An auxiliary mode of apoptotic DNA fragmentation provided by phagocytes (original) (raw)
. 2000 Mar 1;14(5):549–558.
Abstract
CAD (caspase-activated DNase) can cause DNA fragmentation in apoptotic cells. Transgenic mice that ubiquitously express a caspase-resistant form of the CAD inhibitor (ICAD) were generated. Thymocytes prepared from the mice were resistant to DNA fragmentation induced by a variety of stimuli. However, similar numbers of TUNEL-positive cells were present in adult tissues of transgenic and wild-type mice. Exposure to γ-irradiation caused a striking increase in the number of TUNEL-positive cells in the thymus of wild-type, but not transgenic, mice. TUNEL-positive nuclei in transgenic mice were confined to thymic macrophages. When apoptotic thymocytes from the transgenic mice were cocultured with macrophages, the thymocytes underwent phagocytosis and their chromosomal DNA underwent fragmentation. This DNA fragmentation was sensitive to inhibitors that block the acidification of lysosomes. Hence, we conclude that the DNA fragmentation that occurs during apoptosis not only can result cell-autonomously from CAD activity but can also be attributed to a lysosomal acid DNase(s), most likely DNase II, after the apoptotic cells are engulfed.
Keywords: Apoptosis; TUNEL; DNase, caspase; CAD
Programmed cell death, or apoptosis, is the principal mechanism by which superfluous or potentially harmful cells are eliminated in metazoans (Jacobson et al. 1997; Vaux and Korsmeyer 1999). Signals that can induce apoptosis are relatively diverse and include the ligation of cell death receptors at the cell surface, growth factor starvation, DNA damage, and oxidative stress (Nagata 1997; Ashkenazi and Dixit 1998; Green and Reed 1998; Raff 1998). Regardless of the origin of the apoptotic stimulus, commitment to apoptosis occurs through the activation of caspases, a family of cysteine proteases present in growing cells as inactive precursors (Thornberry and Lazebnik 1998). The initiator caspases are activated by forming heteromeric complexes with accessory molecules. For example, caspase 8 combines with Fas and FADD to form the DISC (death-inducing signaling complex), whereas caspase 9 becomes active when complexed with cytochrome c and Apaf1 in a structure termed the apoptosome. This is followed by the cleavage and subsequent activation of downstream caspases such as 3 and 6. Cleavage of a select group of substrates by downstream caspases (Stroh and Schulze-Osthoff 1998) is responsible for the dismantling of essential cell components, which results in the morphological and biochemical changes that characterize apoptotic cell death: cytoskeletal rearrangement, cell membrane disruption and blebbing, nuclear condensation, and DNA fragmentation.
The degradation of nuclear DNA into nucleosomal units is one of the best-characterized biochemical features of apoptotic cell death (Wyllie 1980; Earnshaw 1995). It occurs in a wide variety of cell types and serves as the basis for the techniques that are most commonly used to detect apoptotic cells in situ (Gavrieli et al. 1992). The relationship between caspase activation and nuclear DNA fragmentation was elucidated by the purification and cloning of a caspase-activated DNase (CAD), and its inhibitor (ICAD) (Enari et al. 1998). The full-length form of mouse ICAD (ICAD-L), also called DNA fragmentation factor (DFF-45) (Liu et al. 1997), is a 45-kD protein composed of 331 amino acids. A short form (ICAD-S), which consists of amino acids 1–265 of ICAD-L, is generated by alternative splicing and is also expressed in a number of different cells (Enari et al. 1998; Sabol et al. 1998; Gu et al. 1999; Kawane et al. 1999). CAD has an intrinsic DNase activity but is present in growing cells as a complex with ICAD-L (Enari et al. 1998; Liu et al. 1998; Sakahira et al. 1999a). Caspase 3-mediated cleavage of ICAD at amino acids Asp-117 and Asp-224 dissociates the CAD:ICAD-L complex and allows CAD to cleave chromosomal DNA (Enari et al. 1998). The expression of caspase-resistant mutant ICAD inhibits the DNA fragmentation induced by diverse apoptotic stimuli (Sakahira et al. 1998; McIlroy et al. 1999) and inhibits DNA cleavage into both large-scale and nucleosomal fragments (Sakahira et al. 1999b), confirming that CAD is the major DNase responsible for the cell-autonomous nuclear DNA fragmentation that occurs in apoptotic cells.
To determine the role of apoptotic DNA fragmentation in vivo, and thereby shed light on the biological raison d'etre of the CAD:ICAD system, we established a line of transgenic mice that express caspase-resistant mutant ICAD. The cell-autonomous DNA fragmentation in apoptotic cells was blocked in the cells from transgenic mice. However, DNA fragmentation in situ, revealed by the TUNEL reaction, was still observed in these mice. Coculturing apoptotic thymocytes with macrophages indicated that the lysosomal DNase of engulfing cells could cause DNA fragmentation in apoptotic cells that had undergone phagocytosis. This result agrees with previous reports suggesting that the nuc-1 gene product involved in DNA fragmentation during programmed cell death of Caenorhabditis elegans works after engulfment of dying cells (Hedgecock et al. 1983; Ellis and Horvitz 1986).
Results
Generation of ICAD–Sdm transgenic mice
Previously we showed that the transformant cell lines expressing caspase-resistant ICAD-L or ICAD-S protein are resistant to DNA fragmentation during apoptosis (Sakahira et al. 1998; McIlroy et al. 1999). To test the effects of a similar block in CAD function in whole animals, we generated transgenic mice expressing caspase-resistant ICAD–Sdm protein under the control of the human EF-1α promoter (Fig. 1A), which drives ubiquitous gene expression (Hanaoka et al. 1991). Independent lines with germ-line transmission were identified by PCR (as described in Materials and Methods) and screened for transgene expression by Western blotting of thymus and liver extracts using the M2 anti-Flag antibody. The line showing the highest level of transgene expression (line ICAD–Sdm) was selected for further study. ICAD–Sdm transgenic mice developed normally, and both sexes were fertile when crossed with wild-type B6 mice.
Figure 1.
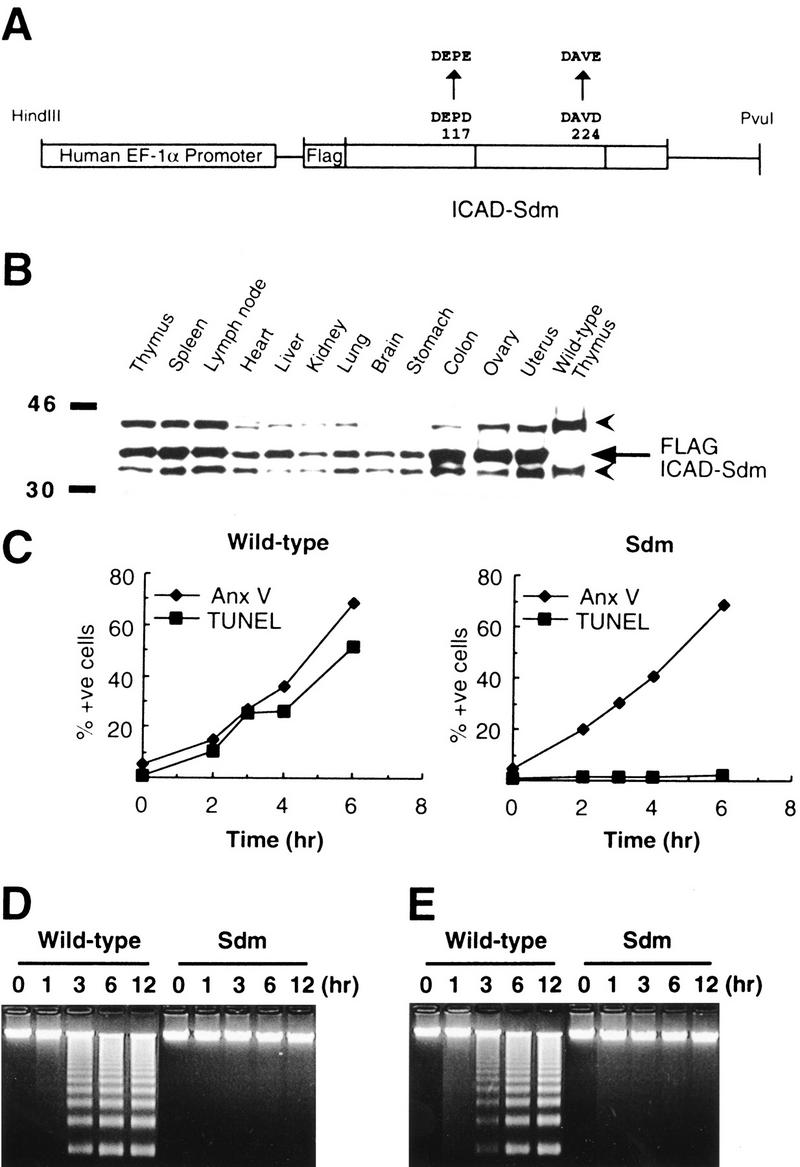
Ubiquitous expression of caspase-resistant ICAD-S in transgenic mice and its effect on in vitro apoptosis. (A) A schematic view of the expression plasmid used to produce the ICAD–Sdm transgenic mice. A 0.8-kb ICAD-S cDNA (open boxes) carrying the D117E and D224E mutations was placed under the control of a 2.4-kb DNA fragment that carries the 5′-flanking sequence, exon 1, and intron 1 of the human EF-1α gene. (B) Ubiquitous expression of the ICAD–Sdm protein. The cell lysates were prepared from the indicated tissues of the transgenic mice and the thymus of wild-type mice. The lysates (20-μg protein) were analyzed by Western blotting with a rabbit antibody against mouse ICAD. The Flag-tagged ICAD–Sdm is indicated by an arrow; arrowheads represent the endogenous ICAD-L and ICAD-S. (C) No DNA degradation in the transgenic thymocytes by γ-irradiation. Mouse thymocytes from the ICAD–Sdm transgenic (right) or wild-type (left) littermate mice were exposed for 10 min to γ-rays (1.2 Gy/min) and cultured for the indicated periods of time. Cells were then stained with FITC-labeled annexin V for apoptotic cells (♦) or with ApopTag fluorescein for TUNEL-positive cells (█) and analyzed by flow cytometry. The results are plotted as percentages of total cells. (_D_-E) No DNA fragmentation in the transgenic thymocytes by treatment with dexamethasone or anti-Fas antibody. Mouse thymocytes (1 × 107 cells) from the ICAD–Sdm transgenic mice or wild-type littermates were treated for the indicated periods of time with either 10 μm dexamethasone (D) or 1 μg/ml of anti-Fas antibody in the presence of 30 μm cycloheximide (E). The chromosomal DNA was analyzed by electrophoresis on an agarose gel.
To confirm the expression of the ICAD–Sdm protein in transgenic mice, cell lysates were prepared from various tissues from adult mice and analyzed by Western blotting using an anti-mouse ICAD antibody. As shown in Figure 1B, all tissues expressed the endogenous ICAD-S and ICAD-L proteins (35- and 45-kD proteins), which agrees with the observation that ICAD-L and ICAD-S mRNAs are ubiquitously expressed in mouse tissues (Kawane et al. 1999). The cell lysates from all tissues of transgenic mice, but not from wild-type mice, showed an extra band at 38 kD. This band was also recognized by the anti-Flag antibody (data not shown), indicating that the 38-kD protein was the transgene product. The expression level of ICAD–Sdm was slightly higher than that of the endogenous ICAD (ICAD-L and ICAD-S) in all tissues.
DNA fragmentation in vitro
Thymocytes undergo apoptosis accompanied by DNA fragmentation in response to various stimuli. To examine the effect of ICAD–Sdm on these events, thymocytes were prepared from ICAD–Sdm transgenic and wild-type mice, and exposed to γ-irradiation. As shown in Figure 1C, the irradiated wild-type thymocytes became annexin V and TUNEL positive with the same time course. On the other hand, no TUNEL-positive population was found in the thymocytes from ICAD–Sdm mice at 6 hr after γ-irradiation, even though 70% of the thymocyte population was annexin V positive at this time point. A similar blockage of apoptotic DNA fragmentation in thymocytes in these mice was observed after treatment with dexamethasone or with anti-Fas antibody (Fig. 1D,E). That is, both stimuli caused DNA fragmentation within 6 hr in wild-type thymocytes but not in thymocytes expressing ICAD–Sdm (Fig. 1D,E). Furthermore, large-scale chromosomal degradation, assessed by pulse-field gel electrophoresis, was barely detectable within 6 hr in glucocorticoid-treated thymocytes from ICAD–Sdm mice, although it occurred within 1 hr in wild-type thymocytes (data not shown). The inhibitory effect of ICAD–Sdm on apoptotic DNA fragmentation was dose dependent. That is, the thymocytes from other transgenic mouse lines that expressed a lower level of caspase-resistant ICAD showed significant DNA fragmentation in response to apoptotic stimuli (data not shown).
DNA fragmentation in vivo
DNA fragmentation in vivo was assessed next by TUNEL staining of tissues in which apoptosis occurs under normal conditions (Surh and Sprent 1994; Umemura et al. 1996). In adult ICAD–Sdm transgenic mice, TUNEL-positive cells were found in the thymic cortex and medulla and in both the corpus luteum and the granulosa cells of ovarian follicles (Fig. 2). The number of TUNEL-positive cells in these adult tissues did not differ between ICAD–Sdm transgenic mice and nontransgenic littermates. Hence, apoptosis accompanied by DNA fragmentation in two different tissues appeared to be normal in vivo in ICAD–Sdm transgenic mice.
Figure 2.
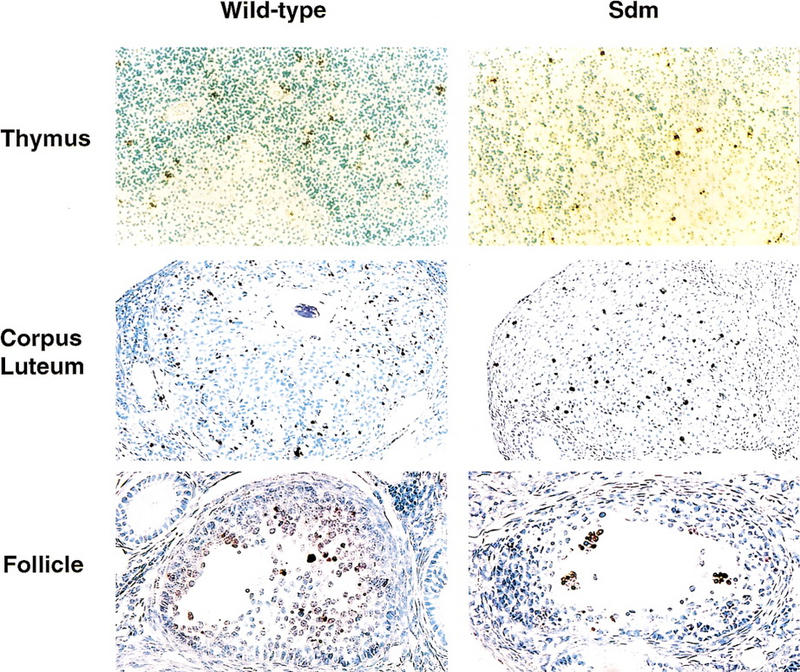
TUNEL-positive cells in various tissues of wild-type and ICAD–Sdm transgenic mice. Paraffin sections were prepared from the thymus and ovary of adult wild-type or ICAD–Sdm transgenic mice. TUNEL-positive cells were detected using an Apotag kit, and DAB-black as a peroxidase substrate. Nuclei were counterstained with hematoxylin. Original magnifications: thymus, 100×; corpus luteum, 100×; ovarian follicules, 150×.
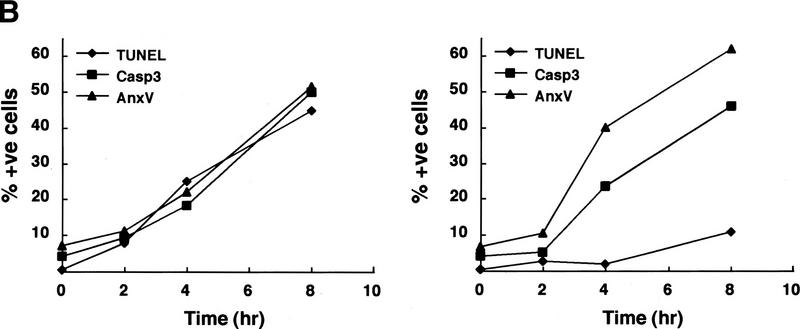
However, when the mice were exposed to γ-irradiation to induce thymocyte apoptosis, a clear difference emerged between ICAD–Sdm transgenic mice and their wild-type littermates. In wild-type mice, 4 hr after exposure to 13 Gy γ-irradiation, large numbers of cells became TUNEL positive in the thymus with extremely high numbers in the cortex (Fig. 3A), in agreement with a previous report (Surh and Sprent 1994). In contrast, the same treatment of ICAD–Sdm transgenic mice generated markedly fewer TUNEL-positive cells in the thymus. The positive cells were clustered together, and the nuclei outside these clusters were uniformly TUNEL negative until 8 hr after irradiation. Thymocytes were then prepared from the mice at different time points after irradiation and analyzed by flow cytometry for TUNEL, annexin V binding and activated caspase 3. As shown in Figure 3B, the proportion of thymocytes containing activated caspase 3 and that binding annexin V increased steadily after irradiation in both wild-type and ICAD–Sdm transgenic mice, reaching 45%–60% by 8 hr. The TUNEL-positive cell population also increased in wild-type mice, whereas that from the transgenic mice remained at a low level (<10%).
Figure 3.
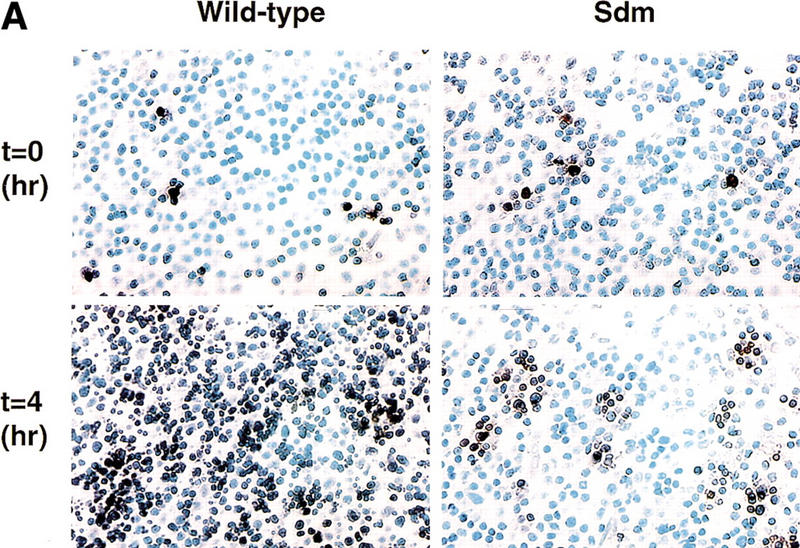
Effect of γ-irradiation on TUNEL-positive cells in the thymus. (A) In situ detection of TUNEL-positive cells in the thymus. Wild-type and ICAD–Sdm transgenic mice were left untreated or exposed for 11 min to γ-rays (1.2 Gy/min). After 4 hr, the mice were sacrificed, and paraffin sections of the thymic cortex were immunohistochemically stained for TUNEL reaction, using an Apotag kit. The sections were then counterstained with methyl green. Original magnifications, 400×. (B) Apoptosis of thymocytes after γ-irradiation. Wild-type (left) and ICAD–Sdm (right) mice were irradiated as above. Thymocytes were prepared from the irradiated mice at the indicated time. Aliquots of 1 × 105 cells were stained with FITC-conjugated annexin V (▴), ApopTag-fluorescein for TUNEL-positive cells (♦), or PhiPhiLux-G1D2 for the activated caspase 3 (█), and analyzed by flow cytometry. The results are expressed as percentages of total cells.
Involvement of phagocytes in DNA degradation in apoptotic cells
A close examination of thymus sections from nonirradiated and irradiated ICAD–Sdm transgenic mice indicated that TUNEL-positive cells were engulfed by phagocytic cells. Double staining of thymic sections for TUNEL and for macrophage-specific F4/80 antigen (Austyn and Gordon 1981) indicated that all TUNEL-positive cells in the thymic cortex of untreated and γ-irradiated ICAD–Sdm transgenic mice were inside F4/80-positive macrophages (Fig. 4A; data not shown). In contrast, large numbers of TUNEL-positive thymocytes were located outside F4/80+ macrophages in irradiated wild-type mice.
Figure 4.
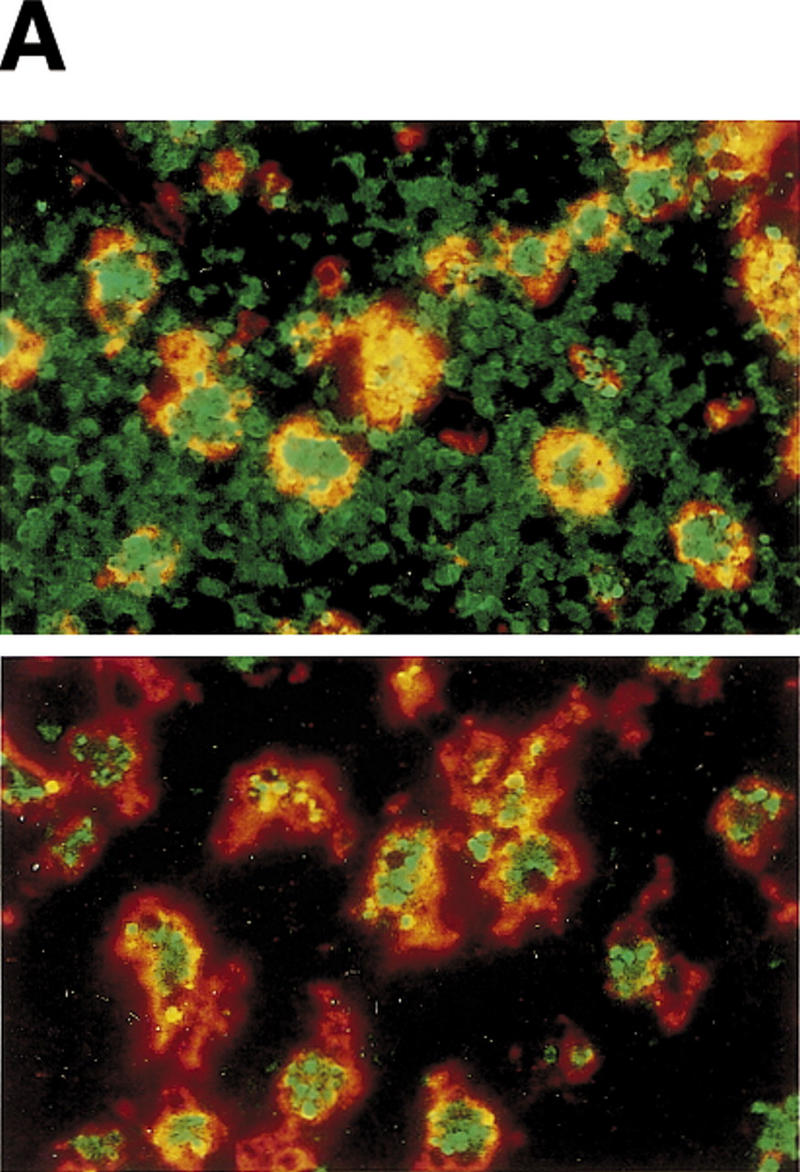
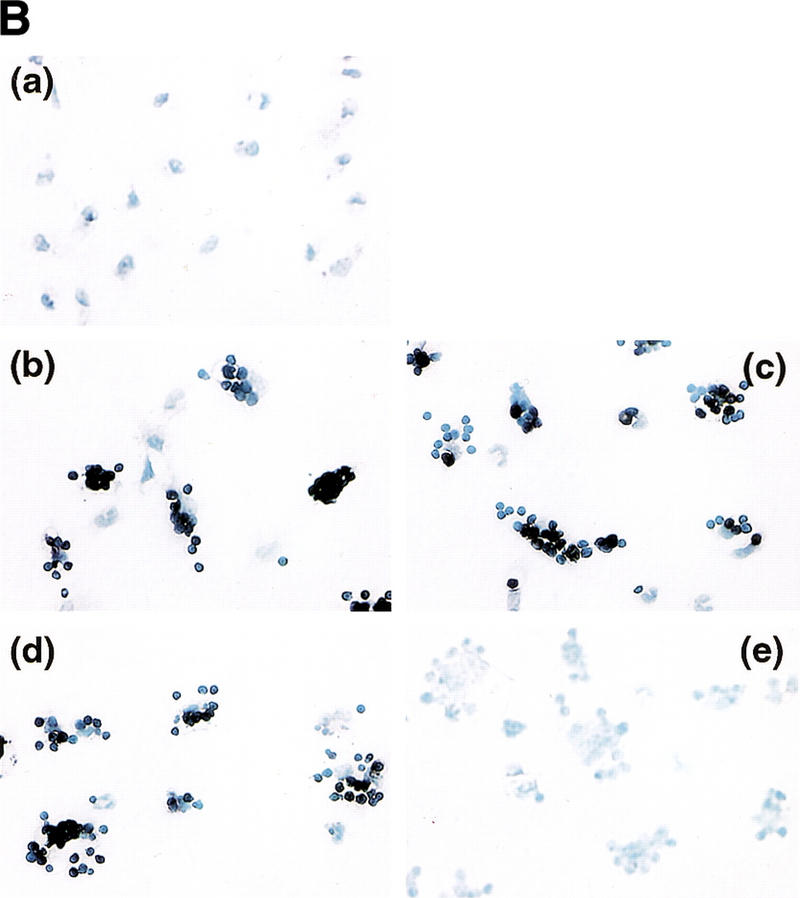
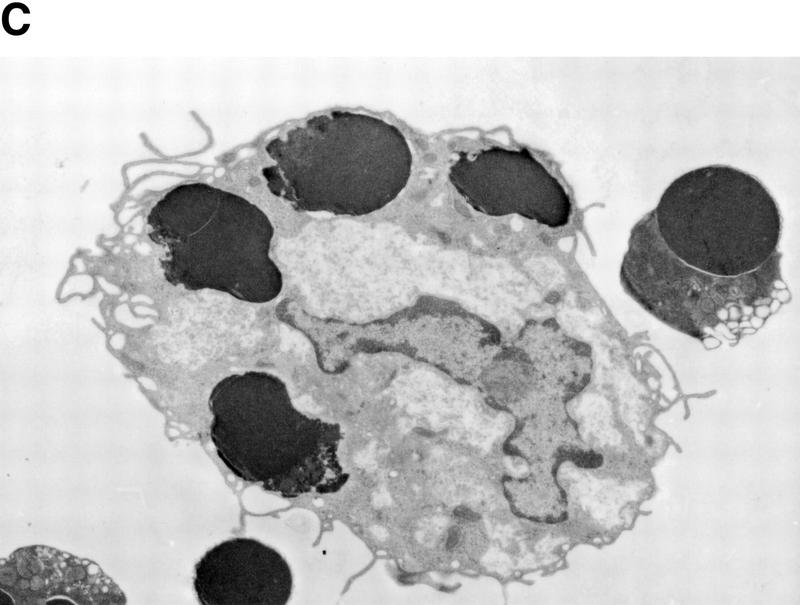
TUNEL-positive thymocytes in macrophages. (A) Ingestion of TUNEL-positive cells in thymuses of ICAD–Sdm transgenic mice by macrophages. Wild-type (top) and ICAD–Sdm transgenic mice (bottom) were exposed for 11 min to γ-rays (1.2 Gy/min). Four hours later, cryosections of the thymus were double-stained for TUNEL-positive cells (green) and F4/80 antigen (red). Original magnifications, 400×. (B) In vitro generation of TUNEL-positive cells by macrophages. Thioglycollate-elicited peritoneal macrophages were resuspended in RPMI medium containing 10% FCS, distributed in eight-well glass chamber slides, and cultured overnight at 37°C. Mouse thymocytes from wild-type (b,d) or ICAD–Sdm (c,e) mice were incubated at 37°C for 3.5 hr with 10 μm dexamethasone in RPMI medium containing 10% FCS. Apoptotic thymocytes were then added to macrophage cultures, and phagocytosis was allowed to proceed for 1 hr in the absence (b,c) or presence (d,e) of 100 μm chloroquine. Cells not undergoing phagocytosis were removed, and the adherent macrophages were fixed with 1% paraformaldehyde, and stained using the TUNEL reaction, and lightly counterstained with Wright's stain. As a control, macrophages without thymocytes were subjected to the TUNEL staining procedure (a). Original magnifications, 400×. (C) Electron micrograph of macrophages carrying thymocytes that underwent phagocytosis. The apoptotic thymocytes from wild-type mice underwent phagocytosis by macrophages as described above. The cells were fixed, stained with uranyl acetate, and examined using a Hitachi H7100 transmission electron microscope.
To examine whether macrophages can cause DNA fragmentation in apoptotic cells, thymocytes were induced to become apoptotic by glucocorticoid treatment and fed to peritoneal macrophages. Phagocytosis was allowed to proceed for 1 hr, during which the apoptotic thymocytes from wild-type and ICAD–Sdm transgenic mice underwent phagocytosis with similar efficiency (data not shown). The transmission electron microscope analysis indicated that macrophages carry three to five apoptotic thymocytes (Fig. 4C). The thymocytes from both wild-type and ICAD–Sdm transgenic mice were TUNEL positive (Fig. 4B, b and c). The appearance of the TUNEL-positive cells was blocked when thymocytes from ICAD–Sdm mice were cocultured with macrophages in the presence of chloroquine (e). Monensin at 30 μm which, like chloroquine, inhibits lysosome acidification, also blocked generation of TUNEL-positive cells from ICAD–Sdm thymocytes (data not shown), indicating that lysosomal acidic DNase was responsible for the DNA fragmentation in these cells. In contrast, the wild-type thymocytes that underwent phagocytosis were TUNEL positive even in the presence of chloroquine (d), as their chromosomal DNA could be independently cleaved by CAD. The same results were obtained using peritoneal macrophages from ICAD–Sdm transgenic mice as phagocytes (data not shown).
Bafilomycin A1 is a specific irreversible inhibitor of vacuolar H+/ATPase and thus inhibits acidification in endosomes, as well as in lysosomes (Bowman et al. 1988; Clague et al. 1994). To determine whether lysosomal DNase from apoptotic thymocytes or phagocytes was responsible for generating TUNEL-positive cells, thymocytes or macrophages were preincubated for 1 hr with 50 nm bafilomycin. After removing bafilomycin, the apoptotic thymocytes were cocultured with macrophages as described above. As shown in Figure 5f, bafilomycin-treated thymocytes from ICAD–Sdm mice became TUNEL-positive after coculture with untreated macrophages. In contrast, treatment of macrophages with bafilomycin inhibited their ability to generate TUNEL-positive ICAD–Sdm thymocytes (Fig. 5e), although the macrophages still efficiently engulfed the apoptotic thymocytes. As expected from the functional CAD system, the engulfed wild-type thymocytes were TUNEL positive irrespective of whether or not macrophages or thymocytes were treated with bafilomycin (Fig. 5a–c). These results indicate that acid lysosomal DNase in macrophages is responsible for the generation of TUNEL-positive cells.
Figure 5.
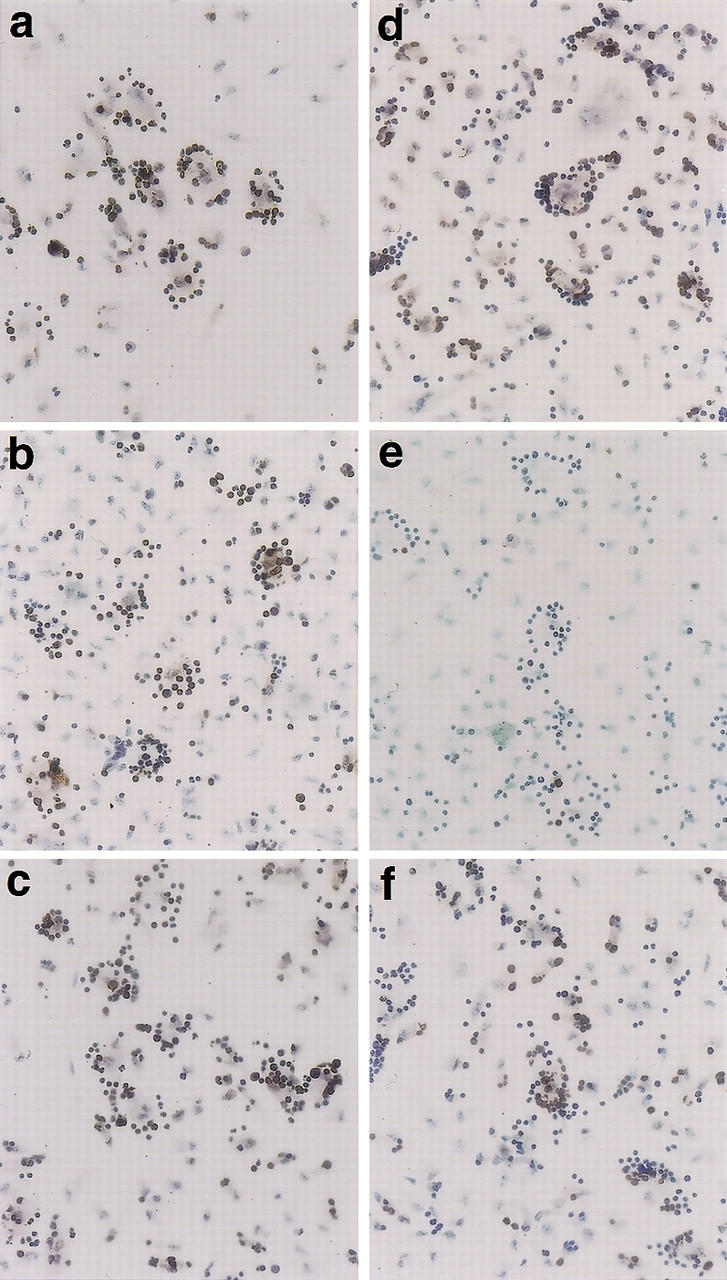
Effect of bafilomycin on macrophage-induced generation of TUNEL-positive cells. Thymocytes from wild-type (a_–_c) or ICAD–Sdm (d_–_f) mice were incubated at 37°C for 3.5 hr with 10 μm dexamethasone in RPMI medium containing 10% FCS. Bafilomycin A1 at 50 nm was added to the culture for the last hour of incubation in c and f. Thioglycollate-elicited peritoneal macrophages were resuspended in RPMI medium containing 10% FCS, distributed in eight-well glass chamber slides, cultured overnight, and incubated at 37°C for 1 hr in the presence (b,e) or absence (a,c,d,e) of 50 nm bafilomycin A1. Thymocytes and macrophages were washed to remove bafilomycin, cocultured at 37°C for 1 hr, and stained using the TUNEL reaction as described in Materials and Methods. Original magnification, 400×.
DNA fragmentation by phagocytes
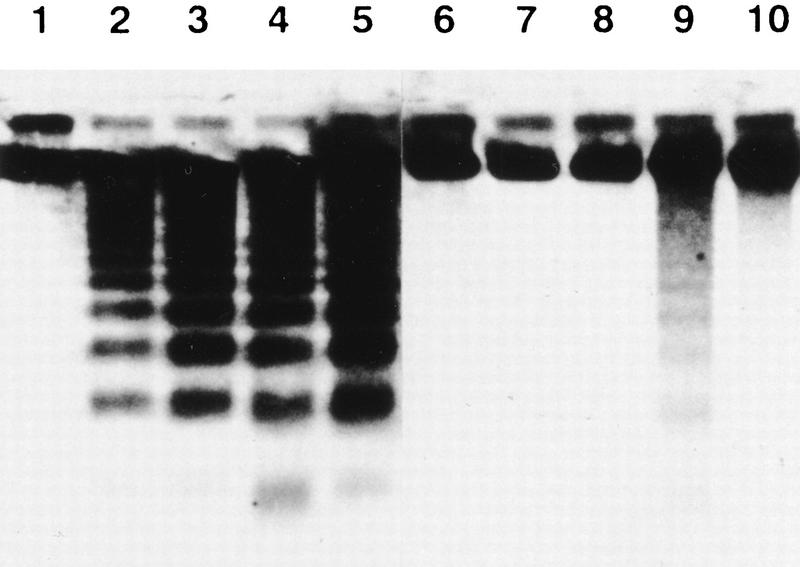
To confirm DNA fragmentation by macrophages, thymocytes whose DNA was labeled in vivo with BrdU were then used as targets. After coculturing these thymocytes with macrophages, the DNA of phagocytosed or nonphagocytosed thymocytes was separated by electrophoresis on an agarose gel and visualized by Western blotting with an anti-BrdU antibody. As shown in Figure 6, the apoptotic thymocytes from wild-type mice showed DNA fragmentation whether or not they underwent phagocytosis (lanes 3,4), and this DNA fragmentation was not inhibited by chloroquine (lane 5). In contrast, when thymocytes from transgenic mice were used as targets, DNA fragmentation was observed only in cells that underwent phagocytosis (lanes 8,9). This DNA fragmentation was blocked when the coculture was carried out in the presence of chloroquine (lane 10). These results indicate that acid DNase provided by phagocytes caused DNA fragmentation in apoptotic thymocytes.
Figure 6.
DNA fragmentation in thymocytes after phagocytosis by macrophages. Wild-type (lanes 1_–_5) and ICAD–Sdm transgenic (lanes 6_–_10) mice were given BrdU for 7 days in drinking water, and their thymocytes were prepared. The thymocytes were left untreated (lanes 1,6) or treated (lanes 2_–_5,7_–_10) with 10 μm dexamethasone as described above. The apoptotic thymocytes were then added to the culture of peritoneal macrophages from wild-type B6 mice, and incubated at 37°C for 1 hr in the absence (lanes 3,4,8,9) or presence (lanes 5,10) of 100 μm chloroquine. DNA was prepared from nonadherent (lanes 3,8) or adherent (lanes 4,5,9,10) cells, separated by electrophoresis on an agarose gel, transferred to a Hybond-N membrane, and probed with a peroxidase-conjugated anti-BrdU monoclonal antibody. Bands were revealed by chemiluminescence.
Discussion
Apoptosis is known to be a cell death process that occurs cell-autonomously. That is, cells have a mechanism for committing suicide or are programmed to die. Various apoptotic stimuli activate caspase cascades that lead to cleavage of >60 death-related substrates (Stroh and Schulze-Osthoff 1998) and to activation of the CAD DNase that causes DNA fragmentation (Enari et al. 1998).In this report we show that DNA fragmentation can be induced not only cell-autonomously by CAD but also by phagocytic cells after apoptotic cells have undergone phagocytosis. This may explain, at least in part, why ICAD-null (Zhang et al. 1998) or ICAD–Sdm transgenic (this report) mice, which cannot produce functional CAD, have no abnormal phenotype during development.
Cell-autonomous DNA fragmentation in wild-type mice generated TUNEL-positive cells and was not inhibited by chloroquine or bafilomycin, which blocks acidification of lysosomes. Terminal transferase used for TUNEL reaction extends 3′-hydroxyl ends of DNA (Roychoudhury et al. 1976). CAD is active at neutral pH (M. Enari and S. Nagata, unpubl.) and cleaves DNA leaving 5′-phosphate and 3′-hydroxyl ends (Liu et al. 1999), confirming that CAD is responsible for the cell autonomous generation of TUNEL-reactive DNA fragments. No TUNEL-positive cells were generated cell-autonomously in thymocytes from ICAD–Sdm mice, indicating that no alternative DNase can cause DNA fragmentation in dying cells under these conditions. DNA fragmentation in apoptotic thymocytes caused by coculture with macrophages was inhibited by agents that block the acidification of lysosomes, suggesting that acid DNase in the lysosomes of phagocytic cells was responsible for DNA fragmentation. The most likely candidate for the acid DNase in lysosomes that causes DNA fragmentation is DNase II (Barry and Eastman 1993; Baker et al. 1998; Krieser and Eastman 1998; Wang et al. 1998; Yasuda et al. 1998). However, it is unlikely that the action of DNase II directly generates TUNEL-positive cells, because cleavage of DNA by DNase II generates 5′-hydroxyl and 3′-phosphate ends that cannot be used as a substrate for terminal transferase. Lysosomes are rich in acid phosphatases, which can remove the phosphate group from the 3′ end of DNA fragments. Thus, it is reasonable to assume that DNase II, in combination with acid phosphatase in lysosomes, generates TUNEL-reactive ends.
Wu et al. (2000) report that Nuc-1, one of the gene products required for DNA degradation during programmed cell death in the C. elegans system, is DNase II. According to them, the apoptotic DNA degradation in C. elegans proceeds in three steps. In apoptotic dying cells, an as yet unidentified CAD-like DNase produces TUNEL-positive DNA ends; the CAD-like DNase is followed by Nuc-1/DNase II, which renders the TUNEL-positive ends TUNEL negative. Finally, DNA is degraded completely by an enzyme(s) provided by engulfing cells. The postulation of CAD-like DNase in the apoptotic cells in C. elegans agrees with our model developed with mammalian cells. However, the function of DNase II in dying cells in C. elegans apparently contradicts our results. It is known that if apoptotic mammalian cells do not undergo phagocytosis, they undergo autophagy and lysosomes rupture (Ohsawa et al. 1998; Neuzil et al. 1999). Under these conditions, DNase II may cause DNA fragmentation in dying cells. We suspect that if the apoptotic cells undergo phagocytosis rapidly (see below), DNase II is provided primarily by engulfing cells to cause DNA fragmentation in dying cells. In any case, we surmise that in both mammalian and C. elegans systems, at least two DNases are involved in apoptotic DNA fragmentation. It is likely that further genetic and cellular analysis will reveal the evolutional conservation of the molecular mechanism of this process in mammalian and C. elegans systems.
The role of engulfment by phagocytes has been thought to be solely to remove unnecessary cell corpses, that is, to act as a passive scavenger. However, all of the TUNEL-positive cells in the thymus were found inside macrophages in both wild-type and ICAD–Sdm transgenic mice, supporting the idea that macrophages are actively involved in the destruction of apoptotic cells (Aliprantis et al. 1996; Diez-Roux and Lang 1997). If macrophages can degrade the chromosomal DNA of the target cells, why do apoptotic cells have a cell-autonomous mechanism (the CAD/ICAD system) for degrading chromosomal DNA? In particular, nucleosomes produced by the cleavage of chromosomal DNA in the apoptotic cells are known to be released from the cells and to activate an immune reaction, which may cause autoimmunity (Bell et al. 1990; Casciola-Rosen et al. 1995). In the developmental setting, phagocytes seem to recognize apoptotic cells efficiently at a very early stage, when no apparent morphological changes or DNA fragmentation have occurred (Durrieu et al. 1998). Thus, all of the apoptotic events, such as DNA fragmentation by CAD and cleavage of the many death-related substrates by caspases, occur inside the phagocytes, and this process could be facilitated by lysosomal enzymes. On the other hand, when the mice were exposed to γ-irradiation, most of the apoptotic cells were not engulfed by macrophages, suggesting that an unphysiologically large load of apoptotic cells exceeded the local phagocytic clearance capacity. In these situations, which may occur not only as a result of γ-irradiation but also during ischemia or acute infection, cell-autonomous cleavage of chromosomal DNA by CAD may be more beneficial for the organisms than releasing intact DNA into the circulation. Alternatively, CAD may be necessary to destroy DNA in apoptotic cells before their engulfment by phagocytes, thereby preventing the transfer of potentially dangerous viral or oncogenic DNA to phagocytes (Holmgren et al. 1999).
Thymocytes treated with glucocorticoids, but not untreated thymocytes, underwent phagocytosis efficiently by macrophages (D. McIlroy, M. Tanaka, and S. Nagata, unpubl.), suggesting that apoptotic cells present “eat me” signal (Platt et al. 1998; Ren and Savill 1998) to phagocytes. DNA fragmentation was initially considered as one such signal. However, the apoptotic thymocytes from wild-type and ICAD–Sdm transgenic mice underwent phagocytosis with the same efficiency, indicating that DNA fragmentation of the apoptotic cells is not a marker used by phagocytes to recognize dying cells.
In summary, we have shown that there is a backup system for cleaving chromosomal DNA during apoptosis. This system seems to be operated by lysosomal enzymes in phagocytic cells. The availability of ICAD–Sdm transgenic mice described in this report and ICAD-null mice established by Zhang et al. (1998), will be useful for elucidating the role of CAD-mediated DNA fragmentation in situations where this backup system becomes overloaded.
Materials and methods
Generation of ICAD–Sdm transgenic mice
The mouse ICAD-S cDNA in which the codons for Asp-117 and Asp-224 were changed to those for Glu has been described previously (Sakahira et al. 1998). A DNA fragment coding for a Flag epitope was ligated in-frame to the 5′ end of the ICAD coding sequence and placed under the promoter of the human EF-1α gene of the pEF321 vector (Kim et al. 1990) to generate pEF3-F-ICADSdm. The plasmid pEF3-F-ICADSdm was digested with _Hin_dIII and _Pvu_I, and the 4.0-kb fragment carrying the ICAD cDNA was microinjected into fertilized mouse eggs collected after mating C57BL/6 with DBA2 mice, as described previously (Yamamura et al. 1984). Transgenic offspring were identified by PCR, using a forward primer (5′-CAAGCCTCAGACAGTGGTTC-3′) carrying a sequence on the EF-1α promoter (Uetsuki et al. 1989) and a reverse primer (5′-TCCACTTCTCATTGCAGGCC-3′) specific for mouse ICAD cDNA (Enari et al. 1998). PCR was carried out with Taq polymerase (Amersham/Pharmacia) as described previously (Adachi et al. 1995). The transgenic mouse lines were maintained by crossing with wild-type C57BL/6 mice, and transgenic offspring and wild-type littermates were used at age 5–12 weeks.
Western blotting
Mouse tissues were homogenized using a Polytron (Kinematica AG) in 50 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl, 1% NP-40, 1 mm PMSF, 2 mm o_-phenanthroline, 3 μg/ml leupeptin, and 3 μg/ml pepstatin. Homogenates were cleared by centrifugation at 30,000_g for 15 min, and mouse IgG was removed with protein A– and protein G–Sepharose beads (Amersham-Pharmacia). The cell lysates were separated by electrophoresis on a 10%–20% gradient polyacrylamide gel and transferred to a PVDF membrane by semidry blotting. Membranes were probed with 0.9 μg/ml rabbit anti-mouse ICAD antibody (Sakahira et al. 1999a), followed by peroxidase-conjugated secondary antibodies (Dako), and bands were revealed by chemiluminescence using a Renaissance kit (GIBCO/NEN).
Induction and detection of apoptosis
To induce apoptosis, mouse thymocytes were suspended at 1 × 107 cells/ml in RPMI medium containing 10% FCS and treated with either dexamethasone, anti-mouse Fas antibody (Jo2) (Ogasawara et al. 1993), or γ-irradiation using a Cs137 source Gammacell 40 Exactor (Nordion International Inc.). Apoptosis of thymocytes was also induced in vivo by exposing the mice to γ-irradiation.
Apoptotic cells were quantified by the binding of FITC-conjugated annexin V using an Apoptosis Detection kit (R+D Systems), followed by flow cytometry. Activation of caspase 3 in the cells was detected using the cell-permeable fluorescent caspase 3 substrate PhiPhiLux-G1D2 (OncoImmunin Inc.) according to the manufacturer's instructions. DNA fragmentation in thymocytes was detected by a modified TUNEL procedure using an ApopTag–fluorescein in situ apoptosis detection kit (Intergen) followed by flow cytometry. The flow cytometry was carried out using a FACS Calibur from Becton-Dickinson. DNA cleavage into nucleosomal fragments was also assessed by fractionating DNA on an agarose gel as described previously (Itoh et al. 1991). In brief, cells were lysed in 100 mm Tris-HCl buffer (pH 8.5) containing 5 mm EDTA, 0.2% SDS, 200 mm NaCl, and 200 μg/ml proteinase K and incubated overnight at 37°C. Nucleic acids were recovered by precipitation with one volume of isopropanol, and DNA equivalent to 1 × 105 cells was fractionated by electrophoresis on a 1.5% agarose gel.
Immunohistochemistry
A rat monoclonal antibody against mouse F4/80 antigen was prepared by growing an F4/80 hybridoma (Austyn and Gordon 1981) in RPMI1640 medium containing 10% FCS. Paraffin tissue sections (4 μm thick) were subjected to the TUNEL reaction using an Apoptag kit (Intergen) and DAB-black (Zymed) as the peroxidase substrate. For double staining for TUNEL and F4/80 antigen, cryosections (5 μm thick) of mouse tissues were first labeled with DIG–dUTP and blocked with 0.5% BSA and 10% rabbit normal serum in PBS. The sections were then incubated with undiluted F4/80 hybridoma supernatant, followed by peroxidase-conjugated rabbit anti-rat secondary antibody (Dako), followed by Cy3-labeled tyramide as the peroxidase substrate. Finally, TUNEL-positive cells were detected with FITC-conjugated anti-DIG antibodies (Intergen).
In vitro phagocytosis assay
Resident or thioglycollate-elicited peritoneal macrophages were resuspended in RPMI medium containing 10% FCS, distributed at 1 × 105 cells per well in eight-well glass chamber slides (Nalge-Nunc), and cultured overnight at 37°C. Mouse thymocytes were incubated at 37°C for 3.5 hr with 10 μm dexamethasone in RPMI medium containing 10% FCS. The apoptotic thymocytes were added to macrophage cultures (2 × 106 thymocytes per well), and phagocytosis was allowed to proceed for 1 hr. Cells not undergoing phagocytosis were removed by washing in PBS containing 0.7 mm Ca2+ and 0.5 mm Mg2+, and the adherent macrophages were fixed in PBS containing 1% paraformaldehyde and stained using the TUNEL technique, as described above.
To detect DNA fragmentation in thymocytes in the mixed culture with macrophages, thymocyte DNA was labeled with BrdU. In brief, mice were given BrdU (Sigma Chemical) for 7 days in their drinking water at a concentration of 0.8 mg/ml. The BrdU was dissolved in sterile water, which was changed daily. After coculturing the BrdU-labeled thymocytes with macrophages, the DNA was extracted and separated by electrophoresis on a 1.5% agarose gel. The DNA was transferred to a Hybond-N membrane (Amersham-Pharmacia) and probed with a peroxidase-conjugated anti-BrdU monoclonal antibody (Boehringer Mannheim). Bands were revealed by chemiluminescence using a Renaissance kit.
Acknowledgments
We thank Ms. C. Ariga for preparation of paraffin sections, Ms. Y. Seto for maintaining the mice, and Ms. S. Kumagai for secretarial assistance. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture in Japan. D.M. is the recipient of a postdoctoral fellowship from the European Union/Japan Society for the Promotion of Science, and H.S. is supported by a Research fellowship of the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nagata@genetic.med.osaka-u.ac.jp; FAX 81-6-6879-3319.
References
- Adachi M, Suematsu S, Kondo T, Ogasawara J, Tanaka T, Yoshida N, Nagata S. Targeted mutation in the Fas gene causes hyperplasia in the peripheral lymphoid organs and liver. Nat Genet. 1995;11:294–300. doi: 10.1038/ng1195-294. [DOI] [PubMed] [Google Scholar]
- Aliprantis AO, Diez-Roux G, Mulder LC, Zychlinsky A, Lang RA. Do macrophages kill through apoptosis? Immunol Today. 1996;17:573–576. doi: 10.1016/s0167-5699(96)10071-2. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- Baker KP, Baron WF, Henzel WJ, Spencer SA. Molecular cloning and characterization of human and murine DNase II. Gene. 1998;215:281–289. doi: 10.1016/s0378-1119(98)00280-7. [DOI] [PubMed] [Google Scholar]
- Barry M, Eastman A. Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch Biochem Biophys. 1993;300:440–450. doi: 10.1006/abbi.1993.1060. [DOI] [PubMed] [Google Scholar]
- Bell DA, Morrison B, VandenBygaart P. Immunogenic DNA-related factors. Nucleosomes spontaneously released from normal murine lymphoid cells stimulate proliferation and immunoglobulin synthesis of normal mouse lymphocytes. J Clin Invest. 1990;85:1487–1496. doi: 10.1172/JCI114595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman EJ, Siebers A, Altendorf K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciola-Rosen LA, Anhalt GJ, Rosen A. DNA-dependent protein kinase is one of a subset of autoantigens specifically cleaved early during apoptosis. J Exp Med. 1995;182:1625–1634. doi: 10.1084/jem.182.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Urbe S, Aniento F, Gruenberg J. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J Biol Chem. 1994;269:21–24. [PubMed] [Google Scholar]
- Diez-Roux G, Lang RA. Macrophages induce apoptosis in normal cells in vivo. Development. 1997;124:3633–3638. doi: 10.1242/dev.124.18.3633. [DOI] [PubMed] [Google Scholar]
- Durrieu F, Belloc F, Lacoste L, Dumain P, Chabrol J, Dachary-Prigent J, Morjani H, Boisseau MR, Reiffers J, Bernard P, Lacombe F. Caspase activation is an early event in anthracycline-induced apoptosis and allows detection of apoptotic cells before they are ingested by phagocytes. Exp Cell Res. 1998;240:165–175. doi: 10.1006/excr.1997.3918. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC. Nuclear changes in apoptosis. Curr Biol. 1995;7:337–343. doi: 10.1016/0955-0674(95)80088-3. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Horvitz H. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Gu J, Dong RP, Zhang C, McLaughlin DF, Wu MX, Schlossman SF. Functional interaction of DFF35 and DFF45 with caspase-activated DNA fragmentation nuclease DFF40. J Biol Chem. 1999;274:20759–20762. doi: 10.1074/jbc.274.30.20759. [DOI] [PubMed] [Google Scholar]
- Hanaoka K, Hayasaka M, Uetsuki T, Fujisawa-Sehara A, Nabeshima Y. A stable cellular marker for the analysis of mouse chimeras: the bacterial chloramphenicol acetyltransferase gene driven by the human elongation factor 1 alpha promoter. Differentiation. 1991;48:183–189. doi: 10.1111/j.1432-0436.1991.tb00256.x. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Sulston JE, Thomson JN. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science. 1983;220:1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- Holmgren L, Szeles A, Rajnavolgyi E, Folkman J, Klein G, Ernberg I, Falk KI. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood. 1999;93:3956–3963. [PubMed] [Google Scholar]
- Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Adachi M, Sakahira H, Copeland NG, Gilbert DJ, Jenkins NA, Nagata S. Structure and promoter analysis of murine CAD and ICAD genes. Cell Death Differ. 1999;6:745–752. doi: 10.1038/sj.cdd.4400547. [DOI] [PubMed] [Google Scholar]
- Kim DW, Uetsuki T, Kaziro Y, Yamaguchi N, Sugano S. Use of the human elongation factor 1 alpha promoter as a versatile and efficient expression system. Gene. 1990;91:217–223. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- Krieser RJ, Eastman A. The cloning and expression of human deoxyribonuclease II. A possible role in apoptosis. J Biol Chem. 1998;273:30909–30914. doi: 10.1074/jbc.273.47.30909. [DOI] [PubMed] [Google Scholar]
- Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, Wang X. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc Natl Acad Sci. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zou H, Widlak P, Garrard W, Wang X. Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1. J Biol Chem. 1999;274:13836–13840. doi: 10.1074/jbc.274.20.13836. [DOI] [PubMed] [Google Scholar]
- McIlroy D, Sakahira H, Talanian RV, Nagata S. Involvement of caspase 3-activated DNase in internucleosomal DNA cleavage induced by diverse apoptotic stimuli. Oncogene. 1999;18:4401–4408. doi: 10.1038/sj.onc.1202868. [DOI] [PubMed] [Google Scholar]
- Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- Neuzil J, Svensson I, Weber T, Weber C, Brunk UT. alpha-tocopheryl succinate-induced apoptosis in Jurkat T cells involves caspase-3 activation, and both lysosomal and mitochondrial destabilisation. FEBS Lett. 1999;445:295–300. doi: 10.1016/s0014-5793(99)00141-6. [DOI] [PubMed] [Google Scholar]
- Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- Ohsawa Y, Isahara K, Kanamori S, Shibata M, Kametaka S, Gotow T, Watanabe T, Kominami E, Uchiyama Y. An ultrastructural and immunohistochemical study of PC12 cells during apoptosis induced by serum deprivation with special reference to autophagy and lysosomal cathepsins. Arch Histol Cytol. 1998;61:395–403. doi: 10.1679/aohc.61.395. [DOI] [PubMed] [Google Scholar]
- Platt N, da Silva RP, Gordon S. Recognizing death: the phagocytosis of apoptotic cells. Trends Cell Biol. 1998;8:365–372. doi: 10.1016/s0962-8924(98)01329-4. [DOI] [PubMed] [Google Scholar]
- Raff M. Cell suicide for beginners. Nature. 1998;396:119–122. doi: 10.1038/24055. [DOI] [PubMed] [Google Scholar]
- Ren Y, Savill J. Apoptosis: The importance of being eaten. Cell Death Differ. 1998;5:563–568. doi: 10.1038/sj.cdd.4400407. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R, Jay E, Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976;3:101–116. doi: 10.1093/nar/3.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabol SL, Li R, Lee TY, Abdul-Khalek R. Inhibition of apoptosis-associated DNA fragmentation activity in nonapoptotic cells: the role of DNA fragmentation factor-45 (DFF45/ICAD) Biochem Biophys Res Commun. 1998;253:151–158. doi: 10.1006/bbrc.1998.9770. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L and ICAD-S. J Biol Chem. 1999a;274:15740–15744. doi: 10.1074/jbc.274.22.15740. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Ohsawa Y, Uchiyama Y, Nagata S. Apoptotic nuclear morphological change without DNA fragmentation. Curr Biol. 1999b;9:543–546. doi: 10.1016/s0960-9822(99)80240-1. [DOI] [PubMed] [Google Scholar]
- Stroh C, Schulze-Osthoff K. Death by a thousand cuts: An ever increasing list of caspase substrates. Cell Death Differ. 1998;5:997–1000. doi: 10.1038/sj.cdd.4400451. [DOI] [PubMed] [Google Scholar]
- Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- Uetsuki T, Naito A, Nagata S, Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor 1α. J Biol Chem. 1989;264:5791–5798. [PubMed] [Google Scholar]
- Umemura S, Yasuda M, Osamura RY, Kawarada Y, Sugiyama T, Tsutsumi Y. Enhancement of TdT-mediated dUTP-biotin nick end-labeling (TUNEL) method using mung bean nuclease, a single-stranded DNA digestion enzyme. J Histochem Cytochem. 1996;44:125–132. doi: 10.1177/44.2.8609368. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Wang CC, Lu SC, Chen HL, Liao TH. Porcine spleen deoxyribonuclease II. Covalent structure, cDNA sequence, molecular cloning, and gene expression. J Biol Chem. 1998;273:17192–17198. doi: 10.1074/jbc.273.27.17192. [DOI] [PubMed] [Google Scholar]
- Wu, Y.C., G.M. Stanfield, and H.R. Horvitz. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes & Dev. (this issue). [PMC free article] [PubMed]
- Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Yamamura K, Kikutani H, Takahashi N, Taga T, Akira S, Kawai K, Fukuchi K, Kumahara Y, Honjo T, Kishimoto T. Introduction of human gamma 1 immunoglobulin genes into fertilized mouse eggs. J Biochem. 1984;96:357–363. doi: 10.1093/oxfordjournals.jbchem.a134845. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Takeshita H, Iida R, Nakajima T, Hosomi O, Nakashima Y, Kishi K. Molecular cloning of the cDNA encoding human deoxyribonuclease II. J Biol Chem. 1998;273:2610–2616. doi: 10.1074/jbc.273.5.2610. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu X, Scherer DC, van Kaer L, Wang X, Xu M. Resistance to DNA fragmentation and chromatin condensation in mice lacking the DNA fragmentation factor 45. Proc Natl Acad Sci. 1998;95:12480–12485. doi: 10.1073/pnas.95.21.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]