Functional selectivity of recombinant mammalian SWI/SNF subunits (original) (raw)
Abstract
The SWI/SNF family of chromatin-remodeling complexes plays a key role in facilitating the binding of specific transcription factors to nucleosomal DNA in diverse organisms from yeast to man. Yet the process by which SWI/SNF and other chromatin-remodeling complexes activate specific subsets of genes is poorly understood. We show that mammalian SWI/SNF regulates transcription from chromatin-assembled genes in a factor-specific manner in vitro. The DNA-binding domains (DBDs) of several zinc finger proteins, including EKLF, interact directly with SWI/SNF to generate DNase I hypersensitivity within the chromatin-assembled β-globin promoter. Interestingly, we find that two SWI/SNF subunits (BRG1 and BAF155) are necessary and sufficient for targeted chromatin remodeling and transcriptional activation by EKLF in vitro. Remodeling is achieved with only the BRG1–BAF155 minimal complex and the EKLF zinc finger DBD, whereas transcription requires, in addition, an activation domain. In contrast, the BRG1–BAF155 complex does not interact or function with two unrelated transcription factors, TFE3 and NF-κB. We conclude that specific domains of certain transcription factors differentially target SWI/SNF complexes to chromatin in a gene-selective manner and that individual SWI/SNF subunits play unique roles in transcription factor–directed nucleosome remodeling.
Keywords: SWI/SNF, zinc fingers, chromatin, transcription, EKLF, β-globin
The selective expression of genes that are packaged into repressive chromatin structures is a fundamental process that controls gene regulation during development. Genetic and biochemical studies have defined several elegant mechanisms that relieve nucleosomal repression and increase accessibility of DNA for protein interactions that establish appropriate patterns of gene expression (for review, see Kadonaga 1998; Kingston and Narlikar 1999). These mechanisms often involve large enzymatic complexes that structurally disrupt or modify histone–DNA contacts within nucleosomes to facilitate transcription factor binding.
One important category of such complexes is the SWI/SNF family of ATP-dependent chromatin-remodeling proteins (for review, see Muchardt and Yaniv 1999). SWI/SNF is a 2 Mda, multisubunit, DNA-dependent ATPase that has been shown genetically to regulate subsets of inducible genes in yeast (for review, see Winston and Carlson 1992) and biochemically to facilitate the interaction of a variety of transcription factors with nucleosomal DNA (Utley et al. 1997; for review, see Workman and Kingston 1998). Mammalian SWI/SNF complexes consist of ∼15 subunits and fall into two broad classes, depending on whether they contain hBRM or BRG1 as the ATPase. BRG1-associated factors (BAFs) tightly bind to either ATPase subunit to form distinct SWI/SNF complexes. These complexes are subject to cell cycle control by changes in phosphorylation (Muchardt et al. 1996; Sif et al. 1998) and can be quite biochemically diverse, which suggests that they may have specialized cellular functions (Wang et al. 1996).
Current studies support the view that SWI/SNF causes the partial unwrapping of DNA from the nucleosome without actual loss of histones, in contrast to the histone acetyl transferase p300 (Ito et al. 2000), and can promote both octamer sliding and transfer to neighboring DNA (for review, see Peterson and Workman 2000). Interestingly, human SWI/SNF (hSWI/SNF) can convert nucleosomal structure from a base state to a remodeled structure in a reversible manner (Schnitzler et al. 1998) and the recombinant ATPase subunit, hBRM or BRG1, is sufficient to disrupt histone–DNA contacts (Phelan et al. 1999).
Although considerable information is available concerning the mechanism by which SWI/SNF alters nucleosomal structure by ATP hydrolysis, little is known about how this complex is targeted to specific promoters to generate transcriptionally active genes. In this regard, SWI/SNF has been found to associate with diverse regulators of gene activation and cell proliferation. These include the glucocorticoid receptors (GRs) and estrogen receptors (Yoshinaga et al. 1992; Muchardt and Yaniv 1993; Ichinose et al. 1997; Ostlund-Farrants et al. 1997; Fryer and Archer 1998), the retinoblastoma tumor suppressor protein, Rb (Dunaief et al. 1994), and cyclin E (Shanahan et al. 1999). The c-MYC proto-oncogene and papillomavirus E1 protein can each associate and function with SWI/SNF, further supporting the notion that this complex participates in cell growth control (Cheng et al. 1999; Lee, D. et al. 1999; for review, see Muchardt and Yaniv 1999). Moreover, activation of the mammalian hsp70 gene in response to certain signaling pathways is also dependent on SWI/SNF components (de La Serna et al. 2000). Mammalian SWI/SNF has functional interactions with tissue-restricted activators such as EKLF (Armstrong et al. 1998) and C/EBPβ (Kowenz-Leutz and Leutz 1999) and cooperates with these proteins to regulate expression of β-globin and myeloid genes, respectively. Involvement of SWI/SNF in the developmental regulation of the human β-globin locus has been demonstrated recently in vivo (Lee, C.H. et al. 1999; O'Neill et al. 1999). Taken together, these studies clearly show that SWI/SNF has a critical role in a wide variety of transcriptional programs and that the specificity of chromatin remodeling must be a highly regulated process.
We have previously shown that a mammalian SWI/SNF complex (E-RC1) regulates transcription of chromatin-assembled human β-globin genes in combination with the erythroid factor EKLF in vitro (Armstrong et al. 1998). SWI/SNF facilitates the targeted interaction of EKLF to its binding site at −90 within the β-globin promoter, resulting in the generation of a DNase hypersensitive region, which is indicative of structurally remodeled chromatin. In contrast, SWI/SNF is unable to activate expression from chromatin-assembled HIV-1 promoters by the E-box binding protein, TFE3, which indicates that remodeling and activation by SWI/SNF is transcription factor selective in vitro. We have examined the basis for this apparent functional selectivity and the role of specific SWI/SNF subunits in factor-directed nucleosomal targeting and gene activation. Here we show that mammalian SWI/SNF cooperates with several proteins containing zinc finger DNA-binding domains (DBDs) to disrupt chromatin structure and to activate transcription. Targeted remodeling by SWI/SNF results from direct interaction with the zinc finger DBDs but not with the activation domains (ADs) of these factors. In contrast, SWI/SNF does not interact or function with two non–zinc finger proteins, TFE3 and NF-κB. We have examined the role of individual SWI/SNF subunits in this process and found that a minimal recombinant complex composed of two proteins is sufficient for factor-directed chromatin disruption and transcription. Thus functional selectivity by mammalian SWI/SNF occurs by direct interactions between specific protein domains and individual SWI/SNF subunits to achieve targeted chromatin remodeling.
Results
Mammalian SWI/SNF functions with several proteins containing distinct zinc finger DBDs
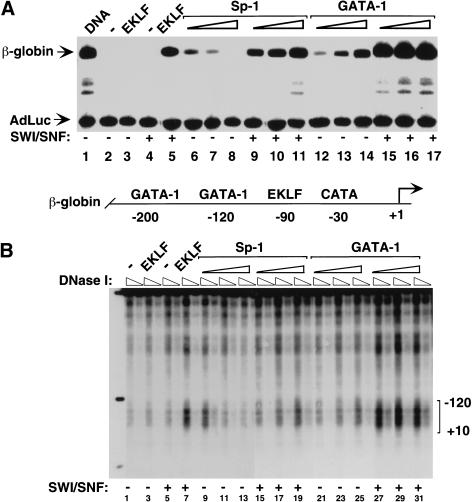
To further assess the specificity of targeted chromatin remodeling, we tested the ability of other zinc finger DBDs in addition to EKLF (Sp1, GATA-1) to cooperate with SWI/SNF and to activate transcription from nucleosome-assembled human β-globin promoters in vitro. The transcription factor Sp1 binds to DNA with the same sequence specificity as EKLF, and recognizes a CACC motif in the β-globin promoter (−90), whereas the GATA-1 factor interacts with two different sites (at −120 and −200) within this region. Sp1 and EKLF share a related zinc finger DBD that is composed of three fingers, which is quite distinct from the two-finger structure of GATA-1 (for review, see Mackay and Crossley 1998). As shown in Figure 1A, the native mammalian SWI/SNF complex strongly activated transcription of the chromatin-assembled β-globin gene by either EKLF (cf. lanes 3 and 5), Sp1 (cf. lanes 6–8 with lanes 9–11), or GATA-1 (cf. lanes 12–14 with lanes 15–17). Activation by each of the three proteins was invariably accompanied by nucleosome structural remodeling, as assessed by the formation of DNase I hypersensitive sites in the β-globin promoter (Figure 1B). Thus SWI/SNF can cooperate with different zinc finger–containing proteins (EKLF, Sp1, and GATA-1) to structurally remodel the nucleosomal β-globin promoter and to activate transcription in vitro.
Figure 1.
Mammalian SWI/SNF selectively functions with several zinc finger DNA-binding proteins to remodel chromatin and activate transcription in vitro. (A) In vitro transcription of chromatin-assembled β-globin plasmids by zinc finger containing transcription factors (EKLF, GATA-1, and Sp-1) in the presence of mammalian SWI/SNF chromatin-remodeling complex. β-globin plasmid templates were assembled into chromatin and incubated in the absence (lanes 1–3, 6–8, 12–14) or presence (lanes 4,5, 9–11,15–17) of SWI/SNF and, where indicated, 37 pmole of recombinant EKLF (lanes 3,5); 30 nmole, 60 nmole, and 150 nmole of an Sp1 fraction (lanes 6–11), and 35 pmole, 45 pmole, or 65 pmole of recombinant GATA-1 (lanes 12–17) per 1 μg of chromatin in a 100 μL of reaction volume. Triangles indicate increasing concentrations of Sp1 (which was inhibitory to transcription at high levels in the absence of SWI/SNF) or GATA-1. All proteins were added to assembled chromatin and incubated for 30 min at 27°C. Chromatin assembly and transcription reactions were conducted as described in Materials and Methods. Primer extension products of the β-globin promoter and the AdLuc internal control gene are indicated by arrows. (B) Analysis of the ability of different zinc finger-containing DNA-binding proteins to generate DNase I hypersensitivity within the β-globin promoter in the presence of SWI/SNF. Assembled chromatin was incubated with SWI/SNF and either EKLF, Sp1, or GATA-1 as in A, and half of the reaction was divided in two and digested with 2 U and 1 U of DNase I as described in Materials and Methods. Triangles indicate increasing amounts of DNase I. Brackets show the −120–+10 region of the β-globin promoter. A schematic diagram of the β-globin promoter is shown between panels A and B.
The zinc finger DBD of EKLF is sufficient for targeted SWI/SNF chromatin remodeling
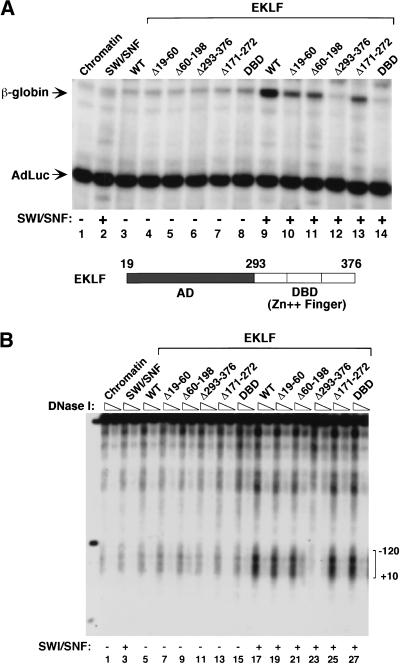
To identify the role of distinct EKLF protein domains in SWI/SNF-mediated chromatin remodeling and transcriptional activation, we examined a series of EKLF-mutant proteins in vitro. As expected, mutations affecting the EKLF AD decreased β-globin transcription (Fig. 2A, cf. lanes 10,11,13 with lane 9), and no activity was observed with the isolated AD (Fig. 2A, lane 12) or the zinc finger DBD (Fig. 2A, lane 14). Importantly, however, the zinc finger DBD was as active as full-length EKLF in its ability to support nucleosome remodeling by SWI/SNF (Fig. 2B, cf. lanes 17 and 27). In contrast, the isolated EKLF activation domain had no ability to support SWI/SNF-dependent remodeling (Fig. 2B, lane 23). These data show that the zinc finger DBD of EKLF is sufficient to generate specific nucleosome remodeling in the presence of native SWI/SNF, whereas transcriptional activation requires, in addition, the EKLF activation domain.
Figure 2.
Distinct protein domains of EKLF are required for SWI/SNF-dependent chromatin remodeling and transcriptional activation. (A) Analysis of different EKLF mutant proteins in β-globin promoter activation in the presence or absence of SWI/SNF in vitro. Assembled chromatin templates were incubated with either wild-type or mutant EKLF proteins (37 pmole/1 μg of chromatin in a 100 μL reaction volume) and SWI/SNF, as indicated for each lane. The reactions were then split, and half was transcribed as in Figure 1A and as described in Materials and Methods. (B) The zinc finger DNA-binding domain of EKLF is sufficient to direct SWI/SNF-dependent DNase I hypersensitivity within the β-globin promoter. After assembly, chromatin was incubated with either wild-type or mutant EKLF protein in the presence or absence of SWI/SNF, as indicated above each lane. Reactions were then split and half was transcribed as shown in A and the remaining chromatin was divided into two tubes with 150 ng chromatin per tube and digested with 1 U and 2 U of DNase I. Triangles indicate increasing amounts of DNase I. Brackets show the −120–+10 region of the promoter. (M) Digested β-globin plasmid as a size marker. Bands represent digests of _Nco_I (4.7 Kb), _Nco_I/_Bsa_AI (841 bp) and _Nco_I/_Eco_RI (301 bp). A schematic diagram of the domain structure of EKLF is shown between panels A and B.
SWI/SNF subunits interact with the distinct zinc finger DBDs of EKLF and GATA-1
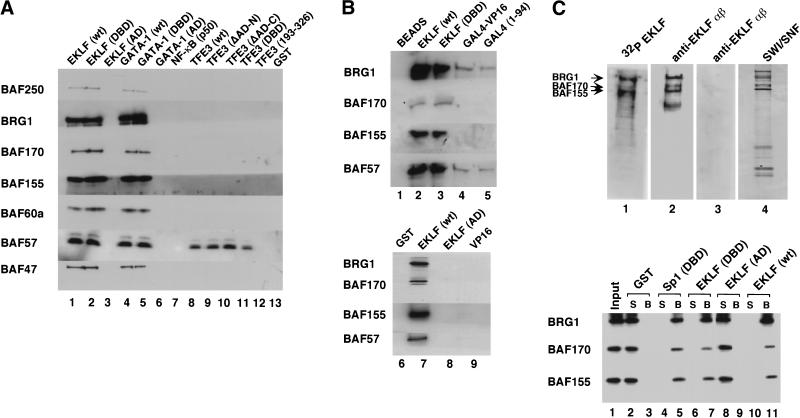
To assess whether EKLF interacts directly with SWI/SNF, recombinant EKLF was incubated with the purified native SWI/SNF complex and examined for associated SWI/SNF subunits following GST pull-down and immunoblotting experiments (Fig. 3A). All SWI/SNF subunits were recovered with full-length EKLF and GATA-1 (Fig. 3A, lanes 1,4) or the distinct zinc finger DBD of either protein (Fig. 3A, lanes 2,5). In contrast, SWI/SNF failed to bind the AD of either EKLF (Fig. 3A, lane 3) or GATA-1 (Fig. 3A, lane 6). Parallel experiments were carried out with recombinant TFE3 and NF-κB (p50). These proteins regulate the HIV-1 promoter and our previous studies have shown that SWI/SNF does not facilitate TFE3-dependent transcription in vitro (Armstrong et al. 1998). Binding analysis of TFE3 and NF-κB (p50) revealed only a weak interaction with one of the SWI/SNF subunits, BAF57 (Fig. 3A, lanes 8–11), which may represent some unincorporated subunit in our SWI/SNF preparation. Further experiments using the chimeric transcription activator, GAL4–VP16, showed only weak association with BRG1 and BAF57 (Fig. 3B, lanes 4,5), but not with the other SWI/SNF subunits. Importantly, SWI/SNF did not bind the ADs of either EKLF (Fig. 3B, lane 8) or VP16 (Fig. 3B, lane 9). Thus three distinct activators (TFE3, NF-κB, and GAL4–VP16) failed to interact significantly with native mammalian SWI/SNF, whereas EKLF and GATA-1 bound SWI/SNF avidly through structurally distinct zinc finger DBDs. Protein overlay (far-Western) experiments using a native flag-tagged hSWI/SNF complex showed that EKLF can interact with BRG1, BAF155, and BAF170 (Fig. 3C, top). Moreover, full-length EKLF and the related zinc finger DBDs of EKLF and Sp1, but not the EKLF AD, also bound to these recombinant SWI/SNF subunits in a GST pull-down assay (Fig. 3C, bottom). These results show that one critical parameter of SWI/SNF functional selectivity is its ability to directly interact with particular protein domains and to be targeted to specific nucleosomal sites. Among the five transcription factors that we have examined, the basis for this selectivity is the presence of structurally distinct zinc finger DBDs (found in either Sp1/EKLF or GATA-1) rather than the other DBDs or ADs represented in this sample.
Figure 3.
SWI/SNF subunits interact specifically with the zinc finger DNA-binding domains (DBDs) of EKLF, GATA-1, and Sp1. (A) Mammalian SWI/SNF interacts with zinc finger DBDs. GST pull-down assays were performed with 3 μg SWI/SNF and 1 μg GST-fused wild-type or mutant EKLF, GATA-1, TFE3, NF-κB (p50). SWI/SNF subunits were detected by Western blot analyses using the antisera indicated on the left as described in Materials and Methods. (B) Acidic and proline-rich activation domains do not interact with mammalian SWI/SNF. GST pull-down assays were performed using 1.5 μg of SWI/SNF and 500 ng each of GST-fused wild-type or mutant EKLF and VP16 proteins. Histidine pull-down assays were performed using 500 ng each of wild-type EKLF, EKLF DBD, wild-type GAL4–VP16, and GAL4 DBD as described in Materials and Methods. Proteins that were pulled down with the beads were analyzed on a 10% SDS-PAGE gel and immunoblotted with antibodies against SWI/SNF subunits, BRG1, BAF170, BAF155, and BAF57. (C) Specific SWI/SNF subunits interact with the EKLF zinc finger DBD. (Top panel) Flag-tagged hSWI/SNF was analyzed by SDS-PAGE, stained with silver (lane 4), blotted onto a PVDF membrane, and processed for far-Western analysis using three different probes: GST–EKLF, followed by anti-EKLF antibody (lane 2), 32P-labeled GST–EKLF (lane 1), or GST–EKLF (AD) followed by anti-EKLF antibody (lane 3) as described in Materials and Methods. The positions of SWI/SNF subunits, BRG1, BAF170, and BAF155 are indicated. (Bottom panel) Wild-type EKLF and the EKLF DBD interact with recombinant SWI/SNF subunits. GST pull-down assays were carried out using 200 ng of purified recombinant F-BRG1, F-BAF170, or F-BAF155 with 200 ng of GST-fused wild-type or mutant EKLF and Sp1 proteins. Equal amounts of supernatants (S) and beads (B) were analyzed on a 10% SDS-PAGE gel and immunoblotted with antibodies against SWI/SNF subunits, BRG1, BAF170, and BAF155.
Recombinant BRG1 and BAF155 subunits cooperate with EKLF to activate the chromatin-assembled β-globin promoter
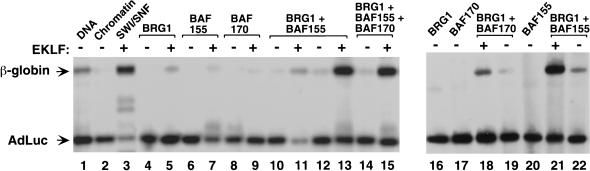
Previous studies have shown that nucleosomal disruption in vitro can be achieved with only a partial SWI/SNF complex or with the ATPase BRG1 alone (Phelan et al. 1999). To define the minimal SWI/SNF complex required for selective regulation by transcriptional activators, we analyzed recombinant SWI/SNF subunits in place of native SWI/SNF complex. As shown in Figure 4, incubation of EKLF with recombinant BRG1 (lane 5), the yeast SWI3 homolog BAF155 (lane 7), or BAF170 (lane 9) did not significantly activate the nucleosome-assembled β-globin gene. In contrast, the combination of BRG1 and BAF155 supported high levels of β-globin transcription (Fig. 4, lane 13), and somewhat weaker transcription was observed when BAF155 was replaced with BAF170 (Fig. 4, cf. lanes 18 and 21). Interestingly, addition of BAF170 into a complex with BRG1 and BAF155 did not further increase transcription (Fig. 4, cf. lanes 13 and 15). Thus a minimal complex composed of two recombinant SWI/SNF subunits is sufficient to facilitate EKLF-dependent transcriptional activation in vitro.
Figure 4.
A minimal recombinant SWI/SNF complex is sufficient for EKLF-dependent transcriptional activation of chromatin-assembled β-globin genes. Recombinant BRG1 and BAF155 or BAF170 cooperate with EKLF to activate chromatin-assembled β-globin genes in vitro. After nucleosome assembly, 100 ng of chromatin was incubated with: 3.7 pmole of EKLF as indicated, 20 ng of F-BRG1 (lanes 4,5,10–16,18–19,21–22), 140 ng of F-BAF170 (lanes 8–9,14–15,17–19), 100 ng of F-BAF155 (lanes 10–11), and 140 ng of F-BAF155 (lanes 12–15,20–22), followed by primer extension analysis of transcripts. As a positive control, an aliquot of 58 ng purified SWI/SNF was incubated with 3.7 pmole of EKLF (lane 3).
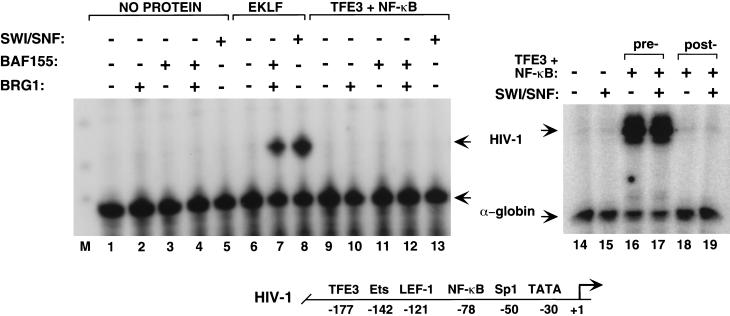
Native and recombinant SWI/SNF function with EKLF but not with TFE3 or NF-κB to activate the chromatin-assembled HIV-1 promoter
To determine whether this minimal SWI/SNF complex still retained its ability to functionally discriminate between classes of transcription factors, we examined whether it could cooperate with TFE3 or NF-κB to activate transcription of chromatin-assembled HIV-1 promoters in vitro. As a positive control, we confirmed that EKLF was able to cooperate with either native SWI/SNF or recombinant BRG1–BAF155 to bind to the Sp1 sites at −50 and to activate transcription from nucleosome-assembled HIV-1 DNA (Fig. 5, lanes 7,8). In contrast, the enhancer factors TFE3 and NF-κB (p50:p65) failed to stimulate transcription from chromatin-assembled HIV-1 templates when incubated with native or recombinant SWI/SNF (Fig. 5, lanes 9–13,19). TFE3 and NF-κB strongly activate HIV-1 transcription when allowed to bind to the template before chromatin assembly (Fig. 5, lanes 16,17), which indicates that the proteins are transcriptionally active. Thus the recombinant BRG1–BAF155 complex retains the functional properties and factor selectivity of the native SWI/SNF complex.
Figure 5.
Recombinant SWI/SNF subunits do not support transcription from chromatin-assembled HIV-1 promoters by TFE3 and NF-κB. Factor-dependent transcriptional activation by native and recombinant SWI/SNF on a chromatin-assembled HIV-1 promoter. (Left ) an aliquot of 100 ng pHIV-1–Luc was assembled into chromatin in the absence of transcription factors (lanes 1–5), or with the following recombinant proteins: 4 pmole EKLF (lanes 6–8) or 5 pmole TFE3 plus one pmole NF-κB subunits (p50:p65; lanes 9–13). Where indicated, 20 ng F-BRG1, 140 ng F-BAF155, and 58 ng native SWI/SNF were added after nucleosome assembly. (Right) A mixture of 5 pmole TFE3 and 1 pmole NF-κB (p50:p65) was incubated with 100 ng HIV-1 chromatin either before (lanes 16–17) or after (lanes 18–19) nucleosome assembly. Transcription reactions contained (lanes 15,17,19) or lacked (lanes 14,16,18) 58 ng native SWI/SNF. The α-globin gene was included as a control for transcription and RNA recovery. A schematic diagram of the HIV-1 promoter is shown below.
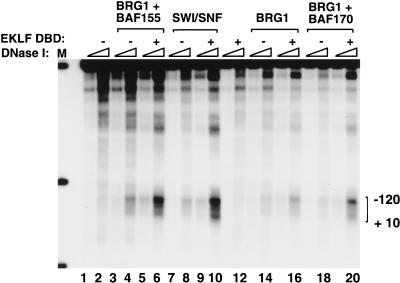
The EKLF zinc finger DBD directs chromatin remodeling by the BRG1–BAF155 minimal complex
We next tested whether the BRG1–BAF155 complex could direct the EKLF zinc finger DBD to specific nucleosomal sites to remodel chromatin in vitro. Importantly, the EKLF DBD was as active as full-length EKLF in its ability to cooperate with native SWI/SNF or the recombinant BRG1–BAF155 complex and generate DNase I hypersensitive sites in the β-globin promoter (Fig. 6, lanes 6,10). Partial remodeling activity was also observed with a recombinant BRG1–BAF170 complex (Fig. 6, lane 20). We conclude that the BRG1–BAF155 minimal complex and a Krüppel-like Sp1/EKLF zinc finger DBD, and presumably the two zinc finger DBD of GATA-1 (Fig. 1B), are together sufficient for targeted chromatin remodeling in vitro.
Figure 6.
The EKLF zinc finger DNA-binding domain (DBD) is sufficient to target chromatin remodeling by the BRG1–BAF155 minimal complex. Analysis of recombinant SWI/SNF subunits to generate DNase I hypersensitive sites within the chromatin-assembled β-globin promoter in the presence of the EKLF DBD. After nucleosome assembly, 300 ng of chromatin was incubated with the following proteins as indicated: recombinant SWI/SNF subunits (60 ng F-BRG1, 400 ng F-BAF155, 400 ng F-BAF170); native SWI/SNF (180 ng); and EKLF DBD (11 pmole). 100 ng of the reaction was transcribed as a control (data not shown) and the remaining chromatin was divided in half and digested with 2 U and 1 U of DNase I as described in Materials and Methods. Triangles indicate increasing amounts of DNase I. (M) Digested β-globin plasmid used as a size marker. Bands represent digests of _Nco_I (4.7 kb), _Nco_I/_Bsa_AI (841 bp) and _Nco_I/_Eco_RI (301 bp).
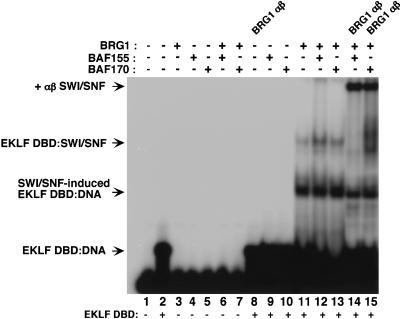
Interaction of SWI/SNF subunits with EKLF DBD–DNA complexes
An important issue is how the interaction of SWI/SNF with a zinc finger DBD affects its ability to bind specific DNA sequences. Incubation of recombinant SWI/SNF subunits with the EKLF DBD and oligonucleotides containing consensus EKLF binding sites generated two novel complexes in gel mobility shift assays. One complex (EKLF DBD–SWI/SNF) was produced by incubation with BRG1 or BRG1 in combination with BAF155 or BAF170 (Fig. 7, lanes 11–13) but not with either BAF subunit alone (Fig. 7, lanes 9,10). This complex was recognized by antisera to BRG1 (Fig. 7, lanes 14,15; BRG1 αβ SWI/SNF) and to the histidine-tag within EKLF (data not shown), indicating that EKLF can bind to DNA simultaneously with this SWI/SNF subunit. Another complex (SWI/SNF-induced EKLF DBD–DNA) was generated upon incubation with BRG1 that contained EKLF but was not recognized by BRG1 antisera (Fig. 7, lanes 11–13). It is possible that the EKLF DBD–DNA complex is stably modified by a transient interaction with BRG1, which does not remain associated under gel mobility shift conditions. The structural nature of the BRG1-associated or -induced EKLF DBD–DNA complexes is unknown. However, these experiments show that the interaction of BRG1 with the EKLF DBD does not prevent its association with DNA, although it may change the nature of EKLF binding.
Figure 7.
Interaction of recombinant SWI/SNF subunits with the EKLF DNA-binding domain (DBD) and DNA. A 60-bp region of the β-globin promoter containing the EKLF binding site was incubated with the following proteins: 10 ng of EKLF DBD (lanes 2,8–15); 20 ng F-BRG1 (lanes 3,6–7,11–15); 140 ng of F-BAF155 (lanes 4,6,9,12,14); 140 ng of F-BAF170 (lanes 5,7,10,13,15) and analyzed by EMSA. One microliter of undiluted antibodies was added as indicated.
Among the SWI/SNF subunits examined, BRG1 is clearly the critical component in recognizing an EKLF DBD–DNA complex and forming a stable association with it or modifying its structure (Fig. 7, lanes 11–13). BAF subunits bind to the EKLF DBD in solution (Fig. 3) but not under gel mobility shift conditions, even in the presence of BRG1. However, the BRG1 ATPase subunit is still not sufficient for EKLF-dependent chromatin remodeling (Fig. 6) or transcription of chromatin templates (Fig. 4) because either BAF155 or BAF170 is required for these processes. Future studies will be directed toward understanding the functional relationship of BRG1 to BAF subunits within a two component SWI/SNF complex in facilitating targeted nucleosome recognition and stable protein interaction.
Discussion
Our studies show that native mammalian SWI/SNF selectively regulates chromatin remodeling through direct interaction with specific DNA-binding proteins. An examination of six transcription factors (EKLF, GATA-1, Sp1, TFE3, NF-κB, and GAL4–VP16) revealed that mammalian SWI/SNF facilitates chromatin remodeling and transcriptional activation of the subset of these factors that contain zinc finger DBDs. Direct protein–protein interactions were observed between mammalian SWI/SNF subunits and the structurally distinct Krüppel-like Sp1/EKLF and GATA-1 zinc finger DBDs. Surprisingly, none of the ADs or other DBDs in this set of six proteins could interact or function with SWI/SNF. We further defined a minimal SWI/SNF complex composed of only two recombinant subunits as being necessary and sufficient for zinc finger–DBD-directed nucleosome remodeling and transcriptional activation in vitro.
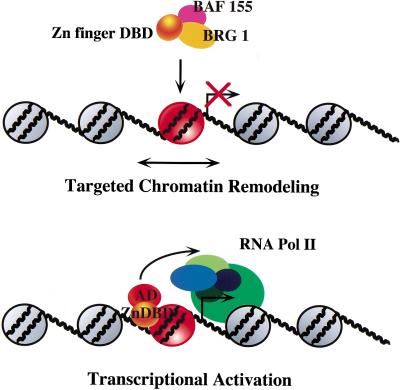
We propose that one mechanism by which mammalian SWI/SNF regulates specific subsets of genes is by interacting with distinct zinc finger DBD structures, through the BRG1 and BAF155 or BAF170 subunits, and targeting the native complex to nucleosomal sites. This results in extended chromatin accessibility (as measured by DNase hypersensitivity) over ∼1 nucleosome and can be achieved in the absence of an AD. Once the critical remodeling step occurs, the AD can recruit other components of the transcription apparatus to promote initiation. A summary of this model is shown in Figure 8. Our experiments support the notion that SWI/SNF does not function with all DNA-binding proteins but only the ones with which it can directly associate. This selectivity may provide the basis for promoter-specific regulation by SWI/SNF in vivo. Importantly, our studies argue against a mechanism by which SWI/SNF transiently disrupts nucleosomes to enable any protein in the vicinity to bind. This is apparent from the inability of native or recombinant mammalian SWI/SNF to facilitate transcriptional activation by TFE3 or NF-κB even when it transiently disrupts nucleosomes on the HIV-1 promoter in vitro. Thus transcriptional regulation by SWI/SNF may require direct association with specific proteins to achieve nucleosome targeting and stable remodeling. Whether SWI/SNF is recruited by proteins that are already bound to the promoter or is directed to specific sites when it is associated with factors in solution is unknown. Our data are consistent with either mechanism because the EKLF DBD interacts with native and recombinant SWI/SNF in solution and when bound to DNA, although we observe negligible EKLF binding to chromatin templates in the absence of SWI/SNF. Through either mechanism, SWI/SNF enhances EKLF binding and remodels adjacent chromatin over at least one nucleosome, which may allow interaction of neighboring transcription factors.
Figure 8.
Model for targeted chromatin remodeling and transcriptional activation, highlighting the interaction between the EKLF zinc finger DNA-binding domain and mammalian SWI/SNF subunits.
A wealth of data show that SWI/SNF can facilitate the binding of many factors to chromatin (Imbalzano et al. 1994; Kwon et al. 1994; Burns and Peterson 1997; Utley et al. 1997; Kingston and Narlikar 1999) and selectively regulate gene expression (Winston and Carlson 1992; Zhao et al. 1998; Dimova et al. 1999). Yeast SWI/SNF complexes appear to be targeted to genes by interaction with specific transactivation domains (Natarajan et al. 1999; Neely et al. 1999; Yudkovsky et al. 1999), similar to chromatin-modifying complexes (Utley et al. 1998). Mammalian SWI/SNF most likely employs several strategies to target transcription factors to nucleosomal sites. For example, recent reports indicate that SWI/SNF may be recruited through interactions between the INI1 subunit and c-MYC DBD (Cheng et al. 1999), or hBRM and the C/EBPβ (Kowenz-Leutz and Leutz 1999) transactivation domain. Our findings highlight an important interaction between structurally distinct zinc finger DBDs and two SWI/SNF subunits, BRG1 and BAF155. In support of this model, the GR zinc finger DBD has been shown to function cooperatively with SWI/SNF in transcription (Yoshinaga et al. 1992; Muchardt and Yaniv 1993), although other reports propose that SWI/SNF is recruited through the GR AD (Wallberg et al. 2000). The observation that chromatin remodeling can occur in the absence of an AD (Pazin et al. 1994; Wong et al. 1997), even though transcription itself is abolished, is consistent with the idea that SWI/SNF can target nucleosome disruption through direct interactions with specific DBDs.
Interactions between the SWI/SNF complex and multiple activators may be important for genes in which SWI/SNF is continuously required for transcription beyond the initial remodeling events (Biggar and Crabtree 1999; Sudarsanam et al. 1999). It is possible that SWI/SNF plays a more direct role by forming a physical bridge between the activator and the transcription machinery that is required to maintain the active state of chromatin (Kingston and Narlikar 1999). SWI/SNF recruitment to a gene may induce a poised state that remains transcriptionally silent, yet competent for initiation when additional cofactors are bound (Struhl 1999). Once recruited to a template, SWI/SNF may also facilitate binding of transcription factors that are otherwise unable to engage remodeling complexes and the interactions may further depend on the environment at the DNA-binding site (Cosma et al. 1999). The observation that the HIV-1 enhancer factors TFE3 and NF-κB cannot bind or function with native or recombinant SWI/SNF raises the possibility that these proteins may recruit distinct classes of chromatin remodeling complexes or depend on other factors to direct remodeling complexes to the HIV-1 promoter. The significance of BAF57 interaction with TFE3 is unclear, and the absence of remaining BAF subunits indicates that some BAF57 may exist as a free subunit in our native SWI/SNF complex.
Much effort is focused on understanding the processes involved in SWI/SNF-dependent regulation using both yeast and human complexes (for review, see Peterson and Workman 2000). Our studies define a minimal mammalian SWI/SNF complex that selectively functions with zinc finger DNA-binding proteins, such as Sp1/EKLF and GATA-1, but not with several unrelated transcription factors, to provide insight on the mechanism and requirements for targeted nucleosome disruption. Further investigations that describe the functional properties of chromatin remodeling complexes and the basis for specificity should help to elaborate the mechanism of gene-selective transcription.
Materials and methods
Plasmid constructions
The β-CAT and HIV-1-Luc plasmids were constructed as described in Jane et al. (1992) and Sheridan et al. (1995), respectively. A full-length EKLF cDNA was cloned into the _Nde_I and _Bam_HI sites of the pET-14b bacterial expression vector (Novagen). GST–EKLF and EKLF mutants (Δ19–60, Δ171–272, Δ293–376, DBD) were constructed as outlined in Bieker and Southwood (1995). The construction of full-length GATA-1, GATA-1 (DBD), and GATA-1 (AD) is described in Hung et al. (1999). Constructs containing full-length TFE3 and the DBDs of TFE3 and Sp1 are outlined in Sheridan et al. (1997) and Merika and Orkin (1995), respectively. TFE3 mutants such as ΔAD-N and ΔAD-C were cloned into the _Xba_I and _Xho_I sites of a pGEX–KG expression vector as described (Guan and Dixon 1991) by introducing amino acids 32–326, 1–193, and 193–326 of TFE3 downstream from GST. NF-κB (P50) was made as described in Pazin et al. (1996).
Protein purification
Histidine-tagged or GST-fusion wild-type and mutant proteins were expressed in Escherichia coli BL21(DE3)pLysS or BL21(DE3) cells, except that 100 μM ZnCl2 was added to the medium during induction for all the zinc finger transcription factors. Histidine-tagged proteins were purified under denaturing conditions on Ni2+-NTA resin (Qiagen) and then renatured by step dialysis. GST-fusion proteins were purified according to the manufacturer's (Pharmacia) protocol, except that 10 μM ZnCl2 was added to all buffers during purification. Human flag-tagged SWI/SNF, F-BRG1, F-hBRM, F-BAF155, and F-BAF170 were purified as described (Phelan et al. 1999). HeLa FL-INI-1–11 cells were grown by the National Cell Culture Center. SWI/SNF from mouse erythroleukemia cells was purified as described (Armstrong et al. 1998) and was functionally equivalent to hSWI/SNF in all assays.
Chromatin assembly and transcription conditions
Chromatin was reconstituted using Drosophila embryonic extracts as described (Armstrong and Emerson 1996). Following assembly, the chromatin template (1 μg in 100 μL) was incubated with wild-type or mutant proteins and SWI/SNF, as described in the Figure legends, for 30 min at 27°C. The reactions were then split in half for either transcription or structural analysis. For transcription, 25 μL nuclear HeLa extract (typically ∼8 mg/mL), prepared as described (Dignam et al. 1983) was added to 0.5 μg of chromatin and incubated on ice for 10 min. Transcription mix was added (20 mM HEPES at pH 7.9; 50 mM KCl; 5 mM MgCl2; 0.2 mg/mL bovine serum albumin; 0.5 mM ATP; 0.5 mM CTP; 0.5 mM UTP; 0.5 mM GTP; 0.7 μg/mL Adeno-luciferase control template [AdLuc]; 1 mM DTT) to a final volume of 150 μL. Transcription proceeded at 30°C for 30 min and was stopped by the addition of 250 μL of transcription stop buffer (1% SDS, 20 mM EDTA). The purified RNA product was analyzed by primer extension analysis.
DNase hypersensitivity analysis
Following incubation of assembled chromatin with transcription factors, 100–150 ng chromatin was digested with DNase I for 1 min at 27°C. The chromatin amounts and nuclease concentrations are given in the Figure legends. Reactions were stopped by the addition of 5× nuclease stop buffer (2.5% sarkosyl, 10 mM EDTA) to a 1× concentration. Purified DNA was digested with _Nco_I and analyzed by Southern blot hybridization (Gene Screen Plus) with a random prime-labeled 301-bp _Eco_RI/_Nco_I fragment from the β–CAT plasmid.
Protein–protein interactions
GST pull-down assays were carried out as described (Garber et al. 1999). Reactions containing 1 μg of wild-type or mutant GST-fusion proteins were incubated with 3 μg of SWI/SNF in 100 μL of binding buffer (50 mM Tris-HCl at pH 8.0, 120 mM NaCl, 0.5% NP-40, 1 mM DTT, and 1 mM PMSF) for 30 min at 27°C. Reactions were then incubated for 1 h at 4°C with 20 μL of glutathione beads and washed three times with binding buffer containing 450 mM NaCl. Histidine-tag pull-down assays were performed using 500 ng of histidine-fusion proteins incubated with 1.5 μg of SWI/SNF in 100 μL of binding buffer (20 mM HEPES at pH 7.9, 20% glycerol, 70 mM KCl, 0.1 mg/ml BSA, and 0.5% Triton-X100) for 30 min at 27°C. Reactions were then incubated with 10 μL of Ni2+ resin and washed three times in buffer (50 mM NaPO4 at pH 6.0, 20% glycerol, 600 mM NaCl, 10 mM β-mercaptoethanol; 0.75% Triton X-100, and 0.75% SDS). The beads were resuspended in 1× loading dye, electrophoresed on a 8% SDS-PAGE gel and blotted onto a polyvinylidene difluoride (PVDF) membrane according to standard procedures. Western blot analyses were carried out using a 1:1000 dilution of primary antisera [anti-BAF250, BRG1, BAF170, BAF155, BAF60a, BAF57, and INI1(BAF47)]. Westerns were developed using Amersham enhanced chemiluminesence reagents.
Far-Western analysis
Far-Western analysis was carried out as described (Edmondson and Roth 1998). Briefly, SWI/SNF was electrophoresed on an 8% SDS-PAGE gel and blotted onto PVDF membranes (Biorad) using standard protocols. The membrane was blocked with 9% milk in 1× TBST (20 mM Tris base at pH 7.6, 137 mM NaCl, 0.1% Tween 20) for 14 hr at 4°C. In three separate experiments, one membrane was probed with 32P-labeled GST–EKLF, followed by autoradiography. The other membranes were probed with GST–EKLF or GST–EKLF (AD) and then incubated with a 1:1000 dilution of monoclonal antibodies directed against EKLF. Western analysis was performed to facilitate alignment with the silver-stained protein gel and identification of interacting SWI/SNF subunits.
Gel shift assays
The 60-bp region of β-globin promoter DNA used for gel shift analyses was generated by polymerase chain reaction amplification from the plasmid β-CAT. The primers (sense 5′-tgtcat cacttagacctca, antisense 5′-tgggagtagattggccaa) used for amplification were 32P end-labeled using T4 polynucleotide kinase. For each 20 μL reaction, 10 ng of EKLF (DBD) was diluted in binding buffer (25 mM HEPES at pH 7.5, 16 mM KCl, 50 mM NaCl, 2 μM ZnCl2, 0.6 mM β-mercaptoethanol, 8% glycerol, 2 mM spermidine) containing 0.25 mg/mL bovine serum albumin (BSA, molecular biology grade, Boehringer Mannheim) and mixed with 200 ng poly(dIdC)·poly(dIdC) competitor DNA (Pharmacia) and 20,000 cpm of 32P-labeled DNA probe (∼0.1 ng). Reactions were incubated for 20 min on ice and electrophoresed on a 5% (39:1 acrylamide:bisacrylamide)/0.25× TBE gel at 150 V (constant voltage) for 6 hr in the cold. Gels were dried and subjected to autoradiography. For the supershift assays, EKLF (DBD) was incubated with the 32P-labeled DNA probe for 10 min on ice, followed by the addition of recombinant SWI/SNF subunits. After an additional 20-min incubation on ice, the samples were electrophoresed on a 5% native polyacrylamide gel. For antibody shift assays, EKLF (DBD) and recombinant SWI/SNF subunits were incubated with 1 μL of undiluted antibody for 30 min on ice before the addition of incubation buffer/BSA, poly(dIdC)·poly(dIdC), and 32P-labeled DNA probe.
Acknowledgments
We thank Drs. James Bieker, Gerd Blobel, Merlin Crossley, Jerry Workman, and Weidong Wang for EKLF, GATA-1, and Sp1 plasmids; GAL4–VP16; and SWI/SNF antisera, respectively. We also acknowledge the services of the National Cell Culture Center. This work was supported by grants from the National Institutes of Health to B.M.E., K.A.J, and R.E.K. and by grants to B.M.E. and K.A.J. from The Mathers Foundation. G.S.M. was supported by the University-wide AIDS Research Program. M.L.P. is a Research Fellow of the National Cancer Institute of Canada.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL emerson@salk.edu; FAX (858) 535-8194.
References
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Emerson BM. NF-E2 disrupts chromatin structure at human β-globin locus control region hypersensitive site 2 in vitro. Mol Cell Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker JJ, Southwood CM. The erythroid Krüppel-like factor transactivation domain is a critical component for cell-specific inducibility of a β-globin promoter. Mol Cell Biol. 1995;15:852–860. doi: 10.1128/mcb.15.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi–Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LG, Peterson CL. The yeast SWI–SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-WG, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle-and developmentally regulated promoter. Cell. 1999;30:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI–SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and co-operate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Roth SY. Interactions of transcriptional regulators with histones. Methods. 1998;15:355–364. doi: 10.1006/meth.1998.0639. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Garber ME, Wei P, Caderas G, Jones KA. Protein and RNA affinity selection techniques using HIV-1 Tat. Methods Enzymol. 1999;306:352–364. doi: 10.1016/s0076-6879(99)06023-1. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J Biol Chem. 1991;266:17026–17030. [PubMed] [Google Scholar]
- Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H, Garnier JM, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. P300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes & Dev. 2000;14:1899–1907. [PMC free article] [PubMed] [Google Scholar]
- Jane SM, Ney PA, Vanin EF, Gumucio DL, Nienhuis AW. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the β-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lee CH, Murphy MR, Lee JS, Chung JH. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc Natl Acad Sci. 1999;96:12311–12315. doi: 10.1073/pnas.96.22.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Sohn H, Kalpana GV, Choe J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature. 1999b;399:487–491. doi: 10.1038/20966. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;1:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Reyes JC, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The mammalian SWI/SNF complex and the control of cell growth. Semin Cell Dev Biol. 1999;10:189–195. doi: 10.1006/scdb.1999.0300. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Zhou H, Winston F, Hinnebusch AG. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright APH, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc Natl Acad Sci. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund-Farrants A-K, Blomquist P, Kwon H, Wrange O. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kamakaka RT, Kadonaga JT. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, Jones KA. NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes & Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI–SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan PL, Mayall TP, Verdin E, Jones KA. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes & Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, Jones KA. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes & Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes & Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Cao Y, Wu L, Laurent BC, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Cote J, Owen-Hughes T, Workman JL. SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Neely KE, Hassan AH, Gustafsson J-A, Workman JL, Wright APH. Recruitment of the SWI–SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol Cell Biol. 2000;20:2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Wong J, Shi Y-B, Wolffe AP. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes & Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]