CCAAT/enhancer binding protein α (C/EBPα)-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation (original) (raw)
Abstract
Earlier work has shown that pre-B cells can be converted into macrophages by the transcription factor CCAAT/enhancer binding protein α at very high frequencies. Using this system, we performed a systematic analysis of whether during transdifferentiation the cells transiently reactivate progenitor-restricted genes or even retrodifferentiate. A transcriptome analysis of transdifferentiating cells showed that most genes are up- or down-regulated continuously, acquiring a macrophage phenotype within 5 d. In addition, we observed the transient reactivation of a subset of immature myeloid markers, as well as low levels of the progenitor markers Kit and FMS-like tyrosine kinase 3 and a few lineage-inappropriate genes. Importantly, however, we were unable to observe the reexpression of cell-surface marker combinations that characterize hematopoietic stem and progenitor cells, including c-Kit and FMS-like tyrosine kinase 3, even when CAAT/enhancer binding protein α was activated in pre-B cells under culture conditions that favor growth of hematopoietic stem and progenitor cells or when the transcription factor was activated in a time-limited fashion. Together, our findings are consistent with the notion that the conversion from pre-B cells to macrophages is mostly direct and does not involve overt retrodifferentiation.
Keywords: cell fate decision, cell reprogramming, hematopoietic differentiation lineage commitment
Transcription factor-induced cell reprogramming has become a major field within stem cell research. Two major types of forced cell-fate changes have been described: the induction of somatic cells into induced pluripotent stem (iPS) cells and the transdifferentiation of cells from one lineage into another (1, 2). The number of transcription factor-mediated lineage conversions has increased steadily in recent years, mostly involving “short jumps” between closely related cell types, although ”long jumps” from mesoderm to either ectoderm or endoderm also have been reported recently (3, 4). It has been suggested that lineage conversions represent direct transitions from one differentiated state into another, with cells “hopping over mountains” within Waddington's epigenetic landscape (5). This argument is based primarily on the findings that transdifferentiation does not require cell divisions (3, 6, 7), that the process is fast, and that no stable intermediates are generated. Consistent with this notion, no reactivation of selected transcription factors characteristic of progenitors could be observed during the conversion of fibroblasts into cardiomyocytes (8) or of exocrine into endocrine pancreatic cells (6), although this question has not been systematically studied. In an interesting example of physiological transdifferentiation in Caenorhabditis elegans, in which a gut epithelial cell transforms into a neuron, the cell transits through an intermediate stage during which it completely erases its identity before redifferentiating into a motoneuron, in a mechanism that requires Uncoordinated family member 3 (unc-3) activity (9).
The transdifferentiation of pre-B cells into macrophages induced by CCAAT/enhancer binding protein α (C/EBPα) constitutes an ideal system to examine whether cells retrodifferentiate or reactivate progenitor genes during the process, because cells can be converted at essentially 100% efficiency within 3–5 d, during which time the population doubles once (10, 11). In addition, the system offers the advantage that hematopoietic stem cells and various intermediate progenitor cells (HSPCs) are defined by specific cell-surface antigen combinations and that expression array databases are available. In this study we asked whether overexpression of C/EBPα induces pre-B cells to “hop over the mountain” or whether it reactivates progenitor traits and markers (Fig. S1_A_). We conclude that, although a few progenitor markers become reactivated, the transdifferentiation process is mostly direct.
Results
C/EBPα Converts the Transcriptome of Pre-B Cells into One Resembling Normal Macrophages and Strongly Represses Cell-Cycle Genes.
To address whether during reprogramming of pre-B cells into macrophage cells transiently reactivates progenitor markers, we analyzed transdifferentiating cells by gene-expression arrays. A prerequisite for these experiments was that the cells studied switch at high frequencies. Earlier work with a pre–B-cell line expressing C/EBPα fused with the estrogen receptor (CEBPαER) had shown that these cells could be converted at 100% efficiency (11), a significant improvement over the 65% conversion observed with primary pre-B cells carrying a wild-type C/EBPα (10). However, a cell line is not suitable for our studies, because as it is not identical to its normal counterparts and is separated from normal progenitors by many population doublings. We therefore tested if primary cells could be switched at high frequencies by preparing bone marrow, sorting primary CD19+ cells, infecting them with C/EBPαER-GFP virus, and seeding them for 2 d on stromal cells (OP9 or S17) in the presence of IL-7. After their expansion, GFP+ cells were sorted again, seeded on stroma under conditions permissive for hematopoietic progenitors, B-lineage cells, and myeloid-lineage cells [stem cell factor (SCF), IL-7, FMS-like tyrosine kinase 3 (Flt3), IL-3, and macrophage colony-stimulating factor (M-CSF)], and induced them with β-estradiol (β-Est). As shown in Fig. S1_B_, the infected cells began to down-regulate CD19 and up-regulate member of AAA family binding CED-4 (Mac-1) at 24 h and turned into fully CD19− Mac-1+ cells within 96–120 h at nearly 100% efficiency. Finally we sorted GFP+ cells from two biological replicates at 0, 3, 12, 48, and 120 h to extract RNA, as well as from bone marrow-derived macrophages cultured under the same conditions.
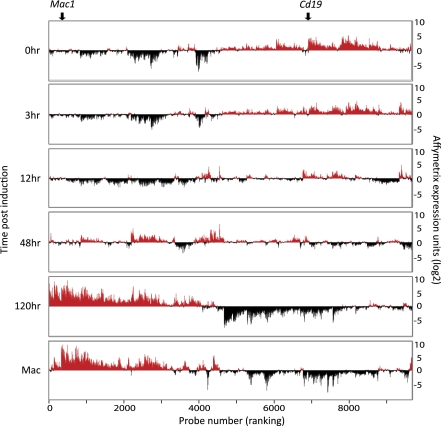
The RNAs were analyzed by Affymetrix 430.2 arrays, containing ∼45,000 gene probes corresponding to 25,000 genes. Of these gene probes, 9,650 changed more than twofold at any time point, with 2,992 probes becoming up-regulated and 3,536 becoming down-regulated (Fig. S1_C_). An unsupervised gene-expression clustering of all genes revealed two predominating groups: genes that became up-regulated (Fig. 1, on the left) and genes that became down-regulated (Fig. 1, on the right). In addition, some genes were transiently up- or down-regulated. The gene-expression pattern obtained 120 h after induction differed dramatically from the starting cells and resembled that of bone marrow-derived macrophages, showing a Pearson correlation coefficient of 0.876 for genes that change and of 0.964 for all genes (Fig. 1 and Fig. S1 D and E). Furthermore, the 120-h expression levels of macrophage and B-cell–associated genes reached levels close to those seen in bone marrow-derived macrophages (Fig. S1_F_). However, many genes became down-regulated to levels below those of normal macrophages (Fig. 1_G_). Functional analysis of these genes using the Database for Annotation, Visualization and Integrated Discovery (DAVID) revealed a high enrichment of genes involved in cell cycle, mitosis, and DNA replication and synthesis. Therefore it is likely that the strong repression of genes in this group is caused mostly by the known cell-cycle inhibitory activity of C/EBPα through its ability to inactivate the transcription factor E2F (12). These findings imply that the majority of genes whose expression changes establish the differentiation phenotype of characteristic macrophages.
Fig 1.
Gene-expression profiles of cells during C/EBPα-induced reprogramming. Unsupervised hierarchical clustering of Affymetrix gene-expression array data of pre-B cells induced for the times indicated. All probes that showed a greater than twofold change in expression at any time during the experiment were included. Negative peaks in black represent probes expressed below the median value; positive peaks in red represent probes expressed above the median value. Affymetrix expression values are indicated in a log2 scale. Positions corresponding to the expression of Mac1 and Cd19 genes are shown above the profiles.
A Small Subset of Myelomonocytic Precursor Genes Becomes Transiently Activated.
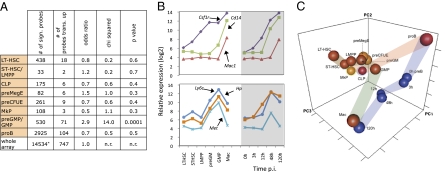
If cells retrodifferentiate during transdifferentiation or diverge into alternative lineages before acquiring their final fate, they should transiently activate hematopoietic precursor or lineage-restricted genes. We therefore compared our array data with gene-expression data from normal hematopoietic progenitors (13, 14). To this end we first determined cell stage-specific signatures consisting of the most highly expressed gene probes in a given cell type relative to all other cell types (sometimes two groups were combined to obtain statistically meaningful numbers). The following stages/groups (Fig. 2_A_) were included: long-term hematopoietic stem cells (LT-HSCs); a combination short-term hematopoietic stem cells (ST-HSCs) and lymphoid-primed multipotential progenitors (LMPPs); common lymphoid progenitors (CPLs); pro-B cells (proBs); granulocyte monocyte precursors (preGMs) plus granulocyte monocyte progenitors (GMPs); megakaryocyte erythroid precursors (preMegEs); erythroid precursors (preCFUEs); and megakaryocyte precursors (MkPs). Among the 14,534 gene probes that showed >1.7-fold changes in expression across all progenitor stages, 747 were transiently up-regulated in at least one time point of the reprogramming process. Of the transiently up-regulated probes only preGM/GMP-specific probes showed a significant enrichment among transiently up-regulated genes (Fig. 2_A_). Specifically, 55 of the 530 signature probes (10.4%) showed their highest expression at 48 h post induction (p.i.), 9 peaked at 12 h p.i., and 5 peaked at 3–12 h p.i. (Table S1 and Fig. S2_A_). Three genes of the 48-h group [lymphocyte antigen 6 complex (Ly6c), Met, and haptoglobin (Hp)] illustrate the concordance of expression with preGM/GMPs (Fig. 2_B_, Lower), whereas three macrophage-specific genes [colony-stimulating factor 1 receptor (Csf1r), Mac1, and _Cd14_] reached their peaks only at 120 h p.i. (Fig. 2_B_, Upper). Ly6C is a granulocyte/monocyte-associated glycosylphosphatidylinositol-linked cell-surface antigen (15), Met corresponds to the hepatocyte growth factor receptor, and haptoglobin is a hormone secreted by granulocytes and hepatocytes (16), but a function during myelopoiesis is not known for any of these proteins. The difference in the timing of expression between Ly6c and Mac1 was confirmed by quantitative RT-PC (qRT-PCR) (Fig. S2_B_) and by FACS analysis (Fig. S2_C_). The observed lack of a global reactivation of progenitor genes also is supported by a principal component analysis showing that the trajectory of the transdifferentiating cells does not deviate substantially toward early progenitors (Fig. 2_C_).
Fig. 2.
Comparative analysis of the transcriptome of transdifferentiating cells with that of normal progenitors reveals enrichment of myeloid progenitor genes. (A) Enrichment of cell stage-specific signatures in transdifferentiating cells. Signature genes were defined as genes more highly expressed in a given cell stage than in all other stages, compiled from 14,534 probesets with a greater than twofold change in expression across all differentiation stages. The table shows the number of signature genes that are transiently up-regulated by more than twofold during reprogramming; the P value indicates the significance of enrichment. (B) Gene-expression values (Affymetrix arrays) of three preGMP/GMP signature genes (Upper) and three macrophage signature genes (Lower) in comparison with transdifferentiating cells. Mac, cultured bone marrow-derived macrophages. (C) Principal component analysis of gene probes that show greater than twofold changes across all samples. Normal lymphoid-myeloid progenitors and differentiated progeny (pro-B cells and macrophages) are shown as red balls, megakaryocyte/erythroid progenitors are shown in orange, and transdifferentiating cells are shown in blue. The ribbons indicate the pathways leading to myeloid differentiation (green), to the B-cell lineage (red), and to transdifferentiation as well as the transition from pro-B to pre-B cells in culture (blue).
Erythroid, Megakaryocytic, and T-Cell Genes Remain Essentially Silent.
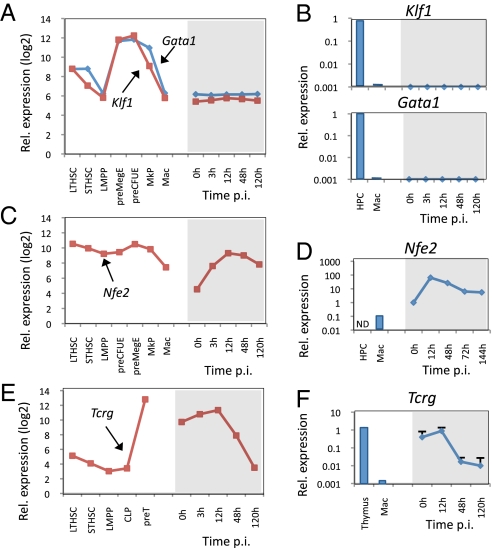
Ablation of the B-cell regulator paired box gene 5 (Pax5) in B cells leads to their dedifferentiation (17), with cells expressing myeloid/erythroid genes such as Csf1r and GATA binding protein 1 (Gata1) as well as T-cell genes including GATA binding protein 3 (Gata3) and pre T-cell antigen receptor α (Ptcra, encoding the pre–T-cell receptor). Because C/EBPα-induced reprogramming involves the rapid down-regulation of Pax5 (Fig. S3_A_), we interrogated our gene-expression database for the reactivation of alternative lineage-restricted genes. We first examined expression of the erythroid lineage-determining transcription factor genes Gata1 and Kruppel-like factor 1 (Klf1) and found that both remained silent (Fig. 3 A and B). We next tested expression of megakaryocyte-specific marker genes. Although Cd41 remained silent, as also was confirmed by FACS, nuclear factor, erythroid derived 2 (Nfe2) showed a peak at 12 h, which also was seen by qRT-PCR (Fig. 3 C and D). We next tested a range of T-cell genes and detected no reactivation of Ptcra or of the T-cell receptor (TCR) genes Tcra, Tcrg-c, and Tcrb-j and the TCR coreceptor genes Cd3e and Cd3g (Fig. S3_B_, Upper). Likewise, genes encoding the T-cell–associated transcription factors Gata3, B-cell leukemia/lymphoma 11B (Bcl11b), and Notch1 remained essentially silent (Fig. S3_B_, Lower). However, Tcrg became transiently activated (Fig. 3 E and F). Together, these data show that, with a few exceptions, alternative lineage markers remain silent during transdifferentiation.
Fig. 3.
Megakaryocyte and T-cell–associated gene expression during transdifferentiation. Comparison of lineage-associated gene expression (Affymetrix arrays and qRT-PCR) in different progenitors/macrophages and transdifferentiating cells. (A and B) Relative expression of the erythroid transcription factor genes Klf1 and Gata1. Hematopoietic progenitors (HPC7 cell line) and macrophages (Mac) were used as controls (blue bars). (C and D) Relative expression of the megakaryocytic transcription factor gene Nfe2. Macrophages (Mac) were used as control. (E and F) Relative expression of the T-cell–associated gene Tcrg. The pre-T (DN3) T-cell line FA2C1 and whole thymus were used as positive controls. Mac, cultured bone marrow-derived macrophages.
Low Levels of Kit and Flt3 mRNAs Become Up-Regulated in a Developmentally Regulated Fashion.
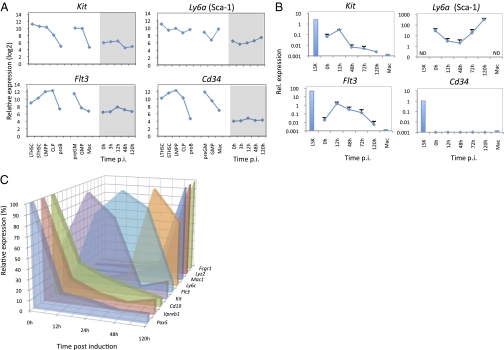
Next we tested the expression of the embryonic stem cell/iPS cell reprogramming genes Oct4, Nanog, SRY-box containing gene 2 (Sox2), Kruppel-like factor 4 (Klf4), and myelocytomatosis oncogene (Myc). No expression of Oct4, Nanog, or Sox2 could be detected (Fig. S4_A_). In contrast, Klf4 became up-regulated, likely reflecting its known function in monocyte differentiation (18), and Myc became down-regulated (Fig. S4_B_), correlating with the gene's role in cell proliferation (19). Next we interrogated our gene-expression database for the transient reactivation of marker genes that phenotypically define HSPCs. As summarized in Fig. S5_A_, LT-HSCs are lineage antigen-negative/low and express Sca-1, c-Kit, and CD150. ST-HSCs and LMPPs are CD150− and CD34+ or Flt3+, respectively (20) (13, 14, 21). CLPs, on the other hand, are also lineage-negative but express Sca-1, Flt3, and IL-7R (22). The expression patterns of Kit, Ly6a (Sca-1), signaling lymphocytic activation molecule family member 1 (Slamf1, CD150), and Cd34 in the various HSPCs (Fig. 4_A_) reflect these progenitor-specific combinatorial codes. During reprogramming Flt3 and Kit became slightly up-regulated at 12 h p.i.; Slamf1, also expressed on non-HSCs, became slightly down-regulated; and Cd34 remained negative at all time points. Finally, Ly6a and Il7r genes were first down-regulated and then up-regulated. qRT-PCR analyses confirmed the transient up-regulation of Kit and Flt3, reaching expression levels at 12 h that were 10- and 45-fold lower, respectively, than in early progenitors (Fig. 4_B_).
Fig. 4.
Progenitor-restricted gene expression during C/EBPα-induced reprogramming. (A) Comparison of Affymetrix gene-expression values in different progenitors and in cells during reprogramming (gene names are shown above the line graphs). (B) qRT-PCR expression values of progenitor genes in cells during reprogramming. Values represent average plus SD of samples from three independent experiments. Positive controls are Lin−/Sca1+/Kit+ progenitors (LSK) and cultured bone marrow-derived macrophages (Mac), indicated by blue bars. (C) Timing of specific genes during reprogramming as determined by Nanostring technology. The highest values obtained in the different time points for each gene (Pax5, CD19, and Vpreb1 at 0 h: 2,046, 10,544, and 37,932, respectively; Kit at 12 h: 156; Flt3 at 24 h: 226; Ly6c at 48 h: 51,786; Mac1, Lyz2, and Fcgr1 at 120 h: 4,787, 36,563, and 23,221, respectively) were normalized to 100%.
To analyze the timing of progenitor markers during transdifferentiation more precisely, we used Nanostring technology, a sensitive and highly reproducible technique for monitoring mRNA expression. We tested the mRNA of Kit, Flt3, and Ly6c and as controls three B-cell markers [Pax5, Cd19, and pre-B lymphocyte gene 1 (Vpreb1)] and three macrophage markers [Mac1 (integrin, alpha M, Itgam), lysozyme 2 (Lyz2), and Fc receptor, IgG, high affinity I (Fcgr1)]. The data in Fig. 4_C_ show that Kit, Flt3, and Ly6c were transiently activated with peaks at 12, 24, and 48 h, respectively, and the B-cell and macrophage markers became down-regulated and up-regulated as expected. This result suggests that the order in which Kit, Flt3, and Ly6c become activated corresponds to their onset during the transition from LT-HSCs to multipotent and myeloid-restricted progenitors.
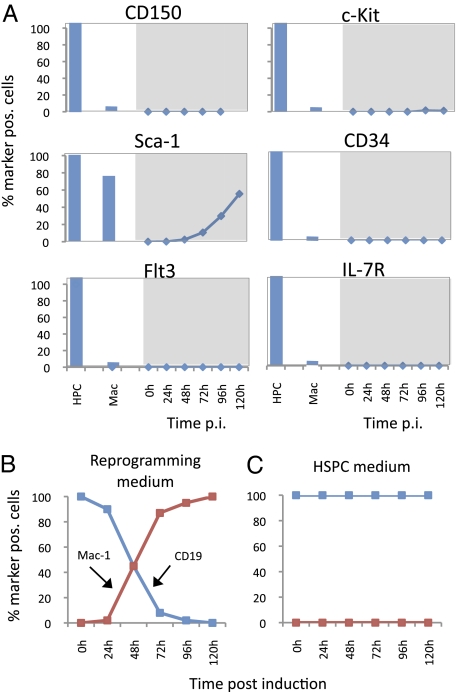
Transitional Stages Do Not Reactivate Cell-Surface Marker Combinations Characteristic of Early Progenitors.
The observed expression of Sca-1, Kit, and Flt3 mRNAs raised the possibility that at least a subset of transdifferentiating cells is positive for the combination of markers characteristic of early hematopoietic progenitors. To test this notion, C/EBPαER-GFP–infected pre-B cells were induced and analyzed by FACS at daily intervals for the expression of the multipotent progenitor antigens CD150, c-Kit, Sca-1, CD34, Flt3, and IL-7R as well as of the differentiation antigens Mac-1 and CD19 as a control. All progenitor markers remained silent, except for Sca-1, which became steadily up-regulated (Fig. 5_A_); Mac-1 and CD19 exhibited the expected reciprocal regulation (Fig. 5_B_). Sca-1 activation was unexpected, but under our culture conditions it acts as a macrophage marker because the antigen also is expressed in cultured bone marrow macrophages. The absence of detectable c-Kit and Flt3 expression suggested that not even a small proportion of the cells expressed these antigens, but it still was possible that proteins were made but were not transported to the membrane. To test this possibility, transdifferentiated cells were permeabilized and stained with c-Kit and Flt3 antibodies. However, neither of the two antigens could be detected by FACS analysis (Fig. S5 C and D). Another possibility is that the B-cell/macrophage culture conditions used were not favorable to capture the transient formation of progenitor-like cells. We therefore repeated the experiments under conditions appropriate for the growth of HSPCs in liquid culture (23) and for the formation of mixed-lineage colonies in semisolid medium. However, we could not observe progenitor cell-surface antigen reactivation under liquid culture conditions favoring HPSC growth (Fig. 5_C_). In addition, no colonies containing more than four cells could be found in methylcellulose cultures containing SCF, IL-3, IL-6, erythropoietin, and TPO (Methocult GF M3434; Stem Cell Technologies) cultures.
Fig. 5.
Progenitor-restricted cell-surface antigen expression during C/EBPα-induced reprogramming. (A) Expression of progenitor cell-surface markers in cells induced with β-Est for various times. Positive controls are as in Fig. 3. (B) Kinetics of differentiation-marker expression of C/EBPαER-infected pre-B cells induced with β-Est under myeloid/B-cell culture conditions. (C) As in B, but induced cells were grown under hematopoietic culture conditions.
Time-Limited Activation of C/EBPα Fails to Induce Progenitor Antigen Expression.
The experiments described so far did not rule out the possibility that progenitor-like cells would form during transdifferentiation if not prevented by continuously active C/EBPα. Earlier work with a C/EBPα-ER–expressing pre–B-cell line had shown that transdifferentiated macrophages, once formed, retain their phenotype even after withdrawal of β-Est, and that a 24-h exposure to the inducer is sufficient to induce an irreversible cell-fate change in the majority of the cells (11). We therefore tested whether primary pre-B cells, maintained B-cell/myeloid cytokines and whether S17 stroma show a similar behavior. For this purpose, cells were infected with C/EBPαER-GFP virus, expanded, treated with β-Est, and, after the inducer was washed out at different times, were analyzed by FACS for the following 5 d (Fig. S6_A_). Although a 12-h β-Est treatment did not cause a stable phenotypic change in the majority of cells, about 90% of the cells treated for 24 h converted into macrophages, and nearly 100% did so after 48 h (Fig. S6_B_). We therefore tested the effect of pulse-inducing the cells for 24 h. As in the previous experiment, CD150, c-Kit, CD34, Flt3, and IL-7R cell-surface antigens remained negative, and Sca-1 became up-regulated (Fig. S6_C_).
Discussion
Our results have shown that C/EBPα-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation, based on gene transcriptome profiling in comparison with normal HSPCs and FACS analyses of cell-surface antigens that define HSCs and various intermediate progenitors. Our results broadly confirm and extend conclusions reached in other systems (6, 8). The lack of retrodifferentiation therefore appears to be a general principle of transdifferentiation that sets it apart from iPS cell reprogramming and from dedifferentiation induced by transcription factor ablation. It will be interesting to determine whether the same principle also applies to transcription factor-induced transdifferentiation between distantly related cell types.
In addition to the majority of genes that become directly up- or down-regulated, we found that a small number of genes become transiently up-regulated. These transiently up-regulated genes fall into three broad classes: (i) Genes that peak at 48 h p.i. (∼10% of the genes most highly expressed in preGMs and GMPs; i.e., the immediate precursors of granulocytes and macrophages). However, none of these genes has been described as being important for myeloid specification, and their relevance for transdifferentiation is questionable. (ii) Selected multipotent progenitor-restricted genes. Here, Kit and Flt3 were found to peak at 12 and 24 h, respectively, their onset recapitulating expression during normal hematopoietic development, where Kit already is expressed on HSCs, and Flt3 becomes expressed from the ST-HSC/LMPP stages onwards (13, 14, 20, 21). However, these genes were not detected at the protein level (see below), and they therefore appear to be irrelevant for transdifferentiation. Of note, CD34, a marker of ST-HSCs, remained negative, whereas Sca-1 became continuously up-regulated at both mRNA and protein levels. However, Sca-1 is expressed on bone marrow-derived macrophages and thus behaves as a myeloid marker under our culture conditions. (iii) Lineage-inappropriate genes. In this category, we observed the transient upregulation of the megakaryocytic regulator Nfe2 and the T cell marker Tcrg at 12–24 h postinjection. Their deregulation might represent a bystander effect resulting from the transition between the B cell and macrophage regulatory networks.
Despite the rapid down-regulation during C/EBPα-induced reprogramming of B-cell master regulators such as Pax5, we observed no reactivation of genes corresponding to the majority of genes restricted to the erythroid and T-cell lineages tested. This absence of reactivation contrasts with the situation when Pax5 is ablated in B-lineage cells (17). A possible explanation is that C/EBPα not only represses B-cell genes but also inhibits erythroid and T-cell genes. Thus, the transcription factor represses erythroid genes in red blood cell lines, and knockout mice exhibit an increase in the number of erythroid cells (24). In addition, it induces the rapid down-regulation of Gata3 and Notch1 in committed T-lineage cells (DN3 and DN4 stages), along with the extinction of the T-cell program (25).
It has been reported that reprogramming of mature B cells by the transcription factors Oct4, Sox2, Klf4, and Myc (OSKM) into iPS cells is enhanced greatly by ectopic expression of C/EBPα (26, 27). Our transcriptome data now offer a possible explanation: C/EBPα-mediated pre–B-cell reprogramming induces the partial up- and down-regulation of many genes, showing that cells coexpress most B-cell– and macrophage-restricted genes at moderate levels 12 and 48 h p.i. (Fig. 1). Because about a fourth of all genes in the genome are involved in this process, it is possible that the chromatin of intermediate-stage cells exhibits a more open configuration than that of cells at either end of the spectrum, without significantly affecting progenitor-restricted genes. The relaxed configuration, in turn, might facilitate the accessibility to OSKM factors, thereby enhancing the frequency with which iPS cells can be obtained. This speculation predicts that in B cells expression of C/EBPα together with alternative lineage-instructive transcription factors might generate cell fates other than macrophages. Such an approach, if feasible, also might be applicable to nonhematopoietic cell types, offering a potential strategy for generating cells desired for cell therapy.
Methods
Cells and Viral Constructs.
B-cell precursors and macrophages were obtained from mouse bone marrow as described (10, 28). The hematopoietic progenitor line HPC7 was kindly provided by L. Carlsson (Umeå Center for Molecular Medicine, Umeå University, Umeå, Sweden) (29). The LSK cell population was sorted as described (30). The construction and production of a murine stem cell virus (MSCV) C/EBPαER internal ribosome entry site (IRES) GFP virus was as described (11).
Cell Reprogramming and FACS Analyses.
Sorted primary B cells were infected for 2 d with C/EBPαER-GFP (infection efficiencies between 30 and 70%), sorted again, induced with 100 nM β-Est (Calbiochem), and grown on S17 cells in special induction medium containing 10 ng/mL IL-7, IL-3, SCF, Flt3 ligand (Peprotech), and human colony-stimulating factor 1 (hCSF-1). For HSC growth conditions, C/EBPαER-infected cells were plated onto OP9 stromal cells plus SCF, TPO, insulin-like growth factor 1 (IGf1), FGF-1, and heparin (23). Antibodies to cell-surface antigens were purchased (BD PharMingen). Cells were analyzed with a FACS LSRII flow cytometer (BD Biosciences), using FlowJo software (Tree Star). For pulse-induction experiments cells were washed thoroughly and incubated with 10 uM of the β-Est antagonist ICI (Tocris Bioscience).
Gene-Expression Profiling by Microarrays and Real-Time RT-PCR.
Biological duplicates of pre-B cells infected with C/EBPαER were induced, and RNA was extracted at various times thereafter, after GFP+ cells were separated from the stromal cells. RNA was extracted with the RNeasy Micro Kit (Qiagen) [quality determined by Bioanalyzer (Agilent 2100)], biotinylated, and amplified in two cycles. The amplified RNAs were hybridized against Affymetrix 430.2 mouse arrays. Gene-expression array data of hematopoietic precursors were from sorted cells (13). qRT-PCR reactions were carried out in triplicate as described (11). Ct values were normalized to glucuronidase beta (GusB), and the relative expression was calculated by the Pfaffl method (31). The Ct values obtained were expressed relative to hematopoietic progenitor cells (Lin−/Kit+) in the case of Cd34, Kit, and Flt3 and to 0-h cells in the case of Ly6c and Met.
Nanostring Gene-Expression Analysis.
Nanostring technology uses molecular barcodes to detect and count mRNA molecules in a digital mode (32). Accuracy and reproducibility were verified by spiking each sample with a dilution series of a known concentration of an RNA control, showing a coefficient of variation of <5% and an essentially linear dynamic range within three orders of magnitude.
Bioinformatics Analyses.
Analysis of the Affymetrix gene-expression data was performed as published (11).
Principal Component Analysis.
Principal component analysis was performed after scaling the values of the data matrix containing all the probes that passed the filter criteria as described above with the stats package in R (version 2.7.0). The image of the three principal components in a 3D scatter plot was generated using the RGL package.
Identification of Cell Type-Specific Genes.
After the raw data from the progenitors were normalized by RMA (Affy v2.7.0), expression values for each probe were averaged among replicates. Probes not reaching 5 (log2 scale) in any cell type were discarded. When a probe's expression value in a specific cell type was greater than 0.8 (log2 scale) as compared with its values in all other cell types, it was considered a signature probe. A signature probe was considered to be transiently up-regulated during transdifferentiation whenever one of its expression values at 3 h, 12 h, and 48 h was more than 1.7-fold higher than its expression value at both 0 h and 120 h. To determine enrichment, we calculated an odds ratio between the transiently up-regulated probes and the total number of signature probes, comparing this enrichment with the enrichment observed in the whole array; we tested the significance of enrichment using the Pearson's χ2 test.
Supplementary Material
Supporting Information
Acknowledgments
We thank S. E. Jacobsen for facilitating the progenitor expression array data, M. Stadtfeld, H. Xie, M. Ye, P. Cosma, and members of the T.G. laboratory for comments, R. Stanley for providing hCSF-1, L. Sumoy, M. Hummel, and G. Castellano for bioinformatics support, and V. Chigancas and L. de Andres for technical assistance. We give special thanks to P. Lundberg, H. Pahl, and R. Roelz for collaborating in the early phase of this work. We received funding from the Ministerio Educación y Ciencia (Grants SAF.2007-63058 and CSD 2006-00049) and from the HEROIC consortium.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data for normal progenitors have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14833). The expression array data for the cells during reprogramming will be submitted to GEO.
This article is a PNAS Direct Submission.
References
- 1.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 2.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 3.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Melton DA. Extreme makeover: Converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinrich C, et al. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richard JP, et al. Direct in vivo cellular reprogramming involves transition through discrete, non-pluripotent steps. Development. 2011;138:1483–1492. doi: 10.1242/dev.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 11.Bussmann LH, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Porse BT, et al. Pedersen TA. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 13.Pronk CJ, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Månsson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 16.Theilgaard-Mönch K, et al. Haptoglobin is synthesized during granulocyte differentiation, stored in specific granules, and released by neutrophils in response to activation. Blood. 2006;108:353–361. doi: 10.1182/blood-2005-09-3890. [DOI] [PubMed] [Google Scholar]
- 17.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg MW, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh AM, Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 21.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhang CC, Lodish HF. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood. 2005;105:4314–4320. doi: 10.1182/blood-2004-11-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suh HC, et al. C/EBPalpha determines hematopoietic cell fate in multipotential progenitor cells by inhibiting erythroid differentiation and inducing myeloid differentiation. Blood. 2006;107:4308–4316. doi: 10.1182/blood-2005-06-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie H, Laiosa CV, Graf T. Reprogramming of committed lymphoid cells by enforced transcription factor expression. Methods Mol Biol. 2010;636:219–232. doi: 10.1007/978-1-60761-691-7_14. [DOI] [PubMed] [Google Scholar]
- 29.Pinto do O P, Richter K, Carlsson L, Pinto do Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood. 2002;99:3939–3946. doi: 10.1182/blood.v99.11.3939. [DOI] [PubMed] [Google Scholar]
- 30.Ye M, et al. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 2003;19:689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- 31.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortina P, Surrey S. Digital mRNA profiling. Nat Biotechnol. 2008;26:293–294. doi: 10.1038/nbt0308-293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information