Molecular mechanisms of long noncoding RNAs (original) (raw)
. Author manuscript; available in PMC: 2012 Sep 16.
Summary
Long noncoding RNAs (lncRNAs) are an important class of pervasive genes involved in a variety of biological functions. Here we discuss the emerging archetypes of molecular functions that lncRNAs execute—as signals, decoys, guides, and scaffolds. For each archetype, examples from several disparate biological contexts illustrate the commonality of the molecular mechanisms, and these mechanistic views provide useful explanations and predictions of biological outcomes. These archetypes of lncRNA function may be a useful framework to consider how lncRNAs acquire useful properties as biological signal transducers, and hint at their possible origins in evolution. As new lncRNAs are being discovered at a rapid pace, the molecular mechanisms of lncRNAs are likely to be enriched and diversified.
Introduction
The conventional view of gene regulation in biology has centered around protein-coding genes via the central dogma of DNA → mRNA → protein. However, over the past decade, evidence from numerous high-throughput genomic platforms suggests that the evolution of developmental processes regulating the complexity of the organism is mainly due to the expansion of regulatory potential of the noncoding portions of the genome (Mattick, 2004). In fact, the portion of the genome responsible for protein coding constitutes approximately 1.5%, while many noncoding regulatory elements are transcribed into noncoding RNA (ncRNA)—the implication being that ncRNAs could play significant regulatory roles in complex organisms. Indeed, the recent explosion in knowledge demonstrating the importance of ncRNAs in the regulation of multiple major biological processes impacting development, differentiation, and metabolism have brought these hereto neglected molecular players to the forefront (Mercer et al., 2009; Ponting et al., 2009; Wilusz et al., 2009).
The complexity of mammalian transcriptome has been highlighted by recent high-throughput studies, which have revealed that tens of thousands of sites are transcribed to produce transcripts with little protein-coding potential—this was most recently demonstrated through ab initio reconstruction (Guttman et al., 2010). In contrast to the small noncoding RNAs (ncRNAs) such as siRNAs, miRNAs, and piRNAs, which are highly conserved and involved in transcriptional and post-transcriptional gene silencing through specific base pairing with their targets, long ncRNAs (lncRNAs)—defined as transcribed RNA molecules greater than 200 nucleotides in length—are poorly conserved and regulate gene expression by diverse mechanisms that are not yet fully understood (Bernstein and Allis, 2005; Bracken and Helin, 2009; Faghihi and Wahlestedt, 2009; Mercer et al., 2009; Whitehead et al., 2009; Wilusz et al., 2009).
Although only a small number of functional lncRNAs have been well characterized to date, they have been shown to control every level of the multi-level regulated gene expression pathway (Wapinski and Chang, 2011). For example, they have long been implicated in post-transcriptional gene regulation through controlling processes like protein synthesis, RNA maturation and transport, and very recently, in transcriptional gene silencing through regulating the chromatin structure (Bernstein and Allis, 2005; Whitehead et al., 2009). Structurally different RNAs engage diverse mechanisms that lead to different regulatory outcomes. Although there is no conservation at the primary sequence level between these RNAs, there are several similarities in their mode of action. Some of the lncRNAs converge on chromatin structure to silence multiple genes located on the overlapping and non-overlapping sides. They interact with DNA and/or chromatin-modifying proteins and recruit them to their target regions. The exact physical association between these lncRNAs and chromatin modifiers and/or gene promoter chromatin remains to be elucidated. Given the large number of lncRNAs whose functions are still to be elucidated, there is clear potential for widespread regulation of chromatin modifications and gene expression.
In this review, we distill the myriad functions of lncRNAs into four archetypes of molecular mechanisms. Each archetype is illustrated with examples drawn from diverse systems and organisms, and we explore similarities and differences between the archetypes to demonstrate the changes in functional complexity. As it will soon become clear, an individual lncRNA may fulfill several archetypes; thus each archetype is not meant to be mutually exclusive. Rather, we aim to illustrate how apparently complex functions can be constructed from combinatorial usage of archetypal molecular mechanisms. By understanding the possible commonalities of the underlying mechanisms, it may facilitate instructive and predictive models of lncRNA function.
Archetype I: Signals
The majority of lncRNAs are transcribed by RNA polymerase II, as evidenced by PolII occupancy, 5‘ caps, histone modifications associated with PolII transcriptional elongation, and polyadenylation (Guttman et al., 2009). LncRNAs show cell type specific expression and respond to diverse stimuli, suggesting that their expression is under considerable transcriptional control. As such, lncRNAs can serve as molecular signals because transcription of individual lncRNAs occurs at very specific time and place to integrate developmental cues, interpret cellular context, or respond to diverse stimuli. Some lncRNAs in this archetype possess regulatory functions, while others are merely by-products of transcription—it is the act of initiation, elongation, or termination that is regulatory. In either case, one can conveniently infer the chromatin state of regulatory elements merely by the expression of their associated lncRNAs. Furthermore, the advantage of using RNA as a medium suggests that potential regulatory functions can be performed quickly without protein translation. Several examples below illustrate lncRNA as signals marking space, time, developmental stage, and expression for gene regulation. Specifically, the lncRNAs in this archetype can act as markers of functionally significant biological events.
Allele specificity: KCNQ1ot1, Air, and Xist
Imprinting is an epigenetic regulatory mechanism that best illustrates the concept of allele specificity. Mammals are diploid organisms carrying two alleles of each autosomal gene, one inherited from the mother and one from the father. Whereas in most cases both parental alleles are expressed equally, a subset of genes show imprinting in which expression is restricted by an epigenetic mechanism to either the maternal or the paternal allele.
Recent emerging evidence indicates that long ncRNAs such as Kcnq1ot1 and Air, which map to the Kcnq1 and Igf2r imprinted gene clusters, respectively, mediate the transcriptional silencing of multiple genes by interacting with chromatin and recruiting the chromatin modifying machinery. For example, in mouse placenta, lncRNAs such as Air and Kcnq1ot1 accumulate at promoter chromatin of silenced alleles and mediate repressive histone modifications in an allele-specific manner (Mohammad et al., 2009). Kcnq1ot1 is a 90-kilobase long noncoding RNA expressed from the paternal allele that directs silencing of a cluster of genes in the imprinted Kcnq1 domain (Pandey et al., 2008). Kcnq1ot1 interacts with the histone methyltransferases G9a and PRC2, effectively forming a repression domain in cis to its transcription site through recruitment of polycomb complexes—the RNA itself seems to play a critical role in the bidirectional silencing of genes in the Kcnq1 domain, thus resembling the mechanisms of action of Xist RNA.
Simiarly, the noncoding RNA Air is imprinted and expressed only from the paternal allele, and its transcription is required for repression of several imprinted genes on the paternal chromosome in a tissue and allele-specific manner. In the placenta, similar to the recruitment of chromatin-modifying activities of Kcnq1ot1, the Air transcription unit initiates in the second intron of the mouse Igf2r gene, and recruits G9a to its target promoter to bring about gene silencing (Nagano et al., 2008). However, in embryonic tissues, Air exerts its effects via a different mechanism, where its own transcription plays a critical role in the silencing of the overlapping gene (Stoger et al., 1993).
X-inactivation (XCI) is a closely related process that equalizes gene expression between mammalian males and females by inactivating one X in female cells. Xist is a well-known lncRNA that plays an essential role in X inactivation (reviewed by Pontier and Gribnau, 2011). During female development, Xist RNA is expressed from the inactive X and “coats” the X chromosome from which it is transcribed, leading to chromosome-wide repression of gene expression. An overlapping antisense lncRNA called Tsix represses Xist expression in cis, while the lncRNA Jpx, whose expression accumulates during XCI, activates Xist on the inactive X (Tian et al., 2010).
In all three examples, presence of the transcribed lncRNA indicates active silencing at their respective genomic locations.
Anatomic specific expression: HOTAIR and HOTTIP
Another example of the tight relationship between time and space is illustrated by two noncoding RNAs from the mammalian Hox loci. In mammals, the homeobox transcription factors (HOX) are organized into four chromosomal clusters, and expressed in a segmental fashion that is collinear between gene position within the cluster and spatial position along the anterior-posterior anatomic axis of the body (reviewed by Wang et al., 2009). Numerous long noncoding RNAs were found to be transcribed from within the human HOX clusters (Rinn et al., 2007) that were expressed in a temporal and site-specific fashion. The lncRNAs were found to be also co-linear with the overall anatomic expression pattern of the HOX loci, and implies that they probably used the same enhancers as the HOX genes. For example, HOTAIR, a lncRNA of the HOXC locus at is expressed in cells with distal and posterior positional identities while Frigidair, another HOXC lncRNA, has an anterior pattern of expression. In contrast, HOTTIP, another lncRNA found at the distal end of the human HOXA cluster, is also expressed in distal cells. In addition to serving as signals of anatomic position, both lncRNAs have additional biological functions, as detailed below.
Induction by DNA damage: linc-p21 and PANDA
Long noncoding RNAs that act to integrate contextual and environmental cues can be found not only during development, but also during times of organismal stress. Huarte et al. (2010) showed that lncRNAs play a key regulatory role in the p53 transcriptional response. One of the direct p53 targets in response to DNA damage, a lncRNA called linc-p21 located upstream of CDKN1A gene, was found to act as a transcriptional repressor in the canonical p53 pathway and to play a role in triggering apoptosis. p53 regulates linc-p21 by directly inducing its expression, likely through direct binding to the linc-p21 promoter, while reduction of lincRNA-p21 increases expression of numerous p53-repressed transcripts. Furthermore, linc-p21 repressed p53 regulated genes through its binding to and modulation of heterogeneous nuclear ribonucleoprotein K (hnRNP-K) localization (Huarte et al., 2010).
Another example of modulation of gene activity in response to external stimuli is found again in the mammalian CDKN1A promoter, where upon DNA damage several lncRNA are transcribed (Hung et al., 2011). One such lncRNA, named PANDA, is also induced in a p53-dependent manner. PANDA cannot be activated by DNA damage in the absence of p53. After DNA damage, p53 directly binds to the CDKN1A locus and activates PANDA; PANDA then interacts with the transcription factor NF-YA to limit expression of pro-apoptotic genes and enables cell cycle arrest, suggesting potentially widespread roles for lncRNAs in cell-growth control.
These results reveal insights into a prototypical transcriptional response and raise the possibility that lncRNAs may act as key regulatory nodes in multiple transcriptional pathways, serving as both a signal and a convenient means of tracking the transcriptional activity of promoters in response to stimuli.
Pluripotency and reprogramming—ROR lncRNA
Pluripotency-associated lincRNAs were initially discovered in mouse embryonic stem cells (ESCs; Guttman et al., 2009), but evidence to support their direct functional relevance has been lacking. Loewer and colleagues show that somatic cell reprogramming to induced pluripotent stem cells (iPSCs) is accompanied by enriched expression of large intergenic non-coding RNAs (lincRNAs; Loewer et al., 2010). One of these reprogramming-induced lincRNAs, named lincRNA-RoR, was shown to be directly targeted by the key pluripotency factors Oct4, Sox2, and Nanog through co-localization of the three factors close its promoter region (Loewer et al., 2010). RoR was downregulated upon Oct4 depletion, as well as during differentiation of iPSCs, implicating co-regulation of specific lincRNAs by key pluripotency factors.
Induction by cold: COLDAIR and COOLAIR
The phenomenon of combinatorial transcriptional regulation by lncRNAs is also found in plants. The transition from vegetative to reproductive development is a highly regulated process that, in many plant species, is sensitive to environmental cues that provide seasonal information to initiate flowering during optimal times of the year. One environmental cue is the cold of winter. Vernalization is an environmentally-induced epigenetic switch in which prolonged exposure to winter cold triggers epigenetic silencing of floral repressors and provides competence to flower in spring (Kim et al., 2009). In the plant Arabidopsis thaliana, winter cold triggers enrichment of trimethylated histone H3 Lys27 at the chromatin of the floral repressor, FLOWERING LOCUS C (FLC), and results in epigenetically stable repression of FLC. This epigenetic change is mediated also through the PRC2 complex. One of the earliest events in this silencing is a large increase in abundance of the antisense transcripts, aptly named COOLAIR, that silences sense FLC transcription and promotes Polycomb occupancy (Swiezewski et al., 2009). The COOLAIR promoter is cold-inducible, and is sufficient to induce cold-dependent silencing of a heterologous reporter construct (Swiezewski et al., 2009). Interestingly, it is apparently the act of 3‘ processing of COOLAIR transcript that triggers FLC silencing (Liu et al., 2010).
More recently, Heo and Sung (2011) showed that a long intronic noncoding RNA termed COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR) is required for the vernalization-mediated epigenetic repression of FLC, via the establishment of stable repressive chromatins at FLC through its physical association with and recruitment of PRC2 to the locus. In contrast to COOLAIR where transcription termination triggers silencing, the COLDAIR lncRNA directly recruits the silencing factors. COLDAIR is transcribed in the sense direction from an intron of its target gene, FLC. FLC harbors a cryptic promoter for COLDAIR ncRNA within its first intron, and this promoter becomes active when FLC is being repressed (Heo and Sung, 2011). The demonstration of a similar interaction of a lncRNA with PRC2 in plants suggests that the noncoding RNA-PRC2 relationship appears to be an evolutionarily conserved mechanism of gene repression. In addition, both COOLAIR and COLDAIR appear to serve as signals of transcriptional activity with spatial and temporal specificity.
Coordinated activity as exemplified by eRNAs
It is well known that regulatory proteins exert their functions by binding stretches of noncoding DNA, either close to a gene's mRNA transcription start site at a promoter, or further away on the genome at an enhancer. Enhancers, in turn, act by helping to recruit the RNA polymerase to the promoter. Recently, a new class of noncoding RNAs—enhancer or eRNAs—have been described that are produced by activity-dependent RNA polymerase II binding of specific enhancers (Kim et al., 2010). The level of eRNA expression at these enhancers positively correlates with the level of messenger RNA synthesis at nearby genes, suggesting that eRNA synthesis occurs specifically at enhancers that are actively engaged in promoting mRNA synthesis (Kim et al., 2010; Wang et al., 2011a). These findings suggest that enhancers have a more active “promoter-like” role in regulating gene expression.
Approaching the same question from a different angle, another group identified a new class of lncRNAs with an enhancer-like function in various human cell lines (Orom et al., 2010). Depletion of these lncRNAs led to decreased expression of their neighboring protein-coding genes, including several master regulators of cellular differentiation. Like classical enhancers, long ncRNAs are orientation-independent and require a minimal promoter in their target genes to enhance their transcription. Although the precise molecular mechanism is yet to be defined, this group of lncRNAs illustrate that eukaryotic transcription is very tightly regulated by overlapping mechanisms. Both examples above provide evidence that RNAs can serve as markers of active regulatory pathways.
Repeat detector: RNA-decay and Staufen
Yet another unforeseen function for lncRNAs was uncovered when a group identified several lncRNAs that are important in downregulating subsets of Staufen 1 (STAU1)-mediated messenger RNA decay (SMD; Gong and Maquat, 2011). SMD regulates diverse classes of mRNAs in mammalian cells that have Staufen 1 binding sites in their 3' un-translated region (3'UTR). It was thought that the Staufen 1 binding site is a cis element with specific secondary structures within the 3'UTR of SMD targets. Interestingly, a group of cytoplasmic lncRNAs, named the half-STAU1-binding site RNAs (1/2-sbsRNAs), was found to facilitate the formation of Staufen 1 binding sites. This was accomplished through imperfect base-pairing, based on Alu repeats, between the lncRNA and the 3'UTR of certain mRNAs, resulting in the degradation of the mRNAs via SMD (Gong and Maquat, 2011). Individual members of this functional class of lncRNAs can downregulate a subset of SMD targets, and distinct lncRNAs can downregulate the same SMD target. These findings reveal a novel role of lncRNAs in mRNA metabolism, specifically, as a mechanism of temporal- and spatial-specific recruitment of proteins to mediate mRNA decay. The idea in which partially complementary lncRNA/mRNA duplexes can form Staufen binding sites is likely to also apply to the regulation by other double-stranded RNA binding protein-dependent pathways.
Thus, the signal archetype of lncRNAs includes not only markers of downstream transcriptional elements but also detectors of transcript abundance/repetition. Taken together, the first archetype of lncRNAs all function as indicators of transcriptional activity, independent of additional functional roles, in a surprisingly straightforward one-to-one relationship.
Archetype II: Decoys
The pervasive transcription of enhancers and promoters (Guenther et al., 2007) hints at a central role for lncRNAs in regulating transcription, both positively and negatively. The means by which such ncRNAs regulate transcription are expanding to encompass a diversity of mechanisms, a major one of which is to act as molecular decoys. This archetype of lncRNAs is transcribed and then binds and titrates away a protein target, but does not exert any additional functions. The RNAs act as a “molecular sink” for RNA-binding proteins (RBPs), which are themselves transcription factors, or chromatin modifiers, or other regulatory factors.
LncRNAs that fit into this functional archetype would presumably act by negatively regulating an effector. Thus, the logical operation is RNA inhibits effector X from executing effector function. Knockdown of the lncRNA should mimick the gain-of-function of the protein partner, and loss of function of both the lncRNA and the effector would be result in a rescue phenotype.
DHFR minor
Alternative promoters within the same gene are a general phenomenon in gene expression (Ayoubi and Van De Ven, 1996). Mechanisms of their selective regulation vary from one gene to another are just beginning to be uncovered. The human dihydrofolate reductase (DHFR) gene is one such locus, and it has been shown to possess an RNA dependent mechanism of transcriptional repression (Martianov et al., 2007). The lncRNA initiated from the upstream minor promoter of the DHFR gene inhibits assembly of the preinitiation complex at the major promoter by forming a stable non-coding RNA-DNA complex with promoter sequences, as well as through direct interactions with the general transcription factor IIB (TFIIB). When the lncRNA was specifically degraded through siRNA knockdown, the occupancy of TFIIB on the major promoter remained high (Martianov et al., 2007). This is a highly dynamic process and presents a specific mechanism that may contribute to promoter targeting and repression, and highlight the importance of intergenic ncRNAs in regulation of neighboring gene expression, by acting as a decoy.
TERRA—telomeres
Telomeres, the DNA-protein complexes located at the physical ends of eukaryotic chromosomes that are essential for chromosome stability, have been found to be transcribed into telomeric repeat containing RNA (TERRA), a lncRNA, which forms an integral part of telomeric heterochromatin (Azzalin et al., 2007). It had been hypothesized that the existence of TERRA RNA hints at a new level of regulation and protection of chromosome ends—this is indeed the case as TERRA has now been demonstrated to physically interact with the telomerase through a repeat sequence complementary to the template sequence of telomerase RNA (Redon et al., 2010). In addition, TERRA also contacts the telomerase reverse transcriptase (TERT) protein subunit independently of the telomerase template RNA moiety. Of note, telomeric heterochromatin bound TERRA is thought to bind and sequester telomerase, in a scenario where TERRA retains telomerase near the telomeric 3‘-end while inhibiting its action (Redon et al., 2010). Additionally, TERRA levels change in a cell cycle-dependent manner—accumulating in early G1, continuously decreases in S phase, and reaches its lowest expression levels at the transition between late S and G2 (Porro et al., 2010). Downregulation of TERRA in S phase might unleash telomerase and allow extension of the telomeric strand in a cell cycle-dependent manner. Thus, telomerase regulation by the telomere substrate may be mediated via its transcription; this is also an example of a natural RNA ligand act as a direct regulator of enzymatic activity, without being a substrate.
PANDA—NF-YA
The lncRNA PANDA, discussed above as a p53-dependent transcript, also appears to possess decoy function. DNA damage can result in apoptosis or cell cycle arrest, and PANDA is very sensitive to DNA damage and its expression is induced temporally ahead of that of CDKN1A. PANDA inhibits expression of apoptotic genes to favor cell cycle arrest, through direct binding to and sequestration of NF-YA, a nuclear transcription factor that activates the apoptotic program upon DNA damage (Hung et al., 2011), resulting in promotion of cell survival (in the context of low level DNA damage) through repression of the apoptotic gene expression program. Depletion of PANDA substantially increased NF-YA occupancy at target genes, while concomitant knockdown of NF-YA and PANDA substantially attenuated induction of apoptotic genes and apoptosis. Interestingly, a subset of human breast cancers overexpress PANDA, and PANDA depletion can sensitize cells to chemotherapeutic agent, hinting at a possible clinical application.
Gas5—Glucocorticoid receptor
Recently, the lncRNA Gas5 (Growth arrest–specific 5) has been identified as a novel mechanism by which cells can create a state of relative glucocorticoid resistance (Kino et al., 2010). Gas5 represses the glucocorticoid receptor through formation of a RNA motif from one of its stem loop structures, mimicking the DNA motif equivalent to that of hormone response elements found in the promoter regions of glucocorticoid-responsive genes. Gas5 then competes for binding to the DNA binding domain of the glucocorticoid receptor, acting as a molecular decoy, and effectively precludes its interaction with the chromosome (Kino et al., 2010). This may turn out to be may be an integral component of the regulatory machinery for modulating steroid hormone activity in target tissue.
LncRNA decoys for miRNAs and splicing factors—pseudogenes, miRNA target mimics, and MALAT1
MicroRNAs, a large class of small non-coding RNAs, have emerged as a critical element in gene regulation by interacting with incompletely complementary sequences in a target messenger RNA (Baek et al., 2008; Bartel, 2009). miRNAs function by annealing to complementary sites on the coding sequences or 3‘ UTRs of target gene transcripts, where they promote recruitment of protein complexes that impair translation and/or decrease the stability of mRNA, leading to a decrease in target protein abundance (Baek et al., 2008; Bartel, 2009). There is now evidence that the converse mechanism may be also in play—i.e., mRNA expression can affect the distribution of miRNAs. Recent work on the tumor suppressor pseudogene PTENP1, previously considered biologically inconsequential, has brought forth the idea that it may have biological function as 'decoys' through sequesteration of miRNAs to affect their regulation of expressed genes (Poliseno et al., 2010). Specifically, the 3‘UTR of PTENP1 RNA was found to bind the same set of regulatory miRNA sequences that normally target the tumor-suppressor gene PTEN, reducing the downregulation of PTEN mRNA and allowing its translation into the tumor-suppressor protein PTEN. In addition to the decoy idea for ncRNAs, this finding nicely illustrates another possible regulatory role for mRNAs in addition to their protein-coding function. Similar relationships likely exist between other cancer-related genes and their pseudogenes (Song et al., 2011). Similarly, in the plant A. thaliana, the noncoding RNA IPS1 binds to and sequester the phosphate-starvation inducible miR-399 through near perfect sequence complementarity, and as a result blunts miR-399 action and alters shoot phosphate content (Franco-Zorrilla et al., 2007).
One of the most abundant nuclear lncRNAs in mammalian cells is MALAT1 (metastasis associated lung adenocarcinoma transcript 1), which is localized in nuclear speckles. MALAT1 binds to and sequesters several serine/arginine (SR) splicing factors to nuclear speckles. Depletion of MALAT1 alters splicing factor localization and activity, leading to altered pattern of alternative splicing for a set of pre-mRNAs (Tripathi et al., 2010). In hippocampal neurons, MALAT1 regulation of SR splicing factors is important for synapse formation (Bernard et al., 2010). Thus, lncRNA decoys can function in nuclear subdomains as well as on chromatin or in the cytoplasm. In sum, these myriad examples illustrate that lncRNA decoys can titrate away proteins and small regulatory RNAs, and likely function in multiple kingdoms of life.
Archetype III: Guides
The third archetype of lncRNA is the guide—RNA binds protein(s), then directs the localization of ribonucleoprotein complex to specific targets. As is evident from the discussions so far, lncRNAs can guide changes in gene expression either in cis (on neighboring genes) or in trans (distantly located genes) in a manner that is not easily predicted based on lncRNA sequence. The myriad of distinctive and likely widespread roles of these RNAs in transcriptional regulation dictates the necessity that certain local changes in chromatin structure can have not only local consequences, but structural repercussions at a distance. Indeed, lncRNAs such as Air and eRNAs appear to exert their effects in cis by spreading from focal sequence elements of transcriptional control such as promoters or enhancers. In contrast, for lncRNAs such as HOTAIR and linc-p21, long range actions of gene regulation require additional ability of components of the interacting partners to be properly localized to their sites of action. In principle, lncRNAs can guide chromatin change in cis in a cotranscriptional manner (tethered by RNA polymerase) or as a complementary target for small regulatory RNAs; guidance in trans can occur by lncRNA binding to target DNA as a RNA:DNA heteroduplex, as RNA:DNA:DNA triplex, or RNA recognition of complex surface of specific chromatin features (reviewed by Hung and Chang, 2010; Bonasio et al., 2010).
The gene regulatory components brought on by the lncRNAs include both repressive (e.g. Polycomb) and activating (MLL) complexes, as well as transcription factors (TFIIB). However, no matter the distance or mechanism (either cis or trans), the principle remains the same: to convey regulatory information across an intervening stretch of DNA to control target gene expression, bringing about changes in the epigenome.
There is further complexity built-in to the archetype in that there are several possible functional classes of effector molecules—activating complexes such as the Trithorax group proteins (TxG), repressive complexes such as the Polycomb group proteins (PcG), as well as the usual collection of transcription factors. Additionally, the effectors can be localized both in cis and in trans. Taking the idea a step further, some lncRNAs may be “tethers” that recruit several chromatin modifications to their sites of synthesis (Lee, 2009), while other lncRNAs may act on distantly located genes as “guides” to affect desired chromatin states. One can then speculate with regards to additional levels of regulation, such as binding of lncRNA by small molecules (e.g. ions, enzyme cofactors, and nucleotide analogs), similar to classic riboswitch interactions (Wachter, 2010), that results in modulation of the RNA-chromatin/RNA-chromatin remodeling enzyme interface.
Key predictions for this archetype of lncRNAs are as follows: knockdown of the lncRNA would change/interfere with the proper localization of the effector molecule, or may phenocopy loss of function of the effector itself; a double knockdown of both the lncRNA and the effector will probably result in exacerbation of the phenotype instead of a rescue as would be expected from the decoy archetype.
Guides in cis: Xist, Air, COLDAIR, rDNA transcript, CCND1, and HOTTIP
Work over the past several years have shed light on the advantages that RNA offers in delivering allelic, cis-limited, and locus-specific control. Perhaps the most intensely studied and best understood cis mechanism of regulation by lncRNAs is the mammalian X inactivation center (Xic), a genetic locus that specifies a number of ncRNAs, including Xist (Plath et al., 2002; Lee, 2010). Xic controls the silencing of one of the two X chromosomes in female mammals, to achieve dosage compensation between the sexes. One of the first changes to occur during the chromosome-wide silencing step of the extra X-chromosome is the recruitment of Polycomb repressive complex 2 (PRC2); PRC2 is brought in cis by RepA RNA, a 1.6 kb ncRNA originating from the 5‘ end of Xist (Wutz et al., 2002). RepA-mediated PRC2 recruitment and Histone 3 lysine 27 trimethylation of the Xist promoter result in the creation of a “heterochromatic state” (Sun et al., 2006) that is required for transcriptional induction of Xist. Spreading of Xist is accompanied by the recruitment of Polycomb and their associated chromatin modifications to the inactive X chromosome (Xi). More recently, a matrix protein hnRNP U was shown to be required for the accumulation of Xist RNA on the Xi (Hasegawa et al., 2010). Xist RNA and hnRNP U interact, and depletion of hnRNP U causes Xist detachment from the Xi and becoming diffusely localized in the nucleoplasm. Thus, X inactivation represents a prominent example of the recruitment of chromatin-modifying activities by lncRNAs and provides a prototype model for a cis localization mechanism.
Similar mechanisms of action appear to be in play for other lncRNAs with transcriptional repression activities. The lncRNA Air silences transcription of its target gene on the paternal chromosome via a specific interaction between the ncRNA and chromatin at its promoter (Nagano et al., 2008); accumulated Air at the promoter recruits G9a and leads to targeted Histone 3 lysine 9 methylation and allelic silencing. The cold-induced plant lncRNA COLDAIR, described above, is required to establish and maintain stable respressive chromatin. COLDAIR plays a critical role in guiding PRC2 complexes to the chromatin of FLC, a strong floral repressor, during vernalization, affecting gene repression through trimethylation of Histone 3 lysine 27 (Heo and Sung, 2011). Similarly in yeast, antisense lncRNAs at numerous gene loci act to silence sense transcription by affecting histone acetylation and methylation states (Camblong et al., 2007; van Dijk et al., 2011). Taken together, these findings suggest a mechanism by which the lncRNAs can function through specific interaction with chromatin to mediate targeted recruitment of repressive histone-modifying activities in cis to epigenetically silence transcription.
The potential of RNA to bind to complementary DNA sequences has led many to hypothesize that RNAs may play important guiding roles in the establishment and transmission of chromatin states. Grummt and colleagues have now shown that this could indeed be a major mechanism by showing that pRNA (promoter-associated RNA), a ncRNA that is complementary to the ribosomal DNA promoter, can form a RNA-DNA triplex structure at the site of binding of TTF-1, the major transcription factor for ribosomal RNA (rRNA) transcription by polymerase I (Schmitz et al., 2010). The triplex was shown to prevent TTF-1 binding while at the same time recruit DNMT3b, a DNA methylase that facilitates promoter methylation and leads to transcriptional silencing of the rRNA gene. This may represent a more general means of epigenetic regulation, by promoter-specific targeting of chromatin modifying enzymes through triplex-forming ncRNAs, with the ncRNAs serving as molecular guides.
lncRNAs are also involved in the regulation of gene expression programs by transcriptional co-activator and co-repressor complexes such as CREB-binding protein (CBP) and p300 histone acetyltransferase (Wang et al., 2008). The RNA-binding protein TLS (translocated in liposarcoma), known to be involved in chromosomal translocations in sarcomas and leukemias, is recruited to chromatin via a tethered lncRNA produced at the cyclin D1 promoter in response to ionizing radiation. Binding of the lncRNA to TLS, in turn, induces a conformation change in TLS that allows its amino terminus to inhibit the histone acetyl transferase activity of p300 and CBP, and thereby inhibit gene expression (Wang et al., 2008). Although the extents to which this particular mechanism is used remains to be established, the identification of highly conserved lncRNAs and the presence of RNA binding domains in a large number of transcriptional co-regulators raise the possibility that promoter-specific lncRNA/coregulator interactions play broad roles in the regulation of gene expression in cis.
The recent identification of HOTTIP lncRNA from the human HoxA cluster (described above; (Wang et al., 2011b) adds an additional dimension to cis regulation by lncRNAs by defining a central role for chromosome looping in delivering a lncRNA to its site of action. By serving as key intermediates that transmit information from higher order chromosomal looping into chromatin modifications, lincRNAs may organize chromatin domains to coordinate long-range gene activation. It is interesting to note that application of genome-wide chromosome conformation capture (3C) technology has recently revealed a vast network of physical interactions between Polycomb group protein target sites (Bantignies et al., 2011). In theory, the chromosome configuration can provide a mechanism by which lncRNAs could regulate transcriptional activity of multiple continguous loci—a phenomenon termed locus control—such as in the HOX clusters.
Guides in trans: HOTAIR, linc-p21, Jpx, other PRC2-boundRNAs
In contrast to the group of cis-regulatory lncRNAs, there are a couple of examples of lncRNAs that exert their transcriptional effects across chromosomes in trans. Expression of the Hox lncRNA HOTAIR has recently been associated with cancer metastasis (Gupta et al., 2010). Elevated expression of HOTAIR is observed in primary and metastatic breast cancer. Furthermore, depletion of HOTAIR from cancer cells leads to a reduced invasiveness of cells that express a high level of Polycomb proteins (PRC2; Gupta et al., 2010). These findings suggest that noncoding RNA-mediated targeting of Polycomb complexes is a crucial event in breast tumorigenesis. Specifically, the implication is that lncRNAs such as HOTAIR are able to alter and regulate epigenetic states in cells through their targeting of chromatin modifying complex occupancy/localization/enzymatic activity in trans. In support of this idea, multiple lncRNAs expressed in various cell types bind PRC2, and that siRNA-mediated depletion of a number of these lncRNAs led to enrichment for genes normally repressed by PRC2, akin to a partial PRC2 knockdown phenotype (Khalil et al., 2009; Zhao et al., 2010).
Jpx, the lncRNA important for activation of Xist RNA on the inactive X, is developmentally regulated and accumulates during XCI (Tian et al., 2010). Deleting Jpx blocks XCI, and post-transcriptional knockdown of Jpx recapitulates the knockout phenotype. Moreover, supplying Jpx in trans rescues the Jpx knockout (Tian et al., 2010). The mechanism by which Jpx transactivates Xist RNA is not yet understood, but perhaps involves loading of the Polycomb complex onto the Xist promoter to create a permissive state for Xist transactivation.
LincRNA-p21 is able to exert its effects on chromatin structure and gene expression across multiple sites in the genome (Huarte et al., 2010),. Ectopic expression of lincRNA-p21 induces gene expression changes and apoptosis, bypassing the upstream regulator p53. It remains to be determined how the repressive complex associated with the p53 induced lncRNA, lincRNA-p21 recognizes targeted loci, or how the complex silences transcription.
Archetype IV: Scaffolds
lncRNAs can serve as central platforms upon which relevant molecular components are assembled; in many diverse biological signaling processes this characteristic of precise control is vital to the precise control of the specificity and dynamics of intermolecular interactions and signaling events (Spitale et al., 2011). Traditionally, proteins were thought to be the major players in various scaffolding complexes (Good et al., 2011). Recent evidence, however, raise the possibility that lncRNAs may also play a similar role.
The fourth archetypal class of lncRNAs is the scaffolds. This is perhaps the functionally most intricate and complex class where the lncRNA possesses different domains that bind distinct effector molecules. The lncRNA would bind its multiple effector partners at the same time, and by doing so brings the effectors, which may have transcriptional activating or repressive activities, together in both time and space. Once a greater understanding of how these scaffolding complexes are assembled and regulated is achieved, it would then be possible to design strategies to selectively utilize specific signaling components and outputs to redirect and reshape cellular behavior. This would be analogous to assembly of an integrated circuit board for molecular scaffolds that functions to dictate flow of information.
Key predictions for this archetype of lncRNAs would include the following: knockdown of the lncRNA would change/interfere with the proper localization of the effector molecule, or may phenocopy loss of function of the component effector itself, through dismantling of the lncRNA-effector scaffold such that the components no longer assemble together. A double knockdown of both the lncRNA and the effector(s) will be expected to result in exacerbation of the phenotype instead of a rescue, as would be expected from the decoy archetype. Finally, disruption of distinct regions of the lncRNA should affect different effector partners and function.
TERC—assembles telomerase
The telomerase is a specialized reverse transcriptase conserved in almost all eukaryotes, and plays a fundamental role in maintenance of genome stability by adding back telomeric DNA repeats lost from chromosome ends. The protein and RNA subunits of telomerase fold and function in a co-dependent manner to establish a high fidelity of telomeric repeat synthesis (Lustig, 2004). Telomerase catalytic activity requires the association of two universal telomerase subunits: an integral RNA subunit, the telomerase RNA (TERC) that provides the template for repeat synthesis, and a catalytic protein subunit, the telomerase reverse transcriptase (TERT), as well as several species-specific accessory proteins. The TERC, in particular, also possesses structures that contribute to TERT binding and catalytic activity, in addition to those that play major roles in stability of the complex (Collins, 2008).
In particular, RNA domains have been identified that affect template usage (Chen and Greider, 2003; Lai et al., 2003) as well as TERT association (Ly et al., 2003). Mutations that alter the equilibrium between different conformational states of TERCs result in disease states such as dyskeratosis congenita (Chen and Greider, 2004), presumably through disruptions of the RNA scaffold structure into which are plugged modular binding sites for telomeric regulatory proteins. Thus, the primary functional role for TERC appears to be that of a scaffold. In an elegant demonstration of this concept, Zappula and Cech engineered functionally active mini-telomerase RNAs that reassembled the telomerase complex in budding yeast, by stitching together minimal RNA motifs (Zappulla and Cech, 2004). This evidence suggests that the telomerase RNA serves as a loosely ordered flexible scaffold for its protein subunits.
HOTAIR—PRC2 and LSD1, resolution of bivalent chromatin
On the basis of their dynamic patterns of expression (Guttman et al., 2009), specific lncRNAs can potentially integrate and direct complex patterns of chromatin states at specific target loci in spatially and temporally specific manner during both organismal development and disease.
As mentioned above, the lncRNA HOTAIR binds the polycomb complex PRC2, which methylates histone H3 on lysine 27 to promote gene repression (Rinn et al., 2007); the fragment responsible for PRC2 binding was recently identified to be the first three hundred nucleotides in the 5‘ end of the lncRNA (Tsai et al., 2010). In addition, the 3‘ seven hundred nucleotides of HOTAIR was found to also interact with a second complex containing LSD1, CoREST, and REST, that demethylates histone H3 on lysine 4 to antagonize gene activation (Tsai et al., 2010). This finding shows that multiple chromatin-modifying complexes are targeted by HOTAIR, suggesting that HOTAIR acts as a scaffold and bridges between PRC2 and the LSD1/CoREST/REST complex—in one package, the HOTAIR/PRC2/LSD1 complex can suppress gene expression via multiple mechanisms at the same time. Indeed HOTAIR expression can induce the interaction of PRC2 and LSD1 complexes, while depletion of HOTAIR led to loss of occupancy of both complexes from target genes. Notably, many additional lincRNAs can interact with both PRC2 and LSD1 complexes in several cell types (Khalil et al., 2009). It is quite possible that other lincRNAs may also contain multiple binding sites where distinct protein complexes can assemble to more specifically bring forth specific combinations of histone modifications on target gene chromatin. KCNQ1ot1 may perform an analogous function for PRC2 and G9a, mediating H3K27me3 and H3K9me3 (Pandey et al., 2008).
ANRIL—PRC2 and PRC1
The molecular interplay between lncRNAs and chromatin modifying complexes can also be found in the transcriptional repression of the well-studied INK4a locus. Expression of the INK4b/ARF/INK4a tumor suppressor locus in normal and cancerous cells is controlled by methylation of histone H3 at lysine 27 as directed by the Polycomb group proteins (Gil and Peters, 2006). The antisense noncoding RNA, ANRIL that emanates from the same INK4b/ARF/INK4a locus is also important for expression of the protein-coding genes in cis. Work over the past few years have demonstrated a direct interaction between ANRIL and components from both PRC1 and PRC2 complexes (Kotake et al., 2011; Yap et al., 2010). Binding to ANRIL contributes to the functions of both PRC1 and PRC2 proteins, and disruption of either interaction impacts transcriptional repression of the target INK4b locus. Thus, similar to HOTAIR, ANRIL represents a prototype of a lncRNA that is always present at the locus, recruits multiple sets of chromatin modifying complexes to the target gene for silencing, serving as a molecular scaffold to dynamically modulate transcriptional activity. In addition, ANRIL provides a powerful system with which to further investigate the biological significance of RNA-mediated targeting of Polycomb. X inactivation may also involve RNA scaffolding of both PRC1 and PRC2 (Bernstein et al., 2006; Zhao et al., 2008).
Pericentric heterochromatin—alpha satellite repeat lncRNA
In recent years, a role for non-coding RNA in heterochromatin establishment has emerged. Heterochromatin at pericentric satellites, characterized by a specific chromatin signature and chromocenter organization, is of paramount importance for genome function (Probst and Almouzni, 2011). Maison and colleagues provide evidence for the presence of long nuclear noncoding transcripts corresponding to major satellite repeats at the periphery of pericentric heterochromatin, and that major transcripts of these lncRNAs in the forward orientation specifically associate with Small Ubiquitin-like Modifier (SUMO)-modified heterochromatin protein 1 (HP1) proteins (Maison et al., 2011). The forward RNA associates with and provides specificity to the initial targeting of the SUMO-HP1 complex at pericentric heterochromatin to seed further HP1 localization. Thus, the lncRNA acts as a molecular scaffold for the targeting and local accumulation of HP1. It would be interesting to investigate if similar principles are exploited in biological situations where major nuclear reorganization occurs.
Integrated function
Not surprisingly, several lncRNAs possess characteristics from multiple archetypes that, in combination, are critical to its eventual biological function. For example, COLDAIR and COOLAIR are transcribed in response to the environmental cue of cold temperature—their transcription serves as a signal of a significant biological event, in this case the preparations for competence to flower after a prolonged winter. Epigenetic repression of floral repressors is then achieved through binding of PRC2 by COLDAIR, with the lncRNA serving as a guide to affect silencing at the FLC locus to bring about the biological effect of vernalization. The lncRNA HOTTIP is another example of a “signal plus guide” combination archetype—it is transcribed in a temporal and spatial manner along with the rest of the distal HOXA genes to convey positional identity, and functions by binding to and targeting the trithorax protein complex Mixed Lineage in Leukemia-1 (MLL-1) to the 5‘ HOXA locus to drive histone methylation and gene transcription.
Another combinatorial archetype is exemplified by HOTAIR. Like HOTTIP, HOTAIR is transcribed in posterior and distal cells, acting as a signal for anatomic specificity. By bindings to both the PRC2 and LSD1 complexes, HOTAIR serves as a modular scaffold, and by targeting PRC2 to its proper genomic locations it acts as a guide. Thus, the desired biological outcome—positional identity and appropriate chromatin modifications leading to proper gene expression—is ultimately achieved through a functional multi-functional lncRNA.
One emerging theme from the analysis of the four lncRNA archetypes is that of stepwise complexity. When one considers each of the archetype classes from an evolutionary perspective, it is a strikingly simple process of incremental modifications that confer alterations in molecular utility. The simple signal archetype lncRNA, such as eRNAs, merely requires the transcription of a regulatory DNA element. If the lncRNA that is produced also binds a protein due to the formation of a RNA motif mimicking its DNA counterpart, as is the case for Gas5, then the lncRNA develops into a molecular decoy. If the lncRNA then gains the ability to target the bound effectors to a specific DNA sequence either in cis or trans, it transitions to become a guide. With nucleic acid duplication, fusion, and recombination events, it is not far-fetched to imagine that lncRNAs may subsequently acquire multiple effector binding sites to turn into a scaffold. This step-wise scenario is potentially quite likely because the regulatory DNA being transcribed, such as enhancers, by definition possess high affinity transcription factor binding sequences, and often in tandem and combinatorial arrangements. Thus, the primordial lncRNA “signals” may often contain functional seeds to become decoys, guides, and scaffolds.
In fact, experimental evolution hints at the feasibility of evolving many new lncRNA regulators of gene expression. Work by Kehayova and Liu (2007) highlights the value of RNA evolution, and argue that the polyanionic characteristic, in combination with the great structural and functional diversity of RNA, makes it especially well suited to mediate processes that involve proteins with cationic patches (Kehayova and Liu, 2007). In fact, RNA-based transcript regulators are relatively easy to evolve, on the order of 104, in contrast to in vitro selections for RNA aptamers for a specific ligand or protein, in which the rate of active RNAs among random library members is approximately 1 in 1010 to 1014 (Kehayova and Liu, 2007).
Polymorphisms and mutations in regulatory regions are increasingly shown to be associated with human disease. However, currently, we are only observing the tip of the iceberg. It is becoming clear that many common disease-association studies are identifying noncoding region variants as the underlying cause of these later onset disorders. It will be exciting, and potentially useful for disease management and treatment, to see what aspects of fine tuning are altered in different anomalies. Areas for future exploration will include the mechanisms through which physiological and environmental changes are translated into altered gene function through lncRNAs and their regulatory networks. We hope that we have provided logic and experimental evidence to support the archetypal classifications of lncRNAs as a useful framework. As more examples of regulation by long ncRNA are uncovered, one might predict that the large transcripts will eventually rival small RNAs and proteins in their versatility as regulators of genetic information.
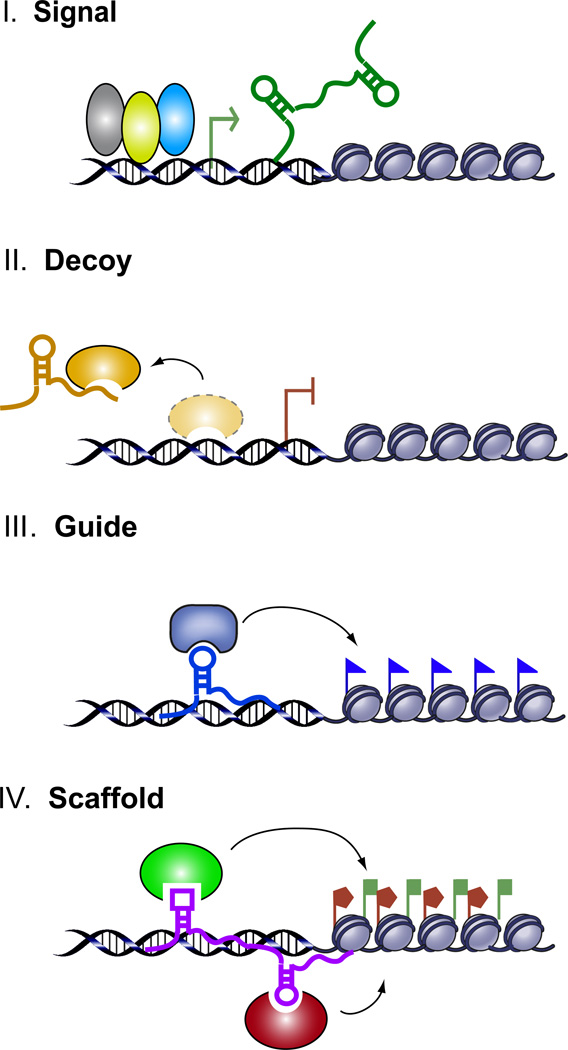
Figure 1.
Schematic diagram of the four archetypes of lncRNA mechanism. Archetype I: As Signals, lncRNA expression can faithfully reflect the combinatorial actions of transcription factors (colored ovals) or signaling pathways to indicate gene regulation in space and time. Archetype II: As Decoys, lncRNAs can titrate away transcription factors and other proteins away from chromatin, or titrate the protein factors into nuclear subdomains. A further example of decoys is lncRNA decoy for miRNA target sites (not shown on schematic). Archetype III: As Guides, lncRNAs can recruit chromatin modifying enzymes to target genes, either in cis (near the site of lncRNA production) or in trans to distant target genes. Archetype IV: As scaffolds, lncRNAs can bring together multiple proteins to form ribonucleoprotein complexes. The lncRNA-RNP may act on chromatin as illustrated to affect histone modifications. In other instances, the lncRNA scaffold is structural and stabilizes nuclear structures or signaling complexes
Acknowledgments
We apologize to colleagues whose work was not discussed due to space constraints. We thank R. Flynn and O. Wapinski for critical reading of the manuscript and assistance with the figure. Supported by NIH and California Institute for Regenerative Medicine (H.Y.C.). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayoubi TA, Van De Ven WJ. Regulation of gene expression by alternative promoters. FASEB J. 1996;10:453–460. [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet.;2011 doi: 10.1038/ng.848. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehayova PD, Liu DR. In vivo evolution of an RNA-based transcriptional silencing domain in S. cerevisiae. Chem Biol. 2007;14:65–74. doi: 10.1016/j.chembiol.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. The X as model for RNA's niche in epigenomic regulation. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a003749. a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ. Telomerase RNA: a flexible RNA scaffold for telomerase biosynthesis. Curr Biol. 2004;14:R565–R567. doi: 10.1016/j.cub.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Ly H, Blackburn EH, Parslow TG. Comprehensive structure-function analysis of the core domain of human telomerase RNA. Mol Cell Biol. 2003;23:6849–6856. doi: 10.1128/MCB.23.19.6849-6856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. NAT Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier DB, Gribnau J. Xist regulation and function eXplored. Hum Genet. 2011 doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Almouzni G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 2011;27:177–185. doi: 10.1016/j.tig.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, Pandolfi PP. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011a doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009;19:268–275. doi: 10.1016/j.tcb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011b;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790:936–947. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]