FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis (original) (raw)
. Author manuscript; available in PMC: 2012 Feb 23.
Published in final edited form as: Cell Stem Cell. 2009 Nov 6;5(5):540–553. doi: 10.1016/j.stem.2009.09.013
Summary
The PI3K-AKT-FoxO pathway is integral to lifespan regulation in lower organisms and essential for the stability of long-lived cells in mammals. Here, we report the impact of combined FoxO1, 3 and 4 deficiencies on mammalian brain physiology with a particular emphasis on the study of the neural stem/progenitor cell (NSC) pool. We show that the FoxO family plays a prominent role in NSC proliferation and renewal. FoxO deficient mice show initial increased brain size and proliferation of neural progenitor cells during early postnatal life, followed by precocious significant decline in the NSC pool and accompanying neurogenesis in adult brains. Mechanistically, integrated transcriptomic, promoter and functional analyses of FoxO deficient NSC cultures identified direct gene targets with known links to the regulation of human brain size and the control of cellular proliferation, differentiation, and oxidative defense. Thus, the FoxO family coordinately regulates diverse genes and pathways to govern key aspects of NSC homeostasis in the mammalian brain.
Introduction
The PI3K-AKT-FoxO signaling pathway plays a central role in diverse physiological processes relevant to aging and cancer. This signaling axis translates extracellular cues into intracellular gene expression changes to maintain cellular homeostasis. In line with genetic studies in C_. elegans and D. melanogaster_ revealing an epistatic relationship across PI3K-AKT-FoxO, the mammalian FoxO family of transcription factors (FoxO1, 3 and 4, hereafter referred collectively as ‘FoxOs’) also act as key downstream effectors of PI3K-AKT pathway and function to control many genes governing various cellular processes such as proliferation, survival, metabolism, differentiation, and oxidative defense (reviewed in Greer and Brunet, 2005). Recent studies from our group have underscored that the mammalian FoxOs function redundantly to promote the stability of long-lived cells such as thymocytes and endothelial cells and to maintain hematopoietic stem cell (HSC) reserves which produce countless blood cells over a life-time. To control these processes across these various systems, the FoxOs have been shown to regulate distinct sets of gene targets depending on cell type, tissue context and developmental stage (Paik et al., 2007; Tothova et al., 2007).
The mammalian brain is a dynamic organ that maintains the capacity to generate new neurons as a function of advancing age and following brain injury. This regenerative capacity derives from resident neural stem cells (NSC) which contribute directly to neurogenesis throughout development and adulthood. Processes including the proliferation, differentiation and fate determination of NSC and progenitor cells, and the survival, maturation, and integration of newborn neurons into existing neural circuits have been shown to be necessary steps in adult neurogenesis. NSC resides within the ventricular zone of the embryonic neural tube and, in adult brain, primarily in the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the hippocampal dentate gyrus (DG). In adult mice, relatively quiescent NSC give rise to transit-amplifying progenitor cells which rapidly divide and contribute to neuroblasts (Morshead et al., 1994; reviewed in Zhao et al., 2008). Newly generated neuroblasts in the SVZ migrate to become granule and periglomerular neurons in the olfactory bulb and neurons born in the SGZ migrate into the granule cell layer of the DG and. become dentate granule cells, contributing to the maintenance and reorganization of the existing circuitry and affecting memory and behavior (Imayoshi et al., 2008; reviewed in Zhao et al., 2008).
Recent studies have identified a number of pathways, including the PI3K-Akt, PTEN, and TSC, governing embryonic and tissue stem cell self-renewal, maintenance and regenerative responses (Gan et al., 2008; Groszer et al., 2006; Groszer et al., 2001; Paling et al., 2004; Yilmaz et al., 2006). Extracellular cues present within the stem cell niche include Wnt, Hedgehog, bone morphogenetic protein (BMP), fibroblast growth factor (FGF), epidermal growth factor (EGF), and the transforming growth factor (TGFβ), all of which have been shown to affect self renewal capacity both in vivo and in vitro (Battista et al., 2006; Doetsch et al., 2002a; Jin et al., 2003; Palma et al., 2005; Reya et al., 2003). Furthermore, several studies have underscored the importance of finely tuned regulation of cell cycle/quiescence, apoptosis, and intracellular redox status in the maintenance and fate determination within tissue stem cell compartment (Cheng et al., 2000a; Cheng et al., 2000b; Doetsch et al., 2002b; Ito et al., 2004; Ito et al., 2006; Kippin et al., 2005; Kozar et al., 2004; Malumbres et al., 2004; Yuan et al., 2004). The molecular mechanisms by which the PI3K-Akt pathway and the FoxO family in particular, regulate NSC self-renewal, proliferation, and differentiation in the developing or mature brain are areas of active investigation.
On the basis of the fact that FoxOs had been reported to influence each of these processes in other systems (reviewed in Greer and Brunet, 2005), and the importance of the FoxOs in the maintenance of HSC reserves (Miyamoto et al., 2007; Tothova et al., 2007), and finally the highly cell type-specific nature of the FoxO action (Paik et al., 2007), we sought to gain an in-depth understanding of the specific functions of the FoxOs in the mammalian central nervous system (CNS) with an emphasis on the growth and renewal potential of neural stem and progenitor cells (hereafter referred to collectively as “NSC”). Here, an integrated computational, molecular, cellular and organismal analysis has identified and validated FoxO direct targets and their linked pathways and biological processes that serve to regulate various aspects of NSC biology including proliferation, survival, renewal and oxidative defense. This multi-level analysis establishes FoxOs as key molecules in processes central to NSC homeostasis and defines the genes through which the FoxOs execute their biological actions in this critical stem cell system.
Results
FoxOs constrain proliferation and sustain renewal of NSC
Immunohistochemical analysis of wild type adult mouse brain shows that FoxO 1 and 3 are broadly expressed in multiple cell types including neurons (NeuN+), astrocytes (GFAP+), and oligodendrocytes (Olig2+), in different locations as reported previously (Supplemental Figure S1a and m-q) (Hoekman et al., 2006). In particular, we documented high levels of FoxO1 and 3 expression in the multipotent progenitors (Sox2+) of the SVZ in the forebrain and the SGZ in the hippocampus (Supplemental Figure S1c, e, i, m, and n). In the embryonic brain (E15.5), FoxO1 and 3 also had broad and partially overlapping patterns of expression particularly within the NSC-rich ventricular zone (VZ). Within the VZ, FoxO1 expression is somewhat restricted with the highest level of expression noted in Nestin-positive radial glial progenitors of the dorso-medial wall of the VZ; in contrast, FoxO3 is expressed more broadly throughout all regions of the VZ examined (Supplemental Figure S1j-l). As further evidence for their potential functional relevance in the NSC, we also detected high level expression of FoxO mRNA and proteins in NSC-containing neurospheres cultured from embryonic and postnatal brains (Supplemental Figures S2). The prominent expression of FoxO 1 and 3 in NSC-rich regions of the developing and adult brain, and FoxO 1, 3 and 4 expression in cultured NSC-containing neurospheres from embryonic and adult brain prompted further study of the FoxO family in these cells.
To circumvent embryonic lethality associated with impaired vascular development due to broad FoxO1 deficiency (Hosaka et al., 2004), we utilized conditional knockout alleles of FoxO1, 3 and 4 (FoxO1/3/4L/L) to achieve somatic inactivation of FoxO in the mouse brain. Here, we selected a particular hGFAP-Cre transgenic line that has been well documented in multiple published studies to be highly active in neural stem/progenitor cells (Supplemental Figure S1g and h)(Zheng et al., 2008; Zhuo et al., 2001; Lim et al., 2009) although it is also active in developing and mature astrocytes. Indeed, nearly complete deletion of FoxO was observed in NSC-rich regions of adult brain, including SVZ, in hGFAP-Cre: FoxO1/3/4L/L mice (Supplemental Figure S1b, d, f, and i).
FoxO1/3L/L allelic combinations with hGFAP-Cre showed significantly increased overall brain size (i.e. thickness of cortices and corpus callosum) and weight relative to hGFAP-Cre negative controls that was evident at 12 weeks of age (Figure 1A and Supplemental Figure S3 b and c). The low expression of FoxO4 in these regions targeted by this particular hGFAP-Cre allele raised the possibility that the increased brain size of hGFAP-Cre: FoxO1/3/4L/L mice was predominantly due to FoxO1 and FoxO3 deficiencies. Indeed, hGFAP-Cre:FoxO4L/L mice, alone or in the triple knockout, exhibited a minimal effect on brain size consistent with its low expression (Supplemental Figure S3b). Consistent with prominent FoxO1 and 3 co-expression patterns in vivo, combined FoxO1 and 3 deficiencies (hGFAP-Cre: FoxO1/3/4L/L) had a more pronounced effect on brain size than individual FoxO1 or FoxO3 deficiency in the context of FoxO4 deficiency (hGFAP-Cre: FoxO3/4L/L or :FoxO1/4L/L). Notably across all of the genotypes, FoxO3 exerts the greatest phenotypic impact on brain size as observed in both conditional and germline knockout mice (Supplemental Figure S3b; data not shown).
Fig. 1.
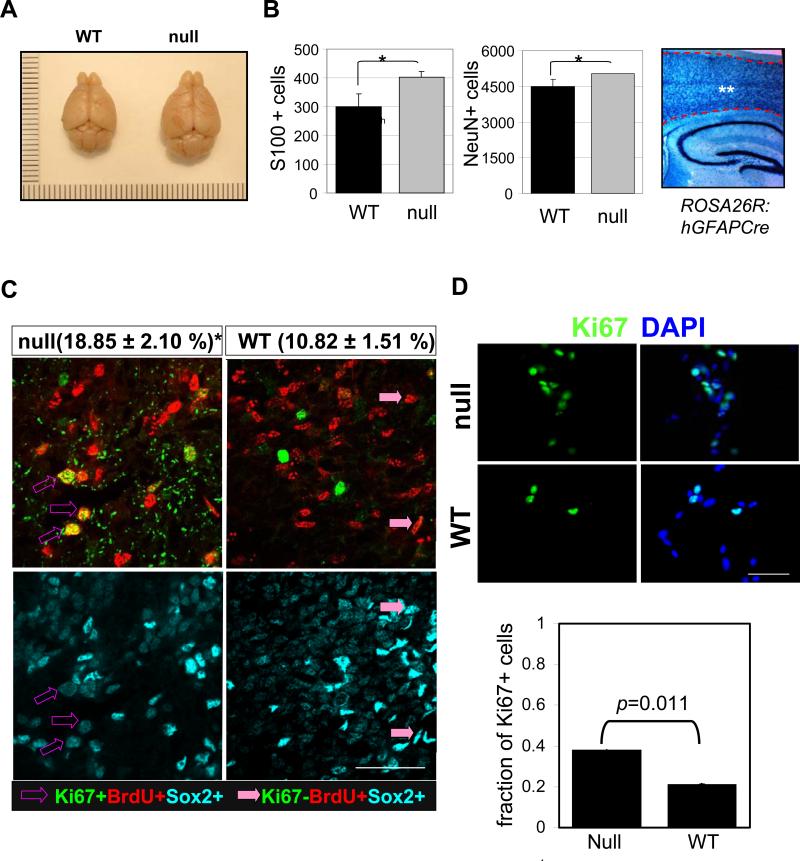
Increased brain size and cycling of NSC in FoxO null mice and their derived culture. (A) Representative photograph of brains from 20 week old FoxO WT (left) and null (right) mice. (B) The number of glia and neorons was estimated by counting S100 or NeuN-positive cells in the cerebral cortex of 10 day old FoxO WT (■) and null ( ) pups. There are more S100 or NeuN-positive cells in FoxO null brain compared with the control (*, p<0.1). ** indicates the region where S100+ and NeuN+ cells were scored. hGFAP-cre mice were crossed with ROSA26R mice, where Cre-mediated recombination drives the constitutive expression of the ß-galactosidase. X-gal staining positice cells are shown in blue. (C) Increase in cell cycle re-entry of FoxO null SVZ cells. Young mice (P8) were single-pulse labeled with BrdU 24hrs prior to the sacrifice and brain sections were stained for BrdU (red), Ki67 (green), and Sox2 (cyan). The fraction of Sox2 positive cells re-entered cell cycle (open arrow, BrdU+/Ki67+/Sox2+ triple positive) increased in FoxO null SVZ and that of no longer dividing (filled arrow, BrdU+/Ki67-/Sox2+) is higher in WT SVZ. Percent mean ±s.d. of Ki67+ cells from BrdU+/Sox2+ cells from 10 WT and 6 FoxO null mice is shown ( *, p<0.001 by two tail t-test). Bar=40μm (D) Ki67 positive NSC of FoxO WT and null cultures (n=5, P2). Bar=20μm.
) pups. There are more S100 or NeuN-positive cells in FoxO null brain compared with the control (*, p<0.1). ** indicates the region where S100+ and NeuN+ cells were scored. hGFAP-cre mice were crossed with ROSA26R mice, where Cre-mediated recombination drives the constitutive expression of the ß-galactosidase. X-gal staining positice cells are shown in blue. (C) Increase in cell cycle re-entry of FoxO null SVZ cells. Young mice (P8) were single-pulse labeled with BrdU 24hrs prior to the sacrifice and brain sections were stained for BrdU (red), Ki67 (green), and Sox2 (cyan). The fraction of Sox2 positive cells re-entered cell cycle (open arrow, BrdU+/Ki67+/Sox2+ triple positive) increased in FoxO null SVZ and that of no longer dividing (filled arrow, BrdU+/Ki67-/Sox2+) is higher in WT SVZ. Percent mean ±s.d. of Ki67+ cells from BrdU+/Sox2+ cells from 10 WT and 6 FoxO null mice is shown ( *, p<0.001 by two tail t-test). Bar=40μm (D) Ki67 positive NSC of FoxO WT and null cultures (n=5, P2). Bar=20μm.
Histological inspection showed no apparent developmental abnormalities or alterations in brain cytoarchitecture prior to early adulthood (3 weeks of age) in any of the genotype combinations (Supplemental Figure. S3a). At 10 days postnatally, the number of both neurons and glia increased in cerebral cortex of FoxO null brain (NeuN+, 4486 vs 5024; S100+, 301 vs 402, p<0.1) where almost all the cells underwent Cre-mediated recombination as measured by reporter activity (Figure 1B). At approximately 15 weeks of age, mild ventricular enlargement became obvious in all three FoxO-deficient brains (Supplemental Figure S4a and b). The above co-expression profiles, coupled with high functional redundancy among FoxO family members in the brain (this study) and other tissues (Paik et al., 2007) prompted us to focus our detailed phenotypic analyses on the triply null FoxO brains (GFAP-Cre: FoxO1/3/4L/L hereafter referred to as “null” and FoxO1/3/4L/L as “WT”) for detailed CNS and NSC analyses.
To understand the basis for increased brain size, we examined cell proliferation and size as a function of FoxO status in the young brain (P8). FoxO deletion was nearly complete in Sox2, NeuN, GFAP, or Olig2 positive cell types in the FoxO null brain with the exception of a few anatomical regions (Supplemental Figure S1i, and o-q). A prominent increase in cell cycle re-entry (i.e., Ki67/BrdU/Sox2 triple-positive cells) among BrdU-labeled population was evident in the NSC-rich regions of the SVZ in FoxO null brain (Figure 1C; 18.85% vs 10.82%, p<0.01). In contrast, detailed morphometric analyses showed no change in the soma size of neurons or astrocytes in FoxO null brains (Supplemental Figure S5a-c). Of note, we detected the presence of hypertrophic astrocytes only in older FoxO null animals (>32 week) and in vitro differentiated culture from late passage FoxO null NSC (Supplemental Figure S5d and e). Together, these observations suggest that FoxOs function to constrain the cycling of NSC and/or progenitor cells, but not their size, an observation in line with drosophila FoxO control of cell proliferation but not cell size (Puig et al., 2003).
Given that the hGFAP-Cre transgene directs FoxO deletion in NSC as well as other CNS lineages (Zhou et al., 2001, Supplemental Figure S1o-q), we sought to further define the specific impact of FoxO deficiency on NSC proliferation and renewal using well-established cell culture-based approaches. Ki67 expression (Figure 1D) and population doubling (Figure 4A) in NSC cultures revealed that early passage (P2) FoxO null NSC cultures had increased proliferation. Thus, FoxOs constrain the proliferative activity of NSC and/or progenitor cells in the young adult brain and in early passage cell cultures.
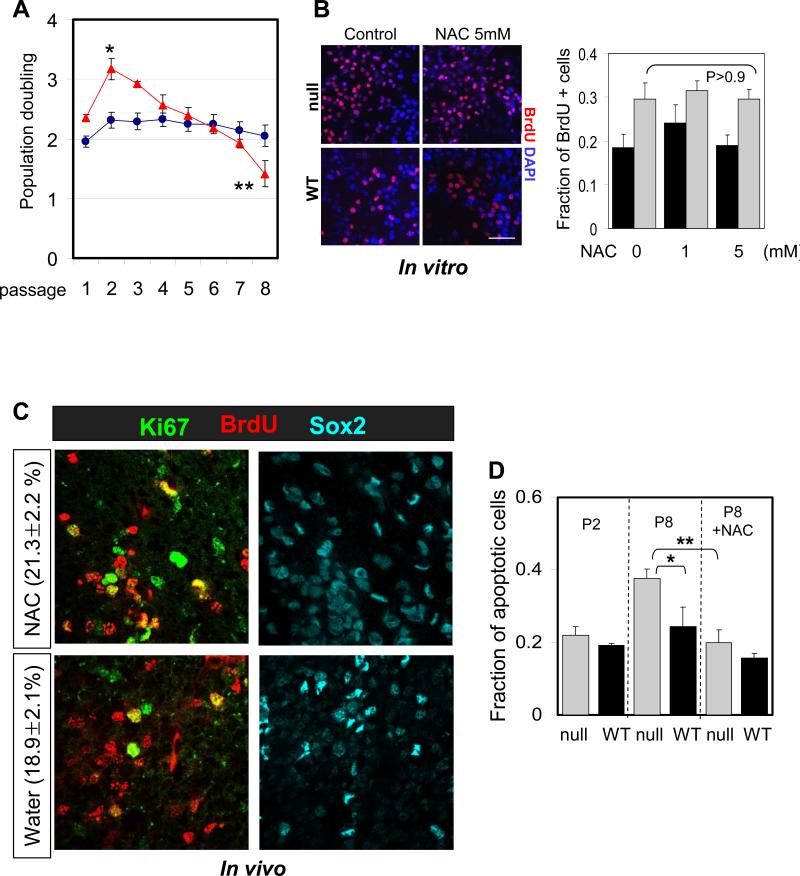
Fig 4.
Suppression of ROS does not inhibit hyper-proliferation of FoxO null NSC. (A) Proliferation of FoxO WT (blue line) and null (red line) NSC (Rosa26-CreERT2+ and -:FoxO1/3/4L/L respectively) over the 8 passages after 4OHT treatment is plotted. Y axis indicates log2 (cells counted after 5 days in culture/cells seeded). Result is representative of three independent cultures (*, p=0.0047; **, p=0.015). (B) NAC treatment does not attenuate proliferation of FoxO null NSC. P2 NSC were treated with up to 5mM NAC for 24hr and fraction of BrdU+ cells was plotted. Bar=30μm (C) NAC treatment does not inhibit increased cycling of Sox2+ cells in 8 day old FoxO null brain. 3 day old FoxO null mice were treated with NAC for 5 days before sacrificed. Percent mean ±s.d. of Ki67+ cells from BrdU+/Sox2+ cells from NAC (n=6) or water (n=6) treated FoxO null mice is shown (p=0.08 by two tail t-test). (D) Increased apoptosis in late passage (P8) FoxO null NSC was reversed by 1mM NAC treatment. No apparent changes in apoptosis were observed in early passage (P2). Cleaved caspase3 staining positive cells were scored and normalized by total number of DAPI positive cells. Fractions of apoptotic cells are plotted. *, p=0.017, **, p=0.021.
FoxO deficient brains exhibit precocious NSC depletion
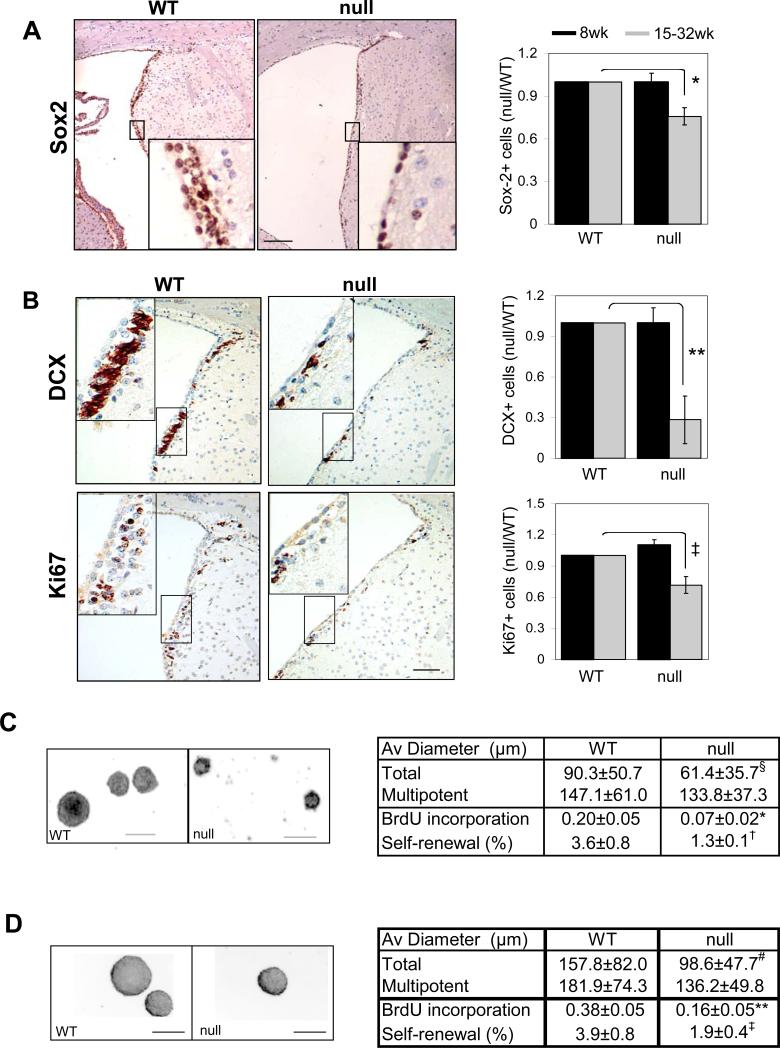
Although increased proliferation of FoxO null NSC cultures was observed initially, continued passage in culture showed exhaustion of neurosphere formation potential with accompanying decreased proliferation and marginally increased apoptosis (Figure 4A and D; note the use of single cell clonogenic assays in Figure 2D, BrdU). This progressive renewal defect in cell culture prompted closer examination of NSC reserves in vivo. Staining brain sections with antibodies to Sox2, a marker for NSC, revealed that 15 week-old FoxO null brain shows a significant 25% decline in Sox2-positive cells in the SVZ (Figure 2A; gray bars, 1 vs 0.757, p=0.004). In addition to the decrease in Sox2 positive SVZ cells to levels well below FoxO WT brains, there was an accompanying decline in doublecortin (DCX, Figure 2B; gray bars, 1 vs 0.283, p=0.0039) and Ki67 (Figure 2B; gray bars, 1 vs 0.717, p=0.026) positive cells and a decline in the number of newborn neurons in the olfactory bulb to numbers below those in FoxO WT brains (1.76% vs 0.92%, p=0.018, Supplemental Figure S4c). Accompanying this decline in NSC and neurogenic populations, the FoxO null brain shows consistent and progressive ventricular dilation and thinning of the lateral ventricular wall and SVZ (Supplemental Figure S4a and b; 100% vs 751.1%, p=0.011). These pathological changes were most evident in more aged brains, while absent in the developing brain, suggesting that the loss of FoxO function in the embryonic and adult brain together impacts most prominently processes controlling the size and function of NSC pool in postnatal mouse brain (Figure 2A and B; black bars).
Fig. 2.
FoxO deficient brain exhibits decrease in NSC reserve and neurogenesis. (A) Decreased Sox2 positive NSC in 32 week old FoxO null (right) brain. Bar=50μm. (B) Decline in neural progenitor proliferation and neurogenesis measured by Ki67 and DCX expression in SVZ of lateral wall from 32 week old FoxO null mice (right). 200x microscopic field of comparable region from WT littermate control mouse is shown (left). Insets are higher magnification view of boxed regions. Bar=50μm. Sox2 (A), DCX (B), Ki67(B) expression levels in young (8 week old, (■)) and relatively old (15-32 week old, ( )) mice SVZ were measured by laser scanning and plotted (4 pairs, *, p=0.004, **, p=0.0039, ‡, p=0.026, by two tail paired t-test). Decreased self-renewal capacity of NSC derived from 18-22 week old FoxO null mice (C) and acutely deleted for FoxOs in vitro (D). For conditional deletion NSC were isolated from SVZ of 4 week old Rosa26-CreERT2+ (null) or -(WT) FoxO1/3/4L/L mice and treated with 400nM 4OHT prior to the assay. Results are representative of three independent experiments with two primary cultures. Both average diameters for all the neurospheres and for multipotential ones are shown as mean±s.d. Proliferation index is shown as fraction of BrdU incorporation from triplicate cultures in two independent experiments. §, p=0.0043, #, p<0.0001, *, p=0.01, **, p=0.03, †, p=0.06, ‡, p=0.096 by two tail t-test. Bar= 100 μm.
)) mice SVZ were measured by laser scanning and plotted (4 pairs, *, p=0.004, **, p=0.0039, ‡, p=0.026, by two tail paired t-test). Decreased self-renewal capacity of NSC derived from 18-22 week old FoxO null mice (C) and acutely deleted for FoxOs in vitro (D). For conditional deletion NSC were isolated from SVZ of 4 week old Rosa26-CreERT2+ (null) or -(WT) FoxO1/3/4L/L mice and treated with 400nM 4OHT prior to the assay. Results are representative of three independent experiments with two primary cultures. Both average diameters for all the neurospheres and for multipotential ones are shown as mean±s.d. Proliferation index is shown as fraction of BrdU incorporation from triplicate cultures in two independent experiments. §, p=0.0043, #, p<0.0001, *, p=0.01, **, p=0.03, †, p=0.06, ‡, p=0.096 by two tail t-test. Bar= 100 μm.
The in vivo marker patterns suggesting a decline in NSC/progenitor cell populations prompted us to perform a series of complementary cell culture-based quantitative studies to measure the self-renewal capacity of cells derived from the forebrain of adult FoxO null mice. The percentage of cultured cells from adult FoxO null mice (18-22 week-old) capable of forming secondary multipotent neurospheres was lower than cells derived from WT littermate controls (Figure 2C; 3.6% vs 1.3%). Specifically, FoxO null cultures exhibited significant reduction in proliferation by BrdU incorporation (Figure 2C; 0.2 vs 0.07, p=0.03) and in average diameter of neurospheres (Figure 2C; Supplemental Figure S6a; 90.3μm vs 61.4μm, p=0.0043). Together, these findings suggest that FoxOs regulate the proliferative and self-renewal potential of NSC and that the loss of FoxO function results in the progressive decline of NSC number.
It is important to emphasize that our experimental strategy deletes FoxO in NSC and other CNS lineages thus potentially affecting NSC biology directly and/or indirectly via alterations in the surrounding microenvironment. Thus, we sought to assess whether deletion of FoxO upon in vitro isolation and culturing would produce the same phenotypes observed in NSC cultures where FoxOs were deleted in vivo. To that end, using single cell clonogenic assays, we tested the self-renewal capacity of NSC from SVZ of young adult mice (4 week old) that was subjected to acute FoxO deletion in vitro using an inducible Cre system. Consistent with in vivo findings, acute in vitro deletion of all three FoxOs using the transiently 4OHT-activated Rosa26-CreERT2 system resulted in a same decrease in self-renewal capacity (Figure 2D; 3.9% vs 1.9%, p=0.096) along with decreased average diameter of neurospheres (Figure 2D; Supplementary Fig. S6b; 157.8μm vs 98.6μm, p<0.0001) and proliferation as per BrdU incorporation (Figure 2D; 0.38 vs 0.16, p=0.010). Furthermore, an accompanying article shows that deletion of FoxO3 alone in the brain via an independent Cre-deletor transgenic line, Nestin-Cre, which allows deletion in NSC (as well as other cell type, also produces similar decline in NSC number both in vivo and in vitro (Renault et al.). The concordant phenotypes in vitro and in vivo, as well as across multiple different experimental systems support the view that precocious depletion of NSC in FoxO deficient brain is, at least in part, a cell-autonomous phenomenon. Thus, we propose that the FoxOs function to support NSC homeostasis in vivo, perhaps by preventing the rapid amplification of progenitors and the consequent exhaustion of the relatively quiescent NSC.
FoxO-deficient NSC have increased ROS and decreased self-renewal potential
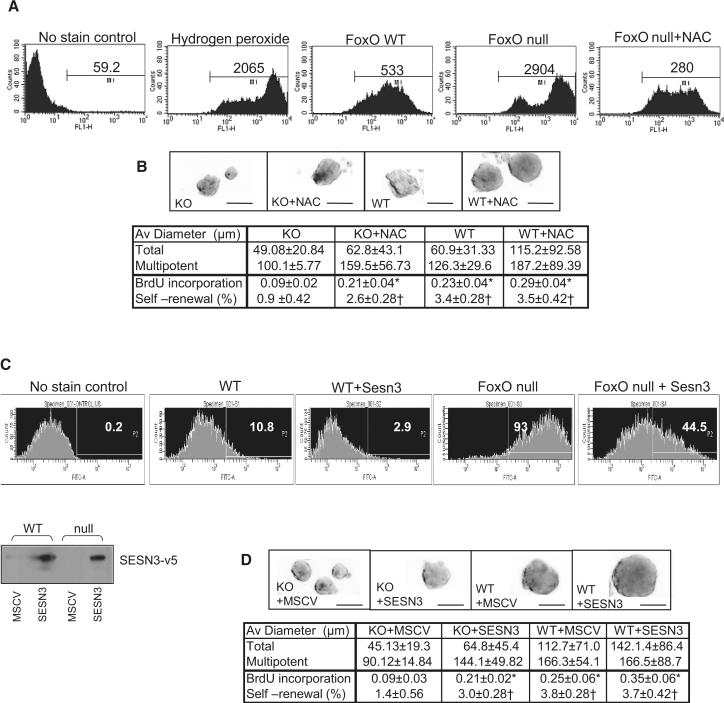
We have previously reported that loss of all three FoxOs in HSC results in decreased expression of ROS-detoxifying enzymes, β-catalase and MnSOD, and that increased intracellular ROS levels in FoxO null HSC are causally linked to enhanced G0 exit and increased apoptosis, resulting in progressive depletion of HSC reserves (Tothova et al., 2007). To determine whether altered regulation of genes involved in oxidative defense contribute to these similar defects in NSC, we performed the DCF-DA ROS measurement assay, which showed an increase in intracellular ROS levels in FoxO null NSC relative to WT control cultures (Figure 3A; mean intensity level of 2904 vs 533). However, contrary to the HSC findings of altered β-catalase and MnSOD expression, the FoxO NSC transcriptome instead shows prominent dysregulation of a distinct set of ROS detoxifying enzymes including peroxiredoxin, glutathione peroxidase1, and sestrin3 (SESN3) (Supplemental Table S3). To examine whether ROS accumulation in NSC also causally contributes to the biphasic phenotypes of hyper-proliferation and/or long-term exhaustion, we reduced ROS levels either by treatment with the anti-oxidant, N-acetyl cysteine (NAC) or by enforcing the expression of SESN3, a cysteine sulfinic acid reductase that reduces peroxiredoxins. Normalized (by NAC) or reduced (by enforced SESN3) intracellular ROS levels (Figure 3A and C) enhanced the self-renewal capacity of FoxO null NSC cultures as measured by neurosphere formation assay (Figure 3B and D; 0.9% vs 2.6% and 1.4% vs 3.0%, respectively, p<0.05). In particular, the late passage (P7) of NAC-treated or SESN3-transduced FoxO null NSC showed improved proliferation as measured by BrdU incorporation (Figure 3B and D; 0.09 vs 0.21, p<0.01), decreased apoptosis (Figure 4D), and increased overall neurosphere diameter (Supplemental Figure S6c and d).
Fig. 3.
Regulation of NSC self-renewal by FoxO through controlling intracellular ROS level. (A) Accumulation of intracellular ROS in FoxO null NSC (P7) is measured by DCF-DA staining and flowcytometry. Numbers indicate mean intensity level (A) and % of total event above the threshold (C). Both NAC treatment (1mM) and enforced expression of SESN3 normalized and suppressed ROS level (A, C) and partially rescued self renewal in FoxO null NSC (B, D). Expression of SESN3-v5 in NSC is shown below by immunobloting of v5 epitope (C). (B, D) Representative images of cultured neurospheres from two independent experiments are shown. Both average diameters for all the neurospheres and for multipotential ones are shown as mean±s.d. Proliferation index is shown as fraction of BrdU incorporation from triplicate cultures in two independent experiments. *, p<0.01, †, p< 0.05 by ANOVA with Bonferroni's post test. Bar= 100 μm
In contrast, in the early passage FoxO null NSC, NAC treatment (up to 5mM) had no effect on the transient hyper-proliferation phenotype nor did it affect the proliferation of FoxO WT NSC (Figure 4B; con vs NAC, 0.295 vs 0.294, p=0.966, P2). Furthermore, in vivo administration of NAC did not suppress increased cycling of Sox2+ cells in postnatal FoxO null brains (Figure 4C; NAC vs PBS, 21.3% vs 18.9%, p>0.05), supporting the view that increased ROS plays no discernable causal role in increased proliferation of FoxO null NSC in the young postnatal brain. Together, these studies suggest that, while FoxO-directed regulation of intracellular ROS level plays an important role in NSC self renewal capacity, it plays no discernable role in controlling the proliferation activity of the NSC.
Identification of FoxO transcriptional targets regulating NSC biology
The lack of complete rescue of the self-renewal defects, even with normalization of ROS by NAC treatment, pointed to additional FoxO regulated pathways governing NSC biology. To identify functionally relevant FoxO direct gene targets and their linked physiological actions, we conducted unbiased transcriptomic profiles in WT and FoxO null NSC and in silico promoter analysis followed by confirmatory biochemical and biological validation studies.
Transcriptomic analysis of multiple independent FoxO WT and null early passage neurosphere cultures revealed differential regulation of 3425 genes by FoxOs as determined by two independent statistical methods (Supplemental Table S1). Among these are known effectors of FoxO and their family members (e.g., Cyclin D1, Inhibitor of DNA binding 1 and 2, and Polo like kinase 2) (Alvarez et al., 2001; Birkenkamp et al., 2007; Schmidt et al., 2002) as well as many potential novel FoxO targets that relate well to our biological observations including cell cycle (e.g., G two and S phase expressed1, Cdk inhibitor 1C, Sal like2) and glial differentiation (e.g., Hes5, Sox10, Myelin basic protein, Plp1, Cystatin C) and that functionally interact with FoxOs (e.g. FoxG1, Sirt2) (Seoane et al., 2004; Wang et al., 2007). Notably, the NSC FoxO transcriptome demonstrates minimal overlap with other cell types including HSCs – a finding in line with previous observations that the FoxOs regulate their targets in a highly cell type-specific manner (i.e., the FoxO transcriptomes of HSC, thymocytes, or endothelial cells show minimal overlap) (Paik et al., 2007; Tothova et al., 2007). Nevertheless, despite these cell type-specific gene target differences, knowledge-based pathway analysis showed convergent regulation of well-known FoxO networks such as IGF-1/PI3K/PTEN, cell cycle and metabolic pathways (data not shown).
Next, we sought to identify direct FoxO targets in order to gain further insights into FoxO's role in NSC biology. To that end, a systematic and validated in silico promoter analysis approach (Paik et al., 2007) was used to identify evolutionarily conserved FoxO consensus binding elements (BE) in the regulatory regions of the top 186 genes (144 downregulated, 42 upregulated in FoxO null) that show the most significant expression differentials (SAM >2; and see Methods) (Supplemental Table S2). To search for FoxO BE, each gene region was surveyed from 10 kb up or downstream of the transcription start or end site. By this approach, 96 of the 186 genes were found to contain putative conserved FoxO BEs. Eighteen of the 96 genes from diverse gene ontology categories were selected and their direct regulation by FoxOs was tested on several levels including (1) differential mRNA expression by RT-qPCR in FoxO null versus WT, (2) adenoviral-transduced expression of active form of FoxO1 and 3 (CA-FoxO1/3) (Supplemental Figure S8a), (3) documentation of physical binding of FoxOs on predicted BE by chromatin immunoprecipitation (ChIP) (Supplemental Figure S8b), and (4) confirmation of target gene differential expression by RISH in FoxO WT and null embryonic brains (data not shown). Of the 18 putative targets tested, these integrated analyses and highly stringent criteria confirmed 8 bona fide novel FoxO direct targets (ASPM, TMEFF2, SIRT2, KITL, TSPAN7, sFRP1/2, and SOST). Several of these FoxO targets have plausible, albeit unexplored, links to NSC biology prompting further functional study.
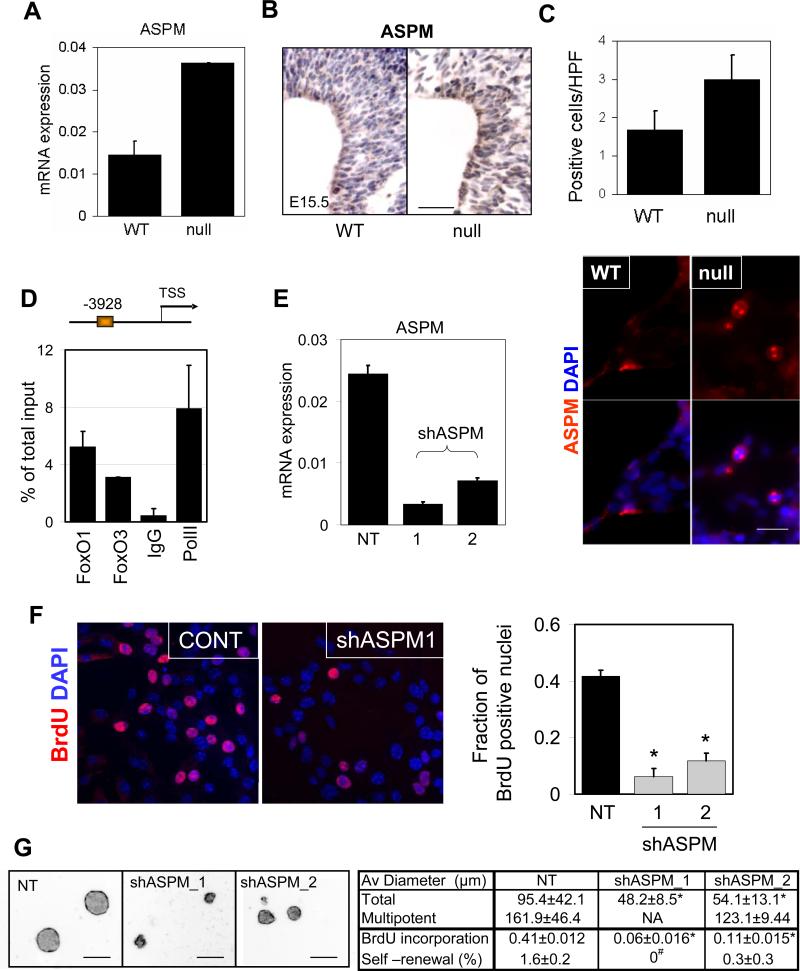
FoxOs directly repress ASPM expression in the regulation of NSC proliferation
The FoxO NSC transcriptome and pathway analyses were notable for alterations in proliferation and differentiation networks, the imbalance of which may provide an explanation for the enlarged brain phenotype and the abnormal cycling that could ultimately lead to exhaustion of NSC. For example, there is increased expression of cyclins (Ccn A2, B1, B2, C, D1, D3, E1 G1, T2), cyclin dependent kinases (Cdk 2, 4, 7), and decrease in specific cyclin dependent kinase inhibitors (p57KIP2) which correlate well with observed short-term hyper-proliferation phenotype in FoxO null NSC (Supplemental table S1). A notable cell cycle gene which may relate to the FoxO null brain size phenotype is ASPM (abnormal spindle-like microcephaly-associated) gene. In humans, ASPM is a determinant of human cerebral cortical size, i.e., individuals with ASPM germline loss-of-function mutations show primary microcephaly (Bond et al., 2002). ASPM expression has been shown to be required for symmetric, proliferative divisions of neural stem/progenitor cells of the mammalian brain (Fish et al., 2006; Horvath et al., 2006).
Consistent with the relevance of the FoxO-ASPM link in NSC biology, we observed (i) significantly increased ASPM mRNA levels in FoxO null NSC by RT-qPCR (Figure 5A), (ii) increased ASPM protein and mRNA in NSC-rich regions of FoxO null embryonic brains (Figure 5B, RISH, not shown), and (iii) increased ASPM-positive cells in FoxO null NSC cultures (Figure 5C). Second, in silico methods (Paik et al., 2007) identified evolutionarily conserved FoxO BEs in the ASPM promoter which were confirmed to bind FoxO by ChIP assay in NSC (Figure 5D). Third, on the functional level, ASPM shRNA-mediated knockdown (two independent hairpins) resulted in a profound decrease in proliferation of FoxO null NSC (Figure 5E, F, and G; 0.41 vs 0.06 and 0.11 for BrdU incorporation, p<0.01). Notably, NSC with ASPM knockdown exhibited significant decrease in size of neurospheres and self-renewal potential in single cell clonogenic assays (Figure 5G and Supplemental Figure S6e; 95.4μm vs 48.2μm and 54.1μm in average diameter, p<0.01 and 1.6% vs 0 and 0.3% for self-renewal, p<0.05), a phenotype associated with increased cell cycle exit and differentiation (data not shown). Thus, ASPM is a physiologically relevant target of FoxO in the regulation of proliferation and the renewal of NSC. We propose that the de-repression of ASPM in FoxO null NSC contributes to the observed short-term hyper-proliferative defects in this progenitor compartment and the enlarged brain phenotype.
Fig.5.
FoxO-dependent expression of ASPM regulates NSC proliferation. Differential expression of ASPM mRNA in FoxO WT and null embryonic NSC measured by rtqPCR (A) and protein by IHC analysis (B) on FoxO WT and null E15.5 embryonic brain. Bar=50μm. (C) Number of mitotic spindle-localized ASPM positive NSC in FoxO WT and null cultures. Representative images are shown. Bar=10μm. (D) The ASPM promoter is occupied by both FoxO1 and FoxO3 in NSC. On the schematic representation of the ASPM promoter FoxO binding sites are depicted relative to the +1 transcriptional start site. ChIP analysis of NSC with FoxO1, FoxO3, IgG, or PolII antibodies. Primers were designed to encompass the FoxO binding sites. (E) Knockdown of ASPM by shRNAs in FoxO null NSC. ASPM mRNA level was measured by rtqPCR after 48hr of shASPM lentiviral transduction. Decreased proliferative potential in ASPM knockdown NSC was measured by BrdU incorporation assay (F) and neurosphere formation (G) *, p<0.01, #, p<0.05 by ANOVA with Bonferroni's post test. NA, not analyzed, NT, non targeting control shRNA infected culture. Bar=100μm.
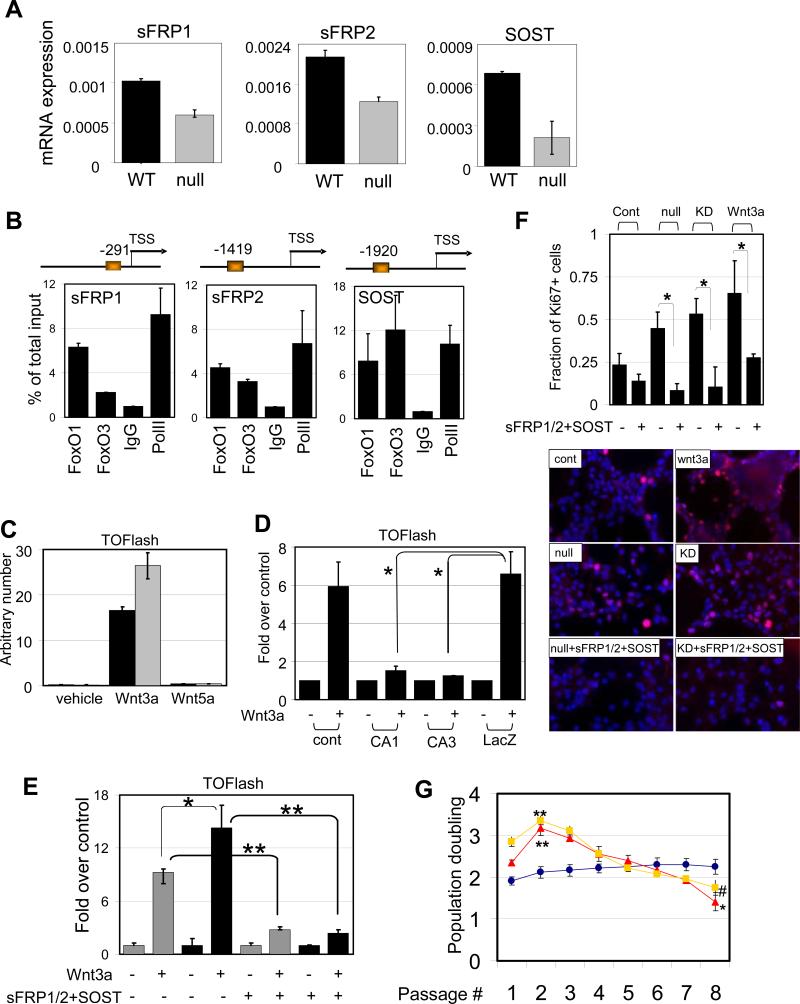
FoxOs attenuate Wnt signaling in NSC
The FoxO transcriptome is also notable for regulatory factors of the Wnt pathway, a pathway whose stringent control is required for the long-term homeostasis of many tissue stem cells. In particular, there was significant downregulation of soluble frizzled receptors (sFRP) 1 and 2 as well as sclerostin (SOST) in FoxO null NSC transcriptome (Supplemental Table S1). First, as above, the link between FoxO status and these factors was confirmed by their higher mRNA expression in FoxO WT relative to FoxO null NSC (Figure 6A) and by the presence of conserved ChIP-validated FoxO binding elements in their promoters (Figure 6B). Second, enforced expression of constitutively active (CA)-FoxO1 or 3 can induce expression of sFRP1/2 and SOST in FoxO null NSC (Supplemental Figure S8a). Third, since these soluble factors act as an antagonist for Wnt ligands, we utilized the TOFlash reporter system to measure the activity of canonical Wnt pathway in our system. Addition of Wnt3a, but not Wnt5a, induced TCF/Lef/β-catenin dependent transcriptional activity confirming the specificity of reporter activity responsive to the canonical Wnt signaling. This response is reproducibly enhanced in FoxO null NSC (Figure 6C). Conversely, enforced expression of CA-FoxO1 or 3 suppressed reporter activity (i.e., dampened Wnt signaling) (Figure 6D). In agreement with the reporter assays, the expression of the targets dependent on Wnt signaling including Cyclin D2 and Myc were upregulated in FoxO null NSC (Supplemental Figure S7a). Fourth, RNAi-mediated knockdown of sFRP1/2 and SOST was performed to examine the role of these factors in FoxO-mediated repression of Wnt signaling in NSC. Combined knockdown of all three sFRP1/2 and SOST resulted in a 1.5-fold increase in Wnt3a-induced reporter activity and this increase was completely reversed by the addition of recombinant sFRP1/2 and SOST (Supplemental Figure S7b and Figure 6E). Knockdown of individual factors did not cause appreciable differences in Wnt3a-induced reporter activity (data not shown), suggesting that these novel FoxO targets act cooperatively as soluble inhibitors of the Wnt pathway to repress Wnt signaling in NSC.
Fig. 6.
FoxOs negatively regulate Wnt signaling in NSC. (A) Expression of soluble antagonists of Wnt. Differential expression of sFRP1/2 and SOST mRNA in FoxO WT and null embryonic NSC measured by rtqPCR. (B) Promoters for sFRP1/2 and SOST are occupied by both FoxO1 and FoxO3 in NSC. On the schematic representation of the promoter FoxO binding sites are depicted relative to the +1 transcriptional start site. ChIP analysis of NSC with FoxO1, FoxO3, IgG, or PolII antibodies. Quantification of ChIP analysis on each promoter was assessed by rtqPCR analysis. (C) Wnt3a-induced canonical Wnt signaling is measured TOFlash reporter assay. Note enhanced signal in FoxO null ( ) NSC compared to WT (■) . *, p=0.08053. (D) Attenuated Wnt3a-dependent canonical signaling by constitutively active FoxOs. CA1, CA3; FoxO1-ADA and FoxO3-AAA mutant respectively. *, p<0.1. (E) Enhanced canonical signaling in sFRP1/2 and SOST knockdown NSC (■) compared to WT (
) NSC compared to WT (■) . *, p=0.08053. (D) Attenuated Wnt3a-dependent canonical signaling by constitutively active FoxOs. CA1, CA3; FoxO1-ADA and FoxO3-AAA mutant respectively. *, p<0.1. (E) Enhanced canonical signaling in sFRP1/2 and SOST knockdown NSC (■) compared to WT ( ) is reversed by the addition of soluble sFRP1/2 and SOST (500ng/ml). *, p=0.068; **, p<0.005 (F) Exogenously added sFRP1/2 and SOST (500ng/ml) reversed increased proliferation of FoxO null NSC. Wnt3a, 50ng/ml of recombinant wnt3a was added for 24hrs. KD, 72 hrs after knockdown by siRNAs for sFRP1/2 and SOST. Fractions of Ki67 positive nuclei are plotted. (G) Decreased long term proliferation potential of NSC by Wnt3a stimulation (100ng/ml). Y axis indicates log2 (cells counted after 5 days in culture/cells seeded). Lines in red, FoxO null; blue, WT; yellow, Wnt3a treated. *, p=0.0041; **, p<0.01; #, p=0.0169.
) is reversed by the addition of soluble sFRP1/2 and SOST (500ng/ml). *, p=0.068; **, p<0.005 (F) Exogenously added sFRP1/2 and SOST (500ng/ml) reversed increased proliferation of FoxO null NSC. Wnt3a, 50ng/ml of recombinant wnt3a was added for 24hrs. KD, 72 hrs after knockdown by siRNAs for sFRP1/2 and SOST. Fractions of Ki67 positive nuclei are plotted. (G) Decreased long term proliferation potential of NSC by Wnt3a stimulation (100ng/ml). Y axis indicates log2 (cells counted after 5 days in culture/cells seeded). Lines in red, FoxO null; blue, WT; yellow, Wnt3a treated. *, p=0.0041; **, p<0.01; #, p=0.0169.
Finally, we examined the biological impact of FoxO-repressed Wnt signaling on NSC proliferation. Exogenous Wnt3a treatment can induce a strong proliferative response in NSC, and correspondingly, both FoxO null and sFRP1/2 and SOST knockdown NSC showed increased basal proliferation which was significantly inhibited by the addition of sFRP1/2 and SOST (Figure 6F). However, it is important to note that the Wnt3a-induced proliferative response is short-lived and is followed by substantial decline in number of cycling cells, resembling the growth kinetics of FoxO null NSC culture (Figure 6G). These observations further reinforce the cross talk between FoxO and Wnt signaling pathways in NSC and the importance of finely-tuned Wnt pathway activation in the long-term homeostasis of NSC.
Discussion
In this study, an integrated molecular, cellular and organismal analysis has established FoxOs as essential regulators of genes governing processes central to NSC biology (Figure 7). Disruption of these FoxO-dependent homeostatic processes in the mouse brain leads to initial deregulated NSC proliferation followed by a decline in NSC pool and accompanying neurogenesis that is only evident in adult mice. The abnormal proliferation kinetics of NSC in vitro and in vivo and the enlarged brains in FoxO-deficient brains, establishes a role for FoxOs in the enforcement of NSC quiescence and control of NSC self-renewal. Mechanistically, a combined transcriptomic, computational, and biochemical analyses have identified a diverse set of direct functional targets and linked pathways through which FoxO directly executes its complex actions on NSC and brain physiology.
Fig. 7.
Model for FoxO-dependent regulation of NSC. FoxOs control NSC biology at multiple levels. FoxOs (A) negatively regulate G0 exit of NSC and keep their quiescence within the niche, (B) support their long-term self-renewal activity, (C) constrain proliferation of lineage-committed neural progenitors, and (D) contribute to continued neurogenesis.
Gradual depletion of NSC in FoxO null animals might be explained a cell autonomous role of FoxOs in regulating adult self-renewal potential. Alternatively increased cell cycling during the development and establishment of NSC pool in the FoxO null brain may lead to later decline in their number by intrinsic mechanism to adjust the size of stem cell pool within the niche. Despite the reinforcing parallel findings of our in vitro analyses showing an impact of FoxO deletion on NSC biology, it is possible that some of the NSC phenotypes derive in part from the broad deletion of FoxOs in cellular compartments other than the NSC in the adult brain. However, it is worth noting that neuron-specific deletion of FoxOs (Syn-Cre: FoxO1/3/4L/L) is not accompanied by NSC abnormalities (data not shown). Nevertheless, we acknowledge that specific deletion of FoxOs only in mature astrocytes or only in adult NSC will be needed to distinguish whether FoxOs exert their actions on NSC homeostasis via altered astrocyte support functions or by acting in NSC. Our validated collection of FoxO targets establishes strong evidence that FoxO regulates ROS detoxification, Wnt signaling, cell cycle, and differentiation processes, which collectively serve to fine tune the balance of growth, survival and differentiation needed to achieve the regulation of NSC throughout life (Figure 7).
The precocious decline in the NSC pool and neurogenesis in FoxO null mice at 6 months of age indicates that FoxOs are important regulators of NSC function and renewal under physiological conditions in the brain. Consistent with functional redundancy among FoxOs in other tissues (Paik et al., 2007), the single and double FoxO null brains show overtly normal patterns of neurogenesis through mid-adulthood (Muller and DePinho, unpublished observations). However, we also acknowledge that it remains possible that single (particularly FoxO3) FoxO null mice may also experience more subtle decline in CNS function. Indeed, accompanying study by Renault et al. demonstrates that lack of FoxO3 alone in the brain is sufficient to cause a defect in NSC number in vivo, a result suggesting a dominant role of FoxO3 relative to other FoxO family member in the brain.
That FoxO deficiency leads to transient increase in cycling cells is not unanticipated based on previous studies with other PI3K pathway components wherein loss of the negative regulator PTEN was shown to result in sustained proliferative potential of NSC (Groszer et al., 2006; Groszer et al., 2001). In contrast, the FoxO null NSC/progenitor proliferation is distinct from that of PTEN null NSC in that it does not show sustained increased proliferation. When viewed through the multiple mechanistic findings of our study, we speculate that increased proliferation and ultimate precocious decline of NSC and associated neurogenesis results from the combined impact of loss of fine-tuned cell cycle/quiescence control by dysregulation of molecules such as the p57KIP2 cell cycle inhibitor, ASPM, and WNT dampeners, among others. Beyond cell cycle regulation, it is possible that altered lineage commitment may also contribute to a decline in neurogenesis as the FoxO null transcriptome shows changes of key developmental regulators (Paik and DePinho, unpublished), a finding in line with previously reported role for FoxOs in cellular fate determination observed in myogenesis and hematopoiesis (Hu et al., 2008; Kitamura et al., 2007; Tothova et al., 2007). Thus, the FoxOs appear to regulate many cellular processes that require fine regulation to maintain the right balance of proliferation, survival and cell fate commitment decisions in tissue stem cell pools, while responding to the regenerative needs of tissues throughout life (Figure 7).
Our analysis of the FoxO null NSC transcriptome, along with analogous studies in the HSC, confirm that FoxOs have conserved roles across different stem cell compartments via regulation of distinct lineage-specific sets of genes governing proliferation and intracellular ROS levels. However, relative to FoxO function in the HSC (Tothova et al., 2007), this report establishes that its function in the NSC shows a greater diversity of activities and pathway engagement. For example, the finding of ASPM as a direct target of FoxOs in NSC is notable given the hypothetical role of ASPM as one of the genetic determinants underlying the positive selection in cerebral cortical size across different species (Bond et al., 2002). Significantly, ASPM deficiency was discovered to be the basis for a human genetic disorder characterized by microcephaly, which presents with reduced neurogenesis. Our findings that ASPM is one of the major targets of FoxOs and its knockdown limits expansion of the FoxO null NSC cultures suggests that ASPM contributes to increased FoxO null brain size phenotype via its known activities in the control of neural progenitor cell biology (Fish et al., 2006).
Our identification of sFRP1/2 and SOST as direct targets of FoxOs also supports the view that the FoxOs regulate NSC cellular responses to the extracellular signals via a coordinated regulation of specific target genes. Here, we establish that FoxOs engage the Wnt pathway on many levels to ensure a tight regulation of NSC. We observe that enhanced Wnt pathway activity in FoxO null NSC leads to an acute short-term increase in their proliferation. While the ensuing depletion of long-term self-renewal potency may relate to the dominance of other dysregulated pathways (see below), hyper-activity of Wnt signaling could paradoxically contribute to this decline as well. This possibility derives from the mechanistic analysis of the accelerated aging phenotype of klotho-deficient mice, a mutant mouse manifesting augmented Wnt signaling (Liu et al., 2007). As exemplified in this study, loss of soluble antagonist for Wnt leads to cellular senescence caused by continuous Wnt signaling contributing to suppression of the number of stem/progenitor cells. Furthermore, consistent with a possible role of elevated Wnt signaling in stem cell maintenance, addition of Wnt has been shown to convert myogenic progenitor culture into fibrogenic lineage (Brack et al., 2007). Thus, while Wnt signaling is clearly essential for tissue stem cell renewal, the loss of finely tuned Wnt pathway activity in a narrow window in the FoxO null NSC could contribute to the defective maintenance via the collective impact of aberrant proliferation activity and presumably altered cell fate.
In summary, this study shows that the mammalian FoxOs have evolved to serve as pro-longevity tumor suppressors designed primarily to maintain the homeostasis and the integrity of long-lived cells including tissue stem cell reserves. This contrasts with other tumor suppressors (e.g., p16Ink4a) which appear to suppress cancer at the cost of tissue resident stem cells and therefore may contribute to decreased regenerative potential during the aging process (Krishnamurthy et al., 2006; Molofsky et al., 2006). The impact of FoxO activity on the CNS health under aging and stress, especially how FoxOs regulate NSC in the aging brain, may prove useful for defining strategies to modulate regenerative response in the setting of acute brain injury or long-term degenerative conditions. Specifically, such insights may inform the development of drugs targeting pathways that may impair or improve CNS health in aging, neurodegenerative diseases, and injury states including those associated with medical treatments such as cancer chemo/radiation therapy.
Methods Summary
Generation of mice
All experimental and control animals were littermates or age matched. An hGFAP-Cre+ male was mated with a FoxO1L/L; FoxO3L/L; FoxO4L/L (FoxO1/3/4L/L) female, and the progeny were then crossed with FoxO1/3/4L/L mice. The resulting offsprings were intercrossed to generate mice of the desired genotypes. For the mapping of Cre activity hGFAP-Cre+ mouse was bred with ROSA26R reporter mouse (Soriano, 1999) and X-gal staining was performed to locate cells expressed Cre. For in vitro deletion of FoxOs embryonic or adult NSC were isolated from Rosa26-CreERT2+ (kindly provided by Dr Anton Berns, The Netherlands Cancer Institute, Amsterdam, The Netherlands);FoxO1/3/4L/L mice (Hameyer et al., 2007). These mice were generated using above mating scheme. All the mice were cared and monitored following DFCI animal research facility guideline.
Neurosphere Formation
Primary neurospheres were dissociated and seeded at 2 cells/μl density in multi well plates. After 7-10 days cultures were monitored for the formation of neurospheres. Alternatively single cells were sorted into individual well on 384 well plate on Dako MoFlo high speed cell sorter and cultured for 3 weeks. Neurospheres were transferred to culture wells coated with poly-L-ornithine (15μg/ml) and fibronectin (1μg/ml) and differentiated in 1% FBS in neurobasal media to measure their multipotentiality. Quantification of neurosphere numbers and diameters were performed by bright field microscopy coupled with an in-house semi-automated segmentation algorithm generated using MATLAB software (The Mathworks, Natick, MA).
Intracellular ROS measurement
FoxO WT and null NSC were incubated with NAC (1mM) or H2O2 (100μM) for entire culture period or 30min respectively. DCF-DA (25 μM , Molecular Probes) was added to cultures and incubated for an additional 30 min at 37°C. Levels of fluoresecent adduct was measured by flowcytometry using excitation at 488nm.
Measuring NSC apoptosis and proliferation
For analysis of apoptosis, NSC were fixed with 4% paraformaldehyde, permeabilized, and stained for cleaved caspase-3 (Calbiochem). For BrdU incorporation BrdU-treated (6μg/ml, 2hr) cells were fixed and treated with 2N HCl for 15 min and permeabilized with 0.2% triton-X100 followed by primary mouse anti-BrdU (1:200, Roche, Bu20a) incubation followed by secondary antibody labeling as below.
In vivo administration of NAC
NAC (500mg/kg, Sigma) and BSO (5mmol/kg, Sigma) were dissolved in PBS and injected subcutaneously for 5 days starting from P3. On day 4 (P7) mice were intraperitonically injected with 1mg of BrdU. 24hr after BrdU injection mice were sacrificed and analyzed.
Reporter assay for Cannonical Wnt pathway
500ng of reporter construct TOPFLASH (kindly provided by Dr. Randall Moon) was transfected into 1×106 FoxO WT and null NSC using Lipofectamine 2000. 50ng of pTk-renilla control vector was cotransfected as an internal control. 50ng/ml of recombinant Wnt3a or Wnt5 (R&D systems) was added to media and 48 hours after transaction cells were lysed and analyzed for luciferase using the Dual Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions.
Quantitation of Neurons and Glia
Brains sections from four littermates WT and FoxO null mice (P10) were stained for NeuN and S100. Positive cells with the cerebral cortex region dorsal to the hippocampal formation were scored.
Laser Scanning Analysis for the quantitation of IHC
Laser Scanning Cytometry quantification of DCX and Sox-2 was performed by iCys Research Imaging Cytometer (Compucyte) as described early (Gorczyca et al., 2001) with a few following modifications. Counts of DCX+ or Sox-2+ (DAB positive) were carried out within the sub-ventricular zones that were predefined by a certified pathologist using the H&E stained brain architecture. The target number for each sample was approximately 500 cells counted. The size of ventricular area and thickness of corpus callosum and cortex was measured by ImageJ software. The paired-t test was used to compare the differences between wild-type and FoxO null brains.
Statistical Analysis
All the data were analyzed by unpaired two tail Student's t-test unless specified in the figure legend.
Supplementary Material
01
02
03
04
5
Supplemental Figure Legends:
Fig. S1. Expression of FoxOs in mouse brain. IHC analysis on the expression of FoxO1 in cortex (a, b) and SVZ (c, d), FoxO3 (e, f), and Cre (h) in NSC-rich SVZ of 10 day old FoxO WT (upper) and null (lower) mice. Red arrows point to neurons that do not express hGFAP-Cre therefore retain FoxO expression. (g) x-gal staining on a brain slice from hGFAP-cre;ROSA26R mice shows blue cells in SVZ underwent Cre-mediated recombination. EP, ependymal cells, LV, lateral ventricle. Bars= 40 and 60μm. (i) Immunofluorescence analysis on the expression of FoxO1 and FoxO3 in Sox2+ neural progenitors of SVZ in P8 mice. OL, overlap image of two separate channel images of marker co-stained panels. Note lack of FoxO1 and 3 staining in Sox2+ cells in FoxO null brain (right panels). Bar=30μm. Expression of FoxO1 (j) and 3 (k) in the brain of E15.5 embryos. High magnification images of boxed regions are shown on the right. Bars=200μm, 40μm. (l) Expression of FoxO1 and 3 in Nestin+ neural progenitors of VZ from boxed region of panel (j-k) is examined. Note that FoxO 1 and 3 are both nucleocytoplasmically present in the Nestin+ cells of VZ and FoxO3 is more broadly and strongly expressed. Bar=40μm. Expression of FoxO1 (m) and FoxO3 (n) in different anatomic regions of WT mouse brain (8 week old). FoxO1 and 3 IHC used Nova red and DAB chromophore, respectively. 400X microscopic field of indicated regions were taken and boxed areas are shown in the right panels. Identification of cell types expressing FoxO1 or 3 by marker co-staining for cortical neurons (o, NeuN+), astrocytes (p, GFAP+) and oligodendrocytes (q, Olig2+) from corpus callosum. Note all three cell types express detectable level of FoxO1 and 3 that disappear in FoxO null brain (right panels). Bars= 30μm.
Fig. S2. mRNA (a) and protein (b) expression of FoxO 1,3, and 4 in Rosa26-Cre ERT2 -(black bar) and + (gray bar) NSC 1 week after 4OHT treatment. NSC were derived from E13.5 embryo (a) and P1 pups (b). (c) Expression and localization of FoxO 1 and 3 in NSC (P1) in vitro. Both WT and FoxO null NSC grown as neurospheres were dissociated, cytospun, and co-stained for FoxOs (green), Nestin (red), Sox2 (magenta), and DAPI (blue). Bar= 40μm.
Fig. S3. (a) FoxO deficiency does not affect overall brain cyotarchitecture and anatomy of young mice. H&E stained coronal brain sections of FoxO WT (right) and null (left) of 3 week old mice. From the top, corpus callosum, SVZ, hippocampus, cerebellum, internal granular layers and Purkinje cell layers are shown. All paired images are under same magnification. Scales bars from top to bottom= 1mm, 1mm, 1mm, 1mm, 200μm. (b) Brain weights of indicated genotypes from 3-24 week old mice were plotted after normalizing with average brain weights of their age-matched WT littermate controls. (c) Thickness of cortices (upper) and corpus callosum (lower) from 24-32 week old mice were plotted. Numbers are normalized by their age-matched WT littermate controls. n=4, **, p=0.0136, *, p=0.0444
Fig. S4. FoxO deficient brain exhibits signs of degeneration in older mice. (a) H&E images of lateral ventricle dilation in 20 week old FoxO null brain (right). Bar=1mm. 600x microscopic field of boxed regions are shown on the side. Arrow points to abnormal thinning of ventricular wall. Bar=60μm. (b) Degree of ventricular dilation was measured by unilateral ventricular surface area from 6 pairs of 15-32 week old mice. Y axis displays % difference. WT vs null is 100% vs 751.1%, **, p=0.011. Bar=200μm. (c) Decrease in olfactory bulb neurogenesis in 32 weeks old FoxO null mice. Mice received BrdU in drinking water for a week and taken off of BrdU for three weeks before sacrificed. BrdU+ cells (upper panels) and colocalization of BrdU+ and NeuN+ cells (lower panels) are shown. Percent of BrdU+ cells among NeuN+ neurons within olfactory bulb is plotted. WT vs null is 1.76% vs 0.92%, **, p=0.018. Bar=60μm
Fig. S5. (a) No significant differences in cell size of FoxO WT (right) and null (left) 15 week old mice. Left panels show GFAP positive astrocytes and right panels are TuJ1 positive neurons in FoxO WT and null brains. Bar= 40 μm. (b) Average volume of dye-loaded cortical neurons isolated from neonatal mice (p1) (n=60, p=0.21 by two tail t-test). Representative z stack view of FoxO WT (right) and null (left) neurons is shown. Heat map shows the distance from the bottom (μm). Bar= 30μm. (c) Size of astrocytes isolated from neonatal mice (P1) is measured by flow cytometry. Representative histogram is shown (median values are 128k and 127k for WT and null respectively). (d) FoxO null astrocyte from old mice shows signs of activation. In vitro differentiated astrocytes from FoxO null NSC derived from adult SVZ shows activated morphology. Bar= 30μm (e) Presence of hypertrophic GFAP-positive astrocytes in the cortex of 32 week old FoxO null brain (top). Corresponding region of wild type control lacks GFAP positive cells (bottom). Bar= 40μm.
Fig. S6. Histograms display size distribution of all the neurospheres. (a, b) WT/KO 1 and 2 are two independent primary cultures of WT and FoxO null NSC. Cultures are from 18-22 week old (a) and 4 week old (b) mice. NAC (1mM) treated (c) or SESN3-retrovirus-transduced (d) NSC culture. (e) NT or shASPM_1 or shASPM_2 lentivirus-infected FoxO null NSC culture.
Fig. S7. (a) Increased mRNA expression of canonical Wnt target genes, CyclinD2 and Myc in FoxO null (gray bar) NSC compared to WT (black bar). (b) Knockdown of sFRP1/2 and SOST by individual (200nM) or mixture (mix, each 100nM) siRNAs. mRNA expression was measured by rtqPCR 24hr after the trasfection.
Fig. S8. Molecular targets of FoxO in NSC. (a) Regulation on putative FoxO target genes. For mRNA expression of each putative target was confirmed by rtqPCR after enforced expression of CA1 or CA3 in FoxO null NSC. Y axis is fold change over control. Mean ± s.e. is shown. (b) ChIP analysis for FoxO1 and FoxO3. Targets bearing conserved FoxO BE were tested for the presence of FoxO binding. Chromatins from FoxO null NSC were used as control. 5% of inputs were used for PCR. ‘-‘ and ‘+’ stand for null and WT respectively.
Supplemental Table S1. List of genes differentially expressed in FoxO null embryonic (E13.5) NSC.
Supplemental Table S2. List of genes differentially expressed with SAM>2.0 from Supplemental Table S1.
Supplemental Table S3. FoxO-dependent changes in expression of genes regulating intracellular ROS levels. List is from selected from Supplemental Table S1. Fold changes are calculated using two different statistical methods.
Supplemental Experimental Procedure:
Isolation and culture of NSC. Embryonic NSC were isolated from E13.5 ganglionic eminence as described (Reynolds and Weiss, 1992). Cells were physically dissociated, and propagated in Dulbecco's Modification of Eagle's Medium/F-12 1:1 mix supplemented with B12 and 20ng/ml of bFGF and recombinant human EGF. Primary neurospheres were raised and passaged by enzymatic dissociation using TrypLE™express (Invitrogen). To measure proliferation of NSC 180,000 cells were plated in 6 well plates and then dissociated, counted, and replated every 5 days. Population doubling was calculated as log2 (cell number counted/plated). Adult NSC were isolated from SVZ of different age of mice. SVZ were dissected out in DMEM/F12 media and mechanically dissociated and filtered through 40 micron mesh. Cells were cultured as above. For differentiation cells were treated with neurobasal media with 1% FBS was added to induce differentiation into neuronal and glial cell types. Cultures were assessed by immunocytochemistry for the presence of different cell lineages after 4-5 days. Embryonic NSC for gene expression profiling study were dissociated and cultured at 100,000 cells/ml with the addition of growth factors every three days. For in vitro deletion of FoxOs freshly isolated _Rosa26-CreERT2+ or - ;FoxO1/3/4L/_L NSC were cultured for 1 week then 400nM 4-hydroxy tamoxifen (4OHT) was added to the culture twice in 48hr interval or for adult NSC cells were grown in the presence of 100nM 4OHT during the formation of primary spheres. All the measurement for mRNA and protein expression was performed on actively growing neurospheres.
Immunohistochemistry and immunofluorescence. Mice were perfused with 4% paraformaldehyde and brains were removed and fixed further and processed for hematoxylin and eosin (H&E) by standard techniques. The entire brain was sectioned in 1–2 mm coronal blocks and submitted in one cassette for paraffin embedding to facilitate analysis of the whole brain. For immunohistochemical analysis, sections were prepared for staining with Paraffin embedded tissues were sectioned at 5 μm thickness. Sections were hydrated in series of ethanol and equilibrated in PBS. Slides were antigen-retrieved using citrate buffer and processed following manufacturer's recommendation (DAKO envision kit). Slides were incubated overnight at 4°C with the following antibodies: 1:400 Goat-anti-Sox-2 (sc-12, Santa Cruz Biotechnology), 1:200 goat-anti-doublecortin (C-18, Santa Cruz Biotechnology), NeuN (Chemicon), S100 (Neomarkers), FoxO1 (clone C29H4, Cell signaling Technology), FoxO3 (H-144, Santa Cruz Biotechnology), Ki67 (DAKO), GFAP (DAKO), ASPM (Bethyl laboratories), Cre (Covance).. Following secondary antibody incubation, sections were then incubated with DAB and then counterstained with hematoxylin. For immunocytochemistry neurospheres were dissociated and cytospun on a slide in 2% BSA/PBS, fixed in 4% paraformaldehyde and permeabilized with 0.2% TritonX-100 and incubated with the primary antibody. The following antibodies were used: rabbit anti-Ki67(1:100), mouse anti-III-tubulin (Tuj1; 1:400, covance), mouse anti-nestin (1:100, Chemicon), and 8-oxoG (Chemicon). Secondary antibodies (Alexa Fluor-conjegated, Invitrogen) were incubated and coverslips were mounted with Prolong gold antifade mounting media (Invitrogen). Images were acquired on Yokogawa spinning disk confocal microscope (Andor). For quantitation several random visual fields from each section were viewed. The percentage of positive cells was calculated in relation to the total number of DAPI-stained nuclei present in the culture. All experiments were independently repeated from three separate cultures.
Microarray analysis and identification of putative FoxO BE. Cells were washed and cultured additional 72hr after 4OHT treatment before the harvest of their RNA using Trizol (Invitrogen) and the RNeasy mini kit (Qiagen). Gene expression profiling was performed utilizing the Affymetrix 430 2.0 chips. dChip (Li and Wong, 2001; Li and Wong, 2003) was used to normalize arrays and to compute expression indices. A list of 186 genes significantly differentially expressed between wild type and FoxO null NSC was generated using the SAM statistic. It was required that the absolute value of the SAM statistic be greater than two. Then the 5kb promoter regions of these genes were isolated and scanned for enrichment of the 550 binding motifs in TRANSFAC. Enrichment was assessed by comparing the target regions to matched control regions at the same distance from the transcription start sites of random genes. Promoter analysis on these gene sets for FoxO BE used the CisGenome software (http://www.biostat.jhsph.edu/~hji/cisgenome/).
Gain or loss of function assay. CA or DBD-FoxO1 (Frescas et al., 2005) and CA-FoxO3 mutant (T32A/S253A/S315A) in adenoviral vectors were generously provided by Domenico Accilli (Columbia University) and purchased from Vector Labs respectively. Viral particles were amplified in NSC medium from HEK293 and titer was determined. shRNAs in pLKO lentiviral vector were acquired from TRC (the RNAi consortium). shRNAs for ASPM are TRCN0000110637-9. Two shRNAs for each gene were selected based on the knockdown efficiency. Vector control or ineffective shRNAs targeting same gene were used as an experimental control. Lentiviral particles were generated in 293T cells by transfecting 1, 6, 6μg of pVSV-G, Δ8,9, and viral vector respectively in 100mm dish using FuGENE6 (Roche) according to the manufacturer's protocol. Supernatant containing viral particles was collected between 48-72hr post transfection and ultracentrifuged (Beckman Coulter) at 23,000rpm for 3hrs. Alternatively On-target plus SMART pool pre designed siRNA mix from Dharmacon were used to knockdown the expression of target genes by Lipofectamine2000 using OptiMEM media for 3hrs. For gain of function experiment human SESN3 ORF was cloned into MSCV-puro-v5 gateway retroviral destination vector and virus was generated by co-transfecting pCL-eco into HEK293T cells. Supernatant was collected and concentrated using centriprep YM-30. NSC were plated on poly-L-ornithine and fibronectin-coated dishes were spin-infected with viral supernatant containing polybrene (8μg/ml). Cells were incubated 96 hrs prior to harvest for verification of knockdown by rtqPCR.
Cell Size Measurement. Primary cortical neurons and astrocytes were isolated from neonatal mice (P1) as described previously (Bachoo et al., 2004). Neurons were cultured in neurobasal media with B27 supplement grown on poly-d-lysine coated coverslips. Neurons were labeled with DiD and scanned though the cell body for collection of Z-axis stacks with 1μm interval using 100x objective on Olympus laser scanning confocal microscope. Collected images were processed for cellular volume in cubic micronmeter using an in-house semi-automated segmentation algorithm. Astrocytes were lifted by gentle trypsinization and dead cells were gated out by propidium iodide staining. Histogram was created with FSC as X-axis and median values were used for the comparison of the cell size.
Quantitation of Olfactory bulb neurogenesis 32 week old mice received BrdU (1mg/ml) in the drinking water for a week and taken off of BrdU for three weeks before sacrificed. Brain tissues were processed and stained with BrdU and NeuN as above and double positive cells were scored from 5 different areas of olfactory bulb.
Chromatin Immunoprecipitation (ChIP) assay. Two million NSC either from FoxO null or WT embryos were crosslinked by addition of 1% formaldehyde to the medium for 10 min followed by quenching with 125mM glycine. The cells were resuspended in lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris (pH 8.1), Protease Inhibitor Cocktail II (Roche)), sonicated 10 times for 30 s with 2 min idle time, the lysates were cleared by centrifugation. One hundred microliters of the sheared DNA was diluted 1:10 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl). Chromatin solution was precleared for 1 h at 4°C with 60 μl of protein G-agarose/salmon sperm DNA. Ten microliters of the precleared chromatin solutions was saved for assessment of input chromatin, and the rest of the precleared chromatin solutions was incubated with 1ug of anti-Rabbit IgG, anti-FoxO1 IgG, or anti-FoxO3 IgG (Cell signaling, Santa Cruz biotechnology, sc-11350, 11351, respectively) overnight at 4°C. Immune complexes were collected on 60 μl of protein A/G Plus-agarose/salmon sperm beads. Precipitates were washed sequentially for 5 min each in Low salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 150 mM NaCl], High salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 500 mM NaCl], LiCl immune wash buffer. Precipitates were then washed twice with 1X TE (pH. 8.0) and extracted two times with 1% SDS, 0.1 M NaHCO3. Elutes were incubated at 65°C with 0.25 M NaCl overnight to reverse cross-linking followed by another 1 hr incubation at 45°C with 10 μM EDTA, 40 μM Tris-HCl (pH 6.8) and 2μg Proteinase K (Sigma). The DNA was purified using a PCR purification kit (Qiagen) with 60 μl of distilled water. One microliter of immunoprecipitated DNA was used for each PCR reaction. Optimal cycle number was determined by Ct values from rtqPCR. Enrichment was calculated and plotted by % of total input (1:100 diluted).
RNA isolation and real-time PCR. Total RNA was extracted using the PureLink Micro-to-Midi kit (invitrogen) and treated with RQ1 RNase-free DNase Set (Promega). First-strand cDNA was synthesized using 1μg of total RNA and SuperscriptII (Invitrogen). rtqPCR was performed in duplicates with a MxPro3000 and SYBR Greener qPCR mix (Invitrogen). The relative amount of specific mRNA was normalized to beta actin. Primer sequences are available upon request.
Acknowledgements
J-H.P is Damon Runyon Fellow was supported by the Damon Runyon Cancer Research Foundation (DRG 1900-06). Grant support includes the Claudia Adam Barr Foundation (J-H.P.), ACS fellowship (F.M.), ABTA fellowship (H.Y.), Ellison Medical Foundation (R.A.D.), and 5P01CA95616 (R.A.D., K.L.L, L.C.). R.A.D. is an American Cancer Society Research Professor and supported by the Robert A. and Renee E. Belfer Foundation Institute for Applied Cancer Science. We thank D. Pellman and S. Yoshida for the deconvolution microscope use and advice, R. Maser for MSCV-puro-v5 gateway vector, W. Hahn for shRNA constructs, J. Ling for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez B, Martinez AC, Burgering BM, Carrera AC. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature. 2001;413:744–747. doi: 10.1038/35099574. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, et al. Molecular diversity of astrocytes with implications for neurological disorders. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista D, Ferrari CC, Gage FH, Pitossi FJ. Neurogenic niche modulation by activated microglia: transforming growth factor beta increases neurogenesis in the adult dentate gyrus. Eur J Neurosci. 2006;23:83–93. doi: 10.1111/j.1460-9568.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- Birkenkamp KU, Essafi A, van der Vos KE, da Costa M, Hui RC, Holstege F, Koenderman L, Lam EW, Coffer PJ. FOXO3a induces differentiation of Bcr-Abl-transformed cells through transcriptional down-regulation of Id1. The Journal of biological chemistry. 2007;282:2211–2220. doi: 10.1074/jbc.M606669200. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. ASPM is a major determinant of cerebral cortical size. Nature genetics. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science (New York, NY. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Dombkowski D, Stier S, Scadden DT. Stem cell repopulation efficiency but not pool size is governed by p27(kip1). Nature medicine. 2000a;6:1235–1240. doi: 10.1038/81335. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science (New York, NY. 2000b;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002a;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Verdugo JM, Caille I, Alvarez-Buylla A, Chao MV, Casaccia-Bonnefil P. Lack of the cell-cycle inhibitor p27Kip1 results in selective increase of transit-amplifying cells for adult neurogenesis. J Neurosci. 2002b;22:2255–2264. doi: 10.1523/JNEUROSCI.22-06-02255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. The Journal of biological chemistry. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca W, Deptala A, Bedner E, Li X, Melamed MR, Darzynkiewicz Z. Analysis of human tumors by laser scanning cytometry. Methods Cell Biol. 2001;64:421–443. doi: 10.1016/s0091-679x(01)64024-x. [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, Zack JA, Geschwind DH, Liu X, Kornblum HI, Wu H. PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science (New York, NY. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiological genomics. 2007;31:32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6:134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Geles KG, Paik JH, DePinho RA, Tjian R. Codependent activators direct myoblast-specific MyoD transcription. Dev Cell. 2008;15:534–546. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Ohtsuka T, Takao K, Miyakawa T, Yamaguchi M, Mori K, Ikeda T, Itohara S, Kageyama R. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nat Neurosci. 2008;11:1153–1161. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nature medicine. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Jin K, Sun Y, Xie L, Batteur S, Mao XO, Smelick C, Logvinova A, Greenberg DA. Neurogenesis and aging: FGF-2 and HB-EGF restore neurogenesis in hippocampus and subventricular zone of aged mice. Aging Cell. 2003;2:175–183. doi: 10.1046/j.1474-9728.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes & development. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. DNA-Chip Analyzer (dChip). In: Parmigiani G, E.S. G, Irizarry R, Zeger SL, editors. The analysis of gene expression data: methods and software. Springer; 2003. [Google Scholar]
- Lim D, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;58:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science (New York, NY. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, Dubus P, Barbacid M. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead CM, Reynolds BA, Craig CG, McBurney MW, Staines WA, Morassutti D, Weiss S, van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paling NR, Wheadon H, Bone HK, Welham MJ. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. The Journal of biological chemistry. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- Palma V, Lim DA, Dahmane N, Sanchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Ruiz i Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes & development. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science (New York, NY. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Shen H, Franklin DS, Scadden DT, Cheng T. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat Cell Biol. 2004;6:436–442. doi: 10.1038/ncb1126. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
5
Supplemental Figure Legends:
Fig. S1. Expression of FoxOs in mouse brain. IHC analysis on the expression of FoxO1 in cortex (a, b) and SVZ (c, d), FoxO3 (e, f), and Cre (h) in NSC-rich SVZ of 10 day old FoxO WT (upper) and null (lower) mice. Red arrows point to neurons that do not express hGFAP-Cre therefore retain FoxO expression. (g) x-gal staining on a brain slice from hGFAP-cre;ROSA26R mice shows blue cells in SVZ underwent Cre-mediated recombination. EP, ependymal cells, LV, lateral ventricle. Bars= 40 and 60μm. (i) Immunofluorescence analysis on the expression of FoxO1 and FoxO3 in Sox2+ neural progenitors of SVZ in P8 mice. OL, overlap image of two separate channel images of marker co-stained panels. Note lack of FoxO1 and 3 staining in Sox2+ cells in FoxO null brain (right panels). Bar=30μm. Expression of FoxO1 (j) and 3 (k) in the brain of E15.5 embryos. High magnification images of boxed regions are shown on the right. Bars=200μm, 40μm. (l) Expression of FoxO1 and 3 in Nestin+ neural progenitors of VZ from boxed region of panel (j-k) is examined. Note that FoxO 1 and 3 are both nucleocytoplasmically present in the Nestin+ cells of VZ and FoxO3 is more broadly and strongly expressed. Bar=40μm. Expression of FoxO1 (m) and FoxO3 (n) in different anatomic regions of WT mouse brain (8 week old). FoxO1 and 3 IHC used Nova red and DAB chromophore, respectively. 400X microscopic field of indicated regions were taken and boxed areas are shown in the right panels. Identification of cell types expressing FoxO1 or 3 by marker co-staining for cortical neurons (o, NeuN+), astrocytes (p, GFAP+) and oligodendrocytes (q, Olig2+) from corpus callosum. Note all three cell types express detectable level of FoxO1 and 3 that disappear in FoxO null brain (right panels). Bars= 30μm.
Fig. S2. mRNA (a) and protein (b) expression of FoxO 1,3, and 4 in Rosa26-Cre ERT2 -(black bar) and + (gray bar) NSC 1 week after 4OHT treatment. NSC were derived from E13.5 embryo (a) and P1 pups (b). (c) Expression and localization of FoxO 1 and 3 in NSC (P1) in vitro. Both WT and FoxO null NSC grown as neurospheres were dissociated, cytospun, and co-stained for FoxOs (green), Nestin (red), Sox2 (magenta), and DAPI (blue). Bar= 40μm.
Fig. S3. (a) FoxO deficiency does not affect overall brain cyotarchitecture and anatomy of young mice. H&E stained coronal brain sections of FoxO WT (right) and null (left) of 3 week old mice. From the top, corpus callosum, SVZ, hippocampus, cerebellum, internal granular layers and Purkinje cell layers are shown. All paired images are under same magnification. Scales bars from top to bottom= 1mm, 1mm, 1mm, 1mm, 200μm. (b) Brain weights of indicated genotypes from 3-24 week old mice were plotted after normalizing with average brain weights of their age-matched WT littermate controls. (c) Thickness of cortices (upper) and corpus callosum (lower) from 24-32 week old mice were plotted. Numbers are normalized by their age-matched WT littermate controls. n=4, **, p=0.0136, *, p=0.0444
Fig. S4. FoxO deficient brain exhibits signs of degeneration in older mice. (a) H&E images of lateral ventricle dilation in 20 week old FoxO null brain (right). Bar=1mm. 600x microscopic field of boxed regions are shown on the side. Arrow points to abnormal thinning of ventricular wall. Bar=60μm. (b) Degree of ventricular dilation was measured by unilateral ventricular surface area from 6 pairs of 15-32 week old mice. Y axis displays % difference. WT vs null is 100% vs 751.1%, **, p=0.011. Bar=200μm. (c) Decrease in olfactory bulb neurogenesis in 32 weeks old FoxO null mice. Mice received BrdU in drinking water for a week and taken off of BrdU for three weeks before sacrificed. BrdU+ cells (upper panels) and colocalization of BrdU+ and NeuN+ cells (lower panels) are shown. Percent of BrdU+ cells among NeuN+ neurons within olfactory bulb is plotted. WT vs null is 1.76% vs 0.92%, **, p=0.018. Bar=60μm
Fig. S5. (a) No significant differences in cell size of FoxO WT (right) and null (left) 15 week old mice. Left panels show GFAP positive astrocytes and right panels are TuJ1 positive neurons in FoxO WT and null brains. Bar= 40 μm. (b) Average volume of dye-loaded cortical neurons isolated from neonatal mice (p1) (n=60, p=0.21 by two tail t-test). Representative z stack view of FoxO WT (right) and null (left) neurons is shown. Heat map shows the distance from the bottom (μm). Bar= 30μm. (c) Size of astrocytes isolated from neonatal mice (P1) is measured by flow cytometry. Representative histogram is shown (median values are 128k and 127k for WT and null respectively). (d) FoxO null astrocyte from old mice shows signs of activation. In vitro differentiated astrocytes from FoxO null NSC derived from adult SVZ shows activated morphology. Bar= 30μm (e) Presence of hypertrophic GFAP-positive astrocytes in the cortex of 32 week old FoxO null brain (top). Corresponding region of wild type control lacks GFAP positive cells (bottom). Bar= 40μm.
Fig. S6. Histograms display size distribution of all the neurospheres. (a, b) WT/KO 1 and 2 are two independent primary cultures of WT and FoxO null NSC. Cultures are from 18-22 week old (a) and 4 week old (b) mice. NAC (1mM) treated (c) or SESN3-retrovirus-transduced (d) NSC culture. (e) NT or shASPM_1 or shASPM_2 lentivirus-infected FoxO null NSC culture.
Fig. S7. (a) Increased mRNA expression of canonical Wnt target genes, CyclinD2 and Myc in FoxO null (gray bar) NSC compared to WT (black bar). (b) Knockdown of sFRP1/2 and SOST by individual (200nM) or mixture (mix, each 100nM) siRNAs. mRNA expression was measured by rtqPCR 24hr after the trasfection.
Fig. S8. Molecular targets of FoxO in NSC. (a) Regulation on putative FoxO target genes. For mRNA expression of each putative target was confirmed by rtqPCR after enforced expression of CA1 or CA3 in FoxO null NSC. Y axis is fold change over control. Mean ± s.e. is shown. (b) ChIP analysis for FoxO1 and FoxO3. Targets bearing conserved FoxO BE were tested for the presence of FoxO binding. Chromatins from FoxO null NSC were used as control. 5% of inputs were used for PCR. ‘-‘ and ‘+’ stand for null and WT respectively.
Supplemental Table S1. List of genes differentially expressed in FoxO null embryonic (E13.5) NSC.
Supplemental Table S2. List of genes differentially expressed with SAM>2.0 from Supplemental Table S1.
Supplemental Table S3. FoxO-dependent changes in expression of genes regulating intracellular ROS levels. List is from selected from Supplemental Table S1. Fold changes are calculated using two different statistical methods.
Supplemental Experimental Procedure:
Isolation and culture of NSC. Embryonic NSC were isolated from E13.5 ganglionic eminence as described (Reynolds and Weiss, 1992). Cells were physically dissociated, and propagated in Dulbecco's Modification of Eagle's Medium/F-12 1:1 mix supplemented with B12 and 20ng/ml of bFGF and recombinant human EGF. Primary neurospheres were raised and passaged by enzymatic dissociation using TrypLE™express (Invitrogen). To measure proliferation of NSC 180,000 cells were plated in 6 well plates and then dissociated, counted, and replated every 5 days. Population doubling was calculated as log2 (cell number counted/plated). Adult NSC were isolated from SVZ of different age of mice. SVZ were dissected out in DMEM/F12 media and mechanically dissociated and filtered through 40 micron mesh. Cells were cultured as above. For differentiation cells were treated with neurobasal media with 1% FBS was added to induce differentiation into neuronal and glial cell types. Cultures were assessed by immunocytochemistry for the presence of different cell lineages after 4-5 days. Embryonic NSC for gene expression profiling study were dissociated and cultured at 100,000 cells/ml with the addition of growth factors every three days. For in vitro deletion of FoxOs freshly isolated _Rosa26-CreERT2+ or - ;FoxO1/3/4L/_L NSC were cultured for 1 week then 400nM 4-hydroxy tamoxifen (4OHT) was added to the culture twice in 48hr interval or for adult NSC cells were grown in the presence of 100nM 4OHT during the formation of primary spheres. All the measurement for mRNA and protein expression was performed on actively growing neurospheres.
Immunohistochemistry and immunofluorescence. Mice were perfused with 4% paraformaldehyde and brains were removed and fixed further and processed for hematoxylin and eosin (H&E) by standard techniques. The entire brain was sectioned in 1–2 mm coronal blocks and submitted in one cassette for paraffin embedding to facilitate analysis of the whole brain. For immunohistochemical analysis, sections were prepared for staining with Paraffin embedded tissues were sectioned at 5 μm thickness. Sections were hydrated in series of ethanol and equilibrated in PBS. Slides were antigen-retrieved using citrate buffer and processed following manufacturer's recommendation (DAKO envision kit). Slides were incubated overnight at 4°C with the following antibodies: 1:400 Goat-anti-Sox-2 (sc-12, Santa Cruz Biotechnology), 1:200 goat-anti-doublecortin (C-18, Santa Cruz Biotechnology), NeuN (Chemicon), S100 (Neomarkers), FoxO1 (clone C29H4, Cell signaling Technology), FoxO3 (H-144, Santa Cruz Biotechnology), Ki67 (DAKO), GFAP (DAKO), ASPM (Bethyl laboratories), Cre (Covance).. Following secondary antibody incubation, sections were then incubated with DAB and then counterstained with hematoxylin. For immunocytochemistry neurospheres were dissociated and cytospun on a slide in 2% BSA/PBS, fixed in 4% paraformaldehyde and permeabilized with 0.2% TritonX-100 and incubated with the primary antibody. The following antibodies were used: rabbit anti-Ki67(1:100), mouse anti-III-tubulin (Tuj1; 1:400, covance), mouse anti-nestin (1:100, Chemicon), and 8-oxoG (Chemicon). Secondary antibodies (Alexa Fluor-conjegated, Invitrogen) were incubated and coverslips were mounted with Prolong gold antifade mounting media (Invitrogen). Images were acquired on Yokogawa spinning disk confocal microscope (Andor). For quantitation several random visual fields from each section were viewed. The percentage of positive cells was calculated in relation to the total number of DAPI-stained nuclei present in the culture. All experiments were independently repeated from three separate cultures.
Microarray analysis and identification of putative FoxO BE. Cells were washed and cultured additional 72hr after 4OHT treatment before the harvest of their RNA using Trizol (Invitrogen) and the RNeasy mini kit (Qiagen). Gene expression profiling was performed utilizing the Affymetrix 430 2.0 chips. dChip (Li and Wong, 2001; Li and Wong, 2003) was used to normalize arrays and to compute expression indices. A list of 186 genes significantly differentially expressed between wild type and FoxO null NSC was generated using the SAM statistic. It was required that the absolute value of the SAM statistic be greater than two. Then the 5kb promoter regions of these genes were isolated and scanned for enrichment of the 550 binding motifs in TRANSFAC. Enrichment was assessed by comparing the target regions to matched control regions at the same distance from the transcription start sites of random genes. Promoter analysis on these gene sets for FoxO BE used the CisGenome software (http://www.biostat.jhsph.edu/~hji/cisgenome/).
Gain or loss of function assay. CA or DBD-FoxO1 (Frescas et al., 2005) and CA-FoxO3 mutant (T32A/S253A/S315A) in adenoviral vectors were generously provided by Domenico Accilli (Columbia University) and purchased from Vector Labs respectively. Viral particles were amplified in NSC medium from HEK293 and titer was determined. shRNAs in pLKO lentiviral vector were acquired from TRC (the RNAi consortium). shRNAs for ASPM are TRCN0000110637-9. Two shRNAs for each gene were selected based on the knockdown efficiency. Vector control or ineffective shRNAs targeting same gene were used as an experimental control. Lentiviral particles were generated in 293T cells by transfecting 1, 6, 6μg of pVSV-G, Δ8,9, and viral vector respectively in 100mm dish using FuGENE6 (Roche) according to the manufacturer's protocol. Supernatant containing viral particles was collected between 48-72hr post transfection and ultracentrifuged (Beckman Coulter) at 23,000rpm for 3hrs. Alternatively On-target plus SMART pool pre designed siRNA mix from Dharmacon were used to knockdown the expression of target genes by Lipofectamine2000 using OptiMEM media for 3hrs. For gain of function experiment human SESN3 ORF was cloned into MSCV-puro-v5 gateway retroviral destination vector and virus was generated by co-transfecting pCL-eco into HEK293T cells. Supernatant was collected and concentrated using centriprep YM-30. NSC were plated on poly-L-ornithine and fibronectin-coated dishes were spin-infected with viral supernatant containing polybrene (8μg/ml). Cells were incubated 96 hrs prior to harvest for verification of knockdown by rtqPCR.
Cell Size Measurement. Primary cortical neurons and astrocytes were isolated from neonatal mice (P1) as described previously (Bachoo et al., 2004). Neurons were cultured in neurobasal media with B27 supplement grown on poly-d-lysine coated coverslips. Neurons were labeled with DiD and scanned though the cell body for collection of Z-axis stacks with 1μm interval using 100x objective on Olympus laser scanning confocal microscope. Collected images were processed for cellular volume in cubic micronmeter using an in-house semi-automated segmentation algorithm. Astrocytes were lifted by gentle trypsinization and dead cells were gated out by propidium iodide staining. Histogram was created with FSC as X-axis and median values were used for the comparison of the cell size.
Quantitation of Olfactory bulb neurogenesis 32 week old mice received BrdU (1mg/ml) in the drinking water for a week and taken off of BrdU for three weeks before sacrificed. Brain tissues were processed and stained with BrdU and NeuN as above and double positive cells were scored from 5 different areas of olfactory bulb.
Chromatin Immunoprecipitation (ChIP) assay. Two million NSC either from FoxO null or WT embryos were crosslinked by addition of 1% formaldehyde to the medium for 10 min followed by quenching with 125mM glycine. The cells were resuspended in lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris (pH 8.1), Protease Inhibitor Cocktail II (Roche)), sonicated 10 times for 30 s with 2 min idle time, the lysates were cleared by centrifugation. One hundred microliters of the sheared DNA was diluted 1:10 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl). Chromatin solution was precleared for 1 h at 4°C with 60 μl of protein G-agarose/salmon sperm DNA. Ten microliters of the precleared chromatin solutions was saved for assessment of input chromatin, and the rest of the precleared chromatin solutions was incubated with 1ug of anti-Rabbit IgG, anti-FoxO1 IgG, or anti-FoxO3 IgG (Cell signaling, Santa Cruz biotechnology, sc-11350, 11351, respectively) overnight at 4°C. Immune complexes were collected on 60 μl of protein A/G Plus-agarose/salmon sperm beads. Precipitates were washed sequentially for 5 min each in Low salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 150 mM NaCl], High salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 500 mM NaCl], LiCl immune wash buffer. Precipitates were then washed twice with 1X TE (pH. 8.0) and extracted two times with 1% SDS, 0.1 M NaHCO3. Elutes were incubated at 65°C with 0.25 M NaCl overnight to reverse cross-linking followed by another 1 hr incubation at 45°C with 10 μM EDTA, 40 μM Tris-HCl (pH 6.8) and 2μg Proteinase K (Sigma). The DNA was purified using a PCR purification kit (Qiagen) with 60 μl of distilled water. One microliter of immunoprecipitated DNA was used for each PCR reaction. Optimal cycle number was determined by Ct values from rtqPCR. Enrichment was calculated and plotted by % of total input (1:100 diluted).
RNA isolation and real-time PCR. Total RNA was extracted using the PureLink Micro-to-Midi kit (invitrogen) and treated with RQ1 RNase-free DNase Set (Promega). First-strand cDNA was synthesized using 1μg of total RNA and SuperscriptII (Invitrogen). rtqPCR was performed in duplicates with a MxPro3000 and SYBR Greener qPCR mix (Invitrogen). The relative amount of specific mRNA was normalized to beta actin. Primer sequences are available upon request.