Circadian changes in long noncoding RNAs in the pineal gland (original) (raw)
Abstract
Long noncoding RNAs (lncRNAs) play a broad range of biological roles, including regulation of expression of genes and chromosomes. Here, we present evidence that lncRNAs are involved in vertebrate circadian biology. Differential night/day expression of 112 lncRNAs (0.3 to >50 kb) occurs in the rat pineal gland, which is the source of melatonin, the hormone of the night. Approximately one-half of these changes reflect nocturnal increases. Studies of eight lncRNAs with 2- to >100-fold daily rhythms indicate that, in most cases, the change results from neural stimulation from the central circadian oscillator in the suprachiasmatic nucleus (doubling time = 0.5–1.3 h). Light exposure at night rapidly reverses (halving time = 9–32 min) levels of some of these lncRNAs. Organ culture studies indicate that expression of these lncRNAs is regulated by norepinephrine acting through cAMP. These findings point to a dynamic role of lncRNAs in the circadian system.
Keywords: RNA sequencing, neuroendocrine regulation, differential expression, chronobiology
Long noncoding RNAs (lncRNAs; >200 bp), including long intergenic noncoding RNAs, are of special interest because of their large and growing numbers and the possibility that they may represent a substantially underrepresented, functionally critical component of the genome (1–3). Current research points to a role in cellular regulation through interaction with proteins and DNA (4–6). The abundance of many lncRNAs changes gradually during development, and many are involved in epigenetic processes impacting gene expression (7–12) [e.g., chromosome inactivation by _XIST_ (13), imprinting by Kcnq1ot1 (14), development and cancer by HOTAIR (15, 16), cancer by PCA3 (17), and disease by PISTR1 (18)]. From this information, it is generally thought that lncRNAs control long-term processes; however, it is also clear that they may play roles in more dynamic processes, including signaling (19–23).
Here, we addressed the broad issue of whether lncRNAs may play a role in vertebrate circadian systems, which seems to be the case in plants (24). We searched for lncRNAs that exhibit daily changes in abundance in the rat pineal gland. The pineal gland is responsible for the daily rhythm in circulating melatonin, the hormone of the night. In this tissue, daily changes in the abundance of protein-coding transcripts (25) are controlled by a neural pathway linking the master mammalian circadian oscillator, the suprachiasmatic nucleus (SCN), to the pineal gland; SCN stimulation of this pathway causes release of norepinephrine (NE) from sympathetic nerve processes. NE acts through an adrenergic receptor/cAMP mechanism to broadly control gene expression. These changes seem to establish optimal conditions for the precise daily rhythm in melatonin production (25). Results presented here provide an indication that lncRNAs play a role in the vertebrate circadian system.
Results
Daily Changes in lncRNA Abundance in the Pineal Gland.
lncRNAs were sought in RNA sequencing (RNA-Seq) results from analysis of rat pineal glands and other tissues. A computer-based search algorithm was used to find lncRNAs with either daily changes in abundance in the rat pineal gland or high relative expression (rEx) compared with a pool of RNA from 15 other tissues (SI Appendix, Table S1, Experiment 1). Each candidate was further evaluated individually by three investigators through analysis of coverage plots.
The coding potential of the lncRNA candidates was evaluated using several criteria. First, transcripts were considered to be lncRNA candidates if a BLASTX search failed to identify an ORF similar to a known protein-coding transcript. Second, coding potential was determined by the Coding Potential Calculator software (26) (http://cpc.cbi.pku.edu.cn/). In addition, other criteria were used (SI Appendix).
This process revealed that the location of the lncRNAs relative to protein-coding genes ranged considerably. Some could be classified as long intergenic noncoding RNAs and were from 10 to >200 kb away; others were located closer. Some were located on the opposite strand from protein-coding genes and either overlapped or encompassed the gene; in some cases, they were included within the gene boundaries (Dataset S1).
This effort identified 112 lncRNAs, given the common identifier lncSN (long noncoding RNA, Section on Neuroendocrinology), with a more than twofold night [Zeitgeber time (ZT) 17]/day (ZT7) differential expression (59% increased at night, and 87% had rEx values > 4) (Dataset S1); 97 of 122 (80%) lncRNAs with rEx > 4 exhibited night/day differential expression, reflecting a high association between differential night/day and pineal-selective expression. These changes were confirmed in subsequent experiments (SI Appendix, Table S1, Experiments 2–4).
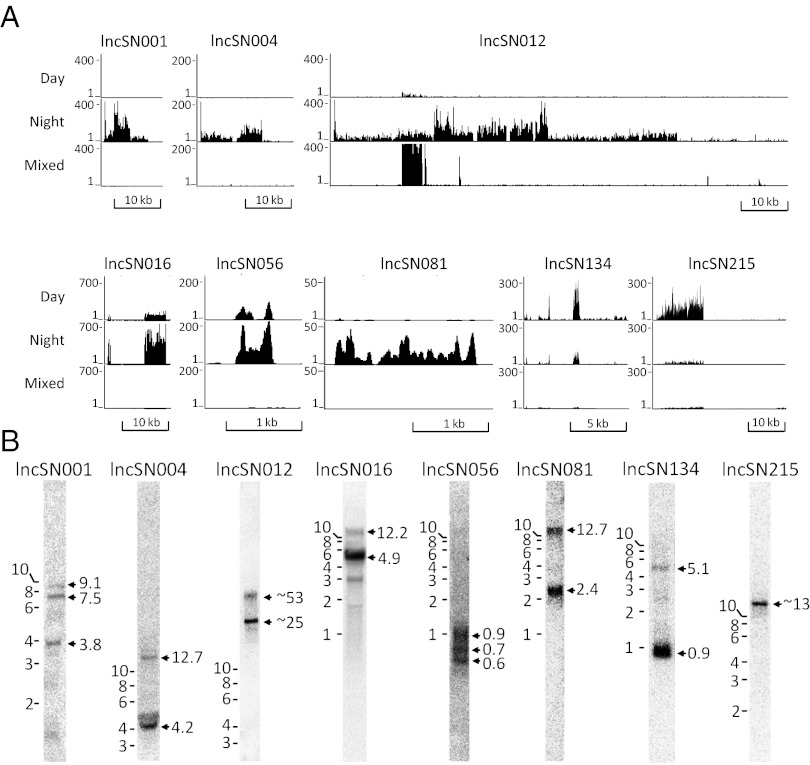
A set of eight lncRNAs with high expression and a more than twofold differential night/day expression (six high at night) was selected for additional study (Fig. 1, Table 1, and SI Appendix). Northern blots (Fig. 1_B_), coverage plots, and splice junction analysis (SI Appendix) indicated that they range in size from <1 to >50 kb, and in most cases, they generate multiple transcripts (Fig. 1_B_): lncSN001 (9.1, 7.5, and 3.8 kb); lncSN004 (12.7 and 4.2 kb); lncSN012 (∼53 and ∼25 kb); lncSN016 (12.2 and 5.1 kb); lncSN056 (0.9, 0.7, and 0.6 kb); lncSN081 (12.7 and 2.4 kb); lncSN134 (0.9 and 5.1 kb); and lncSN215 (13 kb). High evolutionary conservation was not evident, except lncSN016. Flanking genes were not coregulated (Table 1 has links to coverage plots), except in the case of lncSN004 (SI Appendix).
Fig. 1.
Overview of selected lncRNAs. (A) Coverage plots in which data are displayed as reads from RNA-Seq displayed in 10-bp bins on the University of California Santa Cruz (UCSC) Genome Browser. Tracks 1 and 2 represent expression in the pineal gland at day (ZT7) and night (ZT19), respectively. Track 3 represents average expression in a mixture of RNA from 15 tissues (mixed). Scale bars for each plot are shown in the lower right corner. (B) Northern blot analysis of lncRNA transcripts. Total RNA from ZT19 pineal glands was resolved on either 0.5% or 1.5% agarose gels and probed with [32P]-labeled PCR products (300–500 bp). Size markers are on the left of each panel; estimated size of the detected transcripts (arrows) appears on the right side. Northern blot images of ZT7 and mixed tissue RNA are presented in SI Appendix.
Table 1.
Characteristics of eight rhythmic and pineal-selective lncRNAs studied
| lncSN | Chromosomal address | Nominal lncSN length | FPKM day | FPKM night | FPKM mixed | N/D ratio | rEx (max) | Strand |
|---|---|---|---|---|---|---|---|---|
| lncSN001 | chr12: 5,279,390–5,288,650 | 9,260 | 0.26 | 69.78 | 0.17 | 267 | 401 | + |
| lncSN004 | chr15: 106,554,430–106,567,170 | 12,740 | 0.13 | 23.04 | 0.10 | 178 | 227 | + |
| lncSN012 | chr17: 16,296,600–16,393,000 | 96,400 | 0.12 | 5.98 | 2.88 | 50 | 2 | + |
| lncSN016 | chrX: 34,340,700–34,352,700 | 12,000 | 21.68 | 115.57 | 0.48 | 5 | 239 | − |
| lncSN056 | chr5: 22,880,900–22,881,450 | 550 | 14.15 | 81.86 | 0.39 | 6 | 208 | − |
| lncSN081 | chr5: 143,451,600–143,453,500 | 1,900 | 0.39 | 14.30 | 0.05 | 36 | 279 | + |
| lncSN134 | chr13: 47,391,100–47,400,200 | 9,100 | 16.87 | 7.24 | 1.90 | 0.4 | 9 | − |
| lncSN215 | chr8: 44,565,900–44,604,200 | 38,300 | 18.54 | 3.30 | 1.91 | 0.2 | 10 | + |
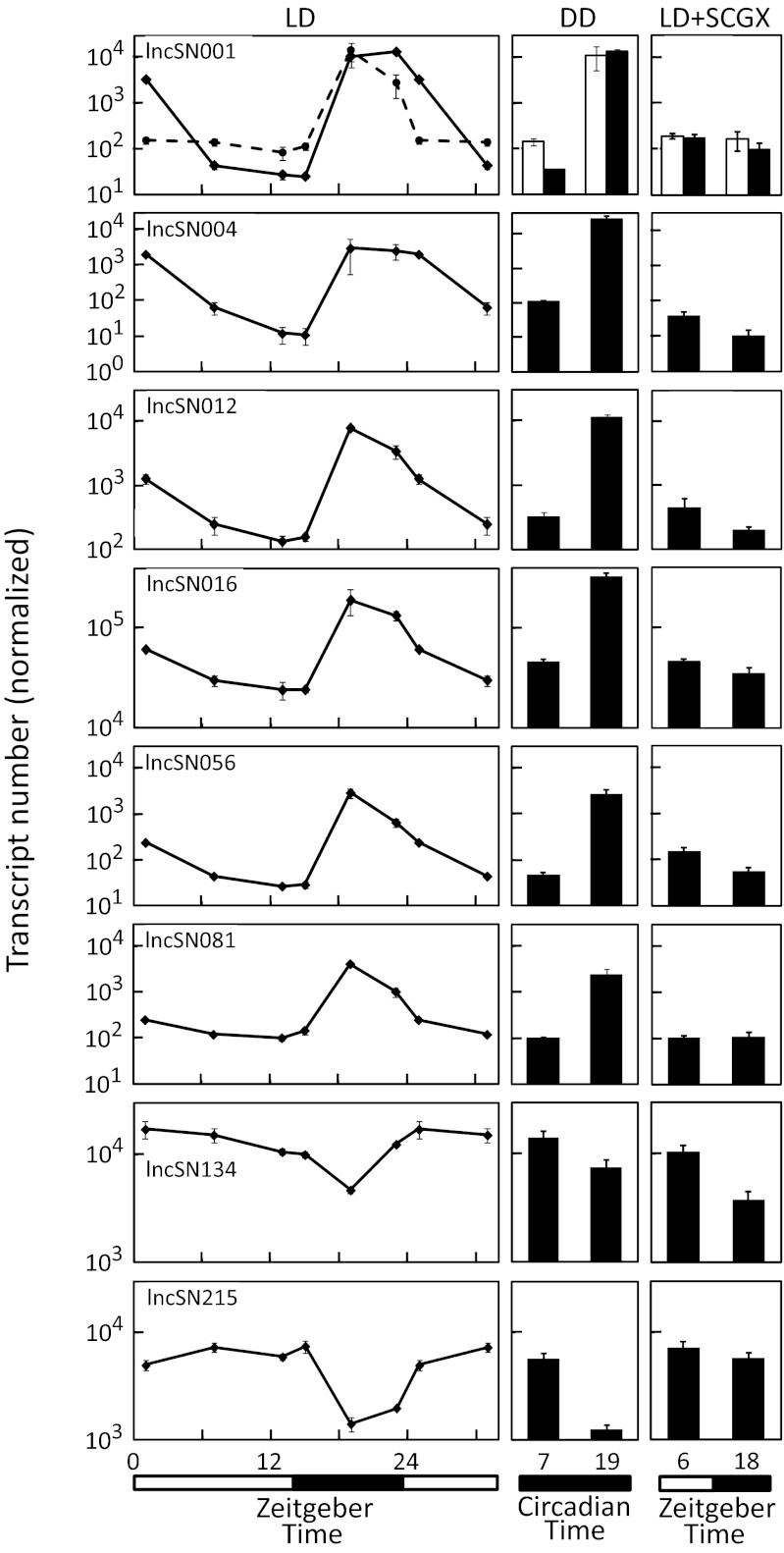
Daily changes ranged from >100-fold increase (lncSN001, lncSN004, and lncSN056) to an approximately fivefold decrease (lncSN215) at night (Fig. 2, Left). Light at midnight reversed some of these changes within 30 min (lncSN001, lncSN056, lncSN081, and lncSN215) (SI Appendix, Fig. S2). The abundance of intron-encoded sequences in the lncSN001 transcript changed more rapidly (sharper peak at night and nearly complete disappearance on light exposure) than the abundance of the lncSN001 exon 2-encoded sequence, consistent with a greater instability of introns (27, 28).
Fig. 2.
Daily rhythm in lncRNA expression is under circadian control. Rat pineal glands were obtained at the indicated times, RNA was extracted, and lncRNA expression was measured by quantitative PCR. The lighting schedule is indicated at the bottom. In cases where an error bar is not visible, it is hidden by the symbol. (Left) A 24-h, six-time point collection. The values at ZT1 and ZT7 have been double-plotted. (Center) Animals were housed in constant darkness for 2 d before being euthanized at subjective circadian times. (Right) Animals were superior cervical ganglionectomized (SCGX) 2 wk before being euthanized at midday (ZT6) or midnight (ZT18). For lncSN001 (row 1), the dashed line and the open bars represent quantitative PCR products of intron 1 primers; the solid line and the filled bars represent quantitative PCR products of exon 2 primers. Transcript number was normalized using a factor calculated from the geometric mean of four reference genes (Rnr1, Gapdh, Actb, and Hprt1) using geNorm. DD, dark:dark; LD, light:dark.
lncRNAs with Daily Changes Are Pineal-Enriched.
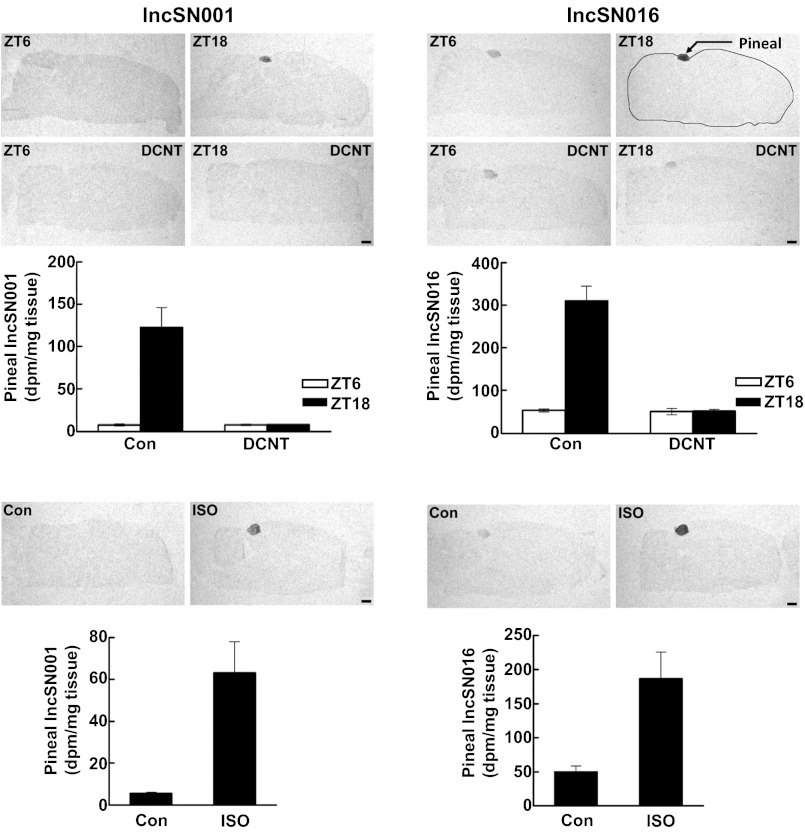
The indication that peak abundance of the six lncRNAs that increased at night were substantially higher than the abundance in the tissue pool (Fig. 1 and Table 1) was confirmed by autoradiographic in situ hybridization histology (Fig. 3).
Fig. 3.
Neural regulation of the expression of lncSN001 and lncSN016. Autoradiographic in situ hybridization histology on saggital sections of the rat brain was used to detect expression of lncSN001 and lncSN016. (Upper) The images of the hybridized sections show the results of surgical decentralization of the superior cervical ganglia (DCNT) on the daily rhythm of the indicated lncRNAs in the pineal gland. (Lower) The images show the effect of midday injection of the β-adrenergic agonist, isoproterenol (ISO), on pineal expression of the indicated lncRNA. The histograms are derived from densitometric in situ data based on three to seven animals in each group. (Scale bar: 1 mm.) Con, control.
Additional analysis (SI Appendix, Table S3) indicated that extrapineal expression in 15 tissues was less than 15% of maximum pineal gland expression in all cases, except for lncSN004 in the small intestine (59%), lncSN012 in the cerebellum (37%), lncSN056 in the pituitary (17%), and lncSN134 in the retina (23%); lncSN215 is highly expressed in many other tissues. In contrast, the daily minimum pineal levels of the eight lncRNAs studied were at or below the levels in other tissues. Accordingly, whereas this set of lncRNAs is expressed in extrapineal tissue, marked night/day differential expression is not seen in these tissues, with the exception of lncSN004 in the small intestine (SI Appendix).
Circadian, Neural, and Transsynaptic Control.
The SCN oscillator drives circadian changes in pineal function and is hard wired to the pineal gland by a multisynaptic neural pathway (29–31). Such SCN-dependent circadian changes persist in constant darkness (32). This finding was the case for the eight lncRNAs (Fig. 2, Center), providing evidence that these changes are truly circadian in nature.
The SCN-pineal neural pathway includes the superior cervical ganglia (SCG), which innervate the pineal gland (29, 32). Surgical interruption of this pathway blocks SCN circadian changes in the pineal gland. Here, rhythmic changes in seven of eight lncRNAs (the exception is lncSN134) were blocked by removal or decentralization of the SCG (Fig. 2, Right and Fig. 3, Upper). These observations, together with the constant darkness results, support the conclusion that the SCN pineal pathway (28) controls expression of these lncRNAs.
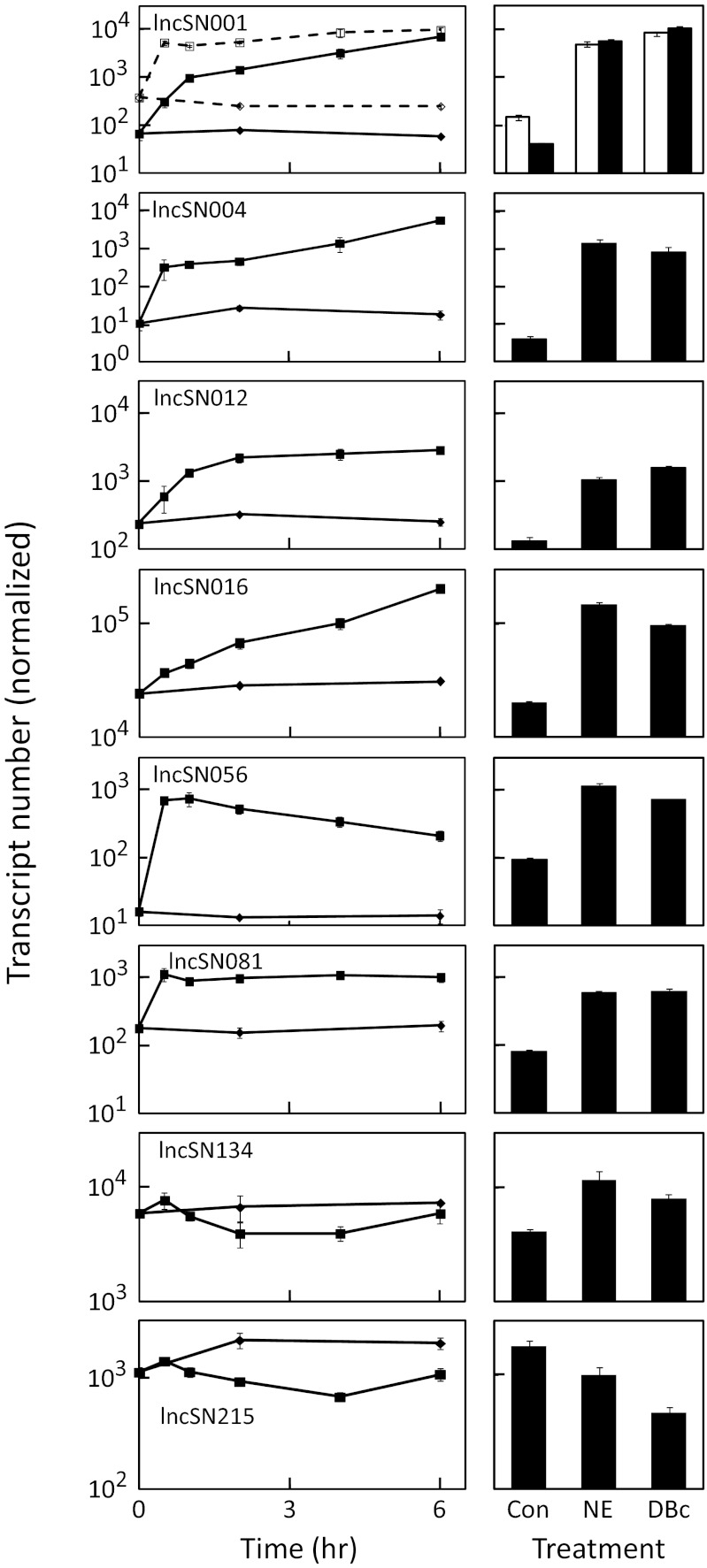
The transsynaptic mechanism controlling expression of the selected lncSNs was studied using in vivo and in vitro experimental designs focused on NE (25, 29, 33–36). Injection of the β-adrenergic agonist isoproterenol during the day elevated lncSN001 and lncSN016 transcripts (Fig. 3, Lower). Organ culture studies (Fig. 4) revealed that NE treatment elevated expression of the lncRNAs that increase in abundance at night; in the case of lncSN001, -004, -056, and -081, the increase was large and rapid (Fig. 4, Left). This finding supports the conclusion that NE mediates neural stimulation of the pineal expression of these lncRNAs that increase at night. The role of NE in controlling daily changes in lncRNAs that decrease at night is not clear, because NE had a weak negative effect on lncSN215 and inconsistent effects of lncSN134 (Fig. 4, Right).
Fig. 4.
Stimulation of lncRNA expression by NE or dibutyryl cAMP (DBc) in cultured pineal glands. Rat pineal glands were incubated in organ culture for 48 h and then treated as indicated. RNA was extracted, and lncRNAs were measured by quantitative PCR. The error bar is not visible where it is smaller than the symbol. (Left) Glands were treated with vehicle (◆) or 1 μM NE (■) for the indicted times. (Right) Glands were treated with vehicle, NE (1 μM), or DBc (500 μM) for 6 h. For lncSN001 (row 1), the dashed line and the open bars represent quantitative PCR products of intron 1 primers; the solid line and the filled bars represent quantitative PCR products of exon 2 primers. Thyroid hormone (100 nM) and 9-_cis_-retinoic acid (100 nM) were added to the culture medium in the experiment in Left but not the experiment in Right. In both cases, NE had the same effect on expression of each lncRNA (except lncSN134), indicating that thyroid hormone and 9-_cis_-retinoic acid are not required for regulation; NE induction requires concurrent thyroid hormone presence for at least one gene (Drd4) in the pineal gland (41). Transcript number was normalized using a factor calculated from the geometric mean of four reference genes (Rnr1, Gapdh, Actb, and Hprt1) using geNorm.
The NE-induced changes in expression of this set of lncRNAs occurred in the presence of cycloheximide (SI Appendix, Fig. S3), a protein synthesis inhibitor, which was also observed in the case of induction of the immediate early genes Fos and Fosl2 (SI Appendix, Fig. S4).
The primary second messenger of NE in the pineal gland is cAMP (29, 36, 37); treatment of cultured pineal glands with the cell-permeable cAMP analog, dibutyryl cAMP, mimicked effects of NE (Fig. 4, Right). This finding establishes that NE acts on these lncRNAs through a cAMP mechanism. This evidence and the evidence that induction occurs without protein synthesis (SI Appendix, Fig. S3) provide reason to suspect that cAMP acts to induce expression of these lncRNAs through a posttranslational mechanism [e.g., phosphorylation of cAMP response element binding proteins (Crebs)], which is the case for protein-coding transcripts (25).
In Silico Analysis of _cis_-Regulation of lncRNAs.
The putative transcription factors controlling expression of the lncRNAs listed in Dataset S1 were investigated in silico (Genomatix) (SI Appendix). Creb binding sites were found in nearly all regions flanking those lncRNA transcripts with a night/day abundance ratio >2 (56 of 57); these sites were overrepresented compared with genomic background. Overrepresented binding sites were also found for transcription factors highly expressed in the pineal gland, including Crx, Otx2, NeuroD1, and Pax4. The flanking regions of eight lncRNAs containing multicomponent Creb/Ets/Bsx modules were more than 20-fold overrepresented in the lncRNA dataset compared with genome-wide promoters, which is of interest, because Bsx expression increases at night in the pineal gland (38). Accordingly, the in silico results are consistent with the hypothesis that Creb sites combined with other sites (e.g., Creb/Ets/Bsx modules) mediate the 24-h lncRNA expression pattern in the pineal gland.
Discussion
The results of these studies provide evidence that lncRNAs play a role in vertebrate circadian biology. The dynamics and magnitude of the daily changes in the abundance of the lncRNAs that were studied in detail are generally similar to the dynamics and magnitude seen for many protein-coding genes expressed in the pineal gland on a circadian basis (25). Moreover, the rhythmically expressed lncRNAs seem to be regulated by the same transsynaptic/cAMP system that controls expression of hundreds of protein-coding genes in the pineal gland, which are thought to be controlled through a Creb-based mechanism.
Additional support for functional involvement of Creb and other factors was derived from analysis of transcription factor binding site (TFBS) frameworks: highly organized patterns of TFBSs directly linked to biological function (in contrast to individual TFBSs; also not prone to overprediction). This approach makes it possible to detect association of frameworks with functional promoters by overrepresentation compared with all promoters in the genome (impossible with individual TFBSs), which in our case, strongly supports the role of Creb combined with one or more other transcription factors.
This similarity in the regulation of the abundance of protein-coding transcripts and lncRNAs supports the suspicion that daily changes in lncRNAs in the rat pineal gland may be an important feature of the circadian regulation. The precise mechanism controlling the dynamic changes in the abundance of each lncRNAs could involve changes in transcription, degradation/clearance, or a combination; the relative importance of each might vary on an individual basis. This finding may explain differences in dynamics seen in the current studies. Independent of the precise mechanism involved in regulating these changes, it is noteworthy that such circadian changes occur, thereby adding a level of complexity to lncRNA biology.
The discovery of 24-h patterns of expression of lncRNAs leads to the compelling question of their function in the pineal gland. The available literature provides a broad range of potential mechanisms through which lncRNAs act in other systems (1, 2, 5, 21). The rapid nature of the changes in lncRNAs seen in the current studies suggests that they are involved in the complex 24-h changes in cell physiology that characterize this tissue that are linked directly and indirectly to changes in melatonin synthesis (25, 39). The multitude of lncRNAs and the potential roles that they play in biology challenge our imagination. The discovery that expression of particular lncRNAs can be dramatically increased through a defined receptor and intracellular signaling pathway raises questions of how these changes impact responses to hormones, transmitters, and immunogens in this tissue and others. Moreover, it is useful to consider that dysregulation of such dynamically expressed lncRNAs may result in disease.
The findings presented here provide additional support for the argument that lncRNAs should be included as mapping targets in RNA-Seq studies (40). Ideally, this inclusion would involve the identification of all lncRNAs expressed in each tissue, which seems essential in understanding the biological roles of lncRNAs.
Materials and Methods
Materials.
[32P] α-Deoxycytidine triphosphate and [35S] α-dATP were purchased from Perkin-Elmer. l-(−)-norepinephrine, dibutyryl cAMP, (−)-isoproterenol, 3,3′,5-triiodo-l-thyronine, and 9-_cis_-retinoic acid were obtained from Sigma-Aldrich.
Animals and Tissues.
Rats (Sprague–Dawley; 150–200 g) used for RNA-Seq and in vivo and organ culture experiments (at the National Institutes of Health) were obtained from Taconic Farms; the rats used for radioactive in situ hybridization, surgical manipulations, or drug injections (at the Panum Institute) were obtained from Charles River. Animals at the National Institutes of Health were housed for 2 wk in light:dark 14:10 lighting cycles, whereas animals at the Panum Institute were housed in light:dark 12:12.
Animal use and care protocols were approved by the local ethical review, and they were in accordance with National Institutes of Health guidelines and the Health Sciences Animal Policy European Union Directive 86/609/EEC (approved by the Danish Council for Animal Experiments).
Tissues removed for analysis during a dark period were obtained from animals euthanized and decapitated under dim red light. A mixed tissue RNA sample was prepared from 15 neural and peripheral tissues (SI Appendix).
Surgery.
Superior cervical ganglionectomy and decentralization of the superior cervical ganglia were performed as described (41).
Drug injection.
Animals were injected with either vehicle or isoproterenol (10 mg/kg for 3 h from ZT5 to ZT8) as described (42).
Light exposure at night experiment.
Rats were exposed to 30 min of light at ZT19 and then killed at ZT19.5. Control rats were kept under normal lighting conditions and killed at ZT19 and ZT19.5.
Organ culture and treatments.
Information is in SI Appendix (43, 44).
Biochemical methods.
Published methods were used for RNA extraction (25), Northern blot analysis (44), and quantitative PCR (25); strandedness was determined biochemically using asymmetric PCR (SI Appendix).
Autoradiographic in situ hybridization.
The probes used are described in SI Appendix (45).
RNA sequence analysis.
RNA-Seq was done using Illumina technology. Mapping and analysis are described in detail in SI Appendix.
Putative transcription factor binding sites.
Transcription factor binding sites were identified using the Genomatix Software package (http://www.genomatix.de). Details are given in SI Appendix.
Supplementary Material
Supporting Information
Acknowledgments
We thank Ms. Tine Thorup Mellergaard for expert technical assistance. This work was supported, in whole or in part, by the Intramural Research Programs of the National Institute of Child Health and Human Development, National Institutes of Health (S.L.C., S.J.H.C., C.F., M.E.O. and D.C.K.), the Center for Information Technology, National Institutes of Health (P.J.M. and Z.R.), and the National Human Genome Research Institute, National Institutes of Health (P.F.C. and J.C.M.); the Biotechnology and Biological Sciences Research Council United Kingdom (D.S.); the Danish Medical Research Council (M.F.R.); the Lundbeck Foundation (M.F.R. and M.M.); and the Novo Nordisk Foundation, the Carlsberg Foundation, and the Simon Fougner Hartmanns Familiefond (M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
1A complete list of the NISC Comparative Sequencing Program can be found in SI Appendix.
References
- 1.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redrup L, et al. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qureshi IA, Mehler MF. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med. 2011;17:337–346. doi: 10.1016/j.molmed.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 11.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 14.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke RA, et al. New genomic structure for prostate cancer specific gene PCA3 within BMCC1: Implications for prostate cancer detection and progression. PLoS One. 2009;4:e4995. doi: 10.1371/journal.pone.0004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’haene B, et al. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: Implications for mutation screening. PLoS Genet. 2009;5:e1000522. doi: 10.1371/journal.pgen.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 20.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuno M, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazen SP, et al. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 2009;10:R17. doi: 10.1186/gb-2009-10-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey MJ, et al. Night/day changes in pineal expression of >600 genes: Central role of adrenergic/cAMP signaling. J Biol Chem. 2009;284:7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L, et al. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Kamitakahara A, Kim AJ, Aguilera G. Cyclic adenosine 3′,5′-monophosphate responsive element binding protein phosphorylation is required but not sufficient for activation of corticotropin-releasing hormone transcription. Endocrinology. 2008;149:3512–3520. doi: 10.1210/en.2008-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponzio TA, Yue C, Gainer H. An intron-based real-time PCR method for measuring vasopressin gene transcription. J Neurosci Methods. 2007;164:149–154. doi: 10.1016/j.jneumeth.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein DC. Photoneural regulation of the mammalian pineal gland. Ciba Found Symp. 1985;117:38–56. doi: 10.1002/9780470720981.ch4. [DOI] [PubMed] [Google Scholar]
- 30.Moore RY. Neural control of the pineal gland. Behav Brain Res. 1996;73:125–130. doi: 10.1016/0166-4328(96)00083-6. [DOI] [PubMed] [Google Scholar]
- 31.Møller M, Baeres FM. The anatomy and innervation of the mammalian pineal gland. Cell Tissue Res. 2002;309:139–150. doi: 10.1007/s00441-002-0580-5. [DOI] [PubMed] [Google Scholar]
- 32.Bustos DM, et al. Global daily dynamics of the pineal transcriptome. Cell Tissue Res. 2011;344:1–11. doi: 10.1007/s00441-010-1094-1. [DOI] [PubMed] [Google Scholar]
- 33.Axelrod J. The pineal gland: A neurochemical transducer. Science. 1974;184:1341–1348. doi: 10.1126/science.184.4144.1341. [DOI] [PubMed] [Google Scholar]
- 34.Sugden D, Klein DC. Rat pineal alpha 1-adrenoceptors: Identification and characterization using [125I]iodo-2-[beta-(4-hydroxyphenyl)-ethylaminomethyl]tetralone. Endocrinology. 1984;114:435–440. doi: 10.1210/endo-114-2-435. [DOI] [PubMed] [Google Scholar]
- 35.Auerbach DA, Klein DC, Woodard C, Aurbach GD. Neonatal rat pinealocytes: Typical and atypical characteristics of [125I]iodohydroxybenzylpindolol binding and adenosine 3′,5′-monophosphate accumulation. Endocrinology. 1981;108:559–567. doi: 10.1210/endo-108-2-559. [DOI] [PubMed] [Google Scholar]
- 36.Vanecek J, Sugden D, Weller J, Klein DC. Atypical synergistic alpha 1- and beta-adrenergic regulation of adenosine 3′,5′-monophosphate and guanosine 3′,5′-monophosphate in rat pinealocytes. Endocrinology. 1985;116:2167–2173. doi: 10.1210/endo-116-6-2167. [DOI] [PubMed] [Google Scholar]
- 37.Maronde E, et al. Signal transduction in the rodent pineal organ. From the membrane to the nucleus. Adv Exp Med Biol. 1999;460:109–131. doi: 10.1007/0-306-46814-x_14. [DOI] [PubMed] [Google Scholar]
- 38.D’Autilia S, Broccoli V, Barsacchi G, Andreazzoli M. Xenopus Bsx links daily cell cycle rhythms and pineal photoreceptor fate. Proc Natl Acad Sci USA. 2010;107:6352–6357. doi: 10.1073/pnas.1000854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stehle JH, von Gall C, Korf HW. Melatonin: A clock-output, a clock-input. J Neuroendocrinol. 2003;15:383–389. doi: 10.1046/j.1365-2826.2003.01001.x. [DOI] [PubMed] [Google Scholar]
- 40.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JS, et al. Thyroid hormone and adrenergic signaling interact to control pineal expression of the dopamine receptor D4 gene (Drd4) Mol Cell Endocrinol. 2010;314:128–135. doi: 10.1016/j.mce.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rath MF, et al. Developmental and diurnal dynamics of Pax4 expression in the mammalian pineal glandNnocturnal down-regulation is mediated by adrenergic-cyclic adenosine 3′,5′-monophosphate signaling. Endocrinology. 2009;150:803–811. doi: 10.1210/en.2008-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein DC, Weller J. Input and output signals in a model neural system: The regulation of melatonin production in the pineal gland. In Vitro. 1970;6:197–204. doi: 10.1007/BF02617764. [DOI] [PubMed] [Google Scholar]
- 44.Kim JS, et al. Methionine adenosyltransferase:adrenergic-cAMP mechanism regulates a daily rhythm in pineal expression. J Biol Chem. 2005;280:677–684. doi: 10.1074/jbc.M408438200. [DOI] [PubMed] [Google Scholar]
- 45.Rath MF, Morin F, Shi Q, Klein DC, Møller M. Ontogenetic expression of the Otx2 and Crx homeobox genes in the retina of the rat. Exp Eye Res. 2007;85:65–73. doi: 10.1016/j.exer.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information