Mouse, but not human STING, binds and signals in response to the vascular disrupting agent DMXAA (original) (raw)
. Author manuscript; available in PMC: 2014 May 15.
Published in final edited form as: J Immunol. 2013 Apr 12;190(10):5216–5225. doi: 10.4049/jimmunol.1300097
Abstract
Vascular disrupting agents (VDAs) such as DMXAA (5,6-dimethylxanthenone-4-acetic acid) represent a novel approach for cancer treatment. DMXAA has potent anti-tumor activity in mice and, despite significant pre-clinical promise, failed human clinical trials. The anti-tumor activity of DMXAA has been linked to its ability to induce type I interferons in macrophages although the molecular mechanisms involved are poorly understood. Here we identify STING as a direct receptor for DMXAA leading to TBK1 and IRF3 signaling. Remarkably, the ability to sense DMXAA was restricted to murine STING. Human STING failed to bind to or signal in response to DMXAA. Human STING also failed to signal in response to cyclic-dinucleotides, conserved bacterial second messengers known to bind and activate murine STING signaling. Collectively, these findings detail an unexpected species-specific role for STING as a receptor for an anti-cancer drug and uncover important insights that may explain the failure of DMXAA in clinical trials for human cancer.
Introduction
Over the last several years, attention has focused on the role of the innate immune system in both pro- and anti-tumor immunity (1, 2). Manipulating immune surveillance and effector mechanisms is important for anti-tumor immunity. Vadimezan or ASA404 (originally called DMXAA) is a xanthenone derivative with potent anti-tumor effects in multiple mouse models (3). In addition to disrupting tumor blood supply, the anti-tumor effects of DMXAA result from the activation of NK cells and the release of cytokines from tumor associated macrophages leading to hemorrhagic necrosis in tumors (4, 5). Additionally, production of chemokines such as MCP-1, IP-10, and RANTES leads to the recruitment of activated tumor-specific CD8+ T-cells that contribute to the disruption of tumors. DMXAA is also a potent inducer of IFN-β (5–7), a cytokine typically induced during infection with viral and bacterial infections. The induction of IFN-β expression by DMXAA slows the growth of tumors in vivo (8, 9). DMXAA showed great promise in initial phase clinical trials (10, 11) but ultimately performed poorly in follow up phase III clinical trials. Understanding the mechanisms by which DMXAA elicits cytokine and interferon production could allow a better understanding of its anti-tumor effects and enable the development of improved anti-tumor agents.
DMXAA treatment of macrophages has been linked to MAP kinase and NFκB signaling (12–16) although activation of these pathways is very modest (6). In contrast, we identified interferon (IFN) regulatory factor 3 (IRF3), a transcription factor important in innate immunity as an important mediator of DMXAA-induced macrophage activation (6). DMXAA is a very strong activator of IRF3 signaling. Normally, IRF3 is present in the cytoplasm and undergoes phosphorylation leading to its dimerization and interaction with the co-activators CBP-p300. TBK1, an IκB kinase-related kinase coordinates the phosphorylation-induced activation of IRF3 leading to transcriptional regulation of immune response genes including type I IFNs and anti-viral interferon stimulated genes (17–19). Despite more than 15 years of research on DMXAA and its parent compound, flavone acetic acid (FAA), the molecular mechanisms responsible for the immune stimulatory effect of DMXAA remains unknown.
Great progress has been made over the last decade in understanding TBK1 activation (20, 21). Several classes of innate sensors including the TLRs and RIG-I like helicases engage TBK1-IRF3 signaling pathways to regulate transcription of type I IFNs. Activation of TBK1 by DMXAA occurs independently of TLRs and RIG-I like receptors (6). DNA sensing receptors such as DAI, IFI16, DDX41 and most recently cGAS have all been shown to couple dsDNA recognition to TBK1 activation (22, 23) (24–28). An ER and/or mitochondrial resident protein called STING is a critical mediator of DNA-induced TBK1 activation (29–32). In addition to this adaptor-like function for STING, STING also acts as a direct innate immune sensor of cyclic di-guanylate monophosphate (c-di-GMP) and cyclic-di-adenylate monophosphate (c-di-AMP), conserved signaling molecules produced by bacteria (33).
Using RNAi in macrophages, as well as using macrophages from mice lacking STING, we recently found that DMXAA-induced IFN production required STING (34). In this present study, we sought to delineate the molecular mechanism of activation of STING by DMXAA. We found that DMXAA directly binds to STING and activates the TBK1-IRF-3 signaling pathway resulting in IFN-β production. Ectopic expression of STING in 293T cells which, themselves are unable to respond to DMXAA facilitated DMXAA-induced TBK1 activation, IRF3 phosphorylation and IFN-β gene induction. STING bound DMXAA directly via its C terminal domain. Remarkably, the ability of STING to mediate DMXAA signaling was seen only with murine STING. When human PBMC or human THP1 cells were treated with DMXAA, no induction of type I IFNs or IFN-stimulated genes was observed. Consistent with these observations, human 293T cells expressing human STING or a chimeric molecule in which the CTD of murine STING was replaced with that of human STING failed to respond to DMXAA. Unlike murine STING, which efficiently binds DMXAA, human STING failed to bind DMXAA. Comparison of DMXAA with cyclic-dinucleotides revealed similar important differences between human and mouse STING for cyclic-dinucleotide signaling. Together, these findings detail a STING pathway critical for DMXAA signaling and reveal a likely explanation for the failure of this molecule to exert its anti-tumor effects in humans.
Materials and Methods
Antibodies and reagents
DMXAA was from Sigma-Aldrich (D5817). Cyclic-di-GMP was from Biolog. The phospho-TBK1 (Ser172) antibody used on the TBK1 IPs was from BD (558397). The total TBK1 antibody used on the TBK1 IPs was from Imgenex (IMG-139A). Anti-Flag Agarose Beads (A2220), mouse M2 anti-Flag (F3165), rabbit anti-Flag (F7425), and mouse anti-β-actin (A5316) were from Sigma-Aldrich. Phospho-TBK1 Ser172 (5483), total TBK1 (3504), and phospho-IRF3 Ser396 (4947) antibodies were from Cell Signaling Technology. STING-deficient mice were from G. Barber (U. Miami).
Plasmids
pOTB7-huSTING (Openbiosystems), pUNO-huSTING (Invivogen) and pCMV-Sport6-moSting (Tmem173)(Openbiosystems) were all subcloned into pEF-Bos-Flag-His. pEF-Bos hu-STING with Arg and His at position 232 behaved similarly in all of our DMXAA and cyclic-di nucleotide assays. pEF-Bos huSTING-Flag-His was generated by using the huSTING N-term forward primer and the huSTING C-term reverse primer to PCR amplify huSTING cDNA (aa 1-379). The resulting PCR product was digested with Esp31 to generate Xho1- and BamH1-compatible ends. The Esp3I-digested PCR product was ligated into BamH1-Xho1-digested pEF-BOS Flag-His. pEF-BOS moSTING-Flag-His was generated using the same strategy and the appropriate primers (moSTING N-term forward primer combined with moSTING C-term reverse primer using pCMV-Sport6-moSTING as template. Plasmids encoding MAVS were originally from Z. Chen (UT Southwestern). Human NOD2 was from Millennium Pharmaceuticals and murine NOD2 from M. Kelliher (Umass Medical School).
Generation of STING chimeras
Primers (Forward: 5′-GCCCGACGTCTCCTCGAGCCACCATGCCCCACTCCAGCCTGC-3′, Reverse: 5′-ATCCGATCGTCTCTGACCATGCCAGCCCATGGGCCACG-3′) were used to PCR amplify cDNA encoding huSTING N-term (aa 1-162). These primers introduce Esp3I sites at both ends of the cDNA. The forward primer also introduced a Xho1 site immediately after the Esp31 site. Another pair of primers (Forward: 5′-CATGGGCGTCTCTGGTCATATTACATCGGATATCTGCGGCTGA-3′, Reverse: 5′-TGGTACCGTCTCGGATCCAGAGAAATCCGTGCGGAGAGGGAGGGGCT-3′) were used to amplify huSTING C-term (aa 163-379). In a fashion similar to the primers for the N-terminal region of huSTING, these primers introduced Esp3I sites at either end of the cDNA. The reverse primer introduced a BamH1 site immediately after the Esp31 site. The same strategy was used to PCR amplify moSTING N-term (aa 1-161; forward primer: 5′-TCCGCGCGTCTCCTCGAGCCACCATGCCATACTCCAACCTGCATCCAGCCA-3′, reverse primer: 5′-CCCAATCGTCTCTGACCAGGCCAGCCCGTGGGCAACATTTAACTTC-3′) and moSTING C-term (aa 162-378; forward primer: 5′-CACGGGCGTCTCTGGTCATACTACATTGGGTACTTGCGGTTGA-3′, reverse primer: 5′-TGGTACCGTCTCGGATCCGATGAGGTCAGTGCGGAGTGGGAGAGGCT-3′). All four PCR products were digested with Esp31. The huSTING N-terminal cDNA fragment and the moSTING C-terminal cDNA fragment were ligated into pEF-BOS Flag-poly His digested with Xho1 and BamH1. The resulting vector encodes the hu-moSTING chimera with a C-terminal Flag-His tag. The moSTING N-terminal fragment and the huSTING C-terminal fragment were combined in a similar fashion to create pEF-BOS mo-huSTING-Flag-His. The fusion site (Ser161/162-Tyr162-163) occurs within helix 1(α1) of the cyclic-di-GMP-binding domain (CBD), the long hydrophobic helix that forms a major part of the dimerization interface (6, 7, 8, 9, 10). This site was chosen because the cDNA sequence encoding these two amino acids is perfectly conserved between mouse and human, creating perfectly complimentary overhangs after digestion with Esp31.
Cell Stimulations
Human embryonic kidney 293T cells were from ATCC (Manassas, VA). Peripheral blood mononuclear cells (PBMC) were freshly isolated by density-gradient centrifugation using Ficoll Hypaque (GE Healthcare). Wild-type immortalized bone marrow-derived macrophages were generated as described (35). Bone marrow derived macrophages were isolated from mice as described (35). 293T cells, mouse macrophages PBMC and THP1 cells were stimulated with DMXAA essentially as previously described (1). Cells were transfected with c-di-GMP or cyclic-di-AMP using Lipofectamine 2000 (Life Technologies) as previously described (5).
Reporter Assays
Reporter assays were carried out as previously described (4). Cells were transfected with reporters plus 1 ng or 50 ng of the indicated STING, DDX41, NOD2 or MyD88. Eighteen hours after transfection, cells were stimulated with DMXAA (.2, 1, 5, 25, or 50 μg/ml) for 6 hours and luciferase activity measured.
In vitro kinase assays
Kinase assays were carried out as previously described (1, 2, 3), with modifications. Whole cell lysates were prepared and endogenous TBK1 was immuno-precipitated using a polyclonal antibody (89246) raised to a C-terminal portion of huTBK1. The IP samples were split into two sets of aliquots: one set was analyzed by in vitro kinase assay using γ-P32-ATP and purified IRF3 (173–427; Qin, Nature Structural and Molecular Biology 2003) as substrates, while the other set was analyzed by immuno-blotting for phospho-TBK1 (Ser172) and total TBK1. Purified IRF3 (aa 173-427, a generous gift from Bill Royer and Kai Lin, 5) was used as a substrate for TBK1.
Quantitative real time PCR
PBMC (2.106cells per 10 cm plate) were stimulated as indicated and RNA isolated using RNeasy (Qiagen, Valencia CA). cDNA were synthetized as previously described (36) using the SuperScript III enzyme (Invitrogen). Quantitative real-time PCR analysis was performed using SYBR green reagent (Invitrogen) using the following primers: IFNβ-F CAGCAATTTTCAGTGTCAGAAGC; IFNβ-R CATCCTGTCCTTGAGGCAGT; βactin-F CCTGGCACCCAGCACAAT; βactin-R GCCGATCCACACGGAGTA. The specificity of amplification was assessed for each sample by melting curve analysis, and the size of the amplicon checked by electrophoresis. All gene expression data were normalized with βactin.
nCounter Nanostring Analysis
Mouse bone marrow derived macrophages or human peripheral blood mononuclear cells were treated as described and RNA was purified using an RNeasy Mini Kit (QIAGEN). 100 ng of total RNA was hybridized to a custom designed mouse or human gene codeset (non-enzymatic RNA profiling using bar-coded fluorescent probes) according to the manufacturer’s protocol (Nanostring technologies) and loaded onto the nCounter prep station, followed by quantification with the nCounter Digital Analyzer. We normalized the data obtained for each sample to the expression of six control mouse or human genes depending on the cell types used. For every sample, we computed the weighted average of the mRNA counts of the six control transcripts and normalized the sample’s values by multiplying each transcript count by the weighted average of the controls. Values were log-transformed and displayed via heat map generated using the ggplot package within the open source R software environment.
Preparation of 293T cell lysates and immunoprecipitations
HEK293T cells were plated at a density of 1×106 cells/well in a 6-well plate. The following day, the cells were transfected as indicated with STING constructs using Lipofectamine 2000 (Invitrogen). The day after that, the cells were rinsed once with PBS and transferred to Eppendorf tubes in PBS containing 1mM EDTA. The cells were pelleted briefly by centrifugation at 1,000_g_ at 4°C. The cell pellet was lysed in an equal volume of digitonin lysis buffer (0.5% digitonin, 20mM Tris-HCl, pH7.4, and 150mM NaCl) containing protease inhibitors (Roche) for 10min on ice. The cell lysates were centrifuged at 10,000_g_ for 10min at 4°C. The protein concentration in the resultant supernatant was measured using the Bradford reagent (Bio-Rad). The cell lysates were subjected to a c-di-GMP binding (crosslinking) assay (see below). The lysates were separated by SDS–PAGE, and the separated proteins were transferred to a nitrocellulose membrane, which was then probed with rat anti-HA antibodies (Roche), to confirm STING–HA expression, and mouse anti-β-actin antibodies (Santa Cruz Biotechnology). To immunoprecipitate HA-tagged STING, the cell lysates were prepared similarly in digitonin lysis buffer and incubated with anti-HA-antibody-conjugated agarose beads (Sigma) for 2h at 4°C. Washed beads were subjected to a c-di-GMP binding assay or separated by SDS–PAGE and stained with colloidal blue protein stain (Thermo Scientific). In other assays, flag tagged STING was detected by immunoblotting with anti-Flag antibody.
c-di-GMP UV crosslinking assay
The c-di-GMP binding assay (also called the crosslinking assay) was carried out as described previously (33). Briefly, 50 μg HEK293T cell lysate at a final concentration of 2 μg/μl, or 1 μg recombinant His6-tagged STING, was incubated with 2 μCi radiolabeled nucleotide in binding buffer (20 mM Tris-HCl, pH7.4, 200 mM NaCl and 1 mM MgCl2) for 15 min at 25° C. The reactions were irradiated at 254 nm for 20 min on ice at a 3 cm distance from a UVG-54 mineral light lamp (UVP). Immediately after crosslinking, the reactions were terminated by the addition of SDS sample buffer (40% glycerol, 8% SDS, 2% 2-mercaptoethanol, 40mM EDTA, 0.05% bromophenol blue, and 250mM Tris-HCl, pH6.8), boiled for 5 min and then separated by SDS–PAGE. The gels were dried, exposed to a phosphor screen and visualized using a Typhoon Trio imager (GE Healthcare).
Thermal shift assay
Both human and mouse STING CTDs were expressed and purified similar to what was reported (33). The thermal shift assay was performed using a ProteoStat® thermal shift stability assay kit (Enzo Life Sciences, Farmingdale, NY) following the manufacturer’s instructions. Thirty-six μM of protein and 200 μM of ligands were mixed and heated using a linear gradient of 20–80° C in 30 min. Fluorescence signal as a function of temperature was recorded using a RealPlex4 real-time PCR cycler (Eppendorf, Hauppauge, NY) with the excitation and emission wavelengths of 470 nm and 605 nm, respectively. Each sample was measured in triplet and fitted with the Boltzmann equation using GraphPad Prism (GraphPad Software, San Diego, CA).
Statistical Analysis
Data were analyzed by one-way or two-way ANOVA followed by Bonferroni’s post-hoc test using Prism software. p values of less than 0.05 were considered significant.
Results
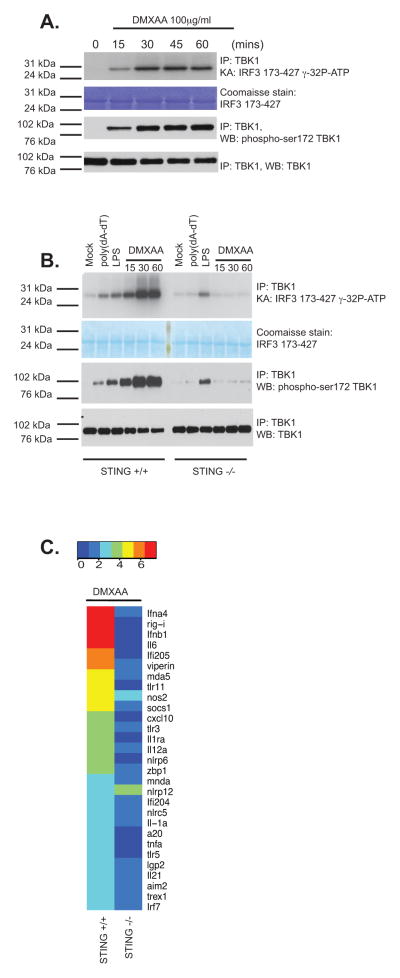
To study TBK1 activation in response to DMXAA treatment of murine bone marrow derived macrophages, we isolated the TBK1 kinase complex using a highly specific anti-TBK1 antibody, and performed an in vitro kinase assay to monitor IRF3 phosphorylation. DMXAA induced rapid TBK1 activation resulting in phosphorylation of IRF3 as early as 15 min post-stimulation (Fig. 1a). We also observed rapid signal-induced phosphorylation of TBK1 in its activation loop (pS172) (lower panels) with similar kinetics. Since activation of TBK1 by DMXAA occurs independently of TLRs and RIG-I like receptors (6), we wanted to examine the role of the STING pathway in DMXAA-induced TBK1 activation by examining the ability of DMXAA to activate TBK1 in wild-type macrophages and in macrophages from STING-deficient mice. The ability of DMXAA to induce TBK1 activation loop phosphorylation and to activate TBK1 kinase activity leading to IRF3 phosphorylation was entirely dependent on STING (Fig. 1b). STING-deficient macrophages displayed TBK1 and IRF3 activation normally following LPS stimulation. We have previously shown that STING-deficient macrophages are impaired in their ability to induce IFN-β expression following DMXAA treatment (34). To understand the immunostimulatory potential of DMXAA, we profiled the expression of a broad panel of inducible immune genes (type I IFNs, as well as a selection of inflammatory cytokines and interferon-stimulated genes (ISGs)) using a non-enzymatic RNA profiling technology that employs bar-coded fluorescent probes (nCounter, Nanostring). Using this technology, we found that DMXAA robustly activated expression of type I IFNs, interferon-stimulated genes as well as inflammatory cytokines, chemokines, and other immune regulators in wild-type cells. In all cases, induction of these genes was largely abrogated in macrophages from mice lacking STING (Fig. 1c). Notably, DMXAA induced IL-6, IL1α, IL12α, IL1ra, TNFα and IL21, as well as A20 in a STING-dependent manner. Since all of these genes are NFκB-dependent these data indicate that in addition to activating the TBK-1 and IRF3 pathway, DMXAA also induced NFκB target genes via STING. These data indicate that DMXAA signals via STING to induce both TBK1-IRF3-dependent IFN production and NFκB-dependent cytokine gene transcription in mouse macrophages.
Figure 1. DMXAA signals via STING in macrophages.
(A) Wild-type immortalized bone marrow-derived macrophages were stimulated with DMXAA (100μg/ml) at the indicated time points. Whole cell lysates were prepared and endogenous TBK1 immunoprecipitated (IP) and analyzed by in vitro kinase assay and western blotted (WB) for phospho-TBK1 (Ser172) and total TBK1. (B) Wild-type or STING−/− bone marrow-derived macrophages were stimulated with poly(dA-dT) (3μg/ml), LPS [10ng/ml] or DMXAA (75μg/ml) as indicated and analyzed as in A. (C) BMDM from wild-type and STING−/− mice were stimulated with DMXAA (75μg/ml) for 4 hours and mRNA levels for a selection of innate immune genes were analyzed by Nanostring analysis. Heatmaps representing differentially regulated genes are presented and scaled by log2(X-min(X) + 1). Data are presented from one experiment which is representative of three experiments (A, B) or two experiments (C).
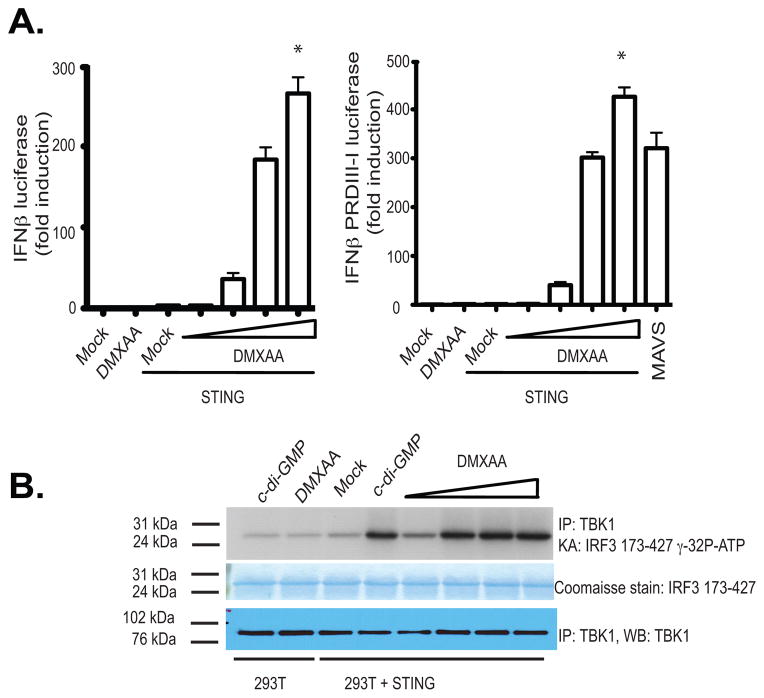
It is currently unclear how DMXAA engages the STING pathway. In an effort to understand these events further, we sought to identify molecules that could reconstitute the IFN response to DMXAA in 293T cells, which do not respond to DMXAA. We expressed mouse STING in 293T cells and monitored IFN-β luciferase reporter gene activation as well as TBK1-induced phosphorylation of IRF3. DMXAA induced IFN-β luciferase reporter gene expression, as well as IRF3 activation, only in cells expressing mouse STING. Low level of mouse STING protein was sufficient to reconstitute the responsiveness of 293T cells to DMXAA (Fig. 2a and 2b). Similar results were obtained when the IFN-β PRD III-I luciferase reporter, which contains a multimerized PRDIII-I element that binds to activated IRFs) was used (Fig. 2a, right panel). Recently, NOD 1 and 2 were shown to contribute to DMXAA induced NFκB signaling using HEK293 cells. NOD 1 and NOD2 are members of the larger NOD-like receptor family and are important in recognizing specific motifs within peptidoglycans of both Gram-negative and Gram-positive bacteria. NOD1 and NOD2 signal via the downstream adaptor serine/threonine kinase RIP2/CARDIAK/RICK to initiate NFκB activation and the release of inflammatory cytokines/chemokines. In light of these findings we next compared the ability of STING and NOD2 to reconstitute the responsiveness of 293T cells to DMXAA. In contrast to murine STING, neither human nor murine NOD2 reconstituted DMXAA induced IFN-β luciferase reporter gene expression (Supplementary Fig. 1a). Although, both human and murine NOD2 constitutively increased basal NFκB activation, similar to what we found with MyD88, neither human nor murine NOD2 reconstituted DMXAA induced NFκB activation. Under these conditions mouse STING facilitated a modest increase in NFκB activation (Supplementary Fig. 1b).
Figure 2. Reconstitution of DMXAA induced IFNβ signaling in STING expressing HEK293 cells.
(A) 293T cells were transfected with either empty vector or mSTING in the presence of an IFN-β luciferase reporter gene (left panel) or a multimerized PRDIII-I reporter gene (right panel) and transfected cells stimulated with DMXAA from 10–100 μg/ml for 18hours and luciferase activity was measured. Data are presented as the mean ± s.e.m of one experiment representative of three experiments. * indicates p <0.05 for the comparison of the highest dose of DMXAA relative to vector control. (B) 293T cells transfected as above were stimulated with cyclic-di-GMP (10μg/ml) or with increasing amounts of DMXAA as in A and endogenous TBK1 activity was analyzed by in vitro kinase assay.
Cyclic di-guanylate monophosphate (c-di-GMP) and cyclic-di-adenylate monophosphate (c-di-AMP), conserved signaling molecules produced by bacteria have been shown to signal via STING (33). Recently DDX41; a member of the DEXD helicase family has also been shown to be important for DNA and cyclic-dinucleotide signaling (37). We therefore also compared the ability of mouse STING and DDX41 to reconstitute the IFN response to DMXAA in 293T cells. Unlike mouse STING, however, ectopic expression of DDX41 did not reconstitute DMXAA signaling (Supplementary Fig. 1b). The ability of mouse STING in 293T cells to facilitate DMXAA-induced signaling is reminiscent of published studies with cyclic-di-AMP and cyclic-di-GMP (33). Collectively, these results reveal that STING expression is sufficient to restore the responsiveness of 293T cells to DMXAA.
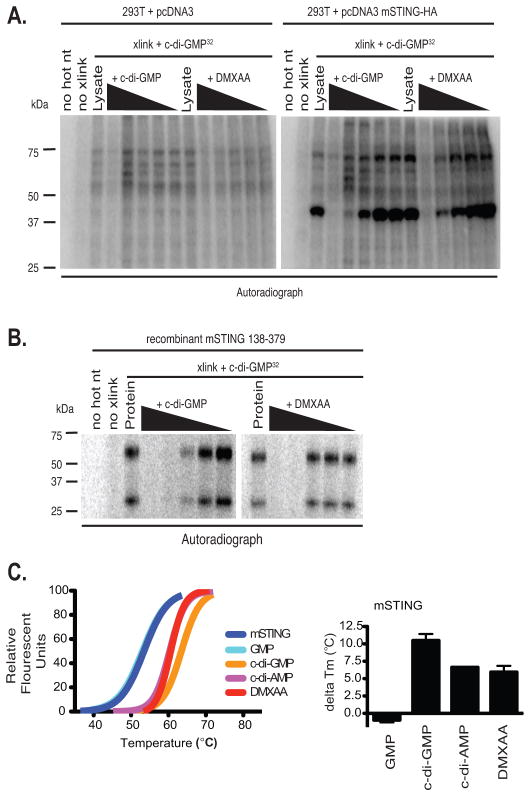
In the case of cyclic-di-nucleotides, STING acts as a sensor by directly binding these small molecules (33). STING encodes an amino-terminal domain with multiple transmembrane segments, followed by a globular carboxy-terminal domain (CTD). To examine if STING can bind to DMXAA, we took advantage of an in vitro ultraviolet radiation crosslinking assay used previously to demonstrate cyclic-di-GMP binding to STING (33). Consistent with published studies (38–41), we observed cyclic-di-GMP binding to the mouse STING CTD after UV crosslinking in both 293T cell transfectants and when a purified C-terminal domain of mouse STING was used in vitro (Fig. 3a and b). In both cases, binding of cyclic-di-GMP to the STING CTD was specifically competed away with cold (unlabeled) c-di-GMP. We reasoned that if DMXAA similarly bound STING, it should also compete with radiolabelled c-di-GMP for STING binding. Similar to what we observed with cold c-di-GMP, binding of labeled c-di-GMP to STING was competed away with DMXAA (Fig. 3b, right panel). These data show that c-di-GMP and DMXAA compete for the mouse STING CTD binding indicating that STING is a direct sensor for DMXAA. We next examined the direct binding of DMXAA to the mouse STING CTD by employing a thermal shift assay. This assay is used to monitor the thermal stability of proteins and investigate factors affecting this stability. Using this assay we found that like c-di-AMP and c-di-GMP, DMXAA increased the melting temperature (Tm) of the STING CTD, an indication of binding and stabilization of the STING CTD structure (Fig. 3c). These data indicate that DMXAA, c-di-GMP and c-di-AMP all bind directly to mouse STING via its CTD.
Figure 3. STING binds to DMXAA and is a direct sensor for DMXAA.
(A) 293T cells were transfected with empty vector or mSTING and cell lysates were UV-crosslinked to c-di-GMP in the presence of cold competing c-di-GMP or DMXAA in 10-fold serial dilutions. (B) One μg His6-mSTING 138–378 was analyzed as described in C**. (C**) Left panel, thermal shift analysis of mouse STING in the presence of GMP, c-di-GMP, c-di-AMP, and DMXAA. The melting temperature shifts in the presence of the ligands are plotted in the graph to the right of the melting curves. Data are representative of two independent experiments (A–C).
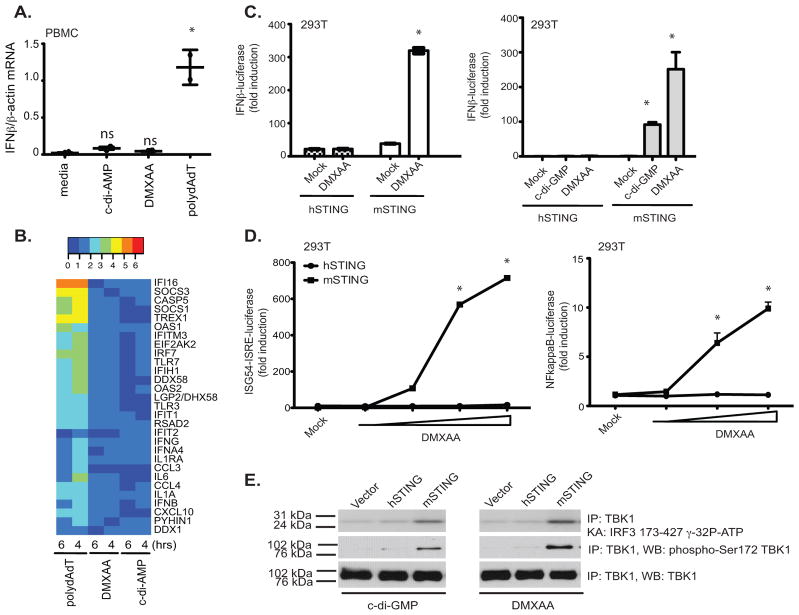
While examining the immune stimulatory activity of DMXAA, we compared responses in human and mouse cells and, to our surprise, found that unlike murine macrophages or dendritic cells (data not shown), peripheral blood mononuclear cells (PBMC) isolated from normal healthy volunteers failed to induce measurable IFN-β responses to DMXAA. This was in contrast to PBMCs treated with polydA-dT, a synthetic B-form dsDNA that elicited a strong IFN-β mRNA induction (Fig. 4a). Surprisingly, there was also a very minimal response to c-di-AMP and c-di-GMP in human PBMC. Therefore, we extended this analysis to include a broader panel of inducible immune response genes, which were all robustly induced by poly(dA-dT). Neither DMXAA nor c-di-AMP turned on expression of this panel of immune genes in human cells (Fig. 4b).
Figure 4. Mouse, but not human, STING signals in response to DMXAA and c-di nucleotides.
**(A)**PBMC from two volunteers were stimulated with DMXAA (10μg/ml), c-di-AMP (5μM) or poly (dA-dT) (4μg/ml) for 6 hours and IFN-β mRNA levels measured by quantitative PCR. * indicates p <0.05 for the comparison of each ligand relative to medium control. (B) RNA isolated from PBMCs stimulated as in A for 4 and 6 hours was subject to Nanostring analysis to monitor expression of 30 innate immune genes. (C–D) 293T cells were transfected with empty vector, pEF-BOS hSTING-Flag-His, or pEF-BOS mSTING-Flag-His, followed by stimulation with either DMXAA or c-di-GMP or with increasing concentrations of DMXAA (D). IFN-β luciferase (C) ISG54-ISRE luciferase, and NF-κB luciferase (D) activities were measured. * indicates p <0.05 for the comparison of DMXAA or c-d-GMP relative to vector control. (E) 293T cells were transfected as above and stimulated with DMXAA and cyclic-di-GMP. Endogenous TBK1 was immunoprecipitated and analyzed as in Figure 1. Data are presented as the mean ± s.e.m of one experiment representative of three experiments (A). (B–E) data is representative of three separate experiments.
Based on these findings, we hypothesized that the differential responses of human and mouse cells to DMXAA might be due to different capacities of human and mouse STING to respond to DMXAA. To test this possibility directly, we directly compared the ability of human and murine STING to reconstitute DMXAA signaling in 293T cells. All of the data presented thus far utilized murine STING in the reconstitution assays and purified murine STING-CTD in the binding assays. Unlike murine STING, expression of human STING in 293T cells failed to render these cells responsive to DMXAA (Fig 4c). This was true not only for DMXAA, but also for cyclic-di-GMP (Fig. 4c, right panel) and cyclic-di-AMP (data not shown). The inability of human STING to restore DMXAA signaling was also observed when an ISG54-ISRE reporter or an NF-κB luciferase reporter gene was utilized (Fig. 4d). Consistent with the differential ability of murine, but not human, STING to enable 293T cells to respond to DMXAA, only 293T cells expressing murine STING elicited TBK1 activation (Fig. 4e). These data clearly indicated that activation of the TBK1-IRF3 pathway in response to DMXAA and other small molecule agonists such as cyclic-di-nucleotides occurs in mouse cells and weakly, if at all, in human cells.
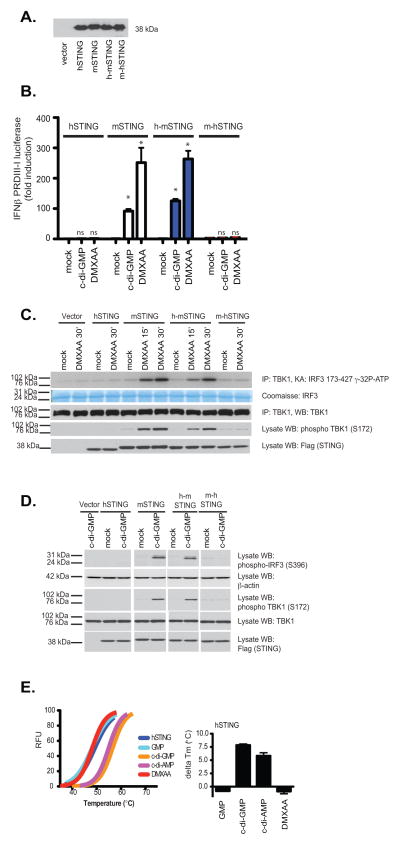
Our binding data indicated that the CTD of STING was important in binding to both DMXAA and c-di-nucleotides, therefore, we next generated chimeric STING molecules in which the CTD of human STING was replaced with that of the mouse and vice versa and examined the ability of these chimeric STING molecules to signal in response to DMXAA and c-di-GMP. We transfected these constructs into 293T cells and found that all were expressed to a similar degree (Fig. 5a). While murine STING conferred DMXAA and c-di-GMP responsiveness to 293T cells, as we had previously shown, expression of human STING or a chimeric mouse-human STING (in which the CTD of the mouse was replaced with that of humans) failed to facilitate DMXAA-induced IFN-β PRDIII-I reporter gene activation (Fig. 5b). In contrast, when the CTD of human STING was replaced with that of mouse STING, this hu-mo-STING chimera responded to both DMXAA and c-di-GMP. We also monitored TBK1 activation as well as phosphorylation of IRF3 in 293T cells expressing human STING, mouse STING, and the STING chimeras. Only the mouse STING and the h-mSTING chimeric molecules facilitated DMXAA-induced TBK1 and IRF3 activation (Fig. 5c).
Figure 5. DMXAA-induced signaling is dependent on the mouse STING C-terminal domain.
293T cells were transfected with human, mouse, or the indicated chimeric STING molecules as well as the IFNβ PRDIII-I luciferase reporter gene. Transfected cells were (A) subjected to immunoblotting with anti-Flag antibody or (B) stimulated with DMXAA or cyclic-di-GMP as indicated and analyzed for IFN-β luciferase reporter gene activation. hu-STING, hu-moSTING, and mo-huSTING were compared to moSTING for each treatment and the statistical significance (p <0.05) was indicated by *. ns=non-significant. (C–D) Cells transfected as above without reporter genes were treated as indicated and analyzed by immunoblotting for phospho-TBK1 (Ser172), phospho-IRF3 (Ser396), total TBK1, β-actin, and Flag (to detect expression of recombinant STING) as indicated. (E) Left panel, thermal shift analysis of human STING CTD in the presence of GMP, c-di-GMP, c-di-AMP, and DMXAA. The melting temperature shifts in the presence of the ligands are plotted in the right panel.
We also conducted similar experiments in cells stimulated with cyclic-di-GMP and found that only mouse STING reconstituted responses to cyclic-di-GMP (Fig. 5d). In light of these data we next examined the binding of DMXAA to purified human STING CTD using the thermal shift assay described above. No significant enhancement of Tm was observed when the human STING CTD was incubated with DMXAA (Fig. 5e). In contrast, c-di-GMP and c-di-AMP both bound human STING, consistent with previous structural studies and the strict sequence conservations for the c-di-GMP-binding residues between human and mouse STING (Suppl. Fig 2. sequence alignment, residues highlighted in red). Collectively, these data indicate that mouse STING binds both cyclic-di-nucleotides and DMXAA and signals TBK1-IRF3 activation as well as NF-κB activation. In contrast, human STING fails to bind DMXAA and activate TBK1. Although human STING still binds cyclic-di-nucleotides, this binding appears to be insufficient to trigger downstream TBK1-IRF3 activation. Taken together these data demonstrate that unlike mouse STING, human STING lacks the ability to respond to DMXAA and cyclic dinucleotides.
Discussion
Vascular Disrupting Agents (VDAs) disrupt blood flow leading to the rapid and irreversible collapse of the established tumor vasculature. The acute ischemia that results leads to widespread tumor necrosis and a significant reduction in tumor burden. Flavone derivatives represent a novel class of VDAs and include FAA and DMXAA. FAA was discovered based on its unexpectedly potent antitumor activity in mice (42). The related compound DMXAA was found to be much more potent in mouse cancer models, and despite showing great promise in early phase l and II trials ultimately failed in phase III clinical trials (43, 44). DMXAA kills tumors through complex mechanisms leading to the induction of tumor hemorrhagic necrosis. DMXAA treatment also activates tumor-associated macrophages to increase local and systemic levels of inflammatory cytokines such as TNF-α, which are also important in anti-tumor activity of this drug (3, 45, 46).
Considerable efforts have focused on the molecular mechanism of action of DMXAA. Early studies provided insights into similarities and differences in gene induction in murine macrophages stimulated by DMXAA when compared with E. coli LPS (7, 46, 47). Compared to LPS, DMXAA potently induced a more limited subset of genes that included IFN-β and interferon-γ-inducible protein (IP-10), but relatively low levels of TNF-α and other proinflammatory cytokine genes (7). Much like TNF-α, the IFNs have been known for decades to possess pleiotropic effects on tumor growth. The anti-tumor properties of IFNs are well known (48) and a number of IFNs are currently used in the clinic to treat more than a dozen malignancies. Notably, type I IFNs represent the first human recombinant proteins to be used successfully in the clinic to treat cancer (49). Although tumor associated macrophages (TAMs) have been shown to promote the growth of tumors by promoting a local cytokine milieu that tends to favor tumor growth (50), in response to DMXAA, TAMs may engender a CD8+ T-cell-mediated attack on the tumor (5). Type I IFNs may play a direct role in mediating these effects (51, 52). In addition to the direct effects of IFN on the protection of the host from cancer, these cytokines also have numerous indirect effects resulting from downstream autocrine and paracrine effects. Type I IFN has also been shown in numerous studies to have anti-angiogenic functions (53, 54), while IFN-inducible genes, such as the CXC chemokine IP-10, have angiostatic properties (55). Roberts et al. showed that the anti-tumor action of DMXAA was dependent on host IFN-β (9).
In our own published studies we identified TBK-1 and IRF3 signaling as key transducers of DMXAA signaling (6). TBK1 represents a point of convergence for multiple innate sensing pathways that activate type I IFN gene transcription. In the present study, we extended these findings by showing that DMXAA treatment of macrophages led to a marked induction of IFNs, ISGs as well as a panel of additional NFκB-dependent inflammatory cytokines, and other regulators of inflammation. In all cases, induction of these genes was abrogated in macrophages lacking STING suggesting that STING mediates both the IRF3 and NFκB signaling pathways. It is important to point out that although DMXAA can turn on NFκB target genes such as IL-1α and IL-6, the induction of these genes is very modest by comparison to IFN and ISG induction. STING was first identified as an ER-residing protein relaying signals to IRF activation and IFN transcription from a variety of stimuli, especially cytosolic dsDNAs (30–32). STING-deficient cells fail to produce type I IFN in response to transfection with dsDNA or infection with herpes simplex virus 1 (HSV-1) (30). STING was also shown to be essential for IFN production in response to c-di-nucleotides such as c-di-GMP and c-di-AMP (56). A mutant mouse strain, Goldenticket, that harbors a missense mutation (I199N) in the STING protein, fails to produce detectable protein and to activate IFN production in response to c-di-GMP (56). Both c-di-GMP and c-di-AMP are second messengers secreted by bacteria (56). Surprisingly, STING was shown to function as a direct sensor for c-di-GMP, and this interaction mapped to the cytosolic C terminal domain and was shown to be critical for cellular responses to c-di-nucleotides (33).
STING contains four transmembrane helices and a globular carboxy-terminal domain (CTD), predicted to be localized in the cytosol. The cytosolic domain does not exhibit significant sequence homology to any proteins with known structures. Within the cytosolic domain, a recent study showed that the very C-terminal tail (CTT) interacts with and activates TBK1 and IRF3 in vitro (57). The CTT of STING is also important for maintaining the autoinhibited state of inactive STING (58). Structural studies suggest that the CTT is involved in stable STING-dimerization and that c-di-nucleotides bind at the cleft/groove between two STING monomers (37–41, 58) It is suspected that the binding of c-di-nucleotides would relieve this autoinhibitory state, expose the CTT, and stabilize STING dimers with c-di-nucleotides buried in the cleft between the interacting protomers (39, 41, 58) (shown in Suppl. Fig. 3).
In this study, we found that DMXAA behaves similarly to c-di-nucleotides and binds directly to the mouse STING CTD. Our 293T reconstitution studies, combined with our binding studies, indicate that mouse STING serves as a direct sensor for this chemotherapeutic drug. Unlike mouse STING, NOD2 and DDX41 both failed to reconstitute signaling to DMXAA. STING binds directly to DMXAA and the competition experiments indicate that DMXAA competes with cyclic-di-GMP for binding. Surprisingly, we found that only murine STING could serve as a direct sensor for DMXAA. Comparison of the ability of mouse and human STING to reconstitute DMXAA signaling in 293T cells revealed a clear difference between mouse and human STING in terms of DMXAA signaling. The failure of DMXAA to induce STING-dependent signaling in humans was mapped to the CTD, since chimeric molecules comprised of the murine STING molecule with the CTD from humans was unresponsive to DMXAA, while a chimeric human STING molecule with the CTD from mouse was fully competent for signaling in response to DMXAA. Binding studies indicate that human STING did not bind to DMXAA, suggesting that non-conserved residues between hSTING and mSTING are part of the DMXAA binding surface. It is possible that DMXAA and c-di-GMP have overlapping but non-identical binding surfaces on mouse STING, and one such model of the STING CTD in complex with DMXAA is shown in Suppl. Fig 4. Another possibility is that DMXAA may bind near the mSTING dimer interface residues D273 or K275, which are not conserved in hSTING (See Suppl. Fig 2 and 3). Such binding may change the mSTING dimer interface and modulate STING autoinhibition. STING is autoinhibited by an intramolecular interaction between the CBD and the C-terminal tail (CTT) and that c-di-GMP releases STING from this autoinhibition by displacing the CTT near the dimer interface. If such an autoinhibition model involving the CTT is correct (58), then DMXAA or c-di-GMP may displace the CTT from a mSTING auto-inhibited state to induce signaling, but they do not necessarily need to bind at the same site.
It is tempting to speculate that the failure of DMXAA in humans might relate to its inability to engage the STING pathway in human cells. While murine STING serves as a sensor for DMXAA, activating a robust IFN and inflammatory resp3onse important for the anti-tumor effect, a defect in this capability could drastically affect the therapeutic efficacy of DMXAA in humans. The inability of DMXAA to bind to human STING and activate STING signaling in humans could, therefore, provide important insights that may help explain the failure of DMXAA in clinical trials for human cancer. The inability of cyclic di-nucleotides to trigger human STING signaling was also surprising. Previously a number of mouse STING mutants were reported to retain their ability to associate with c-di-GMP but failed to induce IFN production (Burdette et al., 2011). Several such mutants that “decouple” c-di-GMP binding and IFN induction are at the non-conserved residues between human and mouse STING (see sequence alignment Suppl. Fig 2, highlighted in yellow). Conceivably, these and perhaps other nonconserved residues may play a role in the species-specific immune responses to c-di-GMP. The inability of these molecules to engage human STING may lessen enthusiasm for the development of these immunostimulatory molecules as immunotherapeutics or vaccine adjuvants for clinical use. In addition these findings might also pave the way to redirect efforts to identify and develop analogs of DMXAA that could bind to human STING and signal and therefore have efficacy in humans.
Supplementary Material
1
Acknowledgments
This work is supported by the NIH (AI067497 to K.A.F, AI063302 and AI080749 to R.E.V. and by a postdoctoral fellowship AI091100 to D.L.B) and by DS Pharma Incorporated (Japan). R.E.V. is supported by an Investigator Award from the Burroughs Wellcome Fund.
The authors would like to thank Anna Cerny, Megan Monroe and Kelly Army for animal husbandry.
References
- 1.Rakoff-Nahoum S, Medzhitov R. Role of toll-like receptors in tissue repair and tumorigenesis. Biochemistry Biokhimiia. 2008;73:555–561. doi: 10.1134/s0006297908050088. [DOI] [PubMed] [Google Scholar]
- 2.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nature reviews Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 3.Baguley BC, Ching LM. Immunomodulatory actions of xanthenone anticancer agents. BioDrugs: clinical immunotherapeutics, biopharmaceuticals and gene therapy. 1997;8:119–127. doi: 10.2165/00063030-199708020-00005. [DOI] [PubMed] [Google Scholar]
- 4.Wallace A, LaRosa DF, Kapoor V, Sun J, Cheng G, Jassar A, Blouin A, Ching LM, Albelda SM. The vascular disrupting agent, DMXAA, directly activates dendritic cells through a MyD88-independent mechanism and generates antitumor cytotoxic T lymphocytes. Cancer research. 2007;67:7011–7019. doi: 10.1158/0008-5472.CAN-06-3757. [DOI] [PubMed] [Google Scholar]
- 5.Jassar AS, Suzuki E, Kapoor V, Sun J, Silverberg MB, Cheung L, Burdick MD, Strieter RM, Ching LM, Kaiser LR, Albelda SM. Activation of tumor-associated macrophages by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid induces an effective CD8+ T-cell-mediated antitumor immune response in murine models of lung cancer and mesothelioma. Cancer research. 2005;65:11752–11761. doi: 10.1158/0008-5472.CAN-05-1658. [DOI] [PubMed] [Google Scholar]
- 6.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. The Journal of experimental medicine. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera PY, Barber SA, Ching LM, Vogel SN. Activation of LPS-inducible genes by the antitumor agent 5,6-dimethylxanthenone-4-acetic acid in primary murine macrophages. Dissection of signaling pathways leading to gene induction and tyrosine phosphorylation. Journal of immunology. 1994;153:4684–4693. [PubMed] [Google Scholar]
- 8.Head M, Jameson MB. The development of the tumor vascular-disrupting agent ASA404 (vadimezan, DMXAA): current status and future opportunities. Expert opinion on investigational drugs. 2010;19:295–304. doi: 10.1517/13543780903540214. [DOI] [PubMed] [Google Scholar]
- 9.Roberts ZJ, Ching LM, Vogel SN. IFN-beta-dependent inhibition of tumor growth by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2008;28:133–139. doi: 10.1089/jir.2007.0992. [DOI] [PubMed] [Google Scholar]
- 10.McKeage MJ, Reck M, Jameson MB, Rosenthal MA, Gibbs D, Mainwaring PN, Freitag L, Sullivan R, Von Pawel J. Phase II study of ASA404 (vadimezan, 5,6-dimethylxanthenone-4-acetic acid/DMXAA) 1800mg/m(2) combined with carboplatin and paclitaxel in previously untreated advanced non-small cell lung cancer. Lung cancer. 2009;65:192–197. doi: 10.1016/j.lungcan.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 11.McKeage MJ. The potential of DMXAA (ASA404) in combination with docetaxel in advanced prostate cancer. Expert opinion on investigational drugs. 2008;17:23–29. doi: 10.1517/13543784.17.1.23. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Wang LC, Fridlender ZG, Kapoor V, Cheng G, Ching LM, Albelda SM. Activation of mitogen-activated protein kinases by 5,6-dimethylxanthenone-4-acetic acid (DMXAA) plays an important role in macrophage stimulation. Biochemical pharmacology. 2011;82:1175–1185. doi: 10.1016/j.bcp.2011.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LC, Thomsen L, Sutherland R, Reddy CB, Tijono SM, Chen CJ, Angel CE, Dunbar PR, Ching LM. Neutrophil influx and chemokine production during the early phases of the antitumor response to the vascular disrupting agent DMXAA (ASA404) Neoplasia. 2009;11:793–803. doi: 10.1593/neo.09506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woon ST, Hung SS, Wu DC, Schooltink MA, Sutherland R, Baguley BC, Chen Q, Chamley LW, Ching LM. NF-kappaB-independent induction of endothelial cell apoptosis by the vascular disrupting agent DMXAA. Anticancer research. 2007;27:327–334. [PubMed] [Google Scholar]
- 15.Wang LC, Woon ST, Baguley BC, Ching LM. Inhibition of DMXAA-induced tumor necrosis factor production in murine splenocyte cultures by NF-kappaB inhibitors. Oncology research. 2006;16:1–14. doi: 10.3727/000000006783981288. [DOI] [PubMed] [Google Scholar]
- 16.Woon ST, Reddy CB, Drummond CJ, Schooltink MA, Baguley BC, Kieda C, Ching LM. A comparison of the ability of DMXAA and xanthenone analogues to activate NF-kappaB in murine and human cell lines. Oncology research. 2005;15:351–364. doi: 10.3727/096504005776449743. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature immunology. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 18.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 20.Uematsu S, Akira S. TLR family and viral infection. Uirusu. 2004;54:145–151. doi: 10.2222/jsv.54.145. [DOI] [PubMed] [Google Scholar]
- 21.Akira S, Saitoh T, Kawai T. Nucleic acids recognition by innate immunity. Uirusu. 2012;62:39–45. doi: 10.2222/jsv.62.39. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 25.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature immunology. 11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature immunology. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barber GN. STING-dependent signaling. Nature immunology. 2011;12:929–930. doi: 10.1038/ni.2118. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cellular and molecular life sciences: CMLS. 2011;68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA, Vogel SN. 5,6-Dimethylxanthenone-4-acetic Acid (DMXAA) Activates Stimulator of Interferon Gene (STING)-dependent Innate Immune Pathways and Is Regulated by Mitochondrial Membrane Potential. The Journal of biological chemistry. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature immunology. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenfusser S, Goutagny N, Diperna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA Helicase Lgp2 Inhibits TLR-Independent Sensing of Viral Replication by Retinoic Acid-Inducible Gene-I. Journal of immunology. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 37.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nature immunology. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 42.Smith GP, Calveley SB, Smith MJ, Baguley BC. Flavone acetic acid (NSC 347512) induces haemorrhagic necrosis of mouse colon 26 and 38 tumours. European journal of cancer & clinical oncology. 1987;23:1209–1211. doi: 10.1016/0277-5379(87)90157-x. [DOI] [PubMed] [Google Scholar]
- 43.Jameson MB, Thompson PI, Baguley BC, Evans BD, Harvey VJ, Porter DJ, McCrystal MR, Small M, Bellenger K, Gumbrell L, Halbert GW, Kestell P IIITCoCRUK Phase. Clinical aspects of a phase I trial of 5,6-dimethylxanthenone-4-acetic acid (DMXAA), a novel antivascular agent. British journal of cancer. 2003;88:1844–1850. doi: 10.1038/sj.bjc.6600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baguley BC. Antivascular therapy of cancer: DMXAA. The lancet oncology. 2003;4:141–148. doi: 10.1016/s1470-2045(03)01018-0. [DOI] [PubMed] [Google Scholar]
- 45.Philpott M, Baguley BC, Ching LM. Induction of tumour necrosis factor-alpha by single and repeated doses of the antitumour agent 5,6-dimethylxanthenone-4-acetic acid. Cancer chemotherapy and pharmacology. 1995;36:143–148. doi: 10.1007/BF00689199. [DOI] [PubMed] [Google Scholar]
- 46.Ching LM, Goldsmith D, Joseph WR, Korner H, Sedgwick JD, Baguley BC. Induction of intratumoral tumor necrosis factor (TNF) synthesis and hemorrhagic necrosis by 5,6-dimethylxanthenone-4-acetic acid (DMXAA) in TNF knockout mice. Cancer research. 1999;59:3304–3307. [PubMed] [Google Scholar]
- 47.Ching LM, Young HA, Eberly K, Yu CR. Induction of STAT and NFkappaB activation by the antitumor agents 5,6-dimethylxanthenone-4-acetic acid and flavone acetic acid in a murine macrophage cell line. Biochemical pharmacology. 1999;58:1173–1181. doi: 10.1016/s0006-2952(99)00194-x. [DOI] [PubMed] [Google Scholar]
- 48.Gresser I, Belardelli F, Maury C, Maunoury MT, Tovey MG. Injection of mice with antibody to interferon enhances the growth of transplantable murine tumors. The Journal of experimental medicine. 1983;158:2095–2107. doi: 10.1084/jem.158.6.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nature reviews Immunology. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 50.Mantovani A, Schioppa T, Porta C, Allavena P, Sica A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer metastasis reviews. 2006;25:315–322. doi: 10.1007/s10555-006-9001-7. [DOI] [PubMed] [Google Scholar]
- 51.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, Murphy KM, Schreiber RD. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. The Journal of experimental medicine. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends in immunology. 2012 doi: 10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dinney CP, Bielenberg DR, Perrotte P, Reich R, Eve BY, Bucana CD, Fidler IJ. Inhibition of basic fibroblast growth factor expression, angiogenesis, and growth of human bladder carcinoma in mice by systemic interferon-alpha administration. Cancer research. 1998;58:808–814. [PubMed] [Google Scholar]
- 54.Singh RK, Gutman M, Bucana CD, Sanchez R, Llansa N, Fidler IJ. Interferons alpha and beta down-regulate the expression of basic fibroblast growth factor in human carcinomas. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4562–4566. doi: 10.1073/pnas.92.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strieter RM, Polverini PJ, Arenberg DA, Walz A, Opdenakker G, Van Damme J, Kunkel SL. Role of C-X-C chemokines as regulators of angiogenesis in lung cancer. Journal of leukocyte biology. 1995;57:752–762. doi: 10.1002/jlb.57.5.752. [DOI] [PubMed] [Google Scholar]
- 56.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infection and immunity. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP Sensing via the Innate Immune Signaling Protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1