The Legionella pneumophila EnhC protein interferes with immunestimulatory muramyl peptide production to evade innate immunity (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 11.
Published in final edited form as: Cell Host Microbe. 2012 Aug 16;12(2):166–176. doi: 10.1016/j.chom.2012.06.004
Abstract
Successful pathogens have evolved to evade innate immune recognition of microbial molecules by pattern recognition receptors (PRR), which control microbial growth in host tissues. Upon Legionella pneumophila infection of macrophages, the cytosolic PRR Nod1 recognizes anhydro-disaccharide-tetrapeptide (anhDSTP) generated by soluble lytic transglycosylase (SltL), the predominant bacterial peptidoglycan degrading enzyme, to activate NF-κB-dependent innate immune responses. We show that L.pneumophila periplasmic protein EnhC, which is uniquely required for bacterial replication within macrophages, interferes with SltL, to lower anhDSTP production . L. pneumophila mutant strains lacking EnhC (ΔenhC) increase Nod1-dependent NF-κB activation in host cells, while reducing SltL activity in a ΔenhC strain restores intracellular bacterial growth. Further, L. pneumophilaΔenhC is specifically rescued in Nod1-, but not Nod2-, deficient macrophages, arguing that EnhC facilitates evasion from Nod1 recognition. These results indicate that a bacterial pathogen regulates peptidoglycan degradation to control the production of PRR ligands and evade innate immune recognition.
Introduction
The Gram-negative intracellular bacterium Legionella pneumophila is the causative agent of Legionnaires’ disease (Fraser et al. 1977; McDade et al. 1977). Fresh water amoebae, which support L. pneumophila intracellular replication, form a natural bacterial reservoir that is thought to result in selective pressure for traits contributing to virulence in the human host (Rowbotham 1980). Disease is caused by inhalation of aerosols from fresh water supplies followed by ingestion of the bacteria by alveolar macrophages (McDade et al. 1977). Within all cell types, L. pneumophila resides and replicates in a membrane-bound compartment surrounded by endoplasmic reticulum (ER), called the Legionella containing vacuole (LCV) (Horwitz 1983; Swanson and Isberg 1995; Kagan and Roy 2002). The L. pneumophila Icm/Dot Type IV protein system is required for formation of the LCV (Berger and Isberg 1993; Segal and Shuman 1997; Segal et al. 1998; Vogel et al. 1998). Over 200 different protein substrates have been identified as having recognition sequences that allow translocation into host cells via the Icm/Dot system (Luo and Isberg 2004; de Felipe et al. 2005; Burstein et al. 2009; Huang et al. 2010).
As most of the selective pressures that gave rise to the pathogenic potential of L. pneumophila resulted from growth within amoebae, L. pneumophila should not express a cadre of proteins specific for survival in macrophages (Rowbotham 1980). The L. pneumophila periplasmic protein EnhC appears to be an exception to this rule (Liu et al. 2008). A L. pneumophila ΔenhC strain is selectively restricted by macrophages previously exposed to bacteria, as the mutant grows with wild type kinetics in the amoebal Dictyostelium discoideum species (Liu et al. 2008). Possibly related to this observation, the absence of EnhC results in a compromised bacterial envelope, which may allow selective sensitivity to cytokine-stimulated macrophages (Liu et al. 2008).
Innate immune recognitions of microorganisms by membrane bound Toll-like receptors (TLR) and cytosolic Nod-like receptors (NLR) are primary strategies for controlling pathogens (Roy and Mocarski 2007; Ishii et al. 2008). At least three classes receptors are involved in recognition and restriction of L. pneumophila in both cultured macrophages and mouse infection models (Archer and Roy 2006; Hawn et al. 2006; Archer et al. 2009; Archer et al. 2010). Intracellular replication of L. pneumophila is restricted in macrophages from C57BL/6 mice, due to coordinate recognition of L. pneumophila flagellin by NLR sensors Naip5/Birc1e and Ipaf (Nlrc4/Card12) (Amer et al. 2006; Molofsky et al. 2006; Ren et al. 2006; Zamboni et al. 2006; Lightfield et al. 2008). In addition, Rip2 and MyD88, critical components of peptidoglycan (PG)-dependent Nod signaling and TLR signaling respectively, modulate host cell responses to L. pneumophila via NF-κB (Losick and Isberg 2006; Shin et al. 2008; Bartfeld et al. 2009). Mice defective for MyD88 function are further impaired for restriction of L. pneumophila if they lack either the Rip2-dependent signaling or the Naip5/Ipaf signaling, indicating that cytosolic flagellin and PG recognition synergize with TLR signaling to allow innate immune clearing of L. pneumophila in mice (Archer et al. 2010). Therefore, the ability of L. pneumophila to modulate production of bacterial products that stimulate pattern recognition should alter host cell recognition of the microorganism.
Soluble lytic murein transglycosylase (Slt), the predominant enzyme involved in PG degradation, generates _N_-acetylglucosamine (GlcNAc)-1,6-anhydro-_N_-acetylmuramic acid (MurNAc)-peptide (G-anhM-peptide) by cleaving the glycosidic bond between MurNAc and GlcNAc (Holtje 1996; Park and Uehara 2008). Slt activity generates pores for insertion of newly synthesized PG fragments or protein complexes, facilitating bacterial growth and division. The action of all PG degradation enzymes must be strictly regulated to maintain a critical balance between PG synthesis and degradation (Scheurwater et al. 2008). The mammalian Nod1 protein, one of the archetypal NLR proteins, recognizes the G-anhM-peptide generated by Slt (Chamaillard et al. 2003a; Girardin et al. 2003b; Girardin et al. 2003a; Inohara et al. 2003). In contrast, the PG-sensing pathway controlled by Nod2 does not recognize this product (Chaput et al. 2006; Nigro et al. 2008).
Here we show that EnhC binds to the L. pneumophila Slt and interferes with its function. The primary consequence of EnhC activity is to facilitate intracellular growth of L. pneumophila by reducing host cell Nod1 innate immune recognition of bacteria
Results
L. pneumophila EnhC directly interacts with soluble lytic murein transglycosylase (Slt)
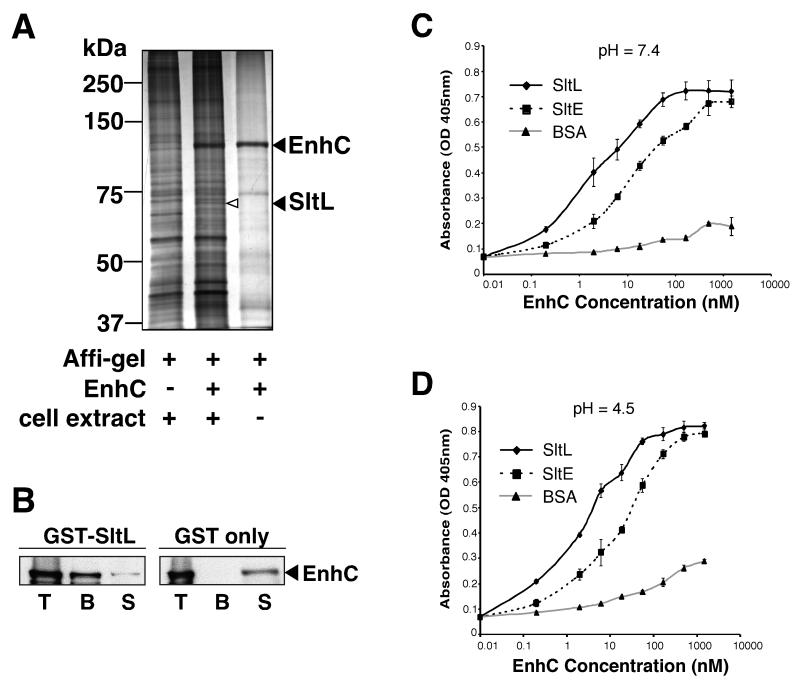
There is little information that suggests a function for EnhC, although strains lacking the protein are osmotically unstable (Liu et al. 2008). To identify binding partners of EnhC, a His-tagged derivative of EnhC was purified (Supplemental Fig. S1A), coupled to Affigel-10 beads, and used to isolate proteins from L. pneumophila lysates (Experimental Procedures). With beads linked to EnhC, we identified a 66 kDa band not present after elution from control beads (Fig. 1A). The band identified by mass spectrometry was a 606 amino acid protein annotated as soluble lytic murein transglycosylase (Slt; Lpg0663) (Chien et al. 2004) because it shares 29% identity to Slt of E. coli. This L. pneumophila ortholog of the E. coli protein will be referred to as SltL and is predicted to have a periplasmic locale, consistent with previous localization data for EnhC (Liu et al. 2008).
Figure 1. L. pneumophila EnhC directly binds L. pneumophila soluble lytic peptidoglycan transglycosylase (SltL).
(A) SltL specifically binds EnhC-coated beads. Lysates of L. pneumophila were incubated with EnhC-Affigel beads. Bound proteins were released from the beads by boiling in SDS, separated by 10 % SDS-PAGE and visualized by silver staining (Experimental Procedures). Left lane: L. pneumophila extract incubated with control Affigel; Middle lane: L. pneumophila extracts incubated with EnhC-Affigel; Right lane: boiled beads from buffer incubated with Affigel-EnhC. (B) EnhC directly binds SltL. GST-SltL- or GST-Glutathione-Sepharose beads were incubated with purified EnhC and proteins associated with the beads were analyzed by Western blot using anti-EnhC. T: Total input of EnhC; B: EnhC bound to beads; S: EnhC in the supernatant after incubation with beads. (C) EnhC binds L. pneumophila Slt (SltL) with higher affinity than E. coli Slt (SltE). ELISA plates coated with SltL or SltE or BSA were probed with increasing concentrations of EnhC. Shown are mean +/− standard deviation (s.d.) of triplicate wells. The experiment was repeated 3 times. (D). High affinity binding of SltL is maintained at pH = 4.5. Experiment performed exactly as in panel C, probing wells coated with proteins noted in the legend. See also Fig. S1.
To test whether SltL directly binds EnhC, we incubated purified EnhC with Glutathione-Sepharose beads bound to GST-SltL and the amount of EnhC retained was analyzed by Western blot (Experimental Procedures). The majority of EnhC was pulled down by GST-SltL, whereas only trace amounts of EnhC were left in the supernatant (Fig. 1B). No such binding of EnhC was observed with GST beads.
The E. coli Slt enzyme, which we will refer to as SltE, shows optimal cell wall hydrolysis at pH = 4.5 (Holtje et al. 1975). As the activity of the enzyme is measured at pH = 4.5, but the protein likely acts in the periplasm at neutral pH, we wanted to ensure that purified SltL (Supplemental Fig. S1B) bound to EnhC at both acidic and neutral pH, using an enzyme-linked immunosorbent assay (ELISA). Half-maximal binding of EnhC to SltL at pH = 7.4 was obtained at an EnhC concentration of 2 nM (Fig. 1C), while the half-maximal binding at pH = 4.5 was similar (Fig. 1D). EnhC did not bind to bovine serum albumin (BSA)-coated plates under both tested pH conditions (Fig. 1C,1D), indicating that the observed interaction between SltL and EnhC was not dependent on the buffer. We also measured the binding of purified E. coli SltE (Supplemental Fig. S1C) to EnhC. Although SltE bound to EnhC at both pH conditions (Fig.1C, 1D, dashed lines), the half-maximal binding of SltE for EnhC was 10 fold lower than that of SltL for EnhC.
L. pneumophila SltL catalyzes transglycosylation of L. pneumophila peptidoglycan (PG)
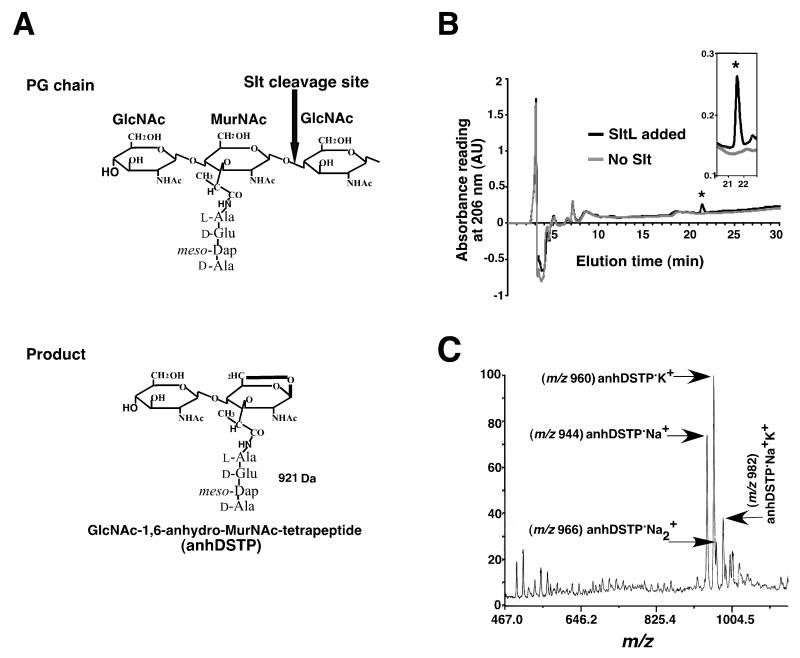
SltE cleaves the β-1,4-glycosidic bond between MurNAc and GlcNAc and generates an intramolecular 1,6-anhydro bond between the C1 and C6 positions in the sugar ring of MurNAc (Fig. 2A). If the short peptide on MurNAc is not cross-linked, then GlcNAc-1,6-anhydro-MurNAc-tetrapeptide (anhydro-disaccharide-tetrapeptide or anhDSTP) is produced, a species of 921 Da (Fig. 2A) (Scheurwater et al. 2008). To determine that L. pneumophila SltL is a lytic murein transglycosylase, PG was prepared from a lysate of L. pneumophila (Experimental Procedures), incubated with purified SltL in pH 4.5 buffer for 16 hr at 37°C and analyzed by high performance liquid chromatography (HPLC) (Experimental Procedures). Incubation of L. pneumophila PG with SltL, yielded a prominent peak that overlapped with the elution peak of pure E. coli anhDSTP. No such peak was observed in absence of added SltL (Fig. 2B). MALDI-TOF mass spectrometry (MS) analysis showed that the peak fraction consisted primarily of anhDSTP•Na+ (m/z 944) and anhDSTP•K+ (m/z 960) (Fig. 2C). Therefore, SltL has lytic transglycosylation activity against L. pneumophila PG.
Figure 2. Transglycosylation of L. pneumophila peptidoglycan (PG) by SltL.
(A) Slt cleaves the β-1,4-glycosidic bond between MurNAc and GlcNAc in the glycan strand and generates an intramolecular 1,6-anhydro linkage between the C1 and C6 positions in the sugar ring of MurNAc, releasing GlcNAc-1,6-anhydro-MurNAc-tetrapeptide (MW = 921Da). (B) Identification of a product generated by SltL incubation with PG. L. pneumophila PG was incubated with SltL and subjected to HPLC (black line). The elution time of purified GlcNAc-1,6-anhydro-MurNAc-tetrapeptide (anhDSTP) is indicated by the asterisk and overlaps with the SltL product (inserted image). (C) Peak in 2(B) was subjected to MALDI-TOF mass spectrometry. Four forms of modified anhDSTP were identified.
L. pneumophila EnhC inhibits the enzymatic activity of L. pneumophila SltL
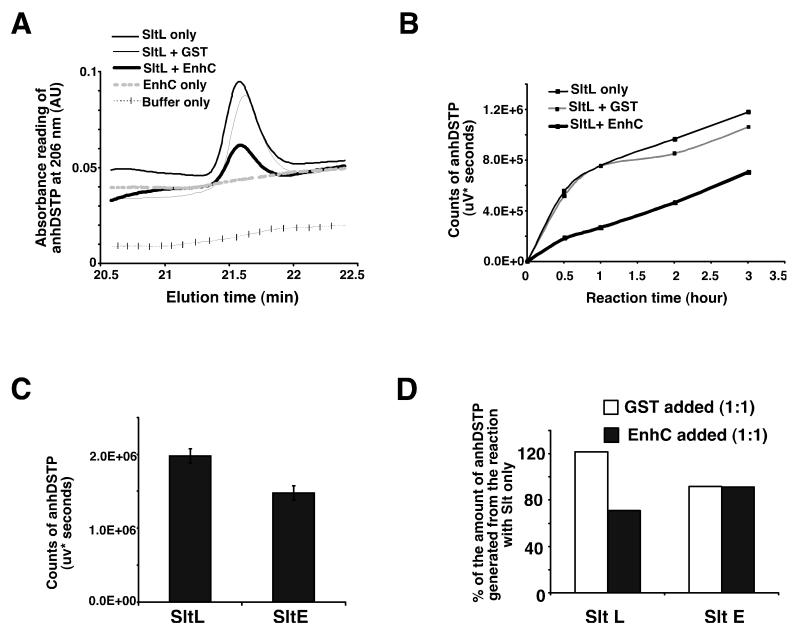
As EnhC directly binds to SltL, we tested whether EnhC controls the production of anhDSTP by SltL. Purified EnhC was added to the mixture of Legionella PG and SltL (Experimental Procedures) at a 1:1 molar ratio of EnhC :SltL and Slt enzymatic activity was measured at pH = 4.5. The addition of equimolar EnhC to SltL interfered with the production of anhDSTP during a 1 hr reaction, whereas adding the same molar amount of GST had no effect (Fig. 3A, Supplemental Fig. S2). To characterize further the effect of EnhC on the activity of SltL, EnhC or GST was added to SltL-catalyzed reactions and the amount of anhDSTP product was determined over time (Experimental Procedures). The product of each reaction was separated by HPLC and the amount of anhDSTP was plotted as a function of the time of reaction (Fig. 3B). After 30 min. of reaction time, equimolar amounts of SltL and EnhC generated only 30% of the product observed when SltL was incubated in the presence of GST (Fig. 3B).
Figure 3. EnhC specifically inhibits the enzymatic activity of L. pneumophila SltL.
(A) EnhC interferes with SltL activity. Equimolar amounts of EnhC and SltL were incubated with L. pneumophila PG at pH = 4.5 and production of anhDSTP was monitored by HPLC fractionation. (B) EnhC causes severe reduction in the level of anhDSTP produced by SltL. EnhC or GST was added to SltL in equimolar amounts and the production of anhDSTP was followed over time. (C) E. coli SltE cleaves L. pneumophila PG. SltE was incubated with L. pneumophila PG for 16 hr and the amount of anhDSTP was quantified based on the HPLC elution peak of anhDSTP. Means +/− s.d. from triplicate reactions are shown. (D) EnhC does not interfere with SltE activity. Displayed is the amount of SltE activity in the presence of EnhC compared to its absence. Production of anhDSTP was determined as in panel A. Data represent the mean value from two independent reactions for a typical experiment. See also Fig. S2.
Although L. pneumophila SltL shares 29% identity and 47% similarity with E. coli SltE, there is no apparent EnhC ortholog in E. coli. To determine whether inhibition by EnhC is specific for SltL, the effect of EnhC on SltE-mediated hydrolysis of L. pneumophila PG was tested. The SltL or SltE preparations were incubated with L. pneumophila PG for 16 hr and the production of anhDSTP by SltL or SltE was quantified (Fig. 3C). As expected, SltE was active on L. pneumophila PG and released anhDSTP from L. pneumophila PG. Nevertheless, the generation of anhDSTP by SltL appeared more robust than that by SltE, suggesting a lower activity of SltE on L. pneumophila PG. To examine whether EnhC inhibits SltE, L. pneumophila PG was incubated with SltE for 1hr with or without EnhC added (Fig. 3D). There was no effect on the production of anhDSTP. On the other hand, adding equimolar amounts of EnhC to SltL still caused about 50% reduction of anhDSTP during a 1hr reaction (Fig. 3D). This result supports the idea that EnhC shows species specificity for inhibition of Slt activity.
Misregulation of SltL results in enhanced NF-κB activation and depressed growth within macrophages
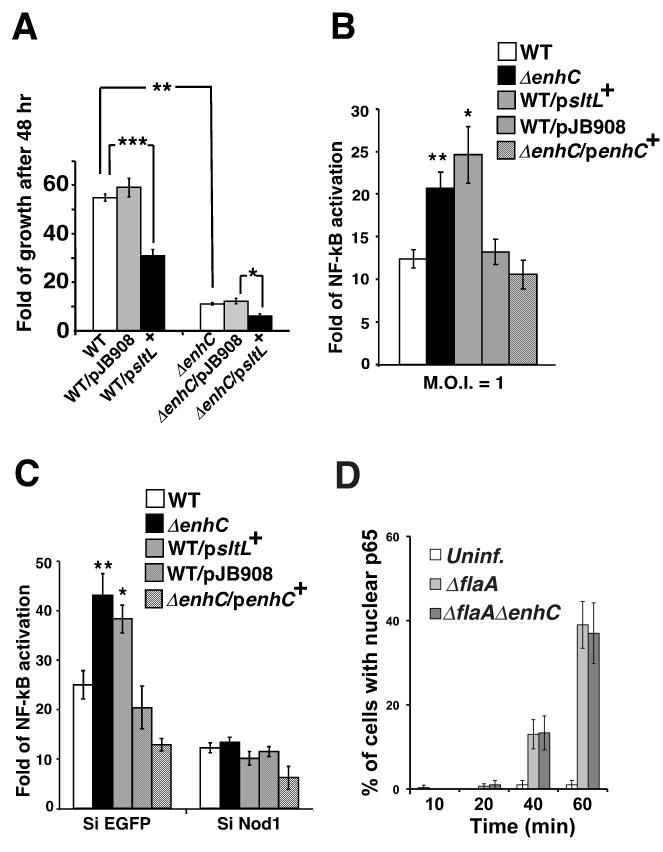
To determine if the observed lowered intracellular growth was the consequence of a hyperactive SltL protein, we constructed deletions of sltL (Supplemental Fig. S3). The ΔsltL mutant was elongated relative to the wild type strain, grew slowly in broth culture and could not reach post-exponential phase, which is required for optimal intracellular infection. This prevented analysis of intracellular growth, so strains overexpressing SltL were analyzed (Supplemental Fig. S3, WT/p_sltL_+). Within mouse bone marrow derived macrophages (BMDM), an SltL-overexpressing strain showed growth defects that were strikingly similar to the ΔenhC mutant, (WT/p_sltL_+; Fig. 4A). Furthermore, over-expression of SltL in the ΔenhC mutant (ΔenhC/p_sltL_+; Fig. 4A) enhanced the intracellular growth defect of ΔenhC, indicating that the depression in intracellular growth was proportional to the level of SltL in the cell.
Figure 4. Overexpression of SltL results in aberrant Nod1-dependent NF-κB activation and defective intracellular growth in host cells.
(A) Overexpression of SltL is detrimental to the intracellular growth of L. pneumophila in mouse bone marrow derived macrophages (BMDMs). Fold growth = (CFU at 48 hr post-infection)/(CFU at 2 hr post-infection). WT: L. pneumophila LP02; ΔenhC: MLL101; WT/p_sltL+_: MLL801; _ΔenhC/p_sltL+: MLL901; pJB908: empty vector. ***: P < 0.0005; **: P < 8.17E-06; *: P < 0.005. (B) Overexpression of SltL stimulates NF-κB activation in HEK293T cells challenged by L. pneumophila. _ΔenhC/p_enhC+: MLL201. NF-κB activation was measured using luciferase fused to NF-κB sensitive promoter (Experimental Procedures). Data are represented as luciferase units from infected samples/ luciferase units in uninfected controls. **: P < 0.003; *: P < 0.003. (C) Overexpression of SltL causes Nod1-dependent NF-κB activation. NF-κB activation was measured as above in HEK293T cells depleted of Nod1 (SiNod1) or with control (SiEGFP). Fold activation = luciferase units from infected samples/ luciferase units of uninfected sample. **: P < 0.002; *: P < 0.004. See also Fig. S3. (D)Time course of p65 translocation. Bone marrow macrophages from C57BL/6 myd88−/− mice were challenged with noted bacterial strains at MOI = 1 and NF-κB p65 nuclear localization was determined (Losick and Isberg 2006). For panels A and D, mean +/− s.d. from 3 samples are shown. For panel B and C, means +/− s.d. from 6 samples are shown. P value was calculated by unpaired two-tailed Student’s _t_-test.
SltL generates the iE-DAP moiety (Fig. 2A), which activates host cell NF-κB response via the Nod1 sensor (Magalhaes et al. ; Chaput et al. 2006; Fritz et al. 2006; Hasegawa et al. 2006; Benko et al. 2008; Nigro et al. 2008). Unregulated SltL should result in excess anhDSTP, consequently triggering a Nod1-dependent NF-κB response (Viala et al. 2004). To test this hypothesis, activation of NF-κB in HEK 293T cells challenged with L. pneumophila was quantified by measuring luminescence produced by an NF-κB driven luciferase reporter (Experimental Procedures). L. pneumophila overexpressing SltL (WT/p_sltL_+; Fig. 4B) or lacking EnhC (ΔenhC, Fig. 4B) caused hyperactivation of NF-κB when compared to controls (WT or WT/pJB908, Fig. 4B). Therefore, overexpression of SltL mimics a strain lacking EnhC in regard to NF-κB activation.
To examine whether hyperactivation of NF-κB was dependent on peptidoglycan sensing by Nod1, siRNA knockdown of Nod1 was performed (Experimental Procedures). Depletion of Nod1 in HEK 293T cells was confirmed by Western blot (Supplemental Figure, Fig. S3, pSiNod1construct2). As shown in Figure 4C, NF-κB activation elicited by the challenge of WT bacteria was reduced by 50% in cells depleted of Nod1 (SiNod1) compared to cells having a control siRNA knockdown (SiEGFP). Therefore, a significant portion of NF-κB activation in response to L. pneumophila was triggered via Nod1. Strikingly, hyper-activation of NF-κB in response to either the ΔenhC strain or the strain overexpressing SltL (WT/p_sltL_+) was abolished in cells knocked down with SiNod1 (Fig. 4C). Hyperactivation by the ΔenhC strain could also be reversed by introducing enhC on a plasmid (ΔenhC/ p_enhC_+; Fig. 4B and 4C). To determine if a similar increase in NF-κB activation could be seen in bone marrow macrophages, activation was measured using a different assay (Losick and Isberg 2006), quantitating the fraction of infected C57BL/6 myd88−/− macrophages that had observable NF-κB p65 nuclear translocation. Bacterial strains used here were ΔflaA (flagellin deficient) to prevent the Naip5/Birc1e signaling that occurs in the C57BL/6 mouse background. This assay did not detect an increase in p65 translocation when macrophages from myd88−/− mice were used to limit the TLR signaling contribution (Fig. 4D). This is either because these two readouts show different levels of sensitivity to changes in NF-κB activation, MyD88 signals that collaborate with Nod1 in HEK 293T cells could not be detected in these macrophages, or else the Nod1-dependent response observed in the HEK 293T is independent of NF-κB activation. The latter model will be treated in the Discussion.
Inhibition of SltL by EnhC is required to suppress Nod1-dependent NF-κB activation induced by L. pneumophila
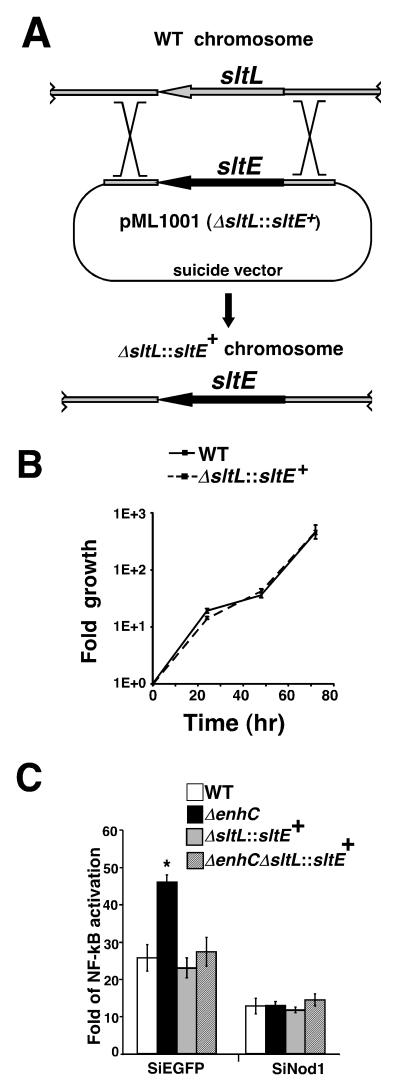
To further demonstrate that lack of inhibition of SltL by EnhC is responsible for the phenotype of the ΔenhC mutant, we tested whether lowering Slt activity in L. pneumophila could bypass the EnhC requirement. As L. pneumophilaΔsltL was defective for growth in broth culture, deleting sltL in a strain lacking EnhC was not feasible. Instead, sltL was substituted with E. coli sltE gene on the L. pneumophila chromosome to generate a new strain predicted to have lowered Slt activity (MLL1001; ΔsltL::sltE+) (Fig. 5A). The SltE protein had lowered activity on L. pneumophila PG (Fig. 3C) and transcription of sltE in L. pneumophila was about 20% that of sltL as monitored by qRT-PCR (Experimental Procedures; Supplemental Fig. S4). When the ΔsltL::sltE+ strain was used to challenge mouse BMDMs, it had the same intracellular growth rate as WT L. pneumophila, indicating that there was sufficient Slt activity to support growth in a number of conditions (Fig. 5B). This predicts that altering Slt activity in L. pneumophila should make EnhC dispensable for intracellular growth and normal level of Nod1-dependent NF-κB activation. To test this hypothesis, the ΔsltL::sltE+ replacement was moved into a ΔenhC background (Supplemental Fig. S4). Nod1-dependent NF-κB activation in response to the ΔenhC ΔsltL::sltE+ strain was similar to that observed for either WT bacteria or the ΔsltL::sltE+ strain (Fig. 5C), indicating that hyperactivation resulting from loss of EnhC was dependent on having a wild type sltL allele.
Figure 5. The ΔenhC phenotype requires SltL function.
(A) Construction of L. pneumophila strain with sltL replaced by sltE (MLL1001; ΔsltL::sltE+). A DNA fragment containing the ORF of sltE flanked by L. pneumophila chromosomal DNA was cloned into suicide vector pSR47S. Selection for integration and recombination is as described (Experimental Procedures). (B) MLL1001 (ΔsltL::sltE+) grows as well as WT in mouse bone marrow derived macrophages (BMDMs). The intracellular growth of L. pneumophila was monitored by measuring CFU every 24 hr during a 3 day incubation (Experimental Procedures). (C) Replacement of sltL with sltE bypasses the ΔenhC defect. NF-κB activation was determined by luciferase reporter assay. WT: LP02. ΔenhC: MLL101. ΔsltL::sltE+: MLL1001. ΔenhC ΔsltL::sltE+: MLL1101. *: P < 0.001. For panel B and C, a representative experiment is shown; experiment was repeated 3 times. For panel B, means +/− s.d. from 3 samples are shown. For panel C, means +/− s.d. from 6 samples are shown. P value was calculated by unpaired two-tailed Student’s _t_-test. See also Fig. S4
Nod1 and products of soluble lytic transglycosylase are responsible for the intracellular growth defect of the ΔenhC mutant
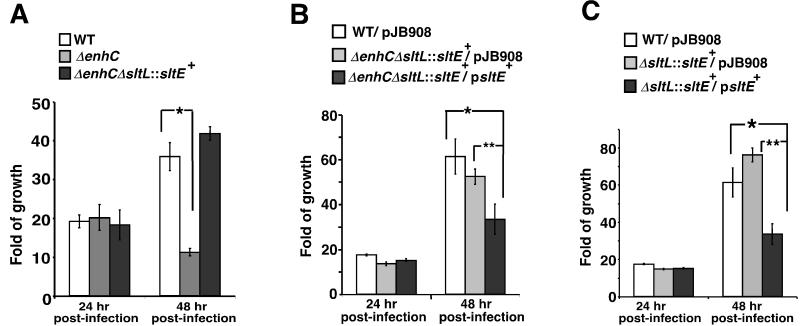
To evaluate the importance of EnhC and the reduced Nod1 sensing resulting from the presence of EnhC, we monitored the intracellular growth of a variety of Legionella strains. Intracellular growth of the ΔenhC mutant is known to be restricted at a time point roughly corresponding to 24-48 hr post-infection (hpi) (Liu et al. 2008). To test whether the restriction of ΔenhC strain was due to an inability to regulate the activity of SltL, intracellular growth in BMDMs of both the ΔenhC and the ΔenhCΔsltL::sltE+ strains was monitored (Fig. 6A). The ΔenhC mutant replicated at the same rate as WT in the first round of intracellular growth (24 hpi) and showed a defect in growth at 48 hpi, consistent with previous observations (Liu et al. 2008). In contrast, the ΔenhC strain harboring the ΔsltL::sltE+ replacement grew as well as WT at 48 hpi (Fig. 6A). This result is consistent with the dependence of the intracellular growth defect of ΔenhC on the slt allele.
Figure 6. The inhibition of SltL by EnhC promotes intracellular growth of L. pneumophila.
(A) Replacement of sltL bypasses the ΔenhC defect for intracellular growth. WT: LP02; ΔenhC: MLL101; ΔenhC ΔsltL::sltE+: MLL1101. *: P < 0.003. (B) Overexpression of SltE in the absence of EnhC and SltL causes an intracellular growth defect. pJB908: empty vector. WT/pJB908: MLL221; ΔenhC ΔsltL::sltE+/pJB908: MLL1101/pJB908; ΔenhC ΔsltL::sltE+/p_sltE_+: MLL1301. *: P < 0.01; **: P < 0.012. (C) Overexpression of SltE in the presence of EnhC causes an intracellular growth defect. ΔsltL::sltE+/pJB908: MLL 1001/pJB908; ΔsltL::sltE+/p_sltE_+: MLL1201. *: P < 0.008; **: P < 0.0004. For all panels, a representative experiment is shown and the experiments were repeated 3 times. Means +/− s.d. from triplicate samples are shown. P value was calculated by unpaired two-tailed Student’s _t_-test.
Results from Figs 5C and 6A indicated that reduced production of anhDSTP caused by a low level of Slt was responsible for rescue of the ΔenhC mutant. To examine this further, we tested whether overexpression of sltE had a similar phenotype to overproduction of SltL. Plasmid-borne sltE was introduced into the ΔenhC ΔsltL::sltE+ replacement strain, and overexpression of sltE was confirmed by qRT-PCR (Supplemental Fig. S4). Overexpression of sltE reduced intracellular growth at 48 hpi (Fig. 6B; ΔenhC ΔsltL::sltE+/p_sltE_+). The presence of an intact enhC gene did not alter these results, as the EnhC+ replacement strain overproducing SltE also showed a significantly reduced intracellular growth at 48 hpi (Fig. 6C; ΔsltL::sltE+/p_sltE_+). These results are consistent with unregulated production of anhDSTP being responsible for the intracellular growth defect of ΔenhC. Furthermore, it is unlikely that the mutant phenotype of a ΔenhC strain is due to SltL having a unique activity not catalyzed by SltE.
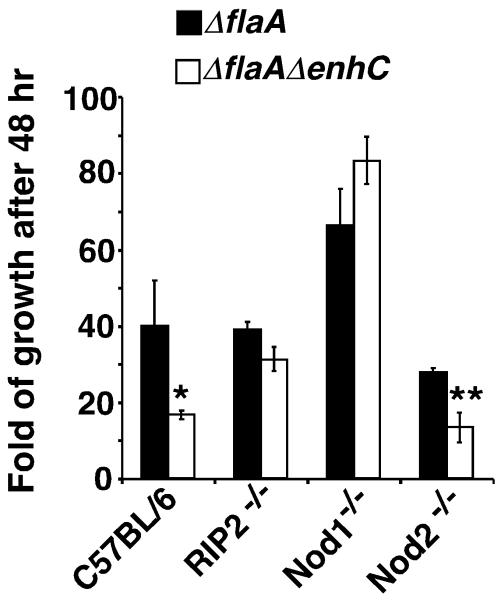
As Slt activity appears tightly linked to the phenoytpe of a ΔenhC strain, we tested whether EnhC promotes intracellular growth of L. pneumophila by limiting Nod1 sensing of anhDSTP. Therefore, the intracellular growth of the mutant lacking EnhC was examined in BMDMs from C57BL/6 mice having the Nod1−/− , Nod2−/− and RIP2−/− (defective for both Nod1 and Nod2 signaling) genotypes. All bacterial strains used in this assay were ΔflaA (flagellin deficient) to prevent the Naip5/Birc1e signaling that occurs in the C57BL/6 mouse background (Supplemental Fig. S5). In macrophages prepared from either RIP2 or Nod1 knockout mice, defective intracellular growth due to the ΔenhC mutation was no longer apparent (Fig. 7; for nod1−/−, P ≤ 0.61; for rip2−/−, P ≤ 0.64 by unpaired Student’s _t_-test). In contrast, the growth defect of ΔenhC was maintained in the Nod2 knockout macrophages, which, unlike Nod1−/− and RIP2−/− macrophages, still retain the ability to recognize anhDSTP (P≤ 0.002). These observations support the model that EnhC promotes intracellular growth of L. pneumophila by interfering with Nod1 signaling.
Figure 7. The absence of Nod1 allows proficient growth of the ΔenhC strain.
C57BL/6J bone marrow derived macrophages from the indicated knockout mice were challenged with L. pneumophila and intracellular growth was determined by plating for CFU after 48 hrs incubation. ΔflaA strains were used to prevent the Naip5/Birc1e signaling that occurs in the C57BL/6 mouse background. ΔflaA: MLL099; ΔflaA ΔenhC: MLL111. **: P < 0.001; *: P < 0.03. A representative experiment is shown and the experiment was repeated 3 times. Data are expressed as mean +/− s.d. from triplicate samples. P value was calculated by unpaired two-tailed Student’s _t_-test. See also Fig. S5.
Discussion
In this study we showed that EnhC modulates the activity of SltL transglycosylase, generating the identical anhydro-disaccharide-tetrapeptide (anhDSTP) produced by E. coli soluble lytic transglycosylase. EnhC binding to L. pneumophila SltL interferes with PG degradation, indicating that mutants lacking EnhC likely increase the amount of anhDSTP available to host cells. Negative regulation of SltL by EnhC provides an explanation for the previous observation that the absence of EnhC results in enhanced sensitivity to a variety of stress-inducing reagents (Liu et al. 2008). Presumably, unregulated SltL results in a loss of PG integrity.
The negative regulation of SltL activity by EnhC allows growth phase control of peptidoglycan degradation. The levels of SltL protein are similar at all phases of L. pneumophila growth (Supplemental Fig. S5), whereas EnhC is highly expressed in post-exponential phase (Liu et al. 2008). During growth and division of L. pneumophila, expression of SltL in the absence of EnhC may allow modulation of PG structure, facilitating septation (Heidrich et al. 2002) and insertion of protein complexes in the cell envelope (Koraimann 2003). When growth slows and bacteria enter post-exponential phase, PG turnover becomes less important, and cell envelope integrity becomes paramount. Although the transglycosylase could be regulated by proteolytic turnover, the presence of an inhibitory protein offers a clear advantage. Proteolysis could remove the entire pool of SltL from the periplasm, reducing enzyme concentration below the basal level necessary for maintaining PG homeostasis. In contrast, modulation by a protein inhibitor allows basal levels of enzyme activity controlled by the concentration of EnhC.
Association of soluble lytic transgycosylase with periplasmic proteins has been demonstrated previously. Slt from Pseudomonas aeruginosa has been found to associate with PG synthase-PBP2 (Legaree and Clarke 2008) and Slt from Brucella suis was found to interact with protein complexes of the Type IV secretion system (Hoppner et al. 2005). Such physical associations could, in the first case, allow coordination of PG degradation and synthesis. In the second example, the association of Slt with the Type IV secretion system may facilitate the assembly of this complex, allowing it to extend through PG layers by generating space for its insertion (Koraimann 2003).
The data from the HEK 293T cell reporter assay are consistent with the Nod1 effects associated with the enhC− mutant being mediated via an NF-κB-dependent pathway. In contrast, the link between NF-κB activation and Nod1 restriction in bone marrow macrophages is not as clear. Although Nod1 was required to selectively restrict the enhC− mutant in bone marrow macrophages, we did not observe enhanced kinetics of p65 translocation in response to the mutant bacterium (Fig. 4D). This is consistent with Nod1-restriction acting via an alternate pathway in macrophages. It is now apparent that restriction of bacterial growth resulting from Nod1 activation can occur independently of NF-κB activation, as Shigella flexneri infection stimulates a Nod1-associated NF-κB-independent antimicrobial response that recruits a subset of cellular autophagy components (Travassos et al. 2010). A similar strategy for restriction of L. pneumophila growth may be occurring in macrophages, so that Nod1 activation results in different responses in different cell types.
The Nod1 response is stimulated by a variety of microorganisms (Girardin et al. 2001; Kim et al. 2004; Viala et al. 2004; Travassos et al. 2005; Nigro et al. 2008; Archer et al. 2010), arguing that EnhC impinges on an important arm of innate immunity. Pathogenic bacteria manage to escape from Nod sensing using a variety of strategies, including modifying PG components, changing the composition of PG and efficiently recycling muramylpeptides back to the bacterial cytosol (Chamaillard et al. 2003b; Psylinakis et al. 2005; Chaput et al. 2006; Boneca et al. 2007; Davis et al. 2008; Nigro et al. 2008). Our data present another strategy, which is to inhibit the major enzyme generating anhydro-muramylpeptides.
Nod1 signaling is associated with immune surveillance in multicellular eukaryotes, so the selective pressures that resulted in the retention of EnhC function are not obvious. It is thought that the pressures that sculpted the L. pneumophila genome selected for a bacterium able to grow in a wide variety of amoebal species. Consistent with this model, there is little evidence for L. pneumophila directly interfering with host innate immune sensing pathways (Shin et al. 2008), and it seems unlikely that the mammalian host immune system exerted selective pressure. Rather, the enhanced stress resistance conferred by EnhC on post-exponential phase bacteria is probably the primary selective advantage for retention of this gene. Alternatively, EnhC may be important for survival in the presence of killing strategies used by an amoebal host that is found in the environment but not interrogated in this work.
For bacteria with access to host cell cytosol, such as Shigella flexneri, muramylpeptides resulting from PG turnover can be shed directly into cytosol and detected by Nod1 (Nigro et al. 2008). For bacteria residing in vacuoles, such as L. pneumophila, it is unclear how hydrophilic muramylpeptides can cross the vacuolar membrane and be sensed by Nod1. Presentation of Helicobacter pylori muramylpeptides to cytosolic Nod1 is dependent on the CagPAI-encoded Type IV secretion system (Viala et al. 2004), so it is possible that muramylpeptides from L. pneumophila could be presented through the Icm/Dot secretion system. Strains defective for Icm/Dot show very low activation of NF-κB in HEK 293T cells (Losick and Isberg 2006), indicating that muramylpeptide stimulation of host cells requires a functional Type IV secretion system. In further support of this model, both NF-κB activation and activation of multiple MAP kinases by the Nod signaling pathway require the Icm/Dot system (Shin et al. 2008). It seems likely that release via Icm/Dot might act as the mechanism for PG presentation.
It is not clear how Nod1 recognition leads to restriction of L. pneumophila. Nod1 recognition can trigger a variety of host immune responses, including macrophage priming and the production of various cytokines, chemokines and antimicrobial peptides, as well as autophagic response described above (Boneca 2005; Chaput and Boneca 2007; Travassos et. 2010). Previous work has shown that Nod1 activity plays a role in interfering with bacterial survival in the presence of a variety of cell types, including fibroblasts (Travassos et al. 2005) and bone marrow-derived dendritic cells (Le Bourhis et al. 2009). In addition, Nod1 controls the expression of a subset of β-defensin anti-microbial peptides (Grubman et al. 2010). Strategies such as these, combined with the inherent increased instability of the bacterial cell wall, could contribute to the reduced yield of the enhC− mutants when exposed to macrophages.
In summary, our work uncovers a mechanism for regulating the activity of soluble lytic transglycosylase. This mechanism allows high viability of L. pneumophila in the presence of increased stress and allows the integrity of the bacterial envelope to be maintained during post-exponential phase in which a large number of bacterial products are poised to be deposited in host cells.
Experimental Procedures
Bacterial strains, plasmids, reagents and cell culture
All bacterial strains and plasmids used in this work are listed in Table 1. All PCR primers are listed in Supplementary Table S1. Charcoal Yeast Extract agar (CYE) and ACES buffered Yeast Extract (AYE) broth were used to cultivate Legionella pneumophila in culture as described (Feeley et al. 1979; Berger and Isberg 1993). Thymidine was added at a concentration of 100 μg/ml when needed.
Table 1.
Bacterial strains and plasmids used in this work
| Strain/plasmid | Genotype/ relevant characteristics | Reference |
|---|---|---|
| pSR47s oriTRP4 | oriTRP4 oriPR6k sacB KanR suicide vector | (Rankin et al. 2002) |
| pJB908 | pMMB66EH oriPRSF1010 ΔoriT bla+ tdΔI | (Laguna et al. 2006) |
| pKB5 | pMMB66EH oriPRSF1010 tdΔi bla+ | (Berger and Isberg 1993) |
| pGE148 | pQE-32-His-tagged EnhC | (Liu et al. 2008) |
| pML701 | pGEX-6P-1-GST-tagged L. pneumophila | Slt this study |
| pMLD204 | pET-His-tagged E. coli Slt | (Stenbak et al. 2004) |
| pML101 | pSR47s ΔenhC | (Liu et al. 2008) |
| pML201 | pJB908_enhC_ + | (Liu et al. 2008) |
| pML301 | pSR47s ΔsltL | this study |
| pML601 | pKB5_sltL_+ | this study |
| pML801 | pJB908_sltL_+ | this study |
| pML1001 | pSR47s ΔsltL :: sltE+ | this study |
| pML1201 | pJB908_sltE_+ | this study |
| LP02 | Philadelphia-1 _rpsL hsdR thyA_− | (Berger and Isberg 1993) |
| LP03 | LP02 dotA03 | (Berger et al. 1994) |
| MLL099 | LP02_ΔflaA_ | (Ren et al. 2006) |
| MLL101 | LP02_ΔenhC_ | (Liu et al. 2008) |
| MLL111 | LP02_ΔflaAΔenhC_ | this study |
| MLL201 | LP02_ΔenhC_/pML201 | (Liu et al. 2008) |
| MLL211 | LP02_ΔenhC_/pJB908 | (Liu et al. 2008) |
| MLL221 | LP02/pJB908 | (Liu et al. 2008) |
| MLL301 | LP02_ΔsltL_ | this study |
| MLL601 | LP02_ΔsltL_ / pML601 | this study |
| MLL611 | LP02_ΔsltL_ / pKB5 | this study |
| MLL801 | LP02/pML801 | this study |
| MLL901 | LP02_ΔenhC_/pML801 | this study |
| MLL1001 | LP02 ΔsltL :: sltE+ | this study |
| MLL1001/pJB908 | LP02 ΔsltL :: sltE+/pJB908 | this study |
| MLL1101 | LP02_ΔenhC ΔsltL_ :: sltE+ | this study |
| MLL1101/pJB908 | LP02_ΔenhC ΔsltL_ :: sltE+/pJB908 | this study |
| MLL1201 | LP02 ΔsltL :: sltE+/pML1201 | this study |
| MLL1301 | LP02_ΔenhC ΔsltL_ :: sltE+/pML1201 | this study |
Plasmids were constructed as described in Supplemental Data. pHis-SltE (pMLD204) was a kind gift of Dr. Dominique Mengin-Lecreulx (University Paris-Sud, Orsay, France).
Mouse bone marrow derived macrophages (BMDM) were prepared from the femurs of female A/J mice (Jackson Laboratories) or mouse strains-RIP2−/−, Nod-1 −/−, and Nod-2 −/− (generous gifts of Dr. Koichi S. Kobayashi, Dana Farber Cancer Center, Boston, MA) (Dietrich et al. 1995) and cultivated in RPMI 1640 (Invitrogen-Gibco) with 10% heat-inactivated fetal bovine serum(FBS) (Gibco). HEK 293T cells (ATCC CRL-11268) were passaged in high glucose DMEM media containing 4.5g/L D-Glucose (Cat. 11995, Invitrogen-Gibco) supplemented with 10% heat-inactivated Hyclone FBS (Thermo Scientific).
Affinity Chromatography from Legionella lysates
200 ml of post-exponential phase (A600 = 3.7~3.9) motile Legionella pneumophila culture was pelleted at 9,000 × g for 15 min at 4 °C, then resuspended in 10 ml ice-cold lysis buffer (PBS, 1 mM β-mercaptoethanol [β-ME], protease inhibitor cocktail [Roche]) and lysed by French press at 1000 psi (Thermo Electron) and cleared by 2x centrifugation at 10,000 × g. Lysate from approximately 7.4 × 1010 bacteria, was incubated with 75 μl Affigel-10 beads linked to approximately 90 μg EnhC for 12 hr at 4°C. Beads were washed 4 or 5 times with ice-cold lysis buffer, resuspended in 12 μl SDS sample buffer, and boiled for 5 min and analyzed by 10 % SDS-PAGE and silver staining (Invitrogen). Proteins were isolated from gels and analyzed by high-performance liquid chromatography/mass spectrometry (HPLC/MS) (Tufts University proteomics core facility).
Recombinant protein purification, anti-SltL antibody production and assay for binding of purified proteins
Details of protein purification are found in Supplemental Data. Purified SltL was injected into rabbits to raise anti-SltL serum (Pocono Rabbit Farm and Laboratory Inc.). Anti-SltL antibody recognized a single protein band with predicted size from LP02 (WT) extract using dilution of 1: 5000 when 5 × 107 bacteria were loaded, but did not recognize any protein from ΔsltL extracts. To analyze binding to SltL, purified EnhC was incubated with GST-SltL bound to GS beads for 12 hr at 4°C. Beads were washed 4 times with PBS, then resuspended and boiled in SDS sample buffer before analysis by Western blot with polyclonal antibody specific for EnhC (1: 10,000 dilution) (Liu et al. 2008).
Quantitative measurement of Slt and EnhC interaction
Interactions between purified EnhC and either purified SltL or purified SltE were measured using ELISA. Each well of ELISA plates (Linbro) was coated with 100 μl of 100 nM SltL or SltE or BSA in PBS at 4°C for over night. Wells were washed three times with PBS, then were blocked with 3% BSA in PBS at 4°C for over night. Wells were washed once with PBS before probing with sequential 3-fold dilutions of EnhC starting at a concentration of 1500 nM in either PBS buffer (pH =7.4) or PBS supplemented with 0.01 M sodium acetate (pH =4.5) and 0.05 M MgCl2 for 2 hr at room temperature. Wells were washed six times with PBS. EnhC bound to wells was detected by probing wells with rabbit anti-EnhC polyclonal antibody (Liu et al., 2008) for 90 min at room temperature. Wells were subjected to 6 washes with PBS before probing with goat anti-rabbit IgG conjugated to alkaline phosphatase (Zymed-Invitrogen) for 90 min at room temperature. Wells were washed 6 times with PBS before phosphatase activity was detected with 3 mM PNPP (Sigma 104 phosphatase substrate, Sigma) in 0.05 M Na2CO3 (pH = 8.5) and 0.05 mM MgCl2. The reactions were stopped with 0.2 M NaOH after about 25 min.
Preparation of peptidoglycan (PG) from Legionella pneumophila
Peptidoglycan was prepared as described (Uehara and Park 2003). Further details on its isolation can be found in Supplemental Materials.
Digestion of L. pneumophila peptidoglycan (PG) by soluble lytic transglycosylase (Slt) and high performance liquid chromatography (HPLC) analysis
50 μg of L. pneumophila PG was incubated with about 3 μg of purified L. pneumophila or E. coli Slt in 100 μl of reaction buffer (10 mM NaAc [pH=4.5], 50 mM MgCl2) at 37°C. The undigested PG was cleared by ultracentrifugation at 130, 000 × g for 55 min and the supernatant was separated by Sunfire™ C18 reverse-phase column (4.6 × 150 mm; particle size, 5 μm; Waters Co., Milford, MA) using a Waters 1525 binary HPLC pump. The column was equilibrated by solvent A at a flow rate of 0.5 ml/min for 15 min. Then samples were injected and separated at a flow rate of 0.5 ml/min with a linear gradient of 0 to 10% solvent B over a period of 5 min, followed by a linear gradient of 10 to 50% solvent B over a period of 25 min. Solvent A: 25mM formic acid (HCOOH), 12.5mM ammonium hydroxide (NH4OH), pH = 3.75. Solvent B: 25mM HCOOH, 12.5mM NH4OH and 80% Methanol (MeOH). For mass spectrometry (MS), 150 μg of PG instead of 50 μg was digested for 16 hr and separated by HPLC. The main fraction was collected, lyophilized, and identified by MS with a Voyager DE Pro matrix-assisted laser desorption ionization—time-of-flight (MALDI-TOF) mass spectrometer (Applied Biosystems) at Tufts University Core Facility.
NF-κB activation assays
For NF-κB activation, assays in HEK293T cells were performed as described, challenging cells with bacteria for 8 hours at MOI = 0.05 or = 1 (Losick et al. 2010). For NF-κB assays in siRNA-treated HEK293T cells, cells were transfected with 100ng endotoxin-free psiRNA (ie SiEGFP or SiNod1) per well using 0.25μl Lipofectamine 2000 in total 50μl OPTI-MEM. After 20 hrs, cells were observed under fluorescence microscope to ensure there was efficient transfection by determining that greater than 75% of the cells showed GFP fluorescence. Media were changed and cells were transfected with endotoxin-free pNF-κB-luciferase reporter plasmid (Losick et al. 2010). 18 hrs later, cells were infected with bacteria at M.O.I. = 1. L. pneumophila uptake into HEK293T cells was monitored in all NF-κB assays to ensure that the NF-κB activation measurements were not affected by bacterial load. For p65 translocation assays, bone marrow macrophages were challenged with bacteria at M.O.I. = 1.0 and p65 nuclear translocation was determined by indirect immunofluorescence microscopy, probing fixed cells with anti-p65 (Losick and Isberg 2006). 100 cells per coverslip were counted visually.
Nod1 psiRNA constructs and assay for testing the efficiency of knockdown constructs
Construction of siRNA plasmids can be found in Supplemental Data. To test the knockdown efficiency of psiNod1, HEK293T cells were cotransfected with 10ng pmyc-Nod1 and 200ng of psiEGFP or psiNod1. 48 hrs later, cells were lysed and analyzed by Western blot with anti-myc to determine steady state protein level of myc-Nod1.
Quantitative Real-time PCR (QRT-PCR) assay of slt transcripts in L. pneumophila
Primers for QRT-PCR are listed in Supplemental Table 2. RNA and genomic DNA (gDNA) from L. pneumophila were extracted using the RNeasy and DNeasy miniprep kits (Qiagen). QRT-PCR reactions were set up in 96 well polypropylene plates (Strategene) using real-time PCR machine Mx3005 model (Strategene). DNA-free RNA samples in the absence of reverse transcriptase incubation were also included in QRT-PCR to ensure that there was no gDNA contamination in cDNA samples. All QRT-PCR reactions were normalized with 16s ribosome RNA as the internal control. Means ± s.d. were generated from triplicate QRT-PCR reactions.
Supplementary Material
01
HIGHLIGHTS.
- L. pneumophila EnhC directly inhibits soluble lytic peptidoglycan transglycosylase (Slt)
- SltL overexpression causes enhanced NF-κB activation and depressed intracellular growth
- SltL inhibition by EnhC is required to suppress Nod1-dependent NF-κB activation
- Growth of the L. pneumophila ΔenhC strain is rescued in Nod1 deficient macrophages
Acknowledgements
We want to thank Dr. Koichi S. Kobayashi for providing bone marrows of RIP2−/−, Nod1−/− and Nod2−/− mice, Dr. Dominique Mengin-Lecreulx for providing the E. coli SltE expression plasmid pMLD204 and Dr. Waldemar Vollmer for the E. coli ΔsltE strain MUF16, respectively, as well as Dr. Tao Ren for providing the L. pneumophila ΔflaA strain. We would also like to thank Drs. Edward Geisinger, Dervla Isaac, Eva Haenssler, Kerri Sheahan, Elizabeth Creasey, Sina Mohammadi and Greg Crimmins for comments on the text. RI is an Investigator of the Howard Hughes Medical Institute (HHMI). This work was supported by HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281(46):35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- Archer KA, Roy CR. MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires’ disease. Infect Immun. 2006;74:3325–3333. doi: 10.1128/IAI.02049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KA, Alexopoulou L, Flavell RA, Roy CR. Multiple MyD88-dependent responses contribute to pulmonary clearance of Legionella pneumophila. Cell Microbiol. 2009;11:21–36. doi: 10.1111/j.1462-5822.2008.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KA, Ader F, Kobayashi KS, Flavell RA, Roy CR. Cooperation between multiple microbial pattern recognition systems is important for host protection against the intracellular pathogen Legionella pneumophila. Infect Immun. 2010;78:2477–2487. doi: 10.1128/IAI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, Engels C, Bauer B, Aurass P, Flieger A, et al. Temporal resolution of two-tracked NF-κB activation by Legionella pneumophila. Cell Microbiol. 2009;11(11):1638–1651. doi: 10.1111/j.1462-5822.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- Benko S, Philpott DJ, Girardin SE. The microbial and danger signals that activate Nod-like receptors. Cytokine. 2008;43:368–373. doi: 10.1016/j.cyto.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Berger KH, Merriam JJ, Isberg RR. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- Boneca IG. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol. 2005;8:46–53. doi: 10.1016/j.mib.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Zusman T, Degtyar E, Viner R, Segal G, et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 2003a;5:581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003b;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Chaput C, Boneca IG. Peptidoglycan detection by mammals and flies. Microbes Infect. 2007;9:637–647. doi: 10.1016/j.micinf.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Chaput C, Ecobichon C, Cayet N, Girardin SE, Werts C, et al. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2006;2:e97. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M, Morozova I, Shi S, Sheng H, Chen J, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- Davis KM, Akinbi HT, Standish AJ, Weiser JN. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 2008;4:e1000241. doi: 10.1371/journal.ppat.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Damron DM, Isberg RR, Lander ES, Swanson MS. Lgn1, a gene that determines susceptibility to Legionella pneumophila, maps to mouse chromosome 13. Genomics. 1995;26:443–450. doi: 10.1016/0888-7543(95)80161-e. [DOI] [PubMed] [Google Scholar]
- Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, et al. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, et al. Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003a;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Tournebize R, Mavris M, Page AL, Li X, et al. CARD4/Nod1 mediates NF-κB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003b;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- Grubman A, Kaparakis M, Viala J, Allison C, Badea L, et al. The innate immune molecule, NOD1, regulates direct killing of Helicobacter pylori by antimicrobial peptides. Cell Microbiol. 12:626–639. doi: 10.1111/j.1462-5822.2009.01421.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Yang K, Hashimoto M, Park JH, Kim YG, et al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281:29054–29063. doi: 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- Hawn TR, Smith KD, Aderem A, Skerrett SJ. Myeloid differentiation primary response gene (88)- and toll-like receptor 2-deficient mice are susceptible to infection with aerosolized Legionella pneumophila. J Infect Dis. 2006;193:1693–1702. doi: 10.1086/504525. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV. Lytic transglycosylases. EXS. 1996;75:425–429. doi: 10.1007/978-3-0348-9225-4_21. [DOI] [PubMed] [Google Scholar]
- Holtje JV, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology. 2005;151:3469–3482. doi: 10.1099/mic.0.28326-0. [DOI] [PubMed] [Google Scholar]
- Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, et al. The E-Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2010;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- Kim JG, Lee SJ, Kagnoff MF. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by toll-like receptors. Infect Immun. 2004;72:1487–1495. doi: 10.1128/IAI.72.3.1487-1495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci. 2003;60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci U S A. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L, Magalhaes JG, Selvanantham T, Travassos LH, Geddes K, et al. Role of Nod1 in mucosal dendritic cells during Salmonella pathogenicity island 1-independent Salmonella enterica serovar Typhimurium infection. Infect Immun. 2009;77:4480–4486. doi: 10.1128/IAI.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaree BA, Clarke AJ. Interaction of penicillin-binding protein 2 with soluble lytic transglycosylase B1 in Pseudomonas aeruginosa. J Bacteriol. 2008;190:6922–6926. doi: 10.1128/JB.00934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Conover GM, Isberg RR. Legionella pneumophila EnhC is required for efficient replication in tumor necrosis factor alpha-stimulated macrophages. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Isberg RR. NF-κB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Haenssler E, Moy MY, Isberg RR. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 2010;12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- Magalhaes JG, Sorbara MT, Girardin SE, Philpott DJ. What is new with Nods? Curr Opin Immunol. 23:29–34. doi: 10.1016/j.coi.2010.12.003. [DOI] [PubMed] [Google Scholar]
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, et al. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro G, Fazio LL, Martino MC, Rossi G, Tattoli I, et al. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell Microbiol. 2008;10:682–695. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan) Microbiol Mol Biol Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psylinakis E, Boneca IG, Mavromatis K, Deli A, Hayhurst E, et al. Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J Biol Chem. 2005;280:30856–30863. doi: 10.1074/jbc.M407426200. [DOI] [PubMed] [Google Scholar]
- Rankin S, Li Z, Isberg RR. Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect Immun. 2002;70:3637–3648. doi: 10.1128/IAI.70.7.3637-3648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Segal G, Shuman HA. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun. 1997;65:5057–5066. doi: 10.1128/iai.65.12.5057-5066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, et al. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbak CR, Ryu JH, Leulier F, Pili-Floury S, Parquet C, et al. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Girardin SE, Boneca IG, Lemos R, et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem. 2005;280:36714–36718. doi: 10.1074/jbc.M501649200. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Uehara T, Park JT. Identification of MpaA, an amidase in Escherichia coli that hydrolyzes the gamma-D-glutamyl-meso-diaminopimelate bond in murein peptides. J Bacteriol. 2003;185:679–682. doi: 10.1128/JB.185.2.679-682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01