Subgroup specific structural variation across 1,000 medulloblastoma genomes (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 17.
Published in final edited form as: Nature. 2012 Aug 2;488(7409):49–56. doi: 10.1038/nature11327
Summary
Medulloblastoma, the most common malignant pediatric brain tumour, is currently treated with non-specific cytotoxic therapies including surgery, whole brain radiation, and aggressive chemotherapy. As medulloblastoma exhibits marked intertumoural heterogeneity, with at least four distinct molecular variants, prior attempts to identify targets for therapy have been underpowered due to small samples sizes. Here we report somatic copy number aberrations (SCNAs) in 1087 unique medulloblastomas. SCNAs are common in medulloblastoma, and are predominantly subgroup enriched. The most common region of focal copy number gain is a tandem duplication of the Parkinson’s disease gene SNCAIP, which is exquisitely restricted to Group 4α. Recurrent translocations of PVT1, including PVT1-MYC and PVT1-NDRG1 that arise through chromothripsis are restricted to Group 3. Numerous targetable SCNAs, including recurrent events targeting TGFβ signaling in Group 3, and NF-κB signaling in Group 4 suggest future avenues for rational, targeted therapy.
Brain tumours are the most common cause of childhood oncological death, and medulloblastoma is the most common malignant pediatric brain tumour. Current medulloblastoma therapy including surgical resection, whole brain and spinal cord radiation, and aggressive chemotherapy supplemented by bone marrow transplant yields five-year survival rates of 60–70%1. Survivors are often left with significant neurological, intellectual, and physical disabilities secondary to the effects of these non-specific cytotoxic therapies on the developing brain2.
Recent evidence suggests that medulloblastoma actually comprises multiple molecularly distinct entities whose clinical and genetic differences may require separate therapeutic strategies3–6. Four principal subgroups of medulloblastoma have been identified: WNT, SHH, Group 3, and Group 47, and there is preliminary evidence for clinically significant subdivisions of the subgroups3,7,8. Rational, targeted therapies based on genetics are not currently in use for medulloblastoma, although inhibitors of the Sonic Hedgehog pathway protein Smoothened have shown early promise9. Actionable targets for WNT, Group 3, and Group 4 tumours have not been identified4,10. Sanger sequencing of 22 medulloblastoma exomes revealed on average only 8 SNVs per tumour11. Some SNVs were subgroup restricted (PTCH1, CTNNB1), while others occurred across subgroups (TP53, MLL2). We hypothesized that the observed intertumoural heterogeneity might have underpowered prior attempts to discover targets for rational therapy.
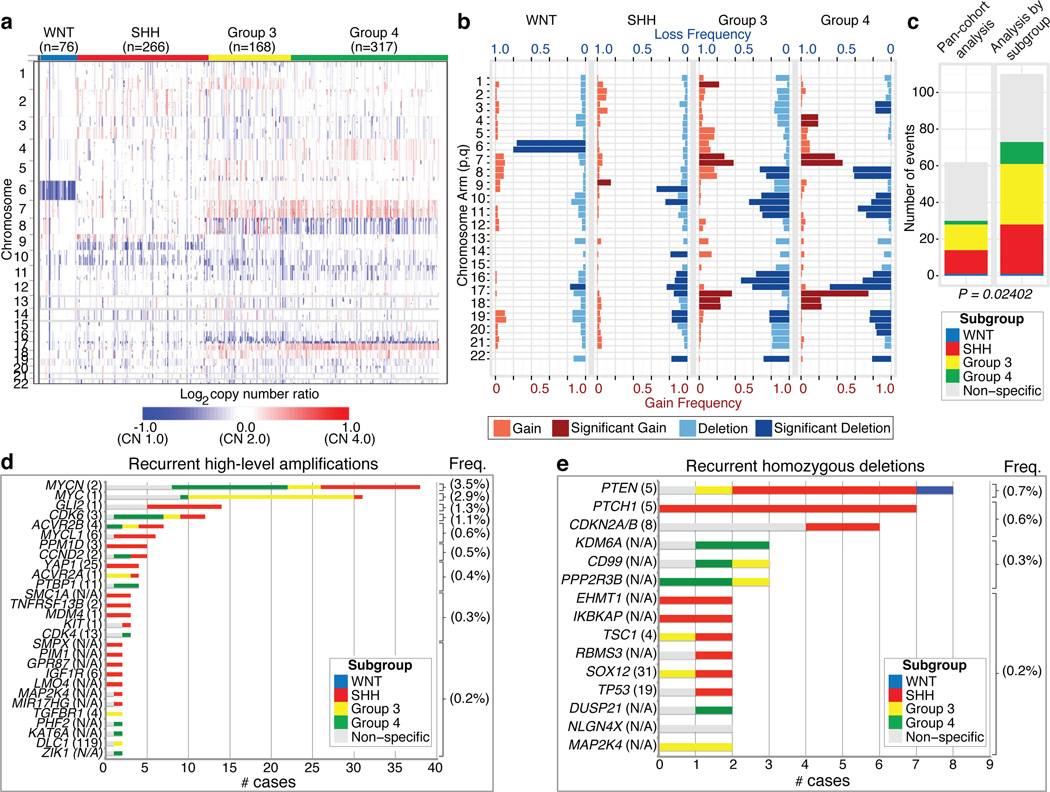
The Medulloblastoma Advanced Genomics International Consortium (MAGIC) consisting of scientists and physicians from 46 cities across the globe gathered >1200 medulloblastomas which were studied by SNP arrays (n=1239; Figure 1a; Supplementary Figure 1; Supplementary Tables 1–3). Medulloblastoma subgroup affiliation of 827 cases was determined using a custom nanoString-based RNA assay (Supplementary Figure 2)12. Disparate patterns of broad cytogenetic gain and loss were observed across the subgroups (Figure 1b; Supplementary Figures 3, 7, 8, 10, 11). Analysis of the entire cohort using GISTIC213 to discover significant ‘driver’ events delineated 62 regions of recurrent SCNA (Figure 1c; Supplementary Figure 4; Supplementary Tables 4–5); analysis by subgroup increased sensitivity such that 110 candidate ‘driver’ SCNAs were identified, most of which are subgroup enriched (Figure 1c–e; Supplementary Table 6).
Figure 1. Genomic heterogeneity of medulloblastoma subgroups.
a, The medulloblastoma genome classified by subgroup. b, Frequency and significance (_q_-value≤ 0.1) of broad cytogenetic events across medulloblastoma subgroups. c, Significant regions of focal SCNA identified by GISTIC2 in either pan-cohort or subgroup-specific analyses. d, e, Recurrent high-level amplifications (d; segmented CN≥5) and homozygous deletions (e; segmented CN≤0.7) in medulloblastoma. The number of genes mapping to the GISTIC2 peak region (where applicable) is listed in brackets after the suspected driver gene, as is the frequency of each event.
Twenty-eight regions of recurrent high-level amplification (copy number ≥5) were identified (Figure 1d; Supplementary Table 7). The most prevalent amplifications affected members of the MYC family with MYCN predominantly amplified in SHH and Group 4, MYC in Group 3, and MYCL1 in SHH medulloblastomas. Multiple genes/regions were exclusively amplified in SHH, including GLI2, MYCL1, PPM1D, YAP1, and MDM4 (Figure 1d). Recurrent homozygous deletions were exceedingly rare, with only 15 detected across 1087 tumours (Figure 1e). Homozygous deletions targeting known tumour suppressors PTEN, PTCH1, and CDKN2A/B were the most common, all enriched in SHH cases (Figure 1e; Supplementary Table 7). Novel homozygous deletions included KDM6A, a histone-lysine demethylase deleted in Group 4. A custom nanoString CodeSet was used to verify 24 significant regions of gain across 192 MAGIC cases, resulting in a verification rate of 90.9% (Supplementary Figure 5). We conclude that SCNAs in medulloblastoma are common, and are predominantly subgroup enriched.
Subgroup-specific SCNAs in medulloblastoma
WNT medulloblastoma genomes are impoverished of recurrent focal regions of SCNA, exhibiting no significant regions of deletion and only a small subset of focal gains found at comparable frequencies in non-WNT tumours (Supplementary Figures 4, 6; Supplementary Table 8). CTNNB1 mutational screening confirmed canonical exon 3 mutations in 63/71 (88.7%) WNT tumours, whereas monosomy 6 was detected in 58/76 (76.3%) (Supplementary Figure 6; Supplementary Table 9). Four WNT tumours (4/71; 5.6%) had neither CTNNB1 mutation nor monosomy 6, but maintained typical WNT expression signatures. Given the size of our cohort and the resolution of the platform, we conclude that there are no frequent, targetable SCNAs for WNT medulloblastoma.
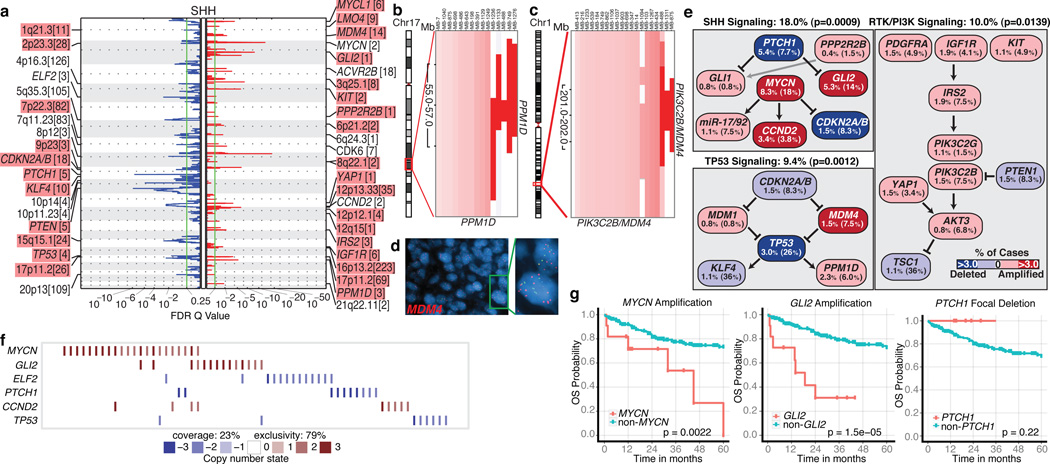
SHH tumours exhibit multiple significant focal SCNAs (Figure 2a; Supplementary Figures 12, 15, 16; Supplementary Tables 10–11). SHH enriched/restricted SCNAs included amplification of GLI2 and deletion of PTCH1 (Figure 2a, e, f)10. MYCN and CCND2 were among the most frequently amplified genes in SHH (Supplementary Table 6), but were also altered in non-SHH cases. Genes up-regulated in SHH tumours (i.e. SHH signature genes) are significantly over-represented among the genes focally amplified in SHH tumours (P=0.001–0.02, permutation tests; Supplementary Figure 9). Recurrent amplification of SHH signature genes has clinical implications, as amplification of downstream transcriptional targets could mediate resistance to upstream SHH pathway inhibitors14.
Figure 2. Genomic alterations affect core signaling pathways in SHH medulloblastoma.
a, GISTIC2 significance plot of amplifications (red) and deletions (blue) observed in SHH. The number of genes mapping to each significant region are included in brackets and regions enriched in SHH are shaded red. b, c, Recurrent amplifications of PPM1D (b) and PIK3C2B/MDM4 (c) are restricted to SHH. d, FISH validation of MDM4 amplification. e, SHH signaling, TP53 signaling, and RTK/PI3K signaling represent the core pathways genomically targeted in SHH. P-values indicate the prevalence with which the respective pathway is targeted in SHH vs. non-SHH cases (Fisher’s exact test). Frequencies of focal and broad (parentheses) SCNAs are listed. f, Mutual exclusivity analysis of focal SCNAs in SHH. g, Clinical implications of SCNAs affecting MYCN, GLI2, or PTCH1 in SHH (log-rank tests).
Novel, SHH-enriched SCNAs included components of TP53 signaling, including amplifications of MDM4 and PPM1D, and focal deletions of TP53 (Figure 2a–e). Targetable events, including amplifications of IGF signaling genes IGF1R and IRS2, PI3K genes PIK3C2G and PIK3C2B, and deletion of PTEN were restricted to SHH tumours (Figure 2a, c, e). Importantly, focal events affecting genes in the SHH pathway were largely mutually exclusive and prognostically significant (Figure 2f, g). Many of the recurrent, targetable SCNAs identified in SHH medulloblastoma (IGF1R, KIT, MDM4, PDGFRA, PIK3C2G, PIK2C2B, and PTEN) have already been targeted with small molecules for treatment of other malignancies, which might allow rapid translation for targeted therapy of subsets of SHH patients (Supplementary Table 16). Novel SHH targets identified here are excellent candidates for combinatorial therapy with Smoothened inhibitors, in order to avoid the resistance encountered in both humans and mice9,14,15.
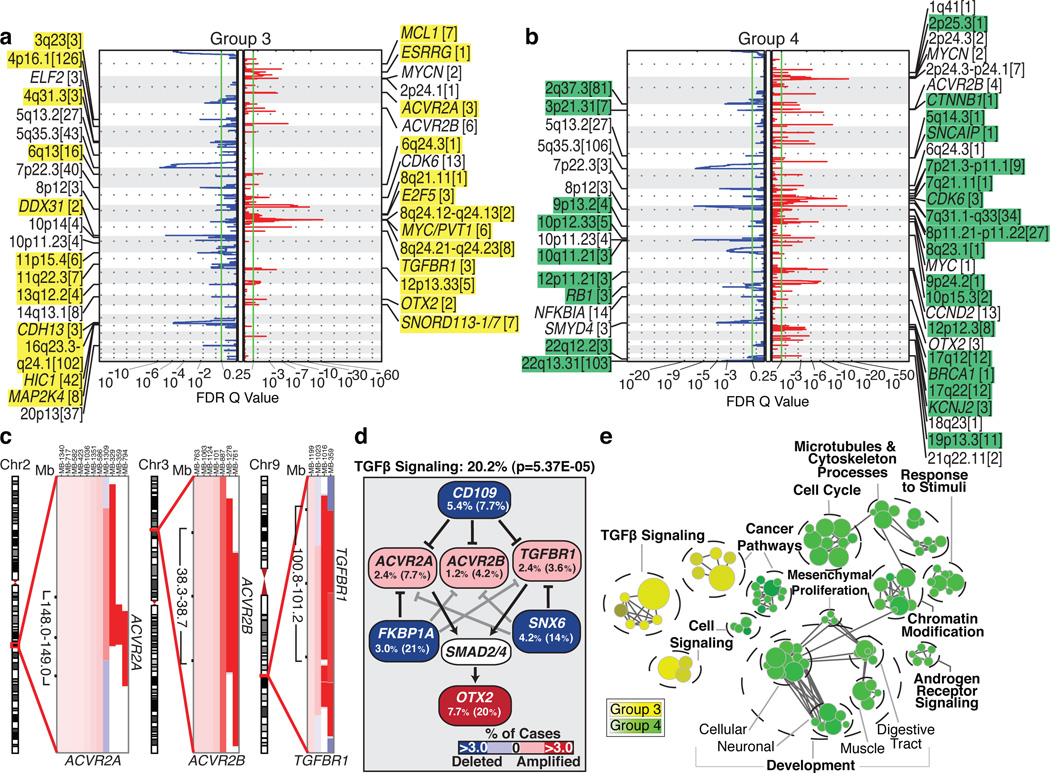
Group 3 and Group 4 medulloblastomas have generic names as comparatively little is known about their genetic basis, and no targets for rational therapy have been identified7. MYC amplicons are largely restricted to Group 3, while MYCN amplicons are seen in Group 4 and SHH tumours (Figure 1d)3,4. Indeed, MYC and MYCN loci comprise the most significant regions of amplification observed in Group 3 and Group 4, respectively (Figure 3a, b; Supplementary Figures 13, 14, 17–20; Supplementary Tables 12–15). Group 3 MYC amplicons were mutually exclusive from those affecting the known medulloblastoma oncogene OTX216 and were highly prognostic (Supplementary Figure 21)3,16. Type II activin receptors, ACVR2A and ACVR2B and family member TGFBR1 are highly amplified in Group 3 tumours, suggesting deregulation of TGFβ signaling as a driver event in Group 3 (Figure 3c–e; Supplementary Figure 22). The Group 3-enriched medulloblastoma oncogene OTX2 is a prominent target of TGFβ signaling in the developing nervous system17 and TGFβ pathway inhibitors, CD10918, FKBP1A19,20, and SNX620 are recurrently deleted in Group 3 (Figure 3a, d). SCNAs in TGFβ pathway genes were heavily enriched in Group 3 (P=5.37E-05, Fisher’s exact test) and found in at least 20.2% of cases, suggesting that TGFβ signaling represents the first rational target for this poor prognosis subgroup (Figure 3d). Similarly, novel deletions affecting regulators of the NF-κB pathway, including NFKBIA21 and USP422 were identified in Group 4 (Supplementary Figure 23), proposing that NF-κB signaling may represent a rational Group 4 therapeutic target.
Figure 3. The genomic landscape of Group 3 and Group 4 medulloblastoma.
a, b, GISTIC2 plots depicting significant SCNAs in Group 3 (a) and Group 4 (b) with subgroup-enriched regions shaded in yellow and green, respectively. c, Recurrent amplifications targeting type II (ACVR2A and ACVR2B) and type I (TGFBR1) activin receptors in Group 3. d, Recurrent SCNAs affecting the TGFβ pathway in Group 3 (P=5.73E-05, Fisher’s exact test). Frequencies of focal and broad (parentheses) SCNAs are listed. e, Enrichment plot of gene sets affected by SCNAs in Group 3 vs. Group 4.
Network analysis of Group 3 and Group 4 SCNAs illustrates the different pathways over-represented in each subgroup. Only TGFβ signaling is unique to Group 3 (Figure 3e). In contrast, cell cycle control, chromatin modification, and neuronal development are all Group 4-enriched. Cumulatively, the dismal prognosis of Group 3 patients, the lack of published targets for rationale therapy, and the prior targeting of TGFβ signaling in other diseases suggest that TGFβ may represent an appealing target for Group 3 rational therapies (Supplementary Table 16).
SNCAIP tandem duplication is common in Group 4
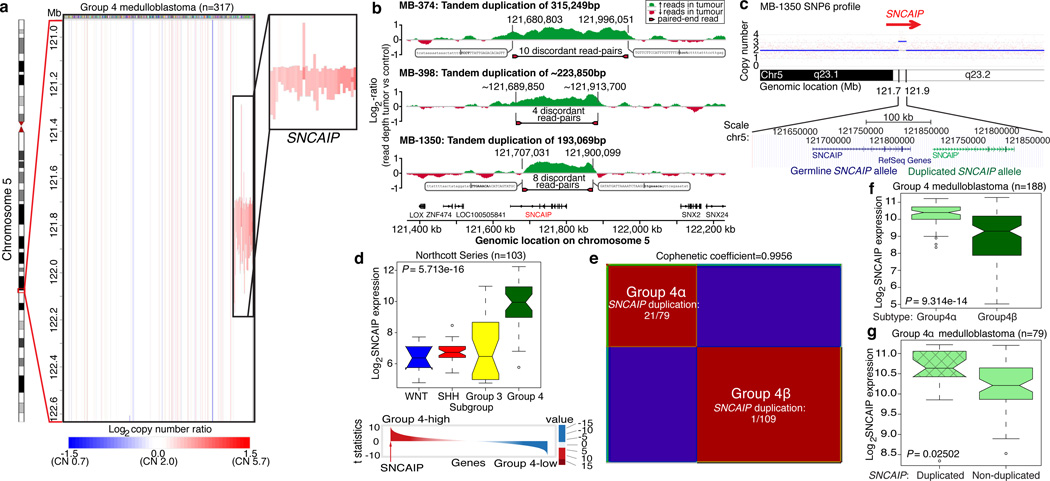
Although Group 4 is the most prevalent medulloblastoma subgroup its pathogenesis remains poorly understood. The most frequent SCNA observed in Group 4 (33/317; 10.4%) is a recurrent region of single copy gain on chr5q23.2 targeting a single gene – SNCAIP (synuclein, alpha interacting protein) (Figure 4a; Supplementary Figure 24). SNCAIP, encodes SYNPHILIN-1, which binds to α-SYNUCLEIN to promote the formation of Lewy bodies in the brains of patients with Parkinson’s disease23,24. Additionally, rare germline mutations of SNCAIP have been described in Parkinson’s families25. Large insert, mate-pair, whole genome sequencing (WGS) demonstrates that SNCAIP copy number gains arise from tandem duplication of a truncated SNCAIP (lacking non-coding exon 1), inserted telomeric to the germline SNCAIP allele (Figure 4b, c; Supplementary Figure 25). SNP6 profiling of patient-matched germline material confirmed that SNCAIP duplications are somatic (Supplementary Figure 26) and subsequent whole transcriptome sequencing (RNASeq) of select Group 4 cases (n=5) verified that SNCAIP is the only gene expressed in the duplicated region (Supplementary Figure 27). Analysis of published copy number profiles for 3131 primary tumours26 and 947 cancer cell lines27 (total of 4078 cases) revealed only four cases with apparent duplication of SNCAIP, all of which were inferred as Group 4 medulloblastomas (data not shown). We conclude that SNCAIP duplication is a somatic event highly specific to Group 4 medulloblastoma.
Figure 4. Tandem duplication of SNCAIP defines a novel subtype of Group 4.
a, Highly recurrent, focal, single copy gain of SNCAIP in Group 4. b, Paired-end mapping verifies recurrent tandem duplication of SNCAIP in Group 4. c, Schematic representation of SNCAIP tandem duplication. d, SNCAIP is a Group 4 signature gene. Upper panel. SNCAIP expression across subgroups in a published series of 103 primary medulloblastomas. Lower panel. SNCAIP ranks among the top 1% (rank=39/16,758) of highly expressed genes in Group 4. e, NMF consensus clustering of 188 expression-profiled Group 4s supports two transcriptionally distinct subtypes designated 4α and 4β (Cophenetic coefficient=0.9956). 21/22 SNCAIP duplicated cases belong to Group 4α (P=3.12E-08, Fisher’s exact test). f, SNCAIP expression is significantly elevated in Group 4α vs. 4β (P=9.31E-14, Mann-Whitney test). g, Group 4α cases harboring SNCAIP duplication exhibit a ~1.5-fold increase in SNCAIP expression.
Re-analysis of 499 published medulloblastoma expression profiles confirmed that SNCAIP is one of the most highly up-regulated Group 4 signature genes (Figure 4d; Supplementary Figure 28). Profiling of 188 Group 4 tumours on expression microarrays followed by consensus non-negative matrix factorization (NMF) clustering delineates two subtypes of Group 4 (4α and 4β; Figure 4e; Supplementary Figure 29). Strikingly, 21/22 SNCAIP duplicated cases belonged to Group 4α (P=3.12E-08, Fisher’s exact test). SNCAIP is more highly expressed in Group 4α than 4β (Figure 4f), and 4α samples with tandem duplication showed ~1.5-fold increased expression, consistent with gene dosage (Figure 4g; Supplementary Figures 35, 36). Group 4α exhibits a relatively balanced genome compared to 4β (Supplementary Figures 30–32), and several 4α cases harbour SNCAIP duplication in conjunction with i17q and no other SCNAs (Supplementary Figure 33). Importantly, SNCAIP duplications are mutually exclusive from other prominent SCNAs in Group 4, including MYCN and CDK6 amplifications (Supplementary Figure 34).
PVT1 fusions arise via chromothripsis in Group 3
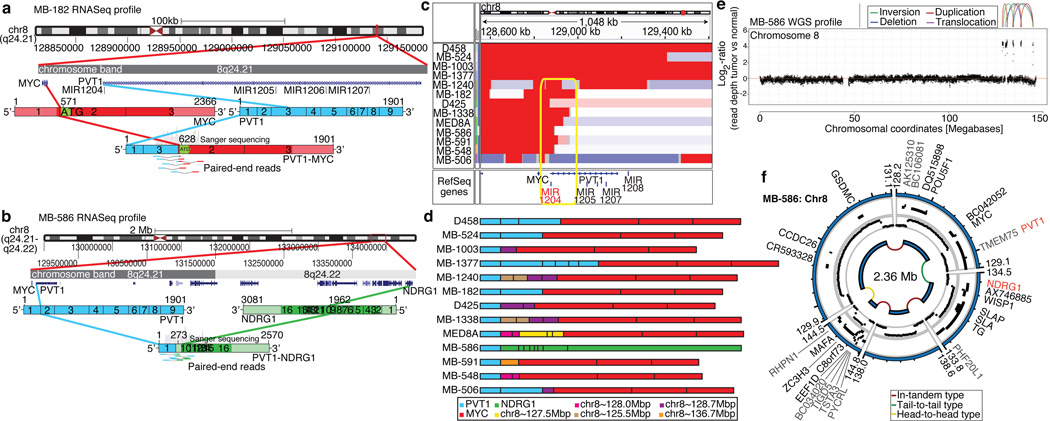
Although recurrent gene fusions have recently been discovered in solid tumours, none have been reported in medulloblastoma. RNASeq of Group 3 tumours (n=13) identified two independent gene fusions in two different tumours (MB-182 and MB-586, both involving the 5’ end of PVT1 - a non-coding gene frequently co-amplified with MYC in Group 3 (Figure 5a, b; Supplementary Figure 37; Supplementary Tables 17–18). Sanger sequencing confirmed a fusion transcript consisting of exons 1 and 3 of PVT1 fused to the coding sequence of MYC (exons 2 & 3) in MB-182, and a fusion involving PVT1 exon 1 fused to the 3’ end of NDRG1 in MB-586 (Figure 5a, b).
Figure 5. Identification of frequent PVT1-MYC fusion genes in Group 3.
a, b, RNASeq identifies multiple fusion transcripts driven by PVT1 in Group 3. Schematics depict the structures of verified PVT1-MYC (b) and PVT1-NDRG1 (c) fusion genes. c, Heatmap of the MYC/PVT1 locus showing a subset of 13 _MYC_-amplified Group 3 cases subsequently verified to exhibit PVT1 gene fusions (shown in d). Yellow box highlights the common breakpoint affecting the first exon/intron of PVT1, including miR-1204. d, Summary of PVT1 fusion transcripts identified in Group 3. e, f, WGS confirms complex patterns of rearrangement on chr8q24 in PVT1 fusion(+) Group 3.
Group 3 copy number data at the MYC/PVT1 locus suggested that additional samples might harbour PVT1 gene fusions (Figure 5c). RT-PCR profiling of select Group 3 cases confirmed PVT1-MYC fusions in at least 60% (12/20) _MYC_-amplified cases (Figure 5d; Supplementary Table 19). Fusion transcripts included many other portions of chr8q, with up to four different genomic loci mapping to a single transcript, a pattern reminiscent of chromothripsis28,29 (Figure 5d). WGS performed on four _MYC_-amplified Group 3s harbouring PVT1 fusion transcripts identified a series of complex genomic rearrangements on chr8q (Figure 5e, f; Supplementary Figure 38; Supplementary Tables 20–21). Chromosome 8 copy number profile for MB-586 (PVT1-NDRG1) derived from WGS showed that PVT1 and NDRG1 are structurally linked as predicted by RNASeq, and several adjacent regions of 8q24 were extensively rearranged (Figure 5e, f; Supplementary Table 21). Monte Carlo simulation suggests that this fragmented 8q amplicon arose through chromothripsis, a process of erroneous DNA repair following a single catastrophic event in which a chromosome is shattered into many pieces (Supplementary Figure 39). Further examination of our copy number dataset revealed rare examples of chromothripsis across subgroups (Supplementary Figure 40), with only chr8 in Group 3 demonstrating statistically significant, region-specific chromothripsis (q=0.0004, FDR-corrected Fisher’s exact test). Among Group 3 tumours, the occurrence of chr8q chromothripsis is correlated with deletion of chr17p (location of TP53; data not shown), in keeping with the association of loss of TP53 and chromothripsis recently described in medulloblastoma (P=0.0199, Fisher’s exact test)28. While the PVT1 locus has been suggested to be a genomically fragile site, we observe that the majority of _MYC_-amplified Group 3 tumours harbour PVT1 fusions that arise through a process consistent with chromothripsis.
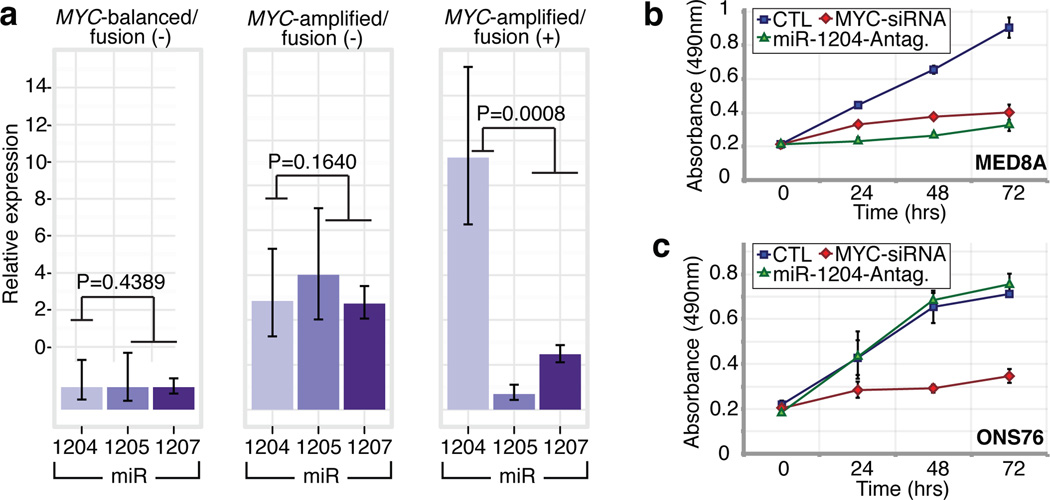
PVT1 is a non-coding host gene for 4 miRNAs – miR-1204–1207. Previous studies have implicated miR-1204 as a candidate oncogene that enhances oncogenesis in combination with MYC30,31. PVT1 fusions identified in this study involve only PVT1 exon 1 and miR-1204. Importantly, miR-1204, but not the adjacent miR-1205 and miR-1206, is expressed at a higher level in PVT1-MYC fusion(+) Group 3 tumours compared to fusion(−) cases (P=0.0008, Mann-Whitney test; Figure 6a). To evaluate whether aberrant expression of miR-1204 contributes to the malignant phenotype, we inhibited miR-1204 in MED8A cells, a Group 3 medulloblastoma cell line with a confirmed PVT1-MYC fusion (Figure 5d). Antagomir-mediated RNAi of miR-1204 had a pronounced effect on MED8A growth (Figure 6b). A comparable reduction in proliferative capacity was achieved with knockdown of MYC. Conversely, the medulloblastoma cell line ONS76 exhibits neither MYC amplification, nor a detectable PVT1-MYC fusion gene and knockdown of miR-1204 had no effect in this line (Figure 6c).
Figure 6. Functional synergy between miR-1204 and MYC secondary to PVT1-MYC fusion.
a, qRT-PCR of PVT1-encoded microRNAs confirms up-regulation of miR-1204 in PVT1-MYC fusion(+) Group 3s: _MYC_-balanced/fusion(−), n=4; _MYC_-amplified/fusion(−), n=6; _MYC_-amplified/fusion(+), n=8. Error bars represent standard error of the mean (SEM) and reflect variability among samples. b, c Knockdown of miR-1204 attenuates the proliferative capacity of PVT1-MYC fusion(+) MED8A medulloblastoma cells (b) but has no effect on fusion(−) ONS76 cells (c). Error bars represent the standard deviation (SD) of triplicate experiments.
PVT1 has been reported previously in fusion transcripts with a number of partners30,32,33. The most prevalent form of the PVT1-MYC fusion in Group 3 tumours lacks the first, non-coding exon of MYC, similar to forms of MYC that have been described in Burkitt’s lymphoma34 (Figure 5a, d). The PVT1 promoter contains two non-canonical E-boxes and can be activated by MYC31. This suggests a positive feedback model where MYC can reinforce its own expression from the PVT1 promoter in PVT1-MYC fusion(+) tumours. Indeed, knockdown of MYC alone in MED8A cells resulted in diminished expression of both MYC and miR-1204, suggesting MYC may positively regulate PVT1 (i.e. miR-1204) expression in medulloblastoma cells (Supplementary Figure 41).
Discussion
Medulloblastomas have few SNVs as compared to many adult epithelial malignancies11, while SCNAs appear to be quite common. Medulloblastoma is a heterogeneous disease7, there-by requiring large cohorts to detect subgroup specific events. Through the accumulation of >1200 medulloblastomas in MAGIC, we have identified novel and significant SCNAs. Many of the significant SCNAs are subgroup restricted, highly supporting their role as driver events in their respective subgroups.
Expression of SYNPHILIN-1 in neuronal cells results in decreased cell doubling time35, decreased caspase-3 activation36, decreased TP53 transcriptional activity and mRNA levels, and decreased apoptosis37. SYNPHILIN-1 is ubiquitinated by PARKIN, which is encoded by the hereditary Parkinson’s disease gene PARK224, a candidate tumour suppressor gene38. While patients with Parkinson’s disease have an overall decreased risk of cancer, they may have an increased incidence of brain tumours39,40. As tandem duplications of SNCAIP are highly recurrent, stereotypical, subgroup restricted, affect only a single gene, and as _SNCAIP_-duplicated tumours have few if any other SCNAs, SNCAIP is a probable driver gene, and merits investigation as a target for therapy of Group 4α. Similarly, PVT1 fusion genes are highly recurrent, restricted to Group 3, arise through a chromothripsis-like process, and are the first recurrent translocation reported in medulloblastoma.
We identify a number of highly targetable, recurrent, subgroup-specific SCNAs that could form the basis for future clinical trials (i.e. PI3K signaling in SHH, TGFβ signaling in Group 3, and NF-κB signaling in Group 4). Activation of these pathways through alternative, currently unknown genetic and epigenetic events could increase the percentage of patients amenable to targeted therapy. We also identify a number of highly ‘druggable’ events that occur in a minority of cases. The co-operative, global approach of the MAGIC consortium has allowed us to overcome the barrier of intertumoural heterogeneity in an uncommon pediatric tumour, and to identify the relevant and targetable SCNAs for the affected children.
Methods Summary
All patient samples were obtained with consent as outlined by individual institutional review boards. Genomic DNA was prepared, processed, and hybridized to Affymetrix SNP6 arrays according to manufacturer’s instructions. Raw copy number estimates were obtained in dChip, followed by CBS segmentation in R. SCNAs were identified using GISTIC213. Driver genes within SCNAs were inferred by integrating matched expressions, literature evidence, and other datasets. Pathway enrichment of SCNAs was analyzed with g:Profiler and visualized in Cytoscape by enrichment mapping. FISH was performed as described previously8,10. Medulloblastoma subgroup was assigned using a custom nanoString CodeSet as described12. Tandem duplication of SNCAIP was confirmed by paired-end mapping as previously reported28. RNA was extracted, processed and hybridized to Affymetrix Gene 1.1 ST Arrays as recommended by the manufacturer. Consensus NMF clustering was performed in GenePattern. Gene fusions were identified from RNASeq data using Trans-ABySS. Medulloblastoma cell lines were maintained as described10. Proliferation assays were performed with the Promega CellTiter 96 Assay. Additional methods are detailed in full in Supplementary Methods available online at Nature.com.
Supplementary Material
Supplementary Information
Acknowledgements
MDT is the recipient of a CIHR Clinician-Scientist Phase II award, and was formerly a Sontag Distinguished Scholar with funds from the Sontag Foundation. Funding is acknowledged from the Pediatric Brain Tumour Foundation (MDT and JTR), and the National Institutes of Health (CA159859 to MDT, RWR, and BW), Genome Canada, Genome BC, Terry Fox Research Institute, Ontario Institute for Cancer Research, Pediatric Oncology Group Ontario, Funds from ‘The Family of Kathleen Lorette’ and the Clark H. Smith Brain Tumour Centre, Montreal Children’s Hospital Foundation, Hospital for Sick Children: Sonia and Arthur Labatt Brain Tumour Research Centre, Chief of Research Fund, Cancer Genetics Program, Garron Family Cancer Centre, B.R.A.I.N. Child, CIHR (grant # ATE-110814); the University of Toronto McLaughlin Centre, CIHR Institute of Cancer Research (grant # AT1 – 112286) and C17 through the Advancing Technology Innovation through Discovery competition (Project Title: The Canadian Pediatric Cancer Genome Consortium: Translating next-generation sequencing technologies into improved therapies for high-risk childhood cancer). Canada's Michael Smith Genome Sciences Centre is supported by the BC Cancer Foundation. JR is supported by The Children's Discovery Institute. PAN was supported by a Restracomp Fellowship (Hospital for Sick Children) and is currently a Roman-Herzog Postdoctoral Fellow (Hertie Foundation). Salary support for LG was provided by the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. EVGM is supported by NIH grants CA86335, CA116804, CA138292, NCI contracts 28XS100 and 29XS193, the Southeastern Brain Tumour Foundation, and the Brain Tumour Foundation for Children. This study includes samples provided by the UK Children’s Cancer and Leukaemia Group (CCLG) as part of CCLG-approved biological study BS-2007-04. JK and SP were supported by a grant from the German Cancer Aid (109252). We thank Dr. Chao Lu, Kozue Otaka, and The Centre for Applied Genomics for excellent technical assistance. We thank Narra S Devi and Zhaobin Zhang for excellent technical assistance. We thank Dominik Stoll for expert project management, Susan Archer for technical writing, and Paul Paroutis for artwork. The MAGIC project is part of the International Cancer Genome Consortium.
Footnotes
Author Contributions
PAN and MDT co-conceived the study. PAN, MAM, and MDT led the study. PAN planned and executed experiments and analyses, supervised data acquisition, performed bioinformatic analyses, and extracted nucleic acids for the MAGIC cohort. DJHS led the bioinformatics and performed analyses. JP performed quantitative RT-PCR and Sanger sequencing of PVT1 fusions, expression profiled PVT1-encoded miRNAs, and generated schematics for PVT1 fusion genes. LG performed the MYC and miR-1204 knockdown experiments. SM supervised the RNASeq and WGS experiments and performed data analysis. TZ, AMS, and JOK performed the large insert paired-end sequencing and PCR verification of SNCAIP duplication samples. AK performed interphase FISH and IHC for candidate genes. JR and GBD led the pathway analyses and generated enrichment plots. SES and RB provided technical support with the GISTIC2 bioinformatic platform. DWE performed interphase FISH for candidate genes. CRW, ACL, and SWS performed the SNP6 genotyping analysis, provided a database of normal copy number variants, and the control dataset used to infer copy number in the tumour samples. SM, AD, FC, MK, DTWJ, and HW performed bioinformatic analyses and provided technical advice. YY sequenced CTNNB1 in the WNT tumours. VR, DK, MFR, TA, and PD performed functional assays for candidate genes. BL extracted nucleic acids, managed biobanking, and maintained the patient database. SM and AR performed the drug database analysis. Xin W, Xiaochong W, and MR provided technical support. RYBC, AC, EC, RDC, GRH, SDJ, YL, AL, KLM, KMN, JQQ, AGJR, NT, RJV, IB, RAM, AJM, RH, and SJMJ led the RNASeq and WGS experiments and performed data analyses. AFL and AMK provided the database of Shh-responsive genes. RJWR, WAG, MPP, CCH, OD, SSR, FFD, SSPF, BKC, SKK, KCW, WS, CGE, MFM, AJ, IFP, XF, KMM, GYG, CDR, LM, EMCM, NKN, PJF, JMK, JMO, RGE, KZ, LK, RCT, MKC, BL, REM, DDB, AF, SA, NJ, JCL, SB, NG, WAW, LB, AK, TEVM, TK, TT, SKE, JRL, JBR, LML, EGVM, MF, HN, GC, MG, PH, AGS, AI, SJ, CGC, RV, YSR, SR, MZ, CCF, JAC, MLL, YJC, UT, CEH, EB, SCC, and SMP provided the patient samples and clinical details that made the study possible. PHBS, MM, SLP, YJC, UT, CEH, EB, SWS, JTR, DM, SCC, SJMJ, JOK, SMP, and MAM provided valuable input regarding study design, data analysis, and interpretation of results. PAN, DJHS, JP, LG, SM, and MDT wrote the manuscript. MAM and MDT provided financial and technical infrastructure and oversaw the study. MAM and MDT are joint senior authors and project co-leaders.
Author information
SNP6 copy number and gene expression array data have been deposited at the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) as a GEO SuperSeries under accession number GSE37385. Whole genome and transcriptome sequencing data have been deposited at the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/) hosted by the EBI, under accession number EGAD00001000158.
The authors declare no competing financial interests.
References
- 1.Gajjar A, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 2.Mabbott DJ, et al. Serial evaluation of academic and behavioral outcome after treatment with cranial radiation in childhood. J Clin Oncol. 2005;23:2256–2263. doi: 10.1200/JCO.2005.01.158. [DOI] [PubMed] [Google Scholar]
- 3.Cho YJ, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remke M, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. 2011;29:3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 6.Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012 doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MD, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta neuropathologica. 2011 doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Northcott PA, et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 2011;122:231–240. doi: 10.1007/s00401-011-0846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudin CM, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. The New England journal of medicine. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Northcott PA, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buonamici S, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Science translational medicine. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson DC, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia S, Wu D, Xing C, Meng A. Smad2/3 activities are required for induction and patterning of the neuroectoderm in zebrafish. Developmental biology. 2009;333:273–284. doi: 10.1016/j.ydbio.2009.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Bizet AA, et al. The TGF-beta co-receptor, CD109, promotes internalization and degradation of TGF-beta receptors. Biochimica et biophysica acta. 2011;1813:742–753. doi: 10.1016/j.bbamcr.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 19.Wang T, Donahoe PK, Zervos AS. Specific interaction of type I receptors of the TGF-beta family with the immunophilin FKBP-12. Science. 1994;265:674–676. doi: 10.1126/science.7518616. [DOI] [PubMed] [Google Scholar]
- 20.Parks WT, et al. Sorting nexin 6, a novel SNX, interacts with the transforming growth factor-beta family of receptor serine-threonine kinases. The Journal of biological chemistry. 2001;276:19332–19339. doi: 10.1074/jbc.M100606200. [DOI] [PubMed] [Google Scholar]
- 21.Bredel M, et al. NFKBIA deletion in glioblastomas. The New England journal of medicine. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao N, et al. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFalpha-induced cancer cell migration. Biochem J. 2012;441:979–986. doi: 10.1042/BJ20111358. [DOI] [PubMed] [Google Scholar]
- 23.Engelender S, et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nature genetics. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- 24.Chung KK, et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nature medicine. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 25.Marx FP, et al. Identification and functional characterization of a novel R621C mutation in the synphilin-1 gene in Parkinson's disease. Human molecular genetics. 2003;12:1223–1231. doi: 10.1093/hmg/ddg134. [DOI] [PubMed] [Google Scholar]
- 26.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shtivelman E, Bishop JM. The PVT gene frequently amplifies with MYC in tumor cells. Molecular and cellular biology. 1989;9:1148–1154. doi: 10.1128/mcb.9.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carramusa L, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. Journal of cellular physiology. 2007;213:511–518. doi: 10.1002/jcp.21133. [DOI] [PubMed] [Google Scholar]
- 32.Shtivelman E, Bishop JM. Effects of translocations on transcription from PVT. Molecular and cellular biology. 1990;10:1835–1839. doi: 10.1128/mcb.10.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 35.Li X, et al. Synphilin-1 exhibits trophic and protective effects against Rotenone toxicity. Neuroscience. 2010;165:455–462. doi: 10.1016/j.neuroscience.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith WW, et al. Synphilin-1 attenuates neuronal degeneration in the A53T alpha-synuclein transgenic mouse model. Human molecular genetics. 2010;19:2087–2098. doi: 10.1093/hmg/ddq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giaime E, et al. Caspase-3-derived C-terminal product of synphilin-1 displays antiapoptotic function via modulation of the p53-dependent cell death pathway. The Journal of biological chemistry. 2006;281:11515–11522. doi: 10.1074/jbc.M508619200. [DOI] [PubMed] [Google Scholar]
- 38.Veeriah S, et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nature genetics. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen JH, et al. Atypical cancer pattern in patients with Parkinson's disease. British journal of cancer. 2005;92:201–205. doi: 10.1038/sj.bjc.6602279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moller H, Mellemkjaer L, McLaughlin JK, Olsen JH. Occurrence of different cancers in patients with Parkinson's disease. Bmj. 1995;310:1500–1501. doi: 10.1136/bmj.310.6993.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information