Higher-Order Assemblies in a New Paradigm of Signal Transduction (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 20.
Abstract
Recent studies have revealed that multiple intracellular signaling proteins may assemble into structured, yet sometimes infinite, higher-order signaling machines for transmission of receptor activation information to cellular responses. These studies advance our understanding of cell signaling and implicate new molecular mechanisms in proximity-driven enzyme activation, threshold behavior, signal amplification, reduction of biological noise, and temporal and spatial control of signal transduction.
A Classical Concept of Signal Transduction
We have come a long way since Paul Ehrlich proposed more than 100 years ago the concept of receptors as cellular communicators in the context of toxin action. Receptors are sensing components in signal transduction pathways; they may reside on the cell surface to recognize extracellular ligands or exist inside the cell to interact with phagocytosed or cell-permeable signals. By triggering intracellular events in response to environmental changes, receptor signal transduction influences nearly every physiological reaction in multicellular organisms. We have learned much about the mechanism of signal transduction in different receptor systems. These studies led to a general view of signal transduction as a chain reaction in which ligand binding sequentially induces conformational changes to receptors, activation of enzymes and second messengers, and generation of transcriptional and nontranscriptional effects to complete signal transmission and amplification (Figure 1A).
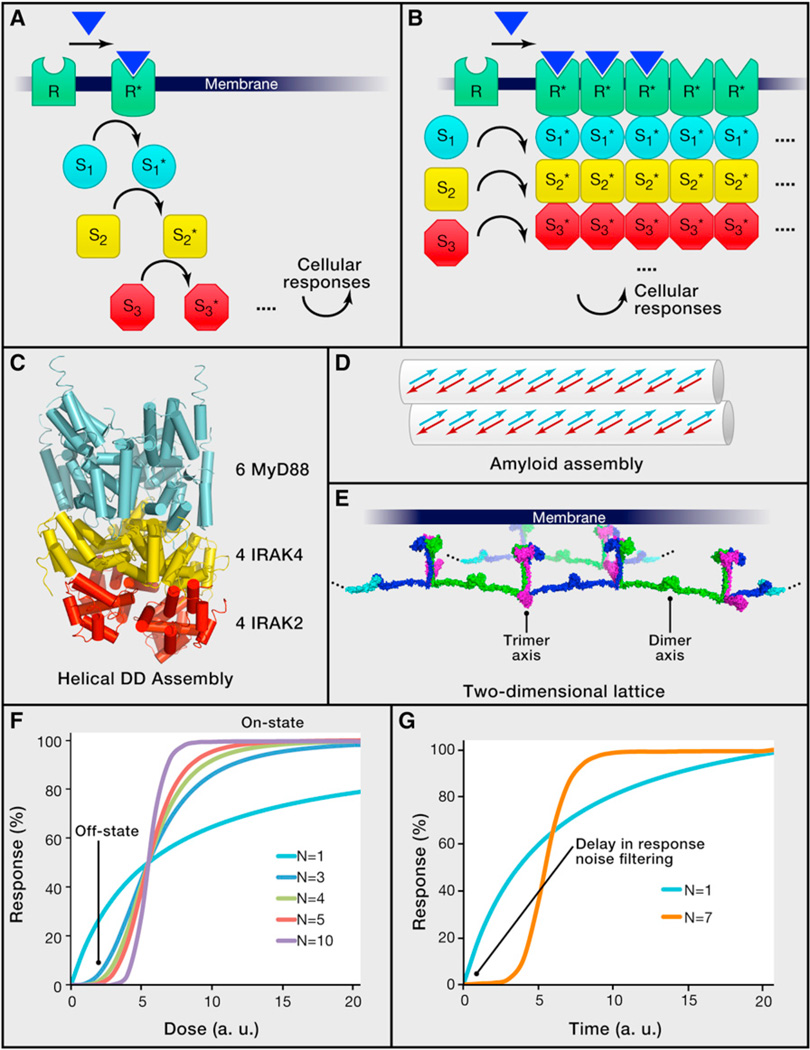
Figure 1. Classical and Higher-Order Assembly Modes of Signal Transduction.
(A) A classical view of receptor signal transduction involving successive steps of activation of intracellular signaling proteins (denoted S1 to S3 as examples). Asterisks are used to indicate activated forms of receptors and signaling proteins.
(B) A view of receptor signal transduction involving formation of higher-order oligomeric signalosomes between receptors and signaling proteins, with implications in proximity-induced enzyme activation, threshold behavior, signal amplification, and spatial regulation. Two potential amplification mechanisms are illustrated respectively by incorporation of unliganded receptors and recruitment of overstoichiometric number of signaling proteins (dotted lines) into the signalosome. It should be noted that, although the extracellular domains of many such receptors form defined oligomers, extensive evidence suggests higher-order lateral clustering of full-length receptors.
(C) Crystal structure of a higher-order signalosome assembly, showing the formation using helical symmetry of a 14 subunit complex containing the DD of six MyD88 (an adaptor protein, in cyan), four IRAK4 (a kinase, in yellow), and four IRAK2 (a kinase, in red) molecules (Lin et al., 2010). Similar helical symmetries may mediate the formation of infinite filamentous structures.
(D) Schematic diagram of an amyloid assembly in signaling (Li et al., 2012a). Arrows indicate βstrands.
(E) A proposed two-dimensional lattice structure of TRAF6 formed by alternating dimerization and trimerization (Yin et al., 2009).
(F) Simulated response curves as a function of dose of stimulation in arbitrary units (a.u.) for differentially cooperative processes. N: Hill coefficients. The off-state and on-state locations of highly cooperative processes are indicated.
(G) Simulated response curves as a function of time in arbitrary units (a.u.). N: Hill coefficients. A delay in response for a highly cooperative process may act as a noise filter in response.
Some well-studied examples illustrate these basic concepts. For G-protein-coupled receptors (GPCRs) such as the β-adrenergic receptor, ligand binding induces a conformational change that leads to recruitment of heterotrimeric G-proteins and promotion of the exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP) (Granier and Kobilka, 2012). The GTP-bound α subunits of G proteins (Gα) dissociate from the β and γ subunits (Gβγ), each effecting target proteins for production of second messengers such as cAMP and for regulation of channel activities, respectively. For receptor tyrosine kinases (RTKs) such as the epidermal growth factor receptor (EGFR), ligand binding at the extracellular domain induces formation of signalingcompetent receptor dimers to allow one kinase domain (activator) to allosterically activate the other kinase domain (acceptor) (Endres et al., 2011). Autophosphorylation of tyrosine residues in the C-terminal tail of EGFR leads to its association with phosphotyrosine-binding domain-containing proteins, eliciting MAP kinase and AKT pathways to promote cell proliferation. In both GPCR and RTK signaling pathways, the strength of the signal transduction may be graded and determined by the lifetime of activated effectors and counter-enzyme-mediated deactivation kinetics.
Higher-Order Assemblies as Intracellular Signalosomes
Our initial hypothesis paints a similar picture of signal transduction in innate immune receptors such as those in the tumor necrosis factor (TNF) receptor (TNFR) superfamily and the Toll-like receptor/ interleukin-1 receptor (TLR/IL-1R) superfamily. Ligand-induced conformational changes through formation of proper receptor trimers or dimers appear to be key events for the transmission of receptor activation signals across the membrane. Unlike GPCRs and RTKs, receptors in the TNFR and TLR/IL-1R superfamilies do not contain enzymatic activities or directly couple to intracellular enzymes. They use adaptor proteins to connect to enzymatic activation in the pathways, culminating in alterations of cell fates through nuclear factor κB (NF-κB) and MAP kinase activation and programmed cell death.
Unexpectedly, through structural studies of intracellular signaling complexes in these pathways, we began to see a different scenario that involves formation of higher-order signaling machines, or signalosomes (Figure 1B). The picture first emerged from our pursuit of complexes in the death domain (DD) fold superfamily, which consists of four subfamilies—DD, death effector domain (DED), caspase recruitment domain (CARD), and Pyrin domain (PYD). From crystal structures of three oligomeric DD complexes, the 5:7 PIDD/RAIDD complex for caspase-2 activation in the PIDDosome (Park et al., 2007), the 5:5 Fas/FADD complex in caspase-8 activation by the TNFR family member Fas (Wang et al., 2010), and the 6:4:4 MyD88/IRAK4/IRAK2 complex in the Myddosome for kinase activation in the TLR/IL-1R pathway (Lin et al., 2010), we showed the surprising mode of assembly through helical symmetry (Figure 1C). Because helical symmetry is the basis of most open-ended filamentous structures, the discovery begins to explain the capability of some of these proteins to form filaments, such as those reported for FADD and caspase-8 DED in apoptosis (Siegel et al., 1998) and MAVS CARD for RIG-I and MDA5 signaling (Hou et al., 2011). Many other filaments of the DD superfamily have also been observed in our lab (H.W., unpublished data).
We have identified two additional types of open-ended higher-order complexes in innate immune pathways. One is a filamentous amyloid complex between the RIP1 and RIP3 kinases formed in TNFR-induced necrosis (Li et al., 2012a). This complex is assembled through stacking of short β strands into long β sheets (Li et al., 2012a) (Figure 1D), like the amyloid plaques formed in diseases such as Alzheimer’s and Parkinson’s. The other is a proposed two-dimensional lattice structure of the ubiquitin ligase TRAF6 formed through alternating dimerization of its N-terminal region and trimerization of its C-terminal region (Yin et al., 2009) (Figure 1E). Together with Lys63-linked polyubiquitin chains it promotes to assemble, TRAF6 forms a higher-order signaling assembly that acts as the platform for NF-κB activation. Collectively, these three types of higher-order intracellular assemblies are present in almost every signaling cascade in innate immunity.
Higher-Order Assemblies, Punctates, and Proximity-Facilitated Molecular Machines
The realization that higher-order assemblies, rather than dimers or trimers, are formed during receptor signaling immediately explained frequent observations of punctate morphologies of receptor signaling complexes in cells. For example, upon ligand binding, the TNFR superfamily member Fas has been shown to aggregate progressively from micropuncta to large clusters at the cell surface in a manner that is dependent on recruitment of the adaptor protein FADD and procaspase-8 (Algeciras-Schimnich et al., 2002). This phenomenon is consistent with formation of large oligomeric helical complexes between the DD of Fas and FADD and between the DED of FADD and procaspase-8. Similarly, we showed that, upon stimulation of TLR4 by bacterial lipopolysaccharide (LPS), the intracellular signaling protein TRAF6 relocates from the cytosol to the TLR4 signaling complex to form clustered patches on the cell surface in a manner that is consistent with an infinite two-dimensional lattice structure (Yin et al., 2009). In both cases, the fluorescent spots on the micrometer scale may be composed of thousands of signaling molecules, each in the nanometer scale organized in an orderly fashion.
A common feature of these higher-order signalosomes is that the domains responsible for assembly are either part of or bound to an enzyme such as a caspase or a protein kinase. Therefore, the spatial clustering within the higher-order assemblies would greatly increase the local concentration of the bound enzyme domains to facilitate proximity-driven autoactivation that would not have been possible because of the intrinsic low affinity of the interactions. Consistently, earlier studies have shown that only the high-molecular-weight fractions of certain signaling proteins are active in reconstituted pathways of these systems (Sun et al., 2004). It is well established that caspases are activated by allosteric changes upon dimerization. Many kinases are activated by proximity such as RTKs (Endres et al., 2011). Furthermore, substrates may be recruited to the same signalosomes such as in the Myddosome in which IRAK2 is a substrate of IRAK4 to enable more efficient enzymatic reactions due to increased local concentration. Therefore, these higher-order signalosomes may be molecular machines analogous to mechanical machines in which all parts are integrated into one to increase efficiency.
The concept of intracellular higher-order signaling machines may be distinct from receptor clustering per se. For example, activated GPCRs and RTKs all appear to form oligomers on the cell surface without clear evidence for the simultaneous assembly of intracellular signalosomes. In the innate immune systems that we study, although the isolated ligand-binding domains of many receptors form defined oligomers such as dimers and trimers with their respective ligands, lateral higher-order clustering of full-length receptors seems to exist and cooperates with assembly of multiple intracellular signaling proteins into supramolecular complexes. Therefore, it appears that, in these systems, ligand/receptor complexes at the extracellular region, the transmembrane helices of the receptors, and the intracellular signaling complexes together reinforce a precise molecular machine that transmits the receptor activation signal across the cellular membrane.
Higher-Order Assemblies, Cooperativity, and Threshold Responses
Cooperativity in classical biophysics originates from nonlinear behavior due to molecular interactions (Qian, 2012). Formation of oligomeric complexes may be cooperative if a productive encounter between two subunits enhances the success of the next encounter within the complex. For an infinite higher-order assembly, the cooperativity may reflect formation of the minimal stable multimeric seed complex required for further polymerization. The cooperativity leads to a sigmoidal binding curve as a function of concentration, which may be fit with a Hill equation (Figure 1F). Although cooperativity in oligomerization is due to the requirement for simultaneous interactions with multiple binding partners, its essence of facilitation is equivalent to cooperativity due to allosteric changes such as in oxygen binding to tetrameric hemoglobin.
The intrinsic cooperativity in the assembly of large signalosomes predicts that a cell could exhibit a sharp transition in its response as a function of the dose of ligand stimulation (Figure 1F), leading to an all-or-none digital reaction, also called a threshold response. A number of previous studies have indicated such threshold behavior of cells, for example, in apoptosis induced by the death receptor Fas as a function of ligand concentration (Bentele et al., 2004). More recently, detailed, high-throughput single-cell studies have been conducted on TNF-induced NF-κB activation under TNF doses covering four orders of magnitude (Tay et al., 2010). These studies showed that, although cells exhibit different threshold to TNF doses due to both preexisting variations in sensitivity and stochastic elements, the total NF-κB nuclear activity was equal among cells during the primary response to the signal regardless of TNF doses, indicating a digital activation. At lowers doses, fewer cells respond, whereas at higher doses, more cells switch from nonresponders to responders. This digital characteristic in TNF response is consistent with the nature of the higher-order assemblies in the signal transduction.
The biophysical properties of higher-order assemblies would also predict cooperativity in the temporal response to stimuli (Figure 1G). An intuitive understanding may arise from the slow kinetics in complex formation due to the requirement for assembly of oligomeric seeds. Therefore, the thermodynamic and the kinetic characteristics of higher-order assemblies may result in threshold responses on both temporal and dose scales. A sampling of receptor signaling rates shows that GPCRs mediate effects within seconds (Jensen et al., 2009), RTKs become phosphorylated within seconds but take minutes to accumulate phosphorylation due to antagonism from phosphatases (Kleiman et al., 2011), and innate immune receptors that require higher-order assemblies such as the TNF receptor generate effects in ~20 and ~50 min for high and low ligand doses, respectively (Tay et al., 2010) (Table 1). Because mounting an immune response in the absence of significant danger would generate the damaging effects of tissue injury, a failsafe mechanism might have evolved in signal transduction in the immune system. By providing time delay (Figure 1G) and dose threshold in activation, higher-order assemblies likely overcome transient and stochastic variations of the stimulation to initiate a response only when there is sufficient danger with a persistent and high dose of stimulation. In single-cell studies, TNF-induced NF-κB activation showed dose dependence in time delay of activation (Tay et al., 2010). The longer delay of activation at lower doses is consistent with the concentration dependence of rate constants and may serve to fine-tune the urgency of the response depending on the strength of the external stimulation.
Table 1.
A Sampling of Receptor Signaling Kinetics
| Receptors | Downstream Effects | TimeConstants | References | |
|---|---|---|---|---|
| GPCRs | M1 muscarinic receptor | channel closure | ~5 s | (Jensen et al., 2009) |
| adrenergic and purinergic receptors | cAMP accumulation | ~20–50 s | (Lohse et al., 2008) | |
| RTKs | insulin receptor | substrate phosphorylation | ~1 min | (Flores-Riveros et al., 1989) |
| EGFRs | receptor phosphorylationa | a few seconds | (Kleiman et al., 2011) | |
| accumulation of receptor phosphorylation | ~2–10 min | (Kleiman et al., 2011) | ||
| Innate Immune Receptors | Fas | clustering and caspase-8 activation | ~10–15 min | (Siegel et al., 2004) |
| TNF receptor, high ligand doses | NF-κB translocation | ~20 min | (Tay et al., 2010) | |
| TNF receptor, low ligand doses | NF-κB translocation | ~50 min | (Tay et al., 2010) | |
| Dectin-1 receptor | p38 activation and ROS production | ~15 min | (Goodridge et al., 2011) |
Higher-Order Assemblies in Signal Amplification
In signal transduction, the ligand-binding-induced receptor activation must be amplified to generate a sufficient response from cells. Different receptors may have evolved distinct mechanisms for signal amplification. In GPCR, each ligand-bound, activated receptor can sustain multiple rounds of G protein activation during its lifetime, resulting in considerable signal amplification. Unlike GPCRs, most innate immune receptors do not activate signaling enzymes through multiple rounds of catalytic actions. Instead, a single round of proximity-induced allosteric changes activates the signaling enzymes such as caspases and kinases in the TNFR and TLR/IL-1R pathways. We propose that formation of higher-order, infinite assemblies provides the potential of signal amplification by incorporating an overstoichiometric number of signaling enzymes into the complexes. For example, the death receptor signaling complex is composed of the DD interaction between the receptor (R) and the bifunctional adaptor (A) FADD and of the DED interaction between FADD and caspase-8, an enzyme (E). It has been elucidated that the DED-containing caspases are present in vast overstoichiometry in the signaling complexes to amplify caspase activation (Schleich et al., 2012), which is consistent with helical assembly of DED filaments,
| R*nAn(seed)+mE⇒R*nAnEm(m≫n) | (amplification mechanism 1) |
|---|
In addition, in some cases, unliganded receptors (R) may also have a low but appreciable propensity to assume the active conformation (R*) (Figure 1B). Once the receptor signaling complex is nucleated upon ligand binding, the seeded complex may draw transiently activated receptors (R*) into the existing complex to amplify the signaling process,
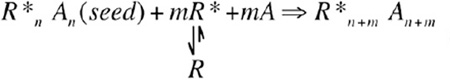 |
(amplification mechanism 2) |
|---|
These amplification processes may continue until all available enzymes are activated and therefore control the sensitivity and magnitude of the response. In an extreme case, bacterial chemotaxis receptors form clustered patches on the cell surface via two-dimensional close-packed lattice structures (Zhang et al., 2007), similar to our proposed structure of TRAF6 (Figure 1E). Lateral propagation of conformational changes upon ligand binding among clustered receptors has been attributed as the reason of high sensitivity of bacterial chemotaxis reactions in detecting a minute amount of attractants.
Higher-Order Assemblies in Reduction of Biological Noise
Erwin Schrödinger reminded us in ”What Is Life?” that physical laws are only approximate, and their precision is based on the statistical power of large numbers. He went on to predict that the master code of a living organism also has to depend on large numbers due to the orderly nature of life. However, many studies have now shown that cell behavior has a strong stochastic component, not only due to low copy numbers and their temporal variation from bursts in gene expression but also because of macromolecular conformational fluctuations and randomness in Brownian diffusion (Qian, 2012). Therefore, instead of adopting a precise, deterministic system, evolution appears to favor a balance between stochasticity and precision either because a deterministic system would be too large and costly or because some imprecision may provide advantage in meeting the challenge of survival in a changing environment.
In this context of stochasticity, we propose that higher-order assemblies may reduce biological noise in signal transduction both kinetically and thermodynamically. The slow association kinetics of these assemblies may filter out transient fluctuations in concentration, diffusion, and conformation so that cells only respond when the stimulation is persistent and strong (Figure 1G). Thermodynamically, because a sharp sigmoidal response provided by higher-order assemblies as a function of ligand concentration essentially only allows the occupation of either the off state or the on state (Figure 1F), the variance in response due to stochastic fluctuations should be intuitively smaller in comparison with less cooperative transitions. Additionally, the capacity for signal amplification by higher-order assemblies may ensure that, as long as the threshold is reached, a cell achieves its maximal magnitude of the response, which is essentially equal among cells in a population as shown for primary NF-κB response to TNF (Tay et al., 2010), further reducing the biological noise in the signal transduction.
Higher-Order Assemblies, Spatial Regulation, and Signal Termination
Numerous studies have now established that cells are not containers of fully diffusible components but rather consist of countless compartments such as organelles and temporary membrane-bound structures for enhancing the specificity of cellular biochemical processes. This spatial regulation is of fundamental importance in signaling transduction. We propose that higher-order assemblies may provide transient spatial compartmentalization without the use of membrane partitions because of their large size and slow-or non-diffusibility. Therefore, a signaling process initiated locally in a cell may stay there without generating unnecessary and undesirable cross-reactions.
To add an additional layer of complexity, previous experimental evidence has suggested that oligomerization may strongly promote phase separation as a result of steric effects of aggregation. In a detailed analysis of assembly of signaling proteins into large, dynamic supramolecular polymers via multivalent interactions, it was found that such polymers produce sharp liquid-liquid demixing phase transitions, generating micron-sized liquid droplets in aqueous solution (Li et al., 2012b). Similarly, low-complexity sequence-containing proteins within cytoplasmic RNA granules have been shown to assemble into amyloid-like fibers that undergo a concentration-dependent phase transition to a hydrogel-like state (Kato et al., 2012). Therefore, higher-order assemblies formed during signal transduction may further partition together into highly concentrated phases or transient organelles, within which the biochemical processes are greatly facilitated by proximity.
How do cells terminate signaling from higher-order assemblies? We know very little about the answer to this question. Biophysics would predict that higher-order assemblies exhibit a slow kinetics of dissociation, and signal termination may require active counter forces such as protein degradation by proteasomes and autophagosomes. In cases in which signalosome assembly is reinforced by phosphorylation or ubiquitination, feedback activation of phosphatases and deubiquitinases may promote disassembly and conclusion of the active cycle of signal transduction.
Conclusions
Higher-order oligomerization appears to exist everywhere in the signaling cascades we have studied. Although it is well established that cells may assemble defined oligomers in signaling such as formation of the apoptosome to activate caspase-9, the discovery that large, infinite assemblies have acquired signaling functions opens up new insights beyond proximity-driven activation. These supramolecular assemblies may allow unique mechanisms of signal amplification, impart threshold response and reduction of biological noise, and render temporal and spatial control of signaling. Certainly, cell signaling is much more complex than depicted here, and we have only begun to see the tip of this signaling iceberg. So many questions are raised by higher-order assemblies, which need to be explored rigorously with experimental and theoretical approaches. Elucidating the biophysical principles governing the assembly and disassembly of higher-order signalosomes in a dynamic, regulated fashion may uncover the structural basis of clustering and reveal new paradigms in cell signaling.
Is it possible that formation of higher-order assemblies is not only widely used in signal transduction in the immune system but also a general mechanism cells use for numerous other biological functions? Many such anecdotal examples exist, such as helical oligomerization of the endoplasmic reticulum (ER) unfolded response protein Ire1 in promoting its allosteric activation, SAM domain-mediated polymerization in inducing DNA packing and transcription silencing, and helical polymers formed on nucleic acids in homologous recombination and DNA replication. Therefore, higher-order assemblies may be an important aspect of many biological processes because they enable formation of precisely organized-molecular machines from constituents present in inactive states at low concentrations to promote biochemical reactions in cells.
ACKNOWLEDGMENTS
I apologize for incomplete citations due to space limitation and thank my colleagues and lab members for many insightful discussions, in particular Kai Wucherpfennig, Hong Qian, Qi Qiao, Qian Yin, and Neer Asherie. This work is supported by the National Institutes of Health.
REFERENCES
- Algeciras-Schimnich A, Shen L, Barnhart BC, Murmann AE, Burkhardt JK, Peter ME. Molecular ordering of the initial signaling events of CD95. Mol. Cell. Biol. 2002;22:207–220. doi: 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H, Krammer PH, Eils R. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J. Cell Biol. 2004;166:839–851. doi: 10.1083/jcb.200404158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres NF, Engel K, Das R, Kovacs E, Kuriyan J. Regulation of the catalytic activity of the EGF receptor. Curr. Opin. Struct. Biol. 2011;21:777–784. doi: 10.1016/j.sbi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Riveros JR, Sibley E, Kastelic T, Lane MD. Substrate phosphorylation catalyzed by the insulin receptor tyrosine kinase. Kinetic correlation to autophosphorylation of specific sites in the beta subunit. J. Biol. Chem. 1989;264:21557–21572. [PubMed] [Google Scholar]
- Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, et al. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature. 2011;472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nat. Chem. Biol. 2012;8:670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JB, Lyssand JS, Hague C, Hille B. Fluorescence changes reveal kinetic steps of muscarinic receptor-mediated modulation of phosphoinositides and Kv7.2/7.3 K+ channels. J. Gen. Physiol. 2009;133:347–359. doi: 10.1085/jgp.200810075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman LB, Maiwald T, Conzelmann H, Lauffenburger DA, Sorger PK. Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol. Cell. 2011;43:723–737. doi: 10.1016/j.molcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012a;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012b;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ, Hein P, Hoffmann C, Nikolaev VO, Vilardaga JP, Bünemann M. Kinetics of G-protein-coupled receptor signals in intact cells. Br. J. Pharmacol. 2008;153(Suppl 1):S125–S132. doi: 10.1038/sj.bjp.0707656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H. Cooperativity in cellular biochemical processes: noise-enhanced sensitivity, fluctuating enzyme, bistability with nonlinear feedback, and other mechanisms for sigmoidal responses. Annu. Rev. Biophys. 2012;41:179–204. doi: 10.1146/annurev-biophys-050511-102240. [DOI] [PubMed] [Google Scholar]
- Schleich K, Warnken U, Fricker N, Oztürk S, Richter P, Kammerer K, Schnölzer M, Krammer PH, Lavrik IN. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol. Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J, Lenardo MJ. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J. Cell Biol. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RM, Muppidi JR, Sarker M, Lobito A, Jen M, Martin D, Straus SE, Lenardo MJ. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J. Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat. Struct. Mol. Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]