Prolonged Exposure to HIV Reinforces a Poised Epigenetic Program for PD-1 Expression in Virus-specific CD8 T Cells (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 15.
Published in final edited form as: J Immunol. 2013 Jun 14;191(2):540–544. doi: 10.4049/jimmunol.1203161
Abstract
Antigen-specific CD8 T cells play a critical role in controlling HIV infection but eventually lose antiviral functions in part because of expression and signaling through the inhibitory PD-1 receptor. To better understand the impact of prolonged TCR ligation on regulation of PD-1 expression in HIV-specific CD8 T cells we investigated the capacity of virus-specific CD8 T cells to modify the PD-1 epigenetic program following reduction in viral load. We observed that the transcriptional regulatory region was unmethylated in the PD-1hi HIV-specific CD8 T cells while it remained methylated in donor matched naïve cells at acute and chronic stages of infection. Surprisingly, the PD-1 promoter remained unmethylated in HIV-specific CD8 T cells from subjects with a viral load controlled by antiviral therapy for greater than 2 years or from elite controllers. Together these data demonstrate that the epigenetic program at the PD-1 locus becomes fixed following prolonged exposure to HIV virus.
INTRODUCTION
Persistent antigen-presentation during chronic viral infections results in a progressive deterioration in T-cell function known as T-cell exhaustion (1–4). Expression of the inhibitory PD-1 receptor on antigen-specific T cells has been associated with T cell dysfunction and lack of viral control in animal models, in HIV infection and other chronic viral diseases. Importantly, expression of PD-1 on virus-specific and total memory T cells correlates with disease progression in HIV-infected subjects (5). Specifically, signaling through PD-1 results in reduced cytokine production and impaired proliferation of HIV-specific CD8 T cells that can be reversed upon in vitro and in vivo exposure of HIV-specific CD8 T cells to antibodies that block the PD-1 - PD-1 ligand interaction (6–8). The therapeutic potential of PD-1 blockade has been demonstrated by administration of blocking antibodies to SIV infected macaques, which results in significant improvement in CTL function and reduction in SIV viral load (9). Recently, therapeutic PD-1 blockade in humans has proven to be an effective strategy for controlling several types of cancer (10, 11).
In light of the tremendous potential that PD-1 blockade strategies have to alleviate human chronic infections and cancer, current efforts are focused on understanding the mechanisms(s) that initiate and sustain PD-1 expression during chronic infection. It is now known that persistent TCR ligation during chronic viral infection maintains elevated levels or PD-1 transcription in nonfunctional antigen-specific CD8 T cells, whereas PD-1 transcription is rapidly downregulated in functional antigen-specific CD8 T cells that develop during an acute infection (4, 12) or when T cells are not exposed to their cognate peptide, such as when viral escape mutations have occurred (13). We have centered our investigation on epigenetic regulation of PD-1 expression as epigenetic modification of transcriptional regulatory regions, including DNA methylation, constitutes an important mechanism for the regulation of tissue and gene specific transcription (14–20). Here we provide data on DNA methylation of the PD-1 regulatory regions in HIV-specific CD8 T cells at different stages of infection and different levels of viral control. This study represents the first report on epigenetic program stability of HIV-specific CD8 T cells during the course of HIV infection in HIV progressors and elite controllers. Our results highlight the negative impact of prolonged exposure to HIV antigen on the ability of virus-specific CD8 T cells to progressively adapt their transcriptional programs to a changing environment.
MATERIALS AND METHODS
Study Subjects and Isolation of HIV-specific T cells
Written informed consent, approved by the University of Montreal Health Center ethics review board (CRCHUM) and Partners Human Research Committee of the Massachusetts General Hospital, was provided and signed by study participants prior to enrollment in the study. Research conformed to ethical guidelines established by the ethics committee of the Massachusetts General Hospital, University of Montreal Health Center and Vaccine and Gene Therapy Institute Florida Inc. institutional review board. Total CD8+ T cells were purified from PBMCs by negative selection using magnetic beads separation (Stem Cell). Cells were then stained with Live/dead aqua dye (Invitrogen) according to the manufacturer instruction and then labeled with phycoerythrin (PE) or allophycocyanin (APC) tetramers at 37° for 15 min as described previously (6, 7). Subsequently, cells were incubated at 4°C for 20 min with cell surface antibodies: CD3 (BD), PD-1 (Biolegend), CD8 (BD), CD45RA (BD), CD27 (Invitrogen), CCR7 (BD). PBMCs were sorted for Naïve (CD8+CD45RA+CCR7+CD27+), CM (CD8+CD45RACCR7+CD27+), TM (CD8+CD45RA−CCR7+CD27−),EM (CD8+CD45RA+CCR7−CD27−), and HIV- specific cells (CD8+ CD3+ tetramer+) using the BD ARIAII flow cytometer (BD Biosciences).
Genomic methylation analysis
Sodium bisulfite induced deamination of cytosine was performed using the Zymo Research EZ DNA methylation kit. The bisulfite modified DNA was PCR amplified with locus specific primers as previously described (18). Statistical significance of CpG site methylation was determined with a two-tailed unpaired Students t test using Prism 4 software.
RESULTS
DNA demethylation of the PD-1 locus during acute and chronic HIV infection
We sought to determine if the PD-1 transcriptional regulatory region in HIV-specific CD8 T cells acquired an “active” epigenetic program at the acute and chronic stages of HIV infection in viremic subjects. Donor matched naïve and HIV-specific CD8 T cells were isolated at the acute (0–4 months post infection) and chronic stages of infection (>4 months post infection) (Supplemental Table 1). Bisulfite sequencing methylation analysis of the PD-1 conserved regulatory region (CR) was performed using genomic DNA from FACS purified cells (Figure 1a & b). Consistent with our previous report (18), naïve CD8 T cells have a methylated transcriptional regulatory region, regardless of the disease stage. In contrast, PD-1hi HIV-specific CD8 T cells had a completely unmethylated regulatory region during conditions of high viremia at the acute stage of infection. Furthermore, virus-specific CD8 T cells isolated at the chronic stage of the infection retained an unmethylated PD-1 regulatory region, whereas donor matched naïve cells retained a methylated regulatory region. These data indicate that exposure to HIV results in a loss of the DNA methylation program at the PD-1 regulatory region in virus-specific CD8 T cells and suggest that the unmethylated state persists with the sustained engagement of TCR by its cognate Ag.
Figure 1. The PD-1 locus transcriptional regulatory region is demethylated in virus-specific CD8 T cells at the acute stage of HIV infection and remains unmethylated during the chronic stage of infection.
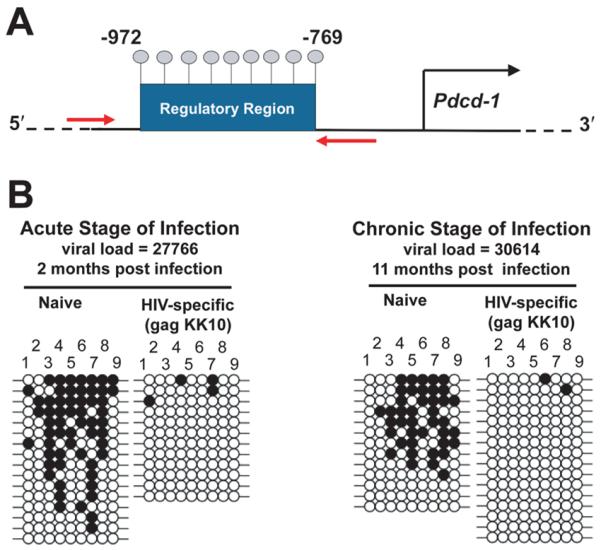
A) Cartoon representation of the PD-1 transcriptional regulatory region. Gray lollipops indicate the location of CpG sites ranging between −769 and −972 base pairs from the start codon. The red arrows indicate the approximate location of the primers used for bisulfite sequencing methylation analysis. B) Representative bisulfite sequencing DNA methylation analysis. Bisulfite sequencing of the PD-1 conserved regulatory (CR) region was performed on purified (>95% purity) naïve and HIV-specific CD8 T cell genomic DNA from donors at the acute (n=2) and chronic (n=3) stages of infection. Each line represents an individual clone picked for sequencing. Filled circles = methylated cytosine. Open circles = non-methylated cytosine. CpG sites 18–26 are shown for clarity.
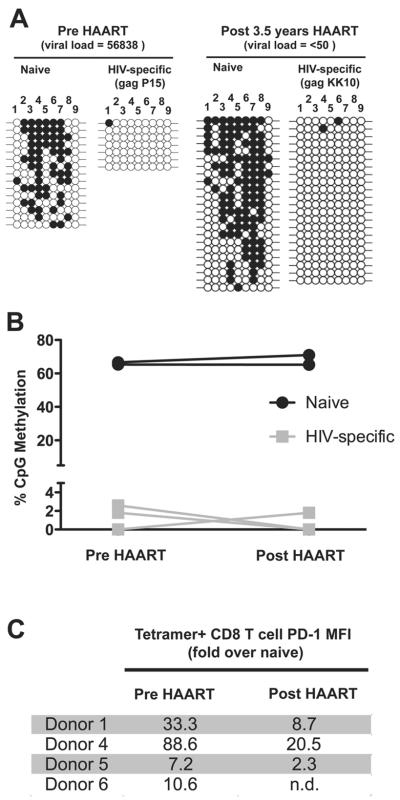
PD-1 locus demethylation persists following HAART mediated reduction in HIV viral load
Given the striking reduction in DNA methylation at the PD-1 locus in HIV-specific CD8 T cells relative to donor matched naïve cells, we next sought to determine if reduction in antigen would result in remethylation of the locus since it is also known that PD-1 levels decrease upon antiviral therapy concomitantly with a decreases viral load to undetectable levels (21). Virus-specific CD8 T cells were isolated from donors prior to HAART therapy and following HAART therapy after viral loads of <50 copies / ml were maintained for more than 1 year (Supplemental Table 1). Akin to the virus-specific CD8 T cells analyzed at the acute and chronic stages from untreated donors, virus-specific CD8 T cells were unmethylated at the PD-1 locus prior to HAART therapy (Figure 2a & b). Methylation analysis of naïve and HIV-specific CD8 T cells were performed in a longitudinal manner with cells isolated from the same donor pre and post (1–4 years) HAART (Figure 2c). Surprisingly, these cells remained unmethylated at the PD-1 regulatory region following HAART therapy despite undetectable levels of viral load for up to 3 years (Figure 2a & b) and despite the fact that PD-1 cell surface expression was reduced on virus-specific CD8 T cells in donors 1, 4, and 5 following HAART therapy (Figure 2d).
Figure 2. Reduced viremia in HAART treated donors does not result in DNA remethylation of the PD-1 locus in virus-specific CD8 T cells.
A) Representative bisulfite sequencing DNA methylation analysis of naïve and HIV-specific CD8 T cells from donors pre- and post-HAART therapy. B) Longitudinal analysis of percent CpG methylation at the PD-1 regulatory region in donor matched naïve and HIV-specific CD8 T cells pre and post HAART. C) PD-1 MFI on HIV-specific CD8 T cells used for methylation studies. PD-1 MFI on tetramer+ CD8 T cells represented as a fold over PD-1 MFI on donor matched naïve CD8 T cells.
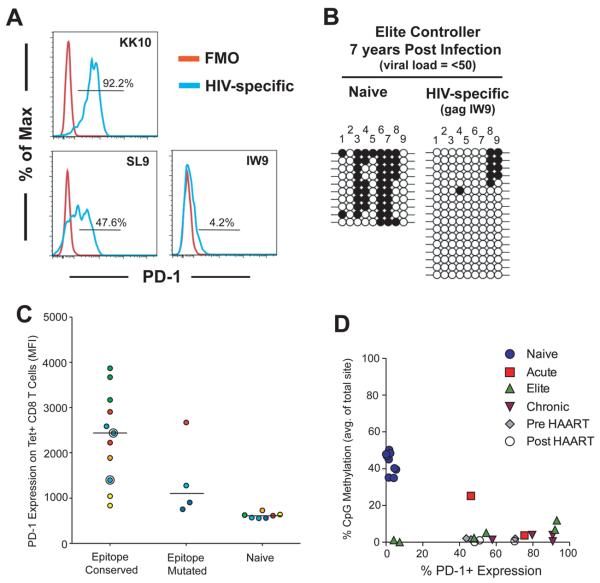
Elite Controller HIV-specific CD8 T cells retain a demethylated PD-1 locus
The lack of PD-1 regulatory region DNA methylation in CD8 T cells from HAART treated donors suggests that prolonged TCR ligation preserves the epigenetic program acquired at the acute stage of the infection even after reduction of viral load by antiviral therapy (ART). To further examine the impact of transient high viremia on the plasticity of the PD-1 epigenetic program, we measured the level of DNA methylation at the PD-1 regulatory region in HIV-specific CD8 T cells from elite controllers (EC). Elite controllers can initially have elevated levels of viremia, but eventually control viral infection to <50 copies of HIV RNA / ml (22). PD-1 expression varied amongst different epitope-specificities within the total population of HIV-specific CD8 T cells (Figure 3a & Supplemental Figure 1d). DNA methylation analyses were performed on virus-specific CD8 T cells isolated from donors with undetectable viral loads for more than 7 years (Supplemental Table 1). Again we observed that HIV-specific CD8 T cells retained an unmethylated PD-1 transcriptional regulatory region in an environment of reduced TCR ligation (Figure 3b). These data suggest that modification of the PD-1 epigenetic program that occurs during naïve to effector differentiation becomes permanent during chronic HIV infection even under conditions where viremia was naturally controlled after the onset of infection.
Figure 3. Chronic HIV infection uncouples PD-1 locus remethylation from CD8 T cell activation.
A) Histogram analysis of PD-1 expression on HIV-specific CD8 T cells KK10, SL9, and IW9 (blue) relative to fluorescence minus one (red). B) Representative bisulfite sequencing DNA methylation analysis of naïve and HIV-specific CD8 T cells from elite controllers. C) PD-1 levels were measured on HIV tetramer-specific CD8 T cells (n=15) from HIV-infected subjects (n=7) with viral loads <50 copies/mL. PD-1 expression is plotted according to the autologous viral sequence of the epitopes corresponding to the tetramers used: “conserved”: autologous epitope sequence identical to the tetramer; “mutated”: autologous epitope harboring a known escape mutation. Each color corresponds to a single individual. D) Graph of average CpG methylation among all sites vs % PD-1+ expression on naïve and HIV-specific CD8 T cells. The difference in PD-1 locus DNA methylation between naïve and HIV-specific CD8 T cells is highly significant; p-value < 0.0001. Pearson analysis of PD-1 expression vs DNA methylation results in R2 values of 0.32 and 0.04 for naïve and HIV-specific CD8 T cells respectively.
In particular, two of the antigen-specific CD8 T cell populations from elite controllers had less than 10% positive PD-1 expression (Figure 3c), specifically IW9-specific CD8 T cells isolated from donor #10 for the DNA methylation analysis were only ~4% positive for PD-1 expression (Figure 3b). MFI analysis of PD-1 expression on the HIV-specific CD8 T cells from HAART treated donors and elite controllers also demonstrated that the degree of PD-1 downregulation was variable among different populations of antigen-specific CD8 T cells when normalized to donor matched naïve cells (Supplemental Figure 1d). The heterogeneity in PD-1 expression among different virus-specific CD8 T cell populations in low viremic conditions may suggest that the antigen-specific CD8 T cells have lost the ability to fully suppress PD-1 transcription.
To further explore the relationship of PD-1 downregulation and antigen persistence we next assessed the level of PD-1 expression on virus-specific CD8 T cells from donors who recognized a conserved HIV epitope in the persisting virus versus PD-1 expression on virus-specific CD8 T cells from donors who have a mutated HIV epitope. As expected the level of PD-1 expression (median MFI from 4 responses) was lower on the antigen-specific CD8 T cells in the presence of the mutated epitope (Figure 3c). Interestingly, PD-1 expression (MFI) on the mutated epitope-specific CD8 T cells was still significantly higher than the donor matched naïve cells. These data suggest that the mechanism for downregulation of PD-1 expression is altered following TCR ligation.
To assess the association between PD-1 expression and DNA methylation at the PD-1 regulatory region prior to and following exposure to HIV antigen, we compared the average % of DNA methylation from the cumulated sites in the PD-1 transcriptional regulatory region relative to the % of PD-1+ expression from all cells used for the DNA methylation study. All naïve CD8 T cells isolated from HIV-infected donors retained a clear inverse correlation between DNA methylation and PD-1 expression. In contrast, the level of PD-1 expression was uncoupled from remethylation of the PD-1 regulatory region (Figure 3d) in HIV specific T cells from elite controllers. Taken together our data reveal that significant reduction in HIV viral load is not sufficient to further modify the DNA methylation program at the PD-1 locus in antigen-specific CD8 T cells. To further determine the impact of antigen induced T cell differentiation on PD-1 transcriptional programming we proceeded to measure the methylation status at the PD-1 promoter in the polyclonal memory populations in HIV infected individuals. Indeed, the level of DNA methylation at all CpG positions in each of the memory subsets was significantly lower than naïve CD8 T cells (Supplemental Figure 1f). Importantly, a population of cells within the Tcm compartment of the CD8 T cells retained a level of methylation that is significantly higher than the HIV-specific CD8 T cells and also had the lowest level of PD-1 expression relative to the other polyclonal memory subset (Supplemental Figure 1e).
DISCUSSION
Persistent exposure to antigen drives T cells towards a state of functional impairment and terminal differentiation (23, 24). Our previous report on epigenetic regulation of PD-1 expression in antigen-specific CD8 T cells differentiating in response to acute and chronic viral infections in mice and humans revealed that the transient upregulation of PD-1 expression in both mouse and human functional virus-specific CD8 T cells was coupled to acquisition of an unmethylated transcriptional regulatory region during the peak of viremia (18). Following clearance of the acute viral infection, the PD-1 transcriptional regulatory region regained the DNA methylation program in functional memory CD8 T cells from both the murine LCMV model system of acute viral infection and yellow fever vaccination of humans. Importantly, the repressive transcriptional program was not reacquired in human EBV and CMV-specific CD8 T cells (18). In the current study analysis of the PD-1 epigenetic program was performed on virus-specific CD8 T cells from HIV infected individuals that are well defined in there date of initial exposure, peak viremia, and reduction in viral load. The results from our current study suggest that the pliable quality of epigenetic mechanisms in naïve and functional memory CD8 T cells is lost during chronic HIV infection, even in HIV-specific CD8 T cells from elite controllers. Although the exact mechanism for fixation of epigenetic programs is not well understood, it is clear that it involves altered expression of de novo DNA methyltransferase variants, as well as lineage defining transcription factors (25).
It is important to note that although reduction in PD-1 expression is likely due to lack of TCR engagement, virus-specific cells could re-circulate from other anatomical sites where HIV replication is still persistent at levels higher than those detected in plasma. Thus, it is possible that limited or intermittent TCR ligation resulting from low-level persistent viremia may reinforce an unmethylated state through periodic activation of the virus-specific cells. Interestingly, our data and those of others demonstrated that PD-1 expression is quite heterogeneous amongst different tetramer+ CD8 T cells following the reduction in viral load. Recently it has been reported that viral escape mutations can result in reduced PD-1 expression on CD8 T cells specific to the WT epitope (13). Thus, mutation in viral epitopes may contribute to the wide range of PD-1 expression observed. Indeed, in the current manuscript we observed that there was a trend for lower PD-1 expression on antigen-specific CD8 T cells targeting a mutated epitope compared to responses targeting conserved viral sequences. However, since the absence of methylation at the PD-1 locus leaves it poised for transcriptional activation, non-TCR stimulatory signals may be sufficient to induce PD-1 expression, including γ–C receptor cytokines and type I interferons (26–30). Such alternative mechanisms are strongly suggested by the broad range in PD-1 expression seen on HIV-specific CD8 T cells targeting conserved epitopes in individuals with similar viral loads. It will be of great interest to determine if the inflammatory environment that is characteristic of HIV infection can induce PD-1 expression on the poised PD-1lo HIV-specific CD8 T cells.
The present study on epigenetic regulation of PD-1 reveals that even though HIV-specific CD8 T cells have reduced or no PD-1 expression when isolated from donors who have undergone successful HAART treatment or from elite controllers, the cells retain a transcriptional program that is poised for PD-1 expression. This first analysis of epigenetic programs in HIV-specific CD8 T cells during infection represent a basic model for acquisition of heritable transcriptional regulation of PD-1 in the context of chronic HIV infection that invokes preservation of the demethylated regulatory regions during persistent stimulation throughout naïve to effector to memory differentiation (Supplemental Figure 1g). In the proposed model, acquired transcriptional programs are maintained during cell division via duplication of the epigenetic modifications from the parental DNA onto the newly synthesized strand of DNA. Therefore antigen-experienced PD-1lo HIV-specific CD8 T cells are poised to rapidly upregulate PD-1 expression and are then susceptible to PD-1 mediated termination of an effective recall response. Further, these results emphasize the need for PD-1 signaling blockade in conjunction with therapeutic strategies that attempt to restore the immune response in order to purge the latent HIV reservoir, and indicate that an alternative approach to PD-1 antibody blockade may involve directed reprogramming of epigenetic modifications.
Supplementary Material
1
2
3
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants P01 AI080192-04 (to R.A., J.M.B, R.P.S, D.E.K.), R37 AI30048-17 (to R.A.), American Cancer Society (A.C.S) postdoctoral fellowship PF-09-134-01-MPC (to B.A.Y.), and NIH R01 HL092565 (to D.E.K)
Footnotes
Conflict of interest statement: R.A. holds a patent for the PD-1 inhibitory pathway.
REFERENCES
- 1.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annual review of immunology. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. International immunology. 2007;19:813–824. doi: 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 3.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nature immunology. 2007;8:239–245. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 4.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 5.Zhang JY, Zhang Z, Wang X, Fu JL, Yao J, Jiao Y, Chen L, Zhang H, Wei J, Jin L, Shi M, Gao GF, Wu H, Wang FS. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood. 2007;109:4671–4678. doi: 10.1182/blood-2006-09-044826. [DOI] [PubMed] [Google Scholar]
- 6.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 7.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, Routy JP, Haddad EK, Sekaly RP. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nature medicine. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 8.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, Precopio ML, Schacker T, Roederer M, Douek DC, Koup RA. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. The Journal of experimental medicine. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velu V, Titanji K, Zhu B, Husain S, Pladevega A, Lai L, Vanderford TH, Chennareddi L, Silvestri G, Freeman GJ, Ahmed R, Amara RR. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 2009;458:206–210. doi: 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brahmer JR, T. S. L., Powderly J, Wollner I, Picus J, Drake CG, Stankevich E, Korman A, Pardoll D, Lowy I. Phase II experience with MDX-1106 (Ono-4538), an anti-PD-1 monoclonal antibody, in patients with selected refractory or relapsed malignancies. Journal of Clinical Oncology. 2009 [Google Scholar]
- 11.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Janbazian L, Price DA, Canderan G, Filali-Mouhim A, Asher TE, Ambrozak DR, Scheinberg P, Boulassel MR, Routy JP, Koup RA, Douek DC, Sekaly RP, Trautmann L. Clonotype and repertoire changes drive the functional improvement of HIV-specific CD8 T cell populations under conditions of limited antigenic stimulation. J Immunol. 2012;188:1156–1167. doi: 10.4049/jimmunol.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 15.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 16.Kersh EN. Impaired memory CD8 T cell development in the absence of methyl-CpG-binding domain protein 2. J Immunol. 2006;177:3821–3826. doi: 10.4049/jimmunol.177.6.3821. [DOI] [PubMed] [Google Scholar]
- 17.Kersh EN, Fitzpatrick DR, Murali-Krishna K, Shires J, Speck SH, Boss JM, Ahmed R. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J.Immunol. 2006;176:4083–4093. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 18.Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, Boss JM, Ahmed R. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8+ T cells. Immunity. 2011;35:13. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youngblood B, Wherry EJ, Ahmed R. Acquired transcriptional programming in functional and exhausted virus-specific CD8 T cells. Current opinion in HIV and AIDS. 2012;7:50–57. doi: 10.1097/COH.0b013e32834ddcf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner SL. Epigenetic control in the immune response. Human molecular genetics. 2005;14(Spec No 1):R41–46. doi: 10.1093/hmg/ddi115. [DOI] [PubMed] [Google Scholar]
- 21.Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. Journal of virology. 2012;86:8161–8170. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura T, Brumme ZL, Brockman MA, Rosato P, Sela J, Brumme CJ, Pereyra F, Kaufmann DE, Trocha A, Block BL, Daar ES, Connick E, Jessen H, Kelleher AD, Rosenberg E, Markowitz M, Schafer K, Vaida F, Iwamoto A, Little S, Walker BD. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. Journal of virology. 84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wherry EJ. T cell exhaustion. Nature immunology. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 24.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. Journal of virology. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 26.Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, O'Shea MA, Hallahan CW, Daucher M, Ward DJ, Moir S, Mullins JI, Kovacs C, Fauci AS. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. The Journal of clinical investigation. 2005;115:3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, Ward DJ, Kovacs JA, Mannon PJ, Fauci AS. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. The Journal of infectious diseases. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 28.Kinter AL, Godbout EJ, McNally JP, Sereti I, Roby GA, O'Shea MA, Fauci AS. The common gamma-chain cytokines IL-2, IL-7, IL-15, and IL-21 induce the expression of programmed death-1 and its ligands. J Immunol. 2008;181:6738–6746. doi: 10.4049/jimmunol.181.10.6738. [DOI] [PubMed] [Google Scholar]
- 29.Terawaki S, Chikuma S, Shibayama S, Hayashi T, Yoshida T, Okazaki T, Honjo T. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol. 2011;186:2772–2779. doi: 10.4049/jimmunol.1003208. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson AS, Korber BT, Markowitz M, Ho DD. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. The New England journal of medicine. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3