The ALS8 protein VAPB interacts with the ER–Golgi recycling protein YIF1A and regulates membrane delivery into dendrites (original) (raw)
Abstract
The vesicle-associated membrane protein (VAMP) associated protein B (VAPB) is an integral membrane protein localized to the endoplasmic reticulum (ER). The P56S mutation in VAPB has been linked to motor neuron degeneration in amyotrophic lateral sclerosis type 8 (ALS8) and forms ER-like inclusions in various model systems. However, the role of wild-type and mutant VAPB in neurons is poorly understood. Here, we identified Yip1-interacting factor homologue A (YIF1A) as a new VAPB binding partner and important component in the early secretory pathway. YIF1A interacts with VAPB via its transmembrane regions, recycles between the ER and Golgi and is mainly localized to the ER–Golgi intermediate compartments (ERGICs) in rat hippocampal neurons. VAPB strongly affects the distribution of YIF1A and is required for intracellular membrane trafficking into dendrites and normal dendritic morphology. When VAPB-P56S is present, YIF1A is recruited to the VAPB-P56S clusters and loses its ERGIC localization. These data suggest that both VAPB and YIF1A are important for ER-to-Golgi transport and that missorting of YIF1A may contribute to VAPB-associated motor neuron disease.
Keywords: amyotrophic lateral sclerosis (ALS), dendrite morphology, secretory pathway, VAMP-associated protein (VAP), Yip1 interacting factor (YIF1)
Introduction
The secretory pathway is responsible for the delivery of a variety of protein and lipid cargo, such as ion channels, receptors and lipid membranes, to their proper location. In the early secretory pathway, newly synthesized proteins that pass the quality control of the endoplasmic reticulum (ER) concentrate at specialized ER subdomains called the ER exit sites (ERES) (Lee et al, 2004; Dancourt and Barlowe, 2010). From here, cargo leaves the ER and is transported to the Golgi compartments to undergo further processing, sorting and subsequent delivery to the plasma membrane or other cellular organelles. ER-derived vesicles deliver their cargo either directly to the cis-Golgi membrane or pass a specialized membrane compartment called the ER–Golgi intermediate compartment (ERGIC) (Watson and Stephens, 2005; Appenzeller-Herzog and Hauri, 2006; Saraste et al, 2009; Lorente-Rodriguez and Barlowe, 2011). Whereas the early secretory pathway and its organization is well studied in a variety of cellular model systems such as yeast and fibroblasts, little information is available about this trafficking pathway in neuronal cells. A number of studies have demonstrated the presence of ER-to-Golgi transport components in neurons (Krijnse-Locker et al, 1995; Pierce et al, 2001; Horton and Ehlers, 2003) and indicated the importance of the early secretory trafficking for neuronal development and morphogenesis (Hanus and Ehlers, 2008; Tang, 2008; Aridor and Fish, 2009; Jan and Jan, 2010).
Members of the highly conserved VAP family, including the mammalian vesicle-associated membrane protein associated protein A (VAPA) and VAPB proteins, are present in the ER and have been proposed to play a role in maintaining Golgi complex identity, ER morphology, lipid transfer and in regulating ER and Golgi transport (Soussan et al, 1999; Skehel et al, 2000; Pennetta et al, 2002; Amarilio et al, 2005; Teuling et al, 2007; Lev et al, 2008; Peretti et al, 2008; Tsuda et al, 2008; Han et al, 2012). VAP proteins contain an N-terminal domain, which is highly homologous to the nematode major sperm protein (MSP), a central domain that forms a coiled-coil structure and the C-terminal transmembrane tail domain (Nishimura et al, 1999; Kaiser et al, 2005). Interest in VAP proteins has greatly increased after the discovery of a dominant missense mutation P56S in the MSP domain of VAPB, which causes a familial motor neuron disease, designated amyotrophic lateral sclerosis type 8 (ALS8) (Nishimura et al, 2004). P56S mutant VAPB accumulates in inclusions that contain abnormal organized ER (Teuling et al, 2007; Papiani et al, 2012) and recruit wild-type VAPA/B in these inclusions, suggesting that VAPB-P56S could have a dominant-negative effect on VAP function (Teuling et al, 2007). Indeed, a loss-of-function mechanism has been observed in several model systems (Chai et al, 2008; Ratnaparkhi et al, 2008; Tsuda et al, 2008; Suzuki et al, 2009; Forrest et al, 2013). Moreover, a reduction in VAP protein levels has been described in sporadic ALS patients, SOD1 mutant mice as well as ALS-derived motor neurons (Teuling et al, 2007; Anagnostou et al, 2010; Mitne-Neto et al, 2011). However, mechanisms leading to VAPB-linked motor neuron degeneration are still poorly understood.
To gain more insight into the role of VAPB in neurons, we searched for new binding partners of VAPB. We identified Yip1-interacting factor homologue A (YIF1A) as a new VAPB binding partner that interacts with both wild-type VAPB and mutant VAPB-P56S. YIF1A is the human orthologue of the budding yeast Yif1p, a transmembrane protein that plays an important role in early secretory transport in yeast and mammalian cells (Matern et al, 2000; Barrowman et al, 2003; Jin et al, 2005; Yoshida et al, 2008). We find that YIF1A recycles between the ER and Golgi and is mainly localized to the ERGIC in hippocampal neurons. Both VAPB and YIF1A are required for membrane trafficking to dendrites and proper dendrite morphology. In addition, we show that ALS8 mutant VAPB-P56S strongly disrupts the localization of YIF1A to ERGIC. Our data suggest an important role for YIF1A and VAPB in the early secretory pathway and we propose that the missorting of YIF1A plays a role in VAPB-associated motor neuron disease. These findings advance the knowledge of fundamental trafficking processes in neuronal cells and have important implications for our understanding of neuronal degeneration.
Results
YIF1A interacts with both wild-type and mutant VAPB
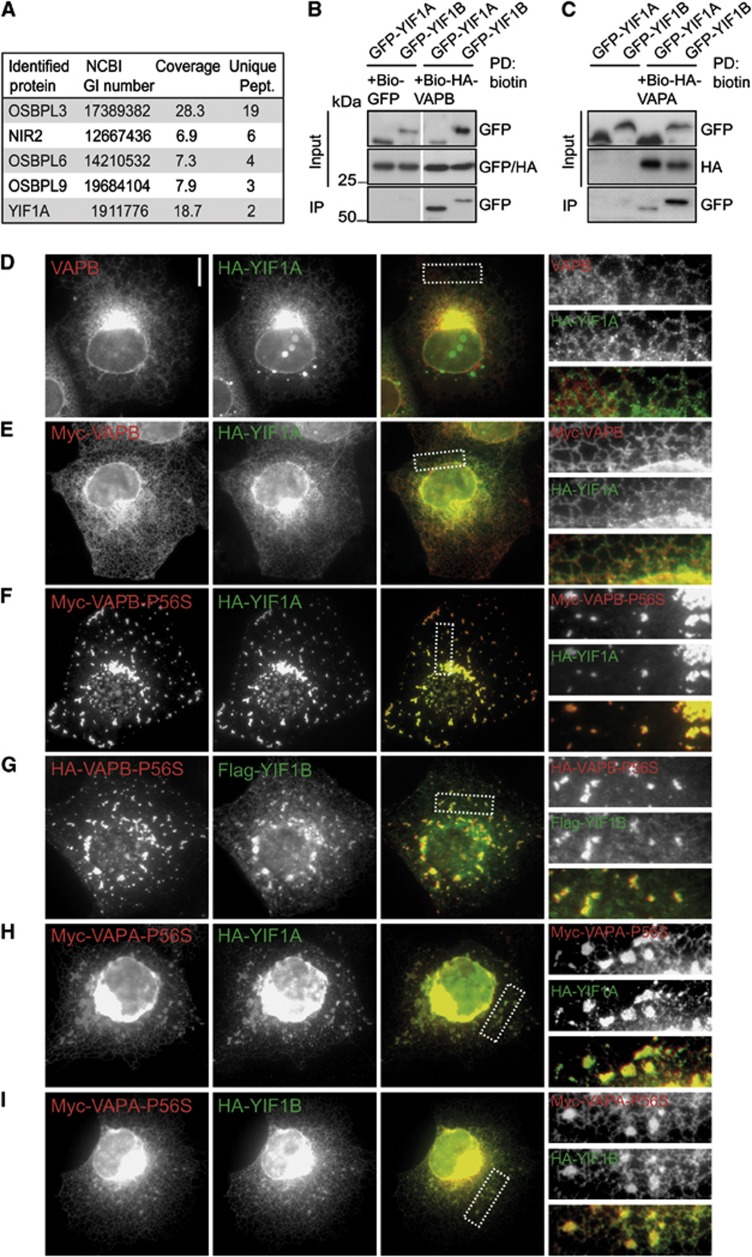
To identify new VAPB interacting proteins, we performed pull-down assays combined with mass spectrometry. Biotinylated and GFP-tagged VAPB (bio-GFP-VAPB) and control bio-GFP constructs were transiently co-expressed in HeLa cells together with the protein-biotin ligase BirA, isolated with streptavidin beads and the proteins were analysed by mass spectrometry. Bio-GFP-VAPB bound to several previously identified VAPB binding partners, such as the FFAT-motif containing proteins NIR2, OSBPL3, OSBPL6 and OSBPL9 (Wyles and Ridgway, 2004; Amarilio et al, 2005; Lehto et al, 2005; Teuling et al, 2007) (Figure 1A). In addition, a novel potential VAPB binding partner was identified, YIF1A, a transmembrane protein of 293 amino acids and mammalian orthologue of the Saccharomyces cerevisiae protein Yif1p (Yip1p-interacting factor 1) (Matern et al, 2000). YIF1A and its close homologue YIF1B are members of a large protein family, named FinGERs, which share a common structure with an N-terminal hydrophilic region, followed by conserved transmembrane regions (Shakoori et al, 2003; Pfeffer and Aivazian, 2004).
Figure 1.
Interaction of YIF1A with wild-type and mutant VAPB. (A) Identification of wild-type VAPB binding partners by mass spectrometry in HeLa cell extract. The table shows proteins identified with a significant Mascot score in the pull-down with streptavidin beads from an extract of HeLa cells co-expressing Bio-GFP-VAPB and biotin ligase BirA. The list is corrected for background proteins, which were identified in a control pull-down from HeLa cells expressing bio-GFP. Abbreviations used in the table to indicate the identified proteins: OSBPL, oxysterol binding protein-like; NIR, N-terminal domain-interacting receptor. (B) Biotin pull-downs (PD) from HEK293T extract transfected with Bio-HA-VAPB and GFP-YIF1A, GFP-YIF1B or control bio-GFP and probed for GFP and HA. (C) Biotin pull-downs from HEK293T extract transfected with Bio-HA-VAPA and GFP-YIF1A or GFP-YIF1B and probed for GFP and HA. The ratio input/pellet is 2–5% for all pull-down and immunoprecipitation experiments. (D) COS-7 cells transfected with HA-YIF1A and stained with anti-HA (green) and anti-VAPB (red) antibodies. (E, F) COS-7 cells double transfected with HA-YIF1A and myc-VAPB (D) or myc-VAPB-P56S (E) stained with anti-HA (green) and anti-myc (red) antibodies. (G) COS-7 cells double transfected with HA-VAPB-P56S and Flag-YIF1B, fixed and stained with anti-HA (green) and anti-Flag (red) antibodies. (H, I) COS-7 cells double transfected with myc-VAPA-P56S and HA-YIF1A (H) or HA-YIF1B (I) stained with anti-HA (green) and anti-myc (red) antibodies. Panels on the right side show enlargements of the boxed regions. Scale bar, 10 μm.
The interaction of VAPB and YIF1A was confirmed by biotin pull-down experiments using extracts of HEK293T cells overexpressing GFP-YIF1A and bio-HA-VAPB (Figure 1B). Pull-down experiments also revealed binding between YIF1B and VAPB (Figure 1B) and between VAPA and both YIF homologues (Figure 1C). To further confirm the interaction between VAPB and YIF1A, we performed immunofluorescence experiments in COS-7 cells. HA-YIF1A co-localized with both endogenous VAPB and co-transfected myc-VAPB, which as previously demonstrated localize to the ER (Nishimura et al, 2004; Kanekura et al, 2006; Teuling et al, 2007; Kim et al, 2010; Papiani et al, 2012) (Figure 1D and E). Significantly, HA-YIF1A also co-distributes with ALS-linked mutant VAPB-P56S and VAPA-P56S (Figure 1F and H), which accumulates in small spherical inclusions (Nishimura et al, 2004; Kanekura et al, 2006; Teuling et al, 2007; Kim et al, 2010; Papiani et al, 2012). Likewise also YIF1B was recruited to mutant VAPA/B inclusion (Figure 1G and I). Together, these results show that YIF1A/B interacts with VAPA/B family proteins.
The transmembrane domains of both VAPB and YIF1A are required for their interaction
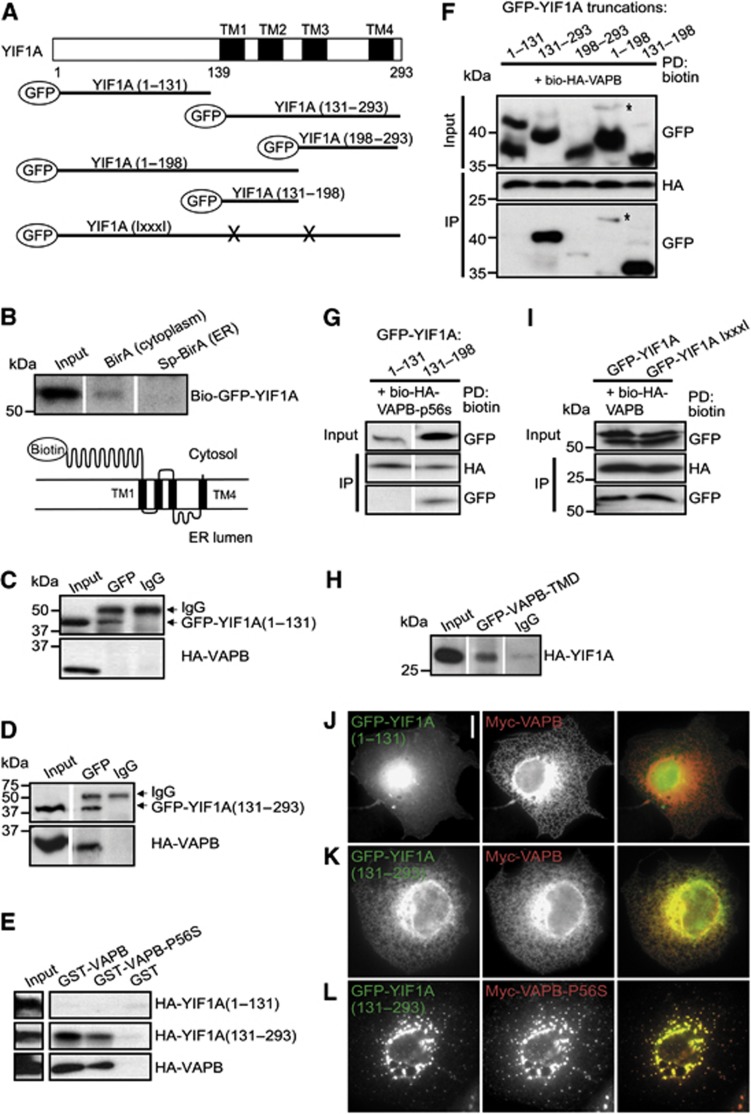
Secondary structure predictions indicate that YIF1A contains four transmembrane domains at the C-terminus (Figure 2A) (Altschul et al, 1997; Hirokawa et al, 1998), while the N-terminus of the yeast homologue Yif1p has been shown to face the cytosol (Matern et al, 2000). To confirm that the N-terminus of YIF1A faces the cytosol, we generated a YIF1A construct with a biotinylation tag at the N-terminus (Figure 2B). Pull-down experiments showed that this construct was biotinylated when the biotinylating enzyme BirA was localized in the cytoplasm, but not by a variant BirA that is localized in the ER lumen (Figure 2B).
Figure 2.
The transmembrane domain of YIF1A interacts with VAPB. (A) YIF1A deletion constructs were made containing amino acids 1–131 of YIF1A, amino acids 131–293, 198–293, 1–198 and amino acids 131–198. GxxxG motifs in transmembrane domain one and three were mutated by replacing the glycine residues with isoleucine. The predicted transmembrane domains are labelled with TM. (B) Biotin pull-down to determine the topology of YIF1A using HEK293T extracts transfected with bio-GFP-YIF1A and BirA (cytoplasm) or SP-BirA (ER lumen). Bio-GFP-YIF1A binds to streptavidin beads in the presence of cytoplasmic BirA but not in the presence of luminal BirA. Samples were immunoblotted using anti-GFP antibodies. (C, D) Analysis of YIF1A binding domain by co-immunoprecipitation. HEK293T cells co-transfected with (C) GFP-YIF1A(1–131) or (D) GFP-YIF1A(131-293) and HA-VAPB were immunoprecipitated with anti-GFP or IgG (control) antibodies. (E) Binding domain analysis by GST pull-down assay using lysates of HEK293T cells expressing HA-YIF1A truncated constructs and GST-VAPB or GST-VAPB-P56S. Samples were immunoblotted using anti-HA antibodies. (F) Biotin pull-downs (PD) from HEK293T extracts transfected with GFP-YIF1A truncated constructs and bio-HA-VAPB. Probed for GFP and HA. The asterisk denotes a band corresponding to the YIF1A(1–198) protein. (G) Biotin pull-down (PD) from HEK293T extracts transfected with GFP-YIF1A truncated constructs and bio-HA-VAPB-P56S and probed for GFP and HA. (H) Immunoprecipitation from extract of HEK293T cells co-expressing GFP-VAPB-TMD and HA-YIF1A. Immunoblot is probed for HA. (I) Biotin pull-down from HEK293T extracts transfected with GFP-YIF1A IxxxI and bio-HA-VAPB and probed for GFP and HA. The ratio input/pellet is 2–5% for all pull-down and immunoprecipitation experiments. (J–L) COS-7 cells double transfected with myc-VAPB (J, K) or myc-VAPB-P56S (L) and GFP-YIF1A truncation constructs, fixed and stained with anti-myc (red) antibodies. Scale bar, 10 μm.
To identify the regions through which VAPB interacts with YIF1A, we first made truncated GFP-YIF1A constructs encoding either the N-terminus (1–131) or the C-terminal transmembrane region (131–293) of YIF1A (Figure 2A). Pull-down experiments using HEK293T cells co-expressing HA-VAPB and the YIF1A deletion constructs showed that VAPB only co-precipitates with YIF1A construct that contained the transmembrane region (Figure 2C and D). These data were confirmed by a glutathione S-transferase (GST) pull-down assay with cell lysates expressing truncated YIF1A constructs using VAPB and VAPB-P56S immobilized on GST beads. The C-terminal part, but not the N-terminal part of YIF1A co-precipitated with VAPB and VAPB-P56S (Figure 2E). To characterize in more detail the YIF1A domain that binds VAPB, we made additional truncated YIF1A constructs, which contain only the first two transmembrane domains (131–198), the last two transmembrane domains (198–298) or the N-terminal cytosolic domain with the first two transmembrane domains (1–198) (Figure 2A). Pull-down experiments with biotinylated VAPB showed that all transmembrane containing YIF1A constructs co-precipitated with VAPB (Figure 2F). However, VAPB brings down considerable higher amounts of truncated GFP-YIF1A proteins that contain the first and second transmembrane domain (131–198) (Figure 2F), indicating that the VAPB interaction is strongest with this region. The marked difference between binding of the first two transmembrane domains compared to the last two transmembrane domains suggests a specific interaction between YIF1A and VAPB. GFP-YIF1A (131–198) also efficiently co-precipitated with VAPB-P56S (Figure 2G). Moreover, the GFP-tagged single transmembrane domain of VAPB co-immunoprecipitates with YIF1A, suggesting that the transmembrane domain of VAPB is important for binding YIF1A (Figure 2H). To further explore the binding of YIF1A with VAPB, we mutated two GxxxG motifs that are present in the first and third transmembrane domain of YIF1A (Figure 2A). GxxxG motifs are known to facilitate interactions between transmembrane helices and mediate the assembly of the transmembrane helices in VAPB (Kim et al, 2010). However, disruption of these motifs in YIF1A did not interfere with VAPB binding (Figure 2I). These data were confirmed by immunofluorescence in COS-7 cells (Figure 2J–L; Supplementary Figure S1). The results show that the first two transmembrane domains of YIF1A are important for VAPB binding but that this binding does not depend on GxxxG motifs. Together, these data indicate that the transmembrane domains of both VAPB and YIF1A are important for their interaction.
YIF1A recycles between the Golgi and ER and localizes to the ERGIC in hippocampal neurons
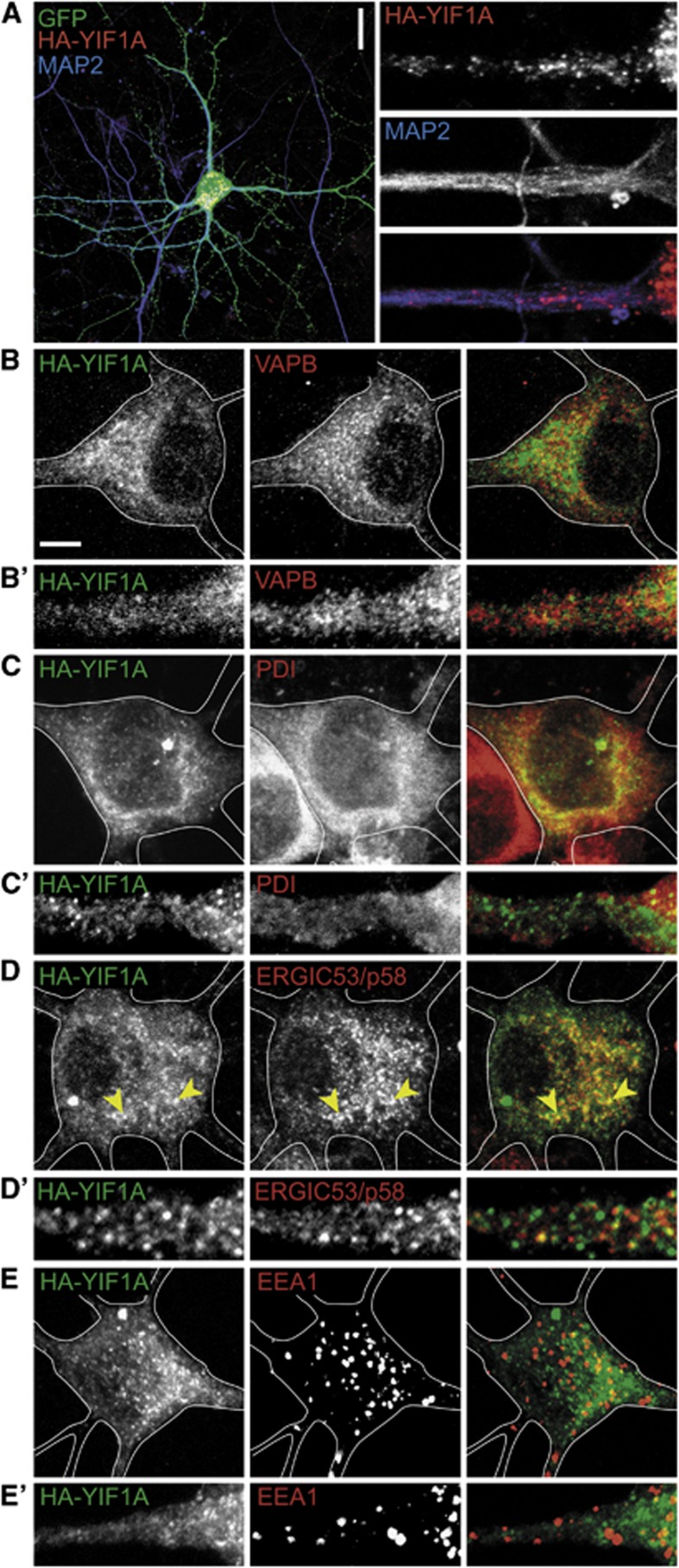
Next, we investigated the function of the VAPB–YIF1A interaction using primary cultured rat hippocampal neurons as a model system. VAPB is mainly localized to the ER in neuronal cells (Teuling et al, 2007), whereas the distribution of YIF1A in neurons is not clear. Immunofluorescent stainings of HA-YIF1A expressing neurons at 16 days in vitro (DIV16) showed that YIF1A is present in a reticular network throughout the neuron and localizes to discrete puncta in the cell body (Figure 3A). The reticular YIF1A staining partially co-localizes with endogenous VAPB and the ER marker protein disulphide isomerase (PDI) (Figure 3B and C). Double labelling with theERGIC marker ERGIC53/p58 showed partial co-localization in the cell body, indicating that the YIF1A-positive puncta coincide with the ERGIC (Figure 3D). In contrast, the endosome marker early endosomal autoantigen 1 (EEA1) showed no co-localization with HA-YIF1A (Figure 3E).
Figure 3.
YIF1A localization in cultured hippocampal neurons.(A) Representative images of rat hippocampal neurons (DIV16) co-transfected with HA-YIF1A and GFP to visualize morphology and labelled with anti-HA (red) and anti-MAP2 (blue) antibodies. Scale bar, 20 μm. In the right panel, a dendritic segment is enlarged to show the presence of HA-YIF1A in proximal dendrites. (B–E) Representative images of rat hippocampal neurons transfected with HA-YIF1A and labelled with anti-HA (green) and anti-VAPB (red in B), anti-PDI (red in C), anti-ERGIC53/p58 (red in D) or anti-EEA1 (red in E). Solid lines indicate the cell edge and arrows showco-localization. Scale bars represent 20 μm in (A) and 5 μm in (B). (B’–E’) Enlargement of dendritic segments to show localization of endogenous proteins and overexpressed YIF1A.
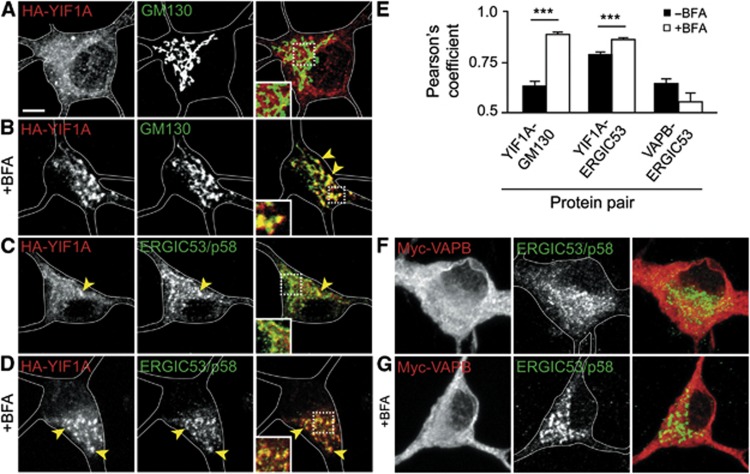
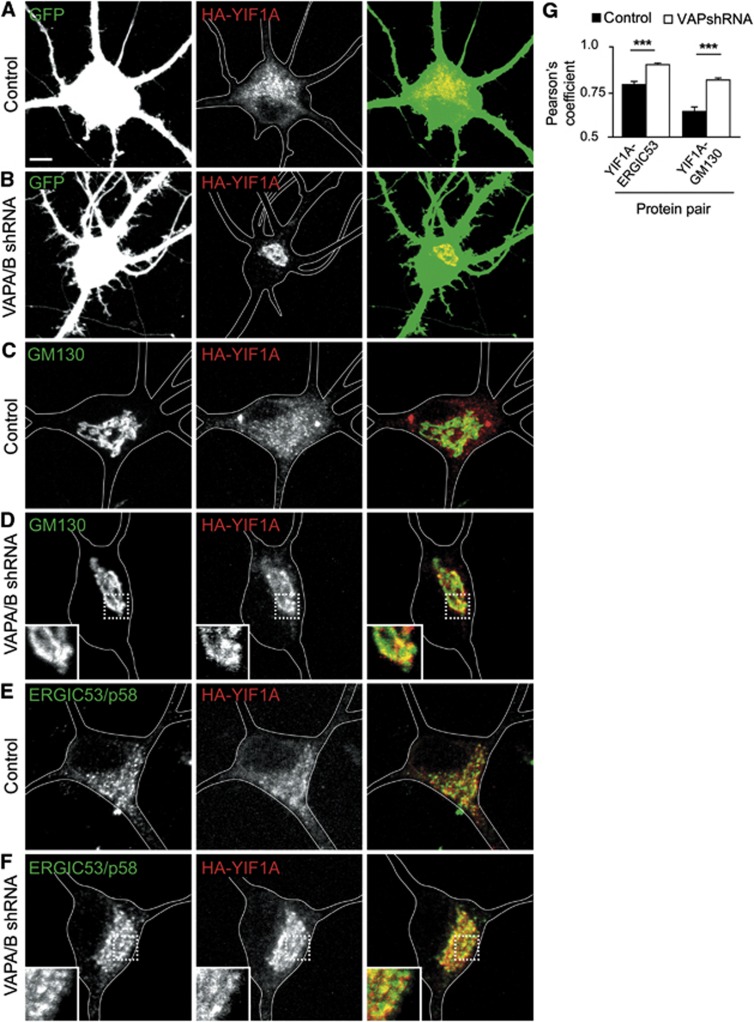
To further verify the co-localization of YIF1A with the ERGIC, we blocked vesicular trafficking between ER and Golgi using brefeldin A (BFA). Co-stainings with cis-Golgi marker GM130 and ERGIC marker ERGIC53/p58 showrespectively little and moderate co-localization with YIF1A in control neurons (Figure 4A, C and E). However, after blocking vesicular trafficking using BFA, YIF1A accumulates in the perinuclear region of the cell body where it stronglyco-localizes with Golgi and ERGIC markers (Figure 4B and D). Analysis of the Pearson’s correlation coefficients of fluorescent signals confirmed that BFA treatment significantlyincreases the co-localization of YIF1A with Golgi and ERGIC (Figure 4E). In contrast, VAPB is present in the ER, not localized to Golgi and ERGIC structures and its localization is unaffected by BFA treatment (Figure 4F, G and E). These results indicate that while VAPB is restricted to the ER, YIF1A recycles between the Golgi and ER and is predominantly present in the ERGIC.
Figure 4.
YIF1A localizes to the ER–Golgi intermediate compartment (ERGIC). (A) Image of the cell body of a hippocampal neuron transfected with HA-YIF1A and stained with anti-HA (red) and anti-GM130 (green) antibodies. (B) Redistribution of HA-YIF1A by BFA treatment. Neurons were transfected with HA-YIF1A, treated with BFA (5 μg/ml) for 15 min fixed and labelled with anti-HA (red) and anti-GM130 (green).(C) Representative image of the cell body of a hippocampal neuron transfected with HA-YIF1A and labelled with anti-HA (red) and anti-ERGIC (green). Redistribution of HA-YIF1A after BFA treatment is shown in (D). (E) Summary of co-localization experiments. Pearson’s coefficient (_r_p) for YIF1A versus ERGIC53/p58, YIF1A versus GM130 and VAPB versus ERGIC53/p58 in control (black bars) and BFA-treated cells (white bars). Twenty-three to twenty-five ROIs were analysed for each condition. Error bars indicate s.e.m., ***P<0.001. (F, G) Images of cell bodies of hippocampal neurons transfected with myc-VAPB and labelled with anti-myc (red) and anti-ERGIC53/p58 (green). BFA treatment has no effect on myc-VAPB distribution (G). Solid lines indicate the cell edges; the insets show magnifications of boxed areas and arrows indicateco-localization. Scale bar, 5 μm.
VAPB retains YIF1A in the ER and inhibits its recycling into ERGIC and Golgi
We next determined whether the specific distribution of YIF1A and VAPB depends on each other’s subcellular localization. We generated YIF1A, YIF1B, VAPA and VAPB shRNAs based on the previously published siRNA sequences to perform knockdown experiments in neurons (Teuling et al, 2007; Carrel et al, 2008). Both VAPA and VAPB shRNAs showed a significant reduction in immunostaining for the respective VAP, indicating an effective knockdown for all VAP-shRNA constructs (Supplementary Figure S2). First, we tested if the VAPB localization in the ER depends on YIF1. In DIV16 neurons, absence of YIF1A/B did not affect the localization of VAPB. Second, we tested if YIF localization depends on VAP using VAPA and VAPB shRNAs. The VAPA/B-shRNA expressing neurons at DIV16 showed a consistent change in YIF1A localization. Whereas control cells have a widespread reticular and punctate YIF1A staining (Figure 5A), VAPA/B knockdown neurons displayed a strong accumulation of YIF1A in the perinuclear region of the cell body (Figure 5B) and a marked co-distribution of YIF1A with cis-Golgi marker GM130 and ERGIC marker ERGIC53/p58. Quantification revealed that the co-localization of YIF1A with GM130 and ERGIC53/p58 is strongly increased in the absence of VAP (Figure 5C–G). These data indicate that knockdown ofVAP in neurons leads to a translocation of YIF1A in post-ER structures.
Figure 5.
Effect of VAP knockdown on YIF1A localization in cultured hippocampal neurons. (A, B) Representative images of cell bodies of hippocampal neurons co-transfected at DIV16 for 4 days with HA-YIF1A (red), GFP and pSuper control vector (A) or pSuper-VAPA and pSuper-VAPB shRNAs (B). VAP knockdown results in relocalization of HA-YIF1A. (C–F) Cell bodies of neurons co-transfected with HA-YIF1A (red) and pSuper control vector (C, E) or pSuper-VAPA and pSuper-VAPB shRNAs (D, F). Neurons were stained for either GM130 (C, D) or ERGIC53/p58 (E, F). Solid lines indicate the cell edges. Scale bar, 5 μm. (G) Summary of co-localization experiments. Pearson’s coefficient (_r_p) for YIF1A versus ERGIC53/p58 and GM130 in control (black bars) and VAP knockdown neurons (white bars). Seventeen to twenty-three ROIs were analysed for each condition. Error bars indicate s.e.m., ***P<0.001.
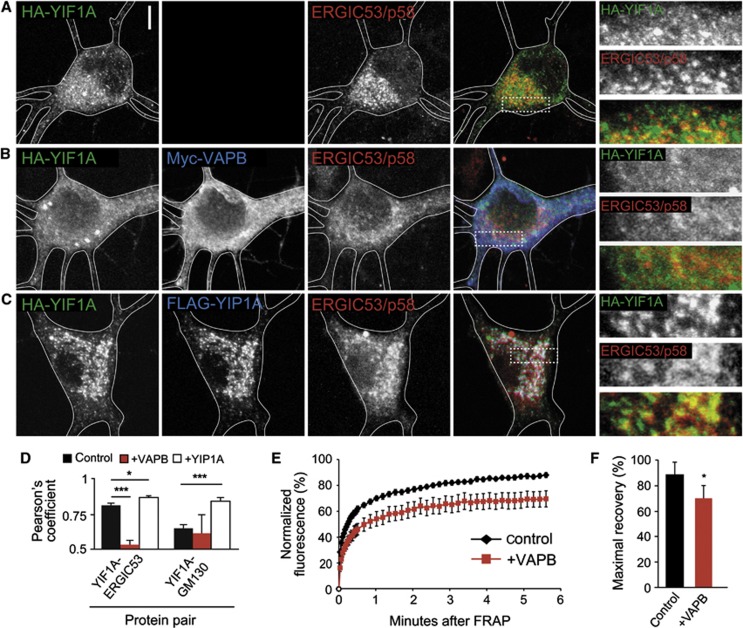
The above data imply that VAPB is important to retain YIF1A in the ER and might control the recycling of YIF1A from the ER to the ERGIC and Golgi. To test this hypothesis, we overexpressed myc-VAPB together with HA-YIF1A. Indeed, YIF1A shows a strong reticular distribution throughout the neuron and loses its characteristic ERGIC localization (Figure 6A, B and D). Next, we tested whether a previously characterized interacting partner of YIF1A, YIP1, also retains YIF1A in the ER. YIP1 has been shown to localize to ER and Golgi membranes as well as coat protein complex II (COPII) transport vesicles in yeast (Yang et al, 1998; Matern et al, 2000; Heidtman et al, 2005; Jin et al, 2005). In hippocampal neurons, Flag-YIP1A has moderate Golgi and a pronounced ERGIC localization (Figure 6C). In contrast to VAPB, Flag-YIP1A overexpression increased YIF1A in the Golgi and ERGIC, while reducing its ER localization, thus having the opposite effect of VAPB (Figure 6C and D). Next, we used fluorescence recovery after photobleaching (FRAP) to examine the mobility of GFP-YIF1A molecules under the influence of increased VAPB levels. A 3-by-3 μm region of the cell soma was bleached by high laser power and fluorescence intensity was measured over a period of ∼5 min (Figure 6E). GFP-YIF1A fluorescence recovered rapidly in control neurons and reached a maximal recovery of ∼90% within this time frame. In neurons co-expressing myc-VAPB the maximal recovery of GFP-YIF1A is decreased to ∼70% (Figure 6E and F), indicating an increase in the immobile fraction of YIF1A molecules in the ER. Taken together, these data suggest that the ER-resident protein VAPB indeed binds YIF1A in the ER and thereby inhibits its recycling to the ERGIC and Golgi.
Figure 6.
VAPB overexpression relocalizes YIF1A. (A–C) Hippocampal neurons transfected with HA-YIF1A and stained with anti-HA (green) and anti-ERGIC53/p58 (blue). Co-transfection with either myc-VAPB (B) or FLAG-YIP1A (C) results in relocalization of HA-YIF1A. Solid lines indicate the cell edges and panels on the right side show enlargements of the boxed regions. Scale bar, 5 μm. (D) Summary of co-localization experiments. Pearson’s coefficient (_r_p) for YIF1A versus ERGIC53/p58 and GM130 in control (black bars), VAPB overexpressing (red bars) and YIP1A overexpressing neurons (white bars). Thirteen to twenty-three ROIs were analysed for each condition. (E) Fluorescent recovery plots showing the rates of GFP-YIF1A recovery in cell bodies of control neurons and neurons overexpressing VAPB. Fluorescent intensity was normalized to intensity before bleaching. _P_=0.008; repeated measures ANOVA. (F) Histogram representing the maximal recovery of fluorescence (estimated mobile fraction) in hippocampal neurons expressing GFP-YIF1A with (_n_=11) or without VAPB overexpression (_n_=10). Data are presented as means±s.e.m., *P<0.05, ***P<0.001.
YIF and VAP are required for normal dendrite morphology
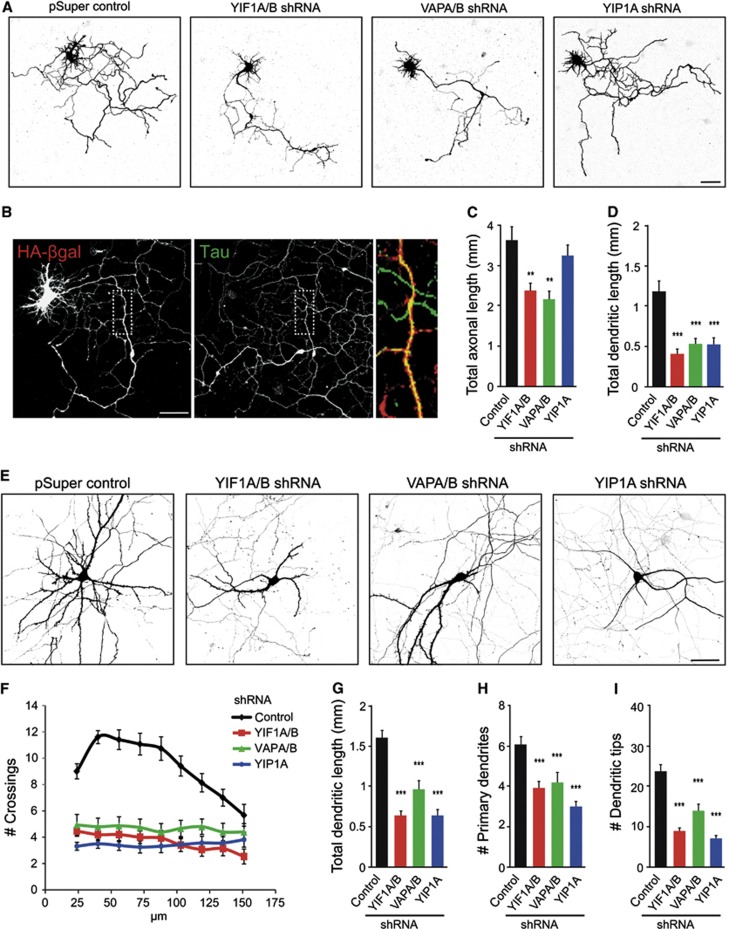
Studies in Drosophila neurons and recently in zebrafish suggest that VAPB has an important role in the transport of proteins to the axon (Yang et al, 2012; Forrest et al, 2013) and that loss of VAP proteins affects neuron morphology (Pennetta et al, 2002; Tsuda et al, 2008; Forrest et al, 2013). To assess the morphological effects of depleting YIF1, YIP1 and VAP proteins, we analysed the length of axons and dendrites in hippocampal neurons transfected at DIV1 with VAPA/B-shRNAs, YIF1A/B-shRNAs or YIP1A-shRNA, together with β-galactosidase to visualize neuron morphology (Figure 7A). Neurons were fixed at DIV5 and immunofluorescent staining with antibody against tau was used to distinguish the axon from the dendrites (Figure 7B). Depletion of YIF1 and VAP, but not YIP1, caused a significant decrease in axonal length (Figure 7C). Previous data showed that ER-to-Golgi transport is particularly important for dendrite morphology (Ye et al, 2007). Since YIF1, YIP1 and VAP proteins localize to the early secretory pathway in hippocampal neurons we wondered whether depletion of these proteins had any effect on dendrite morphology. Indeed, DIV5 neurons showed a significant decrease in dendritic length when transfected with VAPA/B-shRNAs, YIF1A/B-shRNAs and YIP1A-shRNA (Figure 7D). Because in DIV5 neurons dendrites are short and hardly branched we used mature neurons to examine more closely the effect of VAP, YIF1 and YIP1 depletion on dendrite morphology. At DIV19, VAPA/B-shRNAs, YIF1A/B-shRNAs and YIP1A-shRNA transfected neurons showed a striking dendritic phenotype (Figure 7E). Quantitative analysis of the pattern of dendritic branching by Sholl analysis (Sholl, 1953) revealed a simplified dendritic tree with reduced branching in proximal dendrites in neurons transfected with VAPA/B-shRNAs, YIF1A/B-shRNAs and YIP1A-shRNA (Figure 7F). The reduced dendritic complexity in the VAPA/B, YIF1A/B and YIP1A knock-down neurons was also revealed by measuring the total dendritic length, counting the number of primary dendrites directly emanating from the soma, and analysing the total number of dendritic tips (Figure 7G–I). Similar results were obtained with independent VAPA, VAPB, YIF1A and YIF1B shRNA sequences (Supplementary Figure S3). In summary, the early secretory pathway components VAPA/B, YIF1A/B and YIP1A are required for normal dendrite morphology in early and later stages of dendrite development.
Figure 7.
YIF1 and VAP are required for normal dendrite morphology. (A) Hippocampal neurons co-transfected at DIV1 with indicated constructs and β-galactosidase to visualize morphology. (B) Representative image of a hippocampal neurons (DIV5) co-transfected with empty pSuper and β-galactosidase and co-stained with Tau (green) to highlight the axon. (C, D) Quantification of total axonal length and dendritic length after 4 days overexpression of knockdown constructs or empty pSuper as control (14–16 cells were analysed for each condition). (E) Hippocampal neurons co-transfected at DIV15 with indicated constructs and β-galactosidase to visualize morphology. (F) Sholl analysis and quantification of the total dendritic length (G), number of primary dendrites (H) and dendritic tips (I) after 4 days overexpression of knockdown constructs or empty pSuper as control (15–17 cells were analysed for each condition). Error bars indicate s.e.m., **P<0.01, ***P<0.001. Scale bars represent 50 μm.
YIF and VAP are required for intracellular membrane delivery into dendrites
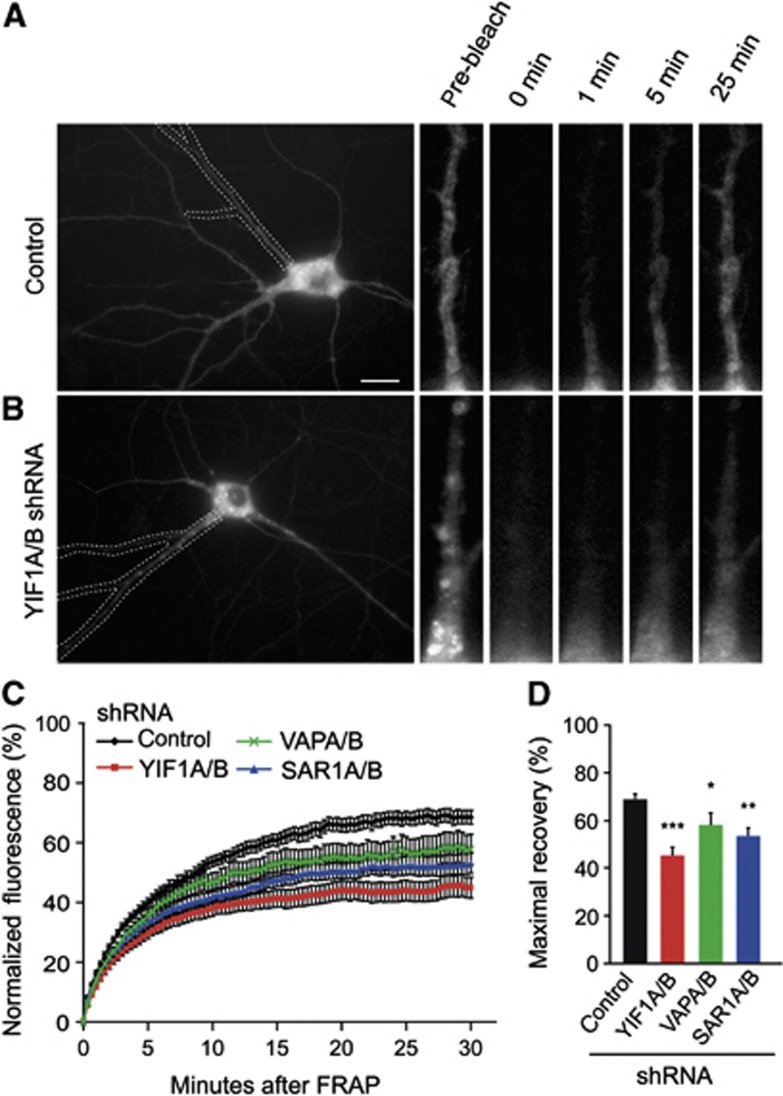
The trafficking of new membrane proteins is essential for normal dendritic growth and morphology (Lecuit and Pilot, 2003). We next determined whether YIF1 and VAP proteins play a role in the delivery of vesicles or cellular material to dendrites. Recent papers show that YIF1B mediates the transport of the serotonin G-coupled receptor (5-HT1A) to dendrites (Carrel et al, 2008; Al Awabdh et al, 2012), however after transfection with VAPA/B-shRNAs or YIF1A/B-shRNAs we do not find changes in YFP-5HT1A receptor localizationto mature dendrites (Supplementary Figure S4). Previous studies showed that the small GTPase Sar1, a critical regulator of ER-to-Golgi transport, controls dendritic growth through the delivery of membrane vesicles to dendrites (Ye et al, 2007). We therefore hypothesized that the observed morphological phenotype is caused by a decrease in membrane supply to the dendrites. To see whether VAP and YIF knockdown affect membrane trafficking in primary hippocampal neurons, we employed FRAP experiments. To examine the membrane supply from the soma to the dendrites, we marked membrane with the fluorescent transmembrane protein CD8-GFP (Hoogenraad et al, 2005; Ye et al, 2007). After photo-bleaching one entire dendrite the fluorescence recovery in the same dendrite was monitored immediately over a time period of 20 or 30 min. The fluorescence recovery, due to supply from the soma, in the proximal dendrite (35 μm from the soma) was used for quantifications. In control neurons, CD8-GFP fluorescence recovered to ∼70% (Figure 8A, C and D; Supplementary Figure S5). The recovery could be fit with a double exponent (f) with 2 time constants (τ) of 36±5 s and 476±15 s (Supplementary Figure S5). To determine the source of GFP-CD8 recovery, we conducted several control experiments that indicate that the majority of GFP-CD8 recovery depends on intracellular trafficking in the secretory pathway but not on surface diffusion or local protein synthesis (Supplementary Figure S5). Next, we depleted VAP, YIF1 or SAR1 to test whether there is an effect on membrane delivery to dendrites. Sar1-shRNA as well as VAPA/B and YIF1A/B-shRNAs significantly reduced the delivery of CD8-GFP from the soma to the dendrites to ∼40–60%, as evident from the recovery curve (Figure 8B and C) as well as the maximal recovery (Figure 8D). These results show that VAP and YIF are required for the intracellular delivery of membrane, labelled by CD8-GFP, from soma to dendrites.
Figure 8.
YIF1 and VAP play a role in membrane trafficking in primary hippocampal neurons. (A, B) Representative images of membrane-bound GFP (CD8-GFP) moving from soma into dendrites in hippocampal neurons (DIV16–19) co-expressing CD8-GFP with either empty pSuper control vector (A) or YIF1A and YIF1B shRNAs (B). Boxed area indicates photobleached dendritic region. In the right panel, proximal part of photobleached dendrite is shown. Scale bar, 20 μm. (C) Fluorescent recovery plots showing the rates of CD8-GFP recovery in photobleached dendrites of control and knockdown neurons. Fluorescent intensity was normalized to intensity before bleaching. P<0.001 for YIF1A/B versus control, _P_=0.033 for VAPA/B versus control and _P_=0.008 for SAR1A/B versus control (repeated measures ANOVA followed by Tukey’s post hoc test). (D) Maximal fluorescence recovery. Nine to fifteen cells were analysed for each condition. Data are presented as means±s.e.m., *P<0.05, **P<0.01, ***P<0.001; one-way ANOVA, Tukey’s post hoc test.
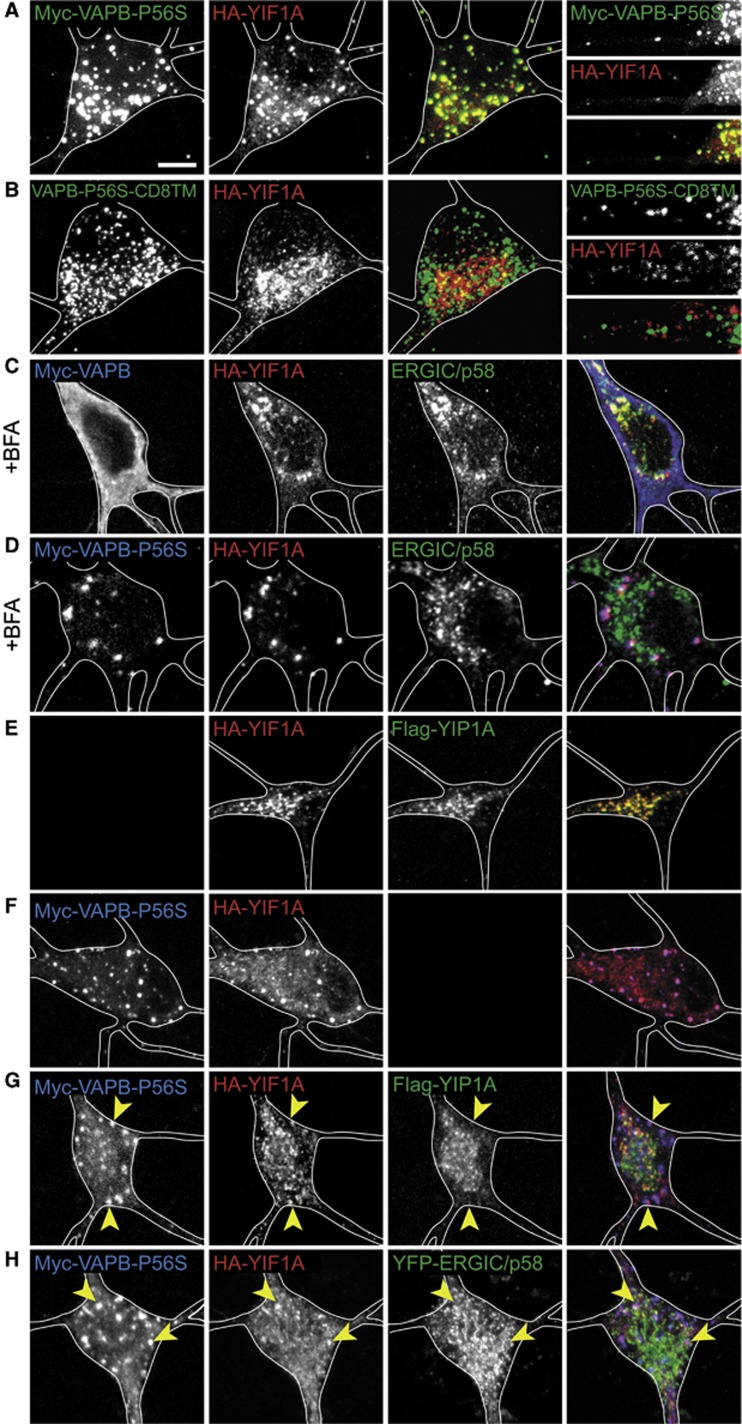
VAPB-P56S traps YIF1A in aggregates and thereby affects its localization
The P56S mutant form of VAPB is associated with motor neuron disorder ALS8. VAPB-P56S proteins aggregate and form multiple spherical inclusions in cellular and invertebrate model systems. As in COS-7 cells (Figure 1F), HA-YIF1A is recruited to the myc-VAPB-P56S inclusions in neurons (Figure 9A). Consistent with the data that the interaction depends on the transmembrane domain of VAPB-P56S,replacement of this region with a different, unrelated transmembrane domain prevents YIF1A recruitment to VAPB inclusions (Figure 9B). In cells transfected with wild-type VAPB, YIF1A strongly co-localized to the ERGIC after BFA treatment (Figures 4D and 9C). Interestingly, in VAPB-P56S expressing cells, YIF1A is absent from the ERGIC structures even after BFA treatment (Figure 9D). These data suggest that when VAPBP-P56S is present, YIF1A loses its localization to the ERGIC. In contrast, the YIF1A binding protein Flag-YIP1A and YPF-ERGIC53/p58 do not co-localize with these VAPB-P56S/YIF1A-positive aggregates (Figure 9E–H).
Figure 9.
Mutant VAPB expression results in recruitment of YIF1A to clusters. (A) Co-transfection of hippocampal neurons with HA-YIF1A (red) and myc-VAPB-P56S (green) shows that YIF1A is recruited to the VAPB-P56S clusters. The far right panel shows enlargement of a dendrite. (B) Hippocampal neuron co-transfected with VAPB-P56S-CD8TM (green) and HA-YIF1A (red). (C, D) Images show that in the presence of overexpressed wild-type VAPB, HA-YIF1A localizes to ERGIC after BFA treatment. However, when VAPBP-P56S is expressed YIF1A loses its localization to the ERGIC. (E–G) Neurons co-transfected with HA-YIF1A (green) and Flag-YIP1A (D), myc-VAPB-P56S (E) or both (F). (H) Hippocampal neuron co-transfected with myc-VAPB-P56S (blue), YFP-ERGIC (green) and HA-YIF1A (red). Scale bar, 5 μm.
The MSP domain in VAP proteins binds to the ‘two phenylalanines in an acid tract’, or ‘FFAT’ motif found in several cytoplasmic lipid-binding proteins, including OSBPL9 and NIR2 (Kaiser et al, 2005; Loewen and Levine, 2005; Teuling et al, 2007). In this way, VAP proteins may act as docking sites for cytoplasmic factors to interact with the ER, coordinate lipid transfer between ER and Golgi apparatus or maintain the structure of the ER by interacting with the cytoskeleton (Amarilio et al, 2005; Peretti et al, 2008; Ngo and Ridgway, 2009). It was recently shown that the P56S substitution in VAPB induces conformational changes within the MSP domain and perturbs FFAT-motif binding (Teuling et al, 2007; Kim et al, 2010). Consistent with these results, FFAT-motif containing proteins OSBPL9 and NIR2 are absent from YIF1A-positive VAPB-P56S aggregates in neurons (Supplementary Figure S6). Together, these results suggest that both perturbation of FFAT-dependent association with the ER and mislocalization of YIF1A could contribute to the pathological mechanisms observed in ALS8.
Discussion
In this study, we identified YIF1A as a new VAPB binding partner and we showed that, in contrast to other VAPB interaction partners, YIFIA does not bind to the MSP domain of VAPB, but via its transmembrane domain interacts with the transmembrane domain of VAPB. In hippocampal neurons, we showed that VAPB–YIF1A interaction controls the shuttling of YIF1A between the ERGIC and the ER and plays an important role in early secretory events in neurons, promoting membrane trafficking and normal dendritic growth. In addition, we show that YIF1A in contrast to other VAPB interaction partners also binds ALS8-mutant VAPB andco-accumulates in abnormal ER-derived structures. We propose that the mislocalization of YIF1A into VAPB-P56S aggregates could contribute to pathological mechanisms observed in VAPB-associated motor neuron diseases.
YIF1A interacts with VAPB in the early secretory pathway
VAPB has been implicated in multiple cellular functions, either as a conserved ER anchoring protein or as a secreted signalling molecule in invertebrates (Lev et al, 2008; Han et al, 2012). Proteins with an FFAT or an FFAT-like motif that can bind the FFAT-motif binding site in the MSP domain of VAPA and VAPB represent a major group of VAPB interacting proteins. FFAT-motif proteins include oxysterol binding proteins (OSBPs), OSBP-related proteins (ORPs) and NIR proteins (Wyles and Ridgway, 2004; Amarilio et al, 2005; Loewen and Levine, 2005; Kawano et al, 2006) and have been shown to play a role in ER structure (Wyles et al, 2002; Amarilio et al, 2005; Lehto et al, 2005) and the transport of membrane lipids (Kawano et al, 2006; Peretti et al, 2008). Other proteins with FFAT-like motifs include for instance PKA anchoring proteins that could play a role in cAMP signalling on the ER (Mikitova and Levine, 2012). In addition, an outer mitochondrial membrane protein (PTPIP51) was shown to interact with VAPB (De Vos et al, 2012) putatively via FFAT-like motifs, although the basis for this interaction was not studied (Mikitova and Levine, 2012). In contrast to the previously described VAP binding partners, the interaction with YIF1 occurs via the transmembrane domain, although we cannot fully exclude a role for the cytosolic VAPB domain. During submission of this paper, another study reported a similar transmembrane mediated interaction between Drosophila VAP and Sac1, a phosphoinositide phosphatase (Forrest et al, 2013). YIF1A and YIF1B in mammals are homologous to Yif1p in yeast. Yif1p is located to the Golgi membrane and COPII vesicles, forms a tight complex with its family member Yip1p and plays an important role in early secretory transport in yeast (Entian et al, 1999; Matern et al, 2000; Otte et al, 2001). Cell-free assays and thermosensitive yeast strains demonstrated that Yif1p and Yip1p play a critical role in the biogenesis of ER-derived COPII transport vesicles (Barrowman et al, 2003; Heidtman et al, 2005). Recent genome-wide RNA interference screens in Drosophila S2 and HeLa cells found that depletion of YIF1 or YIP1 inhibits secretion, indicating that YIF1/YIP1 are functionally conserved components of the secretory pathway (Wendler et al, 2010; Simpson et al, 2012). We find that YIF1A is predominantly localized to the ERGIC and recycles between the ER and Golgi in hippocampal neurons. In agreement with these results, proteomics analysis identified YIF1A in ERGIC-enriched membranes (Breuza et al, 2004) and BFA treatment of HeLa cells results in YIF1A accumulations in the ERGIC (Yoshida et al, 2008). Our data indicate that both VAPB and YIF1A are needed for an efficient transition of membrane cargo through the early secretory pathway in neurons. Furthermore, we show that, while YIF1A is cycling between ER and Golgi, VAPB is restricted to the ER and is not present in ERGIC or Golgi, indicating that YIF1A-VAPB binding is most likely to occur in the ER membrane (Supplementary Figure S7). We hypothesize that YIF1A is retained in the ER membrane by its interaction with VAPB which subsequently allows specific membrane lipids and/or cargo proteins to concentrate at ERES before budding off in carriers destined for further transport towards the ERGIC and cis-Golgi membrane (Gurkan et al, 2006; Jensen and Schekman, 2011). Moreover, interactome analyses in yeast revealed that the Yip1p/Yif1p family proteins associate with a large range of gene products that function in the vesicular transport pathway, including SNAREs, Rab GTPases, ARFGAP and sorting nexins (Ito et al, 2000; Uetz et al, 2000). Future studies using cell-free assays driven by purified proteins that recapitulate protein transport between the ER and the Golgi complex will help to elucidate the mechanisms regulating the interplay between VAPB, YIF1A and the other components of the ER-to-Golgi trafficking pathway and advance our understanding of the biogenesis of transport carriers in the early secretory transport.
YIF and VAP are required for intracellular membrane delivery into dendrites
The early secretory pathway is fundamentally important for neuronal development and function (Hanus and Ehlers, 2008; Tang, 2008; Aridor and Fish, 2009; Jan and Jan, 2010), and some lines of evidence suggest that the organization of secretory trafficking in neurons differs in several ways from the pathways in non-neuronal cells (Nelson and Yeaman, 2001; Stephens and Pepperkok, 2001; Tang, 2008). In particular components of the ER, ERGIC and Golgi have been identified in dendrites, specifically localizing at branch points and close to dendritic spines (Krijnse-Locker et al, 1995; Pierce et al, 2001; Horton et al, 2005; Appenzeller-Herzog and Hauri, 2006). Here, we identify VAPB and YIF1A as early secretory trafficking components that are required for membrane transport and important for normal dendrite morphology. First, we showed that VAPB is able to retain YIF1A in the ER thereby regulating its recycling to ERGIC and Golgi. Depletion of VAP proteins results in a relocalization of ERGIC protein YIF1A to the Golgi, indicating that VAPB regulates trafficking of YIF1A through the ERGIC. Recent studies showed that disruption of VAPB affects ER-to-Golgi transport and anterograde VSVG trafficking in HeLa cells (Peretti et al, 2008; Rao et al, 2012). Second, depletion of VAP, YIF1 as well as YIP1 affects dendrite morphology. This is consistent with the observed dendritic phenotype in secretory trafficking deficient Drosophila mutants (Ye et al, 2007). Third, we show that both VAPA/B and YIF are required for a normal membrane supply to dendrites, indicating that the observed effect on dendrite morphology is caused by a defect in early secretory trafficking. The fact that other proteins of the ER-to-Golgi trafficking pathway, such as Sar1, also affect dendritic growth and membrane trafficking (Ye et al, 2007) strengthens the model that VAPB and YIF1A have an important role in the secretory transport. Interestingly, YIF1B has been shown to play a role in the neuronal ER–Golgi trafficking machinery by specific targeting the serotonin G-coupled receptor (5-HT1A) to dendrites (Carrel et al, 2008). A recent follow-up study proposed that YIF1B acts as a scaffolding complex that recruits 5-HT1A together with Yip1A and Rab6 in dendritic transport vesicles (Al Awabdh et al, 2012). These results imply that at least YIF1B and possibly other Yip1p/Yif1p family proteins play a role in the anterograde transport of a limited subset of cargo molecules from the ER to the Golgi. Although in our study we do not find evidence that YIF1 or VAP plays a role in the transport of the serotonin receptor we believe that defects in the transport of specific proteins under the influence of VAP or YIF impairments are difficult to measure as other, unconventionally, transport routes can be used as default (Grieve and Rabouille, 2011).
YIF1A and ALS8
The P56S mutation in the gene encoding VAPB causes ALS8 and some other related forms of motor neuron disease (Nishimura et al, 2004). Several studies already showed that VAPB-P56S induces the formation of abnormal ER-derived inclusions, perturbs ER–Golgi trafficking and triggers ER stress (Nishimura et al, 2004; Teuling et al, 2007; Tsuda et al, 2008; Suzuki et al, 2009; Chen et al, 2010; Fasana et al, 2010; Moumen et al, 2011). Ubiquitin (Moumen et al, 2011) and BAP31 (Fasana et al, 2010) have been described to accumulate in the VAPB-P56S clusters, suggesting that mutant VAPB-induced aggregation is part of the ER-associated degradation pathway. Mutant VAPB-P56S also induces the co-aggregation of wild-type VAPB, suggesting a dominant-negative mode of pathogenesis (Teuling et al, 2007). Disruption of ER and Golgi structure and function has previously been suggested as a possible pathological mechanism for neurodegenerative diseases (Mourelatos et al, 1996; Lehotsky et al, 2003; Paschen and Mengesdorf, 2005; Vlug et al, 2005; Yoshida, 2007). How VAPB-P56S disrupts protein trafficking between the ER and Golgi and how this may lead to the pathogenesis of ALS8 is not yet understood. The identification of the VAPB–YIF1A interaction and its role in early secretory transport raises the possibility that YIF1A might be involved in the development of neurodegenerative diseases. Both YIF1A and YIF1B are ubiquitously expressed in the CNS (Carrel et al, 2008) and a microarray analysis showed downregulation of YIF1A in motor neurons isolated from the spinal cord of SOD1-G93A transgenic mice, a model for familial ALS (Ferraiuolo et al, 2007). This suggests that YIF1A could be a potential factor involved in the multi-factorial causes leading to ALS. Although the relevance of our observations to heterozygous patients remains uncertain (as our observations are under overexpression conditions), our model suggests that mislocalization of YIF1A to aggregates leads to a defect in secretory trafficking and a subsequent inhibition of dendritic growth and maintenance. Defects in neuronal secretory trafficking already have been shown to affect dendritic growth (Horton et al, 2005; Ye et al, 2007). Dendritic alterations have been documented in mouse models for motor neuron dysfunction (Wiggins et al, 2012) and in ALS-affected patients (Liu et al, 2011). In patients, dendrites in anterior horn cells are shorter and thinner, or are lost (Kato et al, 1987; Sasaki and Iwata, 1996). Moreover, loss and atrophy of dendrites was found in motor neurons in the spinal cord (Karpati et al, 1988). Similarly, in pre-symptomatic SOD1-G93A transgenic mice dendrites in the motor cortex show signs of degeneration and in the prefrontal cortex a reduction in the length of basal dendrites and branch points was observed (Sgobio et al, 2008; Jara et al, 2012). Changes in dendritic architecture are indicative of defects in neuronal function and connectivity, and could be contributing to the development of sites of degeneration. Though there is overwhelming evidence for axonal transport dysfunctions in the pathogenesis of ALS (Chevalier-Larsen and Holzbaur, 2006; Sau et al, 2011), our study indicates that alterations in dendritic structure and trafficking may also play an important role in neurodegeneration. A recent study has reported mild late onset motor deficits in VAPB-deficient mice in the absence of axonal and neuromuscular junction abnormalities (Forrest et al, 2013). Whether these mice develop dendritic abnormalities and the degree of compensation of VAPB deficiency by VAPA in these mice requires further investigation.
In conclusion, we have identified YIF1A as a novel binding partner of VAPB and established a key role for these proteins in the neuronal early secretory pathway. We found that both VAP and YIF1 proteins are important for proper membrane trafficking and normal dendrite growth. Moreover, the ALS-linked mutant VAPB-P56S recruits YIF1A to aggregates and disrupts its normal localization to ERGICs. Understanding the cellular function of VAPB may indicate what molecular and cellular events are associated with the disease process of ALS8. Our current findings provide new molecular targets to investigate VAPB-linked neurodegeneration. It is likely that this information will be of relevance to both the inherited condition and the more common sporadic forms of disease.
Materials and methods
Expression constructs and shRNA
The following mammalian expression plasmids have beendescribed previously: bio-HA-VAPB, GFP-VAPB-TMD, GFP-VAPB, HA- and myc-tagged VAPA, VAPB and VAPA/B-P56S constructs (Teuling et al, 2007), protein-biotin ligase BirA (Lansbergen et al, 2006), flag-YIF1B (Carrel et al, 2008), flag-YIP1A (Dykstra et al, 2010), ERGIC/p58-YFP (Ward et al, 2001) and CD8-GFP (Hoogenraad et al, 2005). Bio-GFP-VAPB, Bio-HA-VAPB-P56S and Bio-HA-VAPA were generated by incorporating a biotinylation-tag (MSGLNDIFEAQKIEWHE) before the GFP- or HA-tagged mutant or wild-type VAPA/B construct. VAPB-P56S-CD8TM was made by removing the transmembrane domain of VAPB-P56S and adding the transmembrane domain of CD8 with a PCR-based strategy using HA-VAPB-P56S and GFP-CD8 as a template and subcloned into a pβactin expression vector. Full-length and truncated human YIF1A constructs were generated by PCR using IMAGE clone 3451489 as template and cloned into HA- and GFP-tagged pGW1-expression vectors. For bio-GFP-YIF1A, a biotinylation tag was inserted in front of the pEGFP-C2 (Clontech) and the YIF1A openreading frame subsequently subcloned in the biotin-tag-GFP vector. GFP-YIF1A IxxxI was generated from GFP-YIF1A by introducing G154I, G158I, G222I and G226I mutations using a PCR-based strategy. GFP-YIF1B was generated by a PCR-based strategy using flag-YIF1B and subcloned into a GFP-tagged pGW1-expression vector. Sp-BirA was made by incorporating the Ig kappa light chain leader sequence (METDTLLLWLLLLWVPGSTG) at the N-terminus of BirA and myc tag at its C-terminus in a pSCT expression vector. The following shRNA sequences were used in this study:VAPA#1 and #2 and VAPB#1 shRNA constructs (Teuling et al, 2007) and rat VAPB#2 (5′-GTTTATGGTTCAGTCTATG-3′), YIF1A#1 (5′-CCATGGCCTTCATCACATA-3′), rat YIF1B#1 (5′-gtactcatgtactggctca-3′), rat YIF1B#2 (5′-GCCATGGCTTTCATAACCT-3′), rat SAR1A (5′-AACCACTCTTCTTCACAT-3′) and rat SAR1B (5′-AACTACCTTCCTGCTATCA-3′) sequences were designed based on previously published sequences (Kanekura et al, 2006; Ye et al,2007; Carrel et al, 2008). The targeting sequences for rat YIP1A (5′-GCAGTATGCTGGCTGTGAG-3′) and YIF1A#2 (5′-gaagctagggctattggtc-3′) were designed by using the siRNA selection program at the Whitehead Institute for Biomedical Research ( http://jura.wi.mit.edu/bioc/siRNAext/home.php) (Yuan et al, 2004). The complementary oligonucleotides were annealed and inserted into a pSuper vector (Brummelkamp et al, 2002). Unless otherwise specified, all VAP and YIF shRNAs presented in the paper refer to shRNAs#1.
Antibodies and reagents
The following antibodies were used for immunocytochemistry: rabbit-anti-VAPB and rabbit-anti-VAPA (1:500) (Teuling et al, 2007); NIR2 (1:500) (Litvak et al, 2004), OSBPL9 (1:600) (Ngo and Ridgway, 2009); human-anti-EEA1 (1:500) (Selak et al, 2000). The following antibodies were obtained from commercial sources: mouse-anti-tau (1:500, Chemicon); mouse-anti-disulphide isomerase (PDI; 1:300, Affinity BioReagents); mouse-anti-GM130 (1:1000, BD Biosciences); mouse-anti-HA (1:500, Roche); mouse-anti-MAP2 (1:2000, Sigma); mouse-anti-myc (1:200, Santa Cruz Biotechnology); mouse-anti-flag (1:2000, Sigma); rabbit-anti-ERGIC53/p58 (1:200, Sigma); rabbit-anti-HA (1:500; Santa Cruz Biotechnology); rabbit-anti-flag (1:2000, Sigma); rabbit-anti-myc (1:200; Cell Signaling Technology); rabbit-anti-β-galactosidase (1:2000, MP Biomedicals); rat-anti-HA (1:200, Roche); mouse-anti-CD8 (1:20, Mabtech); Alexa Fluor 488-, Alexa Fluor 598-, and Alexa Fluor 633-conjugated secondary antibodies (1:400, Invitrogen) and Cy5-conjugated secondary antibody (1:400, Jackson ImmunoResearch Labs). The following antibodies were used for western blot analysis: rabbit anti-GFP (1:1000, Abcam); rabbit anti-HA (1:500, Santa Cruz); mouse anti-HA (1:500, Covance) and HRP-conjugated secondary antibodies (1:5000, Dako). BFA and cycloheximide were obtained from Sigma.
Transfection and immunofluorescence of cultured COS-7 cells
COS-7 cells were cultured in DMEM/Ham’s F10 (50/50%) medium containing 10% FCS and 1% penicillin/streptomycin. Two days before transfection, cells were plated at 1:30 in Lab-tek chamber slides (Nunc). Cells were transfected with Fugene6 (Roche) according to manufacturer’s protocol and incubated overnight. Cells were fixed in 4% paraformaldehyde for 10 min at room temperature followed by 5 min in 0.1% Triton X-100 in PBS. Slides were blocked in 0.5% BSA/0.02% glycine in PBS and labelled with primary antibody for 2 h at room temperature. Slides were washed three times with 0.05% Tween-20 in PBS, labelled with secondary antibodies for 1 h at room temperature, washed three times with 0.05% Tween-20 in PBS and mounted using Vectashield mounting medium (Vector laboratories). Images were acquired using a Leica DMRBE microscope equipped with × 40 and × 100 oil objectives.
Animals
All animal experiments were performed in compliance with the guidelines for the welfare of experimental animals issued by the Federal Government of The Netherlands. All animal experiments were approved by the Animal Ethical Review Committee (DEC) of the Erasmus Medical Center and Utrecht University.
Hippocampal neuron cultures, transfection and immunohistochemistry
Primary hippocampal cultures were prepared from embryonic day 18 (E18) rat brains (Banker and Goslin, 1988; Kapitein et al, 2010). Cells were plated on coverslips coated with poly-L-lysine (35 μg/ml) and laminin (5 μg/ml) at a density of 75 000/well. Hippocampal cultures were grown in Neurobasal medium (NB) supplemented with B27, 0.5 mM glutamine, 12.5 μM glutamate and penicillin/streptomycin. Hippocampal neurons were transfected using Lipofectamine 2000 (Invitrogen). Briefly, DNA (3.6 μg/well) was mixed with 3 μl of Lipofectamine 2000 in 200 μl of NB, incubated for 30 min, and then added to the neurons in NB at 37°C in 5% CO2 for 45 min. Next, neurons were washed with NB and transferred in the original medium at 37°C in 5% CO2. After 2–4 days of transfection, neurons were fixed with 4% paraformaldehyde/4% sucrose inPBS, washed three times in PBS for 10 min and incubatedwith the indicated primary antibodies in GDB buffer (0.2% BSA, 0.8 M NaCl, 0.5% Triton X-100, 30 mM phosphate buffer, pH 7.4) overnight at 4°C. Neurons were then washed three times in PBSfor 30 min, incubated with secondary antibodies in GDB for 1 hat room temperature and washed three times in PBS for 30 min. Slides were mounted using Vectashield mounting medium (Vector Laboratories). Images for co-localization measurements were acquired using a Nikon microscope equipped with a × 100 oil objective. Confocal images were acquired using a LSM510 confocal microscope (Zeiss) with a × 40 or × 63 oil objective.
Immunoprecipitation
HEK293T cells were cultured in DMEM/Hams-F10 (50/50%) medium containing 10% FCS and 1% penicillin/streptomycin and were transfected using Lipofectamine2000 (Invitrogen). Cells were harvested 24 h after transfection, by scraping the cells in ice-cold PBS and lysing cell pellets in lysis buffer (50 mM Tris–HCl, 100 mM NaCl, 1.0% Triton X-100 and protease inhibitors (Roche). Supernatant and pellet fractions were separated by centrifugation at 13 200 r.p.m. for 5 min. Supernatants were mixed with an equal amount of lysis buffer, protein-A-agarose beads (GE Healthcare), and 3 μg of rabbit anti-GFP, mouse anti-HA or control IgG (Sigma). Samples were incubated 4 h while rotating at 4°C, centrifuged at 2000, r.p.m. and pellets were washed three times with lysis buffer. Samples were mixed with 4 × Sample Buffer (8% SDS, 25% Glycerol, 0.05 M Tris pH 6.8, 200 mM DTT, 40 mg/l Bromophenol Blue) and boiled. Equal amounts of protein were loaded onto SDS–PAGE gels and subjected to western blotting on polyvinylidene difluoride membrane. Blots were blocked with 2% bovine serum albumin/0.05% Tween-20 in PBS and incubated with primary antibodies at 4°C overnight. Blots were washed with 0.05% Tween-20 in PBS three times for 10 min at room temperature and incubated with secondary antibodies conjugated to horseradish peroxidase (Dako). Blots were developed with enhanced chemiluminescent Western blotting substrate (Pierce).
GST pull-down
Full-length wild-type and mutant VAPB GST fusion proteins were obtained as described earlier (Teuling et al, 2007). HEK293T cells were transfected as described before with HA-YIF1A, HA-YIF1A(1–131), HA-YIF1A (131–193) and HA-YIF1A (198–293) and lysed in 50 mm Tris–HCl, 100 mm NaCl, and 1% Triton X-100 containing protease inhibitors (Roche). Lysates were incubated with GST beads (GE Healthcare Bio-Sciences) for 2 h at 4°C, washed four times with lysis buffer, and analysed by SDS–PAGE and western blotting as described before.
Biotin-streptavidin pull-down and mass spectrometry
For biotin-streptavidin pull-down assays, HeLa or HEK293T cells were transfected with biotin-tagged VAPB using Lipofectamine-2000 (Invitrogen) transfection reagent according to manufacturer’s instructions. Cells were lysed 16 h later in 20 mm Tris–HCl, pH 8.0, 150 mm KCl, 1% Triton X-100, and protease inhibitors (Roche). To increase the solubility of VAPB-P56S, transfected cells were incubated for 2 h at 20°C before lysis. Cell lysates were centrifuged at 13 000 r.p.m. for 15 min and the supernatants were incubated with Dynabeads M-280 streptavidin (Dynal; Invitrogen) for 45 min. Beads were separated by using a magnet (Dynal; Invitrogen) and washed five times in lysis buffer. For protein elution, the beads were boiled in NuPAGE LDS 4 sample buffer (Invitrogen), separated, and supernatants were run on a 10% NuPAGE Bis-Tris gel (Invitrogen). The gel was stained with the Colloidal Blue staining kit (Invitrogen) and analysed by western blotting. Mass spectrometry was performed as described previously (Lansbergen et al, 2006). The Mascot score cutoff value for a positive protein hit was set to 100. Individual peptide tandem mass spectrometry spectra with Mascot scores below 100 were checked manually and either interpreted as valid identifications or discarded. Proteins present in the negative controls (pull-down assays withbio-GFP alone) were regarded as background.
Photobleaching experiments
For quantitative FRAP experiments, neurons were transfected as described before, and imaged on a Nikon Eclipse TE2000E (Nikon) equipped with an incubation chamber (INUG2-ZILCS-H2; Tokai Hit) mounted on a motorized stage (Prior) (Jaworski et al, 2009). Coverslips (24 mm) were mounted in metal rings and maintained at 37°C and 5% CO2. A dendrite or a 3 × 3 μm regions of interest (ROI) in the cell body was photobleached with high laser power. Immediately after photobleaching, images of GFP fluorescence were acquired using a × 40 objective (Nikon) and a Coolsnap HQ camera (Photometrics). Intensity of GFP signal in dendrites was measured over a length of 30 μm from the soma with MetaMorph image analysis software (Universal Imaging). Background intensity was subtracted and fluorescence intensity after photobleaching was calculated relative to the intensity before bleaching. Sigmaplot 12.3 software was used to perform curve fitting.
Image analysis and quantification
Measurement of neurite outgrowth
To measure neurite length, we used β-galactosidase as an unbiased cell-fill. Hippocampal neurons were transfected with the indicated constructs, fixed at the appropriate time point and subjected to immunofluorescent staining. Confocal images were obtained at 1024 × 1024 pixel resolution using a LSM510 confocal microscope (Zeiss) with a × 40 oil objective (0.7 digital zoom). Each image was a z-series of images; the obtained stack was ‘flattened’ into a single image using maximum projection. Morphometric analysis and quantification were performed using MetaMorph image analysis software (Universal Imaging). For measurement of total dendrite or axonal length, all dendrites or axons of individual neurons were traced. All non-axonal protrusions initiating from the cell soma longer than 10 μm were defined as primary dendrites. For dendrite tip number, tips of all non-axonal protrusions longer than 10 μm were counted. For Sholl analysis, concentric circles with 16 μm differences in diameter were automatically drawn around the cell body, and the number of dendrites crossing each circle was counted.
Co-localization of two fluorescent signals
Co-localization of two fluorescent signals was indicated by the Pearson's coefficient (_r_p), determined using the JACoP plugin (Bolte and Cordelieres, 2006) for ImageJ. For each cell, three ROIs were selected; n was defined as the number of ROIs.
Statistical analyses
Statistical analyses were performed with MS Excel or SPSS software. Data were averaged over multiple cells and statistical analysis was performed with Student’s _t_-test, one-way ANOVA or repeated measures ANOVA and Tukey’s post hoc test. P<0.05 was considered as significant.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Dr N Ridgway for the OSBP9 antibody, Dr S Lev for the NIR2 antibody, Dr T Lee for the Flag-YIP1A construct, Dr M Darmon for the Flag-YIF1B and YFP-5HT1A plasmid, Dr. JF Presley for the p58-YFP construct and Jeroen Demmers and Karel Bezstarosti for help with mass spectrometry analyses. This work was supported by Het Prinses Beatrix Spierfonds grant (DJ and CCH), Netherlands Organization for Scientific Research (NWO-ALW-VICI, AA and CCH), the Netherlands Organization for Health Research and Development (ZonMW-TOP, CCH), the European Science Foundation (EURYI, CCH), EMBO Young Investigators Program (YIP, CCH) and the Human Frontier Science Program (HFSP-CDA, CCH).
Author contributions: MK, DJ, AA and CCH designed research; MK, KLY and ET performed research; MK analysed the data; MK and CCH wrote the paper; CCH supervised the project.
Footnotes
The authors declare that they have no conflict of interest.
References
- Al Awabdh S, Miserey-Lenkei S, Bouceba T, Masson J, Kano F, Marinach-Patrice C, Hamon M, Emerit MB, Darmon M (2012) A new vesicular scaffolding complex mediates the G-protein-coupled 5-HT1A receptor targeting to neuronal dendrites. J Neurosci 32: 14227–14241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarilio R, Ramachandran S, Sabanay H, Lev S (2005) Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem 280: 5934–5944 [DOI] [PubMed] [Google Scholar]
- Anagnostou G, Akbar MT, Paul P, Angelinetta C, Steiner TJ, de Belleroche J (2010) Vesicle associated membrane protein B (VAPB) is decreased in ALS spinal cord. Neurobiol Aging 31: 969–985 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Hauri HP (2006) The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci 119: 2173–2183 [DOI] [PubMed] [Google Scholar]
- Aridor M, Fish KN (2009) Selective targeting of ER exit sites supports axon development. Traffic 10: 1669–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K (1988) Developments in neuronal cell culture. Nature 336: 185–186 [DOI] [PubMed] [Google Scholar]
- Barrowman J, Wang W, Zhang Y, Ferro-Novick S (2003) The Yip1p.Yif1p complex is required for the fusion competence of endoplasmic reticulum-derived vesicles. J Biol Chem 278: 19878–19884 [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Breuza L, Halbeisen R, Jeno P, Otte S, Barlowe C, Hong W, Hauri HP (2004) Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem 279: 47242–47253 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Carrel D, Masson J, Al Awabdh S, Capra CB, Lenkei Z, Hamon M, Emerit MB, Darmon M (2008) Targeting of the 5-HT1A serotonin receptor to neuronal dendrites is mediated by Yif1B. J Neurosci 28: 8063–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai A, Withers J, Koh YH, Parry K, Bao H, Zhang B, Budnik V, Pennetta G (2008) hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum Mol Genet 17: 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Anagnostou G, Chai A, Withers J, Morris A, Adhikaree J, Pennetta G, de Belleroche JS (2010) Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J Biol Chem 285: 40266–40281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen E, Holzbaur EL (2006) Axonal transport and neurodegenerative disease. Biochim Biophys Acta 1762: 1094–1108 [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C (2010) Protein sorting receptors in the early secretory pathway. Annu Rev Biochem 79: 777–802 [DOI] [PubMed] [Google Scholar]
- De Vos KJ, Morotz GM, Stoica R, Tudor EL, Lau KF, Ackerley S, Warley A, Shaw CE, Miller CC (2012) VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet 21: 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra KM, Pokusa JE, Suhan J, Lee TH (2010) Yip1A structures the mammalian endoplasmic reticulum. Mol Biol Cell 21: 1556–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian KD, Schuster T, Hegemann JH, Becher D, Feldmann H, Guldener U, Gotz R, Hansen M, Hollenberg CP, Jansen G, Kramer W, Klein S, Kotter P, Kricke J, Launhardt H, Mannhaupt G, Maierl A, Meyer P, Mewes W, Munder T et al. (1999) Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet 262: 683–702 [DOI] [PubMed] [Google Scholar]
- Fasana E, Fossati M, Ruggiano A, Brambillasca S, Hoogenraad CC, Navone F, Francolini M, Borgese N (2010) A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J 24: 1419–1430 [DOI] [PubMed] [Google Scholar]
- Ferraiuolo L, Heath PR, Holden H, Kasher P, Kirby J, Shaw PJ (2007) Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. J Neurosci 27: 9201–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest S, Chai A, Sanhueza M, Marescotti M, Parry K, Georgiev A, Sahota V, Mendez-Castro R, Pennetta G (2013) Increased levels of phosphoinositides cause neurodegeneration in a Drosophila model of amyotrophic lateral sclerosis. Hum Mol Genet (advance online publication, 29 March 2013; doi:; DOI: 10.1093/hmg/ddt118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieve AG, Rabouille C (2011) Golgi bypass: skirting around the heart of classical secretion. Cold Spring Harb Perspect Biol 3: a005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan C, Stagg SM, Lapointe P, Balch WE (2006) The COPII cage: unifying principles of vesicle coat assembly. Nat Rev Mol Cell Biol 7: 727–738 [DOI] [PubMed] [Google Scholar]
- Han SM, Tsuda H, Yang Y, Vibbert J, Cottee P, Lee SJ, Winek J, Haueter C, Bellen HJ, Miller MA (2012) Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev Cell 22: 348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus C, Ehlers MD (2008) Secretory outposts for the local processing of membrane cargo in neuronal dendrites. Traffic 9: 1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidtman M, Chen CZ, Collins RN, Barlowe C (2005) Yos1p is a novel subunit of the Yip1p-Yif1p complex and is required for transport between the endoplasmic reticulum and the Golgi complex. Mol Biol Cell 16: 1673–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M (2005) GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci 8: 906–915 [DOI] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD (2003) Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci 23: 6188–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD (2005) Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron 48: 757–771 [DOI] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y (2000) Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA 97: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY (2010) Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci 11: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara JH, Villa SR, Khan NA, Bohn MC, Ozdinler PH (2012) AAV2 mediated retrograde transduction of corticospinal motor neurons reveals initial and selective apical dendrite degeneration in ALS. Neurobiol Dis 47: 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC (2009) Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61: 85–100 [DOI] [PubMed] [Google Scholar]
- Jensen D, Schekman R (2011) COPII-mediated vesicle formation at a glance. J Cell Sci 124: 1–4 [DOI] [PubMed] [Google Scholar]
- Jin C, Zhang Y, Zhu H, Ahmed K, Fu C, Yao X (2005) Human Yip1A specifies the localization of Yif1 to the Golgi apparatus. Biochem Biophys Res Commun 334: 16–22 [DOI] [PubMed] [Google Scholar]
- Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT (2005) Structural basis of FFAT motif-mediated ER targeting. Structure 13: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Kanekura K, Nishimoto I, Aiso S, Matsuoka M (2006) Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8). J Biol Chem 281: 30223–30233 [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Yau KW, Hoogenraad CC (2010) Microtubule dynamics in dendritic spines. Methods Cell Biol 97: 111–132 [DOI] [PubMed] [Google Scholar]
- Karpati G, Carpenter S, Durham H (1988) A hypothesis for the pathogenesis of amyotrophic lateral sclerosis. Rev Neurol (Paris) 144: 672–675 [PubMed] [Google Scholar]
- Kato T, Hirano A, Donnenfeld H (1987) A Golgi study of the large anterior horn cells of the lumbar cords in normal spinal cords and in amyotrophic lateral sclerosis. Acta Neuropathol 75: 34–40 [DOI] [PubMed] [Google Scholar]
- Kawano M, Kumagai K, Nishijima M, Hanada K (2006) Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J Biol Chem 281: 30279–30288 [DOI] [PubMed] [Google Scholar]
- Kim S, Leal SS, Ben Halevy D, Gomes CM, Lev S (2010) Structural requirements for VAP-B oligomerization and their implication in amyotrophic lateral sclerosis-associated VAP-B(P56S) neurotoxicity. J Biol Chem 285: 13839–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J, Parton RG, Fuller SD, Griffiths G, Dotti CG (1995) The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol Biol Cell 6: 1315–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, Akhmanova A (2006) CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell 11: 21–32 [DOI] [PubMed] [Google Scholar]
- Lecuit T, Pilot F (2003) Developmental control of cell morphogenesis: a focus on membrane growth. Nat Cell Biol 5: 103–108 [DOI] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R (2004) Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20: 87–123 [DOI] [PubMed] [Google Scholar]
- Lehotsky J, Kaplan P, Babusikova E, Strapkova A, Murin R (2003) Molecular pathways of endoplasmic reticulum dysfunctions: possible cause of cell death in the nervous system. Physiol Res 52: 269–274 [PubMed] [Google Scholar]
- Lehto M, Hynynen R, Karjalainen K, Kuismanen E, Hyvarinen K, Olkkonen VM (2005) Targeting of OSBP-related protein 3 (ORP3) to endoplasmic reticulum and plasma membrane is controlled by multiple determinants. Exp Cell Res 310: 445–462 [DOI] [PubMed] [Google Scholar]
- Lev S, Ben Halevy D, Peretti D, Dahan N (2008) The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol 18: 282–290 [DOI] [PubMed] [Google Scholar]
- Litvak V, Argov R, Dahan N, Ramachandran S, Amarilio R, Shainskaya A, Lev S (2004) Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Mol Cell 14: 319–330 [DOI] [PubMed] [Google Scholar]
- Liu M, Duggan J, Salt TE, Cordeiro MF (2011) Dendritic changes in visual pathways in glaucoma and other neurodegenerative conditions. Exp Eye Res 92: 244–250 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Levine TP (2005) A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem 280: 14097–14104 [DOI] [PubMed] [Google Scholar]
- Lorente-Rodriguez A, Barlowe C (2011) Entry and exit mechanisms at the cis-face of the Golgi complex. Cold Spring Harb perspect Biol 3: a005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern H, Yang X, Andrulis E, Sternglanz R, Trepte HH, Gallwitz D (2000) A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J 19: 4485–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikitova V, Levine TP (2012) Analysis of the key elements of FFAT-like motifs identifies new proteins that potentially bind VAP on the ER, including two AKAPs and FAPP2. PLoS One 7: e30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitne-Neto M, Machado-Costa M, Marchetto MC, Bengtson MH, Joazeiro CA, Tsuda H, Bellen HJ, Silva HC, Oliveira AS, Lazar M, Muotri AR, Zatz M (2011) Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum Mol Genet 20: 3642–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A, Virard I, Raoul C (2011) Accumulation of wildtype and ALS-linked mutated VAPB impairs activity of the proteasome. PLoS ONE 6: e26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Gonatas NK, Stieber A, Gurney ME, Dal Canto MC (1996) The Golgi apparatus of spinal cord motor neurons in transgenic mice expressing mutant Cu,Zn superoxide dismutase becomes fragmented in early, preclinical stages of the disease. Proc Natl Acad Sci USA 93: 5472–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Yeaman C (2001) Protein trafficking in the exocytic pathway of polarized epithelial cells. Trends Cell Biol 11: 483–486 [DOI] [PubMed] [Google Scholar]
- Ngo M, Ridgway ND (2009) Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell 20: 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M (2004) A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 75: 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Hayashi M, Inada H, Tanaka T (1999) Molecular cloning and characterization of mammalian homologues ofvesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem Biophys Res Commun 254: 21–26 [DOI] [PubMed] [Google Scholar]
- Otte S, Belden WJ, Heidtman M, Liu J, Jensen ON, Barlowe C (2001) Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J Cell Biol 152: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papiani G, Ruggiano A, Fossati M, Raimondi A, Bertoni G, Francolini M, Benfante R, Navone F, Borgese N (2012) Restructured endoplasmic reticulum generated by mutant amyotrophic lateral sclerosis-linked VAPB is cleared by the proteasome. J Cell Sci 125: 3601–3611 [DOI] [PubMed] [Google Scholar]
- Paschen W, Mengesdorf T (2005) Cellular abnormalities linked to endoplasmic reticulum dysfunction in cerebrovascular disease--therapeutic potential. Pharmacol Ther 108: 362–375 [DOI] [PubMed] [Google Scholar]
- Pennetta G, Hiesinger PR, Fabian-Fine R, Meinertzhagen IA, Bellen HJ (2002) Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron 35: 291–306 [DOI] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S (2008) Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell 19: 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Aivazian D (2004) Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 5: 886–896 [DOI] [PubMed] [Google Scholar]
- Pierce JP, Mayer T, McCarthy JB (2001) Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol 11: 351–355 [DOI] [PubMed] [Google Scholar]
- Rao M, Song W, Jiang A, Shyr Y, Lev S, Greenstein D, Brantley-Sieders D, Chen J (2012) VAMP-Associated Protein B (VAPB) promotes breast tumor growth by modulation of Akt Activity. PLoS One 7: e46281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaparkhi A, Lawless GM, Schweizer FE, Golshani P, Jackson GR (2008) A Drosophila model of ALS: human ALS-associated mutation in VAP33A suggests a dominant negative mechanism. PloS One 3: e2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, Dale HA, Bazzocco S, Marie M (2009) Emerging new roles of the pre-Golgi intermediate compartment in biosynthetic-secretory trafficking. FEBS Lett 583: 3804–3810 [DOI] [PubMed] [Google Scholar]
- Sasaki S, Iwata M (1996) Dendritic synapses of anterior horn neurons in amyotrophic lateral sclerosis: an ultrastructural study. Acta Neuropathol 91: 278–283 [DOI] [PubMed] [Google Scholar]
- Sau D, Rusmini P, Crippa V, Onesto E, Bolzoni E, Ratti A, Poletti A (2011) Dysregulation of axonal transport and motorneuron diseases. Biol Cell 103: 87–107 [DOI] [PubMed] [Google Scholar]
- Selak S, Woodman RC, Fritzler MJ (2000) Autoantibodies to early endosome antigen (EEA1) produce a staining pattern resembling cytoplasmic anti-neutrophil cytoplasmic antibodies (C-ANCA). Clin Exp Immunol 122: 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgobio C, Trabalza A, Spalloni A, Zona C, Carunchio I, Longone P, Ammassari-Teule M (2008) Abnormal medial prefrontal cortex connectivity and defective fear extinction in the presymptomatic G93A SOD1 mouse model of ALS. Genes Brain Behav 7: 427–434 [DOI] [PubMed] [Google Scholar]
- Shakoori A, Fujii G, Yoshimura S, Kitamura M, Nakayama K, Ito T, Ohno H, Nakamura N (2003) Identification of a five-pass transmembrane protein family localizing in the Golgi apparatus and the ER. Biochem Biophys Res Commun 312: 850–857 [DOI] [PubMed] [Google Scholar]
- Sholl DA (1953) Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87: 387–406 [PMC free article] [PubMed] [Google Scholar]
- Simpson JC, Joggerst B, Laketa V, Verissimo F, Cetin C, Erfle H, Bexiga MG, Singan VR, Heriche JK, Neumann B, Mateos A, Blake J, Bechtel S, Benes V, Wiemann S, Ellenberg J, Pepperkok R (2012) Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat Cell Biol 14: 764–774 [DOI] [PubMed] [Google Scholar]
- Skehel PA, Fabian-Fine R, Kandel ER (2000) Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc Natl Acad Sci USA 97: 1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussan L, Burakov D, Daniels MP, Toister-Achituv M, Porat A, Yarden Y, Elazar Z (1999) ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles. J Cell Biol 146: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DJ, Pepperkok R (2001) Illuminating the secretory pathway: when do we need vesicles? J Cell Sci 114: 1053–1059 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kanekura K, Levine TP, Kohno K, Olkkonen VM, Aiso S, Matsuoka M (2009) ALS-linked P56S-VAPB, an aggregated loss-of-function mutant of VAPB, predisposes motor neurons to ER stress-related death by inducing aggregation of co-expressed wild-type VAPB. J Neurochem 108: 973–985 [DOI] [PubMed] [Google Scholar]
- Tang BL (2008) Emerging aspects of membrane traffic in neuronal dendrite growth. Biochim Biophys Acta 1783: 169–176 [DOI] [PubMed] [Google Scholar]
- Teuling E, Ahmed S, Haasdijk E, Demmers J, Steinmetz MO, Akhmanova A, Jaarsma D, Hoogenraad CC (2007) Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci 27: 9801–9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Han SM, Yang Y, Tong C, Lin YQ, Mohan K, Haueter C, Zoghbi A, Harati Y, Kwan J, Miller MA, Bellen HJ (2008) The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell 133: 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Vlug AS, Teuling E, Haasdijk ED, French P, Hoogenraad CC, Jaarsma D (2005) ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur J Neurosci 22: 1881–1894 [DOI] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J (2001) Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol 155: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Stephens DJ (2005) ER-to-Golgi transport: form and formation of vesicular and tubular carriers. Biochim Biophys Acta 1744: 304–315 [DOI] [PubMed] [Google Scholar]
- Wendler F, Gillingham AK, Sinka R, Rosa-Ferreira C, Gordon DE, Franch-Marro X, Peden AA, Vincent JP, Munro S (2010) A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J 29: 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins LM, Kuta A, Stevens JC, Fisher EM, von Bartheld CS (2012) A novel phenotype for the dynein heavy chain mutation Loa: Altered dendritic morphology, organelle density, and reduced numbers of trigeminal motoneurons. J Comp Neurol 520: 2757–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles JP, McMaster CR, Ridgway ND (2002) Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem 277: 29908–29918 [DOI] [PubMed] [Google Scholar]
- Wyles JP, Ridgway ND (2004) VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp Cell Res 297: 533–547 [DOI] [PubMed] [Google Scholar]
- Yang X, Matern HT, Gallwitz D (1998) Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J 17: 4954–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Huh SU, Drennan JM, Kathuria H, Martinez JS, Tsuda H, Hall MC, Clemens JC (2012) Drosophila Vap-33 is required for axonal localization of Dscam isoforms. J Neurosci 32: 17241–17250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN (2007) Growing dendrites and axons differ in their reliance on the secretory pathway. Cell 130: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H (2007) ER stress and diseases. FEBS J 274: 630–658 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Suzuki K, Yamamoto A, Sakai N, Bando M, Tanimoto K, Yamaguchi Y, Sakaguchi T, Akhter H, Fujii G, Yoshimura S, Ogata S, Sohda M, Misumi Y, Nakamura N (2008) YIPF5 and YIF1A recycle between the ER and the Golgi apparatus and are involved in the maintenance of the Golgi structure. Exp Cell Res 314: 3427–3443 [DOI] [PubMed] [Google Scholar]
- Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F (2004) siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res 32: W130–W134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File