Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system (original) (raw)
Abstract
A simple and robust method for targeted mutagenesis in zebrafish has long been sought. Previous methods generate monoallelic mutations in the germ line of F0 animals, usually delaying homozygosity for the mutation to the F2 generation. Generation of robust biallelic mutations in the F0 would allow for phenotypic analysis directly in injected animals. Recently the type II prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated proteins (Cas) system has been adapted to serve as a targeted genome mutagenesis tool. Here we report an improved CRISPR/Cas system in zebrafish with custom guide RNAs and a zebrafish codon-optimized Cas9 protein that efficiently targeted a reporter transgene Tg(-5.1mnx1:egfp) and four endogenous loci (tyr, golden, mitfa, and ddx19). Mutagenesis rates reached 75–99%, indicating that most cells contained biallelic mutations. Recessive null-like phenotypes were observed in four of the five targeting cases, supporting high rates of biallelic gene disruption. We also observed efficient germ-line transmission of the Cas9-induced mutations. Finally, five genomic loci can be targeted simultaneously, resulting in multiple loss-of-function phenotypes in the same injected fish. This CRISPR/Cas9 system represents a highly effective and scalable gene knockout method in zebrafish and has the potential for applications in other model organisms.
Keywords: genome engineering, RNA-guided mutagenesis, pigmentation
Recent methodological innovations that exploit zinc finger nucleases (ZFNs) and transcription-activator–like effector nucleases (TALENs) have made it possible to introduce site-specific genomic modifications in cell culture systems and in a wide range of species (1, 2). These designer nucleases target and cleave specific genomic sequences. Error-prone nonhomologous end joining (NHEJ) DNA repair then generates mutations. The engineering of efficient ZFNs requires extensive technical expertise and empirical testing to find efficient enzymes. The TALEN technology provides an attractive alternative to ZFNs, but like ZFNs, it requires the assembly of two relatively large DNA-binding proteins for each target. Most recently, a new class of genome editing tool based on the type II prokaryotic CRISPR (clustered regularly interspaced short palindromic repeats) adaptive immune system has been developed (3).
CRISPR systems in eubacteria and archaea use small RNAs and CRISPR-associated proteins (Cas) proteins to target and cleave invading foreign DNAs (4–6). One of the simplest CRISPR systems is the type II CRISPR system from Streptococcus pyogenes: an endonuclease Cas9 and two small RNAs, CRISPR RNA (crRNA) and a transacting RNA (tracrRNA), are sufficient for RNA-guided cleavage of foreign DNAs (7). Recent studies show that a single guide RNA (gRNA) chimera that mimics the crRNA:tracrRNA complex can guide Cas9 to introduce site-specific DNA double-stranded breaks in vitro (3) and in mammalian cell lines (8–11), bacteria (12, 13), yeast (14), zebrafish (15, 16), and mice (17) with efficiencies similar to or surpassing those obtained by ZFNs and TALENs. However, unlike ZFNs and TALENs, gRNA is the only component that needs customization for each genomic target, thus greatly simplifying the design and lowering the cost of gene targeting compared with the protein-based target recognition platforms.
Here we report a CRISPR/Cas9 system that targets and modifies genomic loci in zebrafish with high efficiencies in somatic and germ cells. In testing one hemizygous egfp reporter gene and four endogenous loci, we found that the mutagenesis efficiency consistently reached 75–99%. In four cases, mosaic, null-like phenotypes were observed in the injected embryos (founders), presumably due to high frequencies of biallelic gene disruptions. Furthermore, we showed that five genomic loci in the same embryos could be disrupted simultaneously and efficiently, resulting in the embryos exhibiting the combination of distinct phenotypes caused by multiple biallelic gene inactivations. Finally, in addition to the high efficiency of Cas9-induced mutagenesis in somatic tissues, we also observed efficient germ-line transmission of the mutations to the F1 generation. In summary, we have developed a simple, economical, and scalable CRISPR/Cas9 system for targeted genome editing in zebrafish with efficiencies generally exceeding what has been achieved by ZFNs, TALENs, and earlier Cas9 technologies.
Results
Design of the CRISPR/Cas9 System Optimized for Zebrafish Genome Editing.
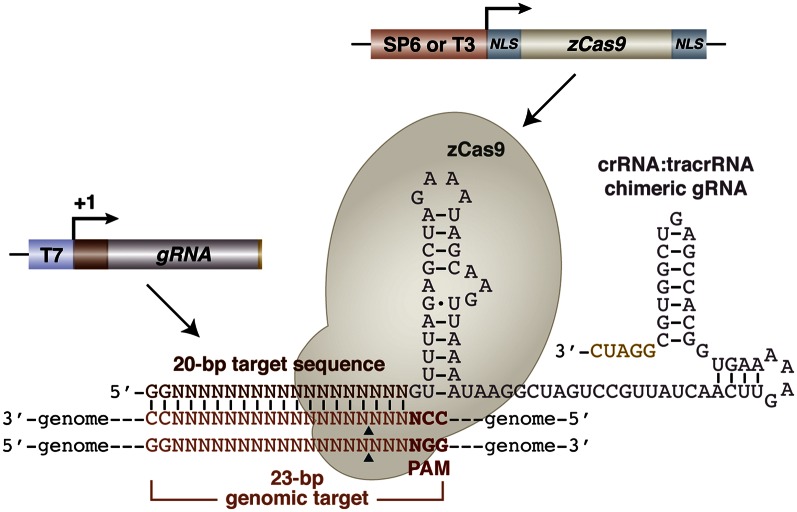
A recent breakthrough using the purified Cas9 protein and designer gRNAs to achieve site-specific DNA double-stranded breaks in vitro (3) prompted us to develop a similar targeted genome editing approach in zebrafish. To optimize the Cas9 expression and its nuclear targeting in zebrafish, we synthesized a zebrafish codon-optimized version of S. pyogenes Cas9 with SV40 large T-antigen nuclear localization signals (nls) at both its amino and carboxyl termini (hereafter referred to as nls-zCas9-nls). The nls-zCas9-nls coding sequence was cloned into vectors for making the capped, polyadenylated mRNA via in vitro transcription from either the SP6 or T3 promoters (Fig. 1). The Cas9 protein sequence (National Center for Biotechnology Information, NCBI RefSeq: YP_005388840) in this study is derived from a recently emerged strain (MGAS1882) and is slightly different from the ones recently reported (8, 15, 16) (Fig. S1_A_).
Fig. 1.
Genome editing in zebrafish using a CRISPR/Cas9 system. The system consists of two components, a dual NLS-tagged zebrafish codon-optimized Cas9 protein and a single crRNA:tracrRNA chimeric gRNA, comprising a 20-bp target sequence (dark red) complementary to the genomic target adjacent to a PAM site of NGG. Both components are first generated as RNAs by in vitro transcription from the SP6 or T3 (for Cas9) or T7 (for gRNA) promoter. The mix of gRNA and Cas9 RNA was then injected into one-cell–stage embryos to induce RNA-guided targeted DNA double-stranded breaks mediated by the Cas9 enzyme. Arrowheads denote putative cleavage sites.
To generate the gRNA that targets the genomic loci, we designed a gRNA cloning vector that harbors the combined features of the crRNA:trascRNA backbone, immediately preceded by a 20-bp “target sequence” (which can be easily added from short annealed oligonucleotides) (Fig. 1, Fig. S1_B_, and SI Materials and Methods). In this system, a potential genomic target is a stretch of 23-bp genomic sequence that starts with two GG residues at the 5′ end for efficient transcription from the T7 promoter and ends with the protospacer-adjacent motif (PAM) sequence (18), NGG, for efficient Cas9 binding (Fig. 1). The desired single targeting gRNA was generated by in vitro transcription from a T7 promoter after linearizing the plasmid. The sequence of the crRNA:tracrRNA chimera in this study is identical to those recently described (8, 15, 16) except that the 3′ end of the gRNA ends with residues GGAUC instead of a string of U residues.
Cas9-Induced Mutagenesis of an egfp Transgene Resulted in Mosaic EGFP Expression in Motoneurons.
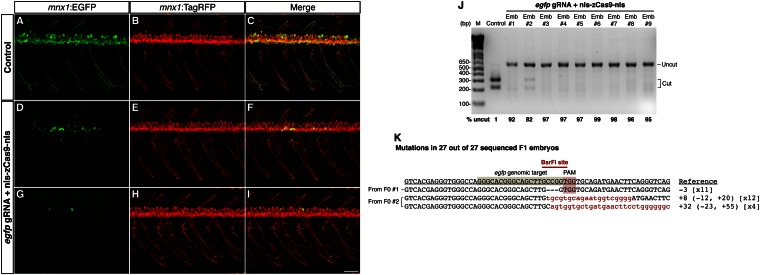
In our initial test, we designed a gRNA for targeting the egfp transgene in Tg(-5.1mnx1:EGFP)/Tg(-5.1mnx1:TagRFP) double transgenic fish (19), in which motoneurons and their processes are labeled by EGFP and TagRFP. In this system, successful targeting was scored as the loss of EGFP expression. We coinjected egfp gRNA (6 or 30 pg) and in vitro transcribed, capped, polyadenylated nls-zCas9-nls mRNA (150 pg) into one-cell–stage embryos. After 1 d postfertilization (dpf), around half of the uninjected siblings (48.2%, n = 83) had their motoneurons labeled with EGFP, a ratio expected from the outcross of a hemizygous transgene. However, only 5.7% (6 pg of egfp gRNA, n = 106) and 2.8% (30 pg of egfp gRNA, n = 141) of normally developed egfp gRNA/_nls-zCas9-nls_–injected embryos showed visible EGFP-positive motoneurons (Fig. S2_A_). Strikingly, all of the injected embryos with EGFP-positive motoneurons exhibited mosaic EGFP expression in the motoneurons (examples in Fig. 2 D and G). The mosaic EGFP expression was not due to the partial loss of motoneurons because the motoneurons developed normally as revealed by the TagRFP labeling (Fig. 2 E and H). These results indicate that using our CRISPR/Cas9 system, the egfp transgene can be targeted efficiently, resulting in sparse motoneuron EGFP expression in the injected embryos (i.e., founders). To further assess the mutagenesis frequencies, the target region was PCR amplified from 20 randomly selected individual founders; 9 of 20 founders were Tg(-5.1mnx1:EGFP) positive. Site-specific mutations would make the region resistant to BsrFI digestion. We observed that 8 had >90% and 1 had >80% mutation rates at the target site (means ± SD: 94.8 ± 5.2%, n = 9) (Fig. 2_J_). Such high mutation rates at the egfp target are why the majority of the injected embryos lost or had sparse EGFP expression in the motoneurons.
Fig. 2.
Efficient disruption of the Tg(-5.1mnx1:EGFP) transgene by Cas9 results in mosaic EGFP expression in the motoneurons. egfp gRNA (6 or 30 pg) and nls-zCas9-nls RNA (150 pg) were injected into Tg(-5.1mnx1:EGFP)/Tg(-5.1mnx1:TagRFP) double transgenic embryos. The control embryos were injected with nls-zCas9-nls RNA and a gRNA lacking the egfp target sequence. (A_–_I) Confocal images of spinal motoneurons of live zebrafish embryos at 3 dpf. In contrast to the uniform motoneuron EGFP expression pattern in the control (A), _egfp_-targeted embryos showed mosaic EGFP expression in the motoneurons (D and G). However, the motoneurons of the _egfp_-targeted embryos developed normally as indicated by the uniform TagRFP labeling (E and H). (J) BsrFI digestion showed high mutagenesis rates at the egfp target site (82–99%) in nine randomly selected _egfp_-targeted embryos. (K) Sequences of egfp mutations in 27 F1 embryos from two _egfp_-targeted _Tg(-5.1mnx1:EGFP)_-positive founders. The wild-type reference sequence is underlined with the target site and PAM highlighted in gray and red, respectively. Note that all 27 F1 siblings had egfp mutations with the BsrFI site (marked by a red line) disrupted. Deletions and insertions are indicated by dashes and lowercase red letters, respectively. The net change of each indel mutation is noted at the Right of each sequence (+, insertion; –, deletion). The number of times a mutant allele was identified is indicated in brackets. (Scale bar: 50 µm.)
Biallelic Cas9-Induced Disruptions of Endogenous Loci Resulted in Null-Like Phenotypes.
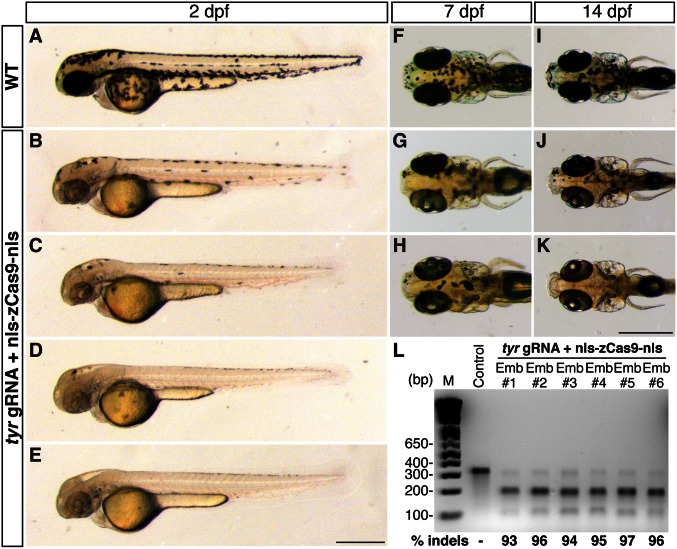
We next tested our CRISPR/Cas9 system in targeting endogenous loci in wild-type zebrafish, where two copies of a given endogenous gene are present. We chose to target three genes involved in body pigmentation because pigmentation defects are a convenient readout for efficient biallelic gene disruption. When we targeted tyrosinase (tyr), the gene required for converting tyrosine into the pigment melanin, we observed that all of the injected founders (n = 66) showed mosaic pigmentation patterns; some of them were indistinguishable from sandy mutants (20) (Fig. 3 A_–_E). As in the case of targeting the egfp transgene, Cas9-induced pigmentation phenotypes are also dose dependent (Fig. S2_B_). We randomly selected six embryos and quantified the mutagenesis rates using the T7 endonuclease I (T7EI) assay; all assayed embryos showed >90% mutagenesis rates (means ± SD: 95.2 ± 1.5%, n = 6) (Fig. 3_L_). The tyr knockout phenotypes persisted to later developmental stages (Fig. 3 F_–_K), indicating that the abrogation of tyrosinase activity was caused by genetic alterations, not by epigenetic changes. Sequencing the tyr target region further confirmed the presence of insertions/deletions (indels), which were likely introduced by the error-prone NHEJ repair following DNA double-stranded breaks (Fig. S3). Consistent with the report that Cas9 cuts target DNA 6 bp upstream of the PAM (3), all mutations occurred at the 3′ end of the target sequence. The fact that all of the sequenced tyr alleles (17 of 17) had indel mutations further confirmed the high mutagenesis rates indicated by the T7EI assay.
Fig. 3.
Biallelic disruption of tyrosinase (tyr) by Cas9 generates mosaic pigmentation phenotypes. tyr gRNA (30 pg) and nls-zCas9-nls RNA (150 pg) were injected into wild-type embryos. (A_–_E) Lateral views of wild-type (A) and _tyr_-targeted embryos (B_–_E) at 2 dpf. _tyr_-targeted embryos showed different degrees of hypopigmentation, some of them were almost unpigmented (e.g., E). (F_–_K) Dorsal view of the wild-type (F and I) and _tyr_-targeted larvae (G, H, J, and K) at 7 dpf (F_–_H) and 14 dpf (I_–_K). Mosaic pigmentation defects in _tyr_-targeted larvae persisted to later stages. (L) T7EI assays showed high mutagenesis rates at the tyr target (93–97%) in six randomly selected _tyr_-targeted embryos. (Scale bars: 500 µm.)
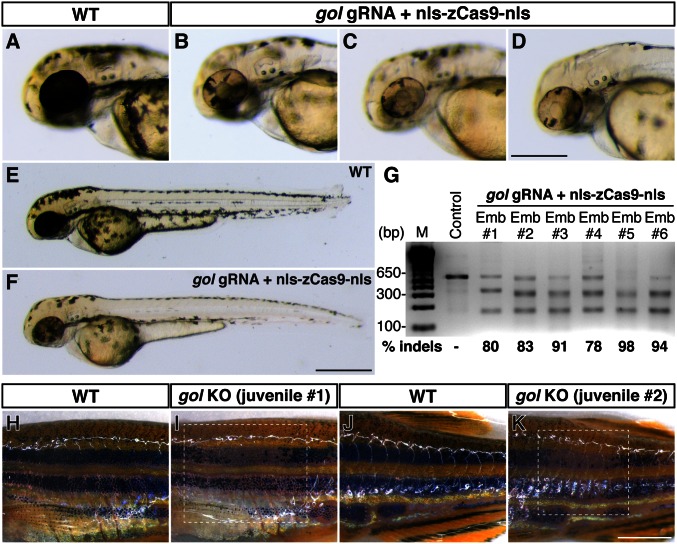
The second pigmentation gene we targeted was golden (gol), which encodes the putative cation exchanger Slc24a5. Homozygous gol mutants show hypopigmentation in retinal pigment epithelium (RPE) and skin melanophores (21). Targeting gol using our CRISPR/Cas9 system in wild-type embryos led to mosaic hypopigmentation in RPE and skin melanophores in 94% of the injected embryos (n = 184) at 2 dpf (Fig. 4 A_–_F). The mutagenesis rates were 78–98% (means ± SD: 87.3 ± 8.1%, n = 6) among six randomly selected embryos (Fig. 4_G_). Sequencing the gol target region further confirmed the Cas9-induced mutations (Fig. S3) The mosaic gol phenotypes also persisted and were observed in juvenile _gol_-targeted founders, wherein patches of pale melanophores were present within the melanophore-rich skin stripes (Fig. 4 H_–_K). We next targeted mitfa, which encodes a basic helix–loop–helix/leucine zipper transcription factor and is required for neural crest-derived melanocyte development (22). Targeting mitfa also yielded high mutagenesis rates (means ± SD: 76.7 ± 8.3%, n = 6) (Fig. S4). However, surprisingly, we did not observe discernible pigmentation defects (see Discussion).
Fig. 4.
Biallelic disruption of golden (gol) by Cas9 generates mosaic pigmentation phenotypes. gol gRNA (50 pg) and nls-zCas9-nls RNA (150 pg) were injected into wild-type embryos. (A_–_F) Lateral views of wild-type (A and E) and _gol_-targeted embryos (B_–_D and F) at 2 dpf. Whereas the wild-type embryo showed darkly pigmented retinal pigment epithelium (RPE), _gol_-targeted embryos had patches of unpigmented cells in RPE (B_–_D). The skin melanophores in _gol_-targeted embryos were also hypopigmented (F). (G) T7EI assays showed high mutagenesis rates at the gol target (80–98%) in six randomly selected _gol_-targeted embryos. (H_–_K) Lateral views of wild-type (H and J) and _gol_-targeted (I and K) juvenile fish at 54 dpf. _gol_-targeted fish had patches of pale melanophores (dashed boxes) among the dark skin stripes. (Scale bars: in D, 300 µm; in F, 500 µm; in K, 2 mm.)
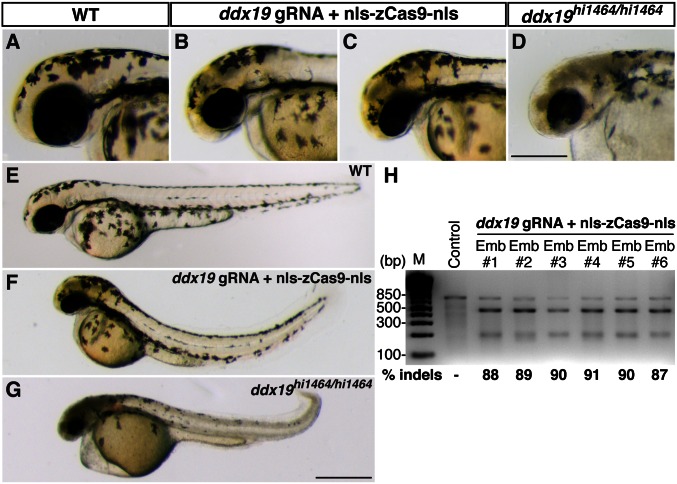
In addition to targeting the genes involved in pigmentation pathways, we also targeted ddx19, which encodes a DEAD-box protein whose Saccharomyces cerevisiae ortholog, Dbp5, is a well-characterized protein essential for mRNA export and protein translation (23, 24). A null insertional allele (ddx19 hi1464) is available in the zebrafish (25). Homozygous ddx19 mutants (ddx19 hi1464/hi1464) showed widespread neuronal cell death, small eyes, and curved body axis at 1 dpf (Fig. 5 D and G). Using our CRISPR/Cas9 system to target ddx19 in the wild-type embryos generated similar gross morphological phenotypes, albeit less severe than the ddx19 hi1464/hi1464 phenotypes (Fig. 5 B, C, and F). In agreement with the induction of null-like phenotypes in _ddx19_-targeted founders, high mutagenesis rates were observed at the ddx19 target site (means ± SD: 89.2 ± 1.5%, n = 6) (Fig. 5_H_). Mutations were further confirmed by sequencing the ddx19 target region (Fig. S3).
Fig. 5.
Biallelic disruption of ddx19 by Cas9 generates null-like phenotypes. ddx19 gRNA (150 pg) and nls-zCas9-nls RNA (150 pg) were injected into wild-type embryos. (A_–_G) Lateral views of the wild-type (A and E), _ddx19_-targeted (B, C, and F), and homozygous ddx19 mutants (ddx19 hi1464/hi1464 ) (D and G) embryos. _ddx19_-targeted embryos recapitulated several ddx19 null phenotypes (e.g., brain necrosis, small eyes, and curved body axis), albeit to a lesser extent. (H) T7EI assays showed high mutagenesis rates at the ddx19 target (87–91%) in six randomly selected _ddx19_-targeted embryos. (Scale bars: in D, 300 µm; in G, 500 µm.)
All of the _ddx19_-targeted founders died before 5 dpf. To establish mutant lines for essential genes (e.g., ddx19), it is necessary to reduce the mutagenesis rates so the founders can survive to adulthood. Thus, we further tested whether Cas9-induced mutagenesis is dose dependent by injecting embryos with different doses of ddx19 gRNA/Cas9 RNA. As in the case of targeting egfp and tyr, we observed a dose-dependent mutagenic activity for ddx19 (Fig. S2_C_). When lowering the ddx19 gRNA/Cas9 concentrations, we obtained increasing numbers of wild-type–like, _ddx19_-targeted embryos, which later survived past larval stages. The T7EI assay indicated that these wild-type–like animals still had relatively high indel rates (∼35–40%) at the ddx19 target (Fig. S2_D_). Therefore, to achieve germ-line transmission of Cas9-induced mutations for essential genes, the mutagenesis rates could be adjusted by simply titrating down the amounts of RNAs injected.
Multiplex Genome Mutagenesis Generated Multiple Phenotypes in the Same Fish.
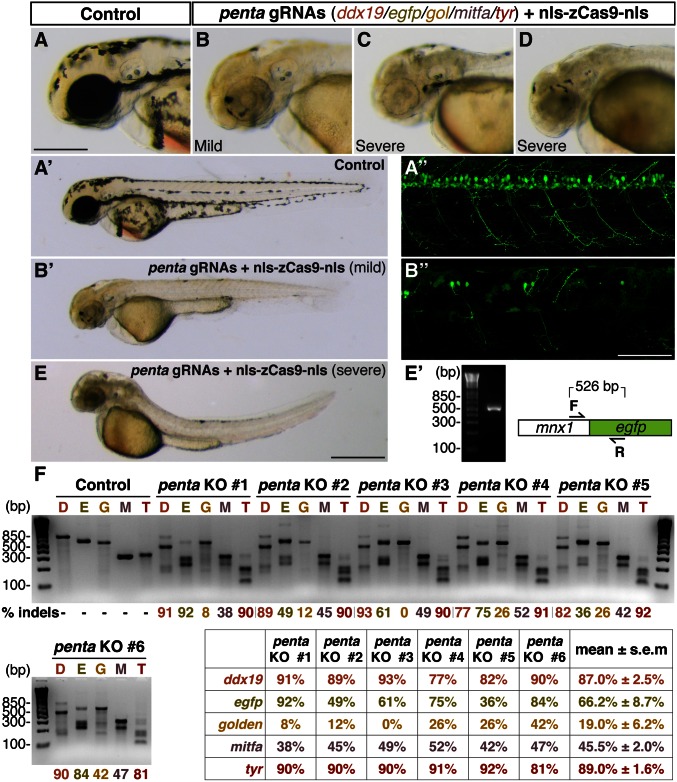
Given that the targeting specificity of the CRISPR/Cas9 system is solely dependent on a single gRNA, we sought to determine whether multiple genomic loci could be targeted simultaneously. We coinjected nls-zCas9-nls RNA with a mix of gRNAs that targeted the five different genomic loci described above. In these embryos, we observed the combination of three distinct phenotypes in the same embryos (Fig. 6 A_–_E): the brain necrosis/small eyes/curved body axis caused by ddx19 disruption, the pigmentation defects caused by tyr and/or golden inactivation, and the mosaic motoneuron EGFP expression caused by mutations in Tg(-5.1mnx1:EGFP). In some _Tg(-5.1mnx1:egfp)_-positive embryos, the motoneuron EGFP signals were not detectable. However, genotyping PCR confirmed the presence of the Tg(-5.1mnx1:EGFP) transgene (Fig. 6_E_′), indicating that the transgene expression could be abrogated in a nonmosaic fashion, similar to what was observed in the case of the single gRNA injection (Fig. 2). Consistent with the induction of pronounced multiple founder phenotypes in these quintuple knockout embryos, we detected efficient genome editing at all five loci (Fig. 6_F_). Sequencing one of the quintuple targeted founders further confirmed that Cas9-induced indels were present at all five genomic target sites in a single animal (Fig. S3). The mutagenesis efficiencies in the quintuple knockout embryos were comparable to that in single knockouts for some gene targets (e.g., ddx19 and tyr), but were lower than that in single knockouts for others (e.g., golden and mitfa). These data indicate that our CRISPR/Cas9 system robustly mediates multiplex, biallelic gene disruptions in a single zebrafish genome, resulting in multiple phenotypes in the same embryos.
Fig. 6.
Multiplex genome editing in zebrafish by Cas9. A mix of five gRNAs (penta gRNAs: ddx19 gRNA, 55 pg; egfp gRNA, 15 pg; gol gRNA, 25 pg; mitfa gRNA, 60 pg; and tyr gRNA, 25 pg) were coinjected with nls-zCas9-nls RNA (150 pg) into Tg(-5.1mnx1:egfp) transgenic embryos. (A_–_E) Lateral views of the wild-type (_A_–_A_′′) and the penta gRNAs/nls-zCas9-nls RNA-injected (_B_–_B_′′ and C_–_E) embryos. The combination of three distinct phenotypes—brain necrosis/small eyes/curved body axis, hypopigmentation, and loss of EGFP expression in the motoneurons—caused by biallelic inactivations of multiple target genes simultaneously were observed in the same embryos (_B_–_B_′′ and E and _E_′). Some quintuple knockout embryos showed no EGFP-positive motoneurons due to the almost nonmosaic disruption of the Tg(-5.1mnx1:egfp) transgene, whose presence was confirmed by PCR (_E_′). (F) T7EI assays showed high mutagenesis rates at five genomic targets in six randomly selected quintuple knockout embryos (penta KO 1–6: D, ddx19; E, egfp; G, gol; M, mitfa; T, tyr). (Scale bars: in A, 300 µm; in _B_′′, 50 µm; in E, 500 µm.)
Efficient Germ-Line Transmission of Cas9-Induced Mutations.
To determine whether the Cas9-induced mutations were heritable, we first outcrossed two of the _egfp_-targeted _Tg(-5.1mnx1:EGFP)_-positive founders to generate F1 progeny. Strikingly, none of the F1 embryos that carried the Tg(-5.1mnx1:EGFP) transgene showed EGFP expression in the motoneurons (n = 27). This result suggested that the Tg(-5.1mnx1:EGFP) loci in the germ line were effectively mutagenized. Sequencing the egfp target region further confirmed the presence of indels in all 27 F1 embryos (Fig. 2_K_).
We next crossed _gol_-targeted founders (two pairs of intercrosses and five outcrosses) to test germ-line transmission of gol mutations. The pigmentation of the F1 progeny from F0 intercrosses was first examined. As shown in Fig. S5, both F0 intercrosses generated F1 transheterozygotes with pigmentation defects. Strikingly, in one of the F0 intercrosses, 82% of the F1 embryos (n = 99) showed null-like pigmentation, and the remaining 18% of embryos were hypopigmented. The lack of F1 progeny with normal pigmentation indicated that close to 100% of germ-line alleles in both founders had biallelic disruptions of gol. Sequencing of the gol target site further confirmed this remarkably high rate of germ-line mutagenesis, as 33 of 33 sequences from 12 F1 embryos had gol mutations (Fig. S5_F_). In addition, the F1 sequence data revealed that both gol alleles in all of the phenotypically null-like F1 embryos had either frameshift or nonsense mutations, whereas all of the hypopigmented F1 had a frameshift mutation in one gol allele and an in-frame small indel in the other. These small in-frame indel alleles likely encoded partial loss-of-function gol products and thus conferred the hypomorphic phenotypes. Finally, for the five gol F0 outcrosses, the T7EI assay was performed to detect gol mutations in the F1 progeny. The results showed that 5 of 5 gol founders transmitted the gol mutations efficiently (Fig. S5_G_). In sum, 9 of 9 _gol_-targeted founders transmitted gol mutations efficiently to the F1 generation.
Off-Target Analyses of Cas9-Induced Mutations.
Three different approaches were used to assess the targeting specificity of our Cas9 system (Fig. S6). First, we searched the genome for potential off-target sites for egfp and tyr gRNAs. Previous studies show that 8–12 bases (“seed sequence”) immediately to the 5′ end of the PAM, NGG, are critical in determining target specificity: single mismatches within this region largely abolish the gRNA/Cas9 target activity, whereas single mismatches beyond the seed sequence toward the 5′ end of the target are tolerated (3, 10, 12). Based on these parameters, four sites with 11-bp perfect matches followed by NGG for the egfp gRNA (egfp OT1–OT4) and two sites with 12-bp perfect matches followed by NGG for the tyr gRNA (tyr OT1 and OT2) exist in the zebrafish genome. Among these six potential off-target sites, the T7EI assay showed that only egfp OT1 exhibited detectable mutagenesis activity by egfp gRNA/Cas9. When further examining the egfp OT1, there were only two mismatches between the egfp OT1 and the perfect 20-bp egfp target. From these data, we conclude that the seed region alone on the gRNA (i.e., 11–12 bases before the PAM) is not sufficient to guide the Cas9 cleavage. The sequence beyond the seed region also confers target specificity and two mismatches are tolerated. This finding is consistent with a recent study in human cells (ref. 26; SI Discussion).
We next tested whether using different gRNAs targeting the same gene would generate the same phenotypes. We found that another gRNA targeting a different location on the gol locus (gol target 2) induced similar pigmentation defects with equivalent efficiencies as the first gol gRNA when coinjected with Cas9 (compare Fig. S7 with Fig. 4).
Last, we tested the specificity of Cas9-induced phenotypes by an RNA rescue assay. When the egfp-ddx19 RNA was coinjected with ddx19 gRNA/Cas9, a full rescue of Cas9-induced ddx19 knockout phenotypes was observed (Fig. S6_D_). T7EI assays on those morphologically wild-type, rescued larvae confirmed that the ddx19 target site in the rescued embryos was mutated as efficiently as in the embryos without egfp-ddx19 RNA (Fig. S6_E_), indicating that the wild-type–like phenotypes were true rescued phenotypes, not due to the failure of Cas9-induced mutagenesis. Taken together, we conclude that our Cas9 mutagenesis system is highly specific. This information should be taken into consideration when designing gRNAs to avoid potential off-target sites. Using independent gRNAs targeting the same gene and performing rescue assays can further validate the specificity of Cas9-induced phenotypes.
Discussion
Here we report a highly efficient and straightforward CRISPR/Cas9 strategy for targeted gene knockouts in zebrafish. Two CRISPR/Cas9 genome editing strategies in zebrafish have been recently reported (15, 16). The mutagenesis efficiencies in these two studies are ∼24–60% (15) and ∼35% (16), respectively. In contrast, the mutagenesis efficiency in our CRISPR/Cas9 system consistently surpasses 75%, often exceeding 90%. This improved CRISPR/Cas9 system represents one of the most efficient targeted vertebrate genome editing systems reported to date.
The higher mutagenesis rates in our system could be partially due to better expression and nuclear localization of the Cas9 protein with the zebrafish codon-optimized Cas9 and dual NLS tags. It is also noteworthy that the Cas9 protein sequence in our system (Fig. S1_A_) is slightly different from that used in vitro (3) and that in recently reported mammalian (8, 10, 11) and zebrafish (15, 16) studies. In addition, the 3′ end of our gRNA is also different from others, ending with the string of GGAUC residues instead of U residues. Future experiments are needed to determine whether these small variations in the Cas9 protein and the gRNA structure contribute to the higher mutagenesis rates observed in our system.
Assuming that disruption of two alleles of a given gene is largely independent, the 75–90% mutagenesis rates induced by our CRISPR/Cas9 system are estimated to cause 56–81% biallelic disruption of the target gene. Consistent with such an estimate, we observe pronounced recessive founder phenotypes in four of five genes targeted. In the cases of targeting tyr (Fig. 3) or ddx19 (Fig. 5), the estimated biallelic disruption could reach 80–90% of all cells in the injected embryos. These exceptionally high efficiencies generate phenotypes nearly equivalent to the null phenotypes. Thus, we expect that for certain genomic targets, the CRISPR/Cas9 system can serve as an alternative to the antisense morpholino knockdown strategy to generate loss-of-function phenotypes. As to the variability between morpholino knockdown and Cas9 knockout approaches, both approaches appear to be dose dependent. Because the Cas9-induced mutagenesis results in variable indels near the target site, some alleles will encode partial loss-of-function products (e.g., in-frame indels). Thus, hypomorphic or mosaic phenotypes can be observed in the highly mutagenized animals due to the generation of hypomorphic alleles. We speculated that at least part of the mosaicism observed in the Cas9-targeted founders is due to generation of hypomorphic clones due to in-frame indels.
It is surprising that targeting mitfa results in ∼75% mutagenesis rates (Fig. S4), and yet shows no discernible phenotype in injected embryos. mitfa is a master transcription regulator required for both specification and differentiation of melanocytes (27). In the absence of mitfa function, melanocyte precursors are thought to die (27), but specific ablation of melanocytes can trigger regeneration of new melanocytes from surrounding unpigmented precursors (28, 29). Therefore, one possible explanation for the lack of _mitfa_-deficient pigmentation phenotypes in the CRISPR/Cas9 system is that the few remaining _mitfa_-positive precursor cells surrounding the _mitfa_-null cells regenerate and mask the mitfa defects. Unless a complete disruption of mitfa is achieved, the compensatory melanocyte regeneration program could make it difficult to generate _mitfa_-deficient phenotypes in the mosaic mitfa disrupted larvae.
Our data also suggest that not all genomic loci are equally accessible for mutagenization by the CRISPR/Cas9 system; a similar observation has also been noted in an earlier study (15). For example, in our studies, targeting the Tg(-5.1mnx1:EGFP), tyr, and ddx19 loci consistently results in higher mutagenesis rates than targeting gol and mitfa. Testing a range of gRNA/Cas9 concentrations for those targets does not alter such a tendency, suggesting that the intrinsic properties of the target sites (e.g., chromatin/epigenetic states, transcription activities, etc.) and/or of the targeting gRNA (e.g., RNA secondary structures, the affinity to Cas9, etc.) could affect the targeting efficiency. An important area awaiting further investigation is to determine the underlying mechanisms governing target site recognition and gRNA–Cas9 interaction.
We observe efficient germ-line transmission of Cas9-induced mutations in the Tg(-5.1mnx1:EGFP) transgene and endogenous gol loci to the F1 progeny. Interestingly, the same mutations are repeatedly identified among individual _egfp_- and _gol_-targeted F1 progeny (Fig. 2_K_ and Fig. S5_F_), suggesting low complexity of germ-line mutations in the founders. In contrast, the Cas9-induced mutations in the somatic cells of the founders are highly diverse. Previous studies have shown that although zebrafish germ plasm can be detected at the first two cleavage planes, they asymmetrically segregate as four subcellular granules during subsequent divisions in the first 4 h of zebrafish development. Consequently, there are only approximately four germ cells by the end of 1,000-cell stage (30, 31). Therefore, if Cas9-induced mutagenesis occurs around this stage, for any given endogenous locus, there will be ≤8 targets in the germ cells but ∼2,000 targets in the somatic cells. Because the Cas9-induced mutation disrupts the target site (unlike the ZFNs and TALENs, in which the cleavage and target sites are separate), each target can only be mutated once. Even if the gRNA/Cas9 activity has a long perdurance, the complexity of mutations will not change. Hence, Cas9-induced mutagenesis during the blastula stage will result in low complexity of germ-line mutations but highly diverse somatic mutations.
One of the most exciting findings in this study is the demonstration of robust multiplex, biallelic somatic gene disruptions in zebrafish. We have shown that five genomic loci can be disrupted simultaneously in a highly efficient manner. This is a unique example of multiplex gene knockouts in zebrafish, accompanied by the generation of founders exhibiting multiple biallelic gene-knockout phenotypes. When this study was under review, another study also demonstrated multiplex gene knockouts in mice (17). The efficient multiplex gene targeting capability of the CRISPR/Cas9 method will be particularly beneficial for reverse genetics applications in zebrafish, wherein ∼20–30% of the genes are duplicated from an ancient genome duplication event (32, 33). In addition to the zebrafish model, this RNA-guided genome editing strategy has the potential for broad applications in other model organisms.
Materials and Methods
Plasmids.
The coding sequence for S. pyogenes MGAS1882 (YP_005388840) was synthesized with codons preferentially used in zebrafish. The sequence also encodes a nuclear localization signal of SV40 T-antigen at both the N- and C termini and contains a Kozak consensus at the translational start. The synthetic gene, named nls-zCas9-nls hence forward (full sequence in Fig. S1), was cloned into pCS2 and pT3TS for SP6 and T3 promoter-driven synthesis of capped sense RNA in vitro, respectively. See SI Materials and Methods for the generation of target gRNA constructs and Tables S1 and S2 for sequences of genomic targets and oligonucleotides for PCR.
RNA Synthesis.
For making nls-zCas9-nls RNA, the template DNA was linearized either by NotI (pCS2-nls-zCas9-nls) or XbaI (pT3TS-nls-zCas9-nls) digestion and purified using a QIAprep column (Qiagen). Capped nls-zCas9-nls RNA was synthesized using mMESSAGE mMACHINE SP6 or T3 kit (Invitrogen) and purified using RNeasy Mini kit (Qiagen). For making gRNA, the template DNA was linearized by BamHI digestion and purified using a QIAprep column. gRNA was generated by in vitro transcription using T7 RNA polymerase (New England Biolabs) or MEGAshortscript T7 kit (Invitrogen). After in vitro transcription, the gRNA (∼100 nucleotides long) was purified using _mir_Vana miRNA isolation kit (Invitrogen). The size and quality of resulting gRNA was confirmed by electrophoresis through a 2% (wt/vol) formaldehyde agarose gel.
Microinjection.
The mix of gRNA and nls-zCas9-nls RNA was injected directly into one-cell–stage embryos as previously described (19). Embryos were either derived from the wild-type AB cross or from the outcross of Tg(-5.1mnx1:EGFP) and/or Tg(-5.1mnx1:TagRFP) fish. All animal research was approved by the Institutional Animal Care and Use Committee, Office of Animal Welfare Assurance, Vanderbilt University.
Supplementary Material
Supporting Information
Acknowledgments
The authors thank Koichi Kawakami for the Tol2 transposon system, Chi-Bin Chien for Gateway plasmids, members of S.R.W. and W.C. laboratories for discussions, and Patrick Page-McCaw for critical reading of the manuscript. This work was supported by FY-10-360 (to S.R.W.) from the March of Dimes Foundation and R01DK088686 (to W.C.) from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Joung JK, Sander JD. TALENs: A widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 3.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 5.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14(3):321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 7.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 12.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23(4):465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 19.Jao LE, Appel B, Wente SR. A zebrafish model of lethal congenital contracture syndrome 1 reveals Gle1 function in spinal neural precursor survival and motor axon arborization. Development. 2012;139(7):1316–1326. doi: 10.1242/dev.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page-McCaw PS, et al. Retinal network adaptation to bright light requires tyrosinase. Nat Neurosci. 2004;7(12):1329–1336. doi: 10.1038/nn1344. [DOI] [PubMed] [Google Scholar]
- 21.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310(5755):1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 22.Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126(17):3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 23.Noble KN, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export II: Nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25(10):1065–1077. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross T, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315(5812):646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 25.Amsterdam A, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101(35):12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013 doi: 10.1038/nbt.2623. dx.doi.org/10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson SL, Nguyen AN, Lister JA. mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev Biol. 2011;350(2):405–413. doi: 10.1016/j.ydbio.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang CT, Sengelmann RD, Johnson SL. Larval melanocyte regeneration following laser ablation in zebrafish. J Invest Dermatol. 2004;123(5):924–929. doi: 10.1111/j.0022-202X.2004.23475.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang CT, Johnson SL. Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development. 2006;133(18):3563–3573. doi: 10.1242/dev.02533. [DOI] [PubMed] [Google Scholar]
- 30.Raz E. Primordial germ-cell development: The zebrafish perspective. Nat Rev Genet. 2003;4(9):690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- 31.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124(16):3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 32.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282(5394):1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 33.Postlethwait JH, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18(4):345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information