High-Resolution Patterns of Meiotic Recombination across the Human Major Histocompatibility Complex (original) (raw)
Abstract
Definitive characteristics of meiotic recombination events over large (i.e., >1 Mb) segments of the human genome remain obscure, yet they are essential for establishing the haplotypic structure of the genome and for efficient mapping of complex traits. We present a high-resolution map of recombination at the kilobase level across a 3.3-Mb interval encompassing the major histocompatibility complex (MHC). Genotyping of 20,031 single sperm from 12 individuals resulted in the identification and fine mapping of 325 recombinant chromosomes within genomic intervals as small as 7 kb. Several principal characteristics of recombination in this region were observed: (1) rates of recombination can differ significantly between individuals; (2) intense hot spots of recombination occur at least every 0.8 Mb but are not necessarily evenly spaced; (3) distribution in the location of recombination events can differ significantly among individuals; (4) between hot spots, low levels of recombination occur fairly evenly across 100-kb segments, suggesting the presence of warm spots of recombination; and (5) specific sequence motifs associate significantly with recombination distribution. These data provide a plausible model for recombination patterns of the human genome overall.
Introduction
Continual development of the human genome sequence has led to a growing interest in defining haplotypic blocks that will assist in the mapping of genes involved in complex diseases (Daly et al. 2001; Goldstein 2001; Reich et al. 2001; Rioux et al. 2001). A number of molecular genetic mechanisms, including mutation, meiotic recombination (gene conversion and crossing over), selection, and demographic history, are likely to dictate the length of haplotypic blocks and the extent of linkage disequilibrium (LD) within each block. Analyses of recombination hot spots have been reported for a number of genomic regions in humans (Lebo et al. 1983; Chakravarti et al. 1984, 1986; Rouyer et al. 1986; Bowcock et al. 1988; Grimm et al. 1989; Charmley et al. 1990; Benger et al. 1991; Oudet et al. 1992; Pentao et al. 1992; Hubert et al. 1994; Reiter et al. 1996; Jeffreys et al. 1998; Lopes et al. 1998; Smith et al. 1998; Yip et al. 1999; Badge et al. 2000; Schneider et al. 2002), the most detailed of which have involved the major histocompatibility complex (MHC) class II region (Cullen et al. 1995, 1997; Jeffreys et al. 2000, 2001). Recent data have indicated a convincing inverse correlation between rate of meiotic recombination and strength of LD within a 200-kb segment of the MHC class II region (Jeffreys et al. 2001), providing concrete evidence that recombination plays a primary role in defining the boundaries of strongly associated haplotypic blocks.
The MHC spans >3 Mb on the short arm of chromosome 6 and contains the most polymorphic loci (and, consequently, haplotypes) known in humans. About 40% of the 128 genes in this region participate in immune responses (MHC Sequencing Consortium 1999), and it is likely that beneficial interactions among specific allelic products of these loci result in selective pressure to secure their close linkage relationship (Beck and Trowsdale 2000) and maintain particular haplotypes (Kwok et al. 1993). On the other hand, a constant—though perhaps low-level—shuffling of haplotypes may protect against novel human pathogens such as HIV-1. In this case, haplotypes that are particularly beneficial against a novel pathogen may arise through recombination and would not necessarily already be present in high frequencies if they were not effective against pathogens that have existed in the population over long periods of time. Measurements of linkage disequilibrium among HLA class I and II genes in the CEPH have emerged over the past several years (Begovich et al. 1992; Klitz et al. 1995; Cullen et al. 1997; Bugawan et al. 2000; Sanchez-Mazas et al. 2000), revealing significant associations between specific alleles of loci separated by distances >1 Mb. However, the dynamic interplay between the generation of new haplotypes through recombination and the maintenance of those that are under selective pressure to remain in the population is poorly understood, in part because of a lack of detailed information regarding recombination across the entire region.
Previous studies of recombination in families have suggested that recombination across the MHC overall is lower than expected (Martin et al. 1995), although the use of family material for the generation of such information is severely limited in power. Fairly extensive mapping of recombination hot spots and cold spots in the class II region has been performed (Cullen et al. 1995, 1997; Jeffreys et al. 2000, 2001), the most detailed of which involved sperm typing of a 200-kb segment encompassing the genes DNA and DMB (Jeffreys et al. 2001).
Here we use single-sperm typing (Li et al. 1988; Cui et al. 1989; Goradia et al. 1991; Schmitt et al. 1994; Yu et al. 1996; Brown et al. 1998; Lien et al. 2000) to generate a reliable estimate of the frequency and distribution of recombination events across a large segment of DNA (3.3 Mb), using a strategy virtually identical to that employed by Hubert et al. (1994) for high-resolution mapping of recombination events in single sperm. The recombination characteristics described here for the MHC provide reasonable expectations of recombination patterns for the genome as a whole. Furthermore, these data will enhance our ability to assess putative selection processes in the MHC, by considering LD values in light of recombination intensity between the loci in question.
Subjects and Methods
Single-Sperm Isolation
All donations of semen for this study were provided on an anonymous basis, as specified in the National Institutes of Health institutional review board–approved protocol OH97-C-N070. All donors were white, with the exception of the MZ twins X and Y, who were of African American descent. Identity was confirmed for twin sib pairs X-Y and V-W on the basis of genetic identity at 13 and 10 unlinked STRs, respectively, as described elsewhere (Hammond et al. 1994). Donor ages ranged from 18 to 49 years.
Sample preparation began within 4 h of semen collection. Fresh cells were washed repeatedly in 1× PBS and were stained in Hoechst 33342. Single sperm were isolated in each well of a microtiter plate by fluorescence-activated cell sorting followed by alkaline lysis release of one haploid genome per well (Li et al. 1991; Cullen and Carrington 2001).
Primer Extension Preamplification (PEP)
PEP, or whole-genome amplification (Zhang et al. 1992), was performed on each haploid sample in a total volume of 60 μl containing 10 mM Tris-HCl (pH 8.3); 2.5 mM MgCl2; 200 μM each of dATP, dCTP, dGTP, and dTTP; 40 μM random 15-mer oligonucleotide primer; 5 U of Taq DNA polymerase; and 10 μl of neutralized sample. PEP reactions were covered with an overlay of mineral oil and proceeded under the following conditions: 95°C for 5 min, 50 cycles of denaturation at 92°C for 1 min, annealing at 37°C for 2 min, and extension at 55°C for 4 min (3-min ramp time from 37°C to 55°C), followed by final extension at 72°C for 10 min. A 60-μl post–PEP-amplified sperm DNA sample provides sufficient material for ⩾12 individual PCRs. Multiplexing several STRs in a single PCR dramatically increased the number of markers typed per sample. Markers exhibiting robust amplification characteristics were successfully amplified with as little as 2 μl of PEP-DNA.
PCR Amplification Using PEP Products
PCR amplification was performed in a total volume of 20 μl containing 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 2.5 mM MgCl2; 200 μM each of dATP, dCTP, dGTP, and dTTP; 0.2–1.6 μM forward and reverse primers; 1.25 U of Taq DNA polymerase; and a 5-μl aliquot of PEP-amplified haploid DNA. The PCR proceeded under the following conditions: 95°C for 9 min, 10 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 4 min, 15 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 3 min, followed by final extension at 72°C for 10 min. The internal primer used in the second round of nested PCR was end-labeled with γ(32P)-ATP. The second round of nested PCR amplification was performed in a total volume of 20 μl containing 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 2.5 mM MgCl2; 200 μM each of dATP, dCTP, dGTP, and dTTP; 0.9 μM internal primer; 1 μM complementary primer; 1.25 U of Taq DNA polymerase; 0.016 μM γ(32P)-ATP–labeled internal primer; and 1/10 the contents of the completed first-round amplification reaction. The PCR proceeded under the following conditions: 94°C for 9 min, 5 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, 25 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, followed by final extension at 72°C for 30 min. Alleles of STRs were resolved by electrophoresis through a 6% denaturing polyacrylamide gel. Further details are available elsewhere (Cullen and Carrington 2001)
Single-Sperm Typing Errors
Accurate scoring of single-sperm genotypes is critical for estimating the recombination fraction between two polymorphic markers. Ideally, estimates are generated by simply dividing the number of two-marker haplotypes tested (recombinant + nonrecombinant) into the number of recombinant haplotypes identified. However, genotyping errors resulting from lack of amplification, PCR contamination, or the presence of zero or two sperm per well are not uncommon. As a result, data generated from a two-marker analysis consist of 16 potential outcomes: two types of nonrecombinant haplotypes, two types of recombinant haplotypes, and 12 erroneous genotypes resulting from experimental error (table 1). Estimates of the recombination fraction calculated without consideration for the errors inherent in single-sperm typing may be imprecise when recombination events cannot be confirmed by typing of additional markers and when the order of the markers along the chromosome is not known. In this situation, the number of obvious errors (e.g., blank wells, typing of only one marker, etc.; see table 1) would correlate positively with errors that are not obvious (i.e., errors that occurred in a situation where only one allele at each of the two delineating markers is typed), and the use of a statistical program that provides maximum-likelihood estimates of the recombination fraction and associated standard errors is necessary (Lazzeroni et al. 1994). On the other hand, if estimates of the recombination fraction between pairs of tightly linked markers of known chromosomal order are calculated for the purpose of comparing recombination rates between different genomic intervals, and recombination events are confirmed using two or more pairs of markers, the program is not necessary (Leeflang et al. 1994). This point is exemplified in the study by Hubert et al. (1994), in which 25 recombinant sperm were fine mapped in a 1-Mb region near the Huntington disease gene, without requiring the generation of likelihood estimates of recombination fractions and error parameters. Our mapping strategy is virtually identical to that developed by Hubert et al. (1994). Furthermore, 91% of recombinants identified in the initial survey and fine mapped within the MHC were confirmed with at least two and usually more than three markers on either side of the crossover. No convincing cases of multiple crossovers were identified.
Table 1.
Genotypes Observed from Single-Sperm Typing of Markers Flanking the MHC[Note]
| No. of Sperm Identified for Genotype in Donor | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Two-Marker Genotypea | A | B | C | G | L | E | S | T | X | Y | V | W |
| Informative meiosesb: | ||||||||||||
| L/S | 1,492 | 16 | 412 | 862 | 683 | 865 | 619 | 529 | 16 | 14 | 896 | 955 |
| S/L | 1,425 | 15 | 405 | 673 | 588 | 820 | 583 | 535 | 36 | 31 | 875 | 952 |
| L/L | 29 | 856 | 12 | 13 | 17 | 29 | 12 | 15 | 854 | 793 | 32 | 25 |
| S/S | 46 | 919 | 13 | 57 | 62 | 19 | 17 | 7 | 969 | 895 | 25 | 18 |
| Blank wells: | ||||||||||||
| Null/null | 168 | 253 | 55 | 267 | 315 | 130 | 244 | 173 | 60 | 142 | 94 | 184 |
| Only one locus amplifies: | ||||||||||||
| Null/S | 94 | 96 | 28 | 63 | 80 | 60 | 51 | 30 | 48 | 70 | 36 | 14 |
| Null/L | 112 | 94 | 20 | 53 | 72 | 54 | 42 | 25 | 49 | 101 | 28 | 18 |
| S/null | 100 | 81 | 67 | 24 | 57 | 31 | 107 | 61 | 41 | 64 | 36 | 25 |
| L/null | 72 | 88 | 39 | 20 | 43 | 40 | 72 | 59 | 18 | 32 | 39 | 29 |
| Two-allele genotype at only one locus: | ||||||||||||
| B/null | 1 | 0 | 0 | 0 | 1 | 1 | 3 | 2 | 2 | 1 | 2 | 1 |
| Null/B | 0 | 2 | 0 | 0 | 1 | 4 | 2 | 1 | 3 | 4 | 2 | 0 |
| Three-allele genotypes: | ||||||||||||
| S/B | 24 | 3 | 7 | 29 | 16 | 49 | 7 | 7 | 10 | 9 | 26 | 12 |
| L/B | 5 | 2 | 11 | 65 | 11 | 36 | 7 | 6 | 35 | 10 | 10 | 9 |
| B/S | 17 | 2 | 5 | 7 | 17 | 29 | 14 | 9 | 5 | 3 | 25 | 15 |
| B/L | 1 | 3 | 3 | 0 | 2 | 9 | 5 | 1 | 63 | 16 | 28 | 20 |
| Four-allele genotypes: | ||||||||||||
| B/B | 26 | 5 | 8 | 36 | 40 | 69 | 33 | 49 | 24 | 14 | 38 | 9 |
| Anomalous genotypesc | 1 | 2 | 7 | 16 | 22 | 8 | 14 | 9 | 35 | 9 | 16 | 14 |
| Total | 3,612 | 2,435 | 1,084 | 2,169 | 2,005 | 2,245 | 1,818 | 1,509 | 2,233 | 2,199 | 2,192 | 2,286 |
Identification of STRs and Recombination Sequence Motifs within the MHC
A total of 3.3 Mb of MHC sequence data was screened for STRs through use of the DNA analysis program SPUTNIK (SPUTNIK Web site). The exact positions of all di-, tri-, tetra-, and pentanucleotide repeats containing ⩾12, 8, 6, and 5 repeats per locus, respectively, were mapped on a 3.3-Mb genomic sequence retrieved through a public MHC database (Sanger Institute Web site) as a contig of 79 overlapping sequences edited to remove sequence overlap. Polymorphic STRs were identified by PCR amplification of DNA from a reference panel of 15 cell lines (homozygous typing cells [HTCs]) (Yang et al. 1989). Sequence information for oligonucleotide primers used in the study is available in table A (online only)table A (online only). In addition, the number and distribution of 21 candidate recombination sequence motifs (table 2) were mapped within the MHC using the sequence analysis program FINDPATTERN (Wisconsin Package Version 10.2; Genetics Computer Group).
Table A.
Oligonucleotide Primer Sequences for the First- and Second-Round Amplification of 49 STRs[Note]
| Locusa | Sequence (5′→3′) | Location | Nucleotide Positionb |
|---|---|---|---|
| *D6S291: | |||
| 5o′ | AGCTCAGAGGATGCCATGTC | ||
| 5i′ | GTCTAAAATATCCATCCGGC | ||
| 3′ | GGGGATGACGAATTATTCACTAACT | 2.4 Mb centromeric to DPB1 | Centromeric to DPB1 |
| *D6S439: | |||
| 5o′ | GTCCACATTGCATGGGTCTG | ||
| 5i′ | GATGATTTAAGTTTCCTGTGGACC | ||
| 3′ | TTCAAGGACAGCCTCAGGG | .85 Mb centromeric to DPB1 | Centromeric to DPB1 |
| DPB2: | |||
| 1720 | TTACAGGAGGACATAAGTCC | ||
| 2000 | TTTAGGCAAGGTCAGCAACC | DPB2 intron 5 | 221554 |
| DPB2A2: | |||
| 5′ | TGGGCTTGAGCTAAAGACTG | ||
| 3i′ | GGCAGGAGAATGGCATAAAC | ||
| 3o′ | TAGTGCCAGCTACTAGGAAG | Centromeric to DPB1 | 254315 |
| DPB1: | |||
| 14251 | TGTCGTCTAAGTCTGCATTC | ||
| 14528 | TGCTGTGCACTGATTGTGTG | DPB1 3′ UTR | 263644 |
| DPB1tetra: | |||
| 5o′ | ATGCTGCCTGGGTAGAAATC | ||
| 5i′ | TCTTGGAGGGGGAAACATTC | ||
| 3′ | TGCATTCCAGACTTGGCAAC | DPB1 | 266072 |
| DNA: | |||
| 5200 | CCTCAGTGGAGTCAGTCATG | ||
| 5446 | CCCAAGCAGTTCTTATGTGC | DN-A 3′ UTR | 344577 |
| DNRNGCA: | |||
| 5o′ | GGTGACATACATCAACTTGTG | ||
| 5i′ | AGGAATCTAGTGCTCTCTCC | ||
| 3′ | CTCTAGCAAAAGGAAGAGCC | Between HLA-DNA and RING3 | 355527 |
| RING3CA: | |||
| 5′ | GCATACACTCTGAAGCAGCC | ||
| 3i′ | GATGGGAAGTTTCCAGAGTG | ||
| 3o′ | GAGGTAATGTCACAGGATGGG | RING3 | 375386 |
| G3.64842: | |||
| 5o′ | GAGACAGAAGTAACAAAGGC | ||
| 5i′ | TGATGGCAGAAGTAATCCAG | ||
| 3′ | GTATAAGCCATTGCCACCAG | Between HLA-DMB and LMP2 | 461218 |
| *TAP1CA: | |||
| 5o′ | CGCCATGTACTGTTCATATC | ||
| 5i′ | GCTTTGATCTCCCCCCTC | ||
| 3′ | GGACAATATTTTGCTCCTGAGG | TAP1 | 498684 |
| 2-7A: | |||
| 5o′ | GGGGAAGGATTCTAAATAGG | ||
| 5i′ | GTATTTCTGTATGACTCTGG | ||
| 3′ | AACCCAGCATTCTGGAGGTG | Between DQA2 and DQB3 | 601210 |
| G4-96: | |||
| 5o′ | AGTCCCAGCTACTACTCAGG | ||
| 5i′ | TCACACCACTACACTCCATG | ||
| 3′ | CAACGGCATGTCAATTAGTGC | Between DQA2 and DQB3 | 612822 |
| 2-8: | |||
| 5′ | ATGAGATTGCACCACTGCAC | ||
| 3i′ | TACCTCCAACACTTTCTGGC | ||
| 3o′ | TATACGTCCCATGAGGACAG | Telomeric to DQB3 | 634333 |
| G5-11525: | |||
| 5o′ | GGTAAAATTCCTGACTGGCC | ||
| 5i′ | ATTTGCCCTGAGGTCTATGC | ||
| 3′ | GACAGCTCTTCTTAACCTGC | Between DQB3 and DQB1 | 648774 |
| *DQCARII: | |||
| 5o′ | TCTTGAGGACTGAGTCTGAG | ||
| 5i′ | GGGCAGCATTTGTAGATTTC | ||
| 3′ | GGCAAGAATCCAGCATTTTGG | Telomeric to DQB1 | 700000 |
| DRB1CA: | |||
| 5o′ | TAGATTGTGGTACTATCAGG | ||
| 5i′ | TTTGAGTTAACTTCCACACG | ||
| 3′ | GAGCTAGTCATTCCTTATTC | DRB1 | 767929 |
| DRA-CA: | |||
| 5o′ | ACATTATGTTCTGTTGCATG | ||
| 5i′ | TGGAATCTCATCAAGGTCAG | ||
| 3′ | TACTTTCCTAATTCTCCTCC | DRA | 890718 |
| 2-10: | |||
| 5o′ | AGGTGACCTGGACCTTACTG | ||
| 5i′ | TTGTTGGAGGGGCAAAAGT | ||
| 3′ | ACACTATGCTAGTCTGTGCC | Between BTL-II and TSBP | 952064 |
| 2-11: | |||
| 5o′ | TTCAGTATCCACTCACCAGG | ||
| 5i′ | CTGCCTATAGATGTTCCCTC | ||
| 3′ | TGTTGAGTTCTGGAGATAGG | Between TSBP and NOTCH4 | 959038 |
| 3-3A: | |||
| 5o′ | AAGATCACCCTACCACACTC | ||
| 5i′ | AGAGTGAGAGACTCTGTCTC | ||
| 3′ | AGGGAGAAGGATAGATAAGG | Between TSBP and NOTCH4 | 995315 |
| 3-2A: | |||
| 5′ | GGCAAAGTCAAGAACTTTCC | ||
| 3i′ | AGCTGAATAGTATTCCACTAC | ||
| 3o′ | CCAGGTTCATCCATGTTGTC | Between TSBP and NOTCH4 | 1049652 |
| 3-1: | |||
| 5′ | AAGCAACTGCTCTCGTAGAG | ||
| 3i′ | CCCAACACCTGCTCATTTTC | ||
| 3o′ | TTCAGTGACTTTGCTCTCAGC | Between NOTCH4 and G18 | 1111584 |
| 3-1A: | |||
| 5′ | AAAGGCCTCTACACCCAGAG | ||
| 3i′ | TGACAGACTGGGACTCCATC | ||
| 3o′ | AGGAGGTGAAGGTTGCAGTG | Between NOTCH4 and G18 | 1114474 |
| 3-3: | |||
| 5′ | CCTGACCACAAAGCTTTCTC | ||
| 3i′ | GGCAACAGAGCTCAAAAAAC | ||
| 3o′ | AAAGGTTGCAGTGAGCCAAG | Between TNXB and CYP21A2 | 1247950 |
| 3-4: | |||
| 5o′ | AGTTCGCACCACTGTAGTCC | ||
| 5i′ | ATAGAGTGAGACCCTGTCTC | ||
| 3′ | ATGAGGATACCCACTCTACC | Between BF and C2 | 1354878 |
| 3-4d: | |||
| 5o′ | ACAAAGTTCTGGCGAGGAAC | ||
| 5i′ | ACCAGTGGCTTCCAAACTTC | ||
| 3′ | GAGATCACGCCATTGCACTC | Between BF and C2 | 1358331 |
| 3-5: | |||
| 5o′ | CATGGAGAGAGCATGTGTTC | ||
| 5i′ | CTGATTGTGAGGTCACTGTG | ||
| 3 | AGATTGTGCCACTGCACTCC | Between HSPA1L and G7b | 1483675 |
| *D6S273: | |||
| 5o′ | CTTGCTAAATCAAATAAAGTC | ||
| 5i′ | GGAGAAGTTGAGTATTTCTGC | ||
| 3′ | ACCAAACTTCAAATTTTCGG | Between G6d and G6e | 1577288 |
| BAT2CA: | |||
| 5o′ | AAAGCTCCCTTCACTTTATC | ||
| 5i′ | TACAAGGGCTTTAGGAGGTC | ||
| 3′ | AGCCTGGATAACAGAACGAG | Telomeric to BAT2 | 1683589 |
| 3-7: | |||
| 5o′ | AGATTTCATCCAGCCACAGG | ||
| 5i′ | GCCTTTAAAGGTCGTCAGAG | ||
| 3′ | ATCCAATCACCTCTGCTCAC | Between LTA and NFKBIL1 | 1725566 |
| MICB-CA: | |||
| 5o′ | ATGGGCAAGACTTCAATGGC | ||
| 5i′ | CCACAACGAGATACCACTTC | ||
| 3′ | CTACCTCCTTGCCAAACTTG | MICB | 1789221 |
| 3-11: | |||
| 5′ | TAGCTGGGACTACAGACTGG | ||
| 3i′ | ACAAAGCAAGACCCAGTCTC | ||
| 3o′ | AAGTTCAAGACCAGCATGGG | Between MICA and HLA-B | 1895528 |
| *MIB: | |||
| 5o′ | CCCATGCTGAATTAGCTGAC | ||
| 5i′ | GCTTCACCCGATCAGTAGAAGAC | ||
| 3′ | CGTACCACAGTCTCTATCAGTCCAG | Between MICA and HLA-B | 1909387 |
| HLABC-CA: | |||
| 5o′ | CCCAGTAGTAAGCCAGAAGC | ||
| 5i′ | TGGGCAATGAGTCCTATGAC | ||
| 3′ | TGCCATTTGGCCCTAAATGC | Between HLA-B and HLA-C | 1999074 |
| 1-1k: | |||
| 5o′ | CCAAGATTGTGCCATTGCAC | ||
| 5i′ | GCGACAAGAGAGAAACTCTG | ||
| 3′ | GGTGTTGGTTTTGTAACTCC | Telomeric to CDSN | 2183701 |
| 1-1g: | |||
| 5o′ | TACTCGGAAAGCTGAGGCAC | ||
| 5i′ | CTGAGATCGCACTACTGCAC | ||
| 3′ | GTCTTTCCCCTGAACCAAAC | Telomeric to CDSN | 2189182 |
| 1-2A: | |||
| 5o′ | TATTGTCCTATGACCCTGCC | ||
| 5i′ | CTCTGCGAGAAACACCCAAG | ||
| 3′ | CAGCGGAAAAATTCAGATCG | Centromeric to P52 | 2307351 |
| 1-3: | |||
| 5o′ | AACCTAAGCCTTCACCTCTC | ||
| 5i′ | CTCAGCAGGAACCAGACAAC | ||
| 3′ | TTGGGACTACAGGTGTGAGC | Between DDR and IER3 | 2419106 |
| 1-3A: | |||
| 5′ | CAGCAGACCTACCCTAAAAG | ||
| 3i′ | TGAGATCCTGCTTCTACACG | ||
| 3o′ | AGGAGTTTGAGAGTAGCCTG | Between DDR and IER3 | 2420460 |
| 1-3B: | |||
| 5′ | AGAGCGAGACTCCTTCTAAG | ||
| 3i′ | TTGCACTTACTGGACCCCTG | ||
| 3o′ | TTTCTGACTCCACCTCACAG | Between DDX16 and PPP1R10 | 2554452 |
| 1-3F: | |||
| 5o′ | ATTACTTGAACCCAGGAGGC | ||
| 5i′ | CTGGCAACAGAGTAAGACTC | ||
| 3′ | CTTCACAGGGTTATTAGGGG | Between PPP1R10 and ABCF1 | 2623029 |
| 1-6: | |||
| 5′ | CTGAGGTCTGTGGTCATAAC | ||
| 3i′ | ATATTTTCCTGCTGCTGCCC | ||
| 3o′ | TCCCATGATCCTTACAGCAG | Between RANBP1 and GT257 | 2776550 |
| 1-7A: | |||
| 5′ | TACATTCTGTAGGCATTTTCAAGC | ||
| 3i′ | GGGCAACAAGATCAAAACTC | ||
| 3o′ | ATTGTGGTAAGCCGAGATTG | Between CAT75X and ZNF173 | 2947999 |
| 1-8: | |||
| 5′ | CTTACATGAATCGCAGAGGG | ||
| 3i′ | AATTTGGACCCACTCCGCTG | ||
| 3o′ | TTCTGGGTAATTCGAGACGC | Between ZNFB7 and RNF9 | 3052507 |
| 1-8A: | |||
| 5o′ | TGTAAGGATTGCTCATCGGG | ||
| 5i′ | CCCTCCTACGGTCCCAAATC | ||
| 3′ | TCTTGCCTTCTCCCCGCTAC | Between ZNFB7 and RNF9 | 3053094 |
| 1-8e: | |||
| 5′ | CGTGCAAGCCGAAAGTCTTC | ||
| 3i′ | CACTGTCCCTGGTTTATAGC | ||
| 3o′ | ACCACAACCTCACTTGATCC | Between HCG1 and ZNRD1 | 3121184 |
| *D6S265: | |||
| 5′ | AGTCACCCTACTGTGCTATC | ||
| 3i′ | ATCGAGGTAAACAGCAGAAAG | ||
| 3o′ | AAAGGTTGATCTAATCGAGG | Between HTEX4 and HCGVIII-1 | 3164186 |
| 1-9: | |||
| 5o′ | CCATCCTGAGTCTTGTGATC | ||
| 5i′ | TTAAAGATTTGCACAATGAG | ||
| 3′ | GGAAGCTTAAAAACCAGCAG | Between HCGIV-6 and HCGII-7 | 3300981 |
| 1-9d: | |||
| 5′ | AGTCTATCATGGGACTTCTC | ||
| 3i′ | GCCTCCATAATTGTGTGAGC | ||
| 3o′ | ATCAGTGATACTATGACAGG | Between HCGII-7 and HLA-G | 3375510 |
| *RF: | |||
| 5′ | CTGTCCTATTTCATATGCTCAGG | ||
| 3i′ | CTGAGAATGAAGGTCTAGAG | ||
| 3o′ | ATGAACTTGTCCTGAGAATGAAG | HLA-F | 3490957 |
| *MOG-CA: | |||
| 5o′ | GAAATGTGAGAATAAAGGAGA | ||
| 5i′ | CACTCTCTAAGATCTCTTCC | ||
| 3′ | GATAAAGGGGAACTACTACA | Telomeric to HLA-F | 3541259 |
Table 2.
Correlation between Recombination Intensity and Motif Density[Note]
| Motif and Reference | Size(bp) | Observed in MHC | Correlation _R_2 (P value) | Sequencea |
|---|---|---|---|---|
| Bacterial: | ||||
| χ (Smith et al. 1981) | 8 | 198 | .082 (.12) | 5′-GCTGGTGG |
| RuvC mediated Holliday resolution motif (Shah et al. 1994) | 4 | ∼175,000 | Not tested | 5′-(A/T)TT(G/C) |
| Yeast: | ||||
| Ade6-M26 heptamer (Schuchert et al. 1991) | 7 | 80 | .018 (.48) | 5′-ATGACGT |
| CRE related sequence (Fox et al. 2000) | 8 | 114 | .072 (.15) | 5′-(C/T/G)TGACGT(A/C) |
| Autonomously replicating sequences 1 (Gale et al. 1992) | 11 | 177 | .013 (.55) | 5′-WTTTATRTTTW |
| Autonomously replicating sequences 2 (Gale et al. 1992) | 11 | ∼650 | Not tested | 5′-WRTTTATTTAW |
| Eukaryotic: | ||||
| Topoisomerase II binding site (Sander and Hsieh 1985) | 15 | 33 | .021 (.44) | 5′-GTNWAYATTNATNNR |
| Rodent: | ||||
| Mouse LTR-IS (Edelmann et al. 1989) | 11 | 4 | .113 (.07) | 5′-TGGAAATCCCC |
| Mouse retrotransposon LTR (Zimmerer and Passmore 1991) | 27 | Absent | NA | 5′-TCATACACCACGCAGGGGTAGAGGACT |
| Chinese hamster scaffold attachment sites 1, 2, 3, & 4 (Gale et al. 1992) | 10 | ∼.8k, ∼9k, ∼12k, ∼20k | Not tested | 5′-AATAAAYAAA, 5′-TTWTWTTWTT, 5′-WADAWAYAWW, 5′-TWWTDTTWWW |
| Human: | ||||
| Protein binding site translin 1 (Aoki et al. 1995) | 16–18 | 20 | .049 (.24) | 5′-GCNCWSSWN(0–2)GCCCWSSW |
| Protein binding site translin 2 (Aoki et al. 1995) | 14–18 | 12 | .091 (.10) | 5′-MTGCAGN(0–4)GCCCWSSW |
| Minisatellite core sequence (Jeffreys et al. 1985) | 10 | 95 | .046 (.25) | 5′-GGGCAGGARG |
| Hypervariable minisatellite 1 (Jeffreys et al. 1985) | 16 | Absent | NA | 5′-GGAGGTGGGCAGGARG |
| Hypervariable minisatellite 2 (Jeffreys et al. 1985) | 16 | 1 | .013 (.55) | 5′-AGAGGTGGGCAGGTGG |
| Protein binding site pur (Bergemann and Johnson 1992; Smith et al. 1998) | 16 | 54 | .066 (.17) | 5′-GGNNGAGGGAGARRRR |
| Replication origin (Dobbs et al. 1994) | 21 | 9 | .006 (.67) | 5′-WAWTTDDWWWDHWGWHMAWTT |
| XY32 homopurine-pyrimidine H-palindrome (Rooney and Moore 1995) | 30 | Absent | NA | 5′-AAGGGAGAARGGGTATAGGGRAAGAGGGAA |
| STRs: | ||||
| (Pentanucleotide)n | ⩾25 | 30 | .017 (.49) | n based on published sequence |
| (Tetranucleotide)n | ⩾24 | 115 | .050 (.23) | n based on published sequence |
| (Trinucleotide)n | ⩾24 | 22 | .028 (.37) | n based on published sequence |
| (non-GT Dinucleotide)n | ⩾24 | 37 | .016 (.50) | n based on published sequence |
| (GT Dinucleotide)n | ⩾24 | 58 | .160 (.03) | n based on published sequence |
| (GT Dinucleotide)n | ⩾24 | 56 | .141 (.04) | n based on the most prevalent allele present in a population of 36 HTCs |
| (GT Dinucleotide)n | ⩾40 | 19 | .203 (.01) | n based on the most prevalent allele present in a population of 36 HTCs |
Statistical Analysis
Contingency tables were evaluated with the log-likelihood ratio statistic (G), Pearson's χ2 statistic, or exact tests (Sokal and Rohlf 1995). The testing of levels of heterogeneity in the rates of recombination across donors as well as genomic segments was performed by the G test for goodness of fit (Sokal and Rohlf 1995). Differences in crossover location between individuals were tested using a Monte Carlo estimation of Fisher’s exact test, with 106 iterations (107 for the 6 × 8 table), using PROC FREQ in SAS.
To compare the recombination rate differences between the three pairs of HLA-identical individuals and pairs of unrelated individuals, a (nonparametric) rank test was performed on the differences between pairs. The three identical pairs were ranked among all 66 comparisons possible among 12 individuals, and their rank sum was computed. Since the 66 comparisons are nonindependent, the usual methods for evaluating the significance of the rank scores are not valid. Therefore, we evaluated the significance of the observed rank sum for the three MHC-identical pairs by specifically testing the null hypothesis that the differences between the identical pairs do not differ significantly from the differences between any three separate (nonoverlapping) pairs chosen from among the 12 individuals. In fact, there are 13,860 ways to choose three pairs; a numerical tabulation shows that 703 (5.07%) have a rank sum less than or equal to the rank sum for the actual identical pairs.
Potential correlations between recombination intensity (cM/Mb) and motif density (number observed/Mb) were tested within each segment independently, using linear regression analysis as described by Majewski and Ott (2000). Correlations were determined by PROC REG in SAS, with variables significant in the multivariate regression chosen by stepwise selection and confirmed by best-subset selection.
Results
Comprehensive Rate of Recombination in the MHC
Martin et al. (1995) reported a sex-averaged recombination rate for the MHC (defined as DPB1 to HLA-A, totaling 3 Mb) of 0.63 cM/Mb, based on 952 informative meioses in 59 CEPH families. The class I region (HLA-B to HLA-A; 1.34 Mb) was responsible for diminishing the recombination rates within the MHC overall, since the rate in this segment was only 0.15 cM/Mb (Martin et al. 1995), representing a recombination desert (i.e., <0.3 cM/Mb), as proposed elsewhere (Broman et al. 1998).
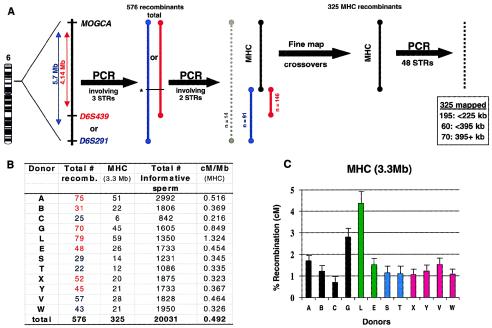
To provide a more accurate estimate of recombination rates, single-sperm typing representing thousands of informative meioses was used to generate large numbers of recombination events from 12 donors. DNA from single sperm was isolated, and the haploid genome of each was amplified using PEP (Zhang et al. 1992). Approximately 26,000 sperm from 12 donors were genotyped for STRs flanking the MHC (fig. 1_A_). A total of 20,031 sperm (the informative meioses) were successfully typed for both telomeric and centromeric markers. The remaining sperm showed 1 of 12 possible erroneous genotypes resulting from experimental error associated with a two-marker analysis of haploid DNA (see table 1 and the “Subjects and Methods” section). Crossovers occurring within a 4.14-Mb (MOGCA to D6S439) segment or an overlapping 5.7-Mb (MOGCA to D6S291) segment were identified in 576 sperm. PEP-DNA from recombinant sperm was genotyped again, using two STRs located in the vicinity of the MHC class II gene DPB1 at the centromeric boundary of the MHC. Among the 576 recombinants, 325 were localized within the MHC (DPB1 to MOGCA; 3.3 Mb), and 91% of these were confirmed by typing at least two markers on either side of the crossover breakpoint. The remaining 251 recombinants mapped within a 0.85-Mb segment (_n_=146) or an overlapping 2.4-Mb segment (_n_=91) centromeric to the class II region (fig. 1_A_) or remain unresolved (_n_=14). An estimate of 0.49 cM/Mb across the MHC, based on 20,031 informative meioses from 12 donors, fell below the male-specific estimate of 0.92 cM/Mb for the entire genome and 0.71 cM/Mb for chromosome 6 based on previously reported genetic (Broman et al. 1998) and physical (Human Genome at NCBI) distances. The extensive polymorphism within the MHC (Hughes and Hughes 1995; MHC Sequencing Consortium 1999) may account, to some extent, for the low rate of recombination in this genomic region.
Figure 1.
Summary of steps involved in recombination fine mapping. A, The MHC was surveyed for recombinants through use of PEP-amplified sperm DNA and the polymorphic markers MOGCA at the telomeric end and either D6S439 (red) or D6S291 (blue) at the centromeric end (depending on heterozygosity in each donor). Four hundred sperm containing a single crossover in the 4.14-Mb segment from MOGCA to D6S439 and 176 sperm containing a single crossover within an overlapping 5.7-Mb segment from MOGCA to D6S291 were identified. Of the 576 recombinants, 325 mapped within the MHC, as determined by typing all recombinants with two additional closely spaced markers (indicated with an asterisk [*]). Forty-eight STRs were used to localize 325 MHC crossovers within the smallest possible interval bordered by adjacent markers. Limited amounts of PEP-DNA restricted the number of markers typed for each recombinant to only those markers that would narrow the segment in which the recombinant occurred (4–11 markers typed per recombinant). B, Numbers representing recombinants identified over the entire region initially screened (red and blue indicate recombinants from MOGCA to D6S439 and MOGCA to D6S291, respectively), recombinants within the MHC, number of informative sperm, and the distance-normalized recombination rate within the MHC. C, Histogram of individual variation in the rates of recombination among 12 donors. Donors A, B, C, and G are unrelated (black). Donors L and E share one parental HLA haplotype (green), donors S and T are _HLA_-identical siblings (blue), and donors X and Y and donors V and W are two sets of MZ twins (red). Frequency of recombination plus 1 SD is shown.
Significant differences in the numbers of the two nonrecombinant single-sperm haplotypes were observed for donors G (P<10-5), L (P<.01), X (P<.01), and Y (P<.05) (table 1), corresponding with a significant difference in the numbers of the two recombinant haplotypes in these same donors (G, P<10-6; L, P<10-6; X, P<10-2; and Y, P<.05). Whether the observed departure from the expected one-to-one transmission ratio of haplotypes is due to experimental error or meiotic drive working upon particular haplotypes remains to be determined.
Variation in Recombination Rates among Individuals
Significant heterogeneity in recombination rates within the MHC (0.22–1.32 cM/Mb; fig. 1_B_ and 1_C_) was observed among the donors tested in the present study, where data from members of each sib pair were averaged and treated as a single value in a G test for heterogeneity (_G_=52.2, 7 df, P<10-8). Donors C and L showed a sixfold difference in recombination frequency over the entire MHC (0.71% and 4.3%, respectively), representing the greatest disparity. These data concur with the previous observation of up to twofold differences in recombination frequency (5.1%–11.2%) among five donors screened for recombination in a genomic segment of 20–25 Mb, which encompassed the MHC (Yu et al. 1996).
To address more specifically whether variability in the rate of recombination is due to random occurrence or genomic sequence variation (either within or outside of the MHC) among donors, single-sperm typing of male siblings who share one or two parental MHC haplotypes was performed (fig. 1_C_). Analyses of data derived from two pairs of MZ twins (X and Y, and V and W) and one pair of MHC-identical DZ brothers (S and T) indicated no significant difference in the level of recombination within the MHC between members of any MHC-identical sib pair. Furthermore, when recombination rates across the entire segment (i.e., MOGCA to D6S439 or D6S291) were compared between members of any MHC-identical pair, no significant differences were observed. However, a highly significant (P<10-5, using a 2×2 exact test) difference in recombination rate within the MHC (2.9-fold) was observed between siblings L and E, who shared only one parental MHC haplotype.
Having established that recombination rates across the MHC vary significantly among members of our donor pool but not between the two members of each MHC-identical sib pair, we specifically compared the extent of variability in recombination rates between MHC-identical sib pairs with that between nonidentical donor pairs. The difference in recombination rates between two individuals was determined for all of the 66 possible pairs of individuals. Recombination rate differences for the identical pairs were at the lower end of the distribution of recombination rate differences. Significant differences between the observed rank sum for the three MHC-identical pairs and any three separate (nonoverlapping) pairs chosen from all 12 individuals was tested. Only 703 of the 13,860 possible unique pairs of three have a rank sum less than or equal to the rank sum for the identical pairs (_P_=.0507). This analysis indicates a possible genetic influence on overall recombination rates within the MHC itself.
Donor age did not correlate with recombination frequency; furthermore, no significant variation in recombination rates was observed over the course of 15 mo among three samples derived from a single donor (data not shown).
Identification of Recombination Hot Spots across the MHC
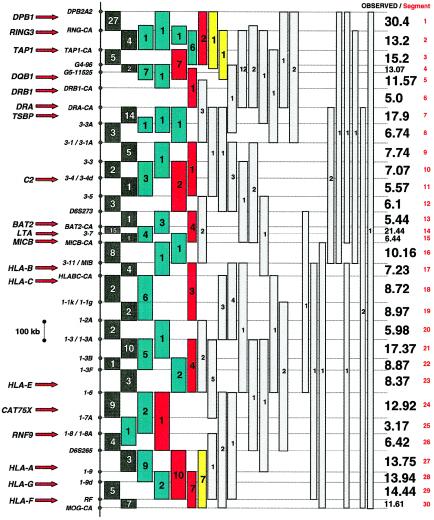
The presence of recombination hot spots in various regions of the human genome indicates a preference for crossing over to take place at discreet sites (Lebo et al. 1983; Chakravarti et al. 1984, 1986; Rouyer et al. 1986; Bowcock et al. 1988; Grimm et al. 1989; Charmley et al. 1990; Benger et al. 1991; Oudet et al. 1992; Pentao et al. 1992; Hubert et al. 1994; Cullen et al. 1995, 1997; Reiter et al. 1996; Jeffreys et al. 1998; Lopes et al. 1998; Smith et al. 1998; Yip et al. 1999; Badge et al. 2000; Jeffreys et al. 2000, 2001; Schneider et al. 2002). To identify potential hot spots for recombination over the entire MHC, 48 polymorphic markers distributed across the MHC were used to fine map the 325 MHC recombinants identified among the 12 donors studied. Recombination breakpoints were resolved to regions of 7–395 kb for 255 of the 325 recombinants identified, 195 of which were mapped to segments of ⩽225 kb (fig. 2). Seventy recombinants, which were distributed evenly among the 12 donors, could not be mapped to regions <395 kb, because of insufficient PEP-DNA or lack of informative markers in the corresponding donor. Crossovers spanning uninformative markers were partitioned equally to the corresponding segments. The observed number of recombination breakpoints identified in each of 30 segments was then compared to the expected number based on segment size and the assumption of an even distribution of events across the MHC (i.e., one crossover per 10.1 kb, given 325 crossovers identified within 3,300 kb of the MHC). Complete genotyping data are available in table B (online only). (To view this very large table properly, readers should be sure to use version 5.0 or higher of Microsoft Internet Explorer or Netscape.)
Figure 2.
Map of the MHC in which 325 recombinant chromosomes were fine mapped by single-sperm typing. The relative positions of 19 MHC genes (red arrows) and 37 polymorphic STRs used to partition the MHC into 30 segments, illustrated as the region between the fine horizontal lines, are shown. The location in which each recombination breakpoint occurred is depicted as a colored box. The number in each box indicates the number of recombinants fine mapped within the area covered by the box. Black boxes represent those recombinants that occurred within a single segment; green, within two adjacent segments (each segment is therefore credited one-half the number of recombinants); red, within three adjacent segments (each segment is credited one-third the number of recombinants); yellow, within four adjacent segments (each segment is credited one-fourth the number of recombinants); and gray, within a segment >395 kb (each segment is therefore credited 1/n the number of recombinants, where n is the number of segments spanned by the recombinant). The total number of potential recombination breakpoints identified in each of the 30 segments (numbered) is listed to the right in boldface type.
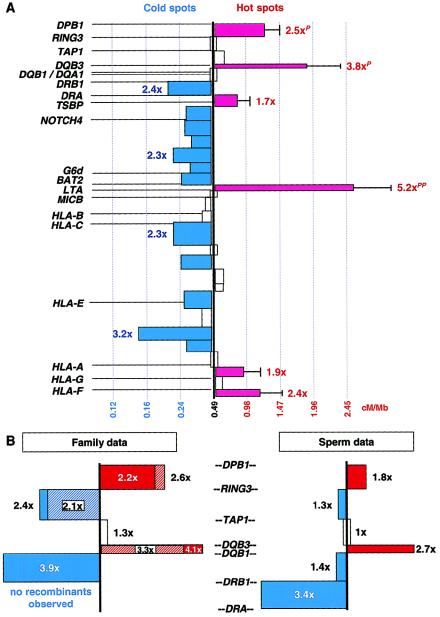
A highly significant difference from the expected frequency of one crossover per 10.1 kb was observed in the analysis of recombinant events from all donors combined (_G_=139, 29 df, P<10-15). Six segments, the total length of which accounts for 13% of the MHC (428 kb), showed levels of recombination that were 1.7–5.2× that expected (fig. 3_A_). Three of these were highly significant (DPB1 to RING3, DQB3 to DQB1, and BAT2 to LTA; P<10-5, 10−5, and 10−15, respectively, after correction for multiple tests). None of the classical class I or II genes (HLA-A, -B, -C, DRB1, DQB1, DQA1, or DPB1) fell within any of the six segments containing hot spots.
Figure 3.
Recombination intensity across the MHC. A, The MHC was subdivided into 30 segments. Segments displaying an excess of recombination are represented as a red bar, which is proportional in length to the increase in recombination over the expected (0.49 cM/Mb). One SD is shown for the six segments where excess recombination was observed. Numbers adjacent to the bars indicate the increase in recombination over the expected. Three segments displayed significantly higher rates of recombination over that expected, after correction for multiple tests (where a superscript “P_” is <10−5 and a superscript “_PP_” is <10−15). Blue bars to the left represent segments displaying a reduction in recombination over that expected, none of which were statistically significant. A distance-normalized scale of recombination rates (in cM/Mb), which allows direct comparison of rates between segments of varied size, is shown at the bottom. White bars straddling the center line represent a 1–1.1× deviation in recombination from the expected. White bars flanking the center line represent a 1.1–1.35× deviation from the expected. B, Recombination intensity across the MHC class II region was compared using data from 88 sperm-based class II recombinants (present study) and previously reported class II recombinants identified in families (Cullen et al. 1997); 22 crossovers of maternal origin (hatched bars) and the complete data set of 31 maternally and paternally derived crossovers (solid bars). The expected number of recombinants for the sperm-based class II recombination profile was based on 88 recombinants evenly distributed across a 635-kb genomic segment and therefore is not identical to the class II profile shown in 3_A. No crossovers were identified between DRB1 and DRA in the family data. However, crossovers of paternal origin were identified between DQB1 and DRB1 in the family data.
The human and mouse class III regions have high sequence similarity and conserved gene organization, relative to the class I and II regions of these species (Yu et al. 2000), allowing a fairly direct comparison of recombination hot spots between the species in class III. The human _BAT2_-to-LTA segment overlaps with the 70-kb segment from LTA to H2-D in the mouse MHC, which also contains a hot spot for recombination (Heine et al. 1994), suggesting that sequences conserved between mouse and human in this segment may promote high frequencies of recombination. On the other hand, regions in humans that are syntenic with the murine class III hot spots Int3 to Tnx and Hsp70 to G7c (Yoshino et al. 1994; Snoek et al. 1998) exhibited 1.7- and 1.5-fold decreases in recombination, respectively, compared with the expected rate.
Although much of the MHC was associated with low levels of recombination, no significant decrease over the expected was observed for any segment. Nevertheless, fine mapping of 255 recombinants indicated the presence of two regions, DRB1 to DRA and CAT75X to RNF9 (i.e., DRB1-CA to_DRA-CA_ and I-7A to I-8A in fig. 2, respectively) where recombination occurred rarely, if ever.
Recombination Distribution Patterns Determined Using Sperm Typing versus Family Data
The distribution of recombination events shown in fig. 3_A_ is indicative of patterns observed in males and does not necessarily reflect that occurring in females. To test whether recombination patterns may be similar in both sexes, the distribution of 88 class II recombinant haplotypes derived from sperm typing was compared with 22 previously reported recombinants of maternal origin derived from segregation analysis in 20 families (Cullen et al. 1997). Similar patterns of recombination distribution between the two groups were observed, the most striking of which are the segments from DPB1 to RING3 and DQB3 to DQB1, where high frequencies of recombination were observed in both groups (fig. 3_B_). Likewise, the recombination distribution pattern determined for all 31 family-derived class II recombinants (22 maternal and 9 paternal) described elsewhere (Cullen et al. 1997) was not significantly different from that observed using single-sperm typing. Thus, comparisons of female versus male and family versus sperm class II recombination data indicate a pattern that is common to both sexes and is reproducible using different methods.
Warm Spots of Recombination
Of the 255 fine-mapped MHC recombinants (i.e., regions of <395 kb), 38% (n_=98) occurred within the six hot spots identified in fig. 3_A_. The remaining 157 fine-mapped recombinants were scattered over 24 additional segments that, together with the six segments incorporating hot spots, constitute the entire 3.3 Mb of the MHC. Twenty-six of the 30 segments shown in figure 2 definitely contained at least one crossover, and the absence of recombination could not be determined absolutely for any of the 30 segments. Rather than suggesting a fairly high level of random dispersal of recombination events throughout the MHC, these data may indicate the presence of many closely spaced “warm spots” of recombination (i.e., one or more warm spots every 100–200 kb). Theoretically, warm spots could be due to intermediate levels of recombination in most individuals or to high levels of recombination in a relatively limited number of individuals in a population. Defining a warm spot would require further fine mapping of the recombinants to a few kilobases or less, such as that performed previously in the TAP2 gene (Cullen et al. 1995; Jeffreys et al. 2000) and in a neighboring 200-kb segment of class II (Jeffreys et al. 2001). This is not feasible in the present study, given the limited amount of PEP-DNA generated from a single sperm. Other methods designed to scrutinize recombination in short DNA fragments (6–8 kb [Jeffreys et al. 2001]), rather than those designed to study a broad view across the entire MHC, would be necessary for identifying warm spots with confidence. A model invoking recombination hot spots and warm spots would explain the identification, in the present study, of the highly active DNA hot spot(s) (Jeffreys et al. 2001; previously referred to as the “_DNA to _RING3_” hot spot [Cullen et al. 1997]) and the inability to identify either of the less active TAP2 or DMB recombination sites (putative warm spots) reported elsewhere (Cullen et al. 1995, 1997; Jeffreys et al. 2000, 2001). The presence of one or more warm spots within each ∼110-kb segment would correspond well with previously estimated lengths of haplotypic blocks (<100 kb) (Daly et al. 2001; Jeffreys et al. 2001; Reich et al. 2001).
Variation among Individuals in the Distribution of Recombination Events
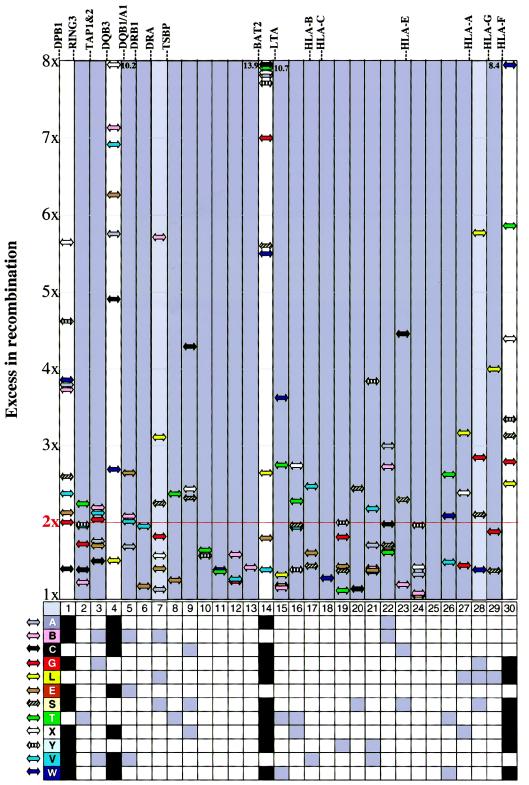
To examine the extent to which multiple individuals contributed to a particular hot spot, recombination intensity at each of the 30 MHC segments was plotted for individual donors (fig. 4) (crossover distribution data available in table C [online only]table C [online only]). These profiles indicate that for certain segments (DPB1 to RING3, DQB3 to DQB1, BAT2 to LTA, and telomeric to HLA-F), recombination occurred at a high rate (at least two times that expected, ranging from 2× to 13.9×) in seven to nine unrelated individuals. The analysis of all recombinants pooled (fig. 3_A_) also detected these four hot spots (segments 1, 4, 14, and 30 in fig. 4), although only three (DPB1 to RING3, DQB3 to DQB1, and BAT2 to LTA) were significant after correction for multiple tests. High levels of recombination in 18 other segments appeared to be specific to only one to three individuals (⩽25% of those surveyed). In two of these segments (28 and 7), recombination was particularly high (more than five times the expected) for single individuals, and removal of those individuals from each analysis resulted in recombination levels that were reduced to 1× and 1.4× the expected, in segments 28 and 7, respectively. Overall, the data support a model in which recombination frequencies vary among segments, on the basis of both rates and fraction of individuals contributing, the extremes of which are the most reliable statistically.
Figure 4.
Variation among individuals in the distribution of recombination events. Individual levels of recombination exceeding that expected, given an overall MHC rate of 0.49 cM/Mb (1×), were plotted for each of 30 segments as excess in recombination activity (range = 1–8×). The magnitude of the increase in recombination is represented as an arrow relative to the scale on the ordinate, and each donor is represented by a unique color. The minimum threshold rate for a hot spot (red horizontal line) is twice the baseline rate of 0.49 cM/Mb. The six peaks in recombination intensity identified in the pooled donor recombination data (fig. 3_A_) corresponded with segments 1, 4, 7, 14, 28, and 30. Only segments 1, 4, and 14 exhibited highly significant increases in recombination after correction for multiple tests (see fig. 3_A_). Segments 7 and 28 (light gray) were not as robust, with only 3 of 12 donors exceeding the 2× threshold. Although segment 30 did not undergo significant levels of recombination over the expected after correction for multiple tests, 7 of 12 donors exhibited hot spot levels in this segment. The relative locations of 18 classical and nonclassical HLA genes are shown, and the three gray segments (2–3, 5–13, and 15–29) represent three major haplotypic blocks. Gray and black boxes in the lower 12×30 grid represent MHC segments that exhibit hot spot (⩾2×) levels of recombination for each individual.
Table C.
Crossover Distribution (No. of Crossovers Per Donor Per Segment)
| No. of Crossovers in Donor | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Segment | A | B | C | G | L | E | S | T | X | Y | V | W | Total | Segment size(kb) |
| 325 MHC recombinants: | ||||||||||||||
| 1 | 7.125 | 3 | .3125 | 3.3955 | 0 | 2.0333 | 1.333 | 0 | 4.1666 | 3.5666 | 2.458 | 3 | 30.3905 | 121 |
| 2 | 1.125 | 1 | .3125 | 2.8955 | 2.116 | .0333 | .333 | 1 | .1666 | 1.5666 | 2.058 | .6 | 13.2065 | 123 |
| 3 | 3.125 | 1.666 | .3125 | 3.2285 | 2.116 | 1.5329 | .333 | 0 | .1666 | .0666 | 2.058 | .6 | 15.2051 | 114 |
| 4 | 3.125 | 1.666 | .3125 | .3955 | .949 | 1.7329 | 0 | 0 | 2.1666 | .0666 | 2.058 | .6 | 13.0721 | 35 |
| 5 | 3.125 | 1.666 | .0625 | .3955 | .949 | 2.482 | 0 | 0 | .1666 | .0666 | 2.058 | .6 | 11.5712 | 119 |
| 6 | .125 | 0 | .0625 | .0625 | .6999 | 1.1499 | 0 | 0 | .1666 | .0666 | 2.058 | .6 | 4.991 | 123 |
| 7 | 1.8666 | 4 | .0625 | 2.604 | 5.832 | 1.1499 | 1 | 0 | 1 | .0666 | .325 | 0 | 17.9066 | 105 |
| 8 | 1.8666 | 0 | .0625 | 1.6041 | .666 | 1.1499 | 0 | 1 | 0 | .0666 | .325 | 0 | 6.7407 | 116 |
| 9 | .49166 | 0 | 1.0625 | .6041 | .166 | 1.0333 | 1.333 | .1428 | 2 | .0666 | .7 | .1428 | 7.74276 | 136 |
| 10 | 1.157 | 0 | .0625 | .6041 | 1.832 | .5333 | .333 | .6428 | 0 | 1.0666 | .7 | .1428 | 7.0741 | 107 |
| 11 | 1.157 | 0 | .0625 | .6041 | .8326 | .5333 | .333 | .6428 | 0 | .0666 | .2 | 1.1428 | 5.5747 | 129 |
| 12 | 1.157 | 1 | .0625 | 1.6041 | .3326 | .599 | 0 | .1428 | 0 | .0666 | 1 | .1428 | 6.1074 | 94 |
| 13 | 1.0746 | 1 | .0625 | 1.028 | .999 | .599 | 0 | .1428 | 0 | .0666 | 0 | .4758 | 5.4483 | 106 |
| 14 | 5.0746 | 0 | 1.0625 | 4.027 | 1.999 | .599 | 1 | 1.6428 | 2 | 2.0666 | .5 | 1.4758 | 21.4473 | 42 |
| 15 | .5746 | .5 | .0625 | .528 | 1.499 | .599 | 0 | .6428 | 0 | .0666 | .5 | 1.4758 | 6.4483 | 64 |
| 16 | .74166 | .5 | .0625 | .195 | 1 | .599 | 1 | 1 | 2 | 1.0666 | 2 | 0 | 10.16476 | 120 |
| 17 | .54166 | 0 | 0 | 1.132 | 1.416 | 1.199 | .583 | .0714 | 0 | .2863 | 2 | 0 | 7.22936 | 95 |
| 18 | 1.0416 | 0 | 0 | .4655 | 1.416 | .866 | .583 | .4044 | 0 | 1.285 | 1 | 1.666 | 8.7275 | 202 |
| 19 | 1.0416 | 0 | 0 | 2.4655 | 1.416 | 1.116 | .583 | .4044 | 0 | 1.285 | 0 | .666 | 8.9775 | 100 |
| 20 | .0416 | 0 | .25 | 1.714 | .75 | .6163 | 1.25 | .4044 | 0 | .2863 | .5 | .1666 | 5.9792 | 120 |
| 21 | 3.5416 | 1.25 | .25 | 1.524 | 2 | 1.449 | 0 | .4044 | 0 | 3.285 | 2.5 | 1.1666 | 17.3706 | 134 |
| 22 | 3.5416 | 1.25 | .25 | 1.524 | .5 | .449 | .5 | .4044 | 0 | .2863 | 0 | .1666 | 8.8719 | 69 |
| 23 | 2.0416 | 1.25 | 1.25 | .5249 | .5 | .449 | 1.5 | .4044 | 0 | .2863 | 0 | .1666 | 8.3728 | 154 |
| 24 | 3.5416 | 1.25 | 0 | .47 | 2.333 | 1.449 | 0 | .0714 | 1.5 | 2.1435 | .166 | 0 | 12.9245 | 172 |
| 25 | .5416 | 0 | 0 | .47 | .333 | .449 | 0 | .0714 | .5 | .1435 | .166 | .5 | 3.1745 | 104 |
| 26 | 1.0416 | 0 | 0 | .47 | .333 | .449 | 0 | 1.0714 | 0 | .1435 | 1.416 | 1.5 | 6.4245 | 112 |
| 27 | .3746 | 0 | 0 | 2.7188 | 7.75 | .283 | 0 | .0714 | 2 | .1435 | .416 | 0 | 13.7573 | 137 |
| 28 | .3746 | .333 | 0 | 2.908 | 7.75 | .283 | .666 | .0714 | .333 | .1435 | .416 | .666 | 13.9445 | 75 |
| 29 | .3746 | .333 | 0 | 2.908 | 8.25 | .283 | .666 | .0714 | .333 | .1435 | .416 | .666 | 14.4445 | 115 |
| 30 | .0416 | .333 | 0 | 1.908 | 2.25 | .283 | .666 | 1.0714 | 1.333 | 1.0666 | 0 | 2.666 | 11.6186 | 50 |
| Total | 50.99278 | 21.997 | 6 | 44.9782 | 58.9851 | 25.9823 | 13.995 | 11.9972 | 19.9986 | 20.9934 | 27.994 | 20.995 | ||
| 255 MHC recombinants mapped within segments ⩽395 kb: | ||||||||||||||
| 1 | 7 | 3 | .25 | 3.333 | 0 | 2 | 1.333 | 0 | 4 | 3.5 | 2 | 3 | 29.416 | 121 |
| 2 | 1 | 1 | .25 | 2.833 | 1.75 | 0 | .333 | 1 | 0 | 1.5 | 0 | 0 | 9.666 | 123 |
| 3 | 3 | 1.666 | .25 | 3.166 | 1.75 | 1.333 | .333 | 0 | 0 | 0 | 0 | 0 | 11.498 | 114 |
| 4 | 3 | 1.666 | .25 | .333 | .583 | 1.333 | 0 | 0 | 2 | 0 | 0 | 0 | 9.165 | 35 |
| 5 | 3 | 1.666 | 0 | .333 | .583 | 1.333 | 0 | 0 | 0 | 0 | 0 | 0 | 6.915 | 119 |
| 6 | 0 | 0 | 0 | 0 | .333 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .333 | 123 |
| 7 | 1.5 | 4 | 0 | 2.5 | 5.5 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 15.5 | 105 |
| 8 | 1.5 | 0 | 0 | 1.5 | .5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 4.5 | 116 |
| 9 | 0 | 0 | 1 | 0 | 0 | 1 | 1.333 | 0 | 2 | 0 | .5 | 0 | 5.833 | 136 |
| 10 | .666 | 0 | 0 | 0 | 1.5 | .5 | .333 | .5 | 0 | 1 | .5 | 0 | 4.999 | 107 |
| 11 | .666 | 0 | 0 | 0 | .5 | .5 | .333 | .5 | 0 | 0 | 0 | 1 | 3.499 | 129 |
| 12 | .666 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3.666 | 94 |
| 13 | .833 | 1 | 0 | .833 | .833 | 0 | 0 | 0 | 0 | 0 | 0 | .333 | 3.832 | 106 |
| 14 | 4.833 | 0 | 1 | 3.833 | 1.833 | 0 | 1 | 1.5 | 2 | 2 | .5 | 1.333 | 19.832 | 42 |
| 15 | .333 | .5 | 0 | .333 | 1.333 | 0 | 0 | .5 | 0 | 0 | .5 | 1.333 | 4.832 | 64 |
| 16 | .5 | .5 | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 1 | 2 | 0 | 9 | 120 |
| 17 | .5 | 0 | 0 | 1 | .666 | 1 | .333 | 0 | 0 | 0 | 2 | 0 | 5.499 | 95 |
| 18 | 1 | 0 | 0 | 0 | .666 | .5 | .333 | 0 | 0 | 1 | 1 | 1.5 | 5.999 | 202 |
| 19 | 1 | 0 | 0 | 2 | .666 | .5 | .333 | 0 | 0 | 1 | 0 | .5 | 5.999 | 100 |
| 20 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | .5 | 0 | 2.5 | 120 |
| 21 | 3.5 | 0 | 0 | 1 | 2 | 1 | 0 | .333 | 0 | 3 | 2.5 | 1 | 14.333 | 134 |
| 22 | 3.5 | 0 | 0 | 1 | .5 | 0 | .5 | .333 | 0 | 0 | 0 | 0 | 5.833 | 69 |
| 23 | 2 | 0 | 1 | 0 | .5 | 0 | 1.5 | .333 | 0 | 0 | 0 | 0 | 5.333 | 154 |
| 24 | 3.5 | 0 | 0 | 0 | 2.333 | 1 | 0 | 0 | 1.5 | 2 | 0 | 0 | 10.333 | 172 |
| 25 | .5 | 0 | 0 | 0 | .333 | 0 | 0 | 0 | .5 | 0 | 0 | .5 | 1.833 | 104 |
| 26 | 1 | 0 | 0 | 0 | .333 | 0 | 0 | 1 | 0 | 0 | 1 | 1.5 | 4.833 | 112 |
| 27 | .333 | 0 | 0 | 2.25 | 7.75 | .25 | 0 | 0 | 2 | 0 | 0 | 0 | 12.583 | 137 |
| 28 | .333 | .333 | 0 | 2.583 | 7.75 | .25 | .666 | 0 | .333 | 0 | 0 | .666 | 12.914 | 75 |
| 29 | .333 | .333 | 0 | 2.583 | 8.25 | .25 | .666 | 0 | .333 | 0 | 0 | .666 | 13.414 | 115 |
| 30 | 0 | .333 | 0 | 1.583 | 2.25 | .25 | .666 | 1 | 1.333 | 1 | 0 | 2.666 | 11.081 | 50 |
| Total | 45.996 | 16.997 | 4 | 34.996 | 51.995 | 12.999 | 12.995 | 8.999 | 18.999 | 17 | 14 | 15.997 | ||
| 143 MHC recombinants mapped within individual segments: | ||||||||||||||
| 1 | 6 | 3 | 0 | 3 | 0 | 2 | 1 | 0 | 4 | 3 | 2 | 3 | 27 | 121 |
| 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 4 | 123 |
| 3 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 114 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 35 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 119 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 123 |
| 7 | 1 | 4 | 0 | 2 | 5 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 14 | 105 |
| 8 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 116 |
| 9 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 5 | 136 |
| 10 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 107 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 129 |
| 12 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 94 |
| 13 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 106 |
| 14 | 4 | 0 | 1 | 3 | 1 | 0 | 1 | 1 | 2 | 2 | 0 | 0 | 15 | 42 |
| 15 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 64 |
| 16 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 1 | 2 | 0 | 8 | 120 |
| 17 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 4 | 95 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 202 |
| 19 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 100 |
| 20 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 120 |
| 21 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 3 | 2 | 1 | 10 | 134 |
| 22 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 69 |
| 23 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 154 |
| 24 | 3 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 9 | 172 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 104 |
| 26 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 4 | 112 |
| 27 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 137 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 75 |
| 29 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 115 |
| 30 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 7 | 50 |
| Total | 20 | 10 | 3 | 19 | 20 | 7 | 7 | 6 | 17 | 14 | 11 | 9 |
High-frequency recombination occurred within three to eight MHC segments per donor, and 22/30 segments exceeded two times the expected recombination frequency in at least one donor (fig. 4). Differences in recombination frequency at specific sites among various donors could potentially be explained by sequence polymorphisms located at or near the hot spot that influences the recombination process. Thus, hot spots common to most individuals, such as that located within the _BAT2_-to-LTA segment (segment 14), may exist because of the presence of a recombination-promoting sequence motif (allele) at this locus that is present on most, but not all, MHC haplotypes. Correspondingly, relatively low-frequency recombination-promoting alleles may also exist at certain polymorphic loci, an example of which may be the _DRA_-to-TSBP segment (segment 7), where a high frequency of recombination was observed in only a single individual.
Determining whether variation in recombination distribution (locations where recombination occurred) differed significantly among the 12 donors across all 30 segments treated individually was computationally unfeasible. Furthermore, <30 recombination events were identified for most donors, limiting the power to detect differences in patterns of recombination distribution between pairs of individuals across the 3.3-Mb region. However, a Fisher’s exact test was performed on the three individuals (A, G, and L) for whom >40 recombination events were observed, considering all 30 segments. The test indicated that these three individuals differed significantly in the distribution pattern of their respective recombination events (_P_=.01). A second Fisher’s exact test, analyzing 11 of the 12 individuals (C had only one recombinant observed within a hot spot and was excluded from the analysis), was performed to determine whether there were differences in recombinant numbers for each of the six hot spots. This test used data from the eight genetically independent sources (individuals A, B, G, L, and E and the summed values for each of the three HLA-identical sib pairs), and it indicated significant heterogeneity in recombination distribution patterns (_P_=.0007), supporting results from the initial test. These data suggest that genetic variation affects not only the rate but also the distribution pattern of recombination in unrelated individuals. On the other hand, no significant difference was observed in the comparisons between the two members of any of the MHCidentical pairs (_P_>.5 in all three analyses); however, small sample sizes for the pairs severely limit the power of this analysis.
Haplotypic Blocks within the MHC
The identification of four intense hot spots within the MHC (segments 1, 4, 14, and 30 in fig. 4) allows the assignment of three major haplotypic blocks across the complex. Major haplotypic blocks (shown as grey segments in fig. 4) are defined here as contiguous regions in which no more than three individuals have recombination rates greater than two times that expected on the basis of the 0.49 cM/Mb relationship determined for the MHC overall. We propose that each major block is composed of many smaller haplotypic blocks (in agreement with Daly et al. 2001), which are defined by warm spots of recombination that vary in the degree of their recombination intensity. Gaudieri et al. (1997) proposed the existence of six strongly associated “frozen” haplotypic blocks within the MHC, which were defined as 100–300-kb stretches of DNA, primarily in regions containing the extremely polymorphic classical HLA class I and II genes. However, recombination occurs in most if not all of the 30 MHC segments defined herein, and segments containing the highly polymorphic class I and II loci generally have average levels of recombination (0.49 cM/Mb). Overall, the recombination data suggest that (1) on the basis of recombination patterns, there are likely to be many more than six strongly associated haplotypic blocks within the MHC; (2) strongly associated blocks are short (probably <100 kb); (3) such blocks are ubiquitous throughout the MHC and are not necessarily defined by high levels of polymorphism; and (4) there is no apparent bilateral pattern of steadily decreasing recombination culminating in a cold spot at each polymorphic class I and class II gene.
Sequence Motifs Involved in Recombination
Specific sequence motifs have been studied for their potential role as recombination-enhancement elements in prokaryotes (Smith et al. 1981; Shah et al. 1994) and eukaryotes (Sander and Hsieh 1985), including yeast (Schuchert et al. 1991; Gale et al. 1992; Fox et al. 2000) and rodents (Edelmann et al. 1989; Zimmerer and Passmore 1991; Gale et al. 1992), and have been proposed to exist in human DNA, as well (Jeffreys et al. 1985; Bergemann and Johnson 1992; Dobbs et al. 1994; Aoki et al. 1995; Rooney and Moore 1995; Smith et al. 1998). Some of these sequence motifs directly bind molecules involved in the recombination process and/or alter DNA/chromatin structure (Wahls 1998), whereas others have been implicated as participants in the process simply as a result of their close proximity to known hot spots. A screen of the MHC for precise sequence motifs previously implicated as potential recombination signal sequences (Smith et al. 1981; Jeffreys et al. 1985; Sander and Hsieh 1985; Edelmann et al. 1989; Schuchert et al. 1991; Zimmerer and Passmore 1991; Bergemann and Johnson 1992; Gale et al. 1992; Dobbs et al. 1994; Shah et al. 1994; Aoki et al. 1995; Rooney and Moore 1995; Smith et al. 1998; Fox et al. 2000) identified 18 of the 21 non-STR motifs considered (table 2). Six of those observed were found ubiquitously throughout the MHC (>600 times) and were excluded from further analysis, since there is no indication from previous studies of a correlation between recombination rate and the number of times these sequences are repeated in a given segment. Linear regression analysis of the remaining 12 motifs was used to detect potential correlations between recombination frequency and motif density (i.e., frequency of the motif within a segment divided by segment size). No significant correlations were observed using the linear regression analysis of motifs independently, although both mouse LTR-IS and translin consensus 2 sites had P values of .07 and .1, respectively, suggesting a weak association (see below). Translin is a single-strand DNA binding protein that associates with breakpoint junctions in some chromosomal translocations and has a potential role in Ig/TCR gene rearrangement (Aoki et al. 1995). LTR-IS has been shown to enhance recombination by 13× in an in vitro system using nuclear extracts from mouse cells (Edelmann et al. 1989).
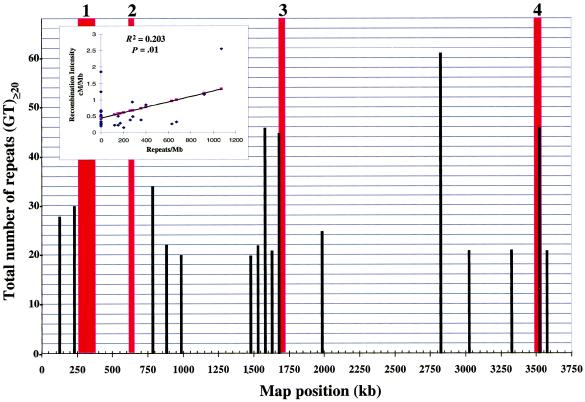
The presence of STRs of GT has been associated with regions of enhanced recombination for the entire chromosome 22 (Majewski and Ott 2000), as well as with increased recombination between two nonreplicating plasmid substrates in human cells in culture (Wahls et al. 1990). It was proposed that left-handed Z-DNA formed by long tracts of GT repeats stabilize the paranemic joint formed during synapsis (Wahls et al. 1990), influence resolution of the Holliday junction (Dutreix 1997), and promote the binding of recombination machinery (Tracy et al. 1997). We tested for potential correlations between the density of various STRs and recombination intensity across the 30 segments and observed a significant positive correlation with GT dinucleotide repeats that were ⩾24 bp (_R_2=0.14, _P_=.04) or ⩾40 bp (_R_2=0.20, _P_=.01) in length (fig. 5). Although this association was not as strong as that reported for chromosome 22 (Majewski and Ott 2000), it was stronger than the association reported for the entire genome (Yu et al. 2001). We further tested the influence of all motifs and repeats (see table 2) on recombination through use of a multivariate regression analysis. Interestingly, the mouse LTR-IS (_P_=.007), translin consensus 2 (_P_=.05), and GT repeats that were >40 bp long (_P_=.006) were all significantly associated with recombination intensity (_R_2=0.44 for the multivariate analysis). This analysis suggests that a portion of the variation in recombination distribution can be explained by these three motifs/repeats, and their role as recombination signal sequences warrants further investigation.
Figure 5.
Relationship between recombination intensity and (GT)⩾20 repeat density. The four major hot spots are labeled 1–4 and are illustrated by red bars. The MHC extends from map position 250 kb to 3550 kb on the _X_-axis. The total number of (GT)⩾20 repeat loci was based on the sum of all tandem (GT)n or (CA)n repeat units (_n_⩾20) per marker per segment, as determined from MHC sequence data (available on the MHC sequence database) and illustrated as black bars. The number of tandem repeats per marker used to determine the repeat length plotted in the histogram reflects the most common allele size within a population of 36 HTCs (M. Cullen, A. Harding, and M. Carrington, unpublished data). The graph insert shows a plot of recombination intensity by (GT) repeat intensity used in the linear regression analysis.
Discussion
The data described in the present article indicate complex patterns of meiotic recombination involving sites that vary significantly in levels of recombination intensity. The presence of four highly active hot spots (three of which were significant even after correction for multiple tests) across a region of 3.3 Mb suggests a density of at least one hot spot every 0.8 Mb of DNA, or potentially 3,000–4,000 intense hot spots throughout the human genome. This range may underestimate the total number of intense hot spots present in the genome and does not include weaker hot spots, which may be undetectable when our approach is used. Theoretically, identification of intense hot spots would define limits across which linkage equilibrium can be expected. Within regions defined by intense hot spots, the level of disequilibrium between variants will vary substantially and, in particular, will be strongly dependent on the distance between the two variants being considered. On the basis of the recombination patterns, we propose that segments defined by intense hot spots are composed of smaller haplotypic units separated by the warm spots of recombination and that these smaller units contain variants that are uniformly in strong LD. Indeed, the presence of strongly associated haplotypes ranging from tens to hundreds of kilobases has been observed previously (Daly et al. 2001; Reich et al. 2001). Furthermore, the existence of warm spots of recombination occurring one or more times every 100 kb is consistent with data described by Jeffreys et al. (2001), as well as with the even distribution of low-level recombination that occurred within nearly all 30 segments of the MHC observed in the present study.
The fine mapping of crossover sites to only one of the 30 MHC segments analyzed was accomplished for 143 of the 325 recombinant haplotypes identified. The remaining 182 crossovers were mapped across two or more segments, because of marker homozygosity and limited amounts of PEP-DNA, which reduces the degree of heterogeneity in recombination frequencies among the 30 segments. An analysis of only those crossovers that mapped within a single segment indicated significant levels of recombination (after correction for multiple tests) in the following segments: DPB1 to RING3, DRA to TSBP, BAT2 to LTA, and the segment telomeric to HLA-F. This result supports that which was obtained when all recombinants were included in the analysis (fig. 3A), with the exception that the _DQB3_-to-DQB1 hot spot was identified only when the complete data set was employed. Additional studies are necessary to determine whether the difference in significance between the two data sets for the _DQB3_-to-DQB1 segment can be attributed to overestimation of recombination due to insufficient fine mapping when the entire data set is used or to lack of power when only 143 recombinants are analyzed. We suspect the latter, since family-derived recombination data also indicate a hot spot for recombination in this region. Higher marker density will refine crossover sites substantially, pinpointing dramatically localized peaks of recombination intensity, as have been observed previously for the DNA, DMB, and TAP2 hot spots (Jeffreys et al. 2000, 2001).
Rates of recombination and distribution of recombination events across the complex varied significantly among individuals. Therefore, haplotypic blocks are likely to vary across individuals, potentially confounding gene mapping of particular traits. Although more than half of the 12 individuals tested had recombination rates more than double the expected in each of the four intense hot spots (fig. 4; DPB1 to RING3, DQB3 to DQB1, BAT2 to LTA, and telomeric to HLA-F), some individuals had lower rates than expected in these hot spot regions. By their nature, differences among individuals in recombination frequency at warm spots are likely to vary even more extensively. Several observations suggest that genetic variation dictates, to some extent, where and how frequently recombination occurs: (1) rates of recombination did not differ significantly between members of any MHC-identical pair, but significant differences were observed between individuals who were not MHC identical; (2) recombination occurred at a significantly greater frequency than expected in some segments, suggesting that sequences local to those hot spots take on a configuration that is particularly amenable to the molecular machinery of the recombination process; and (3) the presence of specific sequence motifs (i.e., [GT]⩾12) are significantly associated with recombination hot spots in the MHC. These data support studies of recombination using other model organisms, such as bacteria, yeast, and mouse, in which sequences local to sites of crossover are thought to promote crossing over (Smith et al. 1981; Sander and Hsieh 1985; Edelmann et al. 1989; Schuchert et al. 1991; Zimmerer and Passmore 1991; Gale et al. 1992; Shah et al. 1994; Fox et al. 2000). Thus, the variability in rate and distribution of recombination among regions of the genome, such as the MHC, and more uniform across a population in conserved genomic segments.
The data described herein can be used directly in gaining a better understanding of MHC evolution and selective processes occurring at this locus. Regions of increased recombination intensity identified in this study will allow proper selection of markers for use in surveys of disease gene associations within the MHC. Disease associations with two or more markers located on opposite sides of a recombination hot spot could potentially indicate an epistatic interaction between two MHC loci, which is entirely plausible given the abundance of genes in this region that encode molecules with related functions. Similarly, LD between specific alleles at two loci that map to opposite sides of a hot spot might indicate selection for that particular haplotype, due to some beneficial effect conferred by that combination. It will be of interest to type loci located on the boundaries of the intense hot spots defined here, to determine the extent to which LD patterns correlate with recombination patterns across the MHC.
Acknowledgments
The authors wish to thank Dr. David Reich, for his careful review of the manuscript; Gary Smythers and Dr. Doug Oman, for assistance with computer programming; and Kathleen Noer, Gilbert McCrary, Michael Malasky, Darlene Marti, and Dr. Xiaojiang Gao, for technical assistance. This publication has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-C0-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Human Genome at NCBI, http://www.ncbi.nlm.nih.gov/genome/guide/human/
- Sanger Institute: Human Chromosome 6 Home, http://www.sanger.ac.uk/HGP/Chr6/ (for the MHC sequence database)
- SPUTNIK, http://abajian.net/sputnik/index.html
References
- Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M (1995) A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet 10:167–174 [DOI] [PubMed] [Google Scholar]
- Badge RM, Yardley J, Jeffreys AJ, Armour JA (2000) Crossover breakpoint mapping identifies a subtelomeric hotspot for male meiotic recombination. Hum Mol Genet 9:1239–1244 [DOI] [PubMed] [Google Scholar]
- Beck S, Trowsdale J (2000) The human major histocompatibility complex: lessons from the DNA sequence. Annu Rev Genomics Hum Genet 1:117–137 [DOI] [PubMed] [Google Scholar]
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W (1992) Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 148:249–258 [PubMed] [Google Scholar]
- Benger JC, Teshima I, Walter MA, Brubacher MG, Daouk GH, Cox DW (1991) Localization and genetic linkage of the human immunoglobulin heavy chain genes and the creatine kinase brain (CKB) gene: identification of a hot spot for recombination. Genomics 9:614–622 [DOI] [PubMed] [Google Scholar]
- Bergemann AD, Johnson EM (1992) The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol 12:1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Hebert JM, Wijsman E, Gadi I, Cavalli-Sforza LL, Boyd CD (1988) High recombination between two physically close human basement membrane collagen genes at the distal end of chromosome 13q. Proc Natl Acad Sci USA 85:2701–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GM, Leversha M, Hulten M, Ferguson-Smith MA, Affara NA, Furlong RA (1998) Genetic analysis of meiotic recombination in humans by use of sperm typing: reduced recombination within a heterozygous paracentric inversion of chromosome 9q32-q34.3. Am J Hum Genet 62:1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugawan TL, Klitz W, Blair A, Erlich HA (2000) High-resolution HLA class I typing in the CEPH families: analysis of linkage disequilibrium among HLA loci. Tissue Antigens 56:392–404 [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Buetow KH, Antonarakis SE, Waber PG, Boehm CD, Kazazian HH (1984) Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet 36:1239–1258 [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A, Elbein SC, Permutt MA (1986) Evidence for increased recombination near the human insulin gene: implication for disease association studies. Proc Natl Acad Sci USA 83:1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmley P, Chao A, Concannon P, Hood L, Gatti RA (1990) Haplotyping the human T-cell receptor beta-chain gene complex by use of restriction fragment length polymorphisms. Proc Natl Acad Sci USA 87:4823–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XF, Li HH, Goradia TM, Lange K, Kazazian HH, Jr., Galas D, Arnheim N (1989) Single-sperm typing: determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci USA 86:9389–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Carrington M (2001) Single-sperm typing: a rapid alternative to family-based linkage analysis. In: Carrington M, Hoelzel AR (eds) Molecular epidemiology: the practical approach series. Oxford University Press, Oxford, pp 145–179 [Google Scholar]
- Cullen M, Erlich H, Klitz W, Carrington M (1995) Molecular mapping of a recombination hotspot located in the second intron of the human TAP2 locus. Am J Hum Genet 56:1350–1358 [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Noble J, Erlich H, Thorpe K, Beck S, Klitz W, Trowsdale J, Carrington M (1997) Characterization of recombination in the HLA class II region. Am J Hum Genet 60:397–407 [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Dobbs DL, Shaiu WL, Benbow RM (1994) Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res 22:2479–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutreix M (1997) (GT)n repetitive tracts affect several stages of RecA-promoted recombination. J Mol Biol 273:105–113 [DOI] [PubMed] [Google Scholar]
- Edelmann W, Kroger B, Goller M, Horak I (1989) A recombination hotspot in the LTR of a mouse retrotransposon identified in an in vitro system. Cell 57:937–946 [DOI] [PubMed] [Google Scholar]
- Fox ME, Yamada T, Ohta K, Smith GR (2000) A family of cAMP-response-element-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics 156:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale JM, Tobey RA, D'Anna JA (1992) Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J Mol Biol 224:343–358 [DOI] [PubMed] [Google Scholar]
- Gaudieri S, Leelayuwat C, Tay GK, Townend DC, Dawkins RL (1997) The major histocompatability complex (MHC) contains conserved polymorphic genomic sequences that are shuffled by recombination to form ethnic-specific haplotypes. J Mol Evol 45:17–23 [DOI] [PubMed] [Google Scholar]
- Goldstein DB (2001) Islands of linkage disequilibrium. Nat Genet 29:109–111 [DOI] [PubMed] [Google Scholar]
- Goradia TM, Stanton VP Jr, Cui XF, Aburatani H, Li HH, Lange K, Housman DE, Arnheim N (1991) Ordering three DNA polymorphisms on human chromosome 3 by sperm typing. Genomics 10:748–755 [DOI] [PubMed] [Google Scholar]
- Grimm T, Muller B, Dreier M, Kind E, Bettecken T, Meng G, Muller CR (1989) Hot spot of recombination within DXS164 in the Duchenne muscular dystrophy gene. Am J Hum Genet 45:368–372 [PMC free article] [PubMed] [Google Scholar]
- Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55:175–189 [PMC free article] [PubMed] [Google Scholar]
- Heine D, Khambata S, Wydner KS, Passmore HC (1994) Analysis of recombinational hot spots associated with the p haplotype of the mouse MHC. Genomics 23:168–177 [DOI] [PubMed] [Google Scholar]
- Hubert R, MacDonald M, Gusella J, Arnheim N (1994) High resolution localization of recombination hot spots using sperm typing. Nat Genet 7:420–424 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MK (1995) Natural selection on the peptide-binding regions of major histocompatibility complex molecules. Immunogenetics 42:233–243 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Murray J, Neumann R (1998) High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell 2:267–273 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Ritchie A, Neumann R (2000) High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet 9:725–733 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL (1985) Hypervariable ‘minisatellite' regions in human DNA. Nature 314:67–73 [DOI] [PubMed] [Google Scholar]
- Klitz W, Stephens JC, Grote M, Carrington M (1995) Discordant patterns of linkage disequilibrium of the peptide-transporter loci within the HLA class II region. Am J Hum Genet 57:1436–1444 [PMC free article] [PubMed] [Google Scholar]
- Kwok WW, Kovats S, Thurtle P, Nepom GT (1993) HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J Immunol 150:2263–2272 [PubMed] [Google Scholar]
- Lazzeroni LC, Arnheim N, Schmitt K, Lange K (1994) Multipoint mapping calculations for sperm-typing data. Am J Hum Genet 55:431–436 [PMC free article] [PubMed] [Google Scholar]
- Lebo RV, Chakravarti A, Buetow KH, Cheung MC, Cann H, Cordell B, Goodman H (1983) Recombination within and between the human insulin and beta-globin gene loci. Proc Natl Acad Sci USA 80:4808–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang EP, Hubert R, Zhang L, Schmitt K, Arnheim N (1994) Single-sperm typing. In: Dracopoli N, Haines J, Korf B, Moir D, Morton C, Seidman C, Seidman J, Smith D (eds) Current protocols in human genetics (Suppl 3). Vol 1. John Wiley & Sons, New York, pp 1.6.1–1.6.15 [Google Scholar]
- Li H, Cui X, Arnheim N (1991) Analysis of DNA sequence variation in single cells. In: Arnheim N (ed) Methods—a companion to methods in enzymology. Vol 2. Academic Press, San Diego, pp 49–59 [Google Scholar]
- Li HH, Gyllensten UB, Cui XF, Saiki RK, Erlich HA, Arnheim N (1988) Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335:414–417 [DOI] [PubMed] [Google Scholar]
- Lien S, Szyda J, Schechinger B, Rappold G, Arnheim N (2000) Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am J Hum Genet 66:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Ravise N, Vandenberghe A, Palau F, Ionasescu V, Mayer M, Levy N, Wood N, Tachi N, Bouche P, Latour P, Ruberg M, Brice A, LeGuern E (1998) Fine mapping of de novo CMT1A and HNPP rearrangements within CMT1A-REPs evidences two distinct sex-dependent mechanisms and candidate sequences involved in recombination. Hum Mol Genet 7:141–148 [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J (2000) GT repeats are associated with recombination on human chromosome 22. Genome Res 10:1108–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Mann D, Carrington M (1995) Recombination rates across the HLA complex: use of microsatellites as a rapid screen for recombinant chromosomes. Hum Mol Genet 4:423–428 [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Nomenclature Committee of the International Union of Biochemistry (1986) Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. J Biol Chem 261:13–17 [PubMed] [Google Scholar]
- Oudet C, Hanauer A, Clemens P, Caskey T, Mandel JL (1992) Two hot spots of recombination in the DMD gene correlate with the deletion prone regions. Hum Mol Genet 1:599–603 [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300 [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228 [DOI] [PubMed] [Google Scholar]
- Rooney SM, Moore PD (1995) Antiparallel, intramolecular triplex DNA stimulates homologous recombination in human cells. Proc Natl Acad Sci USA 92:2141–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F, Simmler MC, Johnsson C, Vergnaud G, Cooke HJ, Weissenbach J (1986) A gradient of sex linkage in the pseudoautosomal region of the human sex chromosomes. Nature 319:291–295 [DOI] [PubMed] [Google Scholar]
- Sanchez-Mazas A, Djoulah S, Busson M, Le Monnier de Gouville I, Poirier JC, Dehay C, Charron D, Excoffier L, Schneider S, Langaney A, Dausset J, Hors J (2000) A linkage disequilibrium map of the MHC region based on the analysis of 14 loci haplotypes in 50 French families. Eur J Hum Genet 8:33–41 [DOI] [PubMed] [Google Scholar]
- Sander M, Hsieh TS (1985) Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res 13:1057–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt K, Lazzeroni LC, Foote S, Vollrath D, Fisher EM, Goradia TM, Lange K, Page DC, Arnheim N (1994) Multipoint linkage map of the human pseudoautosomal region, based on single-sperm typing: do double crossovers occur during male meiosis? Am J Hum Genet 55:423–430 [PMC free article] [PubMed] [Google Scholar]
- Schneider JA, Peto TE, Boone RA, Boyce AJ, Clegg JB (2002) Direct measurement of the male recombination fraction in the human beta- globin hot spot. Hum Mol Genet 11:207–215 [DOI] [PubMed] [Google Scholar]
- Schuchert P, Langsford M, Kaslin E, Kohli J (1991) A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J 10:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Bennett RJ, West SC (1994) Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell 79:853–864 [DOI] [PubMed] [Google Scholar]
- Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL (1981) Structure of chi hotspots of generalized recombination. Cell 24:429–436 [DOI] [PubMed] [Google Scholar]
- Smith RA, Ho PJ, Clegg JB, Kidd JR, Thein SL (1998) Recombination breakpoints in the human beta-globin gene cluster. Blood 92:4415–4421 [PubMed] [Google Scholar]
- Snoek M, Teuscher C, van Vugt H (1998) Molecular analysis of the major MHC recombinational hot spot located within the G7c gene of the murine class III region that is involved in disease susceptibility. J Immunol 160:266–272 [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ (1995) Biometry: the principles and practice of statistics in biological research. W. H. Freeman and Company, New York [Google Scholar]
- Tracy RB, Baumohl JK, Kowalczykowski SC (1997) The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev 11:3423–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP (1998) Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr Top Dev Biol 37:37–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD (1990) The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol 10:785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Milford E, Hammerling V, Dupont B (1989) Description of the reference panel of B-lymphoblastoid cell lines for factors of the HLA system: the B-cell line panel designed for the Tenth International Histocompatibility Workshop. In: Dupont B (ed) Immunobiology of HLA. Vol 1. Springer, New York [Google Scholar]
- Yip SP, Lovegrove JU, Rana NA, Hopkinson DA, Whitehouse DB (1999) Mapping recombination hotspots in human phosphoglucomutase (PGM1). Hum Mol Genet 8:1699–1706 [DOI] [PubMed] [Google Scholar]
- Yoshino M, Sagai T, Lindahl KF, Toyoda Y, Shirayoshi Y, Matsumoto K, Sugaya K, Ikemura T, Moriwaki K, Shiroishi T (1994) Recombination in the class III region of the mouse major histocompatibility complex. Immunogenetics 40:280–286 [DOI] [PubMed] [Google Scholar]
- Yu A, Zhao C, Fan Y, Jang W, Mungall AJ, Deloukas P, Olsen A, Doggett NA, Ghebranious N, Broman KW, Weber JL (2001) Comparison of human genetic and sequence-based physical maps. Nature 409:951–953 [DOI] [PubMed] [Google Scholar]
- Yu CY, Yang Z, Blanchong CA, Miller W (2000) The human and mouse MHC class III region: a parade of 21 genes at the centromeric segment. Immunol Today 21:320–328 [DOI] [PubMed] [Google Scholar]
- Yu J, Lazzeroni L, Qin J, Huang MM, Navidi W, Erlich H, Arnheim N (1996) Individual variation in recombination among human males. Am J Hum Genet 59:1186–1192 [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N (1992) Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA 89:5847–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerer EJ, Passmore HC (1991) Structural and genetic properties of the Eb recombinational hotspot in the mouse. Immunogenetics 33:132–140 [DOI] [PubMed] [Google Scholar]