The M1 and M2 paradigm of macrophage activation: time for reassessment (original) (raw)
Abstract
Macrophages are endowed with a variety of receptors for lineage-determining growth factors, T helper (Th) cell cytokines, and B cell, host, and microbial products. In tissues, macrophages mature and are activated in a dynamic response to combinations of these stimuli to acquire specialized functional phenotypes. As for the lymphocyte system, a dichotomy has been proposed for macrophage activation: classic vs. alternative, also M1 and M2, respectively. In view of recent research about macrophage functions and the increasing number of immune-relevant ligands, a revision of the model is needed. Here, we assess how cytokines and pathogen signals influence their functional phenotypes and the evidence for M1 and M2 functions and revisit a paradigm initially based on the role of a restricted set of selected ligands in the immune response.
Introduction
The concept of classic and alternative activation, also termed M1 and M2 to mimic Th cell nomenclature, has become increasingly broad and overinterpreted, hindering the understanding of pathogenesis and possible manipulation. Although there is evidence that many stimuli combine to determine the phenotype of macrophages, our view of this complex process has become too bipolar.
Macrophages evolved in simple multicellular organisms to perform phagocytic clearance of dying cells in development and adult life, and to protect the host through innate immunity, both as resident tissue macrophages and monocyte-derived recruited cells during inflammation. The development of acquired immunity with reciprocal interactions between macrophages and activated T and B lymphocytes provided novel levels of regulation and acquisition of enhanced antimicrobial resistance. The role of Th1-derived interferon-gamma (IFN-γ) in cell-mediated immunity to intracellular infection and of interleukin-4 (IL-4) (Th2) in extracellular parasitic infection gave rise to the concept of analogous M1 and M2 macrophages, now extended to a wider range of immunomodulatory agents and trophic functions.
In this review, we discuss signaling and genetic and functional signatures acquired during maturation and activation and consider how they fit the current M1/M2 model of macrophage polarization. Growing information indicates that recognition receptors, cytokines, and the signaling and genetic programs behind them control every aspect of cell activation, pointing to the need to recognize a broader functional repertoire for macrophages.
M1-M2 concept: background
Because macrophages are key modulator and effector cells in the immune response, their activation influences and responds to other arms of the immune system. In 1986, Mosmann, Coffman and colleagues put forward the hypothesis that two subsets of helper T cells could be distinguished by the cytokines secreted after T lymphocyte activation, mediating distinct regulatory and effector functions [1]. Coffman recounts that the hypothesis derived from separate studies to answer the following questions: “are there T helper cells analogous to the classes of antibody made by B cells?” and “how are allergic responses, especially the immunoglobulin E (IgE) class of antibody, regulated?” [2]. These questions are implicitly relevant for infective diseases, in which intracellular and extracellular pathogens induce IgG vs. IgE responses, respectively, and macrophages deal with the infection, but also in type I and type II immune diseases in which macrophages contribute to tissue damage and pathology.
The term macrophage activation (classical activation) was introduced by Mackaness in the 1960s in an infection context to describe the antigen-dependent, but non-specific enhanced, microbicidal activity of macrophages toward BCG (bacillus Calmette-Guerin) and Listeria upon secondary exposure to the pathogens [3]. The enhancement was later linked with Th1 responses and IFN-γ production by antigen-activated immune cells [4] and extended to cytotoxic and antitumoral properties [5,6]. At the time, the effect on the macrophages of the Th2 arm of immunity leading to IgE and extracellular parasite protection and allergic responses remained unclear. The discovery that the mannose receptor was selectively enhanced by the Th2 IL-4 and IL-13 in murine macrophages, and induced high endocytic clearance of mannosylated ligands, increased major histocompatibility complex (MHC) class II antigen expression, and reduced pro-inflammatory cytokine secretion, led Stein, Doyle, and colleagues to propose that IL-4 and IL-13 induced an alternative activation phenotype, a state altogether different from IFN-γ activation but far from deactivation [7,8].
While investigating the factors that regulate macrophage arginine metabolism, Mills and colleagues found that macrophages activated in mouse strains with Th1 and Th2 backgrounds differed qualitatively in their ability to respond to the classic stimuli IFN-γ or lipopolysaccharide (LPS) or both and defined an important metabolic difference in the pathway: M1 macrophages made the toxic nitric oxide (NO), whereas M2 macrophages made the trophic polyamines [9]. They proposed that these be termed M1 and M2 macrophage responses, although this model dealt more with the predisposition of macrophages to develop specific phenotypes, it relied on the transforming growth factor-beta (TGF-β)-mediated inhibition of inducible nitric oxide synthase (iNOS) and was independent of T and B cells.
Bona fide evidence of in vivo macrophage alternative activation, equivalent to the observation of Mackaness for intracellular pathogens, came from work done by Allen, de Baetselier, Brombacher, and colleagues in parasite infection, which elicits a strong IgE and Th2 response; a recent review provides a more comprehensive functional perspective revealing heterogeneity in the response, depending on the nematode, the tissue, and type of macrophage [10].
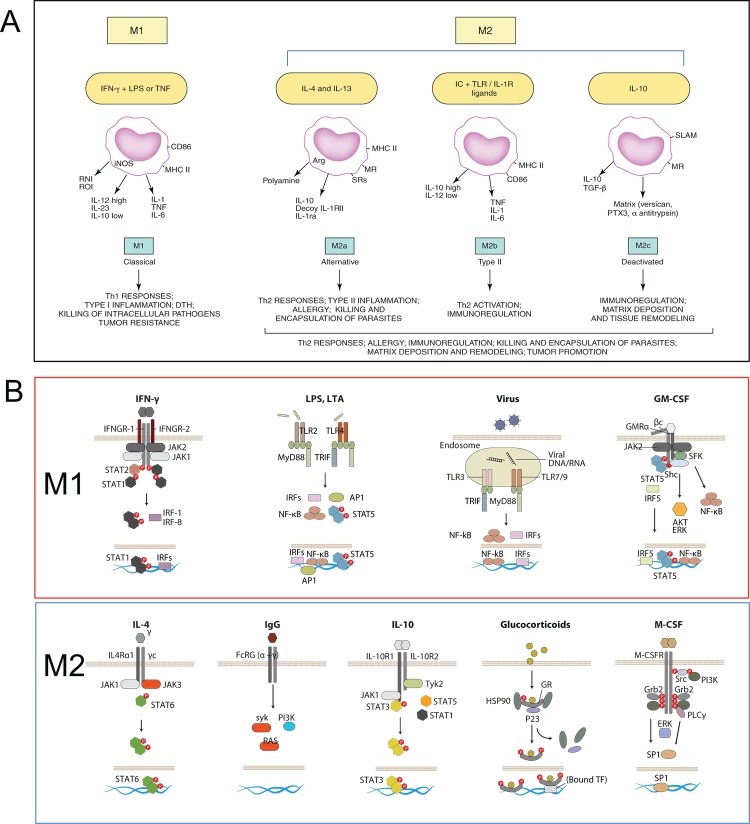
Until here, the questions were in tune with the Th1 and Th2 context. However, other cytokines and factors, such as IL-10, TGF-β (today recognized regulatory T [Treg] products), and glucocorticoids did not fit clearly in the context of the Th1 Th2 response and nonetheless seemed to elicit similar phenotypes in macrophages, with reports showing upregulation of mannose receptor, induction of IL-10 itself, and apparent antagonism to classic stimuli with downregulation of inflammatory cytokines and dampening of reactive nitrogen intermediate and reactive oxygen intermediate killing mechanisms. To integrate the phenotypic similarities and differences, Mantovani and colleagues grouped the stimuli in a continuum between two functionally polarized states, based on their effects on selected macrophage markers, termed M1 (IFN-γ combined with LPS or tumor necrosis factor [TNF]) and M2 (IL-4 [M2a], IL-10, and GCs [M2c]) (Figure 1A) [11]; activation induced by Fc receptors and immune-complexes, described by Mosser, was termed M2b. This careful categorization also made distinctions between M2 groups, such as “product of Th2 activation”, “pro-Th2 activation”, and “immunoregulation”. Later, findings regarding granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) effects in macrophages led to the independent inclusion of these as M1 and M2 stimuli, respectively [12].
Figure 1. The M1/M2 paradigm, origin, and molecular basis.
(A) Mantovani and colleagues [11] proposed an M1-M2 macrophage model, in which M1 included interferon-gamma (IFN-γ) + lipopolysaccharide (LPS) or tumor necrosis factor (TNF) and M2 was subdivided to accommodate similarities and differences between interleukin-4 (IL-4) (M2a), immune complex + Toll-like receptor (TLR) ligands (M2b), and IL-10 and glucocorticoids (M2c). Diagram reproduced with the permission of Elsevier. (B) The signaling behind the effects of M1 and M2 stimuli in macrophages has gained clarity in recent years. Here, we highlight receptors and key signaling mediators in common and distinct pathways, explained in the text. The diagram includes granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) as M1 and M2 stimuli.
The current classification of macrophage immune activation is challenging because two very distinct aspects are considered: the in vitro effects of selected immune-related ligands on the phenotype of macrophages and in vivo evidence for distinct subsets of macrophages in disease, comparable to polarized B- and T-cell responses. The main limitations of the current view are, first, it ignores the source and context of the stimuli; second, the M1 and M2 stimuli do not exist alone in tissues; and, third, macrophages may not form clear-cut activation subsets nor expand clonally. Next, we discuss these and other aspects of the macrophage activation paradigm based on published evidence.
Grouping of M1 and M2 stimuli: need for immunological contextualization
The definition of M1 and M2 macrophage polarity derives from the pre-genomic era, when a few markers were considered to establish differences and similarities in macrophage responses to stimuli. The original definition took into account the possible context of the stimuli, but current generalized views lack this perspective. Furthermore, updated knowledge of cytokine signaling, the role of cytokines in the development of the hematopoietic system and in disease models with genetically modified mice and transcriptomic and proteomic analysis reveal a far more complex picture and challenge the current grouping. In the next section, we discuss the different role of stimuli considered in the M1 and M2 paradigm and highlight signaling pathways and gene expression singularities between the M1 and M2 stimuli.
M1 stimuli
The M1 stimuli are grouped according to their ability to induce prototypic inflammatory responses and markers, but their source, role, receptors, and signaling pathways differ substantially. We discuss, as examples, three of the main M1 stimuli recognized today. IFN-γ is the main cytokine associated with M1 activation and the main Th1 cell product. Other cells, such as natural killer (NK) cells and macrophages, themselves have been shown to produce the cytokine. The IFNGR-1 and IFNGR-2 chains form IFN-γ receptor (Figure 1B). The receptor recruits Janus kinase (Jak)1 and Jak2 adaptors that activate STAT1 (signal transducers and activators of transcription1) and interferon regulatory factors (IRF), such as IRF-1 and IRF-8; for a recent comprehensive review, see [13]. IFN-γ controls specific gene expression programs involving cytokine receptors (CSF2RB, IL15 receptor alpha [RA], IL2RA, and IL6R), cell activation markers (CD36, CD38, CD69, and CD97), and a number of cell adhesion molecules (intercellular adhesion molecule 1 [ICAM1], integrin alpha L [ITGAL], ITGA4, ITGbeta-7 [B7], mucin 1 [MUC1], and ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 [SIAT1]). The major mediators of IFN-γ-induced signaling, STAT1, JAK2, and IRF1, and regulators cytokine inducible SH2-containing protein (CISH), N-myc-interactor (NMI), protein tyrosine phosphatase, receptor type, C PTPRC, protein tyrosine phosphatase, receptor type, O (PTPRO), and suppressor of cytokine signaling 1 (SOCS1) are also under the control of the cytokine [14]. IFN-γ is included in combination with LPS in the M1/M2 paradigm, and gene expression profiles of the combination are different from LPS or IFN-γ profiles alone [15,16]. Mice lacking IFN-γ or its receptors are viable and fertile and their steady-state macrophage numbers are normal [17,18]. Macrophages, however, show impaired production of antimicrobial products, and mice are susceptible to Mycobacterium bovis and Listeria monocytogenes. This defect is not important for prototypical Th1/M1 responses only: knockout (KO) mice are susceptible to protozoa, such as Trypanosoma cruzi [19], Leishmania amazonensis and Leishmania major [20], and Cryptosporidium parvum [21], as well as defense against some nematodes (e.g. Litomosoides sigmodontis [22], Schistosoma mansoni [23], and Schistosoma japonicum [24]). In humans, mutations resulting in the lack of expression of the receptor drive severe immunodeficiency (e.g. susceptibility to mycobacteria M. avium; M. kansasii; M. chelonei, Salmonella typhimurium and S. paratyphi) in patients with familial disseminated atypical mycobacterial disease [25].
Pathogens are recognized by pattern recognition receptors. The activation induced is part of the M1 group and also defined as “innate” activation [26]. Full bacteria induce gene programs similar to those of isolated Toll-like receptors (TLRs), and major parts of the pathogen profiles can be ascribed to TLR ligands, such as LPS, muramyl dipeptide, and lipoteichoic acid [16]. LPS is the best-studied M1 macrophage signal and is recognized by TLR4 (Figure 1B), although recent evidence shows that LPS can also be recognized by TLR4-independent mechanisms leading to inflammasome activation [27,28]. Conventionally, TLR4 activation induces MyD88 and MaL/Tirap (Toll-interleukin 1 receptor domain containing adaptor protein)-dependent pathways that lead to strong pro-inflammatory cytokine profiles (e.g. IFN-β, IL-12, TNF, IL-6, and IL-1β), chemokines (e.g. chemokine [C-C motif] ligand 2 CCL2, chemokine [C-X-C motif] ligand 10 [CXCL10], and CXCL11), and antigen presentation molecules, such as MHC members, co-stimulatory molecules, and antigen-processing peptidases. The profiles are controlled by nuclear factor of kappa light polypeptide gene enhancer (NF-κB), activator protein 1 (AP-1), IRFs, STAT1, and EGR (early growth response) family members, many of which participate in the IFN response [13]. Although there is a degree of overlap between LPS and IFN-γ gene profiles, similarities are not enough to consider the stimuli to be homologous. As for IFN-γ, the numbers of macrophages in TLR KO animals are normal, but their activation is defective and therefore survival to infection is severely impaired; for seminal and recent views, see [29-32]. In humans, genetic mutations in the TLR family have gained clarity, and as for mice, there is evidence for susceptibility to infection with mycobacteria, pneumococci, meningococci, malaria, and susceptibility to develop bacteremia [33].
Granulocyte macrophage colony-stimulating factor (GM-CSF) is the latest addition to the M1 category of stimuli. GM-CSF is produced by a variety of cells, including macrophages and parenchyma cells. The GM-CSF receptor forms a dodecamer structure [34] and recruits JAK2, leading to the activation of STAT5, extracellular signal-regulated kinase (ERK), and V-Akt murine thymoma viral oncogene homolog 1 (AKT) as well as the nuclear translocation of NF-κB and IRF5 upon binding (Figure 1B) [35]. Many of these regulators are part of the IFN-γ and TLR signaling pathways. GM-CSF enhances antigen presentation, complement- and antibody-mediated phagocytosis, microbicidal capacity, leukocyte chemotaxis, and adhesion. GM-CSF induces monocyte and macrophage cytokine production of IL-6, IL-8, G-CSF, M-CSF, TNF, and IL-1β, but less than, for example, LPS. GM-CSF transcriptome analysis shows that GM-CSF regulates several known cell surface molecules (e.g. CD14, Fc fragment of IgG, high affinity Ia (FCγR1A), CD163, and nuclear receptor subfamily 1, group H, member 3 [NR1H3]) [36]. The GM-CSF KO has normal numbers of macrophages in some tissues but has defects in the maturation of alveolar macrophages and develops pulmonary alveolar proteinosis [37]. In humans, mutations in the GM-CSF receptor, especially in the common beta chain, lead to alveolar macrophage defects and proteinosis but also to malignancy [38,39]. As such, the main functions proposed for GM-CSF include regulation of hematopoietic cell proliferation and differentiation, and modulation of the function of mature hematopoietic cells. Other stimuli that share pro-inflammatory properties have been termed M1 (e.g. TNF, IL-1ß, and IL-6). This adds further heterogeneity to a group that already comprises T cells, bacterial products, and a lineage-determining cytokine.
M2 stimuli
The M2 group of stimuli arose from the initial IL-4 observations, and they are grouped mainly due to their ability to antagonize prototypic inflammatory responses and markers; however, as for M1 stimuli, their source, role, receptors, and signaling pathways differ. We discuss, as examples, five of the main M2 stimuli. IL-4 is produced by the Th2 cells, eosinophils, basophils, or macrophages themselves and is recognized by three different receptor pairs. IL-4Rα1 can pair with the common gamma chain (γc), enabling IL-4 binding, and with the IL13Rα1 chain, enabling IL-4 or IL-13 binding (Figure 1B). In addition, IL-13 binds to the IL13Rα2 chain, a controversial signaling receptor. Receptor binding of IL-4 activates JAK1 and JAK3. JAK activation leads to STAT6 activation and translocation. Other transcription factors involved include c-Myc and IRF4. IL-4 induces macrophage fusion and decreases phagocytosis. The IL-4 multispecies transcriptome includes transglutaminase 2 (TGM2), mannose receptor (MRC1), cholesterol hydroxylase CH25H, and the prostaglandin-endoperoxide synthase PTGS1 (prostaglandin G/H synthase 1), the transcription factors IRF4, Krüppel-like factor 4 (KLF4), and the signaling modulators CISH and SOCS1 [40]. IL-13 signatures are similar to IL-4 signatures but are not totally overlapping [41]. In IL-4 KO animals, the numbers of macrophages and maturation are normal, and defects appear in the immune response against nematodes and some viral infections; for recent reviews, see [42,43]. In humans, polymorphisms in the IL-4R have been associated with the development of asthma and atopy [44,45].
Another M2 category is the type II-activated macrophage defined by Mosser and classified M2b by Mantovani and colleagues. This represents the only example of crosstalk with the B cell. This is another combined state, similar to the M1 combination of IFN-γ + LPS, in which ligation of FcγRs on LPS-activated macrophages turns off IL-12 and induces IL-10 secretion in addition to upregulating antigen presentation and, importantly, promoting Th2 responses [46,47]. IgGs are recognized by the Fc gamma receptor family that includes the activatory FcγRI (CD64), the inhibitory FcγRIIA (CD32), FcγRIIB (CD32), and the activatory FcγRIIIA (CD16a) and FcγRIIIB (CD16b). CD32 seems to be crucial for the type II activation in human monocytes and macrophages [48]. FcR signaling involves spleen tyrosine kinase (Syk) and phosphoinositide 3-kinase (PI3K) activation [49], but details in macrophages and its interplay with Myd88 pathways need further study. Type II-activated macrophages are distinct from IL-4-activated macrophages and their gene expression profiles overlap only partially. FcR KO animals have normal macrophage numbers, but their opsonic phagocytic capacity is highly impaired [50]. In humans, genetic differences in FcRs contribute to autoimmune diseases, such as systemic lupus erythematosus, rheumatoid arthritis, and multiple sclerosis [50]. Curiously, IgE, more relevant for Th2 and antiparasitic responses, has not been implicated in this phenotype.
Glucocorticoids and IL-10 are included in the current M2 category, although they represent a very different type of stimulus. Glucocorticoid hormones secreted by the adrenal glands are metabolized by cellular enzymes in macrophages. Active glucocorticoids are lipophilic and diffuse through the membrane to bind the glucocorticoid receptor (GCR) alpha, leading to nuclear translocation of the complex (Figure 1B). The GCR complex binds DNA directly to promote/repress gene transcription or indirectly by interacting with transcription factors, such as NF-κB or AP-1. Expression analysis of glucocorticoid-stimulated monocytes showed induction of complement component 1 subunit A (C1QA), TSC22 domain family, member 3 (DSIPI), MRC1, thrombospondin 1 (THBS1), IL-10, IL1R2, and CD163 [51]. Long-term exposure drives different gene expression programs that interact with LPS and IFN-γ pathways in a complex and non-exclusively antagonistic manner [52]. The profiles are altogether different from those induced by IL-4. Glucocorticoids affect monocyte adherence, spreading, phagocytosis, and apoptosis. Mice with GCR deficiency do not survive long after birth, because of respiratory failure, and alterations in receptor dimerization induce susceptibility to sepsis [53]. In humans, GCR polymorphisms in the genes are pleiotropic and have been involved in a variety of malignancies and inflammatory and autoimmune disorders [54].
IL-10 binds the IL-10 receptor, a dimer of IL10R1 and IL10R2 (Figure 1B). Receptor autophosphorylation leads to the activation of the transcription factor STAT3 and its binding mediates inhibition of pro-inflammatory cytokine expression. IL-10 is a Th2 product and potent inhibitor of Th1 cells [55]. IL-10 is produced by virtually all leukocytes. In macrophages, IL-10 is elicited in response to TLR activation, glucocorticoids, and C-type lectin signaling (e.g. DC-SIGN [CD209 molecule] and dectin 1 ligation). The macrophage transcriptome induced by IL-10 includes selected Fc receptors, the chemoattractants CXCL13 and CXCL4, and the recognition receptors formyl peptide receptor 1 (FPR1), TLR1, TLR8, and macrophage receptor with collagenous domain (MARCO) [56]. IL-10-deficient mice have normal macrophage numbers but develop inflammatory bowel disease following colonization of the gut with resident enteric bacteria [57] and show exaggerated inflammatory responses to parasites [58]. In humans, defects in the cytokine receptors are similar and involve colitis and exacerbated inflammation [59].
M-CSF, like GM-CSF, is a late addition to the paradigm and has been classified as an M2 stimulus. The M-CSF receptor is a tyrosine kinase transmembrane receptor (Figure 1B). M-CSF binding leads to receptor dimerization, autophosphorylation, activation of ERK, phosphatidylinositol 3-kinase, phospholipase C, and eventually Sp1 transcription factor nuclear localization. The transcriptional response to M-CSF includes transient gene clusters with overrepresentation of cell cycle genes (e.g. cyclins A2, B1, D1, and E1) and downregulation of human leukocyte antigen (HLA) members and stable gene clusters, including TLR7 and the complement C1QA/B/C subunits [15].
Reports generally focus on the differences between M-CSF and GM-CSF; a recent and comprehensive comparison of the response to GM-CSF and M-CSF by human and mouse macrophages shows 530 genes regulated in the same direction in both human and murine models [60,61]. M-CSF mutant mice show reduced levels of monocytes and selected macrophages and osteopetrosis [62]. Mutations in the M-CSF receptor in humans lead to myelodysplastic syndromes or acute myeloid leukemia [63], and the mutations have been associated with hereditary diffuse leukoencephalopathy [64], but no human patients with osteopetrosis secondary to M-CSF deficiency have been identified.
The M2 group includes very different stimuli that span four levels of recognition/response: a level in which the macrophage acquires matured phenotypes, a level in which the macrophage interacts with immune cells (eosinophils, basophils, and Th2 cells), a level in which the macrophage actually deals with the pathogen, and a resolution level.
We discussed signaling cascades elicited by current M1 and M2 stimuli, which are complex and include transient and stable gene signatures. Importantly, activation of macrophages is controlled not only by intracellular kinases and transcription factors. Other mechanisms, such as microRNAs (miRNAs) [65-69], enhancer RNAs [70-72], and epigenetic enzymes, control the activation landscape [73-78]. These are beginning to be elucidated in macrophages, and more information exists for the mouse. The research done on epigenetics does not cover the full spectrum of macrophage activation but does provide messages similar to the signaling pathways discussed above (i.e. specific mechanisms control different forms of activation, and these mechanisms are not fixed; for every acetylase or methylase, there is a counterpart ready to be activated). Before discussing our view of macrophage activation, we briefly discuss the evidence for M1/M2 activation in disease.
M1 and M2 in disease: lack of defined subsets
In vitro studies have contributed to our understanding of macrophage activation, and as discussed, KO animals for key cytokines and receptors have established a role for some of these in the development and maturation of macrophages, whereas others regulate activation and the tuning of the response. The activation signatures defined in vitro are highly influenced by factors that are often overlooked but important in vivo (e.g. maturation of the cell, adhesion, extracellular matrix composition, and chemoattractants). Translating in vitro results to disease poses a major problem because of the complexity of in vivo systems and the failure to mimic these conditions in vitro [79]. Defining specific M1 and M2 functions with cytokines or receptor KO is difficult because the genes are pleiotropic and expressed at different stages of macrophage development or in other cell types; development of conditional, macrophage-specific KOs will help to illuminate these functions. Whole genome studies have shown substantial differences between M1 and M2 activation programs in humans and mice, indicative of evolutionary plasticity among macrophages, yet adding difficulty to translation. When it comes to infection, it is clear that macrophage responses to different pathogens are affected by virulence and evasion mechanisms. Non-infectious diseases in humans are not as homogeneous as in mouse models, and thus we are often looking at a collection of tissue and systemic conditions that lead to a common syndrome but with different macrophage phenotypes.
Because of the relationship with the Th1 and Th2 paradigm, macrophage M1 and M2 markers have been investigated in prototypic diseases (e.g. Th2 Asthma [80] and Th1 chronic obstructive pulmonary disease (COPD) [81]). Another area where macrophage profiles have been investigated is atherosclerosis [82] and tumors [83]. For a recent review, see [84]. The emphasis has always been to fit the profile of tissue macrophages in diseases to in vitro predictions, but the message is clear: in vitro models are unable to mimic the complex profiles observed in disease and, as such, the numbers of genes that can be confirmed is limited. Our feeling is that when it comes to tissue macrophages, we need to start with a fresh view. Importantly, because macrophages can develop mixed M1/M2 phenotypes in pathological conditions, we need to focus not only on populations but also at the single-cell level [85,86].
From a functional view, the main properties of macrophages are phagocytosis, endocytosis, secretion, and microbial killing, but chemotaxis, adhesion, and trophic functions are an integral part of their activation [11]. Because macrophages are able to perform all these activities in the steady state, M1 and M2 contribution to disease is, for the most part, modulation and tuning. As before with markers, functions are complex (e.g. phagocytosis involves a collaboration of multiple receptors as well as interactions with different particles). The discrimination and killing of microbes and host target cells are also incompletely understood. To date, M1- or M2-specific functions beyond M1-enhanced microbicidal and M2 antiparasitic defense have expanded to encompass metabolic, thermoregulatory, healing, and antiviral effects [10,42,43,87-90]. Recent evidence shows that M1 and M2 activation display differences but also overlapping effects that need clarification and a more dynamic appreciation of the activation process. In Table 1, we have summarized a few findings that exemplify the increasing complexity of the M1/M2 landscape.
Table 1. Selection of M1 and M2 effects in macrophages.
| M1 (IFN-γ) | M2 (IL-4/IL-13) | |
|---|---|---|
| Functions elicited in macrophages | ||
| Phagocytosis / endocytosis | -Increases phagocytosis of C. albicans [95]-Decreases Fc-mediated phagocytosis [96]-Decreases complement-mediated phagocytosis [97] | -Decreases phagocytosis of particles while increasing inflammatory cytokine production [98] |
| Autophagy | -Induces autophagy in TB infection [99] | -Decreases autophagy in TB infection [100] |
| Macrophage Fusion | -Increases fusion in combination with concanavalin A [101]-Induces fusion in alveolar macrophages [102] | -Induces fusion [103]-Inhibits IFN-γ-induced fusion [101] |
| Nitric Oxide | -Induces Mycobacteria killing via NO [104] | -Favours Arginase-1 vs. i-NOS, Arg1+ macrophages suppress Th2 inflammation and fibrosis [105] |
| Parasite killing and expulsion | -Mediates parasite killing via NO [106, 107] | -Although the cytokine is important for worm expulsion, the effect does not depend on macrophages [108] |
| Virus replication | -Inhibits replication of HIV at early pre-integration steps [109] | -Inhibits HIV replication at post-integration level [109] |
| Markers | ||
| Human | CD64, IDO, SOCS1, CXCL10 | MRC1, TGM2, CD23, CCL22 |
| Mouse | CXCL9, CXCL10, CXCL11, NOS2 | Mrc1, tgm2, Fizz1, Ym1/2, Arg1 |
Detailing M1 and M2 marker studies in particular human disease processes is beyond the scope of this review; we highlight some considerations:
(a) Rather than distinct macrophage populations, M1 and M2 signatures do not necessarily exclude each other and often coexist; the resultant mixed phenotype then depends on the balance of activatory and inhibitory activities and the tissue environment.
(b) Pleiotropism of stimuli and lack of cell specificity of markers indicate that macrophage specialization rests in part on their ability to migrate and deliver specific functions where barriers have failed to stop infection.
(c) The role of M1 or M2 stimuli needs to be considered in their dynamic complexity, beyond the current bipolar dogma of IFN-γ as exclusively important for intracellular pathogens or IL-4 for allergy and extracellular parasitic defense. The multipolar interplay between these and many other signals requires further studies in vivo and in vitro.
Conclusions
Today, we know that, in addition to Th1 and Th2 cells, Treg cells and Th17 cells participate in the pathogenesis and resolution of disease and that these cells are able to display shades of activation within the general categories [91]. The Th1/Th2 paradigm has remained valuable in T lymphocyte heterogeneity and, to an extent, in the field of macrophage activation, but a reassessment is required to accommodate current findings.
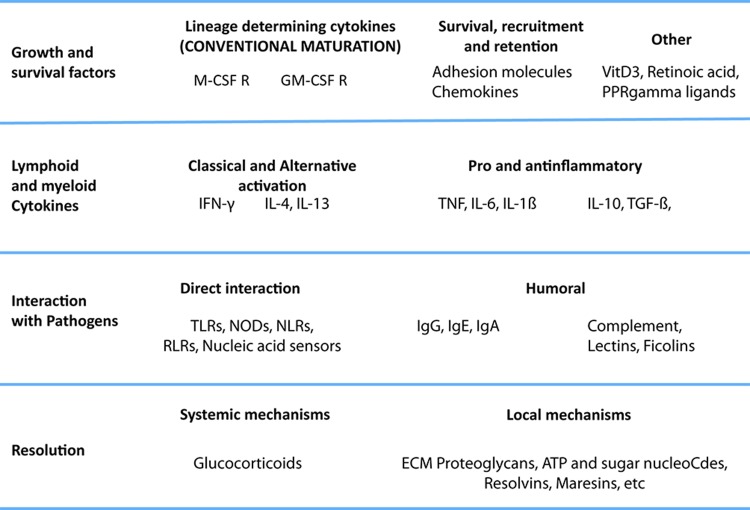
We do not here propose a revised model or nomenclature for macrophage activation but present a view on what needs to be taken into account (Figure 2). Our opinion is that macrophages do not form stable subsets but respond to a combination of factors present in the tissue; we have, rather than subsets of macrophages, pathways that interact to form a complex, even mixed, phenotypes. A satisfactory paradigm needs to take into account at least four levels of recognition/response: first, one in which the monocyte survives and acquires matured phenotypes; second, a level in which the macrophage interacts with immune cells (NK and Th cells, eosinophils, and basophils); third, one in which the macrophage deals with the pathogen, and a final level, of resolution. Also, the M1-M2 paradigm is commonly associated with properties of mature macrophages, but activation takes place in the extended macrophage family, including monocytes [41,48,51,92], myeloid-derived dendritic cells [93,94] and multinucleated giant cells. In tissues, all these events combine to produce a resultant phenotype, and, though useful for the sake of understanding, any sort of hierarchy or order does not represent the biology of the cells. We require a dynamic view of this process to take into account the multiple elements in their systemic and local milieu and to define the kinetics, plasticity, reversibility, and memory of their responses in order to encompass the full functional range of activated macrophages. The process is highly complex, and for an improved understanding, considerably more information is required about macrophages in vivo and at the population level as well as at the single-cell level. Epigenetic, gene expression, and functional studies will help to elucidate these matters.
Figure 2. A multipolar view of the macrophage activation paradigm from an immunological perspective.
The cytokines and stimuli that we call M1 and M2 play different roles in the development, maturation, and activation of macrophages. Integration of these isolated stimuli is necessary to represent the complex changes that macrophages undergo during full activation. Lessons from basic immunology suggest that many of the mechanisms that affect innate and acquired immunity are underappreciated (for example, humoral pattern recognition receptors, inflammatory cytokines other than interferon-gamma (IFN-γ) and tumor necrosis factor (TNF)). Here, we focus on the levels of activation imposed by the immune system rather than proposing a new classification of macrophage phenotypes, which is currently under discussion internationally. We propose that the stimuli governing macrophage activation should be organized according to their role in the immune response, rather than in groups that overlook their individual differences, and highlight the fact that complex combinations should be assessed to understand the full repertoire of macrophages. We identify at least four levels: growth and survival factors, interaction with lymphoid and myeloid cytokines, interaction with pathogens, and resolution. In the first level, in addition to prototypic maturation signals, we add stimuli known to promote survival of monocytes. A second level is interaction with cytokines. Here, we place interleukin-4 (IL-4), and IFN-γ and other cytokines produced by lymphoid and myeloid cells. However, the contribution of non-hematopoietic cells cannot be ignored. The next level is that of interaction with pathogens directly or through humoral recognition receptors, such as lectins, ficolins, and the B cell-derived immunoglobulins. A final level of resolution we classify as systemic (e.g. glucocorticoids) or local, such as ATP, resolvins, and other mediators with general anti-inflammatory properties. The combinations of stimuli commonly associated with extreme phenotypes of cells are IFN-γ + lipopolysaccharide (LPS) or TNF, immune complexes + Myd88, and granulocyte macrophage colony-stimulating factor (GM-CSF) + IL-4, which in monocytes induce a dendritic cell-like phenotype. However, there are many other possible combinations that are not considered special cases. We plan to extend the potential of macrophages, taking into consideration other functionally relevant combinations.
At present, the M1/M2 paradigm has provided a useful framework, especially for selected immune responses, but a more comprehensive classification is clearly required. This should guide an iterative research strategy from in vitro to in vivo studies and back to disease models in genetically defined mice, for example, to establish mechanisms and possible therapeutic targets for manipulation in human disease.
Acknowledgments
The authors acknowledge Ronny Milde for their help with Figure 1B and Elsevier for Figure 1A.
Abbreviations
AP-1
activator protein 1
C1QA/B/C
complement component 1 subunit A/B/C
CISH
cytokine inducible SH2-containing protein
c-Myc
V-Myc avian myelocytomatosis viral oncogene homolog
CXCL
chemokine (C-X-C motif) ligand
EGR
early growth response
ERK
extracellular signal-regulated kinase
FCγR
Fc fragment of IgG, high affinity
GCR
glucocorticoid receptor
GM-CSF
granulocyte macrophage colony-stimulating factor
IFN
interferon
IFNGR
interferon gamma receptor
Ig
immunoglobulin
IL
interleukin
IL-4Rα1
interleukin 4 receptor alpha 1
IRF
interferon regulatory factor
ITGB7
integrin beta-7
Jak
Janus kinase
KO
knockout
LPS
lipopolysaccharide
M-CSF
macrophage colony-stimulating factor
MHC
major histocompatibility complex
MRC1
mannose receptor
MyD88
myeloid differentiation primary response 88
NF-κB
nuclear factor of kappa light polypeptide gene enhancer
NK
natural killer
SOCS1
suppressor of cytokine signaling 1
STAT
signal transducers and activators of transcription
TGF-β
transforming growth factor-beta
Th
T-helper
TLR
Toll-like receptor
TNF
tumor necrosis factor
Treg
T regulatory
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/6/13
Contributor Information
Fernando O. Martinez, Email: fernando.martinezestrada@kennedy.ox.ac.uk.
Siamon Gordon, Email: siamon.gordon@path.ox.ac.uk.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–57. [PubMed] [Google Scholar]
- 2.Coffman RL. Origins of the T(H)1-T(H)2 model: a personal perspective. Nat Immunol. 2006;7:539–41. doi: 10.1038/ni0606-539. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718270380
- 3.Mackaness GB. Cellular resistance to infection. J Exp Med. 1962;116:381–406. doi: 10.1084/jem.116.3.381. [DOI] [PubMed] [Google Scholar]
- 4.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158:670–89. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pace JL, Russell SW, Schreiber RD, Altman A, Katz DH. Macrophage activation: priming activity from a T-cell hybridoma is attributable to interferon-gamma. Proc Natl Acad Sci USA. 1983;80:3782–6. doi: 10.1073/pnas.80.12.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celada A, Gray PW, Rinderknecht E, Schreiber RD. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J Exp Med. 1984;160:55–74. doi: 10.1084/jem.160.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–92. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle AG, Herbein G, Montaner LJ, Minty AJ, Caput D, Ferrara P, Gordon S. Interleukin-13 alters the activation state of murine macrophages in vitro: comparison with interleukin-4 and interferon-gamma. Eur J Immunol. 1994;24:1441–5. doi: 10.1002/eji.1830240630. [DOI] [PubMed] [Google Scholar]
- 9.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–73. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins SJ, Allen JE. Similarity and diversity in macrophage activation by nematodes, trematodes, and cestodes. J Biomed Biotechnol. 2010;2010:262609. doi: 10.1155/2010/262609. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718270383
- 11.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–86. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Verreck FAW, Boer T de, Langenberg DML, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, Waal-Malefyt R de, Ottenhoff THM. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA. 2004;101:4560–5. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–50. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, Levin M, Relman DA. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS ONE. 2010;5:e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1050430
- 16.Nau GJ, Richmond JFL, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA. 2002;99:1503–8. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1003974
- 17.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–5. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 18.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–42. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 19.Marinho CRF, Nuñez-Apaza LN, Martins-Santos R, Bastos KRB, Bombeiro AL, Bucci DZ, Sardinha LR, Lima MRD, Alvarez JM. IFN-gamma, but not nitric oxide or specific IgG, is essential for the in vivo control of low-virulence Sylvio X10/4 Trypanosoma cruzi parasites. Scand J Immunol. 2007;66:297–308. doi: 10.1111/j.1365-3083.2007.01958.x. [DOI] [PubMed] [Google Scholar]
- 20.Pinheiro RO, Rossi-Bergmann B. Interferon-gamma is required for the late but not early control of Leishmania amazonensis infection in C57Bl/6 mice. Mem Inst Oswaldo Cruz. 2007;102:79–82. doi: 10.1590/S0074-02762007000100013. [DOI] [PubMed] [Google Scholar]
- 21.Jakobi V, Petry F. Humoral immune response in IL-12 and IFN-gamma deficient mice after infection with Cryptosporidium parvum. Parasite Immunol. 2008;30:151–61. doi: 10.1111/j.1365-3024.2007.01013.x. [DOI] [PubMed] [Google Scholar]
- 22.Saeftel M, Arndt M, Specht S, Volkmann L, Hoerauf A. Synergism of gamma interferon and interleukin-5 in the control of murine filariasis. Infect Immun. 2003;71:6978–85. doi: 10.1128/IAI.71.12.6978-6985.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rezende SA, Oliveira VR, Silva AM, Alves JB, Goes AM, Reis LF. Mice lacking the gamma interferon receptor have an impaired granulomatous reaction to Schistosoma mansoni infection. Infect Immun. 1997;65:3457–61. doi: 10.1128/iai.65.8.3457-3461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du X, Wu J, Zhang M, Gao Y, Zhang D, Hou M, Ji M, Wu G. Upregulated expression of cytotoxicity-related genes in IFN-γ knockout mice with Schistosoma japonicum infection. J Biomed Biotechnol. 2011;2011:864945. doi: 10.1155/2011/864945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorman SE, Holland SM. Interferon-gamma and interleukin-12 pathway defects and human disease. Cytokine Growth Factor Rev. 2000;11:321–33. doi: 10.1016/S1359-6101(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S, Chen Y, Sankala M, Peiser L, Pikkarainen T, Kraal G, Tryggvason K, Gordon S. MARCO, an innate activation marker of macrophages, is a class A scavenger receptor for Neisseria meningitidis. Eur J Immunol. 2006;36:940–9. doi: 10.1002/eji.200535389. [DOI] [PubMed] [Google Scholar]
- 27.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, MuszyŃski A, Forsberg LS, Carlson RW, Dixit VM. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–9. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718047661
- 28.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–3. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718106716
- 29.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto M, Takeda K. Current views of toll-like receptor signaling pathways. Gastroenterol Res Pract. 2010;2010:240365. doi: 10.1155/2010/240365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Netea MG, Wijmenga C, O’Neill LAJ. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol. 2012;13:535–42. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 33.Casanova J, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol. 2011;29:447–91. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 34.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, McKinstry WJ, Lopez AF, Parker MW. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1119465
- 35.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–8. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/9382957
- 36.Lehtonen A, Ahlfors H, Veckman V, Miettinen M, Lahesmaa R, Julkunen I. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol. 2007;82:710–20. doi: 10.1189/jlb.0307194. [DOI] [PubMed] [Google Scholar]
- 37.Dranoff G, Mulligan RC. Activities of granulocyte-macrophage colony-stimulating factor revealed by gene transfer and gene knockout studies. Stem Cells. 1994;12 (Suppl 1):173–82. discussion 182-4. [PubMed] [Google Scholar]
- 38.Dirksen U, Nishinakamura R, Groneck P, Hattenhorst U, Nogee L, Murray R, Burdach S. Human pulmonary alveolar proteinosis associated with a defect in GM-CSF/IL-3/IL-5 receptor common beta chain expression. J Clin Invest. 1997;100:2211–7. doi: 10.1172/JCI119758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dirksen U, Hattenhorst U, Schneider P, Schroten H, Göbel U, Böcking A, Müller KM, Murray R, Burdach S. Defective expression of granulocyte-macrophage colony-stimulating factor/interleukin-3/interleukin-5 receptor common beta chain in children with acute myeloid leukemia associated with respiratory failure. Blood. 1998;92:1097–103. [PubMed] [Google Scholar]
- 40.Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, Thomas B, Fabbri M, Crawshaw A, Ho LP, Hacken NH ten, Cobos Jiménez V, Kootstra NA, Hamann J, Greaves DR, Locati M, Mantovani A, Gordon S. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- 41.Scotton CJ, Martinez FO, Smelt MJ, Sironi M, Locati M, Mantovani A, Sozzani S. Transcriptional profiling reveals complex regulation of the monocyte IL-1 beta system by IL-13. J Immunol. 2005;174:834–45. doi: 10.4049/jimmunol.174.2.834. [DOI] [PubMed] [Google Scholar]
- 42.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–83. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/717147858
- 43.van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–43. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ford AQ, Heller NM, Stephenson L, Boothby MR, Keegan AD. An atopy-associated polymorphism in the ectodomain of the IL-4R(alpha) chain (V50) regulates the persistence of STAT6 phosphorylation. J Immunol. 2009;183:1607–16. doi: 10.4049/jimmunol.0803266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beghé B, Barton S, Rorke S, Peng Q, Sayers I, Gaunt T, Keith TP, Clough JB, Holgate ST, Holloway JW. Polymorphisms in the interleukin-4 and interleukin-4 receptor alpha chain genes confer susceptibility to asthma and atopy in a Caucasian population. Clin Exp Allergy. 2003;33:1111–7. doi: 10.1046/j.1365-2222.2003.01731.x. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1015264
- 46.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80:1298–307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson CF, Mosser DM. A novel phenotype for an activated macrophage: the type 2 activated macrophage. J Leukoc Biol. 2002;72:101–6. [PubMed] [Google Scholar]
- 48.Sironi M, Martinez FO, D’Ambrosio D, Gattorno M, Polentarutti N, Locati M, Gregorio A, Iellem A, Cassatella MA, van Damme J, Sozzani S, Martini A, Sinigaglia F, Vecchi A, Mantovani A. Differential regulation of chemokine production by Fcgamma receptor engagement in human monocytes: association of CCL1 with a distinct form of M2 monocyte activation (M2b, Type 2) J Leukoc Biol. 2006;80:342–9. doi: 10.1189/jlb.1005586. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez-Mejorada G, Rosales C. Signal transduction by immunoglobulin Fc receptors. J Leukoc Biol. 1998;63:521–33. doi: 10.1002/jlb.63.5.521. [DOI] [PubMed] [Google Scholar]
- 50.Takai T. Roles of Fc receptors in autoimmunity. Nat Rev Immunol. 2002;2:580–92. doi: 10.1038/nri856. [DOI] [PubMed] [Google Scholar]
- 51.Ehrchen J, Steinmüller L, Barczyk K, Tenbrock K, Nacken W, Eisenacher M, Nordhues U, Sorg C, Sunderkötter C, Roth J. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood. 2007;109:1265–74. doi: 10.1182/blood-2006-02-001115. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1097620
- 52.van de Garde MDB, Martinez FO, Melgert BN, Hylkema MN, Jonkers RE, Hamann J. Chronic exposure to glucocorticoids shapes gene expression and modulates innate and adaptive activation pathways in macrophages with distinct changes in leukocyte attraction. J Immunol. 2014;192:1196–208. doi: 10.4049/jimmunol.1302138. [DOI] [PubMed] [Google Scholar]
- 53.Kleiman A, Hübner S, Rodriguez Parkitna JM, Neumann A, Hofer S, Weigand MA, Bauer M, Schmid W, Schütz G, Libert C, Reichardt HM, Tuckermann JP. Glucocorticoid receptor dimerization is required for survival in septic shock via suppression of interleukin-1 in macrophages. FASEB J. 2012;26:722–9. doi: 10.1096/fj.11-192112. [DOI] [PubMed] [Google Scholar]
- 54.Bray PJ, Cotton RGH. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum Mutat. 2003;21:557–68. doi: 10.1002/humu.10213. [DOI] [PubMed] [Google Scholar]
- 55.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–95. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park-Min K, Antoniv TT, Ivashkiv LB. Regulation of macrophage phenotype by long-term exposure to IL-10. Immunobiology. 2005;210:77–86. doi: 10.1016/j.imbio.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 57.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gazzinelli RT, Wysocka M, Hieny S, Scharton-Kersten T, Cheever A, Kühn R, Müller W, Trinchieri G, Sher A. In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J Immunol. 1996;157:798–805. [PubMed] [Google Scholar]
- 59.Glocker E, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hätscher N, Pfeifer D, Sykora K, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashimoto S, Suzuki T, Dong HY, Yamazaki N, Matsushima K. Serial analysis of gene expression in human monocytes and macrophages. Blood. 1999;94:837–44. [PubMed] [Google Scholar]
- 61.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook AD, Hamilton JA. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–65. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/716398013
- 62.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW, Ahmed-Ansari A, Sell KW, Pollard JW, Stanley ER. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci USA. 1990;87:4828–32. doi: 10.1073/pnas.87.12.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tobal K, Pagliuca A, Bhatt B, Bailey N, Layton DM, Mufti GJ. Mutation of the human FMS gene (M-CSF receptor) in myelodysplastic syndromes and acute myeloid leukemia. Leukemia. 1990;4:486–9. [PubMed] [Google Scholar]
- 64.Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, Adamson J, Kouri N, Sundal C, Shuster EA, Aasly J, MacKenzie J, Roeber S, Kretzschmar HA, Boeve BF, Knopman DS, Petersen RC, Cairns NJ, Ghetti B, Spina S, Garbern J, Tselis AC, Uitti R, Das P, van Gerpen JA, Meschia JF, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet. 2012;44:200–5. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13623956
- 65.Etzrodt M, Cortez-Retamozo V, Newton A, Zhao J, Ng A, Wildgruber M, Romero P, Wurdinger T, Xavier R, Geissmann F, Meylan E, Nahrendorf M, Swirski FK, Baltimore D, Weissleder R, Pittet MJ. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. 2012;1:317–24. doi: 10.1016/j.celrep.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–25. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lagrange B, Martin RZ, Droin N, Aucagne R, Paggetti J, Largeot A, Itzykson R, Solary E, Delva L, Bastie J. A role for miR-142-3p in colony-stimulating factor 1-induced monocyte differentiation into macrophages. Biochim Biophys Acta. 2013;1833:1936–46. doi: 10.1016/j.bbamcr.2013.04.007. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718270396
- 68.Mildner A, Chapnik E, Manor O, Yona S, Kim K, Aychek T, Varol D, Beck G, Itzhaki ZB, Feldmesser E, Amit I, Hornstein E, Jung S. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood. 2013;121:1016–27. doi: 10.1182/blood-2012-07-445999. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718270398
- 69.Swaminathan S, Hu X, Zheng X, Kriga Y, Shetty J, Zhao Y, Stephens R, Tran B, Baseler MW, Yang J, Lempicki RA, Huang D, Lane HC, Imamichi T. Interleukin-27 treated human macrophages induce the expression of novel microRNAs which may mediate anti-viral properties. Biochem Biophys Res Commun. 2013;434:228–34. doi: 10.1016/j.bbrc.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santa F de, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/3267958
- 71.Kaikkonen MU, Lam MTY, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–40. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718270403
- 72.Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–5. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718026148
- 73.Bowdridge S, Gause WC. Regulation of alternative macrophage activation by chromatin remodeling. Nat Immunol. 2010;11:879–81. doi: 10.1038/ni1010-879. [DOI] [PubMed] [Google Scholar]
- 74.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–61. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718270404
- 75.Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34:216–23. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718270418
- 76.Mukhopadhyay S, Ramadass AS, Akoulitchev A, Gordon S. Formation of distinct chromatin conformation signatures epigenetically regulate macrophage activation. Int Immunopharmacol. 2013;18:7–11. doi: 10.1016/j.intimp.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 77.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–44. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/6283956
- 78.Kruidenier L, Chung C, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–8. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717952568
- 79.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–95. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718114022
- 80.Madore A, Perron S, Turmel V, Laviolette M, Bissonnette EY, Laprise C. Alveolar macrophages in allergic asthma: an expression signature characterized by heat shock protein pathways. Hum Immunol. 2010;71:144–50. doi: 10.1016/j.humimm.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaykhiev R, Krause A, Salit J, Strulovici-Barel Y, Harvey B, O’Connor TP, Crystal RG. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009;183:2867–83. doi: 10.4049/jimmunol.0900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feig JE, Vengrenyuk Y, Reiser V, Wu C, Statnikov A, Aliferis CF, Garabedian MJ, Fisher EA, Puig O. Regression of atherosclerosis is characterized by broad changes in the plaque macrophage transcriptome. PLoS ONE. 2012;7:e39790. doi: 10.1371/journal.pone.0039790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ojalvo LS, Whittaker CA, Condeelis JS, Pollard JW. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J Immunol. 2010;184:702–12. doi: 10.4049/jimmunol.0902360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pettersen JS, Fuentes-Duculan J, Suárez-Fariñas M, Pierson KC, Pitts-Kiefer A, Fan L, Belkin DA, Wang CQF, Bhuvanendran S, Johnson-Huang LM, Bluth MJ, Krueger JG, Lowes MA, Carucci JA. Tumor-associated macrophages in the cutaneous SCC microenvironment are heterogeneously activated. J Invest Dermatol. 2011;131:1322–30. doi: 10.1038/jid.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718270426
- 86.Vogel DYS, Vereyken EJF, Glim JE, Heijnen PDAM, Moeton M, van der Valk P, Amor S, Teunissen CE, van Horssen J, Dijkstra CD. Macrophages in inflammatory multiple sclerosis lesions have an intermediate activation status. J Neuroinflammation. 2013;10:35. doi: 10.1186/1742-2094-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu Rev Pathol. 2011;6:275–97. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ricote M, Valledor AF, Glass CK. Decoding transcriptional programs regulated by PPARs and LXRs in the macrophage: effects on lipid homeostasis, inflammation, and atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:230–9. doi: 10.1161/01.ATV.0000103951.67680.B1. [DOI] [PubMed] [Google Scholar]
- 89.Cunard R, Ricote M, DiCampli D, Archer DC, Kahn DA, Glass CK, Kelly CJ. Regulation of cytokine expression by ligands of peroxisome proliferator activated receptors. J Immunol. 2002;168:2795–802. doi: 10.4049/jimmunol.168.6.2795. [DOI] [PubMed] [Google Scholar]
- 90.Palsson-McDermott EM, O’Neill LAJ. The Warburg effect then and now: from cancer to inflammatory diseases. Bioessays. 2013;35:965–73. doi: 10.1002/bies.201300084. [DOI] [PubMed] [Google Scholar]
- 91.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamassia N, Le Moigne V, Calzetti F, Donini M, Gasperini S, Ear T, Cloutier A, Martinez FO, Fabbri M, Locati M, Mantovani A, McDonald PP, Cassatella MA. The MyD88-independent pathway is not mobilized in human neutrophils stimulated via TLR4. J Immunol. 2007;178:7344–56. doi: 10.4049/jimmunol.178.11.7344. [DOI] [PubMed] [Google Scholar]
- 93.Mazzoni A, Segal DM. Controlling the Toll road to dendritic cell polarization. J Leukoc Biol. 2004;75:721–30. doi: 10.1189/jlb.1003482. [DOI] [PubMed] [Google Scholar]
- 94.Kapsenberg ML. Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol. 2003;3:984–93. doi: 10.1038/nri1246. [DOI] [PubMed] [Google Scholar]
- 95.Maródi L, Schreiber S, Anderson DC, MacDermott RP, Korchak HM, Johnston RB. Enhancement of macrophage candidacidal activity by interferon-gamma. Increased phagocytosis, killing, and calcium signal mediated by a decreased number of mannose receptors. J Clin Invest. 1993;91:2596–601. doi: 10.1172/JCI116498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frausto-Del-Río D, Soto-Cruz I, Garay-Canales C, Ambriz X, Soldevila G, Carretero-Ortega J, Vázquez-Prado J, Ortega E. Interferon gamma induces actin polymerization, Rac1 activation and down regulates phagocytosis in human monocytic cells. Cytokine. 2012;57:158–68. doi: 10.1016/j.cyto.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 97.Schlesinger LS, Horwitz MA. Phagocytosis of Mycobacterium leprae by human monocyte-derived macrophages is mediated by complement receptors CR1 (CD35), CR3 (CD11b/CD18), and CR4 (CD11c/CD18) and IFN-gamma activation inhibits complement receptor function and phagocytosis of this bacterium. J Immunol. 1991;147:1983–94. [PubMed] [Google Scholar]
- 98.Varin A, Mukhopadhyay S, Herbein G, Gordon S. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood. 2010;115:353–62. doi: 10.1182/blood-2009-08-236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matsuzawa T, Kim B, Shenoy AR, Kamitani S, Miyake M, Macmicking JD. IFN-γ elicits macrophage autophagy via the p38 MAPK signaling pathway. J Immunol. 2012;189:813–8. doi: 10.4049/jimmunol.1102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harris J, Haro SA de, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–17. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1092128
- 101.Takashima T, Ohnishi K, Tsuyuguchi I, Kishimoto S. Differential regulation of formation of multinucleated giant cells from concanavalin A-stimulated human blood monocytes by IFN-gamma and IL-4. J Immunol. 1993;150:3002–10. [PubMed] [Google Scholar]
- 102.Nagasawa H, Miyaura C, Abe E, Suda T, Horiguchi M. Fusion and activation of human alveolar macrophages induced by recombinant interferon-gamma and their suppression by dexamethasone. Am Rev Respir Dis. 1987;136:916–21. doi: 10.1164/ajrccm/136.4.916. [DOI] [PubMed] [Google Scholar]
- 103.Helming L, Gordon S. Macrophage fusion induced by IL-4 alternative activation is a multistage process involving multiple target molecules. Eur J Immunol. 2007;37:33–42. doi: 10.1002/eji.200636788. [DOI] [PubMed] [Google Scholar]
- 104.Herbst S, Schaible UE, Schneider BE. Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS ONE. 2011;6:e19105. doi: 10.1371/journal.pone.0019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thomas GR, McCrossan M, Selkirk ME. Cytostatic and cytotoxic effects of activated macrophages and nitric oxide donors on Brugia malayi. Infect Immun. 1997;65:2732–9. doi: 10.1128/iai.65.7.2732-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Piedrafita D, Parsons JC, Sandeman RM, Wood PR, Estuningsih SE, Partoutomo S, Spithill TW. Antibody-dependent cell-mediated cytotoxicity to newly excysted juvenile Fasciola hepatica in vitro is mediated by reactive nitrogen intermediates. Parasite Immunol. 2001;23:473–82. doi: 10.1046/j.1365-3024.2001.00404.x. [DOI] [PubMed] [Google Scholar]
- 108.Urban JF, Noben-Trauth N, Schopf L, Madden KB, Finkelman FD. Cutting edge: IL-4 receptor expression by non-bone marrow-derived cells is required to expel gastrointestinal nematode parasites. J Immunol. 2001;167:6078–81. doi: 10.4049/jimmunol.167.11.6078. [DOI] [PubMed] [Google Scholar]
- 109.Cassol E, Cassetta L, Rizzi C, Alfano M, Poli G. M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. J Immunol. 2009;182:6237–46. doi: 10.4049/jimmunol.0803447. [DOI] [PubMed] [Google Scholar]