The Dispersed Archaeal Eukaryome and the Complex Archaeal Ancestor of Eukaryotes (original) (raw)
Abstract
The ancestral set of eukaryotic genes is a chimera composed of genes of archaeal and bacterial origins thanks to the endosymbiosis event that gave rise to the mitochondria and apparently antedated the last common ancestor of the extant eukaryotes. The proto-mitochondrial endosymbiont is confidently identified as an α-proteobacterium. In contrast, the archaeal ancestor of eukaryotes remains elusive, although evidence is accumulating that it could have belonged to a deep lineage within the TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) superphylum of the Archaea. Recent surveys of archaeal genomes show that the apparent ancestors of several key functional systems of eukaryotes, the components of the archaeal “eukaryome,” such as ubiquitin signaling, RNA interference, and actin-based and tubulin-based cytoskeleton structures, are identifiable in different archaeal groups. We suggest that the archaeal ancestor of eukaryotes was a complex form, rooted deeply within the TACK superphylum, that already possessed some quintessential eukaryotic features, in particular, a cytoskeleton, and perhaps was capable of a primitive form of phagocytosis that would facilitate the engulfment of potential symbionts. This putative group of Archaea could have existed for a relatively short time before going extinct or undergoing genome streamlining, resulting in the dispersion of the eukaryome. This scenario might explain the difficulty with the identification of the archaeal ancestor of eukaryotes despite the straightforward detection of apparent ancestors to many signature eukaryotic functional systems.
The apparent ancestors of key eukaryotic features (e.g., ubiquitin signaling, RNA interference, and cytoskeletal structures) are identifiable in different Archaea. But the specific archaeal ancestor of eukaryotes remains elusive.
The origin of the eukaryotes obviously is one of the key problems in the entire study of the history of life on Earth. The extreme complexity of the eukaryotic cellular organization compared with that of archaeal and bacterial (collectively, prokaryotic) cells cries for an evolutionary explanation and even has brought the specter of “irreducible complexity” into the scientific debate (Kurland et al. 2006; Martin et al. 2007). The “standard model” derived primarily from the classic phylogenetic analysis of 16S RNA by Woese and coworkers has eukaryotes as the sister group of Archaea, to the exclusion of Bacteria (Woese et al. 1990; Pace 1997, 2006, 2009). However, advances of comparative genomics along with discoveries of cell biology precipitated a major shift in our understanding of the origin of eukaryotes by showing that eukaryotes are Archaeo–Bacterial chimeras. First, because it has been shown that all extant eukaryotes possess mitochondria or their degraded relatives, hydrogenosomes and mitosomes, by inference the Last Common Eukaryotic Ancestor (LECA), already harbored the α-proteobacterial endosymbiont that gave rise to the mitochondria (Embley and Martin 2006; van der Giezen 2009; Shiflett and Johnson 2010). Second, comparative genomic analysis indicates that eukaryotes possess two distinct sets of genes, one of which shows apparent phylogenetic affinity with homologs from Archaea, whereas the other one is more closely related to bacterial homologs (obviously, not all eukaryotic genes belong to these two classes because many are of uncertain origin, and many more appear to be unique to eukaryotes). The eukaryotic genes of apparent archaeal descent encode, primarily, proteins involved in information processing (translation, transcription, replication, repair), and the genes of inferred bacterial origin encode mostly proteins with “operational” functions (metabolic enzymes, components of membranes and other cellular structures, etc.) (Esser et al. 2004; Rivera and Lake 2004; Pisani et al. 2007; Thiergart et al. 2012). Some of the informational and operational systems of eukaryotes show qualitative separation between “archaeal” and “bacterial” genes: thus, the key proteins involved in DNA replication in Archaea and eukaryotes are not homologous to the functionally analogous proteins of Bacteria (Leipe et al. 1999), and conversely, some of the principal enzymes of membrane biogenesis are homologous in eukaryotes and Bacteria but not in Archaea (Pereto et al. 2004).
The parsimonious scenario of eukaryogenesis that takes into account the ancestral presence of mitochondria in eukaryotes and the hybrid composition of the eukaryotic gene complement seems to be that the first eukaryote emerged through the invasion of an archaeon by an α-proteobacterium (Martin and Muller 1998; Rivera and Lake 2004; Martin and Koonin 2006; Martin et al. 2007). Under this scenario, a plausible chain of events has been proposed leading from the endosymbiosis to the evolution of eukaryotic innovations such as the endomembrane system including the nucleus and the cytoskeleton (Koonin 2006; Martin and Koonin 2006). Furthermore, argument has been presented that the energy requirements for the functioning of a eukaryotic cell that is orders of magnitude bigger than typical prokaryotic cells cannot be met by means other than utilization of multiple “power stations” such as the mitochondria (Lane and Martin 2010; Lane 2011).
Despite the plausibility of the symbiogenetic scenario, alternatives are still seriously considered (Embley and Martin 2006; Poole and Penny 2007a,b). Conceptually, these alternatives converge to the archezoan hypothesis under which the host of the α-proteobacterial endosymbiont was not an archaeon but a primitive, amitochondrial proto-eukaryote that already possessed the hallmarks of the eukaryotic cellular architecture (including the nucleus) and was capable of phagocytosis, a function that is sometimes considered essential for the acquisition of endosymbionts (Cavalier-Smith 1991, 2009; Kurland et al. 2006; Poole and Penny 2007a,b). The two scenarios of eukaryogenesis substantially differ with respect to the level of complexity that is attributed to the host of the mitochondrial endosymbiont. Under the symbiotic hypothesis, the host was a “garden-variety” archaeon, and the dramatic complexification of the cellular organization was triggered by the symbiosis. In contrast, the archezoan hypothesis posits that at least some major features of the eukaryotic organizational complexity, such as the endomembrane system and the cytoskeleton, evolved before and independent of the symbiosis, and were already in place in the organism that hosted the mitochondrion. Under the archezoan scenario, the presence of genes of apparent archaeal origin in the ancestral Eukaryotic gene set is explained by postulating that the proto-eukaryotic lineage was a sister group of Archaea or possibly even a derivative of one of the archaeal lineages. The archezoan hypothesis was severely damaged by the demonstration that all unicellular eukaryotes previously thought to be primitively amitochondrial actually possess derived organelles of α-proteobacterial descent. Nevertheless, the archezoan scenario has not been abandoned but, rather, was amended with the additional proposition that the ancestral Archezoan lineage had gone extinct (Poole and Penny 2007a,b). A purported key argument in support of the archezoan hypothesis is that the complex organization of the eukaryotic cell, in particular, the cytoskeleton, which provides for the capability of phagocytosis, would enable engulfment of potential endosymbionts (Cavalier-Smith 2009).
Another aspect of eukaryogenesis that received considerable attention ever since the discovery of the Archaea is the exact nature of the evolutionary relationship between Archaea and eukaryotes. The “standard model” of Woese and colleagues clearly defines these two domains of cellular life as sister groups (Woese et al. 1990; Pace 1997, 2006). However, very shortly after the publication of the three-domain tree of life, an alternative relationship has been inferred from phylogenetic trees of the same 16S rRNA that were constructed using a different method. The topology of these alternative trees underlies the eocyte hypothesis according to which eukaryotes evolved from within the Archaea and are a sister group to the “eocytes,” that is, the branch that is currently known as Crenarchaeota (Lake et al. 1984; Lake 1988, 1998; Rivera and Lake 1992). Support for the eocyte hypothesis has been subsequently claimed from comparative analysis of ribosomal protein sequences (Vishwanath et al. 2004) and from a novel phylogenomic approach (Rivera and Lake 2004). Most convincingly, a subsequent phylogenetic analysis of multiple conserved genes that used the technique that eliminates fast-evolving alignment columns and is supposed to minimize the effect of several common artifacts of phylogenetic analysis (long-branch attraction and others) supported the affinity of eukaryotes with Crenarchaeota (Cox et al. 2008). The investigators of this study suggested that the alternative topologies including the standard three-domain phylogeny resulted from such undetected artifacts.
However, the eocytes are by no means the only proposed candidate for the role of the archaeal ancestor of eukaryotes. The origin of the “archaeal” genes of eukaryotes from Euryarchaea and, specifically, from methanogens was proposed on the basis of biological considerations, within the framework of the hydrogen hypothesis (Martin and Muller 1998) and one of the so-called syntrophic hypotheses (Lopez-Garcia and Moreira 2006), which postulate different forms of metabolic cooperation between the archaeal and bacterial partners of the primary endosymbiosis. The origin of these genes from methanogens also is compatible with the results of some phylogenetic analyses (Moreira and Lopez-Garcia 1998; Horiike et al. 2004). Others have argued that the archaeal ancestor of eukaryotes lies outside the known diversity of Archaea, in accord with the standard model, on the basis of biological considerations (Vellai et al. 1998) or phylogenetic analysis results (Hedges et al. 2001; Tekaia and Yeramian 2005; Ciccarelli et al. 2006; Fukami-Kobayashi et al. 2007).
Most of the aforementioned phylogenetic studies either used a relatively small number of concatenated, highly conserved protein sequences (e.g., those of ribosomal proteins) for phylogenetic tree construction or built trees on the basis of gene repertoire comparisons (phyletic patterns), or else used other features, such as domain architectures of multidomain proteins, as phylogenetic characters. An extensive phylogenomic study that analyzed nearly 6000 gene sets from 185 genomes using a supertree approach has suggested phylogenetic affinity between eukaryotes and Thermoplasma albeit with limited statistical support (Pisani et al. 2007). Thermoplasma or a related, wall-less archaeon also has been proposed as the likely archaeal ancestor of eukaryotes on the basis of biochemical and cytological considerations (Searcy et al. 1978; Margulis and Stolz 1984; Hixon and Searcy 1993; Margulis 1996; Margulis et al. 2000).
An exhaustive phylogenetic analysis of individual eukaryotic genes of apparent archaeal origin suggested a deep (i.e., outside of the known archaeal diversity) origin for most of these genes but also identified a considerable number of genes with a crenarchaeal affinity and a small number of genes with a euryarchaeal affinity (Yutin et al. 2008). In this work, the possibility was introduced that these results did not necessarily result from artifacts of phylogenetic analysis but, rather, might reflect diverse sources of eukaryotic genes due to horizontal gene transfer (HGT) between different Archaea.
Such was the state of the art in the study of the archaeal roots of eukaryotes—far from certainty, even if leaning toward the crenarchaeal (eocyte) origin—when genome analysis of several poorly characterized Archaea suggested the existence of multiple new phyla and yielded unexpected discoveries that seem to call for a change of perspective on the origin of eukaryotes. In this article, we examine the scattered distribution of apparent ancestors of signature eukaryotic genes among Archaea and develop the “dispersed eukaryome” scenario for the origin of eukaryotes.
NEW ARCHAEAL PHYLA, THE TACK SUPERPHYLUM, AND THE COMPLEX ARCHAEAL ANCESTOR
The new outlook of archaeal diversity and evolution emerged in 2008, with the sequencing of the genome of Candidatus Korarchaeum cryptofilum, the first (and so far the only) representative of a poorly characterized group of uncultured Archaea known as Korarchaeota (Elkins et al. 2008) and the almost simultaneous demonstration that mesophilic marine Archaea, previously included in the Crenarchaeota, represented a distinct lineage that was roughly equidistant from Crenarchaeota and Euryarchaeota, and was denoted Thaumarchaeota (Brochier-Armanet et al. 2008). Subsequent studies have shown that Thaumarchaeota are a widespread microbial group of major geochemical importance that includes the key ammonia oxidizers in marine and soil habitats (Pester et al. 2011; Brochier-Armanet et al. 2012; Lloyd et al. 2013).
Phylogenomic analysis enabled by the sequencing of the first representative genome of the Korarchaeota also suggested a new phylum that either is distantly related to Crenarchaeota or branched off before the Euryarchaeota–Crenarchaeota split (Elkins et al. 2008). Subsequent studies have supported these observations and revealed a substantial diversity of Korarchaeota in various marine and terrestrial habitats (Reigstad et al. 2010; Miller-Coleman et al. 2012). The more recent sequencing of the uncultivated archaeon Candidatus Caldiarchaeum subterraneum pointed to the existence of yet another archaeal phylum, dubbed Aigarchaeota (Nunoura et al. 2011). Additional archaeal phyla are likely to lurk within the uncultured microbial diversity, as suggested by 16S RNA phylogenies (Robertson et al. 2005; Teske and Sorensen 2008; Pace 2009; Brochier-Armanet et al. 2011; Rajendhran and Gunasekaran 2011) and the emerging results of single-cell genomics (Lloyd et al. 2013), but, even with the currently available genomes that provide for a more satisfactory phylogenomic analysis, the complex picture of archaeal evolution is starting to take shape.
Several phylogenomic analyses of multiple conserved genes seem to consistently reveal the existence of a deeply rooted archaeal “TACK” superphylum that encompasses Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota (Fig. 1) (Guy and Ettema 2011; Williams et al. 2012; Wolf et al. 2012; Yutin et al. 2012; Martijn and Ettema 2013). The validity of the TACK superphylum is further supported by the reconstruction of the evolution of the archaeal gene repertoire, which assigned a massive gene gain to the base of the TACK (Fig. 1) (Wolf et al. 2012). This reconstruction, in an extension of previous analyses (Makarova et al. 2007; Csurös and Miklos 2009), reveals a striking trend in the evolution of Archaea that might reflect a general tendency of genome evolution, namely, the preponderance of genome reduction that is punctuated with episodes of explosive genome expansion (Wolf and Koonin 2013). According to these reconstructions, each of the major archaeal lineages underwent some degree of genome reduction, and the gene complement of the last archaeal common ancestor (LACA) was at least as complex as that of most of the extant Archaea. The reductive evolution associated with genome streamlining appears to be a general feature of successful lineages that reach large effective population sizes and are subject to strong selective pressure. However, evolution of the Archaea might have involved additional factors leading to genome reduction such as the adaptation to the thermophilic lifestyle (Forterre 1995; Boussau et al. 2008).
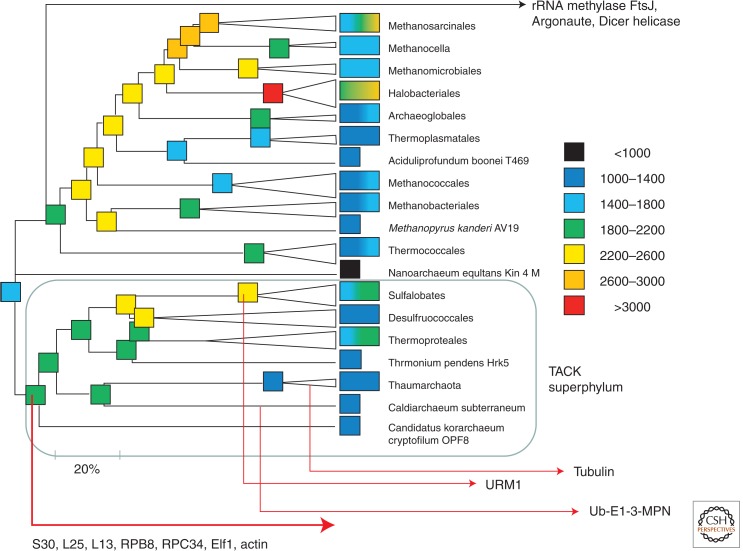
Figure 1.
A reconstruction of the evolution of archaeal gene complements and the inferred origins of the eukaryome components. The boxes show the approximate maximum-likelihood estimates for the number of genes in extant and reconstructed ancestral genomes according to the color code shown on the right. The reconstruction was performed using the COUNT software (Csurös 2010). The tree topology is from the phylogenetic tree of concatenated ribosomal proteins (Wolf et al. 2012; Yutin et al. 2012). The inferred origins of some key eukaryotic genes and functional systems are indicated by arrows (see text); S30, L25, and L13 are ribosomal proteins. (From Wolf et al. 2012; modified, with permission, from the author.)
The delineation of new archaeal phyla and the putative TACK superphylum stimulated further phylogenomic effort aimed at the identification of the archaeal ancestor of eukaryotes. One such study has suggested an affinity of eukaryotes with Thaumarchaeota (Kelly et al. 2011). However, the two most detailed to date, state-of-the-art phylogenetic analyses of rRNA and universal protein-coding genes provided significant support for the monophyly of eukaryotes with the TACK superphylum but failed to recover a specific relationship between eukaryotes and any lineage within the TACK (Williams et al. 2012; Lasek-Nesselquist and Gogarten 2013). Thus, the current last word on the evolutionary relationship between the cores of the information-processing systems of the Archaea and eukaryotes seems to be that the ancestors of eukaryotes are not a sister group to all Archaea but are more likely to comprise a deep archaeal branch sharing a common ancestry with the TACK superphylum (Fig. 1). This assignment is compatible with the identification of several ancestral genes that eukaryotes exclusively shared with the TACK superphylum, in contrast to a smaller number of such derived shared characters in Euryarchaeota (Fig. 1). Notably, the shared derived characters of eukaryotes and the TACK superphylum include components of the information system core such as three ribosomal proteins (Yutin et al. 2012), the RNA polymerase subunits RPB8 (Koonin et al. 2007) and RPC34 (Blombach et al. 2009), and the transcription factor Elf1 (Daniels et al. 2009).
THE DISPERSED ARCHAEAL EUKARYOME
Perhaps, the most remarkable discoveries coming from the recent comparative analysis of archaeal and eukaryotic genomes go beyond the identification of the specific eukaryotic ancestry within Archaea. The unexpected outcome of these comparisons is that, apart from the core of the information systems, the apparent archaeal ancestors of essential eukaryotic genes and entire functional systems—the set of genes that we here denote the “eukaryome”—are often found in different groups of Archaea.
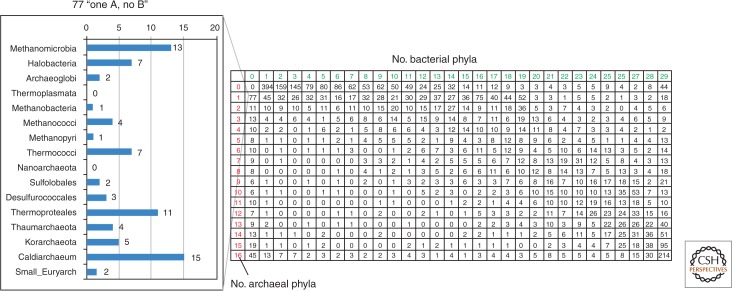
Figure 2 shows the breakdown of the (putative) prokaryotic homologs of ancestral eukaryotic genes by the number of major groups of Archaea and Bacteria in which the respective genes are represented. Focusing on the archaeal homologs of eukaryotic genes, an enrichment in genes that are represented in only one archaeal lineage (and to a lesser extent, in two to four lineages) is notable. A further classification of the 77 putative ancestral genes that were identified in a single group of Archaea shows a wide spread across the entire archaeal domain (Fig. 2). Of special interest seems to be the enrichment of lineage-specific archaeal ancestral genes in Ca. Caldiarchaeum subterraneum, the only known representative of Aigarchaeota. Below we discuss some of the most striking, biologically relevant cases of the dispersal of eukaryome components among Archaea.
Figure 2.
The patterns of the evolutionary relationships between Archaea and eukaryotes. A total of 8440 eukaryotic protein families derived from the KOD (cluster of eukaryotic orthologous domains) data set (Yutin et al. 2008) were used for a PSI-BLAST search (Altschul et al. 1997) against 113 archaeal and 1285 bacterial genomes. Then, 5365 of the 8440 families that had prokaryotic hits with an _e_-value better than 0.01 were subjected to the detailed phyletic analysis. Each number in a cell represents the number of eukaryotic protein families that had putative homologs (hits) in the given number of bacterial or archaeal taxonomic groups. Prokaryotic genomes were grouped into taxonomic groups as follows (numbers in parentheses represent the numbers of the genomes in the respective group). Bacteria: Alpha(proteobacteria) (143); Gamma(proteobacteria) (292); Beta(proteobacteria) (98); Delta(proteobacteria) (41); Nitrospirae (2); Deferribacteres (4); Epsilon (36); Acidobacteria (5); Elusimicrobia (2); Spirochaetes (28); Planctomycetes (5); Chlamydiae_Verrucomicrobia (22); Bacteroidetes_Chlorobi (64); Gemmatimonadetes (1); Fibrobacteres (1); Deinococci (13); Actinobacteria (132); Chloroflexi (16); Cyanobacteria (40); Fusobacteria (5); Mollicutes (36); Firmicutes (272); Synergistetes (2); Coprothermobact (1); Dictyoglomi (2); Thermotogae (11); Aquificae (9); unclassified_Cloacamonas (1); and Chrysiogenetes (1). Archaea: Methanomicrobia (16); Halobacteria (14); Archaeoglobi (4); Thermoplasmata (4); Methanobacteria (8); Methanococci (14); Methanopyri (1); Thermococci (9); Nanoarchaeota (1); Sulfolobales (13); Desulfurococcales (10); Thermoproteales (10); Thaumarchaeota (2); Korarchaeota (1); Caldiarchaeum (1); and “small Euryarchaeota” (5). “Small Euryarchaeota” consist of Candidatus Micrarchaeum acidiphilum ARMAN_2, Candidatus Parvarchaeum acidiphilum ARMAN_4, Candidatus Parvarchaeum acidiphilus ARMAN_5, Candidatus Nanosalinarum sp. J07AB56, and Candidatus Nanosalina sp. J07AB43.
The Ubiquitin Signaling System
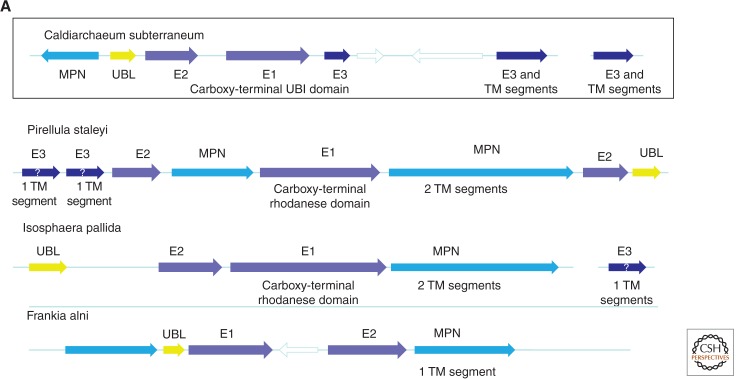
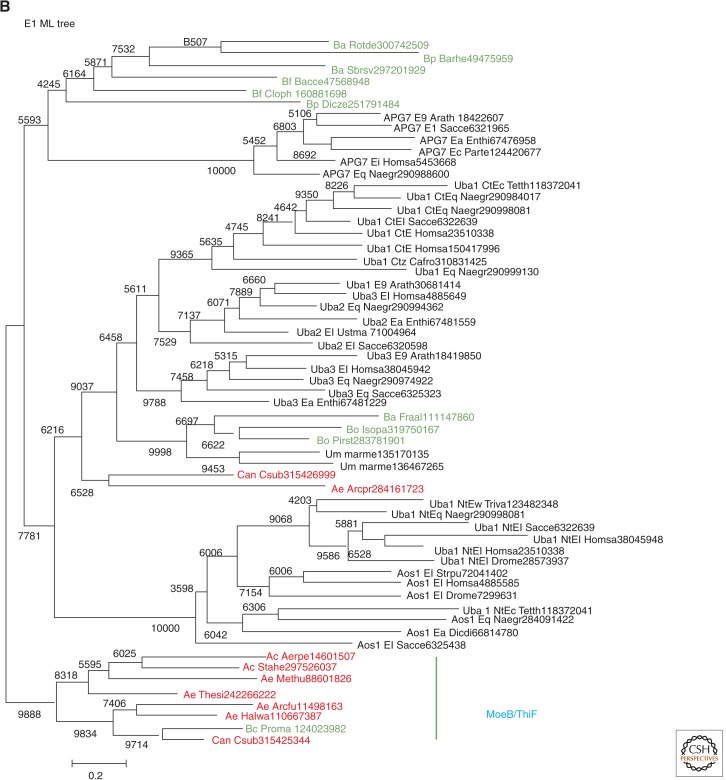
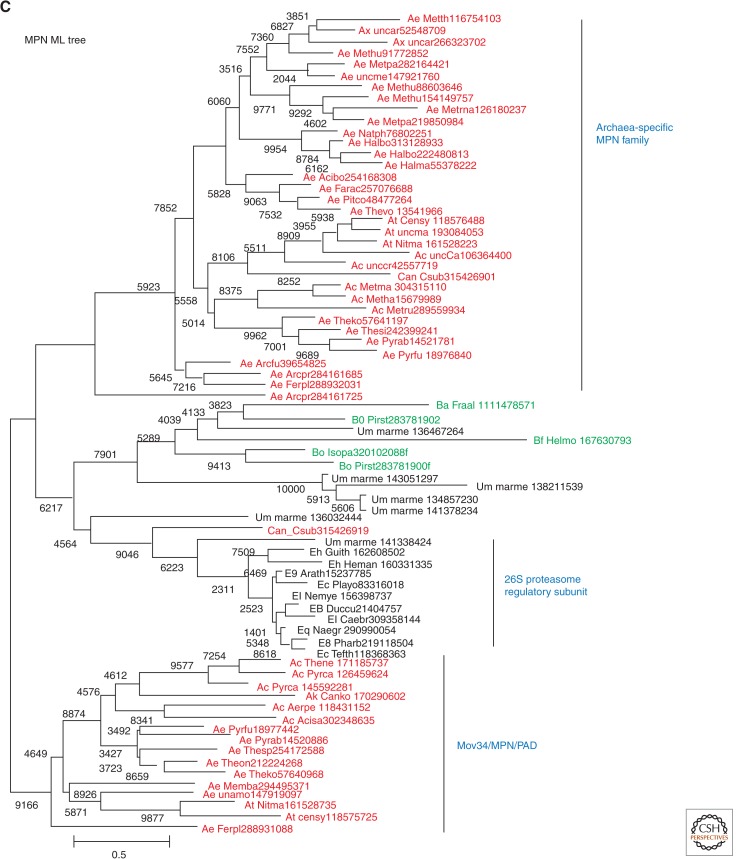
The ubiquitin (Ub) system is the extremely complex machinery that regulates protein degradation, topogenesis, and function in all eukaryotes through modification of proteins by conjugation with various forms of (poly)Ub and its paralogs (Burroughs et al. 2012a,b; van der Veen and Ploegh 2012). For many years, the Ub system was viewed as a eukaryotic innovation that seemed to have evolved from prokaryotic enzymes involved in coenzyme biosynthesis (Hochstrasser 2000). More recently, it has been shown that, with the participation of a homolog of the E1 subunit of eukaryotic Ub ligases, some of the archaeal homologs of Ub are conjugated with proteins and target them for degradation (Humbard et al. 2010; Miranda et al. 2011; Maupin-Furlow 2013). However, these are distant homologies, so the origin of the E2 and E3 subunits of the Ub ligases remained elusive. Analysis of the genome of Ca. Caldiarchaeum subterraneum dramatically transformed the entire story of the origin of the Ub system by revealing a predicted operon encoding a Ub-like protein and homologs of all three Ub ligase subunits, along with a key deubiquitinating enzyme (Nunoura et al. 2011); operons with a similar organization of Ub-related genes are detectable also in several Bacteria, suggestive of horizontal dissemination of these operons among prokaryotes (Fig. 3A). Unlike the homologs of Ub system components detected previously in other Archaea, the homologs from Ca. Caldiarchaeum subterraneum show high sequence similarity to eukaryotic counterparts, and phylogenetic trees for the E1 subunit of Ub ligase (Fig. 3B) and the deubiquitinating enzyme MPN (Fig. 3C) place Ca. Caldiarchaeum subterraneum in the midst of eukaryotes. The possibility of acquisition of the Ub system by Ca. Caldiarchaeum subterraneum via HGT from a eukaryotic source can be effectively discarded given the operonic organization of the archaeal genes. Thus, these findings indicate that, to date, Ca. Caldiarchaeum subterraneum encodes the best candidate for the ancestor of the eukaryotic Ub system.
Figure 3.
The apparent archaeal ancestor of the ubiquitin system. The operon organization of the Ub system components in Ca. Caldiarchaeum subterraneum and similar operon structures in Bacteria. (A) A maximum-likelihood tree for the E1 component of the Ub ligase. (B) A maximum-likelihood tree for the MPN deubiquitinating enzyme. The sequences for the tree construction were retrieved from GenBank nr and env_nr databases (NCBI, NIH) and aligned using the MUSCLE program (Edgar 2004). Positions that included gaps in more than one-third of the sequences and positions with low information content were removed before tree computation (Yutin et al. 2008), which left 133 unambitious positions in the E1 alignment and 103 positions in the MPN alignment. Maximum-likelihood trees were constructed using the TreeFinder program (WAG matrix,G[Optimum]:4, 1000 replicates, Search Depth 2) (Jobb et al. 2004). The bootstrap values (shown for selected branches) represent expected-likelihood weights from 1000 local rearrangements. Branches with bootstrap support <0.5 were collapsed. For each sequence, the species name abbreviation and the gene identification numbers are indicated; (env) marine metagenome. Species: Acibo, Aciduliprofundum boonei T469; Acisa, Acidilobus saccharovorans 345-15; Aerpe, Aeropyrum pernix K1; Arath, Arabidopsis thaliana; Arcfu, Archaeoglobus fulgidus DSM 4304; Arcpr, Archaeoglobus profundus DSM 5631; Bacce, Bacillus cereus G9241; Barhe, Bartonella henselae str. Houston-1; Caebr, Caenorhabditis briggsae AF16; Cafro, Methanocella arvoryzae MRE50; Can_Csub, Candidatus Caldiarchaeum subterraneum; CanKo, Candidatus Korarchaeum cryptofilum OPF8; Censy, Cenarchaeum symbiosum A; Cloph, Clostridium phytofermentans ISDg; Dicdi, Dictyostelium discoideum AX4; Dicze, Dickeya zeae Ech1591; Drome, Drosophila melanogaster; Enthi, Entamoeba histolytica HM-1:IMSS; Ferac, Ferroplasma acidarmanus fer1; Ferpl, Ferroglobus placidus DSM 10642; Fraal, Frankia alni ACN14a; Guith, Guillardia theta; Halbo, Halogeometricum borinquense DSM 11551; Halla, Halorubrum lacusprofundi ATCC 49239; Halma, Haloarcula marismortui ATCC 43049; Halwa, Haloquadratum walsbyi DSM 16790; Helmo, Heliobacterium modesticaldum Ice1; Heman, Hemiselmis andersenii; Homsa, Homo sapiens; Isopa, Isosphaera pallida ATCC 43644; Metar, Methanocella arvoryzae MRE50; Metbo, Methanoregula boonei 6A8; Metbu, Methanococcoides burtonii DSM 6242; Methu, Methanospirillum hungatei JF-1; Metma, Methanohalophilus mahii DSM 5219; Metpa, Methanosphaerula palustris E1-9c; Metru, Methanobrevibacter ruminantium M1; Metth, Methanosaeta thermophila PT; Naegr, Naegleria gruberi strain NEG-M; Natph, Natronomonas pharaonis DSM 2160; Nemve, Nematostella vectensis; Nitma, Nitrosopumilus maritimus SCM1; Parte, Paramecium tetraurelia; Phatr, Phaeodactylum tricornutum CCAP 1055/1; Picto, Picrophilus torridus DSM 9790; Pirst, Pirellula staleyi DSM 6068; Playo, Plasmodium yoelii yoelii 17XNL; Proma, Prochlorococcus marinus str. MIT 9303; Pyrab, Pyrococcus abyssi GE5; Pyrar, Pyrobaculum arsenaticum DSM 13514; Pyrca, Pyrobaculum calidifontis JCM 11548; Pyrfu, Pyrococcus furiosus DSM 3638; Rotde, Rothia dentocariosa M567; Sacce, Saccharomyces cerevisiae S288c; Stahe, Staphylothermus hellenicus DSM 12710; Strpu, Strongylocentrotus purpuratus; Strsv, Streptomyces sviceus ATCC 29083; Tetth, Tetrahymena thermophila; Theko, Thermococcus kodakarensis KOD1; Thene, Pyrobaculum neutrophilum V24Sta; Theon, Thermococcus onnurineus NA1; Thesi, Thermococcus sibiricus MM 739; Thesp, Thermococcus sp. AM4; Thevo, Thermoplasma volcanium GSS1; Triva, Trichomonas vaginalis G3; uncar, uncultured archaeon GZfos18C8; uncCa, uncultured Candidatus Nitrosocaldus sp.; unccr, uncultured crenarchaeote; uncma, uncultured marine crenarchaeote KM3-86-C1; Ustma, Ustilago maydis 521. Taxa: Ac, Crenarchaeota; Ae, Euryarchaeota; Ak, Korarchaeota; At, Taumarchaeota; Ax, environmental samples; Ba, Actinobacteria; Bc, Cyanobacteria; Bf, Firmicutes; Bo, Planctomycetes; Bp, Proteobacteria; E8, Stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalia.
Notably, apparent archaeal orthologs of the distant Ub homolog Urm1, which is conserved in all eukaryotes and performs a dual function as a sulfur carrier in tRNA thiolation and in protein modification (Van der Veen et al. 2011; Wang et al. 2011), are detectable only among the members of the order Sulfolobales of the Crenarchaeota (Fig. 1) (Makarova and Koonin 2010). Thus, within the broadly defined Ub system, two different archaeal ancestors of essential eukaryotic functional modules are detectable.
The Cytoskeleton
All eukaryotic cells possess an advanced, extremely elaborate cytoskeleton that consists of two major types of structural elements, actin-based filaments, and tubulin-based microtubules (Lowe and Amos 2009; Aylett et al. 2011). The cytoskeleton looms large in the discussion of the origin of eukaryotes, primarily because actin filaments play the central role in phagocytosis, the process that is thought to be critical for the engulfment of the proto-mitochondrial endosymbiont by its elusive future host (Cavalier-Smith 2009). For years, Bacteria and Archaea have been thought to encode only distant homologs of actin and tubulin, the proteins of the MreB/FtsA and FtsZ families, respectively, that perform essential functions in the septation of bacterial and some archaeal cells (Cabeen and Jacobs-Wagner 2010; Celler et al. 2013). However, analysis of the expanding archaeal genome collection overturned this perspective. Proteins with high sequence similarity and phylogenetic affinity to eukaryotic actins were discovered in the order of Thermoproteales of the Crenarchaeota, Korarchaeum and Can. C. subterraneum, suggesting that actin was already present in the last common ancestor of the TACK superphylum of the Archaea (Yutin et al. 2009; Makarova et al. 2010). Following this comparative-genomic finding, it has been shown that these archaeal proteins, dubbed crenactins, indeed form helical filaments resembling typical eukaryotic actin filaments (Bernander et al. 2011; Ettema et al. 2011).
Highly conserved orthologs of tubulins so far have been discovered only in the genomes of two Thaumarchaeota. In this case, there is no operon organization to rule out HGT from eukaryotes to Archaea, but the results of phylogenetic analysis appear to be best compatible with these “artubulins” being the ancestors of the eukaryotic tubulins (Yutin and Koonin 2012). The functions of the artubulins remain to be characterized.
Thus, contrary to recent beliefs, both major forms of the eukaryotic cytoskeleton apparently constitute part of the archaeal heritage although the apparent ancestral forms were detected in different extant Archaea.
Cell Division and Membrane-Remodeling Systems
Obviously, cell division is an essential process in all cellular life forms. Nevertheless, there are fundamental mechanistic differences between the division mechanisms in Bacteria and at least some Archaea compared with eukaryotes. In Bacteria, cell division is coupled to DNA replication whereby the replicating DNA is immediately pumped into the daughter cells, and the division is completed through the formation of the septum, which starts with the Z-ring that consists of the FtsZ protein, a distant homolog of eukaryotic tubulins (Adams and Errington 2009; Margolin 2009). The FtsZ-based division system is present in most of the Euryarchaeota, Thaumarchaeota, and Korarchaeota, with the implication that these archaeal cells divide similarly to Bacteria (Makarova et al. 2010). Recently, however, a distinct division system homologous to the eukaryotic ESCRT-III membrane-remodeling complex has been experimentally characterized in the crenarchaeon Sulfolobus (Lindas et al. 2008; Samson et al. 2008; Ettema and Bernander 2009; Samson and Bell 2009) and subsequently identified with comparative-genomic methods in all Sulfolobales and Desulfurococcales (two of the three crenarchaeal orders) as well as some Euryarchaeota (Makarova et al. 2010). A follow-up study has shown that the ESCRT-III-like complex is the primary cell division system in the thaumarchaeon Nitrosopumilus maritimus (Pelve et al. 2011). The broad, even if patchy, distribution of the FtsZ-based and ESCRT-III-like division systems in Archaea implies that both were present in the LACA, with subsequent differential losses in multiple lineages. However, the Crenarchaeota of the order Thermoproteales lack both of these division systems, implying the existence of a third one, conceivably centered around the crenactin cytoskeleton (Makarova et al. 2010; Bernander et al. 2011). Given that the origin of crenactin can be mapped to the base of the TACK superphylum (Fig. 1; see above), one comes to the striking conclusion that the common ancestor of the TACK possessed all three cell division systems that are scattered among the extant Archaea.
The RNA Interference System (RNAi)
The RNA interference system (RNAi) is a major eukaryotic hallmark system that is involved in both antivirus defense and regulation of gene expression and apparently was already in place in the LECA (Cerutti and Casas-Mollano 2006; Shabalina and Koonin 2008; Carthew and Sontheimer 2009). The multiple RNAi mechanisms are centered around proteins of two families, the Dicers that are responsible for the processing of small interfering RNAs and microRNAs, and the Argonautes, some of which are nucleases directly attacking the RNA target (slicers), whereas others bind microRNA and guide it to the target, cleaving the latter (Tolia and Joshua-Tor 2007; Hock and Meister 2008; Jaskiewicz and Filipowicz 2008). The Argonautes have numerous homologs in Bacteria and Archaea, primarily Euryarchaeota, and phylogenetic analysis clearly points to the euryarchaeal origin of the eukaryotic Argonaute family (Makarova et al. 2009). The Dicers have no direct counterparts in Bacteria or Archaea. These proteins comprise a fusion of a helicase domain and two RNase III domains that is unique to eukaryotes. The RNase III domain is of obvious bacterial provenance, whereas the helicase domain of Dicer has been traced to a euryarchaeal ancestor, the Hef helicase that is involved in DNA replication (Shabalina and Koonin 2008). Thus, the RNAi system, a hallmark eukaryotic machinery that has no direct counterpart in Archaea or Bacteria, has mixed archaeal and bacterial origins, with the archaeal components apparently derived from Euryarchaeota.
THE ARCHAEAL ANCESTOR OF EUKARYOTES: A COMPLEX, ANCIENT LINEAGE IN THE TACK SUPERPHYLUM?
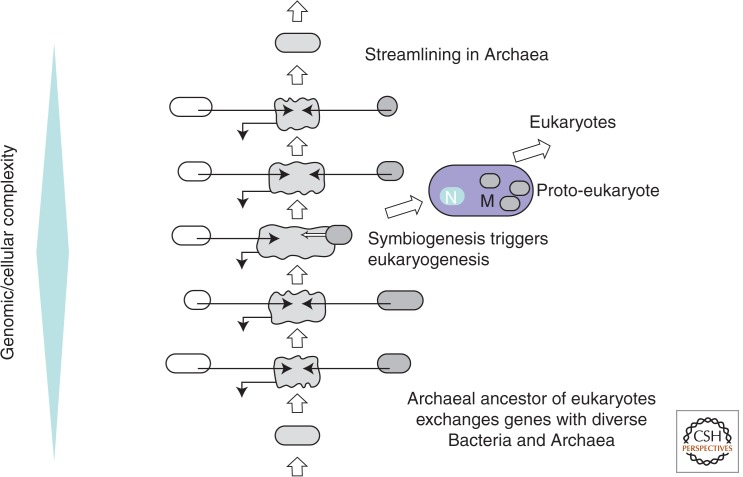
Combined with the results of phylogenomic analysis, the discovery of the dispersed archaeal eukaryome implies a complex archaeal ancestor of eukaryotes, which encoded the ancestors of various eukaryotic functional systems that are currently found in different lineages of extant Archaea (Fig. 1). Specifically, the hypothetical eukaryotic ancestor probably possessed a fairly advanced cytoskeleton that encompassed both actin filaments and tubulin microtubules and could provide for a primitive phagocytic capability (Yutin et al. 2009; Martijn and Ettema 2013). It stands to reason that the eukaryotic ancestor was a wall-less, mesophilic organism that coexisted with diverse Bacteria, so even a limited capacity for phagocytosis would greatly facilitate the capture of prospective endosymbionts. Extant mesophilic Archaea such as Methanosarcina or Halobacteria clearly have acquired numerous genes via HGT (Koonin et al. 2001; Deppenmeier et al. 2002; Nelson-Sathi et al. 2012; Wolf et al. 2012). When it comes to the ancestor of eukaryotes, this gene-gaining capacity might have been further enhanced by the primitive phagocytosis, conceivably through transient engulfment of other Archaea and Bacteria (Fig. 4). Such a lifestyle could result both in the accumulation of the eukaryome components that are currently found in diverse Archaea (see above) and in the acquisition of genes from diverse bacterial sources, possibly explaining the weakness of the α-proteobacterial signal among the eukaryotic genes of apparent bacterial descent (Esser et al. 2007).
Figure 4.
Evolutionary scenario for the origin of the proto-eukaryote from a complex archaeal ancestor. M, Mitochondria; N, nucleus.
As discussed above, phylogenomic analysis suggests that the ancestor of eukaryotes could have been a deep branch within the TACK superphylum that radiated from the other phyla shortly after the emergence of the TACK (Williams et al. 2012; Lasek-Nesselquist and Gogarten 2013). Thus, the hypothetical lineage of the eukaryotic ancestor would have the same evolutionary status as the currently known phyla, Crenarchaeota, Thaumarchaeota, Korarchaeota, and Aigarchaeota. Evolutionary reconstructions suggest that archaeal evolution was dominated by genome reduction on a path from complex ancestors to streamlined extant genomes (Wolf et al. 2012; Wolf and Koonin 2013). The ancestor of eukaryotes could have been an exceptionally complex ancient archaeon. Hence, a notable possibility: one of the extant phyla within the TACK, for example, Aigarchaeota or more likely another group lurking among the still uncultured Archaea, could be the ancestral lineage of the eukaryotes. We have difficulty ascertaining this relationship because of the contrasting trends of genome reduction in Archaea and complexification in eukaryotes, the likely compressed cladogenesis (Rokas and Carroll 2006; Puigbo et al. 2009) at the base of the TACK superphylum and acceleration of evolution during eukaryogenesis. It is interesting to point out the discovery of giant Thaumarchaeota associated with bacterial ectoparasites (Muller et al. 2010). Such forms might resemble the ancestor of the eukaryotes, and genome sequences of these and other “exotic” Archaea are eagerly anticipated.
The proposition that the archaeal ancestor of eukaryotes (the host of the mitochondrial endosymbiont) was a complex organism that possessed some signature features of eukaryotic cells (such as cytoskeleton) begs the question: Was the eukaryotic ancestor an archaeon or actually an Archezoan? This question potentially could be considered semantic, but we are inclined to take it as a substantial one given the qualitative differences between the cellular organizations in eukaryotes versus Archaea and Bacteria (prokaryotes). Given the apparent unsustainability of the eukaryotic genome expansion and cell organization in the absence of multiple energy-converting organelles (such as mitochondria) (Lane and Martin 2010) and the lack of a plausible function for the nucleus in the context of the prokaryotic cell (Koonin 2006; Lopez-Garcia and Moreira 2006; Martin and Koonin 2006), we find it overwhelmingly more likely that the host of the mitochondrial endosymbiont was a bona fide archaeon.
CONCLUSIONS
Deep phylogenies are notoriously artifact-prone, and deriving definitive topology of the major branches of organisms is extremely hard; the results of independent analysis are often in conflict, leading to the notion of a “phylogenomic impasse” with regard to the origin of eukaryotes (Gribaldo et al. 2010). However, these legitimate concerns notwithstanding, there seems to be some emerging clarity with respect to the nature of the archaeal ancestor of eukaryotes. The two key observations are the apparent deep phylogenetic affinity of the core of the eukaryotic information-processing machinery with the archaeal TACK superphylum and the dispersal of the eukaryome components across Archaea. The combination of these findings implies a highly complex archaeal ancestor of eukaryotes that possessed certain signature eukaryotic features, such as the cytoskeleton and the Ub system, while remaining a typical archaeon in terms of overall cellular organization and genome structure. The presence of a well-developed cytoskeleton could facilitate the engulfment of Bacteria, creating the conditions for the evolution of endosymbiosis. The complexity of the archaeal ancestor was apparently fixed in the emerging eukaryotes thanks to endosymbiosis. In contrast, the proto-eukaryotic features were differentially lost in archaeal lineages in the course of reductive evolution, resulting in the currently observed dispersed eukaryome. Given the dispersed eukaryome, extensive sampling of the archaeal diversity by genome sequencing is essential to advance our understanding of eukaryogenesis.
ACKNOWLEDGMENTS
We thank Thijs Ettema, Kira Makarova, and Bill Martin for useful discussions. Our research is supported by intramural funds of the U.S. Department of Health and Human Services (to the National Library of Medicine, NIH).
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
- Adams DW, Errington J 2009. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Lowe J, Amos LA 2011. New insights into the mechanisms of cytomotive actin and tubulin filaments. Int Rev Cell Mol Biol 292: 1–71 [DOI] [PubMed] [Google Scholar]
- Bernander R, Lind AE, Ettema TJ 2011. An archaeal origin for the actin cytoskeleton: Implications for eukaryogenesis. Commun Integr Biol 4: 664–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der Oost J 2009. Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol Direct 4: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456: 942–945 [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P 2008. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6: 245–252 [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P, Gribaldo S 2011. Phylogeny and evolution of the Archaea: One hundred genomes later. Curr Opin Microbiol 14: 274–281 [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Gribaldo S, Forterre P 2012. Spotlight on the Thaumarchaeota. ISME J 6: 227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L 2012a. The natural history of ubiquitin and ubiquitin-related domains. Front Biosci 17: 1433–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L 2012b. Structure and evolution of ubiquitin and ubiquitin-related domains. Methods Mol Biol 832: 15–63 [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C 2010. The bacterial cytoskeleton. Annu Rev Genet 44: 365–392 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1991. Archamoebae: The ancestral eukaryotes? Biosystems 25: 25–38 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2009. Predation and eukaryote cell origins: A coevolutionary perspective. Int J Biochem Cell Biol 41: 307–322 [DOI] [PubMed] [Google Scholar]
- Celler K, Koning RI, Koster AJ, van Wezel GP 2013. Multidimensional view of the bacterial cytoskeleton. J Bacteriol 195: 1627–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA 2006. On the origin and functions of RNA-mediated silencing: From protists to man. Curr Genet 50: 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM 2008. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci 105: 20356–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csurös M 2010. Count: Evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26: 1910–1912 [DOI] [PubMed] [Google Scholar]
- Csurös M, Miklos I 2009. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol 26: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels JP, Kelly S, Wickstead B, Gull K 2009. Identification of a crenarchaeal orthologue of Elf1: Implications for chromatin and transcription in Archaea. Biol Direct 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Baumer S, Jacobi C, et al. 2002. The genome of Methanosarcina mazei: Evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol 4: 453–461 [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, et al. 2008. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci 105: 8102–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W 2006. Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, Gelius-Dietrich G, Henze K, Kretschmann, Richly E, Leister D, et al. 2004. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21: 1643–1660 [DOI] [PubMed] [Google Scholar]
- Esser C, Martin W, Dagan T 2007. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett 3: 180–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJ, Bernander R 2009. Cell division and the ESCRT complex: A surprise from the Archaea. Commun Integr Biol 2: 86–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJ, Lindas AC, Bernander R 2011. An actin-based cytoskeleton in Archaea. Mol Microbiol 80: 1052–1061 [DOI] [PubMed] [Google Scholar]
- Forterre P 1995. Thermoreduction, a hypothesis for the origin of prokaryotes. CR Acad Sci III 318: 415–422 [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Minezaki Y, Tateno Y, Nishikawa K 2007. A tree of life based on protein domain organizations. Mol Biol Evol 24: 1181–1189 [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C 2010. The origin of eukaryotes and their relationship with the Archaea: Are we at a phylogenomic impasse? Nat Rev Microbiol 8: 743–752 [DOI] [PubMed] [Google Scholar]
- Guy L, Ettema TJ 2011. The archaeal “TACK” superphylum and the origin of eukaryotes. Trends Microbiol 19: 580–587 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Chen H, Kumar S, Wang DY, Thompson AS, Watanabe H 2001. A genomic timescale for the origin of eukaryotes. BMC Evol Biol 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hixon WG, Searcy DG 1993. Cytoskeleton in the archaebacterium Thermoplasma acidophilum? Viscosity increase in soluble extracts. Biosystems 29: 151–160 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol 2: E15–E17 [DOI] [PubMed] [Google Scholar]
- Hock J, Meister G 2008. The Argonaute protein family. Genome Biol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiike T, Hamada K, Miyata D, Shinozawa T 2004. The origin of eukaryotes is suggested as the symbiosis of pyrococcus into γ-proteobacteria by phylogenetic tree based on gene content. J Mol Evol 59: 606–619 [DOI] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA 2010. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W 2008. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol 320: 77–97 [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K 2004. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kelly S, Wickstead B, Gull K 2011. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc Biol Sci 278: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2006. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early versus introns-late debate? Biol Direct 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L 2001. Horizontal gene transfer in prokaryotes—Quantification and classification. Annu Rev Microbiol 55: 709–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Elkins JG 2007. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and “Korarchaeota.” Biol Direct 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny 2006. Genomics and the irreducible nature of eukaryote cells. Science 312: 1011–1104 [DOI] [PubMed] [Google Scholar]
- Lake JA 1988. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature 331: 184–186 [DOI] [PubMed] [Google Scholar]
- Lake JA 1998. Optimally recovering rate variation information from genomes and sequences: Pattern filtering. Mol Biol Evol 15: 1224–1231 [DOI] [PubMed] [Google Scholar]
- Lake JA, Henderson E, Oakes M, Clark MW 1984. Eocytes: A new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci 81: 3786–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N 2011. Energetics and genetics across the prokaryote–eukaryote divide. Biol Direct 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W 2010. The energetics of genome complexity. Nature 467: 929–934 [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Gogarten JP 2013. The effects of model choice and mitigating bias on the ribosomal tree of life. Mol Phylogenet Evol 69: 17–38 [DOI] [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Koonin EV 1999. Did DNA replication evolve twice independently? Nucleic Acids Res 27: 3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R 2008. A unique cell division machinery in the Archaea. Proc Natl Acad Sci 105: 18942–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, et al. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496: 215–218 [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D 2006. Selective forces for the origin of the eukaryotic nucleus. Bioessays 28: 525–533 [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA 2009. Evolution of cytomotive filaments: The cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol 41: 323–329 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV 2010. Archaeal ubiquitin-like proteins: Functional versatility and putative ancestral involvement in tRNA modification revealed by comparative genomic analysis. Archaea pii: 710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV 2007. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol Direct 2: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, van der Oost J, Koonin EV 2009. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol 8: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W 2009. Sculpting the bacterial cell. Curr Biol 19: R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L 1996. Archaeal–eubacterial mergers in the origin of Eukarya: Phylogenetic classification of life. Proc Natl Acad Sci 93: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Stolz JF 1984. Cell symbiosis theory: Status and implications for the fossil record. Adv Space Res 4: 195–201 [DOI] [PubMed] [Google Scholar]
- Margulis L, Dolan MF, Guerrero R 2000. The chimeric eukaryote: Origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci 97: 6954–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Ettema TJ 2013. From archaeon to eukaryote: The evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans 41: 451–457 [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus–cytosol compartmentation. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Martin W, Muller M 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Martin W, Dagan T, Koonin EV, Dipippo JL, Gogarten JP, Lake JA 2007. The evolution of eukaryotes. Science 316: 542–543 [DOI] [PubMed] [Google Scholar]
- Maupin-Furlow JA 2013. Ubiquitin-like proteins and their roles in Archaea. Trends Microbiol 21: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Coleman RL, Dodsworth JA, Ross CA, Shock EL, Williams AJ, Hartnett HE, McDonald AI, Havig JR, Hedlund BP 2012. Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning. Plos ONE 7: e35964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Soll D, Maupin-Furlow JA 2011. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in Archaea. Proc Natl Acad Sci 108: 4417–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P 1998. Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: The syntrophic hypothesis. J Mol Evol 47: 517–530 [DOI] [PubMed] [Google Scholar]
- Muller F, Brissac T, Le Bris N, Felbeck H, Gros O 2010. First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol 12: 2371–2383 [DOI] [PubMed] [Google Scholar]
- Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF 2012. Acquisition of 1000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci 109: 20537–20542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, et al. 2011. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39: 3204–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR 1997. A molecular view of microbial diversity and the biosphere. Science 276: 734–740 [DOI] [PubMed] [Google Scholar]
- Pace NR 2006. Time for a change. Nature 441: 289. [DOI] [PubMed] [Google Scholar]
- Pace NR 2009. Mapping the tree of life: Progress and prospects. Microbiol Mol Biol Rev 73: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelve EA, Lindas AC, Martens-Habbena W, de la Torre JR, Stahl DA, Bernander R 2011. Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol 82: 555–566 [DOI] [PubMed] [Google Scholar]
- Pereto J, Lopez-Garcia P, Moreira D 2004. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci 29: 469–477 [DOI] [PubMed] [Google Scholar]
- Pester M, Schleper C, Wagner M 2011. The Thaumarchaeota: An emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14: 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO 2007. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol 24: 1752–1760 [DOI] [PubMed] [Google Scholar]
- Poole A, Penny D 2007a. Eukaryote evolution: Engulfed by speculation. Nature 447: 913. [DOI] [PubMed] [Google Scholar]
- Poole AM, Penny D 2007b. Evaluating hypotheses for the origin of eukaryotes. Bioessays 29: 74–84 [DOI] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV 2009. Search for a tree of life in the thicket of the phylogenetic forest. J Biol 8: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendhran J, Gunasekaran P 2011. Microbial phylogeny and diversity: Small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res 166: 99–110 [DOI] [PubMed] [Google Scholar]
- Reigstad LJ, Jorgensen SL, Schleper C 2010. Diversity and abundance of Korarchaeota in terrestrial hot springs of Iceland and Kamchatka. ISME J 4: 346–356 [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA 1992. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257: 74–76 [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431: 152–155 [DOI] [PubMed] [Google Scholar]
- Robertson CE, Harris JK, Spear JR, Pace NR 2005. Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol 8: 638–6342 [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB 2006. Bushes in the tree of life. PLoS Biol 4: e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Bell SD 2009. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol 17: 507–513 [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD 2008. A role for the ESCRT system in cell division in Archaea. Science 322: 1710–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searcy DG, Stein DB, Green GR 1978. Phylogenetic affinities between eukaryotic cells and a thermophilic mycoplasma. Biosystems 10: 19–28 [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23: 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett AM, Johnson PJ 2010. Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol 64: 409–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F, Yeramian E 2005. Genome trees from conservation profiles. PLoS Comput Biol 1: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A, Sorensen KB 2008. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J 2: 3–18 [DOI] [PubMed] [Google Scholar]
- Thiergart T, Landan G, Schenk M, Dagan T, Martin WF 2012. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol 4: 466–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L 2007. Slicer and the Argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- van der Giezen M 2009. Hydrogenosomes and mitosomes: Conservation and evolution of functions. J Eukaryot Microbiol 56: 221–231 [DOI] [PubMed] [Google Scholar]
- Van der Veen AG, Ploegh HL 2012. Ubiquitin-like proteins. Annu Rev Biochem 81: 323–357 [DOI] [PubMed] [Google Scholar]
- Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S 2011. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci 108: 1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs K, Vida G 1998. A new aspect to the origin and evolution of eukaryotes. J Mol Evol 46: 499–507 [DOI] [PubMed] [Google Scholar]
- Vishwanath P, Favaretto P, Hartman H, Mohr SC, Smith TF 2004. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol Phylogenet Evol 33: 615–625 [DOI] [PubMed] [Google Scholar]
- Wang F, Liu M, Qiu R, Ji C 2011. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell 2: 612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM 2012. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci 279: 4870–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 87: 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV 2013. Genome reduction as the dominant mode of evolution. BioEssays 35: 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Makarova KS, Yutin N, Koonin EV 2012. Updated clusters of orthologous genes for Archaea: A complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV 2012. Archaeal origin of tubulin. Biol Direct 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol 25: 1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf MY, Wolf YI, Koonin EV 2009. The origins of phagocytosis and eukaryogenesis. Biol Direct 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Puigbo P, Koonin EV, Wolf YI 2012. Phylogenomics of prokaryotic ribosomal proteins. Plos ONE 7: e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]