TGFβ signalling in context (original) (raw)
. Author manuscript; available in PMC: 2014 May 20.
Published in final edited form as: Nat Rev Mol Cell Biol. 2012 Sep 20;13(10):616–630. doi: 10.1038/nrm3434
Abstract
The basic elements of the transforming growth factor-β (TGFβ) pathway were revealed more than a decade ago. Since then, the concept of how the TGFβ signal travels from the membrane to the nucleus has been enriched with additional findings, and its multifunctional nature and medical relevance have relentlessly come to light. However, an old mystery has endured: how does the context determine the cellular response to TGFβ? Solving this question is key to understanding TGFβ biology and its many malfunctions. Recent progress is pointing at answers.
Transforming growth factor-β (TGFβ) signalling provides animal cells with a versatile means of driving developmental programmes and controlling cell behaviour, a role that is evident in the many effects of TGFβ-related cytokines on cell proliferation, differentiation, morphogenesis, tissue homeostasis and regeneration, and the severe diseases that result from their malfunctions.
Pioneers in this field in the 1980s welcomed the multifunctional nature of TGFβ with mixed feelings. Up to that point, endocrinology was forged on the principle that, by and large, a hormone has one main role and this role only. With TGFβ it was clear from the beginning that this paradigm did not quite fit. The effects of TGFβ were different, even opposite, depending on the cell type and the conditions. The cellular context more than the cytokine dictated the nature of the response. To those intent on elucidating the TGFβ pathway, this contextual functionality was sobering news, as it raised the specter of an impossibly complicated signal transduction process. Yet, the work uncovered a pathway that is relatively simple (in hindsight) and with the power to mediate most effects of any TGFβ family member in any cell type. The TGFβ receptor system, its activation mechanism and SMAD proteins, which function both as substrates for TGFβ receptors and signal transducers, came to light in quick succession. Disease-causing mutations in these components stressed the medical relevance of the new findings. Regulators and complementary pathways were also found. TGFβ target genes that trigger differentiation in stem cells, cell cycle arrest in epithelial cells or homeostatic constraint in immune and vascular cells were identified. Crystal structures of the pathway components were emerging in the blink of an eye as the century was drawing to a close1. The TGFβ pathway had been solved, to a first approximation at least.
Writing on this subject at the time, the phrase “How cells read TGFβ signals” was picked as a title for two reasons2. It was an affirmation that a molecular framework for the exploration of TGFβ biology was firmly in hand. Fleshing out the newly defined pathway became the next task, and the field keenly obliged by identifying additional components, regulators, ancillary pathways and biological effects of the TGFβ family. However, that phrase also implied a challenging question: how does the cellular context determine the response to TGFβ? It was not clear how TGFβ can inhibit cell proliferation but also promote cell growth, enhance stem cell pluripotency but also differentiation, regulate muscle genes in myoblasts and neural genes in neuroblasts, or suppress pre-malignant cells but encourage metastatic ones. These paradoxes suggested that cells read TGFβ signals in ways that could not be explained. Non-canonical TGFβ pathways and malignant switches were explored as alternatives but yielded no answers either. Fifty thousand TGFβ papers later, the old enigma carries on.
Interest in solving this puzzle is growing, and it is driven by the importance of TGFβ signalling in medically relevant processes of immunity, inflammation, cancer and fibrosis, as well as bone, muscle, adipose, vascular and haematopoietic homeostasis. At last, recent progress is pointing to a resolution. To cover this progress, the present article provides an overview of the contextual determinants of TGFβ action followed by an update on the signalling, transcriptional and genomic elements of the pathway. Building on this, the final section covers, in broad strokes, the mode of action of TGFβ in various contexts, including embryonic stem (ES) cells, lineage-committed progenitors, cells undergoing epithelial–mesenchymal transition (EMT), induced pluripotent stem (iPS) cells, differentiated cells and cells at various stages of malignancy.
Contextual determinants of TGFβ action
The TGFβ pathway seems to have emerged with the first animal species3, ostensibly to control multicellular life in metazoans. It does so largely by regulating gene expression. The number of target genes for a given TGFβ family member can range from just a few in pluripotent ES cells4,5 to hundreds in differentiated cells4–8. The effects of TGFβ on transcription can be positive or negative depending on the targeted gene and the cell ular context. For example, TGFβ represses inhibitor of differentiation 1 (ID1) in mammary epithelial cells9 but induces this gene in metastatic breast cancer cells10. Few TGFβ target genes are common to all cell types, such exceptions being the negative feedback regulators SMAD7 and SKIL (SKI-like oncogene; also known as SNON)11,12. The SMAD signalling pathway is a central `conduit' for all these gene responses and, as such, it recapitulates the context-dependent nature of TGFβ action.
Three types of contextual determinants shape the TGFβ transcriptional response in a cell (FIG. 1). Within a given epigenetic and transcriptional context the response of a cell is determined by the extracellular and intracellular composition of the TGFβ signal transduction system. The abundance and activity of different TGFβ ligands, receptors and regulators determine the nature and intensity of the TGFβ signal in the nucleus, and coexisting cues further shape the response by regulating SMAD function or activating non-canonical pathways13–15. Inputs that affect the intensity of the TGFβ signal may qualitatively influence the cellular response. A dramatic example is the different cell fates that emerge from finely tuned gradients of bone morphogenetic protein (BMP) acting alongside WNT and Hedgehog signals during development16–18.
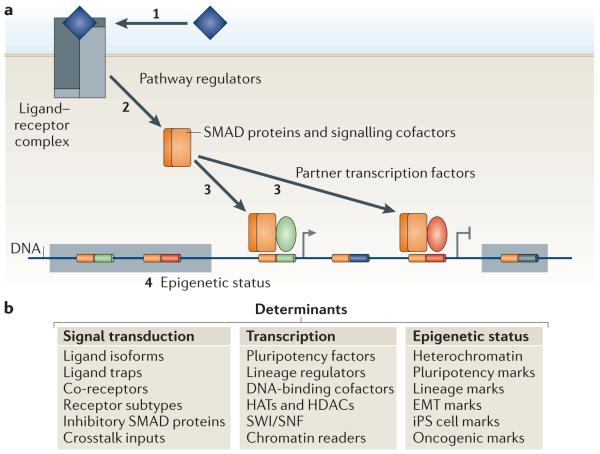
Figure 1. Contextual determinants of TGFβ action.
a | Three types of contextual determinants shape the transforming growth factor-β (TGFβ)-mediated transcriptional response in a cell. First, a large number of signal transduction regulatory factors determine the access of TGFβ ligands to signalling receptors (1), and of receptors to SMAD proteins and other signal delivery factors (2). Second, transcription factors, histone readers and modifiers and chromatin remodellers that bind to activated SMAD proteins determine what genes will be targeted by the signal transduction complexes and whether expression of the target genes will be positively or negatively regulated (3). The third type of contextual determinants is presented by the epigenetic status of the cell. The epigenetic state dictates whether genes are in an active `open' chromatin conformation (and are therefore accessible for SMAD complexes), in a repressive chromatin conformation (and therefore in a silenced `closed' state that is not accessible for transcriptional regulation), or whether these genes are in a poised chromatin state that is silenced yet responsive to TGFβ signalling and the appropriate chromatin readers (4). Composite enhancer elements are represented as bi-coloured segments on the DNA, inside a shaded box if the genes are in repressive chromatin, and unoccupied if the cell does not express the required DNA-binding SMAD cofactor. b | A list of the contextual determinants that affect the signal transduction and transcription steps and the regulators of the epigenetic status. EMT, epithelial–mesenchymal transition; HATs, histone acetyl transferases; HDACs, histone deacetylases; iPS cell, induced pluripotent stem cell; SWI/SNF, Switch/sucrose nonfermentable.
A second set of determinants is the factors that co operate with SMAD proteins to regulate transcription (FIG. 1). The role of such factors was originally highlighted with the identification of forkhead box H1 (FOXH1; also known as FAST1) as a factor that allows SMAD proteins to recognize `activin response elements' (AREs) in promoters of genes involved in mesoderm differentiation19. This principle was extended to the TGFβ subfamily of BMPs with the identification of zinc-finger protein 423 (ZFP423; also known as OAZ), which is a SMAD cofactor for the activation of ventral mesoderm homeobox genes20. Lineage-specific transcription factors direct TGFβ- and BMP-activated SMAD proteins to specific loci genome-wide in myoblasts, pro-B cells, myeloid precursors and erythroid precursors4,8. In differentiated cells, diverse transcription factors guide SMAD proteins to distinct subsets of target genes21. The resulting SMAD complexes then recruit chromatin readers, modifiers and remodellers to regulate transcription. The availability of all these SMAD partners determines what genes will be targeted and whether they will be activated or repressed.
Last, but not least, the epigenetic landscape of the cell, including DNA methylation marks, histone modifications, nucleosome positioning, non-coding RNAs and other components, shapes the chromatin and dictates what genes are open for expression and thus susceptible to regulation (FIG. 1). For example, under conditions that favour self-renewal, ES cells keep pluripotency-enforcing genes in an open conformation that permits transcriptional activation downstream of TGFβ signals, whereas genes involved in differentiation remain repressed and refractory to these inputs. When conditions are permissive for ES cell differentiation, specific chromatin marks open master differentiation genes to activation driven by Nodal, which is a TGFβ family member that regulates stem cell pluripotency and differentiation4,5,8,22,23. EMT and iPS cell generation also incur epigenetic changes that influence TGFβ-mediated gene regulation22,24,25.
Collectively, these three classes of contextual determinants channel, skew or switch the pleiotropic capacity of TGFβ signalling, thereby giving rise to specific response programmes that are archetypical of particular cell types and conditions.
Activating TGFβ signalling
The TGFβ family includes over 30 members in humans, and orthologues are found in the most primitive metazoan genome sequenced, Trichoplax adhaerens. In this organism, the TGFβ pathway is already equipped with four SMAD proteins and four receptors3. Sequence similarities define two ligand subfamilies: the TGFβ–activin–Nodal subfamily and the BMP subfamily1. The ligands are disulphide-linked dimers, and dimerization is essential for receptor activation (FIG. 2). Most family members act as paracrine factors on cells near the source.
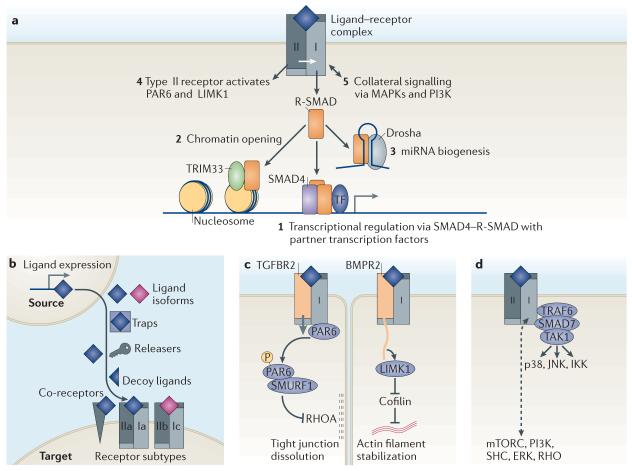
Figure 2. TGFβ receptors and signal transducers.
a | Transforming growth factor-β (TGFβ) family ligands signal by assembling a hetero-tetrameric receptor complex with two type I (which are the main signal propagators) receptor components (shown in light grey) and two type II (which are activators) components (shown in dark grey). Signalling is mediated via a cytoplasmic Ser/Thr kinase domain. Signalling modality 1 is the canonical SMAD pathway, and modality 2 its companion. Signalling modalities 3–5 are considered as non-canonical TGFβ signalling pathways. Receptor-phosphorylated SMAD proteins (R-SMAD proteins) form transcriptional complexes that pair with other context-dependent transcription factors to regulate hundreds of genes (1) (for details, see FIGS 3,5). Activated R-SMAD proteins can also form a complex with TRIM33 (tripartite motif containing 33) that recognizes certain histone marks and disables their repressive action, which results in chromatin opening and thereby allows access for canonical SMAD complexes (2) (for details, see FIG. 4). R-SMAD proteins can participate in microRNA (miRNA) processing by Drosha complexes for the biogenesis of a subset of SMAD-binding miRNA precursors (3). TGFβ and bone morphogenetic protein (BMP) type II receptors signal by directly activating partitioning defective 6 (PAR6) and LIM kinase 1 (LIMK1), respectively (4). TGFβ and BMP receptors can activate various mitogen-activated protein kinase (MAPKs) and phosphoinositide 3-kinase (PI3K) pathways (5). b | Seven classes of determinants regulate the access of ligand to TGFβ receptors. These include: the level of expression of a ligand in source cells, the type of ligand isoforms that is available, factors that can sequester the ligands (termed traps) or release them (termed releasers), decoy ligands that occupy the receptors or co-receptors without triggering signalling and the type of receptor and co-receptor that is expressed in target cells. c | Direct signalling by type II receptor kinases. TGFβ receptor type II (TGFBR2) phosphorylates the tight junction regulator PAR6 to recruit the E3 ubiquitin ligase SMURF1 and target RHOA for degradation. This leads to dissolution of tight junctions in epithelial cells. BMP receptor type II (BMPR2) contains a long carboxy-terminal tail that binds and activates LIMK1, thereby inhibiting the actin-disassembling protein cofilin. This results in stabilization of actin filaments. d | TRAF6 (tumour necrosis factor receptor-associated factor 6) acts together with SMAD7 and both are known binding partners for TGFβ receptors. The interaction between TRAF6 and SMAD7 is implicated in the activation of TAK1 (TGFβ-activated kinase 1), which is a protein kinase upstream of the signal transduction kinases p38, JNK (Jun amino-terminal kinase) and IKK (inhibitor of κB kinase). TGFβ and BMP receptors can activate several other signal transducers (such as mTORC (mammalian target of rapamycin), PI3K, SHC (SH2 domain-containing transforming protein), ERK (extracellular signal-regulated kinase) and RHO), although the biochemical and structural bases for many of these links remain unknown.
Receptor combinations
Ligand binding assembles a complex consisting of two type I (that is, signal-propagating) receptor components and two type II (that is, activator) components (FIG. 2a). Both components are Ser/Thr protein kinases. A short cytoplasmic segment is followed by the kinase domain and, only exceptionally, a carboxy-terminal extension (for example, in the type II BMP receptor BMPR2). In the complex, type II receptors phosphorylate the type I components, which then propagate the signal26. Phosphorylation switches a region in the type I receptor from a site that binds 12 kDa FK506-binding protein (FKBP12), which silences kinase activity, into a site that binds substrate SMAD proteins for their phosphorylation27. These are the sole cell surface receptor Ser/Thr kinases known in humans. Why TGFβ uses receptor Ser/Thr kinases and not Tyr kinases as all other kinase-activating cytokines do remains a mystery.
Seven type I receptors and five type II receptors exist in humans. Interactions with contiguous or non-contiguous molecular surfaces determine the specificity of ligand–receptor pairings28,29. Detailed ligand–receptor combinations are reviewed elsewhere1,30. Briefly, TGFβ binds exclusively to the type I receptor TGFBR1 (also known as ALK5 and TβRI) and the type II receptor TGFBR2. Activin, Nodal and BMPs share the type II receptors activin receptor type 2A (ACVR2A) and ACVR2B. Activin and Nodal, but not BMPs, share the type I receptors ACVR1 (also known as ALK2), ACVR1B (also known as ALK4) and ACVR1C (also known as ALK7), whereas BMPR1A (also known as ALK3) and BMPR1B (also known as ALK6) primarily act as type I receptors for BMPs and anti-Muellerian hormone (AMH). BMPR2 is another type II receptor for BMPs, and AMHR2 is the type II receptor for AMH. Activin receptor-like 1 (ACVRL1; also known as ALK1) is a BMP9 (also known as GDF2) and BMP10 type I receptor, but it can be collaterally engaged by high TGFβ concentrations31.
Extracellular regulators
Seven variables outside the target cell determine the extent of stimulation by a TGFβ cytokine (FIG. 2b). First is the level of ligand expression by the source, which is highly regulated by many contextual elements. A good example is Drosophila melanogaster BMP Decapentaplegic16,17. Second, the ligand subtype (for example, TGFβ1 and TGFβ2) also has a role, as subtype s differ in receptor affinity32.
Next, various ligand-trapping proteins control the formation of ligand gradients in embryogenesis and depots in adult tissues1,33,34. Some traps, for example the BMP trap Noggin, occlude crucial receptor contacting residues in the ligand35. Fusion of phalangeal joints in individuals with NOG mutations36 and the involvement of the BMP inhibitor gremlin 1 (GREM1) in basal cell carcinoma37 highlight the importance of the traps. Fourth, the mediators of ligand release from these traps are relevant, and their action can be nuanced. Latent TGFβ complex activation involves conformational motions driven by contacts with cell surface integrins38. Inadequate anchoring of LTBP1 (latent TGFβ-binding protein 1) owing to mutations in the gene encoding fibrillin 1 in individuals with Marfan syndrome is thought to cause faulty TGFβ release in the aortic wall, which can cause aneurysms39.
A fifth variable at play is antagonistic ligands. Lefty (also known as left–right determination factor) inhibits Nodal binding to receptors and is essential for the establishment of left–right asymmetry in embryos40,41. Inhibin prevents activin binding to its receptors42,43. Furthermore, a sixth variable is the presentation of ligands to the signalling receptors by accessory receptors. For example, β-glycan presents TGFβ to its receptors44 and inhibin to activin receptors42,43. Cripto (also known as TDGF1) is essential for Nodal binding to activin receptors41,45. Mutations in the accessory receptor endoglin, like mutations in its client receptor ACVRL1, cause haemorrhagic telangectasia46,47. Last, the combination of expressed signalling receptors is another determinant of how cells receive TGFβ signals. These seven variables tightly control the initiation of TGFβ signalling.
The SMAD signalling cycle
TGFβ family members regulate gene expression by receptor-mediated activation of SMAD transcription factors (FIG. 2a). Activated SMAD proteins regulate the transcriptional output of active genes and can also open repressive chromatin. Additionally, SMAD proteins serve as hubs for the integration of regulatory inputs and context-dependent modulation of TGFβ signalling.
SMAD activation by TGFβ receptors
SMAD proteins consist of two globular domains (termed MH1 and MH2 domains) coupled by an unstructured linker1 (FIG. 3a). The amino-terminal MH1 domain contains a hairpin structure with DNA-binding ability. The MH2 domain has a series of hydrophobic surface patches for versatile interactions with cytoplasmic adaptor proteins, activated TGFβ receptors, various partner DNA-binding cofactors, co-activators and co-repressors. BMP type I receptors phosphorylate SMAD1, SMAD5 and SMAD8, and BMP type I receptors for TGFβ, activin and Nodal mainly phosphorylate SMAD2 and SMAD3. Receptor-mediated phosphorylation of this set of SMAD proteins (collectively known as receptor-regulated SMAD proteins (R-SMAD proteins)) targets two C-terminal Ser residues, creating an acidic knob that binds to homologous MH2 domains or to the MH2 domain of SMAD4. SMAD4 itself is not a receptor substrate but functions as a shared partner of all R-SMAD proteins (FIG. 3b). Trimers with two R-SMAD molecules and one SMAD4 are thought to be the principal functional units1.
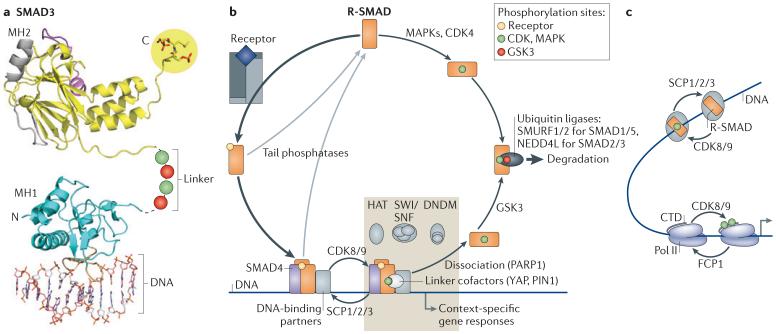
Figure 3. The SMAD signalling cycle.
a | Structure of SMAD3 as a representative of receptor-phosphorylated SMAD proteins (R-SMAD proteins). SMAD proteins consist of two globular domains (termed MH1 and MH2) that are coupled by a linker region. The signalling receptors phosphorylate R-SMAD proteins at the carboxy-terminal sequence Ser-X-Ser (where X can be any amino acid), creating an acidic tail that allows binding to SMAD4 (not shown). The linker is phosphorylated by cyclin-dependent kinases (CDKs), mitogen-activated protein kinases (MAPKs) (green) and glycogen synthase kinase 3 (GSK3) (red). The phosphorylated linker creates a docking sites for positive and negative regulators of SMAD function. b | Following phosphorylation by receptors (yellow), R-SMAD proteins bind SMAD4 and form a hetero-trimeric complex that binds to DNA with partner transcription factors. The transcriptional kinases CDK8 and CDK9 phosphorylate the linker region for peak activation (green). Key participants in the SMAD transcriptional complex include the SWI/SNF nucleosome positioning complex, the HATs (histone acetyl transferases) p300 and CBP (cyclic AMP response element-binding protein) and in certain target genes a DNA demethylating complex (DNDM). Repressive SMAD complexes recruit histone deacetylases (HDACs) through co-repressors (not shown). Various linker-bound factors also participate. Small C-terminal domain (CTD) phosphatases (SCPs) dephosphorylate the linker, allowing repeated utilization of the activated SMAD complex. Poly(ADP-ribose) polymerase 1 (PARP1) and other factors mediate the dissociation of this complex, and tail phosphatases return R-SMAD proteins to the basal state. If R-SMAD proteins are not dephosphorylated, GSK3 recognizes R-SMAD proteins phosphorylated by CDK8 and CDK9 and further increases R-SMAD phosphorylation (red), thereby marking R-SMAD proteins for recognition by E3 ubiquitin ligases and proteasome-mediated degradation. In this manner, SMAD transcriptional action becomes coupled to SMAD turnover. Mitogens and stresses acting through MAPKs, and the cell cycle acting through CDK4, can also phosphorylate the linker to limit the availability of R-SMAD proteins for TGFβ or bone morphogenetic protein (BMP) signalling. c | Intriguing parallels exist between the SMAD andRNA polymerase II (Pol II) transcriptional cycles. R-SMAD proteins and Pol II are phosphorylated (green) by the same kinases (which are CDK8 and CDK9) for peak activation and dephosphorylated by structurally related phosphatases (SCP1, SCP2 and SCP3) that reset the basal state. FCP1, transcription factor IIF-associated CTD phosphatase 1; NEDD4L, neural precursor cell expressed developmentally downregulated protein 4-like; PIN1, peptidylprolyl cis/trans isomerase, NIMA-interacting 1; SMURF, SMAD-specific E3 ubiquitin protein ligase 1; YAP, Yes-associated protein. Image in part a is reproduced, with permission, from REF. 63 © (2011) Cold Spring Harbor Laboratory Press.
In the basal state, SMAD proteins constantly shuttle between the cytoplasm and the nucleus via contact with nucleoporins for transit through the nuclear pore48. However, in the receptor-induced oligomeric state, SMAD proteins require nuclear import and export factors49–51. In the nucleus, R-SMAD proteins in activated SMAD4–R-SMAD complexes bind other DNA-binding transcription factors as partners for target gene recognition and transcriptional regulation4,7,8,19,20 (FIG. 3b).
The DNA-binding hairpins in SMAD1, SMAD2, SMAD3 and SMAD5 and are identical in amino acid sequence and all recognize the DNA motif CAGAC (which is the SMAD-binding element (SBE))1. However, the positioning of the hairpin in SMAD1 (REF. 52) differs from that in SMAD3 (REF. 53), and this perhaps explains why SMAD1 also bind to GC-rich sequences54. These differences are manifest in the genome-wide binding patterns: TGFβ and Nodal-activated SMAD2 and SMAD3 predominantly bind canonical SBEs55,56, whereas BMP-activated SMAD1 and SMAD5 bind GC-rich elements in addition to canonical SBEs57,58. Thus, recognition of specific DNA sequences by different SMAD complexes is dictated by preferences for different SBEs and different DNA-binding partners.
The SMAD action turnover switch
SMAD molecules that are engaged in transcription become rapidly phosphorylated in the linker region by cyclin C–cyclin-dependent kinase 8 (CDK8) and cyclin T–CDK9 (REFS 59,60) (FIG. 3a,b). The phosphorylated motifs recruit factors like YAP (Yes-associated protein) for transcriptional action59 but also prime the linker for sub-sequent phosphorylation by glycogen synthase kinase 3 (GSK3)61,62. GSK3 creates binding sites for the E3 ubiquitin protein ligases SMURF1 (SMAD-specific E3 ubiquitin protein ligase 1) or NEDD4L (neural precursor cell expressed developmentally downregulated protein 4-like), which target SMAD proteins for polyubiquitylation and proteasome-mediated degradation59,60. Binding of YAP, SMURF1 and NEDD4L involves two WW domains on these proteins; one WW domain contacts the SMAD phosphorylated linker motif, and the other WW domain interacts with a vicinal Pro/Tyr motif. The order of events, which is SMAD action first and degradation later, is controlled by a GSK3-driven switch in the SMAD motifs that bind WW domains63. This SMAD phospho-Ser code and the set of WW code readers provide an efficient solution to the problem of coupling TGFβ signal delivery to turnover of SMAD messenger molecules.
Links to the RNA polymerase II cycle
CDK8 and CDK9 are components of the transcriptional mediator complex and elongation complex, respectively. As such, CDK8 and CDK9 phosphorylate Ser/Pro motifs in the C-terminal domain (CTD) of RNA polymerase II (Pol II) to recruit proteins for DNA transcription, transcript capping and splicing64. SMAD linker phosphorylation is therefore coordinated by CDKs that also orchestrate Pol II action. Moreover, the SMAD linker is dephosphorylated by small CTD phosphatases (SCP1, SCP2 and SCP3) that prolong the participation of activated SMAD proteins in transcription before SMURF1 and NEDD4L target them for destruction65,66 (FIG. 3b). SCPs are structurally related to FCP1 (transcription factor IIF-associated CTD phosphatase 1; also known as CTDP1), which dephosphorylates the Pol II CTD for repeated cycles of transcription67. These provocative parallels suggest that SMAD and Pol II transcriptional cycles are coordinated by a common team of linker and CTD kinases and phosphatases (FIG. 3c).
Ending SMAD action
Additional factors have roles in limiting the pool of activated SMAD protein or turning over SMAD molecules that are engaged in transcription (FIG. 3b). Phosphatases remove SMAD C-terminal phosphorylation; however, which of these phosphatases acts in the nucleus needs clarification65,68. The co-activators p300 and CBP (cyclic AMP response element-binding protein) acetylate SMAD proteins on Lys residues that reside in the MH1 domain to enhance SMAD binding to DNA69, whereas poly(ADP)-ribosylation of this domain inhibits binding to DNA70. In addition, SMAD proteins are sumoylated, however, the functional implications of this post-translational modification are still unresolved30. Negative feedback is provided by SMAD-induced expression of SMAD7, which recruits SMURF to TGFβ and BMP receptors for polyubiquitylation and degradative endocytosis11,71. Receptor ubiquitylation is countered by the deubiquitinases USP4 (ubiquitin-specific processing protease 4), USP11 and USP15 (REFS 72–74). SMAD7 simultaneously recruits USP15 and SMURF2 to TGFβ receptor complexes, and these the two enzymes compete at this site73. USP15 can also deubiquitylate R-SMAD proteins75. The decoy receptor BAMBI (BMP and activin membrane-bound inhibitor)76 and SKIL also provide negative feedback, which disrupts SMAD transcriptional complexes12,77. Arkadia, a RING-domain E3 ubiquitin ligase, counters SMAD7 and SKIL by ubiquitylating these inhibitors78,79. SMAD phosphorylations at other sites provide added control30.
SMAD proteins as a hub for regulation and integration
The SMAD linker region is a hotspot for the integration of regulatory inputs (FIG. 3). The CDK target sites in SMAD linker regions are phosphorylated by mitogen-activated protein kinases (MAPKs) in response to growth factors (for example, fibroblast growth factor and epithelial growth factor) and stress signals, as well as by CDK4 during cell cycle progression62,80,81. GSK3 phosphorylation of CDK-primed or MAPK-primed SMAD linker is a key in this regard. By inhibiting GSK3, WNT augments the useful life of activated SMAD1, thereby providing an entry point for the cooperation between BMP and WNT pathways61.
WNT also cooperates with BMP and TGFβ through co-occupancy of SMAD target enhancers by WNT-activated LEF1 (lymphoid enhancer-binding factor 1; also known as TCF1α) and TCF7L2 transcription factors82–84. SMAD1 and TCF7L2 co-occupy the genome with lineage-identity transcription factors to implement differentiation during regenerative haematopoiesis8. Integration with the AKT pathway is provided by FOXO factors as SMAD partners85 and targets86. Pathway integration is also achieved by SMAD target genes, in particular by their protein products that interact with SMAD proteins to regulate other genes. For example, in epithelial cells TGFβ-activated SMAD proteins stimulate the expression of activating transcription factor 3 (ATF3) and SNAIL (also known as SNAI1) and then cooperate with these proteins to repress, respectively, ID1 and CDH1 (which encodes epithelial cadherin (E-cadherin))9,87. Many other roles of SMAD proteins as a hub for signal regulation and integration are known (reviewed in REFS 14,15,18,88).
SMAD proteins and chromatin
The transcriptional action of signal-activated SMAD proteins involves close interactions with chromatin. Recent insights have shed light on how SMAD proteins remodel chromatin or gain access to loci that are secluded by repressive histone marks.
SMAD-recruited chromatin and DNA modifiers
SMAD proteins recruit the histone acetyl transferases (HATs) p300 and CBP to stimulate transcription7, leading to acetylation of Lys9, Lys14, Lys18 and Lys23 on histone H3 (REFS 5,89,90). SMAD inhibition of gene expression depends on binding partners that in turn recruit histone deacetylases (HDACs)7. Among these partners, TGIF1 (5′-TG-3′-interacting factor 1) and TGIF2 recruit the co-repressor CTBP (C-terminal-binding protein), which in turn binds HDAC1 to limit TGFβ–Nodal signalling7. TGIF1 mutations in humans are associated with holoprosencephaly, which is a devastating defect of craniofacial development. In mice, knockout of Tgif1 causes a persistent increase in Nodal and TGFβ responses91 and the occurrence of holoprosencephaly92.
Recently, studies on TGFβ signalling provided a remarkable example for signal-directed, locus-specific DNA demethylation during gene activation93. TGFβ induces expression of CDKN2B, which encodes the CDK4 inhibitor p15Ink4b. This results in an antiproliferative effect that is mediated by promoter binding of a SMAD4–SMAD2/3–FOXO complex and dissociation of a MYC–MIZ1 (MYC-interacting zinc-finger protein 1; also known as ZBT17) complex94,95. In the absence of TGFβ, a complex consisting of zinc-finger protein 217 (ZFN217), co-repressor of RE1-silencing transcription factor (CoREST; also known as RCOR1) and the DNA methyltransferase 3A (DNMT3A) binds to the promoter and methylates a CpG island to mediate repression93. Following TGFβ stimulation, SMAD4–SMAD2/3–FOXO binds to the promoter and recruits a DNA excision repair complex that demethylates the promoter, which leads to CDNK2B expression. When TGFβ stimulation ends, ZFN217–CoREST–DNMT3A returns the promoter to a methylated, inactive state93.
Gene activation or repression by TGFβ additionally require, in most cases, a SWI/SNF nucleosome repositioning complex96. SWI/SNF complexes mediate nucleosome sliding in an ATP-dependent manner to allow access for the transcriptional machinery to DNA97. BRG1 (also known as SMARCA4) is one98 of two mutually exclusive ATPase subunits that nucleate SWI/SNF complexes. BRG1 binds directly to SMAD2 and SMAD3 (REFS 90,99) and recruits BAF250B (also known as ARID1B), BAF170 and BAF155 to the SMAD complex96.
A SMAD-associated histone reader for silent chromatin
In ES cells and progenitor cells, signal-activated SMAD2 and SMAD3 bind to the histone-binding protein TRIM33 (tripartite motif containing 33; also known as TIF1γ)5,99 (FIG. 2b). TRIM proteins contain a RING-finger and various protein interactions domains. TRIM33 additionally contains a plant homeodomain (PHD) and a bromodomain, which are two histone-binding domains essential for its role in TGFβ signal transduction5. TRIM33–SMAD2/3 and trimeric SMAD4–SMAD2/3 complexes form with similar abundance and kinetics in response to TGFβ or Nodal stimulation5,99. Although TRIM33 was implicated to have a role in inhibitory SMAD4 mono-ubiquitylation89,100, TRIM33-deficient mouse embryos lack a mesoderm101, and TRIM33 is essential for Nodal activation of mesendoderm genes in ES cells5. TRIM33 mediates erythroid differentiation in human haematopoietic progenitors in response to TGFβ99. Trim33 knockout in the mouse pancreas pheno copies the tumorigenic effect of Smad4 knockout, arguing that TRIM33 and SMAD4 converge on tumour suppression102,103.
The role of TRIM33 in TGFβ signalling was recently elucidated by genetic ablation experiments in ES cells coupled with X-ray crystal structure analysis5 (FIG. 4a). The TRIM33 PHD–bromodomain cassette binds to N-terminal tails of histone H3 that contain unmodified Lys4 residues, trimethylated Lys9 (H3K9me3) and acetylated Lys18 (H3K18ac). Notably, H3K9me3 is a chromatin mark that represses yet `poises' master differentiation genes for activation23. Driven by Nodal, TRIM33–SMAD2/3 recognizes these histone marks and promotes a transition from poise to open chromatin, thereby allowing access for SMAD4–SMAD2/3-dependent transcriptional activation (see below)5. However, other target genes, such as SMAD7 and SKIL in ES cells and most of TGFβ target genes in skin and breast epithelial cells5,99 do not require TRIM33 for their activation. These genes must already be in an active, open chromatin state for SMAD4–SMAD2/3 to bind.
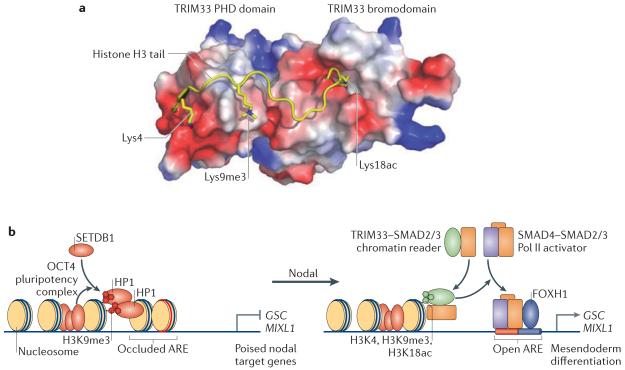
Figure 4. SMAD access to chromatin.
a | TRIM33 (tripartite motif containing 33) is a chromatin reader that recognizes a triple feature on histone H3: unmodified Lys4, trimethylated Lys9 (Lys9me3) and acetylated Lys18 (Lys18ac), as seen in the crystal structure of the TRIM33 plant homeodomain PHD-bromo cassette bound to the cognate histone peptide. b | The OCT4 pluripotency complex (which contains OCT4, SOX2 and NANOG) in embryonic stem cells prompts SETDB1 (SET-domain binding 1) to trimethylate histone H3 at Lys9 (H3K9me3) at the promoters of the mesendoderm differentiation genes goosecoid homeobox (GSC), mix paired-like homeobox (MIXL1) and other genes. H3K9me3 recruits the chromatin compacting factor heterochromatin protein 1 (HP1) to implement repression. Under differentiation conditions, GSC and MIXL1 are silent but poised for activation by Nodal signals. The gene promoters present cognate histone marks that are recognized by TRIM33. Nodal drives the formation of two companion complexes, TRIM33–SMAD2/3 and SMAD4–SMAD2/3. The TRIM33–SMAD2/3 complex binds to the poised nucleosomes, displacing HP1, thus enabling SMAD4–SMAD2/3 and their partner in this context, FOXH1 (forkhead box H1), to co-occupy cognate DNA elements (termed activin response elements (AREs)) and activate transcription. Image in part a is courtesy of Z. Wang and D. J. Patel, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Non-canonical signalling
Although TGFβ signals mainly via the the SMAD pathway, TGFβ can also activate other pathways that are collectively referred to as `non-canonical' TGFβ signalling and complement SMAD action (FIG. 2c,d).
Signalling by type II receptors
Various forms of signal transduction emanate directly from type II receptors. In cells undergoing EMT, ligand-bound TGFBR2 directly phosphorylates the cell polarity regulator partitioning defective 6 (PAR6) (FIG. 2c). Phosphorylated PAR6 recruits SMURF1 to target RHOA GTPase at tight junctions, thereby causing dissolution of the junctions and polarized migration104,105. In addition, in neocortical neurons in the developing brain, TGFβ turns naive neurites into axons through TGFBR2-mediated phosphorylation of PAR6 (REF. 106).
BMP type II receptor BMPR2 has a long C-terminal extension that is not required for SMAD signalling, but mutations that truncate this domain cause primary pulmonary hypertension107. The C-terminal domain of BMPR2 binds to LIM kinase 1 (LIMK1), which is an inhibitor of the actin-depolymerizing factor cofilin (FIG. 2c). BMP binding prompts LIMK1 to inhibit cofilin, thereby stabilizing actin filaments108. BMPR2-bound LIMK1 at the tips of neurites synergizes with the RHO GTPase CDC42 to induce the den-dritic arbor in cortical neurons in response to BMP109. Moreover, the BMPR2–LIMK2–cofilin pathway mediates loss of epithelial identity during EMT of the neural crest in the chick embryo110.
Links to MAPK and PI3K outputs
Besides the negative impact of MAPKs on TGFβ and BMP signalling through phosphorylation of SMAD linker sites (see above), reports over the past two decades have described effects of TGFβ and BMP on the activity of various MAPKs and also on the phosphoinositide 3-kinase (PI3K) pathway. These effects can be either immediate and transient or delayed and secondary, depending on the cell type and the culture conditions14,15,30. MAPK and PI3K activation has been proposed to complement and converge with SMAD signalling14,15,30,111, although these pathways can also antagonize SMAD signalling in other contexts.
The biochemical links between TGFβ receptors and the MAPK and PI3K pathways are complex and involve TRAF6 (tumour necrosis factor receptor-associated fac-tor 6), mTORC (mammalian target of rapamycin) and other mediators30,31,112 (FIG. 2d). Due to a lack of structural information it is not known whether activation of the MAPK and PI3K pathways is directly coupled to TGFβ receptors, whether it is the result of collateral activation of other receptors or whether it is a consequence of network-wide signalling crosstalk. Regardless, the MAPK and PI3K pathways on their own are major signalling routes for receptor tyrosine kinases, metabolic inputs and environmental stresses, and in cancer these pathways are often activated by key oncoproteins. These considerations raise questions about what an extra TGFβ input into PI3K or MAPKs can accomplish in this context. It has been suggested that pro-tumorigenic effects of TGFβ such as induction of EMT or enhancement of metastasis involve a pathologic switch of TGFβ signalling from SMAD-mediated tumour suppression to non-canonical malignant pathways. However, canonical SMAD signalling in cancer cells does drive EMT24,113, tumour-initiating cell stemness114–116 and pro-metastatic gene expression10,117.
SMAD proteins as Drosha components in microRNA biogenesis
SMAD proteins have other functions besides their role in transcriptional regulation. A remarkable example is their involvement in micro-RNA (miRNA) biogenesis118,119. In human vascular smooth muscle cells, activated R-SMAD proteins stimulate the maturation of a specific set of miRNAs in a SMAD4-independent manner (FIG. 2a). TGFβ-activated SMAD3 or BMP-activated SMAD1 are recruited to the Drosha miRNA processing complex by the RNA helicase DDX5 (DEAD box protein 5), which is a component of the Drosha complex. In the case of miR-21, the R-SMAD proteins promote the processing of primary transcripts (pri-miR-21) into precursor miR-21 (pre-miR-21). The resulting increase in miR-21 levels downregulates PDCD4 (programmed cell death 4) and induce a contractile phenotype in vascular smooth muscle cells. Most R-SMAD-regulated miRNAs contain a consensus SBE within the stem region of the primary transcript, and this element is required for the incorporation and regulatory effect of R-SMAD proteins in the Drosha complex118.
TGFβ action in context
TGFβ and BMP regulate pluripotency and differentiation in ES cells and lineage-committed progenitors, reprogramming in EMT and iPS cells, homeostasis in differentiated cells and altered versions of these processes in cancer. The signalling engine in these various contexts is essentially the same. The context more than the proteins involved in the signalling pathway is what shapes the response.
Contexts for ES cell self-renewal and differentiation
The core transcriptional regulators OCT4 (also known as POU5F1 and OCT3), SOX2 and NANOG form an interactive, self-sustaining protein network that induces pluripotency in ES cells22,23. This triad directs chromatin-modifying complexes to establish repressive marks on differentiation genes (FIG. 4b). Polycomb repressive complex 2 (PRC2) promotes accumulation of H3K27me3, which promotes the recruitment of PRC1, and SETDB1 (SET-domain binding 1; also known as KMT1E) catalyses the formation of repressive H3K9me3 marks23. These repressive marks may coexist with activating marks that poise chromatin for abrupt transcriptional activation by appropriate signals23.
Recent work on the genome-wide integration of signalling pathways with the OCT4 network provided fresh insights into the function of SMAD proteins in the context of this network4,120. BMP signalling stimulates ES cell self-renewal121 by directing SMAD1 to co-occupy the genome with leukaemia inhibitory factor (LIF)-activated signal transducer and activator of transcription 3 (STAT3), OCT4, SOX2 and NANOG at sites that contain the H3K4me3 mark of active transcription. The targeted genes include Oct4, Sox2, Nanog and Id3 (REF. 120), implementing a feed-forward circuit. The OCT4 complex also directs Nodal-activated SMAD3 to neighbouring sites4, although this results in the activation of only a few genes, including Nanog and the Nodal negative feedback regulators, Lefty1, Lefty2 and Smad7 (REF. 4).
How does this scenario change when conditions are permissive for ES cell differentiation? In the absence of the pluripotency enforcing factor LIF (FIG. 5), as yet unknown changes enable ES cells to respond to autocrine signals and differentiate into mesendodermal cells of the primitive streak and ectodermal cells. Nodal drives mesendodermal differentiation by inducing the expression of the homeobox transcription factors goosecoid homeobox (GSC) and mix paired-like homeobox (MIXL1)5. Certain sites in the regulatory regions of GSC and MIXL1 have H3K9me3 and acetylated Lys18 of histone H3 (H3K18ac)5 (FIGS 4b,5a). The chromatin compacting protein HP1γ (heterochromatin protein 1γ) bound to H3K9me3 prevents binding of Nodal-activated SMAD4–SMAD2/3 to AREs. The activation of MIXL1 and GSC in response to Nodal and ensuing differentiation require the previous action of a Nodal-activated TRIM33–SMAD2/3 complex. TRIM33 recognizes the dual histone mark H3K9me3–H3K18ac and binds with high affinity, thereby displacing bound HP1γ and opening the downstream AREs to allow access by SMAD4–SMAD2/3 complexes5. FOXH1 acts as a primitive mesendoderm identity factor that recruits SMAD4–SMAD2/3 complexes to co-occupy AREs19,122. The resulting induction of GSC and MIXL1 commits primitive embryo cells to mesendodermal fates5 (FIG. 4b). In other words, BMP-activated SMAD1 stimulates ES cell self-renewal by co-occupying the genome with LIF-activated STAT3 and the core pluripotency triad OCT4–SOX2–NANOG. When self-renewal signals fade, the poised chromatin marks that were established by the OCT4 complex provide an entry point for SMAD3 complexes to activate differentiation genes.
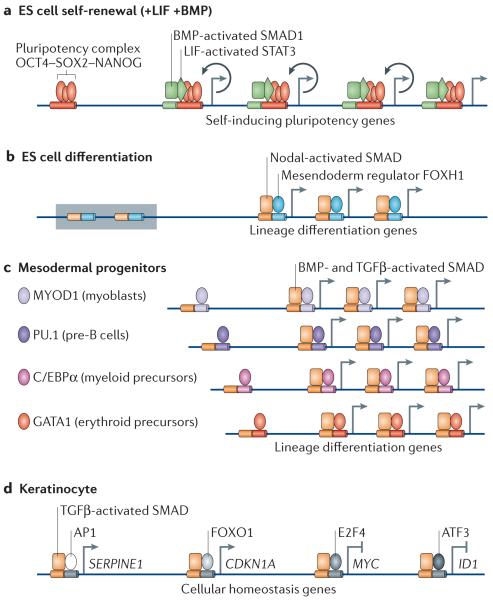
Figure 5. TGFβ action in ES cells, lineage progenitors and differentiated cells.
a | In embryonic stem (ES) cells, OCT4, SOX2 and NANOG form the core of a self-renewal network that is stimulated by the bone morphogenetic protein (BMP) mediator SMAD1 and the leukaemia inhibitory factor (LIF) mediator STAT3. SMAD1, STAT3 and the OCT4 complex co-occupy many active sites throughout the genome, including OCT4, SOX2 and NANOG themselves, to enforce self-renewal (indicated by circular arrows). b | In ES cells that lack self-renewal signals, the Nodal-activated SMAD4–SMAD2/3 complex, together with the TRIM33 (tripartite motif containing 33)–SMAD2/3 complex (see FIG. 4), activates mesendoderm differentiation genes in poised chromatin (indicated by a shaded box). Forkhead box H1 (FOXH1) is a mesendoderm lineage factor that recruits the SMAD4 complex to multiple differentiation genes. c | In lineage-restricted progenitors, lineage identity factors are dominant partners of transforming growth factor-β (TGFβ)- or BMP-activated SMAD4 complexes and co-occupy the genome to implement differentiation. d | In differentiated cells, TGFβ activated SMAD4–SMAD2/3 complexes are recruited by different partner transcription factors to different subsets of target genes (each subset is represented in the figure by only one gene). The combination of these pathways that lead to transcriptional regulation constitutes the overall TGFβ-mediated transcriptional response for regulation of cell proliferation, adhesion, extracellular matrix properties, the secretome and other cell homeostasis functions in any cell type. The diagram shows the factors that regulate homeostasis of a keratinocyte (that is, a differentiated ectodermal derivative). ATF3, activating transcription factor 3; AP1, adaptor protein 1; C/EBPα, CCAAT/enhancer-binding protein-α; CDKN1A, cyclin-dependent kinase inhibitor 1A; ID1, inhibitor of DNA binding 1; MYOD1, myoblast determination protein 1.
Context for lineage regulation in committed progenitors
Recent studies in different types of progenitor cells have shown that lineage identity factors recruit SMAD proteins to many sites in the genome to implement specific differentiation programmes4,8 (FIG. 5b). TGFβ activated SMAD3 co-occupies the genome with the myogenic identity facto r myoblast determination protein 1 (MYOD1) in mesenchymal progenitors and with the lymphoid identity factor PU.1 (also known as SPI1) in pro-B cells4. BMP-activated SMAD1 colocalizes at target genome sites with the myeloid lineage regulator CCAAT/enhancer-binding protein-α (C/EBPα) or the erythroid lineage regulators GATA1 and GATA2 in haematopoietic progenitors to prompt differentiation towards these two lineages8. Ectopic expression of these lineage regulators can redirect SMAD proteins to the corresponding lineage specific loci. Similarly to FOXH1 in ES cells, the master regulators MYOD1, PU.1, C/EBPα and GATA1 and GATA2 in lineage-committed progenitors recruit SMAD4–R-SMAD complexes to enhancer elements in order to implement differentiation. Taken together, core pluripotency factors and line-age identity factors direct signal-driven SMAD complexes to different sets of chosen genes, where SMAD proteins orchestrate transcriptional activation for self-renewal or differentiation.
Context for EMT
EMT, which is the switch of epithelial cells into a mesenchymal migratory phenotype, is a crucial morphogenetic event in gastrulation, embryonic tissue formation and regeneration24,113. Pathological forms of EMT participate in fibrosis and cancer. Enforced expression of EMT transcriptional regulators can turn mammary epithelial cells into stem-like cells (FIG. 6a) and provide breast cancer cells with a tumour-initiating phenotype25. EMT is driven by an interactive network of transcriptional repressors including SNAIL1, SNAIL2 (also known as SLUG), ZEB1 (zinc-finger E-box binding factor), ZEB2, KLF4 (Krueppel-like factor 4), TCF3 (also known as E47) and TWIST24,113. A negative feedback loop is established by miR-200, miR-205 and miR-183, which suppress ZEB1 and ZEB2, whereas ZEB1 represses miR-200 (REFS 24,123) (FIG. 6b).
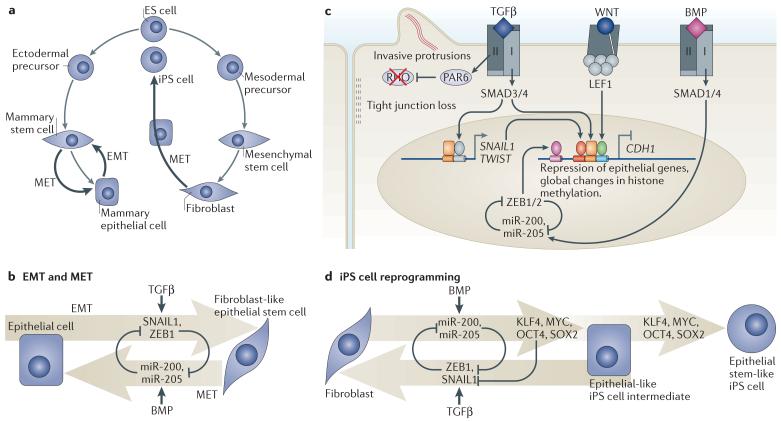
Figure 6. TGFβ action in EMT, MET and iPS cell transition.
a | Cell reprogramming processes regulated by transforming growth factor-β (TGFβ) and bone morphogenetic protein (BMP) include epithelial–mesenchymal transition (EMT) in mammary epithelial cells, which can generate epithelial stem-like cells, and the reverse process, mesenchymal–epithelial transition (MET). Reprogramming of fibroblasts into induced pluripotent stem (iPS) cells involves an intermediate MET-like process that is also regulated by BMP and TGFβ. b | EMT is driven by the core regulators SNAIL1, SNAIL2, ZEB1 (zinc-finger E-box binding factor 1) and ZEB2 (SNAIL2 and ZEB2 are not shown for simplicity) and other factors. EMT is inhibited by the pro-epithelial microRNAs miR-200 and miR-205, which in turn are suppressed by ZEB1. In mammary epithelial cells, EMT is stimulated by TGFβ and opposed by BMP. c | WNT primes epithelial cells to undergo EMT in response to TGFβ. TGFβ triggers SMAD-dependent induction of SNAIL1 and TWIST. SMAD3 and SMAD4 then join SNAIL1 and WNT-activated LEF1 (lymphoid enhancer-binding factor 1) to co-occupy the promoter of CDH1 (which is the gene encoding epithelial cadherin (E-cadherin)) for repression of this key epithelial gene. This and an associated genome-wide chromatin changes implement the mesenchymal phenotype. In parallel, TGFβ receptor type II (TGFBR2) phosphorylates partitioning defective 6 (PAR6) to mediate dissolution of tight junctions and foster migration (see FIG. 2c). BMP signalling through SMAD1 and SMAD4 upregulates miR-200 and miR-205 to oppose EMT. d | Fibroblast reprogramming into iPS cells by ectopic expression of OCT4, Krueppel-like factor 4 (KLF4), SOX2 and MYC requires MET, which is facilitated by BMP-induced miR-200 and miR-205 expression. By interfering with MET, TGFβ suppresses the generation of iPS cells. The use of TGFBR1 inhibitors increases the efficiency of iPS cell generation and reduces the requirement for MYC and SOX2.
TGFβ triggers EMT during heart development, palate fusion and renal fibrosis, as well as in breast and hepatic epithelial cells124. WNT acts as a competence factor that creates a favourable environment for TGFβ-induced EMT113 (FIG. 6c), as otherwise epithelial cells would undergo growth arrest in response to TGFβ. In mammary epithelial cells, TGFβ-activated SMAD proteins directly stimulate the expression of SNAIL1 and TWIST1 (TWIST-related protein 1)125,126. This is followed by the recruitment of SMAD proteins and SNAIL1 to the CDH1 promoter to repress the expression of this key epithelial cell junction gene87 (FIG. 6c). The WNT effector LEF1 also co-occupies the CDH1 promoter with SMAD proteins127, providing a molecular mechanism for the observed cooperation of WNT and TGFβ in EMT25. BMPs, which are known to induce miR-200 and miR-205 expression128, antagonize EMT and favour mesenchymal–epithelial transition (MET)25.
Downregulation of E-cadherin and upregulation of neural cadherin (N-cadherin) and vimentin are markers of EMT and mediators of distinct morphological changes. However, TGFβ-induced EMT proceeds with epigenetic events, which is suggestive of genome-wide reprogramming. These events include a global increase in H3K4me3 and H3K36me3, and a reduction in H3K9me2 within large, organized heterochromatin Lys9 modification regions that depends on the Lys-specific deacetylase LSD1 (REF. 129). The transcriptional effects of TGFβ are complemented by TGFBR2-mediated phosphorylation of PAR6 to dissolve tight junctions and promote a protrusive and invasive phenotype104,105 (see above) (FIG. 6c). Therefore, TGFβ seems to trigger EMT via a two-pronged TGFBR1–SMAD and TGFBR2–PAR6 pathways, with WNT–TCF providing the right transcriptional context.
iPS cell reprogramming
Ectopic expression of OCT4, Krueppel-like factor 4 (KLF4), SOX2 and MYC (which is termed the `OKSM cocktail') reprogrammes fibroblasts and other differentiated cells into pluripotent iPS cells130. As in ES cell self-renewal and EMT transitions, TGFβ and BMP affect iPS cells by signalling to the core reprogramming network22 (FIG. 6a). In fact, OKSM-driven reprogramming of mouse embryonic fibroblasts is a multistep process that passes first through an MET. BMP facilitates OKSM-driven iPS cell formation by inducing expression of miR-200 and miR-205, which facilitate MET128 (FIG. 6d). Moreover, OCT4 and SOX2 seem to suppress SNAIL1 expression, and KLF4 enhances E-cadherin expression131. TGFβ interferes with iPS cell reprogramming by favouring EMT131. Inclusion of TGFBR1 kinase inhibitors in the media eliminates the requirement of ectopic SOX2 and MYC (endogenous SOX2 and MYC may suffice in this context) and increases the rate of iPS cells generation132,133.
Regulation of homeostasis in differentiated cells
The TGFβ family exerts broad control over many aspects of the biology of differentiated cells, including their proliferation, migration and adhesion, the secretome, extracellular matrix (ECM) production and other functions. The negative regulation of cell cycle progression by TGFβ in epithelial, haematopoietic and neural cells is also of long-standing interest for its implications in cancer. It is largely mediated by the induction of CDK inhibitors (for example, CDKN1A, CDKN1C and CDKN2B) and repression of MYC14. TGFβ regulates the expression of components of the ECM, including collagens, fibronectin, tenascins, proteoglycans and their transmembrane receptor integrins134. The profound effects of TGFβ on ECM and inflammatory cytokine production and their implications in fibrosis and cancer have long spurred the consideration of targeting TGFβ pathways to treat these conditions. TGFβ is a major inducer of stromal chemokines in wound healing and tumour microenvironments135. Its colossal role as a regulator of immune and inflammatory functions depends on balancing acts between positive and negative effects on gene expression136. This extends to the regulation of tolerogenic and immunogenic forces in TH17 lymphocytes, which is a recently identified lineage of intestinal mucosal immunity mediators and suspected culprits in autoimmune diseases137–139. BMPs, activins, myostatin and other family members have important roles in the homeostasis of muscle, bone and adipose tissues, blood vessels and haematopoiesis. Such biological impact makes them potential therapeutical targets, which is prompting brisk activity in the pharmaceutical sector.
These diverse effects involve different transcription factors acting as DNA binding partners of SMAD proteins for the recognition of different sets of target genes (FIG. 5c). The sum of these partnerships defines the overall response of a particular cell type to the signal. For example, in skin keratinocytes, the TGFβ-activated SMAD4–SMAD2/3 complex cooperates with FOXO, the transcription factor ETS1 and the adaptor protein AP2 to bind to enhancers in CDKN1A and CDNK2B21,85,140 and with E2F4 and E2F5 to bind to an inhibitory element in MYC94,141 to induce antiproliferative gene responses. This distribution of activated SMAD among distinct effector complexes is in contrast with the dedicated interaction of SMAD with a few regulators to enforce differentiation in ES cells and progenitors.
Tumour suppression and tumour progression contexts
TGFβ is a powerful tumour suppressor in the context of pre-malignant cells but an enhancer of invasion and metastasis in the context of more advanced carcinoma cells14. This dichotomy may seem paradoxical, but it has its logic. TGFβ has cytostatic effects that are important in tissue regeneration and homeostasis142,143 (FIG. 7a). When cells incur oncogenic mutations and reach a pre-malignant state TGFβ-mediated suppressive effects become more dramatic, as in this context TGFβ triggers apoptosis14,103,143 (FIG. 7b). Exactly how pre-malignant cells become programmed to undergo apoptotic cell death is not clear, but the context forces them to avert TGFβ action. In some tumour types, this pressure selects for mutations that eliminate TGFβ signalling altogether (FIG. 7c). Inactivating mutations in TGFBR2 and SMAD4 are frequent in the carcinoma transition of gastro-intestinal and pancreatic tumours14,144. With the TGFβ pathway eliminated, cancer cells can create with impunity a TGFβ-rich microenvironment that favours tumour progression via effects on the stroma135.
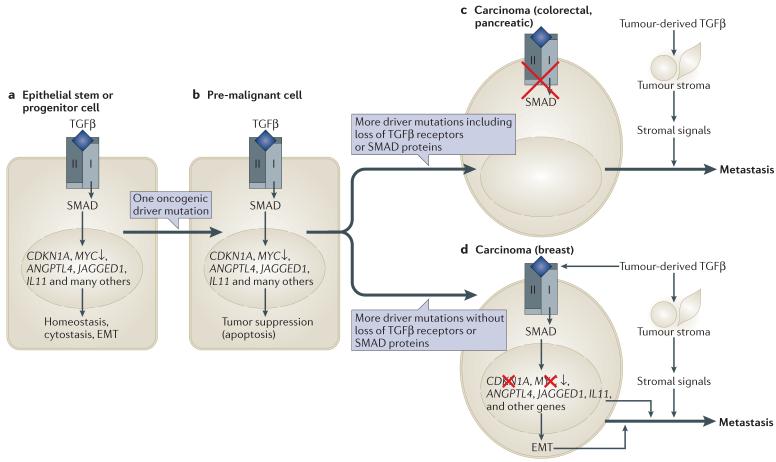
Figure 7. TGFβ action in tumour suppression and tumour progression.
a | Transforming growth factor-β (TGFβ)-activated SMAD proteins regulate hundreds of genes in normal epithelial cells, including cytostatic genes and genes involved in homeostasis. The examples include genes that are upregulated by TGFβ, except MYC, which is downregulated. b | When a stem or progenitor cell incurs an oncogenic mutation (for example, loss of APC (adenomatous polyposis coli) tumour suppressor, activation of KRAS or amplification of HER2) it becomes liable to undergo apoptosis if exposed to TGFβ. To advance in the tumorigenic path, this cell must accumulate additional alterations that, among other achievements, disable the tumour suppressive responsiveness of the cell to TGFβ. c | One path to tumour formation involves the selection of malignant clones that have lost TGFβ signalling owing to mutations in TGFBR1 (TGF beta receptor type II), TGFBR2 or SMAD4. This outcome is frequent in colorectal and pancreatic carcinomas. As a result, cancer cells can withstand a TGFβ-rich tumour stroma and benefit from pro-tumorigenic effects such as TGFβ induction of stroma-derived cytokines that promote cell survival. d | Another path involves the selection of clones that have lost tumour suppressive TGFβ responses but retain an intact SMAD signalling machinery. This outcome is frequent in breast carcinomas, gliomas and melanomas. As a result, cancer cells not only withstand a TGFβ-rich microenvironment but also respond to TGFβ, resulting in SMAD-dependent gene responses that in this context are profitable for metastasis. Examples include the induction of angiopoietin-like 4 (ANGPTL4) that primes breast cancer cells for extravasation, increased expression of interleukin 11 (IL11) and NOTCH ligand JAGGED1 that allows cancer cells in the bone marrow to activate osteoclasts for osteolytic metastasis, and epithelial–mesenchymal transition (EMT) that provides an invasive and tumour initiating phenotype.
Things are more devious in breast cancer, melanomas and gliomas. The cancer cell clones that prevail in these tumours retain an intact SMAD signalling machinery. These clones are disabled for TGFβ tumour suppressive responses, but in contrast to their colonic or pancreatic counterparts, the loss of the tumour suppressive responses occurs downstream of SMAD signalling (FIG. 7d). Genetic loss of CDKN2B rids cancer cells of this tumour suppressive SMAD target gene, and oncogenic drivers such as human epidermal growth factor receptor 2 (HER2) in breast carcinomas or PI3K in gliomas weaken tumour suppressor SMAD cofactors such as FOXO and C/EBPβ94,114,145. Under such conditions, cancer cells can use the remaining signalling capacity of the SMAD pathway to their advantage. Many SMAD-dependent pro-metastatic effects have been identified in such contexts. SMAD-driven EMT enhances stemness and metastatic seeding in breast cancer cells25,105,146. TGFβ signalling in breast tumours is associated with lung relapse, partly due to SMAD-dependent expression of angiopoietin-like 4, which enhances extravasation of circulating tumour cells10. SMAD-dependent induction of PTHRP (parathyroid hormone-like hormone; also known as PTHLH), IL11 (interleukin 11), CTGF (connective tissue growth factor) and JAGGED1 enhances osteolytic metastasis in breast carcinoma117,147 and melanoma cells148. SMAD-dependent activation of SOX2, PDGFB (platelet-derived growth factor beta polypeptide) and LIF supports glioblastoma stem cells114–116, and these findings provide a rationale for the clinical development of TGFβ inhibitors against glioma149.
Such SMAD-mediated gene responses would not be oncogenic in a normal cellular context, but in cancer cells they become mediators of malignancy. Aberrations in the TGFβ signalling machinery could certainly also participate in tumorigenesis, as cancer cells will use all the help they can get. However, in cancer cells that retain a functional SMAD pathway, it is the context that uses this pathway to promote cancer.
Outlook
These recent advances have further exposed the logic and power of TGFβ signalling in physiology and disease and moved the field closer to an unambiguous understanding of the context-dependent nature of TGFβ action. Looking ahead, some specific problems seem particularly worthy of future research. Structural analysis of ligand interactions with traps and co-receptors would aid the pharmaceutical development of mimics and blockers of these interactions. Moreover, structural analysis of receptor interactions with non-canonical mediators, would solve long-standing questions. Similarly, solving the structure of SMAD proteins with their partners bound to DNA would shed light on outstanding questions. A further investigation of SMAD-associated chromatin readers, DNA modifiers and miRNA processors would take our understanding beyond the recent pioneering findings in these promising areas, and quantitative analysis of SMAD dynamics and interactions in live cells would add robustness to this knowledge. It has been known that pathways crosstalk, but the increasingly apparent close interaction between the TGFβ and WNT pathways deserves special attention. Defining the biochemical links between the SMAD cycle and the Pol II cycle could reveal new principles of signal-driven transcriptional action. Genome-wide analysis of SMAD-binding sites, the co-occupying partners and the associated transcriptional outputs, would bring clarity to TGFβ response programmes in contexts of interest. A more vigorous scrutiny of non-transcriptional TGFβ effects on cell contacts, the cytoskeleton and mechanotransduction is warranted, as cell behaviour constantly relies on these processes. It would also be important to elucidate how the responsiveness to TGFβ changes as cells transit from one context to the next (that is, from ES cells to progenitors, from progenitors to differentiated cells, from these to EMT and iPS cells and from pre-malignant cells to metastatic cells). Finally, unravelling how TGFβ signalling activates cell death pathways in aspiring malignant cells would benefit in the development of cancer drugs.
Acknowledgements
The author would like to thank the members of his laboratory for their contributions and critical reading the manuscript. The authors' research in this field is supported by grants from the US National Institutes of Health (NIH).
Glossary
Myoblasts
Mesenchymal progenitor cells that are committed to differentiate into muscle cells.
Epithelial–mesenchymal transition
(EMT). A phenotypic change that is characteristic of some developing tissues and certain forms of cancer. During EMT, cells lose intercellular junctions and apical–basal polarity, become migratory and, in the case of cancer, become invasive.
Homeobox genes
A family of genes encoding transcription factors that are essential for patterning along the anterior–posterior body axis.
Pro-B cells
Cells in the earliest stage of B cell development in the bone marrow. They are characterized by incomplete immunoglobulin heavy-chain rearrangements and are defined as CD19+ cytoplasmic immunoglobulin M (IgM)– or, sometimes, as B220+CD43+ (according the Hardy classification scheme).
Latent TGFβ complex
A complex that includes a bioactive transforming growth factor-β (TGFβ) dimer non-covalently bound to the cleaved propetide of the TGFβ biosynthetic precursor. This cleavage product in turn covalently binds to latent TGFβ-binding protein (LTBP).
Nucleoporins
Family of proteins that constitute the nuclear pore complex, which is a structure that spans the nuclear envelope in eukaryotic cells.
Polyubiquitylation
Post-translational modification of proteins that involves the covalent attachment and polymerization of ubiquitin moieties to Lys chain amino groups.
WW domains
Protein interaction domains that are found in many proteins. The WW domain is characterized by a pair of Trp residues 20–22 amino acids apart, and an invariant Pro residue within a region of 40 amino acids. WW domains interact with Pro-rich regions, including those containing phospho-Ser or phospho-Thr.
Mediator complex
A multiprotein complex that functions as a transcriptional co-activator and binds to the carboxy-terminal domain of the RNA polymerase II (Pol II) holoenzyme. This complex acts as a bridge between the Pol II and transcription factors.
SWI/SNF
(Switch/sucrose nonfermentable). A chromatin-remodelling complex family that was first identified genetically in yeast as a group of genes required for mating type switching and growth on alternative sugar sources to sucrose. This complex is required for the transcriptional activation of~7% of the genome. SWI/SNF complexes exist in multiple forms made up proteins referred to as BRGl-associated factors (BAFs).
MicroRNA
(miRNA). An approximately 21–22 ribonucleotide RNA that arises from the action of the Dicer double-stranded ribonucleases on short stem–loop precursors. miRNAs initiate blocking of the targeted mRNAs, which have nucleotide sequences that are complementary to the miRNA.
Drosha
A ribonuclease III enzyme that initiates processing of microRNAs.
Autocrine
Autocrine signalling refers to when the target cell is the signal-releasing cell itself.
Primitive streak
Structure that forms during early stages of embryonic development. It establishes the first axis of symmetry and marks the beginning of gastrulation.
Melanomas
Malignant tumours derived from melanocyte precursors.
Gliomas
The most common types of malignant tumour in the brain.
Footnotes
Competing interests statement The author declares no competing financial interests.
References
- 1.Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 2.Massagué J. How cells read TGF-β signals. Nature Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 3.Huminiecki L, et al. Emergence, development and diversification of the TGF-β signalling pathway within the animal kingdom. BMC Evol. Biol. 2009;9:28. doi: 10.1186/1471-2148-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi Q, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies TRIM33 as a partner of Nodal-activated SMAD3 that binds to and disables repressive histone marks in master regulators of ES cell differentiation.
- 6.Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 7.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 8.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates, together with reference 4, that master regulators of pluripotency in the context of ES cells and of lineage determination in progenitor cells direct signal-activated SMAD proteins to many sites in the genome.
- 9.Kang Y, Chen CR, Massagué J. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 10.Padua D, et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFβ receptor for degradation. Mol. Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 12.Wu JW, et al. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-β signaling. Cell. 2002;111:357–367. doi: 10.1016/s0092-8674(02)01006-1. [DOI] [PubMed] [Google Scholar]
- 13.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial–mesenchymal transition. Curr. Opin. Cell Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Massagué J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu MY, Hill CS. Tgf-β superfamily signaling in embryonic development and homeostasis. Dev. Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nature Rev. Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 17.Kicheva A, Gonzalez-Gaitan M. The Decapentaplegic morphogen gradient: a precise definition. Curr. Opin. Cell Biol. 2008;20:137–143. doi: 10.1016/j.ceb.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Plouhinec JL, Zakin L, De Robertis EM. Systems control of BMP morphogen flow in vertebrate embryos. Curr. Opin. Genet. Dev. 2011;21:696–703. doi: 10.1016/j.gde.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, et al. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 20.Hata A, et al. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP–Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 21.Gomis RR, et al. A FoxO–Smad synexpression group in human keratinocytes. Proc. Natl Acad. Sci. USA. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 25.Scheel C, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 27.Huse M, et al. The TGFβ receptor activation process: an inhibitor- to substrate-binding switch. Mol. Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 28.Greenwald J, et al. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol. Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 29.Groppe J, et al. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 30.Moustakas A, Heldin CH. The regulation of TGFβ signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 31.Pardali E, Goumans MJ, ten Dijke P. Signaling by members of the TGF-β family in vascular morphogenesis and disease. Trends Cell Biol. 2010;20:556–567. doi: 10.1016/j.tcb.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Cheifetz S, et al. The transforming growth factor-β system, a complex pattern of cross-reactive ligands and receptors. Cell. 1987;48:409–415. doi: 10.1016/0092-8674(87)90192-9. [DOI] [PubMed] [Google Scholar]
- 33.Zakin L, De Robertis EM. Extracellular regulation of BMP signaling. Curr. Biol. 2010;20:R89–R92. doi: 10.1016/j.cub.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zilberberg L, et al. Specificity of latent TGF-β binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J. Cell Physiol. 2012;227:3828–3836. doi: 10.1002/jcp.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groppe J, et al. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- 36.Potti TA, Petty EM, Lesperance MM. A comprehensive review of reported heritable noggin-associated syndromes and proposed clinical utility of one broadly inclusive diagnostic term: NOG-related-symphalangism spectrum disorder (NOG-SSD) Hum. Mutat. 2011;32:877–886. doi: 10.1002/humu.21515. [DOI] [PubMed] [Google Scholar]
- 37.Sneddon JB, et al. Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc. Natl Acad. Sci. USA. 2006;103:14842–14847. doi: 10.1073/pnas.0606857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi M, et al. Latent TGF-β structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindsay ME, Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. doi: 10.1038/nature10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller P, et al. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science. 2012;336:721–724. doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schier AF. Nodal morphogens. Cold Spring Harb. Perspect. Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis KA, et al. β-glycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- 43.Wiater E, Harrison CA, Lewis KA, Gray PC, Vale WW. Identification of distinct inhibin and transforming growth factor β-binding sites on β-glycan: functional separation of β-glycan co-receptor actions. J. Biol. Chem. 2006;281:17011–17022. doi: 10.1074/jbc.M601459200. [DOI] [PubMed] [Google Scholar]
- 44.López-Casillas F, Wrana JL, Massagué J. β-glycan presents ligand to the TGFβ signaling receptor. Cell. 1993;73:1435–1444. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 45.Gritsman K, et al. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- 46.Gallione C, et al. Overlapping spectra of SMAD4 mutations in juvenile polyposis (JP) and JP-HHT syndrome. Am. J. Med. Genet. A. 2010;152A:333–339. doi: 10.1002/ajmg.a.33206. [DOI] [PubMed] [Google Scholar]
- 47.Marchuk DA. Genetic abnormalities in hereditary hemorrhagic telangiectasia. Curr. Opin. Hematol. 1998;5:332–338. doi: 10.1097/00062752-199809000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Xu L, Kang Y, Col S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- 49.Chen X, Xu L. Specific nucleoporin requirement for Smad nuclear translocation. Mol. Cell. Biol. 2010;30:4022–4034. doi: 10.1128/MCB.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill CS. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 2009;19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 51.Dai F, Lin X, Chang C, Feng XH. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-β signaling. Dev. Cell. 2009;16:345–357. doi: 10.1016/j.devcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.BabuRajendran N, et al. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-β effectors. Nucleic Acids Res. 2010;38:3477–3488. doi: 10.1093/nar/gkq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, et al. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 54.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 55.Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011;25:1654–1661. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koinuma D, et al. Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci. 2009;100:2133–2142. doi: 10.1111/j.1349-7006.2009.01299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fei T, et al. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 2010;20:36–44. doi: 10.1101/gr.092114.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morikawa M, et al. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 2011;39:8712–8727. doi: 10.1093/nar/gkr572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alarcón C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao S, et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuentealba LC, et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows the integration of dorsoventral BMP and anteroposterior WNT signal gradients at the level of SMAD1 phosphorylation during embryonic pattern formation.
- 62.Sapkota G, Alarcón C, Spagnoli FM, Brivanlou AH, Massagué J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 63.Aragón E, et al. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows, together with reference 59, how the nuclear kinases CDK8, CDK9 and GSK3 drive a cycle of SMAD utilization and disposal that is an integral part of canonical BMP and TGFβ pathways.
- 64.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Briochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 65.Bruce DL, Sapkota GP. Phosphatases in SMAD regulation. FEBS Lett. 2012;586:1897–1905. doi: 10.1016/j.febslet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 66.Sapkota G, et al. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-β pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- 67.Ghosh A, Shuman S, Lima CD. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol. Cell. 2008;32:478–490. doi: 10.1016/j.molcel.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin X, et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simonsson M, Kanduri M, Gronroos E, Heldin CH, Ericsson J. The DNA binding activities of Smad2 and Smad3 are regulated by coactivator-mediated acetylation. J. Biol. Chem. 2006;281:39870–39880. doi: 10.1074/jbc.M607868200. [DOI] [PubMed] [Google Scholar]
- 70.Lonn P, et al. PARP-1 attenuates Smad-mediated transcription. Mol. Cell. 2010;40:521–532. doi: 10.1016/j.molcel.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 71.Ebisawa T, et al. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 72.Al-Salihi MA, Herhaus L, Maccartney T, Sapkota G. USP11 augments TGFb signalling by deubiquitylating ALK5. Open Biol. 2010;2:120063. doi: 10.1098/rsob.120063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eichhorn PJ, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nature Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nature Cell Biol. 2012;14:717–726. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 75.Inui M, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nature Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 76.Paulsen M, Legewie S, Eils R, Karaulanov E, Niehrs C. Negative feedback in the bone morphogenetic protein 4 (BMP4) synexpression group governs its dynamic signaling range and canalizes development. Proc. Natl Acad. Sci. USA. 2011;108:10202–10207. doi: 10.1073/pnas.1100179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 78.Koinuma D, et al. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. EMBO J. 2003;22:6458–6470. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levy L, et al. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 2007;27:6068–6083. doi: 10.1128/MCB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 81.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 82.Labbé E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and wnt pathways. Proc. Natl Acad. Sci. USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Labbé E, et al. Transcriptional cooperation between the transforming growth factor-β and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 84.Nakano N, et al. Requirement of TCF7L2 for TGF-β-dependent transcriptional activation of the TMEPAI gene. J. Biol. Chem. 2010;285:38023–38033. doi: 10.1074/jbc.M110.132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 86.Naka K, et al. TGF-β–FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 87.Vincent T, et al. A SNAIL1–SMAD3/4 transcriptional repressor complex promotes TGF-β mediated epithelial–mesenchymal transition. Nature Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that formation of a SNAIL1–SMAD3–SMAD4 transcriptional complex provides a mechanism for repression of epithelial genes in the context of EMT.
- 88.Varelas X, Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 2012;22:88–96. doi: 10.1016/j.tcb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 89.Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1γ to chromatin via its PHD finger–bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol. Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 90.Ross S, et al. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J. 2006;25:4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zerlanko BJ, Bartholin L, Melhuish TA, Wotton D. Premature senescence and increased TGFβ signaling in the absence of Tgif1. PLoS ONE. 2012;7:e35460. doi: 10.1371/journal.pone.0035460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taniguchi K, Anderson AE, Sutherland AE, Wotton D. Loss of Tgif function causes holoprosencephaly by disrupting the SHH signaling pathway. PLoS Genet. 2012;8:e1002524. doi: 10.1371/journal.pgen.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thillainadesan G, et al. TGF-β-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol. Cell. 2012;46:636–649. doi: 10.1016/j.molcel.2012.03.027. [DOI] [PubMed] [Google Scholar]; Demonstrates a remarkable case in which a SMAD transcriptional complex recruits a base excision repair complex to remove repressive DNA methylation from a TGFβ target gene.
- 94.Gomis RR, Alarcón C, Nadal C, Van Poznak C, Massagué J. C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 95.Seoane J, et al. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nature Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 96.Xi Q, He W, Zhang XH, Le HV, Massagué J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor-β transcriptional program. J. Biol. Chem. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 98.Hatakeyama S. TRIM proteins and cancer. Nature Rev. Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 99.He W, et al. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 100.Dupont S, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 101.Morsut L, et al. Negative control of Smad activity by ectodermin/Tif1γ patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 102.Vincent DF, et al. Inactivation of TIF1γ cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5:e1000575. doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ozdamar B, et al. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 105.Viloria-Petit AM, et al. A role for the TGFβ–Par6 polarity pathway in breast cancer progression. Proc. Natl Acad. Sci. USA. 2009;106:14028–14033. doi: 10.1073/pnas.0906796106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows a non-canonical mode of TGFβ signalling that involves direct phosphorylation of a cell polarity regulator by the TGFβ type II receptor and that facilitates cancer cell invasion.
- 106.Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-β signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morrell NW. Pulmonary hypertension due to BMPR2 mutation: a new paradigm for tissue remodeling? Proc. Am. Thorac. Soc. 2006;3:680–686. doi: 10.1513/pats.200605-118SF. [DOI] [PubMed] [Google Scholar]
- 108.Foletta VC, et al. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J. Cell Biol. 2003;162:1089–1098. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee-Hoeflich ST, et al. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J. 2004;23:4792–4801. doi: 10.1038/sj.emboj.7600418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park KS, Gumbiner BM. Cadherin-6B stimulates an epithelial mesenchymal transition and the delamination of cells from the neural ectoderm via LIMK/cofilin mediated non-canonical BMP receptor signaling. Dev. Biol. 2012;366:232–243. doi: 10.1016/j.ydbio.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holm TM, et al. Noncanonical TGFβ signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science. 2011;332:358–361. doi: 10.1126/science.1192149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res. 2012;347:11–20. doi: 10.1007/s00441-011-1201-y. [DOI] [PubMed] [Google Scholar]
- 113.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 114.Bruna A, et al. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 115.Ikushima H, et al. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]; Shows that canonical TGFβ–SMAD signalling in the context of glioma stem cells drives expression of the pluripotency factor SOX2 to promote self-renewal.
- 116.Peñuelas S, et al. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]; Shows that canonical SMAD signalling in the context of glioma cells drives expression of pluripotency cytokine LIF and that this promotes tumour reinitiation.
- 117.Kang Y, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl Acad. Sci. USA. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovers, together with reference 118, a non-canonical mode of TGFβ signalling that involves a direct interaction of signal-activated SMAD proteins with a subset of miRNA precursors in the Drosha maturation complex.
- 120.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 121.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 122.Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop – a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-β family proteins in development and disease. Nature Cell Biol. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 125.Tan EJ, et al. Regulation of transcription factor Twist expression by the DNA architectural protein high mobility group A2 during epithelial-to-mesenchymal transition. J. Biol. Chem. 2012;287:7134–7145. doi: 10.1074/jbc.M111.291385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thuault S, et al. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial- to-mesenchymal transition. J. Biol. Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nawshad A, Medici D, Liu CC, Hay ED. TGFβ3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2–Smad4–LEF1 transcription complex. J. Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 129.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature Struct. Mol. Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 131.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]; Reports, together with reference 128, the opposing roles of BMP-induced MET and TGFβ-induced EMT during somatic cell reprogramming.
- 132.Ichida JK, et al. A small-molecule inhibitor of tgf-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maherali N, Hochedlinger K. Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roberts AB, et al. Smad3 is key to TGF-β-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 135.Yang L, Pang Y, Moses HL. TGF-β and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 138.Yang L, et al. IL-21 and TGF-β are required for differentiation of human TH17 cells. Nature. 2008;454:350–352. doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou L, et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Koinuma D, et al. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol. Cell. Biol. 2009;29:172–186. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen CR, Kang Y, Siegel PM, Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 142.Arany PR, et al. Smad3 deficiency alters key structural elements of the extracellular matrix and mechanotransduction of wound closure. Proc. Natl Acad. Sci. USA. 2006;103:9250–9255. doi: 10.1073/pnas.0602473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guasch G, et al. Loss of TGFβ signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that in the context of normal skin epithelial cells TGFβ induces growth inhibition, but in the context of pre-malignant (that is, a KRAS mutant) cells TGFβ drives apoptosis.
- 144.Levy L, Hill CS. Alterations in components of the TGF-β superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 145.Arnal-Estapé A, et al. HER2 silences tumor suppression in breast cancer cells by switching expression of C/EBPβ isoforms. Cancer Res. 2010;70:9927–9936. doi: 10.1158/0008-5472.CAN-10-0869. [DOI] [PubMed] [Google Scholar]
- 146.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial–mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mohammad KS, et al. TGF-β-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011;71:175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Anido J, et al. TGF-β receptor inhibitors target the CD44high/Id1high glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]