An Assembly-incompetent Mutant Establishes a Requirement for Dynamin Self-assembly in Clathrin-mediated Endocytosis In Vivo (original) (raw)
Abstract
Dynamin GTPase activity is required for its biological function in clathrin-mediated endocytosis; however, the role of self-assembly has not been unambiguously established. Indeed, overexpression of a dynamin mutant, Dyn1-K694A, with impaired ability to self-assemble has been shown to stimulate endocytosis in HeLa cells (Sever et al., Nature 1999, 398, 481). To identify new, assembly-incompetent mutants of dynamin 1, we made point mutations in the GTPase effector/assembly domain (GED) and tested for their effects on self-assembly and clathrin-mediated endocytosis. Mutation of three residues, I690, K694, and I697, suggests that interactions with an amphipathic helix in GED are required for self-assembly. In particular, Dyn1-I690K failed to exhibit detectable assembly-stimulated GTPase activity under all assay conditions. Overexpression of this assembly-incompetent mutant inhibited transferrin endocytosis as potently as the GTPase-defective dominant-negative mutant, Dyn1-K44A. However, worm-like endocytic intermediates accumulated in cells expressing Dyn1-I690K that were structurally distinct from long tubules that accumulated in cells expressing Dyn1-K44A. Together these results provide new structural insight into the role of GED in self-assembly and assembly-stimulated GTPase activity and establish that dynamin self-assembly is essential for clathrin-mediated endocytosis.
INTRODUCTION
The GTPase dynamin is a central player in clathrin-mediated endocytosis and synaptic vesicle recycling (Hinshaw, 2000). Dynamin is distinguished from regulatory GTPases by its low affinity for GTP and high turnover rate of GTP hydrolysis (Sever et al., 2000b; Song and Schmid, 2003). In addition, dynamin self-assembles to form higher order structures such as rings and spirals at low salt concentrations (<50 mM NaCl; Hinshaw and Schmid, 1995; Warnock et al., 1996) or at physiological salt concentrations (e.g., 150 mM NaCl) in the presence of templates including phosphatidylinositol-4,5-bisphosphate (PI4,5P2)-containing liposomes or lipid tubules (Warnock et al., 1996; Stowell et al., 1999). Self-assembly can stimulate dynamin GTPase activity as much as 100-fold (Sever et al., 1999; Stowell et al., 1999).
Dynamin is organized into multiple domains. The N-terminal GTPase domain is structurally related to other GTPases (Niemann et al., 2001), and mutations that impair GTP binding and/or hydrolysis block endocytosis (Herskovits et al., 1993; Marks et al., 2001). The C-terminal proline/arginine-rich domain (PRD) and the pleckstrin homology (PH) domain mediate dynamin's interactions with SH3 domain–containing proteins and PI4,5P2, respectively. The GTPase effector domain (GED), which interacts with the GTPase domain, the middle domain, and with itself, is believed to be involved in both self-assembly and assembly-stimulated GTPase activity. It has been proposed that GED functions as an assembly-dependent GTPase-activating protein (GAP; Sever et al., 1999). GED's role as a GAP, however, was brought into question when mutation of a proposed catalytic arginine residue, R725, within GED was found not to affect stimulated GTPase activity measured in the presence of lipid tubules (Marks et al., 2001). Thus, although it is clear that GED functions in self-assembly, it remains unclear as to whether it also plays a more direct role in stimulating dynamin's GTPase activity.
Dynamin self-assembly and assembly-stimulated GTPase activity are thought to be directly involved in severing the neck of a coated pit. Dynamin rings and spirals have dimensions that could allow them to encircle the neck of a clathrin-coated pit, and concerted conformational changes of assembled dynamin upon GTP hydrolysis could provide mechanical forces that directly promote membrane fission (Hinshaw and Schmid, 1995; Takei et al., 1995; Stowell et al., 1999). The possible role for dynamin in endocytosis was initially proposed based on analyses of mutations in shibire, the dynamin homologue (van der Bliek and Meyerowitz, 1991). Shibirets flies are paralyzed at the nonpermissive temperature because synaptic vesicle recycling is blocked (Koenig and Ikeda, 1989). In addition, morphological analysis revealed electron-dense collars around the necks of endocytic pits. The collars are reminiscent of dynamin rings and spirals produced in vitro and are believed to consist of dynamin, although this has not been directly established. Indeed, collars are not detected on the endocytic intermediates that accumulated in other tissues of shibire flies after shift to the nonpermissive temperature, suggesting that other neuron-specific proteins might be needed to generate these structures. Moreover, collars remain in place and membrane appears to flow around these constrictions for long periods after return to the permissive temperature in shibire flies (Koenig and Ikeda, 1989), bringing into question their role as “pinchases” (Roos and Kelly, 1997).
Indeed, there are several lines of evidence suggesting that dynamin alone is insufficient to drive membrane fission. For example, collar-like structures are not observed in thin-section electron micrographs when dynamin alone assembles onto PI4,5P2-containing liposomes, but addition of amphiphysin triggers the formation of electron-dense rings composed of both dynamin and amphiphysin (Takei et al., 1999). Moreover, GTP-dependent fragmentation of liposomes by dynamin is greatly enhanced by amphiphysin (Takei et al., 1999). More recently, cells expressing antisense RNA for clathrin heavy chain were shown to accumulate clathrin-coated pits with elongated necks wrapped by collar-like structures that label for endogenous dynamin. Clathrin-mediated endocytosis was severely inhibited in these cells, suggesting that dynamin assembly around the necks of endocytic intermediates is not, in itself, sufficient for severing the neck of coated pits (Iversen et al., 2003). Finally, dynamin mutants with an impaired ability to self-assemble were found to stimulate the rate-limiting step in endocytosis when overexpressed in HeLa cells (Sever et al., 1999, 2000a).
Although dynamin's self-assembly and mechanochemical properties have been clearly established in vitro, it remains an important objective to establish that these activities are required for dynamin function in vivo. By analogy, the dynamin-related MxA protein that has antiviral activity in vivo, has in vitro characteristics similar to dynamin: it can self-assemble, has assembly-stimulated GTPase activity, and can tubulate liposomes (Richter et al., 1995; Accola et al., 2002). However, although GTPase domain mutants of MxA that are defective in GTP binding and hydrolysis have no antiviral activity (Pitossi et al., 1993), MxA bearing a point mutation in the functionally conserved GED region of MxA that disrupts its ability to self-assemble and abrogates assembly-stimulated GTPase activity is fully functional in vivo (Janzen et al., 2000). Therefore, despite its in vitro properties, self-assembly is not required for the biological function of MxA in vivo. Thus, we have generated new mutations in GED that abrogate dynamin's ability to self-assemble so as to directly test the role of dynamin self-assembly in clathrin-mediated endocytosis.
MATERIALS AND METHODS
Dynamin Constructs and Generation of Recombinant Baculoviruses and Adenoviruses
Point mutations were performed on the gene for human dynamin 1 in a pBluescript vector by the QuickChange method (Stratagene, La Jolla, CA) using the oligonucleotides (Operon, Alameda, CA) indicated in Table 1. The _Kas_I-_Nhe_I fragment containing each mutation was subcloned into pVL1393 vector containing wild-type dynamin 1. To generate wild-type Dyn1-EGFP, the 3′ end of dynamin 1 (pcDNA3.1 HA-dynamin 1) was amplified by PCR (bp 1402–2595) to remove the stop codon and introduce an extra _Age_I restriction site. This fragment was digested with _Eco_RV and _Age_I and subcloned together with the 5′end of HA-dynamin 1 (_Bam_HI-_Eco_RV) into pEGFP-N1 (CLONTECH, Palo Alto, CA). Dyn1-K44A-EGFP was generated by subcloning the _Bam_HI-_Eco_RV fragment from pcDNA3.1 HA-Dyn1-K44A into the wild-type Dyn1-EGFP construct. Dyn1-I690K-EGFP was generated by amplification of the 3′ end of pBluescript vector containing Dyn1-I690K by PCR and subcloning into the _Eco_RV and _Age_I sites of the wild-type-Dyn1-EGFP construct. All constructs were confirmed by sequencing.
Table 1.
Oligonucleotide sequences used for site-directed mutagenesis
| Oligonucleotide | Sequence |
|---|---|
| I690K-f | 5′-GACCATCATGCACCTCATGAAGAACAATACCAAGGAGTTCATC-3′ |
| I690K-r | 5′-GATGAACTCCTTGGTATTGTTCTTCATGAGGTGCATGATGGTC-3′ |
| K694E-f | 5′-CTCATGATTAACAATACCGAGGAGTTCATCTTCTCGGAG-3′ |
| K694E-r | 5′-CTCCGAGAAGATGAACTCCTCGGTATTGTTAATCATGAG-3′ |
| I697K-f | 5′-GATTAACAATACCAAGGAGTTCAAATTCTCGGAGCTGCTGGCC-3′ |
| I697K-r | 5′-GGCCAGCAGCTCCGAGAATTTGAACTCCTTGGTATTGTTAATC-3′ |
Recombinant baculoviruses were generated by using BaculoGold linearized baculovirus DNA according to the manufacturer's instructions (PharMingen, San Diego, CA). For recombinant adenoviruses, the _Nhe_I-_Hin_dIII fragment with an appropriate mutation was first subcloned into pADTetT3T7 vector containing the gene for human dynamin 1 wild type. HEK293-cre4 cells were transfected with the resulting pADTetT3T7 DNA as described (Hardy et al., 1997).
Dynamin Expression and Purification
Dynamin was expressed in Tn5 insect cells and purified either by ion-exchange chromatography after ammonium sulfate precipitation (Warnock et al., 1996; Muhlberg et al., 1997) or by affinity chromatography using amphiphysin II SH3 domain conjugated to glutathione _S_-transferase (Marks et al., 2001). Purified dynamin was dialyzed against storage buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT) and stored at –80°C after freezing in liquid nitrogen. Protein concentration was determined by OD280 using the extinction coefficient of 56170 for dynamin.
Liposomes and Lipid Nanotubules Containing PI4,5P2
All lipids were dissolved in chloroform at the following concentrations: dodecanoic phosphatidylcholine (PC, 10 mg/ml, Calbiochem, La Jolla, CA), synthetic galactoceramide (Galcer, 10 mg/ml, Avanti, Birmingham, AL), cholesterol (1 mg/ml, Calbiochem), and phosphatidylinositol-4,5-bisphosphate (PI4,5P2, 1 mg/ml, Calbiochem or Avanti). For liposome preparation, 2.5 ml of PC and 0.1 ml of PI4,5P2 were mixed, and chloroform was evaporated with nitrogen gas. The lipids were resuspended in 3.25 ml of degassed buffer containing 50 mM HEPES, pH 7.5, and 150 mM KCl. Aliquots of 250 μl were subjected to repeated (five times) freeze-thawing by transferring between liquid nitrogen and a water bath at 37°C followed by vortexing and then stored at –80°C. Right before use, each aliquot was thawed and extruded through a 0.4-μm polycarbonated filter with pressure. For preparation of lipid nanotubules, PC (4 μl), Galcer (4 μl), cholesterol (3 μl), and PI4,5P2 (25 μl) were mixed. After evaporation of chloroform with nitrogen gas, lipids were resuspended in 100 μl of 20 mM HEPES, pH 7.5, and 100 mM NaCl with vortexing followed by sonication for 2 min. Formation of lipid nanotubules was confirmed by electron microscopy. Final concentration of PI4,5P2 is 250 μM.
GTPase Assay
GTP hydrolysis was followed by monitoring released Pi, which was quantitated by a colorimetric method. The color reagent was prepared by dissolving malachite green (50 mg) and ammonium molybdate tetrahydrate (500 mg) in 50 ml of 1 N hydrochloric acid followed by filtration through 0.45-μm membrane. The GTPase reaction was initiated by mixing a solution (10 μl) of dynamin (0.1–1.0 μM) in 20 mM HEPES, pH 7.5, 150 mM KCl, 2 mM MgCl2 with or without templates (typically 2 μl of liposomes, 1 μl of lipid nanotubules) with a solution (10 μl) of GTP (0–1.0 mM) in the same buffer, and the reaction mixture was incubated for increasing time at 37°C. The reaction was stopped by adding 5 μl of 0.5 M EDTA, pH 8.0. Optical density at 650 nm was measured against the standard solution of Pi after addition of malachite-green reagent (150 μl). This method agrees well with product separation of [α-32P]GDP from [α-32P]GTP by TLC. However, in some cases contamination of nonreacting radioactive species with similar mobility to GTP gave lower rates of GTP hydrolysis in the radioactivity assay. A representative data set of several repeated measurements was presented for each experiment.
Velocity Sedimentation Assay
Dynamin self-assembly at low salt (10 mM KCl) or at high salt (150 mM KCl) in the presence of lipid nanotubules was tested by sedimentation after highspeed centrifugation. Dynamin (1–2 μM) was incubated 10–20 min at room temperature in the GTPase assay buffer. The reaction mixture (50 μl) was spun for 10 min at 14,000 rpm in a microfuge at 4°C. The pellet was dissolved in 20 μl of solubilization buffer (4 M urea, 0.5% SDS, 100 mM Tris·HCl, pH 7.0). The supernatant fraction was precipitated by adding 1% Triton X-100 (1 μl) and 10% trichloroacetic acid (10 μl). The precipitates were collected by spinning 10 min in a microfuge at 4°C, rinsed with ice-cold acetone (300 μl), dried in the air after removal of acetone, and dissolved in 1× SDS-PAGE buffer (20 μl). The relative amount of protein for each fraction was analyzed by SDS-PAGE followed by Coomassie staining.
Cross-linking
Cross-linking was performed as described previously with minor modifications (Muhlberg et al., 1997). Cross-linking was initiated by adding 5 μl of 100 mM fresh Bis[sulfonylsuccinimidy]suberate (BS3) to 95 μl of 4.0 μM dynamin in 20 HEPES, pH 7.5, 2 mM MgCl2, and 10 or 150 mM KCl. The reaction mixture was incubated 5 min at room temperature, stopped by adding 5 μl of 2.0 M glycine. Protein was precipitated after addition of 1% Triton X-100 (2 μl) with 10% trichloroacetic acid. The suspension was spun in a microfuge tube for 10 min at 4°C. The pellet was rinsed with ice-cold acetone (0.3 ml), spun 10 min at 4°C, and resuspended in 100 μl of gel loading buffer. The reaction products were analyzed by SDS-PAGE on 6.5% polyacrylamide gel.
Immunoprecipitation of Dynamin 1/Dynamin 2 Heterooligomers
tTA-HeLa cells (∼50% confluent in a 15-cm dish) infected for 16 h with adenovirus encoding HA-Dyn1 (WT or I690K) were harvested in 1 ml of 5 mM EDTA/PBS, pelleted, and resuspended in 1.2 ml of NP-40 buffer (50 mM Tris·Cl, pH 8.0, 150 mM NaCl, 1% NP-40) containing the cocktail of protease inhibitors from Sigma (St. Louis, MO). Cells were incubated 30 min on ice for lysis and spun 10 min in a microfuge at 14,000 rpm at 4°C. The supernatant (0.3 ml), mixed with 0.6 ml of 150 mM NaCl in 50 mM Tris·Cl, pH 8.0, was transferred to protein A-Sepharose (20 μl) rinsed with 150 mM NaCl in 50 mM Tris·Cl, pH 8.0, and gently mixed for 4 h at 4°C. The resulting precleared supernatant was incubated overnight with protein A-Sepharose CL-4B (20 μl) together with pAb 1441, a dynamin 2-specific rabbit antisera, which was generated against the sequence (KDQAENEDGAQE) between the PH domain and GED not conserved between Dyn1, Dyn2, and Dyn3. Western blot analysis against purified Dyn1 and Dyn2 confirmed the antibody specificity (D. E. Warnock and S. L. Schmid, unpublished data). The beads were then washed three times with 200 μl of 150 mM NaCl in 50 mM Tris·Cl, pH 8.0, and proteins on the beads were analyzed on a 10% denaturing polyacrylamide gel followed by Western blot with 12CA5, an antibody recognizing HA-tag of dynamin 1.
Internalization Assays
Internalization assays were performed exactly as described (van der Bliek et al., 1993) using biotinylated transferrin as ligand and assessing its internalization into an avidin-inaccessible compartment. For chronic infections, tTA-HeLa cells (1.0 × 106 in a 35-mm dish) were infected for 16–18 h with 10 μl of recombinant adenovirus encoding wt or mutant dynamin, which gave uniform infection and maximum protein expression determined by immunofluorescence. The relative protein expression level was monitored by Western analysis.
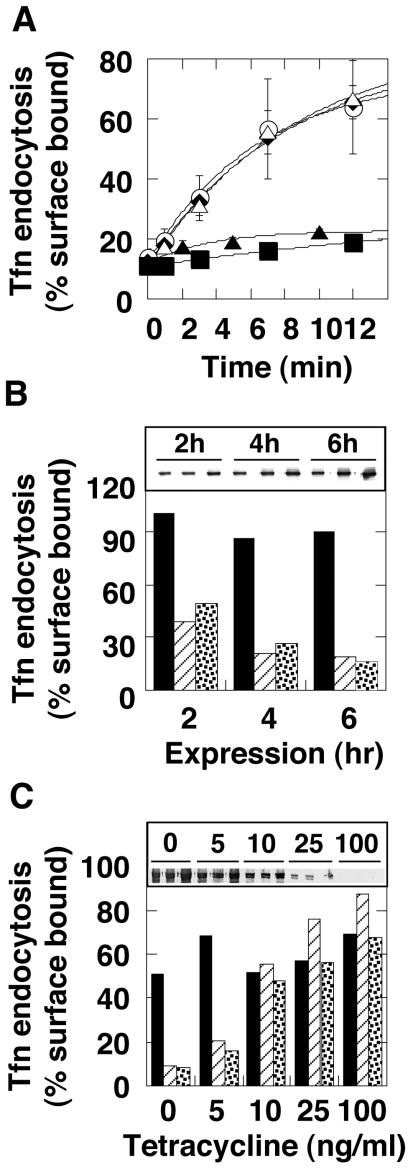
To examine the kinetics of inhibition after bolus protein expression (Ceresa et al., 2001), cells were infected with excess recombinant adenoviruses (100 μl) for 2 h at 37°C. The medium was replaced by medium containing 10 μg/ml cycloheximide and the cells were incubated 3 h. After removal of cycloheximide, cells were incubated for varying lengths of time (2, 4, and 6 h) to allow protein expression. To examine the relative potency of Dyn1-I690K compared with Dyn1-K44A, the amount of protein expression was controlled by adding tetracycline (0, 5, 10, 25, 100, 1000 ng/ml, final) together with recombinant adenovirus and transferrin uptake assay was performed at 16 h postinfection.
TIR-FM and Immunofluorescence Microscopy
Swiss 3T3 cells were grown to 70% confluency in 100-mm tissue culture dishes in DMEM + 10% FCS and transfected with 20 μg DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Cells were allowed to recover for 2 h in DMEM + 10% FCS and then detached from the dishes with trypsin-EDTA, replated on acid-washed glass coverslips (Corning, Corning, NY), and allowed to attach overnight. The following day, the medium was removed and replaced with DMEM without phenol red, 10% FCS, 10 mM HEPES, pH 7.5. Coverslips were then mounted on a slide, sealed with valap (1:1:1 vasoline, lanolin, paraffin), and imaged at 37°C.
Cells were viewed with a 100× 1.45 NA objective using an inverted Nikon TE2000U microscope with through-the-objective TIR fluorescence microscropy (Garden City, NY). The wavelength and the intensity of light from a KrAr laser were controlled with a polychromatic acousto-optical modulator (PCAOM), and light was passed through a dual-bandpass dichromatic mirror. Images were obtained using an Orca II-ER camera (Hamamatsu, Malvern, PA) using Metamorph software (Universal Imaging, Downington, PA) at 2-s intervals.
For indirect immunofluorescence, cells were transfected as described above and fixed for 3 min in methanol at –20°C. Cells were permeabilized with PBS containing 0.1% Triton-X 100 and 2% BSA and stained with the X22 antibody that recognizes clathrin heavy chain (3 μg/ml) and rhodamine-conjugated secondary antibody (Molecular Probes, Eugene, OR). Images were obtained by wide-field epifluorescence microscopy using a Zeiss Axiophot microscope, 63×, 1.4 NA objective, and Axiocam camera (Zeiss, Thornwood, NY).
Subcellular Fractionation of Cells Overexpressing Dynamins
tTA-HeLa cells (1.0 × 106 cells) infected with adenoviruses encoding dynamins were harvested and resuspended in 200 μl of buffer A (20 mM HEPES, pH 7.5, 1 mM MgCl2, 2 mM EGTA, 0.25 M sucrose). Cells were lysed by freeze-thawing five times, followed by centrifugation in a microfuge at 14,000 rpm for 10 min at 4°C. The supernatant was saved and the pellet was resuspended in 200 μl of buffer A. Samples were analyzed on a 10% denaturing polyacrylamide gel and Western blotted with a 12CA5 antibody recognizing HA-tag fused to dynamin 1 and an antitubulin α antibody from Sigma.
RESULTS
Dyn1-I690K Is Assembly Defective
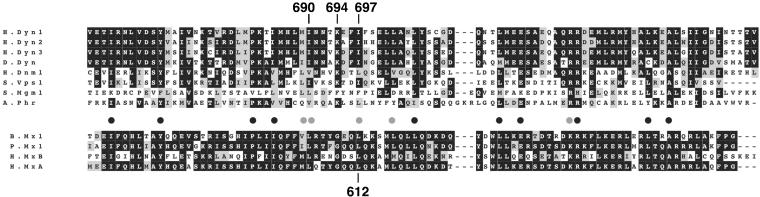
Previous mutations in the GED region of dynamin, Dyn1-K694A and Dyn1-R725A, exhibited impaired assembly activity both in vitro (Sever et al., 1999) and in vivo (Marks et al., 2001); however, neither mutation completely abrogated dynamin's ability to self-assemble. We therefore sought to identify other amino acids in dynamin that might play an essential role in self-assembly. We focused on three residues in the GTPase effector domain (GED) of dynamin, Ile 690, Lys 694, and Ile 697. Ile 697 corresponds to Leu 612 in the GED region of MxA (Figure 1). Mutation of Leu 612 to Lys disrupted MxA self-assembly and abolished its assembly stimulated GTPase activity, but did not affect its antiviral activity (Janzen et al., 2000). Lys 694 was chosen because its mutation to Ala, a hydrophobic residue, impaired, but did not abrogate dynamin self-assembly (Sever et al., 1999). It was mutated to Glu with an expectation that the opposite charge would more severely impair dynamin self-assembly. Finally, Ile 690 was also mutated to lysine because it is conserved among dynamins (Figure 1) and could be part of an amphipathic α-helix involved in dynamin self-assembly.
Figure 1.
Sequence alignment of dynamin's GTPase effector domain. Alignment was performed using the BCM Search Launcher: H.Dyn1, human dynamin 1 (aa 659–750; van der Bliek et al., 1993); H.Dyn2, human dynamin 2 (aa 649–740; Diatloff-Zito et al., 1995); H.Dyn3, human dynamin 3 (aa 649–740; Nagase et al., 1998); D.Dyn, D. melanogaster dynamin (aa 656–747; Chen et al., 1991; van der Bliek and Meyerowitz, 1991); H.Dnm1, human dynamin-related protein (aa 645–736; Shin et al., 1997); S.Vps1, S. cerevisiae vacuolar sorting protein (aa 618–704; Rothman et al., 1990); S.Mgm1, S. cerevisiae mitochondrial structure protein (aa 801–893; Jones and Fangman, 1992); A.Phr, A. thaliana phragmoplastin (aa 520–613; Gu and Verma, 1996); B.Mx1, bovine interferon-induced GTP-binding protein Mx1 (aa 566–654; Ellinwood et al., 1998); P.Mx1, porcine Mx1 protein (aa 575–663; Muller et al., 1992); H.MxB, human MxB protein (aa 623–714; Aebi et al., 1989); H.MxA, human MxA protein (aa 574–662; Aebi et al., 1989). Black boxes represent identical sequences and gray boxes amino acid residues with similar biochemical properties. Similarly, black and gray dots represent residues conserved or similar between the Mx proteins and dynamin-related proteins. Numbers above the alignment indicate amino acid residues of human dynamin 1 that were mutated to Lys, Glu, and Lys, respectively. The number below the alignment indicates the amino acid residue of MxA that abolished its self-assembly when mutated to Lys.
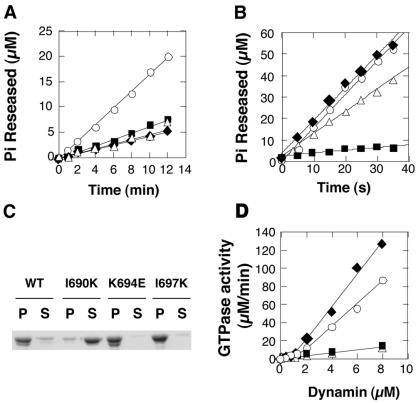
Dynamin's GTPase activity is diagnostic of self-assembly. Dynamin's basal GTPase activity is measured at physiological salt concentration (150 mM KCl) and at low concentrations of dynamin, conditions that disfavor self-assembly (Warnock et al., 1996). In contrast, at low salt concentrations (<50 mM KCl), dynamin can self-assemble into higher order structures, which hydrolyze GTP at a stimulated rate compared with basal activity. Therefore, the absence of GTPase stimulation at lower salt concentrations is indicative of a defect in self-assembly. The rates of GTP hydrolysis for individual mutants were examined at a physiological GTP concentration (100 μM) under both low and high salt conditions. At 150 mM KCl, each of the three mutations in GED showed small (about twofold), but significant effects on basal GTPase activity (Figure 2A, Table 2), suggesting some functional interactions between GED and the GTPase domain, even in dynamin's unassembled state. Incubation of wild-type dynamin (Dyn1-WT) under low salt conditions (5 mM KCl) that favor self-assembly, results in an ∼50-fold stimulation of GTPase activity over the basal rate (Figure 2B, Table 2). Under these conditions Dyn1-K694E exhibited an assembly-stimulated GTPase activity that was comparable to Dyn1-WT, and Dyn1-I697K was slightly impaired (Table 2). In contrast, Dyn1-I690K showed only minimally enhanced GTPase activity when assayed under low salt assembly conditions (Figure 2B, Table 2). Furthermore, unlike Dyn1-WT, K694E, or I697K, Dyn1-I690K was not sedimentable at low salt by centrifugation, suggesting that Dyn1-I690K is assembly-defective (Figure 2C).
Figure 2.
Dyn1-I690K lacks assembly-stimulated GTPase activity. Basal (A) and assembly-stimulated GTPase activity (B) was measured at high (150 mM KCl) and low (5 mM KCl) salt concentrations, respectively, as described in MATERIALS AND METHODS. The concentration of Pi released was plotted as a function of time: wt (○), I690K (▪), K694E (♦), I697K (▵). (C) Self-assembly at 10 mM KCl was examined by velocity sedimentation, followed by the SDS-PAGE analysis of the pellet (P) and supernatant fractions (S). (D) Dependence of GTPase activity on dynamin concentration. The rate of GTP hydrolysis was measured at high GTP concentration (1.0 mM) and plotted as a function of dynamin concentration: wt (○), I690K (▪), K694E (♦), I697K (▵). The GTPase activity of dynI690K is linearly proportional to its concentration.
Table 2.
Relative GTPase activity of dynamin mutants
| wt | I690K | K694E | I697K | |
|---|---|---|---|---|
| Basal | 1.75 ± 0.24 | 0.65 ± 0.20 | 0.61 ± 0.19 | 0.59 ± 0.10 |
| Low Salt | 103 ± 9.2 | 6.81 ± 0.89 | 100 ± 8.4 | 71.6 ± 15.9 |
| Liposome | 2.98 ± 0.59 | 0.69 ± 0.29 | 6.27 ± 1.19 | 0.74 ± 0.28 |
| Lipid Tubule | 115 ± 24 | 3.01 ± 0.61 | 150 ± 41 | 122 ± 42 |
Another indication of dynamin self-assembly is the observation that dynamin GTPase activity shows positive cooperativity with increasing protein concentration (Warnock et al., 1996, 1997). Thus, at physiological salt concentrations and protein concentrations higher than 2.0 μM, the GTPase activity of wild-type dynamin exhibits an abrupt increase reflecting higher-order protein-protein interactions that disproportionately enhance GTPase activity (Figure 2D, ○). A similar cooperative behavior is observed with Dyn1-K694E, whereas the specific activities for Dyn1-I690K and Dyn1-I697K remained constant even up to 8.0 μM dynamin. From these data we conclude that the Dyn1-K694E mutation, in contrast to the Dyn-K694A mutation (Sever et al., 1999), does not affect dynamin's ability to self-assemble in solution, whereas mutations of both I690K and I697K alter dynamin's self-assembly properties. The effects of the I697K mutation are most strongly manifested under physiological salt conditions, but are largely overcome by incubation in low salt. Strikingly, self-assembly of the I690K mutation appears to be severely impaired under both conditions.
Templates Cannot Induce Dyn1-I690K to Self-assemble
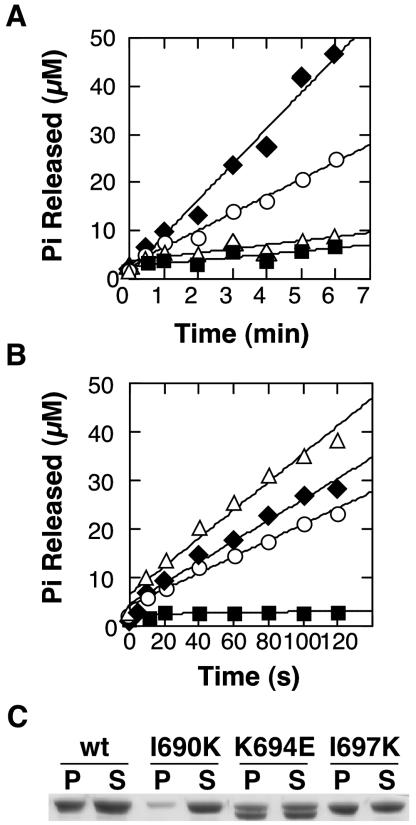
Previous studies have shown that the assembly defect in Dyn1-K694A observed at low salt can be overcome by incubation in the presence of templates that drive dynamin self-assembly (Sever et al., 1999; Marks et al., 2001). We therefore examined dynamin's assembly-stimulated GTPase activity, at physiological salt, in the presence of PI4,5P2-containing liposomes and lipid tubules (LTs). Liposomes were prepared by passing lipid mixtures through 0.4-μm filters and have diameters greater than that of the LTs, which are ∼50 nm in diameter (Stowell et al., 1999). Consistent with findings of others (Stowell et al., 1999; Barylko et al., 2001), we found that liposomes were significantly less effective as templates for assembly-stimulated GTPase activity than LTs (Table 2). Nevertheless, consistent with our previous experiments, the liposome-stimulated GTPase activity of Dyn1-K694E was equal to or greater than that for Dyn1-WT (Figure 3A, Table 2). In contrast, neither Dyn1-I697K nor Dyn1-I690K exhibited enhanced GTPase activity in the presence of liposomes, suggesting a defect in self-assembly on this template.
Figure 3.
Defective self-assembly of DynI690K is not rescued by templates. GTPase activity was initiated by mixing dynamin [1.0 μM for liposomes (A), and 0.1 μM for lipid tubules (B)] with GTP (100 μM): wt (○), I690K (▪), K694E (♦), I697K (▵). (C) Self-assembly on lipid tubules at physiological salt concentrations (150 mM KCl) was followed by a velocity sedimentation assay and SDS-PAGE analysis of the recovered pellets (P) and supernatants (S).
A different result was obtained when LTs, whose diameter is comparable to that of dynamin collars, were used as templates for self-assembly. Assembly of Dyn1-WT around LTs stimulates GTPase activity ∼100-fold and elicits its maximum GTPase rate (Table 2). The LT-stimulated GTPase activity of Dyn1-K694E was comparable to Dyn1-WT (Figure 3B, ♦, ○), confirming our previous results that this mutation does not have an adverse effect on dynamin self-assembly. The GTPase activity of Dyn1-I697K (▵) was also fully stimulated by LTs, suggesting that the assembly defect of this mutation can be overcome in the presence of an ideal assembly template. In contrast, the Dyn1-I690K mutant (▪) was severely impaired in assembly-stimulated GTPase activity, even in the presence of LTs, and exhibited only ∼3% of Dyn1-WT activity in this assay (Table 2). These data suggest that Dyn1-I690K cannot form higher-order structures, even in the presence of templates. This was confirmed by a velocity sedimentation assay. Dyn1-WT, Dyn1-K694E, and Dyn1-I697K bound to and were cosedimented with PI4,5P2-containing LTs, suggesting that these proteins form higher-order structures on lipid tubules (Figure 3C). Strikingly, Dyn1-I690K did not bind to or cosediment with LTs after centrifugation. When examined by electron microscopy highly ordered helical structures, which were observed for Dyn1-WT, were not detected for Dyn1-I690K (unpublished data). From these results, we conclude that the I690K mutation in GED potently blocks dynamin self-assembly with or without templates.
The Effect of I690K GED Mutation on Dynamin Oligomerization
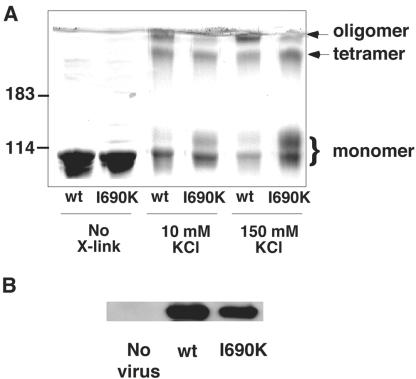
Unassembled dynamin exists in equilibrium between monomeric and tetrameric states (Muhlberg et al., 1997; Binns et al., 2000), which can be detected by chemical cross-linking (Hinshaw and Schmid, 1995; Muhlberg et al., 1997). Cross-linking studies performed under both low-salt (assembled) and high-salt (unassembled) conditions revealed interactions between GED and the GTPase domain (Muhlberg et al., 1997). To test whether the I690K mutation disrupts tetramer formation as well as higher-order assembly, we performed cross-linking reactions with bis[sulfonylsuccinimidyl]suberate (BS3) at low-(10 mM KCl) and high-(150 mM KCl) salt concentrations (Figure 4). PAGE followed by Coomassie staining revealed three major products for both Dyn1-WT and Dyn1-I690K: one migrating slightly slower than untreated proteins, reflecting intramolecular cross-linking, one migrating with an apparent molecular weight of ∼400 kDa, corresponding to dynamin tetramers, and a third band trapped at the stacking gel interface, corresponding to higher molecular weight oligomers (Figure 4). Consistent with our previous observations, the largest molecular weight species was significantly reduced in Dyn1-I690K compared with Dyn1-WT. In contrast, cross-linking of Dyn1-I690K tetramers appeared to be as efficient as Dyn1-WT.
Figure 4.
I690K mutation does not inhibit tetramer formation of dynamin. (A) Dyn1-WT and Dyn1-I690K and Dyn1-WT were cross-linked, as indicated, with BS3 at low-(10 mM KCl) and high-(150 mM KCl) salt concentrations, and their products were analyzed by SDS-PAGE on a 6.5% polyacrylamide gel. Cross-linked species are identified on the right, based on molecule weight markers shown on the left. (B) Coimmunoprecipitation of exogenously expressed HA-Dyn1 with endogenously expressed dynamin 2. The soluble fraction of cell extracts from cells overexpressing HA-Dyn1-WT, or I690K was immunoprecipitated with 1441, an antidynamin 2 antibody, and Western blotted with 12CA5, an anti-HA antibody.
The formation of dynamin tetramers in vivo can be detected by coimmunoprecipitation of antigenically distinct (e.g., myc- and HA-tagged) populations of exogenously expressed dynamin 1 (Okamoto et al., 1999). Similarly, exogenously expressed dynamin 1 will form heterotetramers with endogenous dynamin 2 (Altschuler et al., 1998). Thus, the ability of Dyn1-I690K to form tetramers in vivo could be tested by selective immunoprecipitation of endogenous dynamin 2 followed by Western blotting with a 12CA5 antibody that specifically recognizes the HA-tag of exogenously expressed HA-Dyn1-WT and HA-Dyn1-I690K. The data in Figure 4B show that Dyn1-I690K and Dyn1-WT were efficiently coprecipitated with endogenous dynamin 2. Thus, we conclude that the I690K mutation in GED disrupts formation of higher-order structures such as rings and spirals, but does not block formation of dynamin tetramers.
Assembly Defective Dynamin Inhibits Receptor-mediated Endocytosis
Our data establish that the I690K mutation in GED disrupts dynamin self-assembly regardless of templates or ionic strength. To examine the role of dynamin self-assembly in clathrin-mediated endocytosis, we performed transferrin internalization in tTA-HeLa cells expressing mutant dynamins (I690K, K694E, I697K). As expected, internalization of transferrin was indistinguishable in cells expressing either Dyn1-WT or the Dyn1-K694E mutant, which had wild-type self-assembly activity (Figure 5A). Perhaps less expected, endocytosis in cells overexpressing Dyn1-I697K was also unaffected compared with wild-type controls, even though this mutant exhibited impaired self-assembly and assembly-stimulated GTPase activity under some assay conditions. In contrast, the assembly-defective Dyn1-I690K mutant inhibited transferrin internalization as potently as Dyn1-K44A, a GTPase domain mutant known to dominantly interfere with endogenous dynamin (Damke et al., 1994; Figure 5A).
Figure 5.
Assembly-defective dynamin inhibits clathrin-mediated endocytosis. (A) Effects of chronic expression. Transferrin uptake was followed in tTA-HeLa cells infected for 16 ± 2 h with recombinant adenovirus encoding Dyn1-WT (○), Dyn1-K44A (▴), Dyn1-I690K (▪), Dyn1-K694E (♦), or Dyn1-I697K (▵). (B) Effects of bolus protein expression. Cells were infected with 10 times more virus than used in A in the presence of cycloheximide to accumulate dynamin mRNA. Transferrin uptake was measured 2, 4, and 6 h after removal of the cycloheximide block. Dyn1-WT (solid bars), Dyn1-K44A (striped bars), Dyn1-I690K (dotted bars). Protein expression was confirmed by Western analysis (inset). (C) Dose-response curves for relative potency. The level of protein expression was controlled by tetracycline during chronic infection as in A and the relative extent of transferrin uptake was monitored. Dyn1-WT (solid bars), Dyn1-K44A (striped bars), Dyn1-I690K (dotted bars). The level of protein expression at varying tetracycline (ng/ml) was monitored by Western analysis (inset).
The internalization assay was performed 14–16 h postinfection, and we observed significant changes in cell morphology, including membrane blebbing and infolding in cells infected with recombinant adenovirus containing Dyn1-I690K, by both electron microscopy and light microscopy (unpublished data). It has been proposed that dynamin may function as a regulatory GTPase and that the assembly-dependent GAP activity encoded by GED plays a role in negatively regulating dynamin signaling (Sever et al., 1999). Therefore, it was possible that the morphological changes induced by Dyn1-I690K reflected the activity of an unregulated signaling molecule. Indeed, overexpression of the ubiquitously expressed isoform of dynamin 2 can activate signaling pathways that trigger apoptosis (Fish et al., 2000). Therefore, to rule out possible indirect effects of the mutant protein on endocytosis due to long-term expression, we used a protocol to acutely induce expression of mutant dynamins followed by immediate analysis of endocytosis (Ceresa et al., 2001). For these experiments, cells were infected with a high multiplicity of infection in the presence of cycloheximide and in the absence of tetracycline to allow accumulation of virally encoded dynamin mRNA, but not protein. Cycloheximide was then removed to induce protein expression (Figure 5B, inset), and transferrin internalization was examined after 2, 4, and 6 h, before any changes in cell morphology were observed. The kinetics of inhibition of endocytosis by Dyn1-I690K were identical to those of Dyn1-K44A, suggesting that both are similarly potent inhibitors of clathrin-mediated endocytosis (Figure 5B) and that the inhibitory effects on endocytosis are direct.
GTPase defective dynamin 2 mutants are believed to inhibit endocytosis in HeLa cells by coassembling with endogenous dynamin 2 and interfering with its function (Herskovits et al., 1993). Thus, the potency of inhibition is thought to reflect the ability of the exogenous mutant dynamin to bind to and sequester endogenous dynamin. Indeed Dyn2-K44A mutants are about fourfold more potent than Dyn1-K44A mutants suggesting that homo-oligomerizion of the two isoforms is more efficient than the formation of Dyn2-Dyn1 hetero-oligomers (Altschuler et al., 1998). Dyn1-I690K is defective in higher order self-assembly; hence, it was possible that its mechanism of dominant interference might be different. For example, given its defect in assembly-stimulated GTPase activity, Dyn1-I690K might remain in the GTP-bound form and inhibit endocytosis by sequestering dynamin-binding partners necessary for vesicle formation. To determine whether Dyn1-I690K also inhibited endocytosis by interference with endogenous dynamin 2, we compared the relative potency of the dominant interference of Dyn1-I690K with that of Dyn1-K44A. For this experiment, the level of protein expression was varied in a controlled manner using tetracycline (Figure 5C, inset). As can be seen (Figure 5C), the inhibitory effect of Dyn1-I690K showed a dose dependence similar to that of Dyn1-K44A. These data are consistent with the coimmunoprecipitation results (Figure 4B) and suggest that the mechanism of dominant-interference by Dyn1-I690K is the coassembly of mutant dynamin 2 with endogenous dynamin 2 into tetramers and the inability of the resultant hetero-tetramers to self-assemble.
Dynamin Self-assembly Is Required for Membrane Morphogenesis in Endocytosis
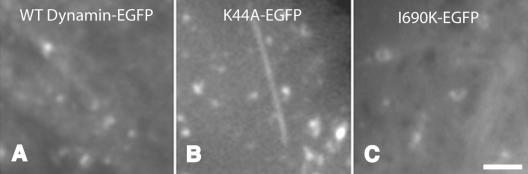
Our data establish Dyn1-I690K as a new class of dominant negative dynamin mutant that inhibits endocytosis because of its inability to self-assemble into higher-order structures, presumably rings and spirals. To determine whether the endocytic intermediates trapped upon overexpression of this new class of mutant dynamins differ from those that accumulate in the presence of assembly competent, GTPase domain mutants, we generated dynamin-EGFP fusion proteins and examined the nature of dynamin-associated endocytic intermediates in transfected cells using total internal reflection fluorescence microscopy (TIR-FM).
Consistent with previous observations (Merrifield et al., 2002; Rappoport and Simon, 2003), live cell imaging by TIR-FM of wild-type Dyn1-EGFP–expressing cells revealed the transient appearance of punctate structures at the cell surface (Figure 6A, Movie 1). In contrast, Dyn1-K44A-EGFP overexpression induced the accumulation of long dynamin-positive tubules, in addition to short, rod-like structures (Figure 6B, Movie 2). The structures observed in Dyn1-I690K-EGFP–expressing cells were dramatically different. Dyn1-I690K-EGFP expression caused the formation of consistently shorter, worm-like structures (Figure 6C, Movie 3). Thus, although the effect of overexpression of the two dominant-negative dynamin mutants on transferrin internalization is biochemically indistinguishable, the endocytic intermediates that accumulate in cells expressing these two classes of mutants are structurally distinct.
Figure 6.
Structurally distinct endocytic intermediates accumulate in cells expressing dyn1-K44A and dyn1-I690K. Shown are individual frames from Movies 1–3 taken using total internal reflection fluorescence microscopy of Swiss 3T3 cells expressing Dyn1-WT-EGFP, Dyn1-K44A-EGFP, or Dyn1-I690K-EGFP, as indicated. Movies are taken 1 frame/2 s. Scale bar, 2 μm.
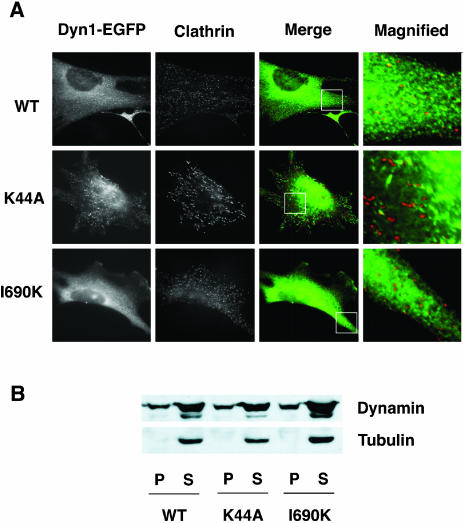
In fixed cells, overexpression of wild-type Dyn1-EGFP, Dyn1-K44A-EGFP, and Dyn1-I690K-EGFP each show partial colocalization with clathrin, consistent with the their association with coated pits (Merrifield et al., 2002; Figure 7A). Subcellular fractionation of cells infected with adenoviruses encoding the dynamins revealed that a fraction of overexpressed Dyn1-I690K similar to Dyn1-WT was localized to particulate membrane fractions (Figure 7B) This confirms that despite the inability of Dyn1-I690K to bind to lipid tubules in vitro, it can be targeted to the plasma membrane in vivo, presumably through interactions with SH3-domain–containing dynamin partners or through hetero-dimerization with endogenous dynamin. Together, these data suggest that dynamin self-assembly is necessary to stabilize and elongate the neck regions of invaginated coated pits.
Figure 7.
Assembly defective Dyn1-I690K is recruited to the endocytic site. (A) WT-dynamin, K44A dynamin, and 1690K-dynamin1 all partially colocalize with clathrin. The images show indirect immunofluorescence of endogenous clathrin in Swiss 3T3 cells transiently transfected with either Dyn1-EGFP wild-type (top), K44A (middle), or I690K (bottom). The columns of panels show the localization of (left-to-right) dynamin-EGFP alleles, clathrin (X22 antibody), merge of dynamin (green) and clathrin (red), and an enlargement of the region within merge panel. (B) Similar to Dyn1-wt, a fraction of assembly defective Dyn1-I690K is localized to the plasma membrane.
DISCUSSION
Dynamin GTPase activity is controlled in an assembly-dependent manner. Our results confirm the proposed role of dynamin's GED in assembly-stimulated GTPase activity and provide new structural insight into GED interactions in self-assembly. Previous studies established that the K694A mutation in GED impaired dynamin's ability to self-assemble in low salt (Sever et al., 1999), although assembly onto a microtubule template at low salt or onto lipid tubules at physiological salt was unaffected (Sever et al., 1999; Marks et al., 2001). We hypothesized that the positive charge on K694 might be important for protein interactions involved in self-assembly and that replacement of the basic residue with an acidic residue would generate a more severely impaired mutant. Instead, we found that the K694E mutant was, if anything, more effective at self-assembly than wild-type dynamin. Nevertheless, we identified two additional mutations in this region of GED, I690K, and I697K, that impaired dynamin's ability to self-assemble. Of these, the I690K mutation was the most severe and completely abrogated dynamin's ability to self-assemble and its assembly-stimulated GTPase activity, as assessed both in solution and on PI4,5P2-containing templates.
Our data suggest that an amphipathic α-helix in GED, defined by well-conserved hydrophobic residues at 690 and 697 and a charged residue at 694 (Figure 1), is involved in higher-order dynamin self-assembly. Mutations in this region of GED appear not to perturb tetramerization of dynamin. Nonetheless, each of the mutations in GED described here have minor, but significant effects on dynamin's basal rate of GTP hydrolysis, suggesting a role for GED-GTPase interactions in regulating GTPase activity, even in the unassembled state. Previous work had identified, by deletion analysis, two adjacent α-helical segments in GED (residues 654–681 and 712–740 of dynamin 2) that are required for the formation of dynamin tetramers in the unassembled state (Okamoto et al., 1999). Thus GED interactions are critical for both tetramerization and higher-order assembly.
Structural studies of dynamin's GTPase domain have revealed a hydrophobic pocket along its surface and it has been suggested that GED might occupy this region (Niemann et al., 2001). In contrast, other studies have suggested that GED interacts with an α-helical region in the middle domain for tetramerization (Okamoto et al., 1999). Studies on the related MxA protein suggest that GED makes extensive contacts with both the middle and the GTPase domain. For example, two-hybrid analysis has localized the target for interaction with L612 in MxA to within the middle domain, yet deletion of the last seven amino acids of MxA (residues 656–662) severely impaired MxA GTPase activity (Schwemmle et al., 1995). Together, these data suggest that extensive interactions between GED and dynamin contribute to its overall structure and activity in both assembled and unassembled states.
Dynamin self-assembly stimulates its GTPase activity as much as 100-fold and the I690K mutation completely abrogates dynamin's assembly-stimulated GTPase activity. These data are consistent with our hypothesis that GED functions as an assembly-dependent GAP; (Sever et al., 1999). GAPs are accessory factors for regulatory GTPases that stimulate their GTPase activity and function to switch them from the GTP-bound active state to the GDP-bound inactive state. Many GAPs function by providing a catalytic arginine in trans into the active site of the GTPase (Scheffzek et al., 1998), and it was proposed that R725 served this function in GED (Sever et al., 1999). However, more recent studies have shown that the LT-stimulated GTPase activity of Dyn1-R725A is fully active (Marks et al., 2001) and that Dyn1-R725K behaves indistinguishably from the wild-type protein (S. Sholly and S. L. Schmid, unpublished results). Given that dynamin's basal rate of GTP hydrolysis is rapid compared with Ras-related GTPases, and more comparable to trimeric G proteins (Song and Schmid, 2003), it is more likely that GED functions like the RGS-type GAPs that stimulate Gα subunits by stabilizing and optimally positioning catalytic residues already in place in the dynamin active site. Further studies will be necessary to determine the mechanism of GED-stimulated GTPase activity.
Dyn1-I690K is defective in binding to and assembly around PI4,5P2-containing lipid tubules even though its PH domain is unaltered. The isolated PH domain binds to PI4,5P2 with low affinity, does not localize to the membrane when expressed in mammalian cells, and requires oligomerization for high-affinity binding to membrane phosphoinositides (Kavran et al., 1998; Klein et al., 1998). Here we show that higher-order assembly of full-length dynamin tetramers through lateral protein-protein interactions is also required for binding to PI4,5P2-containing lipid tubules or liposomes. Despite its inability to bind to PI4,5P2-containing liposomes in vitro, Dyn1-I690K was targeted to the plasma membrane in vivo. This suggests that protein-protein interactions, presumably involving SH3 domain–containing partners of dynamin, are involved in membrane targeting.
Our data provide the first direct evidence that dynamin self-assembly is essential for clathrin-mediated endocytosis. However, because stimulated GTPase activity requires dynamin self-assembly, it is not clear whether it is dynamin self-assembly, assembly-stimulated GTPase activity, or both that are important for endocytosis. New mutants that uncouple dynamin self-assembly from assembly-stimulated GTPase activity will be necessary in order to distinguish these possibilities.
The assembly-activity of Dyn1-I697K was also impaired, as this mutant failed to show positive cooperativity even at high protein concentrations and failed to be stimulated by PI4,5P2-containing liposomes under physiological salt conditions. Dyn1-I697K was fully active, however, when assembly-stimulated GTPase activity was measured in the presence of lipid tubules. Overexpression of Dyn1-I697K did not inhibit endocytosis; thus, the in vivo phenotype correlates with the in vitro activity measured on lipid tubules. From these data, we speculate that the I697K mutation impairs lateral associations between dynamin molecules such that the strength of dynamin interactions are not sufficient to generate the force needed to deform a large liposome, but are sufficient to self-assemble around a lipid template having dimensions similar to the diameter of the neck of a coated pits (Koenig and Ikeda, 1989). Given that Dyn1-I697K fully supports endocytosis, these data suggest that dynamin-dynamin interactions in themselves may not be the driving force for reducing membrane curvature in coated vesicle formation and that other factors, such as the clathrin coat and/or accessory proteins such as endophilin (Schmidt et al., 1999) and epsin (Ford et al., 2002) may help to generate the narrow neck upon which dynamin self-assembles. Consistent with this interpretation are our observations of the worm-like endocytic intermediates seen in cells overexpressing Dyn1-I690K-EGFP. It appears that in the absence of dynamin self-assembly, narrow necks can form. In contrast, in cells expressing the assembly-competent, GTPase domain mutant, Dyn1-K44A-EGFP, these necks appear to become stabilized and/or elongated. Similar tubules were seen in cells expressing the Dyn1-T65A GTPase-defective mutant (Marks et al., 2001) and around the TGN in cells expressing Dyn2-K44A-GFP (Kreitzer et al., 2000).
As function follows from structure, it is perhaps not surprising that dynamin's ability to self-assemble into helical arrays and/or its dramatic assembly-stimulated GTPase activity are required for endocytosis. However, a similar assumption would have applied equally to the MxA protein, a member of the dynamin subfamily of GTPases, that like dynamin, self-assembles, tubulates liposomes and displays dramatic assembly-stimulated GTPase activity (Richter et al., 1995; Accola et al., 2002; Kochs et al., 2002). However, neither self-assembly nor assembly-stimulated GTPase activity, which are abrogated by the L612K mutation in its GED, are required for the antiviral activity of MxA (Janzen et al., 2000). The in vivo functions of the self-assembly and assembly-stimulated GTPase activities of MxA remain to be established. Similarly, although self-assembly and/or assembly-stimulated GTPase activity are clearly required for dynamin function in vivo, their exact roles remain to be established. There remain at least three possibilities, which are not mutually exclusive: 1) assembled dynamin may play a structural role in stabilizing transient narrow necks and/or recruiting fission factors to these structures, 2) assembled dynamin may be a mechanochemical device such that conformational changes driven by GTP hydrolysis directly constrict and/or sever the membranous necks, and 3) assembled dynamin may function as a geometric sensor, whose assembly is triggered by the formation of a narrow neck to activate GTP hydrolysis and terminate dynamin's interactions with itself and with its fission partners. More work is needed to fully understand the function of this complex GTPase in clathrin-mediated endocytosis.
Supplementary Material
MBC Videos
Acknowledgments
We are grateful to Alisa Jones, Tricia Glenn, and Marilyn Leonard for their technical support; Clare Waterman-Storer for her invaluable expertise and assistance in fluorescence video microscopy; Elizabeth Kubalek-Wilson for help with lipid nanotubule preparation; Dale Warnock for preparation of 1441 Dyn2-specific antisera; and members in the Schmid lab for critically reading the manuscript. This work was supported by National Institutes of Health Grants GM42455 and MH61345 to S.L.S. and training grant 5T32HL07695 to D.Y. This is TSRI manuscript No 16137-CB.
V⃞
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Accola, M.A., Huang, B., Al Masri, A., and McNiven, M.A. (2002). The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 277, 21829–21835. [DOI] [PubMed] [Google Scholar]
- Aebi, M., Fah, J., Hurt, N., Samuel, C.E., Thomis, D., Bazzigher, L., Pavlovic, J., Haller, O., and Staeheli, P. (1989). cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler, Y., Barbas, S.M., Terlecky, L.J., Tang, K., Hardy, S., Mostov, K.E., and Schmid, S.L. (1998). Redundant and distinct functions for dynamin-1 and dynamin-2 isoforms. J. Cell Biol. 143, 1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko, B., Binns, D.D., and Albanesi, J.P. (2001). Activation of dynamin GTPase activity by phosphoinositides and SH3 domain-containing proteins. Methods Enzymol. 329, 486–496. [DOI] [PubMed] [Google Scholar]
- Binns, D.D., Helms, M.K., Barylko, B., Davis, C.T., Jameson, D.M., Albanesi, J.P., and Eccleston, J.F. (2000). The mechanism of GTP hydrolysis by dynamin II: a transient kinetic study. Biochemistry 39, 7188–7196. [DOI] [PubMed] [Google Scholar]
- Ceresa, B.P., Lotscher, M., and Schmid, S.L. (2001). Receptor and membrane recycling can occur with unaltered efficiency despite dramatic Rab5(Q79L)-induced changes in endosome geometry. J. Biol. Chem. 276, 9649–9654. [DOI] [PubMed] [Google Scholar]
- Chen, M.S., Ober, R.A., Schroeder, C.C., Austin, T.W., Poodry, C.A., Wadsworth, S.C., and Vallee, R.B. (1991). Multiple forms of dynamin are encoded by Shibire, a Drosophila gene involved in endocytosis. Nature 351, 583–586. [DOI] [PubMed] [Google Scholar]
- Damke, H., Baba, T., Warnock, D.E., and Schmid, S.L. (1994). Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 127, 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatloff-Zito, C., Gordon, A.J., Duchaud, E., and Merlin, G. (1995). Isolation of an ubiquitously expressed cDNA encoding human dynamin II, a member of the large GTP binding protein family. Gene 163, 301–306. [DOI] [PubMed] [Google Scholar]
- Ellinwood, N.M., McCue, J.M., Gordy, P.W., and Bowen, R.A. (1998). Cloning and characterization of cDNAs for a bovine (Bos taurus) Mx protein. J. Interferon Cytokine Res. 18, 745–755. [DOI] [PubMed] [Google Scholar]
- Fish, K.N., Schmid, S.L., and Damke, H. (2000). Evidence that dynamin-2 functions as a signal-transducing GTPase. J. Cell Biol. 150, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, M.G., Mills, I.G., Peter, B.J., Vallis, Y., Praefcke, G.J., Evans, P.R., and McMahon, H.T. (2002). Curvature of clathrin-coated pits driven by epsin. Nature 419, 361–366. [DOI] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695–704. [PMC free article] [PubMed] [Google Scholar]
- Hardy, S., Kitamura, M., Harris-Stansil, T., Dai, Y., and Phipps, M.L. (1997). Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71, 1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits, J.S., Burgess, C.C., Obar, R.A., and Vallee, R.B. (1993). Effects of mutant rat dynamin on endocytosis. J. Cell Biol. 122, 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw, J.E. (2000). Dynamin and Its Role in Membrane Fission. Annu. Rev. Cell Dev. Biol. 16, 483–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw, J.E., and Schmid, S.L. (1995). Dynamin self assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Iversen, T.-G., Skretting, G., and Sandvig, K. (2003). Clathrin-coated pits with long, dynamin-wrapped necks upon expression of a clathrin antisense RNA. Proc. Natl. Acad. Sci. USA 100, 5175–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen, C., Kochs, G., and Haller, O. (2000). A monomeric GTPase-negative MxA mutant with antiviral activity. J. Virol. 74, 8202–8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, B.A., and Fangman, W.L. (1992). Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP binding domain of dynamin. Genes Dev. 6, 380–389. [DOI] [PubMed] [Google Scholar]
- Kavran, J.M., Klein, D.E., Lee, A., Falasca, M., Isakoff, S.J., Skolnik, E.Y., and Lemmon, M.A. (1998). Specificity and promiscuity in phosphoinositide binding by pleckstrin homology (PH) domains. J. Biol. Chem. 273, 30497–30508. [DOI] [PubMed] [Google Scholar]
- Klein, D.E., Lee, A., Frank, D.W., Marks, M.S., and Lemmon, M.A. (1998). The PH-domains of dynamin isoforms require oligomerization for high-affinity phosphoinositide binding. J. Biol. Chem. 273, 27725–27733. [DOI] [PubMed] [Google Scholar]
- Kochs, G., Haener, M., Aebi, U., and Haller, O. (2002). Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 277, 14172–14176. [DOI] [PubMed] [Google Scholar]
- Koenig, J.H., and Ikeda, K. (1989). Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J. Neurosci. 11, 3844–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer, G., Marmorstein, A., Okamoto, P., Vallee, R., and Rodriguez-Boulan, E. (2000). Kinesin and dynamin are required for post-Golgi transport of a plasma-membrane protein. Nat. Cell Biol. 2, 125–127. [DOI] [PubMed] [Google Scholar]
- Marks, B., Stowell, M.H., Vallis, Y., Mills, I.G., Gibson, A., Hopkins, C.R., and McMahon, H.T. (2001). GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231–235. [DOI] [PubMed] [Google Scholar]
- Merrifield, C., Feldman, M.E., Wan, L., and Almers, W. (2002). Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4, 691–698. [DOI] [PubMed] [Google Scholar]
- Muhlberg, A.B., Warnock, D.E., and Schmid, S.L. (1997). Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 16, 6676–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, M., Winnacker, E.L., and Brem, G. (1992). Molecular cloning of porcine Mx cDNAs: new members of a family of interferon-inducible proteins with homology to GTP-binding proteins. J. Interferon Res. 12, 119–129. [DOI] [PubMed] [Google Scholar]
- Nagase, T. et al. (1998). Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 5, 355–364. [DOI] [PubMed] [Google Scholar]
- Niemann, H.H., Knetsch, M.L., Scherer, A., Manstein, D.J., and Kull, F.J. (2001). Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 20, 5813–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, P.M., Tripet, B., Litowski, J., Hodges, R.S., and Vallee, R.B. (1999). Multiple distinct coiled-coils are involved in dynamin self-assembly. J. Biol. Chem. 274, 10277–10286. [DOI] [PubMed] [Google Scholar]
- Pitossi, F., Blank, A., Schroder, A., Schwartz, A., Hussi, P., Schwemmle, M., Pavlovic, J., and Staeheli, P. (1993). A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 67, 6726–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport, J.Z., and Simon, S.M. (2003). Real-time analysis of clathrin-mediated endocytosis during cell migration. J. Cell Sci. 116, 847–855. [DOI] [PubMed] [Google Scholar]
- Richter, M.F., Schwemmle, M., Herrmann, C., Wittinghofer, A., and P. Staeheli. (1995). Interferon-induced MxA protein: GTP binding and GTP hydrolysis properties. J. Biol. Chem. 270, 13512–13517. [PubMed] [Google Scholar]
- Rothman, J.H., Raymond, C.K., Gilbert, T., O'Hara, P.J., and Stevens, T.H. (1990). A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell 61, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Roos, J., and Kelly, R. (1997). Is dynamin really a `pinchase'. Trends Cell Biol. 7, 257–259. [DOI] [PubMed] [Google Scholar]
- Scheffzek, K., Ahmadian, R., and Wittinghofer, A. (1998). GTPase-activating proteins: helping hands to complement active site. Trends Biochem. Sci. 23, 257–262. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., Wolde, M., Thiele, C., Fest, W., Kratzin, H., Podtelejnikov, A.V., Witke, W., Huttner, W.B., and Soling, H.D. (1999). Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature 401, 133–141. [DOI] [PubMed] [Google Scholar]
- Schwemmle, M., Richter, M.F., Hermann, C., Nassar, N., and Staeheli, P. (1995). Unexpected structural requirements for GTPase activity of the interferon-induced MxA protein. J Biol. Chem. 270, 13518–13523. [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000a). Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 150, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000b). Garrotes, springs, ratchets and whips: Putting dynamin models to the test. Traffic 1, 385–392. [DOI] [PubMed] [Google Scholar]
- Sever, S., Muhlberg, A.B., and Schmid, S.L. (1999). Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature 398, 481–486. [DOI] [PubMed] [Google Scholar]
- Shin, H.W., Shinotsuka, C., Torii, S., Murakami, K., and Nakayama, K. (1997). Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J. Biochem. 122, 525–530. [DOI] [PubMed] [Google Scholar]
- Song, B.D., and Schmid, S.L. (2003). A molecular motor or a regulator? Dynamin's in class of its own. Biochemistry 42, 1369–1376. [DOI] [PubMed] [Google Scholar]
- Stowell, M.H.B., Marks, B., Wigge, P., and McMahon, H.T. (1999). Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat. Cell Biol. 1, 27–32. [DOI] [PubMed] [Google Scholar]
- Takei, K., McPherson, P.S., Schmid, S.L., and De Camilli, P. (1995). Tubular membrane invaginations coated by dynamin rings are induced by GTPγS in nerve terminals. Nature 374, 186–190. [DOI] [PubMed] [Google Scholar]
- Takei, K., Slepnev, V.I., Haucke, V., and De Camilli, P. (1999). Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1, 33–39. [DOI] [PubMed] [Google Scholar]
- van der Bliek, A.M., and Meyerowitz, E.M. (1991). Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature 351, 411–414. [DOI] [PubMed] [Google Scholar]
- van der Bliek, A.M., Redelmeier, T.E., Damke, H., Tisdale, E.J., Meyerowitz, E.M., and Schmid, S.L. (1993). Mutations in human dynamin block an intermediate stage in coated vesicle formation. J. Cell Biol. 122, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock, D.E., Baba, T., and Schmid, S.L. (1997). Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol. Biol. Cell 8, 2553–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock, D.E., Hinshaw, J.E., and Schmid, S.L. (1996). Dynamin self assembly stimulates its GTPase activity. J. Biol. Chem. 271, 22310–22314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MBC Videos