RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 1.
Published in final edited form as: Nat Methods. 2014 Jul 13;11(9):959–965. doi: 10.1038/nmeth.3029
Abstract
Many biological processes are RNA-mediated, but higher-order structures for most RNAs are unknown, making it difficult to understand how RNA structure governs function. Here we describe SHAPE mutational profiling (SHAPE-MaP) that makes possible de novo and large-scale identification of RNA functional motifs. Sites of 2’-hydroxyl acylation by SHAPE are encoded as non-complementary nucleotides during cDNA synthesis, as measured by massively parallel sequencing. SHAPE-MaP-guided modeling identified greater than 90% of accepted base pairs in complex RNAs of known structure and was used to define a second-generation model for the HIV-1 RNA genome. The HIV-1 model contains all known structured motifs and previously unknown elements, including experimentally validated pseudoknots. SHAPE-MaP yields accurate and high-resolution secondary structure models, enables analysis of low abundance RNAs, disentangles sequence polymorphisms in single experiments, and will ultimately democratize RNA structure analysis.
Introduction
Higher-order structures govern most aspects of RNA function, modulating interactions with small molecule ligands, individual proteins, large multi-component complexes, and other small and large RNAs1,2. There are numerous features of RNA structure that are difficult or impossible to determine from sequence-based analysis alone. Inclusion of data from chemical probing experiments, in which an RNA reacts with diagnostic chemical reagents in a structure-selective way, dramatically improves the accuracy of RNA structure modeling3.
Substantial effort has therefore been directed toward developing high-throughput approaches to analyze RNA secondary structure. Recently reported approaches for RNA structure analysis that use massively parallel sequencing to read out the results of enzymatic, SHAPE, or DMS probing have provided comprehensive support for large-scale comparative trends in transcript structure but have not been shown to yield accurate secondary structure models or enable novel motif discovery4–11. In general, these "-seq" approaches are not suited to recovering RNA structure probing information because they require complex RNA ligation and library preparation steps that result in substantial nucleobase and local structure biases. In addition, there is no known pathway for using enzyme or DMS probing data, which report on only a subset of nucleotides, to model complex RNAs accurately. Moreover, understanding many critical features of RNA folding mechanisms12, RNA-protein interactions13,14, and in-cell effects on RNA folding and structure15,16 require that all four RNA nucleotides be interrogated simultaneously.
Results
The MaP Strategy
Selective 2"-hydroxyl acylation analyzed by primer extension (SHAPE)17–19 experiments use 2"-hydroxyl-selective reagents that react to form covalent 2"-_O_-adducts at conformationally flexible RNA nucleotides, both under simplified solution conditions13,20 and in cells15,16,21. Recent innovations that include SHAPE data as restraints in RNA structure prediction algorithms consistently yield highly accurate secondary structure models for structurally complex RNAs19,22. Here we quantify SHAPE chemical modifications17,19,20,23 in RNA in a single direct step by massively parallel sequencing (Fig. 1). The approach exploits conditions that cause reverse transcriptase to misread SHAPE-modified nucleotides and incorporate a nucleotide non-complementary to the original sequence in the newly synthesized cDNA. The positions and relative frequencies of SHAPE adducts are thus immediately, directly, and permanently recorded as mutations in the cDNA primary sequence, thereby creating a SHAPE mutational profile (SHAPE-MaP). In a SHAPE-MaP experiment, the RNA is treated with a SHAPE reagent or treated with solvent only, and the RNA is modified under denaturing conditions to control for sequence-specific biases in detection of adduct-induced mutations (Fig. 2a). RNA from each experimental condition is subjected to reverse transcription, and the resulting cDNAs are then prepared for massively parallel sequencing. Reactive positions are identified by subtracting data from the treated sample from data obtained from the untreated sample and by normalizing to data from the denatured control (Figs. 1 and 2b and Supplementary Figs. 1–3).
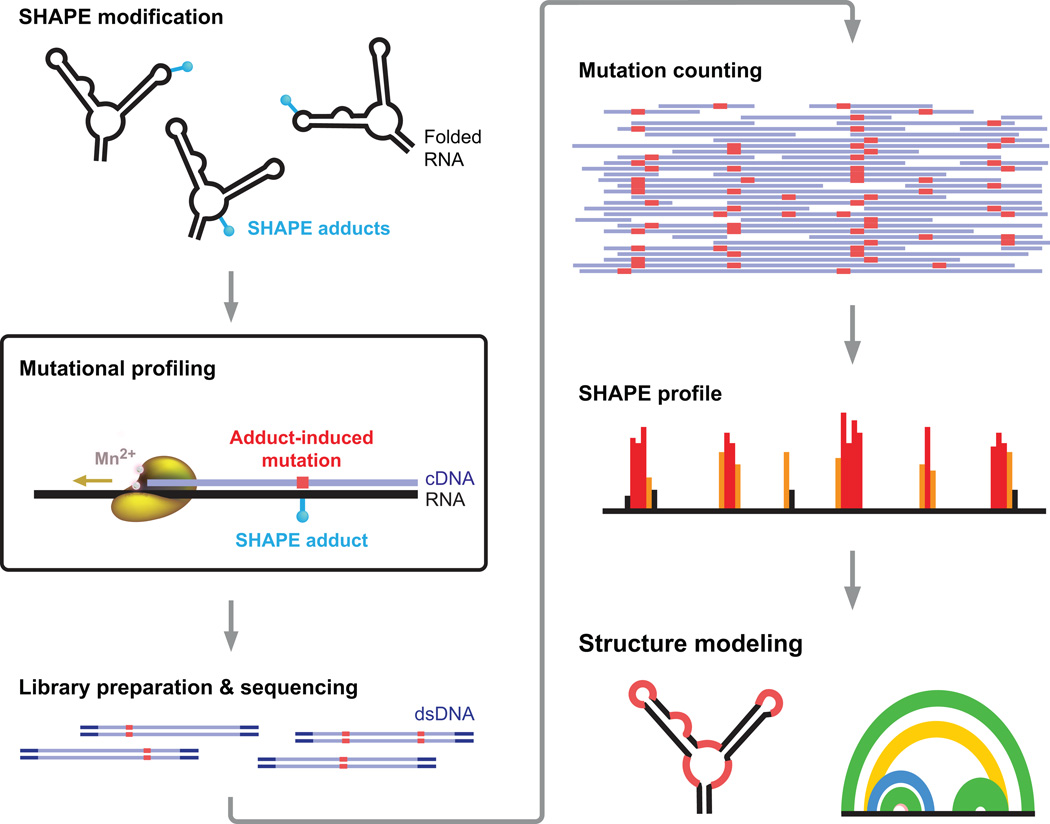
Figure 1. SHAPE-MaP Overview.
RNA is treated with a SHAPE reagent that reacts at conformationally dynamic nucleotides. Reverse transcription is carried out under conditions such that the polymerase reads through chemical adducts in the RNA and incorporates a nucleotide non-complementary to the original sequence (in red) into the cDNA. The resulting cDNA is sequenced using any massively parallel approach to create mutational profiles (MaP). Sequencing reads are aligned to a reference sequence, and nucleotide-resolution mutation rates are calculated, corrected for background and normalized, producing a standard SHAPE reactivity profile. SHAPE reactivities can then be used to model secondary structures, visualize competing and alternative structures, or quantify any process or function that modulates local nucleotide RNA dynamics.
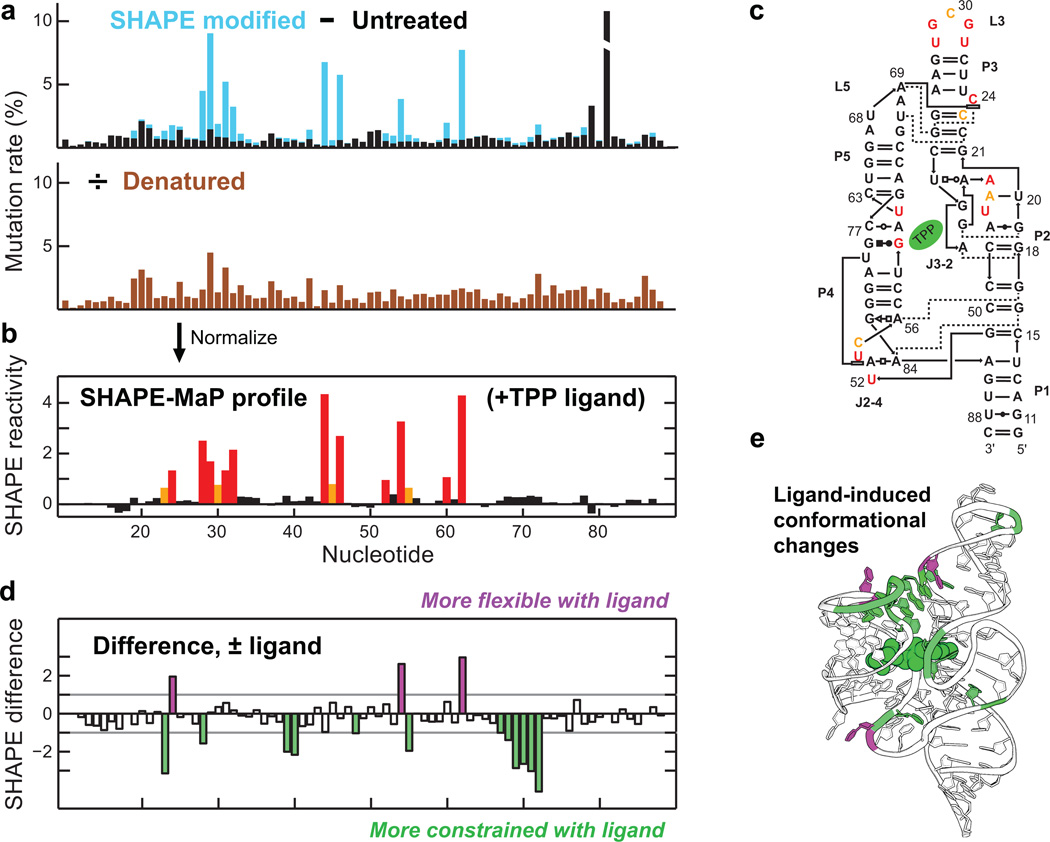
Figure 2. Nucleotide-resolution interrogation of RNA structure and ligand-induced conformational changes.
(a) Mutation rate profiles for the SHAPE modified and untreated TPP riboswitch RNA in the presence of ligand (top) and for SHAPE modification performed under denaturing conditions (bottom). (b) Quantitative SHAPE profile obtained after subtracting the data from the untreated sample from data for the treated sample and normalizing by the denatured control. (c) SHAPE reactivities plotted on the accepted secondary structure of the ligand-bound TPP riboswitch. Red, orange, and black correspond to high, moderate, and low reactivities, respectively. (d) Difference SHAPE profile showing conformational changes in the TPP riboswitch upon ligand binding. (e) Superposition of ligand-induced conformational changes on the TPP riboswitch structure.
Structure Modeling: Validation
We initially examined the structure of the E. coli thiamine pyrophosphate (TPP) riboswitch aptamer domain in the presence and absence of saturating concentrations of the TPP ligand (Fig. 2). SHAPE-MaP profiles recapitulated the known reactivity pattern for the folded, ligand-bound RNA (Figs. 2b, 2c) and accurately reported nucleotide-resolution reactivity differences that occur upon ligand binding (Figs. 2d, 2e). These results, and an analysis of the 1542-nt E. coli 16S rRNA (Supplementary Figs. 4, 5), demonstrate the ability of SHAPE-MaP to capture fine structural details for distinct RNA conformations at nucleotide resolution, accurately and reproducibly, and independently of nucleotide type. Because the SHAPE profiles are reconstructed from mutation frequencies derived from all sequencing reads, uncertainties in SHAPE reactivities can be estimated from the Poisson distribution of mutation events (Methods, Supplementary Figs. 5, 6).
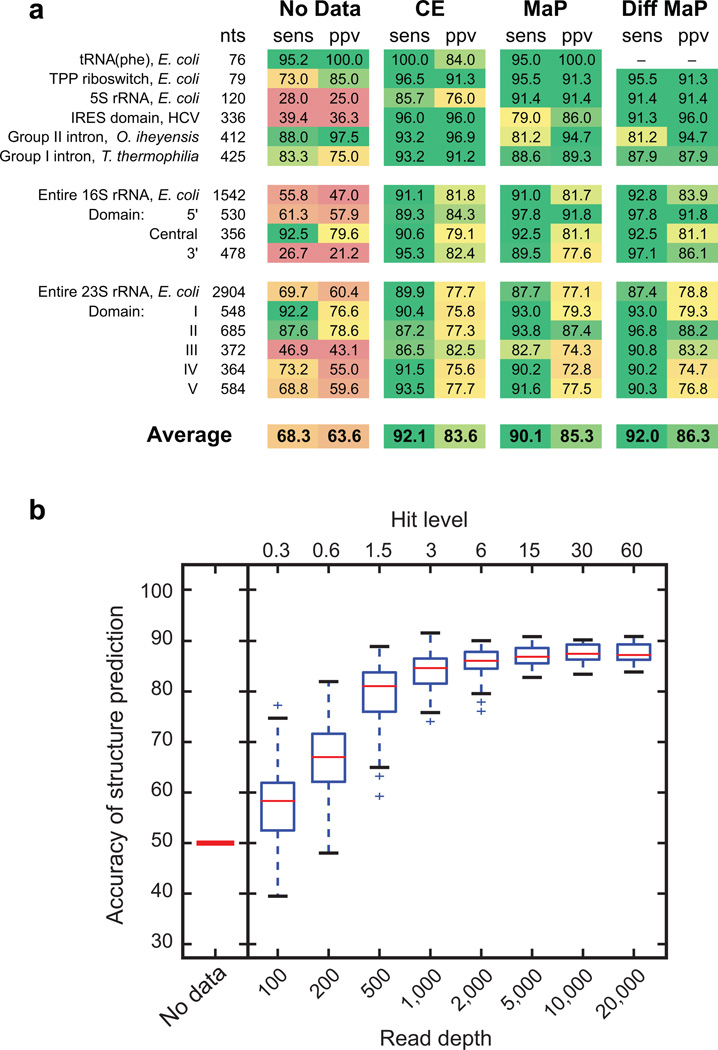
Use of SHAPE data as pseudo-free energy change terms to constrain secondary structure modeling has been extensively benchmarked using RNA test sets specifically chosen to be challenging to conventional secondary structure modeling19,22. To assess the accuracy of SHAPE-MaP, we probed a subset of these RNAs, ranging in size from 78 to 2,904 nucleotides, with the well-validated 1M7 reagent17 We also evaluated the "differential" SHAPE experiment that uses two additional reagents – 1M6 and NMIA – to detect non-canonical and tertiary interactions and yields RNA structural models with consistent high accuracy, even for especially challenging RNAs19,23. The overall accuracy of SHAPE-MaP directed RNA structure modeling using differential reactivities, measured in terms of sensitivity (sens) and positive predictive value (ppv), was similar to and often superior to that of conventional SHAPE reactivities based on adduct-mediated termination of primer extension detected by capillary electrophoresis. The accuracy for recovery of accepted, canonical base pairs exceeded 90% (Fig. 3a).
Figure 3. Accuracy of SHAPE-MaP-directed secondary structure modeling.
(a) Secondary structure modeling accuracies reported as a function of sensitivity (sens) and positive predictive value (ppv) for calculations performed without experimental constraints, with conventional capillary electrophoresis (CE) data, and with SHAPE-MaP data obtained with the 1M7 reagent22,38 or with three-reagent differential (Diff) data19. Results are colored on a scale to reflect low (red) to high (green) modeling accuracy. (b) Relationship between sequencing read depth, hit level, and accuracy of RNA structure modeling. Structure prediction accuracy (vertical axis) is shown as the geometric average of the sens and ppv of predicted structures with respect to the accepted model38. For the 16S rRNA, this accuracy ranges from 50% in the absence of experimental data to 89% for single-reagent SHAPE (shown), and to 91% for the three-reagent “differential”19 experiment. Boxplots summarize modeling the secondary structure of the 16S ribosomal RNA as a function of simulated SHAPE-MaP read depth. At each depth, 100 folding trajectories were sampled. The line at the center of the box indicates the median value and boxes indicate the interquartile range. Whiskers contain data points that are within 1.5 times the interquartile range and outliers are indicated with (+) marks. Hit level is the total signal above normalized background per transcript nucleotide.
SHAPE reactivities obtained using the MaP strategy are measured as many individual events by massively parallel sequencing. Reliability depends on adequate measurement of mutation rates. Accurate modeling of the 16S rRNA structure was achieved using a per nucleotide read depth of 2,000–5,000. This corresponds to 6 to 15 modifications above background per ribosomal nucleotide on average (the hit level; Fig. 3b and Online Methods). Although several prior studies have been performed in which all of the RNAs in a given transcriptome were physically present during the probing phase of the experiment, only a few thousand nucleotides in each case were sampled at a depth that would allow full recovery of the underlying structural information (see Online Methods for detailed analysis). Importantly, accurate SHAPE-MaP directed modeling was achieved using the same parameters originally defined for capillary electrophoresis-based experiments and comparable high accuracies were obtained using both RNA-specific and randomly primed experiments (Supplementary Fig. 3). Data were highly reproducible between experimental replicates performed months apart by different individuals (Supplementary Fig. 5), emphasizing the robustness of SHAPE-MaP.
A Second-Generation Model for an HIV-1 RNA Genome
We obtained single-nucleotide resolution structural information for the entire authentic HIV-1NL4-3 genomic RNA (~9,200 nts) in experiments and data analysis performed over roughly 2 weeks. The 1M7 and differential SHAPE-MaP data were processed to yield SHAPE reactivity profiles and secondary structure models using efficient and fully automated algorithms (Fig. 4, Supplementary Figs. 1–3, 7). Since our report in 2009 of a model for the HIV-1 RNA genome, we have made multiple, fundamental advances in SHAPE-directed RNA structure modeling. These innovations include improved energy models, the ability to model pseudoknots, and concise strategies for detecting tertiary and non-canonical interactions19,22. The MaP approach, implemented in this work, yields nucleotide-resolution reactivity data for large RNAs that are equal or are superior to the prior gold standard capillary electrophoresis data (Fig. 3a). Thus, the HIV-1 genome structure presented here represents a higher resolution, second-generation model for well-defined elements in this RNA.
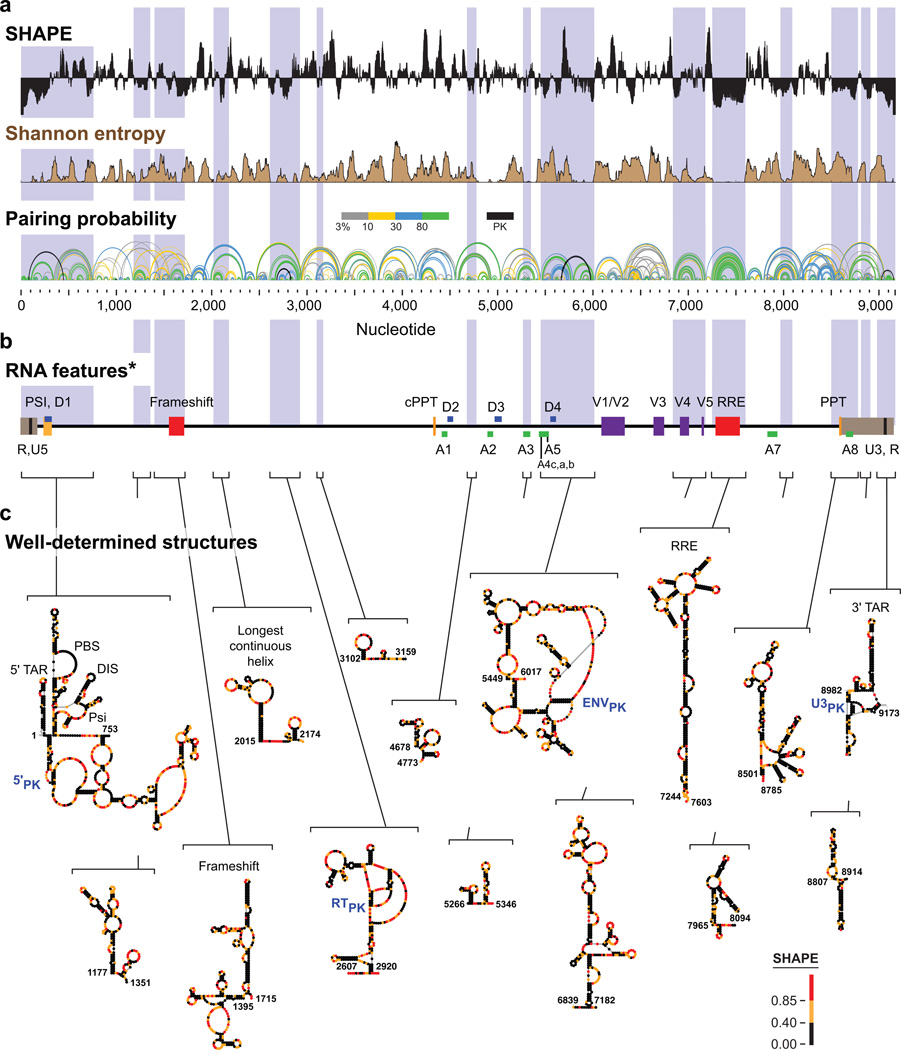
Figure 4. SHAPE-MaP analysis of the HIV-1 NL4-3 genome.
(a) SHAPE reactivities for the NL4-3 HIV-1 genomic RNA. Reactivities are shown as the centered 55-nt median window, relative to the global median; regions above or below the line are more flexible or constrained than the median, respectively. Shannon entropy values for 55-nt windows were calculated by considering the pairing probability of a nucleotide over all structures in a 1M7 and differential SHAPE19 reactivity data-constrained Boltzmann ensemble and reflect how well determined the secondary structure model is for each nucleotide region. Arcs representing base pairs are colored by their respective pairing probabilities, with green arcs indicating highly probable helices. Areas with many overlapping arcs have multiple potential structures. Pseudoknots (PK) are indicated by black arcs. (b) RNA regions identified as having biological functions. Brackets enclose well-determined regions and are drawn to emphasize locations of these regions relative to known RNA features in the context of the viral genome. Regions correspond to low SHAPE-low Shannon entropy domains and are extended to include all intersecting helices from the lowest predicted free-energy secondary structure. 5’ and 3’ UTRs are brown; splice acceptors and donors are green and blue, respectively; polypurine tracts are yellow; variable domains are purple; and the frameshift and RRE domains are red. These elements fall within regions with low SHAPE and low Shannon entropy much more frequently than expected by chance (p = 0.002; see Online Methods). (c) Secondary structure models for regions, identified de novo, with low SHAPE reactivities and low Shannon entropies. Nucleotides are colored by SHAPE reactivity and pseudoknotted structures are labeled in blue. Larger figure images, showing nucleotide identities, are provided in Supplementary Fig. 7.
De Novo Identification of Well-Determined Structures
Almost any long RNA sequence will form some secondary structures24, but not all of these structures are biologically important or well-defined. Therefore, we used SHAPE-directed modeling, whose underlying energy function yields highly accurate models for RNAs with well-defined secondary structures (Fig. 3), to calculate a probability for each base pair across all possible structures in the Boltzmann ensemble of structures predicted for the HIV-1 RNA. These probabilities were used, in turn, to calculate Shannon entropies25,26 (Fig. 4). Regions with higher Shannon entropies are likely to form alternative structures, and those with low Shannon entropies correspond to regions with well-defined RNA structures or persistent single-strandedness, as determined by SHAPE reactivity.
The plot of pairing probability across the entire HIV-1 genome reveals both well-determined and variable RNA structures in the HIV-1 genomic RNA (Fig. 4a). Previously characterized structured regions such as the 5'-UTR, Rev response element (RRE), frameshift element, and polypurine tract (PPT) are well determined in the model (represented by green arcs). In contrast, there are also large regions – for example, from nucleotides 3200 to 4500 and from nucleotides 6100 to 6800 – that have high SHAPE reactivities and high Shannon entropy and are therefore likely to sample many conformations (shown as blue, yellow, and gray arcs). This visualization approach highlights regions with unique, likely stable structures and those regions where multiple structures are likely to be in equilibrium.
Critically, analysis of Shannon entropies and SHAPE reactivities provides an approach for de novo discovery of regions with well-defined structure in long RNAs. Fifteen regions in the HIV-1 genomic RNA had both low SHAPE reactivity values (indicating a high degree of RNA structure) and low Shannon entropies (providing confidence in a single predominant secondary structure) (Figs. 4a, 4b, shaded in purple). We created nucleotide-resolution structure models for each of these regions (Fig. 4c). The models of known, functionally important regulatory structures – RRE, 5' TAR, primer binding site (PBS), packaging element PSI structures, ribosomal frameshift element, and 3' TAR – agreed closely with previously proposed models for these regions. In addition, the longest continuous helix, the hairpins flanking the polypurine tract, and other features remain consistent between the prior27 and this second-generation model (Supplementary Table 1). We next assembled a list of all regulatory elements likely to function via an RNA motif (Fig. 4b; Supplementary Table 2). We then compared the locations of these RNA structural elements with the highly structured and low entropy regions identified de novo by SHAPE-MaP. Functional RNA elements occur overwhelmingly in low SHAPE, low Shannon entropy regions (_p_-value = 0.002; Fig. 4 and Online Methods), indicating that most RNA-mediated functions operate in the context of an underlying RNA structure. Several low SHAPE, low Shannon entropy regions in the HIV-1 genome occur in regions not previously associated with known RNA functional elements: These regions are high-value targets for discovery of new RNA motifs.
Motif Discovery and Deconvolution of Structural Polymorphism
Pseudoknots appear to be rare in large RNAs and are difficult to identify; however, these motifs appear to be overrepresented in functionally important regions of many RNAs28,29. As a rigorous test of the current cumulative advances in SHAPE-directed structure modeling and of the high-throughput SHAPE-MaP data itself, we searched22 for novel pseudoknots in the HIV-1 RNA genome. In our model, there are four pseudoknots in regions of low SHAPE reactivity and low Shannon entropy (Fig. 4c). The pseudoknot adjacent to the 5' polyadenylation signal in the HIV-1 RNA (5'PK) was previously validated13,30. The three additional, novel pseudoknots are predicted to form in the reverse transcriptase coding region (RTPK), at the beginning of env (ENVPK), and in the U3 region adjacent to the 3' polyadenylation signal (U3PK). An additional pseudoknot predicted by the ShapeKnots algorithm that lies in a region of high SHAPE reactivity and Shannon entropy (CAPK, nt 961–1014, Supplementary Fig. 8) was analyzed as a negative control.
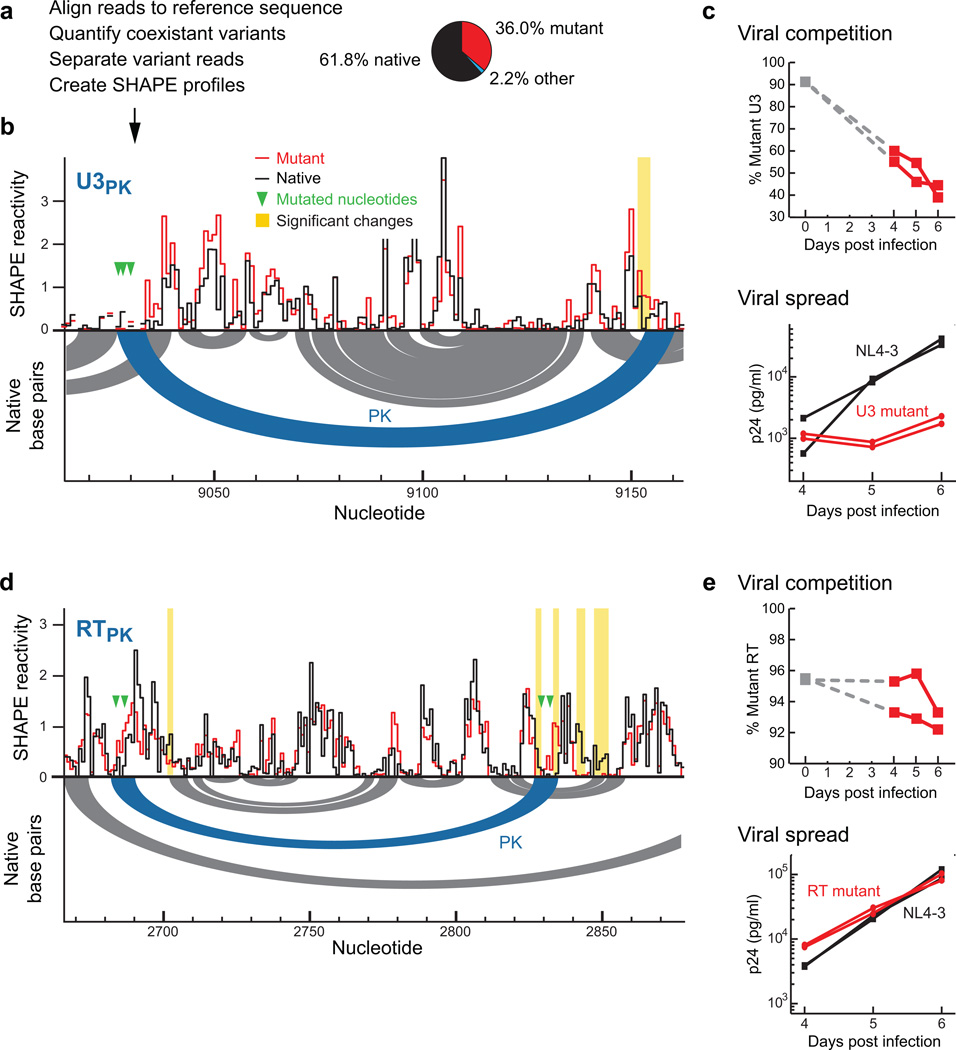
We introduced silent mutations designed to disrupt each pseudoknot into the full-length HIV-1 genome (Supplementary Fig. 8). Special features of the U3PK region illustrate the power of the MaP approach. U3 sequences occur at both the 5' and 3' ends of the viral genome in proviral HIV-1 DNA but only at the 3' end in the viral RNA. During transfection of the provirus-encoding plasmid, these sequences can undergo recombination. When we introduced a single mutant copy of the U3 sequence (at the 3' end) into the pNL4-3 provirus, we observed partial recombination with the native sequence U3 at the 5' end of the proviral DNA. SHAPE-MaP experiments revealed that both native and mutant sequences were present at the 3' ends of individual genomic RNAs in the mutant U3PK sample. Critically, because nucleotides are analyzed in the context of unfragmented RNA regions in the MaP approach, we were able to independently monitor both alleles in the same experiment, computationally separate them, and construct native and mutant SHAPE profiles (Figs. 5a, 5b, Supplementary Fig. 9, and Online Methods). Notable SHAPE reactivity differences between native and mutant U3 were observed, produced by viruses in direct competition with each other and consistent with precise disruption of the U3PK structure (Figs. 5b and Supplementary Fig. 9). Strikingly, mutations introduced in the 5' side of the U3PK pseudoknot helix induced changes in the predicted 3' pairing partner, located over 100 nucleotides away (Fig. 5b). SHAPE-MaP is thus uniquely useful for structural analysis and motif discovery in systems that contain complex mixtures of RNAs and for detecting and deconvoluting structural consequences of single-nucleotide and other allelic polymorphisms.
Figure 5. Functional and structural validation of newly discovered HIV-1 RNA motifs.
(a) Scheme for simultaneous deconvolution and structural analysis of a mixture of native sequence and U3PK mutant genomes.(b) SHAPE profiles for the U3PK pseudoknot bridging U3 and R. The experiment simultaneously probed a mixture of viruses with native sequence and mutant U3PK RNAs. Secondary structure for the native sequence is shown as arcs below the y-axis intercept. Significant SHAPE reactivity differences are emphasized with yellow vertical lines (see Online Methods). (c) Direct growth competition and viral spread for U3PK mutant and native sequence NL4-3 HIV-1 virions in Jurkat cells. Percentage of mutant in the initial inoculum is presented as a grey square at day 0. p24 levels correspond to the amount of HIV-1 capsid protein. (d) SHAPE profiles for the RTPK pseudoknot within the reverse transcriptase coding region. In this case, SHAPE data were obtained in separate experiments for each virus. (e) Viral spread and direct growth competition for RTPK mutant and native sequence NL4-3 HIV-1 virions in Jurkat cells. For the competition data, y-axes are shown on an expanded scale for clarity.
All mutant constructs were analyzed using SHAPE-MaP and in cell-based assays for viral fitness. Mutations in U3PK reduced viral spread in Jurkat cells by ~10-fold relative to NL4-3 and reduced viral fitness in direct competition with NL4-3, with a mean relative fitness difference of –0.32 relative to NL4-3 (Fig. 5c)31. This large effect on viral fitness by mutations in the U3PK is consistent with the general importance of 3'-UTRs in regulating mRNA stability and translation32 and, more specifically, with a role for specific higher-order spatial organization of the poly(A) signal and upstream sequence elements in assembly of the polyadenylation machinery33,34. SHAPE changes in the RTPK mutant were also located directly in or immediately adjacent to the pseudoknotted helix (Fig. 5d). Mutations in RTPK showed a smaller, but reproducible, decrease in viral spread and viral fitness, with a mean relative fitness of –0.14, compared to NL4-3 (Fig. 5e). We also observed changes in SHAPE reactivities at both the 5' and 3' sequences for the long-distance ENVPK mutant, including changes extending 5' from the pseudoknotted helix, suggestive of local refolding caused by disruption of this pseudoknot (Supplementary Fig. 10). Viral spread and viral fitness were not reduced for the ENVPK mutant, which may reflect the challenge of detecting some features of HIV-1 replication in cell culture. The mutations in CAPK (Supplementary Fig. 10), which we analyzed as a negative control, did not support existence of a pseudoknotted structure at this location by SHAPE-MaP analysis, in agreement with its high Shannon entropy profile.
Discussion
This work defines an alternative strategy for reading out nucleic acid structure probing experiments by massively parallel sequencing. With mutational profiling, or MaP, nucleic acid structural information is directly and concisely recorded in the sequence of the complementary cDNA and rendered insensitive to biases in library preparation and sequencing. MaP thus converts reverse transcription or DNA synthesis into a direct engine for nucleic acid structure discovery. MaP is fully independent of sequencing strategy and can therefore be used in any sequencing approach with a sufficiently low base call error rate to quantify chemical modifications in any low-abundance RNA detectable by reverse transcription. Detection of chemical adducts in RNA and DNA via direct read-through can be coupled with strategies for polymerase selection35,36 to record, as mutational profiles or MaPs, a wide variety of post-transcriptional and epigenetic modifications. SHAPE-MaP data contain error estimates and are readily integrated into fully automated, vetted, algorithms for structure modeling (Online Methods). SHAPE-MaP yielded accurate models for RNAs of known structure (Fig. 3a) and of individual functional motifs in large RNA (Fig. 4) and makes possible transcript-wide motif discovery (Figs. 4 and 5).
In large- and genome-scale RNA structural studies, true functional elements must be identified in the background of the complex ensemble of structures that form in any large RNA. The combination of SHAPE-MaP analysis with analysis of pairing probabilities, calculated across large RNA regions, identified almost all known large-scale functional elements within the HIV-1 genome, with the exception of the central polypurine tract (cPPT; Fig. 4), which appears to have a conserved structure37. Thus, the sensitivity of functional element detection by SHAPE-MaP is very high. Moreover, despite the fact that the HIV-1 genome is one of most intensively studied RNAs in scientific history, quantitative and high-resolution SHAPE-MaP analysis nonetheless allowed rapid, de novo discovery and direct validation of new functional motifs, specifically three pseudoknots – a motif that has traditionally been challenging to predict (Figs. 4 and 5). The positive predictive value of the approaches developed here is thus also correspondingly high. SHAPE-MaP is unique in its experimental simplicity and structural accuracy and can be scaled to RNA systems of any size and complexity.
Online Methods
SHAPE-MaP experimental overview
SHAPE-MaP experiments use specialized conditions for reverse transcription that promote incorporation of nucleotides non-complementary to the RNA into the nascent cDNA at the locations of SHAPE adducts. Sites of RNA adducts thus correspond to internal mutations or deletions in the cDNA, relative to comparison with cDNAs transcribed from RNA not treated with SHAPE reagent. Reverse transcription can be carried out using gene-specific or random primers (Supplementary Fig. 3); both approaches are described below. Once cDNA synthesis is complete, RNA structural information is essentially permanently recorded in the sequence and thus independent of biases introduced during any multi-step library construction scheme. Library preparation is similar to that of an RNA-seq experiment, can be readily tailored to any sequencing platform, and allows multiplexing using sequence barcodes. Single-stranded breaks and background degradation do not intrinsically interfere with SHAPE-MaP experiments (in contrast to conventional SHAPE and other reverse transcriptase stop-dependent assays), as these are not detected during read-through sequencing. There is also no signal decay or drop-off in the MaP approach, which otherwise requires complex, partially heuristic, correction.
SHAPE-MaP development and efficiency
Reverse transcriptase enzymes are, in some cases, able to read through unusual 2'-_O_-linkages and adducts, following enzyme pausing39,40. We hypothesize that read-through causes, or results from, structural distortion in the reverse transcriptase active site, that results in a higher rate of nucleotide misincorporation at the location of a pause-inducing SHAPE adduct. We screened multiple reverse transcriptase enzymes for use in SHAPE-MaP as a function of nucleotide concentration, reaction time, buffer conditions, and divalent metal ion identity. We searched for enzyme conditions that produced minimal adduct-induced reverse transcription stops and maximal full-length cDNA products. Of the divalent metal ions tested (including magnesium, manganese, copper, cobalt, nickel, and lead), Mn2+ most effectively promoted enzyme read-through at the sites of bulky 2'-_O_-adducts, particularly using a Moloney murine leukemia virus reverse transcriptase (Superscript II, Invitrogen). This observation is consistent with the high activity of the Moloney reverse transcriptase in Mn2+ 41 and the ability of this ion to promote mutagenic behavior in DNA polymerases42.
We determined the precise classes of adduct-induced misincorporation events by comparing substitution and deletion rates at non-paired and paired nucleotide positions in the 16S rRNA. Misincorporation trends were similar between all three SHAPE reagents (1M717 and the "differential" reagents NMIA and 1M619). Generally, the presence of a SHAPE adduct causes nucleotides to be misread as A, T, or deletion events, although there is significant information content in other misincorporation events (Supplementary Fig. 11). Flexible nucleotides in a dinucleotide model substrate with a single reactive position (AddC)17 are modified with an efficiency of ~2% by NMIA or 1M7 under conditions similar to those used here. Mutation rates above background at flexible positions in the 16S rRNA are ≥0.5%, with many of the most reactive positions above 2% (Supplementary Fig. 4). Given these boundary values, we estimate that the MaP strategy detects SHAPE adducts with an efficiency of ≥50%.
RNA folding and SHAPE probing of model RNAs
DNA templates (IDT) were synthesized for tRNAPhe, TPP riboswitch, E. coli 5S, hepatitis C virus IRES domain, T. thermophila group I intron, or O. iheyensis group II intron RNAs in the context of flanking 5' and 3' structure cassettes. Templates were amplified by PCR and transcribed into RNA using T7 RNA polymerase43. RNAs were purified by denaturing polyacrylamide gel electrophoresis, appropriate regions excised, and RNAs passively eluted from the gel overnight at 4 °C. 16S and 23S rRNAs were isolated from DH5α cells during mid-log phase using non-denaturing conditions38. For each sample, 5 pM of RNA was refolded in 100 mM HEPES, pH 8.0, 100 mM NaCl, and 10 mM MgCl2 in a final volume of 10 µL. After folding, RNAs were modified in the presence of 10 mM SHAPE reagent and incubated at 37 °C for 3 min (1M6 and 1M7) or 22 min (NMIA). No-reagent controls, containing neat DMSO rather than SHAPE reagent, were performed in parallel. To account for sequence-specific biases in adduct detection, RNAs were modified using NMIA, 1M7, or 1M6 under strongly denaturing conditions in 50 mM HEPES (pH 8.0), 4 mM EDTA, and 50% formamide at 95 °C. Following modification, RNAs were isolated using either RNA affinity columns (RNeasy MinElute; Qiagen) or G-50 spin columns (GE Healthcare).
RNA folding and SHAPE probing of the HIV-1 genomic RNA
For whole-genome SHAPE-MaP of HIV-1 (strain NL4-3; group M, subtype B), virus was produced and purified as described27. Viral RNA was gently extracted and purified from protein then precipitated with ethanol from a solution containing 300 mM NaCl. Approximately 30% of genomic RNA is full length when prepared in this manner27; the fragmented nature of native HIV-1 genome samples resulted in decreased sample recovery during column purifications (RNeasy MinElute, Qiagen). Therefore, ~1 µg of HIV-1 RNA was used per sample, more RNA than the 250 ng required for SHAPE-MaP experiments of more intact RNAs.
Mutant viruses were produced by transfection of 293T cells using FuGene6 (Promega) or XtremeGene HP (Roche). Viral supernatants were concentrated using centrifugal concentrators (Vivaspin 20, Sartorius), followed by precipitation (Lenti-X Concentrator, Clontech) to concentrate virions. Pelleted virions were resuspended in viral lysis buffer [50 mM HEPES (pH 8.0), 200 mM NaCl, 3 mM MgCl2]27, and lysed with 1% (w/v) SDS and 100 µg/mL proteinase K (25 °C, 30 min). RNA was extracted with phenol:chloroform:isoamyl alcohol at least three times, followed by two extractions with chloroform and precipitation with ethanol.
Approximately 1 µg of HIV-1 genomic RNA was suspended in modification buffer [50 mM HEPES (pH 8.0), 200 mM potassium acetate (pH 8.0), 3 mM MgCl2] and incubated at 37 °C for 15 min (for SHAPE modified and untreated samples) or in denaturing buffer [50 mM HEPES (pH 8.0), 4 mM EDTA, and 50% formamide] and incubated at 95 °C for 2 minutes. Samples were then treated with SHAPE reagent (10 mM final) or neat solvent.
SHAPE-MaP using fragmented samples
Following SHAPE modification and purification, HIV-1, group II intron, HCV IRES, and ribosomal RNA samples were fragmented (yielding lengths of ~250–350 nts) by a 4 min incubation at 94 °C in a buffer containing 9 mM MgCl2, 225 mM KCl, 150 mM Tris HCl (pH 8.3). RNA fragments were desalted using G-50 spin-columns. Fragmented samples (250–500 ng total mass) were subjected to reverse transcription for 3 hours at 42 °C (using SuperScript II, Invitrogen). Reactions were primed using 200 ng random nonamer primers (NEB) for the ribosome, group II intron, and HCV IRES RNA or with custom LNA primers (Supplementary Fig. 12) for HIV-1 RNA genomes. Reverse transcriptase buffer contained 0.7 mM premixed dNTPs, 50 mM Tris HCl (pH 8.0), 75 mM KCl, 6 mM MnCl2, and 14 mM DTT. Following reverse transcription, reactions were desalted using G-50 spin columns (GE Healthcare). Under these conditions (long incubation times and using 6 mM Mn2+ as the only divalent ion) the reverse transcriptase reads through sites of 2'-_O_-modification by a SHAPE reagent, incorporating a non-complementary nucleotide at the site of the adduct.
Double-stranded DNA libraries for massively parallel sequencing were generated using NEBNext sample preparation modules for Illumina. Second-strand synthesis (NEB E6111) of the cDNA library was performed using 100 ng input DNA, and the library was purified using a PureLink Micro PCR cleanup kit (Invitrogen K310250). End repair of the double-stranded DNA libraries was performed using the NEBNext End Repair Module (NEB E6050). Reaction volumes were adjusted to 100 µL, subjected to a cleanup step (Agencourt AMPure XP beads A63880, 1.6:1 beads-to-sample ratio), dA tailed (NEB E6053), and ligated with Illumina-compatible forked adapters (TruSeq) with a quick ligation module (NEB M2200). Emulsion PCR44 (30 cycles) using Q5 hot-start, high-fidelity polymerase (NEB M0493) was performed to maintain library sample diversity. Resulting libraries were quantified (Qubit fluorimeter; Life Technologies), verified using a Bioanalyzer (Agilent), pooled, and subjected to sequencing using the Illumina MiSeq or HiSeq platform.
SHAPE-MaP using targeted gene-specific primers
The tRNAPhe, TPP riboswitch, 5S rRNA, group I intron, and mutant HIV-1 construct RNAs were subjected to reverse transcription using a DNA primer specific to either the 3' structure cassette (5'-GAA CCG GAC CGA AGC CCG-3') for the small RNAs or to specific HIV-1 sequences flanking a pseudoknot using buffer and reaction conditions described in the previous section. Sequencing libraries were generated using a modular, targeted, two-step PCR approach that makes it possible to inexpensively and efficiently generate data for many different RNA targets. PCR reactions were performed using Q5 hot-start, high-fidelity DNA polymerase. The forward PCR primer (5'-GAC TGG AGT TCA GAC GTG TGC TCT TCC GATC NNNNN-gene-specific primer-3') includes an Illumina-specific region at the 5' end, followed by five random nucleotides to optimize cluster identification on the MiSeq instrument, and ends with a sequence complementary to the 5' end of the target RNA. The reverse primer (5'-CCC TAC ACG ACG CTC TTC CGA TCT NNNNN-gene-specific primer-3') includes an Illumina-specific region followed by five random nucleotides and a sequence that is the reverse complement of the 3' end of the target RNA. The cDNA library was ‘tagged’ by limited, 5-cycle PCR for amplicons or a longer 25 cycle PCR reaction when very low RNA concentrations were used. Excess primer, not used in the first few cycles, was removed (PureLink Micro PCR cleanup kit; Invitrogen). The second round of PCR added the remaining Illumina-specific sequences needed for on-flow cell amplification and barcoded the samples for multiplexing. The forward primer (CAA GCA GAA GAC GGC ATA CGA GAT [Barcode] GT GAC TGG AGT TCA GAC) contains a barcode and targets sequence in the forward primer from PCR 1. The reverse primer (AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT T CCC TAC AC GAC GCT CTT CCG) contains an Illumina-specific sequence and targets the reverse primer from PCR 1. PCR 2 was performed for 25 or 5 cycles to generate the final library for sequencing (not exceeding 30 total cycles). Typical SHAPE-MaP experiments of mutant viruses used ~150 to 200 ng of RNA per experimental condition. However, when material is limiting, as little as 50 ng input RNA is sufficient.
SHAPE-MaP data analysis pipeline
We created a data analysis pipeline, called ShapeMapper, that can be executed on most Unix-based platforms and accepts as input sequencing read files in FASTQ format, reference sequences in FASTA format, and a user-edited configuration file. Without further user intervention, the software creates a SHAPE reactivity profile and standard error estimates for each reference sequence. Other useful outputs are provided including mutation counts, sequencing depths, and predicted secondary structures. The analysis software incorporates several third-party programs. Python 2.7 is required (www.python.org); Bowtie 2 is used for read alignment45; reactivity profiles are generated using the python library matplotlib46; secondary structure prediction uses RNAstructure47; and secondary structure drawing uses the Pseudoviewer web service48.
Configuration
A configuration file is used to specify the reference sequences present in each sample and which samples should be combined to create reactivity profiles. The format is flexible, allowing the alignment of each sample to multiple sequence targets as well as the treatment of multiple samples in unified analyses. Important parameters for each stage of analysis may also be customized.
Quality trimming
Input reads were separated into files by sequencing barcode (this step is integrated into most sequencing platforms). The first analysis stage trims reads by base-call quality. Each read was trimmed downstream of the first base-call with a phred quality score below 10, corresponding to 90% expected accuracy. Reads with 25 or more remaining nucleotides were copied to new FASTQ files for alignment.
Read alignment
Reads were locally aligned to reference sequences using Bowtie 245; parameters were chosen to provide high sensitivity, to detect single nucleotide mismatches, and to allow deletions of up to about 200 nucleotides. Seed length (-L) was 15 nucleotides. One mismatch was allowed per seed (-N). Maximum seed attempts (-D) was set at 20. Maximum “re-seed” attempts (-R) was set at 3. Dynamic programming padding (--dpad) was set at 100 nucleotides. The match bonus (--ma) was 2. The maximum and minimum mismatch penalties (--mp) were 6 and 2, respectively. Gap open and extend parameters (--rdg, --rfg) were 5 and 1, respectively. The default minimum alignment score function was used. Soft-clipping was turned on. Paired-end alignment was used by default. Bowtie 2 outputs aligned reads as SAM files.
Alignment parsing, ambiguous alignment removal, and mutation counting
Paired-end reads in SAM files were combined, and higher-quality base-calls were selected where read pairs disagreed. Mismatches and deletions contribute to mutation counts; insertions were ignored. Since error-prone reverse transcription generates most of the mutations in each read, we treated a sequence change covering multiple adjacent nucleotides as a single mutation event located at the 3´-most nucleotide. If random primers were used, a region one nucleotide longer than the length of the primer was excluded from the 3´ end of each read. Reads with mapping qualities less than 30 were excluded. Deletions are an important part of the mutation signal, but deletions that are ambiguously aligned can blur this signal, preventing single-nucleotide resolution. To resolve this problem, a simple local realignment was performed to identify and remove ambiguously aligned deletions. The reference sequence surrounding a deletion was stored. The deletion was then slid upstream or downstream one nucleotide at a time to a maximum offset equal to the deletion length. At each offset, the surrounding reference sequence was compared to the stored sequence. If any offset sequence matched, this indicated a possible alternate alignment, and the deletion was excluded. This algorithm correctly identified ambiguous deletions in homopolymeric regions as well as repeated sequences.
Reactivity profile creation
The mutation rate (mutr) at a given nucleotide is simply the mutation count (mismatches and unambiguously aligned deletions) divided by the read count at that location. Raw reactivities were generated for each nucleotide using the following expression, where S corresponds to a SHAPE modified sample, U to untreated, and D to reaction under denaturing conditions:
The standard error (stderr) associated with the mutation rate at a given nucleotide in the S, U, or D samples was calculated as:
The final standard error of the reactivity at a given nucleotide is:
| =(stderrSmutrD)2+(stderrUmutrD)2+(stderrD×(mutrS−mutrU)mutrD2)2 | (3) |
|---|
Reactivities were normalized to a standard scale that spanned zero (no reactivity) to ~2 (high SHAPE reactivity) as described22. Nucleotides with mutation rates greater than 5% in no-reagent control samples were excluded from analysis, as were nucleotides with sequencing depths less than 10 in any sample. A much greater depth is required for high-quality data and structure modeling (see Fig. 3).
Final data output
SHAPE reactivity profiles (.shape) were output as tab-delimited text files with the first column indicating nucleotide number and the second reactivity. A SHAPE-MaP reactivity file was also output (.map). This file is in the SHAPE file format with the addition of two columns: standard error and nucleotide sequence. Another file (.csv) containing mutation counts, read depths, mutation rates, raw reactivities, normalized reactivities, and standard errors for SHAPE modified, untreated, and denatured samples was also created. Files containing figures showing mutation rate histograms, sequencing depths, and reactivity profiles may be generated (.pdf). These are useful in diagnosing potential experimental problems (including insufficient sequencing depth or low mutagenesis efficiency).
Automatic RNA folding and structure drawing by the SHAPE-MaP pipeline
For sequences shorter than ~4000 nucleotides and with sufficient read depth, the automated pipeline allows secondary structures to be automatically modeled using RNAstructure, although this capability was not used for the RNAs in this work. FASTA sequence files are converted to SEQ files required by RNAstructure. SHAPE reactivities are incorporated into RNAstructure as pseudo-free energies using standard parameters for the 1M7 reagent22 [slope (-sm) 1.8, intercept (-si) −0.6]. Differential SHAPE reagents are supported by RNAstructure, but have not been incorporated into the automated pipeline at the time of submission. Predicted structures are written to .ct files. The lowest energy predicted secondary structures can be drawn and annotated by SHAPE reactivity. This stage queries the Pseudoviewer web service48 over an active internet connection. A custom client (pvclient.py) submits server requests and retrieves responses. This client also handles coloring of nucleotides by reactivity. Colored structure drawings are vector .eps files. Structures are also automatically converted to .xrna files (http://rna.ucsc.edu/rnacenter/xrna/) for optional manual editing.
Filtering by Z-factor for differential SHAPE data
SHAPE-MaP allows errors in SHAPE reactivity measurements to be estimated from a Poisson distribution describing the measured mutation rates at each nucleotide. The Poisson-estimated SHAPE reactivity error can be used to evaluate statistical significance when comparing two SHAPE signals. Significant differences between NMIA and 1M6 reactivity were identified using a Z-factor test49. This nucleotide-resolution test compares the absolute difference of the means with the associated measurement error:
| Zfactor=1−3(σNMIA+σ1M6)|μNMIA−μ1M6| | (4) | | ------------------------------------- | --- |
Each nucleotide in a SHAPE-MaP experiment has a calculated reactivity µ and an associated standard error σ. The significance threshold for Z-factors was set at Z > 0, equivalent to a SHAPE reactivity difference for 1M6 and NMIA of at least three standard deviations. Differential nucleotide reactivities not meeting this significance criterion were set to zero.
Structure modeling
Secondary structure modeling for RNAs less than 700 nts in length was performed as described19,22; differential SHAPE data were incorporated after filtering by Z-factor. For the HIV-1 RNA genome, we developed an automated windowed modeling approach, implemented in a SHAPE-MaP Folding Pipeline, in which structure calculations were broken into stages designed to increase computational efficiency, generate realistic RNA structures, and reduce end-effects caused by selecting a false 5' or 3' end from an internal fold in a window. This approach facilitated pseudoknot discovery, identification of probable base pairs, and generation of minimum free energy structures (Supplementary Fig. 2). Representative calculations for folding of ribosomal subunits, performed using both one-step and windowed folding showed comparable, high degrees of accuracy and substantial reductions in computation ‘wall time’ using a typical desktop workstation for the windowed folding approach. For shorter RNAs, such as the 16S rRNA, there is a modest performance penalty for breaking the RNA into smaller windows. However, for RNAs longer than ~2000 nucleotides, computation time scales approximately linearly with length.
Nearly all known and well-validated functional RNA structures are modeled identically in the current study and the prior 2009 investigation27 (Table S1). Substantial improvements in digital (MaP) data acquisition, improved SHAPE-based energy functions19,22 and automated data analysis (Supplementary Figs. 1–3) favor the the current second-generatation HIV-1 structure models over previous models in regions of disagreement. This work also reflects other innovations and analysis, notably that not all regions of an RNA are likely to form a single well-defined structure. As a result, an important component of the current work is the identification of regions in the HIV-1 RNA genome that do not form single well-defined structures.
Pseudoknot prediction
During the first stage, the full-length HIV-1 RNA genome was folded in 600-nt sliding windows moved in 100-nt increments using ShapeKnots with slope, intercept, P1, and P2 parameters set to previously defined values (1.8, −0.6, 0.35, 0.65) using 1M7- SHAPE data19,22. Additional folds were computed at the ends of the genome to increase the number of windows that cover terminal sequences. Predicted pseudoknots were retained if the structure appeared in a majority of windows and had low SHAPE reactivity on both sides of the pseudoknotted helix. This list of pseudoknots was used for all later stages of modeling.
Partition function modeling
The partition function was calculated using Partition26,47 and included both 1M7 and differential SHAPE data in the free energy penalty. The max pairing distance was set to 500 nts. Partition was run in 1600-nt windows with a step size of 375 nts. Two extra windows (lengths of 1550 and 1500 nts) were run on the 5' and 3' end sequences to increase sampling at the true ends and to reduce the effect of non-optimal cut site selection. Six sequences (the primer binding site, dimerization sequence, and four pseudoknots known to be involved in unusual or special interactions) were constrained as single stranded during partition function calculations. From the individual partition function files, the Shannon entropy of base pairing was calculated as:
| Hi=−∑j=1Jpi,jlog10pi,j | (5) |
|---|
Where _p_i,j is the probability of pairing for nucleotides i and j over all potential J partners25. Following this calculation, 300 nts were trimmed from the 5' and 3' ends of each window that did not flank the true 5' and 3' ends of the RNA. This calculation retained more consistent internal values and discarded values skewed by end effects. Shannon entropy windows were combined by averaging, creating a single entropy file.
Individual probable pairs from each window were then trimmed using the same approach outlined for the Shannon entropy. Base pairs that formed with a probability less than 10−4 were removed to decrease computation time. Windows were combined, and all remaining pairs were averaged over all of the windows in which they could have appeared. A heuristic color scale was developed from the combined partition file to indicate relative likelihood of a pair appearing in the final structure. The resulting pairs were plotted as arcs (Fig. 4). Base pairs with a probability greater than 0.99 were used as double-stranded constraints in the next step.
Minimum free-energy modeling
A minimum free energy structure was generated using Fold47, 1M7 SHAPE data, and differential SHAPE data. A window size of 3,000 nts with a step size of 300 nts was used to generate potential structures over each window. Four folds (3100, 3050, 2950, and 2900 nts from the ends) were also generated to increase the number of structure models at the termini. These folds from overlapping windows were then combined into a complete structure by comparing base pairs common to each window and requiring that pairs in the final structure appear in a majority of potential windows. As a final step, pseudoknotted helices were incorporated (Supplementary Fig. 2).
Error analysis and determining a minimum number of reads required for accurate RNA structure modeling
The mutation rates for each of the contributing signals (SHAPE modified, untreated, denatured) were modeled using a Poisson distribution because discrete events from individual reads contribute to the overall signal. The variance of a Poisson distribution is equal to the number of observations; thus, the standard error of a ‘true’ rate can be modeled as:
| SErate=λreads=ratsreads | (6) |
|---|
where λ is the number of events (mutations observed), reads is the read depth at the modeled nucleotide (both mutations and non-mutations), and rate is the number of events per read. As expected, bootstrapping of the standard error of SHAPE reactivity showed an _x_−1/2 power relationship as a function the read depth (Supplementary Fig. 6).
Using a deeply sequenced RNA (greater than 50,000 reads for each nucleotide), the number of expected mutation events at much lower read depths is known with high precision. Mutation events can be sampled from a Poisson distribution across the entire RNA to create profiles of plausible SHAPE data. To determine a minimum threshold for number of reads necessary for accurate SHAPE-directed secondary structure modeling, we examined the 16S rRNA because it is modeled poorly in the absence of experimental data (~50% sensitivity). For each simulated read depth, we created 100 SHAPE trajectories based on the expected Poisson variance at the simulated read depth and modeled it using RNAstructure Fold (Fig. 3b). As expected, modeling accuracy improved as read depth increased. For accurate nucleotide resolution structure modeling, we recommend at least 5000 reads; however, even at 500 reads, the measurement is useful for structure modeling (Fig. 3b).
Hit level calculation and comparison with other reports
SHAPE-MaP structure analysis as read out by massively parallel sequencing presents a valuable tool for structural interrogation of RNA at a single nucleotide level. Several other approaches have been developed with similar goals. To compare the read depth requirement of SHAPE-MaP (and its mutational profiling readout) with other approaches, we calculated a “hit level”. The hit level metric quantifies the total background-subtracted signal per nucleotide of transcript:
| hit level=total eventsS−read depthSread depthB×total eventsBtranscript length | (7) |
|---|
where the subscripts S and B indicate the experimental sample and background control, respectively; events are either ligation-detected sequence stops or mutations, depending on readout method, and read depth corresponds to the median number of reads overlapping each nucleotide in the transcript. We obtained a hit level of 160 for the 16S rRNA. Since mutation counts in SHAPE-MaP are proportional to read depths, we estimated the relationship between our sequencing read depth and hit level by dividing our observed hit level by the median read depth in an experimental condition. A hit level of ~15 is required to fully recover RNA structure information as interrogated by SHAPE, although highly useful structure models were consistently obtained at hit levels as low as 5 (Fig. 3b).
High-resolution RNA structure probing and modeling requires that most or all of an RNA be interrogated at a high hit level. Individual regions probed at low hit levels, even if the overall average hit level is ≥5, are likely to contain notable errors. In PARS experiments, a minimum threshold of 1 average read stop per nucleotide of transcript was required5,50, corresponding to hit level of 1, assuming zero background for enzymatic cleavage data. Similarly, a report describing DMS chemical probing, structure-seq, used a similar threshold of ≥1 average stop per A or C nucleotide10; this corresponds to an estimated hit level (by our definition) of 0.2, assuming a signal:background ratio of 1.7 (estimated from Extended Data Fig. 1, panel d in ref. 10) and that half of all transcript nucleotides are A or C. A minimum of 15 reads per A or C on average was required by the creators of DMS-seq11. This corresponds to a hit level of 3.3, assuming a signal:background ratio of 1.8 (estimated from Fig. 1, panel c in ref. 11). The benchmarking and bootstrapping analysis for modeling accuracy reported here (Fig. 3b) has not been implemented in prior massively parallel sequencing-based RNA structure analyses5,7,8,10,11,50; the authors of SHAPE-seq8 and Mod-seq51 have independently noted the importance of read depth in obtaining quantitative RNA structure probing information.
This hit level analysis emphasizes that, although several prior studies have been performed in which the full complement of RNAs in a given transcriptome were present during the probing phase of the experiment, only a few thousand nucleotides in each case were sampled at a depth consistent with recovery of the underlying structure information obtainable using DMS or enzyme probes.
Algorithmic discovery of HIV-1 regions with low Shannon entropy and low SHAPE reactivity
Overlaps of regions with both low SHAPE reactivity and low Shannon entropy were used to identify regions likely to have a single well-determined structure. First, local median SHAPE reactivity and Shannon entropy were calculated over centered sliding 55-nt windows. Next, we selected regions in which the local median fell below the global median for more than 40 nts in both Shannon entropy and SHAPE reactivity. Regions were combined if they were separated by fewer than 10 nts. Finally, regions were expanded to include nested secondary structures from the minimum predicted free-energy model.
To exclude the possibility that the algorithmically discovered structured regions overlapped known RNA elements merely by chance, we generated a randomized pool of segments and calculated the expected distribution of overlapping nucleotides (Table S2). We maintained the same number and length of segments but randomized their locations within the 9173-nt genome. Out of 105 trials, only 219 showed a larger overlap than we observed, corresponding to a _p_-value of 0.002.
HIV-1 mutagenesis
Mutations were introduced into HIV-1 pNL4-3 subclones spanning the region of interest by site-directed mutagenesis (QuikChange XL, Agilent) and verified by sequencing. The mutated subclone fragment was re-introduced into the full-length pNL4-3 plasmid52. The full-length mutant genome sequence was verified by conventional automated sequencing using 16 or more overlapping primers. Viruses from mutant and wild-type NL4-3 plasmids were produced by transfection as outlined above (to yield ~12 ng viral RNA per mL viral supernatant). Virion production was measured by p24 assay (AlphaLISA HIV p24 kit, PerkinElmer AL207C). Viruses were measured for infectivity on TZM-bl indicator cells53 (using Glo Lysis buffer and the Luciferase Assay System; Promega).
Mutations were designed to disrupt the primary pseudoknot sequence but maintain amino acid identity in coding sequences (Supplementary Fig. 8). The primary pseudoknotted helix in U3PK partially overlaps a binding site for transcription factor SP1. A total of three consecutive SP1 binding sites exist in HIV-1. Two point mutations introduced into the U3PK construct overlap the third SP1 binding site. Previous work has demonstrated that only a single binding site is required for complete viral function54. The SP1 protein tolerates variation at several places in the binding consensus sequence and the mutations introduced here maintained a canonical SP1 binding site. To analyze effects on virus production due to the SP1 mutation, the same U3 mutations were introduced into the 5' U3 region of the pNL4-3 clone (with and without concomitant 3' mutations). The resulting viruses had either no phenotype (the 5' U3 mutations alone) or the same phenotype as the original U3PK mutant (those containing both 5' and 3' U3 mutations), suggesting that alteration of the SP1 binding site did not disrupt viral RNA production. The double mutant species was used for viral spread and competition assays as described below.
Separation of mutant U3PK and wild-type NL4-3 SHAPE-MaP data
Gene-specific primer SHAPE-MaP data for U3PK (for the construct in which the 3' U3 alone was mutated) revealed that the three nucleotides targeted for mutagenesis showed unusually high mutation rates when aligned to the mutant sequence, suggesting the presence of multiple sequence populations. Quantifying the relative abundance of each variant sequence showed that 61.8% of reads contained the native sequence, 36.0% contained the designed mutant sequence, and a small fraction (2.2%) contained other sequences (Fig. 5a, Supplementary Fig. 10). These ratios suggest that recombination between the native sequence and mutant U3 regions occurred during transfection producing fitter HIV-1 virus that grew more rapidly than the mutant virus during virus culture. This mutant virus was grown in H9 cells (ATCC) for 3 weeks prior to RNA extraction and SHAPE-MaP testing. Reactivity profiles for the designed mutant and the wild-type sequence were created by computationally separating the reads after alignment (Fig. 5b). In addition, SHAPE-MaP reactivity data for the three mutated nucleotides were obtained by selecting reads targeting two mutated nucleotides at a time to assign wild-type or mutant membership, allowing variation at the third nucleotide to determine the mutation rate. This approach is widely applicable to chemically probing populations of RNAs in which each sequence fraction is larger than the expected reverse transcription-induced per-nucleotide mutation rate (~1% in this work).
HIV replication assays
Viruses were tested for cell-to-cell spread in Jurkat (ATCC) and H9 T-cell lines. Virus inocula were normalized by TZM-bl infectivity at a low multiplicity of infection (less than 0.01) prior to infection and used to infect 5 × 105 cells in 1 mL RPMI-1640 medium in 12-well plates; infections were carried out in duplicate. A full medium change was performed 3 days post-infection (dpi), and the medium in each well was harvested and replaced 4, 5, and 6 dpi. Viral concentrations were quantified by p24 assay (AlphaLISA HIV; PerkinElmer).
HIV competition assays
Mutant and native sequence virus were mixed at a 10:1 ratio, respectively, and used to infect 5 × 105 Jurkat cells in 1 mL total volume in 12-well plates. Infections were performed using half as much mutant and 20-fold less wild-type virus relative to the viral replication assays. Competition experiments were carried out in duplicate. Medium was initially harvested at 2 dpi to represent the initial inoculum. The medium was harvested at 3, 4, 5, and 6 dpi, and p24 (capsid protein) was quantified in medium (AlphaLISA HIV p24 kit). We required that p24 levels increase exponentially through day 6 to ensure that uninfected cells were in excess through the infections. Viral RNA was purified from medium (QIAamp viral RNA mini kit, Qiagen) and reverse transcription using SuperScript III (Life Technologies) was carried out using Primer ID primers55 to barcode each cDNA produced and eliminate population biases introduced during PCR. Subsequent sample preparation was performed as described above for SHAPE-MaP using targeted gene-specific primers.
After sequencing, paired-end reads were merged into longer synthetic reads using FLASh (Fast Length Adjustment of Short reads)56. Next, synthetic reads were aligned to the expected NL4-3 sequence for the targeted regions using Bowtie 245 (using default parameters). A consensus read was built for each PrimerID based on a Phred score voting metric. IDs matching either native or mutant sequences were required to have the expected point mutations in all locations in order to be considered. The fraction of mutant IDs was expressed as the number of mutant IDs out of the sum of mutant and native IDs. Relative fitness of mutant viruses was determined from the rate of change of the ratio of mutant to NL4-3 measured over time31.
Calculation of differences in SHAPE reactivities in pseudoknot mutants
Standard error measurements of SHAPE reactivities, estimated from the Poisson distribution, are dependent on the number of reads obtained for each sample. The observation that standard error decreases with the inverse square of read depth (Supplementary Fig. 6) was used to derive a scaling equation that normalizes to a common depth of 8000 reads to account for differences in sequencing depth between samples. The standard error scaling factor, _f_0, was calculated for each sample based on the average read depth, _r_ave, of the lowest sequenced component (SHAPE modified, untreated, and denaturing conditions) contributing to the SHAPE reactivity profile:
| f0=(rave)−12(8000)−12 | (8) |
|---|
After scaling standard errors to a common read depth, significance for each point was calculated using a modified z-factor test49 requiring differences to be greater than 1.96 times the sum of the standard errors. Z-factor scores greater than zero were considered significant:
| Zfactor=1−1.96(σPK+σWT)|μPK−μWT| | (9) | | ---------------------------------- | --- |
Isolated reactivity changes can be viewed as noise in the context of a global structure shift resulting from disruption of a pseudoknot. Therefore, in addition to the z-factor test, differences were required to be consecutive.
Supplementary Material
1
2
Acknowledgements
We are indebted to R.J. Gorelick for expert preparation of HIV-1 genomic RNA and many insightful discussions, to C.E. Hajdin for extensive discussion and for initial pseudoknot mutant design, to R. Swanstrom for critical discussions, to K. Compliment for expert technical assistance, and to W. Resch for competition calculations. TZM-bl cells were obtained from J. Kappes and X. Wu (Tranzyme Inc.) via the NIH AIDS Reagent Program. This work was supported by the NIH (AI068462 to K.M.W.) and the UNC CFAR (P30 AI50410). N.A.S. is a Lineberger Postdoctoral Fellow in the Basic Sciences and a recipient of a Ruth L. Kirschstein NRSA Fellowship (F32 GM010169). G.M.R. was supported in part by an NIH training grant in molecular and cellular biophysics (T32 GM08570).
Footnotes
Software and Data Availablity and Accession Codes
All software and processed massively parallel sequencing data are freely available as Supporting Information and from the website of the corresponding author. Sequencing files have been uploaded to the NCBI short read archive, accession SRX554885.
Author Contributions
The SHAPE-MaP strategy was conceived and designed by N.A.S. and K.M.W. SHAPE experiments were performed by N.A.S. and G.M.R. HIV-1 replication assays were designed and performed by N.A.S. and J.A.E.N. The SHAPE-MaP data analysis pipeline was created by S.B. RNA folding and motif discovery analyses were conceived and created by G.M.R. and K.M.W. All authors collaborated in interpreting the experiments and writing the manuscript.
Competing Financial Interests
N.A.S, S.B., and K.M.W. are listed as inventors on a provisional patent application based on elements of this work.
References
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mauger DM, Weeks KM. Toward global RNA structure analysis. Nat. Biotechnol. 2010;28:1178–1179. doi: 10.1038/nbt1110-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Meth. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weeks KM. RNA structure probing dash seq. Proc. Natl. Acad. Sci. 2011;108:10933–10934. doi: 10.1073/pnas.1107835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 11.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grohman JK, et al. A guanosine-centric mechanism for RNA chaperone function. Science. 2013;340:190–195. doi: 10.1126/science.1230715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gherghe C, et al. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGinnis JL, Weeks KM. Ribosome RNA assembly intermediates visualized in living cells. Biochemistry. 2014;53:3237–3247. doi: 10.1021/bi500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 18.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20:846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 21.Spitale RC, et al. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steen K-A, Rice GM, Weeks KM. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J. Am. Chem. Soc. 2012;134:13160–13163. doi: 10.1021/ja304027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doty P, Boedtker H, Fresco JR, Haselkorn R, Litt M. Secondary structure in ribonucleic acids. Proc. Natl. Acad. Sci. 1959;45:482–499. doi: 10.1073/pnas.45.4.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynen M, Gutell R, Konings D. Assessing the reliability of RNA folding using statistical mechanics. J. Mol. Biol. 1997;267:1104–1112. doi: 10.1006/jmbi.1997.0889. [DOI] [PubMed] [Google Scholar]
- 26.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brierley I, Pennell S, Gilbert RJC. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Micro. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillart J-C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5’-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 31.Resch W, Ziermann R, Parkin N, Gamarnik A, Swanstrom R. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 2002;76:8659–8666. doi: 10.1128/JVI.76.17.8659-8666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3’ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 33.Gilmartin GM, Fleming ES, Oetjen J. Activation of HIV-1 pre-mRNA 3' processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klasens BI, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghadessy FJ, Holliger P. Compartmentalized self-replication: a novel method for the directed evolution of polymerases and other enzymes. Methods Mol. Biol. 2007;352:237–248. doi: 10.1385/1-59745-187-8:237. [DOI] [PubMed] [Google Scholar]
- 36.Chen T, Romesberg FE. Directed polymerase evolution. FEBS Lett. 2014;588:219–229. doi: 10.1016/j.febslet.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollom E, et al. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 2013;9:e1003294. doi: 10.1371/journal.ppat.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 39.Lorsch JR, Bartel DP, Szostak JW. Reverse transcriptase reads through a 2′-5′ linkage and a 2′-thiphosphate in a template. Nucleic Acids Res. 1995;23:2811–2814. doi: 10.1093/nar/23.15.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson JT, Nickens DG, Burke DH. HIV-1 reverse transcriptase pausing at bulky 2’adducts is relieved by deletion of the RNase H domain. RNA Biol. 2006;3:163. doi: 10.4161/rna.3.4.4020. [DOI] [PubMed] [Google Scholar]
- 41.Matathias A, Fox D, Crouse J. SuperScript II RNase H reverse transcriptase. 18064-3. Focus On, Life Technologies; 1999. [Google Scholar]
- 42.Beckman RA, Mildvan AS, Loeb LA. On the fidelity of DNA replication: manganese mutagenesis in vitro. Biochemistry. 1985;24:5810–5817. doi: 10.1021/bi00342a019. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 44.Williams R, et al. Amplification of complex gene libraries by emulsion PCR. Nat. Meth. 2006;3:545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- 45.Staple DW, et al. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter JD. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007:90–95. [Google Scholar]
- 47.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Byun Y, Han K. PseudoViewer: web application and web service for visualizing RNA pseudoknots and secondary structures. Nucleic Acids Res. 2006;34:W416–22. doi: 10.1093/nar/gkl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Chung T, Oldenburg K. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 50.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talkish J, May G, Lin Y, Woolford JL, McManus CJ. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA. 2014;20:713–720. doi: 10.1261/rna.042218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Derdeyn CA, et al. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrich D, et al. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 1989;63:2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2