Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans (original) (raw)
Abstract
RNA granules have been likened to liquid droplets whose dynamics depend on the controlled dissolution and condensation of internal components. The molecules and reactions that drive these dynamics in vivo are not well understood. In this study, we present evidence that a group of intrinsically disordered, serine-rich proteins regulate the dynamics of P granules in C. elegans embryos. The MEG (maternal-effect germline defective) proteins are germ plasm components that are required redundantly for fertility. We demonstrate that MEG-1 and MEG-3 are substrates of the kinase MBK-2/DYRK and the phosphatase PP2APPTR−½. Phosphorylation of the MEGs promotes granule disassembly and dephosphorylation promotes granule assembly. Using lattice light sheet microscopy on live embryos, we show that GFP-tagged MEG-3 localizes to a dynamic domain that surrounds and penetrates each granule. We conclude that, despite their liquid-like behavior, P granules are non-homogeneous structures whose assembly in embryos is regulated by phosphorylation.
DOI: http://dx.doi.org/10.7554/eLife.04591.001
Research organism: C. elegans
eLife digest
For a gene to be expressed as a protein, its DNA is first used as a template to make a molecule of RNA, which is then translated to make the protein. In most cells, RNA molecules concentrate into aggregates called RNA granules. These granules contain both RNA and proteins that bind to RNA and are used to transport, store, and regulate the translation and breakdown of RNA molecules. Unlike many other structures within cells, RNA granules are not surrounded by a membrane; and the molecules that hold RNA granules together are not known.
P granules are a type of RNA granule that is found in the germ cells (the cells that go on to form eggs and sperm) of a microscopic worm called C. elegans. When a C. elegans embryo is still a single cell, P granules move throughout the cell and the P granules at the front of the cell dissolve, while those at the back condense. As such, when the single-celled embryo divides, the front forms a cell without P granules (that will go on to form the tissues of the worm's body) and the back becomes a P granule-containing germ cell.
Two proteins called MBK-2 and PPTR-1 have opposite effects on P granules: MBK-2 causes P granules to dissolve, while PPTR-1 makes them form. MBK-2 is an enzyme that adds phosphate groups onto other proteins, whereas PPTR-1 is part of an enzyme that removes such groups. Wang et al. have now searched for proteins that interact with MBK-2 and PPTR-1 in order to identify the molecules that regulate the assembly of P granules. They found that a group of proteins, known as MEG proteins, are acted upon by both of these proteins. Wang et al. found that MBK-2 adds phosphate groups to MEG proteins, which encourages granules to disassemble, while PPTR-1 removes these groups to promote granule assembly.
Wang et al. generated mutant worms that lacked each of the MEG proteins. These mutant worms had fewer and smaller P granules than normal worms. Without MEG proteins, P granules failed to assemble or disassemble normally and the worms were infertile. Using high resolution microscopy, Wang et al. observed that the MEG proteins wrap around the P granules and that one of the MEG proteins—called MEG-3—follows an almost ribbon-like path that surrounds and enters each granule. These observations suggest that the MEG proteins stabilize RNA granules by forming a cage-like scaffold around each granule. How the MEG proteins—which are predicted to lack a fixed or ordered three-dimensional structure and show no similarity to proteins with known functions—assemble into a scaffold will be the focus of future studies.
DOI: http://dx.doi.org/10.7554/eLife.04591.002
Introduction
RNA granules are ubiquitous cytoplasmic organelles that contain RNA and RNA-binding proteins (Kedersha et al., 2013; Buchan, 2014). Several types of RNA granules have been described, including germ granules in germ cells, neuronal granules in neurons, and stress granules and P bodies in somatic cells. Their functions include mRNA transport and storage and the regulation of mRNA degradation and translation. Unlike other organelles, RNA granules are not bound by limiting membranes and their internal components are in constant flux with the surrounding cytoplasm. RNA granules assemble and disassemble in response to developmental or environmental cues (Kedersha et al., 2013; Buchan, 2014). Live imaging studies in Caenorhabditis elegans zygotes have suggested that RNA granules behave like liquid droplets that undergo phase transitions (Brangwynne et al., 2009). Granule components exist in a condensed liquid or gel-like phase in the granule and a dispersed phase in the cytoplasm (Weber and Brangwynne, 2012; Toretsky and Wright, 2014). In vitro studies have lent support to this hypothesis by demonstrating that purified proteins can undergo phase transitions in aqueous solutions. For example, proteins that contain weak, multivalent protein-binding domains undergo liquid–liquid demixing in concentrated solutions to form micron-sized droplets (Li et al., 2012). Similarly, RNA-binding proteins that contain low sequence-complexity domains assemble into ordered fibers that coalesce into hydrogels when maintained at low temperature (Frey et al., 2006; Kato et al., 2012). The proteins that drive the phase transitions in vivo, however, are not known. Several proteins and RNAs are required for granule integrity (Buchan, 2014), and recently the kinase DYRK3 has been implicated in the regulation of stress granule dissolution and condensation (Wippich et al., 2013). In the present study, we identify the substrates of the C. elegans DYRK3 homolog MBK-2 and demonstrate that phosphorylation and dephosphorylation of these substrates drive the dissolution and condensation of P granules in embryos.
P granules are the germ granules of C. elegans (Strome and Wood, 1982; Updike and Strome, 2010). P granules are perinuclear during most of the germline development. During the oocyte-to-embryo transition, P granules detach from nuclei and disperse in the cytoplasm for asymmetric segregation to the nascent embryonic germline. Preferential dissolution of the granules in the anterior and condensation in the posterior of the zygote cause the granules to accumulate in the cytoplasm destined for the germline blastomere P1 (Figure 1) (Brangwynne et al., 2009; Gallo et al., 2010). P granules segregate asymmetrically for three more divisions until the P granules are uniquely concentrated in the germline founder cell P4 (Strome and Wood, 1983). Two groups of RNA-binding proteins form the core components of P granules: the RGG domain proteins PGL-1 and PGL-3 and the Vasa-related helicases GLH-1, 2, 3, and 4 (Updike and Strome, 2010). A self-association domain in the PGL family is essential for granule nucleation (Hanazawa et al., 2011), and FG repeats in GLH-1 are required for P granules to associate with nuclei (Updike et al., 2011).
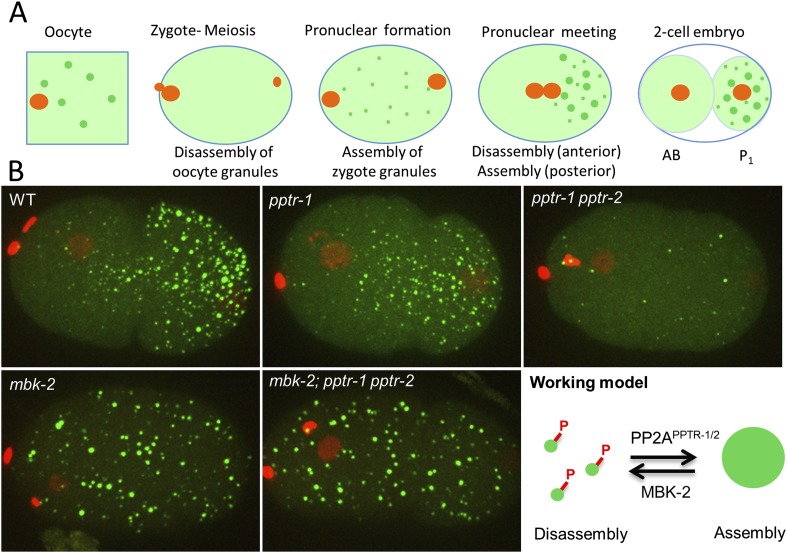
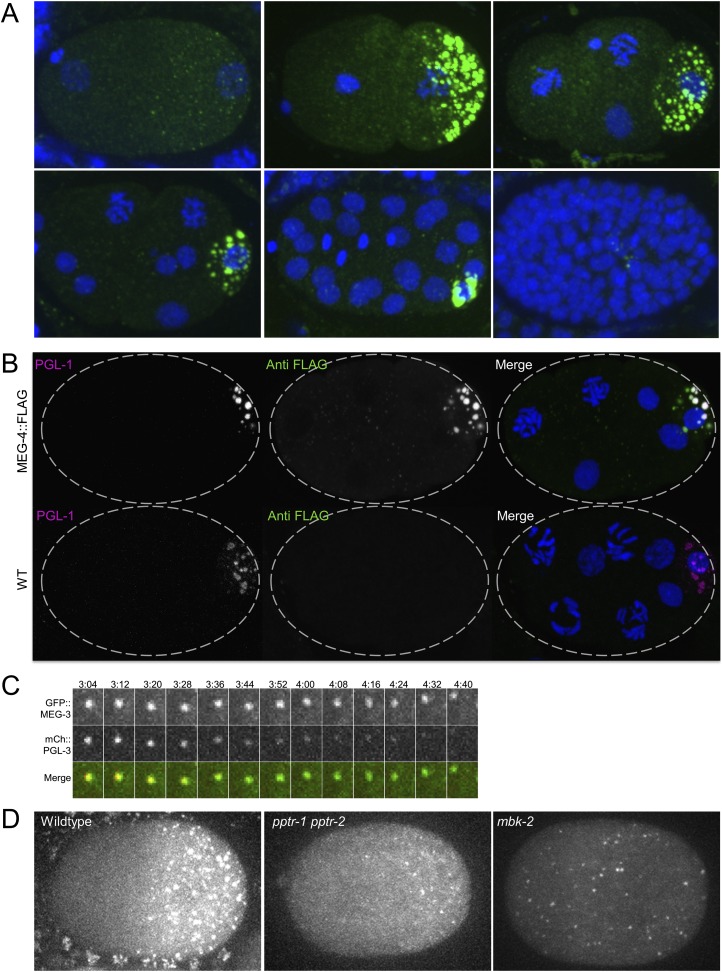
Figure 1. MBK-2 and PP2APPTR−1/2 are an opposing kinase/phosphatase pair.
(A) P granule dynamics during the oocyte to embryo transition. Green puncta are P granules, pale green color represents P granule components that diffuse into the cytoplasm. Orange represents pronuclei. P1 is the germline blastomere. (B) Zygotes of the indicated genotypes expressing GFP::PGL-1 and mCherry::Histone 2B during pronuclear migration. Full genotypes are pptr-1(tm3103), pptr-1(tm3103) pptr-2(RNAi), mbk-2(RNAi), and mbk-2(RNAi);pptr-1(tm3103) pptr-2(RNAi). Right: working model. Phosphorylation by MBK-2 disassembles granules. Dephosphorylation by PP2APPTR−1/2 assembles granules. Each embryo is 50 μm in length. Anterior is to the left, posterior is to the right.
DOI: http://dx.doi.org/10.7554/eLife.04591.003
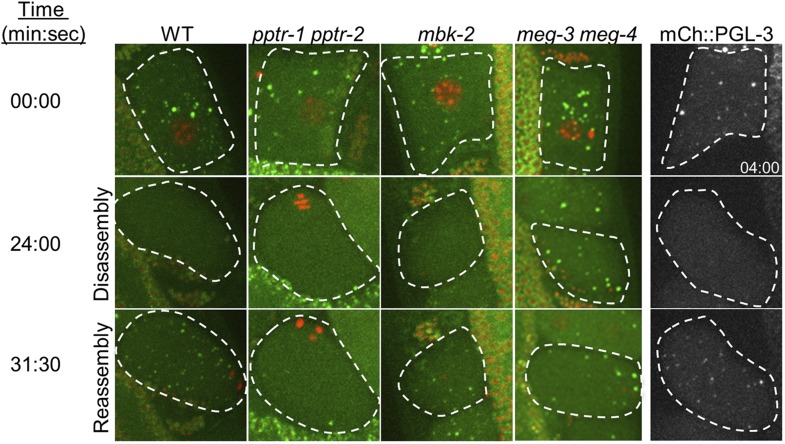
Figure 1—figure supplement 1. Oocyte P granule disassembly during the oocyte-to-zygote transition.
First four columns: time points from movies of eggs (highlighted by dashed lines) undergoing the oocyte-to-embryo transition (top to bottom) and expressing GFP::PGL-1 (green) and mCherry::H2B (red). At time point zero, the oocytes are still in oviduct and not yet fertilized. At 24 min, fertilization has occurred and the zygotes are undergoing the first meiotic division. The oocyte granules have disassembled, except in meg-3 meg-4 zygotes. At 31 min, the zygotes are finishing the second meiotic division, and the zygote granules are forming. Zygote granule assembly is impaired in pptr-1 pptr-2 and meg-3 meg-4 zygotes. Full genotypes are: pptr-1(tm3103) pptr-2(RNAi), mbk-2(RNAi), and meg-3(RNAi) meg-4(RNAi). Fifth column: time points from movies of eggs expressing mCherry::PGL-3. Note that the top image was taken at 4:00. All other images are time-matched to those taken in GFP::PGL-1 mCherry::H2B strains.
In previous studies, we identified two potential regulators of P granule dynamics: MBK-2, the C. elegans DYRK3 homolog (Aranda et al., 2011; Wippich et al., 2013) and PPTR-1, a regulatory subunit of the heterotrimeric phosphatase PP2A (Padmanabhan et al., 2009). Loss of mbk-2 and pptr-1 has opposite effects on P granule dynamics. All P granules remain condensed in mbk-2 zygotes and all P granules disperse in pptr-1 zygotes (Pellettieri et al., 2003; Quintin et al., 2003; Gallo et al., 2010). These phenotypes suggest that MBK-2 and PPTR-1 proteins may share substrates whose phosphorylation regulates the phase of P granules. To investigate this possibility, we searched for MBK-2 and PPTR-1 substrates and identified a group of intrinsically disordered proteins that regulate P granule dynamics.
Results
PPTR-1 functions redundantly with PPTR-2 to stabilize P granules and is antagonized by MBK-2
To determine the earliest time point at which P granules become dynamic during the oocyte-to-zygote transition, we monitored P granules in the oviduct and uteri of live hermaphrodites from ovulation through the first embryonic divisions. We used a GFP::PGL-1 transgene to mark P granules and an mCherry::histone transgene to mark chromosomes. Shortly after ovulation, P granules that were present in the oocyte cytoplasm (oocyte granules) disassemble and new granules (zygote granules) form throughout the cytoplasm as the maternal pronucleus completes the second meiotic division (Video 1, Figure 1A, Figure 1—figure supplement 1). After meiosis, as the zygote becomes polarized along the anterior/posterior axis, granules in the anterior cytoplasm are quickly disassembled and granules in the posterior cytoplasm continue to grow and fuse to form large (≥1 micron) granules (Figure 1, Video 2). We observed the same dynamics with a mCherry fusion to a second P granule component (PGL-3) (Figure 1—figure supplement 1).
Video 1. P granule dynamics during the oocyte-to-zygote transition.
Time-lapse of eggs undergoing the oocyte-to-embryo transition in the gonad of a hermaphrodite expressing GFP::PGL-1 and mCherry::H2B. An oocyte in the oviduct, several sperm in the spermatheca, and a zygote in the uterus are highlighted in the first frame of the movie. Key stages in the P granule disassembly and assembly process are also highlighted in later frames. Images are maximum intensity projections of 11 z stacks separated by 1 µm steps. Stacks were taken every 30 s, total movie time is 30 min 30 s, movie is played back in 60× real time.
DOI: http://dx.doi.org/10.7554/eLife.04591.005
Video 2. P granule dynamics in wild-type and meg-3 meg-4 zygotes.
Time-lapse of embryos expressing GFP::PGL-1 and mCherry::H2B. Images are maximum intensity projections of 8 z planes separated by 1 μm steps. Stacks were taken every 8 s, total movie time is 17 min 52 s, movie is played back in 80× real time. Full genotype of mutant is meg-3(tm4259) meg-4(ax2026). Very small granules form transiently in the posterior of the meg-3 meg-4 embryo at 7:20. The bright red puncta in the anterior (left) side of the embryos are polar bodies. In the wild-type movie, there are also red puncta above the embryo, these are sperm outside of the egg shell.
DOI: http://dx.doi.org/10.7554/eLife.04591.006
The kinase MBK-2 is required for P granule asymmetry in zygotes (Pellettieri et al., 2003; Quintin et al., 2003). We found that in zygotes derived from mothers lacking mbk-2 (hereafter referred to as mbk-2 zygotes), oocyte granule disassembly and zygote granule assembly proceeded as in wild-type during meiosis (Figure 1—figure supplement 1). The zygote granules, however, failed to disassemble in the anterior cytoplasm during zygote polarization (Figure 1B).
PPTR-1 is a B′/B56 regulatory subunit of PP2A that is required for P granule maintenance during mitosis (Gallo et al., 2010). In pptr-1 mutant zygotes, disassembly of oocyte granules and assembly of zygotic granules proceeded normally during meiosis. After meiosis, however, granules in the posterior did not grow (Figure 1B) and all granules eventually disassembled by the onset of mitosis, as previously reported (Gallo et al., 2010). PPTR-1 is 58% identical at the amino acid level to PPTR-2, another predicted B′/B56 regulatory subunit of PP2A (WormBase; [Harris et al., 2014]). To determine whether PPTR-2 might also regulate P granule dynamics, we compared zygotes derived from pptr-1 mothers and pptr-1 pptr-2 mothers_._ In pptr-1 pptr-2, zygote granule assembly during meiosis was less robust and disassembly began even earlier during pronuclear migration (Figure 1B and Figure 1—figure supplement 1). pptr-2 single mutants appeared as wild-type (data not shown). We conclude that pptr-1 and pptr-2 contribute redundantly to the assembly/stabilization of P granules in zygotes.
Intriguingly, when we depleted mbk-2 in pptr-1 pptr-2, we observed the same phenotype as in mbk-2: new P granules assembled in the zygote and remained stable through the first division (Figure 1B). mbk-2 therefore is epistatic to pptr-1 pptr-2, as might be expected in a ‘no phosphorylation’ scenario. These observations suggest that MBK-2 and PP2APPTR−1/2 work in opposition to promote P granule disassembly (MBK-2) and re-assembly (PP2APPTR−1/2). One possibility is that MBK-2 and PP2APPTR−1/2 share substrates that, when phosphorylated, destabilize P granules in zygotes (working model in Figure 1B).
Three intrinsically disordered proteins bind to PPTR-1
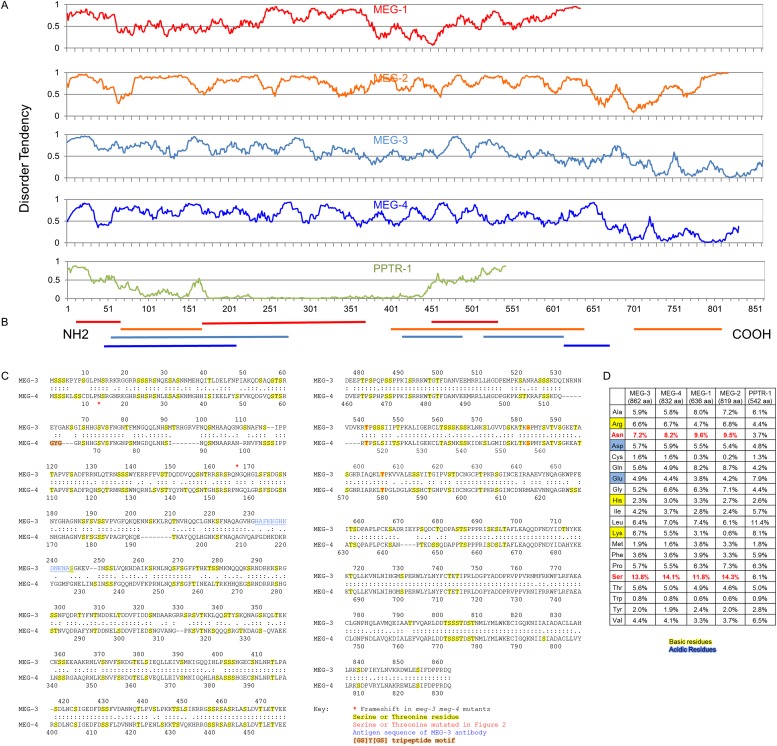
To identify potential substrates of PP2APPTR−1/PPTR−2, we screened a LexA-PPTR-1 fusion against cDNAs from a mixed-stage C. elegans library fused to the GAL-4 activation domain. From 1.2 × 107 transformants, we obtained ∼4500 positive colonies. We sequenced plasmids from 111 colonies and identified 32 unique genes (Figure 2—source data 1). We tested each gene by RNAi (1) for defects in P granule distribution in wild-type embryos, or (2) for suppression of the pptr-1 hyper-granule disassembly phenotype and identified three genes: meg-1, F52D2.4, and C36C9.1 (Figure 2—source data 1). meg-1 (and its paralog meg-2) was identified previously as a P granule component specific to early embryos (Leacock and Reinke, 2008). F52D2.4 was identified previously in a yeast two-hybrid screen as a gex-3 interactor and named gei-12 (Tsuboi et al., 2002). Based on our findings (see below), we have renamed this gene meg-3. C36C9.1 is 71% identical to F52D2.4/meg-3 at the amino acid level, and we have named it meg-4 (Figure 2). meg-3 and meg-4 were identified previously as pptr-2 interactors in a genome-wide yeast two-hybrid screen (Simonis et al., 2009). All four meg genes are X-linked and code for 70–95.8 kDa proteins that contain 11.8–14.3% serine and 7.2–9.6% asparagine residues and >60% residues predicted to be in disordered regions (Figure 2). meg-1/2, however, are not homologous to meg-3/4 (WormBase; [Harris et al., 2014]).
Figure 2. The MEG proteins contain sequences predicted to be intrinsically disordered.
(A) Graphs showing the disorder tendency of sequences along each protein calculated using IUPRED (http://iupred.enzim.hu/) using ‘long disorder’ parameters (Dosztányi et al., 2005). Scores above 0.5 indicate disorder. Number of predicted disordered residues: MEG-1: 420 of 636 aa (66%), MEG-2: 715 of 819 aa (87%), MEG-3: 542 of 862 aa (63%), MEG-4: 570 of 832 aa (69%), PPTR-1: 97 of 542 aa (18%). PPTR-1 is included here as an example of a mostly ordered protein. (B) Lines below graph: regions of low complexity sequence as defined using SEG (http://mendel.imp.ac.at/METHODS/seg.server.html) (Wootton, 1994). Using the default SEG parameters (SEG 12 2.2 2.5) as used in Kato et al. (2012), there are no low complexity sequences greater than 36 residues in the MEG proteins. Using a larger window size (SEG 45 3.4 3.75), all MEGs have predicted low complexity regions as shown, but PPTR-1 does not. (C) Protein sequence alignment of MEG-3 and MEG-4, which are 71% identical to each other. Yellow highlight: serine and threonine residues. Red: predicted MBK-2 phosphorylation site mutated in Figure 3 kinase assay. Blue: antigen sequence of MEG-3 antibody. Orange highlight: [GS]Y[GS] tripeptide motif (Kato et al., 2012). There is only 1 instance of this sequence in MEG-4 and none in MEG-3. Red star: position of the frameshift in the meg-3(tm4259) and meg-4(ax2026) mutants. meg-3(tm4259) is a deletion of 623 nucleotides starting at nucleotide 543 (amino acid 165), followed by an insertion of ‘TACGA’. The frameshift inserts the sequence ‘KQGRH’ at amino acid 165 followed by a premature stop. meg-4(ax2026) is a deletion of 7 nucleotides starting at nucleotide 37 (amino acid 13). The frameshift inserts the amino acids ‘EETEKVTALIVEAIWKVHRKIWDTTLVLKNCFIRS’ at amino acid 13, followed by a premature stop. See Table 2 for full allele descriptions. (D) Amino acid composition of indicated proteins. Basic residues are highlighted in yellow, acidic in blue. Red residues are overrepresented in MEG proteins.
DOI: http://dx.doi.org/10.7554/eLife.04591.007
Figure 2—source data 1. Candidates from yeast two-hybrid screen.
DNA was extracted and sequenced from 111 colonies grown on –Trp–Leu–Ura–His plates. RNAi feeding vectors for each candidate were obtained from the Ahringer or OpenBiosystems RNAi banks, or if unavailable were PCR amplified from genomic DNA and cloned into pL4440. *meg-3(RNAi) also knocks out meg-4 and vice-versa. + in the pptr-1 suppression column means that the RNAi treatment restores P granules in P blastomeres. Pleiotropic indicates a P granule phenotype accompanied by additional cellular defects. Entries in red are the MEG proteins, which uniquely affect P granules in the zygote stage (meg-3 and meg-4) or suppress the pptr-1 phenotype (meg-1) without inducing other phenotypes. Other phenotypes were as follows: emb = embryonic lethal, dpy = dumpy, egl = egg laying defect.
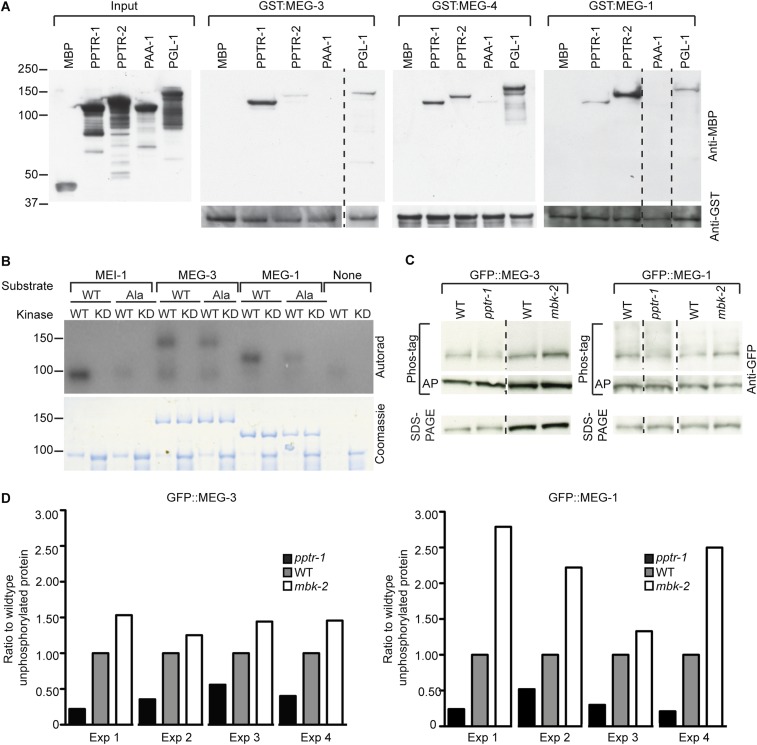
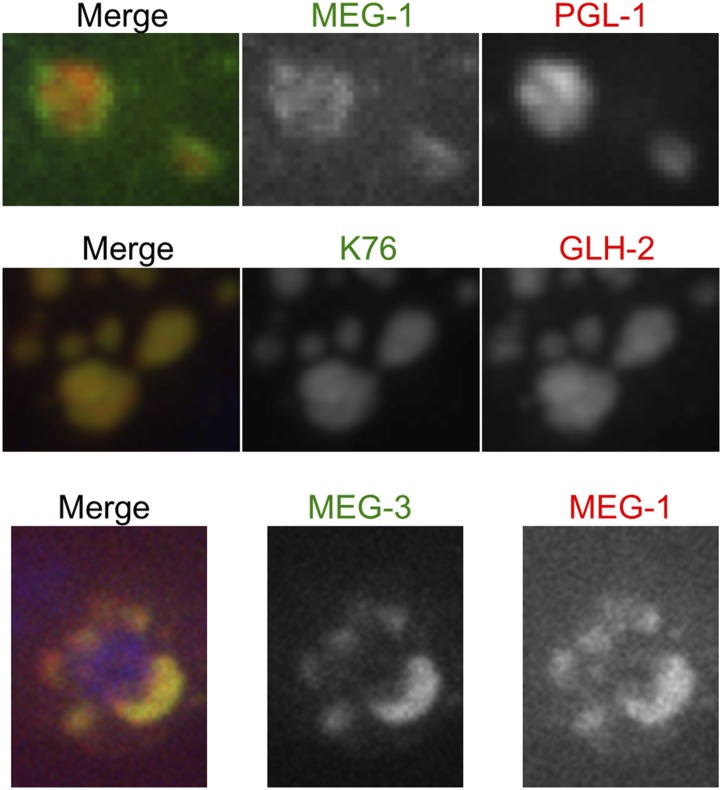
To test that the MEG-1, MEG-3, and MEG-4 proteins interact directly with PPTR-1 and PPTR-2, we performed GST pull down assays (Figure 3A). We found that all three bound to PPTR-1 and/or PPTR-2 but not to maltose-binding protein (MBP) or to PAA-1, the scaffolding subunit of PP2A. MEG-1, MEG-3, and MEG-4 also interacted with the P granule component PGL-1 (Figure 3A).
Figure 3. MEG-1 and MEG-3 are substrates of MBK-2 and PP2APPTR−1/2.
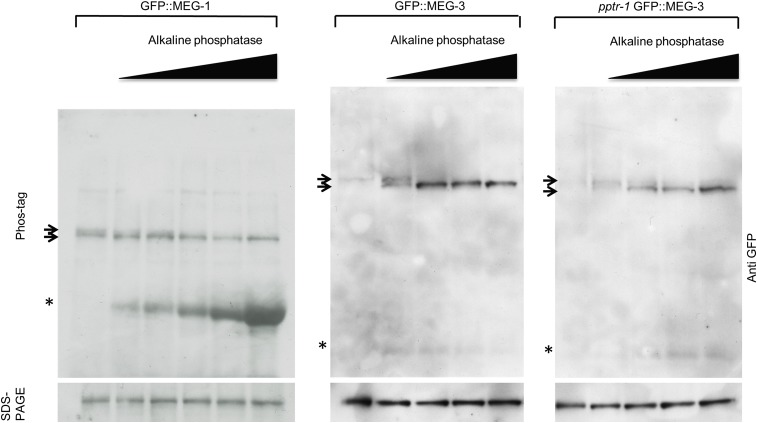
(A) Westerns of E. coli lysates expressing the indicated MBP fusions before (input) and after immobilization on columns containing the indicated GST fusions. MBP and PAA-1 are negative controls. PAA-1 is the scaffolding subunit of PP2A. Dashed lines separate individual gels. (B) In vitro kinase assay. MBP fused to wild-type or kinase-dead (KD) MBK-2 was incubated with gamma32P-ATP and MBP-fused substrates: MEI-1 (94 kDa); MEI-1(S92A) (94 kDa); MEG-3 (138 kDa with degradation band at ∼100 kDa); MEG-3(T541A S582A T605A) (138 kDa with degradation band at ∼100 kDa); MEG-1 (112 kDa); MEG-1(S574A) (112 kDa). MBP::MBK-2 is 99 kDa and autophosphorylates. MEI-1 is a previously known substrate of MBK-2 (Stitzel et al., 2006). Coomassie staining to control for loading is shown below. Phosphorylation is diminished in MEG-3(T541A S582A T605A) (64% of wild-type phosphorylation) and MEG-1 (S574A) (88% of wild-type phosphorylation). (C) Anti-GFP westerns of C. elegans embryonic lysates run on SDS-PAGE gels with (top) or without Phos-tag (bottom). Wild-type, pptr-1(tm3103), and mbk-2(RNAi) embryonic lysates expressing GFP::MEG-3 or wild-type, pptr-1(RNAi), and mbk-2(RNAi) embryonic lysates expressing GFP::MEG-1 were treated with or without alkaline phosphatase (AP) and equal amounts were loaded on gels with or without Phos-tag. Dashed lines separate individual gels. Loading control was protein run on SDS-PAGE without Phos-tag. (D) Graphs showing % unphosphorylated protein relative to wild-type in four Phos-tag experimental replicates. % unphosphorylated was calculated as the ratio of the intensity of the band corresponding to unphosphorylated protein on a Phos-tag gel over the intensity of the band on a non-Phos-tag gel (total protein) and normalized to wild-type.
DOI: http://dx.doi.org/10.7554/eLife.04591.009
Figure 3—figure supplement 1. MEG-1 and MEG-3 are phosphorylated in vivo.
Anti-GFP western of embryonic lysates expressing GFP::MEG-1 or GFP::MEG-3 treated with increasing amounts of alkaline phosphatase and run on Phos-tag SDS-PAGE (top) and SDS-PAGE without Phos-tag (bottom). Equivalent amounts of total cell lysates were loaded in each lane. Top arrow indicates position of phosphorylated band, bottom arrow indicates position of unphosphorylated band. Asterisk indicates alkaline phosphatase, which is detected to varying extents using these Western blotting conditions.
MEG-3 and MEG-1 are MBK-2 and PPTR-1 substrates
To test whether the MEGs are also MBK-2 substrates, we expressed MEG-1 and MEG-3 as recombinant MBP fusions in Escherichia coli and performed in vitro kinase assays with recombinant MBK-2 as in Cheng et al. (2009). MBK-2 could phosphorylate MBP::MEG-1 and MBP::MEG-3, but not MBP alone (Figure 3B). Using the DYRK consensus site K/R X1–3 S/T P (Himpel et al., 2000; Campbell and Proud, 2002), we identified putative consensus sites in MEG-1 (S574) and MEG-3 (T541, S582, T605). S574 in MEG-1 is reported as a phosphorylated site in Phosida (Gnad et al., 2007). We mutated each consensus site to alanine and repeated the kinase assays. Phosphorylation was reduced but not eliminated in the alanine mutants (Figure 3B). We conclude that recombinant MBK-2 phosphorylates MEG-1 and MEG-3 on consensus sites in vitro, as well as other sites that remain to be determined.
To examine whether MEG-1 and MEG-3 are phosphoproteins in vivo, we ran lysates from embryos expressing GFP::MEG-1 or GFP::MEG-3 on Phos-tag SDS-PAGE. Phos-tag retards the migration of phosphorylated proteins (Kinoshita et al., 2006). We detected one primary species for both GFP::MEG-1 and GFP::MEG-3 and several higher molecular weight species that did not resolve well into specific bands. Treatment with alkaline phosphatase eliminated the higher molecular weight species and increased the intensity of the lower species (unphosphorylated isoform, Figure 3C, Figure 3—figure supplement 1). The relative abundance of the unphosphorylated isoform increased in lysates depleted of mbk-2 and decreased in lysates depleted of pptr-1 (Figure 3C,D). We conclude that MEG-1 and MEG-3 are phosphoproteins in vivo and that MBK-2 and PPTR-1 promote and antagonize, respectively, their phosphorylation. Under all conditions, GFP::MEG-1 and GFP::MEG-3 still migrated as a mixture of phosphorylated and unphosphorylated isoforms. Because our assays were done in lysates that were depleted for MBK-2 by RNAi, and lysates that lacked PPTR-1 but retained PPTR-2, we do not know whether MBK-2 and PP2APPTR−1/2 are the only regulators of MEG-1 and MEG-3 phosphorylation or whether other kinases and phosphatases are also involved.
MEG-3 and MEG-4 are required for granule assembly in embryos
To determine the loss of function phenotype of meg-3, we obtained a deletion allele of meg-3 from the National Bioresource Project (Mitani, 2009). tm4259 is an out-of-frame, 623-nucleotide deletion that creates a frameshift at amino acid 165 followed by premature stop at amino acid 170 and therefore is a likely null (Figure 2). Since RNA-mediated interference experiments suggested that meg-3 and meg-4 function redundantly (Figure 4—figure supplement 1), we generated a meg-4 allele in the meg-3(tm4259) background by non-homologous end joining repair of CRISPR/Cas9-induced cuts in meg-4 (Paix et al., 2014). meg-4(ax2026) is a 7 bp deletion which causes a frameshift at amino acid 13, followed by a premature stop at amino acid 48 (Figure 2). We also generated a second meg-4 allele in the wild-type background by non-homologous end joining repair of two CRISPR/Cas9 cuts flanking the meg-4 ORF (Paix et al., 2014). meg-4(ax2081) is a 3.2 kb deletion that removes 733 bases upstream of the meg-4 ATG and 2565 bases of the meg-4 coding sequence (total length: 2589 bases).
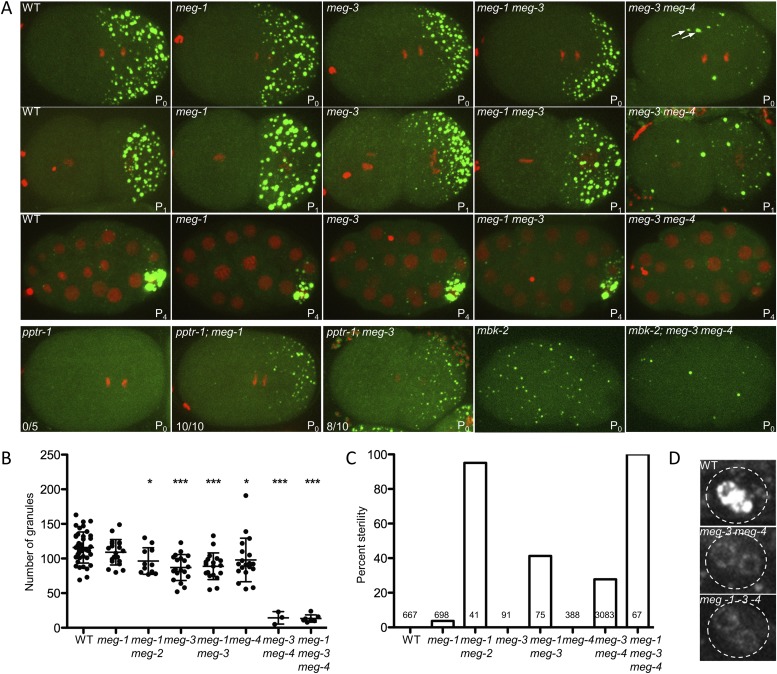
To determine whether meg-3 and meg-4 are required for P granule dynamics, we crossed in GFP::PGL-1 and mCherry::histone transgenes to analyze P granule dynamics in the meg-3, meg-4 single and double mutants (Figure 4). We observed the strongest phenotype in the double mutant. First, we noticed that disassembly of oocyte granules was incomplete in meg-3 meg-4 zygotes (Video 2, Figure 1—figure supplement 1), causing a few oocyte granules to persist through the first mitotic division (arrows in Figure 4A). During mitosis, a few new granules appeared in the posterior cytoplasm (Figure 4A), but these remained small (<1 micron) and most were not maintained during the division (Video 2). As a result, the total number of granules (including oocyte granules) in meg-3 meg-4 zygotes in mitosis was ∼11% that of wild-type (Figure 4B). In the 2-cell stage, new small granules formed again transiently in P1, but again these were fewer and smaller than in wild-type (Figure 4A). This pattern was repeated at each division such that by the 28-cell stage, only a few scattered granules were observed throughout the embryo, with no detectable enrichment in the germline founder cell P4 (Figure 4A). We observed the same P granule dynamics when staining meg-3 meg-4 zygotes with an antibody against another P granule component, the Vasa-homolog GLH-2 (Figure 4—figure supplement 1). meg-3 meg-4 zygotes showed similar levels of overall GFP:PGL-1 fluorescence compared to wild-type and similar levels of PGL-3 by western blot analysis (Figure 4—figure supplement 5). We conclude that the loss of meg-3 and meg-4 causes a dramatic reduction in the number of P granules but does not affect PGL-1 and PGL-3 levels significantly.
Figure 4. meg-1, meg-3, and meg-4 regulate P granule dynamics.
(A) Live embryos of the indicated genotypes and stages expressing GFP::PGL-1 (green) and mCherry::H2B (red) or GFP::PGL-1 only (last two). Arrows in meg-3 meg-4 point to oocyte granules. For pptr-1 zygotes, the number of zygotes with visible posterior P granules is indicated. P0 is 1-cell stage, P1 is 2-cell stage, P4 is 28-cell stage. (B) Number of GFP::PGL-1 granules in zygotes in anaphase of the first mitosis. Each dot represents a different zygote, and mean and standard deviation are shown. Asterisks indicate data that are statistically significantly different from wild-type (one asterisk: p < 0.005, three asterisks: p < 0.0001). (C) Percent sterility of adult hermaphrodites of the indicated genotypes grown at 20°C. Number of hermaphrodites scored (n) is written above the x axis. (D) Primordial germ cells in the L1 larval stage expressing GFP::PGL-1. By this stage, PGCs express P granule components (Kawasaki et al., 2004). Perinuclear P granules form in all three genotypes. GFP::PGL-levels are lower in the meg mutants due to the lack of P granules inherited from the early embryonic stages. Full genotypes are meg-1(vr10), meg-1(vr10) meg-2(RNAi), meg-3(tm4259), meg-1(vr10) meg-3(tm4259), meg-4(ax2081), meg-3(tm4259) meg-4(ax2026), pptr-1(tm3103), pptr-1(tm3103);meg-1(vr10), pptr-1(tm3103);meg-3(tm4259), mbk-2(pk1427) and mbk-2(pk1427); meg-3(RNAi) meg-4(RNAi).
DOI: http://dx.doi.org/10.7554/eLife.04591.011
Figure 4—figure supplement 1. meg-3 and meg-4 are required redundantly for P granule assembly in zygote.
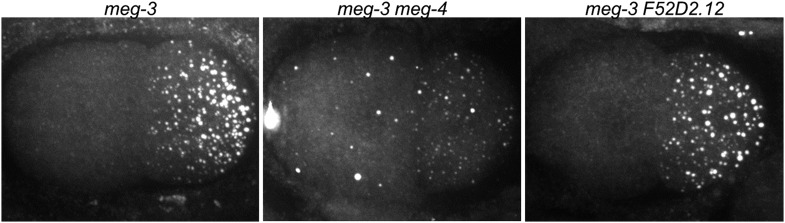
2-cell embryos stained for the P granule component GLH-2 (Gruidl et al., 1996). meg-3(tm4259) hermaphrodites were treated with meg-4 or F52D2.12 RNAi. The RNAi constructs were designed to target regions in meg-4 and F52D2.12 corresponding to the region deleted in meg-3(tm4259) to avoid any possible cross-silencing. Genotypes are meg-3(tm4259), meg-3(tm4259) meg-4(RNAi), and meg-3(tm4259) F52D2.12(RNAi). F52D2.12 is a more distantly related homolog of meg-3 and meg-4 (48% amino acid identity between meg-3 and F52D2.12, compared to 71% amino acid identity between meg-3 and meg-4). No phenotype was observed in meg-3(tm4259) F52D2.12(RNAi) nor in wild-type animals treated with meg-4(RNAi) or F52D2.12(RNAi).
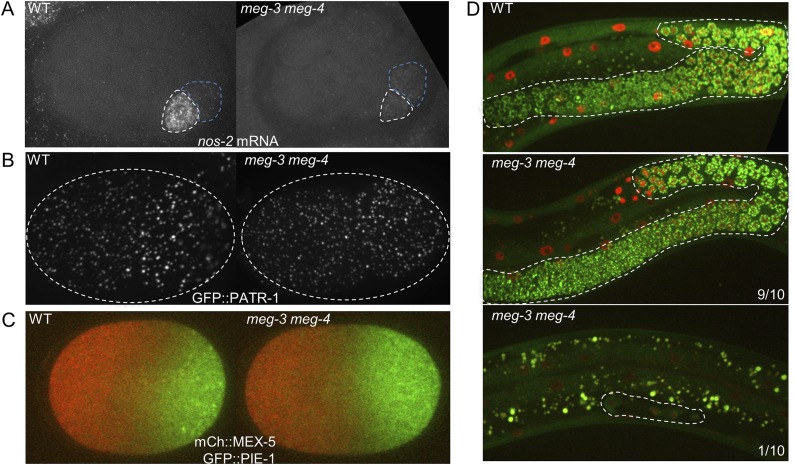
Figure 4—figure supplement 2. Comparison of wild-type and meg-3 meg-4 mutants.
(A) 28-cell stage embryos hybridized to a probe for nos-2 RNA. In wild-type embryos, nos-2 is enriched in the P blastomere (white outline). In meg3 meg-4, nos-2 RNA is segregated equally to the P blastomere (white outline) and its sister somatic blastomere (blue outline). Older somatic blastomeres have already turned over nos-2 RNA. (B) Live zygotes expressing GFP::PATR-1, a marker for P bodies. Assembly of P bodies is not affected in meg-3(RNAi) meg-4(RNAi) zygotes. (C) Live zygotes expressing GFP::PIE-1 and mCherry::MEX-5. Localizations are indistinguishable, except for the lack of PIE-1 on granules in meg-3(RNAi) meg-4(RNAi) zygotes. (D) L4 larvae expressing GFP::PGL-1 and mCherry::H2B. The germline is highlighted in white dashed lines. 9 of 10 meg-3 meg-4 L4 larvae developed a full germline, as in wild-type. The remaining one meg-3 meg-4 L4 larva only contained 4 germ cells.
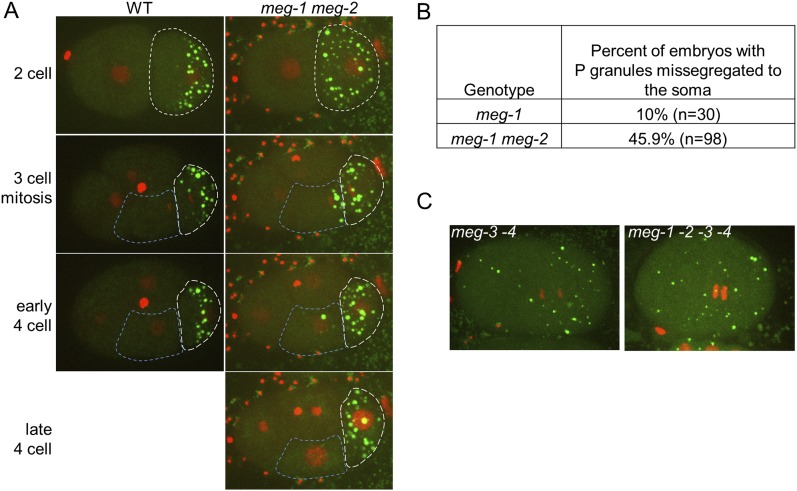
Figure 4—figure supplement 3. meg-1 meg-2 embryos exhibit defects in P granule disassembly in 2-cell and later embryos leading to missegregation of P granules to somatic blastomeres.
(A) Time points from movies of a wild-type and a meg-1(RNAi) meg-2(RNAi) embryo expressing GFP::PGL-1 and mCherry::H2B. The embryos are precisely aged-matched. White dashed lines outline the P blastomeres P1 (2-cell stage) and P2 (3-cell and later stages), blue dashed lines outline the somatic blastomere EMS. In wild-type, P granules are disassembled in the anterior cytoplasm of P1 and segregate only to P2. In meg-1 meg-2, P granule disassembly is defective in P1, resulting in some P granules segregating into EMS. These ectopic P granules are disassembled by the late 4-cell stage. (B) Table showing the number of 4-cell and later stage embryos with missegregated P granules in meg-1(vr10) and meg-1(vr10) meg-2(RNAi). (C) Live meg-3(tm4259) meg-4(ax2026) and meg-1(vr10) meg-2(RNAi) meg-3(tm4259) meg-4(RNAi) zygotes expressing GFP::PGL-1 and mCherry::H2B. Phenotypes are indistinguishable.
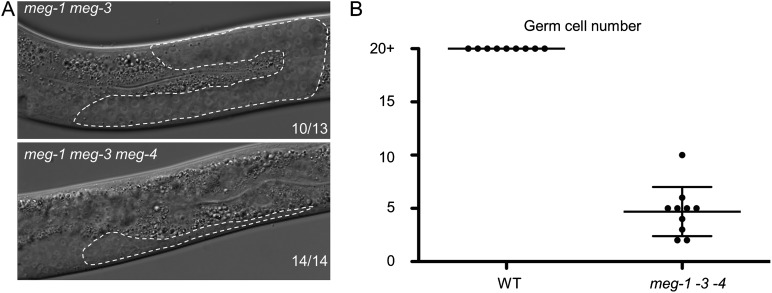
Figure 4—figure supplement 4. MEG proteins are required for germ cell development.
(A) Nomarski images of live L3 larvae. Worms were staged according to vulval development. Germline is outlined. 10 of 13 meg-1 meg-3 animals had wild-type germlines as shown, and 3 of 13 had underdeveloped germlines similar to meg-1 meg-3 meg-4. Full genotypes are: meg-3(tm4259) meg-4(ax2026) and meg-1(vr10) meg-3(tm4259) meg-4(RNAi). (B) Graph showing the total number of germ cells in wild-type and meg-1(vr10) meg-3(tm4259) meg-4(RNAi) L4 larvae. Each dot represents a single larva. Mean and standard deviation are shown.
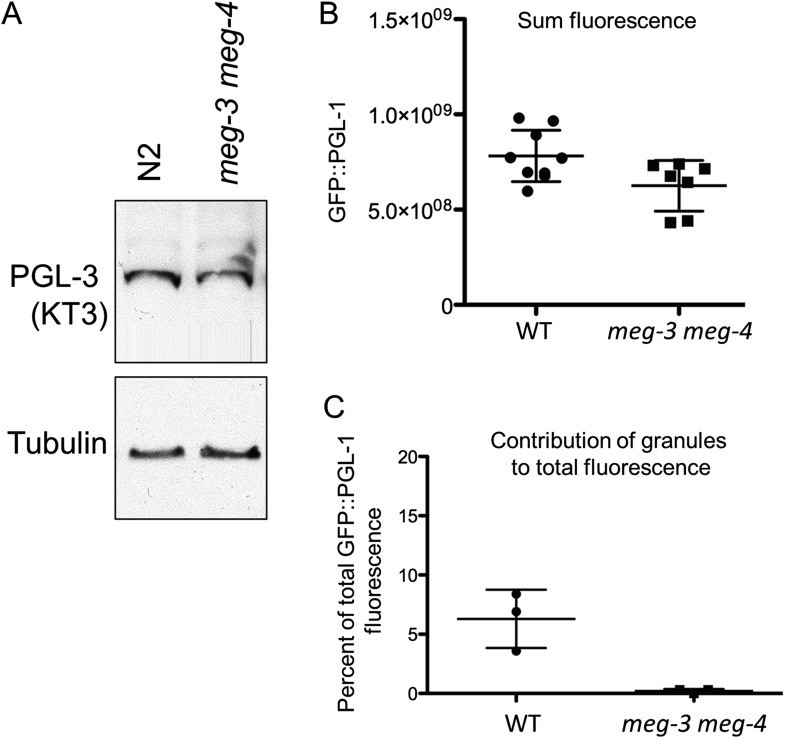
Figure 4—figure supplement 5. meg-3 meg-4 do not affect PGL-1 and PGL-3 levels significantly.
(A) Western blot for endogenous PGL-3 (detected using KT3 antibody) and tubulin in wild-type (N2) embryos and meg-3(tm4259)meg-4(RNAi) embryos. (B) Total GFP::PGL-1 fluorescence in wildtype and meg-3(tm4259) meg-4(ax2026) zygotes. Difference between wildtype and meg-3 meg-4 is not statistically significant (p = 0.3). Mean and standard deviation are shown. (C) Percent GFP::PGL-1 fluorescence present in granules in wild-type and meg-3(tm4259)meg-4(ax2026) zygotes at mitosis. Mean and standard deviation are shown.
meg-3 single mutants exhibited the same phenotypes as the meg-3 meg-4 double mutant but with lower expressivity. In meg-3 zygotes, oocyte granule disassembly was delayed but was eventually completed before the first division (Video 3). Zygote granule assembly was also impaired, but less than in meg-3 meg-4 zygotes: we observed a 30% reduction in the number of P granules in meg-3 zygotes compared to the 89% reduction for meg-3 meg-4 zygotes (Figure 4B). We also observed only a slight reduction in P granule number in meg-4 zygotes (Figure 4B). We conclude that meg-3 and meg-4 contribute redundantly to the assembly of zygote granules, with meg-3 providing the greater contribution.
Video 3. P granule dynamics in wild-type and meg-3.
Time-lapse of embryos expressing GFP::PGL-1 and mCherry::H2B. Wild-type embryo is same one as shown in Video 2. Images are maximum intensity projections of 8 z planes separated by 1 μm steps. Stacks were taken every 8 s, total movie time is 17 min 20 s, movie is played back in 80× real time. Full genotype of mutant is meg-3(tm4259). The bright red puncta in the anterior (left) side of the embryos are polar bodies. In the wild-type movie, there are also red puncta above the embryo, these are sperm outside of the egg shell.
DOI: http://dx.doi.org/10.7554/eLife.04591.017
We next tested whether RNA components of P granules are affected in meg-3 meg-4 embryos. In wild-type, during each asymmetric division of the germline P blastomeres, the P granule-associated RNA nos-2 segregates preferentially with the P granules (asymmetric segregation) (Subramaniam and Seydoux, 1999). The lower levels inherited by the somatic daughter are turned over after division (degradation). In meg-3 meg-4 zygotes, we found that nos-2 RNA was distributed equally to P blastomeres and their somatic sisters, consistent with the lack of stable P granules. nos-2 RNA, however, was still quickly degraded in the somatic blastomeres after division as in wild-type (Figure 4—figure supplement 2A). Thus, as previously reported for pptr-1 mutants (Gallo et al., 2010), despite symmetric segregation of nos-2 RNA, an inherent asymmetry between somatic and germline blastomeres in _meg_-3 meg-4 embryos causes nos-2 RNA to be maintained only in the germline blastomeres. Loss of meg-3 and meg-4 also did not affect the formation of P bodies (Figure 4—figure supplement 2B) nor the distribution of asymmetric proteins that segregate independently of P granules (PIE-1 and MEX-5) (Figure 4—figure supplement 2C). By the L1 larval stage, when new P granule components are synthesized in the PGCs (Kawasaki et al., 2004), low levels of perinuclear P granules became visible in the primordial germ cells of meg-3 meg-4 double mutants (Figure 4D). By the L4 larval stage, P granule levels were indistinguishable from wild-type in fertile meg-3 meg-4 mutants (Figure 4—figure supplement 2D). We conclude that MEG-3 and MEG-4 are only required for P granule assembly in pre-gastrulation embryos and are not required for other soma/germline asymmetries, or for the assembly of P bodies or perinuclear P granules.
MEG-1 contributes to granule assembly and disassembly and is required redundantly with MEG-3 and MEG-4 for fertility
To determine the role of meg-1 and meg-2 in P granule assembly and disassembly, we again used the GFP::PGL-1 and mCherry::H2B transgenes to follow granule dynamics live. Leacock and Reinke (2008) reported that meg-1 and meg-1 meg-2 mutants assemble P granules, but occasionally missegregate P granules to somatic blastomeres in 4-cell and later stage embryos. Consistent with those findings, we observed only a modest reduction in the number of P granules in meg-1 meg-2 zygotes (Figure 4A,B). In 2-cell stage meg-1 mutants, P granules failed to disassemble in the anterior cytoplasm of the P1 blastomere (Figure 4A), causing a few P granules to be inherited by the EMS blastomere (Figure 4—figure supplement 3A). meg-2(RNAi) on meg-1 mutants enhanced the percent of P granule missegregation to somatic blastomeres (Figure 4—figure supplement 3B), as reported previously (Leacock and Reinke, 2008). We conclude that, unlike meg-3 and meg-4, meg-1 and meg-2 contribute only weakly to granule assembly and contribute primarily to granule disassembly.
To determine whether the meg genes act redundantly in granule assembly, we compared double, triple, and quadruple loss-of-function zygotes, combining mutations and RNAi (see Figure 4 legend for complete genotypes). We found that the P granule assembly defects observed in meg-3 and meg-3 meg-4 zygotes were not affected by the additional loss of meg-1 or meg-1 and meg-2 (Figure 4A and Figure 4—figure supplement 3C). In the L1 stage, we observed low levels of perinuclear P granules in the PGCs of meg-1 meg-3 meg-4 triple mutants as seen in meg-3 meg-4 double mutants (Figure 4D). We conclude that meg-3 and meg-4 are the primary contributors to granule assembly in early embryos, and that none of the _meg_s are required for the assembly of perinuclear granules in PGCs later in development.
meg-1 meg-2 double mutants are 100% sterile (Leacock and Reinke, 2008). To examine whether other meg mutant combinations are also sterile, we determined the percent of adult animals with empty uteri (sterile animals). We found that 27% of meg-3 meg-4 mutants and 100% of meg-1 meg-3 meg-4 mutants were sterile (Figure 4C). In wild-type animals, the two PGCs begin to divide in the first larval stage (L1) to generate >1000 germ cells by adulthood. In contrast, we observed <10 germ cells in meg-1 meg-3 meg-4 larvae and no further growth by the adult stage, despite the presence of a somatic gonad (Figure 4—figure supplement 4). This phenotype is identical to that reported for meg-1 meg-2 mutants (Leacock and Reinke, 2008). We conclude that meg-1, meg-2, meg-3, and meg-4 contribute redundantly to germ cell proliferation during larval development.
The MEGs function downstream of MBK-2 and PPTR-1 to regulate P granule dynamics
To test whether the meg genes are epistatic to pptr-1 and mbk-2, we examined the effect of loss of meg-1 and meg-3 in pptr-1 and mbk-2 zygotes. In pptr-1 mutants, all P granules disassemble during mitosis. If this defect were due to hyper-phosphorylation of a PPTR-1 target, then elimination of that target might suppress the pptr-1 phenotype and restore some P granules. Consistent with this possibility, we found that the loss of meg-1 suppressed the granule hyper-disassembly phenotype of pptr-1 zygotes (Figure 4A). Interestingly, the loss of meg-3 also partially suppressed pptr-1 (Figure 4A), indicating that meg-3 also contributes to disassembly.
In mbk-2 mutants, P granule disassembly does not occur during mitosis. We found that in mbk-2, meg-3 meg-4 zygotes, P granules failed to assemble as is observed in meg-3 meg-4 zygotes (Figure 4A). This finding indicates that meg-3 and meg-4 are essential for granule assembly even when disassembly is inhibited. Together with the finding that mbk-2 is epistatic to pptr-1/2 (Figure 1), these genetic interactions are consistent with the meg genes functioning downstream of mbk-2 and pptr-1/2, to regulate the balance of granule assembly and disassembly in embryos.
MEG-3 and MEG-4 localize to embryonic P granules
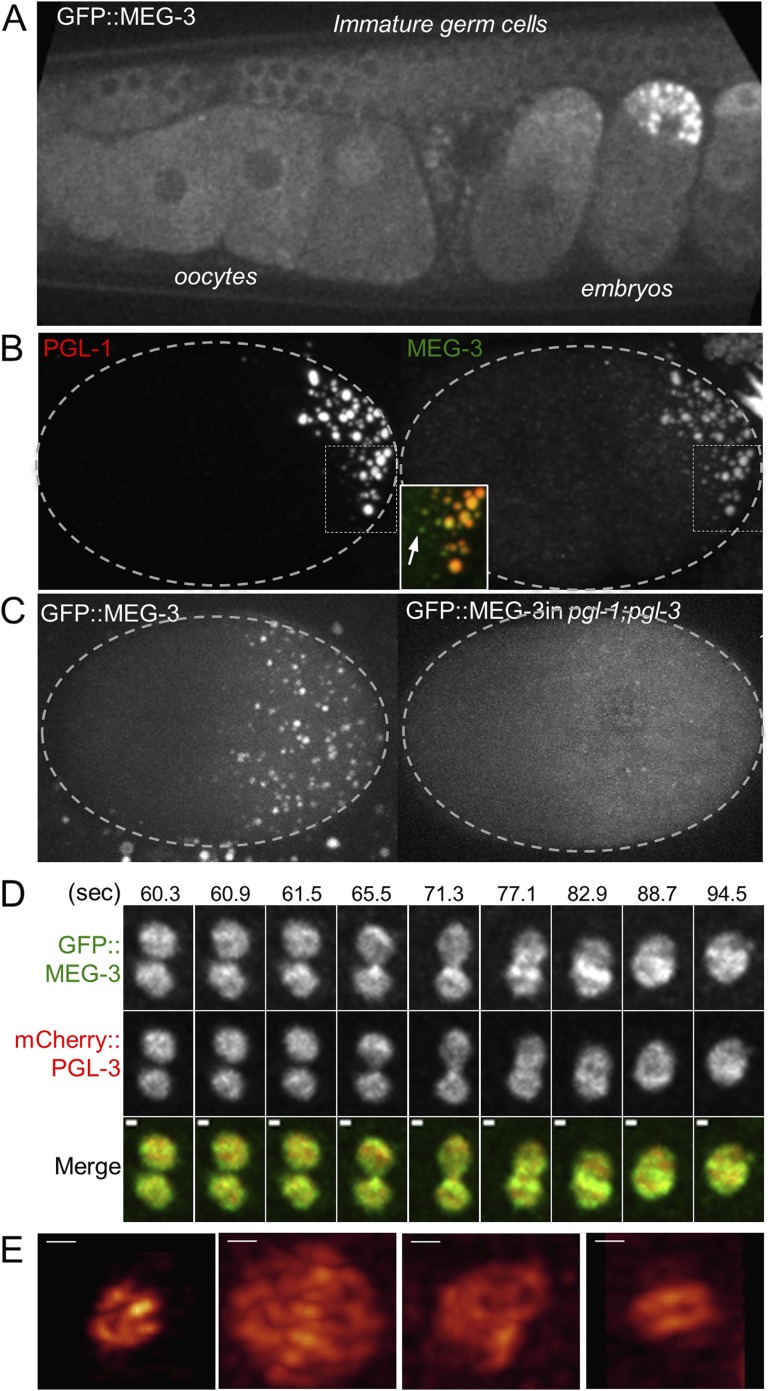
The experiments above suggest a direct role for the MEGs in disassembling and assembling P granules. MEG-1 was previously reported to localize to embryonic P granules from the 4-cell stage to the 100-cell stage (Leacock and Reinke, 2008). To determine the localization of MEG-3, we generated a rescuing GFP-tagged transgene and a polyclonal serum raised against a MEG-3 peptide (‘Materials and methods’ and Figure 5—figure supplement 1). For MEG-4, we inserted a 3× FLAG tag at the carboxy terminus of the meg-4 open reading frame by homology dependent repair of a CRISPR/Cas9-induced cut (Paix et al., 2014). We found that both MEG-3 and MEG-4 localize to P granules from the 1-cell stage to ∼100-cell stage (Figure 5 and Figure 5—figure supplement 2A). After the birth of the primordial germ cells Z2/Z3 (100-cell stage), when P granules are fully perinuclear and no longer dynamic, MEG-3 and MEG-4 levels quickly faded (Figure 5, Figure 5—figure supplement 2 and data not shown).
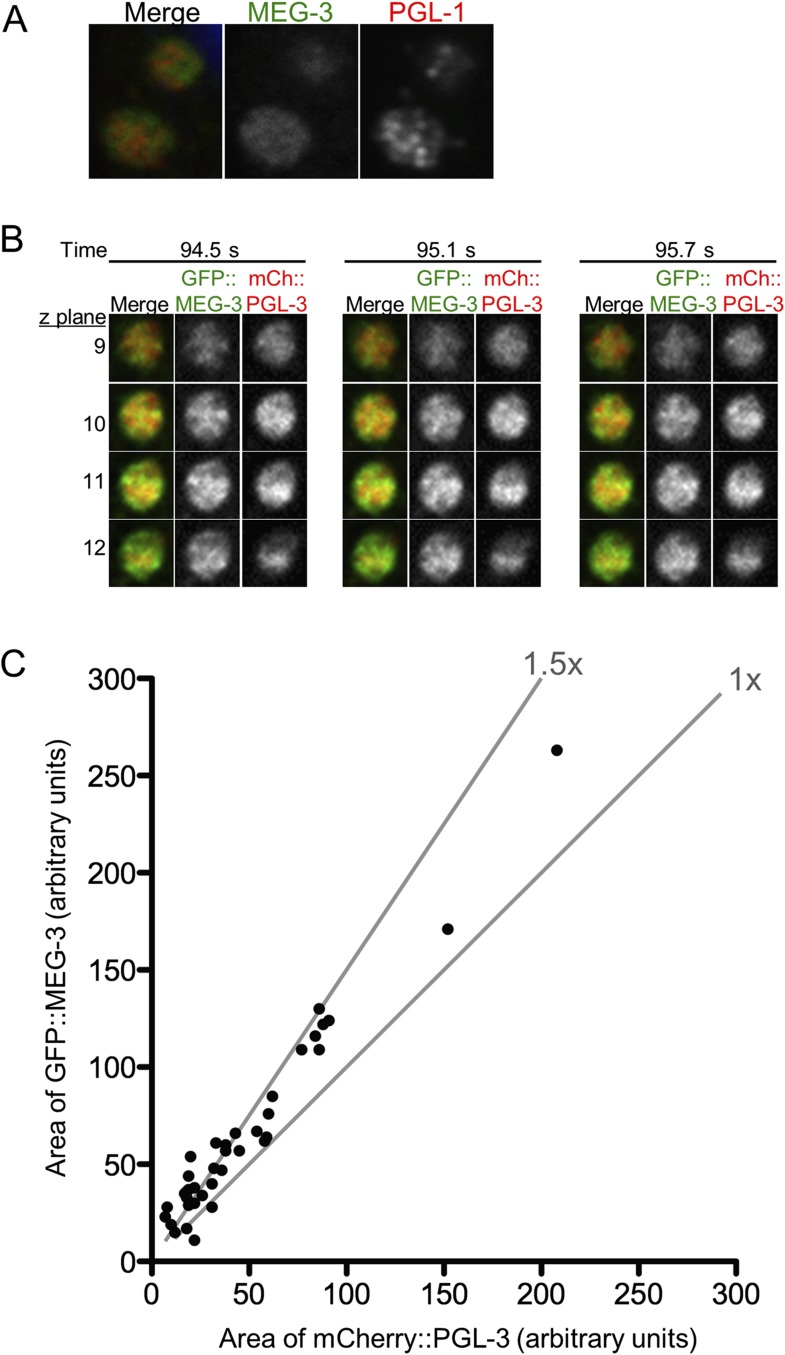
Figure 5. MEG-3 localizes to a dynamic domain that surrounds and penetrates the P granules.
(A) Gonad of an adult hermaphrodite expressing the GFP::MEG-3 transgene under the control of the meg-3 promoter and 3′ UTR. GFP::MEG-3 associates strongly with P granules only in embryos. (B) Fixed wild-type zygote stained with anti:MEG-3 and anti-PGL-1 sera. Inset: magnification of merged image. Arrow points to a MEG-3-positive/PGL-1-negative granule. (C) Live wild-type and pgl-1(RNAi);pgl-3(bn104) zygotes expressing GFP::MEG-3. GFP::MEG-3 localizes to a cytoplasmic gradient and P granules. P granule localization is lost in pgl-1(RNAi);pgl-3(bn104) zygotes, but the cytoplasmic gradient remains. (D) Still images from a movie acquired using lattice light sheet microscopy (Video 5). Max intensity projection of a Z stack through a pair of fusing granules. Time in seconds (Video 5) is indicated above each panel. GFP::MEG-3 and mCherry:PGL-3 domains are not completely co-localized. Also see Figure 5—figure supplement 3B. Resolution is 238 nm × 238 nm × 500 nm. Scale bars: upper left = 500 nm. (E) Lattice light sheet 3D-SIM mode reconstruction of GFP::MEG-3 in P granules in a living zygote (also see Video 6). Scale bars: upper left = 500 nm. First granule on the left: acquisition time is 1 s (Video 6). Subsequent granules: acquisition time is 1.7 s (Video 7).
DOI: http://dx.doi.org/10.7554/eLife.04591.018
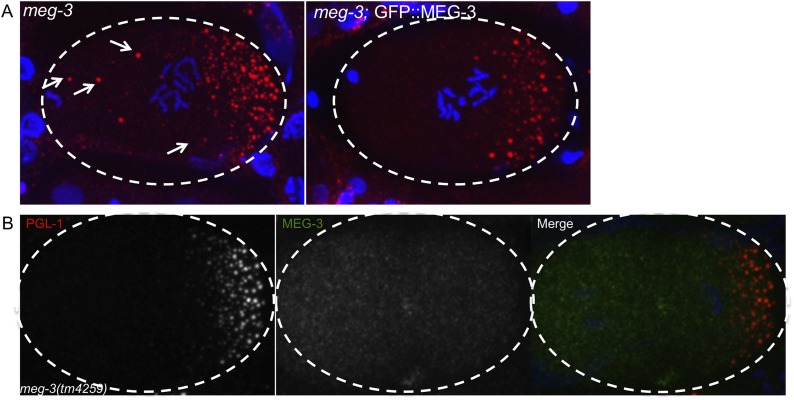
Figure 5—figure supplement 1. GFP::MEG-3 rescues meg-3 mutant and the MEG-3 antibody is specific.
(A) Fixed zygotes stained for P granules (K76 antibody, red). Note the oocyte granules (arrows) in meg-3(tm4259) which are absent in the rescued strain meg-3(tm4259);GFP::MEG-3. At least 20 mothers were stained for each genotype and these images are representative. We could not test whether GFP::MEG-3 also rescues the P granule assembly defect of meg-3(tm4259) mutants, since this defect can only be scored using GFP::PGL-1 (antibody staining of P granules is not reliable enough for accurate counts at this stage). (B) Fixed meg-3(tm4259) zygote stained for P granules (K76 antibody, red) and with anti-MEG-3 serum. Anti-MEG-3 serum does not stain granules in meg-3(tm4259) embryos, but does so in wild-type (Figure 5B), demonstrating specificity of the serum.
Figure 5—figure supplement 2. MEG-4 localizes to P granules and MEG-3 and MEG-4 assemblies persist longer in disassembling granules than PGL-1.
(A) Embryos expressing MEG-4::FLAG (endogenous locus) and stained with anti-FLAG antibody. (B) Top row: maximum projection image of embryo expressing MEG-4::FLAG (endogenous locus) and stained with anti-FLAG antibody and anti-PGL-1 (K76) antibody. Note the MEG-4:FLAG-positive/PGL-1 negative granules in the anterior part of the cell (where P granules disassemble). Bottom row: maximum projection of image of wild-type embryo (no MEG-4::FLAG) stained with anti-FLAG antibody and anti-PGL-1 (K76) antibody. Note the lack of FLAG staining confirming the specificity of the FLAG antibody. (C) Time-lapse of a disassembling P granule from an embryo expressing GFP::MEG-3 and mCherry::PGL-3 granule. During granule disassembly, GFP::MEG-3 persists for longer than mCherry::PGL-3. Also see Video 4. (D) Zygotes expressing GFP::MEG-3 imaged at mitosis. Genotypes are: pptr-1(RNAi) pptr-2(RNAi) and mbk-2(RNAi). Note the perduring GFP::MEG-3 clusters in the pptr-1pptr-2 zygotes which disassemble all PGL clusters by this stage (Figure 4A).
Figure 5—figure supplement 3. MEG-3 and PGL-1/3 overlap only partially in P granules.
(A) Confocal images of two P granules in a fixed zygote expressing GFP::MEG-3. Endogenous PGL-1 is visualized using the K76 antibody. In all fixed preparations, GFP::MEG-3 never co-localized perfectly with PGL-1, but the appearance of the PGL-1 signal varied between experimental replicates. (B) Single plane images of GFP::MEG-3 and mCherry::PGL-3 in fused granule is also shown in Figure 5D. Time points are indicated above in seconds. Four consecutive z planes are shown with 0.3 μm z steps. (C) Graph plotting area of mCherry::PGL-3 signal (x axis) vs area of GFP::MEG-3 signal (y axis) for 37 granules from two embryos visualized by lattice light sheet microscopy (dithered mode). Each dot represents an individual granule. Diagonal lines indicate 1× and 1.5× fold increase of GFP::MEG-3 area over mCherry::PGL-3 area. In 34 of 37 granules, the GFP::MEG-3 signal occupied a larger area than the mCherry::PGL-3 signal (1.57× increase on average). The area occupied by the mCherry::PGL-3 signal was always entirely contained within the area occupied by the GFP::MEG-3 signal (37/37 granules analyzed).
Figure 5—figure supplement 4. MEG-1 only partially co-localizes with PGL-1.
Fixed embryos imaged using confocal spinning disk microscopy. Top: endogenous MEG-1 antibody (green), K76 (anti-PGL-1, red). Middle: K76 (anti-PGL-1, green), GLH-2 (red). Bottom: GFP::MEG-3 (green), endogenous MEG-1 antibody (red).
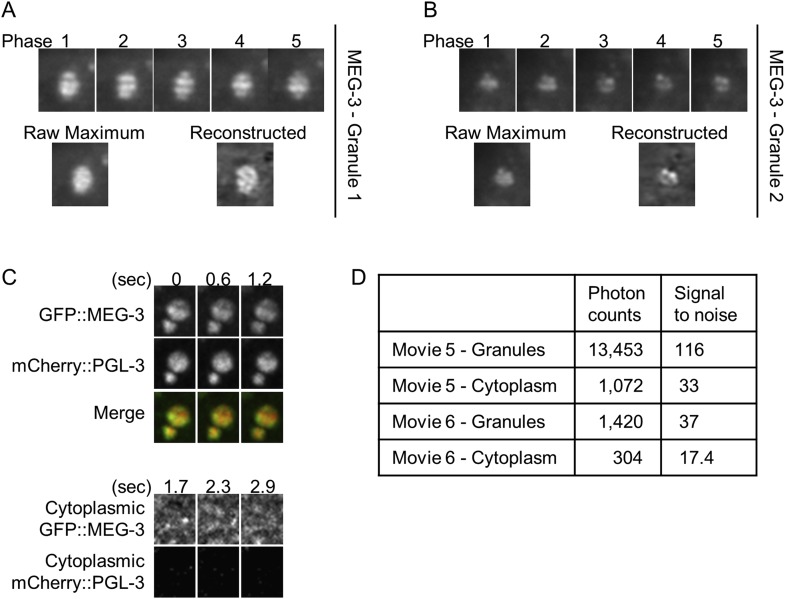
Figure 5—figure supplement 5. Lattice light sheet microscopy identifies sub-granular MEG-3 domain.
(A and B) Single plane images of granules from live embryos expressing GFP::MEG-3 imaged by lattice light sheet 3D-SIM. Granule in B is the same as in Video 6 and Figure 5E. Top row: each structured illumination phase is shown. Bottom row, left: maximum intensity image of the raw phase data. Bottom row, right: image after reconstruction. Note that the structure seen in the reconstructed images is present in the raw maximum images. (C) Top: maximum projections of lattice light sheet images of GFP::MEG-3 and mCherry::PGL-3 granules. MEG-3 sub-granular structure is preserved over successive frames. Bottom: maximum projections of lattice light sheet images of cytoplasmic GFP::MEG-3 and mCherry::PGL-3. Image contrast was enhanced to the same extent for both channels. mCherry::PGL-3 is not visible in the cytoplasm under these conditions. In contrast, GFP::MEG-3 is easily detected and is distributed non-uniformly. The partial pattern repetition seen in the three successive time points and the high signal-to-noise ratio (D) suggest that the cytoplasmic MEG-3 pattern represents true structure and not noise. (D) Photon counts and signal to noise for each movie.
We did not detect MEG-3 or MEG-4 in the perinuclear P granules of adult gonads (Figure 5A for MEG-3 and data not shown). We detected GFP::MEG-3 in the cytoplasm of immature germ cells and oocytes (Figure 5A). In zygotes, cytoplasmic GFP::MEG-3 formed an anterior-to-posterior gradient with highest levels in the posterior (Figure 5C). In embryos depleted for pgl-1 and pgl-3, which do not form P granules (Hanazawa et al., 2011), GFP::MEG-3 was still present in an anterior-to-posterior gradient but no longer localized to large granules (Figure 5C). We conclude that MEG-3 and MEG-4 are maternally provided proteins that segregate with the P lineage and associate with P granules during the embryonic period where P granules are most dynamic.
Live-imaging of zygotes expressing GFP::MEG-3 and mCherry::PGL-3 revealed that, during granule disassembly, GFP::MEG-3 perdures longer in the granules than mCherry::PGL-3 (Video 4 and Figure 5—figure supplement 2C). Consistent with that observation, in fixed zygotes, we observed MEG-4-positive/PGL-1-negative and MEG-3-positive/PGL-1-negative granules at the anterior most edge of the P granule domain (Figure 5B and Figure 5—figure supplement 2B). We also observed GFP::MEG-3-positive/PGL-1-negative granules in pptr-1 zygotes where disassembly is enhanced (Figure 5—figure supplement 2D). We conclude that MEG-3/4 and PGL-1 exhibit different dynamics during disassembly.
Video 4. MEG-3 and PGL-3 dynamics in the anterior of P1 cell.
Time-lapse of embryos expressing GFP::MEG-3 and mCherry::PGL-3. Images are maximum intensity projections of 8 z planes separated by 1 μm steps. Stacks were taken every 8 s, total movie time is 9 min 12 s, movie is played back in 80× real time.
DOI: http://dx.doi.org/10.7554/eLife.04591.024
MEG-3 localizes to a highly dynamic peri-granular domain
Consistent with the different MEG and PGL dynamics, immunostaining experiments in fixed embryos suggested that MEG-1 and MEG-3 do not co-localize precisely with PGL-1 in P granules (Figure 5—figure supplement 3A and Figure 5—figure supplement 4). To avoid potential artifacts from fixation, we turned to lattice light sheet microscopy, a new method that uses ultra-thin light sheets derived from optical lattices to image 3D volumes with high temporal and spatial resolution (Chen et al., 2014). Imaging of live zygotes co-expressing GFP::MEG-3 and mCherry:PGL-3 revealed that both were non-uniformly distributed and constantly rearranged within the granules (Figure 5D, Video 5). The GFP::MEG-3 and mCherry::PGL-3 signals overlapped but were never perfectly coincident, even during granule fusion (Figure 5D, Figure 5—figure supplement 3B). In 34 of 37 granules analyzed, the GFP::MEG-3 signal extended over a larger area than the mCherry::PGL-3 signal (Figure 5—figure supplement 3C), with GFP::MEG-3 extending further out at the periphery of each granule compared to mCherry::PGL-3.
Video 5. Lattice light sheet movie of an embryo expressing GFP::MEG-3 and mCherry::PGL-3.
Time-lapse of the posterior cytoplasm of a zygote (anterior to the left, posterior to the right) acquired using lattice light sheet microscopy in dithered mode. Two fusing granules are highlighted (also shown in Figure 5D). Images are maximum intensity projections of 11 z planes separated by 0.3 μm steps, capturing the entire depth of the fusing pair. Stacks were taken every 580 ms, total movie time is 34 s, movie is played back in 5× real time.
DOI: http://dx.doi.org/10.7554/eLife.04591.025
Images acquired every 580 ms revealed changes in the distribution of GFP::MEG-3 and GFP::PGL-3 at every time point (Figure 5—figure supplement 3B). Overall granule shape was also dynamic, with none of the granules maintaining a perfectly spherical shape (Figure 5D, Video 5). Fusion between granules was comparatively slow, on the order of tens of seconds as documented previously (Brangwynne et al., 2009). Granules remained close to each other for several seconds before initiating fusion. In the typical example shown in Figure 5D and Video 5, the fusing granules took 25 s to return to a quasi-spherical shape. We also detected smaller, dynamic GFP::MEG-3 assemblies in the cytoplasm away from the granules (Video 5, Figure 5—figure supplement 5).
To examine the distribution of GFP::MEG-3 at higher resolution, we collected single-color, single time point light sheet images in the structured illumination (SIM) mode. In this mode, individual images are collected as the lattice light sheet is stepped along the x axis and reconstructed into a 3D image with resolution beyond the diffraction limit in the x and z directions (194 nm × 238 nm in xz; [Chen et al., 2014]). The single time point SIM images confirmed that GFP::MEG-3 is not uniformly distributed in the granules. The non-uniform pattern of GFP::MEG-3 was also visible in raw, unprocessed images (Figure 5—figure supplement 5). In 5 out of 5 granules reconstructed in SIM mode, GFP::MEG-3 localized to a discontinuous ribbon-like domain that surrounded and penetrated the granule (Figure 5E and Videos 6 and 7). We conclude that P granules are not homogeneous assemblies and that MEG-3 localizes to dynamic sub-domains within each granule that only partially overlap with PGL-3.
Video 6. 3D reconstruction of GFP::MEG-3 granule.
Single time point SI images acquired with lattice light sheet microscopy of an individual P granule in an embryo expressing GFP::MEG-3. Acquisition time was 1 s. Resolution is 194 nm × 238 nm × 419 nm. Anterior is to the right, posterior to the left.
DOI: http://dx.doi.org/10.7554/eLife.04591.026
Video 7. 3D reconstructions of GFP::MEG-3 granules.
Single time point SI images acquired with lattice light sheet microscopy of individual P granules in an embryo expressing GFP::MEG-3. Four examples are shown. Acquisition time was 1.75 s. Anterior is to the right, posterior to the left.
DOI: http://dx.doi.org/10.7554/eLife.04591.027
Discussion
In this study, we identify the substrates of MBK-2/DYRK that regulate the condensation and dissolution of P granules in C. elegans embryos. Our findings indicate that the MEGs are required to stabilize the condensed phase of P granules. Phosphorylation of the MEGs by the anteriorly enriched kinase MBK-2/DYRK and dephosphorylation by PP2APPTR−1/PPTR2 antagonizes and promotes, respectively, the stabilizing function of the MEGs. The MEGs also contribute redundantly to a second activity required for germline proliferation during larval development. We discuss each aspect of this model below.
The MEGs stabilize P granules in embryos: possible scaffolding function
During the oocyte-to-zygote transition, oocyte granules disassemble and are replaced by smaller and much more dynamic granules (zygote granules). We suggest that this transition depends on scaffolding of new granules by MEG assemblies throughout the cytoplasm. We propose that, shortly after fertilization, MEG assemblies promote PGL condensation, depleting the cytoplasmic PGL-1 pool available to maintain oocyte granules and causing PGL-1 to redistribute from oocyte granules to MEG-scaffolded zygote granules. In meg mutants, this competition does not take place, allowing oocyte granules to persist longer. The alternative possibility that the primary role of the MEGs is to disassemble oocyte granules, which indirectly is required for zygote granule assembly, is unlikely since (1) the meg-3 single mutant disassembles oocyte granules, yet is still partially defective in zygote granule assembly (Figure 4), (2) mbk-2, which is required for the disassembly of zygote granules, is not required for the disassembly of oocyte granules (Figure 1—figure supplement 1), and (3) oocyte granules represent less than 1% of the total PGL-1 pool available in meg-3 meg-4 zygotes, so their mere presence is unlikely to interfere with the assembly of new granules (Figure 4—figure supplement 5). We detected GFP::MEG-3 in the cytoplasm of oocytes before fertilization, yet loss of meg-3 does not affect oocyte granules until after fertilization. We do not know what cue activates the scaffolding function of MEG proteins specifically in zygotes.
How do MEG proteins stimulate PGL-1 condensation? Unlike the RNA-binding proteins that associate with hydrogels in vitro (Frey et al., 2006; Kato et al., 2012), the MEGs do not contain a recognizable RNA-binding domain or short repeated motifs (Figure 2). The MEGs contain, however, extended regions with predicted high disorder (Figure 2). Intrinsically disordered proteins have been shown to oligomerize and organize scaffolds that template supramolecular structures (Turoverov et al., 2010; Toretsky and Wright, 2014). For example, ameloblastin self-assembles in vitro into ribbons that can extend to hundreds of nanometers in length (Wald et al., 2013). In vivo, ameloblastin is essential for the organization of enamel matrix proteins during teeth biomineralization. MEG-1 and MEG-3 bind directly to PGL-1 in vitro, consistent with a role in organizing PGL-1 assemblies. When over-expressed in mammalian cells, PGL-1 can assemble granules on its own (Hanazawa et al., 2011). In meg-3 meg-4 zygotes, a few small PGL-1 granules form transiently in the posterior cytoplasm but are not maintained. We suggest that, in zygotes, the self-assembly properties of PGL-1 are only sufficient to form small, unstable granules, which must be stabilized by a MEG-dependent scaffold. Our observations using light sheet microscopy indicate that MEG-3 localizes to small assemblies in the cytoplasm and larger assemblies surrounding a PGL core in the P granules. During granule disassembly, MEG-3 and MEG-4 assemblies persist longer than the PGL-1 core, consistent with occupying a distinct domain. Together these observations suggest the following working model: the MEGs promote granule assembly by forming dynamic scaffolds in the cytoplasm that stabilize and wrap around PGL condensates.
By electron microscopy, embryonic P granules were reported to appear as homogenous, fibrillar granular bodies (Wolf et al., 1983). Our observations suggest that embryonic P granules in fact contain distinct surface and internal zones, as also reported for post-embryonic, perinuclear P granules (Pitt et al., 2000; Sheth et al., 2010). Nucleoli, which also behave like liquid droplets, also contain a distinct surface shell (Brangwynne et al., 2011). Nucleoli, however, are perfect spheres (Brangwynne et al., 2011), whereas P granules are imperfect spheres with irregular, asymmetric contours that are constantly in flux. We suggest that this difference is due to the fact that P granules are undergoing continuous exchange with smaller assemblies in the surrounding cytoplasm. Consistent with this possibility, photobleaching experiments have demonstrated rapid exchange of GFP::PGL-1 and GFP::MEG-3 between granules and cytoplasm (Brangwynne et al., 2009 and data not shown). We conclude that P granules in embryos are not stable, homogeneous droplets, but dynamic assemblies of two (or more) condensates that mix only partially and are constantly in flux with smaller assemblies in the cytoplasm.
Phosphorylation of the MEGs by MBK-2/DYRK promotes granule disassembly
MEG-1 and MEG-3 are direct targets of MBK-2/DYRK and the PP2A phosphatase. The MEGs are rich in serines (75 serines in MEG-1 and 119 serines in MEG-3), and their migration on Phos-tag gels suggests a complex pattern of phosphorylation with many phosphoisoforms. Initial attempts to mutate individual serines in MEG-1 and MEG-3 have not yielded detectable phenotypes, suggesting that the serines may function cumulatively (data not shown). Accumulation of negatively charged phosphates could interfere with the interactions that stabilize the MEG scaffold or its interactions with PGL-1. Consistent with this possibility, phosphorylation of the low complexity domain of the FUS stress granule protein interferes with its incorporation into hydrogels in vitro (Han et al., 2012). Our genetic analyses suggest that all MEGs contribute to assembly and disassembly but to different extents, with MEG-3 and MEG-4 together making the most important contribution to assembly. The predicted pI (isoelectric point) of MEG-1 and MEG-2 in the unphosphorylated state is 6.63 and 6.04, compared to 9.74 and 9.33 for MEG-3 and MEG-4. We speculate that the intrinsic negative charge of MEG-1 and MEG-2, which is increased further by phosphorylation, favors a role for these proteins in disassembly. In contrast, the intrinsic weak positive charge of MEG-3 and MEG-4 allows them to contribute robustly to both disassembly and assembly, as they switch to negative and back to positive upon phosphorylation and dephosphorylation.
How are disassembly and assembly spatially segregated? At the onset of disassembly, MBK-2/DYRK is enriched in the anterior half of the zygote (Pellettieri et al., 2003). If dephosphorylation by PP2A continuously antagonizes phosphorylation by MBK-2, even small changes in the level of MBK-2 levels along the anterior/posterior axis could bias MEG phosphorylation and P granule disassembly. Paradoxically, MBK-2/DYRK also accumulates in P granules, especially late in the first cell cycle as the granules continue to grow in the posterior cytoplasm (Pellettieri et al., 2003). Interestingly, DYRK3, the vertebrate homolog of MBK-2 that regulates stress granule dynamics, also localizes in the granules (Wippich et al., 2013). Drug inhibition studies suggest that DYRK3 enhances granule condensation when its kinase activity is inhibited and promotes granule dissolution when activated (Wippich et al., 2013). An attractive possibility is that, as P granules increase in size, MBK-2 activity decreases within the granule, perhaps due to the accumulation of an inhibitor. Interestingly, GFP::PPTR-1 also becomes enriched in P granules in zygotes (data not shown). There are also other asymmetries in the zygote cytoplasm that could bias P granule dynamics. GFP::MEG-3 forms an anterior-low/posterior-high gradient along the length of the zygote, and the RNA-binding protein MEX-5, which is required for full disassembly, forms an opposing gradient with higher levels in the anterior (Schubert et al., 2000). Modeling studies have already demonstrated that, in principle, even weak gradients of phase transition regulators are sufficient to segregate P granules (Lee et al., 2013). It will be important to determine which asymmetry in the zygote cytoplasm is directly responsible for patterning P granule dynamics.
MEG proteins contribute to an essential germ plasm activity that does not require P granules
Analysis of single, double, and triple meg combinations indicate that the _meg_s display synthetic sterility. For example, meg-1 and meg-3 meg-4 mutants are ∼4% and ∼30% sterile, respectively, whereas meg-1 meg-3 meg-4 mutants are 100% sterile (this work and Leacock and Reinke, 2008). In the sterile meg-1 meg-3 meg-4 worms, the primordial germ cells stop dividing soon after initiating divisions in the gonad, a phenotype previously reported for meg-1 meg-2 mutants (Meg phenotype: maternal-effect germ cell defective) (Leacock and Reinke, 2008). We conclude that the MEGs contribute redundantly to an activity essential for germ cell viability and/or proliferation.
The MEGs are components of the C. elegans germ plasm, the specialized maternal cytoplasm that segregates with the embryonic germline and specifies germ cell fate. Germ plasm is thought to have evolved independently several times during metazoan evolution (Extavour and Akam, 2003), and the molecules that initiate germ plasm and/or germ granule assembly in eggs are not conserved between different species, although some contain intrinsically disordered regions (e.g., Bucky Ball/Xvelo in vertebrates, MEGs in C. elegans) (Marlow and Mullins, 2008; Bontems et al., 2009; Nijjar and Woodland, 2013). In all species examined, the germ plasm contains microscopically visible P granule-like structures that are enriched for conserved mRNAs and RNA-binding proteins required for germ cell development (e.g., nanos RNA and VASA-related helicases) (Voronina et al., 2011). Despite this conservation, our genetic analyses suggest that the MEGs' contribution to fertility is not linked to their effects on P granules, as also suggested by Leacock and Reinke (2008). meg-1 meg-2 embryos assemble embryonic P granules and meg-1 meg-3 meg-4 embryos do not, yet both display the same fully penetrant Meg sterility phenotype (Figure 4 and Leacock and Reinke, 2008). Furthermore, meg-3 meg-4 embryos show the same dramatic defect in P granule assembly as meg-1 meg-3 meg-4 embryos, yet most meg-3 meg-4 animals (70%) are fertile (Figure 4). We conclude that P granule assembly in the germ plasm of early embryos is neither required nor sufficient for fertility in C. elegans. In contrast, the perinuclear P granules that form in primordial germ cells and their descendents are required for fertility (Updike and Strome, 2010). Consistent with that view, fertile meg mutants that do not assemble P granules in the germ plasm assemble perinuclear P granules de novo later in development. De novo assembly of perinuclear germ granules is also observed in animals that do not inherit germ plasm and specify their primordial germ cells through inductive mechanisms later in development (Voronina et al., 2011).
Previously, our analyses of pptr-1 mutants led us to conclude that asymmetric segregation of core P granule components is not essential to distinguish germline from soma in embryos (Gallo et al., 2010). Our results here support this view and suggest that the essential activity of the germ plasm resides not in the assembly of P granules per se but in the MEG proteins that regulate granule assembly. How the maternally provided MEGs contribute to the health and proliferating potential of the PGCs will be an interesting area for future exploration.
Materials and methods
Worm handling, transgenics, CRISPR-mediated editing, RNAi, sterility counts
C. elegans was cultured according to standard methods (Brenner, 1974). Transgene plasmids were generated by InFusion cloning (Clontech, Mountain View, CA) and introduced into unc-119 worms by microparticle bombardment (Praitis et al., 2001). GFP::MEG-3 is driven by its endogenous promoter and 3′UTR and consist of 1128 bp of genomic DNA sequence upstream of the MEG-3 ATG (up to but not including the ATG of the next gene Y40A1A.2), GFP from pCM1.53 (Merritt et al., 2008) and 1872 bp of genomic DNA downstream of the stop codon, past both polyA sites annotated in WormBase (WBsf257432 and WBsf216670). GFP::MEG-1 is described in Leacock and Reinke (2008) and is driven by the pie-1 promoter and pie-1 3′UTR. CRISPR/Cas9 experiments to generate meg-3 and meg-4 alleles were performed as described in Paix et al. (2014). See Tables 1 and 2 for list of strains, allele descriptions, and sgRNA and repair templates sequences.
Table 1.
| Name | Description | Genotype | Reference |
|---|---|---|---|
| JH2842 | pie-1 prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | unc-119(ed3) III; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | Gallo et al., 2010 |
| JH2843 | pptr-1 mutant; pie-1 prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | pptr-1(tm3103) V; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | Gallo et al., 2010 |
| JH3055 | meg-3 mutant | meg-3(tm4259) X | This study |
| JH3147 | meg-3 mutant_; pie-1_ prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | meg-3(tm4259) X; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | This study |
| JH3225 | meg-3 meg-4 mutant—see Table 2 | meg-3(tm4259) meg-4(ax2026) X | This study |
| JH3149 | meg-3 meg-4 mutant_; pie-1_ prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | meg-3(tm4259) meg-4(ax2026) X; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | This study |
| JH3148 | meg-1 mutant_; pie-1_ prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | meg-1(vr10) X; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | This study |
| JH3229 | meg-1 meg-3 mutant | meg-1(vr10) meg-3(tm4259) X | This study |
| JH3150 | meg-1 meg-3 mutant; pie-1 prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | meg-1(vr10) meg-3(tm4259) X; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | This study |
| JH3156 | pptr-1; meg-1 mutant; pie-1 prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | pptr-1(tm3103) V; meg-1(vr10) X; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | This study |
| JH3155 | pptr-1; meg-3 mutant; pie-1 prom::GFP::PGL-1-pgl-1 3′UTR; pie-1 prom::mCherry::H2B::pie-1 3′UTR | pptr-1(tm3103) V; meg-3(tm4259) X; axIs1522[pCM4.11]; ltIs37 [pAA64] IV | This study |
| JH2932 | mbk-2 null mutant; pgl-1::TY1::eGFP::3xFLAG992C12 | unc-24(e1172) mbk-2(pk1427) IV / nT1[let-?(m435)](IV;V); ddEX16 | This study |
| YL183 | meg-1 mutant; pie-1 prom::GFP::MEG-1::pie-1 3′UTR | meg-1(vr10) X; GFP::MEG-1 | Leacock and Reinke, 2007 |
| JH3016 | meg-3 prom::GFP::MEG-3::meg-3 3′UTR | unc-119(ed3); axIS2076[pJW6.01] | This study |
| JH3064 | pptr-1 mutant; meg-3 prom::GFP::MEG-3::meg-3 3′UTR | pptr-1(tm3103) V; axIS2076[pJW6.01] | This study |
| JH3019 | meg-3 prom::GFP::MEG-3::meg-3 3'UTR; pie-1 prom::GFP::PGL-3::pie-1 3′UTR | unc-119(ed3) III; axIS2076[pJW6.01]; axIS2077[pJW2.03] | This study |
| JH3230 | pgl-3 mutant; meg-3 prom::GFP::MEG-3::meg-3 3′UTR | pgl-3(bn104) V; axIS2076[pJW6.01] | This study |
| JH3153 | meg-3 mutant_; meg-3_ prom::GFP::MEG-3::meg-3 3′UTR | meg-3(tm4259) X; axIS2076[pJW6.01] | This study |
| JH3247 | C-terminal FLAG insertion in genomic meg-4 locus—see Table 2 | meg-4(ax2080) | This study |
| JH3248 | meg-4 mutant—see Table 2 | meg-4(ax2081) | This study |
Table 2.
| Strain | Allele | Edit | Method | sgRNA (PAM sequence) | Genomic site of edit |
|---|---|---|---|---|---|
| JH3247 | meg-4 (ax2080) | C-terminal 3xFLAG insertion in meg-4 | Homology directed repair with ssODN. Sequence: (homology arms) (cctgtcagatacttgaatgcaaaacgagaatggctgga atctatttttgacccaccgagagatcaa)gactacaaaga ccatgacggtgattataaagatcatgatatcgattacaa ggatgacgatgacaag(tgattgtactgatatatatctatt tcatgtcgagtattttgtattttattcttgttcattgacc) | caatcattgatctctgggt (ggg) | X:1686208 |
| JH3248 | meg-4 (ax2081) | Deletion removing 733 base pairs upstream of the meg-4 start and the first 2565 bases of the gene | NHEJ | gagcgcgaaatagtgtgtg (ggg) tgggaccaaaaagcaagaa (tgg) atttatttatggtctgccc (agg) ctgcccaggaacttgtaac (ggg) | X:1682223..1685521 |
| JH3325 | meg-3 (tm4259) meg-4 (ax2026) | Deletion of 7 nucleotides starting at nucleotide 37 of meg-4(amino acid 13). The frameshift inserts the amino acids ‘EETEKVTALIVEAIWKVHRKIWDTTLVLKNCFIRS’ at amino acid 13 followed by a premature stop | NHEJ for meg-4(ax2026) in meg-3(tm4259) | tctctgtttcctctggagtt (tgg) caagttccgttgattccagc(tgg) | X:1682992 |
RNAi was performed by feeding (Timmons and Fire, 1998). Feeding constructs were obtained from the Ahringer or Openbiosystems libraries and sequenced or newly cloned from C. elegans cDNA. HT115 bacteria transformed with feeding vectors were grown at 37°C in LB + ampicillin (100 µg/ml) for 5 hr, induced with 5 mM IPTG for 45 min, plated on NNGM (nematode nutritional growth media) + ampicillin (100 µg/ml) + IPTG (1 mM), and grown overnight at room temperature before adding L4 worms at 24°C for 24–30 hr. For Phos-tag gel experiments, worms were fed starting in the L1 stage and scored for embryonic lethality (mbk-2(RNAi)) or P granule phenotype (pptr-1(RNAi)).
For sterility counts, at least eight gravid adult worms were allowed to lay embryos for 1 to 2 hr. Adult progeny were scored for empty uteri (‘white sterile’ phenotype) on a dissecting microscope. To determine the number of germ cells in meg larvae, we counted the number of nuclei in GFP::PGL-1-expressing cells, using vulval morphology to stage the larvae.
Yeast two-hybrid
Yeast two-hybrid assays were performed using DUALhybrid kit (P01004, Dualsystems Biotech, Switzerland). All plasmids were converted to Gateway-compatible vectors (Invitrogen, Carlsbad, CA) and Gateway recombination was used to create N-terminally tagged pLexA-PPTR-1 bait vector, pY3H-PAA-1 bridge vector, and candidate prey vectors. For library screening, yeast transformed with PPTR-1 bait and PAA-1 bridge were used with prey library from Dualsystems consisting of polyA+ cDNA from mixed stage C. elegans with 5.7 × 106 independent clones (28 μg of prey library). Total transformation efficiency was 4.3 × 105 clones per μg DNA, 1.2 × 107 total transformants, with 2.1× library coverage.
GST pull-down
GST fusion proteins were cloned into pGEX6p1 (GE Healthcare, Pittsburgh, PA). MBP fusion proteins were cloned into pJP1.09, a Gateway-compatible pMAL-c2x (Pellettieri et al., 2003). Proteins were expressed in E. coli BL21 cells overnight at 16°C, following induction with 0.4 mM IPTG. 200 mg of bacterial pellet of GST fusion proteins was resuspended in 10 mM EGTA, 10 mM EDTA, 500 mM NaCl, 0.1% Tween, PBS pH 7.4 with protease and phosphatase inhibitors, lysed by sonication, and bound to GST beads. Beads were washed and incubated with MBP fusion proteins at 4°C for 1 hr in 50 mM Hepes pH 7.4, 1 mM EGTA, 1 mM MgCl2, 500 mM KCl, 10% glycerol, 0.05% NP-40, pH 7.4 with protease and phosphatase inhibitors. After washing, beads were eluted with 10 mM reduced glutathione and eluates were loaded on SDS-PAGE.
Protein purification and in vitro kinase assay
MBP fusion proteins were cloned into pJP1.09, expressed and partially purified as previously described (Griffin et al., 2011). In vitro kinase assays were performed as described (Cheng et al., 2009).
Phos-tag gel
Embryos were harvested from synchronized young adult worms and sonicated in 2% SDS, 65 mM Tris pH 7, 10% glycerol with protease and phosphatase inhibitors. Lysates were spun at 14,000 rpm for 30 min and cleared supernatants treated with 100 U alkaline phosphatase (Roche, Indianapolis, IN). Samples were run in parallel on Phos-tag gels (7% SDS-PAGE with 25 μM Phos-tag and 50 μM MnCl2, Phos-tag from Wako Chemicals, Japan) and 7% SDS-PAGE at 30 mA for 2.5 hr. Gels were washed in transfer buffer with 1 mM EDTA twice for 10 min each and washed in transfer buffer without EDTA twice for 10 min each. Western blot transfer was performed for 1 hr at 4°C onto nitrocellulose membranes. Membranes were blocked and washed in 5% milk, 0.1% Tween-20 in PBS and probed with JL-8 antibody (1:240 dilutions, Clontech).
Antibody production
Peptide antibodies against MEG-3 were made by Covance (Princeton, NJ) using KLH conjugation in rabbits against the sequence Ac-HAFKKGHKDNKNASC-amide.
In situ hybridization
In situ hybridization of nos-2 mRNA was performed using fluorescent oligonucleotide probes as described previously (Voronina et al., 2012).
Immunostaining
Gravid adult hermaphrodites were laid on a slide coated with 0.01% poly-L-lysine, and embryos were extruded by squashing with a coverslip. Embryos were frozen on pre-chilled aluminum blocks. Coverslips were removed and slides were incubated in −20°C methanol for 15 min, followed by −20°C acetone for 10 min. Slides were pre-blocked in PBS/0.1% Tween/0.1% BSA (PBT) for 30 min and incubated with primary antibody overnight at 4°C. Primary antibodies were diluted in PBT as follows: K76 (1:10, DSHB, Iowa City, IA), rabbit anti-MEG-3 (1:250, Covance), rabbit anti-MEG-1 (1:100, gift from Valerie Reinke), chicken anti-GLH-2 (1:200, gift from Karen Bennett), anti-FLAG (1:500, Sigma, St. Louis, MO). Secondary antibodies were applied for 2 hr at room temperature.
Spinning-disc confocal microscopy
Images were acquired using a Zeiss Axio Imager fitted with a Yokogawa spinning disc confocal scanner with Slidebook software (Intelligent Imaging Innovations, Denver, CO) using a 63× objective (embryos) or 40× objective (in utero movies). Embryos were dissected from gravid mothers and mounted on 3% agarose pads in M9 solution at room temperature.
For single time point images, 10 z planes with a z step size of 1 μm, spanning 9 μm, were acquired. Exposure time was 100 ms per plane per color with one exposure acquired in each color before proceeding to the next plane. Granule counts were performed with Slidebook using Mask Segmentation tools and manually verified for each embryo.
For time-lapse movies of embryos, z-stacks (8 z planes, step size 1 μm) were acquired every 8 s. Exposure time was 100 ms per plane per color with one exposure acquired in each color before proceeding to the next plane.
For in utero movies, young adult mothers were anesthetized in 0.3 mM levamisole for 15 min prior to mounting on 3% agarose pads. Z-stacks (11 z planes with a z step size of 1 μm) were acquired every 30 s. Exposure time was 100 ms per plane per color with one exposure acquired in each color before proceeding to the next plane.
Lattice light sheet microscopy
Embryos were dissected from gravid mothers and mounted on 5-mm diameter coverslips in M9 solution at room temperature. Coverslips were pre-cleaned and coated with 2 μl of BD Cell-Tak (BD Biosciences, San Jose, CA) and 2 μl of poly-D-lysine. In the time-lapse dithered mode, 21 z planes with a z step size 0.3 μm, spanning 6 μm, were acquired. Exposure time was 10 ms per plane per color with one exposure acquired in each color before proceeding to the next plane, for a total exposure time of 21 × 10 × 2 = 420 ms for each 3D stack. A 150 ms pause was added between each time point. Dithered-mode stacks were deconvolved in 3D using the Richardson–Lucy algorithm with experimentally measured point spread functions for each color. Granule cross-sectional area was calculated using ImageJ using maximum-projection images from two embryos.
3D-SIM mode images were acquired as in Chen et al. (2014), with 5 phase SIM. For the first image in Figure 5E, 71 z planes were acquired with 50 ms exposure per phase and a 0.15 μm z step between planes. For the other images in Figure 5E, 71 z planes were acquired with 20 ms exposure per phase and a 0.2 μm z step. SIM images were reconstructed as in Gao et al. (2012). Note that the granules shown in Figure 5E span only 7–10 z planes, corresponding to 1–1.75 s acquisition time. We confirmed that during this time, the granules were stationary on the scale of the SIM excitation pattern, allaying any concerns of motion-induced artifacts in the SIM reconstructions.
Photon counts were calculated according to the formula P(photo) = CF × (C − off)/Q(lambda), where CF is the conversion factor (electron/count), C is output count of the selected pixel, off is the background of the camera, and Q(lambda) is a curve based on the wavelength. Signal to noise ratio was calculated as sqrt(P).
Acknowledgements
We thank Valerie Reinke, Sam Kapelle, Karen Bennett, the National Bioresource Center (Japan), and the CGC (USA) for strains and reagents, the Janelia Farm Visitors Program for hosting Jenn Wang, and Cliff Brangwynne and the Seydoux lab for discussions. Research in the Seydoux lab is supported by R01HD37047 from the National Institutes of Health. G Seydoux is an Investigator of the Howard Hughes Medical Institute. Research in the Betzig lab is wholly supported by the Howard Hughes Medical Institute.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
- National Institute of Child Health and Human Development R01HD37047 to Jennifer T Wang, Jarrett Smith, Alexandre Paix, Bramwell G Lambrus.
- Howard Hughes Medical Institute to Jarrett Smith, Bi-Chang Chen, Helen Schmidt, Dominique Rasoloson, Deepika Calidas, Eric Betzig, Geraldine Seydoux.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
JTW, All other figures, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
JS, Figure 3C, Figure 3D, Figure 3 supplement 1 left panel (GFP::MEG-1), Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
B-CC, Videos 5, 6, 7, which were used to make Figure 5D, 5E, Figure 5 - supplements 3B, 3C, 5., Acquisition of data, Analysis and interpretation of data.
HS, Figure 4 meg-4 data, Figure 5 supplement 2A and 2B, Table 2, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
DR, Figure 3A, Acquisition of data, Analysis and interpretation of data.
AP, Figure 4—figure supplement 2A, Acquisition of data, Analysis and interpretation of data.
BGL, Generation of strain JH3225, Acquisition of data, Analysis and interpretation of data.
DC, Generation of strain JH3247, Acquisition of data, Analysis and interpretation of data.
EB, Analysis and interpretation of data, Drafting or revising the article.
GS, Corresponding author, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
References
- Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB Journal. 2011;25:449–462. doi: 10.1096/fj.10-165837. [DOI] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Current Biology. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences of USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. mRNP granules: assembly, function, and connections with disease. RNA Biology. 2014;11 doi: 10.4161/rna.29034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LE, Proud CG. Differing substrate specificities of members of the DYRK family of arginine-directed protein kinases. FEBS Letters. 2002;510:31–36. doi: 10.1016/S0014-5793(01)03221-5. [DOI] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, III, Liu Z, English BP, Mimori-Kiyosue Y, Romero DP, Ritter AT, Lippincott-Schwartz J, Fritz-Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann AC, Böhme R, Grill SW, Wang JT, Seydoux G, Tulu US, Kiehart DP, Betzig E. Lattice light sheet microscopy: imaging molecules, cells, and embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KC, Klancer R, Singson A, Seydoux G. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell. 2009;139:560–572. doi: 10.1016/j.cell.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmók V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Shao L, Higgins CD, Poulton JS, Peifer M, Davidson MW, Wu X, Goldstein B, Betzig E. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biology. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Odde DJ, Seydoux G. Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell. 2011;146:955–968. doi: 10.1016/j.cell.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of USA. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hanazawa M, Yonetani M, Sugimoto A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. Journal of Cell Biology. 2011;192:929–937. doi: 10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW. WormBase 2014: new views of curated biology. Nucleic Acids Research. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. Journal of Biological Chemistry. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, Motohashi T, Karashima T, Bossinger O, Strome S. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167:645–661. doi: 10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends in Biochemical Sciences. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Molecular & Cellular Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Leacock SW, Reinke V. MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics. 2008;178:295–306. doi: 10.1534/genetics.107.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CF, Brangwynne CP, Gharakhani J, Hyman AA, Julicher F. Spatial organization of the cell cytoplasm by position-dependent phase separation. Physical Review Letters. 2013;111:088101. doi: 10.1103/PhysRevLett.111.088101. [DOI] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Developmental Biology. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3' UTRs are the primary regulators of gene expression in the C. elegans germline. Current Biology. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S. Nematode, an experimental animal in the national BioResource project. Experimental Animals. 2009;58:351–356. doi: 10.1538/expanim.58.351. [DOI] [PubMed] [Google Scholar]
- Nijjar S, Woodland HR. Protein interactions in Xenopus germ plasm RNP particles. PLOS ONE. 2013;8:e80077. doi: 10.1371/journal.pone.0080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, Tissenbaum HA. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Wang Y, Smith HE, Lee CY, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, Seydoux G. Scalable and versatile genome editing using linear DNAs with micro-homology to Cas9 sites in Caenorhabditis elegans. Genetics. 2014;198:1347–1356. doi: 10.1534/genetics.114.170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Reinke V, Kim SK, Seydoux G. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Developmental Cell. 2003;5:451–462. doi: 10.1016/S1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Developmental Biology. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin S, Mains PE, Zinke A, Hyman AA. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO Reports. 2003;4:1175–1181. doi: 10.1038/sj.embor.7400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Molecular Cell. 2000;5:671–682. doi: 10.1016/S1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, Ayivi-Guedehoussou N, Dann E, Bertin N, Szeto D, Dricot A, Yildirim MA, Lin C, de Smet AS, Kao HL, Simon C, Smolyar A, Ahn JS, Tewari M, Boxem M, Milstein S, Yu H, Dreze M, Vandenhaute J, Gunsalus KC, Cusick ME, Hill DE, Tavernier J, Roth FP, Vidal M. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nature Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Pellettieri J, Seydoux G. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic Maturation. Current Biology. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of USA. 1982;79:1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. The Journal of Cell Biology. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi D, Qadota H, Kasuya K, Amano M, Kaibuchi K. Isolation of the interacting molecules with GEX-3 by a novel functional screening. Biochemical and Biophysical Research Communications. 2002;292:697–701. doi: 10.1006/bbrc.2002.6717. [DOI] [PubMed] [Google Scholar]
- Turoverov KK, Kuznetsova IM, Uversky VN. The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Progress in Biophysics and Molecular Biology. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. Journal of Andrology. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Hachey SJ, Kreher J, Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. Journal of Cell Biology. 2011;192:939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Paix A, Seydoux G. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development. 2012;139:3732–3740. doi: 10.1242/dev.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in the germline. In: Paolo Sassone-Corsi MTF, Braun Robert, editors. CSH Perspectives. Cold Spring Harbor: Cold Spring Harbor Press; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald T, Osickova A, Sulc M, Benada O, Semeradtova A, Rezabkova L, Veverka V, Bednarova L, Maly J, Macek P, Sebo P, Slaby I, Vondrasek J, Osicka R. Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. Journal of Biological Chemistry. 2013;288:22333–22345. doi: 10.1074/jbc.M113.456012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Wolf N, Priess J, Hirsh D. Segregation of germline granules in early embryos of Caenorhabditis elegans: an electron microscopic analysis. Journal of Embryology and Experimental Morphology. 1983;73:297–306. [PubMed] [Google Scholar]
- Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Computers & Chemistry. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown. Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Thank you for sending your work entitled “Regulation of RNA granule dynamics by a phosphorylation-sensitive scaffold in _C. elegans_” for consideration at eLife. Your article has been favorably evaluated by James Manley (Senior editor), a Reviewing editor, and 4 reviewers, one of whom has agreed to be identified: Roy Parker (Reviewer #1).
The Reviewing editor and the other reviewers discussed their comments before we reached this decision, and the Reviewing editor has assembled the following comments to help you prepare a revised submission. As you will see, all felt that the work was potentially quite interesting but all also raise a number of points that need to be dealt with via revision. In particular all of the reviewers question the conclusion that the MEG proteins serve as a scaffold for P granules. It is essential that you provide convincing evidence in support of this conclusion. Please address this issue as well as the other concerns of the reviewers as thoroughly as possible.
Although we ordinarily collate the reviews into a single narrative, the reviewers felt that you would benefit by a full disclosure of their comments:
Reviewer #1:
This manuscript addresses the mechanisms by which P-granules (maternal mRNA-protein assemblies in C. elegans) assemble and disassemble in a temporal and spatially regulated manner. The main contribution of the work is to demonstrate that a phosphorylation-dephosphorylation cycle of the Meg1-4 proteins plays a role in modulating the assembly and disassembly of P-granules. The kinase and phosphatase for this process are identified (building on earlier genetics) and the data generally supports the authors' conclusions, although there are some issues as detailed below.
This review is from Roy Parker and I would be happy to directly clarify the comments for the authors if desired.
A major point of the manuscript (in title and abstract) is that the Meg proteins form a scaffold that modulates P-granule assembly/disassembly. While it is clear that Meg proteins modulate P-granule assembly/disassembly in a phosphorylation dependent manner, the evidence that they form a scaffold is solely that by microscopy they are dynamic and non-uniform within P-granules. Although they could form a scaffold, the current evidence is also open to other interpretations. I suggest they a) change the tone of the manuscript to emphasize that how the Meg proteins affect P-granule structurally is not yet known, and b) remove the scaffold conclusion from the title and the abstract. Alternatively, if the authors want to argue for a scaffolding function per se, they would need additional data to support such a conclusion.
Reviewer #2:
The Wang et. al. paper advances our understanding of regulation of P granule dynamics and partitioning by a kinase, a phosphatase, and cycles of substrate phosphorylation and dephosphorylation. Most of the players have been previously described: MBK-2 kinase is needed for P granule disassembly, PP2A phosphatase (PPTR-1, 2) is needed for P granule assembly/stability, and MEG-1 and MEG-2 are needed for P granule segregation in early embryos and for fertility. By screening for proteins that interact with PPTR-1 in yeast 2-hybrid, Wang et al. additionally identified GEI-11/MEG-3 and MEG-4. They showed that a) MEG-1,3,4 directly interact with PPTR-1,2 and also with the P granule component PGL-1, b) MEG-1 and MEG-3 are phosphorylated by MBK-2 in vitro, c) MEG-1 and MEG-3 are phospho-proteins in vivo and the abundance of their phosphorylated forms is influenced by MBK-2 and PPTR-1. Analysis of double and triple mutants showed that MBK-2 is epistatic to PPTR-1, 2, and the MEGs are epistatic to both. MEG-3, 4 function redundantly to promote P granule assembly. The model is that MBK-2 and MEG phosphorylation promote P granule disassembly in the anterior cytoplasm, and PPTR-1, 2 and MEG dephosphorylation promote P granule assembly in the posterior cytoplasm destined for the germline blastomeres. The MEG proteins are predicted to be largely disordered and based on high resolution imaging of MEG-3 in live embryos using lattice light sheet microscopy, the authors propose the MEGs form a scaffold that surrounds and penetrates the P granules and that regulates P granule dynamics.
This is an impressive “stitch it all together” paper. The genetics are especially compelling. The biochemistry contributes important molecular details, but some of it needs strengthening. The lattice light sheet microscopy images in the figures convey that P granules are heterogeneous and dynamic, and the movies support that MEG-3 forms ribbon-like structures around each granule. Whether that is a “scaffold” is speculative, and should be presented as such.
Comments:
Testing molecular epistasis depends on full knock-out of MBK-2 and PPTR-1, 2 functions. Are the authors confident that double RNAi of mbk-2 and pptr-2 in the mbk-2(RNAi); pptr-1(tm3103) pptr-2(RNAi) triple worked? Might observing an mbk-2 phenotype in the triple be due to poor knock-down of pptr-2? If the authors are confident, I suggest mentioning that mbk-2 is epistatic to pptr-1, 2, as might be expected in a “no phosphorylation” scenario.
Did the authors try to generate any phospho-mimetic aa changes in the MEGs, to see if those cause P granule disassembly, even in the absence of MBK-2?
GFP::PGL-1 is the sole reporter of P granule dynamics in wild type. Does mCherry::PGL-3 display the same dynamics as GFP::PGL-1? Are there other tagged P granule reporters that confirm that the dynamics reported are P granules and not just PGL proteins? Similarly, PGL-1 is the sole reporter of P granule integrity in meg-3 meg-4 mutants. Especially since MEG-3 and MEG-4 interact directly with PGL-1, the authors should verify that P granules are absent in meg-3 meg-4 mutants using additional markers as well.
The lattice light sheet microscopy images in the figures certainly convey that P granules are heterogeneous (the distributions of both MEG-3 and PGL-3) and dynamic, but don't strongly support that MEG-3 forms a scaffold around each granule. Videos 5 and 6 best convey the ribbon-like distribution around the perimeter. Can the authors generate still images that better capture the movies than shown in e.g. Figure 5E? How many movies were generated and analyzed? Also, can the authors analyze mCherry::PGL-3 along with GFP::MEG-3 in single time-point images (e.g. Videos 5 and 6) to determine if PGL-3 is located in the spaces between the MEG-3 ribbon zones? Calling the MEGs a scaffold predicts that they would assemble into “granules” in the absence of other P granule components, but in fact MEG granules are disrupted in pgl-1; pgl-3 mutants. I would soften the language from concluding to speculating that the MEGs serve as P granule scaffolds.
There is room for improvement of the analysis of phosphorylation of MEG-3 and MEG-1 shown in Figure 3.
The last two paragraphs of Discussion need to be clarified to accurately reflect the results of this study. In the second to last paragraph, the fact that meg-3 meg-4 adults are 70% fertile, even though these embryos fail to assemble large P granules in germline blastomeres, is used to suggest that P granule assembly is not required for fertility. This is followed by acknowledging that normal looking P granules do actually reform in larval and adult meg-3 meg-4 animals. A more appropriate conclusion would be that P granules ARE required for fertility in adults, but that assembly of large P granules in early embryos is not required to specify the primordial germ cells, similar to their conclusions for pptr-1. In the last paragraph, the authors “suggest that the essential activity of the germ plasm resides not in the germ granules, but in the proteins that scaffold the granules.” It is unclear which of their findings distinguish germ granules from germ granule scaffolds and support their statement.
Reviewer #3:
The manuscript by Wang et. al. “Regulation of RNA granule dynamics by a phosphorylation-sensitive scaffold in _C. elegans_” focuses on the molecular regulation of the assembly and disassembly of C. elegans germline P granules. This study reveals that the phosphorylation state of the proteins MEG-1 and MEG-3 is regulated by activity of MBK-2, a kinase homologous to the DYRK kinase shown to regulate stress granule assembly, and the phosphatase PPTR-1/2 PP2A. The authors perform a detailed molecular mechanistic analysis using mutants and/or RNAi, revealing important insights into the epistastic relationships governing P granule assembly. Moreover, the authors utilize a novel super-resolution imaging technique to gain insights into the molecular distribution within P granules. This is an interesting and important study that makes an important contribution to our molecular understanding of P granule assembly, and should therefore definitely be published in eLife.
Despite my enthusiasm for much of the work in this paper, I nevertheless have a problem with what I think are over-interpretations of the data, particularly the imaging data.
One of the major concerns I have can be seen clearly in Video 5 (and other movies/images). There is a fair amount of “structure” apparent within the P granules. This is particularly obvious in the largest granule at the center of the image, which looks a bit like popcorn. There is apparently structure within the granules, which exists on a characteristic length scale of ∼200 nm. The problem is that if one looks at regions of the cytoplasm that does not contain granules, there is also structure on this length scale. Why is that? The mottled pattern seen both in granules and in the cytoplasm is quite reminiscent of the patterns one gets after running e.g. a Gaussian bandpass filter through unstructured white noise images, or after running deconvolution algorithms on unstructured images (I just did this for a random, white noise image and the result of applying a Gaussian bandpass filter looks, qualitatively, very similar to the mottled patterns you show; however, I can't seem to figure out how to upload an image with this review...). This is very relevant because for at least some of the light sheet images shown, there is a deconvolution algorithm being employed, where care needs to be taken to avoid artifacts. Do you see structure like that seen in the P granules when you use this technique to image a homogenous solution of dye? If you use this technique to image micron-sized fluorescent spherical particles, for which the embedded dye is presumably uniformly distributed within the particle, does it come out looking like popcorn? (is this also true if the particle is “pre-bleached” so that the fluorophores density is comparable to what one might expect for an average GFP density in the granule?). For these reasons, it is difficult for me to put any weight on arguments made about both the internal structure of the granules, and the idea that they are highly non-spherical on these length scales. Since the authors make such a big deal about the non-homogenity of the granules (e.g. last sentence of the abstract), this seems important to nail down.
As far as I can tell from the movies shown, the mottled patterns are highly dynamic, as one would expect if they were random intensity distributions that were amplified by the imaging technique and post-processing. Together with the observation that the MEG “scaffold” does not form in pgl-1/pgl-3 zygotes, the case for referring to this as a “scaffold”, which implies a static structure on which the rest of the P granule components hang, is a bit of a weak proposition.
Reviewer #4:
This manuscript provides new insight into the process of P granule assembly and disassembly by identifying a set of proteins, the MEGs, whose phosphorylation and dephosphorylation cycle may explain the assembly and disassembly of P granules in the oocyte and early stages of embryonic development. The work is intriguing and most of the data presented are convincing, but the study seems incomplete in several respects. First, it is not clear why the authors chose to analyze some mutants or combinations of mutants and not others. For example, why is the meg-4 mutant omitted? In the analysis of triple mutants, why isn't the phenotype of meg-1/2/3 or meg-2/3/4 shown? The localization analysis is one of the strengths of the paper but is limited to MEG-3 protein; the conclusions would be greatly enhanced by the analysis of other MEGs and, ideally, a combination of two different MEGs differentially labeled with GFP and Cherry. Second, the paper seems to stop short of a critical experiment, analyzing the dynamics of MEG-3 (or other MEG protein) in mbk-2 and pptr-1 mutants or even the distribution in wildtype animals during disassembly. A second major concern is that meg-3 meg-4 mutant animals are fertile despite the fact P1 has very few P granules and none are apparent in P4 (Figure 4), which makes me question the importance of these genes and the P granule assembly process.
Additional issues:
Protein-protein interactions are only demonstrated in vitro; the data would be significantly strengthened by demonstration of in vivo interactions through co-IP experiments.
The Phos-tag gel data are not particularly convincing. The authors say that they detected “one primary species for both GFP:MEG1 and GFP:MEG3 and several higher mw species that did not resolve well into bands.” These are not at all apparent on the gels shown (or at least there are no higher mw species that appear different between wt and pptr-1 or mbk-2), although a single higher mw isoform of GFP:MEG-3 is visible in the supplemental figure. Also, treatment of MEG-3 by AP in both the mbk-2 mutant, where presumably the protein is not phosphorylated, results in an increase in the intensity of the band comparable to that in the wt. This would suggest that the protein is primarily phosphorylated by one or more kinases other than mbk-2 but the authors seem to ignore this.
What are the data that allow the conclusion that the MEGs are scaffolds? They aren't sufficient for P granule assembly and it seems like an over interpretation of the localization data.
Reviewer #1:
A major point of the manuscript (in title and abstract) is that the Meg proteins form a scaffold that modulates P-granule assembly/disassembly. While it is clear that Meg proteins modulate P-granule assembly/disassembly in a phosphorylation dependent manner, the evidence that they form a scaffold is solely that by microscopy they are dynamic and non-uniform within P-granules. Although they could form a scaffold, the current evidence is also open to other interpretations. I suggest they a) change the tone of the manuscript to emphasize that how the Meg proteins affect P-granule structurally is not yet known, and b) remove the scaffold conclusion from the title and the abstract. Alternatively, if the authors want to argue for a scaffolding function per se, they would need additional data to support such a conclusion.
We agree with the reviewer that the use of the word scaffold was too speculative.
Accordingly, we have removed this reference from most of the manuscript except for the discussion, where we present this concept clearly as a hypothesis that will need to be tested further. The hypothesis of a “scaffold” is based on the Following observations:1) the MEGs are required to assemble large PGL granules from smaller unstable PGL granules and 2) MEG‐3 localizes to small assemblies in the cytoplasm and larger assemblies surrounding a PGL core in the P granules and 3) (new data!) during granule disassembly, MEG‐3 and MEG‐4 assemblies Persist longer than the PGL‐1 core. Together these observations suggest the following working model: the MEGs promote granule assembly by forming dynamic scaffolds in the cytoplasm that stabilize and wrap around PGL condensates. See new text in Discussion.
Reviewer #2:
Testing molecular epistasis depends on full knock-out of MBK-2 and PPTR-1, 2 functions. Are the authors confident that double RNAi of mbk-2 and pptr-2 in the mbk-2(RNAi); pptr-1(tm3103) pptr-2(RNAi) triple worked? Might observing an mbk-2 phenotype in the triple be due to poor knock-down of pptr-2? If the authors are confident, I suggest mentioning that mbk-2 is epistatic to pptr-1, 2, as might be expected in a “no phosphorylation” scenario.
We are confident that the RNAi treatments worked for three reasons: 1) mbk‐2(RNAi) pptr‐2(RNAi) was applied to pptr‐1(tm3103) animals and resulted in a Mbk‐2 phenotype consistent with mbk‐2(RNAi) working and 2) we applied in parallel control RNAi; pptr‐2(RNAi) to pptr‐1(tm3103) and obtained the expected Pptr‐1/2 phenotype confirming that the pptr‐2(RNAi) treatment also worked. 3) pptr‐1/2(RNAi) on the mbk‐2 null gave the same result. We have clarified this conclusion in the text.
Did the authors try to generate any phospho-mimetic aa changes in the MEGs, to see if those cause P granule disassembly, even in the absence of MBK-2?
We did not make phospho‐mimetic mutants because of the large numbers of potential phosphorylation sites and the difficulty in testing for partial suppression of the mbk‐2 phenotype (granule number is variable in mbk‐2 zygotes). We did mutate S574 in MEG‐1 (site shown to be phosphorylated by
MBK‐2 in vitro and in vivo) to alanine and did not observe a robust phenotype, consistent with the idea that the regulation involves more than one phosphorylation site.
GFP::PGL-1 is the sole reporter of P granule dynamics in wild type. Does mCherry::PGL-3 display the same dynamics as GFP::PGL-1? Are there other tagged P granule reporters that confirm that the dynamics reported are P granules and not just PGL proteins? Similarly, PGL-1 is the sole reporter of P granule integrity in meg-3 meg-4 mutants. Especially since MEG-3 and MEG-4 interact directly with PGL-1, the authors should verify that P granules are absent in meg-3 meg-4 mutants using additional markers as well.
We agree with the reviewer that it is important to confirm that the phenotypes we report are not just specific to PGL‐1, but extend to other P granule components. Accordingly, we have now added the following data to the revision:
mCherry::PGL‐3 undergoes the same dynamics as GFP::PGL‐1 in wild‐type embryos (Figure 1–figure supplement 1, column 5). GLH‐2 behaves like PGL‐1 in meg‐3meg‐4 zygotes (Figure 4–figure supplement 1). We have also confirmed that P granules are absent in meg‐3meg‐4 zygotes by observing the distribution of nos‐2 RNA (RNA component of P granules) and PIE‐1 (another RNA binding protein that associates with P granules) (Figure 4–figure supplement 2).
The lattice light sheet microscopy images in the figures certainly convey that P granules are heterogeneous (the distributions of both MEG-3 and PGL-3) and dynamic, but don't strongly support that MEG-3 forms a scaffold around each granule. Videos 5 and 6 best convey the ribbon-like distribution around the perimeter. Can the authors generate still images that better capture the movies than shown in e.g. Figure 5E? How many movies were generated and analyzed? Also, can the authors analyze mCherry::PGL-3 along with GFP::MEG-3 in single time-point images (e.g. Videos 5 and 6) to determine if PGL-3 is located in the spaces between the MEG-3 ribbon zones?
The strongest evidence that MEG‐3 is present at the perimeter comes from our analysis of 37 granules, where we compared the area occupied by MEG‐3 and
PGL‐3, and found that MEG‐3 occupied a larger area in 34 of the 37 granules, with PGL‐3 entirely contained within the MEG‐3 area in all cases (Figure 5–figure supplement 3).
Unfortunately we were not able to generate SI (highest resolution) images to examine MEG‐3 and PGL‐3 in the same granule, so we do not know whether PGL‐3 is localized in the spaces between the MEG‐3 domains. Our confocal and lower resolution lattice‐sheet (dithered mode) images, however, suggest that this is the case (Figure 5–figure supplement 3).
Calling the MEGs a scaffold predicts that they would assemble into “granules” in the absence of other P granule components, but in fact MEG granules are disrupted in pgl-1; pgl-3 mutants. I would soften the language from concluding to speculating that the MEGs serve as P granule scaffolds.
We agree that we have used the word “scaffold” too freely and in the revision we now mention this concept only in the discussion. The observation that the MEGs no longer localize to large granules in the absence of PGL‐1 and PGL‐3 is actually consistent with our model that dynamic MEG scaffolds present throughout the cytoplasm associate with and stabilize PGL condensates. Without the PGL condensates, the dynamic MEG scaffolds remain distributed throughout the cytoplasm.
There is room for improvement of the analysis of phosphorylation of MEG-3 and MEG-1 shown in Figure 3.
We have expanded Figure 3–figure supplement 1 to show the complete migration pattern of MEG‐1 and MEG‐3 isolated from wild‐type embryos with and without alkaline phosphatase treatment. We have also included data for MEG‐3 in the pptr‐1 mutant. At equivalent concentrations of protein and alkaline phosphatase, MEG‐3 shows greater resistance to alkaline phosphatase treatment in the pptr‐1 background compared to wild‐type.
The last two paragraphs of Discussion need to be clarified to accurately reflect the results of this study. In the second to last paragraph, the fact that meg-3 meg-4 adults are 70% fertile, even though these embryos fail to assemble large P granules in germline blastomeres, is used to suggest that P granule assembly is not required for fertility. This is followed by acknowledging that normal looking P granules do actually reform in larval and adult meg-3 meg-4 animals. A more appropriate conclusion would be that P granules ARE required for fertility in adults, but that assembly of large P granules in early embryos is not required to specify the primordial germ cells, similar to their conclusions for pptr-1. In the last paragraph, the authors “suggest that the essential activity of the germ plasm resides not in the germ granules, but in the proteins that scaffold the granules.” It is unclear which of their findings distinguish germ granules from germ granule scaffolds and support their statement.
We completely agree with the reviewer and have clarified our writing to reflect these points. We do not dispute that the perinuclear P granules that form later in development are required for fertility, as has been shown by many studies. Our analyses only address the function of the cytoplasmic P granules that form within the germ plasm of early embryos. We argue that granule assembly in the context of germ plasm of early embryos is NOT essential. This is an important conclusion as many studies assume that RNA granules perform the critical function of the germ plasm. We have reworded the discussion to make this point clearer.
The sentence in the last paragraph is meant to remind readers that, although the MEGs’ role in granule assembly does not appear essential, the MEGs as a group are essential for fertility. We have reworded this statement.
Reviewer #3:
One of the major concerns I have can be seen clearly in Video 5 (and other movies/images). There is a fair amount of “structure” apparent within the P granules. This is particularly obvious in the largest granule at the center of the image, which looks a bit like popcorn. There is apparently structure within the granules, which exists on a characteristic length scale of ∼200 nm. The problem is that if one looks at regions of the cytoplasm that does not contain granules, there is also structure on this length scale. Why is that? The mottled pattern seen both in granules and in the cytoplasm is quite reminiscent of the patterns one gets after running e.g. a Gaussian bandpass filter through unstructured white noise images, or after running deconvolution algorithms on unstructured images (I just did this for a random, white noise image and the result of applying a Gaussian bandpass filter looks, qualitatively, very similar to the mottled patterns you show; however, I can't seem to figure out how to upload an image with this review...). This is very relevant because for at least some of the light sheet images shown, there is a deconvolution algorithm being employed, where care needs to be taken to avoid artifacts. Do you see structure like that seen in the P granules when you use this technique to image a homogenous solution of dye? If you use this technique to image micron-sized fluorescent spherical particles, for which the embedded dye is presumably uniformly distributed within the particle, does it come out looking like popcorn? (is this also true if the particle is “pre-bleached” so that the fluorophores density is comparable to what one might expect for an average GFP density in the granule?). For these reasons, it is difficult for me to put any weight on arguments made about both the internal structure of the granules, and the idea that they are highly non-spherical on these length scales. Since the authors make such a big deal about the non-homogenity of the granules (e.g. last sentence of the abstract), this seems important to nail down.
As far as I can tell from the movies shown, the mottled patterns are highly dynamic, as one would expect if they were random intensity distributions that were amplified by the imaging technique and post-processing.
We understand the reviewer’s concerns and now include raw data from the SI lattice light sheet analyses in a supplementary figure (Figure 5–figure supplement 5). The new figure shows that:
The GFP::MEG‐3 ribbon‐like structure is visible before the reconstruction algorithm is applied (Figure 5–figure supplement 5A and B), showing that reconstruction does not impose structure at the scale of MEG‐3 ribbons.
The GFP::MEG‐3 structure is maintained between successive time frames (Figure 5–figure supplement 5E), even though movement can also be detected. If the structure were white noise, it would be uncorrelated between frames.
The photon counts are at least an order of magnitude greater than noise, ruling out the possibility that the signal is just filtered white noise. (Figure 5–figure supplement 5F)
Together with the observation that the MEG “scaffold” does not form in pgl-1/pgl-3 zygotes, the case for referring to this as a “scaffold”, which implies a static structure on which the rest of the P granule components hang, is a bit of a weak proposition.
We agree and have removed the mention of a scaffold throughout the text, except
For the discussion where we present this idea as a working hypothesis. We also present new data showing that the MEGs perdure longer in disassembling granules than PGL‐1, consistent with a scaffolding function. We describe our model more clearly in the Discussion.
As mentioned above, we note that the observations that the MEGs no longer localize to large granules in the absence of PGL‐1 and PGL‐3 is actually consistent with our model that dynamic MEG scaffolds present throughout the cytoplasm associate with and stabilize PGL condensates. Without the PGL condensates, the smaller, dynamic MEG scaffolds remain distributed throughout the cytoplasm.
Reviewer #4:
This manuscript provides new insight into the process of P granule assembly and disassembly by identifying a set of proteins, the MEGs, whose phosphorylation and dephosphorylation cycle may explain the assembly and disassembly of P granules in the oocyte and early stages of embryonic development. The work is intriguing and most of the data presented are convincing, but the study seems incomplete in several respects. First, it is not clear why the authors chose to analyze some mutants or combinations of mutants and not others. For example, why is the meg-4 mutant omitted?
We have now added analysis of the meg‐4 mutant (Figure 4). Our initial RNAi analyses indicated that loss of meg‐4 alone is not sufficient to cause a P granule phenotype in wildtype, but enhances the phenotype of meg‐3 mutants. For this reason, we first created a meg‐4 mutant in the background of the meg‐3 mutant to create the double (the two genes are closely linked on X). Since then, we have generated a second meg‐4 mutant in the wild‐type background.
As expected, this mutant has no P granule phenotype. These new data are now included in Figure 4.
In the analysis of triple mutants, why isn't the phenotype of meg-1/2/3 or meg-2/3/4 shown?
Because all the _meg_s are linked on the X chromosome, it is not trivial to create all the different combinations. Analysis of meg‐1 and meg‐2 by Leacock and Reinke has shown that, of the two, meg‐1 plays the primary role, and therefore we used meg‐1 for the triple mutant combination. We did analyze the quadruple combination and found that loss of meg‐1 and meg‐_2 does not affect the meg‐_3 meg‐4 mutant phenotype (Figure 4–figure supplement 3).
The localization analysis is one of the strengths of the paper but is limited to MEG-3 protein; the conclusions would be greatly enhanced by the analysis of other MEGs and, ideally, a combination of two different MEGs differentially labeled with GFP and Cherry.
We have done the most extensive analyses on MEG‐3 because it is the only single meg that shows a strong P granule phenotype in zygotes. In the revision, we now add new localization data for MEG‐1 and MEG‐4. The data confirm that MEG‐4, like MEG‐1, 2 and 3, localizes to P granules specifically in embryos. The data also demonstrate that MEG‐1, like MEG‐3, only partially overlaps with PGL‐1. See new Figure 5–figure supplement 2 and Figure 5–figure supplement 4.
Second, the paper seems to stop short of a critical experiment, analyzing the dynamics of MEG-3 (or other MEG protein) in mbk-2 and pptr-1 mutants or even the distribution in wildtype animals during disassembly.
We have done these analyses (see Figure 5–figure supplement 2). These data indicate that MEG‐3 and MEG‐4 persist longer than PGL‐1 in disassembling granules in wild‐type. We also show that this is the case for GFP::MEG‐3 in ppt‐1 zygotes. These results indicate that the MEGs exhibit different dynamics than PGL‐1 in P granules. Together with the fact that the MEGs are required for the assembly of large stable PGL granules, these observations are consistent with a possible scaffolding function, but do not prove it. For this reason, while we include the new data in the revision, we have removed mention of “scaffold” from the title and text and only mention this idea as a hypothesis in the Discussion.
A second major concern is that meg-3 meg-4 mutant animals are fertile despite the fact P1 has very few P granules and none are apparent in P4 (Figure 4), which makes me question the importance of these genes and the P granule assembly process.
We agree with the reviewer and address this point in the last section of the discussion. Our conclusion is that P granule assembly in the germ plasm of early embryo is NOT essential for fertility. MEG activity, however, is required for fertility since meg‐1meg‐2 and meg‐1meg‐3meg‐4 mutants are 100% sterile.
We suggest therefore that MEGs performs a function required for fertility that is NOT dependent on P granule assembly. This is an important conclusion as granule assembly is often assumed to be an essential function of the germ plasm.
Additional issues:
1) Protein-protein interactions are only demonstrated in vitro; the data would be significantly strengthened by demonstration of in vivo interactions through co-IP experiments.
The proposed interaction between the MEGs and PPTR‐1 is based on four independent lines of evidence: interaction in the yeast two hybrid system, GST pull downs using partially purified proteins, in vivo phosphorylation patterns visualized on Phostag gels, and genetic epistasis.
We also showed that the MEGs interact with PGL‐1 in GST pull‐down assay. As indicated by the reviewer, the MEG‐PGL interaction will need to be confirmed in vivo, since we do not provide any additional evidence for that interaction_._ However, that interaction is not the main focus of this study and none of our conclusions depend on that finding. We can remove those data if the editor prefers.
2) The Phos-tag gel data are not particularly convincing. The authors say that they detected “one primary species for both GFP:MEG1 and GFP:MEG3 and several higher mw species that did not resolve well into bands.” These are not at all apparent on the gels shown (or at least there are no higher mw species that appear different between wt and pptr-1 or mbk-2), although a single higher mw isoform of GFP:MEG-3 is visible in the supplemental figure.
We agree with the reviewer that it is difficult on the Phos‐tag gels to see all the phosphorylated forms of MEG‐1 and MEG‐3. This is consistent with the fact that these proteins have large numbers of serines and threonines and likely exist in many different isoforms, each with a unique migration behavior on the Phos‐tag gels.
We include larger gels in Figure 3–figure supplement 1 that clearly show the presence of at least one phosphoisoform. Since we cannot visualize all isoforms on gels, we based all of our quantitative analyses on the non‐phosphorylated form of MEG‐1 and MEG‐3, which runs as a clear single band in all the experiments. By comparing the abundance of this band on Phos‐tag versus non‐Phos‐tag gels, we estimate the degree of phosphorylation in the different extracts and use these estimates to COMPARE degrees of phosphorylation between wild‐type and mbk‐2 and pptr‐1 mutants.
These experiments were repeated at least 4 times, and always showed the same results: the degree of phosphorylation is highest in pptr‐1 lysates and lowest in mbk‐2 lysates.
Also, treatment of MEG-3 by AP in both the mbk-2 mutant, where presumably the protein is not phosphorylated, results in an increase in the intensity of the band comparable to that in the wt.
That experiment was not done with a mbk‐2 mutant (which is lethal and cannot be grown in large enough quantities for biochemistry), but with mbk‐2(RNAi) embryos which may retain some mbk‐2 activity and therefore some phosphorylation.
Also, as we note in the text, it is very possible that other kinases phosphorylate the MEGs. Our only claim is that MBK‐2 is one of the kinases that influence the degree of MEG phosphorylation.
This would suggest that the protein is primarily phosphorylated by one or more kinases other than mbk-2 but the authors seem to ignore this.
We agree and have included the following in the text: “we do not know whether MBK‐2 and PP2A PPTR‐1/2 are the only regulators of MEG‐1 and MEG‐3 phosphorylation, or whether other kinases and phosphatases are also involved.”
3) What are the data that allow the conclusion that the MEGs are scaffolds? They aren't sufficient for P granule assembly and it seems like an over interpretation of the localization data.
We agree and now only mention the scaffold idea as a working hypothesis in the Discussion.
Supplementary Materials
Figure 2—source data 1. Candidates from yeast two-hybrid screen.
DNA was extracted and sequenced from 111 colonies grown on –Trp–Leu–Ura–His plates. RNAi feeding vectors for each candidate were obtained from the Ahringer or OpenBiosystems RNAi banks, or if unavailable were PCR amplified from genomic DNA and cloned into pL4440. *meg-3(RNAi) also knocks out meg-4 and vice-versa. + in the pptr-1 suppression column means that the RNAi treatment restores P granules in P blastomeres. Pleiotropic indicates a P granule phenotype accompanied by additional cellular defects. Entries in red are the MEG proteins, which uniquely affect P granules in the zygote stage (meg-3 and meg-4) or suppress the pptr-1 phenotype (meg-1) without inducing other phenotypes. Other phenotypes were as follows: emb = embryonic lethal, dpy = dumpy, egl = egg laying defect.