Three-dimensional organotypic culture: experimental models of mammalian biology and disease (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 7.
Published in final edited form as: Nat Rev Mol Cell Biol. 2014 Sep 17;15(10):647–664. doi: 10.1038/nrm3873
Abstract
Mammalian organs are challenging to study as they are fairly inaccessible to experimental manipulation and optical observation. Recent advances in three-dimensional (3D) culture techniques, coupled with the ability to independently manipulate genetic and microenvironmental factors, have enabled the real-time study of mammalian tissues. These systems have been used to visualize the cellular basis of epithelial morphogenesis, to test the roles of specific genes in regulating cell behaviours within epithelial tissues and to elucidate the contribution of microenvironmental factors to normal and disease processes. Collectively, these novel models can be used to answer fundamental biological questions and generate replacement human tissues, and they enable testing of novel therapeutic approaches, often using patient-derived cells.
The anatomical basis of life was first studied by natural historians who identified and named organs across species. A crucial simplification came when Bichat recognized that organs represented combinations of a few fundamental tissues1. Compound microscopes enabled Virchow to define epithelium, connective tissu e, nerve, muscle and blood as the universal tissues2, and by 1900, the microscopic anatomy of humans was well known3. However, it remains difficult at a cellular and molecular level to understand how mammalian organs form during development and how they change during disease.
Compared with the transparent embryos of externally developing species, mammalian tissues and organs are fairly inaccessible to experimental manipulation and optical observation. Furthermore, mammalian development occurs over the time range of days to years. These limitations led Harrison et al. to develop twodimensional (2D) culture techniques in 1907 (REF. 4). 2D culture enabled biologists to observe and manipulate mammalian cells and laid the foundation for cell and molecular biology. However, 2D cultures do not completely recapitulate the three-dimensional (3D) organization of cells and extracellular matrix (ECM) within tissues and organs. Consequently, there is a large gap between our detailed knowledge of sub cellular processes and our incomplete understanding of mammalian biology at the tissue level. Dynamic analyses of organogenesis have instead relied on model systems, such as Caenorhabditis elegans, Drosophilam elanogaster, Xenopus laevis and zebrafish.
The goal of reconstituting organ function ex vivo is broadly shared, and there are successful examples for most tissues and organs (TABLE 1). In pursuit of this goal, a wide range of techniques has been developed that are referred to as 3D culture, organotypic culture or organoid culture. Various subfields use these terms either interchangeably or distinctly; for example, in the field of mammary gland biology, the term organoids refers to primary explants of epithelial ducts into 3D ECM gels5. Conversely, in studies of intestinal biology, organoids can refer to clonal derivatives of primary epithelial stem cells that are grown without mesenchyme6 or can refer to epithelial–mesenchymal co-cultures that are derived from embryonic stem (ES) cells or induced pluripotent stem cells (iPS cells)7.
Table 1.
Cellular and molecular techniques for three-dimensional culture
| Organ | Cellular input | Culture format | Refs* |
|---|---|---|---|
| Mammary gland | Cell line (for example, MCF-10A) | 2.5D culture | 51,111,161 |
| Whole organ | Mechanically supported | 15,162 | |
| Tissue organoid | 3D-embedded culture | 5,27,66,72,163 | |
| Primary cells | 2.5D culture | 57,164 | |
| Salivary gland | Embryonic whole organ | Mechanically supported | 92,99,115 |
| Ex vivo epithelial-mesenchymal recombination | Mechanically supported | 100,165 | |
| Tissue organoid | 3D-embedded culture | 33,63,95 | |
| Primary cells | 3D-embedded culture | 166,167 | |
| Kidney | Cell line (for example, MDCK) | 2.5D culture | 58,105,168,169 |
| Embryonic whole organ | Mechanically supported | 96,170,171 | |
| Ex vivo epithelial-mesenchymal recombination | Mechanically supported or 3D-embedded | 172,173 | |
| Tissue organoid | 3D-embedded culture | 34,64,174 | |
| Stem cell organoid (ES cells) | 2.5D culture | 175 | |
| Stem cell organoid (ES cells and iPS cells) | Mechanically supported | 176,177 | |
| Primary embryonic cells | Mechanically supported | 55,56 | |
| Lung | Normal or neoplastic lung slice | Mechanically supported | 17,156,178 |
| Embryonic whole organ | Mechanically supported | 14,116,179 | |
| Tissue organoid | 3D-embedded culture | 35,179 | |
| Stem cell organoid (tissue stem cells) | 3D-embedded culture | 128,180 | |
| Primary cells (human alveolar cells) | 2.5D culture | 59 | |
| Primary cells (foetal pulmonary cells) | 3D-embedded culture | 181 | |
| Small intestine | Cell line (for example, Caco-2) | 2.5D culture | 182,183 |
| Organ slice | Mechanically supported | 18,19 | |
| Tissue organoid | 3D-embedded culture | 6,28,36,102,184 | |
| Stem cell organoid (LGR5+) | 3D-embedded culture | 6,143 | |
| Stem cell organoid (iPS cells) | 3D-embedded culture | 185 | |
| Colon | Organ slice | Mechanically supported | 19,20 |
| Tissue organoid | 3D-embedded culture | 36,37,144,184,186 | |
| Stem cell organoid (LGR5+) | 3D-embedded culture | 37,144 | |
| Liver | Whole organ and organ slice | Mechanically supported | 14,153,178 |
| Tissue organoid | 3D-embedded culture | 38 | |
| Stem cell organoid (LGR5+) | 3D-embedded culture | 38 | |
| Stem cell organoid (iPS cells) | 2.5D culture | 152 | |
| Stomach | Tissue organoid | 3D-embedded culture | 39 |
| Stem cell organoid (LGR5+ and Troy+) | 3D-embedded culture | 39,187 | |
| Pancreas | Embryonic whole organ | Mechanically supported | 14,84 |
| Tissue organoid | 3D-embedded culture | 40 | |
| Stem cell organoid (LGR5+) | 3D-embedded culture | 40 | |
| Primary pancreatic ductal cells | 3D culture | 65,188 | |
| Oesophagus | Primary cells (oesophageal keratinocytes) | Mechanically supported | 54,89,127 |
| Skin | Cell line (for example, HaCaT) | 3D-embedded culture | 189 |
| Primary cells (epidermal keratinocytes) | Mechanically supported | 52,53,73,114 | |
| Prostate | Tissue organoid | 3D-embedded culture | 41 |
| Primary cells (human prostatic epithelium) | 3D-embedded culture | 190 | |
| Optic cup | Stem cell organoid (ES cells) | 3D-suspension or 3D-embedded culture | 44,45 |
| Brain | Organ slice | Mechanically supported | 21,22 |
| Stem cell organoid (ES cells) | 3D-suspension culture | 46 | |
| Stem cell organoid (iPS cells) | 3D-embedded culture, spinning bioreactor | 47 | |
| Blood vessels | Primary cells (for example, HUVEC) | 2.5D culture | 191,192 |
| Primary cells (for example, HUVEC) | 3D-embedded culture | 193 | |
| Organ slice (aorta) | 3D-embedded culture | 23 | |
| Primary cells (for example, HUVEC) | 3D bioengineered platform | 11,12,151 ,194 |
In this Review, we first provide an overview of the commonly used cellular inputs and culture formats. We then discuss how these experimental systems have been used to visualize the cellular mechanisms that drive epithelial tissue development, to study the genetic regulation of cell behaviours in epithelial tissues and to evaluate the role of microenvironmental factors in normal development and disease. Finally, we provide examples of how 3D culture techniques can be used to build complex organs, to generate replacement human tissues and to advance therapeutic approaches.
Cellular inputs into 3D culture
To understand how mammalian organs can be cultured ex vivo, it is useful to consider their constituent parts. The external surfaces of the body and the linings of organs are built from epithelial tissues8. Epithelial cells are connected to each other by intercellular junctions and are located within a specialized ECM, which is known as the basement membrane. These cell–cell and cell–ECM interactions are not completely modelled in 2D culture. Epithelial tissues are avascular and exist in close proximity to vascularized connective tissue8. In contrast to the epithelium, connective tissue contains an abundance of ECM and a diverse population of stromal cells, including fibroblasts, immune cells and adipocytes8. Epithelium and connective tissue are functionally interdependent units within organs and are integrated with nerves and muscle to varying degrees depending on their organ-specific function.
The first crucial design choice in 3D culture is the extent to which the full in vivo complexity of the organ is recapitulated. Organ function results from cooperation among different tissues, but it can be difficult to isolate the roles of specific genes or cell behaviours in vivo. One approach is to deconstruct organs into their parts (for example, epithelium, stromal cells, vasculature or ECM) and then selectively recombine these parts in 3D culture. Embedding epithelial cells within an ECM gel enables the cells to self-assemble into tissues and to both interpret and remodel the ECM9. Similarly, endothelial cells and perivascular cells can be patterned into functional, perfused networks within ECM gels to model vascular development within connective tissue10–12. Alternatively, multiple tissue components can be combined into the same culture.
Whole-organ and organ-slice cultures
Conceptually, the simplest unit that can be explanted into 3D culture is the whole organ (FIG. 1a). This approach was successfully used to study skeletal development as early as 1929 (REF. 13) and was extended to the kidney, lung, salivary gland, liver, pancreas and mammary gland by the mid-twentieth century14–16. However, the limited diffusion of extracellular molecules into thick tissues restricts these studies to embryonic or thin organs. Alternatively, organs such as the lung17, small intestine18,19, colon19,20, brain21,22 and aorta23 can be sectioned into tissue ‘slices’ and mounted onto siliconized filter paper24 or porous culture membranes25 for mechanical support. Slices from the same organ can be subjected to different experimental conditions, which enables the evaluation of both matrix-bound and soluble paracrine signals; for example, the lactogenic hormones were defined by their ability to induce milk production in whole mammary gland cultures derived from virgin mice26. Although these culture formats enable optical access to the tissue and experimental interventions within native stromal tissues, it can be challenging to maintain tissue viability, and the interpretation of experimental manipulations is more difficult within complex organs.
Figure 1. Cellular inputs to organotypic cultures.
a Whole-organ and organ-slice cultures. Tissues that develop during embryogenesis, such as the salivary gland, kidney and lung, are commonly explanted as whole organs; for example, explants of metanephric kidney that have been isolated from the embryonic urogenital ridge will undergo vigorous branching morphogenesis in three-dimensional (3D) culture. Tissues that develop postnatally, such as the mammary gland, intestine, brain and aorta, can be sectioned into tissue ‘slices’ owing to their large size. b Tissue organoid cultures. Primary organs can be harvested and processed by mechanical disruption and enzymatic digestion into tissue fragments (known as tissue organoids). The native stromal cells and extracellular matrix are typically removed, which enables isolated culture of the epithelial tissues. The resulting organoids contain diverse epithelial cell types organized in their normal spatial configuration and are typically explanted into commercial extracellular matrices, such as Matrigel or collagen I; for example, mammary epithelial organoids will undergo branching morphogenesis in 3D Matrigel. c Stem cell organoid cultures. Stem cells can be used to generate organoids that model the architecture and cellular composition of a larger organ. Both embryonic and adult induced pluripotent stem (iPS) cells have been used to generate organoids of the kidney, lung, intestine, liver, optic cup and brain; for example, embryonic stem (ES) cells that are cultured in the presence of Matrigel and differentiation factors will aggregate and self-organize into optic cup-like structures. Alternatively, single tissue stem cells that have been isolated from an adult organ can be used; for example, Leu-rich repeat-containing G protein-coupled receptor (LGR5) - expressing (LGR5+) tissue stem cells that are embedded within Matrigel will generate many tissues of the digestive tract. d Primary cell cultures. Primary keratinocytes from the skin and oesophagus have been cultured on cell culture inserts to organize into stratified epithelium. In addition, primary epithelial cells from the salivary gland, lung, kidney and pancreas, as well as endothelial cells, have been used in two and one-half-dimensional (2.5D) or 3D culture.
Tissue organoids
A fundamental developmental question is whether epithelial cells determine organ pattern or whether the pattern emerges from a dialogue between the epithelium and the connective tissue or mesenchyme. To answer this question, epithelial tissues are isolated and cultured without their corresponding stromal cells; this approach is known as mesenchyme-free or organoid culture (FIG. 1b). Tissue organoids are freshly isolated from primary mammalian organs for every experiment. Each tissue organoid contains several hundred cells, which are accessible to signalling molecules and can be genetically modified using robust lentivirus- and adenovirus-based techniques27–31.
Organoid protocols have been developed for the mam-mary gland5,32, salivary gland33, kidney34, lung35, small intestine6,36, colon36,37, liver38, stomach39, pancreas40 and prostate41. Tissue organoids are typically explanted into commercial matrices, such as Matrigel42 or collagen I43, which enable optical imaging. As pieces of tissue are explanted intact into culture, the resulting organoids contain diverse epithelial cell types that are organized in their normal spatial configurations as observed in vivo. This culture format has been used to study the cell movements that drive organogenesis and to model the cell and tissue consequences of genetic changes.
Stem cell organoids
Organoids can also be expanded from ES cells, from iPS cells or from primary stem cells that have been purified from organs (FIG. 1c); for example, primary tissue-derived, intestinal stem cells that express Leu-rich repeat-containing G protein-coupled receptor 5 (LGR5) clonally generate crypt–villus architecture in 3D culture6. This approach was extended to the stomach39, colon37, pancreas40 and liver38. Investigators have also developed robust techniques to generate stem cell organoids from ES cells and iPS cells (TABLE 1). An advantage of iPS cell-derived organoids is that they can be gener ated from cells from the patient. ES cell- or iPS cell-derived organoids have also been used to demonstrate the self-organization of the retina44,45, cerebral cortex46,47 and pituitary48. However, in vivo organs do not expand from single isolated stem cells, and therefore the mechanisms that drive the formation of stem cell organoids may be distinct from organogenesis in vivo. For example, cortical organoids that are formed from ES cells generate stratified structures with layer-specific neuronal differentiation46; however, the inside-out pattern of layer formation that is observed in vivo is reversed in 3D culture46. Nonetheless, the extent to which brain anatomy can be recapitulated from defined cellular and molecular starting materials is remarkeable46,47. An additional issue is the timing of molecular interventions in tissues compared with that in single cells, as differences in timing could easily change phenotypes.
Reaggregated single-cell suspensions
Clonal expansion from a single ES cell or iPS cell requires many rounds of cell division to generate organoids. Accordingly, many 3D culture assays start from suspensions of single cells, such as cell lines, stem cells or freshly isolated primary cells (FIG. 1d). Classic amphibian embryology experiments showed that disaggregated single cells would spontaneously reaggregate and recapitulate their normal tissue architecture49. Similarly, mammalian kidney50 or mammary51 cell lines readily form acini from single cells when cultured on top of Matrigel. These epithelial models were used to dissect the molecular basis of epithelial adhesion and polarity50,51. A single-cell suspension can also generate more complex tissues; for example, isolated epidermal or oesophageal keratinocytes organize into a stratified epithelium with highly realistic tissue architecture52–54. Similarly, dissociated cells from mouse embryonic kidneys reaggregate to form renal structures that contain epithelial-derived ureteric buds and mesenchyme-derived nephrons55,56. These approaches facilitate the formation of chimeric tissues that consist of different cell types or cells with distinct genetic modifications.
Culture formats for 3D culture
In addition to the cellular inputs, culture formats can be varied independently in order to answer specific biological questions (TABLE 1).
2.5D cultures
The addition of basement membrane proteins to the medium in 2D cultures is sufficient to induce tissue-specific differentiation of diverse epithelial cells, including mammary57, kidney58 and lung59 (FIG. 2a). Most experiments rely on a commercial basement membrane protein source, such as Matrigel42. Conversely, epithelial tissues often lose their differentiated state and migrate individually when cultured in a stromal matrix such as collagen I (REF. 60). These observations led to the development of diverse assays in which single cells are plated on top of Matrigel, with additional Matrigel in the medium50,51,61. This technique is frequently referred to as two and one-half-dimensional (2.5D) culture or ‘drip’ culture, in reference to the ECM in the medium (FIG. 2b). This format does not perfectly model the in vivo environment, as the cells contact a large fluid reservoir that they would not encounter in vivo. As a result, fluid-facing surfaces of the cell lack contact with the ECM, and cell-generated paracrine factors are diluted. Despite these limitations, 2.5D assays are experimentally convenient, they induce cells to form a more physiological tissue architecture than 2D assays and the cells remain accessible for molecular analysis51.
Figure 2. The major categories of cell culture.
a Two-dimensional (2D) culture. Cells are typically cultured directly on a highly rigid substrate such as glass or plastic. The medium can be supplemented with extracellular matrix (ECM) proteins to induce a more differentiated cell state; for example, addition of laminin I will induce the differentiation of mammary epithelial cells. b Two and one-half-dimensional (2.5D) culture (also known as drip culture). Cells are cultured on top of a thin, organized layer of ECM, and diluted ECM proteins (such as laminins) are present in the medium. This format is ideal for imaging and antibody staining and is sufficient for epithelial acinar formation (for example, in MCF-10A and MDCK (Madin–Darby canine kidney) cell lines). 2.5D cultures have also been used to generate endothelial networks. The mechanical or structural properties of the ECM layer can be varied, and microfluidics can be used to generate flow-over gradients. c Three-dimensional (3D)-embedded culture. Cells are cultured within a gel of ECM proteins. Cells are uniformly exposed to an organized ECM and can further remodel and restructure the ECM over time; for example, mammary tissue organoids will undergo branching morphogenesis in 3D Matrigel. This format requires that the ECM solution is cell-compatible both in liquid and in gel form, and it enables the incorporation of different ECM components, multicellular tissues and stromal cells. If constructed within microfluidic devices, these cultures can be subjected to in- or through-gel gradients. ECMs can also be precisely patterned in 3D. d Mechanically supported culture. Cells, organ slices or whole organs are cultured on a tissue culture insert that is either submerged in medium or maintained at an air–liquid interface. Histologically realistic epithelial tissues can be constructed in stages, with initial assembly of keratinocytes into an epithelial cell layer on a submerged culture insert, followed by exposure of these cells to an air–liquid interface to induce the formation of stratified epidermis. Stromal cells can be co-cultured with the epithelial cells or added to a separate compartment within the culture dish to study paracrine effects without direct physical contact between cell types. Slices of large organs, such as the brain, can be cultured on these inserts.
3D-embedded cultures
To better model in vivo tissue organization, cells can be fully embedded within 3D ECM gels (FIG. 2c). Tissue organoids that are embedded in 3D ECM have been used to study the branching morphogenesis of the mammary5,32,62, salivary33,63, kidne y64, lung35 and pancreatic ductal65 epithelium. In this format, both the composition and mechanics of the ECM environment can be varied66–71. Collagen I models the ECM of connective tissue, basement membrane extracts (such as Matrigel) recapitulate the tissue context of epithelial ducts, and fibroblast-conditioned matrix, which is rich in fibronectin, models the microenvironment during wound healing. Embedded cultures also support a broader and more complex range of tissue architectures than is typically observed in 2.5D formats. However, owing to the location of the cells within a thick ECM gel, both optical imaging72 and the recovery of cells are more complex than in 2.5D cultures.
Mechanically supported cultures
Both whole-organ explant cultures and stratified epithelial cultures that have been reconstituted from single cells are typically cultured on top of mechanical supports. These cultures were historically grown on top of siliconized paper14,16,24 and, more recently, have been grown on cell culture inserts with semipermeable membranes (FIG. 2d; TABLE 1). For organ explants, these membranes provide flexible support and enable the delivery of nutrients and signalling molecules to the bottom surface of the tissue, and optionally to the top surface. Epidermal keratinocytes that are seeded on ECM-coated culture inserts form a monolayer when submerged, and they stratify and differentiate into epidermis when exposed to an air–liquid interface52,53. The addition of fibroblasts to a floating collagen lattice enables the formation of an underlying dermis and the self-organization of full-thickness human skin53. Similar approaches generate histologically realistic normal and neoplastic models of oral, oesophageal and cervical epithelium54,73 (TABLE 1). These reconstituted tissues are used to study normal development and disease processes, and they can also be used for toxicity assays.
Bioengineering-inspired culture systems
Although we cannot review them in detail here, a diverse range of engineered culture assays has also been developed, both to answer fundamental biological questions and to function as platforms for constructing replacement tissues. Important examples include synthetic polymer systems for 2.5D and 3D cultures74–76, integration of microfluidic systems in complex cultures77–79, cell surface-modification techniques to pattern the assembly of epithelial tissues80, microfabricated 3D environments to control tissue geometry and mechanics81 and 3D pattern ing of perfused vascular networks10,11.
Imaging in 3D cultures
Imaging in 3D cultures enables a continuous cellular and molecular description of tissue-level development over a time period of days to weeks72. Imaging is typically carried out using an inverted microscope and requires the robust control of temperature, humidity, CO2 and evaporation. A major obstacle is the scattering of light in thick 3D cultures. One strategy to minimize scattering is to simplify the culture to its core components, such as by using tissue organoids rather than whole organs62,82. A second approach is to match the index of refraction of the immersion medium to that of the culture medium. Finally, the working distance of the lens must be sufficient to image structures throughout the culture.
Different microscope systems can be used in a complementary way. Differential interference contrast enables optical sectioning in thick ECM gels and label-free visualization of collagen I fibres. Confocal imaging enables multicolour 3D time-lapse imaging up to 70–100 μm deep within the tissue82–85, and two-photon microscopy facilitates deeper imaging and visualization of fibrillar ECM using second harmonic signals43,86. However, limitations of two-photon microscopy include increased cost and high levels of energy deposition at the plane of focus. Finally, 3D culture samples can be prepared for transmission electron microscopy and scanning electron microscopy27,59,87,88.
Tailoring the culture format
3D culture assays would ideally be perfectly representative of the in vivo situation, easy to manipulate and inexpensive. However, in practice, trade-offs must typically be made, especially with primary human tissues. It is useful to identify the main experimental goal and then tailor the 3D culture to achieve that goal; for example, replacement human tissues need to be large in size and incorporate into the host vasculature, and the host response to the graft needs to be minimized. By contrast, 3D cultures that are designed for drug screening should ideally fit within 384-well plates, have a low cost per well and predict the results of testing in preclinical animal models and human clinical trials. Important factors to consider include the throughput that is required in the assay, the ease of molecular manipulations and molecular readouts, the necessary recapitulation of the in vivo histology and the availability of primary cells and tissues for that specific organ. For a close match to in vivo histology, the more complex embedded-culture or culture-insert models are most suitable. For genome-scale molecular-interference approaches, the least complex culture system enables the most rapid analysis. Efficient use of limited starting materials is required if the cultures involve primary human tissues. If optical imaging is crucial, the 3D culture needs to be relatively thin and transparent. Finally, the goals of the experiment will determine which cell types need to be incorporated. Many published assays are essentially monocultures of epithelial cells, endothelial cells or neurons. However, each assay format can incorporate additional cell types; for example, cancer cells can be incorporated to study the interactions between normal and neoplastic cells, and stromal cells can be in corporated to study the interactions between epithelial and stromal cells54,89.
The great variety in 3D culture techniques reflects both the various requirements of the different organs and the distinct goals of the investigators. A large body of work has been published on 3D, organotypic or organoid culture, the results of which cannot be comprehensively reviewed here. However, a few common themes emerge that both capture the challenges that motivated the development of 3D culture techniques and illustrate the various biological questions that can be uniquely answered using these approaches.
Organ morphogenesis
Understanding how cells build tissues and organs is of fundamental interest. However, the long timescale of mammalian development and its location deep within an opaque animal limit most organogenesis studies to comparisons of fixed sections from different animals. Owing to the large number of cells and the morpho logical heterogeneity of developing organs, these studies often cannot distinguish among several mechanistic possi bilities of organ formation. Furthermore, an imprecise understanding of the normal developmental trajectory necessarily limits our ability to identify the molecular and cellular differences that define the mutant phenotype. Accordingly, many 3D culture techniques were initially developed to enable the direct observation of developmental processes. In the examples below, we mainly focus on epithelial tissues, as their formation is extensively described in the literature, and highlight common mechanistic themes as well as species- and organ-specific differences.
Mechanisms that drive epithelial tube elongation
A good example of the application of 3D cultures in developmental biology is the study of epithelial tubulogenesis. This process involves an increase in epithelial surface area and changes in tissue shape (FIG. 3a). Tube elongation can, in principle, be accomplished through various combinations of changes in cell number, cell shape and cell size. Terminal branching of the D. melanogaster trachea occurs by subcellular branching of the leading cell90, without proliferation, whereas mammalian tubulo-genesis in the mammary gland, salivary gland, kidney and lung involves a large increase in epithelial tissue size (FIG. 3b–e). The iterative branched structure of these tubular networks and the simple epithelial organization of the resulting ducts initially led to the concept of a conserved process of branching morphogenesis91.
Figure 3. The cellular basis of epithelial tube elongation.
a Schematic diagram showing epithelial bud initiation and tube elongation. Although the process is conceptually similar across the various organs, it was unclear whether tube elongation was accomplished by conserved cellular mechanisms. b Mammary epithelium elongates from a mammary placode into a surrounding fat pad starting at 3 weeks after birth. Branching morphogenesis involves transitions from simple to stratified to simple epithelium. The terminal end bud initiates and elongates as a multilayered structure at the growing front and eventually repolarizes into a simple bilayer62 (inset). c Salivary gland epithelium develops embryonically from a single stratified bud that undergoes successive clefting and extracellular matrix remodelling to form a branched network with simple architecture. At the single bud stage (at embryonic day 13.5), the epithelium already contains a morphologically distinct outer layer of columnar cells, which form the acinar epithelium of the gland, and many inner rounded cells, which form the ductal epithelium160 (inset). d Kidney branching morphogenesis initiates embryonically when the Wolffian duct evaginates into the surrounding metanephric mesenchyme as the ureteric bud. The epithelial bud transitions from simple to pseudostratified to simple architecture94 (inset). e Lung development occurs embryonically. Avian lung maintains simple organization throughout branching morphogenesis and initiates new buds via apical constriction85 (inset). f Salivary gland epithelium requires fibroblast growth factor 10 (FGF10) for branching morphogenesis, and heparan sulphate increases the affinity of FGF10 for its receptor. Specific sulphation patterns of heparan sulphate regulate FGF10-mediated morphogenetic events, such as proliferation, end-bud expansion and duct elongation95. g In branching ureteric bud tips, pre-mitotic epithelial cells delaminate into the lumen to undergo cell division while maintaining a thin basal process at the site of origin. One daughter cell (blue) reinserts at the original site and the second daughter cell (green) inserts at a position one to three cell diameters away96. h In the chick lung, treatment with the proliferation inhibitor aphidicolin does not block bud formation, which shows that cell proliferation is dispensable for bud initiation85. i During mouse lung development, domain branching is characterized by a localized increase in cell division within the incipient bud relative to adjacent trunk cells. Within the bifurcating bud at the end of the tube, there is both an enrichment of proliferation relative to the trunk and a polarization of the plane of cell division within the future cleft region97. Arrows indicate the orientation of cell division.
However, it remained an open question whether conceptually related processes of ductal elongation and bifurcation occurred via a common sequence of cellular events across these organs. Answering this question was challenging as it was difficult to be certain that a duct was elongating at the moment it was fixed. 3D confocal imaging of 3D cultures of the mammary gland, salivary gland, kidney and lung has in fact shown that these organs have distinct cellular mechanisms of branching morphogenesis. Mammary ducts first undergo a simple to stratified transition, elongate as a stratified epithelium and then polarize to re-establish simple ductal architecture62,82,88 (FIG. 3b). Salivary glands start as a stratified epithelium that first clefts and then progressively polarizes to form a network of simple ducts29,92 (FIG. 3c). The kidney ureteric bud transitions from simple to pseudo-stratified, elongates as a pseudostratified epithelium and then polarizes to a simple epithelium93,94 (FIG. 3d). By contrast, epithelial buds initiate and elongate as a polarized simple epithelium during monopodial branching in the chick lung85 (FIG. 3e). Therefore, these tubes have distinct tissue architectures and use different cellular mechanisms during elongation.
Cell proliferation in tubulogenesis
The imaging analyses that are discussed above generated a general framework for branching morphogenesis that enabled researchers to identify the underlying cellular mechanisms. In turn, cell division was shown to have distinct roles in different epithelial organs; for example, asymmetric divisions within mammary luminal epithelial cells initiate stratification and lead to a loss of polarity during both development and ERBB2-induced oncogenic stratification82. Salivary gland epithelial cell proliferation is increased at ductal tips and involves heparan sulphate-mediated regulation of FGF10–FGFR2 signalling; disruption of heparin signalling results in an expanded zone of proliferation and abnormal branching95 (FIG. 3f). In the ureteric bud, proliferating cells delaminate into the luminal space, and daughter cells reintegrate at non-adjacent locations, which leads to mixing of different cellular populations96 (FIG. 3g). The requirement for proliferation in organ development also varies across species. In the chick lung, monopodial branching occurs even in the absence of proliferation85 (FIG. 3h), whereas in the mouse lung, both domain branching and bud bifurcation involve polarized cell divisions97 (FIG. 3i). These species-specific and tissue-specific differences in cell proliferation during tubulo-genesis highlight the importance of real-time analysis as a framework for interpreting molecular interventions. Understanding the role of cell proliferation in development is also relevant in cancer, as oncogenic activation of proliferation might have different consequences in different organs.
Gene regulation of cell behaviour
3D culture techniques have also been developed to elucidate the molecular mechanisms that regulate tissue organization and function in developmental and disease states. Although in vivo studies can test whether a gene is required for organ development, they cannot always determine how the gene regulates cell behaviours to change tissue architecture and function. Conversely, the roles of individual proteins in biochemical pathways have mostly been elucidated in 2D culture studies. Howeve r, these molecular interactions can differ in a 3D tissue context; for example, the molecular composition and phosphorylation status of focal adhesions within fibroblasts are different on 2D surfaces compared with 3D ECM gels98. RNAi, Cre–_lox_-based recombination and lentiviral short hairpin RNA (shRNA) approaches have all recently been adapted to 3D culture, which has enabled the evaluation of single genes and genome-scale screening in tissues, including mammary27, intestinal28 and salivary99,100 epithelium. The application of CRISPR–Cas9 (clustered, regularly interspaced short palindromic repeats– CRISPR-associated protein 9)-based genome editing101 further provides the potential for the rapid introduction and functional correction of disease mutations102.
Tissue-specific genetics
MDCK (Madin–Darby canine kidney) cell acinar formation in 2.5D culture provided an early model of a generic epithelium that was readily accessible to genetic manipulation50,58. The molecular machineries that guide polarity initiation, the specification of an apical membrane and lumen formation were identified in this system and provided a conceptual foundation for our current understanding of polarity in more complex tissues58,103–106. Analogous studies in 2.5D cultures of MCF-10A mammary epithelial cells enabled the detailed dissection of the role of growth factor receptors and polarity proteins in normal development and cancer107–110. Both systems have facilitated high-resolution, real-time studies of the role of oncogenes in disrupting normal epithelial cell behaviours111,112. Nevertheless, these cell lines lack important features of epithelial tissues; for example, MCF-10A cells lack tight junctions87.
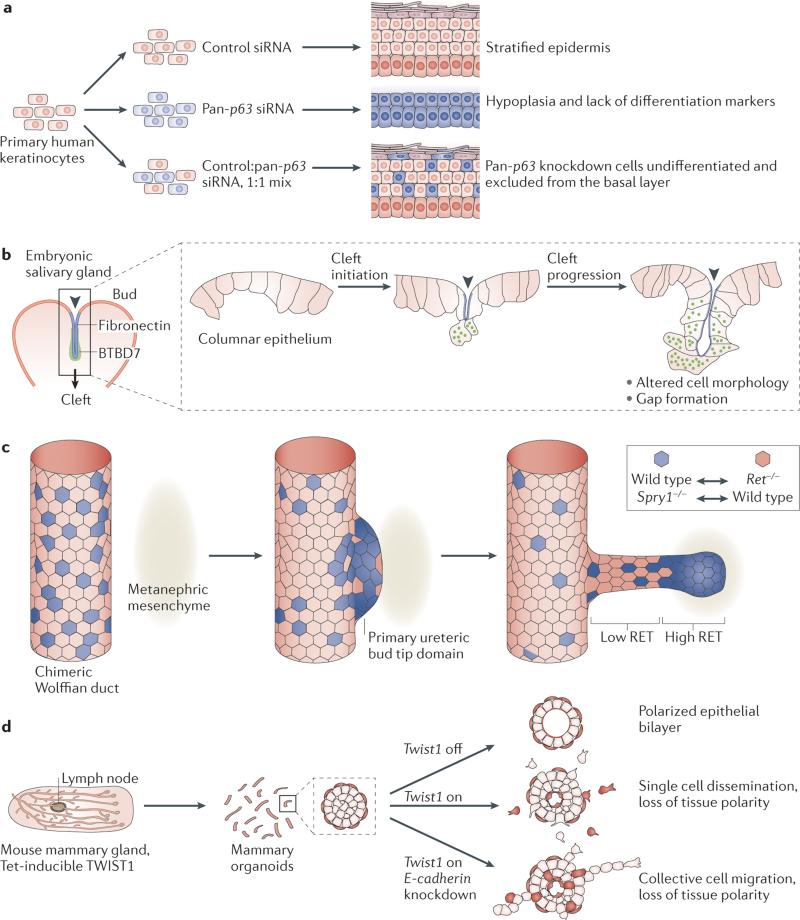
Ultimately, the molecular pathways that have been defined in cell line-based assays need to be validated in more physiological contexts; however, many genes are required during early mammalian embryonic development, which prevents the study of their role in later develop mental processes. This challenge has been overcome by the use of tissue-specific promoters to induce gene deletion or overexpression at later stages. However, many promoters are mosaically expressed within organs, which leads to complex mixtures of wild-type and genetically modified cells. Depending on the pheno type, this heterogeneity can greatly complicate the interpretation of gene function. This challenge can be overcome by combining ex vivo gene manipulation and 3D culture of primary tissues. Knockdown of p63 using siRNA showed that this transcription factor (which is essential for neonatal survival) is required for both proliferation and differentiation in regenerating organotypic epidermis, and distinct contributions are made by different p63 isoforms113 (FIG. 4a). Depletion of p63 leads to tissue hypoplasi a, defects in epidermal stratification and the induction of simple epithelial markers113. In genetic mosaic epidermis, retrovirally delivered LacZ and haemag glutinin epitope-tagged keratin 14 labels were used to distinguish between wild-type and _p63_−/− populations and to show that control of differentiation by p63 is cell-autonomous113. Additional regulators of epidermal differentiation, such as the long non-coding RNA TINCR, have been identified by further combining human organotypic skin cultures with transcriptome sequencing, RNAi, protein micro arrays and RIA–Seq (RNA interactome analysis, followed by deep sequencing)114.
Figure 4. Genetic regulation of cell behaviours in mammalian tissues.
a siRNA-mediated knockdown of p63 showed that this transcription factor is required for both proliferation and differentiation in regenerating organotypic postnatal epidermis. Depletion of p63 in all cells leads to tissue hypoplasia, defects in epidermal stratification and differentiation, and loss of simple epithelial markers113. Mosaic mixtures of control cells and p63 siRNA-treated cells leads to a cell-autonomous failure of differentiation in the p63 knockdown cells113. b Fibronectin was known to accumulate within the forming clefts in the salivary gland115. Gene expression analysis showed that fibronectin binding induces expression of Btbd7 in epithelial cells within the clefts29. BTBD7 regulates cleft progression by reducing cell–cell adhesion and promoting the formation of transient intercellular gaps29. c Labelled cells within chimeric embryonic kidneys compete for contribution to the ureteric bud, depending on their individual level of RET signalling94. In a chimeric Wolffian duct, labelled epithelial cells that lack the receptor tyrosine kinase (RTK) RET (Ret−/− cells) are excluded from the tips of elongating ureteric buds in favour of wild-type cells94. By contrast, cells that are depleted of Sprouty1 (_Spry1_−/−), which is a repressor of RTK signalling, have increased levels of RET and accumulate at the ureteric bud tip domain instead of wild-type cells. d Tet-inducible Twist1 expression leads to loss of tissue polarity and the rapid dissemination of otherwise normal mammary epithelial cells27. Disseminated cells retain epithelial gene expression (for example, cytokeratin 8), localize epithelial cadherin (E-cadherin) and β-catenin to the membrane and require E-cadherin protein to disseminate as single cells27. Part b of the figure from Onodera, T. et al. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science 329, 562–565 (2010). Adapted with permission from AAAS. Part c of the figure adapted from Dev. Cell, 17, Chi, X. et al., Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis, 199–209, Copyright (2009), with permission from Elsevier.
Incorporation of fluorescent reporters simplifies the interpretation of mosaic tissues, as it enables real-time comparisons between genetically modified cells and wild-type cells; for example, deletion of epithelial cadherin (E-cadherin) in a large fraction (>80%) of mammary epithelial cells in tissue organoids does not prevent the initiation of new epithelial ducts27. This result suggests that E-cadherin is dispensable for mam-mary morphogenesis; however, real-time imaging using Cre biosensors showed that these new ducts are elon-gated by E-cadherin-expressing cells27. E-cadherin is in fact required for the maintenance of epithelial architecture and branching morphogenesis27. Importantly, this is consistent with in vivo findings, and therefore, the combination of ex vivo gene manipulation and fluorescent biosensors enables the rapid evaluation of gene requirement in primary tissues.
Real-time genetic analysis
Real-time tracking can be used to determine the effects of genetic modifications on specific cell behaviours; for example, studies using fluorescent labelling, siRNA knockdown and time-lapse imaging identified the signals that are required for salivary gland clefting29,92,115. Gene expression analysis revealed increased levels of BTBD7 in epithelial cells located within clefts29 (FIG. 4b). Moreover, it was shown that BTBD7 promotes cleft progression by inducing Snai2 (also known as Slug) expression and reducing E-cadherin levels29. Live-cell imaging has also been used in embryonic whole-lung explants to demonstrate the requirement for the polarity protein Scribble in lumen morphogenesis in the mammalian lung but not in bud bifurcation116; the depletion of Scribble in lungs leads to the mislocalization of junction proteins and misalignment of distal epithelial cells. Importantly, the finding that Scribble is required for epithelial cell–cell contacts and lumen morphogenesis is consistent with the in vivo Scribble (Scrib) loss-of-function pheno-type (that is, small, misshapen lungs, reduced airway number and defects in epithelial architecture)116. Thus, genetic manipulation, together with live-cell imaging in 3D culture, identified the specific genes that have a key role in salivary gland clefting and lung morphogenesis. In the future, this approach will aid in identifying the genes that are involved in other cellular processes in additional organs.
A major advantage of 3D culture is the ability to observe and follow the same cells over time. This approach was used in the mouse kidney to identify the cells that require the receptor tyrosine kinase (RTK) RET during branching morphogenesis94. Imaging in embryonic whole-organ chimeras showed the dynamic exclusion of epithelial cells that lack RET (Ret−/− cells) from the tips of elongating ureteric buds94 (FIG. 4c). The authors then increased RET signalling by deleting Sprouty1 (Spry1), which is a repressor of RTK signalling. In chimeras with wild-type cells, Spry1−/− cells are enriched in the ureteric bud tip domain. Importantly, normal Wolffian duct cells display heterogeneous RETsignalling in vivo and therefore probably compete on this basis for contribution to the ureteric bud94. Taken together, these results reveal a crucial role for RET signalling, specifically in the cells that are most involved in tube elongation.
Inducible gene expression
Recent advances also enable inducible gene expression and precisely timed molecular analyses in 3D cultures27; for example, expression of the transcription factor Twist1 in mammary epithelial cells induces rapid dissemination of cells into the ECM. Surprisingly, _Twist1_-expressing disseminated cells retain epithelial gene expression, and dissemination requires E-cadherin27 (FIG. 4d). The fraction of cells within a tissue that expresses a gene can also regulate tissue architecture. Mosaic, but not ubiquitous, expression of an activated form of the GTPase HRAS induces multicellular protrusions in MCF-10A aggregates117. In a separate study, activated HRAS was sufficient to drive tissue overgrowth when expressed in myoepithelial or luminal mammary epithelial cells118. However, inhibitor studies demonstrated the involvement of different RAS effectors, which showed that different pathways were used in the two cell types to induce proliferation downstream of the same oncogene118. Taken together, these studies show that 3D culture enables elucidation of the cellular and molecular effects of gene activity in distinct cell populations within a tissue.
The microenvironment and cell behaviour
The examples in the previous section highlight biological insights from the study of isolated epithelial tissues ex vivo. However, in vivo, epithelial cells are surrounded by connective tissue, which contains immune cells, blood vessels, fibroblasts and ECM-bound signalling molecules (FIG. 5a). These components can regulate adjacent epithelial and neuronal tissues and contribute to disease progression, particularly in cancer119,120. However, it is difficult to specifically alter either stromal cell or ECM composition in vivo. By contrast, 3D culture systems enable the precise manipulation of components of the microenvironment and the analysis of how they affect the structure and function of a cell or tissue69,121,122.
Figure 5. The role of the microenvironment in regulating epithelial function.
a Schematic overview of different components of the tissue microenvironment, including immune cells (for example, macrophages), blood vessels, fibroblasts and extracellular matrix (for example, collagen I). The components and properties of the microenvironment can be readily modified in three-dimensional (3D) culture. b Co-cultures of lung or bone marrow stroma mixed with endothelial cells were used to generate a 3D organotypic microvascular niche. The angiogenesis inhibitor thrombospondin 1 (TSP1) induces breast tumour cell dormancy in mature endothelium, whereas transforming growth factor-β1 (TGFβ1) and periostin promote tumour cell growth in neovascular tips, which lack TSP1 (REF. 129). c Direct comparisons of the same tissue in different microenvironments (that is, Matrigel or collagen I) shows that the composition of the ECM regulates invasive and disseminative behaviours of both normal and malignant mammary epithelium66. d The mechanical properties of the microenvironment can affect cell and tissue function. High rigidity (which is achieved by crosslinking poly(ethylene) glycol (PEG) networks within Matrigel scaffolds) suppresses the growth of both normal and neoplastic tissue but does not induce invasion or dissemination121. The addition of adhesive peptides promotes dissemination of both normal and tumour cells121. Part b of the figure adapted from REF. 129, Nature Publishing Group. Part c of the figure adapted from Nguyen-Ngoc, K.-V. et al. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl Acad. Sci. USA 109, E2595–E2604 (2012).
Epithelial–stromal interactions
The ability to add or remove stromal cells in 3D co-cultures has been particularly exploited in cancer invasion studies. It was known that macrophages regulate breast cancer cell invasion and metastasis in vivo123. To study the underlying molecular mechanisms, various immune cells and immune cell-derived soluble factors have been added to 3D cultures of breast tumour organoids124. Macrophages, T cells that express interleukin-4 (IL-4) or exogenous IL-4 can each promote tumour invasion124. Similarly, squamous cell carcinoma invasion into collagen I is strongly promoted by co-culture with fibroblasts125, which interact with both cancer cells and the ECM125. Importantly, if the fibroblasts are allowed to remodel the ECM and are then removed, their tracks within the matrix are sufficient to promote invasion125. Fibroblasts can therefore create lasting, proinvasive changes in the tumour microenvironment125. In breast cancer, fibroblasts use Hippo signalling to promote matrix stiffening and invasion126. In oesophageal cancer, fibroblasts promote invasion via hepatocyte growth factor (HGF) signalling89 and alter the differentiation state of cancer cells127.
The vasculature is another potent source of regulatory signals. Recent studies have implicated thrombospondin 1 (TSP1), which is an angiogenesis inhibitor, in normal epithelial differentiation and tumour growth. A clonal 3D co-culture model was used to show that lung but not liver endothelial cells can direct lineage specification in distal lung stem cells128, and TSP1 promotes differentiation into an alveolar fate following injury128. In a different co-culture model, 3D niches that are composed of lung or bone marrow stroma mixed with endothelial cells were used to show that TSP1 regulates tumour dormancy129 (FIG. 5b). This study revealed that TSP1 induces breast tumour cell dormancy in mature endothelium, whereas transforming growth factor-β1 (TGFβ1) and periostin promote tumour cell growth in neovascular tips, which lack TSP1 (REF. 129). Neovascularization therefore promotes disseminated tumour cells to develop into macrometastases.
3D models of the perivascular niche have also been used to study interactions between brain endothelial cells and cancer stem cells (CSCs) in glioblastoma130. Vascular networks within 3D-engineered scaffolds increase CSC maintenance, growth and migration via paracrine IL-8 signalling, and co-implantation of endothelial cells and CSCs in vivo promotes tumour formation via IL-8 receptor signalling in CSCs130. Importantly, conditioned medium that has been collected from 3D cultures of endothelial cells promotes the maintenance of stem cell markers in CSCs, whereas conditioned medium from 2D cultures does not130. Taken together, these data suggest that the tumour stroma is a potent regulator of primary tumour growth and metastasis.
Mesenchymal components can also regulate the regenerative capacity of an organ. In the salivary gland, removal of the parasympathetic ganglion decreases branching morphogenesis and depletes the pool of keratin 5-expressing progenitor cells131. Morphogenesis and proliferation of these progenitors can be rescued by activation of muscarinic receptors with an acetylcho-line analogue131. Parasympathetic innervation therefore maintains progenitors in an undifferentiated state, and this signalling axis may enable therapeutic intervention to promote organ repair132.
Epithelial–epithelial interactions
Tumours are increasingly recognized to contain cancer cells with different genetic and phenotypic characteristics133. 3D cultures of cancer cells have revealed subtype-specific differences in invasive behaviour and in the capacity of fibroblasts to promote invasion134,135. Interestingly, cell lines that have greater migratory capacity are observed to lead collective invasion of less migratory cell lines via direct contact between cancer cells134. A recent study developed an assay to prospectively identify the most invasive cells within primary mouse and human tumours by embedding tumour-derived organoids into 3D collagen I gels30. Cancer cells that express basal epithelial markers were found to lead collective invasion in mouse breast cancer models as well as in patient samples, in 3D culture and in vivo, across breast cancer subtypes30. Cancer cells that express basal markers directly adhere to, and lead the collective invasion of, cancer cells that express luminal markers30. Thus, interactions between epithelial cancer cells in different differentiation states are crucial for collective invasion. These techniques can be readily generalized to any epithelial malignancy and enable unbiased identification of the cells that are most responsible for invasion and systemic spread. Interestingly, inter actions between normal basal and luminal epithelial cells have also been shown to regulate tissue architecture and polarity in the mammary gland136,137.
ECM composition
In both 2.5D and 3D culture, cells from the same source can be explanted into micro-environments that have varied ECM composition or mechanics. In the MDCK kidney acini model, the composition of the ECM regulates the efficiency and mechanism of lumen formation104. In collagen I gels, cell polarization is slow and lumen formation requires apoptosis104. By contrast, in Matrigel, cell polarization is rapid and apoptosis is dispensable for lumen formation104. Direct comparisons of mammary epithelium in different microenvironments showed that the composition of the ECM regulates invasive and disseminative behaviours of both normal and malignant tissues66 (FIG. 5c). Importantly, the consequences of gene deletion at the tissue level also depend on the ECM, as deletion of placental cadherin (P-cadherin) results in mammary epithelial hyperplasia in Matrigel and increased dissemination in collagen I (REF. 66). Finally, the organization of the ECM can modulate tissue architecture. Radially aligned collagen I fibres promote breast cancer invasion70 and correlate with poor patient outcomes138. Interestingly, aligned collagen I fibres also promote directional growth of normal epithelial cells67,68, which suggests that structural cues can pattern normal morphogenesis.
Mechanical cues
The mechanical properties of the microenvironment also change during development and disease; for example, epithelial tumours increase in rigidity owing to both ECM stiffening and increased cytoskeletal tension139. 3D culture experiments showed that stiff matrices induce integrin clustering, cytoskeletal tension and focal adhesion assembly139. Increased cytoskeletal tension, whether induced by matrix stiffness or by ERK signalling, promotes malignant progression139. Matrix stiffness can also contribute to cell fate specification, as mesenchymal stem cells commit to distinct lineages when cultured on gels with varied rigidity122. Soft matrices promote neurogenic differentiation, intermediate matrices promote myogenic differentiation and rigid matrices promote osteogenic differentiation122.
Recent studies have focused on identifying the signals that cells respond to within these microenvironments; for example, lysyl oxidase-mediated collagen I crosslinking increases ECM rigidity and promotes metastatic progression via stimulation of PI3K signalling69. Another study reported the effect of rigidity on cell behaviour by crosslinking poly(ethylene) glycol (PEG) networks within Matrigel scaffolds121. High rigidity alone suppresses the growth of both normal and neoplastic tissue but does not induce invasion or dissemination121 (FIG. 5d); however, the addition of adhesive peptides to the PEG network promotes the dissemination of both normal and tumour cells121. Collectively, these studies show that stromal cells, ECM composition and microenvironmental mechanics can independently regulate cell and tissue function140.
Stem cell-derived tissues and therapeutics
3D culture can be used to define the necessary and sufficient components to replicate the structure and function of an organ. This approach, which is sometimes termed synthetic tissue biology141, can guide our understanding of normal development and generate replacement tissues142; for example, single LGR5+ epithelial cells that have been isolated from the small intestine are sufficient to generate organoids with crypt–villus architecture, without mesenchymal cells6. However, the efficiency of generating intestinal organoids from LGR5+ cells is greatly increased by co-culture with Paneth cells143. Organoid cultures can also be passaged indefinitely to produce sufficient cells for transplantation37,38,144. In principle, stem cells from the injured organ of a patient could be used to generate organoids to repair the damage. Support ing this concept, a recent study induced experimental colitis in mice and introduced LGR5+ organoids into the colon144. The organoids engrafted and formed crypts with barrier function144. In cases in which the disease state is caused by a known mutation, combinations of patient-derived iPS cells with genome-editing techniques, such as CRISPR–Cas9, could enable the generation of replacement cells and tissues in which the mutation has been corrected102. This strategy has been validated in organoids derived from patients with cystic fibrosis, in which correction of the disease-inducing mutation in the cystic fibrosis transmembrane conductor receptor (CFTR) restored channel function in 3D culture102.
3D cultures of ES cells or iPS cells can also assemble into specific tissues and organs. A striking example is the formation of the optic cup from ES cell aggregates, with the stratified neural tissue containing all six cell types in their appropriate spatial arrangement44. Improved culture conditions have enabled the routine production of both rod and cone photoreceptors and cryogenic storage of sheets of stratified human neural retina45. Adaptation of these techniques for the mouse retina enabled transplantation and functional engraftment into retinal degenerative mice145. This therapeutic approach has also been used for endocrine glands: mouse iPS cells were first differentiated into immature pancreatic cells and then differentiated into islets in 3D culture146. Following transplantation into hyperglycaemic mice, these islets incorporated into the pancreas and improved blood glucose levels146. A recent study further showed that co-culture with mesenchyme increases the efficiency of expansion of human endodermal progenitors more than 106-fold and preserves the capacity of the progenitors to give rise to glucose-sensing, insulin-secreting cells following transplantation in vivo147. Similarly, transient expression of two transcription factors enables the generation of thyroid follicular cells from ES cells in 3D culture, and following transplantation into athymic mice, these cells rescue plasma levels of thyroid hormones148. Taken together, these studies reveal that 3D culture is a crucial method to generate replacement tissues for therapeutic purposes149.
An additional strength of these approaches emerges when studying organs that diverge considerably between mice and humans. ES cell- and iPS cell-based methods enable the real-time analysis of anatomically complex human organs such as the brain. A recent study developed cerebral organoids from human iPS cells in 3D culture and replicated the anatomical organization of multi ple human brain regions, including cerebral cortex47. In turn, organoids derived from iPS cells that had been isolated from a patient with severe microcephaly displayed premature neuronal differentiation and reduced amounts of neuroepithelium, which shows that even complex disorders of the central nervous system can be modelled using 3D culture47.
Future directions
A major goal of biomedical science is to translate our understanding of the fundamental principles that govern biological systems to improve patient outcomes. 3D culture can function as an integration point for modelling human disease in experimental systems and also yields precise answers to biological questions. Achieving the full potential of these methods will involve the expertise of scientists and clinicians and may require new funding mechanisms to sustain the interdisciplinary work that is required.
We anticipate that recent advances in 3D culture will accelerate our understanding of the cellular and molecular basis of mammalian organogenesis, both via the study of primary mouse tissues and in artificially generated human organs. Equally important, real-time analysis is expected to yield insights into the dynamic cellular basis of disease processes, especially when robust mouse models have been developed. This combined knowledge of normal development and pathobiology should enable the identification of molecular signals that promote tissue regeneration, restore tissue function and resist disease progression.
A particularly exciting new development is the ability to explant the living tissue of a patient into 3D culture to obtain individualized predictive or prognostic information, which is the ultimate goal of personalized medicine (BOX 1). It is now possible to culture primary human cells or tissues using excess material from surgery30,37,73,102. These human cell cultures could be used to test novel therapeutic strategies, to guide the selection of therapies for a specific patient or to grow replacement cells or tissues from the cells of the patient. The success of this therapeutic approach will require a high degree of standardization and reproducibility across experiments. In addition, patient tissue in 3D culture could be analysed for its dynamic molecular responses to standardized cellular and molecular cues, such as targeted therapies150.
The development of robust vascular networks in 3D culture10,11 should enable the generation of larger and more complex tissue constructs151, improve transplant efficiency10 and catalyse a transition in 3D culture from the self-organization of simple tissues to the synthetic assembly of multicomponent organs. This transition in complexity should enable an increased focus on the recapitulation of organ function152,153. Recent efforts in this direction suggest that functional properties of an organ can be replicated in microfluidic assays that do not closely model the normal in vivo tissue architecture78. Finally, the successful functional incorporation of transplanted organoids into mouse colon144, retina145, thyroid148, liver38,152 and pancreas146,147 suggest that organ function can be restored in human patients by the transfer of functional units of tissue rather than by whole-organ transplantation.
Box 1 Therapeutic applications of three-dimensional culture.
Predictive assays
The vast number of potential cancer drugs and drug combinations cannot be serially evaluated in individual patients. This challenge has motivated efforts to develop chemotherapy-sensitivity and -resistance assays in which the therapeutic response of the tumour of a patient could be directly assessed154,155. Although no current assays are ready for routine clinical practice154,155, emerging techniques based on three-dimensional (3D) culture and microfluidic systems can potentially provide personalized information on response to therapy (see the figure, part a). For example, ex vivo slice cultures of tumours have been used to identify heterogeneous responses to small-molecule compounds such as PI3K inhibitors156. Similarly, primary glioblastoma specimens show variable cell death rates and DNA damage responses following treatment with the chemotherapeutic drug temozolomide after irradiation157. Primary samples can also be challenged with molecular therapeutic agents and assayed for downstream signalling responses150. Validation of the predictive value of such assays will require clinical trials that test whether assay-guided clinical management results in better patient outcomes than the standard of care154,155.
Prognostic assays
Prognostic assays could be developed to measure functionally based outputs, such as morphological criteria (for example, the extent of invasion into a defined microenvironment30) (see the figure, part b) or molecular criteria (for example, expression of components of the MAPK pathway150). These assays can potentially provide information that is independent of that derived from static phenotypic analyses that have been conducted on formalin-fixed paraffin-embedded sections. If such assays correlate with patient outcomes, they could provide a means to generate individualized prognoses based on tissue from the patient.
Preclinical therapeutic testing
Organotypic models of disease can provide efficient proof-of-principle tests of novel molecular approaches to disease management (see the figure, part c).
Genome editing. Primary intestinal organoids that are derived from patients with cystic fibrosis carry mutations in the gene that encodes the cystic fibrosis transmembrane conductance regulator (CFTR) (for example, CFTRΔF508)102. Forskolin induces rapid swelling in organoids from healthy individuals but not in organoids from patients with cystic fibrosis. Correction of the CFTRΔF508 mutation by CRISPR–Cas9 (clustered, regularly interspaced short palindromic repeats– CRISPR-associated protein 9) in patient-derived stem cells that express Leu-rich repeat-containing G protein-coupled receptor 5 (LGR5+) results in the restoration of CFTR function and in forskolin-induced swelling in the organoids102. Notably, CFTR function improves more with gene editing than with small-molecule drugs158.
Rational therapeutic design. The survival of a cancer cell in the presence of a therapeutic agent can depend on its 3D localization within a tissue. Dual inhibition of PI3K and mTOR in ovarian cancer spheroids leads to the death of inner matrix-deprived cells but the upregulation of pro-survival programmes in matrix-attached cells159. Combined inhibition of PI3K, mTOR and BCL-2 eliminates both matrix-deprived and matrix-attached cells by apoptosis in tumour xenografts and in primary ovarian and breast cancer samples from patients.
Epithelium
A type of animal tissue derived from ectoderm or endoderm that lines all cavities and body surfaces and consists of one or more layers of polarized, tightly connected cells.
Connective tissue
A type of animal tissue derived from mesenchyme that provides structural and nutritional support and connectivity among other tissues; it consists of individual cells, ground substance and fibres.
Extracellular matrix (ECM). The non-cellular component of tissues that provides both structural support and signalling cues to cells; it is composed of a network of proteins such as collagen, fibronectin and laminin.
Mesenchyme
Loosely organized, undifferentiated cells derived from embryonic mesoderm that give rise to the connective tissues of the body and the lymphatic and circulatory systems.
Induced pluripotent stem cells (iPS cells). Adult somatic cells that are genetically reprogrammed to generate embryonic-like pluripotent stem cells.
Basement membrane
An organized thin layer of extracellular matrix proteins that separates the epithelium from the surrounding connective tissue.
Stromal cells
The connective tissue cells of an organ (for example, fibroblasts), which support the function of the parenchymal cells of the organ.
Tissue stem cells
Adult stem cells that can give rise to some or all of the specialized cells of the tissue or organ from which they originate.
Matrigel
A gelatinous basement membrane matrix derived from Engelbreth-Holm-Swarm mouse sarcoma cells; it promotes cell differentiation and models the in vivo microenvironment of many tissues.
Self-organization
In tissues, the spontaneous formation of a highly ordered structure from a population of cells in the absence of pre-patterns.
Stratified epithelium
An epithelium that is composed of two or more layers of cells; it is often found in locations that require increased protection, such as exterior body surfaces.
Microfluidic systems
Devices that comprise submillimetre channels, pumps and valves that enable controlled, reproducible analysis of small samples of cells in nanolitre or picolitre volumes.
Asymmetric divisions
Cell divisions that result in two daughter cells with different fates, such as localization into distinct epithelial cell layers with unequal inheritance of polarity proteins.
Cre-lox
A site-specific recombination tool that uses the enzyme Cre recombinase to induce deletions, translocations or inversions in segments of genomic DNA that are flanked by loxP sites.
CRISPR-Cas9
(Clustered, regularly interspaced short palindromic repeats–CRISPR-associated protein 9). A genome-editing tool that uses the microbial RNA-guided Cas9 nuclease to make targeted changes in the DNA of eukaryotic cells.
Hypoplasia
Abnormal tissue or organ development owing to a deficient number of cells.
Hyperplasia
Abnormal tissue or organ development owing to an excess number of cells.
Tumour xenografts
Human tumours that are implanted into immunocompromised animal hosts.
Acknowledgements
The authors apologize to the many scientists whose outstanding work could not be cited owing to space limitations. A.J.E. and E.R.S. were supported by a Research Scholar Grant (RSG-12-141-01-CSM) from the American Cancer Society. A.J.E. was also supported in part by funds from the National Institutes of Health National Cancer Institute (NIH–NCI) (U01 CA155758), by a Jerome L. Greene Foundation Discovery Project, by a grant from the Mary Kay Ash Foundation (036-13), by funds from the Cindy Rosencrans Fund for Triple Negative Breast Cancer Research and by a grant from the Breast Cancer Research Foundation.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Bichat X. General Anatomy, Applied to Physiology and Medicine. (Richardson and Lord. 1822 [Google Scholar]
- 2.Virchow R. De Witt RM, editor. Cellular Pathology, as Based upon Physiological and Pathological Histology. Twenty Lectures Delivered in the Pathological Institute of Berlin During the Months of February, March and April, 1858. 1860 [Google Scholar]
- 3.Sobotta J, Huber GC, De Witt LMB. Atlas and Epitome of Human Histology and Microscopic Anatomy. W. B. Saunders & company; 1903. [Google Scholar]
- 4.Harrison RG, Greenman MJ, Mall FP, Jackson CM. Observations on the living developing nerve fiber. Anat. Rec. 1907;1:116–128. [Google Scholar]
- 5.Simian M, et al. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato T, et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 7.Finkbeiner SR, Spence JR. A gutsy task: generating intestinal tissue from human pluripotent stem cells. Dig. Dis. Sci. 2013;58:1176–1184. doi: 10.1007/s10620-013-2620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberts B. Molecular Biology of the Cell. 4th edn. Garland Science; 2002. Ch. 19. [Google Scholar]
- 9.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baranski JD, et al. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc. Natl Acad. Sci. USA. 2013;110:7586–7591. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl Acad. Sci. USA. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen D-HT, et al. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl Acad. Sci. USA. 2013;110:6712–6717. doi: 10.1073/pnas.1221526110. [Introduces a 3D in vitro model of angiogenic sprouting from preformed vessels to define the morphogenetic and molecular requirements for neovascularization.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fell HB, Robison R. The growth, development and phosphatase activity of embryonic avian femora and limb-buds cultivated in vitro. Biochem. J. 1929;23:767–784. doi: 10.1042/bj0230767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JM. The cultivation in fluid medium of organised liver, pancreas and other tissues of foetal rats. Exp. Cell Res. 1954;7:518–529. doi: 10.1016/s0014-4827(54)80096-6. [DOI] [PubMed] [Google Scholar]
- 15.Ichinose RR, Nandi S. Lobuloalveolar differentiation in mouse mammary tissues in vitro. Science. 1964;145:496–497. doi: 10.1126/science.145.3631.496. [DOI] [PubMed] [Google Scholar]
- 16.Waymouth C. In: Biology of the Laboratory Mouse. Green Earl L., editor. Dover Publications; 1966. [Google Scholar]
- 17.Guerrero RR, Rounds DE, Booher J. An improved organ culture method for adult mammalian lung. In Vitro. 1977;13:517–524. doi: 10.1007/BF02615145. [DOI] [PubMed] [Google Scholar]
- 18.Browning TH, Trier JS. Organ culture of mucosal biopsies of human small intestine. J. Clin. Invest. 1969;48:1423–1432. doi: 10.1172/JCI106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randall KJ, Turton J, Foster JR. Explant culture of gastrointestinal tissue: a review of methods and applications. Cell Biol. Toxicol. 2011;27:267–284. doi: 10.1007/s10565-011-9187-5. [DOI] [PubMed] [Google Scholar]
- 20.Autrup H, et al. Explant culture of rat colon: a model system for studying metabolism of chemical carcinogens. In Vitro. 1978;14:868–877. doi: 10.1007/BF02616157. [DOI] [PubMed] [Google Scholar]
- 21.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 22.Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends Neurosci. 1997;20:471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- 23.Aplin AC, Fogel E, Zorzi P, Nicosia RF. The aortic ring model of angiogenesis. Methods Enzymol. 2008;443:119–136. doi: 10.1016/S0076-6879(08)02007-7. [DOI] [PubMed] [Google Scholar]
- 24.Topper RJ, Oka T, Vonderhaar BK. Techniques for studying development of normal mammary epithelial cells in organ culture. Methods Enzymol. 1975;39:443–454. doi: 10.1016/s0076-6879(75)39039-3. [DOI] [PubMed] [Google Scholar]
- 25.Hardman P, Klement BJ, Spooner BS. Growth and morphogenesis of embryonic mouse organs on non-coated and extracellular matrix-coated Biopore membrane. Dev. Growth Differ. 1993;35:683–690. doi: 10.1111/j.1440-169x.1993.00683.x. [DOI] [PubMed] [Google Scholar]
- 26.Trott JF, Vonderhaar BK, Hovey RC. Historical perspectives of prolactin and growth hormone as mammogens, lactogens and galactagogues — agog for the future! J. Mammary Gland Biol. Neoplasia. 2008;13:3–11. doi: 10.1007/s10911-008-9064-x. [DOI] [PubMed] [Google Scholar]
- 27.Shamir ER, et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 2014;204:839–856. doi: 10.1083/jcb.201306088. [Demonstrates, using genetic manipulation of primary normal mammary tissue, that epithelial cells can disseminate while retaining epithelial- specific proteins and gene expression. Shows that E-cadherin is required for efficient single-cell dissemination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koo BK, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nature Methods. 2011;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 29.Onodera T, et al. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329:562–565. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [Uses organotypic culture of primary tumour organoids to identify a common subpopulation of cells that leads collective invasion across distinct breast cancer subtypes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daley WP, Gulfo KM, Sequeira SJ, Larsen M. Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev. Biol. 2009;336:169–182. doi: 10.1016/j.ydbio.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fata JE, et al. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFα and FGF7 in morphogenesis of mouse mammary epithelium. Dev. Biol. 2007;306:193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinberg Z, et al. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 2005;132:1223–1234. doi: 10.1242/dev.01690. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Bush KT, Nigam SK. In vitro culture of embryonic kidney rudiments and isolated ureteric buds. Methods Mol. Biol. 2012;886:13–21. doi: 10.1007/978-1-61779-851-1_2. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, et al. Novel role for Netrins in regulating epithelial behavior during lung branching morphogenesis. Curr. Biol. 2004;14:897–905. doi: 10.1016/j.cub.2004.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nature Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 38.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barker N, et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/ R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh S, et al. PI3K/mTOR signaling regulates prostatic branching morphogenesis. Dev. Biol. 2011;360:329–342. doi: 10.1016/j.ydbio.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Wolf K, et al. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eiraku M, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 45.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [References 44 and 45 demonstrate that retinal development can be mostly recapitulated in vitrovia the self-organization of ES cell-derived retinal epithelia.] [DOI] [PubMed] [Google Scholar]
- 46.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;6:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [The first in vitro model of whole brain tissue, which was derived from human iPS cells, with discrete but interdependent brain domains.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suga H, et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- 49.Townes PL, Holtfreter J. Directed movements and selective adhesion of embryonic amphibian cells. J. Exp. Zool. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- 50.O'Brien LE, Zegers MM, Mostov KE. Building epithelial architecture: insights from three-dimensional culture models. Nature Rev. Mol. Cell Biol. 2002;3:531–537. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- 51.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 52.Green H, Kehinde O, Thomas J. Growth of cultured human epidermal cells into multiple epithelia suitable for grafting. Proc. Natl Acad. Sci. USA. 1979;76:5665–5668. doi: 10.1073/pnas.76.11.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuchs E. Epidermal differentiation: the bare essentials. J. Cell Biol. 1990;111:2807–2814. doi: 10.1083/jcb.111.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalabis J, et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nature Protoc. 2012;7:235–246. doi: 10.1038/nprot.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unbekandt M, Davies JA. Dissociation of embryonic kidneys followed by reaggregation allows the formation of renal tissues. Kidney Int. 2009;77:407–416. doi: 10.1038/ki.2009.482. [DOI] [PubMed] [Google Scholar]
- 56.Ganeva V, Unbekandt M, Davies JA. An improved kidney dissociation and reaggregation culture system results in nephrons arranged organotypically around a single collecting duct system. Organogenesis. 2011;7:83–87. doi: 10.4161/org.7.2.14881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Streuli CH, Bailey N, Bissell MJ. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell–cell interaction and morphological polarity. J. Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Brien LE, et al. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nature Cell Biol. 2001;3:831–838. doi: 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- 59.Yu W, et al. Formation of cysts by alveolar type II cells in three-dimensional culture reveals a novel mechanism for epithelial morphogenesis. Mol. Biol. Cell. 2007;18:1693–1700. doi: 10.1091/mbc.E06-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenburg G, Hay ED. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J. Cell Biol. 1982;95:333–339. doi: 10.1083/jcb.95.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 62.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morita K, Nogawa H. EGF-dependent lobule formation and FGF7-dependent stalk elongation in branching morphogenesis of mouse salivary epithelium in vitro. Dev. Dyn. 1999;215:148–154. doi: 10.1002/(SICI)1097-0177(199906)215:2<148::AID-DVDY7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 64.Qiao J, Sakurai H, Nigam SK. Branching morphogenesis independent of mesenchymal– epithelial contact in the developing kidney. Proc. Natl Acad. Sci. USA. 1999;96:7330–7335. doi: 10.1073/pnas.96.13.7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wescott MP, et al. Pancreatic ductal morphogenesis and the Pdx1 homeodomain transcription factor. Mol. Biol. Cell. 2009;20:4838–4844. doi: 10.1091/mbc.E09-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen-Ngoc K-V, et al. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl Acad. Sci. USA. 2012;109:E2595–E2604. doi: 10.1073/pnas.1212834109. [Demonstrates that the composition of the ECM determines the migration strategy and disseminative behaviour of both normal and tumour mammary organoids and can regulate the phenotypic consequences of molecular perturbations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen-Ngoc KV, Ewald AJ. Mammary ductal elongation and myoepithelial migration are regulated by the composition of the extracellular matrix. J. Microsc. 2013;251:212–223. doi: 10.1111/jmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brownfield DG, et al. Patterned collagen fibers orient branching mammary epithelium through distinct signaling modules. Curr. Biol. 2013;23:703–709. doi: 10.1016/j.cub.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Provenzano PP, et al. Collagen reorganization at the tumor–stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Provenzano PP, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ewald AJ. Practical considerations for long-term time-lapse imaging of epithelial morphogenesis in three-dimensional organotypic cultures. Cold Spring Harb. Protoc. 2013;2013:100–117. doi: 10.1101/pdb.top072884. [DOI] [PubMed] [Google Scholar]
- 73.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nature Med. 2010;16:1450–1455. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nature Nanotechnol. 2010;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singh A, Elisseeff J. Biomaterials for stem cell differentiation. J. Mater. Chem. 2010;20:8832–8847. [Google Scholar]
- 77.Young EW, Beebe DJ. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010;39:1036–1048. doi: 10.1039/b909900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [Introduces the concept of using microfabrication and microfluidics to construct biomimetic microsystems with tissue–tissue interfaces, efficient nutrient delivery, mechanical integrity and organ functionality.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stroock AD, Fischbach C. Microfluidic culture models of tumor angiogenesis. Tissue Engineer. Part A. 2010;16:2143–2146. doi: 10.1089/ten.tea.2009.0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl Acad. Sci. USA. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varner VD, Nelson CM. Let’s push things forward: disruptive technologies and the mechanics of tissue assembly. Integr. Biol. 2013;5:1162–1173. doi: 10.1039/c3ib40080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huebner RJ, Lechler T, Ewald AJ. Developmental stratification of the mammary epithelium occurs through symmetry-breaking vertical divisions of apically positioned luminal cells. Development. 2014;141:1085–1094. doi: 10.1242/dev.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larsen M, et al. Role of PI 3-kinase and PIP3 in submandibular gland branching morphogenesis. Dev. Biol. 2003;255:178–191. doi: 10.1016/S0012-1606(02)00047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Puri S, Hebrok M. Dynamics of embryonic pancreas development using real-time imaging. Dev. Biol. 2007;306:82–93. doi: 10.1016/j.ydbio.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim HY, Varner VD, Nelson CM. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development. 2013;140:3146–3155. doi: 10.1242/dev.093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Provenzano PP, et al. Nonlinear optical imaging of cellular processes in breast cancer. Microsc. Microanal. 2008;14:532–548. doi: 10.1017/S1431927608080884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Underwood JM, et al. The ultrastructure of MCF-10A acini. J. Cell. Physiol. 2006;208:141–148. doi: 10.1002/jcp.20639. [DOI] [PubMed] [Google Scholar]
- 88.Ewald AJ, et al. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J. Cell Sci. 2012;125:2638–2654. doi: 10.1242/jcs.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grugan KD, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc. Natl Acad. Sci. USA. 2010;107:11026–11031. doi: 10.1073/pnas.0914295107. [Uses organotypic culture and independent genetic manipulation of epithelial and stromal compartments to implicate fibroblast-secreted HGF and its epithelial receptor MET in the invasion of transformed oesophageal epithelial cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- 91.Lu P, Werb Z. Patterning mechanisms of branched organs. Science. 2008;322:1506–1509. doi: 10.1126/science.1162783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J. Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 93.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev. Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 94.Chi X, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [Demonstrates, using an elegant series of chimeric embryonic kidneys, that levels of RET signalling dictate cellular contribution to the ureteric bud tip domain.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Patel VN, et al. Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. J. Biol. Chem. 2008;283:9308–9317. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Packard A, et al. Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev. Cell. 2013;27:319–330. doi: 10.1016/j.devcel.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schnatwinkel C, Niswander L. Multiparametric image analysis of lung-branching morphogenesis. Dev. Dyn. 2013;242:622–637. doi: 10.1002/dvdy.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 99.Hsu JC, et al. Viral gene transfer to developing mouse salivary glands. J. Dent. Res. 2012;91:197–202. doi: 10.1177/0022034511429346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sequeira SJ, Gervais EM, Ray S, Larsen M. Genetic modification and recombination of salivary gland organ cultures. J. Vis. Exp. 2013;28:e50060. doi: 10.3791/50060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [Demonstrates that CRISPR–Cas9 and organoid culture can be coupled to correct disease mutations in patient-derived cells and assay for restored tissue function.] [DOI] [PubMed] [Google Scholar]
- 103.Yu W, et al. β1-integrin orients epithelial polarity via Rac1 and laminin. Mol. Biol. Cell. 2005;16:433–445. doi: 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martin-Belmonte F, et al. Cell-polarity dynamics controls the mechanism of lumen formation in epithelial morphogenesis. Curr. Biol. 2008;18:507–513. doi: 10.1016/j.cub.2008.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bryant DM, et al. A molecular network for de novo generation of the apical surface and lumen. Nature Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gálvez-Santisteban M, et al. Synaptotagmin-like proteins control the formation of a single apical membrane domain in epithelial cells. Nature Cell Biol. 2012;14:838–849. doi: 10.1038/ncb2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aranda V, et al. Par6–aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nature Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 109.Zhan L, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell–cell cohesion. Nature Cell Biol. 2013;15:1–14. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leung CT, Brugge JS. Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature. 2012;482:410–413. doi: 10.1038/nature10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sakurai A, Matsuda M, Kiyokawa E. Activated Ras protein accelerates cell cycle progression to perturb Madin–Darby canine kidney cystogenesis. J. Biol. Chem. 2012;287:31703–31711. doi: 10.1074/jbc.M112.377804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 116.Yates LL, et al. Scribble is required for normal epithelial cell–cell contacts and lumen morphogenesis in the mammalian lung. Dev. Biol. 2013;373:267–280. doi: 10.1016/j.ydbio.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu JS, Farlow JT, Paulson AK, Labarge MA, Gartner ZJ. Programmed cell-to-cell variability in Ras activity triggers emergent behaviors during mammary epithelial morphogenesis. Cell Rep. 2012;2:1461–1470. doi: 10.1016/j.celrep.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plichta KA, Mathers JL, Gestl SA, Glick AB, Gunther EJ. Basal but not luminal mammary epithelial cells require PI3K/mTOR signaling for Ras-driven overgrowth. Cancer Res. 2012;72:5856–5866. doi: 10.1158/0008-5472.CAN-12-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev. Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beck JN, Singh A, Rothenberg AR, Elisseeff JH, Ewald AJ. The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination. Biomaterials. 2013;34:9486–9495. doi: 10.1016/j.biomaterials.2013.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 123.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 124.DeNardo DG, et al. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nature Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 126.Calvo F, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nature Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Okawa T, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee J-H, et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4–NFATc1–thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ghajar CM, et al. The perivascular niche regulates breast tumour dormancy. Nature Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [References 128 and 129 define a role for endothelial-derived TSP1 in regulating epithelial differentiation and tumour growth.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Infanger DW, et al. Glioblastoma stem cells are regulated by interleukin-8 signaling in a tumoral perivascular niche. Cancer Res. 2013;73:7079–7089. doi: 10.1158/0008-5472.CAN-13-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Knox SM, et al. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 2010;329:1645–1647. doi: 10.1126/science.1192046. [Demonstrates, using a combination of 3D-embedded culture and whole-organ culture, that parasympathetic innervation maintains salivary epithelial progenitors and offers a therapeutic strategy for organ repair.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Knox SM, et al. Parasympathetic stimulation improves epithelial organ regeneration. Nature Commun. 2013;4:1494. doi: 10.1038/ncomms2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nature Rev. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 134.Carey SP, Starchenko A, McGregor AL, Reinhart-King CA. Leading malignant cells initiate collective epithelial cell invasion in a three-dimensional heterotypic tumor spheroid model. Clin. Exp. Metastasis. 2013;30:615–630. doi: 10.1007/s10585-013-9565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dang TT, Prechtl AM, Pearson GW. Breast cancer subtype-specific interactions with the microenvironment dictate mechanisms of invasion. Cancer Res. 2011;71:6857–6866. doi: 10.1158/0008-5472.CAN-11-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gudjonsson T, et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J. Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chanson L, et al. Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proc. Natl Acad. Sci. USA. 2011;108:3264–3269. doi: 10.1073/pnas.1019556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Conklin MW, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 140.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 141.Sia SK, Gillette BM, Yang GJ. Synthetic tissue biology: tissue engineering meets synthetic biology. Birth Defects Res. C Embryo. Today. 2007;81:354–361. doi: 10.1002/bdrc.20105. [DOI] [PubMed] [Google Scholar]
- 142.Elliott MJ, et al. Stem-cell-based, tissue engineered tracheal replacement in a child: a 2-year follow-up study. Lancet. 2012;380:994–1000. doi: 10.1016/S0140-6736(12)60737-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [Demonstrates that Paneth cells, which are a differentiated stem cell progeny, function as an essential part of the stem cell niche in intestinal crypts and significantly increase the ability of LGR5+ stem cells to form long-lived organoids in vitro.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yui S, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nature Med. 2012;18:618–623. doi: 10.1038/nm.2695. [Demonstrates the potential of stem cell organoids to repair experimental injuries to the colon in small-animal models and suggests that organoids could be used therapeutically in human patients.] [DOI] [PubMed] [Google Scholar]
- 145.Assawachananont J, et al. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014;2:662–674. doi: 10.1016/j.stemcr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Saito H, Takeuchi M, Chida K, Miyajima A. Generation of glucose-responsive functional islets with a three-dimensional structure from mouse fetal pancreatic cells and iPS cells in vitro. PLoS ONE. 2011;6:e28209. doi: 10.1371/journal.pone.0028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sneddon JB, Borowiak M, Melton DA. Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature. 2012;491:765–768. doi: 10.1038/nature11463. [Develops techniques to efficiently differentiate ES cells into endodermal progenitors and then, using co-culture with mesenchyme and transplantation, differentiate these progenitors into glucose-sensitive, insulin-secreting cells in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Antonica F, et al. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491:66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 150.Schayowitz A, et al. Functional profiling of live melanoma samples using a novel automated platform. PLoS ONE. 2013;7:e52760. doi: 10.1371/journal.pone.0052760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Miller JS, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nature Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [Develops a novel model for vascularized human liver from iPS cells and demonstrates the functional engraftment of these liver buds into mice. Notably, the transplanted tissue had characteristics of human liver at the level of protein production and drug metabolism.] [DOI] [PubMed] [Google Scholar]
- 153.Sudo R. Multiscale tissue engineering for liver reconstruction. Organogenesis. 2014 doi: 10.4161/org.27968. http://dx.doi.org/10.4161/org.27968. [DOI] [PMC free article] [PubMed]
- 154.Schrag D, et al. American society of clinical oncology technology assessment: chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2004;22:3631–3638. doi: 10.1200/JCO.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 155.Burstein HJ, et al. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2011;29:3328–3330. doi: 10.1200/JCO.2011.36.0354. [DOI] [PubMed] [Google Scholar]
- 156.Vaira V, et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl Acad. Sci. USA. 2010;107:8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Merz F, et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro. Oncol. 2013;15:670–681. doi: 10.1093/neuonc/not003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 159.Muranen T, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [Shows that the dimensionality of cancer spheroids is relevant for the rational design of drug combinations owing to distinct responses in matrix-attached and matrix-deprived cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Walker JL, et al. Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: insights into the formation of acinar and ductal structures. Dev. Dyn. 2008;237:3128–3141. doi: 10.1002/dvdy.21717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ginsburg E, Vonderhaar BK. In: Methods in Mammary Gland Biology and Breast Cancer Research. Ip MM, Asch BB, editors. Springer; US: 2000. pp. 147–154. [Google Scholar]
- 163.Nguyen-Ngoc KV, et al. In: Tissue Morphogenesis: Methods and Protocols Vol. 1189 Methods in Molecular Biology. Nelson CM, editor. Springer Science and Business Media; 2014. [Google Scholar]
- 164.Akhtar N, Streuli CH. An integrin–ILK–microtubule network orients cell polarity and lumen formation in glandular epithelium. Nature Cell Biol. 2012;15:17–27. doi: 10.1038/ncb2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Daley WP, et al. ROCK1-directed basement membrane positioning coordinates epithelial tissue polarity. Development. 2011;139:411–422. doi: 10.1242/dev.075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pradhan-Bhatt S, et al. Implantable three-dimensional salivary spheroid assemblies demonstrate fluid and protein secretory responses to neurotransmitters. Tissue Eng. Part A. 2013;19:1610–1620. doi: 10.1089/ten.tea.2012.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wei C, Larsen M, Hoffman MP, Yamada KM. Self-organization and branching morphogenesis of primary salivary epithelial cells. Tissue Eng. 2007;13:721–735. doi: 10.1089/ten.2006.0123. [DOI] [PubMed] [Google Scholar]
- 168.O'Brien LE, et al. Morphological and biochemical analysis of Rac1 in three-dimensional epithelial cell cultures. Methods Enzymol. 2006;406:676–691. doi: 10.1016/S0076-6879(06)06053-8. [DOI] [PubMed] [Google Scholar]
- 169.Yagi S, Matsuda M, Kiyokawa E. Suppression of Rac1 activity at the apical membrane of MDCK cells is essential for cyst structure maintenance. EMBO Rep. 2012;13:237–243. doi: 10.1038/embor.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Srinivas S, et al. Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev. Genet. 1999;24:241–251. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<241::AID-DVG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 171.Costantini F, Watanabe T, Lu B, Chi X, Srinivas S. Dissection of embryonic mouse kidney, culture in vitro, and imaging of the developing organ. Cold Spring Harb. Protoc. 20112011 doi: 10.1101/pdb.prot5613. http://dx.doi.org/10.1101/pdb.prot5613. [DOI] [PubMed]
- 172.Rosines E, et al. Constructing kidney-like tissues from cells based on programs for organ development: toward a method of in vitro tissue engineering of the kidney. Tissue Eng. Part A. 2010;16:2441–2455. doi: 10.1089/ten.tea.2009.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Steer DL, Bush KT, Meyer TN, Schwesinger C, Nigam SK. A strategy for in vitro propagation of rat nephrons. Kidney Int. 2002;62:1958–1965. doi: 10.1046/j.1523-1755.2002.00694.x. [DOI] [PubMed] [Google Scholar]
- 174.Taub M, Wang Y, Szczesny TM, Kleinman HK. Epidermal growth factor or transforming growth factor α is required for kidney tubulogenesis in matrigel cultures in serum-free medium. Proc. Natl Acad. Sci. USA. 1990;87:4002–4006. doi: 10.1073/pnas.87.10.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Morizane R, et al. Kidney specific protein-positive cells derived from embryonic stem cells reproduce tubular structures in vitro and differentiate into renal tubular cells. PLoS ONE. 2013;8:e64843. doi: 10.1371/journal.pone.0064843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Taguchi A, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 177.Takasato M, Little MH, Elefanty AG. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nature Cell Biol. 2013;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 178.Parrish AR, Gandolfi AJ, Brendel K. Precision-cut tissue slices: applications in pharmacology and toxicology. Life Sci. 1995;57:1887–1901. doi: 10.1016/0024-3205(95)02176-j. [DOI] [PubMed] [Google Scholar]
- 179.del Moral P-M, Warburton D. Explant culture of mouse embryonic whole lung, isolated epithelium, or mesenchyme under chemically defined conditions as a system to evaluate the molecular mechanism of branching morphogenesis and cellular differentiation. Methods Mol. Biol. 2010;633:71–79. doi: 10.1007/978-1-59745-019-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl Acad. Sci. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mondrinos MJ, et al. Engineering three-dimensional pulmonary tissue constructs. Tissue Eng. 2006;12:717–728. doi: 10.1089/ten.2006.12.717. [DOI] [PubMed] [Google Scholar]
- 182.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Magudia K, Lahoz A, Hall A. K-Ras and B-Raf oncogenes inhibit colon epithelial polarity establishment through up-regulation of c-myc. J. Cell Biol. 2012;198:185–194. doi: 10.1083/jcb.201202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kovbasnjuk O, et al. Human enteroids: preclinical models of non-inflammatory diarrhea. Stem Cell Res. Ther. 2014;4:S3. doi: 10.1186/scrt364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li X, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nature Med. 2014;20:769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Stange DE, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Reichert M, et al. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nature Protoc. 2013;8:1354–1365. doi: 10.1038/nprot.2013.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Okugawa YA. Novel three-dimensional cell culture method to analyze epidermal cell differentiation in vitro. Methods Mol. Biol. 2013 doi: 10.1007/7651_2013_45. [DOI] [PubMed] [Google Scholar]
- 190.Lang SH, et al. Experimental prostate epithelial morphogenesis in response to stroma and three-dimensional matrigel culture. Cell Growth Differ. 2001;12:631–640. [PubMed] [Google Scholar]
- 191.Kubota Y, Kleinman HK, Martin GR, Lawley TJ. Role of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary-like structures. J. Cell Biol. 1988;107:1589–1598. doi: 10.1083/jcb.107.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Arnaoutova I, Kleinman HK. In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nature Protoc. 2010;5:628–635. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 193.Davis GE, et al. Control of vascular tube morphogenesis and maturation in 3D extracellular matrices by endothelial cells and pericytes. Methods Mol. Biol. 2013;1066:17–28. doi: 10.1007/978-1-62703-604-7_2. [DOI] [PubMed] [Google Scholar]
- 194.Morgan JP, et al. Formation of microvascular networks in vitro. Nature Protocols. 2013;8:1820–1836. doi: 10.1038/nprot.2013.110. [DOI] [PubMed] [Google Scholar]