Recurring use of a gene regulatory network to orchestrate innate and adaptive cell fates in the immune system (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 25.
Abstract
The transcription factor PU.1 functions in a graded manner to regulate macrophage versus B cell generation; its higher concentration favors the macrophage fate. We now demonstrate that Gfi-1 reciprocally promotes B cell fate choice at the expense of myeloid progeny. Gfi-1−/− MPPs are unable to constrain the expression of PU.1 as Gfi-1 functions to repress the PU.1 gene by displacing PU.1 from positive auto-regulatory elements. Attenuating a transcriptional module comprised of PU.1 and Egr’s suppresses the B lineage developmental defects of Gfi-1−/− MPPs. Finally Ikaros, a transcription factor required for B cell development, functions to activate Gfi-1 and antagonize PU.1 expression in MPPs. Our results reveal that a core transcriptional regulatory network used to direct cell fate choice in the innate immune system has been co-opted by Ikaros to orchestrate the generation of B-lymphocytes. These findings have important implications for the evolution of the adaptive immune system.
Keywords: Gfi-1, PU.1, Ikaros, hematopoiesis, cell fate choice, innate and adaptive immune cells
INTRODUCTION
An emerging developmental framework for hematopoiesis posits that cells of the innate and adaptive immune system arise from a common lymphoid-primed multipotent progenitor (LMPP) lacking erythrocytic and megakaryocytic potential (Adolfsson et al., 2005; Laiosa et al., 2006a). Genetic and molecular analyses of various lineage-determining transcription factors have enabled the assembly of contingent gene regulatory networks that promote the generation of either myeloid or lymphoid progeny from multipotent hematopoietic progenitors (Laiosa et al., 2006a; Laslo et al., 2008; Rothenberg and Taghon, 2005; Singh et al., 2005). While the transcription factors PU.1, Ikaros, Mef2c, E2A, and EBF-1 have been implicated in regulating myeloid versus lymphoid cell fate choice (DeKoter and Singh, 2000; Dias et al., 2008; Pongubala et al., 2008; Reynaud et al., 2008; Stehling-Sun et al., 2009), the nature of the molecular circuits that underlie the onset of cell fate determination in MPPs remain to be elucidated. A deeper understanding of such transcriptional networks may provide insight into the molecular evolution of the adaptive immune cells from their primordial innate counterparts.
The existence of multipotent progenitors restricted to the generation of myeloid and lymphoid progeny was previously proposed based on genetic analysis of the PU.1 (Sfpi1) gene (Singh, 1996). PU.1 is a member of the Ets family of transcription factors and is expressed in multiple lineages of the hematopoietic system, including MPPs (Nutt et al., 2005). In the absence of PU.1, the development of myeloid and lymphoid lineages is severely impaired, while the generation of erythrocytes and megakaryocytes is largely unaffected (Scott et al., 1994). The expression of many myeloid (DeKoter et al., 1998) and lymphoid-specific genes (DeKoter et al., 2002) is abolished in PU.1−/− hematopoietic progenitors. Graded levels of PU.1 appear to regulate the development of myeloid and B lineage progeny, as a low concentration of PU.1 induces the B cell fate, while a higher concentration promotes macrophage development at the expense of B cell generation (DeKoter and Singh, 2000). In addition, higher levels of PU.1 have been shown to inhibit early T cell development (Anderson et al., 2002) and promote macrophage differentiation (Laiosa et al., 2006b). These results suggest that PU.1 expression needs to be constrained in MPPs in order to enable B lymphopoiesis in the bone marrow and T lymphopoiesis in the thymus. The molecular means by which this is achieved remains to be elucidated.
In myeloid progenitors, PU.1 has been shown to induce and resolve a mixed lineage pattern of gene expression resulting in the generation of macrophages and neutrophils (Laslo et al., 2006). In this cellular context, PU.1 is a component of a transcriptional regulatory circuit comprised of the myeloid determinant C/EBPα and the counteracting repressors Egr1,2/Nab2 and Gfi-1. High levels of PU.1 induce Egr2 and Nab2. Importantly, Egr2 functions in a feed forward loop with PU.1 to activate macrophage-specific genes and with Nab2 to repress alternate lineage neutrophil genes, including Gfi-1. Conversely, Gfi-1 promotes neutrophil differentiation by antagonizing PU.1 and Egr activity, the former, presumably, via direct protein-protein interactions (Dahl et al., 2007) and the latter via transcriptional repression (Laslo et al., 2006). Since PU.1 expression appears to be regulated by a positive auto-regulatory loop (Okuno et al., 2005), these results raised the possibility that Gfi-1 could attenuate the expression of PU.1 by antagonizing PU.1 activity in MPPs, thereby lowering its levels to promote the generation of lymphocytes at the expense of myeloid progeny.
Like PU.1, Gfi-1 is expressed in multiple hematopoietic lineages, including MPPs (Zeng et al., 2004). However, Gfi-1 levels appear to be inversely correlated with those of PU.1 in hematopoietic cells. Gfi-1−/− animals are neutropenic; the granulocytic intermediates that develop in the bone marrow mis-express PU.1 regulated genes such as c-fms (Hock et al., 2003). Interestingly, the frequency of myeloid progeny is increased in the bone marrow of Gfi-1−/− mice while the number of B-lineage cells in the bone marrow and T-lineage cells in the thymus are significantly reduced (Hock et al., 2003; Yucel et al., 2003). Since high levels of PU.1 function to induce myeloid development and Gfi-1 activity is critical in early lymphocyte development, we considered whether PU.1 and Gfi-1 might function in an antagonistic manner to regulate innate versus adaptive immune cell fate choice in MPPs as they do in orchestrating macrophage versus neutrophil development. As high levels of PU.1 are inhibitory for early B and T cell development, we reasoned that the underlying basis might involve PU.1 mediated induction of the Egr’s that could directly repress Gfi-1 expression. Consistent with this possibility, the loss of Egr1 results in increased T-lineage precursors in the thymus (Bettini et al., 2002). Given the aforementioned findings, we hypothesized that a network comprised of PU.1, Egr’s and Gfi-1 might function in a recurring manner to regulate myeloid versus lymphoid cell fate choice in MPPs.
As with Gfi-1, the loss of the zinc-finger transcription factor Ikaros has profound consequences on the development of both B and T lineage cells (Wang et al., 1996). Additionally, Ikaros has been implicated in the generation of LMPPs (Yoshida et al., 2006). Interestingly, Ikaros−/− pro-B cells retain myeloid developmental potential and mis-express multiple myeloid-specific genes, including c-fms (Reynaud et al., 2008). These findings suggest similar roles for Ikaros and Gfi-1 in promoting early B and T cell development and in repressing myeloid developmental potential. We therefore considered the possibility that Ikaros and Gfi-1 may represent components of a regulatory network in which they collaborate to promote adaptive immune cell fates and repress innate immune cell fates in MPPs.
We designed a set of genetic and molecular experiments to test the existence of the proposed regulatory network controlling innate versus adaptive immune cell fates. We demonstrate that Gfi-1 promotes B cell fate choice by antagonizing the expression of the PU.1 gene and aspects of the myeloid gene expression program in MPPs. Molecular analyses reveal that Gfi-1 directly represses the PU.1 gene by targeting the PU.1 promoter and a distal upstream regulatory element (URE). Consistent with our hypothesis, Egr’s are shown to function in an opposing manner to Gfi-1, as the Egr’s inhibit B cell development while enabling myelopoiesis. Finally, we show that Ikaros positively regulates Gfi-1 and antagonizes the expression of PU.1 in MPPs. These data are consistent with a model whereby Ikaros and Gfi-1 function within MPPs to inhibit myeloid lineage potential by attenuating PU.1 and Egr activity, thereby facilitating the specification of lymphoid cell fates.
RESULTS
Gfi-1 promotes B cell fate choice
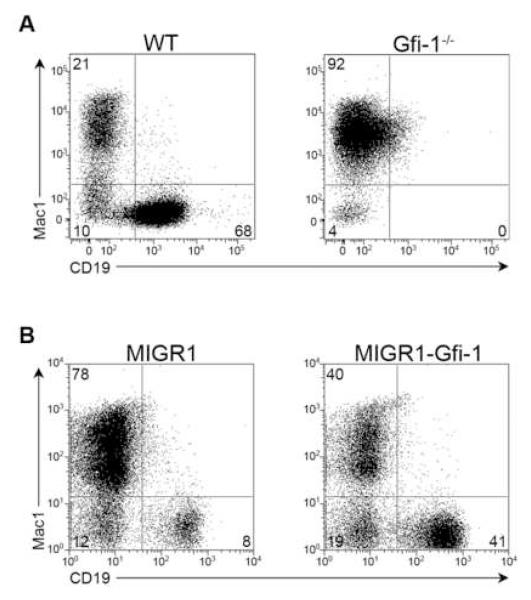
Since Gfi-1−/− animals exhibit a decrease in the frequency of common lymphoid progenitors and an increase in granulocyte/macrophage progenitors (Zeng et al., 2004), we examined if Gfi-1 functions to promote B cell fate specification and repress myeloid development in the context of a MPP. We isolated MPPs from wildtype (WT) and Gfi-1−/− mice (Figure S1A) and plated equivalent numbers on OP9 stroma under conditions that support the generation of both myeloid (Mac1+) and B-lineage (CD19+) progeny. WT progenitors gave rise to both myeloid (21%) and B lymphoid cells (68%). In contrast, MPPs lacking Gfi-1 were severely defective for B cell development in vitro (Figure 1A and S1B). We note that the loss of Gfi-1 in vivo impairs, but does not eliminate B cell development (Hock et al., 2003). We also performed gain-of-function analyses to test whether an increased concentration of Gfi-1 in MPPs enhanced the generation of B lineage progeny at the expense of myeloid precursors. Transduction of WT MPPs with a control vector (MIGR1) via co-culture primarily generated Mac1+ cells. In contrast, MPPs transduced with a Gfi-1 vector (MIGR1-Gfi-1) generated a higher proportion of CD19+ progeny (Figure 1B). Using limiting dilution assays, MPPs transduced with Gfi-1 were seen to give rise to B lineage progeny at a frequency nearly 3.5 times higher than their control counterparts (Figure S1C). These results demonstrate that Gfi-1 promotes B cell fate choice at the expense of myeloid options in the context of MPPs.
Figure 1. Gfi-1 regulates B versus myeloid cell fate choice.
(A) MPPs (Lin−Sca-1+c-Kithi) were isolated from the bone marrow of WT or _Gfi-1−/−_animals. Cells were directly plated on OP9 stroma and analyzed by flow cytometry at day 12. Data are representative of at least three independent experiments. (B) WT MPPs were transduced with a control vector (MIGR1) or one expressing Gfi-1 (MIGR1-Gfi-1). GFP+ transductants were plated on OP9 stroma and examined for the generation of myeloid and B-lineage progeny after 7 days. Data are representative of at least three independent experiments.
Gfi-1 antagonizes PU.1 expression in MPPs
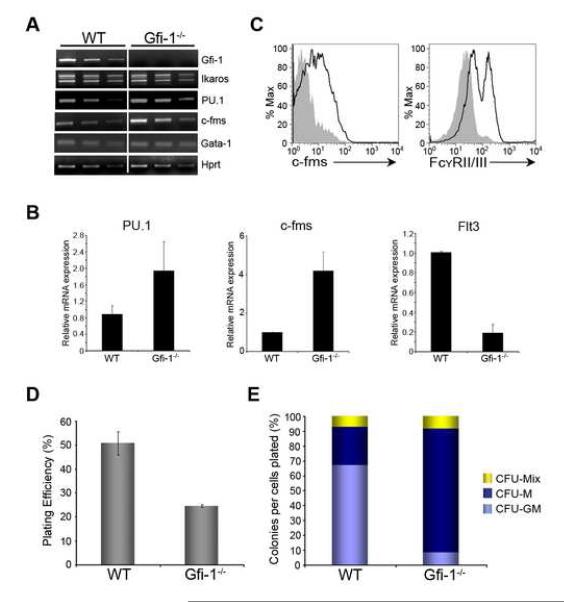
PU.1 expression is elevated in total bone marrow cells lacking Gfi-1 (Hock et al., 2003). We specifically analyzed the expression of PU.1 and other hematopoietic determinants in WT and Gfi-1−/− MPPs. While the expression of Ikaros and Gata1 were unaffected in Gfi-1−/− MPPs, PU.1 transcripts were elevated (Figure 2A and B). Moreover, the expression of the c-fms gene, which is directly activated by PU.1 (Krysinska et al., 2007), was increased in Gfi-1−/− MPPs (Figure 2A and B). We note that Flt3 transcripts are reduced in Gfi-1−/− MPPs (Figure 2B), consistent with reduced Flt3 expression on the surface of Gfi-1−/− MPPs (Zeng et al., 2004). We also assessed expression of c-fms and FcγRII/III on Gfi-1−/− MPPs using flow cytometry. Like c-fms, the FcγRII/III receptors are expressed on myeloid precursors and their genes are directly activated by PU.1. The loss of Gfi-1 resulted in increased expression of c-fms and FcγRII/III in MPPs (Figure 2C). We note that the expression of Ly6G and Mac1, two cell surface markers that are expressed on more mature myeloid cells, was not affected (Figure S2A). Thus, loss of Gfi-1 results in elevated expression of PU.1 and its myeloid target genes in MPPs.
Figure 2. Gfi-1−/− MPPs express increased levels of PU.1 and its target genes.
(A) Semi-quantitative RT-PCR analysis of cDNA (3X serial dilution) from WT or Gfi-1−/− MPPs. Transcript levels were normalized to Hprt. (B) qPCR analysis of PU.1, c-fms and Flt3 transcripts in WT or Gfi-1−/− MPPs. (C) Histograms represent c-fms or FcγRII/III expression on the surface of WT (gray shaded area) or Gfi-1−/− (black line) MPPs. (D) and (E) WT or Gfi-1−/− MPPs were plated in methylcellulose containing erythropoietin, stem cell factor, IL-3, IL-6, and Flt3 ligand. Colony numbers were scored 8 days after plating and analyzed for the generation of CFU-Mix (erythroid/granulocyte/macrophage), CFU-M (macrophage) or CFU-GM (granulocyte/macrophage) by Wright staining of cytospins. Data is from two independent experiments.
We evaluated the developmental potential of c-fms expressing MPPs to test if they were biased towards myeloid fates (Figure S2B). Gfi-1−/− MPPs that were c-fms− had considerably lower B-lineage developmental potential than WT MPPs (Figure S2B). Importantly, c-fms+ MPPs lacking Gfi-1 were further impaired in their ability to give rise to B lineage progeny. Therefore, B lineage potential may be impaired in Gfi-1−/− MPPs as a consequence of enhanced myeloid potential.
We predicted that augmented PU.1 expression in Gfi-1−/− MPPs should result in enhanced macrophage potential. We analyzed Gfi-1−/− MPPs for their ability to undergo multilineage erythro-myeloid differentiation. The absence of Gfi-1 resulted in ~50% reduction in plating efficiency of MPPs relative to WT counterparts (Figure 2D). WT MPPs preferentially gave rise to granulocyte/macrophage colonies (~70%) and, to a lesser extent, macrophage (25%), and myelo/erythroid colonies (5%) (Figure 2E). In contrast, the preponderance of colonies generated from Gfi-1−/− MPPs contained macrophages (85%). No discernable difference was noted between WT and Gfi-1−/− MPPs in the generation of myelo-erythroid colonies, suggesting a role for Gfi-1 in lineage restriction subsequent to the specification of megakaryocyte and erythrocyte fates. Taken together, these data suggest a critical role for Gfi-1 in constraining the expression of PU.1 in the context of two distinct cell fate choices: i) macrophage versus neutrophil and ii) myeloid versus B cell.
PU.1 heterozygosity partially rescues B cell development in Gfi-1−/− mice
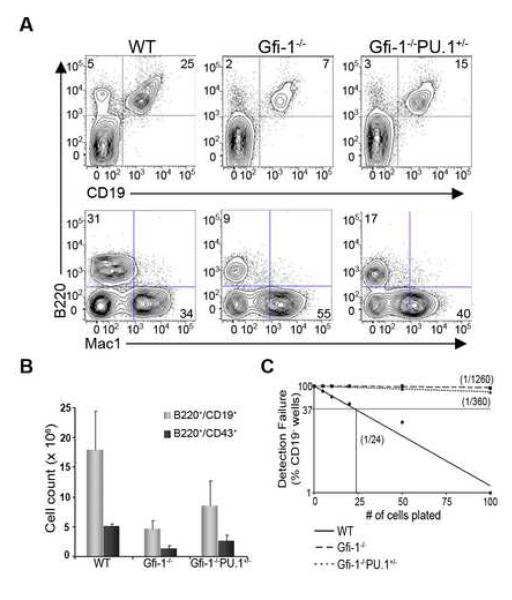
Since the loss of Gfi-1 results in enhanced expression of PU.1, we reasoned that this may partially account for the reduced B cell developmental potential of Gfi-1−/− MPPs. To test this possibility, we determined if reducing the dosage of PU.1 would suppress the B cell developmental defect caused by the loss of Gfi-1. As previously described (Hock et al., 2003), loss of Gfi-1 resulted in fewer committed B lineage cells (B220+CD19+) in the bone marrow (Figure 3A). Combining PU.1 heterozygosity with the Gfi-1 mutation led to a two-fold increase in the frequency as well as the absolute numbers of committed B lineage cells (Figure 3A and B). In addition, there was a two-fold increase in the number of pro-B cells (B220+CD43+) in Gfi-1−/−PU.1+/− mice in comparison to Gfi-1−/− controls (Figure 3B). These results demonstrate that B lineage defects in Gfi-1−/− bone marrow can be partially suppressed by reducing PU.1 gene dosage.
Figure 3. PU.1 heterozygosity promotes B cell development in Gfi-1−/− mice.
(A) Bone marrow cells from WT, Gfi-1−/− and Gfi-1−/−PU.1+/− animals were analyzed by flow cytometry for B-lineage (B220+CD19+) and myeloid (Mac1+) precursors. (B) Absolute numbers of B-lineage (B220+CD19+ or B220+CD43+) cells in the bone marrow of WT (n=5), Gfi-1−/− (n=5), and Gfi-1−/−PU.1+/− (n=7) animals. (C) Quantitative analysis of B-lineage potential in MPPs from WT (solid line), Gfi-1−/− (long dashed line), or Gfi-1−/−PU.1+/− (short dashed line) mice. MPPs were plated on OP9 stroma in limiting dilution as described in Fig. 1. Cultures were analyzed for wells containing CD19+ cells at day 12. Data are representative of two independent experiments.
To determine whether reduced PU.1 gene dosage increases the B-lineage developmental potential of Gfi-1−/− MPPs, we quantitatively analyzed their cell fate outputs in vitro. We note that loss of one allele of PU.1 was associated with a decrease in c-fms and FcγRII/III expression on the cell surface of Gfi-1−/−PU.1+/− MPPs relative to their Gfi-1−/− counterparts (Figure S3). WT MPPs gave rise to B lineage cells at a frequency of ~1/24, while loss of Gfi-1 severely impaired the generation of such cells (1/1260) (Figure 3C). Importantly, we observed ~ 3.5 fold increase (1/360) in the frequency of B lineage cells generated from _Gfi-1−/−/PU.1+/−_MPPs compared with their Gfi-1−/− counterparts. These data demonstrate that Gfi-1 mediated antagonism of PU.1 expression in MPPs plays an important role in restraining myeloid differentiation and, in turn, promoting specification of the B cell fate.
Knockdown of PU.1 in Gfi-1−/− MPPs promotes B cell development
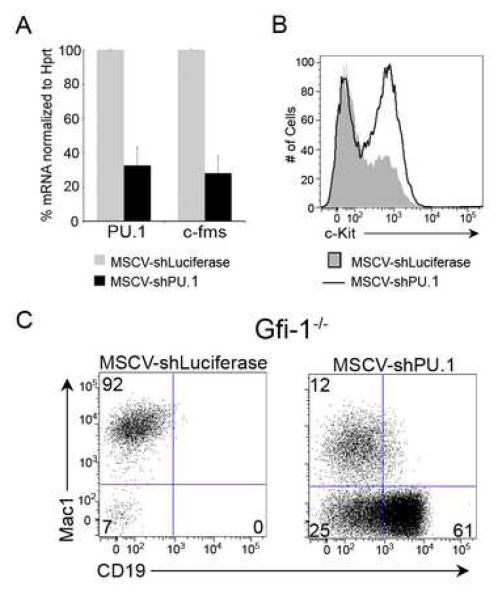
We utilized an shRNA approach to rigorously test if Gfi-1−/− MPPs are impaired in their B-lineage potential due to increased expression of PU.1. Transduction of WT MPPs with a vector targeting PU.1 mRNA (MSCV-shPU.1) resulted in ~70% reduction in PU.1 transcripts and its target gene c-fms (Figure 4A). Reduced PU.1 expression impaired terminal myeloid differentiation, as evidenced by the accumulation of c-Kit+Mac1+ myeloid precursors (Figure 4B). This resembled the accumulation of similar myeloid precursors in the bone marrow of mice in which PU.1 expression is reduced by targeting an upstream distal enhancer (Rosenbauer et al., 2004). Importantly, knocking-down PU.1 expression in WT MPPs resulted in a higher frequency of committed B lineage progeny (27%) and diminished Mac1+ precursors (52%) compared to control transductants (Figure S4). Thus, lowering PU.1 expression in MPPs promoted the generation of B lineage cells at the expense of myeloid precursors.
Figure 4. Attenuation of PU.1 activity promotes B cell fate specification.
(A) shRNA-mediated knockdown of PU.1 in MPPs. WT MPPs were transduced with a MSCV-shLuciferase or MSCV-shPU.1 vector. GFP+ transductants were analyzed for the expression of PU.1 and c-fms transcripts. Data are representative of two independent experiments. (B) WT MPPs transduced with MSCV-shLuciferase (gray shaded area) or MSCV-shPU.1 (black line) were plated on OP9 stroma and analyzed for the presence of c-Kit+ precursors (gated on Mac1+ cells) 7 days post-sort. Data are representative of two independent experiments. (C) Gfi-1−/− MPPs were transduced with MSCV-shLuciferase or MSCV-shPU.1 and analyzed by flow cytometry for the generation of Mac1+ and CD19+ precursors after 10 days.
We next examined whether attenuating PU.1 expression in Gfi-1−/− MPPs restored their ability to give rise to B-lineage progeny. Gfi-1−/− MPPs were transduced with the control or MSCV-shPU.1 construct. While Gfi-1−/− MPPs primarily gave rise to Mac1+ precursors, knockdown of PU.1 strongly induced the generation of CD19+ B lineage cells (Figure 4C). These data establish that Gfi-1 mediated antagonism of PU.1 expression in MPPs is used to regulate B versus myeloid cell fate choice.
Inhibition of Egr activity promotes B cell fate choice
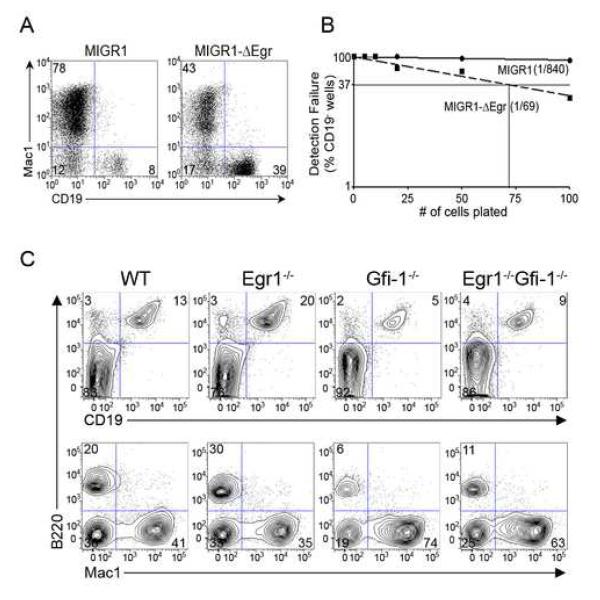
We have previously proposed that higher levels of PU.1 antagonize Gfi-1 expression via induction of the Egr’s which directly bind to the Gfi-1 promoter and repress its activity (Laslo et al., 2006). Conversely, Gfi-1 represses the transcription of Egr genes. Thus perturbing Egr activity would be predicted to have the opposite consequence to that of manipulating Gfi-1 on early B cell development. Furthermore, the Egr’s should promote myeloid vs. B cell fate choice in MPPs as is the case with higher levels of PU.1. To test this prediction, WT MPPs were transduced with a control vector (MIGR1) or one expressing a dominant-negative version of Egr-2 (MIGR1-ΔEgr). While control transductants preferentially give rise to Mac1+ precursors, inhibition of Egr function in MPPs strongly promoted the development of B lineage progeny (Figure 5A). Limiting dilution analyses revealed that blocking Egr activity in MPPs enhanced B cell development by more than an order of magnitude (Figure 5B). Thus, blocking Egr activity in MPPs has the same consequence on B cell development as enhancing Gfi-1 expression (Figure 1B).
Figure 5. Egr activity in MPPs antagonizes B cell development.
(A) WT MPPs transduced with a control vector (MIGR1) or one expressing a dominant-negative version of Egr-2 (MIGR1-ΔEgr) were plated on OP9 stroma and analyzed for Mac1+ and CD19+ precursors after 7 days. (B) GFP+ transductants plated in limiting dilution were analyzed for the generation of CD19+ progeny by flow cytometry 7 days post-sort. Data are representative of at least three independent experiments. (C) Total bone marrow cells from WT, Egr1−/−, Gfi-1−/− and Egr1−/−Gfi-1−/− animals were analyzed for B-lineage (B220+CD19+) and myeloid (Mac1+) cells by flow cytometry. Data is representative of three mice for each genotype.
We next used genetic interaction analyses to test for the functional cross-antagonism between the Egr’s and Gfi-1 in B cell development. Interestingly, we observed that the removal of Egr1 partially suppressed the B cell developmental defect observed in Gfi-1−/− mice (Figure 5C and S5A). This paralleled the partial ‘rescue’ of B cell development in Gfi-1−/− mice by reducing the dosage of PU.1 (Figure 3A and B). Egr1 and Egr2 function redundantly during macrophage differentiation (Laslo et al., 2006). To test if Egr1 and Egr2 also function redundantly in antagonizing the generation of B cells, we generated mice lacking both Egr1 and Egr2 using a conditional Egr2 allele (Taillebourg et al., 2002). This was necessitated as Egr2−/− mice die soon after birth (Swiatek and Gridley, 1993) and loss of Egr1 and Egr2 results in embryonic lethality. Using the Mx1-Cre transgene, we observed efficient excision of the Egr2 allele in total bone marrow cells 4 weeks after polyIC treatment (Figure S5B). Combined loss of Egr-1 and Egr-2 resulted in a ~2-fold increase in the percentage of B lineage progeny in the bone marrow and a comparable reduction in myeloid precursors (Figures S5B and C). These results provide compelling genetic evidence in support of the hypothesis that Egr’s antagonize B cell fate choice at the expense of myeloid options in MPPs.
Gfi-1 targets the PU.1 promoter and URE
Given that Gfi-1 functions as a transcriptional repressor, we examined whether Gfi-1 targets the PU.1 gene. Bioinformatic analysis revealed conserved putative Gfi-1 binding sites in the PU.1 promoter and the URE (Figure 6A). Intriguingly, these presumptive Gfi-1 binding sites were in close proximity to PU.1 binding sites. PU.1 binding sites in the promoter and URE of the PU.1 gene have been implicated in a positive auto-regulatory feedback loop (Okuno et al., 2005). We considered the possibility that Gfi-1 could repress the PU.1 gene by displacing PU.1 from its own promoter and URE.
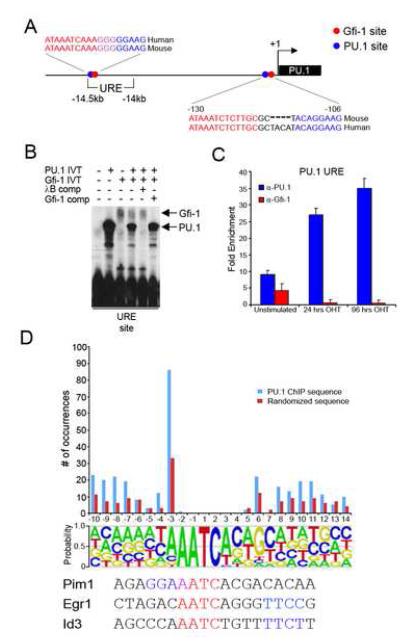
Figure 6. Gfi-1 targets the PU.1 locus.
(A) Schematic structure of the murine PU.1 locus. Sequence alignment between the mouse and human orthologs showing putative PU.1 (blue) and Gfi-1 (red) binding sites in the PU.1 promoter and URE. (B) PU.1 and Gfi-1 compete for overlapping sites within the PU.1 URE. Binding reactions containing either PU.1 and/or Gfi-1 in vitro translation extracts with radiolabelled oligonucleotides were analyzed by EMSA. Unlabelled PU.1 (λB) and Gfi-1 binding site oligonucleotides were used as competitor DNAs. Arrows indicate migration positions of PU.1 or Gfi-1 protein-DNA complexes. (C) ChIP analysis of PU.1 (blue bars) and Gfi-1 (red bars) to the URE in unstimulated and OHT-treated PUER cells. Fold enrichment was assessed by qPCR after normalization to α-actin. Data are representative of three experiments. (D) ChIP-on-chip analysis of PU.1 target promoters in OHT-treated PUER cells. Putative Gfi-1 binding sites were identified in PU.1 target promoters (see Experimental Procedures). The histogram displays the distribution of the various configurations of overlapping presumptive PU.1 and Gfi-1 binding sites in the PU.1 ChIP sequences (blue bars). A randomized set of DNA sequences (see Experimental Procedures) was used to generate a control distribution (red bars). The Gfi-1 binding site matrix is displayed below the frequency distributions. The configurations of the overlapping presumptive PU.1 and Gfi-1 binding sites are tabulated based on the distance (bp) from the center of the Gfi-1 core AATC to the center of the PU.1 core GGAA. PU.1 core sequences that reside either upstream or downstream of the Gfi-1 core sequence are denoted by negative or positive coordinates, respectively.
We performed electrophoretic mobility-shift assays (EMSA) to determine whether PU.1 and Gfi-1 recognize the predicted binding site motifs in the promoter and URE. PU.1 and Gfi-1 bound to their respective motifs in the URE with higher affinity than to the corresponding motifs in the promoter (Figure S6A). Since the Gfi-1 and PU.1 sites overlap within the URE, we tested if the proteins compete for DNA binding. Competition reactions revealed PU.1 and Gfi-1 displace each other when binding to the composite element in the URE (Figure 6B).
To determine if Gfi-1 and PU.1 bind to the URE in a reciprocal manner in vivo, we performed chromatin immunoprecipitation (ChIP) assays using a cell line that expresses an inducible PU.1 protein (PUER) (Walsh et al., 2002). In the absence of tamoxifen (OHT), these cells express Gfi-1 (Laslo et al., 2006) which could be seen to crosslink to the URE (Figure 6C) as well as the PU.1 promoter (Figure S6B). A basal level of crosslinking of PU.1 was also detectable at these regions (Figure 6C and S6B). Upon stimulation with OHT, PU.1 binding at the URE and promoter increased whereas Gfi-1 crosslinking was diminished. Increased PU.1 binding accompanied by the loss of Gfi-1 was also observed at the c-fms promoter (Figure S6C), consistent with recent findings implicating Gfi-1 in the repression of the c-fms gene (Zarebski et al., 2008). Although induction of PU.1 activity resulted in downregulation of Gfi-1, we did not observe reduced Gfi-1 crosslinking to an auto-regulatory site in the Gfi-1 promoter (data not shown, Yucel et al., 2004). Thus loss of Gfi-1 binding at the URE is likely a consequence of displacement by PU.1. Collectively, these data demonstrate that Gfi-1 targets multiple regulatory regions within the PU.1 locus and suggest that it restrains PU.1 expression by interrupting an auto-regulatory loop.
To explore whether the molecular antagonism of PU.1 action by Gfi-1 via apposed binding sites may occur at other loci, we performed ChIP-on-chip assays using a promoter array to identify PU.1 target genes in OHT-induced PUER cells. Bioinformatic analysis (see Experimental Procedures) revealed that 3,170 PU.1 target sequences contained presumptive Gfi-1 binding sites. Of these, 19% contained a putative PU.1 site within 10bp of a Gfi-1 site (Figure S6D). Interestingly, PU.1 and Gfi-1 motifs were seen to overlap in nearly half of this subset of ChIP sequences. As a control, we generated a set of randomized DNA sequences in silico with the same base composition and length as the PU.1 target set. Comparison between PU.1-bound and randomized DNA sequences revealed a statistically significant difference given that only 9% (SD=0.44) of randomized sequences contained presumptive binding sites for the two transcription factors separated by 10bp or less.
Strikingly, the most notable arrangement of presumptive PU.1 and Gfi-1 binding sites consisted of ones in which the core motif of PU.1 (GGAA) and Gfi-1 (AATC) overlapped by a single nucleotide (Figure 6D, see Pim1). The fact that the same configuration was retained as a predominant feature in the randomized sequences, albeit at a significantly lower frequency, led us to consider the possibility that the large frequency of overlapping sites might be explained by compatibilities within the extended binding motifs of PU.1 and Gfi-1. In fact, inspection of the Gfi-1 binding site matrix revealed preferences for specific nucleotides flanking the core base pairs of the Gfi-1 site that potentially encode PU.1 sites (Figure 6D). Detailed analysis of the Gfi-1 binding matrix revealed a number of preferred PU.1 site configurations in relation to the Gfi-1 core, as evidenced in both the PU.1 ChIP-on-chip and randomized DNA analysis (Figure 6D). These analyses suggest that a large number of PU.1 target genes may be repressed via competitive binding or proximal action of Gfi-1.
Ikaros regulates Gfi-1 and constrains PU.1 expression
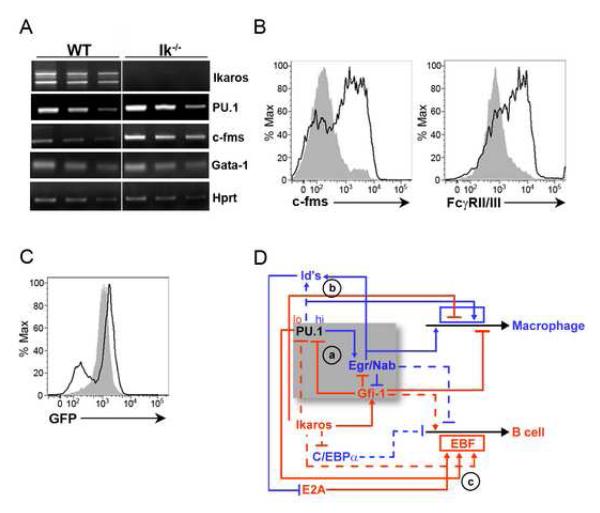
Given that Ikaros is required for B cell development and repression of myeloid potential, we examined whether its loss, like that of Gfi-1, also results in the enhanced expression of PU.1 in MPPs. PU.1 and c-fms transcripts were increased in Ik−/− MPPs (Figure 7A and S7A). In addition, the loss of Ikaros resulted in a significant increase in the expression of both c-fms and FcγRII/III (Figure 7B). The similarities in the myeloid signature observed in Gfi-1−/− and Ik−/− MPPs suggested a potential relationship between Ikaros and Gfi-1 activity in restraining myelopoiesis. Therefore, we sought to determine whether Ikaros activity was required for the expression of Gfi-1 in MPPs. Utilizing a Gfi-1-GFP reporter allele (Yucel et al., 2004), we observed that the loss of Ikaros was associated with the generation of a subset of MPPs expressing lower levels of GFP (Figure 7C). We note that a larger subset of Ik−/− MPPs expressed higher levels of GFP relative to their control counterparts. These cells also expressed higher levels of c-fms (data not shown) and may represent MPPs that are primed to the myeloid lineage (Ng et al., 2009). Bioinformatic analysis revealed several putative Ikaros binding sites in the Gfi-1 locus. Interestingly, two such sites were positioned downstream of the Gfi-1 gene in an ~170bp region that is duplicated (Figure S7B). EMSA and ChIP assays demonstrated that Ikaros bound these sites in vitro and in vivo (Figure S7C and S7D). These data suggest that Ikaros directly promotes Gfi-1 expression in a subset of MPPs.
Figure 7. Ikaros promotes Gfi-1 expression and represses PU.1.
(A) RT-PCR analysis of the indicated transcripts in WT and Ik−/− MPPs. (B) Flow cytometric analysis of c-fms and FcγRII/III expression on WT (gray shaded area) and Ik−/− (black line) MPPs. Data are representative of three independent experiments. (C) Expression of the Gfi-1-GFP knock-in allele in MPPs from Gfi-1+/− (gray shaded area) or Ik−/−Gfi-1+/− (black line) mice by flow cytometry. Data are representative of two independent experiments. (D) A proposed gene regulatory network that dictates myeloid versus B cell fate choice in the context of a MPP. Arrows represent gene activation and barred lines represent gene repression. Hatched lines represent regulatory connections whose molecular basis remains to be elucidated. The shaded region encompasses a core gene regulatory network that is used in a recurring manner to orchestrate innate as well as adaptive immune cell fates.
Discussion
Using PU.1−/− hematopoietic progenitors, we have previously reported that a graded concentration of PU.1 regulates B-lymphocyte versus macrophage cell fate choice (DeKoter and Singh, 2000). Consistent with these findings, it has been reported that lowering the levels of PU.1 in mouse embryonic stem cells using siRNA enhances their B-lineage developmental potential (Zou et al., 2005). However, demonstrating a role for PU.1 dosage in innate versus adaptive cell fate ‘choice’ in vivo has proven to be elusive. Using a sensitized genetic background, a null mutation in the Gfi-1 locus, we show that reducing PU.1 levels via a gene dosage strategy results in enhanced B lymphopoiesis. Importantly, these results establish a role for PU.1 levels in the regulation of myeloid versus B lymphocyte cell fate choice in vivo in the context of a MPP.
Recent studies have revealed that reduced PU.1 expression by removal of a distal PU.1 enhancer resulted in a decrease, rather than an increase, in bone-marrow derived B lineage progeny in vivo (Rosenbauer et al., 2006). In these animals, removal of the distal enhancer reduced PU.1 transcript levels at least 10-fold in MPPs. It may be that reducing PU.1 concentrations to the levels observed in PU.1 enhancer knockout animals is suboptimal for initiating the B cell program in MPPs. It is also possible that impaired B-lymphopoiesis in these knockout mice is a consequence of enhanced generation of neutrophil precursors (Rosenbauer et al., 2004). Lowering the levels of PU.1 in relation to C/EBPα has been shown to promote the generation of neutrophils (Dahl et al., 2003). Furthermore, since PU.1 can negatively regulate the expression of Gfi-1 via the Egr’s (Laslo et al., 2006), Gfi-1 expression is likely to be increased in MPPs of PU.1 enhancer knockout animals. In this context, increased Gfi-1 levels would function in concert with C/EBPα to promote neutrophil development. Nonetheless, our results demonstrate that PU.1 and Gfi-1 have opposing roles in promoting myeloid versus B cell development and establish that these counter acting regulators function in a recurring manner to regulate cell fate choice in the innate as well as the adaptive immune systems.
How does Gfi-1 restrain the expression of PU.1 in MPPs to specify the B cell fate? Previously, Gfi-1 has been shown to interact with the PU.1 protein and this protein-protein interaction has been suggested to inhibit PU.1 mediated transactivation (Dahl et al., 2007). While we cannot rule out the involvement of this protein interaction in antagonism of PU.1 gene expression by Gfi-1, we show that Gfi-1 directly competes for PU.1 binding in vitro and in vivo to auto-regulatory sites in the PU.1 locus. We therefore propose that Gfi-1 represses PU.1 gene activation by binding to DNA sites in the locus and disrupting a PU.1-dependent auto-regulatory loop. Importantly, increased expression of PU.1 can induce Egr expression and the latter represses Gfi-1. Finally, Gfi-1 has been shown to bind to its own promoter and function in a negative auto-regulatory loop (Yucel et al., 2004). Thus, the two auto-regulatory feedback loops and the cross-antagonism between PU.1 and Gfi-1 may generate alternate gene expression states within this network that are driven by either higher PU.1 and Egr activity or higher Gfi-1 activity.
Gfi-1−/− MPPs are poised to differentiate along the myeloid lineage at the expense of the B lineage as a consequence of mis-expression of PU.1 and other myeloid genes. In addition to the PU.1 locus, Gfi-1 targets many myeloid genes including c-fms (Zarebski et al., 2008). Accordingly, the absence of Gfi-1 in MPPs may augment the expression of PU.1 target genes, such as c-fms, via de-repression. Our analysis of PU.1 target sequences has revealed an intriguing feature; a large number of PU.1 core motifs are embedded within the extended binding matrix for Gfi-1. Therefore, robust activation of such PU.1 target genes would be dependent on displacement of Gfi-1 repressor complexes with PU.1 activator complexes at these composite elements, and vice versa. We note that the predicted composite sites are found in a varied set of genes, including: growth factors and their receptors, lineage-determining transcription factors and histone modifying enzymes. We envision that the antagonistic regulation of a large battery of genes by PU.1 and Gfi-1 would have evolved more readily by selecting for favorable composite binding sites for the two factors rather than independent selective events that generate two separated sites. This may represent a general strategy for rapidly evolving counteracting regulatory modules by utilizing transcription factors whose individual binding motifs are compatible with the generation of overlapping composite elements.
Based on the aforementioned data, we propose a transcriptional regulatory network that appears to function in a recurring manner to govern cell fate choice in the immune system (Figure 7D). In MPPs, PU.1 is proposed to function in a graded manner to regulate B-lymphoid versus macrophage cell fates. Higher levels of PU.1 represent a primary input to the macrophage developmental program and can induce the Egr’s (module a). These regulators activate both myeloid gene expression and the Id genes which inhibit E2A activity (module b) and consequently the priming of B-lymphoid developmental potential in MPPs (Dias et al., 2008). Lower levels of PU.1 along with Ikaros and E2A function as primary inputs to activate the B-lymphoid program. These factors induce the B cell fate determinant EBF (module c) (Laslo et al., 2008). In our model, the reduced levels of PU.1 that promote B-lymphoid development are achieved by Ikaros, in part, through the induction of Gfi-1. Ikaros and Gfi-1 constrain the levels of PU.1 while promoting the expression of B-lymphoid genes. We note that Ikaros has been implicated in repression of the myeloid determinant C/EBPα (Reynaud et al., 2008; Ng et al., 2009). This repressive activity of Ikaros is likely to be important for B cell development, as C/EBPα can reprogram committed B cells into macrophages (Laoisa et al., 2006a).
An additional consequence of Ikaros and Gfi-1-mediated repression of the myeloid program in MPPs could be to promote T cell developmental potential. Along these lines, Gfi-1 activity appears to be critical for the generation of the earliest T-lineage progenitors in the thymus (Yucel et al., 2003). It is noteworthy that the loss of Ikaros and Gfi-1 is associated with the increased expression of Id genes (Yucel et al., 2003; Ng et al., 2009). These genes encode proteins that inhibit E2A family transcription factors which are required for early B and T cell development (Quong et al., 2002). Conversely, we have demonstrated that high levels of PU.1 induce Id2 during macrophage differentiation (Laslo et al., 2006). Similarly, increasing PU.1 levels in fetal thymic progenitors induces Id2 and aspects of the myeloid program (Franco et al., 2006). These findings suggest a general mechanism by which Gfi-1 and PU.1 could function in a counteracting and recurring manner to promote B- and T-lymphoid versus myeloid cell fates via the antagonistic regulation of Id genes. Consistent with our model, Egr1 has been identified as a positive regulator of Id gene expression (Quong et al., 2002). Loss of Egr1 results in an increase in the absolute numbers of the earliest T-lineage progenitors (Bettini et al., 2002). We note that multiple defects observed in Gfi-1−/− mice during T cell development, including reduced cellularity and impaired progression through the double negative and double positive stages of development are partially rescued upon the loss of Egr1 in vivo (C.J.S. and H.S. unpublished data). These findings are also in keeping with our proposal that the Egr’s and Gfi-1 comprise a counteracting regulatory module that directs cell fate options or developmental transitions in multiple cellular contexts (Laslo et al., 2006). Based on the above results, the Id’s would represent an additional node within this module whose expression would be counteracted by Gfi-1 to promote lymphopoiesis or induced by PU.1 and the Egr’s to inhibit B and T cell potential.
Our proposed core transcriptional network for lymphoid versus myeloid cell fate determination is derived from one that regulates macrophage versus neutrophil cell fate choice (Laslo et al., 2006). In the former, Ikaros has replaced C/EBPα as a pivotal primary determinant, but the remaining circuit is conserved both with respect to the nature of the regulatory molecules and their connectivity. The modified network architecture has two important evolutionary implications for the emergence of the adaptive immune system from an innate primordial precursor. First, it suggests that the conserved core network comprising of the counteracting PU.1/Egr and Gfi-1 modules predates the origin of lymphocytes and was co-opted by Ikaros. Second, that Ikaros has played a key role in the evolutionary emergence of adaptive immune cells (i.e., lymphocytes). Ikaros manifests two major regulatory functions that are consistent with this proposition: 1) it is able to restrain myeloid developmental potential by antagonizing expression of PU.1 and other myeloid genes and 2) it directly activates Rag gene expression and promotes the recombination of antigen receptor gene segments, a hallmark of adaptive immune cells (Reynaud et al., 2008). Molecular phylogenetic analysis of our network components and their connectivity will be necessary to test this evolutionary proposition.
Experimental Procedures
Mice
PU.1+/− (Scott et al., 1994), Egr1+/− (Swiatek and Gridley, 1993), Egr2+/fl (Taillebourg et al., 2002), Ik+/− (Wang et al., 1996) and Gfi-1+/− (Yucel et al., 2004) mice have been previously described. The Gfi-1 mutant allele represents a GFP knock-in and the mutant mice are phenotypically indistinguishable from a previously described Gfi-1 knockout animal (Karsunky et al., 2002). Therefore, Gfi-1GFP/GFP mice are referred to Gfi-1−/− in these studies. Wildtype C57BL/6 mice were purchased from The Jackson Laboratory. Mice were maintained in pathogen-free conditions in accordance with guidelines approved by the Institutional Animal Care and Use Committees of the University of Chicago.
Flow Cytometry and Cell Culture
Bone marrow single cell suspensions washed in PBS containing 5mM EDTA and 0.5% BSA were analyzed by flow cytometry using an LSRII (Becton Dickinson) and flowjo software. The following antibodies conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), PerCPCy5.5, phycoerythrin-Cy7 (PECy7), and allophycocyanin (APC) were used against cell surface molecules outlined in Supplementary Table 5. DAPI was used to discriminate between viable and dead cells. MPPs were isolated and cultured as previously described (Medina et al., 2004) and analyzed for the presence of Mac1+ and CD19+ cells between days 7-12 by flow cytometry.
Retroviral Transduction
WT MPPs were isolated as described above and transduced by co-culture with GFP (MIGR1), Gfi-1 (MIGR1-Gfi-1) or ΔEgr2 (MIGR1-ΔEgr) as previously described (DeKoter and Singh, 2000). After 2 days, GFP+ transductants were sorted and cultured on OP9 stroma in previously described B-lineage conditions (Medina et al., 2004) and analyzed for the presence of Mac1+ and CD19+ cells by flow cytometry 7 days post-sort.
Plat-E packaging cells were transiently transfected with shRNA retroviral constructs targeting luciferase (MSCV-shLuciferase) or PU.1 (MSCV-shPU.1) with Fugene 6 reagent (Roche). WT or Gfi-1−/− MPPs were isolated as described above and transduced as previously described (Reynaud et al., 2008) and analyzed for the presence of Mac1+ and CD19+ cells by flow cytometry 10 days post-sort. shRNA targeting sequences are available upon request.
Gene Expression Analysis
RNA was isolated with TRIzol reagent (Invitrogen) and reverse-transcribed with First-strand cDNA synthesis kit (GE Healthcare) using manufacturers instructions. Reverse-transcription products were serially diluted and amplified by PCR for semiquantitative analyses. Brilliant SYBR Green was used for quantitative PCR analyses on a Mx4000 system (Stratagene). Expression was normalized relative to the expression of Hprt. Primer sequences used for semiquantitative and quantitative PCR analyses are listed in Supplementary Tables 1 and 2.
Colony-forming assays
Clonogenic assays were performed by sorting ~300 MPPs from WT or Gfi-1−/− mice in 3ml of Methocult M3434 (Stem Cell Technologies) supplemented with 25ng/ml of Flt3 ligand (R&D Systems). ~100 MPPs were plated in duplicate in 35 mm petri dishes. After 8 days in culture, individual colonies were counted, subjected to cytospin and analyzed for the presence of erythroid and/or myeloid colonies by Wright staining.
EMSA
PU.1 and Gfi-1 proteins were generated using the TNT coupled reticulocyte lysate system (Promega). IVT extracts were incubated for 30 min. at RT with α-32P-labeled double-stranded oligonucleotides representing an optimal Gfi-1 (Gfi-1 consensus), a high affinity PU.1 (λB) binding site or sites in the PU.1 promoter and URE in a final volume of 20μl of binding reaction buffer (10mM Tris pH7.5, 50mM NaCl, 3% Ficoll, 1 mM EDTA, 1mM DTT, and 1μg/ml of polydI:dC). Ikaros binding reactions were performed as previously described (Reynaud et al., 2008). EMSA probes are listed in Supplementary Table 4.
ChIP
ChIP assays were performed in PUER cells treated with tamoxifen for 24 or 96 hours as previously described (Laslo et al., 2006) or EBF−/− cells (Pongubala et al., 2008) with some modifications. Briefly, 1 × 107 cells were crosslinked with 1% paraformaldehyde, sonicated, precleared, and incubated with 2 μg of α-IgG (sc-2027; Santa Cruz), α-PU.1 (sc-352; Santa Cruz), α-Gfi-1 (sc-8558; Santa Cruz), pre-immune or α-Ikaros antiserum (Smale lab). Complexes were washed with low and high salt buffers, eluted, reverse-crosslinked and the DNA was precipitated. Immunoprecipitated DNA sequences were analyzed by qPCR (primer sequences used for ChIP analyses are listed in Supplementary Table 3). For ChIP-on-chip, chromatin DNA samples were amplified by ligation-mediated PCR and hybridized on a MM8 RefSeq promoter chip (NimbleGen Systems, Inc.).
Computational Analysis of PU.1 and Gfi-1 Binding Sites
Genomic coordinates for all fragments bound by PU.1 with high confidence as determined by NimbleScan (FDR score < 0.2) were extended by 500bp upstream and downstream and these sequences were retrieved. MotifLocator was used to analyze all ChIP sequences to predict PU.1 (M01172) and Gfi1 (M00250) binding sites of quality _t_>0.85. Algorithms were developed in Perl to determine the arrangement and distribution of PU.1 and Gfi-1 binding sites. Randomized sets of sequences were generated by matching the length and base composition of the DNA sequences obtained from the ChIP-on-chip data. Statistical analyses were based on 100 independent randomized runs.
Supplementary Material
01
Acknowledgements
We are grateful to H. Schjerven and S. Smale (U.C.L.A) for providing α-Ikaros serum and valuable technical advice. We thank D. Leclerc and members of the U of C Immunology Applications Core Facility for performing cell sorting. We thank T. Möröy (ICRM), P. Charnay (INSERM) and S. Winandy (Northwestern Univ.) for kindly providing Gfi-1+/−, Egr2+/fl and Ik+/− mice, respectively. E.P. was a trainee of the Program in Physical and Chemical Biology (T90 DK070076). H.S. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- Bettini M, Xi H, Milbrandt J, Kersh GJ. Thymocyte development in early growth response gene 1-deficient mice. J Immunol. 2002;169:1713–1720. doi: 10.4049/jimmunol.169.4.1713. [DOI] [PubMed] [Google Scholar]
- Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J Biol. Chem. 2007;282:6473–6483. doi: 10.1074/jbc.M607613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat. Imm. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science (New York, NY. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Walsh JC, Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO. 1998;17:4456–4468. doi: 10.1093/emboj/17.15.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc. Nat. Acad. Sci. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Duhrsen U, Moroy T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Gen. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- Krysinska H, Hoogenkamp M, Ingram R, Wilson N, Tagoh H, Laslo P, Singh H, Bonifer C. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol. Cell. Biol. 2007;27:878–887. doi: 10.1128/MCB.01915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid myeloid lineage diversification. Ann. Rev. Imm. 2006a;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006b;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Laslo P, Pongubala JM, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Sem. Imm. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp. Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, Akashi K, Moreau-Gachelin F, Li Y, Zhang P, et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 2005;25:2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Imm. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Ann. Rev. Imm. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Imm. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Gen. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Gen. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. Molecular genetics of T cell development. Ann. Rev. Imm. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science (New York, NY. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Singh H. Gene targeting reveals a hierarchy of transcription factors regulating specification of lymphoid cell fates. Curr. Opin. Imm. 1996;8:160–165. doi: 10.1016/s0952-7915(96)80053-7. [DOI] [PubMed] [Google Scholar]
- Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc. Nat. Acad. Sci. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling-Sun S, Dade J, Nutt SL, DeKoter RP, Camargo FD. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat. Imm. 2009;10:289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Gen. Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Taillebourg E, Buart S, Charnay P. Conditional, floxed allele of the Krox20 gene. Genesis. 2002;32:112–113. doi: 10.1002/gene.10062. [DOI] [PubMed] [Google Scholar]
- Thal MA, Carvalho TL, He T, Kim HG, Gao H, Hagman J, Klug CA. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc. Nat. Acad. Sci. 2009;106:552–557. doi: 10.1073/pnas.0802550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Imm. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp. Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel R, Kosan C, Heyd F, Moroy T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol. Chem. 2004;279:40906–40917. doi: 10.1074/jbc.M400808200. [DOI] [PubMed] [Google Scholar]
- Zarebski A, Velu CS, Baktula AM, Bourdeau T, Horman SR, Basu S, Bertolone SJ, Horwitz M, Hildeman DA, Trent JO, et al. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28:370–380. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GM, Chen JJ, Yoder MC, Wu W, Rowley JD. Knockdown of Pu.1 by small interfering RNA in CD34+ embryoid body cells derived from mouse ES cells turns cell fate determination to pro-B cells. Proc. Nat. Acad. Sci. 2005;102:13236–13241. doi: 10.1073/pnas.0506218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01