Quantifying memory CD8 T cells reveals regionalization of immunosurveillance (original) (raw)
. Author manuscript; available in PMC: 2016 May 7.
Summary
Memory CD8 T cells protect against intracellular pathogens by scanning host cell surfaces, thus infection detection rates depend on memory cell number and distribution. Population analyses rely on isolation from whole organs and interpretation is predicated on presumptions of near complete cell recovery. Paradigmatically, memory is parsed into central, effector, and resident subsets, ostensibly defined by immunosurveillance patterns, but in practice identified by phenotypic markers. Because isolation methods ultimately inform models of memory T cell differentiation, protection, and vaccine translation, we tested their validity via parabiosis and quantitative immunofluorescence microscopy of a mouse memory CD8 T cell population. We report three major findings: lymphocyte isolation fails to recover most cells and biases against certain subsets, residents greatly outnumber recirculating cells within nonlymphoid tissues, and memory subset homing to inflammation does not conform to previously hypothesized migration patterns. These results indicate that most host cells are surveyed for reinfection by segregated residents rather than by recirculating cells that migrate throughout the blood and body.
Introduction
A cardinal feature of the vertebrate adaptive immune system is the retention of a memory of past infections that enhances protective immunity in the event of reinfection. CD8 T cells are a principle component of this process, and protect against those pathogens that invade intracellular compartments. Mechanistically, vertebrates maintain memory CD8 T cells that scan MHC I on the surface of host cells for the presence of pathogen-derived peptides. Recognition triggers infection control. The efficiency achieved by this immunosurveillance depends upon the memory CD8 T cell population 1) magnitude relative to host cells and 2) location.
Quantification of the immune response is essential for our understanding of protective immunity and for evaluating vaccines. Limiting dilution assays suggested that pathogen-specific CD8 T cells were exceedingly rare among responding cells. However, technical innovations, such as the development of MHC I tetramers (Altman et al., 1996), revealed that antigen specific CD8 T cell responses were 10-100 fold bigger than initially thought, precipitating a substantial revision in conceptualization of the immune response (Murali-Krishna et al., 1998).
Memory CD8 T cells are present within secondary lymphoid organs (SLO), blood, and the rest of the organism (nonlymphoid tissues, NLT, as well as primary lymphoid organs such as thymus and bone marrow). Landmark work, based on analysis of human blood, proposed that memory CD8 T cells could be parsed into two subsets based on their patterns of immunosurveillance. Central memory T cells (TCM), defined by expression of lymph node homing molecules, putatively limit surveillance to SLO and are specialized for longevity and proliferation upon reinfection. Effector memory T cells (TEM), defined by the absence of lymph node homing molecules, were thought to recirculate between blood, NLT, and lymph, thus surveying body surfaces and visceral organs that are often the initial portals of reinfection (Sallusto et al., 1999).
However, the (TCM/TEM model failed to capture the true complexity of memory T cell diversity. It recently became clear that a third subset, termed tissue resident memory T cells (TRM), resides in NLT without recirculating (Masopust and Schenkel, 2013; Mueller et al., 2013). Shortly after activation in SLO, this population seeds tissues, then differentiates in response to local environmental cues to adopt unique lineage specific signatures (Casey et al., 2012, Mackay et al., 2013; Masopust et al., 2006). Importantly, the presence of TRM at NLT sites of reinfection can accelerate pathogen elimination (Gebhardt et al., 2009; Jiang et al., 2012; Teijaro et al., 2011; Wu et al., 2014). Fundamentally, TRM are defined by migration: they remain confined to one tissue without leaving and re-entering. Practically, cell migration patterns are laborious or impractical to define in animal models or humans, so phenotypic surface markers have been substituted. The markers CD 103 and CD69 are used to infer TRM status, whereas the absence of both CD62L and CD69 expression defines NLT recirculating TEM (Farber et al., 2014; Masopust and Schenkel, 2013). However, the fidelity of these markers has not been validated.
The emergence of TRM has complicated the longstanding paradigm of T cell-mediated immunosurveillance. It is no longer clear to what degree CD8+ TEM recirculate through NLT, and how immunological memories are apportioned between TRM, TEM, and TCM as each subset has not been quantified throughout the host. Previous identification of significant recirculation through major NLT (Klonowski et al., 2004) requires reassessment in light of recent discoveries of bloodborne populations contaminating even perfused tissues (Anderson et al., 2014). Moreover, while quantitative analyses typically depend on ex vivo isolation to determine memory CD8 T cell subset and phenotype, the accuracy of this approach has not been validated (Peaudecerf and Rocha, 2011; Selby et al., 1984). To address these gaps in the field, we performed a stringent and comprehensive quantitative analysis using migration properties to identify TRM, TEM, and TCM populations. Our findings redress fundamental presumptions that inform models of immunosurveillance, T cell subsets, and protective immunity.
Results
Isolations underestimate total memory CD8 T cells and distort distribution
Memory CD8 T cells are broadly distributed throughout the host organism, but the overall magnitude and anatomic apportionment of this population remain unclear and controversial (Ganusov and De Boer, 2007; Masopust et al., 2001; Peaudecerf and Rocha, 2011; Reinhardt et al., 2001, Rocha et al., 1991). To address this gap, we enumerated a single trackable memory CD8 T cell population established by a well-studied infection model in mice. To this end, we transferred naїve lymphocytic choriomeningitis virus (LCMV)-specific Thy1.1 + P14 transgenic CD8 T cells into naive C57BI/6J mice, which were then infected with LCMV (Armstrong strain). Animals were sacrificed 120-150 days later. These mice, referred to as P14 immune chimeras, were injected with α-CD8α antibody (Ab) i.v. prior to sacrifice. The intravascular injection of α-CD8α antibody was used in each experiment to distinguish i.v. Ab+ cells in vascular contiguous compartments (e.g., peripheral blood, spleen red pulp, RP, liver sinusoids, and lung capillaries) from i.v. Ab- CD8 T cells in the stroma and parenchyma of NLT and SLO (Anderson et al., 2014; Galkina et al., 2005). Cells were isolated from tissues by ex vivo dissociation (see methods), and then analyzed by flow cytometry.
Consistent with previous reports, we isolated ∼6,000 P14 CD8 T cells from the female reproductive tract (FRT) (Nakanishi et al., 2009; Suvas et al., 2007). We also performed immunohistochemistry, taking advantage of the fact that the P14 LCMV system allows for identification of LCMV-specific cells in tissue sections via α-Thy1.1 Ab. Because ∼240 7μm coronal sections could be acquired from the FRT, flow cytometry data predicted ∼25 P14 in a single section. But, we counted ∼1750 P14 per tissue section, suggesting discordance between flow cytometry and immunohistochemistry (data not shown).
For this reason, we developed an image-based quantitative immunofluorescence microscopy (QIM) strategy to compare the recovery of P14 memory CD8 T cells to what was actually present within the tissue (Figure 1A). For QIM, organ volumes of age-matched mice were determined by displacement. These values were consistent with available estimates from previous reports using a variety of methods (Doctor et al., 2010; Nutter et al., 1980; Scheller et al., 1994). Organs from P14 immune chimeras were also frozen, sectioned and stained. Whole sections or large representative regions were imaged by immunofluorescence microscopy (see methods). Image size and section thickness were used to determine the portion of the whole organ represented in each image. This factor was used to extrapolate enumerations from large individual images to whole organs. Cell enumerations were then multiplied by 11/19 to correct for those cells that would be counted twice because they straddle two sections (Figure 1A&B). Importantly, the total number of nucleated cells in a given organ as determined by QIM was similar to that estimated by whole organ DNA content, assuming 6pg DNA per diploid cell (Anjos Pires M et al., 2001), thus independently validating QIM accuracy (Figure 1B and Table 1).
FIGURE 1. Isolations underestimate total memory CD8 T cells and distort distribution.
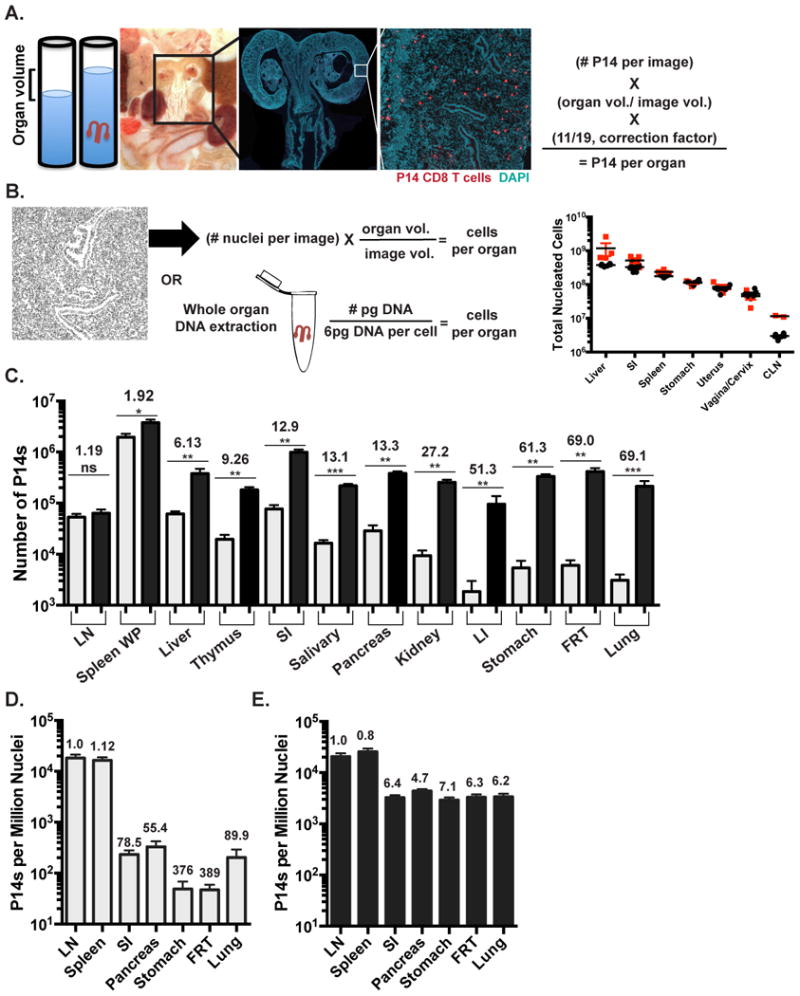
(A&B) Quantitative Immunofluorescence Microscopy (QIM) methodology. (A) Organ volumes were determined by displacement. Tissue sections were stained for Thy1.1 (red) and DAPI (teal) to identify memory P14 CD8 T cells and nucleated cells 120-150 days after LCMV infection of C57BI/6J mice. P14 counts per section were extrapolated to total organ volume and corrected to eliminate double counting. Whole FRT image scale bar=2000μm, cropped close up of FRT image scale bar= 250μm.(B) Total DAPI+ nucleated cells by QIM were extrapolated to total organ volume (black circles) and validated independently by DNA extraction (red squares), n=4. (C) Comparison of α-CD8α i.V.- P14 per tissue determined by cell isolation and flow cytometry (grey) or QIM (black). Total P14 frequency determined by (D) flow cytometry or (E) QIM relative to DAPI+ nucleated cells per organ as determined by QIM. Fold differences shown are relative to LN. n≥ 6, graphs show mean and SEM. *p<0.05, **p<0.01, ***p<0.001, Mann-Whitney-Wilcoxon test, (See also Figure S1, S2).
Table I. Enumeration of memory P14 CD8 T cells by cell isolation and flow cytometry and QIM.
5×104 naїve Thy1.1+ P14 CD8 T cells were transferred to C57BI/6J mice, which were infected one day later with 2×105 LCMV Armstrong i.p. 120-150 days later, 3min prior to sacrifice mice were injected i.v. with α-CD8α antibody to discriminate the blood and marginated pool (i.v.+) from parenchymal P14 (i.v.-). Bold values indicate the average number of P14 or total nucleated cells per tissue derived from cell isolation and flow cytometry or QIM. Kidney accounts for both kidneys, salivary gland reports for both lobes, uterus includes both uterine horns, and mandibular lymph node enumerates a Single unpaired lymph node. Peripheral blood enumeration is extrapolated to 1.74mL of blood, based on average body weight of mice used in this study. Data from 6 or more mice. Underlined italic numbers indicate number of nucleated cells (± SD) as determined by DNA extraction. BD= below detection, NA= not available, LP= lamina propria, IEL= intraepithelial lymphocytes, M.E.= muscularis externa, ILF= isolated lymphoid follicle.
| Tissue | Fold Difference | Flow Cytometry | QIM | QIM & DNA | |||
|---|---|---|---|---|---|---|---|
| QIM/Flow | Total P14 ×104±SD ×104 | % of total P14 i.v.- | Total P14 ×104±SD ×104 | % of total P14 i.v.- | P14 ×103±SD ×103 Per 106 nuclei | Total Nucleated Cells ×106±SD×106 | |
| Spleen (White Pulp) | 1.92 | 195 ±79.7 | 66.6 | 375 ±142 | 82.1 | 25.7 ±9.41 | 178 ±10.7_237_ ±30.2 |
| Spleen i.v.+ (Red Pulp) | 0.84 | 93.2 ±32.7 | 78.5 ±20.5 | ||||
| Mandibular lymph node | 1.19 | 5.31 ±2.00 | 97.7 | 6.32 ±2.83 | BD | 20.7 ±7.74 | 2.98 ±0.48_11.5_ ±0.705 |
| Thymus | 9.26 | 1.96 ±1.11 | 96.6 | 99.7% IV-= 18.1 ±5.61 | 99.7 | 2.86 ±0.32 | 77.3 ±25.7 |
| Liver | 6.13 | 6.12 ±2.05 | 14.8 | 37.5 ±22.0 | 16.9 | 5.42 ±1.17 | 378 ±29.9_1180_±973 |
| Liver i.v.+ | 4.58 | 37.1 ±75.9 | 170 ±36.1 | ||||
| Lung | 69.1 | 0.31 ±0.24 | 9.06 | 21.3±15.3 | 25 | 3.36 ±1.27 | 282 ±45.2 |
| Lung i.v.+ | 13.8 | 5.43 ±6.42 | 75.2±53.7 | ||||
| Kidney | 27.2 | 0.945 ±0.64 | 46.3 | 25.4 ±8.27 | 83.1 | 1.99 ±0.697 | 157 ±25.0 |
| Kidney i.v.+ | 5.02 | 1.03 ±0.76 | 5.17 ±1.95 | ||||
| Pancreas | 13.3 | 2.87 ±2.11 | 94.8 | 99.7% IV-= 37.9 ±9.19 | 99.7 | 4.37 ±0.95 | 86.9 ±11.5 |
| Salivary (serous) | 13.1 | 1.65 ±0.59 | 99.8 | 21.6 ±5.61 | BD | 10.3 ±2.64 | 21.2 ±2.58 |
| Salivary (mucous) | NA | NA | NA | 7.81 ±2.35 | BD | 5.43 ±1.41 | 14.4 ±1.42 |
| Uterus | 69.0 | 0.603 ±0.41 | 90.2 | 25.6 ±4.61 | BD | 3.51 ±0.958 | 75.7 ±15.8_85.4_ ±26.2 |
| Cervix/Vagina | 16.0 ±8.39 | BD | 2.95 ±1.02 | 52.4 ±14.2_45.1_ ±19.8 | |||
| SI (IEL) | 6.10 | 3.81 ±2.02 | 99.8 | 23.2 ±12.7 | BD | 3.24 ±0.91 | 328 ±92.9_517_ ±176 |
| SI (LP) | 18.6 | 4.06 ±1.70 | 99.1 | 74.3 ± 22.5 | BD | ||
| SI (muscle) | 1.04 ±1.05 | BD | |||||
| LI (IEL) | 41.3 | 0.034 ±0.018 | 84.10 | 1.39 ±0.75 | BD | 0.81 ±0.04 | 122 ±12.3 |
| LI (LP) | 68.1 | 0.12 ±0.073 | 82.46 | 7.59 ±3.65 | BD | ||
| LI (ILF) | 0.51 ±0.63 | BD | |||||
| Stomach(IEL) | 17.5 | 0.35E ±0.45 | 91.9 | 6.17 ±2.27 | BD | 2.91 ±0.92 | 118 ±14.9_113_ ±17.4 |
| Stomach (LP) | 122 | 0.22 ±0.157 | 95.1 | 20.0 ±7.22 | BD | ||
| Stomach (Submucosa) Stomach(M.E) | 3.05 ± 0.92 | BD | |||||
| 3.91 ±1.12 | BD | ||||||
| Peripheral Blood | NA | 13.4 ±6.33 | 100 | NA | NA | NA | NA |
QIM revealed that lymphocyte isolation from the FRT was inefficient, thus we tested whether isolation efficiencies varied among tissues by comparing these methods in many organs (Figure 1C). Many mucosal sites, including the stomach, lung, large intestine (LI) and FRT, contained 50-70 fold more α-CD8α i.v. Ab- memory P14 CD8 T cells when evaluated by QIM as compared to cell isolation methods (Figure 1C & Table I). While the density of memory P14 cells in skin was too low to evaluate (data not shown), QIM of other NLTs resulted in 6-27 fold higher estimates of P14s. Examination of SLOs, including the white pulp (WP) of the spleen and the mandibular lymph node (LN), resulted in the most efficient isolations with less than 2-fold differences observed between the two methods. These results demonstrate a wide discrepancy between cell isolation and QIM, suggesting that the most common method of enumeration (isolation) significantly underestimates the size of the memory CD8 T cell pool in NLT. Similar findings were observed when enumerating endogenous LCMV-specific memory CD8 T cells (without P14 transfers) in mice via in situ MHC I tetramer staining (Supplemental Figure 1A) and also when analyzing CD8β+T cells in human cervix (Supplemental Figure 1B).
As memory CD8 T cells patrol and survey all nucleated cells for the presence of infection, we represented the total number of memory P14 CD8 T cells as determined by cell isolation (Figure 1D) or QIM (Figure 1E) per nucleated host cell (as determined by QIM) in LN, spleen, small intestine (SI), pancreas, stomach, FRT and lung. Based on isolation methods, memory P14 CD8 T cells were calculated to be ∼50-400 fold rarer in tissues than SLOs. QIM enumeration significantly altered this perceived immunosurveillance ratio, and revealed that the density of sentinel memory CD8 T cells in NLT was within 8-fold of SLOs. This refinement in perspective could help explain how memory CD8 T cells within NLT can be sufficiently abundant to be first responders against anamnestic infections (Masopust and Schenkel, 2013; Mueller et al., 2013).
Isolation efficiency is biased by tissue compartment and cell phenotype
Because cell isolation methods failed to capture most cells from NLT, we asked whether isolation efficiency varied among memory CD8 T cells with different phenotypes or between compartments within organs, thus further distorting the representation of the memory CD8 T cell population composition and location. Using intravascular α-CD8α Ab, we found that the blood and marginated pool (BMP) of lymphocytes (i.v. Ab+) within kidney and lung were more readily isolated than those within the tissue (i.v. Ab-) (Figure 2A&B). This was also true of splenic RP (i.v. Ab+) compared to splenic WP (i.v. Ab-) (Table I).
FIGURE 2. Isolation efficiency is biased by tissue compartment and cell phenotype.
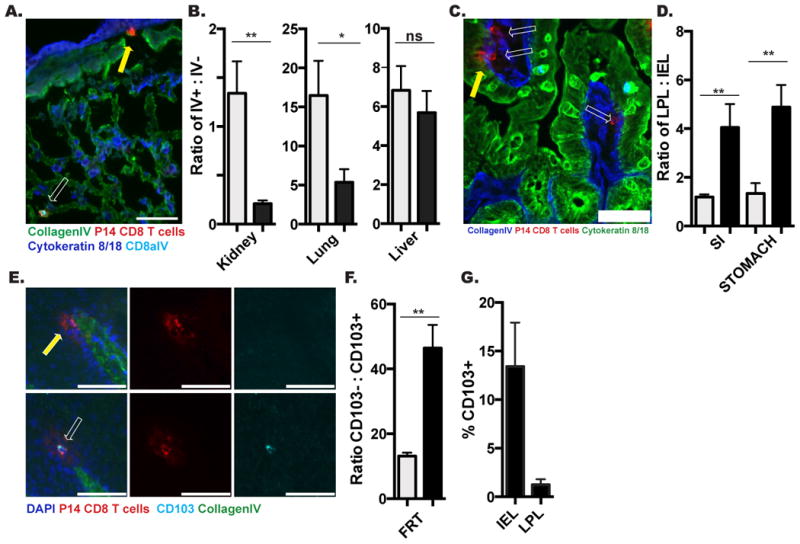
P14 immune chimeras were analyzed 120-150 days after LCMV infection. (A) Representative image of CD8α i.v. Ab+ (white arrow) or CD8α i.v. Ab- (yellow arrow) P14 CD8 T cells in lung. CD8α i.v. Ab (teal), Thy1.1 + P14 (red), Collagen IV (green), Cytokeratin 8/18 (Blue), scale bar= 50μm. (B) Ratio of i.v. Ab+ to i.v. Ab- P14s by flow cytometry (grey) and QIM (black) methodology. (C) Representative image of P14 CD8 T cell in small intestine epithelium (IEL, yellow arrow) and lamina propria (LPL, white arrows). Thy1.1+ P14 (red), Collagen IV (blue), Cytokeratin 8/18 (Green), scale bar= 50μm. (D) Ratio of LPL to IEL P14 by flow cytometry (grey) and QIM (black). (E) Representative image of CD103- (top panels) and CD103+ (bottom panels) P14 CD8 T cells in vaginal epithelium. CD103 (teal). Thy1.1+ P14 (red), Collagen IV (green), DAPI (blue), scale bar = 50μm. (F) Ratio of CD103- to CD103+ P14s by flow cytometry (grey) and QIM (black) in FRT. (G) Percent of vaginal IEL or LPL P14 expressing CD013, determined by QIM. n≥6, graphs show mean and SEM. *p<0.05, **p<0.01, ***p<0.001, Mann-Whitney-Wilcoxon test, (See also Figure S2).
We next investigated if lymphocyte extraction efficiency differed between histologically distinct mucosal compartments. To this end, we separated analyses of memory CD8 T cells isolated or imaged from stomach and SI into fractions localized above the basement membrane (intraepithelial lymphocytes, IEL) or cells contained within the collagen matrix subjacent to the epithelium (lamina propria, LP lymphocytes) (Figure 2C). As shown in figure 2D and Table I, P14 memory CD8 T cells are more efficiently recovered from epithelium than the lamina propria.
We next examined whether lymphocyte isolation misrepresented the proportion of mucosal memory CD8 T cell subsets as defined by phenotype. We focused on the FRT because it contains both CD103+ and CD103- memory P14 CD8 T cells (Figure 2E), and CD103 is one marker used to define TRM. As shown in figure 2F, cell isolation from the FRT over-represents the proportion of P14 memory CD8 T cells that express CD103. This bias may also have an anatomic basis (as in Figure 2D) as CD103+ cells are enriched within epithelium relative to lamina propria (Figure 2G). Taken together, these results indicate that lymphocyte isolation from NLT misrepresents memory CD8 T cell distributions by location and phenotype.
Most memory CD8 T cells in NLT are TRM
A broad and accurate accounting of the anatomic distribution of a memory CD8 T cell population, delineated into resident (TRM) vs. recirculating (TEM and TCM) subsets, has not previously been performed. Moreover, since the identification of TRM as a distinct lineage (previously TRM were conflated with recirculating TEM), it remains unclear what contribution each population makes to the overall NLT memory T cell pool, and how these populations compare numerically with memory T cells positioned within SLOs. We first interrogated this issue by quantifying the proportion of memory CD8 T cells that were resident after LCMV infection. The vasculature of P14 immune chimeras (90 days after infection, generated as in Figure 1) was conjoined to that of naive mice via parabiosis surgery. Thirty days later we tested whether memory P14 CD8 T cells equilibrated between immune and naive parabiont organs, or whether disequilibrium was maintained which indicates residence (Figure 3A). As preliminary evidence indicated that flow cytometry preferentially underestimated TRM as compared to recirculating TEM (data not shown), we utilized the more precise QIM approach for this analysis.
FIGURE 3. The majority of memory CD8 T cells in NLT are TRM.
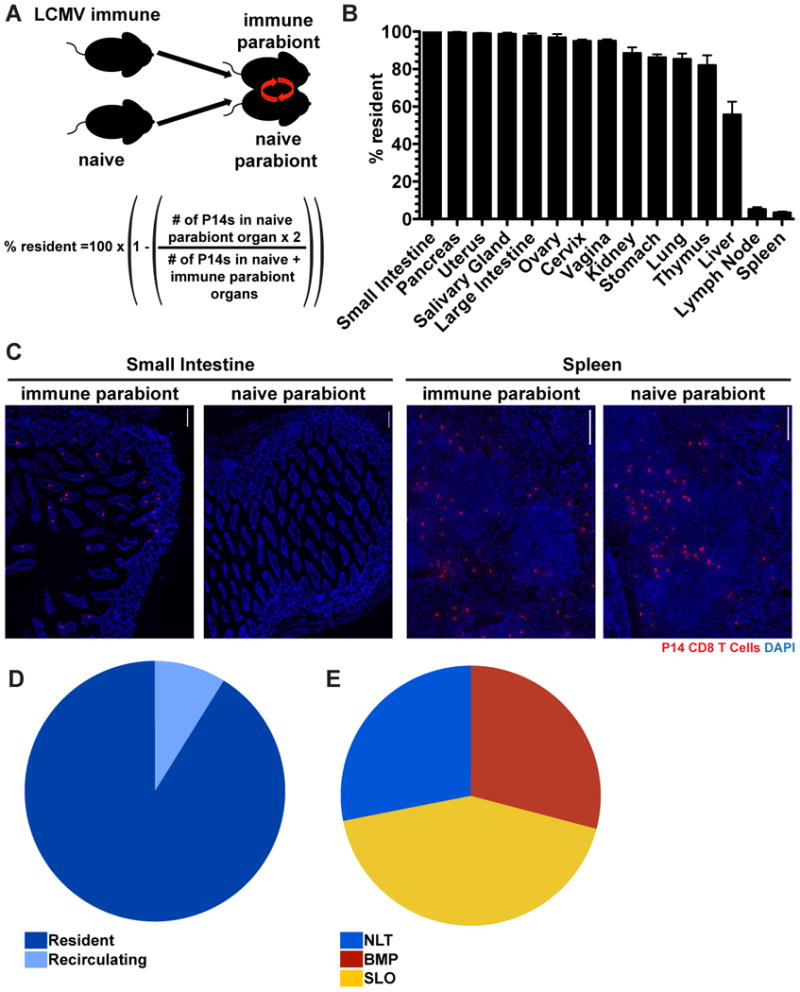
(A) Ninety days after infection with LCMV Armstrong, P14 immune chimeras were conjoined to naive C57BL/6 mice using parabiosis. Thirty days after parabiosis surgery (B) the fraction of resident memory P14 CD8 T cells were calculated for the indicated tissues. n=3, representative of 9 mouse pairs from 3 independent experiments. Graphs show mean and SEM. (C) Representative images of P14 CD8 T cells in the small intestines and spleens of LCMV immune and naive parabionts, P14s (red) and DAPI (blue), scale bar= 50μm. (D) Distribution of resident and recirculating P14 CD8 T cells in nonlymphoid organs calculated by QIM. (E) P14 immune chimeras were analyzed 120-150 days after LCMV infection to determine the distribution of P14 CD8 T cells in secondary lymphoid organs (SLO), nonlymphoid tissues (NLT, including i.v. Ab- cells within liver, lung, kidney, pancreas, salivary gland, uterus, vagina and cervix, small intestine, large intestine, stomach and thymus) and circulating blood and marginated pool (BMP, includes i.v. Ab+ cells from all tissues examined), n≥6. Cell numbers from all tissues were calculated by QIM, except circulating blood, which was enumerated by cell isolation and flow cytometry, (See also Figure S2).
Initially, we restricted analysis to P14 memory CD8 T cells that were not permissive to i.v. Ab staining. SLOs maintained very little disequilibrium between immune and naive parabionts, consistent with the previous observation that they contain only small fractions of TRM after LCMV infection (Schenkel et al., 2014). In contrast, the vast majority of memory P14 CD8 T cells within almost all NLT examined were TRM, as they exhibited little to no evidence of infiltration into the NLTs of naive parabionts (Figure 3B&C). Indeed, liver was the only NLT that supported substantive levels of memory CD8 T cell migration, although even in this case, ∼55% of i.v. Ab- P14 CD8 T cells were resident.
The distribution of T lymphocytes and particular memory subsets remains uncertain and debated, in part due to technical issues of quantifying cell numbers in tissues, identification of antigen-specific populations with a known history of stimulation, and bona fide analyses of cell recirculation. As QIM, parabiosis, and our focus on a single but identifiable population (P14, 120 days after LCMV infection in mice) overcome these hurdles, we summated the parabiosis data from each NLT, revealing that the vast majority of nonlymphoid memory P14 are in fact TRM, not recirculating TEM (Figure 3D). Further, we then leveraged these approaches to generate a global representation of the apportionment of a memory CD8 T cell population throughout the visceral compartments of the organism. These data, shown in figure 3E, support several conclusions. Less than half of the memory P14 pool was localized to SLO; spleen WP and LN (extrapolating mandibular LN data to the 37 macroscopic LNs in mice) (Van den Broeck et al., 2006). This was due to the fact that NLT contained more cells than expected based on previous cell isolation-dependent methods, and also because of the surprising abundance of memory P14 contained within the BMP, a compartment that has not been enumerated in previous studies. Indeed, peripheral blood (from which many estimates of total blood lymphocytes are extrapolated) actually contained less than 4% of the memory P14 within the total bloodborne population, particularly due to the magnitude or increased density of lymphocytes within spleen red pulp, lung and liver vasculature (Table I). These data provide the most extensive quantitative characterization of a single memory CD8 T cell population to date, and revise perceptions of migration and distribution.
Memory CD8 T cell migration is compartmentally restricted within NLT
We next used the advantages of imaging analyses to test whether memory CD8 T cell entry during the memory phase of the response was selective for certain tissues within nonlymphoid organs. As shown in figure 4A, mucosal organs could be segregated into three patterns of memory P14 migration, those in which there was: 1) no migration to mucosal epithelia or LP, 2) no migration to mucosal epithelia but limited migration to LP, submucosa, and muscularis externa, and 3) limited migration to both epithelia and LP. In the thymus, the medulla, but not cortex, was permissive to memory CD8 T cell recirculation (Figure 4B&C). These results suggested that memory CD8 T cell migration differs between compartments within nonlymphoid organs, although TRM dominate all compartments. We next focused our analyses on the i.v. Ab+ BMP in liver and kidney, which includes cells within sinusoids and glomeruli (Anderson et al., 2014). We observed that 35-60% of the marginated pool was TRM even within the vascular compartments of these organs (Figure 4D). These data indicate that migration properties vary by compartment within NLT and that TRM are not exclusively localized to the parenchyma of tissues.
FIGURE 4. Memory CD8 T cell migration is compartmentally restricted within NLT.
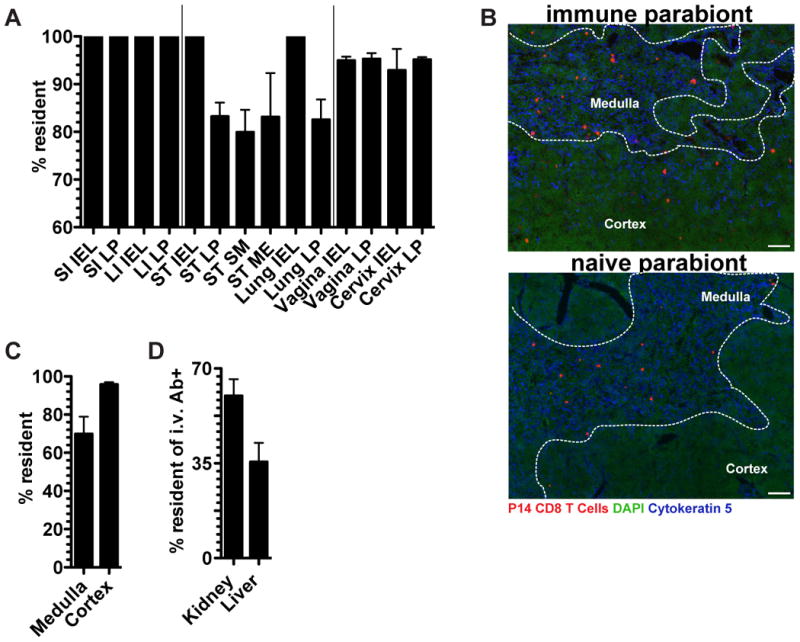
P14 immune chimeras conjoined to naive C57BL/6 mice (as in figure 3) were analyzed 30 days after parabiosis surgery. (A) The fraction of P14 CD8 T cells that are resident in the indicated tissue compartments, small intestine (SI), large intestine (LI), stomach (ST), epithelium (IEL), lamina propria (LP), submucosa (S.M.), muscularis externa (M.E.). (B) Representative thymus images in immune and naive parabionts. P14 CD8 T cells (red), DAPI (green), cytokeratin 5 (blue), scale bar= 50μm. (C) Percent of P14 CD8 T cells that are resident in the thymus medulla and cortex. (D) Percent of i.v. Ab+ P14 CD8 T cells that are resident within the kidney and liver. n=3, representative of 9 mouse pairs from 3 independent experiments. Graphs show mean and SEM, (See also Figure S2).
CD69 is an imperfect marker of tissue residence
Given the impracticality of performing bona fide migration studies, the C-type lectin CD69 has become the defining marker for distinguishing TRM from recirculating TEM because it antagonizes the sphingosine 1-phosphate receptor 1 (SIPR1) which promotes egress via lymphatics and is necessary for TRM maintenance in epidermis (Farber et al., 2014; Mackay et al., 2013). We tested whether CD69 expression was stringently predictive of recirculation properties. Only 25-75% of the memory P14 cells in pancreas, salivary gland (SG), and FRT expressed CD69 (Figure 5A) even though almost all cells from these organs were TRM (Figure 3B). This demonstrates that CD69-cells can also be functionally resident, a result that extends to the vascular compartments of the kidney and liver (Figure 5B-D). Thus, CD69 is not a definitive marker to distinguish recirculating cells from TRM.
FIGURE 5. CD69 is an imperfect marker of tissue residence.
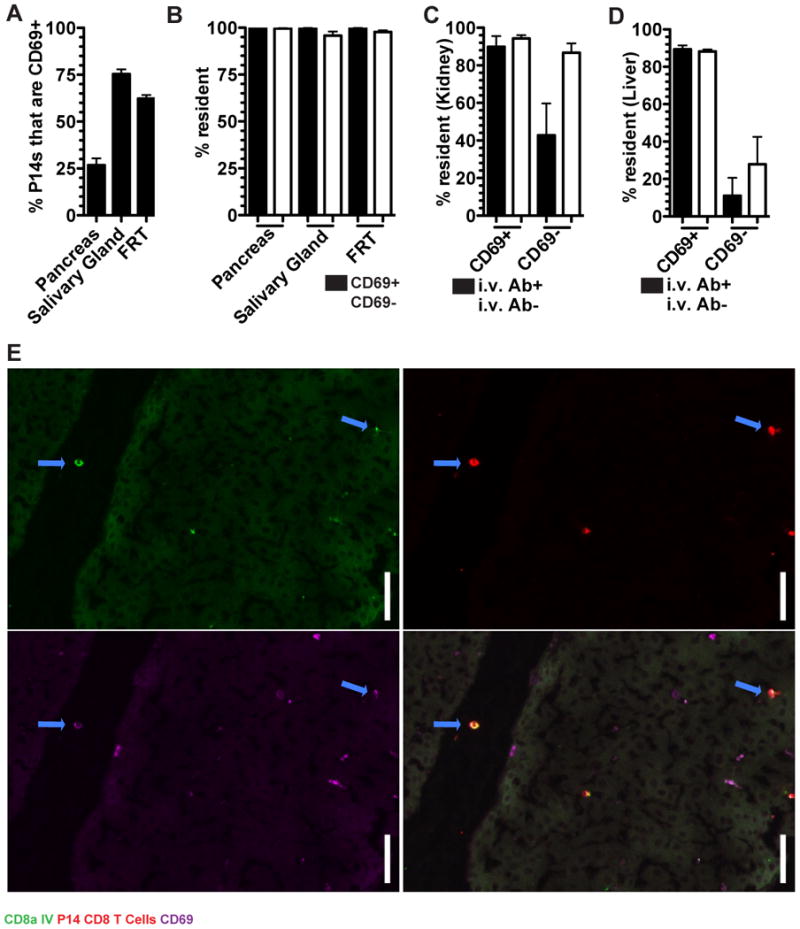
(A) P14 CD8 T cells from immune parabionts were analyzed for the expression of CD69 in the pancreas, salivary gland and FRT by QIM. (B) The fraction of CD69+ and CD69- P14 CD8 T cells that were resident. (C) The percent of P14 CD8 T cells that were resident among i.v. Ab+/- and CD69+/- in the kidney and (D) liver. (E) Representative image of a CD69+ i.v. Ab+ P14 CD8 T cells in a large vessel in the liver. α-CD8α i.v. Ab (green), P14 CD8 T cells (red), and CD69 (purple). Blue arrows indicate α-CD8α i.v. Ab+ CD69+ P14 CD8 T cells, scale bar =20μm. n=3, representative of 9 mouse pairs from 3 independent experiments. Graphs show mean and SEM, (See also Figure S2).
CD69 is known to be induced on TRM precursors upon migration into tissues during the effector phase of immune responses, putatively by tissue-derived instructional cues (Casey et al., 2012; Lee et al., 2011; Masopust et al., 2006;). However, we observed CD69 expression among TRM within the BMP of the liver and kidney, suggesting that parenchymal localization is not a requirement. Indeed, we even detected CD69+ memory P14 CD8 T cells within the large bore vessels of the liver of immune (but not naive) parabionts (Figure 5E). Taken together, in the steady state most CD69+ memory CD8 T cells are TRM, but many TRM are not CD69+.
Migration of memory CD8 T cell subsets
Evidence for equilibration of memory CD8 T cells in non-lymphoid tissues fails to discriminate between bona fide recirculating TEM versus the possibility that a few TEM or TCM continue to seed NLT and form TRM long after immunization (i.e., a one way trip). Because leukocytes use lymphatics to exit tissues, we examined whether we could observe evidence of memory P14 CD8 T cells within lymphatic vessels (visualized by Lyve-1 staining) of naive parabionts. We focused on FRT and SG due to the prominent nature of the lymphatic collecting ducts in these organs (Figure 6A-C). Figure 6C of a representative FRT image shows that P14 memory CD8 T cells could indeed be visualized within lymphatic vessels. In each mouse, we visualized ∼100 lymphatic vessel-bound P14 CD8 T cells in both FRT and SG when 3-4 sections were combined for analysis.
FIGURE 6. Migration of memory CD8 T cell subsets.
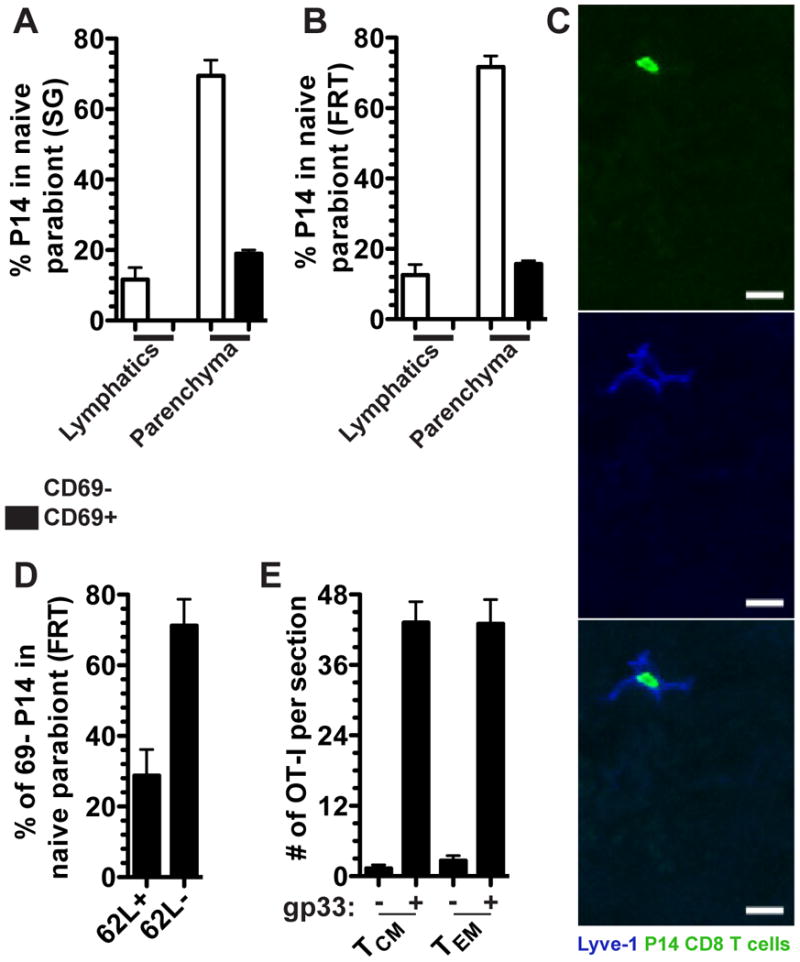
P14 CD8 T cells were analyzed by QIM from naive parabionts were quantified based on their localization within the parenchyma or afferent lymphatic Lyve-1 + vessels and for the expression of CD69 in the (A) salivary gland and (B) female reproductive tract. (C) Representative image of a P14 CD8 T cell in the FRT afferent lymphatics of a naive parabiont. Lyve-1 (blue) and P14 CD8 T cells (green), scale bar =10μm. (D) Fraction of CD69- P14 CD8 T cells in the FRT of the naive parabiont that were CD62L+ or CD62L-. n=3, representative of 9 mice from 3 independent experiments (E) 5×105 CD62L+ or CD62L- memory OT-I CD8 T cells isolated from the spleen of VSV-OVA immune chimeras were transferred into P14 immune chimeras and the next day P14 immune chimeras were challenged transcervically with 50μg gp33 peptide. Two days later total numbers of OT-I CD8 T cells were enumerated in the FRT. n=6, representative of 2 independent experiments. Graphs show mean and SEM, (See also Figure S2).
Quantitative analysis indicated that ∼20% of P14 CD8 T cells that entered SG and FRT of naive parabionts during the memory phase of the response could be localized to lymphatic vessels (Figure 6A&B). These data provide strong evidence that a substantive fraction of P14 CD8 T cells that entered these NLT tissues during the memory phase of the immune response were bona fide TEM that exited these tissues after entry (even though TRM represented the dominant fraction of the overall memory CD8 T cell population in these tissues, see Figures 3&4). Phenotypic analysis indicated that memory P14 CD8 T cells in lymphatic vessels were exclusively CD69- (Figure 6A&B). While this has not previously been reported, we were able to detect a population of CD69+ P14 CD8 T cells that had migrated to the FRT and SG of naive parabionts during the memory phase of the immune response, 90-120 days after infection.
Paradigmatically, TEM recirculate through NLT or respond to NLT sites of inflammation, while TCM limit recirculation to SLO (Sallusto et al., 1999). However, this hypothesis has not been rigorously tested. Parabiosis allowed us to identify bona fide CD69- memory CD8 T cells that had entered the FRT 90-120 days after immunization, thus providing an opportunity to test this model. We found that ∼30% of CD69- migrating memory P14 CD8 T cells in naive parabionts were CD62L+, indicating that much of the NLT recirculating population would conventionally be defined as TCM (Figure 6D).
We next tested whether TEM are in fact specialized to migrate to NLT sites of inflammation compared to TCM. 5×105 CD62L+ (TCM or CD62L- (TEM) memory OT-I CD8 T cells (see methods) were transferred into P14 immune chimeras. The next day, mice were challenged transcervically with gp33 peptide to reactivate P14 TRM in the FRT and precipitate an inflammatory response that recruits circulating memory T cells (Schenkel et al., 2013). As shown in figure 6E, TCM and TEM migrated to NLT inflammation equivalently, revising the current model of how each subset participates in host immunity.
Discussion
This study provides a rigorous and comprehensive analysis of the anatomic distribution of a single memory CD8 T cell population. Preparation of single cell suspensions from tissues recovered as few as 2% of memory CD8 T cells from NLT, and inaccurately represented memory T cell subsets, phenotype and tissue distribution. Similar results were observed in human tissue, suggesting fundamental errors with standard techniques that we rely upon for our basic characterization of the peripheral immune system. These issues may extend to other hematopoietic lineages, evaluation of vaccine responses in tissues, and other clinical investigations.
When the NLT population was summated with the unexpected abundance of memory CD8 T cells observed in BMP, SLO (WP of spleen and the 37 macroscopic LNs in mice) did not contain the majority of memory CD8 T cells (Van den Broeck et al., 2006). And our study likely underestimates NLT memory CD8 T cells because not every tissue was analyzed, including many other locations (heart, bladder, gall bladder, esophagus, trachea, skeletal muscle, etc.) that contain memory CD8 T cells (Casey et al., 2012, data not shown). In particular, skin has been shown to harbor abundant memory T cells in humans, where extraction efficiency is also an important challenge (Clark et al., 2006). This study further highlights the abundance of TRM as well as their broad anatomic distribution, which includes the BMP. Moreover, based on cell isolation and flow cytometry enumerations, cells in mucosal tissues were 50-400 fold more rare than in SLOs. However, QIM revealed that the ratios of memory CD8 T cells relative to potential targets (i.e. host cells) were fairly comparable between SLO and NLT. These observations revise perceptions of immunosurveillance, and may help explain why frontline memory CD8 T cell populations can rapidly detect infections in barrier tissues (Gebhardt et al., 2009; Jiang et al., 2012; Shin and Iwasaki, 2012; Teijaro et al., 2011; Wu et al., 2014).
We focused most analyses on memory resulting from a single infection in order to achieve the depth of characterization described here. However, evidence supports that fundamental observations regarding the abundance of resident memory extend well beyond the context of LCMV. Many infections, whether systemic or local, result in CD8 T cell populations that express peripheral homing molecules and then become broadly distributed throughout multiple nonlymphoid tissues (Masopust et al., 2010, Masopust et al., 2004, Liu et al., 2006, Kaufman et al., 2008). In fact, even lymphopenia-induced proliferation is sufficient to induce widespread CD8 T cell dissemination and acquisition of markers associated with TRM (Casey et al., 2012). These data indicate that TRM development may occur irrespective of local antigen or inflammation. TRM are likely not only widely distributed in a variety of contexts, but also underestimated. Indeed, recent evidence suggests that most CD8 T cells that express markers of antigen-experience also express CD69 when isolated from human tissues, which suggests that most are resident (Thome et al., 2014). We demonstrated that the isolation of CD8 T cells from nonlymphoid tissues was inefficient in both mice and humans, suggesting that memory T cells outside of secondary lymphoid organs are misrepresented regardless of species or pathogen specificity.
This study also raises important caveats with how we define resident and recirculating memory CD8 T cell subsets. CD69 is considered the lineage-defining marker for TRM. It has been shown that CD69 is important for establishing TRM populations in epidermis after HSV-1 infection in mice (Mackay et al., 2013). In accordance with these data, we found that many TRM were CD69+. However, we found that many were not. Moreover, expression of another marker oft used to identify TRM, CD103, was compartment-specific and most TRM lacked CD103. These data define additional complexity amongst TRM and suggest that there is more than one subset. Maintenance of CD69- TRM could be mediated by alternative means such as down regulation of KLF2-dependent S1P receptors (Skon et al., 2013). Our data also reveal that anatomic localization outside (or inside) vasculature is not sufficient to reveal the residence status of a CD8 T lymphocyte. Furthermore, we did detect memory CD8 T cells that had entered certain NLT months after putative clearance of infection. While rare, a substantive proportion of these “latecomers” expressed CD69. It is possible that this represents a one-way trip and that TRM are maintained by a slow matriculation of circulating memory CD8 T cells that convert to TRM, upregulating CD69 post migration.
To what degree do memory CD8 T cells undergo bona fide recirculation through NLT? Leukocytes exit tissues via the afferent lymphatics. Because we identified latecomer memory CD8 T cells in the lymphatics of the FRT and SG, these cells are likely a bona fide NLT recirculating subset in the steady state. In support of this conclusion, this population did not express CD69.
Given the abundance of memory CD8 T cells in the BMP and NLT, and the relative paucity of recirculation through NLT, our data raise questions as to whether most TEM truly survey NLT. Perhaps a more likely scenario is that NLT are surveyed by only a fraction of specialized TEM, and that other TEM serve functions that remain to be fully elucidated. Our data indicate that TCM also contribute to the rare population of NLT recirculating memory CD8 T cells in the steady state, which may also occur in human skin (Clark et al, 2006). Moreover, in the context of inflammation, TCM migrated just as robustly as TEM to the FRT. In contrast to the original and elegant (TCM/TEM model, this may ensure that there is a long-lived pool capable of being recruited because TCM may be maintained longer than CD62L- BMP (Wherry et al., 2003, Marzo et al., 2005).
Supplementary Figure 2 summarizes and contextualizes these observations. Most host cells, which require contact by CD8 T cells for immunosurveillance, are positioned outside of secondary lymphoid organs. These include solid organs and body surfaces such as the gastrointestinal, respiratory, and genitourinary mucosae and skin that represent common primary sites of pathogen exposure. The majority of memory CD8 T cells that patrol these frontlines are segregated populations that confine their surveillance locally and do not migrate between other NLT, SLOs, or blood. Therefore, this major fraction of the memory CD8 T cell pool cannot be captured by sampling blood or SLOs. Indeed, the recirculating populations, which included both CD62L- TEM and CD62L+ TCM actually comprised a small minority of those cells patrolling NLT. The blood and marginated pool (BMP, which includes peripheral blood, the red pulp of the spleen, and vascular compartments within organs such as liver and kidney) also contains a substantial fraction of the overall memory CD8 T cell population. When NLT re-infections are not rapidly eliminated, inflammation recruits both TEM and TCM from the BMP, presumably to contribute to local immunosurveillance and pathogen control. The vascular compartments of certain tissues, including liver and kidney, are also populated by TRM, which may facilitate direct immunosurveillance of the organ via the endothelium, for instance of hepatocytes through sinusoidal fenestrae, or may prevent hematogenous spread of target cells. When infections are not contained within NLTs, pathogens and associated foreign antigens reach the SLOs. Here, TCM (which recirculate between blood and SLOs) can be reactivated to proliferate and provide additional reinforcements that migrate to NLTs.
This revised model highlights the provincial nature of memory CD8 T cell mediated immunosurveillance. Different populations of memory CD8 T cells patrol distinct anatomic niches that form an integrated immunological network to protect the host in the event of reinfection. However, the majority of the host is patrolled by abundant yet discrete regionalized memory CD8 T cell populations that do not recirculate and instead remain confined within single anatomic compartments.
Experimental Procedures
Mice, Adoptive Transfers, Surgeries & Infections
All mice were used in accordance with the Institutional Animal Care and Use Committee at the University of Minnesota. C57BL/6J mice were purchased from The Jackson Laboratory, P14 and OT-I CD8 T cell transgenic mice were maintained in house. P14 immune chimeras were generated by transferring 5×104 P14 CD8 T cells into naive C57BL/6J mice. The following day these mice were infected with 2×105 PFU LCMV Armstrong via intraperitoneal (i.p) injection. For endogenous studies naive C57BL/6J mice were infected with 2×105 PFU LCMV Armstrong i.p. OT-I immune chimeras were generated by transferring 5×104 naїve OT-I CD8 T cells into C57BL/6 mice. The next day, mice were infected with 2×106 PFU Vaccinia Virus expressing chicken ovalbumin. Sixty days after infection, CD62L+ and CD62L- memory OT-I splenocytes were purified using α-CD62L PE and α-PE magnetic beads according to the manufacturers instructions (Miltenyi). 5×105 CD62L+ or CD62L- OT-I cells were transferred into P14 immune chimeras that 60 days previously had been infected with LCMV. The following day animals were transcervically (t.c.) challenged with 50μg gp-33 peptide as previously described (Collins et al.; Schenkel et al., 2013). Parabiosis surgeries were performed as previously described (Schenkel et al., 2013).
Intravascular antibody
To label all CD8 T cells in compartments contiguous with vasculature animals were injected i.v. with 3μg α-CD8α biotinylated antibody (53-6.7, eBioscience) that was allowed to circulate for three minutes prior to sacrifice. For detection of i.v. injected α-CD8α antibody, fluorochrome-conjugated streptavidin (eBioscience) was used for flow cytometry and donkey anti-rat antibodies (Jackson Laboratory) were used for immunofluorescence.
Isolations and Flow Cytometry
Three minutes after in vivo intravascular antibody injection (Anderson et al., 2014), mice were sacrificed and organs of interest were excised. For isolation of SI lELs, the small intestine was removed, Peyer's patches were excised, and the intestine was cut longitudinally and then laterally into 0.5-1 cm2 pieces. Large intestines and stomachs were cut similarly. To remove lELs, small intestine, large intestine and stomach pieces were incubated with 0.154mg/ml dithioerythritol (DTE) in 10% HBSS/HEPES bicarbonate for 30 min at 37 C, stirring at 450rpm. Following IEL isolation, small intestine, large intestine and stomach pieces were further processed to remove lamina propria lymphocytes (LPL), by treatment with 100U/ml type I collagenase (Worthington) in RPMI 1640, 5% FBS, 2mM MgCI2, 2mM CaCI2 for 45 min at 37 C, stirring at 450rpm).
The following tissues were cut into pieces and enzymatically digested with 100U/ml type I collagenase (Worthington) in RPMI 1640, 5% FBS, 2mM MgCI2, 2mM CaCI2 at 37 C, stirring at 450rpm; salivary gland (SG, mucous portion removed, treated for 45 minutes), kidney (treated for 45 minutes), pancreas (treated for 20 minutes), lung (treated for one hour). For isolation of the female reproductive tract, the uterine horns, cervix, and vaginal tissue were resected and cut into small pieces prior to treatment with 0.5mg/ml type IV collagenase (Sigma) RPMI 1640, 5% FBS, 2mM MgCI2, 2mM CaCI2 (treated for one hour) at 37 C, stirring at 450rpm. After enzymatic treatment, the remaining tissue pieces of the stomach LPL, FRT, SG, pancreas, lung, and kidney, were further mechanically disrupted by a gentleMACS Dissociator (setting m_Spleen_01.01). The liver was mechanically dissociated using the back of a syringe over a 70μm nylon cell strainer (Falcon). From single cell suspensions, lymphocytes were separated using a 44/67% Percoll density gradient. Spleen, lymph nodes, and thymus were mechanically dissociated using the back of a syringe against a polystyrene petri dish that had previously been scored in 4 directions with an 18.5 gauge needle. Peripheral blood was treated with ACK lysis buffer. The resulting single cell suspension was stained for acquisition on an LSR II flow cytometer (BD Biosciences).
The following antibodies were used for flow cytometry of mouse cells: α-CD103 (M290) from BD Biosciences; α-CD8α (53-6.7), α-Thy1.1 (HIS51), α-CD44 (IM7), Streptavidin APC, and α-CD45.1 (A20) from eBioscience; and α-Thy1.1 (OX-7) and α-CD8β (YTS156.7.7) from Biolegend.
Quantitative Immunofluorescence Microscopy
To determine volumes of individual organs, mice age-matched to those analyzed for enumeration were sacrificed, and organs were removed and cleared of all fat, connective tissue and fecal matter. Each organ was submerged in PBS, the displaced volume was measured, and this was repeated for each organ 4 times. This displacement procedure was conducted on 6 mice age matched to those used in experiments. For organs too small for accurate volume displacement, including the mandibular lymph nodes, organs were pooled from multiple animals before measuring displacement and dividing the displaced volume by the number of pooled organs. For QIM enumeration, three minutes after in vivo intravascular antibody injection, mice were sacrificed and organs of interest were excised, positioned in plastic cryomolds and snap frozen in optimum cutting temperature (O.C.T) freezing medium. From these frozen tissue blocks slides of 7μm sections were prepared. Slides were stained for acquisition on a Leica DM5500B 4 color fluorescent system with motorized z-focus stage for fully automated image stitching. Enumeration of P14 cells as well as CD103, CD69 and CD62L expression was done manually in Adobe Photoshop. lmageJ64 software was used to enumerate nuclei in each image (as stained by DAPI) as previously described (Schenkel et al., 2013), all counts were manually validated, and these, like the manual enumerations, were extrapolated to whole organs. Area measurements of images were made either in LAS (Leica Acquisition Software) or Adobe Photoshop. Area measurements were multiplied by tissue section thickness (7μm) to determine the volume of enumerated images. Manual and lmageJ64 counts were extrapolated up to whole organ enumerations. We multiplied all enumerations by 11/19 to correct for all cells that would be counted twice because they straddle two adjacent sections. This correction factor is derived because sections are 7μm thick, the diameter of a memory CD8 T lymphocyte is approximately 7μm, and any cell traversing a section by >1 μm would be enumerated (Decoursey et al., 1987). Sections through whole organs or large (∼5mm2) tiled images were counted, no fewer than 100 and up to 3000 P14 were counted per organ per animal, representative tissue sections were sampled which included diverse regions of each organ and non-serial sections (35-70 microns apart) to ensure P14 counts were representative of the entire organ. For example, whole sections of the stomach were counted to ensure anatomical representation of the fundus, body, and antrum regions. The following antibodies were used for immunofluorescence microscopy: α-CD103 (2E7) and α-Thy1.1 (OX-7) from Biolegend; α-CD62L (MEL-14), α-CD8α (53-6.7), α-CD8β (YTS1 56.7.7), α-Ecadherin (DECMA-1), αCD45.1 (A20) from eBioscience; α-CD69 (polyclonal goat), α-Lyve-1 (223322) from R&D; α-Cytokeratin 8 (rabbit polyclonal), α-Cytokeratin 18 (rabbit polyclonal), α-PE (rabbit polyclonal) from novus biologicals; α-Collagen IV (goat polyclonal) from Millipore; α-Cytokeratin5 (PRB-160P) from Covance. DAPI (4,6-diamidino-2-phenylindole) and prolong gold were from Invitrogen. The following secondary antibodies were from Jackson Immunoresearch: donkey α-rabbit (polyclonal), bovine α-goat (polyclonal), donkey α-rat (polyclonal).
DNA extraction
To validate QIM extrapolation, DNA content of whole organs was determined. First organs were dissected, cut into 1mm pieces and digested in tissue digestion buffer (10mM TRIS, 10mM EDTA, 10% SDS, sodium acetate and proteinase K) shaking overnight at 56 C. Phenol-Chloroform-lsoamyl alcohol DNA extraction was then performed on each digested organ. Each DNA sample was resuspended in TE buffer and nucleic acid concentration was determined by a nanodrop spectrophotometer. Each sample was measured 4 times; an average of the 4 was taken to determine the most accurate nucleic acid content of each sample. The total nucleic acid content of each organ was divided by an assumed 6 picograms of DNA per cell to determine total cell number for the organ based on DNA content (Anjos Pires et al., 2001).
Supplementary Material
1
2
3
Acknowledgments
This work was supported by grants from the National Institutes of Health R01-AI084913, and R01-Al11671 to D.M.; T32-AI083196 to E.M.S; F30-5F30DK100159-02 to J.M.S.; and F31-CA183226 to L.S.M.
Footnotes
Author Contributions: E.M.S., J.M.S., L.S.M., and D.M. conceived and designed the experiments, E.M.S., J.M.S., L.B., K.A.F., and B.Z.I. performed the experiments, L.S.M. and P.J.S contributed reagents/materials/analysis tools, and E.M.S., J.M.S., and D.M. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, David MM. Phenotypic analysis o antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Anjos Pires M, Palmeira C, Rodrigues P, Lopes C, Oliveira-Torres F. Establishment of a diploid reference value for DNA ploidy analysis by image cytometry in mouse cells. Anal Quant Cytol Histol. 2001;23:427–432. [PubMed] [Google Scholar]
- Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. Antigen-Independent Differentiation and Maintenance of Effector-like Resident Memory T Cells in Tissues. J Immunol. 2012;188:4866–875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RA, Chong B, Mirchandrani N, Brinster NK, Yamanaka K, Dowgiert RK, Kupper TS. The Vast Majority of CLA+ T cells are Resident in Normal Skin. J Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- Collins MK, Tay CS, Erlebacher A. Dendritic cell entrapment within the pregnant uterus inhibits immune surveillance of the maternal/fetal interface in mice. J Clin Invest. 2009;119:2062–2073. doi: 10.1172/JCI38714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoursey TE, Chandy KG, Gupta S, Cahalan MD. Mitogen induction of ion channels in murine T lymphocytes. J Gen Physiol. 1987;89:405–420. doi: 10.1085/jgp.89.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doctor RB, Serkova NJ, Hasebroock KM, Zafar I, Edelstein CL. Distinct patterns of kidney and liver cyst growth in pkd2(WS25/-) mice. Nephrol Dial Transplant. 2010;25:3496–3504. doi: 10.1093/ndt/gfq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkina E, Thatte J, Dabak V, Williams MB, Ley K, Braciale TJ. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J Clin Invest. 2005;775:3473–3483. doi: 10.1172/JCI24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusov VV, De Boer RJ. Do most lymphocytes in humans really reside in the gut? Trends Immunol. 2007;28:514–518. doi: 10.1016/j.it.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- Horbul JE, Schmechel SC, Miller BR, Rice SA, Southern PJ. Herpes simplex virus-induced epithelial damage and susceptibility to human immunodeficiency virus type 1 infection in human cervical organ culture. PloS ONE. 2011;6(7):e22638. doi: 10.1371/journal.pone.0022638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman DR, Liu J, Carville A, Mansfield KG, Havenga MJE, Goudsmit J, Barouch DH. Trafficking of antigen-specific CD8 T lymphocytes to mucosal surfaces following intramuscular vaccination. J Immunol. 2008;181:4188–4198. doi: 10.4049/jimmunol.181.6.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KM, McNamara JT, Lefrangois L. In situ imaging of the endogenous CD8 T cell response to infection. Science. 2007;318:116–120. doi: 10.1126/science.1146291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonowski KD, Williams KJ, Marzo AL, Blair DA, Lingenheld EG, Lefancois L. Dynamics of Blood-Borne CD8 Memory T cell Migration in Vivo. Immunity. 2004;20:551–562. doi: 10.1016/s1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]
- Lee YT, Suarez-Ramirez JE, Wu T, Redman JM, Bouchard K, Hadley GA, Cauley S. Environmental and antigen receptor-derived signals support sustained surveillance of the lungs by pathogen-specific cytotoxic T lymphocytes. J Virol. 2011;85:4085–4094. doi: 10.1128/JVI.02493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Fuhlbrigge RC, Karibian K, Tian T, Kupper TS. Dynamic programing of CD8+ T cell trafficking after live viral immunization. Immunity. 2006;25:511–520. doi: 10.1016/j.immuni.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. The developmental pathway for CD103+ CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8 T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Schenkel JM. The integration of T cell migration, differentiation and function. Nat Rev Immunol. 2013;13:309–320. doi: 10.1038/nri3442. [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Usherwood EJ, Cauley LS, Olson S, Marzo AL, Ward RL, Wooland DL, Lefrangois L. Activated primary and memory CD8 T cells migrate to nonlymphoid tissues regardless of site of activation or tissue of origin. J Immunol. 2004;172:4875–4882. doi: 10.4049/jimmunol.172.8.4875. [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol. 2006;176:2079–2083. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]
- Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey K, Barber DL, Kawamura KS, Fraser KA, Webby RJ, Brinkmann V, Butcher EC, Newell KA, Ahmed R. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T Cell Subsets, Migration Patterns, and Tissue Residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a revaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462:510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter RL, Gridley DS, Slater JM, McMillan PJ. Responses of mouse spleen morphology to the growth of subcutaneously injected virally transformed cells. Anat Rec. 1980;197:363–368. doi: 10.1002/ar.1091970310. [DOI] [PubMed] [Google Scholar]
- Peaudecerf L, Rocha B. Role of the gut as a primary lymphoid organ. Immunol Lett. 2011;140:1–6. doi: 10.1016/j.imlet.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK. Visualizing the generation of memory CD4 T cells in the whole body. Science. 2001;410:101–105. doi: 10.1038/35065111. [DOI] [PubMed] [Google Scholar]
- Rocha B, Vassalli P, Guy-Grand D. The VB Repertoire of Mouse Gut Homodimeric aCD8+ Intraepithelial T cell Receptor a/b+ Lymphocytes Reveals a Major Extrathymic Pathway of T cell Differentiation. J Exp Med. 1991;173:483–486. doi: 10.1084/jem.173.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999a;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Scheller LF, Wirtz RA, Azad AF. Susceptibility of different strains of mice to hepatic infection with Plasmodium berghei. Infect Immun. 1994;62:4844–4847. doi: 10.1128/iai.62.11.4844-4847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, Fraser KA, Masopust D. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J Immunol. 2014;192:2961–2964. doi: 10.4049/jimmunol.1400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8(+) T cells. Nat Immunol. 2013;14:509–513. doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby WS, Janossy G, Bofill M, Jewell DP. Intestinal lymphocyte subpopulations in inflammatory bowel disease: an analysis by immunohistological and cell isolation techniques. Gut. 1984;25:32–40. doi: 10.1136/gut.25.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012;491:463–467. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skon CN, Lee JY, Anderson KG, Masopust DM, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8 T cells. Nat Immunol. 2013;14:1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvas PK, Dech HM, Sambira F, Zeng J, Onami TM. Systemic and Mucosal Infection Program Protective Memory CD8 T Cells in the Vaginal Mucosa. J Immunol. 2007;179:8122–8127. doi: 10.4049/jimmunol.179.12.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrangois L, Farber DL. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol. 2011;187:5510–5514. doi: 10.4049/jimmunol.1102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome JJC, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, Farber DL. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck W, Derore A, Simoens P. Anatomy and nomenclature of murine lymph nodes: Descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. J Immunol Methods. 2006;312:12–19. doi: 10.1016/j.jim.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Wherry JE, Teichgraber V, Becker TC, Masopust D, Kaech SM, Anita R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, Cauley LS. Lung-resident memory CD8 T cells (trm) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol. 2014;95:215–224. doi: 10.1189/jlb.0313180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3