MIWI2 and MILI Have Differential Effects on piRNA Biogenesis and DNA Methylation (original) (raw)
. Author manuscript; available in PMC: 2016 Aug 25.
Summary
In developing male germ cells, prospermatogonia, two Piwi proteins, MILI and MIWI2, use Piwi-interacting RNA (piRNA) guides to repress transposable element (TE) expression and ensure genome stability and proper gametogenesis. In addition to their roles in post-transcriptional TE repression, both proteins are required for DNA methylation of TE sequences. Here, we analyzed the effect of Miwi2 deficiency on piRNA biogenesis and transposon repression. Miwi2 deficiency had only a minor impact on piRNA biogenesis; however, the piRNA profile of _Miwi2_-knockout mice indicated overexpression of several LINE1 TE families that led to activation of the ping-pong piRNA cycle. Furthermore, we found that MILI and MIWI2 have distinct functions in TE repression in the nucleus. MILI is responsible for DNA methylation of a larger subset of TE families than MIWI2 is, suggesting that the proteins have independent roles in establishing DNA methylation patterns.
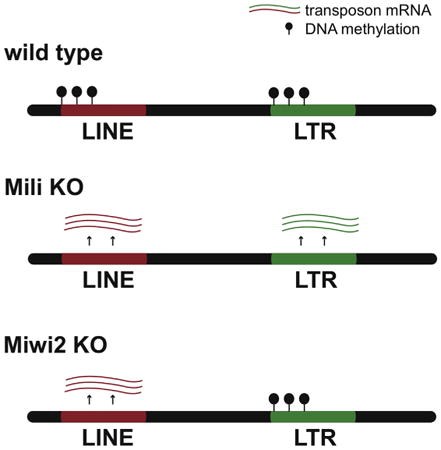
Graphical abstract
Introduction
Piwi proteins and their associated small RNAs, Piwi-interacting RNA (piRNAs), repress transcription of transposable elements (TEs) in germ cells of Metazoa (Siomi et al., 2011). The piRNA pathway is conserved between mouse and flies, and three Piwi proteins are required for proper TE repression in each species. The non-redundant functions of the three Piwi proteins in flies were explained by two different observations. First, only one of the three Piwi proteins, Piwi, is nuclear; it is required for establishment of repressive histone marks on TE sequences (Klenov et al., 2011; Le Thomas et al., 2013; Rozhkov et al., 2013; Sienski et al., 2012). The endonuclease activity of Piwi is not necessary for its function (Darricarrère et al., 2013; Saito et al., 2010; Sienski et al., 2012). The two other proteins, Aub and Ago3, are required for post-transcriptional degradation of TE transcripts (Brennecke et al., 2007). In the cytoplasm, Aub and Ago3 cooperate in an intricate process called the ping-pong cycle that causes destruction of TE transcripts and simultaneous increases in the levels of piRNAs that target active transposons (Brennecke et al., 2007; Gunawardane et al., 2007). Importantly, the ping-pong cycle operates when only one of the two partners, namely Aub, is present (Li et al., 2009). Ago3 is necessary to enrich piRNA populations in sequences that can recognize and target active transposons (Li et al., 2009).
The individual roles of the three Piwi proteins in transposon repression in mice are not as well understood as the functions in flies. One of the three murine Piwi members, MIWI (Piwi-like protein 1, or PIWIL1), associates with pachytene piRNAs that are expressed in spermatocytes during and after meiosis; pachytene piRNAs are not enriched in TE sequences (Deng and Lin, 2002; Girard et al., 2006). The catalytic activity of MIWI is, however, implicated in LINE1 TE silencing (Reuter et al., 2011). The two other proteins mouse Piwi proteins, MILI (Piwi-like protein 2, or PIWIL2) and MIWI2 (Piwi-like protein 4, or PIWIL4), associate with piRNAs enriched in TE-derived sequences, and mutation of either of the two causes derepression of several retrotransposon families (Aravin et al., 2007; Carmell et al., 2007; De Fazio et al., 2011; Kuramochi-Miyagawa et al., 2008; Pezic et al., 2014). Like Drosophila Aub and Ago3, MILI and MIWI2 participate in ping-pong amplification (Aravin et al., 2008); however, the endonuclease activity of MIWI2 is dispensable for ping-pong piRNA processing (De Fazio et al., 2011).
Both MILI and MIWI2 are required for DNA methylation of TE sequences in the nucleus (Aravin et al., 2007; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008). MIWI2 localizes to the nucleus of prospermatogonia in the narrow developmental window when de novo methylation of TE sequences takes place during gametogenesis (Aravin etal., 2008; Kuramochi-Miyagawa et al., 2008). Accordingly, it was proposed that MIWI2 acts as a direct effector of DNA methylation. In contrast to MIWI2, MILI predominantly localizes to the cytoplasm (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2004). MILI is required for loading of MIWI2 with piRNAs and subsequent nuclear localization, (Aravin et al., 2008; De Fazio et al., 2011). These data suggest that MILI induces DNA methylation indirectly by ensuring piRNA loading and proper nuclear localization of MIWI2.
Here, we analyzed individual roles of MILI and MIWI2 in piRNA biogenesis and TE suppression on a genome-wide scale. Our results revealed distinct roles of two proteins in the piRNA pathway. Unlike Mili knockout (KO), which results in a dramatic reduction of piRNA levels (Aravin et al., 2008; De Fazio et al., 2011), Miwi2 deficiency had a smaller effect on piRNA biogenesis. Levels of LINE1-derived piRNAs were reduced in Miwi2 KO mice early during gametogenesis, but their levels were increased later in development due to activation of the ping-pong cycle caused by LINE1 derepression. Analysis of genome-wide DNA methylation patterns in Mili KO and Miwi2 KO mice suggested that functions of MIWI2 and MILI in CpG methylation of TE sequences in the nucleus are at least partially independent.
Results
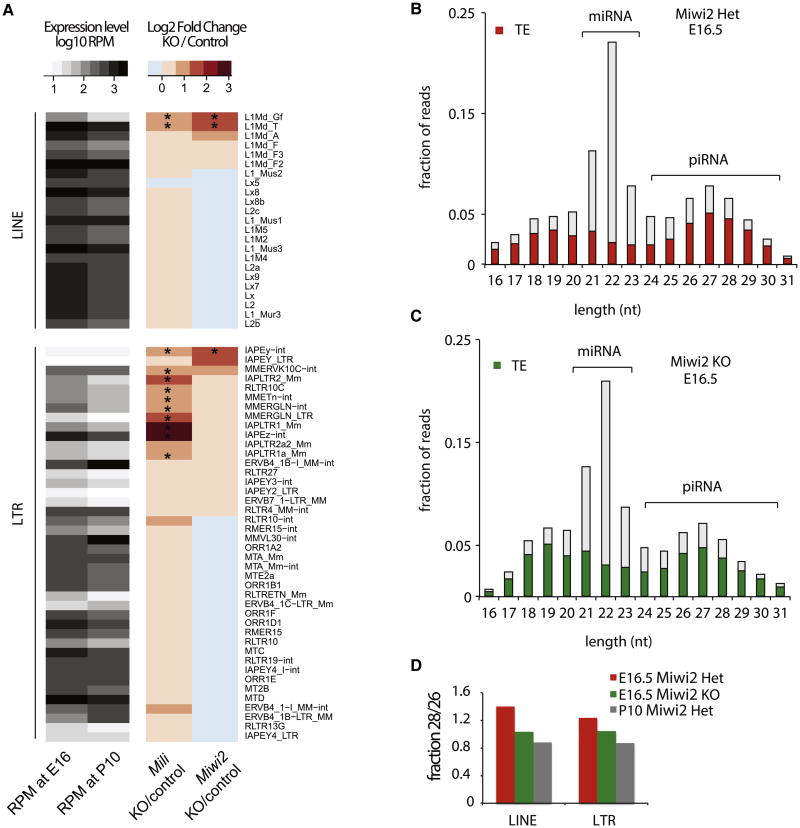
To compare the roles of MILI and MIWI2 in TE repression, we generated and analyzed transcriptome profiles of testes isolated from Mili KO and Miwi2 KO mice and corresponding Heterozygous controls (Het mice) at postnatal day 10 (P10) (Table S1). At least two biological replicates of RNA-seq libraries were generated for each genotype; measurements were highly reproducible (Figures S1A and S1B). Both _Mili_- and _Miwi2_-deficient mice had increased levels of expression of transcripts from several TE families, whereas the vast majority of transcripts of protein-coding genes and many other TE families remained unchanged (Figures S1C and S1D). Importantly, the effects of Mili and Miwi2 deficiencies on transposon expression were not identical: only three families were significantly upregulated in Miwi2 KO mice compared to levels in wild-type mice, whereas 12 families were overexpressed in Mili mutants (Figure 1A). Miwi2 deficiency led to a significant increase in expression of young LINE1 family TEs L1-T and L1-Gf (RepBase names L1Md_T and L1Md_Gf, respectively). Significant but weaker upregulations of L1-T and L1-Gf were also observed in Mili mutants. The levels of transcripts from the old LINE1 families, such as L1-F (RepBase name L1Md_F), were similar in mutants and heterozygote control mice. Miwi2 deficiency also led to overexpression of one specific endogenous retrovirus (ERV) family IAPEy (3.5-fold induction); expression of other ERV elements remained unchanged relative to expression in Heterozygote controls. The same IAPEy family was also upregulated in Mili mutants, but to a lesser extent (∼2-fold). Mili KO led to significant overexpression of many other long terminal repeat (LTR) retrotransposons; those of the IAPEz family showed the strongest upregulation (8-fold) relative to wild-type expression (Figure 1A). Overall, the effect of Mili deficiency on transposon expression was more dramatic than that of Miwi2 KO, although stronger upregulation of L1-T, L1-Gf, and IAPEy in Miwi2 KO indicates that effects of deletions of the genes encoding these Piwi proteins have complex effects. It seems that two Piwi proteins cooperate to suppress LINE families L1-T and L1-Gf and the ERV family IAPEy, and most other LTR elements are predominantly controlled by Mili.
Figure 1. Expression of Transposable Elements and piRNAs in _Mili_- and _Miwi2_-Mutant Mice.
(A) Expression of transposable elements (TEs) in testes of Mili and Miwi2 mutants. The heatmap shows transcript abundance (RPM) of 65 TE families in testis at two different developmental time points (E16.5 and P10) analyzed by RNA-seq. The right panel shows the change in transcript abundance in Mili and _Miwi2_-KO animals relative to corresponding controls at P10. The asterisks mark TEs that are statistically significantly upregulated (p < 0.01 at false discovery rate < 20%). Miwi2 KO induces expression of L1Md_Gf, L1Md_T as well as IAPEy elements, and several additional LTR families are induced in Mili KO.
(B and C) Expression of piRNA in testes of E16.5 _Miwi2_-KO and control (Miwi2 Het) mice. Size profiles of small RNAs indicate that lack of MIWI2 does not grossly disrupt piRNA biogenesis. Only the piRNA fraction of the small RNA population is enriched in TE-derived sequences. The presence of a prominent piRNA peak in _Miwi2_-KO animals indicates that biogenesis of piRNA is not disrupted by Miwi2 deficiency.
(D) The Miwi2 deficiency eliminates a subset of TE-derived piRNAs. The ratio of 28-nt to 26-nt piRNAs (y axis) reflects relative abundance of MIWI2 versus MILI-bound piRNA. The ratio ishigh in E16.5 prospermatogonia of controls animals that express both MIWI2 and MILI; the ratio islower in _Miwi2_-KO samples. The ratio is also low at the later developmental stage (P10) of control animals, since MIWI2 is not expressed after birth.
See also Figure S1.
MILI and MIWI2 repress TEs through recognition of complementary transcripts by associated piRNAs. A deficiency in Mili was reported to eliminate the vast majority or all piRNAs in embryonic testes, including those associated with MIWI2, indicating that MILI is necessary for loading of piRNA into MIWI2 (Aravin et al., 2008; De Fazio et al., 2011). In order to understand differential effect of MILI and MIWI2 on TE expression, we analyzed piRNA expression in Miwi2 KO mice. We generated small RNA libraries from male germ cells at two developmental time points: embryonic day 16.5 (E16.5), when both MILI and MIWI2 proteins are expressed in prospermatogonia of embryonic testes, and P10 (Table S1). MIWI2 expression ceased soon after birth, and the protein was not detected in P10 spermatogonia; thus, MILI was the only Piwi protein that interacts with piRNA in these cells.
The size profile of sequenced small RNA revealed two prominent classes of small RNAs in E16.5 testes: microRNAs (miRNAs) of 19–23 nt and a broader peak of short RNAs 24–29 nt long (Figure 1B). The 24-nt to 29-nt small RNAs have ∼64% bias toward a uridine residue at position 1 and are strongly enriched in TE-derived sequences, suggesting that they are bona fide piRNAs (Figures 1B and S1E). Furthermore, comparison of the 24-nt to 29-nt fraction from total testes RNA and piRNA isolated from immunoprecipitated MILI and MIWI2 complexes (Aravin et al., 2008) showed that majority of these sequences are piRNAs (Table S2). Therefore, the 24-nt to 29-nt fraction of total cellular RNA was used to evaluate the effect of Miwi2 KO on piRNA abundance and biogenesis.
In contrast to Mili KO, which causes dramatic loss of piRNA (Aravin et al., 2008), 24-nt to 29-nt piRNAs were readily detected in Miwi2 KO animals (Figure 1C). miRNAs that do not interact with either MILI or MIWI2 proteins and are not affected by disruption of the piRNA biogenesis were used for normalization of the effect of the Miwi2 mutation on abundance of piRNA. After normalization, we found an approximately 5% decrease in total piRNA abundance in testes of E16.5 Miwi2 KO mice compared to control. The Miwi2 mutation did not have an effect on the fraction of TE-derived piRNA, as ∼67.5% and 69% of all 24-nt to 29-nt small RNA species were derived from TEs in control and mutant libraries, respectively (Figures 1B and 1C). Expression of unistrand and dual-strand piRNA clusters was similar in _Miwi2_-deficient and control samples (Figures S1G and S1H).
Piwi proteins associate with piRNAs of slightly different lengths; the mean length of MILI-bound piRNAs is 26 nt, and that for MIWI2-bound piRNAs is 28 nt (Aravin et al., 2007, 2008). Analysis of the length distribution of small RNA at E16.5 showed that, as expected, the fraction of 28-nt RNA was lower in Miwi2 KO mice than in controls, indicating that MIWI2-bound piRNAs are destabilized in the absence of their protein partner (Figure 1D). Importantly, the length distribution of piRNAs in testes of E16.5 Miwi2 KO animals closely resembled the profile of piRNAs at P10, the time point when MIWI2 protein is not expressed in wild-type mice and only MILI associates with piRNA (Figures 1D and S1F). In summary, our analysis of piRNA profiles showed that MIWI2 deficiency leads to reduction of piRNAs that are normally MIWI2 bound but does not prevent loading of MILI with piRNA. Furthermore, total abundance of piRNAs was not significantly affected by Miwi2 KO, either because MILI protein and its associated piRNAs are more abundant than MIWI2 and associated piRNAs or because MILI is able to compensate for the absence of MIWI2.
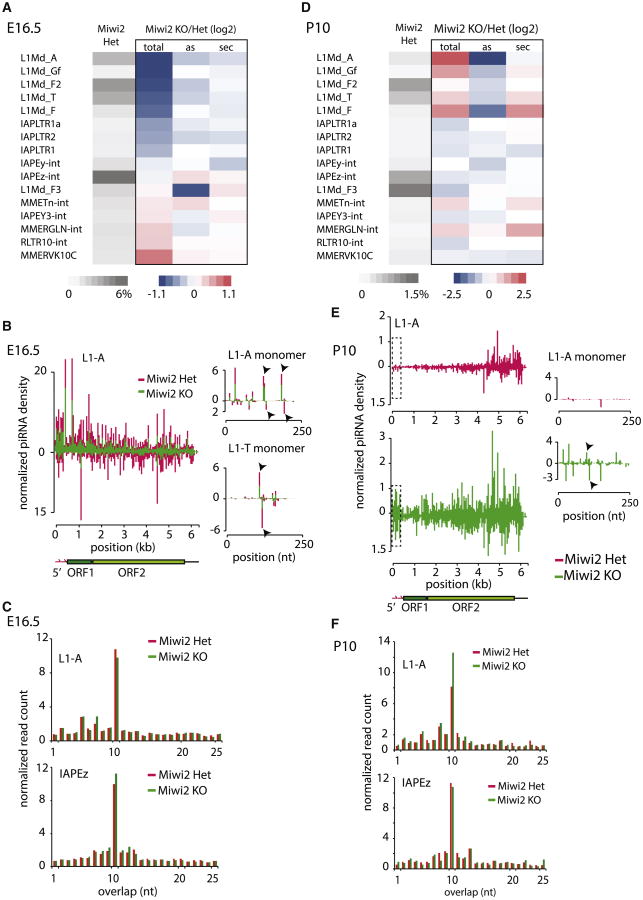
Though total levels of piRNA were not significantly affected by Miwi2 deficiency, further analysis showed a pronounced effect of Miwi2 KO on piRNAs specific for certain TE families. PiRNAs against LINE1 families, such as L1-A, L1-T, and L1-Gf, were reduced approximately 2-fold (Figure 2A). The effect of Miwi2 on piRNA levels was specific for a few LINE1 families, as levels of piRNAs matching IAPEz and other ERV families were not decreased in Miwi2 mutant mice (Figures 2A and S2).For affected L1 families, levels of piRNAs distributed throughout the body of the transposable element were reduced in the Miwi2 KO mice compared to levels in wild-type controls (Figure 2B). The reductions in piRNAs targeting L1-A and L1-T 5′ monomers, which are repeated several times at the 5′ end of LINE and function as a promoter, were also significant (Figure 2B).
Figure 2. The Effect of Miwi2 Knockout on piRNA Populations.
(A) The piRNA levels of young LINE1 families are decreased upon Miwi2 KO in prospermatogonia. The heatmap shows expression of piRNA targeting selected TE families in E16.5 prospermatogonia (gray) and the change in piRNA abundance (“total”) in Miwi2 mutants relative to Miwi2 Het mice. In addition, the heatmap shows the effect of Miwi2 deficiency on the fraction of antisense sequences (“as”) and fraction of secondary piRNA (“sec”, defined as reads that have adenine at position 10 and do not have uridine residue at position one). The differences between KOand Het mice are shown as log2 of ratios of values. Several L1 families show ∼2-fold decrease in piRNA abundance; however, the fractions of antisense and secondary piRNA from these families do not differ significantly when data from _Miwi2_-KO and Het mice are compared.
(B) The distribution of piRNAs along L1-A body in E16.5 prospermatogonia. miRNA-normalized piRNA density shows that Miwi2 deficiency causes a reduction of both sense and antisense piRNA mapping to L1 consensus in E16.5 prospermatogonia. Separately shown are piRNA profiles on 5′ monomer sequences of two L1 families, L1-A and L1-T. Each bar represents a 5′ end of a piRNA mapping to either sense or antisense strand of the element. Arrows indicate the 5′ ends of sense and antisense ping-pong partners (piRNA on two opposite strands those 5′ ends are separated by 10 nt).
(C) The effect of Miwi2 deficiency on the ping-pong piRNA biogenesis in E16.5 prospermatogonia. The E16.5 piRNA mapped to L1 and IAPEz consensuses with up to three mismatches were analyzed to find the signature of the ping-pong processing: complementary piRNA reads those 5′ ends are separated by 10 nt. The graph shows fraction of reads (y axis) whose 5′ ends overlap by a certain number of nucleotides (xaxis). The prominent peak at 10 nt that indicates ping-pong biogenesis is not significantly affected by Miwi2 deficiency.
(D) The levels of L1 piRNAs are increased in Miwi2 mutants in P10 spermatogonia. The analysis and heatmap representation of piRNA at P10 spermatogonia are as described in (A). At P10, Miwi2 deficiency causes increase in total abundance of L1 piRNA as well as increase in the fraction of sense (decrease in antisense) piRNA relative to control.
(E) The distribution of piRNAs along L1-Abody in P10 spermatogonia. miRNA-normalized piRNA densities show that Miwi2 deficiency causes increases in the levels of piRNA mapping to L1 consensus in P10 spermatogonia. More piRNAs are derived from 5′ end of L1-A in _Miwi2_-KO mice than in Het controls. Each bar on L1-A 5′ monomer represents a 5′ end of a piRNA mapping to either sense or antisense strand of the element. Arrows point to the 5′ ends of ping-pong partners, evident only in _Mwi2_-KO animals.
(F) The effect of Miwi2 mutation on the ping-pong piRNA biogenesis in P10 spermatogonia. The analysis was performed as described in (C). The signature of the ping-pong processing, the fraction of piRNA pairs that overlap by 10nt, is increased in _Mwi2_KO mice relative to controls for L1-A, but notfortheIAPEz element.
See also Figure S2.
We next analyzed the effect of Miwi2 deficiency on ping-pong processing. The extent of ping-pong amplification can be measured by quantifying two different parameters: the number of ping-pong pairs (pairs of piRNAs that have 10-nt overlaps between 5′ ends) and the fraction of secondary piRNAs (reads that have A at position 10 and do not have U at 1). Both measurements revealed that ping-pong processing was not strongly affected by the MIWI2 deficiency (Figures 2A and 2C). Even piRNAs with reduced levels, such as those targeting L1 families, preserved the signs of ping-pong processing. Indeed, prominent ping-pong pairs were detected on L1 consensus sequences in Miwi2 KO samples, and quantitative analysis showed only a moderate decrease in ping-pong processing when Miwi2 KO and wild-type samples were compared (Figure 2C). Overall, our data show that MIWI2 is largely dispensable for ping-pong amplification in embryonic testes. Similarly, mutation in one of the two ping-pong partners, Ago3, in Drosophila does not stop ping-pong; however, Ago3 function is required to enrich piRNA populations for antisense molecules that target transposon transcripts (Li et al., 2009). Our analysis shows that, in contrast to the effect of the Ago3 deletion in flies, Miwi2 deficiency did not lead to a significant decrease in fraction of antisense piRNA (Figure 2A). The fraction of antisense piRNA reads did not decrease even for those that target L1 TE families that have reduced abundance of complementary piRNA in the Miwi2 mutants. Taken together, our data reveal similarities and differences between operation of the ping-pong cycle in flies and mice. In both organisms, two Piwi proteins are involved in the ping-pong cycle, but the cycle operates with just one protein (Aub in flies and MILI in mice). The second Piwi protein in flies, Ago3, is required to generate piRNA populations highly enriched in antisense piRNA sequences, whereas MIWI2 in mice does not affect strand bias.
In addition to evaluating effect of Miwi2 deficiency on piRNA populations expressed in embryonic testes (E16.5), we compared piRNA profiles in testes of KO and control mice after birth (P10), the stage of spermatogenesis when only MILI is expressed. In striking contrast to reduction of the L1 piRNAs observed in Miwi2 KO animals in E16.5 prospermatogonia, the piRNA levels for the same L1 families were actually increased in P10 compared to controls (Figures 2D and S2D). The fraction of sense-strand piRNA for several L1 families were increased in Miwi2 KO mice at P10 relative to E16.5, indicating that these piRNAs are processed from L1 mRNAs (Figure 2D).
Further we analyzed the distribution of piRNAs along the body of the L1 transposon. The L1-derived piRNAs in control P10 spermatogonia showed a clear gradient toward the 3′ end of the element (Figure 2E). This gradient likely reflects the fact that a majority of genomic L1 copies are truncated at 5′ ends, and piRNAs are generated from all genomic copies including truncated ones. The truncated copies lack functional promoters indicating that transcription of a substrate for piRNA processing can be initiated from non-TE promoters. In Miwi2 KO spermatogonia the fraction of piRNAs that map to the 5′ end of the L1 copy was higher by ∼5-fold and the 5′ to 3′ gradient almost disappeared, making the profile similar to that observed in wild-type E16.5 prospermatogonia (Figures 2E and 2B). This result indicates that reactivation of full-length L1 copies in Miwi2 KO spermatogonia leads to piRNA generation from these copies, which were silenced in wild-type P10 spermatogonia. In addition to an increase in total abundance and fraction of sense-strand piRNA sequences, we found a moderate increase in ping-pong piRNA processing of upregulated L1 families (Figure 2F). Together, this comprehensive analysis of the piRNA population suggests that Miwi2 deficiency leads to a decrease in L1-derived piRNA abundance early in the development (E16.5). Later during development (P10), derepression of L1 caused by a lack of MIWI2 causes the rise in levels of the sense piRNAs processed from upregulated L1 mRNAs by the ping-pong mechanism.
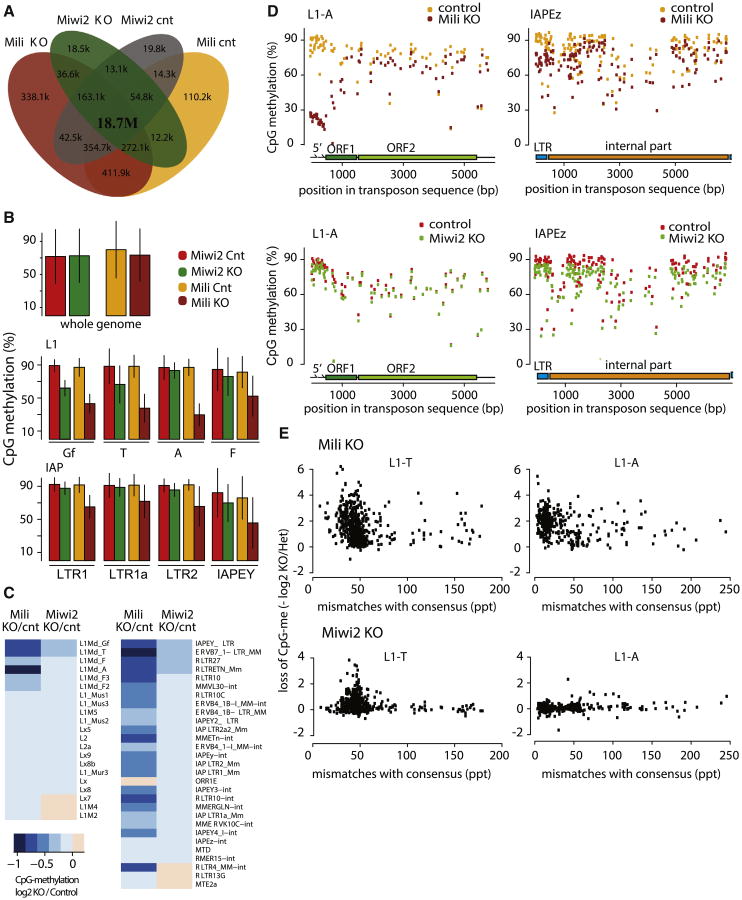
Both Mili and Miwi2 are involved in de novo CpG methylation of several TE families in embryonic prospermatogonia (E15.5 to P1), exactly the time that coincides with MIWI2 expression and nuclear localization (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008). Genome-wide analysis of the effect of Mili KO on DNA methylation was recently reported (Molaro et al., 2014). To obtain similar data for Miwi2-KO mice, we use bisulfite conversion coupled with deep sequencing (BS-seq) for genome-wide profiling of CpG methylation patterns in Miwi2 KO and control spermatogonia isolated from P10 testes. BS-seq libraries were sequenced to a depth comparable with previously published Mili KO data, and datasets were processed using the same bioinformatics pipeline (Figure 3A; Table S3). We first determined the level of CpG methylation on ∼19 million genomic CpG sites (88% of CpG sites in mouse genome) that can be unambiguously detected in the shallowest BS-seq library. Overall, genome-wide methylation levels were very similar among the four samples (Mili KO, Miwi2 KO, and respective controls); ∼70% of CpG sites were methylated in each sample (Figure 3B). As expected, we detected high, and similar, levels of methylation (∼90% mean methylation) on CpG sites within many TE families, including those with expression regulated by MILI and MIWI2, such as L1 and LTR retrotransposons, when control samples were compared (Figure 3B). These results indicate that the efficiency of methylation detection in our experiment was similar to that in the previously published Mili dataset; this allowed us to directly compare the effect of Mili and Miwi2 deficiencies on CpG methylation patterns.
Figure 3. Differential Effect of Mili and Miwi2 Knockouts on CpG Methylation of Transposable Elements.
(A) Most genomic CpG sites are detected in each individual BS-seq library (Mili and Miwi2 KO and corresponding controls). The Venn diagram shows overlap between CpG sites detected by at least one sequencing read in Miwi2 KO and Mili KO BS-seq and respective control datasets. The majority of CpG sites (18.7 million or 85.3%) were detected by at least one uniquely aligned read in each dataset.
(B) The effect of Miwi2 and Mili KO on CpG methylation genome-wide and in specific TE families. The levels of genome-wide CpG methylation are similar (∼70%) in germ cells of each of the four different genotypes (top). CpG methylation of TE families is also similar between the two control datasets. CpG methylation is decreased on both L1 and IAP (LTR) families in the _Mili_-KO sample relative to control. Miwi2 KO only affects L1-Gf and L1-T families and decrease in fraction of methylated CpG sites is slight but significant. The bars represent mean methylation in TE copies belonging to the indicated families and whiskers showSD. For L1 elements only those CpG sites that are located in the promoter regions (i.e., the first 500 bp of a corresponding consensus sequence) were used to evaluate CpG methylation levels.
(C) The effect of Miwi2 and Mili KO on TE methylation. Mili deficiency leads to a decrease in CpG methylation of a greater number of TE families than does Miwi2 KO. The heatmap shows log2 ratio of mean methylation level in Mili and Miwi2 mutants and corresponding control samples.
(D) Effect of Mili mutation on CpG methylation is most pronounced at CpGs in the 5′ end of LINE1 and the LTR portion of IAPEz. BS-seq reads from _Mili_-KO, _Miwi2_-KO, and corresponding control libraries were aligned to L1-A and IAPEz transposon sequences. Each point on the plots represents the mean methylation level at specific CpG site.
(E) Mili and Miwi2 deficiencies affect DNA methylation of young, less divergent TE copies. Each point on the graph represents an individual genomic TE copy that has different level of divergence from the consensus sequence. The x axis reflects the number of mutations in parts per thousand (ppt). The y axis shows loss of CpG methylation in mutants versus controls (negative value of the log2 ratio). L1-T copies that are less divergent from the consensus show loss of CpG methylation in both mutants. CpG methylation of L1-A elements is only affected by Mili KO.
See also Figure S3.
First, our data show that Miwi2 KO does not have a global effect on DNA methylation. As previously reported, Mili deficiency leads to demethylation of many TE families, including both L1 and LTR (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008; Molaro et al., 2014). Remarkably, the effect of Miwi2 KO was modest compared to that of Mili; fewer families were affected in the Miwi2 KO than in the Mili KO, and even in the affected families, the magnitude of methylation loss compared to control was lower (Figure 3C). Nevertheless, our genome-wide profiling of CpG methylation in Miwi2-KO samples revealed an appreciable (15%–20%) decrease of CpG methylation on active L1 families L1-T and L1-Gf (but not L1-A) and LTRs of several ERV families, including IAPEy, relative to levels in control samples. The extent of demethylation was even stronger when assessed by CpG sites located in L1 promoter regions (Figure 3B) Moreover, there is a significant correlation between demethylation and magnitude of derepression of LINE and ERV families in Miwi2 KO P10 spermatogonia (Figure S3).
Next, we analyzed levels of CpG methylation along the body of different transposable elements (Figure 3D). L1 families harbor several CpG-rich ∼250-bp repeats at their 5′ ends that act as a promoter; in the body of L1 elements, CpG density is lower. CpG sites within 5′ repeats were highly methylated (∼90%). CpG sites in L1 bodies had lower levels of methylation; however, methylation exceeded 60% for the majority of the sites, with notable exceptions of a few sites that were methylated at 0%–30%. Supporting bulk analysis of CpG sites, Miwi2 deficiency did not have an effect on methylation of any particular CpG site in the L1-A element. In contrast, our analysis confirmed previously reported result that Mili deficiency led to a dramatic decrease of CpG methylation at 5′ repeats in the promoter of L1-A from ∼90% to ∼20%, with effects in the body much less pronounced. Promoters of ERV elements are situated in LTR regions that have higher concentrations of CpG sites in comparison to the bodies of these elements. Similar to the effect at the L1 element, Mili deficiency had a stronger effect on CpG sites in the LTR promoter region than in the internal region of IAPEz. In contrast, Miwi2 deficiency did not lead to an appreciable decrease of CpG methylation of the LTR or the internal region of IAPEz (Figure 3D).
We next looked at the effect of Miwi2 and Mili KOs on individual copies of a TE within each family. The successful L1 and ERV TE families are represented by thousands of copies scattered throughout the genome. Individual TE copies have different levels of divergence from the active so-called consensus sequence. Less divergent copies of transposons represent recent, potentially active elements, whereas more divergent older copies are potentially damaged inactive elements. We found that Miwi2 as well as Mili deficiency predominantly affected methylation of younger elements within L1-T and other families (Figure 3E). Overall, our data revealed unexpected differences between effects of Mili and Miwi2 on DNA methylation of TE elements: Miwi2 KO had less pronounced effects in comparison to Mili KO, suggesting independent functions of the two in DNA methylation.
Discussion
Our results shed light on the distinct roles of two Piwi proteins, MILI and MIWI2, in TE repression in mammalian germ cells. The proteins have non-redundant roles in silencing of the young and active families of TE, L1-T, L1-Gf, and IAPEy transposons in developing male germ cells. Although MIWI2 affects the biogenesis of far fewer piRNAs compared to MILI, MIWI2 is required to generate high levels of piRNAs targeting several LINE1 families. Indeed, Miwi2 deficiency led to ∼2-fold reduction of embryonic piRNAs against L1-A, L1-T, and L1-Gf families (Figure 2). Both MILI and MIWI2 participate in the ping-pong cycle in the cytoplasm (Aravin et al., 2007, 2008; Shoji et al., 2009). Our results show that in the absence of MIWI2, the ping-pong continues to work. Furthermore, unlike mutation in one of the two ping-pong Piwi partners in flies, Miwi2 deficiency does not lead to decrease in the fraction of antisense piRNAs. It is important to note, however, that in contrast to fly piRNAs that are highly enriched in antisense sequences, piRNAs in mouse do not have a strong antisense bias, even when both Piwi proteins are functional. Overall, our results indicate that MILI alone is sufficient to maintain the proper quality (though not the quantity) of piRNAs in mice. This conclusion corroborates previous findings that the ping-pong cycle operates in the absence of MIWI2 in post-embryonic spermatogonia (Aravin et al., 2007) and that ping-pong amplification occurs in the presence of an MIWI2 point mutant that is impaired in endonuclease activity (De Fazio et al., 2011). It should be stressed that ability of MILI operate the ping-pong processing cycle alone does not mean that MIWI2 is not involved in this process in wild-type cells. MIWI2 clearly participates in the ping-pong processing (Aravin et al., 2008), and our data highlight the high level of redundancy and robustness present in the system that ensures efficient TE silencing.
Investigation of piRNA profiles at two developmental stages (E16.5 and P10) in germ cells of _Miwi2_-KO mice illuminated the existence of a strong feedback loop between piRNA generation and TE expression that adjusts the piRNA population to match expression of specific transposon families. Indeed, the increase in expression of L1 families due to Miwi2 deficiency correlates with increase in ping-pong processing and generation of sense piRNA from these transcripts. These results highlight the essential role of the ping-pong cycle in fine-tuning the piRNA population to match the levels of TE expression.
Comparative analysis of effects of Miwi2 and Mili deficiencies on CpG methylation of transposon sequences unexpectedly revealed independent functions of two proteins in this process. Previously, MILI and MIWI2 were thought to cooperate in linear hierarchical pathway to achieve CpG methylation of their targets. MIWI2 was considered to be directly involved in CpG methylation based on its prominent nuclear localization in prospermatogonia at the time when de novo methylation patterns are established (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008). MILI, concentrated in the cytoplasm of prospermatogonia, was thought to be upstream of MIWI2, as it is required for loading of MIWI2 with piRNAs and subsequent nuclear localization (Aravin et al., 2008; De Fazio et al., 2011). According to this proposed linear pathway, the effect of Mili mutation on DNA methylation was interpreted as indirect and mediated by MIWI2 protein. This model predicts that the effects of Mili and Miwi2 mutations on CpG methylation should be identical. However, our analysis of genome-wide changes in CpG methylation in two mutants showed that MILI has a significant impact on TE methylation that is independent of MIWI2. Indeed, MILI is responsible for DNA methylation of a wider range of TEs than MIWI2, suggesting that MILI can act to induce DNA methylation independently of MIWI2. Similar observations were made by Nagamori and colleagues, which supports our conclusion of differences between functions of MILI and MIWI2 (Nagamori et al., 2015). This result is in line with the observation in the HSP90α mutant, in which transposable elements are upregulated, piRNA levels are decreased and MIWI2 fails to localize to the nucleus, but DNA methylation at a number of elements in not affected (Ichiyanagi et al., 2014). Future studies should reveal whether MILI is present in the nucleus and directly involved in CpG methylation through recognition of nascent TE transcripts or whether it impacts DNA methylation by another mechanism.
In summary, our results demonstrate that MILI and MIWI2, the two mouse Piwi proteins involved in TE silencing in the male germline, do not act in a linear hierarchical pathway but have partially independent functions. MIWI2 is required to silence most active TE families; indeed, these families require both MILI and MIWI2 for silencing.
Experimental Procedures
Animals
The _Miwi2_-KO mouse model has been described previously (Kuramochi-Miyagawa et al., 2008). The _MM_- KO model is described in Kuramochi-Miyagawa et al. (2004). All experiments were conducted in accordance with a protocol approved by the institutional animal care and use committee of the California Institute of Technology.
RNA Isolation, Poly(A)+ Selection, and RNA-Seq Library Cloning
Total RNA was isolated from total testis using Ribozol, and RNA was treated with DNase. For library cloning, 1 μg DNase-treated RNA was poly(A)+ selected and RNA-sequencing (RNA-seq) libraries were cloned using either Illumina TruSeq RNA sample prep kit (version 2, 48 rxns, RS-122-2001) or NEB Next Ultra RNA library prep kit for Illumina (New England Biolabs, E7530). Small RNAs (19–30 nt) were extracted from total RNA by separation on 12% polyacrylamide gel and cloned as described elsewhere (Pastor et al., 2014); for a detailed protocol, see Supplemental Experimental Procedures.
Genomic DNA Isolation, Bisulfite Conversion, and Library Cloning
Cells were isolated from testes using magnetic-activated cell sorting and genomic DNA isolated as described in Pezic et al. (2014) and Supplemental Experimental Procedures. Genomic DNA (200 ng) was sonicated in 100 μl 1× TE and libraries cloned using NuGEN Ovation UltraLow MethylSeq Multiplex System 1-8 kit (NuGEN 0335-32) according to the manufacturer's instructions. Bisulfite conversion of DNA is performed after adaptor ligation using this kit, and it was done using the Active Motif MethylDetector Kit (#55001) per the manufacturer's protocol.
Bioinformatics Analysis of Gene and Transposon Expression
Upon adaptor trimming, RNA-seq reads were aligned to mm10 genome with Bowtie 0.12.7 (Langmead et al., 2009) either in unique alignment mode (to assess gene expression) or permitting reads with multiple valid alignments (to assess expression of TE families). Annotation of gene territories was based on RefSeq transcriptome annotation (Pruitt et al., 2009), and annotation of individual TEs was obtained via RepeatMasker (http://www.repeatmasker.org) track for mm10 genome from the UCSC Genome Bioinformatics Site (Karolchik et al., 2014). Reads with 5′ coordinates intersecting individual TE copies were totaled up for each family weighing each alignment instance by the total number of alignments per read. See Supplemental Experimental Procedures for details.
Bioinformatics Analysis of Bisulfite-Sequencing Libraries to Evaluate CpG Methylation
Upon trimming adaptors, bisulfite-sequencing (BS-seq) libraries were aligned to mm10 genome with Bismark (Krueger and Andrews, 2011). Methylation level of individual CpG sites was determined using MethPipe computational tool suite (Song et al., 2013). To evaluate mean CpG methylation of TE families, we considered TE copies with at least ten CpG sites detected by at least ten sequencing events. Analysis of all libraries was based on a consensus set of ∼19 million CpG sites (88% of genomic sites) that were detected in the shallowest library in this study. See Supplemental Experimental Procedures for details.
Supplementary Material
2
Highlights.
- MILI deficiency induces expression of more transposon families than MIWI2 deficiency does
- Ping-pong piRNA amplification in prospermatogonia can occur without MIWI2
- MILI is responsible for DNA methylation of a larger subset of TE families than MIWI2 is
Acknowledgments
We thank Katalin Fejes Tóth and members of the Aravin lab for helpful discussion and comments on the manuscript. We thank Alyssa Maskell (Caltech) for assistance with mouse work. We thank Igor Antoshechkin (Caltech) for help with RNA-seq and Maria Ninova for help with genome annotation. This work was supported by grants from the NIH (R00 HD057233 and DP2 OD007371A) and by the Searle Scholar Award and Packard Fellowship (to A.A.A.).
Footnotes
Accession Numbers: RNA-seq and BS-seq libraries are available through GEO: GSE70731.
Author Contributions: S.A.M., D.P., and A.A.A. designed the experiments. D.P. performed the experiments, S.A.M. performed computational analysis of all deep-sequencing data, and G.K.M. performed ping-pong analysis. R.S. created bioinformatics tools for small RNA analysis. W.A.P. cloned and sequenced RNA-seq libraries from Mili mutant. S.A.M., D.P., and A.A.A. interpreted the data and wrote the manuscript.
References
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van deKant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Darricarrère N, Liu N, Watanabe T, Lin H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc Natl Acad Sci USA. 2013;110:1297–1302. doi: 10.1073/pnas.1213283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi T, Ichiyanagi K, Ogawa A, Kuramochi-Miyagawa S, Nakano T, Chuma S, Sasaki H, Udono H. HSP90α plays an important role in piRNA biogenesis and retrotransposon repression in mouse. Nucleic Acids Res. 2014;42:11903–11911. doi: 10.1093/nar/gku881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25–R10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 2014;28:1544–1549. doi: 10.1101/gad.244350.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamori I, Kobayashi H, Shiromoto Y, Nishimura T, Kuramochi-Miya-gawa S, Kono T, Nakano T. Comprehensive DNA methylation analysis of retrotransposons in male germ cells. Cell Rep. 2015;12 doi: 10.1016/j.celrep.2015.07.060. Published online August 27, 2015. http://dx.doi.org/10.1016/j.celrep.2015.07.060. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc'his D, et al. MORC1 represses transposable elements in the mouse male germline. Nat Commun. 2014;5:5795. doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezic D, Manakov SA, Sachidanandam R, Aravin AA. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 2014;28:1410–1428. doi: 10.1101/gad.240895.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armit-age and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS ONE. 2013;8:e81148. doi: 10.1371/journal.pone.0081148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2