Phenotypic and Genotypic Characteristics of Small Colony Variants and Their Role in Chronic Infection (original) (raw)
Abstract
Small colony variant (SCV) bacteria arise spontaneously within apparently homogeneous microbial populations, largely in response to environmental stresses, such as antimicrobial treatment. They display unique phenotypic characteristics conferred in part by heritable genetic changes. Characteristically slow growing, SCVs comprise a minor proportion of the population from which they arise but persist by virtue of their inherent resilience and host adaptability. Consequently, SCVs are problematic in chronic infection, where antimicrobial treatment is administered during the acute phase of infection but fails to eradicate SCVs, which remain within the host causing recurrent or chronic infection. This review discusses some of the phenotypic and genotypic changes that enable SCVs to successfully proliferate within the host environment as potential pathogens and strategies that could ameliorate the resolution of infection where SCVs are present.
Keywords: auxotrophy, adhesion, intracellular, biofilm, phenotype
Discovery of Small Colony Variants (SCVs)
Pure bacterial cultures are not genetically homogeneous, and their behavior is determined by genomic characteristics, such as a high degree of plasticity. Slow-growing subpopulations of bacteria in pure culture have been described from as early as 1913; reported to emerge in response to diverse environmental pressures, they were termed SCVs because they formed pin-prick-sized colonies when cultured on solid media.1,2 Initially, SCVs were thought of as morphological variants with a secondary role in infectious disease because of their markedly diminished pathogenicity and impaired production of virulence factors.2 Furthermore, it was believed that the G forms, as they were referred to, may even constitute an ordinary part of the microbial life cycle.3,4 It was not for many decades following their initial phenotypic characterization that the pathogenic potential of SCVs was realized and their presence within a microbial community was regarded as more than a laboratory curiousity.5,6
Early studies clarified the link between environmental stress and the phenotypic changes that became associated with SCVs, including atypical colony morphology, slow growth rate, lack of pigmentation, reduced hemolytic activity, reduced coagulase activity, reduced carbohydrate utilization, low virulence potential, and elevated antibiotic resistance (Fig. 1).7–9 Indeed, the growth rate of SCVs has been estimated to be approximately nine times slower than that of the progenitor organisms.10 As such, SCVs are now better defined as a microbial subpopulation constituting a naturally occurring, slow-growing but diverse bacterial morphotype.7,11 Clinically, this is problematic; the presence of SCVs during infection is correlated with recurrent or chronic infectious disease. A combination of extended incubation time in addition to altered phenotypic and biochemical traits often means that SCVs in patient samples are overlooked by clinical microbiologists utilizing conventional diagnostic tests. This results in the cessation of antimicrobial treatment before SCVs are effectively cleared from an infection; therefore, they persist causing recurrent and chronic infection.7,12
Figure 1.
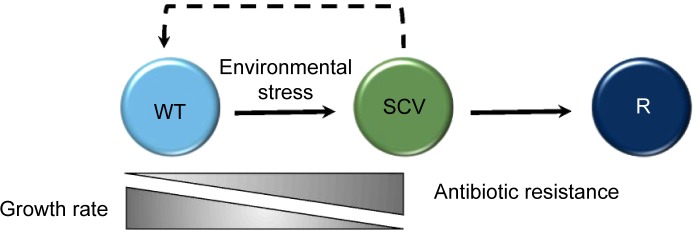
WT organisms undergo a shift to the SCV phenotype under the conditions of stress, where they exhibit a slower growth rate but increased antibiotic resistance. They can revert to a WT-like (indicated by a dashed line to denote that WT-like organisms are not identical to the original WT progenitor) or alternative revertant (R) phenotype when the environmental stress is removed, regaining a faster growth rate but becoming more susceptible to antibiotic treatment.
Various environmental stimuli appear to result in phenotypically distinct varieties of SCV.7,13 Some undergo permanent genetic changes, whereas a subpopulation reverts to a wild-type (WT) phenotype or to a different phenotype that is distinct from both the progenitor and the SCV upon repeated subculture (revertant phenotype) (Fig. 1).14 Phenotypic reversion, where genetic mutations have not occurred, happens rapidly and circumvents any permanent fitness costs.14 The tendency to permanent genetic alteration as compared to phenotypic reversion seems to depend largely on the nature of the original environmental pressure.7 There is not always commonality between phenotypic and genotypic changes within different SCV populations, but there are a number of prevalent auxotrophies, characterized initially in Staphylococcus spp., including hemin, thiamine, menadione, thymidine, or unsaturated fatty acids.7,14–17 Conversely, some SCVs do not demonstrate these auxotrophies,18–21 for example, those of Staphylococcus aureus, selected by Triclosan™ treatment, which revert neither upon supplementation with growth factors nor by repeated subculture.22,23 The reasons why SCVs are phenotypically diverse remain unclear beyond an unproven link to certain environmental contraints.7,24,25
Unique Phenotypic Traits Associated with SCVs
In addition to the auxotrophies described above, there are a number of other phenotypic characteristics typically associated with SCVs that likely contribute to their ability to persist under adverse growth conditions. One such conventional SCV attribute is diminished electron transport, observed in various species of Staphylococcus, Enterococcus, and Pseudomonas. This phenotype arises as a consequence of mutations impairing the function of menadione, hemin, and thiamine, all of which are required for the biosynthesis of components of the electron transport chain (Table 1).7,14–16,26,27 Ordinarily, menadione is isoprenylated to form menaquinone, the acceptor of electrons from nicotinamide adenine dinucleotide (NADH)/flavin adenine dinucleotide (FADH2) in the electron transport chain, which does not occur in some SCVs.10,28,29 Subsequent reduced electron transport results in a decreased electrochemical gradient and therefore reduced synthesis of adenosine triphosphate (ATP). Large amounts of ATP are required for cell wall biosynthesis, and electron transport is directly linked to the biosynthesis of carotenoid pigments, rendering many SCVs of pigmented species colorless.
Table 1.
Characteristics associated with SCVs.
| GENE | PROTEIN | FUNCTION | VARIATION | EXAMPLE ORGANISM(S) | SELECTED REFERENCES |
|---|---|---|---|---|---|
| Altered interaction with the host | |||||
| clfA | Clumping Factor A | Fibrinogen binding | Increased expression in hemB background | Staphylococcus spp. | 57 |
| fnb | Fibronectin Binding Protein | Fibronectin binding | Increased expression in hemB background | 57 | |
| spa | Protein A | Surface protein, inhibits phagocytosis | Transcription reduced in SCV: avoidance of host immunity | 57 | |
| Altered biosynthesis or enzymatic pathways | |||||
| hemL | Glutamate 1-semi-aldehyde aminotransferase | Porphyrin biosynthesis | Gene interruption causing persistent infection | Staphylococcus spp. Pseudomonas aeruginosa | 52 |
| hemB | Porphobilinogen synthase | Porphyrin biosynthesis | Gene interruption causing persistent infection | Escherichia coli Salmonella enterica sv. Typhimurium Enterococcus spp. | 52 |
| menD | 2-succinyl-6-hydroxy-2,4-cyclohexadine-1-carboxylate synthase | Cytochrome biosynthesis | Gene interruption causing persistent infection | Staphylococcus spp. Pseudomonas aeruginosa Enterococcus spp. | 7 |
| ctaB | Haem-O-monooxygenase | Cytochrome biosynthesis | Gene interruption causing persistent infection | Staphylococcus aureus | 7, 39 |
| citB | Aconitase | Catalyses isomerization of citrate to isocitrate in the tricarboxylic acid cycle | Down-regulated in hemB background | Staphylococcus spp. | 13 |
| aroD | 5-dehydroquinate hydrolyase | Menadione biosynthesis | Defective in SCV: increased persistence | Staphylococcus spp. Salmonella enterica sv. Typhimurium | 26 |
| ldh | Lactate dehydrogenase | Converts pyruvate to lactate in hypoxic/anoxic conditions | Defective in SCV: increased persistence | Staphylococcus spp. | 26 |
| thyA | Thymidylate synthase | Catalyses the conversion of deoxyuridine monophosphate (dUMP) to deoxythymydine monophosphate (dTMP) | Varied mutations resulting in thymidine auxotrophy | Stenotrophomonas maltophilia Staphylococcus aureus Pseudomonas aeruginosa Enterococcus spp. | 13, 14 |
| Transcriptional regulation | |||||
| agr | Accessory gene regulator | Global virulence regulator— quorum sensing | Impaired expression in SCV: chronicity | Staphylococcus aureus | 38, 39 |
| sarA | Accessory gene regulator | Global virulence regulator— biofilm formation | Impaired expression in SCV: chronicity | Staphylococcus aureus | 60, 62 |
| sigB | Alternative stress regulator | Alternative stress regulator—intracellular persistence | Down-regulated or silenced in SCV: increased intracellular persistence and resistance to hydrogen peroxide stress | Bacillus cereus Staphylococcus aureus | 58, 59, 61, 63 |
| Miscellaneous function | |||||
| mutL | Member of MutHLS complex | Methyl-directed mismatch repair (MMR) system | Gene truncated due to frameshift mutations in thymidine-dependent SCV isolates: hypermutability | Pseudomonas aeruginosa | 65 |
| nupC | Nucleoside permease | High affinity nucleoside transporter | Gene mutations in thymidine-dependent SCVs | Stenotrophomonas maltophilia Staphylococcus aureus | 7 |
| hla | α-hemolysin | Initiates eukaryotic cell apoptosis and necrosis | Expression impaired in SCV: attenuated virulence and enhanced intracellular persistence | Staphylococcus aureus | 7 |
The unique cell wall structure of SCVs is believed to confer some degree of protection from stress and is allied with aberrant electron transport. Abnormal cell division has been described for SCVs of S. aureus, causing inappropriate cell wall biosynthesis and growth of unusually large cells.21,30–32 SCVs of S. aureus remain to be some of the best characterized, and when examined by electron microscopy are revealed to be a heterogeneous population of differing size, including “empty” cells and substantial amounts of debris.29,30,33 Moreover, while WT S. aureus are spherical, with thin cell walls and a relatively uniform cytoplasm,34,35 their SCV counterparts tend to exhibit much thicker cell walls with irregular cytoplasm of dense granular appearance at the periphery and fine granular materials at the center.34 Additionally, SCVs with incomplete, branched, or multiple cross walls without regular cell separation are often also observed.7,29,30 Ghost or empty cells (as mentioned above and observed also for Enterococcus spp.) devoid of cytoplasmic content and chromosomal or plasmid DNA and with defective cell walls have also been documented; these are categorized as SCVs despite not being viable microorganisms.30–35
Characteristically, in addition to the aforementioned phenotypic changes, the small regulatory RNA molecule, RNAIII, is usually absent from SCVs and has been particularly well defined for SCVs of S. aureus.13,36,37 RNAIII is known to regulate virulence factors, including exoproteins, and cell wall-associated proteins, including adhesins, as well as act as the effector of _agr_-mediated quorum sensing. RNAIII positively regulates the production of toxins and proteases but negatively regulates adhesins, meaning that SCVs tend to be less toxigenic and more prone to adhesion to biotic or abiotic surfaces with enhanced intracellular persistence.38,39 Virulence during infection is reliant on initial colonization of the host; host–pathogen interactions prevail via bridging mechanisms involving bacterial adhesins and corresponding host proteins.40 SCVs adhere to host cells in much the same way as WT microorganisms; the major difference is that SCVs express many more surface adhesins, thus favoring interaction with the host.
Once attached to the host cell, SCVs, like their WT counterparts, induce host-cell changes by actin rearrangement, which mediates internalization, effectively hijacking non-phagocytic cells (including endothelial cells, fibroblasts, osteoblasts, and keratinocytes).41,42 Pathogens that are not categorized as SCVs utilize the same mechanism of internalization, but crucially, SCVs are far more efficient at this process than their progenitors.43 Fundamentally, this intracellular protection affords additional defense against immune clearance or antimicrobial treatment. Owing to their reduced toxicity, the uptake of SCVs in this manner occurs in vitro without damage to host cells.44 Once inside the host cell, intracellular survival is critical to retain protection.40 SCVs characteristically proliferate intracellularly, more successfully than their progenitors, which is a trait that directly contributes to antibiotic treatment failure and poor prognosis in patients.45–47 A marked increase in the expression of member genes of the arginine deaminase pathways in SCVs of S. aureus results in the reduced function of vital host enzymes involved in the immune response and is believed to be key to successful intracellular persistence.36
In addition, SCVs evade the immune response and persist intracellularly by escaping from intracellular phagosomes,43,48 thus avoiding the hydrolytic activity of lysosomes.48 It has been proposed that unlike other intracellular pathogens, once in the cytoplasm, SCVs may no longer disrupt normal actin polymerization of the cells in which they reside, meaning that they do not elicit normal intracellular cytokine and chemokine defense mechanisms.40 Therefore, the ability of SCVs to dampen the proinflammatory response means that the attenuated virulence associated with the SCV phenotype is in fact favorable for their survival and prolonged persistence within the host.46 The recovery of SCVs from the cases of asymptomatic infection supports this theory of persistence through diminished host damage.48,49
Unique Genotypic Features Associated with SCVs
Several genetic mutations can result in the electron transport-defective SCV phenotype described above, including the mutations in menD, hemB, and ctaA.38,44,50,51 MenD encodes for 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, which catalyses the conversion of isochorismate 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate. HemB encodes for porphobilinogen synthase, which is essential for subsequent porphyrin metabolism.36,44,52–54 CtaA encodes haem-O-monooxygenase that converts haem-O to haem-A, which is an essential cofactor for enzymes involved in electron transport,53 and its deficiency inhibits cytochrome biosynthesis.7 Mutations in menD and hemB block the biosynthesis of menadione, which is used in menaquinone biosynthesis. Mutations in menD, hemB, or ctaA can also lead to defective cytochrome biosynthesis.7 Both menD and hemB mutations also impair the biosynthesis of cytochromes.36,44,52,53
During infection, organisms are exposed to high levels of haem that may be toxic because of the accumulation of superoxides. By virtue of the mutations described above, haem stress for SCVs is significantly alleviated, suggesting that a reduction in haem-associated stress may be an additional factor enabling the survival of SCVs during chronic infection.54 Genes governing other aspects of general metabolic pathways associated with energy production and respiration in SCVs often also carry mutations (Table 1). They primarily include genes encoding proteins of the Entner–Doudoroff pathway, reconciling the slower growth rate of SCVs that contributes to their persistence.
Increased adhesion and biofilm formation are correlated with the enhanced expression of surface-bound adhesins and their cognate transcriptional regulators.55–58 Adhesins not only function as a means of binding directly to host proteins prior to colonization but also enable interbacterial aggregation, which is critical to the development of biofilm; SCVs are characteristically prolific biofilm-forming organisms.59 The expression of adhesin genes is often governed by global transcriptional regulators that form part of an intricate transcriptional network that responds to environmental cues, usually involving quorum sensing. Therefore, it is not surprising to find that genes encoding transcriptional regulators, such as agr, sigB, and sarA, are differentially regulated in SCVs of both Bacillus cereus and S. aureus (Table 1).60–65 Indeed, during chronic infection where the SCV phenotype begins to emerge, the expression of these transcriptional regulators is repressed, thus suppressing virulence gene expression, promoting intracellular survival, and dampening the host immune response (Table 1).66
Although differentially expressed or mutated genes tend to be conserved in SCVs, to date no defined core set of SCV genes has been documented. Often SCV-associated phenotypic traits are not the result of permanent genetic mutations but may instead result from genome rearrangements; therefore, identifying SCVs at the genotypic level is potentially as challenging as identifying them based on phenotype alone. Moreover, numerous phenotypic traits can be attributed to epigenetics.67 Where genetic traits are conserved, they usually confer essential adaptations; transient characteristics that are not an absolute requirement for survival, but which confer a competitive advantage, are likely controlled by uncharacterized global transcriptional regulators or alternative sigma factors (Table 1) that form part of a larger and as yet undefined SCV regulon. Since only traits that are conferred by permanent genetic change are heritable, the maintenance of a stable SCV community within the larger microbial consortia is postulated to depend on appropriate regulatory signals. Significantly, DNA mismatch repair systems are often impaired in SCVs, leading to the accumulation of genetic mutations that confer the typical SCV phenotype or, in some cases, result in hypermutability and alternative variant phenotypes.68
SCVs within Microbial Communities
Variants occur at random within microbial populations; most are transitory with only those changes that allow bacteria to remain viable and confer an advantage becoming fixed within a population. Microbial adaptation to a particular environment and competition between the members of a heterogeneous population, comprising a parent (wild type) and progeny (including mutants), are dictated by growth parameters and stresses.69 Numerous laboratory studies have demonstrated that successful microorganisms, namely, those that succeed within a given environment, do so because they exhibit the highest growth rate under prevailing conditions.70 Despite this, SCVs persist within microbial communities, albeit as a minor constituent that is never entirely outcompeted by the parental strain. Given the tenacity of SCVs to survive under stress, it might be expected that they should eventually predominate. Certainly with regard to antimicrobial interventions,71 this is the case, as the more susceptible parent strain is eradicated leaving behind a population of SCVs that can undergo reversion, which results in a recurrence of infection (Fig. 2).72 SCVs that arise during human infection appear better adapted for survival and persistence within the host73 despite their impaired rate of growth and increased host dependency. Selection because of loss or redundancy of metabolic activity is not unusual and under certain growth conditions,74 such as the host environment, might confer a fitness advantage, if not in terms of growth, in terms of survival.75 Many host-adapted and therefore invariably host-dependent pathogens undergo reductive evolution;76 driven by the host habitat and their ability to utilize host metabolites, such organisms become slow growing and nutritionally fastidious, often adopting an intracellular lifestyle.77 This process is ordinarily mediated by the loss of large fragments of genomic DNA. SCVs appear similar to host-adapted pathogens, and the loss of metabolic function renders them reliant on the host to meet their nutritional needs;78 indeed, many SCVs survive intracellularly. Where reductive evolution occurs in pathogens, it is correlated with increased virulence79 that is not observed for SCVs, which conversely seem to exhibit attenuated virulence.80 Significantly, pathogens that undergo reductive evolution do not regain “lost” genetic function81 unlike SCVs that are able to revert to WT or WT like once selective pressures, such as antimicrobial treatment, are removed (Figs. 1 and 2). Therefore, for SCVs, there is apparently a trade-off between virulence and persistence that exploits the ability to revert to a WT or WT-like variant, which is less host dependent and regains virulence. The specific adaptations that the minority SCV population depends on to ensure survival among their faster growing counterparts include the increased expression of surface adhesins as previously described. If two microorganisms are competing for the same human receptor, then those with a binding advantage (ie, more surface adhesins) are more likely to adhere.82,83 Combined with enhanced biofilm formation, such colonizers are less likely to be removed from the host by detachment.84 Intracellular survival provides more than simple protection from immunity, with the cell cytoplasm providing a nutritionally rich habitat for auxotrophic SCVs that is not afforded to the parental strain, but at the same time reduces competition for space at the tissue surface.85 It is proposed that in this way, both SCV and progenitor can coexist.
Figure 2.
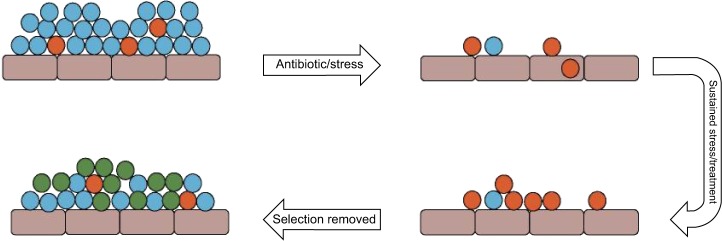
Pure populations of bacteria often comprise WT (major population; blue) and SCV (minor population; orange), which arise spontaneously; under environmental stress, such as antibiotic treatment, the WT population is diminished and SCVs survive; under sustained stress, such as a course of antibiotics to treat an infection, the SCVs become the dominant members of the population. When the selective pressure is removed, WT organisms proliferate and become the dominant members of the population compared to slow-growing SCVs; significantly a proportion of SCVs revert to either the WT phenotype or a WT-like phenotype (green), which regains characteristics that enable faster growth.
SCVs in Chronic Infection
SCVs show enhanced resistance to a range of antibiotics7,25,86–88 and have been directly associated with persistent infections in a number of diseases, including, but not limited to, cystic fibrosis,89,90 chronic obstructive pulmonary disease,91 diabetic foot ulcers,92 chronic rhinosinusitis,93 chronic wound infections,94–96 systemic infections,97 and infections arising from surgical intervention or medical devices,98–102 which can lead to serious and sometimes fatal clinical consequences, such as endocarditis, bacteremia, or meningitis.97,102 It has been proposed that in the case of the ventriculoperitoneal shunt (a medical device used in the treatment of hydrocephalus), SCV-associated meningitis infection can arise from inadequate disinfection of the shunt and failure to identify and treat these persistent variants.103 Chronic infections represent a significant burden to both patients and health-care providers. Where chronicity ensues, biofilm is frequently present.104–108 The presence of SCVs within biofilms has been directly linked to chronic antibiotic-resistant infections, including cystic fibrosis in lung, osteomyelitis, catheter and pacemaker infection, among others as previously described.87–93,108–115 The respiratory tract of cystic fibrosis patients provides a unique environment for the selection of a subgroup of autoaggregative and highly adherent SCVs of Pseudomonas aeruginosa.20,115,116 These SCVs are hyperpiliated and exhibit increased twitching motility as well as have the capacity to emerge and successfully endure in biofilms, thus contributing significantly to the pathogenesis of P. aeruginosa lung infection.115,116 However, it is the hyperaggregative property of these SCVs that is the primary contributing factor to etiology of chronic infection because it enables microorganisms to produce large amounts of polysaccharide intercellular adhesion and highly structured biofilms.117 Significantly, a recent study describing polymicrobial biofilm comprising P. aeruginosa and S. aureus suggests that the cohabitation of these microorganisms not only leads to a more dense and stable biofilm formation but also induces SCV emergence, even in the absence of antibiotics.83 Specifically, SCV of S. aureus emerged following the exposure to 4-hydroxy-2-heptylquinoline-N-oxide, secreted by P. aeruginosa, which is known to impair the growth of S. aureus. Growth impairment was attributed to a shift to the slower growing SCV morphotype.118 This phenomenon has been best studied in cocultured organisms derived from patients with cystic fibrosis who present with chronic infection for which antibiotic treatment is received, where SCVs of S. aureus were identified in 24% of the patients.119 It is believed that for S. aureus, this is a specific survival strategy in the presence of P. aeruginosa, mediating protection from secreted exotoxin A, which targets the electron transport chain.25
The rate of occurrence of SCVs in chronic infection is likely to vary depending on the clinical conditions;7 nonetheless, SCVs are detected in approximately 1% of isolates in a clinical microbiology laboratory and their incidence is the highest in cystic fibrosis and osteomyelitis.7 It is pertinent to highlight that in patients with osteomyelitis, surgical placement of slow-release gentamicin beads along with debridement is a common practice for treatment and may be linked to SCV induction.120 This is of concern as inadvertent iatrogenic-induced SCVs may be formed as a result of the long-term exposure to gentamicin; studies have verified that SCVs can be recovered from patients undergoing treatment with gentamicin beads.120 It has consequently been suggested that routine screening for SCVs should take place for patients treated with gentamicin beads for osteomyelitis.28 Furthermore, given the recalcitrance of SCV-associated infection, it might seem reasonable to screen persons who are predisposed to developing chronic infection following the completion of antimicrobial chemotherapy. Therefore, with regard to efficacious antimicrobial treatments, the identification of SCVs is as important as ensuring an appropriate dose of antimicrobial is administered. However, this approach is confounded by the relatively limited information describing successful treatment of SCV infections. Since aminoglycosides are known to promote the emergence of SCVs in some bacterial species, including P. aeruginosa, they are unlikely to constitute a suitable treatment where such SCVs persist. Vancomycin exhibits a higher degree of efficacy against SCVs than most antibiotics, but its potency is estimated to be approximately half of that typically observed for the treatment of non-SCV organisms.121 It is possible to achieve bactericidal activity against S. aureus hemB mutants using daptomycin, and the effect is concentration dependent, suggesting that at its simplest, SCVs can be effectively treated using higher doses of antibiotics that are normally prescribed to treat infection.122 However, until satisfactory laboratory isolation is achieved, SCVs will remain very difficult to detect in patient samples and will, therefore, remain excluded from the standard antimicrobial testing regimens.
Future Perspectives
Although SCVs have been known to exist for over century, little attention was originally given to them as they were believed to be nonvirulent and therefore not clinically important. However, as more is understood about their role in persistent infections, it has become imperative that mechanisms of SCV persistence and resistance, as well as population dynamics, are thoroughly explored. Recent investigations have proposed a low-cost point-of-care test for the diagnosis of P. aeruginosa in patients at risk of chronic respiratory infection for rapid and economical diagnosis. This method, named electrochemical impedance spectroscopy, has successfully differentiated strains of P. aeruginosa based on their impedance signature, which is influenced by factors such as pyocyanin secretion.123 While this method has not yet been tested using the SCVs of P. aeruginosa, many SCVs exhibit differential pyocyanin production and so could be potentially identified via this means that could replace traditional culture methods. With this in mind, it seems reasonable to suggest that accurate diagnosis of SCV-associated infections will rely on nontraditional diagnostics, including the use of molecular probes, in future. The principle complication for the development of such diagnostic methodology is the varied phenotypic and genotypic traits exhibited by SCVs; without a core set of SCV genes even with new diagnostic techniques, it might prove as easy to misdiagnose SCVs in infection as by traditional culture.
Footnotes
ACADEMIC EDITOR: Raul Rivas, Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1064 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived the concepts: BEJ, KJP, NPT, SEM. Analyzed the data: BEJ, KJP, NPT, SEM. Wrote the first draft of the manuscript: BEJ, KJP, NPT, SEM. Contributed to the writing of the manuscript: BEJ, KJP, NPT, SEM. Agree with manuscript results and conclusions: BEJ, KJP, NPT, SEM. Jointly developed the structure and arguments for the paper: BEJ, KJP, NPT, SEM. Made critical revisions and approved final version: BEJ, KJP, NPT, SEM. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Kolle W, Hetsch H. Die experimentelle Bakteriologie und die Infektionskrankheiten mit besonderer Berücksichtigung der Immunitätslehre. Berlin, Germany: Urban und Schwarzenberg; 1913. [Google Scholar]
- 2.Swingle EL. Studies on small colony variants of Staphylococcus aureus. J Bacteriol. 1935;29(5):467–489. doi: 10.1128/jb.29.5.467-489.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadley P, Delves E, Klimek J. The filterable forms of bacteria: 1. A filterable stage in the life history of the Shiga dysentery bacillus. J Infect Dis. 1931;48(1):1–159. [Google Scholar]
- 4.Hale JH. Studies on Staphylococcus mutation: characteristics of the “G” (Gonidial) variant and factors concerned in its production. Br J Exp Pathol. 1947;28(3):202–210. [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber AU, Craig WA. Aminoglycoside-selected subpopulations of Pseudomonas aeruginosa: characterisation and virulence in normal and leukopenic mice. J Lab Clin Med. 1982;100:671–681. [PubMed] [Google Scholar]
- 6.Musher DM, Baughn RE, Merrell GL. Selection of small-colony variants of Enterobacteriaceae by in vitro exposure to aminoglycosides: pathogenicity for experimental animals. J Infect Dis. 1979;140:209–214. doi: 10.1093/infdis/140.2.209. [DOI] [PubMed] [Google Scholar]
- 7.Proctor RA, von Eiff C, Kahl BC, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4(4):295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 8.McNamara PJ, Proctor RA. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int J Antimicrob Agents. 2000;14(2):117–122. doi: 10.1016/s0924-8579(99)00170-3. [DOI] [PubMed] [Google Scholar]
- 9.Wilson SG, Sanders CC. Selection and characterisation of strains of Staphylococcus aureus displaying unusual resistance to aminoglycosides. Antimicrob Agents Chemother. 1976;10(3):519–525. doi: 10.1128/aac.10.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proctor RA, Kahl B, von Eiff C, Vaudaux PE, Lew DP, Peters G. Staphylococcus small colony variants have novel mechanisms for antibiotic resistance. Clin Infect Dis. 1998;27(suppl 1):S68–S74. doi: 10.1086/514906. [DOI] [PubMed] [Google Scholar]
- 11.Allegrucci M, Sauer K. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol. 2008;190(19):6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Eiff C. Staphylococcus aureus small colony variants: a challenge to microbiologists and clinicians. Int J Antimicrob Agents. 2008;31(6):507–510. doi: 10.1016/j.ijantimicag.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Onyango LA, Dunstan RH, Roberts TK, Macdonald MM, Gottfries J. Phenotypic variants of Staphylococci and their underlying population distributions following exposure to stress. PLoS One. 2013;8(10):e77614. doi: 10.1371/journal.pone.0077614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massey RC, Buckling A, Peacock SJ. Phenotypic switching of antibiotic resistance circumvents permanent costs in Staphylococcus aureus. Curr Biol. 2001;11(22):1810–1814. doi: 10.1016/s0960-9822(01)00507-3. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee I, Herrmann M, Proctor RA, Peters G, Kahl BC. Enhanced post-stationary-phase survival of a clinical thymidine-dependent small-colony variant of Staphylococcus aureus results from lack of a functional tricarboxylic acid cycle. J Bacteriol. 2007;189(7):2936–2940. doi: 10.1128/JB.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besier S, Zander J, Kahl BC, Kraiczy P, Brade V, Wichelhaus TA. The thymidine-dependent small-colony-variant phenotype is associated with hypermutability and antibiotic resistance in clinical Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2008;52(6):2183–2189. doi: 10.1128/AAC.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besier S, Ludwig A, Ohlsen K, Brade V, Wichelhaus TA. Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int J Med Microbiol. 2007;297(4):217–225. doi: 10.1016/j.ijmm.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan ML, Dye W. Growth requirements of some small-colony-forming variants of Staphylococcus aureus. J Clin Microbiol. 1976;4(4):343–348. doi: 10.1128/jcm.4.4.343-348.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan XS, Hamlyn PJ, Talens-Visconti R, Alovero FL, Manzo RH, Fisher LM. Small-colony mutants of Staphylococcus aureus allow selection mediated resistance to dualtarget fluoroquinolones. Antimicrob Agents Chemother. 2002;46(8):2498–2506. doi: 10.1128/AAC.46.8.2498-2506.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Häussler S, Rohde M, Steinmetz I. Highly resistant Burkholderia pseudomallei small colony variants isolated in vitro and in experimental mellioidosis. Med Microbiol Immunol. 1999;188(2):91–97. doi: 10.1007/s004300050110. [DOI] [PubMed] [Google Scholar]
- 21.Häussler S, Ziegler I, Löttel A, et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol. 2003;52(4):295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- 22.Seaman PF, Ochs D, Day MJ. Small-colony variants: a novel mechanism for triclosan resistance in methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2007;59(1):43–50. doi: 10.1093/jac/dkl450. [DOI] [PubMed] [Google Scholar]
- 23.Bayston R, Ashraf W, Smith T. Triclosan resistance in methicillin-resistant Staphylococcus aureus expressed as small colony variants: a novel mode of evasion of susceptibility to antiseptics. J Antimicrob Chemother. 2007;59:848–853. doi: 10.1093/jac/dkm031. [DOI] [PubMed] [Google Scholar]
- 24.Lenhard JR, von Eiff C, Hong IS, et al. Evolution of Staphylococcus aureus under vancomycin selective pressure: the role of the small-colony variant phenotype. Antimicrob Agents Chemother. 2015;59(2):1347–1351. doi: 10.1128/AAC.04508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman LR, Déziel E, D’Argenio DA, et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103(52):19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crompton MJ, Dunstan RH, Macdonald MM, Gottfries J, von Eiff C, Roberts TK. Small changes in environmental parameters lead to alterations in antibiotic resistance, cell morphology and membrane fatty acid composition in Staphylococcus lugdunensis. PLoS One. 2014;9(4):e92296. doi: 10.1371/journal.pone.0092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lannergård J, von Eiff C, Sander G, et al. Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:4017–4022. doi: 10.1128/AAC.00668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dean MA, Olsen RJ, Long SW, Rosato AE, Mausser JM. Identification of point mutations in clinical Staphylococcus aureus strains that produce small colony variants auxotrophic for menadione. Infect Immun. 2014;82:1600–1605. doi: 10.1128/IAI.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley R, Meganathan R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol Rev. 1982;46:241–280. doi: 10.1128/mr.46.3.241-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahl BC, Belling G, Reichelt R, Herrmann M, Proctor RA, Peters G. Thymidine-dependent small-colony variants of Staphylococcus aureus exhibit gross morphological and ultrastructural changes consistent with impaired cell separation. J Clin Microbiol. 2003;41(1):410–413. doi: 10.1128/JCM.41.1.410-413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumert N, von Eiff C, Schaaff F, Peters G, Proctor RA, Sahl HG. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb Drug Resist. 2002;8:253–260. doi: 10.1089/10766290260469507. [DOI] [PubMed] [Google Scholar]
- 32.Mayfield JA, Hammer ND, Kurker RC, et al. The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem. 2013;288(32):23488–23504. doi: 10.1074/jbc.M112.442335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellingausen N, Chatterjee I, Berger A, Niederfuehr A, Proctor RA, Kahl BC. Characterization of clinical Enterococcus faecalis small-colony variants. J Clin Microbiol. 2009;47(9):2802–2811. doi: 10.1128/JCM.00485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki Y, Yamauchi Y, Hayashi H, Takayama Y, Tsuji A. Characterization of small colony variants of methicillin-resistant Staphylococcus aureus regrown in the presence of arbekacin. J Infect Chemother. 1998;4(3):107–111. [Google Scholar]
- 35.Adler H, Schraner EM, Frei R, Wild P. Ultrastructure of a clinical isolate of Staphylococcus aureus small colony variant and its revertant. Microsc Microanal. 2005;11(2):982–983. [Google Scholar]
- 36.Vaudaux P, Francois P, Bisognano C, et al. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect Immun. 2002;70(10):5428–5437. doi: 10.1128/IAI.70.10.5428-5437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohler C, von Eiff C, Peters G, Proctor RA, Hecker M, Engelmann S. Physiological characterisation of a heme-deficient mutant of Staphylococcus aureus by proteomic approach. J Bacteriol. 2003;185(33):6926–6937. doi: 10.1128/JB.185.23.6928-6937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahl BC, Belling G, Becker P, et al. Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect Immun. 2005;73(7):4119–4126. doi: 10.1128/IAI.73.7.4119-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clements MO, Watson SP, Poole RK, Foster SJ. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J Bacteriol. 1999;181(2):501–507. doi: 10.1128/jb.181.2.501-507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atalla H, Gyles C, Mallard B. Staphylococcus aureus small colony variants (SCVs) are their role in disease. Anim Health Res Rev. 2011;12(1):33–45. doi: 10.1017/S1466252311000065. [DOI] [PubMed] [Google Scholar]
- 41.Sendi P, Proctor RA. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 2009;17(2):54–58. doi: 10.1016/j.tim.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, et al. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379(6560):91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 43.Kalinka J, Hachmeister M, Geraci J, et al. Staphylococcus aureus isolates from chronic osteomyelitis are characterized by high host cell invasion and intracellular adaption, but still induce inflammation. Int J Med Microbiol. 2014;304(8):1038–1049. doi: 10.1016/j.ijmm.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Balwit JM, van Langevelde P, Vann JM, Proctor RA. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J Infect Dis. 1994;170:1033–1037. doi: 10.1093/infdis/170.4.1033. [DOI] [PubMed] [Google Scholar]
- 45.von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Gotz F. A site directed Staphylococcus aureus hemB mutant is a small colony variant which persists intracellularly. J Bacteriol. 1997;179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Eiff C, Becker K, Metze D, et al. Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in patients with Darier’s disease. Clin Infect Dis. 2001;32(11):1643–1647. doi: 10.1086/320519. [DOI] [PubMed] [Google Scholar]
- 47.Kahl B, Herrmann M, Everding AS, et al. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J Infect Dis. 1998;177(4):1023–1029. doi: 10.1086/515238. [DOI] [PubMed] [Google Scholar]
- 48.Schröder A, Kland R, Peschel A, von Eiff C, Aepfelbacher M. Live cell imaging of phagosome maturation in Staphylococcus aureus infected human endothelial cells: small colony variants are able to survive in lysosomes. Med Microbiol Immunol. 2006;195(4):185–194. doi: 10.1007/s00430-006-0015-0. [DOI] [PubMed] [Google Scholar]
- 49.Proctor RA, van Langevelde P, Kristjansson M, Maslow JN, Arbeit RD. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin Infect Dis. 1995;20(1):95–102. doi: 10.1093/clinids/20.1.95. [DOI] [PubMed] [Google Scholar]
- 50.Kipp F, Ziebuhr W, Becker K, et al. Detection of Staphylococcus aureus by 16s rRNA directed in situ hybridisation in a patient with a brain abscess caused by small colony variants. J Clin Pathol. 2003;56(10):746. doi: 10.1136/jnnp.74.7.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seggewiss J, Becker K, Kotte O, et al. Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J Bacteriol. 2006;188:7765–7777. doi: 10.1128/JB.00774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bates DM, von Eiff C, McNamara PJ, et al. Staphylococcus aureus mend and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J Infect Dis. 2003;187(10):1654–1661. doi: 10.1086/374642. [DOI] [PubMed] [Google Scholar]
- 53.Wakeman CA, Hammer ND, Stauff DL, et al. Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol Microbiol. 2012;86(6):1376–1392. doi: 10.1111/mmi.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Granick S, Beale SI. Hemes, chlorophylls, and related compounds: biosynthesis and metabolic regulation. Adv Enzymol Relat Areas Mol Biol. 1978;46:33–203. doi: 10.1002/9780470122914.ch2. [DOI] [PubMed] [Google Scholar]
- 55.Ythier M, Resch G, Waridel P, et al. Proteomic and transcriptomic profiling of Staphylococcus aureus surface LPXTG-proteins: correlation with agr genotypes and adherence phenotypes. Mol Cell Proteomics. 2012;11:1123–1139. doi: 10.1074/mcp.M111.014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerca N, Brooks JL, Jefferson KK. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigma B, and IcaR in Staphylococcus aureus. J Bacteriol. 2008;190(19):6530–6533. doi: 10.1128/JB.00482-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Da Re S, Le Quéré B, Ghigo JM, Beloin C. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl Environ Microbiol. 2007;73:3391–3403. doi: 10.1128/AEM.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prigent-Combaret C, Brombacher E, Vidal O, et al. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Eiff C, McNamara P, Becker K, et al. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J Bacteriol. 2006;188(2):687–693. doi: 10.1128/JB.188.2.687-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Möbius K, Arias-Cartin R, Breckau D, et al. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc Natl Acad Sci U S A. 2010;107(23):10436–10441. doi: 10.1073/pnas.1000956107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinha B, François PP, Nüsse O, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasion via fibronectin bridging to integrin to alpha5beta1. Cell Microbiol. 1999;1(2):101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 62.Mitchell G, Fugère A, Pépin Gaudreau K, et al. SigB is a dominant regulator of virulence in Staphylococcus aureus small-colony variants. PLoS One. 2013;8(5):e65018. doi: 10.1371/journal.pone.0065018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moisan H, Brouillette E, Jacob CL, Langlois-Bégin P, Michaud S, Malouin F. Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB. J Bacteriol. 2006;188(1):64–78. doi: 10.1128/JB.188.1.64-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valle J, Toledo-Arana A, Berasain C, et al. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48(4):1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 65.Tuchscherr L, Bischoff M, Lattar SM, et al. Sigma factor SigB is crucial to mediate Staphylococcus aureus adaption during chronic infections. PLoS Pathog. 2015;11(4):e1004870. doi: 10.1371/journal.ppat.1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tormo MA, Martí M, Valle J, et al. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J Bacteriol. 2005;187(7):2348–2358. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casadesús J Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:3830–3885. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Schaik W, Zwietering MH, de Vos WM, Abee T. Deletion of the SigB gene in Bacillus cereus ATCC 14579 leads to hydrogen peroxide hyper resistance. Appl Environ Microbiol. 2005;71(10):6427–6430. doi: 10.1128/AEM.71.10.6427-6430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michelsen CF, Christensen AM, Bojer MS, Hoiby N, Ingmer H, Jelsbak L. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human adapted Pseudomonas aeruginosa lineage. J Bacteriol. 2014;196:3903–3911. doi: 10.1128/JB.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brouillette E, Martinez A, Boyll BJ, Allen NE, Malouin F. Persistence of Staphylococcus aureus small colony variant under antibiotic pressure in vivo. FEMS Immunol Med Microbiol. 2004;41:35–41. doi: 10.1016/j.femsim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Smania AM, Segura I, Pezza RJ, Becerra C, Albesa I, Argarana CE. Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology. 2004;150:1327–1338. doi: 10.1099/mic.0.26751-0. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell G, Grundin G, Bilodeau G, Cantrin AM, Malouin F. Infection of polarized airway epithelial cells by normal and small colony variant strains of Staphylococcus aureus is increased in cells with abnormal cystic fibrosis transmembrane conductive regulator function and is influenced by NF-KB. Infect Immun. 2011;79:3541–3551. doi: 10.1128/IAI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blivn KA, Mavrelli AT. Antivirulence genes: insights into pathogen evolution through gene loss. Infect Immun. 2012;80:4016–4070. doi: 10.1128/IAI.00740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orr HA. The genetic theory of adaptation: a brief history. Nat Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 76.Song H, Hwang J, Yi H, et al. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog. 2010;6:e1000922. doi: 10.1371/journal.ppat.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alder H, Widmer A, Frei R. Emergence of teicoplanin-resistant small colony variant of Staphylococcus epidermidis during vancomycin therapy. Eur J Clin Microbiol Infect Dis. 2003;22:746–748. doi: 10.1007/s10096-003-1029-9. [DOI] [PubMed] [Google Scholar]
- 78.Kriegeskorte A, Grubmüller S, Huber C, et al. Staphylococcus aureus small colony variants show common metabolic features in central metabolism irrespective of underlying auxotrophism. Front Cell Infect Microbiol. 2014;4:141. doi: 10.3389/fcimb.2014.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cole J. Nitrate reduction to ammonia by enteric bacteria: redundancy or a strategy for survival during oxygen starvation? FEMS Lett Microbiol. 1996;136:1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x. [DOI] [PubMed] [Google Scholar]
- 80.Toft C, Andersson SG. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 81.Dagan T, Blekhman R, Graur D. The “domino theory” of gene death: gradual and mass gene extinction events in the lineages of obligate symbiotic bacterial pathogens. Mol Biol Evol. 2006;23(2):310–316. doi: 10.1093/molbev/msj036. [DOI] [PubMed] [Google Scholar]
- 82.Merhej V, Geogiades K, Raoult D. Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors. Brief Funct Genomics. 2013;12:291–304. doi: 10.1093/bfgp/elt015. [DOI] [PubMed] [Google Scholar]
- 83.Kumar A, Ting YP. Presence of Pseudomonas aeruginosa influences biofilm formation and surface protein expression of Staphylococcus aureus. Environ Microbiol. 2015 doi: 10.1111/1462-2920.12890. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 84.Gopal PK, Prasad J, Smart J, Gill HS. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbiol. 2001;67:207–216. doi: 10.1016/s0168-1605(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 85.Bibel DJ, Aly R, Bayles C, Straus WG, Shinefield HR, Maibach HI. Competitive adherence as a mechanism for bacterial interference. Can J Microbiol. 1983;29:700–703. doi: 10.1139/m83-114. [DOI] [PubMed] [Google Scholar]
- 86.Sifri CD, Baresh-Bernal A, Calderwood SB, von Eiff C. Virulence of Staphylococcus aureus small colony variants in the Caenorhabditis elegans infection model. Infect Immun. 2006;74(2):1091–1096. doi: 10.1128/IAI.74.2.1091-1096.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller MH, Wexler MA, Steigbigel NH. Single and combination antibiotic therapy of Staphylococcus aureus experimental endocarditis: emergence of gentamicin-resistant mutants. Antimicrob Agents Chemother. 1978;14(3):336–343. doi: 10.1128/aac.14.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delgado-Valverde M, Fernández-Echauri P, Batista-Diaz N, Pascual-Hernández A. Small-colony variants of Staphylococcus aureus: usefulness of various test [sic] for diagnosis and susceptibility study. Enferm Infecc Microbiol Clin. 2014;32(2):96–98. doi: 10.1016/j.eimc.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 89.Sadowska B, Bonar A, von Eiff C, et al. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol Med Microbiol. 2002;32(3):191–197. doi: 10.1111/j.1574-695X.2002.tb00553.x. [DOI] [PubMed] [Google Scholar]
- 90.Evans TJ. Small colony variants of Pseudomonas aeruginosa in chronic bacterial infection of the lung in cystic fibrosis. Future Microbiol. 2015;10(2):231–239. doi: 10.2217/fmb.14.107. [DOI] [PubMed] [Google Scholar]
- 91.Cullen L, McClean S. Bacterial adaption during chronic respiratory infections. Pathogens. 2015;4(1):66–89. doi: 10.3390/pathogens4010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cervantes-García E, García-Gonzalez R, Reyes-Torres A, Resendiz-Albor AA, Salazar-Schettino PM. Staphylococcus aureus small colony variants in diabetic foot infections. Diabet Foot Ankle. 2015;6:26431. doi: 10.3402/dfa.v6.26431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan NC, Cooksley CM, Roscioli E, et al. Small-colony variants and phenotype switching of intracellular Staphylococcus aureus in chronic rhinosinusitis. Allergy. 2014;69:1364–1371. doi: 10.1111/all.12457. [DOI] [PubMed] [Google Scholar]
- 94.Abele-Horn M, Schupfner B, Emmerling P, Waldner H, Göring H. Persistent wound infection after herniotomy associated with small-colony variants of Staphylococcus aureus. Infection. 2000;28(1):53–54. doi: 10.1007/s150100050014. [DOI] [PubMed] [Google Scholar]
- 95.Posnett J, Franks PJ. The burden of chronic wounds in the UK. Nurs Times. 2008;104(3):44–45. [PubMed] [Google Scholar]
- 96.Siddiqui AR, Bernstein JM. Chronic wound infection: facts and controversies. Clin Dermatol. 2010;28(5):519–526. doi: 10.1016/j.clindermatol.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 97.Agarwal H, Verrall R, Singh SP, Tang YW, Wilson G. Small colony variant Staphylococcus aureus multiorgan infection. Pediatr Infect Dis J. 2007;26(3):269–271. doi: 10.1097/01.inf.0000256749.29244.67. [DOI] [PubMed] [Google Scholar]
- 98.Tande AJ, Osmon DR, Greenwood-Quaintance KE, Mabry TM, Hanssen AD, Patel R. Clinical characteristics and outcomes of prosthetic joint infection caused by small colony variant staphylococci. MBio. 2014;5(5):e1910–e1914. doi: 10.1128/mBio.01910-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roggenkamp A, Sing A, Hornef M, Brunner U, Autenrieth IB, Heesemann J. Chronic prosthetic hip infection caused by a small-colony variant of Escherichia coli. J Clin Microbiol. 1998;36(9):2530–2534. doi: 10.1128/jcm.36.9.2530-2534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Piffaut C, Lustig S, Laurent F, Chidiac C, Ferry T. Small colony variant-producing S aureus prosthesis joint infection highlighted by sonication and treated with prolonged high doses of daptomycin. BMJ Case Rep. 2013;2013 doi: 10.1136/bcr-2013-008637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Idelevich EA, Pogoda CA, Ballhausen B, et al. Pacemaker lead infection and related bacteraemia caused by normal and small colony variant phenotypes of Bacillus licheniformis. J Med Microbiol. 2013;62(6):940–944. doi: 10.1099/jmm.0.051987-0. [DOI] [PubMed] [Google Scholar]
- 102.Maduka-Ezeh A, Seville MT, Kusne S, et al. Thymidine auxotrophic Staphylococcus aureus small-colony variant endocarditis and left ventricular assist device infection. J Clin Microbiol. 2012;50(3):1102–1105. doi: 10.1128/JCM.01170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spanu T, Romano L, D’Inzeo T, et al. Recurrent ventriculoperitoneal shunt infection caused by small-colony variants of Staphylococcus aureus. Clin Infect Dis. 2005;41(5):e48–e52. doi: 10.1086/432577. [DOI] [PubMed] [Google Scholar]
- 104.Maduka-Ezeh AN, Greenwood-Quaintance KE, Karau MJ, et al. Antimicrobial susceptibility and biofilm formation of Staphylococcus epidermidis small colony variants associated with prosthetic join infection. Diagn Microbiol Infect Dis. 2012;74(3):224–229. doi: 10.1016/j.diagmicrobio.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 105.Pradeep Kumar, SS Easwer, HV Maya, Nandkumar A. Multiple drug resistant bacterial biofilms on implanted catheters—a reservoir of infection. J Assoc Physicians India. 2013;61(10):702–707. [PubMed] [Google Scholar]
- 106.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013;136:1–51. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 107.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 108.Bui LM, Turnidge JD, Kidd SP. The induction of Staphylococcus aureus biofilm formation of small colony variants is a strain-specific response to host-generated chemical stresses. Microbes Infect. 2015;17(1):77–82. doi: 10.1016/j.micinf.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 109.Jeukens J, Boyle B, Kukavica-Ibrulj I, et al. Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS One. 2014;9(2):e87611. doi: 10.1371/journal.pone.0087611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rit K. A case report of small colony variant of Staphylococcus aureus isolated from a patient with chronic osteomyelitis [sic] in a tertiary care hospital of eastern India. Adv Biomed Res. 2014;3:32. doi: 10.4103/2277-9175.124683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Häussler S, Tümmler B, Weissbrodt H, Rohde M, Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29(3):621–625. doi: 10.1086/598644. [DOI] [PubMed] [Google Scholar]
- 112.von Eiff C, Vaudaux P, Kahl BC, et al. Bloodstream infections caused by small-colony variants of coagulase-negative staphylococci following pacemaker implantation. Clin Infect Dis. 1999;29(4):932–934. doi: 10.1086/520462. [DOI] [PubMed] [Google Scholar]
- 113.Sendi P, Rohrbach M, Graber P, Frei R, Ochsner PE, Zimmerli W. Staphylococcus aureus small colony variants in prosthetic joint infection. Clin Infect Dis. 2006;43(8):961–967. doi: 10.1086/507633. [DOI] [PubMed] [Google Scholar]
- 114.Singh R, Ray P, Das A, Sharma M. Enhanced production of exopolysaccharide matrix and biofilm by a menadione-auxotrophic Staphylococcus aureus small-colony variant. J Med Microbiol. 2010;59(5):521–527. doi: 10.1099/jmm.0.017046-0. [DOI] [PubMed] [Google Scholar]
- 115.von Götz F, Häussler S, Jordan D, et al. Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. J Bacteriol. 2004;186(12):3837–3847. doi: 10.1128/JB.186.12.3837-3847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ikeno T, Fukuda K, Ogawa M, Honda M, Tanabe T, Taniguchi H. Small and rough colony Pseudomonas aeruginosa with elevated biofilm formation ability isolated in hospitalized patients. Microbiol Immunol. 2007;51(10):929–938. doi: 10.1111/j.1348-0421.2007.tb03989.x. [DOI] [PubMed] [Google Scholar]
- 117.Al Laham N, Rohde H, Sander G, et al. Augmented expression of polysaccharide intercellular adhesin in a defined Staphylococcus epidermidis mutant with small-colony-variant phenotype. J Bacteriol. 2007;189(12):4494–4501. doi: 10.1128/JB.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mitchell G, Séguin DL, Asselin AE, et al. Staphylococcus aureus sigma B dependent emergence of small colony variants and biofilm production following exposure to Pseudomonas aeruginosa4-hydroxy-2-hepylquinoline-N-oxide. BMC Microbiol. 2010;10:33. doi: 10.1186/1471-2180-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schneider M, Muhlemann K, Droz S, Courzinet S, Casualta C, Zimmerli S. Clinical characteristics associated with isolation of small colony variants of Staphylococcus aureus and Pseudomonas aeruginosa from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 2008;46:1832–1834. doi: 10.1128/JCM.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.von Eiff C, Bettin D, Proctor RA, et al. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin Infect Dis. 1997;25(5):1250–1251. doi: 10.1086/516962. [DOI] [PubMed] [Google Scholar]
- 121.Tsuji BT, von Eiff C, Kelchlin PA, Forrest A, Smith PF. Attenuated vancomycin bactericidal activity against Staphylococcus aureus hemB mutants expressing the small-colony-variant phenotype. Antimicrob Agents Chemother. 2008;52(4):1533–1537. doi: 10.1128/AAC.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Begic D, von Eiff C, Tsuji BT. Daptomycin pharmacodynamics against Staphylococcus aureus hemB mutants displaying the small colony variant phenotype. J Antimicrob Chemother. 2009;63(5):977–981. doi: 10.1093/jac/dkp069. [DOI] [PubMed] [Google Scholar]
- 123.Ward AC, Connolly P, Tucker NP. Pseudomonas aeruginosa can be detected in a polymicrobial competition model using impedance spectroscopy with a novel biosensor. PLoS One. 2014;9(3):e91732. doi: 10.1371/journal.pone.0091732. [DOI] [PMC free article] [PubMed] [Google Scholar]