Spatially resolved, highly multiplexed RNA profiling in single cells (original) (raw)
. Author manuscript; available in PMC: 2015 Nov 28.
Published in final edited form as: Science. 2015 Apr 9;348(6233):aaa6090. doi: 10.1126/science.aaa6090
Abstract
Knowledge of the expression profile and spatial landscape of the transcriptome in individual cells is essential for understanding the rich repertoire of cellular behaviors. Here we report multiplexed error-robust fluorescence in situ hybridization (MERFISH), a single-molecule imaging approach that allows the copy numbers and spatial localizations of thousands of RNA species to be determined in single cells. Using error-robust encoding schemes to combat single-molecule labeling and detection errors, we demonstrated the imaging of 100 – 1000 unique RNA species in hundreds of individual cells. Correlation analysis of the ~104 – 106 pairs of genes allowed us to constrain gene regulatory networks, predict novel functions for many unannotated genes, and identify distinct spatial distribution patterns of RNAs that correlate with properties of the encoded proteins.
System-wide analyses of the abundance and spatial organization of RNAs in single cells promise to transform our understanding in many areas of cell and developmental biology, such as the mechanisms of gene regulation, the heterogeneous behavior of cells, and the development and maintenance of cell fate (1). Single-molecule fluorescence in situ hybridization (smFISH) has emerged as a powerful tool for studying the copy number and spatial organization of RNAs in single cells either in isolation or in their native tissue context (2, 3). Taking advantage of its ability to map the spatial distributions of specific RNAs with high resolution, smFISH has revealed the importance of subcellular RNA localization in diverse processes such as cell migration, development, and polarization (4–8). In parallel, the ability of smFISH to precisely measure the copy numbers of specific RNAs without amplification bias has allowed quantitative measurement of the natural fluctuations in gene expression, which has in turn elucidated the regulatory mechanisms that shape such fluctuations and their role in a variety of biological processes (9–13).
Recent advances in imaging and analysis methods have allowed hundreds of smFISH measurements to be performed in an automated manner, substantially expanding our knowledge of the RNA expression profile and spatial organization in different organisms (14, 15). However, application of the smFISH approach to many systems-level questions remains limited by the number of RNA species that can be simultaneously measured in single cells. State-of-the-art efforts using combinatorial labeling by either color-based barcodes or sequential hybridization have enabled simultaneous measurements of 10–30 different RNA species in individual cells (16–19), yet many interesting biological questions would benefit from the measurement of hundreds to thousands of RNAs within a single cell. For example, analysis of how the expression profile of such a large number of RNAs vary from cell to cell and how these variations correlate among different genes could be used to systematically identify co-regulated genes and map regulatory networks; knowledge of the subcellular organizations of numerous RNAs and their correlations could help elucidate molecular mechanisms underlying the establishment and maintenance of many local cellular structures; and RNA profiling of individual cells in native tissues could allow in situ identification of cell type.
Here we report MERFISH, a highly multiplexed smFISH imaging method that substantially increases the number of RNA species that can be simultaneously imaged in single cells by using combinatorial labeling and sequential imaging with error-robust encoding schemes. We demonstrated this transcriptome imaging approach by simultaneously measuring 140 RNA species using an encoding scheme that can both detect and correct errors and 1001 RNA species using an encoding scheme that can detect but not correct errors. Correlation analyses of the copy number variations and spatial distributions of these genes allowed us to identify groups of genes that are co-regulated and groups of genes that share similar spatial distribution patterns inside the cell.
Combinatorial labeling with error-robust encoding schemes
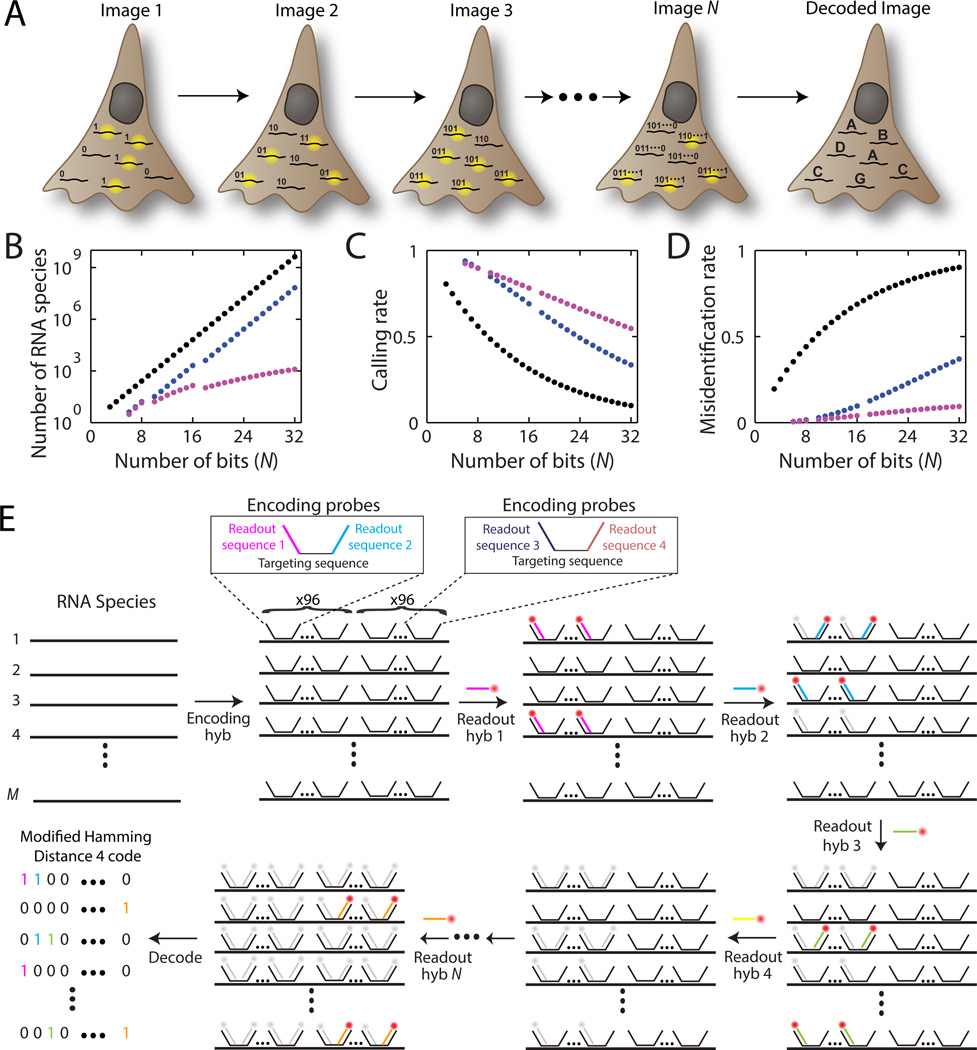
Combinatorial labeling that identifies each RNA species by multiple (N) distinct signals offers a route to rapidly increase the number of RNA species that can be probed simultaneously in individual cells (Fig. 1A). However, this approach to scaling up the throughput of smFISH to the systems scale faces a significant challenge because not only does the number of addressable RNA species increases exponentially with N, but the detection error rates also grow exponentially with N (Fig. 1B–D). Imagine a conceptually simple scheme to implement combinatorial labeling, where each RNA species is encoded with a _N_-bit binary word and the sample is probed with N corresponding rounds of hybridization, each round targeting only the subset of RNAs that should read ‘1’ in the corresponding bit (Fig. S1). N rounds of hybridization would allow 2_N_−1 RNA species to be probed. With just 16 hybridizations, over 64,000 RNA species, which should cover the entire human transcriptome including both messenger RNAs (mRNAs) and non-coding RNAs (20), could be identified (Fig. 1B; black symbols). However, as N increases, the fraction of RNAs properly detected (the calling rate) would rapidly decrease and, more troublingly, the fraction of RNAs that are identified as incorrect species (the misidentification rate) would rapidly increase (Fig. 1C, D; black symbols). With realistic error rates per hybridization (measured below), the majority of RNA molecules would be misidentified after 16 rounds of hybridizations!
Fig. 1. MERFISH: a highly multiplexed smFISH approach enabled by combinatorial labeling and error-robust encoding.
(A) Schematic depiction of the identification of multiple RNA species in N rounds of imaging. Each RNA species is encoded with a _N_-bit binary word and during each round of imaging, only the subset of RNAs that should read ‘1’ in the corresponding bit emit signal. (B–D) The number of addressable RNA species (B), the rate at which these RNAs are properly identified – calling rate (C), and the rate at which RNAs are incorrectly identified as a different RNA species – misidentification rate (D) plotted as a function of the number of bits (N) in the binary words encoding RNA. Black, a simple binary code that includes all 2_N_−1 possible binary words. Blue, the HD4 code where the Hamming distance separating words is 4. Magenta, the modified HD4 (MHD4) code where the number of ‘1’ bits are kept at four. The calling and misidentification rates are calculated with per bit error rates of 10% for the 1→0 error and 4% for the 0→1 error. (E) Schematic diagram of the implementation of a MHD4 code for RNA identification. Each RNA species is first labeled with ~192 encoding probes that convert the RNA into a unique combination of readout sequences (Encoding hyb). These encoding probes each contain a central RNA targeting region flanked by two readout sequences, drawn from a pool of N different sequences, each associated with a specific hybridization round. Encoding probes for a specific RNA species contain a unique combination of four of the N readout sequences, which correspond to the four hybridization rounds where this RNA should read ‘1’. N subsequent rounds of hybridization with the fluorescent readout probes are used to probe the readout sequences (hyb 1, hyb 2, …, hyb N). The bound probes are inactivated by photobleaching between successive rounds of hybridization. For clarity only one possible pairing of the readout sequences is depicted for the encoding probes; however, all possible pairs of the four readout sequences are used at the same frequency and distributed randomly along each cellular RNA in the actual experiments.
To address this challenge, we designed error-robust encoding schemes, in which only a subset of the 2_N_−1 words separated by a certain Hamming distance (21) were used to encode RNAs. In a codebook where the minimum Hamming distance is 4 (HD4 code), at least four bits must be read incorrectly to change one code word into another (Fig. S2A). As a result, every single-bit error produces a word that is uniquely close to a single code word, allowing such errors to be detected and corrected (Fig. S2B). Double-bit errors produce words with an equal Hamming distance of 2 from multiple code words and, thus, can be detected but not corrected (Fig. S2C). Such a code should substantially increase the calling rate and reduce the misidentification rate (Fig. 1C, D, blue symbols). To further account for the fact that it is more likely to miss a hybridization event (an 1→0 error) than to misidentify a background spot as an RNA (an 0→1 error) in smFISH measurements, we designed a modified HD4 (MHD4) code, in which the number of ‘1’ bits were kept both constant and relatively low—only four per word—to reduce error and avoid biased detection. This MHD4 code should further increase the calling rate and reduce the misidentification rate (Fig. 1C, D, magenta symbols).
In addition to the error considerations, several practical challenges have also made it difficult to probe a large number of RNA species, such as the high cost of the massive number of fluorescently labeled FISH probes needed and the long time required to complete many rounds of hybridization. An oligopaint approach has been previously developed to generate a large number of oligonucleotide probes to label chromosome DNA and to introduce non-targeting site for secondary activities (22). Inspired by this approach, we designed a two-step labeling scheme to encode and readout cellular RNAs (Fig. 1E). First, we label cellular RNAs with a set of encoding probes, each probe comprising a RNA targeting sequence and two flanking readout sequences. Four of the N unique readout sequences were assigned to each RNA species based on the MHD4 code word of the RNA. Second, we identified these N readout sequences with complementary FISH probes (the readout probes) via N rounds of hybridization and imaging, each round using a unique readout probe. To increase the signal to background ratio, we labeled every cellular RNA with ~192 encoding probes. Because each encoding probe contained two of the four readout sequences associated with that RNA (Fig. 1E), a maximum of ~96 readout probes can bind to each cellular RNA per hybridization round. To generate the massive number of encoding probes required, we amplified them from array-derived oligonucleotide pools containing tens of thousands of custom sequences using a modified form of the oligopaint protocol comprising in vitro transcription followed by reverse transcription (Fig. S3; Materials and Methods, “Probe Synthesis” section) (22, 23). This two-step labeling approach significantly diminished the total hybridization time for an experiment: we found that efficient hybridization to the readout sequences took only 15 minutes whereas efficient direct hybridization to cellular RNA required more than 10 hours.
Measuring 140 genes with MERFISH using a 16-bit MHD4 Code
To test the feasibility of this error-robust, multiplexed imaging approach, we performed a 140-gene measurement on human fibroblast cells (IMR90) using a 16-bit MHD4 code to encode 130 RNA species while leaving 10 code words as misidentification controls (Table S1). After each round of hybridization with the fluorescent readout probes, cells were imaged by conventional wide-field imaging with an oblique-incidence illumination geometry. Fluorescent spots corresponding to individual RNAs were clearly detected and were then efficiently extinguished via a brief photobleaching step (Fig. 2A). The sample was stable throughout the 16 rounds of iterative labeling and imaging: The change in the number of fluorescent spots from round to round matched the predicted change based on the relative abundances of RNA species targeted in each round derived from bulk sequencing, and we did not observe a systematic decreasing trend with increasing number of hybridization rounds (Fig. S4A). The average brightness of the spots varied from round to round with a standard deviation of 40%, likely due to different binding efficiencies of the readout probes to the different readout sequences on the encoding probes (Fig. S4B). We observed only a small, systematic decreasing trend in the spot brightness with increasing hybridization rounds, which was on average 4% per round (Fig. S4B).
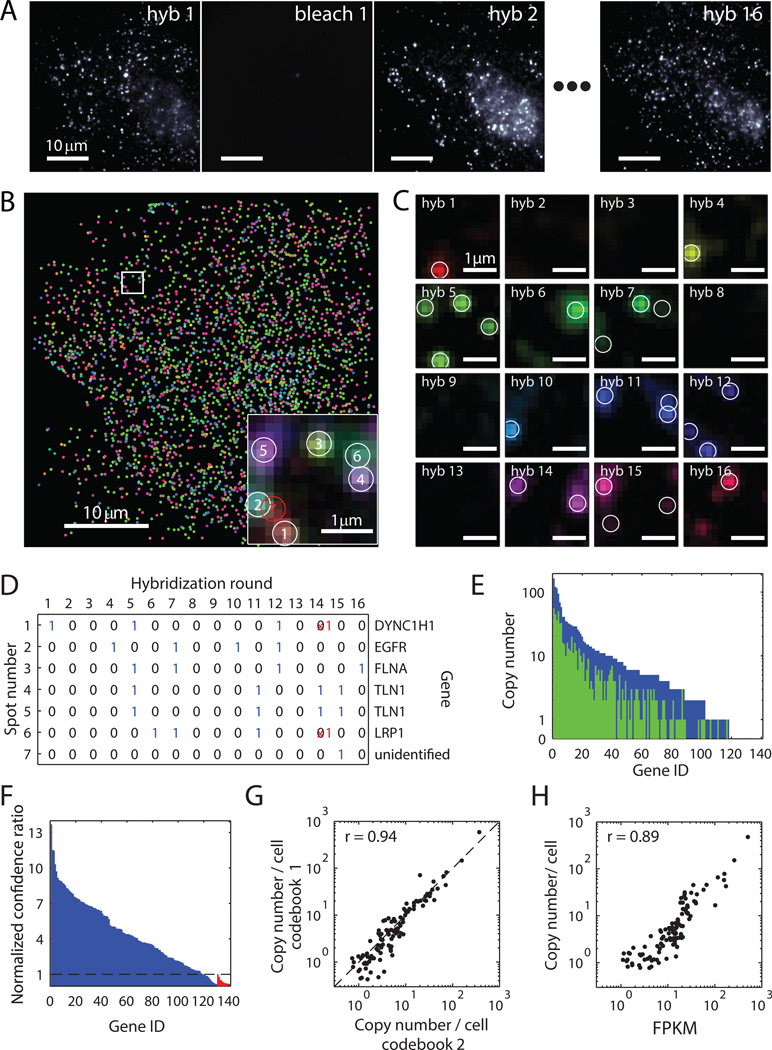
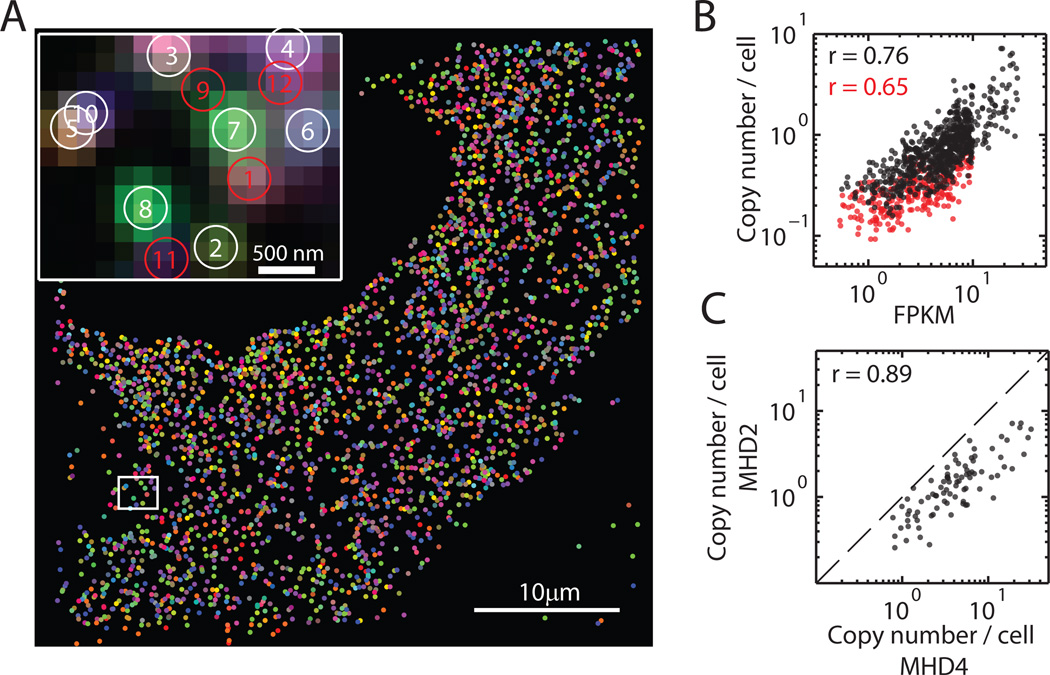
Fig. 2. Simultaneous measurement of 140 RNA species in single cells using MERFISH with a 16-bit MHD4 code.
(A) Images of RNA molecules in an IMR90 cell after each hybridization round (hyb 1 – hyb 16). The image after photobleaching (bleach 1) demonstrates efficient removal of fluorescent signals between hybridizations. (B) The localizations of all detected single molecules in this cell colored based on their measured binary words. Inset: the composite, false-colored fluorescent image of the 16 hybridization rounds for the boxed sub-region with numbered circles indicating potential RNA molecules. A red circle indicates an unidentifiable molecule, the binary word of which does not match any of the 16-bit MHD4 code words even after error correction. (C) Fluorescent images from each round of hybridization for the boxed sub-region in (B) with circles indicating potential RNA molecules. (D) Corresponding words for the spots identified in (C). Red crosses represent the corrected bits. (E) The RNA copy number for each gene observed without (green) or with (blue) error correction in this cell. (F) The confidence ratio measured for the 130 RNA species (blue) and the 10 misidentification control words (red) normalized to the maximum value observed from the misidentification controls (dashed line). (G) Scatter plot of the average copy number of each RNA species per cell measured with two shuffled codebooks of the MHD4 code. The Pearson correlation coefficient is 0.94 with a p-value of 1×10−53. The dashed line corresponds to the y = x line. (H) Scatter plot of the average copy number of each RNA species per cell versus the abundance determined by bulk sequencing in fragments per kilobase per million reads (FPKM). The Pearson correlation coefficient between the logarithmic abundances of the two measurements was 0.89 with a p-value of 3×10−39.
We then constructed binary words from the observed fluorescent spots based on their on-off patterns across the 16 hybridization rounds (Fig. 2B–D). If the word exactly matched one of the 140 MHD4 code words (exact matches) or differed by only one bit (error-correctable matches), we assigned it to the corresponding RNA species (Fig. 2D). Within the single cell depicted in Fig. 2A, B, more than 1500 RNA molecules corresponding to 87% of the 130 encoded RNA species were detected after error correction (Fig. 2E). Similar observations were made in ~400 cells from 7 independent experiments. On average, ~4 times as many RNA molecules and ~2 times as many RNA species were detected per cell after error correction as compared with the values obtained before error correction (Fig. S5).
Two types of errors can occur in the copy number measurement of each RNA species: 1) Some molecules of this RNA species are not detected, leading to a drop in calling rate, and 2) some molecules from other RNA species are misidentified as this RNA species. To assess the extent of misidentification, we utilized the 10 misidentification control words — code words that were not associated with any cellular RNA. Although matches to these control words were observed, they occurred far less frequently than the real RNA-encoding words: 95% of the 130 RNA-encoding words were counted more frequently than the median count for these control words. Moreover, we typically found the ratio of the number of exact matches to the number of matches with one-bit errors for a real RNA-encoding word to be substantially higher than the same ratios observed for the misidentification controls, as expected (Fig. S6A, B). Using this ratio as a measure of the confidence in RNA identification, we found that 91% of the 130 RNA species had a confidence ratio greater than the maximum confidence ratio observed for the misidentification controls (Fig. 2F), demonstrating a high accuracy of RNA identification. Subsequent analyses were conducted only on these 91% of genes.
To estimate the calling rate, we utilized the error-correction ability of the MHD4 code to determine the 1→0 error rates (10% on average) and 0→1 error rates (4% on average) for each hybridization round (Fig. S6C, D). Using these error rates, we estimated an ~80% calling rate for individual RNA species after error correction, i.e. ~80% of the fluorescent spots corresponding to a RNA species were decoded correctly (Fig. S6E). We note that although the remaining 20% of spots contributed to a loss in detection efficiency, most of them did not cause species misidentification because they were decoded as double-bit error words and discarded.
To test for potential technical bias in our measurements, we probed the same 130 RNAs species with a different MHD4 codebook by shuffling the code words among different RNA species (Table S1) and changing the encoding probe sequences. Measurements with this alternative code gave similar misidentification and calling rates (Fig. S7). The copy numbers of individual RNA species per cell measured with these two codebooks showed excellent agreement with a Pearson correlation coefficient of 0.94 (Fig. 2G), indicating that the choice of encoding scheme did not bias the measured counts.
In order to validate the copy numbers derived from our MERFISH experiments, we performed conventional smFISH measurements on 15 of the 130 genes, selected from the full measured abundance range of three orders of magnitude. For each of these genes, both the average copy number and the copy number distribution across many cells agreed quantitatively between our MERFISH and conventional smFISH measurements (Fig. S8A, B). The ratio of the copy numbers determined by these two approaches was 0.82 ± 0.06 (mean ± SEM across the 15 measured RNA species, Fig. S8B), which agrees with the estimated 80% calling rate for our multiplexed imaging approach. The quantitative match between this ratio and our estimated calling rate over the full measured abundance range additionally supports our assessment that the misidentification error was low. Given that the agreement between the MERFISH and conventional smFISH results extended to the genes at the lowest measured abundance (<1 copy per cell, Fig. S8B), we estimate that our measurement sensitivity was at least 1 copy per cell.
As a final validation, we compared the abundance of each RNA species averaged over hundreds of cells to those obtained from a bulk RNA sequencing measurement that we performed on the same cell line. Our imaging results correlated remarkably well with bulk sequencing results with a Pearson correlation coefficient of 0.89 (Fig. 2H).
High-throughput analysis of cell-to-cell variation in gene expression
The MERFISH approach allows parallelization of measurements of many individual RNA species and co-variation analysis between different RNA species. We first illustrated the parallelization aspect by examining the cell-to-cell variation in the expression level of each of the measured genes (Fig. 3A). To quantify the measured variation, we computed the Fano factors, defined as the ratio of the variance to the mean RNA copy number, for all measured RNA species. The Fano factors substantially deviated from 1, the value expected for a simple Poisson process, for many genes and exhibited an increasing trend with the mean RNA abundance (Fig. 3B), consistent with a previous observation for other cell types (24). A simple model for promoter regulation — the promoter stochastically switches between on and off states with global constraints on the kinetic rates — has been previously suggested to rationalize such a trend (24, 25). Based on this model, this trend of increasing Fano factors with mean RNA abundance can be explained by changes in the transcription rate and/or promoter off-switching rates but not by changes in the promoter on-switching rate.
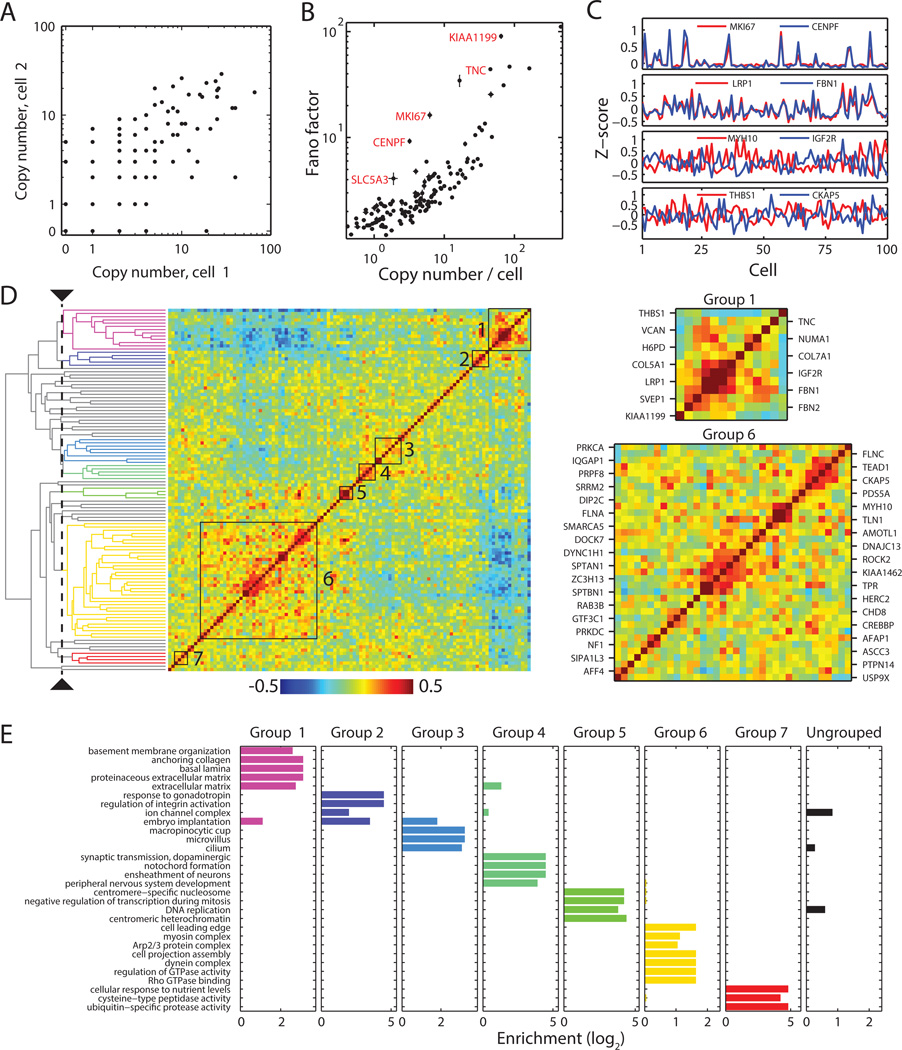
Fig. 3. Cell-to-cell variations and pairwise correlations for the RNA species determined from the 140 gene-measurements.
(A) Comparison of gene expression levels in two individual cells. (B) Fano factors for individual genes. Error bars represent standard error of the mean determined from 7 independent data sets. (C) Z-scores of the expression variations of four example pairs of genes showing correlated (top two) or anti-correlated (bottom two) variation for 100 randomly selected cells. Z-score is defined as the difference from the mean normalized by the standard deviation. (D) Matrix of the pairwise correlation coefficients of the cell-to-cell variation in expression for the measured genes, shown together with the hierarchical clustering tree. The seven groups identified by a specific threshold on the cluster tree (dashed line) are indicated by the black boxes in the matrix and colored lines on the tree, with grey lines on the tree indicating ungrouped genes. Different threshold choices on the cluster tree could be made to select either smaller subgroups with tighter correlations or larger super-groups containing more weakly coupled subgroups. Two of the seven groups are enlarged on the right. (E) Enrichment of 30 selected, statistically significantly enriched GO terms in the seven groups. Enrichment refers to the ratio of the fraction of genes within a group that have the specific GO term to the fraction of all measured genes having that term. Top 10 statistically significantly enriched GO terms for each of the seven groups are shown in Table S2. Not all of the GO terms presented here are in the top 10 list.
Moreover, we identified several RNA species with substantially larger Fano factors than this average trend. For example, we found that SLC5A3, CENPF, MKI67, TNC and KIAA1199 displayed Fano factor values substantially higher than those of the other genes expressed at similar abundance levels. The high variability of some of these genes can be explained by their association with the cell cycle. For example, two of these particularly ‘noisy’ genes MKI67 and CENPF are both annotated as cell-cycle related genes (26), and based on their bimodal expression (Fig. 3C), we propose that their transcription is strongly regulated by the cell cycle. Other high-variability genes did not show the same bimodal expression patterns and are not known to be associated with the cell cycle. Understanding the origin and significance of noisy gene expression is an active topic of current research (24).
Analysis of expression co-variation among different genes
Analysis of co-variations in the expression levels of different genes can reveal which genes are co-regulated and elucidate gene regulatory pathways. At the population level, such analysis often requires the application of external stimuli to drive gene expression variation; hence, correlated expression changes can be observed among genes that share common regulatory elements influenced by the stimuli (27). At the single-cell level, one can take advantage of the natural stochastic fluctuations in gene expression for such analysis and can thus study multiple regulatory networks without having to stimulate each of them individually. Such co-variation analysis can constrain regulatory networks, suggest new regulatory pathways, and predict function for unannotated genes based on associations with co-varying genes (11, 28).
We applied this approach to the 140-gene measurements and examined the ~10,000 pairwise correlation coefficients describing how the expression levels of each pair of genes co-varied from cell to cell. Many of the highly variable genes showed tightly correlated or anti-correlated variations (Fig. 3C). To better understand the correlations for all gene pairs, we adopted a hierarchical clustering approach, commonly used in the analysis of both bulk and single-cell expression data (29, 30), to organize these genes based on their correlation coefficients (Fig. 3D). From the cluster tree structure, we identified seven groups of genes with substantially correlated expression patterns (Fig. 3D and Table S2). Within each of the seven groups, every gene showed significantly stronger average correlation with other members of the group than with genes outside the group (Table S2). To further validate and understand these groups, we identified gene ontology (GO) terms (31) enriched in each of these seven groups. Notably, the enriched GO terms within each group shared similar functions and were largely unique to each group (Fig. 3E and Table S2), validating the notion that the observed co-variation in expression reflects some commonalities in the regulation of these genes.
Here, we describe two of these groups as illustrative examples. The predominant GO terms associated with Group 1 were terms associated with the extracellular matrix (ECM) (Fig. 3D, E and Table S2). Notable members of this group included ECM components, such as FBN1, FBN2, COL5A, COL7A and TNC, and glycoproteins linking the ECM and cell membranes, such as VCAN and THBS1. The group also included an unannotated gene, KIAA1199, which we would predict to play a role in ECM metabolism based on its association with this cluster. Indeed, this gene has recently been identified as an enzyme involved in the regulation of hyaluronan, a major sugar component of the ECM (32).
Group 6 contained many genes that encode vesicle transport proteins and proteins associated with cell motility (Fig. 3D, E and Table S2). The vesicle transport genes included microtubule motors and related genes DYNC1H, CKAP1, and factors associated with vesicle formation and trafficking, like DNAJC13 and RAB3B. Again, we found an unannotated gene, KIAA1462, within this cluster. Based on its strong correlation with DYNC1H1 and DNAJC13, we predict that this gene may be involved in vesicle transport. The cell motility genes in this group included actin-binding proteins like AFAP1, SPTAN1, SPTBN1, and MYH10, and genes involved in the formation of adhesion complexes, like FLNA and FLNC. Several GTPase-associated factors involved in the regulation of cell motility, attachment and contraction also fell into this group, including DOCK7, ROCK2, IQGAP1, PRKCA, and AMOTL1. The observation that some cell motility genes correlated with vesicle transport genes is consistent with the role of vesicle transport in cell migration (33). An additional interesting feature of group 6 is that a subset of these genes, in particular those related to cell motility, were anti-correlated with members of the ECM group discussed above (Fig. 3D). This anti-correlation may reflect regulatory interactions that mediate switching of cells between adherent and migratory states.
Mapping spatial distributions of RNAs
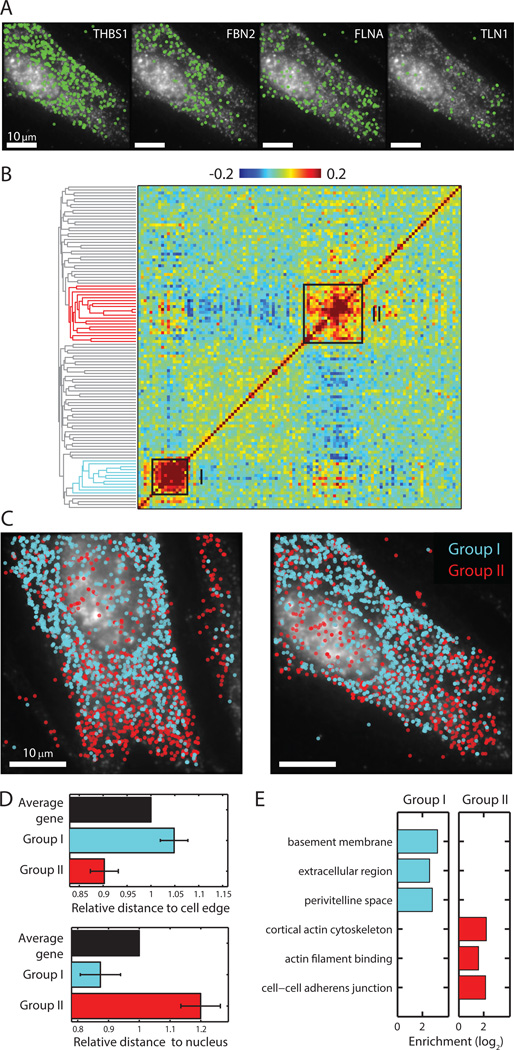
As an imaging based approach, MERFISH also allowed us to investigate the spatial distributions of many RNA species simultaneously. Several patterns emerged from the visual inspection of individual genes, with some RNA transcripts enriched in the perinuclear region, some enriched in the cell periphery, and some scattered throughout the cell (Fig. 4A). To identify genes with similar spatial distributions, we determined the correlation coefficients for the spatial density profiles of all pairs of RNA species and organized these RNAs based on the pairwise correlations again using the hierarchical clustering approach. The correlation coefficient matrix showed groups of genes with correlated spatial organizations, and the two most notable groups with the strongest correlations are indicated in Fig. 4B. Group I RNAs appeared enriched in the perinuclear region whereas group II RNAs appeared enriched near the cell periphery (Fig. 4C). Quantitative analysis of the distances between each RNA molecule and the cell nucleus or the cell periphery indeed confirmed this visual impression (Fig. 4D).
Fig. 4. Distinct spatial distributions of RNAs observed in the 140-gene measurements.
(A) Examples of the spatial distributions observed for four different RNA species in a cell. (B) Matrix of the pairwise correlation coefficients describing the degree with which the spatial distributions of each gene pair is correlated, shown together with the hierarchical clustering tree. Two strongly correlating groups are indicated by the black boxes on the matrix and color on the tree. (C) The spatial distributions of all RNAs in the two groups in two example cells. Cyan symbols: group I genes; Red symbols: group II genes. (D) Average distances for genes in group I and genes in group II to the cell edge or the nucleus normalized to the average distances for all genes. Error bars represent SEM across 7 data sets. (E) Enrichment of GO terms in each of the two groups.
Group I contained genes encoding extracellular proteins such as FBN1, FBN2 and THSB1, secreted proteins such as PAPPA, and integral membrane proteins such as LRP1 and GPR107. These proteins have no obvious commonalities in function. Rather a GO analysis showed significant enrichment for location terms, such as extracellular region, basement membrane, or perivitelline space (Fig. 4E). To reach these locations, proteins must pass through the secretion pathway, which often requires translation of mRNA at the endoplasmic reticulum (ER) (34, 35). Thus, we propose that the spatial pattern that we observed for these mRNAs reflects their co-translational enrichment at the ER. The enrichment of these mRNAs in the perinuclear region (Fig. 4C, D, cyan), where the rough ER resides, supports this conclusion.
Group II contained genes encoding the actin-binding proteins, including filamins FLNA and FLNC, talin TLN1, and spectrins SPTAN1 and SPTBN1; the microtubule-binding protein CKAP5; and the motor proteins MYH10 and DYNC1H1. This group was enriched with GO terms such as cortical actin cytoskeleton, actin filament binding, and cell-cell adherens junction (Fig. 4E). It has been shown previously that beta-actin mRNA is enriched near the cell periphery in fibroblasts as are mRNAs that encode members of the actin-binding Arp2/3 complex (36, 37). The enrichment of group II mRNAs in the peripheral region of the cells (Fig. 4C, D) suggests that the spatial distribution of the Group II genes might be related to the distribution of actin cytoskeleton mRNAs.
Measuring 1001 genes with a 14-bit MHD2 code
Finally, we sought to further increase the throughput of our MERFISH measurement by simultaneously imaging ~1000 RNA species. This increase could be achieved with our MHD4 code by increasing the number of bits per code word to 32 while maintaining the number of ‘1’ bits per word at four (Fig. 1B). While the stability of our samples across many hybridization rounds (Fig. S4) suggests that such an extension is potentially feasible, we pursued an alternative approach that did not require an increase in the number of hybridizations by relaxing the error correction requirement but keeping the error detection capability. For example, by reducing the Hamming distance from 4 to 2, we could use all 14-bit words that contain four ‘1’ bits to encode 1001 genes and probe these RNAs with only 14 rounds of hybridization. However, because a single error can produce a word equally close to two different code words, error correction is no longer possible for this modified Hamming-distance-2 (MHD2) code. Hence we expect the calling rate to be lower and the misidentification rate to be higher with this encoding scheme.
To evaluate the performance of this 14-bit MHD2 code, we set aside 16 of the 1001 possible code words as misidentification controls and used the remaining 985 words to encode cellular RNAs (Table S3). Among these 985 RNAs, we included 107 RNA species probed in the 140-gene experiments as an additional control. We performed the 1001-gene experiments in IMR90 cells using a similar procedure as described above. To allow all encoding probes to be synthesized from a single 100,000-member oligopool, we reduced the number of encoding probes per RNA species to ~94. Fluorescent spots corresponding to individual RNA molecules were again clearly detected in each round of hybridization with the readout probes and, based on their on-off patterns, these spots were decoded into RNA (Fig. 5A and Fig. S9A, B). 430 RNA species were detected in the cell shown in Fig. 5A, and similar results were obtained in ~200 imaged cells in 3 independent experiments.
Fig. 5. Simultaneous measurements of 1001 RNA species in single cells using MERFISH with a 14-bit MHD2 code.
(A) The localizations of all detected single molecules in a cell colored based on their measured binary words. Inset: the composite, false-colored fluorescent image of the 14 hybridization rounds for the boxed sub-region with numbered circles indicating potential RNA molecules. Red circles indicate unidentifiable molecules, the binary words of which do not match any of the 14-bit MHD2 code words. Images of individual hybridization round are shown in Fig. S9A. (B) Scatter plot of the average copy number per cell measured in the 1001-gene experiments versus the abundance measured via bulk sequencing. The black symbols are for the 73% of genes detected with confidence ratios higher than the maximum ratio observed for the misidentification controls. The Pearson correlation coefficient is 0.76 with a p-value of 3×10−133. The red symbols are for the remaining 27% of genes. The Pearson correlation coefficient is 0.65 with a p-value of 3×10−33. (C) Scatter plot of the average copy number for the 107 genes shared in both the 1001-gene measurement with the MHD2 code and the 140-gene measurement with the MHD4 code. The Pearson correlation coefficient is 0.89 with a p-value of 9×10−30. The dashed line is correspond to the y = x line.
As expected, the misidentification rate of this scheme was higher than that of the MHD4 code. 77% of all real RNA words were detected more frequently than the median count for the misidentification controls instead of the 95% value observed in the MHD4 measurements. Using the same confidence ratio analysis as described above, we found that 73% (instead of 91% for the MHD4 measurements) of the 985 RNA species were measured with a confidence ratio larger than the maximum value observed for the misidentification controls (Fig. S9C). RNA copy numbers measured from these 73% RNA species showed excellent correlation with our bulk RNA sequencing results (Pearson correlation coefficient r = 0.76; Fig. 5B, black). It is worth noting that the remaining 27% of the genes still exhibit good, albeit lower, correlation with the bulk RNA sequencing data (r = 0.65; Fig. 5B, red), but we took the conservative measure of excluding them from further analysis.
The lack of an error correction capability also decreased the calling rate of each RNA species: When comparing the 107 RNA species common in both the 1001-gene and 140-gene measurements, we found that the copy numbers per cell of these RNA species were lower in the 1001-gene measurements (Fig. 5C and Fig. S9D). The total count of these RNAs per cell was ~1/3 of that observed in the 140-gene measurements. Thus the lack of error correction in the MHD2 code produced a ~3-fold decrease in the calling rate, which is consistent with the ~4-fold decrease in calling rate observed for the MHD4 code when error correction was not applied. As expected from the quantitative agreement between 140-gene measurements and conventional smFISH results, comparison of the 1001-gene measurements with conventional smFISH results for 10 RNA species also indicated a ~3-fold drop in calling rate (Fig. S8C). Despite the expected reduction in calling rate, the good correlations found between the copy numbers observed in the 1001-gene measurements and those observed in the 140-gene measurements, as well as in conventional smFISH and bulk RNA sequencing measurements, indicates that the relative abundance of these RNAs can be quantified with the MHD2 encoding scheme.
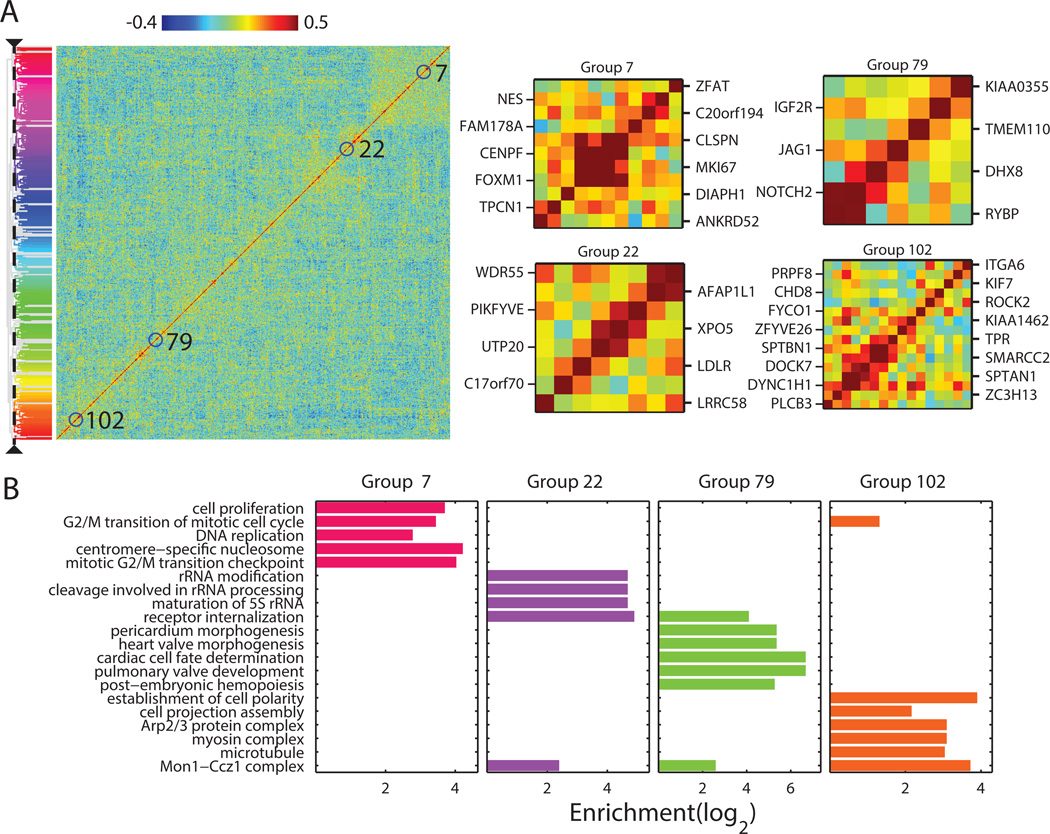
Simultaneously imaging ~1000 genes in individual cells substantially expanded our ability to detect co-regulated genes. Fig. 6A shows the matrix of pairwise correlation coefficients determined from the cell-to-cell variations in the expression levels of these genes. Using the same hierarchical clustering analysis as described above, we identified ~100 groups of genes with correlated expression (Table S4). Remarkably, nearly all of these ~100 groups showed statistically significant enrichment of functionally related GO terms (Fig. 6B; Table S4). These included some of the groups identified in the 140-gene measurements, such as the group associated with cell replication genes and the group associated with cell motility genes (Fig. 6A, B, groups 7 and 102), as well as many new groups. The groups identified here included 46 RNA species lacking any previous GO annotations, for which we can now hypothesize function based on their group association (Table S4). For example, KIAA1462 is part of the cell motility group, as also shown in the 140-gene experiments, suggesting a potential role of this gene in cell motility (Fig. 6A, group 102). Likewise, KIAA0355 is part of a new group enriched in genes associated with heart development (Fig. 6A, group 79), and C17orf70 is part of a group associated with ribosomal RNA processing (Fig. 6A, group 22). Using these groupings, we can also hypothesize cellular functions for 61 transcription factors and other partially annotated proteins of unknown functions (Table S4). For example, the transcription factors Z3CH13 and CHD8 are both members of the cell motility group, suggesting their potential role in the transcriptional regulation of cell motility genes. While these predicted functions based on gene-association analysis require further validation, our co-variation data provide a resource for generating hypotheses on gene function and regulation.
Fig. 6. Co-variation analysis of the RNA species measured in the 1001-gene measurements.
(A) Matrix of all pairwise correlation coefficients of the cell-to-cell variation in expression for the measured genes shown with the hierarchical clustering tree. The ~100 identified groups of correlated genes are indicated by color on the tree. Zoom in of four of the groups described in the text are shown on the right. (B) Enrichment of 20 selected, statistically significantly enriched GO terms in the four groups. The statistically most significantly enriched GO terms (maximum 10) for each of the ~100 groups are shown in Table S4.
Discussion
In summary, we have developed a highly multiplexed detection scheme for transcriptomic-scale RNA imaging in single cells. Using combinatorial labeling, sequential hybridization and imaging, and two different error-robust encoding schemes, we simultaneously imaged either 140 or 1001 genes in hundreds of individual human fibroblast cells. Of the two encoding schemes presented here, the MHD4 code is capable of both error detection and error correction, and hence can provide a higher calling rate and a lower misidentification rate than the MHD2 code, which instead can only detect but cannot correct errors. MHD2, on the other hand, provides a faster scaling of the degree of multiplexing with the number of bits than MHD4. Other error-robust encoding schemes can also be used for such multiplexed imaging, and experimenters can set the balance between detection accuracy and ease of multiplexing based on the specific requirements of the experiments.
By increasing the number of bits in the code words, it should be possible to further increase the number of detectable RNA species using MERFISH with either MHD4 or MHD2 codes. For example, using the MHD4 code with 32 total bits and four or six ‘1’ bits would increase the number of addressable RNA species to 1,240 or 27,776, respectively — the latter is the approximate scale of the human transcriptome. The predicted misidentification and calling rates are still reasonable for the 32-bit MHD4 code (shown in Fig. 1C, D, magenta for the MHD4 code with four ‘1’ bits and similar rates were calculated for the MHD4 code with six ‘1’ bits). If more accurate measurements are desired, an additional increase in the number of bits would allow the use of encoding schemes with a Hamming distance greater than 4, further enhancing the error detection and correction capability. While an increase in the number of bits by adding more hybridization rounds would increase the data collection time and potentially lead to sample degradation, these problems could be mitigated by utilizing multiple colors to readout multiple bits in each round of hybridization.
As the degree of multiplexing is increased, it is important to consider the potential increase in the density of RNAs that need to be resolved in each round of imaging. Based on our imaging and sequencing results, we estimate that including the whole transcriptome of the IMR90 cells would lead to a total RNA density of ~200 molecules/µm3. Using our current imaging and analysis methods, we could resolve 2–3 molecules/µm3 per hybridization round (38), which would reach a total RNA density of ~20 molecules/µm3 after 32 rounds of hybridization. This density should allow all but the top 10% most expressed genes to be imaged simultaneously or a subset of genes with even higher expression levels to be included. By utilizing more advanced image analysis algorithms to better resolve overlapping images of individual molecules, such as compressed sensing (39, 40), it would be possible to extend the resolvable density by ~4-fold and thus allow nearly the entire transcriptome, except for the top 2% most expressed genes, to be imaged all together. Finally, theoretical predictions (17) indicate that the use of super-resolution imaging (41, 42) could increase the resolvable density to ~105 molecules/µm3, which should be ample to address the entire transcriptome even in cell types with RNA densities substantially higher than that of IMR90. However, it is important to note that RNAs in densely packed structures, such as p-bodies and stress granules, may still elude measurement.
We have illustrated the utility of the data derived from highly multiplexed RNA imaging by using co-variation and correlation analysis to reveal distinct sub-cellular distribution patterns of RNAs, to constrain gene regulatory networks, and to predict functions for many previously unannotated or partially annotated genes with unknown functions. We anticipate that many more quantitative analyses could be applied to such data sets that include the spatial localization and copy number information of many RNA species in individual cells. Given its ability to quantify RNAs across a wide range of abundances without amplification bias while preserving native context, we envision that MERFISH will enable many applications of in situ transcriptomic analyses of individual cells in culture or complex tissues.
Materials and Methods
Probe design
Each RNA species in our target set was randomly assigned a binary code word either from all 140 possible code words of the 16-bit MHD4 code or from all 1001 possible code words of the 14-bit MHD2 code, as we describe in the main text. The encoding schemes are provided in Tables S1 and S3.
We used array-synthesized oligopools as templates to make the encoding probes (22, 23). The template molecule for each encoding probe contains three components: i) a central targeting sequence for in situ hybridization to the target RNA, ii) two flanking readout sequences designed to hybridize each of two distinct readout probes, and iii) two flanking primer sequences to allow enzymatic amplification of the probes (Fig. S3). The readout sequences were taken from the 16 possible readout sequences each corresponding to one hybridization round. The readout sequences were assigned to the encoding probes such that for any RNA species each of the 4 readout sequences were distributed uniformly along the length of the target RNA and appeared at the same frequency. Template molecules for the 140-gene library also included a common 20-nucleotide (nt) priming region between the first PCR primer and the first readout sequence. This priming sequence was used for the reverse transcription step described below. All template sequences are provided in Table S5.
We embedded multiple experiments in a single array-synthesized oligopool, and used PCR to selectively amplify only the oligos required for a specific experiment. Primer sequences for this indexed PCR reaction were generated from a set of orthogonal 25-nt sequences (43). These sequences were trimmed to 20 nt and selected for i) a narrow melting temperature range (70 – 80°C), ii) the absence of consecutive repeats of 3 or more identical nucleotides, and iii) the presence of a GC clamp, i.e. one of the two 3’ terminal bases must be G or C. To further improve specificity, these sequences were then screened against the human transcriptome using BLAST+ (44), and primers with 14 or more contiguous bases of homology were eliminated. Finally, BLAST+ was again used to identify and exclude primers that had an 11-nt homology region at the 3’ end of any other primer or a 5-nt homology region at the 3’ end of the T7 promoter. The forward primer sequences (Primer 1) were determined as described above, whereas the reverse primers each contain a 20-nt sequence as described above plus a 20-nt T7 promoter sequence to facilitate amplification via in vitro transcription (Primer 2). The primer sequences used in the 140-gene and 1001-gene experiments are listed below.
| ExperimentName | Primer 1 Sequence(Index Primer 1) | Primer 2 Sequence(T7 promoter plus the reverse complement of IndexPrimer 2) |
|---|---|---|
| 140-gene Codebook 1 | GTTGGTCGGCACTTGGGTGC | TAATACGACTCACTATAGGGAAAGCCGGTTCATCCGGTGG |
| 140-gene Codebook 2 | CGATGCGCCAATTCCGGTTC | TAATACGACTCACTATAGGGTGATCATCGCTCGCGGGTTG |
| 1001-gene | CGCGGGCTATATGCGAACCG | TAATACGACTCACTATAGGGCGTGGAGGGCATACAACGC |
30-nt-long readout sequences were created by concatenating fragments of the same orthogonal primer set generated above by combining one 20-nt primer with a 10-nt fragment of another. These readout sequences were then screened, using BLAST+, for orthogonality with the index primer sequences and other readout sequences (no more than 11 nt of homology) and for potential off-target binding sites in the human genome (no more than 14 nt of homology). Fluorescently labeled readout probes with sequences complementary to the readout sequences were used to probe these readout sequences, one in each hybridization round. All used readout probes sequences are listed below:
| Bit | Readout probes |
|---|---|
| 1 | CGCAACGCTTGGGACGGTTCCAATCGGATC/3Cy5Sp/ |
| 2 | CGAATGCTCTGGCCTCGAACGAACGATAGC/3Cy5Sp/ |
| 3 | ACAAATCCGACCAGATCGGACGATCATGGG/3Cy5Sp/ |
| 4 | CAAGTATGCAGCGCGATTGACCGTCTCGTT/3Cy5Sp/ |
| 5 | GCGGGAAGCACGTGGATTAGGGCATCGACC/3Cy5Sp/ |
| 6 | AAGTCGTACGCCGATGCGCAGCAATTCACT/3Cy5Sp/ |
| 7 | CGAAACATCGGCCACGGTCCCGTTGAACTT/3Cy5Sp/ |
| 8 | ACGAATCCACCGTCCAGCGCGTCAAACAGA/3Cy5Sp/ |
| 9 | CGCGAAATCCCCGTAACGAGCGTCCCTTGC/3Cy5Sp/ |
| 10 | GCATGAGTTGCCTGGCGTTGCGACGACTAA/3Cy5Sp/ |
| 11 | CCGTCGTCTCCGGTCCACCGTTGCGCTTAC/3Cy5Sp/ |
| 12 | GGCCAATGGCCCAGGTCCGTCACGCAATTT/3Cy5Sp/ |
| 13 | TTGATCGAATCGGAGCGTAGCGGAATCTGC/3Cy5Sp/ |
| 14 | CGCGCGGATCCGCTTGTCGGGAACGGATAC/3Cy5Sp/ |
| 15 | GCCTCGATTACGACGGATGTAATTCGGCCG/3Cy5Sp/ |
| 16 | GCCCGTATTCCCGCTTGCGAGTAGGGCAAT/3Cy5Sp/ |
The readout probes used for the 140-gene libraries were probes 1 through 16. The readout probes used for the 1001-gene experiment were probes 1 through 14. /3Cy5Sp/ indicates a 3’ Cy5 modification.
To design the central targeting sequences of the encoding probes, we first compiled the abundance of different transcripts in IMR90 cells using Cufflinks v2.1 (45), total RNA data from the ENCODE project (46), and human genome annotations from Gencode v18 (20). Probes were designed from gene models corresponding to the most abundant isoform using OligoArray2.1 (47) with the following constraints: the target sequence region is 30-nt long; the melting temperatures of the hybridized region of the probe and cellular RNA target is greater than 70 °C; there is no cross hybridization targets with melting temperatures greater than 72 °C; there is no predicted internal secondary structures with melting temperatures greater than 76 °C; and there is no contiguous repeats of 6 or more identical nucleotides. Melting temperatures were adjusted to optimize the specificity of these probes and minimize secondary structure while still producing sufficient numbers of probes for our libraries. To decrease computational cost, isoforms were divided into 1-kb regions for probe design. Using BLAST+, all potential probes that mapped to more than one cellular RNA species were rejected. Probes with multiple targets on the same RNA were kept.
For each gene in the 140-gene experiments, we generated 198 putative encoding probe sequences by concatenating the appropriate index primers, readout sequences, and targeting regions as shown in Fig S3. To address the possibility that concatenation of these sequences introduced new regions of homology to off-target RNAs, we used BLAST+ to screen these putative sequences against all human rRNA and tRNA sequences as well as highly expressed genes (genes with FPKM > 10,000). Probes with greater than 14 nt of homology to rRNAs or tRNAs or greater than 17 nt of homology to highly expressed genes were removed. After these cuts, we had ~192 (with a standard deviation of 2) probes per gene for both MHD4 codebooks used in the 140-gene experiments. We followed the same protocol for the 1001-gene experiments: Starting with 96 putative targeting sequences per gene, we obtained ~94 (with a standard deviation of 6) encoding probes per gene after these additional homology cuts. We decreased the number of encoding probes per RNA for the 1001-gene experiments so that these probes could be synthesized from a single 100,000-member oligopool as opposed to two separate pools. We designed each encoding probe to contain two of the four readout sequences associated with each code word, hence only half of the bound encoding probes can bind readout probe during any given hybridization round. We used ~192 or ~94 encoding probes per RNA to obtain high signal-to-background ratios for individual RNA molecules. The number of encoding probes per RNA could be substantially reduced but still allow single RNA molecules to be identified (17, 48, 49). In addition, increasing the number of readout sequences per encoding probe or using optical sectioning methods to reduce the fluorescence background may allow further reduction in the number of the encoding probes per RNA.
We designed two types of misidentification controls. The first control — blank words — were not represented with encoding probes. The second type of control — no-target words — had encoding probes that were not targeting any cellular RNA. The targeting regions of these probes were composed of random nucleotide sequences subject to the same constraints used to design the RNA targeting sequences described above. Moreover, these random sequences were screened against the human transcriptome to ensure that they contain no significant homology (>14-nt) to any human RNA. The 140-gene measurements contained 5 blank words and 5 no-target words. The 1001-gene measurements contained 11 blank words and 5 no-target words.
Probe synthesis
The encoding probes were synthesized using the following four steps, and this synthesis protocol is illustrated in Fig. S3.
- Step 1: The template oligopool (CustomArray) was amplified via limited-cycle PCR on a Bio-Rad CFX96 using primer sequences specific to the desired probe set. To facilitate subsequent amplification via in vitro transcription, the reverse primer contained the T7 promoter. All primers were synthesized by IDT. This reaction was column purified (Zymo DNA Clean and Concentrator; D4003).
- Step 2: The purified PCR products were then further amplified ~200-fold and converted into RNA via a high yield in vitro transcription according to the manufacturer’s instructions (New England Biolabs, E2040S). Each 20 µL reaction contained ~1 µg of template DNA from above, 10 mM of each NTP, 1× reaction buffer, 1× RNase inhibitor (Promega RNasin, N2611) and 2 µL of the T7 polymerase. This reaction was incubated at 37 °C for 4 hours to maximize yield. This reaction was not purified before the following steps.
- Step 3: The RNA products from the above in vitro transcription reaction were then converted back into DNA via a reverse transcription reaction. Each 50 µL reaction contained the unpurified RNA produce from Step 2 supplemented with 1.6 mM of each dNTP, 2 nmol of a reverse transcription primer, 300 units of Maxima H- reverse transcriptase (Thermo Scientific, EP0751), 60 units of RNasin, and a final 1× concentration of the Maxima RT buffer. This reaction was incubated at 50 °C for 45 minutes, and the reverse transcriptase was inactivated at 85°C for 5 minutes. The templates for the 140-gene libraries contain a common priming region for this reverse transcription step; thus, a single primer was used for this step when creating these probes. Its sequence was CGGGTTTAGCGCCGGAAATG. A common priming region was not included for the 1001-gene library; thus, the reverse transcription was conducted with the forward primer: CGCGGGCTATATGCGAACCG.
- Step 4: To remove the template RNA, 20 µL of 0.25 M EDTA and 0.5 N NaOH was added to the above reaction to selectively hydrolyze RNA, and the sample was incubated at 95 °C for 10 minutes. This reaction was then immediately purified by column purification using a 100-µg-capacity column (Zymo Research, D4030) and the Zymo Oligo Clean and Concentrator protocol. The final probes were eluted in 100 µL of RNase-free deionized water, evaporated in a vacuum concentrator, and then resuspended in 10 µL of encoding hybridization buffer (recipe below). Probes were stored at −20 °C. Denaturing poly-acrylamide gel electrophoresis and absorption spectroscopy were used to confirm the quality of the probes and revealed that this probe synthesis protocol converts 90–100% of the reverse-transcription primer into full length probe and of the probe that is constructed, 70–80% is recovered during the purification step. This protocol is similar to another recently published protocol (23) but provides a substantially larger yield.
Fluorescently labeled readout probes have sequences complementary to the readout sequences described above and a Cy5 dye attached at the 3’ end. These probes were obtained from IDT and HPLC purified.
Sample preparation and labeling with encoding probes
Human primary fibroblasts (American Type Culture Collection, IMR90), a commonly used cell line with a previously determined transcriptome (46), were used in this work. These cells are relatively large and flat, facilitating wide-field imaging without the need for optical sectioning. Cells were cultured with Eagle’s Minimum Essential Medium. Cells were plated on 22-mm, #1.5 coverslips (Bioptechs, 0420-0323-2) at 350,000 cells/coverslip and incubated at 37 °C with 5% CO2 for 48–96 hours within petri dishes. Cells were fixed for 20 minutes in 4% paraformaldehyde (Electron Microscopy Sciences, 15714) in 1× phosphate buffered saline (PBS; Ambion, AM9625) at room temperature, reduced for 5 minutes with 0.1% w/v sodium borohydride (Sigma, 480886) in water to reduce background fluorescence, washed three times with ice-cold 1× PBS, permeabilized for 2 minutes with 0.5% v/v Triton (Sigma, T8787) in 1× PBS at room temperature, and washed three times with ice cold 1× PBS.
Cells were incubated for 5 minutes in encoding wash buffer comprising 2× saline-sodium citrate buffer (SSC) (Ambion, AM9763), 30% v/v formamide (Ambion, AM9342), and 2 mM vanadyl ribonucleoside complex (NEB, S1402S). 10 µL of 100 µM (140-gene experiments) or 200 µM (1001-gene experiments) encoding probes in encoding hybridization buffer was added to the cell-containing coverslip and spread uniformly by placing another coverslip on top of the sample. Samples were then incubated in a humid chamber inside a 37 °C-hybridization oven for 18–36 hours. Encoding hybridization buffer is composed of encoding wash buffer supplemented with 1 mg/mL yeast tRNA (Life technologies, 15401-011) and 10% w/v dextran sulfate (Sigma, D8906-50G).
Cells were then washed with primary encoding wash buffer, incubated at 47 °C for 10 minutes, and this wash was repeated for a total of three times. A 1:1000 dilution of 0.2-µm-diameter carboxylate-modified orange fluorescent beads (Life Technologies, F-8809) in 2×SSC was sonicated for 3 minutes and then incubated with the sample for 5 minutes. The beads were used as fiducial markers to align images obtained from multiple successive rounds of hybridization, as described below. The sample was washed once with 2×SSC, and then post-fixed with 4% v/v paraformaldehyde in 2×SSC at room temperature for 30 minutes. The sample was then washed three times with 2×SSC and either imaged immediately or stored for no longer than 12 hours at 4 °C prior to imaging. All solutions were prepared as RNase-free.
MERFISH imaging with multiple successive rounds of hybridization
The sample coverslip was assembled into a Bioptech’s FCS2 flow chamber, and the flow through this chamber was controlled via a home-built fluidics system composed of three computer-controlled 8-way valves (Hamilton, MVP and HVXM 8-5) and a computer-controlled peristaltic pump (Rainin, Dynamax RP-1). The sample was imaged on a home-built microscope constructed around an Olympus IX-71 body and a 1.45 NA, 100× oil immersion objective and configured for oblique incidence excitation. The objective was heated to 37 °C with a Bioptechs objective heater. Constant focus was maintained throughout the imaging process with a home-built, auto-focusing system. Illumination was provided at 641 nm, 561 nm, and 405 nm using solid state lasers (MPB communications, VFL-P500-642; Coherent, 561-200CWCDRH; and Coherent, 1069413/AT) for excitation of our Cy5-labeled readout probes, the fiducial beads, and nuclear counterstains, respectively. These lines were combined with a custom dichroic (Chroma, zy405/488/561/647/752RP-UF1) and the emission was filtered with a custom dichroic (Chroma, ZET405/488/561/647-656/752m). Fluorescence was separated with a QuadView (Photometrics) using the dichroics T560lpxr, T650lpxr, 750dcxxr (Chroma) and the emission filters ET525/50m, WT59550m-2f, ET700/75m, HQ770lp (Chroma) and imaged with an EMCCD camera (Andor, iXon-897). The camera was configured so that a pixel corresponds to 167 nm in the sample plane. The entire system was fully automated, so that imaging and fluid handling were performed for the entire experiment without user intervention.
Sequential hybridization, imaging, and bleaching proceeded as follows. 1 mL of 10 nM of the appropriate fluorescently labeled readout probe in readout hybridization buffer (2×SSC; 10% v/v formamide; 10% w/v dextran sulfate, and 2 mM vanadyl ribonucleoside complex) was flown across the sample, flow was stopped, and the sample was incubated for 15 minutes. Then 2 mL of readout wash buffer (2×SSC, 20% v/v formamide; and 2 mM vanadyl ribonucleoside complex) was flown across the sample, flow was stopped, and the sample was incubated for 3 minutes. 2 mL of imaging buffer comprising 2×SSC, 50 mM TrisHCl pH 8, 10 % w/v glucose, 2 mM Trolox (Sigma-Aldrich, 238813), 0.5 mg/mL glucose oxidase (Sigma-Aldrich, G2133), and 40 µg/mL catalase (Sigma-Aldrich, C30) was flown across the sample (50). Flow was then stopped, and then approximately 75 to 100 regions were exposed to ~25 mW 642-nm and 1 mW of 561-nm light and imaged. Each region was 40 µm by 40 µm. The laser powers were measured at the microscope backport. Because the imaging buffer is sensitive to oxygen (51), the ~50 mL of imaging buffer used for a single experiment was made fresh at the beginning of the experiment and then stored under a layer of mineral oil throughout the measurement. Buffer stored in this fashion was stable for more than 24 hours.
After imaging, the fluorescence of the readout probes was extinguished via photobleaching. The sample was washed with 2 mL of photobleaching buffer (2×SSC and 2 mM vanadyl ribonucleoside complex), and each imaged region of the sample was exposed to 200 mW of 641-nm light for 3 s. To confirm the efficacy of this photobleaching treatment, imaging buffer was reintroduced, and the sample was imaged as described above.
The above hybridization, imaging, and photobleaching process was repeated either 16 times for the 140-gene measurements using the MHD4 code or 14 times for the 1001-gene measurements using the MHD2 code. An entire experiment was typically completed in ~20 hours.
Following completion of imaging, 2 mL of a 1:1000 dilution of Hoescht (ENZ-52401) in 2×SSC was flown through the chamber to label the nuclei of the cells. The sample was then washed immediately with 2 mL of 2×SSC followed by 2 mL of imaging buffer. Each region of the sample was then imaged once again with ~1 mW of 405-nm light.
Because we imaged cells using wide-field imaging with oblique-incidence illumination, without optical sectioning and z-scanning, we quantified the fraction of individual RNA species that was outside the axial range of our imaging geometry for 6 different RNA species using conventional smFISH. For this purpose, we optically sectioned these cells by collecting stacks of images at different focal depths through the entire depth of the cells. We aligned the images in consecutive focal planes and then computed for each cell the fraction of RNAs that were detected in the three-dimensional stack but not in the basal focal plane. We found that only a small fraction, 15% ± 1% (Mean ± SEM across six different RNA species) of RNA molecules were outside the imaging range of a fixed focal plane without z-scanning. These measurements also confirmed that our excitation geometry illuminated the full depth of our cells. We note that, from an imaging perspective, MERFISH is simply a series of smFISH experiments. Thus, any optical sectioning technique could be employed in MERFISH to allow the imaging of RNAs in thicker cells or tissues.
Construction of measured words
Fluorescent spots were identified and localized in each image using a multi-Gaussian-fitting algorithm (38) assuming a Gaussian with a uniform width of 167 nm. This algorithm was used to allow partially overlapping spots to be distinguished and individually fit. RNA spots were distinguished from background signal, i.e. signal arising from probes bound non-specifically, by setting the intensity threshold required to fit a spot with this software. Due to variation in the brightness of spots between rounds of hybridization, this threshold was adjusted appropriately for each hybridization round to minimize the combined average of the 1→0 and 0→1 error rates across all hybridization rounds (140-gene measurements) or to maximize the ratio of the number of measured words with four ‘1’ bits to those with three or five ‘1’ bits (1001-gene measurements). The location of the fiducial beads was identified in each frame using a faster single-Gaussian fitting algorithm.
Images of the same sample region in different rounds of hybridization were registered by rotating and translating the image to align the two fiducial beads within the same image that were most similar in location after a coarse initial alignment via image correlation. All images were aligned to a coordinate system established by the images collected in the first round of hybridization. The quality of this alignment was determined from the residual distance between five additional fiducial beads, and alignment error was typically ~20 nm.
Fluorescence spots in different hybridization rounds were connected into a single string, corresponding to a potential RNA molecule, if the distance between spots was smaller than 1 pixel (167 nm). For each string of spots, the on-off sequence of fluorescent signals in all hybridization rounds were used to assign a binary word to the potential RNA molecule, in which ‘1’ was assigned to the hybridization rounds that contained a fluorescent signal above threshold and ‘0’ was assigned to the other hybridization rounds. Measured words were then decoded into RNA species using the 16-bit MHD4 code or the 14-bit MHD2 code discussed in the main text. In the case of the 16-bit MHD4 code, if the measured binary word matched the code word of a specific RNA perfectly or differed from the code word by one single bit, it was assigned to that RNA. In the case of the 14-bit MHD2 code, only if the measured binary word matched the code word of a specific RNA perfectly, was it assigned to that RNA. To determine the copy number per cell, the number of each RNA species was counted in individual cells within each 40 µm by 40 µm imaging area. We note that this number accounts for the majority but not all RNA molecules within a cell because a fraction of the cell could be outside the imaging area or focal depth. Tiling images of adjacent areas and adjacent focal planes could be employed to improve the counting accuracy.
In the 140-gene experiments, some regions of the cell nucleus occasionally contained too much fluorescence signal to properly identify individual RNA spots. In the 1001-gene experiments, the cell nucleus generally contained too much fluorescent signal to allow identification of individual RNA molecules. These bright regions were excluded from all subsequent analysis. This work focuses on mRNAs, which are enriched in the cytoplasm. To estimate the fraction of mRNAs missed by excluding the nucleus region, we used conventional smFISH to quantify the fraction of molecules found inside the nucleus for six different mRNAs species. We found that only 5% ± 2% (Mean ± SEM across six RNA species) of these RNA molecules are found in the nucleus. Employment of super-resolution imaging and/or optical sectioning could potentially allow individual molecules in these dense nucleus regions to be identified, which will be particularly useful for probing those non-coding RNAs that are enriched in the nucleus.
smFISH measurements of individual genes
Pools of 48 fluorescently-labeled (Quasar 670) oligonucleotide probes per RNA were purchased from Biosearch Technologies. 30-nt probe sequences were taken directly from a random subset of the targeting regions used for the multiplexed measurements. Cells were fixed and permeabilized as described above. 10 µL of 250 nM oligonucleotide probes in encoding hybridization buffer (described above) was added to the cell-containing coverslip and spread uniformly by placing another coverslip on top of the sample. Samples were then incubated in a humid chamber inside a 37 °C-hybridization oven for 18 hours. Cells were then washed with encoding wash buffer (described above) at 37 °C for 10 minutes, and this wash was repeated for a total of three times. The sample was then washed three times with 2×SSC and imaged in imaging buffer using the same imaging geometry as described above for MERFISH.
Bulk RNA sequencing
Total RNA was extracted from IMR90 cells cultured as above using the Zymo Quick RNA MiniPrep kit (R1054) according to the manufacturer’s instructions. polyA RNA was then selected (NEB; E7490), and a sequencing library was constructed using the NEBNext Ultra RNA library preparation kit (NEB; E7530), amplified with custom oligonucleotides, and 150-bp reads were obtained from on a MiSeq. These sequences were aligned to the human genome (Gencode v18) and isoform abundance was computed with cufflinks (45).
Calculation of the predicted scaling and error properties of different encoding schemes
Analytic expressions were derived for the dependence of the number of possible code words, the calling rate, and the misidentification rate on N. The calling rate is defined as the fraction of RNA molecules that are properly identified. The misidentification rate is defined as the fraction of RNA molecules that are misidentified as a wrong RNA species. For encoding schemes with an error-detection capability, the calling rate and misidentification rate does not add up to 1 because a fraction of the molecules not called properly can be detected as errors and discarded and, hence, not misidentified as a wrong species. These calculations assume that the probability of misreading bits is constant for all hybridization rounds but differs for the 1→0 and 0→1 errors. Experimentally measured average 1→0 and 0→1 error rates (10% and 4% respectively) were used for the estimates shown in Fig.1B–D. For simplicity, the word corresponding to all ‘0’s was not removed from calculations.
For the simple binary encoding scheme in which all possible N_-bit binary words are assigned to unique RNA species, the number of possible code words is 2_N. The number of words that could be used to encode RNA is actually 2_N_ − 1 because the code word ‘00…0’ does not contain detectable fluorescence in any hybridization round, but for simplicity the word corresponding to all ‘0’s was not removed from subsequent calculations. The error introduced by this approximation is negligible. For any given word with m ‘1’s and N_−_m ‘0’s the probability of measuring that word without error — the fraction of RNAs that is properly called — is
where p1 is 1→0 error rate and p0 is 0→1 error rate per bit. Because different words in this simple binary encoding scheme can have different numbers of ‘1’ bits, the calling rate for different words will differ if _p_1 ≠ _p_0. The average calling rate, reported in Fig. 1C, was determined from the weighted average of the value of Eq. (1) for all words. This weighted average is
| 12N∑m=0N(Nm)(1−p1)m(1−p0)N−m, | (2) |
|---|
where (Nm) is the binomial coefficient and corresponds to the number of words with m ‘1’ bits in this encoding scheme. Since in this encoding scheme every error produces a binary word that encodes a different RNA, the average misidentification rate for this encoding scheme, reported in Fig. 1D, follows directly from (2):
| 1−12N∑m=0N(Nm)(1−p1)m(1−p0)N−m. | (3) |
|---|
To calculate the scaling and error properties of the extended Hamming distance 4 (HD4) code, we first created the generator matrix for the desired number of data bits using standard methods (21). The generator matrix determines the specific words that are present in a given encoding scheme and was used to directly determine the number of encoded words as a function of the number of bits. In this encoding scheme, the calling rate corresponds to the fraction of words measured without error as well as the fraction of words measured with a single-bit error. For code words with m ‘1’ bits, this fraction is determined by the following expression:
| (1−p1)m(1−p0)N−m+mp11(1−p1)m−1(1−p0)N−m+(N−m)p01(1−p1)m(1−p0)N−m−1 | (4) |
|---|
where the first term is the probability of not making any errors, the second term corresponds to the total probability of making one 1→0 error at any of the m ‘1’ bits without making any other 0→1 errors, and the final term corresponds to the total probability of making one 0→1 error at any of the N-m ‘0’ bits without making any 1→0 errors. Because the number of ‘1’ bits can differ between words in this encoding scheme, the average calling rate reported in Fig. 1C was computed from a weighted average over Eq. (4) for different values of m. The weight for each term was determined from the number of words that contain m ‘1’ bits as determined from the generator matrix described above.
Because RNA-encoding words are separated by a minimum Hamming distance of 4, at least 4 errors are required to switch one word into another. If error correction is applied, then 3 or 5 errors could also convert one RNA into another. Thus, we estimate the misidentification rate from all possible combinations of 3-bit, 4-bit and 5-bit errors for code words with m ‘1’ bits. Technically, >5-bit errors could also convert one RNA into another, but the probability of making such errors is negligible because of the small per-bit error rate. We approximate this expression with
| ∑i=04(mi)(N−m4−i)p1ip04−i(1−p1)m−i(1−p0)N−m−(4−i)+∑i=03(mi)(N−m3−i)p1ip03−i(1−p1)m−i(1−p0)N−m−(3−i)+∑i=05(mi)(N−m5−i)p1ip05−i(1−p1)m−i(1−p0)N−m−(5−i)+ | (5) |
|---|
The first sum corresponds to all of the ways in which exactly four mistakes can be made. Similarly, the second and third sums correspond to all of the ways in which exactly three or five mistakes can be made. Eq. (5) provides an upper bound for the misidentification rate because not all three, four, or five bit errors produce a word that matches or would be corrected to another legitimate word. Again because the number of ‘1’ bits can differ between words, the average misidentification rate reported in Fig. 1D is calculated as a weighted average of Eq. (5) over the number of words that have m ‘1’ bits.
To generate our MHD4 code where the number of ‘1’ bits for each code word is set to 4, we first generated the HD4 codes as described above, and then removed all code words that did not contain four ‘1’s. The calling rate of this code, reported in Fig. 1C, was directly calculated from Eq. (4) but with m = 4 because all code words in this code have four ‘1’ bits. The misidentification rate of this code, reported in Fig. 1D, was calculated by modifying Eq. (5) with the following considerations: (i) the number of ‘1’ bits, m, was set to 4 and (ii) errors that produce words that do not contain three, four, or five ‘1’ bits were excluded. Thus, the expression in Eq. (5) was simplified to
| (42)(N−42)p12p02(1−p1)2(1−p0)N−6+(41)(N−42)p11p02(1−p1)3(1−p0)N−6+(42)(N−41)p12p01(1−p1)2(1−p0)N−5 | (6) |
|---|
(42)(N−43)p12p03(1−p1)2(1−p0)N−7+(43)(N−42)p13p02(1−p1)1(1−p0)N−6
Again, this expression is an upper bound on the actual misidentification rate because not all words with four ‘1’s are valid code words.
Estimates of the 1→0 and 0→1 error rates for each hybridization round
To compute the probability of misreading a bit at a given hybridization round, we used the error correcting properties of the MHD4 code. Briefly, the probabilities of 1→0 or 0→1 errors were derived in the following way. Let the probability of making an error at the _i_th bit, i.e. _i_th hybridization round, be pi and the actual number of RNA molecules of the given species be A, then the number of exact matches for this RNA will be WE=A∏i=116(1−pi) and the number of one-bit error corrected matches for this RNA corresponding to errors at the _i_th bit will be Wi=Api(1−pi)∏j=116(1−pj). The pi can be directly derived from the ratio: Wi/WE=pi(1−pi). This ratio assumes that the one-bit error-corrected counts were only generated from single-bit errors from the correct word and that multi-error contamination from other RNA words is negligible. Given that our error rate per hybridization round is small and that it takes at least three errors to convert one RNA-encoding word into a word that would be misidentified as another RNA, the above approximation should be a good one.
To compute the average 1→0 or 0→1 error probabilities for each of the 16 hybridization rounds, we use the above approach to calculate the per-bit error rates for each bit of every gene, sort these errors based on whether they correspond to a 1→0 or a 0→1 error, and then take the average of these errors for each bit weighted by the number of counts observed for the corresponding gene.
Estimates of the calling rate for individual RNA species from actual imaging data
With the estimates of the 1→0 or 0→1 error probabilities for each round of hybridization as determined above, it is possible to estimate the calling rate for each RNA based on the specific word used to encoded it. Specifically, the fraction of an RNA species that is called correctly is determined by
| ∏i=1N(1−pi)+∑j=1Npj(1−pj)∏i=1N(1−pi), | (7) |
|---|
where the first term represent the probability of observing an exact match of the code word and the second term represent the probability of observing an error-corrected match (i.e. with one-bit error). The values of the per-bit error rate pi for each RNA species are determined by the specific code word for that RNA and the measured 1→0 or 0→1 error rates for each round of hybridization. If the code word of the RNA contains a ‘1’ in the _i_th bit, then pi is determined from the 1→0 error rate for the _i_th hybridization round; if the word contains a ‘0’ in the _i_th bit, pi is determined from the 0→1 error rate for the _i_th hybridization round.
Hierarchical clustering analysis of the co-variation in RNA abundance
Hierarchical clustering of the co-variation in gene expression for both the 140-gene and 1001-gene experiments was conducted as follows. First, the distance between every pair of genes was determined as 1 minus the Pearson correlation coefficient of the cell-to-cell variation of the measured copy numbers of these two RNA species, both normalized by the total RNA counted in the cell. Thus, highly correlated genes are ‘closer’ to one another and highly anti-correlated genes are ‘further’ apart. An agglomerative hierarchical cluster tree was then constructed from these distances using the Unweighted Pair Group Method with Arithmetic mean (UPGMA). Specifically, starting with individual genes, we constructed hierarchical clusters by identifying the two clusters (or individual genes) that are closest to one another according to the arithmetic mean of the distances between all inter-cluster gene pairs. The pairs of clusters (or individual genes) with the smallest distance are then grouped together and the process is repeated. The matrix of pairwise correlations was then sorted based on the order of the genes within these trees.
Groups of genes with substantial co-variations were identified by selecting a threshold on the hierarchical cluster tree (indicated by the dashed lines in Figs. 3D and 6A) that produced approximately 10 groups of genes each of which contains at least 4 members for the 140-gene experiments or approximately 100 groups each of which contains at least 3 members for the 1001-gene experiments. We note that one can change the threshold in order to identify either more tightly coupled smaller groups or larger groups with relatively loose coupling.
A probability value for the confidence that a gene belongs to a specific group was determined by computing the difference between the average correlation coefficient between that gene and all other members of that group and the average correlation coefficient between that gene and all other measured genes outside that group. The significance (p-value) of this difference was determined with the student’s t-test and is provided in Tables S2 and S4.
Because hierarchical clustering is inherently a one-dimensional analysis, i.e. any given genes can only be a member of a single group, this analysis does not allow all correlated gene groups to be identified. Higher dimension analysis, such as principal component analysis or k-means clustering, could be used to identify more co-varying gene clusters (30).
Analysis of RNA spatial distributions
To identify genes that have similar spatial distributions, we subdivided each of the measured cells into 2×2 regions and calculated the fraction of each RNA species present in each of these bins. To control for the fact that some regions of the cell naturally contain more RNA than others, we calculate the enrichment for each gene — the ratio of the observed fraction in a given region for a given RNA species to the average fraction observed for all genes in that same region. For each pair of RNA species, we then determined the Pearson correlation coefficient of the region-to-region variation in enrichment of these two RNA species for each cell and averaged the correlation coefficients over ~400 cells imaged in 7 independent data sets. We then clustered RNA species based on these average correlation coefficients using the same hierarchical clustering algorithm described above. Because of the large number of cells used for the analysis, we found that the coarse spatial binning (2×2 regions per cell) was sufficient to capture the spatial correlation between genes and finer binning did not produce more significantly correlated groups.
To measure the distances of genes from the nuclei and from the cell edge, we first used brightness thresholds on our cell images to segment the nuclei and identify the cell edge. We then measured the distance from every RNA molecule to the nearest part of the nucleus and nearest part of the cell edge. For each data set, we computed the average distance for each RNA species averaged over all the cells measured. We then averaged these distances for the group I genes, group II genes or all genes. Only those RNA species with at least 10 counts per cell were used in this analysis to minimize statistical error on the distance values.
Gene ontology (GO) analysis
Groups of genes were selected from the hierarchical trees as discussed above. A collection of GO terms (31) was determined for all measured RNA species as well as the RNA species associated with each group from the most recent human GO annotations (http://geneontology.org/page/download-annotations) using both the annotated GO terms and terms immediately upstream or downstream of the found annotations. The enrichment of these annotations was calculated from the ratio of the fraction of genes within each group that have this term to the fraction of all measured genes that have this term and the p-value for this enrichment was calculated via the hypergeometric function. Only statistically significantly enriched GO terms with a p-value less than 0.05 were considered.
Supplementary Material
Supplemental 1
Table 1
Table 2
Table 3
Table 4
Table 5
Acknowledgments
We thank Hazen Babcock for technical advice and help with instrumentation and Bogdan Bintu for aid in error analysis. This work was in part supported by the National Institutes of Health. K.H.C. acknowledges a National Science Scholarship from the Agency for Science, Technology and Research of Singapore. A.N.B. acknowledges support by the Damon Runyon Foundation postdoctoral fellowship. J.R.M. acknowledges support from the Helen Hay Whitney Foundation postdoctoral fellowship. S.W. acknowledges support from Jane Coffins Child Foundation postdoctoral fellowship. X.Z. is a Howard Hughes Medical Institute investigator. X.Z., K.H.C., A.N.B., J.R.M., S.W. are inventors on a patent applied for by Harvard University that covers the MERFISH method described here; X.Z., J.R.M., and A.N.B. are inventors on a patent applied for by Harvard University that covers the probe synthesis method described here.
Footnotes
Online Supplementary Materials:
Figs. S1 to S9
Tables S1 to S5
References
- 1.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 2.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 3.Raj A, van Den Bogaard P, Rifkin S, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr. Opin. Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote Control of Gene Function by Local Translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregor T, Garcia HG, Little SC. The embryo as a laboratory: quantifying transcription in Drosophila. Trends Genet. 2014;30:364–375. doi: 10.1016/j.tig.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2014;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson DR, Singer RH, Zenklusen D. A single molecule view of gene expression. Trends Cell Biol. 2009;19:630–637. doi: 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu. Rev. Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha T. Single-molecule methods leap ahead. Nat. Methods. 2014;11:1015–1018. doi: 10.1038/nmeth.3107. [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods. 2013;10:1127–1133. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- 16.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 17.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levesque MJ, Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods. 2013;10:246–248. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon TK. Error Correction Coding: Mathematical Methods and Algorithms. ed. 1st Wiley: 2005. [Google Scholar]
- 22.Beliveau BJ, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murgha YE, Rouillard J-M, Gulari E. Methods for the Preparation of Large Quantities of Complex Single-Stranded Oligonucleotide Libraries. PLoS One. 2014;9:1–10. doi: 10.1371/journal.pone.0094752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342:1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.So L, et al. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Safran M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolinski K, Botstein D. Changing perspectives in yeast research nearly a decade after the genome sequence. Genome Res. 2005;15:1611–1619. doi: 10.1101/gr.3727505. [DOI] [PubMed] [Google Scholar]
- 28.Padovan-Merhar O, Raj A. Using variability in gene expression as a tool for studying gene regulation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013;5:751–759. doi: 10.1002/wsbm.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehlenborg N, et al. Visualization of omics data for systems biology. Nat. Methods. 2010;7:S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball C, Blake J, Botstein D. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida H, et al. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5612–5617. doi: 10.1073/pnas.1215432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 34.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 35.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521–1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 37.Mingle LA, et al. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci. 2005;118:2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babcock H, Sigal YM, Zhuang X. A high-density 3D localization algorithm for stochastic optical reconstruction microscopy. Opt. Nanoscopy. 2012;1:6. doi: 10.1186/2192-2853-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, Zhang W, Elnatan D, Huang B. Faster STORM using compressed sensing. Nat. Methods. 2012;9:721–723. doi: 10.1038/nmeth.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babcock H, Moffitt J, Cao Y, Zhuang X. Fast compressed sensing analysis for super-resolution imaging using L1-homotopy. Opt. Express. 2013;21:28583–28596. doi: 10.1364/OE.21.028583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hell SW. Microscopy and its focal switch. Nat. Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- 42.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, Schlabach MR, Hannon GJ, Elledge SJ. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouillard J-M, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batish M, Raj A, Tyagi S. In: Methods in molecular biology. Gerst JE, editor. Totowa, NJ: Humana Press; 2011. pp. 3–13. vol. 714 of Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 49.Buxbaum A, Wu B, Singer R. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–423. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 51.Shi X, Lim J, Ha T. Acidification of the oxygen scavenging system in single-molecule fluorescence studies: in situ sensing with a ratiometric dual-emission probe. Anal. Chem. 2010;82:6132–6138. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental 1
Table 1
Table 2
Table 3
Table 4
Table 5