A Structure and Dynamics Continuum of Higher-Order Assemblies: Amyloids, Signalosomes and Granules (original) (raw)
. Author manuscript; available in PMC: 2017 May 19.
Abstract
We here attempt to reach an integrated understanding of the structure and dynamics of a number of higher-order assemblies including amyloids, various kinds of signalosomes, and cellular granules. We propose that the synergy between folded domains, linear motifs and intrinsically disordered regions regulates formation and intrinsic fuzziness of all higher-order assemblies, creating a structural and dynamic continuum. We describe how such regulatory mechanisms could be influenced under pathological conditions.
Introduction
The attainment of the first protein crystal structures approximately half a century ago commenced the structure-function paradigm, in which a folded structure determines protein function. More recently, increasing evidence from various branches of structural biology demonstrated that the function of proteins also originates from their intrinsically disordered regions (IDRs) that lack a well-defined conformation, expanding the classical understanding of how structure determines function (Oldfield and Dunker, 2014; Wright and Dyson, 2015). Some IDR sequences are repetitive and have low information content relative to folded proteins. They are also referred to as low complexity domains (LCDs) (Huntley and Golding, 2002). IDRs may fold into a unique conformation upon binding to another protein or when oligomerized, in order to exert their function. They may also alternatively fold into ensembles of structured conformations or remain largely disordered, exhibiting a fast exchange of conformations even in the complex state, which have both been referred to as fuzzy structures (Tompa and Fuxreiter, 2008).
In parallel with the increasing structural diversity, higher-order complexes have emerged across a wide spectrum of the biological landscape in recent years, including amyloids and prions (Eisenberg and Jucker, 2012), various kinds of signaling complexes generically denoted as signalosomes (Bienz, 2014; Wu, 2013), and nuclear and cytoplasmic granules (Anderson et al., 2015; Hyman et al., 2014; Mitchell and Parker, 2014). They have collectively transformed how the field thinks about cellular organization and signal transduction (Brangwynne et al., 2015; Wu, 2013). These complexes differ from most traditional macromolecular complexes because their component proteins often polymerize or crosslink, leading to variable stoichiometries and heterogeneous conformations. The question is: are there common biophysical principles at play for the different types of higher-order structures? Because lessons learned from one kind of structure may inform the functional mechanisms of others, we have collectively interrogated them in order to reveal mechanistic insights. We found that folded domains, linear motifs and IDRs in higher-order assemblies synergize with each other to form inherently fuzzy structures. These complexes may possess a similar critical concentration of assembly, but exhibit different structural and dynamic properties depending on the underlying assembly mechanism and regulation.
Distinct Structure and Dynamics of Higher-Order Assemblies
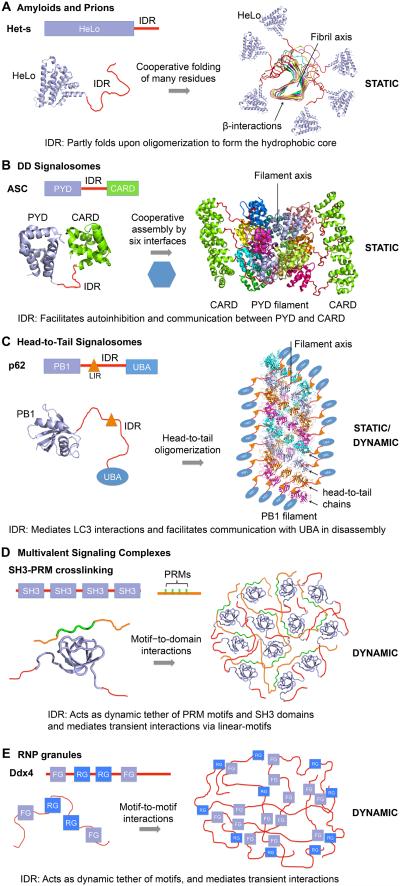
The higher-order structures we discuss in this perspective are formed by distinct interacting elements and with different dynamic properties. Amyloids are the best known higher-order structures, which can be formed from IDRs through folding upon binding. They are fibrous protein aggregates that contain a central cross-β sheet core with solvent-excluded, self-complementing steric zipper interactions (Eisenberg and Jucker, 2012; Sawaya et al., 2007). Amyloids represent one of the most static higher-order structures, with tight packing of adjacent interacting elements, resulting in cooperative contacts and a high barrier of dissociation, and thus irreversible assembly (Table 1). Within each β-sheet, hydrogen-bonding ladders formed from Asn and Gln side chains along the fibrillar axis are often the key stabilizing interactions, which are complemented by hydrophobic interactions between the β-sheets (Alberti et al., 2009; Sawaya et al., 2007). Amyloids and prions have been associated with both human diseases and normal physiology in diverse organisms, including the fungal heterokaryon incompatibility gene product, HET-s, the inherited nonsense codon suppression protein Sup35 (Fowler et al., 2006; Prusiner, 1991; Wickner et al., 2007), and the mammalian necrosome containing RIP1 and RIP3 kinases for necrotic cell death signaling (Li et al., 2012a). The complete amyloid domain structure of HET-s from solid-state NMR depicts a β-solenoid with a triangular hydrophobic core and abundant hydrogen bonds and salt bridges (Wasmer et al., 2008) (Figure 1A).
Table 1.
Relationship of Binding Element Affinity, Valency, IDRs and Material States.
| Complexes | Example | Critical Concentration | Interacting Element | KD of Element | Valency | Dynamic IDR | Material State | Dynamics* | References |
|---|---|---|---|---|---|---|---|---|---|
| Amyloid | HET-s Filament | <15 μM | A residue | > mM | ~46 residues | No | Solid | Static | (Wasmer et al., 2009) |
| RIP1/RIP3 Filament | < 5 μM | A residue | > mM | ~22 residues | No | Solid | Static | (Li et al., 2012a) | |
| DD Signalosomes | ASC PYD Filament | < 2 μM | Each of the Six Interfaces | > mM | 6 interfaces | No | Solid | Static | (Lu et al., 2014b) |
| Head-to-Tail Signalosomes | Dishevelled | ~5-20 μM | One of the two interfaces | ~5-20 μM | 2 interfaces | Yes | Solid | ~Seconds | (Schwarz-Romond et al., 2007; Schwarz-Romond et al., 2005) |
| Multivalent Complex | (SH3)5-(PRM)5 | ~μM | SH3-PRM | 0.35 mM | 5 motifs | Yes | Liquid | ~Seconds | (Li et al., 2012b) |
| Nucleoporin | Nup100 | 0.2 μM | Short motif | 7.5 μM | ~43 motifs | Yes | Hydroge l | ~Minutes | (Schmidt and Gorlich, 2015) |
| RNP Granule | Fus | < 10 μM | Short motif | > mM | 24 motifs | Yes | Liquid | < Seconds | (Burke et al., 2015) |
Figure 1. Examples of higher-order assemblies with indicated modes of interactions, roles of IDRs and dynamic properties.
(A) The yeast prion Het-s assembles into a triangular β-solenoid amyloid fibril by folding-upon-binding via its C-terminal IDR.
(B) The inflammasome adaptor ASC assembles into a filament via the cooperative interactions of its N-terminal PYD. The C-terminal CARDs situate outside the filament to induce downstream caspase-1 recruitment.
(C) The p62 autophagy scaffolding protein assembles head-to-tail via its N-terminal PB1 domain to form the main interactions in the filament. The LC3 interacting region and the UBA domain flank outside to mediate their respective interactions.
(D) The multivalent protein containing four SH3 domains interacts with another multivalent proteins containing four PRMs to induce phase separation.
(E) The RNP granule protein Ddx4 containing multiple short linear motifs undergoes phase separation via weak motif-mediated interactions.
Higher-order structures appear to be especially prevalent in signal transduction, which often contain interactions that are mediated by folded domains. In innate immunity, helical polymerization of death fold domains (DD), which also include the Pyrin domain (PYD) and the caspase recruitment domain (CARD) (Yin et al., 2015), represents a critical signaling mechanism in the Toll-like receptor (TLR) (Lin et al., 2010), inflammasome (Cai et al., 2014; Lu et al., 2014b), and RIG-I pathways (Hou et al., 2011; Wu et al., 2014). These domains share a common six-helix bundle structure, and cooperative interactions among six weak adjacent surfaces lead to stable homo- or hetero-oligomeric assemblies that can be as static as amyloids (Hou et al., 2011; Lu et al., 2014b; Wu et al., 2014) (Table 1, Figure 1B). Owing to their stability, DD fold-mediated signalosomes have been shown to mimic prions in conformational conversion, aggregation, and intercellular propagation (Baroja-Mazo et al., 2014; Cai et al., 2014; Franklin et al., 2014; Hou et al., 2011). Multiple similar domains are often present in the component proteins of these signaling complexes, which are linked by IDRs. These concurrent oligomerization interactions are thought to further compact the helical filaments into punctate structures in cells that are visible under light microscopy (Hornung et al., 2009; Wu et al., 2010).
One of the earliest identified modes of higher-order assembly was head-to-tail polymerization by Phox and Bem1 (PB1), dishevelled (Dvl), homologous (DIX) and sterile alpha motif (SAM) domains, which are involved in many signaling processes, including the Wnt signaling pathway, autophosphorylation of ephrin receptor tyrosine kinases, and induction of autophagy. While the PB1 and DIX domains share a related ubiquitin-like fold (Bienz, 2014), SAM domains possess a fold with largely α-helices (Thanos et al., 1999). PB1, DIX and SAM domains can establish competing homo- and hetero-oligomeric interactions between the acidic residues of one domain and a basic charge cluster on the opposite surface of another domain (Ciuffa et al., 2015; Lamark et al., 2003; Wilson et al., 2003). The interaction dissociation constants (KD) between these domains range from low nM to high μM or mM, resulting in a variety of assemblies, from more ordered stable aggregates to highly dynamic structures with rapid association and dissociation rates (Bienz, 2014; Isono et al., 2013; Schwarz-Romond et al., 2005) (Table 1, Figure 1C). The transient clusters of signal transducers provide a temporary high local concentration for low-affinity effectors and enable a rapid response to activating signals. These domains may further organize the head-to-tail polymerization into a flexible filamentous scaffold, as in the case of the selective autophagy protein p62 (Ciuffa et al., 2015), which exposes the IDR-linked C-terminal protein interaction domains for downstream recruitment (Figure 1C).
Multivalent crosslinking by arrays of domains and their interacting motifs interspersed by IDRs may mediate loosely packed higher-order signalosomes, in which the less cooperative and low-affinity contacts result in a lower barrier for dissociation, and thus more reversible assemblies (Table 1, Figure 1D). In the T-cell receptor (TCR) activation system, engagement of presented antigens initiates a kinase cascade that results in multiple tyrosine phosphorylations Multiplexed crosslinking between SH2 domains and phosphotyrosine (p-Tyr) motifs, and between SH3 domains and proline rich motifs (PRM) is important for assembling and sustaining TCR microclusters in the immunological synapse (Balagopalan et al., 2015; Coussens et al., 2013). In TLR-induced NF-κB activation, the ubiquitin ligase TRAF6 appears to crosslink into higher-order structures through alternating dimerization and trimerization of two interaction domains (Yin et al., 2009). The dependence on the valency of the interacting species for crosslinked higher-order structures was demonstrated for an in vitro signaling system, in which an engineered protein containing multiple flexibly linked SH3 domains formed dynamic liquid droplets when mixed with another engineered protein containing multiple PRMs (Li et al., 2012b) (Table 1, Figure 1D).
The more recently discovered higher-order structures of ribonucleoprotein (RNP) granules and nuclear pore complexes, as phase-separated liquid droplets or hydrogels, are formed from multivalent interactions among motifs in extended IDRs and exhibit highly dynamic properties (Brangwynne et al., 2009; Frey et al., 2006; Hyman et al., 2014) (Table 1). Micron-sized RNP granules in cells are the site of many RNA related processes, including nucleoli for ribosome biogenesis, Cajal bodies for spliceosome assembly, nuage granules in drosophila germlines for selective silencing of genetic elements, processing bodies (P-bodies) for mRNA turnover, and stress granules store translationally arrested mRNAs (Anderson et al., 2015; Hyman et al., 2014; Jain and Parker, 2013; Mitchell and Parker, 2014; Nott et al., 2015). FG-repeat nucleoporins in nuclear pore complexes form a hydrogel that provides a selective permeability barrier for nuclear transport receptors. The IDRs of the component proteins are often LCDs that form higher-order structures using repetitive short motifs for protein-protein interactions, for example [F/Y]G repeats, RG repeats, NQ-rich prion-like sequences, and charge blocks (Ader et al., 2010; Kato et al., 2012; Malinovska et al., 2013; Sun et al., 2011) (Figure 1E). The low-affinity, often transiently interacting elements and the plasticity of the IDRs facilitate a vast number of combinations and rapid exchange of contacts, resulting in a low barrier of dissociation and, thus highly reversible dynamic assembly.
The nature of the interacting elements in granules and nuclear pore complexes is not well defined. Long-range electrostatic interactions among patterned oppositely charged blocks may facilitate initiation of phase-separation (Elbaum-Garfinkle et al., 2015; Lin et al., 2015; Nott et al., 2015). Short-range cation-π, π-π, hydrogen bonding with aromatic rings, dipole-dipole, and hydrophobic interactions may mediate the transient weak contacts among the sequence motifs in LCDs. Among these interactions, cation-π is the strongest, with an upper limit for free energy of about −3.6 kcal/mol, as desolvation penalties of aromatic rings are rather small (Gallivan and Dougherty, 2000). These interactions appear to play an important role in phase separation of the primary nuage granule component Ddx4 (Nott et al., 2015), the P granule protein LAF-1 (Elbaum-Garfinkle et al., 2015), and FUS, whose LCD is required for formation of stress granules and cytoplasmic inclusions in some forms of amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD) (Burke et al., 2015; Sun et al., 2011). The repetitive nature of LCD motifs appears to be crucial, as the combination of multiple polymorphic interactions is required for phase separation (Brangwynne et al., 2015; Burke et al., 2015; Chen et al., 2015; Kato et al., 2012; Lin et al., 2015; Nott et al., 2015).
Fuzzy Structures in Higher-Order Assemblies
A common feature of higher-order assemblies is the involvement of IDRs, either as direct interaction elements, or as largely unstructured linkers and tails in the bound state. Hence, all higher-order assemblies, even the most static ones, exhibit structural multiplicity, polymorphism, and/or dynamic disorder, and can be considered as fuzzy structures. Importantly, conformational diversity of any kind, either static or dynamic, has an impact on the regulated formation and function of higher-order assemblies. In the case of static polymorphism, alternative conformations of the same interacting elements are stabilized within the assembly. In the case of dynamic disorder, the IDRs retain conformational freedom within the assembly and will be referred to as fuzzy regions or dynamic IDRs, to distinguish them from IDRs that fold upon binding. These dynamic IDRs within higher-order structures may link separate binding modules to increase their local concentration, exert transient interactions to influence adjacent binding elements, facilitate allostery, or promote intramolecular autoinhibition (Fuxreiter, 2012; Sharma et al., 2015) via well-characterized mechanisms (see examples in FuzDB, http://protdyn-database.org).
In amyloid formation, a plethora of experimental evidence demonstrates an intrinsic structural polymorphism, which can lead to distinct fibril morphologies, conformational states, and inheritable prion phenotypes (Krishnan and Lindquist, 2005; Tanaka et al., 2004). Different amyloid strains formed under different conditions can exhibit alternative contacts within the same sequence (Sawaya et al., 2007). Yeast prion Sup35 strains show variations in core lengths, intermolecular contacts, and side chain chemical environments or orientations (Frederick et al., 2014; Krishnan and Lindquist, 2005; Reymer et al., 2014). Additionally, the sequence of the Sup35 LCD can be randomized without blocking amyloid formation (Ross et al., 2005).
Albeit surprising, dynamic disorder is also a common feature of many amyloids. Although the aggregates have a well defined, tightly packed core region, a considerable portion of the IDR often stays disordered and flanks the core filament as a solvent-exposed, protease sensitive protein segment (Krishnan and Lindquist, 2005). These fuzzy regions may exert autoinhibition of aggregation before amyloid formation, and/or play active roles in the aggregation mechanism (Tompa, 2009). Dynamic IDRs may also link the amyloid core and a functional domain. In HET-s, the N-terminal HeLo domain purportedly decorates the central amyloid (Greenwald et al., 2010) (Figure 1A). Furthermore, post-translational modifications (PTMs) also regulate amyloid formation via targeting fuzzy regions. In the mammalian functional amyloid necrosome, hyperphosphorylation of the IDRs in RIP1 and RIP3 kinases appears to enhance amyloid formation, as only the hyperphosphorylated forms exist in the insoluble amyloid fraction (Li et al., 2012a). Phosphorylation may create charge repulsion in the IDR to expose the RIP homotypic interaction (RHIM) motifs and initiate assembly.
Polymorphism is also characteristic to many signaling complexes that involve multiple combinations of interacting domains. The AIM2 inflammasome, which recruits the adaptor ASC and the protease caspase-1, possesses at least three such domains with distinct oligomerization mechanisms: the AIM2 HIN domain oligomerizes by wrapping around dsDNA (Jin et al., 2012; Lu et al., 2015), the oligomerized AIM2 PYDs nucleate ASC PYD filaments (Cai et al., 2014; Lu et al., 2014a; Lu et al., 2015), and the clustered ASC CARDs nucleate caspase-1 filaments (Lu et al., 2014a) (Figure 2A). In AIM2 a ~50 residue IDR between the PYD and HIN domains enables combination of the clustered PYDs into variable short helical oligomers. Furthermore, the long linker IDR promotes an intramolecular PYD/HIN interaction to inhibit PYD oligomerization. This autoinhibition can be released by the interaction of the HIN domain with dsDNA to allow PYD oligomerization and signal transduction (Jin et al., 2012; Jin et al., 2013). For full-length ASC, the extended, dynamic linker positions the CARD outside the PYD filament core (Lu et al., 2014b) (Figure 1B) and transmits an allosteric signal between the two domains to align the CARD for oligomerization.
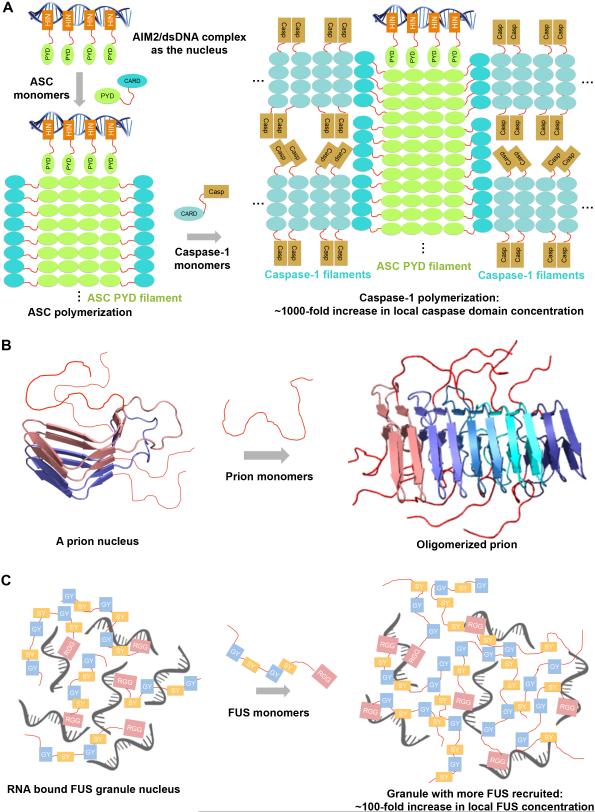
Figure 2. Proposed nucleated polymerization mechanisms in several higher-order systems, with dramatically enhanced local concentrations.
(A) Biochemical and structural studies suggest the involvement of two nucleated polymerization steps in the assembly of the AIM2 inflammasome involving the PYDs and the CARDs respectively. To reduce clutter, ASC CARDs are only shown for the ASC subunits at the periphery of the ASC PYD filament, and Caspase-1 Casp domains are only shown for subunits at the periphery of the Caspase-1 filament.
(B) A prion oligomer acts as the nucleus to induce the polymerization of the prion monomers.
(C) Proposed nucleated polymerization mechanism of a FUS granule in which RNA-bound FUS acts as the nucleus to induce the phase separation of more FUS molecules.
In a similar fashion, the helical filament of the autophagy scaffolding protein p62 also exposes a ~200 residue-long IDR linker to cause the ubiquitin binding UBA domain and the LC3 binding LIR motifs to become fully solvent-accessible (Figure 1C). Structural disorder in the linker is consistent with the observed heterogeneity and flexibility of the polymer (Ciuffa et al., 2015). The dynamic disorder of the linker facilitates binding to the autophagosomes via multiple LC3 interactions and regulates the helical assembly via mediating transient contacts between the UBA domain and the filament.
Intrinsic heterogeneity is a major bottleneck in elucidating the underlying structural features of liquid droplets and hydrogels. Fuzzy structures of liquid granules (Figure 1E) have been recently demonstrated for FUS at high resolution by NMR spectroscopy (Burke et al., 2015). No evidence for static structures on the 10 second or slower timescale was observed (Burke et al., 2015). Phase-separated FUS LCD exhibits only minor chemical shift changes relative to the solution state, which correspond to transient, varying intermolecular contacts distributed all along the chain (Burke et al., 2015). These data may further reflect the similarity of transient intramolecular interactions in the monomeric state to those intermolecular contacts sampled in the phase-separated state. Data from fluorescence recovery after photobleaching (FRAP) are consistent with the presence of dynamic IDRs in RNP granules (Nott et al., 2015; Patel et al., 2015). These fuzzy regions, together with transient non-specific contacts, facilitate shuttling proteins in and out of the assembly and enable dynamic reorganization upon changes in protein and/or nucleic acid content and concentration.
A similar dynamic reorganization is essential for nucleoporins to selectively allow entry of nuclear transport receptors (NTRs) (Ader et al., 2010; Labokha et al., 2013; Schmidt and Gorlich, 2015). Hydrogels, formed by Phe and glycine-rich nucleoporins (FG-Nups), also exploit multiple low-affinity binding motifs that rapidly vary upon interaction with the NTRs (Milles et al., 2015). The individual FG-repeats bind with low affinity and must act in concert for efficient binding. NMR, single-molecule fluorescence, and molecular simulations demonstrated that FG-Nups maintain an unexpectedly high plasticity when bound to NTRs (Milles et al., 2015) allowing for ultrafast association and dissociation rates in agreement with the measured kinetic parameters for nuclear transport (Hough et al., 2015).
Common Themes in Higher-Order Structures: Nucleated Polymerization, Regulation by PTMs, and Local Concentration Increases
One fundamental assembly mechanism of higher-order structures is nucleated polymerization. Conversion from the soluble state to amyloids can be achieved by infection with an aggregated form (Prusiner, 1998), the conformational property of which could be propagated, resulting in strains with distinct phenotypes (Frederick et al., 2014) (Figure 2B). For signalosomes, nucleated polymerization of a downstream protein by a substoichiometric amount of an upstream seed is a mode of recruitment, signal propagation and signal amplification, which leads to threshold responses (Cai et al., 2014; Lu et al., 2014b; Qiao et al., 2013; Schwarz-Romond et al., 2007; Wu, 2013) (Figure 2A). Seeds may induce polymerization via increasing the local concentration of the interacting domains or motifs, relieving autoinhibition, or exerting allosteric effects (Ciuffa et al., 2015; Jin et al., 2013; Lu et al., 2015).
A number of interacting nucleic acids and proteins have been shown to induce phase separation of LCDs in RNP granules (Figure 2C). Phase transition of low complexity IDRs to liquid droplets can be nucleated by interaction partners, such as RNA (Lin et al., 2015; Schwartz et al., 2013; Zhang et al., 2015), poly(ADP-ribose) (Altmeyer et al., 2015), or C-terminal domain of RNA Polymerase II (Kwon et al., 2013). Multivalent binding may crosslink the proteins to form granule seeds, which then grow through recruitment of more monomeric proteins into the granules. In contrast to amyloids and prions, no distinct conformations are propagated in liquid droplets, owing to the dynamic exchange of interactions. Decreasing the dynamics by aging, mutation, or other factors can lead to segregation of structural properties in the assembly (Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015).
In higher-order structures, PTMs represent a powerful mechanism for modulating affinity of the binding elements and dynamics of the intervening IDRs to regulate assembly and disassembly. The Toll-like receptor helical complex dissociates upon kinase activation and phosphorylation (Li et al., 2002), while the RIP1/RIP3 necrosome complex is strengthened by phosphorylation (Li et al., 2012a). In head-to-tail polymerization, phosphorylation of the acidic motifs strengthens electrostatic interactions and facilitates assembly (Wilson et al., 2003), while phosphorylation of positive surfaces blocks polymerization (Matsumoto et al., 2011). Phosphorylation on Tyr increased the avidity of SH2-pTyr interactions in both an in vitro actin polymerization system and in TCR signaling, resulting in enhancement of their respective activities (Balagopalan et al., 2015; Li et al., 2012b). For the Ddx4 granule, modification by protein arginine methyltransferases (PRMT1) weakens electrostatic and cation-π interactions to destabilize the droplets in vivo (Nott et al., 2015). By contrast, PRMT1 depletion decreases the tendency of FUS to associate with cytosolic stress granules (Tradewell et al., 2012). Phosphorylation contributes to dissolution of stress granules when the kinase DYRK3 phosphorylates RNA-binding proteins to release mTORC1 for signaling (Wippich et al., 2013).
Formation of higher-order structures invariably results in a dramatic increase in local concentrations of the component proteins, as well as their linked domains. In the caspase-1 filament, the local concentration of the caspase domain is calculated to be ~3.1 mM (Figure 2A), which is ~1,000-fold higher than the μM cellular concentrations of many signaling proteins. Similar increases in local concentration were reported for Dvl and Axin puncta (Fiedler et al., 2011; Schwarz-Romond et al., 2005), the head-to-tail polymerization of which is required for sequestration of Axin in the β-catenin destruction complex, leading to β-catenin accumulation and transcriptional signaling (Schwarz-Romond et al., 2007). Head-to-tail p62 oligomers formed by PB1 domain interactions increase the local concentration of the C-terminal UBA domains and LIR motifs for recruitment of polyubiquitinated proteins to the LC3-bound membrane to initiate autophagic degradation (Ciuffa et al., 2015). An even more dramatic concentration increase is generated in ordered arrays of the linked functional domains in functional amyloids, which can also provide optimal orientation for their substrates, thereby improving catalytic efficiency (Fowler et al., 2006). Increased local concentration can promote allosteric activation of enzymes, including caspases and kinases, e.g. in caspase-1 activation by inflammasomes, and kinase activation by the TLR pathway, the necrosome, the SAM domain polymer of ephrin receptors, and the TCR signaling complex (Balagopalan et al., 2015; Ferrao et al., 2014; Li et al., 2012a; Lu et al., 2014b; Thanos et al., 1999).
In the crosslinked phase-separated liquid droplets, approximately 100-fold concentration increase of component proteins and their domains, relative to the bulk concentration, has been reported (Li et al., 2012b) (Figure 2C). For RNP granules, selective recruitment of partner proteins per se, due to the high local concentration of granule proteins, may directly modify signaling pathways, metabolic machinery, and stress response programs, with inclusion or exclusion of certain molecules, which can have profound effects on cancer cell viability (Anderson et al., 2015). In a specific case, phase separated LC domains of FUS, EWS, and TAF15 recruit RNA polymerase II via avidity for weak hydrophobic motifs, and when translocated onto a variety of different DNA-binding domains can promote transcriptional activation (Kwon et al., 2013). Taken together, increases in local concentration provide an efficient means to decrease signaling noise, as well as a plethora of mechanisms to promote different functions.
Affinity, Valency and IDR in Determining Dynamics and Material States of Higher-Order Assemblies
We derive the molecular determinants of material states and dynamics of higher-order assemblies using polymer physics approaches, which have been previously applied to chemical polymers. In a microscopic view, a cross-linked system can be described as a system-spanning network, where crosslinks between the monomers are organized into large, networked clusters. In a macroscopic formulation, phase separation is driven by the more favorable association of molecules interacting with themselves than with the solvent. Reversible networks tend to phase separate (Semenov and Rubinstein, 1998), which is promoted by the exchange of intramolecular interactions for intermolecular interactions. This is consistent with the Flory theory (Flory, 1941) and explains the existence of both critical concentrations and surface tension in the liquid droplets (Semenov and Rubinstein, 1998). Within this framework, we propose that the dynamics of higher-order assemblies and their tendency to phase-separate can be deconvoluted to three factors (Table 1): i) the affinity of the interacting elements, which can be motifs or domain interfaces, ii) the frequency and multivalency of the interacting elements, and iii) the dynamics of the connecting regions. In this context, the dynamic nature of phase-separated higher-order structures, such as those mediated by LCDs or multivalent SH3-PRM interactions, originates from the weak affinity between the interacting motifs, their high valency, and the rapid conformational exchange facilitated by the intervening IDRs (Figure 1D-E, Table 1).
A quantitative evaluation of affinity is available for multivalent SH3-PRM systems, in which the individual interactions have KD in the high μM to mM range (Banjade et al., 2015; Li et al., 2012b). From the estimated interaction free energy of −3.6 kcal/mol (Gallivan and Dougherty, 2000), a cation-π interaction in LCDs is equivalent to a weak KD of ~2 mM (Table 1). This supports the requirement of low affinities for liquid-liquid phase separations. The interaction kinetics may be inferred from the relationship between the thermodynamic dissociation constant KD, and the association (ka) and dissociation rates (kd), KD = kd/ka. Assuming a diffusion controlled ka in RNP granules of ~103 M-1s-1, estimated from a ka of ~106 M-1s-1 in water (Cunningham and Wells, 1993), and adjusting for the ~103-fold increased viscosity in the droplets (Brangwynne et al., 2009; Hyman et al., 2014), the kd for a transient interaction of KD = 2 mM is ~2 s−1. While the calculation is extremely approximated, it predicts a dynamic time scale of seconds, which is on the same order of magnitude for FRAP data on in vitro reconstituted and cellular RNP granules (Burke et al., 2015; Lin et al., 2015; Nott et al., 2015; Patel et al., 2015) (Table 1).
The second property required for phase separation is the interaction multivalency, which represents a high degree of redundancy in the possible contacts. As deduced for the engineered SH3-PRM systems, the number of differently crosslinked variants determines the number of microstates, and thus the configurational entropy (Li et al., 2012b). Upon interaction of m number of each type of molecules with n valencies, the number of possible microstates is (n*m)!, a factorial of the product, given that all valencies are saturated. Conformationally heterogeneous IDRs enable the relatively independent binding of n valencies in m interacting molecules to form a vast number of combinatorial interaction patterns that approaches the theoretical limit of possible microstates. Based on the linear dependency between valency and configurational entropy gain (Li et al., 2012b), crosslinking the highly frequent, repetitive linear motifs in LCDs (Elbaum-Garfinkle et al., 2015; Lin et al., 2015; Nott et al., 2015) provides a significant contribution to affinity.
The third property, which impacts phase separation, is the dynamics of the IDRs that connect or flank interacting elements. IDRs regulate assembly via entropic effects; they increase the number of microstates by enabling many different cross-linked geometries, as described above, and decrease the entropic penalty of binding by preserving their dynamic state. These fuzzy regions can also promote the assembly via direct, transient interactions, which may enhance binding affinities by 5-20 fold, as estimated from peptide-protein interactions (Selenko et al., 2003; Yu et al., 1994). In addition, flanking or distant IDRs may contain additional interacting motifs; the linker between the first and second SH3 domains in Nck weakly interacts with the second SH3 domain of another Nck molecule to induce self-association, which promotes higher-order structure formation in the multivalent Nck-N-WASP complex (Banjade et al., 2015).
Static higher-order assemblies, such as amyloids and DD signalosomes, lack dynamic IDRs between their interaction elements (Table 1). While each residue in an amyloid only provides a weak-affinity interaction, their high frequency and proximity results in cooperation with other proximal residues in the β-sheet structure to generate a tightly bound state (Figure 1A). Albeit different amyloid states co-exist owing to interaction polymorphism, they do not exhibit a rapid exchange. A similar cooperative interaction can be observed between six interdependent interfaces upon helical polymerization of PYD and CARD domains (Figure 1B). Simultaneous binding of six interacting elements with low individual affinities results in a helical filament with limited dissociation that can occur only at the ends of the filament. In contrast, divalency, as opposed to multivalency, in head-to-tail polymerization results in more flexible higher-order structures (Figure 1C), where filaments can fragment depending on the single surface dissociation rate.
Not all dynamic higher-order structures undergo phase separation. Albeit head-to-tail polymers can be very dynamic, depending on the affinity between the interaction elements, their lower valency and more constrained intervening IDRs limit conformation exchange and phase separation. Increased valency in TCR signalosomes and nucleopore complexes facilitates a multitude of cross-linking arrangements and phase transition. They however possess slower IDR dynamics as compared to RNP granules, which can generate more stable higher-order assemblies, such as hydrogels. Taken together, the interplay of affinity, valency, and IDR dynamics generates fuzzy higher-order structures that populate a structural and dynamic continuum.
Conversion Between the Different Material States and Relevance to Disease Conditions
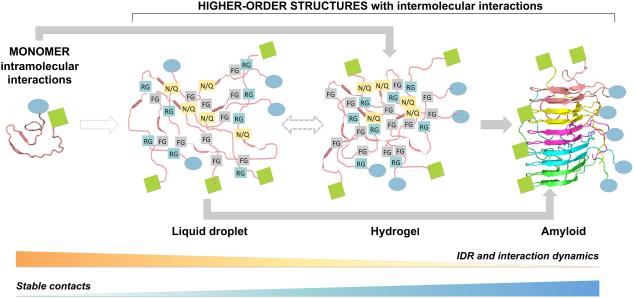
One intriguing observation related to LCD-induced formation of higher-order structures is that the same protein is capable of forming different states under different conditions, ranging from liquid droplets to hydrogels and fibrous aggregates (Elbaum-Garfinkle et al., 2015; Kato et al., 2012; Lin et al., 2015; Nott et al., 2015). Liquid droplets formed by RNP granule component proteins are prone to conversion into the more stable, solid-like fibrous states under pathological conditions (Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015) or simply as a function of time, a phenomenon known as maturation or aging (Lin et al., 2015; Patel et al., 2015). Similarly, polyQ peptides can exhibit two distinct transitions upon an increase in concentration: first forming liquid-like assemblies and then insoluble amyloid fibrils (Crick et al., 2013). These data suggest that alternative, dynamic interactions driven by short, weak motifs in droplets and hydrogels may be intrinsically metastable until fully transformed to the stable amyloid state (Ader et al., 2010; Halfmann et al., 2012). Therefore, on a dynamic continuum, an LCD may proceed from a solution state to liquid droplets, or to hydrogels, and to fibrous aggregates (Figure 3).
Figure 3. A schematic diagram on the conversion between material states.
LCD and embedded linear motifs are shown in pink and two hypothetical folded domains are represented as blue oval and green square respectively.
One outstanding question is whether these different states share any common molecular basis. To shed some light on this question, we compared the conversion between different material states with protein crystallization, in which it is empirically known that liquid-liquid phase separation often precedes the appearance of crystals and its presence bodes well for a successful outcome. Previous numerical simulations of crystal nucleation with short-range attractive interactions explain the phenomenon as a metastable condensed liquid that enormously enhances crystal nucleation by lowering the free energy barrier (ten Wolde and Frenkel, 1997). Hence, it was concluded that the first step towards the critical nucleus is the formation of a liquid-like droplet. Since proteins that can form liquid droplets have a higher tendency to become crystals, we argue that the intrinsic short-range forces in liquid droplets may resemble those in the final stable states of the crystals.
Analogously, extensive evidence at various levels of detail suggests the presence of interaction similarities with regard to the interaction type and residues involved among the different material states of LCDs. First, probing residue solvent exposure using N-acetylimidazole (NAI) chemical footprinting showed comparable patterns of protection for recombinant hnRNPA2 in reconstituted hydrogels and liquid droplets (Xiang et al., 2015). Second, the frequently similar amino acid compositions of amyloids and granules support the similarity of interactions in dynamic liquid droplets and stable amyloids (Malinovska et al., 2013). Hydrogels may already contain dynamic amyloid-like fibers (Kato et al., 2012) or involve amyloid-like interactions (Ader et al., 2010). Third and most intriguingly, NAI protection patterns of monomeric hnRNPA2 are similar to liquid droplet and hydrogel states for key residues that are involved in formation of these assemblies (Xiang et al., 2015). One explanation is that the same key residues in a LCD form transient intramolecular interactions in the absence of oligomerization, which is consistent with the minor NMR chemical shift changes observed between the phase-separated and the solution states of FUS LCD (Burke et al., 2015).
However, the levels of NAI labeling differs significantly among the different states of hnRNPA2, with denatured state > monomeric form > liquid droplet > hydrogel (Xiang et al., 2015). Therefore, labeling pattern similarity cannot be used to deduce dynamics; instead, the level of labeling may correlate with how frequently an interaction exists and protects the protein from labeling in the different material states. Owing to the high redundancy of the contacts in droplets and hydrogels, with regard to not only their number, but also their type (e.g. Tyr can be involved in both cation-π and π-π interactions), different conditions may induce alternative crosslinking arrangements. A main distinction of the fibrillar state is a reduced number of crosslinking microstates with slowed IDR dynamics. To initiate fibrillization, the entropy loss has to reach a certain threshold for commitment to unidirectional propagation. Thus, even if amyloid-like interactions are sampled in the granule, as long as they remain dynamic the conversion to solid aggregates may yet occur.
It is conceivable that the fibrillar state involves a larger number of interactions, comprised of not only motif residues common in the more dynamic assemblies, but also new unique interactions. A detailed study of a hnRNPA1 mutant (D262V) associated with inherited forms of ALS and FTD supports this hypothesis (Molliex et al., 2015). It was shown that the D to V mutation did not significantly impact the forces that drive phase separation. However, the mutant hnRNPA1 produced amyloid fibrils within minutes of droplet formation, suggesting that the mutation significantly potentiated conversion to the amyloid state. This observation may be consistent with the important role of hydrophobic packing in amyloid formation, in comparison with phase separation. No fibrils were formed if the protein was maintained in the soluble state, suggesting that phase separation is required for fibril formation. Seeding by the aggregation-prone disease mutant also destabilized the liquid state of the wild-type protein and triggered fibrillization, resulting in mixed fibrils. Likewise, the patient-derived G156E and R244C FUS mutants formed similar dynamic assemblies as wild-type protein, but had a higher tendency to form fibrils in aging experiments (Patel et al., 2015).
The static and dynamic properties associated with different material states could be directly related to the biological functions of these assemblies. For example, functional amyloids and innate immune signalosomes act as scaffolds to either activate or inactivate the proteins in the complex. For prions, these structures are stable during cell division and act as nuclei to template the conformational transition of newly synthesized monomeric forms. In these cases, stability may be beneficial. In contrast, signaling events occurring on short timescales require higher order assemblies that can rapidly form and dissociate (Bienz, 2014). For RNP granules, it is likely that the observed dynamic properties are important for rapid exchange of recruited proteins in response to stress (Wippich et al., 2013).
Concluding Remarks
In some ways analogous to social constructions like towns and villages, higher-order structures are emerging as the architecture of cellular organelles and signaling complexes that respond to the environment with regulated amplitude, time scale, and spatial distribution. While the different higher-order assemblies that exist on a structural and dynamic continuum offer varying properties to suit their respective functions, their cooperative formation and dramatic increase in local concentration all facilitate protein recruitment and enzyme activation. Redundant motifs and dynamic IDRs in higher-order structures often lead to polymorphic or fuzzy structures, as exemplified in the prion strains of amyloids, heterogeneous crosslinking of DD fold filaments, and dynamic multivalent granules. Remarkably, the more dynamic higher-order structures are often metastable, and can be converted to insoluble fibrillar aggregates as a function of time and environmental conditions, or under pathological circumstances, in which the IDR dynamics decrease with commitment to particular conformations.
The formation of higher-order structures raises many questions, including questions regarding the spatial regulation of their assembly and the molecular mechanism of their disassembly. While it is known that cells have evolved specific recruitment strategies to ensure the induction of certain innate immune pathways from different cellular locations (Kagan et al., 2014), the mechanisms of spatial control remain largely elusive. Once assembled, how do higher-order structures become down-regulated to terminate specific responses? For dynamic signalosomes and granules, it appears that intrinsic mechanisms involving PTMs play an important role in triggering disassembly. For removal of the more static assemblies, such as amyloids and crosslinked DD fold structures, protein disaggregation and degradation machineries involving chaperones, proteasomes, and selective autophagy may be necessary.
Structurally, the existence of alternative contacts in higher-order assemblies, either as transient, weak interactions or as stable, strong interactions, causes polymorphism and heterogeneity, which in turn pose challenges to our methodologies. Traditional structural techniques, especially those that can achieve residue level resolution, such as X-ray crystallography and electron microscopy, generally require structural homogeneity. Divide-and-conquer approaches may generate structural data on the isolated parts suitable for high-resolution structural pursuits. However, characterization of the complete fuzzy ensembles requires more holistic approaches, such as NMR in combination with other spectroscopic methods, as well as computational modeling, to illuminate the exciting, yet still rather unexplored, landscape of higher-order assemblies from different mechanistic angles.
Acknowledgment
We apologize for incomplete citations due to space limitations, and acknowledge support by the National Institutes of Health (to H.W.) and by the Momentum Program of the Hungarian Academy of Sciences and the Hungarian Science Fund (to M.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ader C, Frey S, Maas W, Schmidt HB, Gorlich D, Baldus M. Amyloid-like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci U S A. 2010;107:6281–6285. doi: 10.1073/pnas.0910163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nature communications. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L, Kortum RL, Coussens NP, Barr VA, Samelson LE. The Linker for Activation of T Cells (LAT) Signaling Hub: From Signaling Complexes to Microclusters. J Biol Chem. 2015;290:26422–26429. doi: 10.1074/jbc.R115.665869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, Rosen MK. Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc Natl Acad Sci U S A. 2015;112:E6426–6435. doi: 10.1073/pnas.1508778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Mazo A, Martin-Sanchez F, Gomez AI, Martinez CM, Amores-Iniesta J, Compan V, Barbera-Cremades M, Yague J, Ruiz-Ortiz E, Anton J, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- Bienz M. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci. 2014;39:487–495. doi: 10.1016/j.tibs.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nature Physics. 2015;11:899–904. [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Song J, Chan HS. Theoretical perspectives on nonnative interactions and intrinsic disorder in protein folding and binding. Curr Opin Struct Biol. 2015;30:32–42. doi: 10.1016/j.sbi.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJ, Johansen T, Sachse C. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell reports. 2015;11:748–758. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- Coussens NP, Hayashi R, Brown PH, Balagopalan L, Balbo A, Akpan I, Houtman JC, Barr VA, Schuck P, Appella E, et al. Multipoint binding of the SLP-76 SH2 domain to ADAP is critical for oligomerization of SLP-76 signaling complexes in stimulated T cells. Mol Cell Biol. 2013;33:4140–4151. doi: 10.1128/MCB.00410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2013;110:20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA. Comparison of a structural and a functional epitope. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Zhou H, Shan Y, Liu Q, Li Q, Shaw DE, Li X, Wu H. IRAK4 Dimerization and trans-Autophosphorylation Are Induced by Myddosome Assembly. Mol Cell. 2014;55:891–903. doi: 10.1016/j.molcel.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Mendoza-Topaz C, Rutherford TJ, Mieszczanek J, Bienz M. Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down-regulating beta-catenin. Proc Natl Acad Sci U S A. 2011;108:1937–1942. doi: 10.1073/pnas.1017063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory PJ. Molecular Size Distribution in Three Dimensional Polymers I. Gelation. J Am Chem Soc. 1941;63:3083. [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, et al. The adaptor ASC has extracellular and 'prionoid' activities that propagate inflammation. Nat Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem Biol. 2014;21:295–305. doi: 10.1016/j.chembiol.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M. Fuzziness: linking regulation to protein dynamics. Mol Biosyst. 2012;8:168–177. doi: 10.1039/c1mb05234a. [DOI] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. A computational study of cation-p interaction vs salt bridges in aqueous media: implications for protein engineering. J Am Chem Soc. 2000;122:870–874. [Google Scholar]
- Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein ML, Ness F, Cescau S, Soragni A, Leitz D, et al. The mechanism of prion inhibition by HET-S. Mol Cell. 2010;38:889–899. doi: 10.1016/j.molcel.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Wright JR, Alberti S, Lindquist S, Rexach M. Prion formation by a yeast GLFG nucleoporin. Prion. 2012;6:391–399. doi: 10.4161/pri.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, Cowburn D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 2015;4 doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley MA, Golding GB. Simple sequences are rare in the Protein Data Bank. Proteins. 2002;48:134–140. doi: 10.1002/prot.10150. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J, Ishikura T, Toyoda T, Bernstein BE, Koseki H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Developmental cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Jain S, Parker R. The discovery and analysis of P Bodies. Adv Exp Med Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–13235. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Magupalli VG, Wu H. Supramolecular organizing centres: location-specific higher-order signalling complexes that control innate immunity. Nat Rev Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 Necrosome Forms a Functional Amyloid Signaling Complex Required for Programmed Necrosis. Cell. 2012a;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012b;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci U S A. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol. 2014a;426:1420–1427. doi: 10.1016/j.jmb.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014b;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Yang L, Yin Q, Ruan J, Yu X, Egelman E, Wu H. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discovery. 2015;1:15013. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim Biophys Acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, et al. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, Bracken C, Rosen M, Melnick A, Egelman EH, et al. Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymer A, Frederick KK, Rocha S, Beke-Somfai T, Kitts CC, Lindquist S, Norden B. Orientation of aromatic residues in amyloid cores: structural insights into prion fiber diversity. Proc Natl Acad Sci U S A. 2014;111:17158–17163. doi: 10.1073/pnas.1415663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife. 2015;4 doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14:484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T, Merrifield C, Nichols BJ, Bienz M. The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J Cell Sci. 2005;118:5269–5277. doi: 10.1242/jcs.02646. [DOI] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Krämer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- Semenov AN, Rubinstein M. Thermoreversible Gelation in Solutions of Associative Polymers. 1. Statics. Macromolecules. 1998;31:1373–1385. [Google Scholar]
- Sharma R, Raduly Z, Miskei M, Fuxreiter M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015;589:2533–2542. doi: 10.1016/j.febslet.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- ten Wolde PR, Frenkel D. Enhancement of protein crystal nucleation by critical density fluctuations. Science. 1997;277:1975–1978. doi: 10.1126/science.277.5334.1975. [DOI] [PubMed] [Google Scholar]
- Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. 1999;283:833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- Tompa P. Structural disorder in amyloid fibrils: its implication in dynamic interactions of proteins. The FEBS journal. 2009;276:5406–5415. doi: 10.1111/j.1742-4658.2009.07250.x. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Schutz A, Loquet A, Buhtz C, Greenwald J, Riek R, Bockmann A, Meier BH. The molecular organization of the fungal prion HET-s in its amyloid form. J Mol Biol. 2009;394:119–127. doi: 10.1016/j.jmb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J Clin Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Fu TM, Li J, Wu H. Structural Biology of Innate Immunity Ann Rev Immunology. 2015;33:393–416. doi: 10.1146/annurev-immunol-032414-112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich R, Campos AD, Myszka DG, et al. E2 interaction and dimerization in the crystal structure of TRAF6 Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]