The APOBEC Protein Family: United by Structure, Divergent in Function (original) (raw)
. Author manuscript; available in PMC: 2017 Jul 1.
Published in final edited form as: Trends Biochem Sci. 2016 Jun 6;41(7):578–594. doi: 10.1016/j.tibs.2016.05.001
Summary
The APOBEC (Apolipoprotein B mRNA Editing Catalytic Polypeptide-like) family of proteins has diverse and important functions in human health and disease. These proteins have an intrinsic ability to bind to both RNA and single-stranded (ss)DNA. Both function and tissue-specific expression varies widely for each APOBEC protein. We are beginning to understand that the activity of APOBEC proteins is regulated through genetic alterations, changes in their transcription and mRNA processing, and through their interactions with other macromolecules in the cell. Loss of cellular control of APOBEC activities leads to DNA hypermutation and promiscuous RNA editing associated with the development of cancer or viral drug resistance, underscoring the importance of understanding how APOBEC proteins are regulated.
Keywords: APOBEC, cytidine deaminase, disease mechanisms, DNA mutation, epigenetics, RNA editing
The diversity of the APOBEC family exceeded expectations
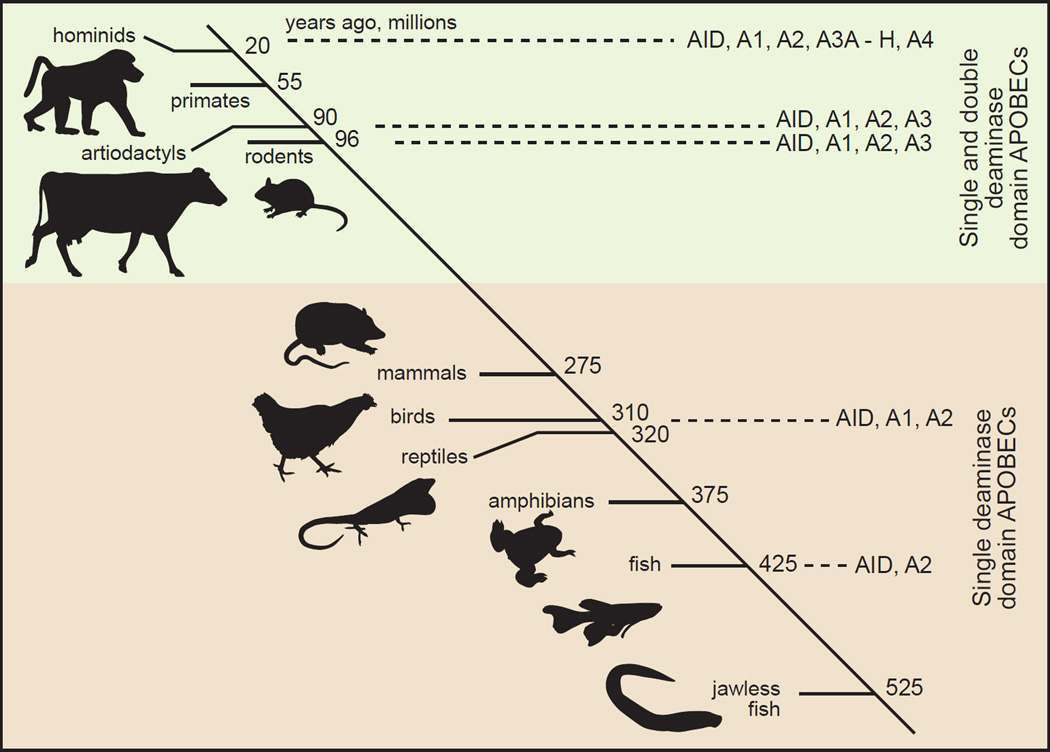
The mid 1980s to early 1990s was a dynamic period of realization during which RNA editing in chloroplasts, mitochondria and the cell nucleus were discovered [1]. Despite the multitude of epigenetic mechanisms becoming apparent, no one predicted the number of APOBEC enzymes that were soon to be discovered or the magnitude of their impact on biological systems. The APOBEC field began with the discovery that apolipoprotein B (apoB) mRNA contained a C to U base-modification that was not genomically encoded [2, 3] [Text Box 1] (Table 1). The APOBEC family in humans consists of 11 primary gene products and alternatively spliced variants that include A1, Activation Induced Deaminase (AID), APOBEC2 (A2), APOBEC3A-H (A3A-H) and APOBEC4 (A4) proteins (Figure 1A). The family was suggested to originate from ancestral AID genes (PmCDA1 & 2) expressed in the lymphocytes of jawless fish over 500 million years ago (Figure 1B). The ancestral AID genes were proposed to have induced up to 1014 somatic recombination events in antigen receptor genes used in adaptive immunity [4]. Gene duplication from AID and A2 and diversification of AID structure and function in bony fish were hypothesized as contributing to the evolutionary process leading to genes that encoded the APOBEC proteins found in amphibians, birds and tetrapods including primates [5, 6].
Text box 1. APOBEC1 function and regulation, the founder of the field.
Apolipoprotein B (ApoB) mRNA editing of cytidine nucleotides to uridines occurs at positions 6666 and 6802 and only in two tissues, the small intestine of all mammals and in the liver of some species (e.g. rodents, dogs and horses) [7, 8]. A long and a short protein variant of apoB carry lipid and cholesterol in circulation from the digestive tract to the rest of the tissues in the body. The short form of apoB is rapidly cleared from the blood by tissues whereas cholesterol associated with the long form of apoB has a longer blood half-life during which it can become oxidized to the 'bad' form of cholesterol which is associated with an elevated risk of atherosclerosis. Protein expressed from C6666-edited mRNA is 48% of full-length apoB protein because a CAA codon is edited to a UAA stop codon. The potential that apoB mRNA editing might mitigate a risk factor for atherogenic disease is why the single nucleotide polymorphisms in ApoB mRNA became of intense interest. The catalytic subunit responsible for deamination of C6666 was discovered in 1993 [9] and named as Apolipoprotein B Editing Catalytic Subunit 1 (APOBEC1 or A1) in 1995 [10] even though no other APOBEC paralogs were known at that time.
A1 alone had no activity on apoB mRNA. Critical to its discovery was the identification of a cis-acting 11 nucleotide sequence required for editing site recognition (the mooring sequence, Table 1) and an RNA binding protein cofactor (A1CF) that binds to A1 and the mooring sequence, thereby targeting A1 to the editing site (reviewed in [11]). A1 complementation can also be provided by RBM47 [12]. Regulation of APOBEC activity through interaction with other proteins or cofactors is an important theme in this field. Potentially complicating the characterization of APOBEC cofactors is the possibility that they may have functions beyond RNA editing. The finding that gene knockouts of A1CF and RBM47 disrupted mouse embryological development [12–14] whereas A1 gene knockouts were not lethal [14] support this possibility.
C to U editing of cytidines proximal to the mooring sequence has been an essential criterion used in the identification of editing targets and editing sites, such as the editing site in neurofibromin (NF1) mRNA [15] and the involvement of A1 in the multiple C to U transitions found in the 3' UTRs of mRNAs [16] (Table 1). The biological significance of 3' UTR editing of mRNA has not been determined but may be relevant to mRNA stability [16, 17]. Editing of NF1 mRNA has been proposed to lead to deregulation of Ras signaling in solid tumors of neurological origin [18, 19] due to the loss of the C-terminal Ras GTPase regulator domain in NF1 through the introduction of a premature stop codon in NF1 mRNA.
Table 1.
Genetic, Cellular and Functional Attributes of APOBEC Proteins
| Name | Genomic Info | Well-DefinedTissue Expression | ActiveDeaminaseDomains | §CellularLocalization | Cofactors/Interactions | Targets | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Location | Exons | BindingActivity | DeaminaseSubstrate | Sequence(C = edit site) | GeneticElements | |||||
| APOBEC1 | 12p13.1 | 5 | GI tract | 1 | n/C | A1CF,RBM47,CRM1 | ssDNA,RNA | ssDNA,RNA | 5’-A C(n4-6)UGAUnnGnnnn-3’(for n, A and U preferred) | apoB & NF1mRNA;3’ UTRs |
| AID | 12p13 | 5 | activated B cells | 1 | n/C | eEF1A,HSP90, CRM1 | ssDNA,RNA | ssDNA,RNA? | 5’-WRC-3’(W = A or T; R = A or G) | immunoglobulingenes |
| APOBEC2 | 6p21 | 3 | heart,skeletal muscle,TNFα activatedliver cells | ? | N/C | ? | ? | ? | ? | Eif4g2 & PtenmRNA |
| APOBEC3A | 22q13.1 | 5 | monocytes/macrophages,non-progenitorcells | 1 | N/C(C inmonocytes) | ? | ssDNA,RNA | ssDNA,RNA | 5’-TC-3’ (preferred) | AAV-2,retroviruses,retroelements;WT1, SDHB &SIN3A mRNAs |
| APOBEC3B | 22q13.1 | 8 | PKC inducedcancer cells, INFαactivated liver cells | 1(C-terminal) | N | ? | ssDNA,RNA | ssDNA | 5’-TC -3’ (preferred) | retroelements,SIV, HBV, HPVproto-oncogenes |
| APOBEC3C | 22q13.1 | 4 | immune centers,peripheral-bloodcells | 1 | N/C | HIV Vif,HIV Gag (MA) | ssDNA,RNA | ssDNA | 5’-TC -3’ (preferred) | SIV,retroelements |
| APOBEC3Dϕ | 22q13.1 | 7 | immune centers,peripheral-bloodcells | 2(both weak) | C | HIV Vif,HIV Gag (NC) | ssDNA,RNA | ssDNA | 5’-TC -3’ (preferred) | HIV,retroelements |
| APOBEC3F | 22q13.1 | 8 | immune centers,peripheral-blood,INFα activatedliver cells | 1(C-terminal) | C | HIV Vif,HIV Gag (NC) | ssDNA,RNA | ssDNA | 5’-TC -3’ (preferred) | manyretroviruses,retroelements,HBV |
| APOBEC3G | 22q13.1 | 8 | immune centers,peripheral-blood,INFα activatedliver cells | 1(C-terminal) | C(restricted) | HIV Vif,HIV Gag (NC) | ssDNA,RNA | ssDNA | 5’-CC -3’ (preferred)5’-TC -3’ (less abundant) | manyretroviruses,retroelements,HBV |
| APOBEC3H | 22q13.1 | 5 | immune centers,peripheral-bloodcells | 1 | N/C(C for hap II) | HIV Vif,HIV Gag (NC) | ssDNA,RNA | ssDNA | 5’-TC -3’ (preferred) | manyretroviruses,retroelements |
| APOBEC4 | 1q25.3 | 2 | ? | 0 | ? | ? | ? | ? | ? | ? |
Figure 1. Alignment of members of the APOBEC family and their predicted phylogeny.
(A) The APOBEC proteins expressed in humans are AID, A2, A1, A3A through A3H and A4. Cartoons of the proteins are shown emphasizing gene duplication and likely divergence from the perspective of cytidine deaminase domain paralogs [Z1 (green), Z2 (orange) and Z3 (blue)] of the ZDD motif. APOBEC protein lengths (amino acid) are depicted to scale with the total length shown at the C terminus and with proteins aligned to the (first) ZDD motif. The canonical amino acid motif of the ZDD and of Z1-, Z2- and Z3-type cytidine deaminase domains are shown at the bottom of the panel. (B) A cartoon depicting the hypothesis for APOBEC paralog evolution. Phylogenetic relationships between several vertebrates and the emergence of APOBEC family members are shown on a timeline in millions of years that is not drawn to scale. AID and A2 are predicted to have emerged in jawless fish whereas A1 may have first appeared in ancestors of reptiles and amphibians. The timing of gene duplications and diversification events that led to the A3 subgroup (brown to green transition) is unclear, but likely occurred around 200 MYA. Humans and other primates express the largest diversity of A3 proteins. The phylogenetic relationship of A4 to other APOBECs is unclear. Though A4 is present in mammals, chickens and frogs, it is absent in fish [35].
A zinc-dependent cytidine deaminase domain of A1 is responsible for apoB mRNA C to U editing [9, 20] (Figure 1). The identification of a homologous cytidine deaminase domain motif within the AID primary amino acid sequence led to the prediction that AID [21] might function as the deoxycytidine deaminase required for the multiple mutations required for immunoglobulin gene diversification and recombination (known as somatic hypermutation (SHM) and class switch recombination (CSR)) [22–25]. It also led to the prediction that A3G [26] was responsible for host cell restriction of HIV infection through extensive dC to dU hypermutation of HIV proviral DNA during reverse transcription [26–29] (Table 1).
Recombination of the V(D)J region within immunogloblin genes gives rise to almost a billion different antibody proteins in a person's lifetime. However, it wasn’t until the discovery of AID and its hypermutational activity that we understood the mechanism for fine tuning that enables specific binding of antibody variable regions to new antigens. Point mutations in the immunoglobulin variable gene region (i.e. SHM) in activated B lymphocytes now is understood to be catalyzed by AID [21]. Moreover, the loss of AID function gives rise to an autosomal recessive, immune compromising disease known as hyper-IgM syndrome type 2 (HIGM2) wherein IgM accumulates due to an inability to perform CSR [30] which otherwise would confer Ig functionality throughout the body by recombining gene segments encoding different constant regions with a given variable region of the heavy chain immunoglobulin gene.
Likewise, A3G was discovered to be the causal factor for the dG to dA hypermutations in HIV proviral DNA that reduced the production of infectious virus [26, 28]. A3F, A3D and A3H likely support A3G through their ability to deaminate cytidines at additional sites in a concerted effort that brings about a catastrophic level of hypermutation of viral genomes; together, they are effectively an armada of host defense [31, 32]. Studies in which sufficient quantities of A3G, A3D, A3F or A3H were stably expressed in T cells demonstrated that each enzyme alone had the ability to mutate HIV during viral replication and inhibited infection in the absence of HIV Vif, the protein expressed by HIV to counteract these host defense factors [Text box 2] [32].
Text box 2. A critical viral defense mechanism is achieved by Vif-binding to A3s.
During HIV-1 infection, A3s can cause viral genomic mutations in ssDNA during reverse transcription that would ablate infection. However, the HIV-1 accessory protein Vif binds with A3C, D, F, G and H and recruits these antiretroviral proteins to an E3 ubiquitin ligase for polyubiquitination and subsequent proteasomal degradation. Vif binding to A3G is localized to the 128-DPD-130 motif at the junction of α4 helix and loop 7, while that for A3F, A3D, and A3C is over a broader conserved region localized mainly to a groove between α2 and α3 helices, opposite the ends that bind zinc and form the catalytic site. The Vif binding interface of A3H is less well understood but involves residues scattered across α3 and α4 helices. These interactions have been reviewed elsewhere [31, 33]. Lentiviral Vif-mediated degradation of A3 proteins is species specific. HIV-1 Vif binds and targets for degradation human A3D, A3F, A3G and to a lesser extent A3H, but not APOBEC of African green monkey (AGM) or rhesus macaque, while AGM simian immunodeficiency virus (SIV) Vif binds both A3G from AGM and rhesus macaque (rh), each coding K128, as opposed to human A3G, which codes for D128 [32, 34]. Likewise, rhA3D, rhA3F, rhA3G and rhA3H are degraded by rhSIV Vif, but only rhA3D is partially sensitive to HIV Vif, while rhA3F, rhA3G, and rhA3H are not [32].
A2 and A4 are the least studied of the APOBECs mainly due to the lack of specific phenotypes linked to either protein’s expression [35, 36]. Emerging evidence suggests that increased expression of A2 was linked to liver cancer and mutations in the genes encoding eukaryotic translation initiation factor 4 gamma 2 (Eif4g2) and phosphatase and tensin homolog (Pten) [37]. A2 may be essential for proper muscle differentiation and development [38], suggesting an as-of-yet unknown function of A2 may exist and may require the binding of a cofactor to regulate activity in certain tissue types.
While APOBEC proteins appear at a cursory level to be very similar because of their common evolutionary origin and a shared basic structural homology, they are very diverse in their impact on human health and disease. This review will consider recent understanding of APOBEC protein expression and its impact on the biology of cells relative to what we have begun to learn of shared structures as well as subtle structural differences that may explain unique functional characteristics. The reader will appreciate through this review that APOBEC genes have diversified greatly throughout evolution and that we are just beginning to contemplate that they have multifaceted roles in biology.
The A3 subfamily targets both foreign and self RNA and DNA
The duplication and divergence of A3 genes leading to expression of seven major homologs in humans (Figure 1A) may have both been driven by and contributed to mutations that promoted the evolution of endogenous and exogenous retroviruses/retroelements that bind to A3 proteins as substrates for editing [Text box 3] [39–42]. In this regard, the primate A3 subfamily of proteins has distinct but fairly lax nucleotide sequence preferences adjacent to edited Cs (Table 1) that may have been selected by A3 interactions that better controlled invasion by diverse retroviruses. Supporting this possibility are the expansion of A3 paralogs from one A3 gene in mice to seven in humans and the comparative decreased presence of active retroelements in humans relative to mice [Text box 3] [43]. Present day differentiation of human myeloid cells and their sensitivity to viral infection from both RNA and DNA viruses appears to be closely tied to the transcriptional regulation of A3 genes [44–50].
Text box 3. A3 proteins limit endogenous retroelement activity.
There are seven A3 proteins in humans (Figure 1A) active against endogenous retroelements (Table 1). In humans there are non-functional endogenous retrovirus (ERV) sequences that utilize long-terminal repeats (LTR) for genomic insertion similar to exogenous retroviruses. A3F and A3G hypermutation footprints have been identified in human ERV-K and ERV-1 elements as imprints of A3s being active against functional ERVs in our ancestral past [51]. There are also both functional and non-functional retroelements in humans that are non-LTR based retroelements (i.e. autonomous retroelements, called long interspersed nuclear elements (LINE) and non-autonomous retroelements that have trans-dependency on LINE replication machinery, called short interspersed nuclear elements (SINE)). To counter this threat to genomic integrity human A3A-H have been implicated in the inhibition of LINE and/or SINE retrotransposition [52]. Most A3 proteins are capable of directly hypermutating retroelement ssDNA and/or inhibiting reverse transcription of the retroelements [52–56]. A3G also may inhibit SINE activity in a deaminase-independent mechanism by binding to SINE RNA (e.g. Alu) and sequestering these RNAs as ribonucleoprotein complexes in the cytoplasm [57, 58]. Conversely, mice have only one A3 gene and their genomes contain active LTR-based and non-LTR based retroelements with 50–60 times more active retroelements in their genomes than humans and the proportion of retroelements causing disease is 35% greater in mice compared to humans [43, 59]. Overall, the variety within A3 genes in humans is a key determinant in combating the genotoxic threat posed by endogenous retroelements. The battle between A3 proteins and endogenous retroelements has clear selective pressure for both A3 genes and retroelement activity. The details of A3 proteins’ specific links to genome evolution through retroelements is a key open question ideally suited for future bioinformatics studies.
RNA viruses
A3G is naturally expressed to significantly higher levels than other A3 family members and contributes most of the APOBEC mutagenic, anti-HIV activity in T cells; A3F and A3H contribute secondarily, while A3D contributes even less [60]. Human A3H is expressed as variants in different populations (haplotypes I-IV) that have a remarkable diversity in DNA mutagenic and antiviral attributes [61–63]. A3H haplotype (hap) II is the only variant with a stable expression profile in humans and its expression correlates with increased anti-HIV activity [64]. Along with A3G and A3F, A3C and A3B are proposed to prevent simian immunodeficiency virus (SIV) from infecting humans, though A3B is weakly expressed in normal T cells [65]. A3C is moderately expressed in T cells, so it is a more likely host defense against SIV, but it lacks anti-HIV activity compared to A3G. This may be determined by its binding to the matrix (MA) region of HIV Gag rather than to the nucleocapsid (NC) region where A3G binds for viral particle encapsidation [66].
DNA viruses
A3B, A3F and A3G are up-regulated by interferon alpha (INFα) in liver cells and this expression has been suggested to protect these cells against hepatitis B virus (HBV) infection [67]. A3A is a potent inhibitor of a parvovirus, adeno-associated virus type 2 (AAV-2), and its activity seems to be related to the presumed DNA binding loops that differ from those found in A3G (Figure 2B, Key Figure) [68]. Interferons also upregulate the expression of two enzymatically active A3A isoforms in monocytes and macrophages, suggesting that A3A may protect monocytes from 'parasitic' DNA (e.g. viruses and retroelements) [69].
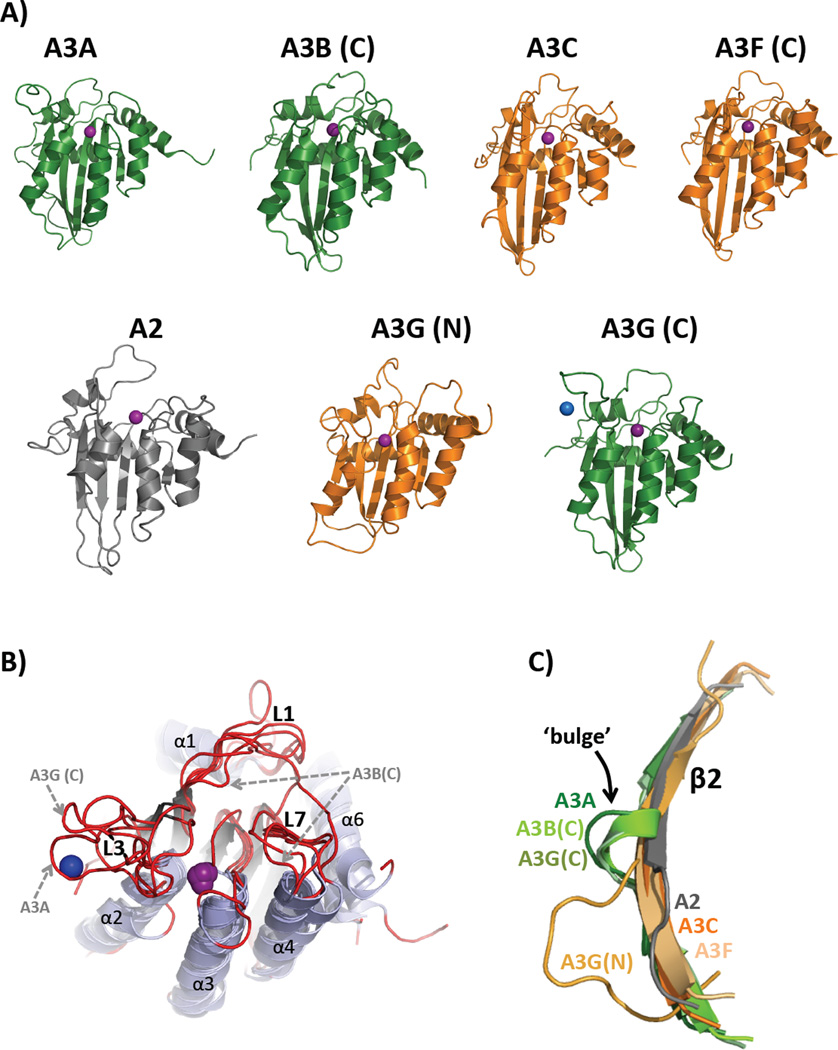
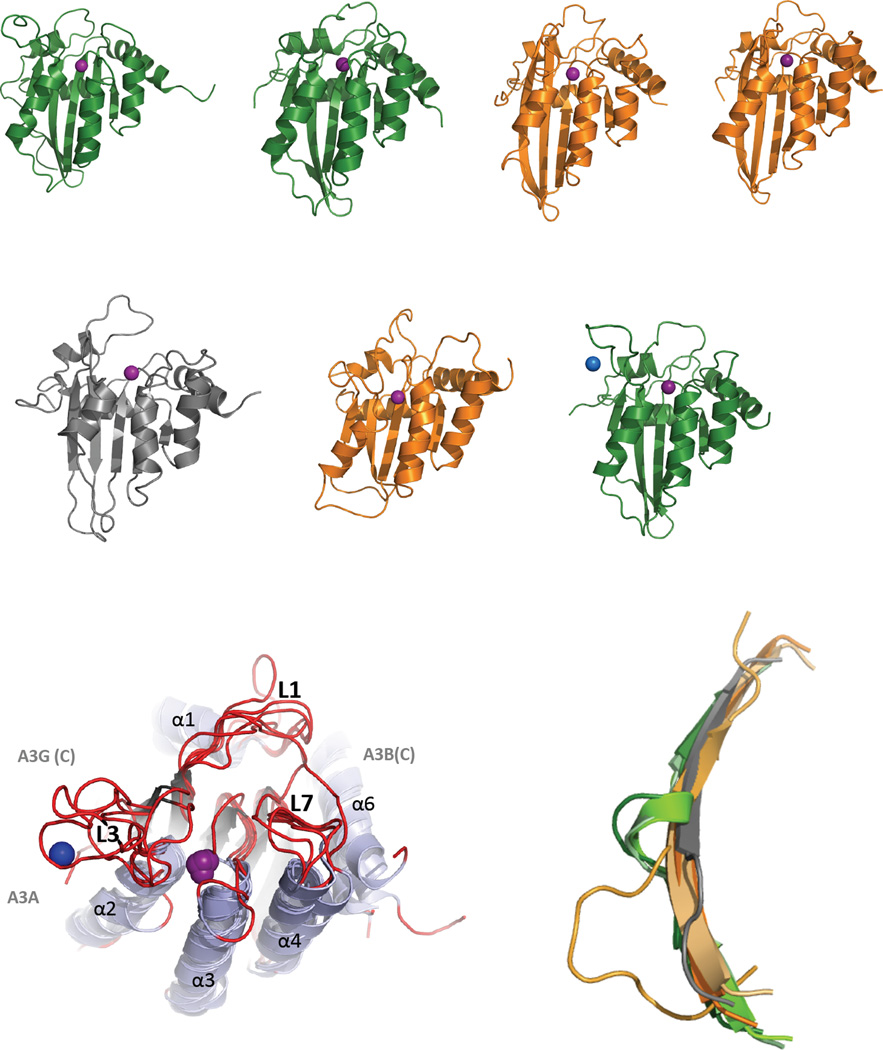
Figure 2, Key Figure. Structural similarities and differences of the APOBEC family of Proteins.
(A) Cartoon representation of the solution NMR structures of A2Δ40 (PDB 2RPZ) [96], A3A (PDB 2M65) [100], and A3G (N-terminal half, PDB 2MZZ [101]) and X-ray crystal structures of A3B (C-terminal half, PDB 5CQI [106]), A3C (PDB 3VOW) [102], A3F (C-terminal half, PDB 4J4J [108]) and A3G (C-terminal half, PDB 3IR2 [104]) are depicted in the same orientation as that shown in [Text Box 4]. A3 Z1-type CDA domains are green and Z2-types are orange. The catalytic zinc ion (or pseudo-catalytic for A3G(N)) is shown as a purple sphere while the blue sphere represents an intermolecular structural zinc ion that is coordinated by two residues from each of the A3G proteins in the asymmetric unit of the crystal mediating a potential zinc-dependent A3G C-terminal oligomeric interface involving loop 3, α2 helix and the β2-bulge-β2’ feature. (B) Alignment of APOBEC structures shown in (A) with a perspective that shows the structural differences among loops 1, 3, and 7 (red), which are predicted to be important for nucleic acid binding, substrate sequence preference and to control access of substrates to the catalytic pocket containing zinc (purple). Variations in loop sequence and structure have been proposed as determinants of ssDNA binding, catalysis and sequence specificity differences among APOBECs. The L3s of A3A and A3G C-term are both significantly longer than those of A2 and the Z2 A3C, A3F C-term and A3G N-term and both bind a secondary zinc ion (shown for A3G C-term structure as a blue sphere) that may allosterically regulate catalysis [114]. L3 of A3B C-term, the other Z1-type CDA domain, was deleted from the construct to facilitate crystallization [104]. L1 and L7 of A3B C-term are significantly shifted toward the catalytic zinc ion that is predicted to block access to the catalytic pocket; suggesting that this structure is a closed conformation (catalytically inactive) of what is known to be a catalytically active CDA domain. (C) Structural alignment of the β2 strands from APOBEC structures in (A) depicting the β2-bulge presence in Z1-type A3s (A3A, A3B C-term and A3G C-term) or absence in A2 and Z2-type A3s (A3C, A3F and A3G N-term).
RNA editing of cellular transcripts
Whereas most A3 proteins only use ssDNA as substrates, A3A (in monocytes), like A1 (in liver and intestine), has the ability to target specific RNAs for C to U editing (Table 1) [70]. The mechanism and phenotypes linked to A3A site-selective mRNA editing are yet to be been determined. Whether there is a phenotype resulting from APOBEC editing of RNA as clearly as seen with A1 editing of apoB mRNA [Text Box 1] is a general open question in the field. An open controversy has been whether AID edits mRNA as part of the mechanism for SHM and CSR [71, 72].
Nine APOBEC proteins with verified activity and a generally lax sequence preference (Table 1) enables the targeting of multiple RNA and DNA substrates by this family. Therefore, the overall number of APOBEC proteins in humans and targets speaks to the necessity of the APOBECs to both protect cells from foreign invaders and to influence cellular gene expression. However, there is an equally crucial need to regulate these proteins in order to prevent possible mutagenic capabilities of APOBEC deaminase activity on off-target cellular RNA and DNA.
Cells regulate APOBEC activity through localization
A1 and AID are regulated by nucleo-cytoplasmic trafficking
A crucial factor regulating APOBEC function is cellular localization and regulation of nucleo-cytoplasmic trafficking. APOBEC proteins that mutate ssDNA may affect genomic integrity and this is likely the reason why their access to the cell nucleus often is regulated. The activities of both A1 on mRNA [Text box 1] and AID on the ssDNA of the immunoglobulin locus take place within the cell nucleus (Table 1) [11]. Both enzymes contain nuclear localization signals (NLS) and CRM1-dependent nuclear export signals (NES) [73, 74]. A1 is maintained in the cytoplasm as an enzymatically inactive 60S complex along with A1CF [Text box 1]. A1 and A1CF then translocate to the cell nucleus and form a 27S active editosome on mRNA following RNA splicing [75]. In addition to NLS and NES trafficking signals, AID is actively retained in the cytoplasm through interactions with eukaryotic Elongation Factor 1-alpha (eEF1A), heat shock protein 90 (HSP90) [76] and possibly RNA [72] (Table 1).
Anti-HIV A3s avoid genomic DNA by staying in the cytoplasm
A3G is restricted to the cytoplasm through a strong cytoplasmic retention signal (CRS) [77]. Even during mitosis, A3G does not come in contact with chromosomal DNA [11]. The A3G CRS region contains an RNA binding domain [78] and RNA binding is involved in A3G association with cytoplasmic RNA processing centers, stress granules and p-bodies [79, 80]. RNA binding to A3G inhibits its ability to hypermutate ssDNA [81]. However, A3G deletion mutations that abrogated RNA binding remained in the cytoplasm, suggesting that RNA binding may only be part of the essential interactions for retention of A3G in the cytoplasm [77]. Interestingly, A3H hap II is the only A3H haplotype that has a predominantly cytoplasmic localization, consistent with other anti-HIV paralogs, A3D, A3F and A3G (Table 1). The localization of A3H in the cytoplasm may also be linked to its RNA binding activity, as nuclear localized haplotypes I,III, and IV did not support RNA binding [78].
Misregulation of APOBECs causes cancer
AID misregulation
Alternatively spliced variants of AID lacking exon 4 retain binding to trafficking and localization cofactors CRM1 and eEF1A, yet they have been specifically linked to misregulation of AID activity and cancer [76, 82]. Tight regulation of AID activity was shown to be important for regulating immunoglobulin gene expression and cMyc oncogenic translocation in B cell tumor development [83, 84]. In these studies off-target translocations were highly sensitive to AID expression, yet on-target SHM and CSR were only mildly affected by AID expression levels [83, 84]. This suggests that there is a regulatory mechanism (e.g. cofactor) that specifically regulates AID on-target hypermutation of the immunoglobulin gene region.
A1 misregulation
Attempts at gene therapy with A1 have been largely abandoned due to studies showing aberrant hypermutations even with regulated expression of A1 in mouse liver [85]. Furthermore over-expression of A1 has been linked to oncogenesis due to increased editing of the novel A1 target mRNA (NAT1), which encodes a widely expressed translational repressor unless the mRNA is edited to contain a premature translation stop codon [86]. A1 has also been shown to have genomic DNA mutagenic activity when expressed in E. coli and an indirect correlation between A1 up-regulation in cells that are precursors to esophageal adenocarcinomas and DNA mutations has been reported; although DNA mutation by A3B, which is also expressed highly in cancerous cells, was not ruled out [87].
A3B links to cancer
A3B is predominantly nuclear due to a NLS that is similar to that of AID [88]. A3B expression is kept at low levels in most normal tissues, however whether this is controlled through transcription, RNA silencing or post-translational regulation is yet to be determined. By contrast, A3B expression is elevated in diverse forms of cancers [89]. Whether A3B mutagenic activity is a cancer driver or a downstream effector remains an open question and the mechanism for A3B up-regulation in cancer cells is not known [90]. A driver of A3B transcriptional up-regulation may be viral infection, as human papillomavirus (HPV) infection in head/neck and cervical cancers increased A3B expression through the viral oncoprotein E6 [91]. That access to genomic DNA seems to be regulated only by limiting the expression of A3B (and not other APOBEC proteins) may explain why A3B expression (among APOBEC proteins) is more commonly associated with cancer.
A3A links to cancer
A3A is cytoplasmic and non-genotoxic in monocytes; however, when A3A was transfected into other cell types it localized throughout the cell and became genotoxic [92]. These findings suggest that either A3A interacts with a nuclear import factor in a foreign cell context or a cytoplasmic retention cofactor for A3A is expressed in greater abundance in monocytes. It is curious that a large proportion of Asian (37%), Amerindian (58%) and Oceania (93%) populations have a deletion in the A3B gene, which is associated with a 20-fold increase in the expression of an A3A from an mRNA variant containing the 3’UTR of A3B. This deletion is associated with an increased risk for breast cancer and hepatocellular carcinoma (HCC) [90]. It is not clear whether the increased risk for cancer is related to the 20-fold increase in expression of the A3A variant, which may be responsible for genomic mutations [93]. However, an analysis of A3A verses A3B signature mutations in cancer cells suggested that perhaps A3A mutations are generally more prevalent than those from A3B in cancer [94].
Overall, the data suggest that regulation of expression and cytoplasmic localization are critical regulatory mechanisms to maintain genomic integrity, but the mechanisms involved appear to differ for each paralog. Localization of APOBECs likely requires binding to a cell type-specific protein or RNA cofactor(s). It is increasingly apparent that misregulation of these processes are associated with cancer [90]. Thus, identifying and characterizing APOBEC cofactors is a key open question in the field.
The structures of APOBEC proteins suggest variations around a common core
The common core of APOBEC structures is the cytidine deaminase domain
Structures of APOBEC proteins have been difficult to obtain. The first APOBEC crystal structure obtained was for A2 with a 40-residue N-terminal deletion [95]. Subsequently, NMR solution structures of full-length A2 [96], the A3G C-terminal half [97–99], A3A [100], and the A3G N-terminal half [101], were reported as well as X-ray structures of A3C [102], the A3G C-terminal half [103–105], the A3B C-terminal half [106], and the A3F C-terminal half [107–109]. A low-resolution small-angle X-ray scattering model of recombinant full-length A3G also has been reported [110]. Representative structures for each APOBEC are depicted as cartoons in Figure 2A.
All 11 APOBECs share a conserved zinc-dependent deaminase sequence motif (ZDD, Figure 1B) within a α-β-α super secondary structural element that forms the core catalytic site of a cytidine deaminase (CDA) domain [Text Box 4]. Variations in the length, composition and spatial location of conserved secondary structural features and in particular the loops adjacent to the catalytic site likely dictate function, substrate selection and regulation of catalytic deamination of (deoxy)cytidine to (deoxy)uridine and oligomerization. The mechanism of deamination is believed to be conserved with that of other CDA superfamily enzymes that act on cytidine, cytosine, or deoxycytidylate substrates [111].
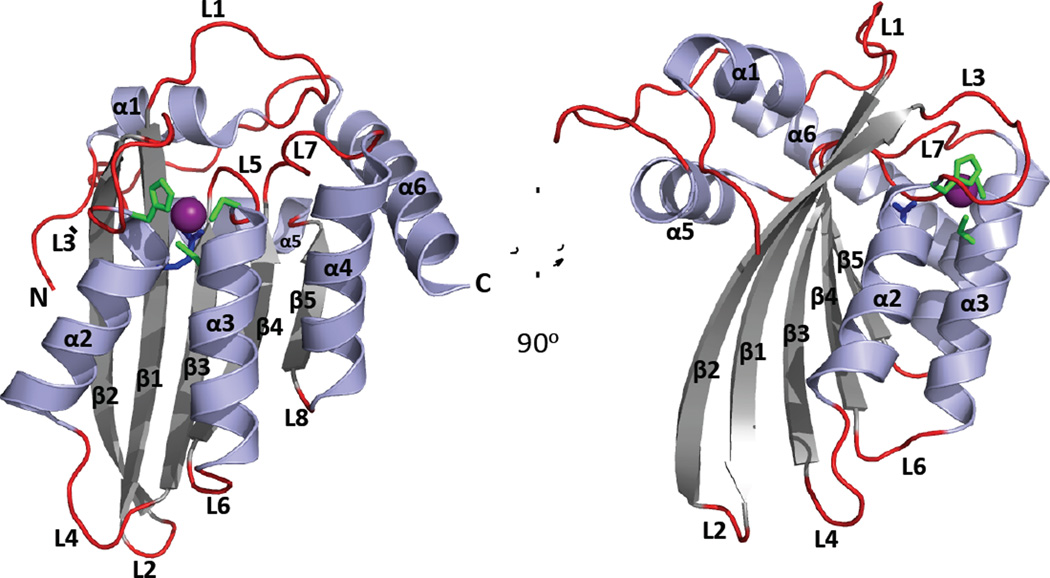
Text Box 4. The APOBEC canonical cytidine deaminase (CDA) fold.
APOBECs share a conserved CDA fold, comprising a five-stranded mixed β-sheet that is stabilized by alpha helices packing against either face and with the secondary structural elements sequentially arranged as α1-β1-β2-α2-β3-α3-β4-α4-β5-α5-α6 (Figure I). The catalytic residues of the ZDD motif (Figure 1A) are found at the N-terminal ends of helices α2 and α3 (Figure I). A zinc ion is coordinated by the conserved Cys and His residues of the ZDD and by a water molecule that is activated by the zinc ion for nucleophilic attack of the C4 atom in the cytidine ring. The conserved Glu residue of the ZDD acts as a proton shuttle to aid the transfer of a proton from water to N3 of the cytidine ring and donates a proton to the leaving ammonia group.
Figure I legend, included with Figure I in Text Box 4.
Ribbon Diagram of an APOBEC Canonical Cytidine Deaminase (CDA) Fold.
A cartoon depiction of the A3F (C-terminal half, PDB 4J4J [108]) crystal structure with a 90 degree counter clockwise rotation illustrating the major secondary structural elements and the canonical CDA fold, that is evolutionarily conserved in the cytidine deaminase super family [36]. The CDA fold comprises a five-stranded mixed β-sheet that is flanked by six α-helices; the conserved catalytic Glu (blue) and zinc binding residues (His and two Cys, green) of the ZDD-motif form the core of the active site and are located at the N-terminal end of α2 and α3 helices. APOBEC proteins comprise either one CDA domain or two CDA domains covalently linked on the same peptide. Many single- and dual-deaminase domain APOBECs oligomerize through an unknown molecular mechanism. Variations in the length, composition and spatial location of conserved secondary structural features and loops dictate cellular localization, function, substrate selection and regulation of catalytic deamination of (deoxy)cytidine to (deoxy)uridine.
The β2 strand bulge is seen in some but not all A3 structures
A3A and the A3G and A3B C-terminal CDAs each possess an interrupted β2 strand (Figure 2A & C). The normally contiguous center of the long β2 strand is replaced with a short, bulging loop or helix (termed the β2 bulge) resulting in a β2-bulge-β2’ topology. This feature is not present in the Z2-type structures of A3C, A3F C-terminal CDA, A3G N-terminal CDA or in A2 as these structures feature continuous β2 strands. While the bulge is missing in the A3G N-terminal structure, the β2 strand is shorter and the preceding loop 2 is longer and less compact, perhaps mimicking the function of the bulge (Figure 2C). The physical proximity of the β2 strand with the beginning of the C-terminal CDA suggests that the β2 strand may be poised to interact with the adjacent N-terminal CDA (for the dual-deaminase A3B and A3G) in a bulge-dependent manner. Structural variability of β2 strands in the APOBEC family may play a role the quaternary organization of single domain A3A. Verification awaits a full-length dual-deaminase domain structure in which the position and interaction of the N-terminal CDA with and C-terminal CDA of dual domain APOBECs are restrained through short and covalent linkages.
Structural foundation of ssDNA and RNA binding by APOBECs
Structural features that regulate APOBEC interactions with nucleic acids
Nucleic acid binding is common to all APOBECs, though in the dual deaminase domains of A3B, A3F, and A3G the N-terminal domains have lost the ability to catalyze cytidine deamination but retained RNA binding activity (Table 1) and, at least for A3G, DNA binding activity as well [112]. The interaction of ssDNA with APOBECs is generally mediated through shallow grooves on the protein surface that lead to the catalytic site and are lined with patches of positively charged (basic) and aromatic (hydrophobic) residues that stabilize interactions with the negatively charged nucleic acid backbone and stack with nucleic acid bases, respectively [31, 99, 104]. While there is strong structural conservation of the CDA architecture among the APOBEC family [Text Box 4], examination of available APOBEC structures reveals a significant amount of structural plasticity and sequence variability that is localized to loops between conserved secondary structural elements (Figure 2B). Functional differences in substrate recognition and deamination activity among APOBECs have been mapped largely to variations in amino acid composition, length and spatial conformation of loops L1, L3 and L7 in particular.
L7 has conserved sequence motifs that determine nearest neighbor editing sequence preference among the APOBECs [107] (Table 1) where L1 together with L7 control substrate access to the catalytic pocket. In most APOBEC structures (the exception being the current A3B C-terminal structure [106]) L1 and L7 are splayed apart enough to grant a deoxycytosine base access to an open catalytic pocket [106] (Figure 2B). L3 is also involved in ssDNA substrate binding and has the greatest length and sequence variation among APOBECs [100, 103, 113]. L3 of A3G and A3A contribute a secondary zinc binding site that is proposed to allosterically regulate dC deaminase activity [114]. Concordantly, a zinc ion with intermolecular coordination is observed between residues in adjacent L3 loops of the asymmetric unit in the A3G(C) crystal structure [104]. Zinc coordination by this loop has also been observed at a dimeric interface in an A3A crystal structure [115].
Models of single-stranded DNA binding to APOBEC3s
Numerous models of ssDNA binding to the C-terminal CDA of A3G have been proposed by a variety of methods. NMR DNA-titration experiments predict a ‘brim‘ model involving a ring of basic, positively charged residues surrounding a concave active site. In this model, a putative ssDNA path navigates between L1, L3 and L7 within extended grooves of the A3G C-terminal surface. These loops are clustered at the confluence of the N-terminal ends of α2 and α3 helices. DNA contacts with residues of L5 and the α4 and α6 helices are also predicted [97] (Figure 2B). A structure-guided mutational assay [103] predicted a similar ‘straight’ path for ssDNA on A3G, while real-time monitoring of the deamination reaction [98] suggested that the ssDNA path is ‘kinked’ and runs through a different surface groove that is perpendicular to the ‘brim’ and ‘straight’ models. Mass spectroscopy (MS) analysis of UV-cross linked ssDNA in complex with full-length A3G suggested that ssDNA bound to both the catalytically active C-terminal CDA (L7 and a region comprising α6) and the catalytically inactive N-terminal CDA, in a region comprising its α6 [112]. Modeling based on this analysis suggested that a large, contiguous surface from both A3G domains engages ssDNA and provides a rationale for why the catalytically active C-terminal CDA of dual deaminase A3s alone is not sufficient for high affinity ssDNA binding [112].
DNA binding models for other A3s have also been proposed. A model of ssDNA binding to the A3F C-terminal half crystal structure [108] based on structure guided mutagenesis predicted a path of ssDNA that is similar to the ‘straight’ and ‘brim’ A3G models. The crystal structure of the A3B C-terminus was solved with a dCMP bound to residues of the α4 and α6 helices that are away from the active site [106]. Modeling of polynucleotide binding based on this structure predicted a similar interaction of ssDNA with A3B as suggested for A3G [106]. In contrast to the aforementioned models, a binding model for A3A suggested ssDNA bends sharply to insert the target dC into a deeper catalytic pocket with fewer localized contacts [100]. Differences in binding configurations may explain why, unlike in A3G and A3F, deamination by A3A is not predicted to be a processive event [116]. Overall, modeling has suggested somewhat similar paths for ssDNA binding to the C-terminal CDA of A3G, A3F, and A3B and the noted exception currently is A3A.
The covalent linkage of tandem CDAs that is inherent to A3B, A3D, A3F, and A3G, and the oligomerization of some single and dual deaminase domain APOBECs, may enable more robust APOBEC interactions with ssDNA. These features also may accommodate additional regulatory mechanisms for ssDNA binding. The dual-domain binding model spanning the N- and C-terminus of A3G predicted by UV-cross linking and MS [112] is supported by: (i) single point mutations in the N-terminal domain ablating nucleic acid binding by full-length A3G [117, 118], (ii) the observation that full-length A3G has ~1000 fold stronger affinity for ssDNA than the C-terminal CDA alone [97, 98, 118], and (iii) results from atomic force microscopy experiments interrogating APOBEC binding to ssDNA which revealed a bimodal distribution of contour lengths for the dual-domain A3G but only a monomodal distribution for the single-domain A3A [119]. In addition, full length A3G forms dimers [120–122] that bind to ssDNA substrates to form catalytically active homo tetramers [121], thus the full extent of a ssDNA interaction with native APOBECs may be underestimated by modeling with single CDA domains.
APOBEC and RNA interactions
A1 requires AC1F for site-specific editing of apoB mRNA but A1 itself, has a low RNA binding sequence-specificity [123]. APOBECs are RNA binding proteins and A3 proteins bind non-selectively to cellular and viral RNAs [124]. Dual deaminase domain APOCECs bind to RNA predominantly through their non-catalytic N-terminal CDA [117, 118, 125, 126] and, at least for A3G, by the C-terminal domain as well [112]. RNA-bound A3s are catalytically inactive on ssDNA [110, 118]. MS analysis of nucleic acid cross-linked to A3G revealed RNA bound to the same sites that DNA bound, which suggests a competitive mechanism for RNA-mediated inhibition of ssDNA deamination [112]. However, the proximity of RNA binding sites in the N-terminal half of A3G are proximal to a DNA-binding site and this raises the question of whether RNA binding might allosterically regulate ssDNA binding. It is currently unclear whether RNA and ssDNA can bind A3G simultaneously. RNA binding to N-terminal and C-terminal CDAs may recruit APOBECs for alternate functions [81, 112, 124].
Catalytic activity, nucleic acid interactions and cellular abundance may be determined by APOBEC homo and hetero protein-protein interactions
A1 and AID
Some members of the APOBEC family form functionally relevant homo or hetero complexes. A1 dimerization [20, 127] and complex formation with A1CF is required for editing activity on apoB mRNA [128, 129]. The dimerization interface utilizes a conserved hydrophobic patch near the C terminus and, though the A1 structure is not known, this patch maps to α6 helix in the CDA structure [130–132]. AID has been reported to form functional homodimers [132, 133] as well as functional monomers [134].
A2
The crystal packing of the initial A2 structure with a 40 residue N-terminal deletion (A2Δ40) [95] revealed a long β2-β2 interface between adjacent A2 molecules, suggesting a potential mechanism for oligomerization of single domain APOBEC proteins such as AID. However, the basis for this mode of oligomerization was refuted by the monomeric NMR solution structure of mouse A2Δ40 (Figure 2A, PDB ID 2RPZ), which demonstrated that native A2 may be a monomer. Further NMR analyses of full-length human A2 showed that the N-terminal 40 residue extension forms 3 dynamic α-helices that pack against the β2 strand, preventing a β2-β2 mediated dimeric interaction [96].
A3 family
The oligomeric state of A3 proteins has effects on catalytic function, nucleic acid binding and cellular distribution, though there is no consensus on the oligomeric state of catalytically active A3s. Factors that have confounded the study of APOBEC oligomerization and hindered a clear understanding of mechanism are that oligomerization occurs through both direct protein-protein interaction and through more complicated, but indirect, nucleic acid-bridged interactions. Fluorescence fluctuation spectroscopy demonstrated that A2, A3A, and A3C were monomeric in cells while A3B, A3D, A3F, A3G, and A3H were multimeric [135]. Interestingly, the ability to form multimeric complexes was correlated with sequestration of A3s and AID in cytoplasmic complexes [75, 76, 136] and p-bodies [79, 80] as well as with packaging in HIV virions in the cytoplasm of cells [78, 137–139]. This suggested that oligomerization, whether through protein-protein interactions or RNA-protein interactions, may contribute to A3 localization and function. Biophysical analyses of purified A3G suggested multiple oligomeric states for catalytically active full-length A3G [110, 120, 140, 141] modeled alternatively as mediated by either the N-[125, 139, 142] or C-terminal [77] CDA. It is likely that the oligomeric state influences the orientation of loops involved in nucleic acid binding and catalytic site access; this could affect and even regulate the kinetics of deamination by providing larger binding surfaces for nucleic acid.
Some members of the A3 family (A3C, D, F, G and H) are bound by HIV-1 Vif protein and subsequently recruited to an E3 ubiquitin ligase complex for polyubiquitination and proteasomal degradation [Text Box 2]. Vif mediated degradation of A3s is essential for a sustainable HIV infection and the Vif-binding A3 surfaces vary among those A3s that are bound by Vif (reviewed in [31, 33]). Recent computational studies suggest how an ancestral lentiviral Vif may have evolved to bind multiple and distinct A3 surfaces from an original host organism with only a single A3 protein [143].
Concluding remarks
Our knowledge of APOBEC proteins, their functions and structures is growing rapidly. In recent years of research, A4 was added to the list of family members and the diversity of A3H haplotypes with differences in antiviral functions were described. A3A recently joined A1 as a deaminase that has mRNA editing activities. In addition, an expanded functional significance for A1 mRNA editing has been suggested by the discovery of 3' UTR mRNA editing and the discovery that the RNA binding protein RBM47 in addition to A1CF is a complementation factor; opening the possibility of yet-to-be identified RNAs that are C to U edited through different auxiliary protein targeting of A1.
Today the role of genetic mutations in A3B and A3A and elevated expression of AID, A3B and A3A leading to a cancer genotype and phenotype is all but certain. We are just beginning to appreciate the extent to which AID, or other APOBECs, contribute to chromosomal translocations in health and disease. We understand that cells have, as part of their innate immunity, an ability to neutralize infection by HIV through A3 proteins and their mutagenic activities on HIV. As the field has made progress in structural characterization of APOBECs and their interactions with substrates and other macromolecules, an overarching focus has become the regulatory processes that limit or enhance APOBEC expression and functions in promotion of healthy cell responses to diseases. Although the timing and reasons why APOBEC proteins evolved may be difficult to prove, a priority will be to understand the function of the present day APOBEC family. If we do come to appreciate the strengths and potential limitations of this family of proteins, there may be opportunities to use these enzymes in cell engineering and for the development of therapeutics for disease intervention.
Outstanding Questions.
- What are the native structures for APOBECs? Notably absent are structures of A1, AID, a full-length dual deaminase domain A3 and structures of substrate- and cofactor-bound APOBECs.
- What are the native functions for APOBECs? The relationships among APOBEC protein expression, cellular localization and function have been driven by native cell-type specific selection pressures. The functions of APOBECs that are inferred by experimentally overexpressing APOBECs in various cell lines have to be evaluated cautiously as cellular regulatory mechanisms may not function under these conditions.
- How do APOBECs bind to ssDNA and RNA and what are the interactions required for deoxycytidine/cytidine deamination?
- What nucleic acid binding or protein cofactor interactions affect APOBEC subcellular compartmentalization, interactions with substrates and regulation of deaminase activity?
- Are the DNA mutagenic activities of proto oncogenic APOBECs that localize to the cell nucleus, like AID, A1, A3A and A3B, involved in the initiation or progression of cancers? The beneficial roles of APOBECs clearly can be compromised through over-expression, production of APOBEC variants and inappropriate retention in the cell nucleus. We do not know what leads to mutation of off-target nucleic acids and which mutations can lead to the induction and progression of cancer.
- Does RNA editing by APOBEC affect the diversity of the protein or RNA function? The technology for transcriptome analysis has enabled the search and identification of modified RNA bases included presumed APOBEC editing sites. However the percentage of RNAs modified and therefore the impact RNAs with modified bases have on cell phenotype has not been addressed. Without quantitative analysis, edited mRNAs identified by high resolution RNA sequencing methods that are the result of over-expressing APOBEC in heterologous cell types raise questions concerning their occurrence in nature.
Trends.
The APOBEC family of proteins does not have a common RNA or DNA substrate for cytidine/deoxycytidine deamination and in fact it appears that not all the predicted cytidine deaminase domains are catalytic activity.
A consensus is emerging that nucleic acid substrates are coordinated for deamination using amino acid residues flanking the catalytic domain.
APOBECs are not essential proteins for development and growth but several play critical roles in the maintenance of long-term health such as in metabolism and immunity (innate and acquired).
APOBECs are controlled through expression of wild type and alternative spliced mRNAs as well as localization, post-translational modification, turnover, and interactions with other proteins or RNAs.
Overexpression of APOBECs and APOBEC variants has been observed in several malignancies suggesting an association with cancer initiation and/or progression.
Acknowledgments
The authors thank Jenny M.L. Smith for the preparation of Figure 1. All authors contributed equally to the planning and writing of this review. The authors wish to acknowledge the numerous contributions to the advancement of science in the APOBEC field made by investigators across the globe whose work may not have been adequately referenced here due to the limitations of space. Preparation of this review was supported in part by a Public Health Services Grant GM110568 and GM110038 awarded to HCS.
Glossary
C to U base-modification
also referred to as cytidine to uridine deamination or editing, this is a reaction in which a coordinated zinc ion in an enzyme active site acts as a Lewis acid to activate a water molecule for hydrolytic, nucleophilic attack of the amide group at the C4 position of cytidine and a conserved glutamic acid acts as a proton shuttle to convert a cytidine base to a uridine in either RNA or DNA with an ammonium leaving group.
Class Switch Recombination (CSR)
an immunoglobulin-specific function in activated B cells and dependent on AID mutagenic activity where non homologous recombination within the Ig gene sequence encoding the immunoglobulin constant region is induced to switch from IgM to new functional classes of antibodies (IgG, IgA or IgE).
Cofactor
a protein/peptide, nucleic acid, lipid or organic small molecule that binds to an enzyme to regulate a specific structure, activity or subcellular distribution.
Hypermutation
when a multitude of nucleotides on the same strand of DNA or RNA are edited.
Interferons
members of the cytokine family of signaling proteins that are synthesized and secreted by cells in response to pathogens (e.g. viruses and bacteria) and that act on immune cells to activate their responses.
Retroelements
the two major classes of autonomous retroelements are long terminal repeat (LTR) based endogenous retroviruses and non-LTR based long interspersed nuclear elements (LINE) that both encode everything needed to reverse transcribe and re-insert their sequence into another location within the cell’s genome. There are also non-autonomous short interspersed nuclear elements (SINE) that are trans-dependent on the LINE encoded machinery for reverse transcription and genomic re-insertion. Endogenous retroelements’ ability to copy themselves into random locations in the genome leads to genomic instability and disease.
Somatic Hypermutation (SHM)
a process specific to immunoglobulin (Ig) genes and dependent on AID mutational activity in activated B-cells in which the gene sequence encoding the Ig variable region is hypermutated to produce Ig gene variants that will encode IgM with the ability to bind to antigens with high affinity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: Dr. HC Smith is a full-time tenured professor at the University of Rochester School of Medicine and Dentistry. He is also the founder and CEO of the University of Rochester spinout company OyaGen, Inc. The company has a financial interest in the development of antiviral and anti-cancer drugs based on APOBEC technology. Drs. JD Salter and RP Bennett are employees of OyaGen, Inc.
References
- 1.Smith HC. RNA and DNA Editing. Hoboken, New Jersey: John Wiley & Sons Inc.; 2008. p. 426. [Google Scholar]
- 2.Chen SH, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 3.Powell LM, et al. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 4.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8(6):647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 5.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9(6):229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. Journal of Virology. 2009;83(2):494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greeve J, et al. Apolipoprotein B mRNA editing in 12 different mammalian species: hepatic expression is reflected in low concentrations of apoB-containing plasma lipoproteins. J Lipid Res. 1993;34(8):1367–1383. [PubMed] [Google Scholar]
- 8.Smith HC, Wedekind JE, Kefang X, Sowden MP. Mammalian C to U Editing. Topics in Current Bioogy. 2005;12:365–400. [Google Scholar]
- 9.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260(5115):1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 10.Davidson NO, et al. Proposed nomenclature for the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme: APOBEC-1 [letter] RNA. 1995;1(1):3. [PMC free article] [PubMed] [Google Scholar]
- 11.Smith HC, et al. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol. 2012;23(3):258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fossat N, et al. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014;15(8):903–910. doi: 10.15252/embr.201438450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanc V, et al. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol Cell Biol. 2005;25(16):7260–7269. doi: 10.1128/MCB.25.16.7260-7269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano K, et al. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J Biol Chem. 1996;271(17):9887–9890. doi: 10.1074/jbc.271.17.9887. [DOI] [PubMed] [Google Scholar]
- 15.Skuse GR, et al. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Research. 1996;24(3):478–485. doi: 10.1093/nar/24.3.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg BR, et al. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nat Struct Mol Biol. 2011;18(2):230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anant S, Davidson NO. An AU-rich sequence element (UUUN[A/U]U) downstream of the edited C in apolipoprotein B mRNA is a high-affinity binding site for Apobec-1: binding of Apobec-1 to this motif in the 3' untranslated region of c-myc increases mRNA stability. Molecular and cellular biology. 2000;20(6):1982–1992. doi: 10.1128/mcb.20.6.1982-1992.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cappione AJ, French BL, Skuse GR. A potential role for NF1 mRNA editing in the pathogenesis of NF1 tumors. Am J Hum Genet. 1997;60(2):305–312. [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay D, et al. C-->U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am J Hum Genet. 2002;70(1):38–50. doi: 10.1086/337952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79(3):285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 21.Muramatsu MSV, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific Expression of Activation-induced Cytidine Deaminase (AID), a Novel Member of the RNA-editing Deaminase Family in Germinal Center B Cells. The Journal of Biological chemistry. 1999;274(26):18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, et al. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18(1):1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 23.Dickerson SK, et al. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197(10):1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20(6):659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell. 2002;109(Suppl):S35–S44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 26.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 27.Smith HC. APOBEC3G: a double agent in defense. Trends Biochem Sci. 2011;36(5):239–244. doi: 10.1016/j.tibs.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Refsland EW, et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38(13):4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 30.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 31.Aydin H, Taylor MW, Lee JE. Structure-Guided Analysis of the Human APOBEC3-HIV Restrictome. Structure. 2014;22(5):668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Hultquist JF, et al. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-deficient HIV-1. Journal of Virology. 2011 doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salter JD, Morales GA, Smith HC. Structural insights for HIV-1 therapeutic strategies targeting Vif. Trends Biochem Sci. 2014 doi: 10.1016/j.tibs.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pery E, et al. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83(5):2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogozin IB, et al. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4(9):1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 36.Conticello SG, et al. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22(2):367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 37.Okuyama S, Marusawa H, Matsumoto T, Ueda Y, Matsumoto Y, Endo Y, Takai A, Chiba T. Excessive activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide 2 (APOBEC2) contributes to liver and lung tumorigenesis. International Journal of Cancer. 2012;130:1294–1301. doi: 10.1002/ijc.26114. [DOI] [PubMed] [Google Scholar]
- 38.Sato Y, et al. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J Biol Chem. 2010;285(10):7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz M, et al. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2(9):E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munk C, Willemsen A, Bravo IG. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol. 2012;12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etienne L, et al. The Role of the Antiviral APOBEC3 Gene Family in Protecting Chimpanzees against Lentiviruses from Monkeys. PLoS Pathog. 2015;11(9):e1005149. doi: 10.1371/journal.ppat.1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35(Pt 3):637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- 44.Koning FA, et al. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83(18):9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng G, et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110(1):393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stopak KS, et al. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem. 2007;282(6):3539–3546. doi: 10.1074/jbc.M610138200. [DOI] [PubMed] [Google Scholar]
- 47.Rose KM, et al. Transcriptional regulation of APOBEC3G, a cytidine deaminase that hypermutates human immunodeficiency virus. J Biol Chem. 2004;279(40):41744–41749. doi: 10.1074/jbc.M406760200. [DOI] [PubMed] [Google Scholar]
- 48.Sarkis PT, et al. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177(7):4530–4540. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- 49.Anderson BD, Harris RS. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-beta/RUNX axis. Sci Adv. 2015;1(8):e1500296. doi: 10.1126/sciadv.1500296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desimmie BA, et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426(6):1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anwar F, Davenport MP, Ebrahimi D. Footprint of APOBEC3 on the genome of human retroelements. J Virol. 2013;87(14):8195–8204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kinomoto M, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Research. 2007;35(9):2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogerd HP, et al. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 55.Muckenfuss H, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. Journal of Biological Chemistry. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 56.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. The Journal of Biological chemistry. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 57.Chiu YL, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hulme AE, et al. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390(1–2):199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Esnault C, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433(7024):430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 60.Chaipan C, et al. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol. 2013;87(1):444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakurai D, et al. APOBEC3H polymorphisms associated with the susceptibility to HIV-1 infection and AIDS progression in Japanese. Immunogenetics. 2015;67(4):253–257. doi: 10.1007/s00251-015-0829-2. [DOI] [PubMed] [Google Scholar]
- 62.Refsland EW, et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10(11):e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naruse TK, et al. APOBEC3H polymorphisms and susceptibility to HIV-1 infection in an Indian population. J Hum Genet. 2015 doi: 10.1038/jhg.2015.136. [DOI] [PubMed] [Google Scholar]
- 64.OhAinle M, et al. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4(3):249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol. Chem. 2004;279(51):53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 66.Wang T, et al. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008;377(1):71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonvin M, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43(6):1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 68.Bulliard Y, et al. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol. 2011;85(4):1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thielen BK, et al. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. The Journal of Biological chemistry. 2010;285(36):27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharma S, et al. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun. 2015;6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bransteitter R, et al. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100(7):4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nonaka T, et al. Carboxy-terminal domain of AID required for its mRNA complex formation in vivo. Proc Natl Acad Sci U S A. 2009;106(8):2747–2751. doi: 10.1073/pnas.0812957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc Natl Acad Sci U S A. 1997;94(24):13075–13080. doi: 10.1073/pnas.94.24.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ito S. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sowden MP, Ballatori N, de Mesy Jensen KL, Hamilton Reed L, Smith HC. The editosome for cytidine to uridine mRNA editing has a native complexity of 27S: identification of intracellular domains containing active and inactive editing factors. J Cell Science. 2002;115(1027 – 1039):1027–1039. doi: 10.1242/jcs.115.5.1027. [DOI] [PubMed] [Google Scholar]
- 76.Methot SP, et al. Consecutive interactions with HSP90 and eEF1A underlie a functional maturation and storage pathway of AID in the cytoplasm. J Exp Med. 2015;212(4):581–596. doi: 10.1084/jem.20141157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bennett RP, et al. Nuclear Exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J Biol Chem. 2008;283(12):7320–7327. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 78.Zhen A, et al. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS One. 2012;7(7):e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallois-Montbrun S, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81(5):2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2(5):e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McDougall WM, Smith HC. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochem Biophys Res Commun. 2011;412(4):612–617. doi: 10.1016/j.bbrc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu X, et al. Alternative splicing regulates activation-induced cytidine deaminase (AID): implications for suppression of AID mutagenic activity in normal and malignant B cells. Blood. 2008;112(12):4675–4682. doi: 10.1182/blood-2008-03-145995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28(5):630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takizawa M, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008 doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hersberger M, et al. Regulatable liver expression of the rabbit apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (APOBEC-1) in mice lacking endogenous APOBEC-1 leads to aberrant hyperediting. Biochem J. 2003;369(Pt 2):255–262. doi: 10.1042/BJ20020694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamanaka S, et al. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11(3):321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 87.Saraconi G, et al. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol. 2014;15(7):417. doi: 10.1186/s13059-014-0417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lackey L, et al. APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol. 2012;419(5):301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494(7437):366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med. 2015;21(5):274–284. doi: 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 91.Vieira VC, et al. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio. 2014;5(6) doi: 10.1128/mBio.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Land AM, et al. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem. 2013;288(24):17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caval V, et al. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 94.Chan K, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47(9):1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prochnow C, et al. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445(7126):447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 96.Krzysiak TC, et al. APOBEC2 is a monomer in solution: implications for APOBEC3G models. Biochemistry. 2012;51(9):2008–2017. doi: 10.1021/bi300021s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452(7183):116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 98.Furukawa A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009;28(4):440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Harjes E, et al. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389(5):819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Byeon IJ, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kouno T, et al. Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat Struct Mol Biol. 2015;22(6):485–491. doi: 10.1038/nsmb.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kitamura S, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19(10):1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 103.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456(7218):121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shandilya SM, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18(1):28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu X, et al. Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA. J Biol Chem. 2015;290(7):4010–4021. doi: 10.1074/jbc.M114.624262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shi K, et al. Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain. J Biol Chem. 2015;290(47):28120–28130. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bohn MF, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21(6):1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Siu KK, et al. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakashima M, et al. Structural Insights into HIV-1 Vif-APOBEC3F Interaction. J Virol. 2015;90(2):1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wedekind JE, et al. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J Biol Chem. 2006;281(50):38122–38126. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Betts L, et al. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994;235(2):635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 112.Polevoda B, et al. RNA binding to APOBEC3G induces the disassembly of functional deaminase complexes by displacing single-stranded DNA substrates. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rathore A, et al. The Local Dinucleotide Preference of APOBEC3G Can Be Altered from 5'-CC to 5'-TC by a Single Amino Acid Substitution. J Mol Biol. 2013;425(22):4442–4454. doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marx A, Galilee M, Alian A. Zinc enhancement of cytidine deaminase activity highlights a potential allosteric role of loop-3 in regulating APOBEC3 enzymes. Sci Rep. 2015;5:18191. doi: 10.1038/srep18191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bohn MF, et al. The ssDNA Mutator APOBEC3A Is Regulated by Cooperative Dimerization. Structure. 2015;23(5):903–911. doi: 10.1016/j.str.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Logue EC, et al. A DNA sequence recognition loop on APOBEC3A controls substrate specificity. PLoS One. 2014;9(5):e97062. doi: 10.1371/journal.pone.0097062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Navarro F, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333(2):374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 118.Iwatani Y, et al. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80(12):5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shlyakhtenko LS, et al. APOBEC3G Interacts with ssDNA by Two Modes: AFM Studies. Sci Rep. 2015;5:15648. doi: 10.1038/srep15648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salter JD, et al. A hydrodynamic analysis of APOBEC3G reveals a monomer-dimer-tetramer self-association that has implications for anti-HIV function. Biochemistry. 2009;48(45):10685–10687. doi: 10.1021/bi901642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McDougall WM, Okany C, Smith HC. Deaminase activity on single-stranded DNA (ssDNA) occurs in vitro when APOBEC3G cytidine deaminase forms homotetramers and higher-order complexes. J Biol Chem. 2011;286(35):30655–30661. doi: 10.1074/jbc.M111.269506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shlyakhtenko LS, et al. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J Biol Chem. 2011;286(5):3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anant S, MacGinnitie AJ, Davidson NO. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270(24):14762–14767. [PubMed] [Google Scholar]
- 124.Prohaska KM, et al. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip Rev RNA. 2014;5(4):493–508. doi: 10.1002/wrna.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Friew YN, et al. Intracellular interactions between APOBEC3G, RNA, HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81(8):3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Navaratnam N, et al. Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J Mol Biol. 1998;275(4):695–714. doi: 10.1006/jmbi.1997.1506. [DOI] [PubMed] [Google Scholar]
- 128.Galloway CA, et al. APOBEC-1 complementation factor (ACF) forms RNA-dependent multimers. Biochem Biophys Res Commun. 2010;398(1):38–43. doi: 10.1016/j.bbrc.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maris C, et al. NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA. 2005;11(2):173–186. doi: 10.1261/rna.7190705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oka K, et al. Tissue-specific inhibition of apolipoprotein B mRNA editing in the liver by adenovirus-mediated transfer of a dominant negative mutant APOBEC-1 leads to increased low density lipoprotein in mice. J Biol Chem. 1997;272(3):1456–1460. doi: 10.1074/jbc.272.3.1456. [DOI] [PubMed] [Google Scholar]
- 131.Lau PP, et al. Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem. 1997;272(3):1452–1455. doi: 10.1074/jbc.272.3.1452. [DOI] [PubMed] [Google Scholar]
- 132.Lau PP, et al. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc Natl Acad Sci U S A. 1994;91(18):8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bhagwat AS, Carpenter MA, Bujnicki JM. Is AID a monomer in solution? DNA Repair (Amst) 2008;7(3):349–350. doi: 10.1016/j.dnarep.2007.10.009. author reply 351-2. [DOI] [PubMed] [Google Scholar]
- 134.Brar SS, et al. Activation-induced deaminase, AID, is catalytically active as a monomer on single-stranded DNA. DNA Repair (Amst) 2008;7(1):77–87. doi: 10.1016/j.dnarep.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li J, et al. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426(6):1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004 doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 137.Svarovskaia ES, et al. Human APOBEC3G is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. The Journal of Biological chemistry. 2004 doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 138.Wang T TC, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. Journal of Molecular Biology. 2008;375(4):1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 139.Huthoff H, et al. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5(3):e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chelico L, et al. A structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J Biol Chem. 2010 doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shlyakhtenko LS, et al. Atomic force microscopy studies of APOBEC3G oligomerization and dynamics. J Struct Biol. 2013 doi: 10.1016/j.jsb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chelico L, Pham P, Goodman MF. Stochastic properties of processive cytidine DNA deaminases AID and APOBEC3G. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):583–593. doi: 10.1098/rstb.2008.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Richards C, et al. The Binding Interface between Human APOBEC3F and HIV-1 Vif Elucidated by Genetic and Computational Approaches. Cell Rep. 2015;13(9):1781–1788. doi: 10.1016/j.celrep.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]