Droplet organelles? (original) (raw)
Abstract
Cells contain numerous, molecularly distinct cellular compartments that are not enclosed by lipid bilayers. These compartments are implicated in a wide range of cellular activities, and they have been variously described as bodies, granules, or organelles. Recent evidence suggests that a liquid–liquid phase separation (LLPS) process may drive their formation, possibly justifying the unifying term “droplet organelle”. A veritable deluge of recent publications points to the importance of low‐complexity proteins and RNA in determining the physical properties of phase‐separated structures. Many of the proteins linked to such structures are implicated in human diseases, such as amyotrophic lateral sclerosis (ALS). We provide an overview of the organizational principles that characterize putative “droplet organelles” in healthy and diseased cells, connecting protein biochemistry with cell physiology.
Keywords: Liquid‐liquid phase separation, low‐complexity domain, nuclear bodies, RNP granules
Subject Categories: Membrane & Intracellular Transport, Neuroscience, RNA Biology
Glossary
Many of the terms used in the recent literature have meanings that are overlapping or refer to subtle differences between concepts. Here, we provide the classical definitions for these terms and comment on their usage.
Liquid–liquid phase separation (LLPS)
It is the phenomenon in which solutes spontaneously separate into a demixed liquid phase suspended within the bulk solvent. Conventionally, the solute is a flexible chain polymer, but the term is also applied to biological macromolecules that may not have a flexible chain‐like tertiary structure.
Low‐complexity domain (LCD)
It is a region within a protein that contains an overrepresentation of a subset of amino acids in the primary sequence. Often this occurs as a repeat motif, but repeats are not a requirement.
Intrinsically disordered regions (IDRs)
These are the protein domains, often containing low‐complexity sequences that appear to lack well‐defined secondary and tertiary structure. Some IDRs have been determined experimentally, while others are inferred and may be structured in certain contexts.
Droplet
It is the spherical fluid morphology adopted by phase‐separated macromolecules in solution. Droplets have measureable surface tension and viscosity. Molecular constituents diffuse within them and can exchange with the bulk solvent.
Hydrogel
It is the hydrated matrix formed by cross‐linked protein polymers. These polymers are best thought of as a stable colloidal solid suspended in water.
Aggregate
It is a solid formation composed of proteins that have precipitated from solution. Precipitation occurs because water is excluded from macromolecular interactions to the extent that the protein mass is no longer stably suspended
Amyloid
It is a class of protein aggregate characterized by a semi‐regular structure formed by the stacking of β sheets among protein monomers in trans. They are experimentally identified by characteristic X‐ray diffraction patterns and staining with the dye, thioflavin T.
Prion‐like domain
It is a protein region characterized by sequence similarity to that of prototypical yeast prion proteins domains. These can be thought of as a special case of low‐complexity domain
Introduction
Cells spatially organize biochemical reactions, a characteristic that is fundamental to life but often evades analysis. Lipid membranes create discrete chemical environments within canonical organelles and achieve separation of constituents from the bulk cytoplasm. Enclosing membrane‐bound compartments requires dedicated machinery to construct and maintain the lipid bilayer and transport substances across them, thus expending energy (Grossman et al, 2012; Neupert, 2015). Many organelles lack lipid bilayers, circumventing these issues and introducing the potential for greater dynamics and alternative mechanisms of regulation. Nuclear structures, such as nucleoli and Cajal bodies, regulate ribonucleoprotein (RNP) assembly in this manner (Mao et al, 2011; Machyna et al, 2013). In the cytoplasm, stress granules and P bodies regulate RNA stability and protein translation in response to cellular stimuli (Buchan & Parker, 2009). Accumulating evidence suggests that such non‐membrane‐bound organelles behave as fluid droplets, which undergo LLPS (see Glossary). The molecular mechanisms that underlie their form and function are currently under intense investigation, and this review focuses on the emergent common principles.
We survey the literature on systems that exhibit LLPS and related behavior in order to bridge the in vivo studies of cellular structures and the molecular understanding afforded by in vitro studies of proteins, RNAs, and their interactions. Our focus will be on the most recent work that has shed light on the molecular mechanisms that lead to LLPS, but we also reference previous key studies to provide a prelude to recent developments. The review closes by returning to the biological consequences of competing models for LLPS systems and discusses how they may relate to diseases characterized by the formation of aberrant protein aggregates. Our goal is to synthesize an outlook for the relevance of LLPS at both molecular and cellular scales—a number of excellent recent reviews provide a more detailed analysis of the individual topics covered (Weber & Brangwynne, 2012; Malinovska et al, 2013; Hyman et al, 2014; Toretsky & Wright, 2014; Uversky, 2015). We emphasize several unresolved issues in the field and attempt to address controversy where it arises.
Nuclear and cytoplasmic bodies are the sites of RNP biogenesis
Many non‐membrane‐bound organelles can be found in the nucleus and were first observed there over a century ago (Gall, 2000). Figure 1 illustrates how a few of these organelles are positioned. Nuclear bodies have been investigated extensively regarding the exchange of constituents with surrounding nucleoplasm. For most components, residence times are on the order of seconds with the most stable constituents exchanging within tens of seconds (Phair & Misteli, 2000; Dundr et al, 2004). The dynamic nature of nuclear bodies likely underlies their function, which the most recent studies of LLPS strive to explain. The most prominent nuclear body is the nucleolus, which forms on active rDNA loci and is the site of pre‐rRNA processing and pre‐ribosomal subunit assembly (Boisvert et al, 2007; Pederson, 2011; Falahati et al, 2016). Similarly, the Cajal body forms on active snRNA loci and is the site of snRNA processing, snRNP assembly, and snRNP surveillance (Frey et al, 1999; Stanek & Neugebauer, 2004; Machyna et al, 2013; Novotny et al, 2015). Nuclear speckles and paraspeckles are more granular in morphology, contain mRNAs and their binding proteins, and form on two long non‐coding RNAs (lncRNAs) MALAT1 and NEAT1, respectively (Mao et al, 2011). Many of the specific functions of these subdomains are still poorly characterized, but at the descriptive level, they are consistent with phase‐separated systems.
Figure 1. In homeostatic cellular conditions, dynamic fluid droplets demix from surrounding nucleoplasm.

(A) Nucleoli, Cajal bodies (CBs), histone locus bodies (HLBs), speckles, and paraspeckles participate in RNA and RNP biogenesis in the nucleus. Associated with chromosomal loci, these nuclear bodies contain specific RNAs and proteins that pass in and out of nuclear bodies during RNP assembly. Unstable RNAs concentrate in P bodies in the cytoplasm, where mRNA decay factors co‐localize. (B) Analogous dynamics and fluid properties are obtained when a purified RNA‐binding protein with a low‐complexity region is incubated in with RNA and observed over time in vitro. (C) Electron micrograph of a droplet showing overall spherical shape with an irregular outline. Micrographs reproduced from Li et al (2012).
Cytoplasmic bodies are more granular in morphology and have functions often related to translational control and/or mRNA stability. The processing body (P body) falls into this second category, in which translation is stalled and transcripts are targeted for degradation by exonucleases (Parker & Sheth, 2007) or selective reactivation of translation (Arribere et al, 2011). Stress granules are related to P bodies, in that they contain translationally repressed mRNA, but form in response to heat, osmotic, and chemical stress stimuli. Figure 2 illustrates changes seen in several bodies during cellular stress including the altered structure of the nucleolus. Stress granules are an example of a phase‐separated structure where the physical properties change in response to a stimulus (Boulon et al, 2010). Others including nuclear speckles, histone locus bodies, and paraspeckles also respond to stress with a variety of morphological and biochemical changes (Carmo‐Fonseca et al, 1992; Lamond & Spector, 2003; Bongiorno‐Borbone et al, 2010; Boulon et al, 2010). Stress granules contain factors that stall translation, as well as a number of RNA‐binding proteins, associated with ALS and other diseases. The normal physiological roles of many stress granule proteins are still unclear (Anderson & Kedersha, 2008; Li et al, 2013).
Figure 2. During cellular stress, hydrogel‐like assemblies form.
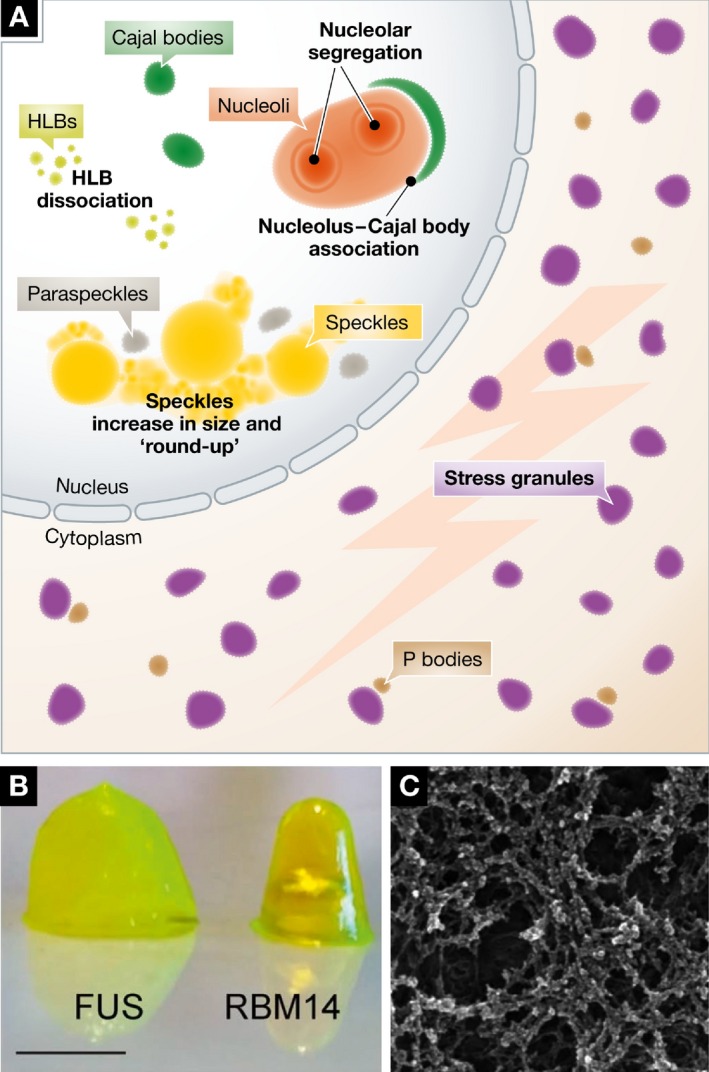
(A) Stressors like heat shock, UV light, and transcription inhibition can cause cellular bodies to disassemble (CBs), change morphology (nucleoli, nuclear speckles), or appear de novo (stress bodies). (B) These granules contain proteins that form hydrogels and ultimately form an intricate proteinaceous network or aggregate (C). Micrographs reproduced from Hennig et al (2015).
Some cytoplasmic RNA granules are developmentally important. In C. elegans, the germ cell lineage is specified by the asymmetric inheritance of P granules during mitosis. The exact function of these RNP granules is unknown, but they have some similarity to P bodies and are functionally implicated in translational control of the germ line (Seydoux & Braun, 2006). In mammals, germ line specification follows a different mechanism, but germ cells still display characteristic RNP structures known as nuage or the chromatoid body. Nuage contains many RNAs as well as several helicases, endonucleases, and proteins involved in miRNA‐mediated degradation of RNA (Kotaja & Sassone‐Corsi, 2007).
RNP bodies and granules display characteristics of phase‐separated liquids
Even as progress has been made on elucidating the biological functions of these nuclear and cytoplasmic bodies, their physical properties have only recently come to light through high‐resolution live‐cell imaging. Like nucleoli, Cajal bodies fuse and split (Platani et al, 2000). In 2005, Gall and colleagues speculated that Cajal bodies were actually “semi‐fluid spheres suspended in semi‐fluid nucleoplasm” (Handwerger et al, 2005). This conclusion was based on observed shape, permeability, and differential protein concentrations between the nucleoplasm and the large Cajal bodies present in Xenopus germinal vesicles. In 2009, seminal work on the P granules of C. elegans demonstrated their liquid‐like properties and that they localize to the future germ cell cytoplasm by dissolving and condensing rather than by moving as discrete objects through the cytoplasm (Brangwynne et al, 2009). Extrachromosomal nucleoli of Xenopus laevis were also shown to exhibit properties expected of liquid droplets, including surface tension and fluidity (Brangwynne et al, 2011). Advanced microscopy techniques, including tracking of subcellular structures over time, turned the conjecture that nuclear and other organelles undergo LLPS into a well‐characterized phenomenon.
These apparent manifestations of LLPS in vivo were striking and harken back to studies of other biomolecules that undergo phase separation in vitro. In 2006, Görlich and colleagues showed that the yeast FG (phenylalanine–glycine) repeat proteins of the nuclear pore complex are capable of condensing into a hydrated gel matrix (Frey et al, 2006). These gels showed no fluorescence recovery after photobleaching (FRAP), suggesting that the hydrogel is not fluid and instead may trap constituent molecules in an immobile meshwork. Cross‐β amyloid‐like interactions are likely to underlie hydrogel formation (see Glossary) and are disrupted by nuclear transport factors. This “melting” of the gel matrix by some proteins but not others effectively constitutes a selectivity filter (Ader et al, 2010; Schmidt & Görlich, 2015). Species other than fungi are less likely to have FG nucleoporins that show strong amyloid character, but all rely on hydrophobic interactions in their formation (Labokha et al, 2013; Schmidt & Görlich, 2015). This in vitro molecular behavior is distinct from that observed for constituents of Cajal bodies and nucleoli in vivo. Specifically, the GFP florescence recovery of coilin and SMN, two core components of Cajal bodies, indicates that they exchange rapidly with bulk nucleoplasm (Phair & Misteli, 2000; Dundr et al, 2004; Brangwynne et al, 2011), which would not be expected of hydrogels. Comparing the findings of in vitro studies of the nuclear pore components and of nuclear bodies in vivo, it would seem that there are two different ways that biological systems can separate from the bulk solvent: either as solid hydrogels or as LLPS droplets.
In vitro systems show fundamental properties of phase separation
New assays have been devised to probe the underlying physical and molecular nature of phase‐separating systems. Using purified protein domains derived from signaling proteins of the N‐WASP pathway, Rosen and colleagues were able to show that multivalency is sufficient to drive the phase separation of concatenated SH3 domains that bind to concatenated proline‐rich motifs (Li et al, 2012). Figure 1B depicts the fusion of droplets containing concatenated SH3 domains (Li et al, 2012), consistent with the proposal that they possess fluid properties. In the same study, RNA with multiple binding motifs was also able to form droplets in combination with the multivalent protein PTB, which exhibit FRAP and form above a critical concentration threshold. Other work has also shown that the properties of phase‐separated proteins depend heavily upon phosphorylation state (Kwon et al, 2013, 2014; Wang et al, 2014; Su et al, 2016), as discussed below. One investigation into these effects on FUS, an RNA‐ and DNA‐binding protein found in both cytoplasm and nucleoplasm, highlighted the importance of post‐translational modification as a means of regulating phase separation and implicated low‐complexity protein domains (LCDs; see Glossary) as fundamental to body formation (Kato et al, 2012).
Proteins with low‐complexity domains are often found in bodies
The bodies so far discussed almost all contain at least one, if not several, disordered proteins that notably contain characteristic LCDs. Many of these proteins are listed in Table 1. It is worth clarifying some of the terminology that has been used to describe LCDs: Several studies have designated LCD sequences based on their sequence similarity to the yeast prion proteins (see Glossary) (Alberti et al, 2009; Lancaster et al, 2014). These so‐called prion‐like domains seem to be related to important diseases and have a propensity to aggregate (King et al, 2012); however, this term is too narrow for the present discussion of phase‐separating systems. “Low‐complexity domain” is currently the most satisfying term available because it includes the prion‐like domains as well as disordered domains with other amino acid compositions that might be important for forming phase‐separated systems like stress granules and P bodies (Decker et al, 2007; Sun et al, 2011).
Table 1.
Protein components of cellular compartments that phase separate in vitro
| Protein (aa) | Low‐complexity motif(s) | Structured domains | Cellular compartment | In vitro morphology | Disease association | References |
|---|---|---|---|---|---|---|
| DDX4 (724) | 19aa R/G‐rich16aa R/G/S9aa P‐rich | DEXDcHELICc | NuageRNP granules | Droplets* | N/A | Nott et al (2015) |
| eIF4GII (914)yeast | 38aa N/K/Y21aa T/P13aa S/R17aa E/A29aa N/S | MIF4G | Stress granules | Droplets* | N/A | Lin et al (2015), Molliex et al (2015) |
| EWS (656) | 19aa Y/A/S82aa A/Q/T85aa Q/S/Y33aa R/G22aa D‐rich60aa R/G/P86aa R/G/P | RRMZnF_RBZ | DNA damage sites | Hydrogel* | ALS | Kwon et al (2013), Altmeyer et al, (2015) |
| Fibrillarin (321) | 72aa R/G | Methyl‐transferase (fibrillarin) | NucleolusCajal body | Droplets | Autoimmunity | Berry et al (2015) |
| FUS (526) | 156aa S/G/Q55aa RGG76aa RGG | RRMZnF | Paraspecklesstress granules | Dropletshydrogel | ALS | Altmeyer et al (2015), Burke et al (2015), Murakami et al (2015), Patel et al (2015) |
| hnRNP A1/A2 (372/353) | 180aa G/S/R/Q | 2 RRMs | Stress granules | Dropletshydrogel | ALS, IBM, Paget's, FTLD | Kim et al (2013), Lin et al (2015), Molliex et al (2015), Xiang et al (2015) |
| LAF‐1 (708) C. elegans | 130aa G/R75aa G/R/Q | DEXDcHELICc | P granules | Droplets | N/A | Elbaum‐Garfinkle et al (2015) |
| Lsm4 (187)yeast | 173aa N/R | Sm | P bodiesstress granules | Droplets* | N/A | Lin et al (2015) |
| PTB (557) | 13aa S/N/A18aa A‐rich30aa A‐rich | 4 RRMs | Nuclear specklesperi‐nucleolar | Droplets | N/A | Li et al (2012), Lin et al (2015) |
| Pub1 (453)yeast | 55aa N/M33aa Q‐rich | 3 RRMs | Stress granules | Droplets* | N/A | Lin et al (2015) |
| RBM14 (669) | 300aa A/R/S/Q/P | 2 RRMs | Paraspeckles | Hydrogel | ALS | Hennig et al (2015) |
| SRSF2 (221) | 113aa R/S | RRM | Nuclear speckles | Hydrogel | MDSleukemias | Kwon et al (2014) |
| TAF15 (589) | 148 S/G/Q/Y39 RGG26aa R/G186aa RGG/YGG | RRMZnF_RBZ | DNA damage sites | Hydrogel* | ALSFTLD | Kwon et al (2013), Altmeyer et al (2015) |
| TDP‐43 (414) | 43aa G/F/N9aa A‐rich17aa Q/N37aa S/G | 2 RRMs | Stress granules | Droplets | ALS | Burke et al (2015), Molliex et al (2015) |
| Tia1 (386) | 23aa S/T/Q | 3 RRMs | Stress granules | Droplets* | Welander distal myopathy | Lin et al (2015) |
| Whi3 (729) A. gossypii | Poly‐Q | RRM | Cytoplasmic RNP granules | Droplets* | N/A | Zhang et al (2015) |
Two proteins containing LCDs have come to the forefront of much recent research due to their relevance to ALS and frontotemporal dementia: FUS and hnRNPA1. FUS is associated with several nuclear and cytoplasmic bodies, including paraspeckles and stress granules. Under normal conditions, FUS is found in the nucleus where it takes part in DNA repair and transcriptional regulation (Wang et al, 2008, 2013). For reasons that are not entirely clear, stress stimuli result in the export of FUS to the cytoplasm where it joins stress granules (Bentmann et al, 2012). These studies have identified the importance of both RNA‐binding properties and the presence of the LCD within FUS. Similar to FUS, nuclear hnRNPA1 has two RNA recognition motifs (RRMs) and an LCD, is recruited to stress granules, and phase separates in vitro (Kim et al, 2013; Kwon et al, 2014). Last year, significant inroads were made into understanding how FUS, hnRNPA1, and other LCD proteins (see Table 1) can generate fluid cellular bodies as outlined in several reviews (Malinovska et al, 2013; Toretsky & Wright, 2014; Uversky, 2015).
Electrostatic interactions and LLPS
What causes an LCD to transition into either a liquid droplet or a solid hydrogel is a matter of debate, and the recent literature considers several contributing factors (Burke et al, 2015; Lin et al, 2015; Xiang et al, 2015). Isolated LCDs from a variety of proteins that scaffold cellular structures are sufficient for phase‐separating behavior. Strikingly, the low‐complexity sequences of eIF4GII, hnRNPA1, and FUS all form liquid droplets without the addition of any other components (Lin et al, 2015). The integrity of the full low‐complexity domain is essential for the formation of paraspeckles, and a tyrosine to serine mutation in the repeat motif in RBM14 clearly alters the morphology of the resulting hydrogel (Hennig et al, 2015). Figure 2B and C shows the hydrogel cross‐linking that may occur during droplet maturation, a process described for numerous intrinsically disordered regions (IDRs) in vitro (see Glossary). Typically, cross‐linking is induced over time by manipulating protein and/or salt concentration, temperature, and molecular crowding (Lin et al, 2015; Nott et al, 2015). This is in contrast to the fluid droplets shown in Fig 1B and C, which have a notably different structure as seen in electron micrographs. Thus, electrostatic interactions may provide an underlying force for LLPS, though the evidence for this is taken from experiments that were performed above physiological protein concentrations or below physiological salt concentrations. It remains to be seen whether hydrogel formation and stability also depend on polar contacts.
By demonstrating that the two‐phase state of these systems requires a low ionic strength, these studies allow us to speculate on the importance of charge and polarity in the composition of LCDs as well as the relevance of post‐translational modifications that can render amino acids either more or less charged. For example, hydrogel recruitment of the SRSF2 LCD, which is serine and arginine‐rich, is blocked by phosphorylation (Kwon et al, 2014). Intriguingly, phosphorylation can also alter the range of structural ensembles and binding interactions for some IDRs (Arai et al, 2015), suggesting that some IDRs may become less disordered or even structured when post‐translationally modified. Experiments employing phosphomimetic or alanine replacements could provide decisive evidence here. While existing data indicate that electrostatic interactions between LCDs are important, we have yet to address the fact that LLPS often coincides with the presence of another charged macromolecule: RNA.
Roles for RNA in vitro and in vivo
Until recently, the role of RNA in droplet formation had been relatively neglected. New studies re‐invigorate considerations of RNA's role in vitro and in vivo. In considering how electrostatic interactions might mediate the formation of liquid droplets, recall that RNA is concentrated in most bodies and granules in vivo. Because of its anionic phosphate backbone, RNA is a highly charged molecule that can potentially contribute to electrostatic interactions with positively charged residues in LCDs. Indeed, RNA enhances the fluid properties of droplets formed by the P granule component LAF1, which contains positively charged arginine–glycine–glycine (RGG) repeats (Elbaum‐Garfinkle et al, 2015). This increase in fluidity might be related to a general promotion of LLPS by RNA, as is also observed in the formation of fibrillarin droplets (Berry et al, 2015). Poly‐(ADP ribose), or PAR, is a polynucleotide synthesized upon DNA damage; PAR acts as a signal in the localization of the DNA repair machinery to sites of DNA damage (Table 1). The chemical structure of PAR strongly resembles RNA and appears to nucleate liquid phase‐separated regions through electrostatic interactions with RGG repeats found in FUS, EWS, and TAF15 (Altmeyer et al, 2015; Patel et al, 2015). Like LAF‐1, these proteins also contain RNA‐binding domains and two regions of RGG repeats, but only the N‐terminal LCD is sufficient for LLPS. Synergistic effects between these regions may contribute to in vivo LLPS by binding multiple proteins and RNAs and associating with other LCDs (Chen et al, 2011; Phan et al, 2011; Burke et al, 2015). That said, the potential for non‐canonical binding of RNA by low‐complexity domains should not be discounted. Indeed, the Cajal body scaffolding protein coilin, which contains RGG repeats as well as other low‐complexity sequences, was recently shown to bind RNA in the absence of an annotated RNA‐binding domain (Machyna et al, 2014). Thus, RNA may play a role as a generic poly‐anion or may regulate LLPS through specific interactions with proteins.
A second way that RNA can participate in LLPS is through protein–RNA interactions mediated by canonical RNA‐binding domains, such as RNA recognition motifs (RRMs), zinc fingers (ZnFs), and KH domains. Indeed, droplet scaffolding proteins frequently contain one or more RNA‐binding domain (Table 1). RNA sequence can afford multivalency to RNA–protein interactions by presenting repeated motifs for protein binding. For example, the highly structured RNA‐binding protein, PTB, forms droplets when added to concatemerized RNA target sequences (Li et al, 2012). More recently, a study of Whi3, a poly‐Q protein that forms cytoplasmic mRNA granules in the fungus Ashbya gossypii, showed that droplet formation depends on a functional RRM and binding to the CLN3 mRNA (Zhang et al, 2015). Full‐length Whi3 never formed fibrous structures in vivo or in vitro, while the LCD alone formed filaments that became less soluble over time. Similar results were obtained in studies of FUS (see below), which was shown to transition from a droplet to filaments over time in vitro (Patel et al, 2015) and to form aggregates in the context of hydrogels (Hennig et al, 2015). The potential role of the FUS RRM in modulating LLPS, aggregate, and/or filament formation has not yet been directly analyzed.
Because RNA‐rich nuclear bodies—Cajal bodies, nucleoli, histone locus bodies, and paraspeckles—can form at transcription sites, nascent RNA likely plays a role in their nucleation (Bond & Fox, 2009; Mao et al, 2011; Shevtsov & Dundr, 2011; Machyna et al, 2014; Falahati et al, 2016). The combination of an RRM with an LCD should be ideal for scaffolding transcriptionally dependent LLPS structures. Indeed, many proteins within the paraspeckle contain N‐terminal RRMs and C‐terminal LCDs (Table 1), and both FUS and RBM14 form droplets and aggregates (Hennig et al, 2015). Recent investigations of hnRNP A1 indicate that RNA binding by N‐terminal RRMs modulates droplet formation promoted by the protein's C‐terminal LCD (Lin et al, 2015; Molliex et al, 2015). Interestingly, fusion of PTB's RRMs to the IDRs of a variety of RNA‐binding proteins rendered these IDRs capable of droplet formation in the presence of RNA, and hnRNP A1's RRM was essential for droplet formation. Thus, nucleic acids may serve in the formation of LLPS cellular structures, by providing a multivalent platform to seed phase separation and by buffering the protein–protein contacts to promote fluidization.
Different molecular interactions create distinct protein states
Why would cells risk the use of systems that promote toxic aggregation? A major focus of the recent work on LLPS has been on determining which LCDs tend to form liquid droplets, hydrogels, or solid amyloid aggregates (Kato et al, 2012; Kwon et al, 2013; Altmeyer et al, 2015; Burke et al, 2015; Courchaine & Neugebauer, 2015; Hennig et al, 2015; Kroschwald et al, 2015; Lin et al, 2015; Molliex et al, 2015; Murakami et al, 2015; Patel et al, 2015). Many neurological diseases include protein aggregation in their pathology. Figure 3 conceptualizes these amyloid‐like aggregates and depicts their formation. The solid stress granules of yeast are controlled by the disaggregase machinery and must be disassembled before the cell may resume growth (Cherkasov et al, 2013; Kroschwald et al, 2015). In addition, yeast stress granules can be disassembled by autophagy in response to cellular conditions (Buchan et al, 2013). In higher organisms, there are likely to be more subtle control processes that determine when LCDs remain liquid and when they aggregate. The idea that proteins have a range of states of matter has been raised previously and is an important concept in the regulation of these processes in vivo (Weber & Brangwynne, 2012).
Figure 3. In disease, amyloid fibers arise from droplets.
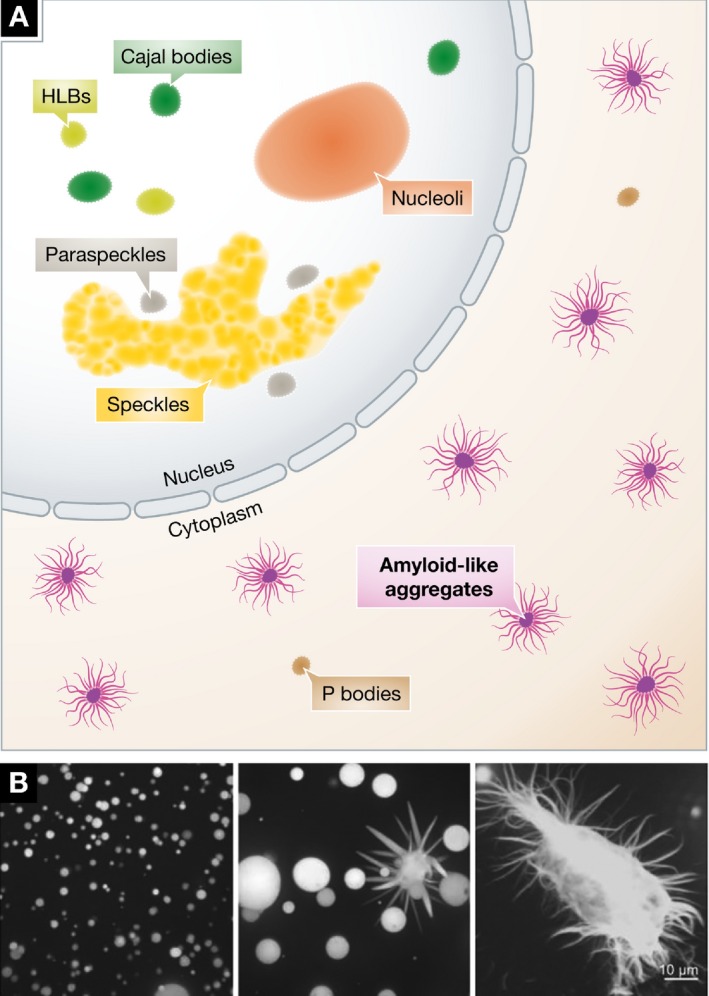
(A) Amyloid fibers can form in the nucleus or cytoplasm when constituent low‐complexity proteins are mutant. Concomitant changes in nuclear morphology have not been reported. (B) In vitro, fiber formation by mutant proteins, such as FUS, can follow droplet formation. Micrographs reproduced from Patel et al (2015).
The dynamic and fluid nature of LLPS systems depends on the attractive potential between the constituent protein and RNA molecules. By buffering ionic strength within a liquid droplet, RNA may be able to maintain the two‐phase state near a theoretical “cloud point”. Two recent papers have demonstrated that droplets formed in LLPS have properties that correspond to those predicted by Flory–Huggins theory (Molliex et al, 2015; Nott et al, 2015). This takes into account the entropic cost of confining large polymers (proteins and RNA) into a localized region, which is counterbalanced by the enthalpy of electrostatic contacts between components that attract (Flory, 1942; Huggins, 1942). This framework from polymer chemistry has broad applicability in biochemistry and has been discussed in detail elsewhere (Hyman et al, 2014). From this perspective, the observed amyloid formation is likely to be a secondary consequence of increased protein concentration rather than LLPS being a consequence of limited amyloid‐like interactions. Therefore, cells can use this property to rapidly respond to changing conditions, with disassembly and assembly occurring on time scales on the order of seconds. Thus, these results substantiate the relevance of LLPS as a framework for understanding droplet formation, but leave open the question of how droplets, hydrogels, and amyloids may be related to one another on a molecular scale.
The cloudy relationship between protein conformation and phase separation
Despite the rapid and substantial progress on this topic, conflicting perspectives on the molecular mechanisms of LLPS have emerged. These will have consequences for understanding how diseases like ALS arise and what therapies might be developed. Three proteins serve as the best examples of this controversy: FUS and the hnRNPs A1 and A2. All contain LCDs and bind nucleic acids (see Table 1). Furthermore, these proteins have been shown to form the droplet, hydrogel, and amyloid aggregate structures seen in Figs 1, 2, 3. In several studies, the formation of these structures has been posited to be the result of the slow formation of cross‐β interactions between LCDs (Hennig et al, 2015; Lin et al, 2015). The intriguing aspect of such a mechanism is that only one interaction causes the formation of both droplets and amyloids, making the differences between each state a matter of kinetics. While wild‐type FUS droplets eventually form fiber structures (Fig 3B), disease mutations drastically shorten the time required for FUS aggregation (Patel et al, 2015). Experiments have also shown that some LCDs, including that of hnRNPA1, undergo an irreversible droplet condensation after a certain time period or after a number of cycles (Lin et al, 2015; Molliex et al, 2015). This lends further support to the idea that cross‐β interactions require time to form hydrogels or aggregates. In an investigation of hnRNPA2 (Xiang et al, 2015) show that a cross‐β structure is found in liquid droplets, hydrogels, and in vivo. This use of a chemical footprinting method would seem to confirm that these manifestations of LCDs lie on a continuum.
Other experiments lend support to an alternative interpretation: that the molecular mechanisms for LLPS and solid aggregation are separable. The fact that droplet condensation is reversible in certain conditions could indicate that a Flory–Huggins phase separation occurs first, but that the high local concentration of an LCD forms cross‐β interactions (Lin et al, 2015; Molliex et al, 2015). This would require that the proteins remain disordered in droplets. This was observed in an NMR study of FUS showing that droplets maintain the disordered state (Burke et al, 2015). Here, we are presented with a conundrum, because the same LCD cannot be both disordered (as described here) and simultaneously participate in cross‐β interactions (as described above). hnRNPA1, which contains a hexapeptide repeat that is frequently mutated in ALS and causes amyloid fibers to form, complicates matters as this repeat is not necessary for LLPS (Molliex et al, 2015). Indeed, this hexapeptide region alone is insufficient to cause precipitation, but when it is inserted into a globular protein like RNase A, it creates robust amyloid fibers (Teng & Eisenberg, 2009; Kim et al, 2013). At minimum, these results indicate that phase separation and precipitation are highly context dependent. More work must be done to determine whether all phase separations are the result of a unified process.
Biological roles for LLPS and amyloids
Molecular mechanisms notwithstanding, the range of properties displayed by liquid droplet systems must be leveraged by the cell. In the Cajal body, the increased local concentration of its components might be a kinetic driver of snRNP assembly, as predicted by simulation (Klingauf et al, 2006). The ability to promote reactions would be a powerful use of LLPS, but would likely require the droplet to remain highly fluid. The maturing droplets observed by Lin et al (2015) might not be as advantageous to reactions, and instead sequester mRNAs to prevent translation during stress, which is the proposed role of the stress granule (Anderson & Kedersha, 2008). LLPS protein mixtures might be involved in vastly unrelated structures as well. The putative spindle matrix has been hypothesized to have such an underlying mechanism requiring the spindle protein BugZ (Jiang et al, 2015). More work must be done on all fronts to determine how LLPS affects function as well as to show how systems with known function might be leveraging a phase separation‐like mechanism.
Even amyloids, which are classically associated with human disease, have important physiological functions (Fowler et al, 2007). Bacteria and fungi have been shown to use fibril formation for its mechanical properties and to promote host invasion processes. Amyloid aggregation is used as a regulatory mechanism for heterokaryon formation in fungi, and amyloid‐like oligomers are essential for long‐term memory formation in Drosophila (Majumdar et al, 2012). Amyloid potentiates mammalian melanin production, and a number of endocrine hormones are stored in an amyloid‐like state before dissociation upon release (Fowler et al, 2006; Maji et al, 2009). More recently, it has been shown that regulated amyloid‐like protein aggregates control gene expression during yeast gametogenesis (Berchowitz et al, 2015). These apparent functions for amyloid suggest that this quaternary structure is not necessarily aberrant, but rather that other regulatory failures cause disease.
Disease states and failure of LLPS regulation
Many of the proteins currently used as models for LLPS and fibril formation are linked to ALS and frontotemporal lobar degeneration (FTLD). The most extensively studied are hnRNP proteins, FUS, and RBM14 (Table 1), which contribute to ALS pathology (Hennig et al, 2015; Molliex et al, 2015; Patel et al, 2015). These proteins all bear known mutations that are either causative in familial disease or correlate with spontaneously arising disease. For FUS, a possible mechanism of neurotoxicity has emerged: ALS‐linked FUS mutants sequester a variety of proteins involved in RNA metabolism, suggesting that the misregulation of LLPS could cause disease through downstream effects on gene expression even if amyloid structures are not toxic on their own (Murakami et al, 2015). While it remains unclear what contexts allow amyloid promoting peptides to induce fibrils, disease‐linked mutations facilitate their formation. Whether the amyloid plaques seen in Alzheimer's and Huntington's diseases also have a relationship to LLPS mechanisms remains to be seen. These diseases often do not manifest until later in life, suggesting that compensatory mechanisms may fail during aging. One such example is the autophagy process, which is responsible for clearing stress granules that are not disassembled by other means (Buchan et al, 2013). Defects in the autophagy receptor in ALS patients are associated with amyloid aggregation of stress granule proteins in neurons (Majcher et al, 2015). What dictates whether a stress granule simply reverses phase separation or must be targeted for total enzymatic degradation is unknown.
Understanding LLPS provides an opportunity to understand how we might intervene in disease. If we consider the formation of toxic amyloids to be a continuous process, the mechanism must be unified in some way. Therefore, any intervention in amyloid formation might also interfere with the normal functions of the phase‐separated body. On the other hand, we might consider a model where amyloid formation occurs independently of LLPS, but is promoted by the high local concentration of certain proteins. In this scenario, we might be able to block amyloid formation without disrupting phase‐separated bodies. Further, these bodies are not just regulated by the phase separation process, but also by disaggregase activity, allowing cells to guard against toxicity by digesting bodies and fibrils that will not dissolve. It will be interesting to learn the fate of RNAs present in granules undergoing disaggregation or autophagy.
Conclusions
While cellular bodies lacking membranes have been known for decades, whether they behave as liquid droplets in vivo has now become a pressing question. Recent advances have brought us a long way toward understanding the intermolecular interactions underlying these phenomena, including the importance of RNA as a structural component and regulator of LLPS. Most of the recent work has focused on in vitro characterization of stress granule proteins. If we are to form a generalizable understanding of LLPS in cells, we must extend these studies to in vitro systems that more faithfully reproduce the molecular complexity of the in vivo situation, by including protein and RNA interactors. Furthermore, we must examine how non‐equilibrium processes mediated by chaperones, helicases, kinases, and proteases alter the dynamics of LLPS and amyloid formation. The field looks forward to these illuminating tests of current hypotheses that may allow us to formally adopt unifying terminology, such as “droplet organelle”, to reflect common mechanisms in the formation and function of all cellular compartments lacking membranes.
Conflict of interest
The authors declare that they have no conflict of interest.
The EMBO Journal (2016) 35: 1603–1612
See the Glossary for abbreviations used in this article.
References
- Ader C, Frey S, Maas W, Schmidt HB, Gorlich D, Baldus M (2010) Amyloid‐like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci USA 107: 6281–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, Lukas J (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP‐ribose). Nat Commun 6: 8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2008) Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Arai M, Sugase K, Dyson HJ, Wright PE (2015) Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci USA 112: 9614–9619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Doudna JA, Gilbert WV (2011) Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol Cell 44: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C (2012) Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA‐binding protein of 43 kDa (TDP‐43). J Biol Chem 287: 23079–23094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A (2015) Regulated formation of an amyloid‐like translational repressor governs gametogenesis. Cell 163: 406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP (2015) RNA transcription modulates phase transition‐driven nuclear body assembly. Proc Natl Acad Sci USA 112: E5237–E5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorno‐Borbone L, De Cola A, Barcaroli D, Knight RA, Di Ilio C, Melino G, De Laurenzi V (2010) FLASH degradation in response to UV‐C results in histone locus bodies disruption and cell‐cycle arrest. Oncogene 29: 802–810 [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010) The nucleolus under stress. Mol Cell 40: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732 [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA (2011) Active liquid‐like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA 108: 4334–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153: 1461–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL (2015) Residue‐by‐residue view of in vitro FUS granules that bind the C‐terminal domain of RNA polymerase II. Mol Cell 60: 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo‐Fonseca M, Pepperkok R, Carvalho MT, Lamond AI (1992) Transcription‐dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 117: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T (2011) Deciphering arginine methylation: tudor tells the tale. Nat Rev Mol Cell Biol 12: 629–642 [DOI] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel‐Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23: 2452–2462 [DOI] [PubMed] [Google Scholar]
- Courchaine E, Neugebauer KM (2015) Paraspeckles: paragons of functional aggregation. J Cell Biol 210: 527–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R (2007) Edc3p and a glutamine/asparagine‐rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T (2004) In vivo kinetics of Cajal body components. J Cell Biol 164: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum‐Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP (2015) The disordered P granule protein LAF‐1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA 112: 7189–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H, Pelham‐Webb B, Blythe S, Wieschaus E (2016) Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 26: 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory PI (1942) Thermodynamics of high polymer solutions. J Chem Phys 10: 51–61 [Google Scholar]
- Fowler DM, Koulov AV, Alory‐Jost C, Marks MS, Balch WE, Kelly JW (2006) Functional amyloid formation within mammalian tissue. PLoS Biol 4: e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW (2007) Functional amyloid–from bacteria to humans. Trends Biochem Sci 32: 217–224 [DOI] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG (1999) Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol 9: 126–135 [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D (2006) FG‐rich repeats of nuclear pore proteins form a three‐dimensional meshwork with hydrogel‐like properties. Science 314: 815–817 [DOI] [PubMed] [Google Scholar]
- Gall JG (2000) Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 16: 273–300 [DOI] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M (2012) Functional architecture of the nuclear pore complex. Annu Rev Biophys 41: 557–584 [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Cordero JA, Gall JG (2005) Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low‐density, sponge‐like structure. Mol Biol Cell 16: 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, Hosoki K, Goshima N, Kawaguchi T, Hatters D, Trinkle‐Mulcahy L, Hirose T, Bond CS, Fox AH (2015) Prion‐like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 210: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins ML (1942) Some properties of solutions of long‐chain compounds. J Phys Chem 46: 151–158 [Google Scholar]
- Hyman AA, Weber CA, Julicher F (2014) Liquid‐liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58 [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, Zheng Y (2015) Phase transition of spindle‐associated protein regulate spindle apparatus assembly. Cell 163: 108–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL (2012) Cell‐free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN_et al_ (2013) Mutations in prion‐like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J (2012) The tip of the iceberg: RNA‐binding proteins with prion‐like domains in neurodegenerative disease. Brain Res 1462: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingauf M, Stanek D, Neugebauer KM (2006) Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 17: 4972–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Sassone‐Corsi P (2007) The chromatoid body: a germ‐cell‐specific RNA‐processing centre. Nat Rev Mol Cell Biol 8: 85–90 [DOI] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S (2015) Promiscuous interactions and protein disaggregases determine the material state of stress‐inducible RNP granules. Elife 4: e06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL (2013) Phosphorylation‐regulated binding of RNA polymerase II to fibrous polymers of low‐complexity domains. Cell 155: 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL (2014) Poly‐dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D (2013) Systematic analysis of barrier‐forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J 32: 204–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Nutter‐Upham A, Lindquist S, King OD (2014) PLAAC: a web and command‐line application to identify proteins with prion‐like amino acid composition. Bioinformatics 30: 2501–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD (2013) Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R (2015) Formation and Maturation of Phase‐Separated Liquid Droplets by RNA‐Binding Proteins. Mol Cell 60: 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM (2013) Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 4: 17–34 [DOI] [PubMed] [Google Scholar]
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM (2014) The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 56: 389–399 [DOI] [PubMed] [Google Scholar]
- Majcher V, Goode A, James V, Layfield R (2015) Autophagy receptor defects and ALS‐FTLD. Mol Cell Neurosci 66: 43–52 [DOI] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale W, Riek R (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White‐Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, Unruh J, Slaughter B, Si K (2012) Critical role of amyloid‐like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148: 515–529 [DOI] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Alberti S (2013) Protein disorder, prion propensities, and self‐organizing macromolecular collectives. Biochim Biophys Acta 1834: 918–931 [DOI] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL (2011) Biogenesis and function of nuclear bodies. Trends Genet 27: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, Kronenberg‐Versteeg D, Li Y, Yang SP, Wakutani Y, Meadows W, Ferry RR, Dong L, Tartaglia GG, Favrin G, Lin WL_et al_ (2015) ALS/FTD Mutation‐Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W (2015) A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol 427: 1135–1158 [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett‐Jones DP, Pawson T, Forman‐Kay JD, Baldwin AJ (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny I, Malinova A, Stejskalova E, Mateju D, Klimesova K, Roithova A, Sveda M, Knejzlik Z, Stanek D (2015) SART3‐dependent accumulation of incomplete spliceosomal snRNPs in cajal bodies. Cell Rep 10: 429–440 [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S (2015) A liquid‐to‐solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Pederson T (2011) The nucleolus. Cold Spring Harb Perspect Biol 3: a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB, Patel DJ (2011) Structure‐function studies of FMRP RGG peptide recognition of an RNA duplex‐quadruplex junction. Nat Struct Mol Biol 18: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI (2000) In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 151: 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Görlich D (2015) Nup98 FG domains from diverse species spontaneously phase‐separate into particles with nuclear pore‐like permselectivity. Elife 4: e04251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE (2006) Pathway to totipotency: lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat Cell Biol 13: 167–173 [DOI] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM (2004) Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 166: 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352: 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD (2011) Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9: e1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng PK, Eisenberg D (2009) Short protein segments can drive a non‐fibrillizing protein into the amyloid state. Protein Eng Des Sel 22: 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky JA, Wright PE (2014) Assemblages: functional units formed by cellular phase separation. J Cell Biol 206: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN (2015) The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett 589: 2498–2506 [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA‐binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 16: 1383–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G (2014) Regulation of RNA granule dynamics by phosphorylation of serine‐rich, intrinsically disordered proteins in C. elegans. Elife 3: e04591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP (2012) Getting RNA and protein in phase. Cell 149: 1188–1191 [DOI] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL (2015) The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid‐like droplets, and nuclei. Cell 163: 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum‐Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS (2015) RNA controls PolyQ protein phase transitions. Mol Cell 60: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]