Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring (original) (raw)
. Author manuscript; available in PMC: 2017 Jun 16.
SUMMARY
Maternal obesity during pregnancy has been associated with increased risk of neurodevelopmental disorders, including autism spectrum disorder (ASD), in offspring. Here we report that maternal high fat diet (MHFD) induces a shift in microbial ecology that negatively impacts offspring social behavior. Social deficits and gut microbiota dysbiosis in MHFD offspring are prevented by co-housing with offspring of mothers on a regular diet (MRD) and transferable to germ-free mice. In addition, social interaction induces synaptic potentiation (LTP) in the ventral tegmental area (VTA) of MRD, but not MHFD offspring. Moreover, MHFD offspring had fewer oxytocin immunoreactive neurons in the hypothalamus. Using metagenomics and precision microbiota reconstitution, we identified a single commensal strain that corrects oxytocin levels, LTP, and social deficits in MHFD offspring. Our findings causally link maternal diet, gut microbial imbalance, VTA plasticity and behavior, and suggest that probiotic treatment may relieve specific behavioral abnormalities associated with neurodevelopmental disorders.
Keywords: neurodevelopmental disorders, autism, dysbiosis, high fat diet (HFD), ventral tegmental area (VTA), long-term potentiation (LTP), probiotic
INTRODUCTION
Recent epidemiological evidence suggests that exposure to maternal obesity_in utero_ increases the risk of neurodevelopmental disorders, such as ASD in children (Connolly et al., 2016; Krakowiak et al., 2012; Sullivan et al., 2014). Given the increase in the prevalence of obesity (Skinner and Skelton, 2014), it is important to understand the neurobiological mechanism by which maternal obesity affects offspring behavior and brain function.
The amount and type of dietary macronutrients strongly influence the intestinal microbiota (Tremaroli and Backhed, 2012), which consists of a vast bacterial community that resides in the lower gut and lives in a symbiotic relationship with the host. Indeed, maternal obesity has been associated with alterations in the gut microbiome in offspring in both human and non-human primates (Galley et al., 2014; Ma et al., 2014). In addition, some individuals with neurodevelopmental disorders including ASD co-present with gastrointestinal problems and dysbiosis of the gut microbiota (Bresnahan et al., 2015; Mayer et al., 2014; Parracho et al., 2005). Given the large body of preclinical literature supporting the notion that a bidirectional communication system between the gut and the brain—known as the gut-brain axis—links gut and brain activities (Cryan and Dinan, 2012; Mayer et al., 2015), it has been speculated that changes in the gut microbiome may be relevant to the development of behavioral symptoms associated with ASD (Hsiao et al., 2013; Mayer et al., 2014). However, how changes in bacteria that inhabit the intestine could influence brain development and function remains unknown.
Here we report that maternal high fat diet (MHFD)-induced obesity in mice is associated with social behavioral deficits, which are mediated by alterations in the offspring gut microbiome. Notably, we also found that MHFD-induced changes in the offspring gut microbiota block long-lasting neural adaptation in the mesolimbic dopamine reward system (ventral tegmental area, VTA). Moreover, oral treatment with a single commensal bacterial species corrects oxytocin levels and synaptic dysfunction in the VTA and reverses social deficits in MHFD offspring.
RESULTS
Social Behaviors are Impaired in MHFD Offspring
To investigate how maternal diet-induced obesity affects offspring neurodevelopment, female mice were fed either regular diet (RD) or high fat diet (HFD) for 8 weeks, a standard period required to reach a state of diet-induced obesity in mice (Aye et al., 2015). Females were then paired with males to produce offspring that were given regular diet after weaning (Figure 1A). As expected, MHFD significantly increased maternal weight (Figures S1A–S1C). Consistent with reports of more frequent spontaneous abortion in obese mothers (King, 2006), the litter size was reduced (Figure S1D) and latency to first litter increased in female mice fed HFD (Figure S1E). It is noteworthy that there was no significant difference in offspring weight between maternal diet cohorts at 7–12 weeks of age (Figures S1F and S1G), the time at which behavioral and electrophysiological experiments were performed.
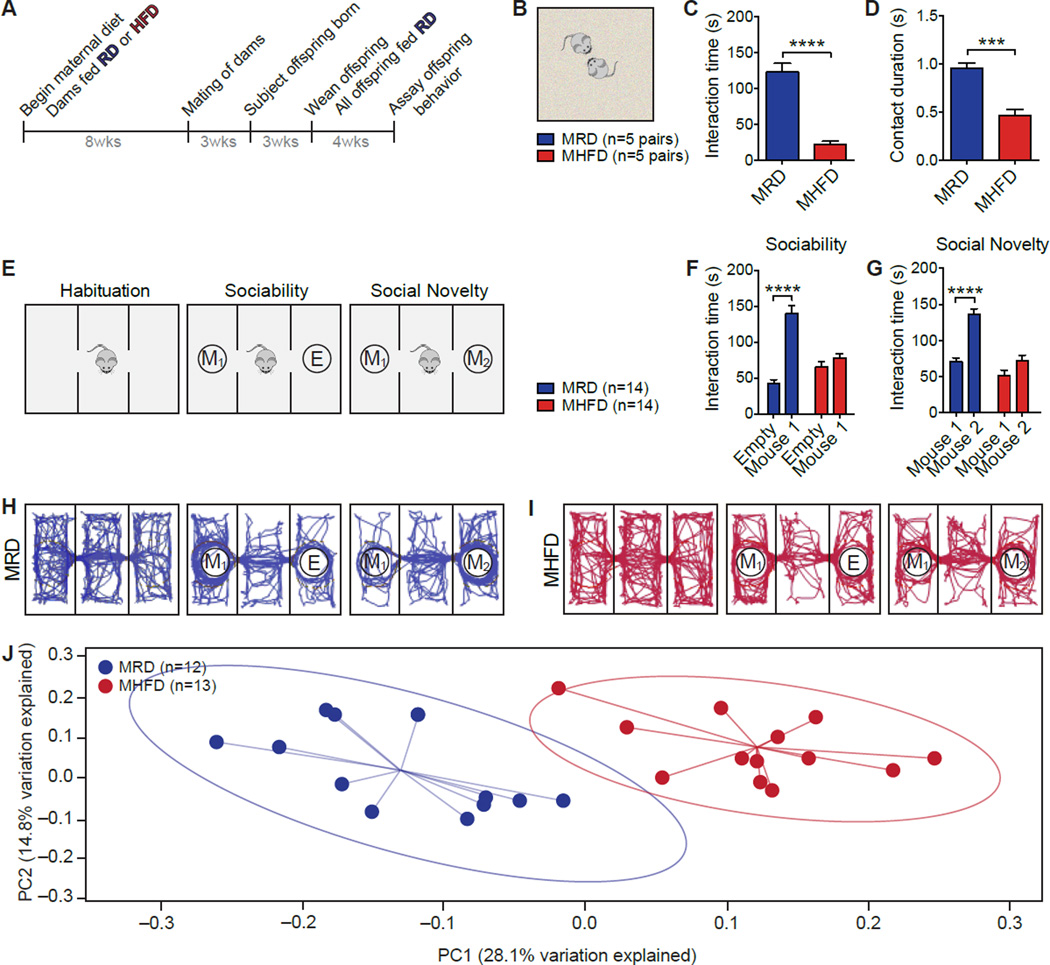
Figure 1. Social Deficits and Dysbiosis of the Gut Microbiota in MHFD Offspring.
A, Schematic of the maternal diet regimen and breeding. B, Schematic of the reciprocal social interaction task. C–D, MHFD offspring showed reduced reciprocal interaction (C, P<0.0001,_t_=7.90; D, P<0.001,_t_=5.89). E, Schematic of the three-chamber social interaction task. F–G, In the sociability test, MRD offspring spent more time interacting with a mouse than with an empty wire cage (F,P<0.0001, _t_=8.817), whereas MHFD offspring showed no preference for the mouse (F, _P_=0.48,_t_=1.19; Maternal diet effect F1,52=6.08,P<0.05). In the social novelty test, unlike MRD (G,P<0.0001, _t_=6.68), MHFD offspring had no preference for interacting with a novel versus a familiar mouse (G,_P_=0.086, _t_=2.08; Maternal diet effect F1,52=34.96, P<0.0001). H–I, Representative exploratory activity of MRD (H) and MHFD (I) offspring in the three-chamber test. J, Principal coordinates analysis (PCoA) of unweighted UniFrac distances from the averaged rarefied 16S rRNA gene dataset (_n_=1,000 rarefactions; 7,617 reads/sample) showed that MRD samples clustered separately from MHFD samples (P<0.001, _R_2=0.37). Plots show mean ± SEM. See also Figures S1–S2.
Given that maternal obesity has been associated with increased risk for neurodevelopmental disorders including ASD in offspring (Bilder et al., 2013; Krakowiak et al., 2012; Moss and Chugani, 2014) and deficient social interactions are a salient behavioral feature of ASD (Mefford et al., 2012), we studied social behavior in MRD and MHFD offspring. First, we assessed reciprocal social interactions by recording the amount of time a pair of mice, unfamiliar with each other, spent interacting in a neutral arena (Figure 1B). When compared to MRD offspring, MHFD offspring had fewer reciprocal interactions (Figures 1C, 1D, S1H, and S1I). Next, we used the three-chamber test (Silverman et al., 2010) to assess a) sociability by comparing the time mice spent interacting with an empty wire cage versus one containing a mouse and b) preference for social novelty by measuring the time mice spent interacting with a familiar versus a stranger mouse (Figure 1E). Consistent with the results from reciprocal social interactions, MHFD offspring had impaired sociability and showed no preference for social novelty (Figures 1F–1I, S1J, and S1K). Taken together these data indicate that MHFD offspring display social deficits.
Dysbiosis of the Gut Microbiota in MHFD Offspring
A variety of factors could contribute to the etiology of MHFD-induced social behavioral abnormalities. However, maternal obesity has been shown to alter the gut microbiome of offspring (Galley et al., 2014; Ma et al., 2014) and individuals diagnosed with ASD can co-present dysbiosis of the gut microbiota (Mayer et al., 2014; Parracho et al., 2005). To examine whether MHFD induces alterations in offspring gut microbiota, we analyzed the bacterial composition and community structure in the feces of MRD and MHFD offspring by 16S ribosomal RNA (rRNA) gene sequencing. The microbial communities in both MRD and MHFD offspring were comprised of a typical mouse gut microbiota, dominated by Bacteroidetes and Firmicutes (Figures S2A–S2D). While bacterial diversity computed based on weighted UniFrac distances [the assessment of community structure by considering abundance of operational taxonomic units (OTUs)] did not differ significantly between the offspring from either diet group (Figure S2E), unweighted analyses of UniFrac distances (assessment of community structure by considering only OTU presence/absence) revealed a marked difference between the structures of the bacterial communities (Figure 1J). Moreover, the diversity of microbiota in MHFD offspring was reduced compared to MRD microbiota (Figure S2F).
Consistent with previous reports (Turnbaugh et al., 2006), an HFD regimen in mothers induced a remarkable change in the maternal microbiome composition and diversity (Figures S2G and S2H), which was similar to that observed in their offspring (Figures 1J and S2F).
Gut Microbiota Mediate MHFD-Induced Social Deficits
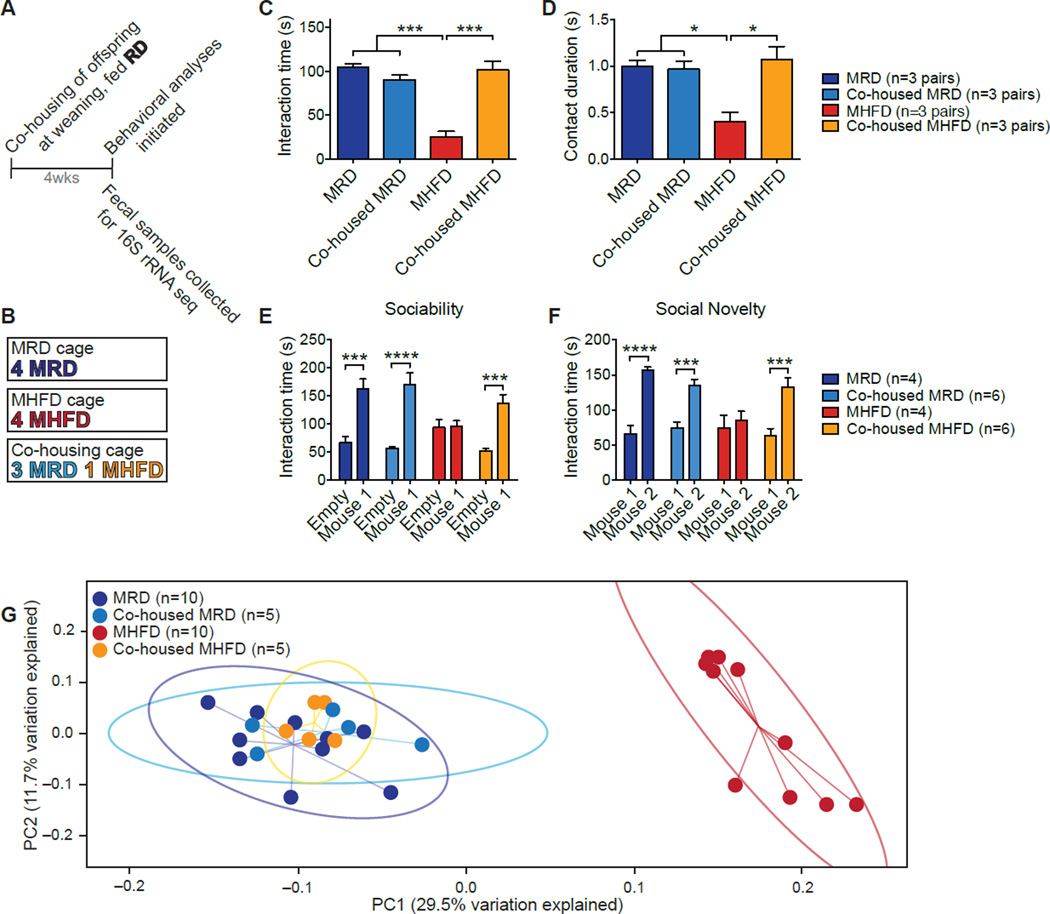
While microbial communities vary across individuals (Yatsunenko et al., 2012), co-housed family members are known to share their microbiota (Song et al., 2013). Since mice are coprophagic and transfer gut microbiota between each other by the fecal-oral route (Ridaura et al., 2013), we examined whether co-housing MHFD with MRD mice prevents the social deficits in MHFD offspring. To this end, at weaning (3 weeks) an MHFD mouse was co-housed with three MRD mice (Figures 2A and 2B). Control groups consisted of individual cages containing either four MHFD mice or four MRD mice (Figure 2B). Fecal samples were collected and social behavior in MRD and MHFD offspring was assessed when mice were 7–8 weeks old. Strikingly, MHFD mice co-housed with MRD mice exhibited normal reciprocal social interactions (Figures 2C, 2D and S3A–S3C), as well as normal sociability and preference for social novelty, as determined by the 3-chamber test (Figures 2E, 2F, S3D and S3E). Thus, co-housing with control mice corrects social deficits in MHFD offspring.
Figure 2. Co-housing MHFD with MRD Offspring Rescues both Social Dysfunction and the Microbiota Phylogenetic Profile of MHFD Mice.
A, Schematic of the co-housing experiment. B, MRD and MHFD offspring were weaned into one of three cage compositions. C–G, Social interaction time (C, MRD vs. MHFD P<0.001, _t_=9.30; MRD vs. co-housed MHFD _P_>0.99, _t_=0.31; MHFD vs. co-housed MHFD P<0.001,_t_=7.99; _P_<0.0001, F3,8=30.51) and contact duration (D, MRD vs. MHFD_P_<0.05, _t_=4.13; MRD vs. co-housed MHFD_P_>0.99, _t_=0.46; MHFD vs. co-housed MHFD P<0.05, _t_=4.59;_P_<0.001, F3,8=9.01) in the reciprocal interaction test; social interaction times in the sociability (E, MRD_P_<0.001, _t_=4.36; MHFD_P_>0.99, t_=0.078; Co-housed MRD_P<0.0001, _t_=6.33; Co-housed MHFD_P_<0.001, _t_=4.78; Maternal diet/Housing/Interaction effect F3,32=6.13,_P_<0.01) and social novelty tests (F, MRD_P_<0.0001, _t_=5.12; MHFD_P_>0.99, t_=0.60; Co-housed MRD_P<0.001, t_=4.20; Co-housed MHFD_P<0.001, _t_=4.76; Maternal diet/Housing/Interaction effect F3,32=4.37,P<0.01), as well as UniFrac-based phylogenetic clustering (G, P<0.001,_R_2=0.552; _n_=1,000 rarefactions; 3,390 reads/sample), are all restored in MHFD offspring co-housed with MRD mice. Plots show mean ± SEM. See also Figure S3.
We next examined whether co-housing also corrected the changes in the microbiota of MHFD offspring. Indeed, co-housing caused a shift in the bacterial phylogenetic profile of MHFD mice to resemble that of MRD or MRD co-housed mice (Figure 2G), thus correcting the MHFD-induced alterations in the commensal microbiota.
In agreement with the idea that the fecal microbiota of MHFD offspring lacks one or more beneficial bacterial species required for normal social behavior, co-housing one MRD with three MHFD offspring was sufficient to rescue both the social behaviors and microbiota phylogenetic profile of MHFD offspring (Figures S3F–S3L).
Colonization of Germ-Free (GF) Mice with the Microbiota from MRD, but not MHFD Mice, Reverses their Deficient Social Behavior
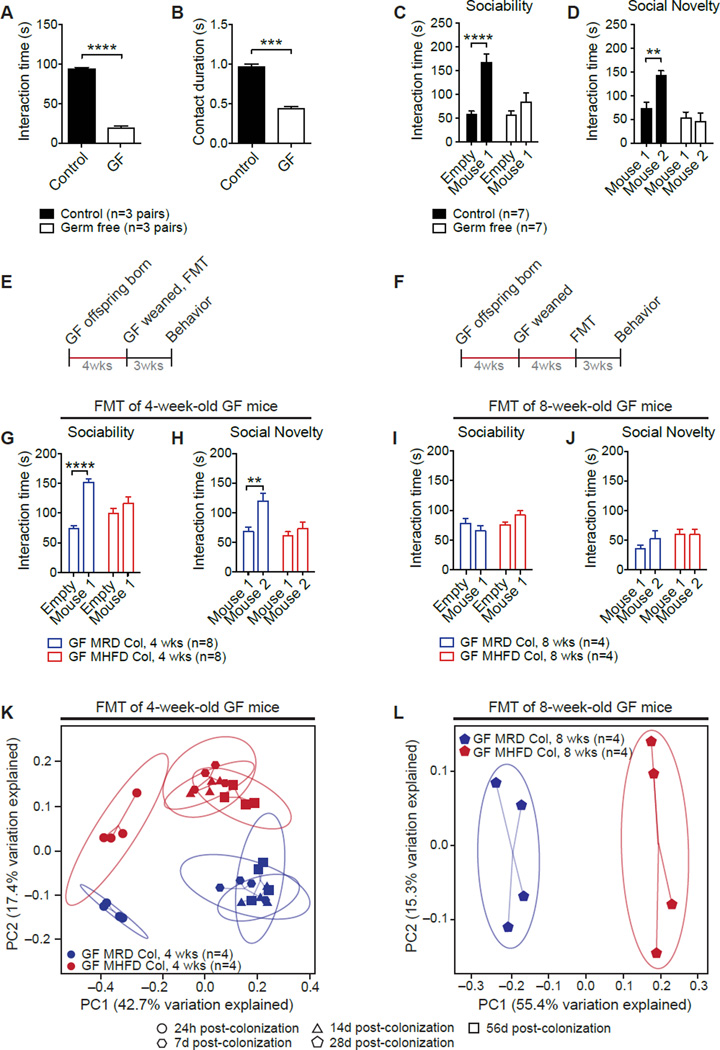
Studies on GF mice have shown that the intestinal microbiota can influence brain development and function (Cryan and Dinan, 2012). We hypothesized that, if the lack of one or more bacterial species in the microbiota of MHFD offspring is responsible for their defective social behavior, GF mice should also be socially deficient. Confirming this hypothesis and in keeping with recent results (Desbonnet et al., 2014), social behaviors were impaired in GF mice (Figures 3A–3D and S4A–S4D).
Figure 3. Fecal Microbiota from MRD, but not MHFD, Offspring Improves Germ-Free (GF) Recipient Social Behavior.
A–D, GF mice show reduced reciprocal social interaction (A,P<0.0001, _t_=22.73; B,_P_<0.001, _t_=11.31) and deficits in sociability (C, Control _P_<0.0001,_t_=5.30, GF _P_>0.99,_t_=0.39; Main group effect F1,24=21.98,P<0.0001) and preference for social novelty (D, Control _P_<0.01, _t_=3.64, GF_P_=0.39, _t_=1.33; Main group effect F1,24=5.29, _P_<0.05). Schematic of fecal microbiota transplant (FMT) at four (E) and eight weeks of age (F). G–H, FMT from MRD, but not MHFD, offspring at weaning restored both GF sociability (G, GFMRDCol _P_<0.0001, _t_=6.66; GFMHFDCol _P_=0.35, _t_=1.40; Donor effect F1,28=32.44, _P_<0.0001) and preference for social novelty (H, GFMRDCol _P_<0.01, _t_=3.60; GFMHFDCol _P_=0.81, _t_=0.84; Donor effect F1,28=9.86, _P_<0.01). I–J, At eight weeks, FMT from either MRD or MHFD donors failed to improve sociability (I, GFMRDCol _P_=0.51, _t_=1.20; GFMHFDCol _P_=0.28, _t_=1.58; Donor effect F1,12=0.07, _P_=0.79) or preference for social novelty in GF mice (J, GFMRDCol _P_=0.48, _t_=1.23; GFMHFDCol _P_>0.99, _t_=0.043; Donor effect F1,12=0.71, _P_=0.42). K–L, PCoA of unweighted UniFrac distances based on the 16S rRNA gene sequencing dataset from GF recipients of stools from either MRD or MHFD donors at four (K,_P_=0.001, _R_2=0.83;_n_=1,000 rarefactions; 4,628 reads/sample) or eight (L,P<0.001, _R_2=0.77;_n_=1,000 rarefactions; 4,805 reads/sample) weeks of age. Plots show mean ± SEM. See also Figure S4.
To identify functional differences between gut microbial communities and determine whether their role is causal, we transplanted (gavaged) fecal microbiota from adult MRD and MHFD offspring into 4 week- (Figure 3E) and 8 week-old (Figure 3F) GF mice. Interestingly, GF mice that received fecal microbiota from MRD offspring at weaning (4 weeks; Figures 3G, 3H, S4E and S4F), but not at 8 weeks (Figures 3I, 3J, S4G, and S4H), showed normal social behavior. By contrast, GF mice that received fecal microbiota from MHFD offspring remained socially impaired, regardless of the age at which the fecal transfer was performed (Figures 3G–3J and S4E–S4H). Moreover, as in the case of the phylogenetic separation of MRD and MHFD microbiota (Figure 1J), the bacterial communities in GF mice receiving feces from MHFD donor offspring clustered separately from those of GF mice receiving feces from MRD donor offspring, irrespective of whether the fecal transplant was performed at 4 or 8 weeks (Figures 3K, 3L, and S4I–S4N). These data reveal a neurodevelopmental window during which microbial reconstitution effectively improves social behavior.
MHFD Negatively Impacts a Subset of Bacteria in the Intestinal Gut and Selective Re-Introduction of Lactobacillus(L.) reuteri Restores Social Deficits in MHFD Offspring
To investigate which bacterial species are absent in the microbiota community of MHFD offspring, we performed metagenomic shotgun sequencing of fecal samples from both MHFD and MRD offspring. Our analysis identified several species whose relative abundance was dramatically reduced in the MHFD offspring microbiota (Table 1). Among these,L. reuteri was the most drastically reduced (> 9-fold) in MHFD microbiota population, compared to the MRD microbiota (Table 1).
Table 1.
Species Whose Abundance Is Reduced in the Gut Microbiota of MHFD Offspring
| Species of interest | MRD Representation | MHFD Representation | Fold Change MRD/MHFD |
|---|---|---|---|
| Lactobacillus reuteri | 7.49 ± 3.0 | 0.879 ± 0.21 | 9.24 ± 0.65 |
| Parabacteroides distasonis | 0.00709 ± 0.0055 | 0.00126 ± 0.0011 | 5.63 ± 1.17 |
| Helicobacter hepaticus | 7.35 ± 2.4 | 2.58 ± 1.3 | 2.84 ± 0.61 |
| Bacteroides uniformis | 5.49 ± 2.2 | 2.07 ± 0.78 | 2.65 ± 0.56 |
| Olsenella unclassified | 0.230 ± 0.064 | 0.121 ± 0.031 | 1.90 ± 0.38 |
| Collinsella unclassified | 0.0866 ± 0.031 | 0.0494 ±0.016 | 1.75 ± 0.48 |
| Bifidobacterium pseudolongum | 19.4 ± 3.3 | 11.3 ± 2.4 | 1.71 ± 0.27 |
| Lactobacillus johnsonii | 24.5 ± 6.2 | 17.1 ± 5.2 | 1.43 ± 0.40 |
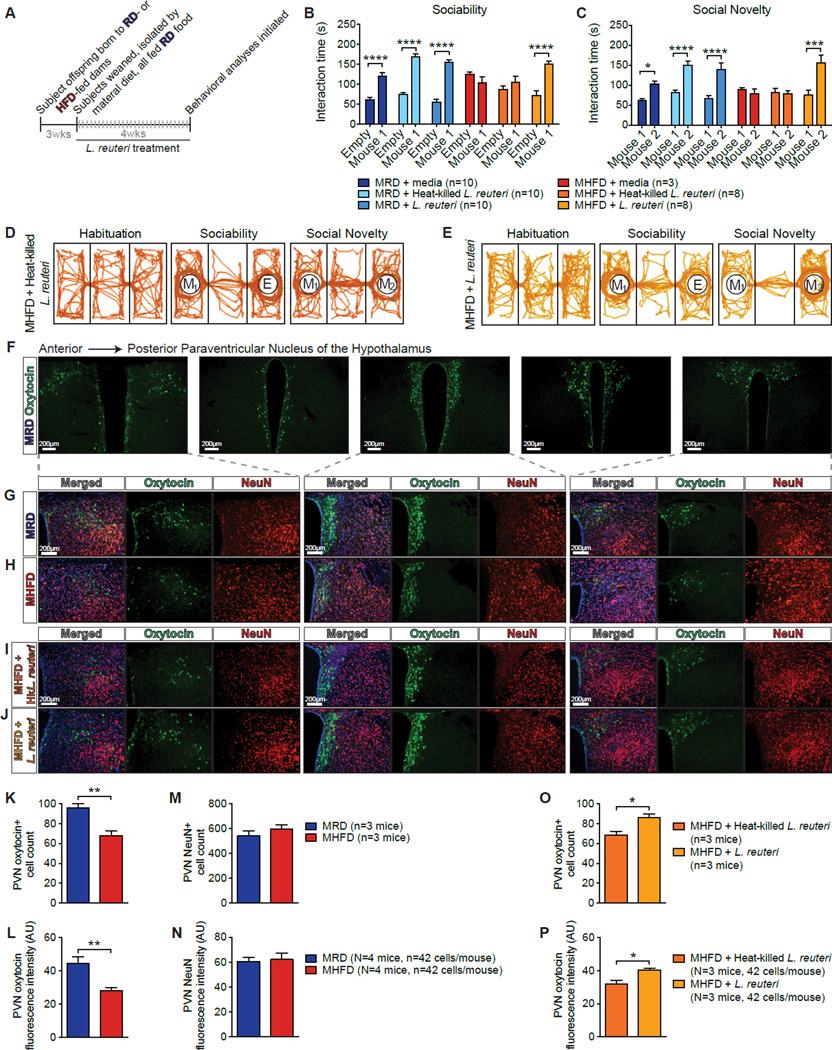
L. reuteri has been shown to promote oxytocin levels (Poutahidis et al., 2013), a hormone that plays a crucial role in social behaviors (Donaldson and Young, 2008). We hypothesized that the selective decrease in L. reuteri in the microbiota of MHFD offspring was causally related to their social deficits. To test this hypothesis, we introduced L. reuteri into the drinking water of MHFD offspring at weaning, for 4 weeks, after which behavior was tested (Figure 4A). Remarkably, treatment with L. reuteri significantly improved sociability and preference for social novelty in MHFD offspring (Figures 4B, 4C, 4E, S5A, and S5B). Results from several control experiments underscore the specificity of L. reuteri_-mediated rescue of social behaviors in MHFD offspring. First, drinking water treated with either resuspension media or heat-killed L. reuteri (80°C for 20 min) failed to restore social behavior in MHFD offspring (Figures 4B–4D, S5A, and S5B).Second, drinking water with live L. reuteri did not change the social behavior of MRD offspring (Figures 4B, 4C, S5A, and S5B), presumably because their gut microbiota already contains ample L. reuteri.Finally, addition of L. reuteri to the drinking water had no major effect on bacterial viability and the heat-killing procedure completely abrogated colony-forming units (Figure S5C). Importantly, the amelioration of the deficient social behavior is specific to L. reuteri since similar treatment with another_Lactobacillus species, L. johnsonii, whose abundance is also reduced in the gut microbiota of MHFD offspring (Table 1), failed to rescue social behaviors in MHFD offspring (Figure S5D–S5M).
Figure 4. Selective Treatment with Lactobacillus (L.) reuteri Restores Social Deficits and Oxytocin Levels in MHFD Offspring.
A, Schematic of _L. reuteri_–treatment. B,C, Unlike resuspension media (B, _P_>0.99,_t_=1.03; c, _P_>0.99,_t_=0.40) or heat-killed L. reuteri (B,_P_>0.99, _t_=1.35; c,_P_>0.99, _t_=0.21), administration of live L. reuteri in the drinking water rescued sociability (B,P<0.0001, _t_=5.98) and preference for social novelty (C, P<0.001,_t_=5.01) in MHFD offspring (B, Treatment effect F1,86=87.53, P<0.0001; C, Treatment effect F1,86=30.24, P<0.0001). D–E, Representative track plots showing exploratory activity. F, Representative images of control oxytocin immunoreactivity at different anteroposterior levels of the PVN. G–J, Oxytocin immunoreactivity in the PVN of MRD (G), MHFD (H), heat-killed _L. reuteri_-treated MHFD (I), and live_L. reuteri_-treated MHFD offspring (J). K–N, Oxytocin immunoreactive cell number (K, P<0.01,_t_=4.76) and oxytocin immunofluorescence intensity (L,P<0.01, _t_=3.80) were reduced in the PVN of MHFD versus MHFD mice. In the PVN of MRD and MHFD offspring, NeuN cell number immunoreactivity (M, _P_=0.34,_t_=1.09) and immunofluorescence intensity (N,_P_=0.79, _t_=0.28) were similar. O–P, Relative to treatment with heat-killed L. reuteri, treatment with live L. reuteri significantly increased oxytocin-positive cell number (O, P<0.05, _t_=2.93) and oxytocin immunofluorescence intensity (P, P<0.05,_t_=3.09) in the PVN of MHFD offspring. AU: arbitrary units. Plots show mean ± SEM. See also Figures S5–S6.
MHFD offspring show other behavioral traits that are associated with ASD, like repetitive behaviors and anxiety (Figures S6). Interestingly, while co-housing MHFD with MRD offspring restores social behavior (Figures 2C and 2D), it failed to rescue marble burying (Figure S6B), a behavioral task reflecting repetitive and perseverative behavior (Thomas et al., 2009). Accordingly, GF mice also showed increased marble burying, and fecal microbial transplants from MRD (or MHFD) offspring into GF mice failed to reverse the repetitive behavior (Figure S6B). Thus, these data suggest that repetitive behaviors in MHFD offspring do not depend on changes in the gut microbiome. In addition,L. reuteri treatment had no effect on anxiety in MHFD offspring (Figure S6C–S6E). Taken together, these data indicate that L. reuteri reconstitution specifically rescues social, but not other behavioral endophenotypes associated with ASD.
Oxytocin Levels are Reduced in the Hypothalamus of MHFD Offspring
There is growing evidence that the neuropeptide oxytocin modulates numerous aspects of social behavior (Donaldson and Young, 2008) and is implicated in ASD (Lerer et al., 2008; Wu et al., 2005). L. reuteri, which rescued social behaviors in MHFD mice (Figures 4B, 4C, and 4E), has been reported to increase oxytocin levels (Poutahidis et al., 2013). Because oxytocin is primarily synthesized in the paraventricular nuclei (PVN) of the hypothalamus, we decided to compare the number of oxytocin-expressing cells in the PVN of MRD and MHFD offspring. Interestingly, MHFD offspring had significantly fewer oxytocin immunoreactive neurons compared to MRD offspring (Figures 4F–4H, 4K, and 4L). The reduction in oxytocin immunoreactivity was not due to an overall decrease in PVN neurons, since the total number of neurons was unchanged (as measured by NeuN staining; Figures 4G, 4H, 4M, and 4N). Notably, in _L. reuteri_-treated MHFD offspring, the number of oxytocin-expressing cells was higher than in control-treated MHFD offspring (Figures 4I, 4J, 4O, and 4P). Thus, the number of oxytocin immunoreactive neurons in the PVN is reduced in MHFD offspring but can be restored by L. reuteri treatment.
Mesolimbic Dopamine Reward System Function is Impaired in MHFD Offspring
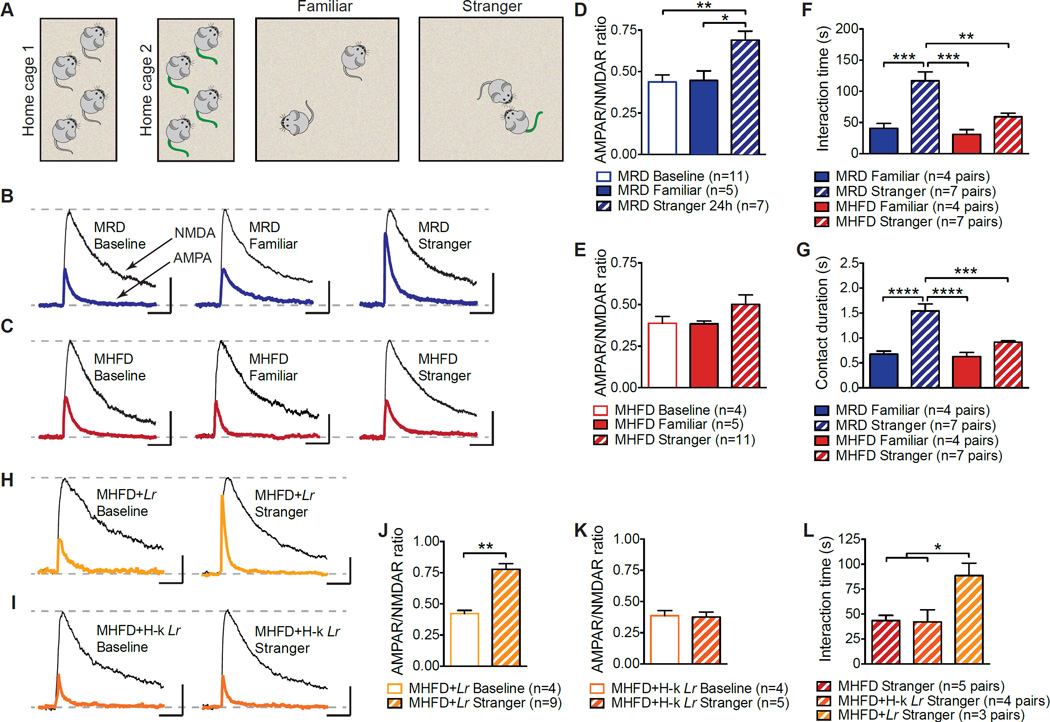
Brain regions that respond to naturally rewarding stimuli, including the ventral tegmental area (VTA) and the nucleus accumbens (NAc), are crucially involved in social behaviors (Dolen et al., 2013; Gunaydin et al., 2014). In addition, oxytocin-expressing neurons in the PVN project to the VTA (Melis et al., 2007). Oxytocin activates VTA neurons in both mice and humans, influencing the processing of socially relevant cues (Groppe et al., 2013; Tang et al., 2014) and oxytocin receptor blockade in the VTA prevents social attachment in rodents (Pedersen et al., 1994). Given that social stimulation can be particularly rewarding and triggers synaptic potentiation in VTA DA neurons of birds (Huang and Hessler, 2008), we examined whether direct social interaction evokes long-term potentiation (LTP) of synaptic inputs to VTA DA neurons (Figure S7A–S7C). To this end, we recorded AMPAR/NMDAR ratios of glutamatergic excitatory postsynaptic currents (EPSCs) in MRD and MHFD offspring 24 hours following a 10 min reciprocal interaction with either a stranger or a familiar mouse (Figure 5A). In control MRD mice, social interaction with a stranger, but not a familiar mouse, triggered LTP in VTA DA neurons, as determined by an increase in AMPAR/NMDAR ratios (Figures 5B and 5D). By contrast, in MHFD offspring, social interaction with a stranger failed to induce LTP in their VTA DA neurons (Figures 5C and 5E). Input-output curves, paired-pulse ratios and miniature EPSCs (mEPSCs) frequency and amplitude show that the impairment of LTP induced by social interaction in MHFD offspring cannot be attributed to changes in basal synaptic transmission (Figure S7D–S7H).
Figure 5. Reciprocal Social Interaction and Social Interaction-Induced LTP in MHFD Offspring VTA DA Neurons are Restored by L. reuteri.
A, Schematic of the experimental design. B–E, LTP was measure 24 hours following reciprocal interaction with either a stranger or a familiar mouse. Only interaction with a stranger mouse induced LTP in MRD VTA DA neurons, as determined by increased AMPAR/NMDAR ratios (B,D, Baseline vs. Familiar_P_>0.99, _t_=0.12, Baseline vs. Stranger P<0.01, _t_=3.79; Familiar vs. Stranger _P_<0.05, _t_=3.03; F=8.03,_P_<0.01). In MHFD offspring, neither stranger nor familiar reciprocal interaction evoked LTP in VTA DA neurons (C,E, Baseline vs. Familiar _P_>0.99, _t_=0.035, Baseline vs. Stranger _P_=0.64, _t_=1.30; Familiar vs. Stranger_P_=0.50, _t_=1.45; F2,15=1.47,_P_=0.26). F–G, Whereas MRD mice spent more time interacting with a stranger than a familiar mouse (Familiar vs. Stranger; F,P<0.001, _t_=4.88; G,_P_<0.0001, _t_=5.87), MHFD mice did not (Familiar vs. Stranger; F, _P_=0.47, _t_=1.87; G, _P_=0.40, _t_=1.96) (F, F3,19=13.8,_P_<0.0001; G, F3,19=18.54,_P_<0.0001). H–K, Live (H,J,_P_<0.01, t=4.95), but not heat-killed _L. reuteri_ (I,K, _P_=0.84, _t_=0.20), restored stranger interaction-evoked LTP in the VTA of MHFD offspring. L, Unlike heat-killed _L. reuteri_ (MHFD vs. MHFD+Hk−_Lr_ _P_>0.99, _t_=0.099), live L. reuteri restored reciprocal social interaction (MHFD vs. MHFD+Lr P<0.05, _t_=3.24; F2,9=6.45,P<0.05). Plots show mean ± SEM. See alsoFigure S7.
Mirroring the electrophysiological results, MRD offspring spent significantly more time interacting with a stranger than a familiar mouse, but MHFD offspring did not (Figures 5F and 5G). Thus, social interaction induces a long-lasting increase in the activity of the dopaminergic reward system of MRD, but not in MHFD, offspring.
Treatment with L. reuteri or Oxytocin Reverses both the LTP in VTA DA Neurons and Social Behavior in MHFD Offspring
We next wondered whether L. reuteri treatment, which restores sociability and preference for social novelty in MHFD offspring (Figures 4B, 4C, and 4E), would also rescue reciprocal social interaction and related changes in synaptic strength in the VTA. Live (Figures 5H and 5J), but not heat-killed (Figures 5I and 5K), L. reuteri rescued stranger interaction-induced LTP in the VTA as well as reciprocal social interactions in MHFD offspring (Figure 5L). Thus, L. reuteri restores social interaction-induced LTP in the VTA of MHFD offspring.
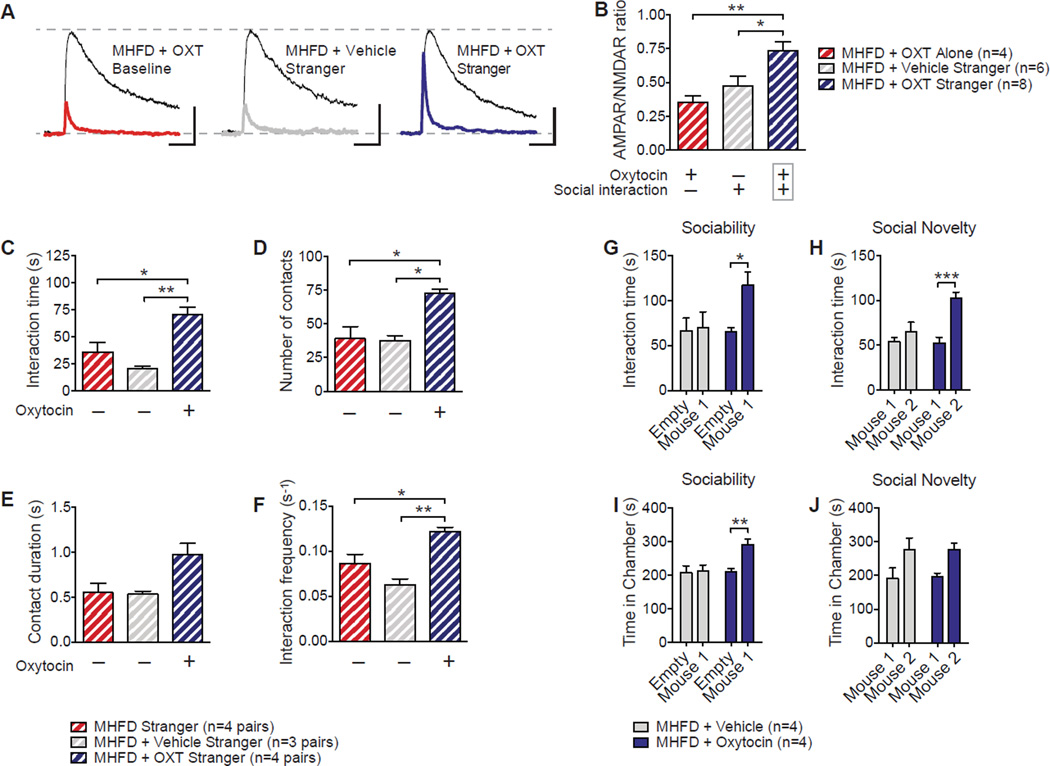
These findings, together with the fact that _L. reuteri_treatment increased oxytocin immunoreactivity in the PVN of MHFD offspring (Figures 4I, 4J 4O and 4P), led us to examine whether direct oxytocin application could also reverse the behavioral and electrophysiological deficits characteristic of MHFD offspring. To test this idea, we administered oxytocin intranasally—a preferred method of administering neuropeptides to the brain bypassing more invasive procedures (Penagarikano et al., 2015)—to MHFD offspring and measured reciprocal social interactions 30 min later. Although either oxytocin alone or social interaction alone failed to rescue social interaction-induced LTP in the VTA, the combination of social interaction and oxytocin treatment restored LTP in the VTA of MHFD offspring (Figures 6A and 6B), supporting prior work implicating a synergistic effect of oxytocin and dopamine in the processing of socially relevant cues (Modi and Young, 2012). Accordingly, oxytocin treatment improved reciprocal social interaction (Figures 6C–6F), as well as sociability and the preference for social novelty (Figures 6G–6J). Thus, oxytocin administration rescues social behavior and related neural adaptations in the VTA of MHFD offspring. Collectively, our data show that MHFD impairs oxytocin-mediated synaptic adaptations in the VTA that underlie social behaviors.
Figure 6. Oxytocin Restores Social Interaction-Induced VTA Plasticity and Social Behavioral Deficits in MHFD Offspring.
A–B, LTP was measure 1–3 hours following a reciprocal interaction. Intranasal oxytocin administration rescued LTP in the VTA of MHFD offspring (B, MHFD+OXT Alone vs. MHFD+OXT Stranger_P_<0.01, _t_=3.66; MHFD+Vehicle Stranger vs. MHFD+OXT Stranger _P_<0.05, _t_=2.86; F2,15=7.97, _P_<0.01). C–F, Oxytocin restored reciprocal social interaction (C, MHFD vs. MHFD+Vehicle_P_=0.55, _t_=1.46; MHFD vs. MHFD+OXT_P_<0.05, _t_=3.62; MHFD+Vehicle vs. MHFD+OXT _P_<0.01, _t_=4.81; F2,8=12.82, _P_<0.01; D, MHFD vs. MHFD+Vehicle _P_>0.99, _t_=0.16; MHFD vs. MHFD+OXT P<0.05, _t_=4.075; MHFD+Vehicle vs. MHFD+OXT _P_<0.05, _t_=3.94; F2,8=10.97, _P_<0.01; E, MHFD vs. MHFD+Vehicle, _P_>0.99, _t_=0.11; MHFD vs. MHFD+OXT _P_=0.052, _t_=2.99; Treatment effect F2,8=5.87, P<0.05; F, MHFD vs. MHFD+Vehicle _P_=0.2, _t_=2.11; MHFD vs. MHFD+OXT_P_<0.05, _t_=3.43; Treatment effect F2,8=14.58, _P_<0.01), sociability (G, MHFD+Vehicle _P_>0.99, _t_=0.44; MHFD+Oxytocin _P_=0.24, _t_=1.74; Treatment effect F1,8=2.37, _P_=0.16; H, MHFD+Vehicle_P_>0.99, t_=0.29; MHFD+Oxytocin_P<0.01, _t_=3.50; Treatment effect F1,12=5.16, P<0.05) and preference for social novelty in MHFD offspring (I, MHFD+Vehicle _P_=0.65,_t_=1.05; MHFD+Oxytocin P<0.05,_t_=3.54; Treatment effect F1,8=10.54,P<0.05; J, MHFD+Vehicle _P_=0.50,_t_=1.25; MHFD+Oxytocin _P_=0.096,_t_=2.34; Treatment effect F(1,8)=6.41,P<0.05). Plots show mean ± SEM.
DISCUSSION
Both genetic and environmental factors, and their interactions, play a crucial role in the etiology of neurodevelopmental disorders including ASD (Hallmayer et al., 2011). There is growing epidemiological evidence that maternal obesity heightens the risk of neuropsychiatric disorders in offspring (Krakowiak et al., 2012; Sullivan et al., 2014). Indeed, a recent study reported that mothers with obesity were 1.5 times more likely to have a child with ASD, and the increased risk of children with ASD was two-fold greater for pregnant mothers with both obesity and gestational diabetes (Connolly et al., 2016).
While most of the focus in the field has been on inflammation (Bolton and Bilbo, 2014) or epigenetic changes (Mathers and McKay, 2009), the biological mechanism by which maternal obesity affects offspring neurodevelopment remains to be determined Here we show that that the behavioral dysfunction associated with MHFD-induced obesity is induced by alterations in the offspring gut microbiota. Several lines of evidence support this idea. First, some individuals diagnosed with ASD present dysbiosis of the gut microbiota and gastrointestinal issues (Bresnahan et al., 2015; Mayer et al., 2014; Parracho et al., 2005).Second, maternal obesity leads to alterations in the offspring’s gut microbiome in humans and non-human primates (Galley et al., 2014; Ma et al., 2014). Third, in mice, the gut microbiota of MHFD offspring is altered (Figure 1J) by the reduction in specific bacterial species (Table 1). Fourth, manipulation of the microbiome community by co-housing MHFD with MRD offspring rescues MHFD-induced social deficits and corrects their microbial phylogenetic profile (Figures 2 and S3). Fifth, GF mice are socially impaired and fecal microbiota transplanted from MRD (but not MHFD) offspring rescues GF social behavior (Figures 3 and S4).Finally, treatment with a single bacterial species, L. reuteri, which is dramatically reduced in MHFD offspring (Table 1), selectively restores social behavior in MHFD mice (Figures 4 and S5A–C).
We propose a model in which L. reuteri improves social behavior by promoting oxytocin-mediated functions. Consistent with this model,_L. reuteri_-treatment enhances oxytocin levels in the PVN of MHFD mice (Figures 4I and 4J) and direct oxytocin-treatment normalizes the social behavior of MHFD offspring (Figure 6). Although the precise mechanism by which L. reuteri promotes oxytocin in the brain remains to be determined, we favor the idea that the vagus nerve (Davari et al., 2013) could be the main pathway of communication between the gut/L. reuteri and changes in oxytocin in the PVN. It is known that vagal nerve fibers project to the PVN (Sabatier et al., 2013; Uvnas-Moberg et al., 2014). In addition, neuronal activity in the PVN induced by bacterial colonization is blocked by subdiaphragmatic vagotomy (Wang et al., 2002). Especially relevant are the reports that the _L. reuteri_–mediated increase in oxytocin depends on the vagus nerve (Poutahidis et al., 2013) and that another Lactobacillus species, L. rhamnosus, reduced stress-induced anxiety in mice in a vagus-dependent manner (Bravo et al., 2011).
Our results provide new insight into the mechanism by which a marked shift in microbial ecology, caused by MHFD, can negatively impact social behaviors and related neuronal changes in offspring. These neuronal adaptations, which underlie social behavior by enhancing the salience and rewarding value of social stimuli, are surprisingly impaired by maternal diet-induced changes in the gut microbiome (Figure 5). Interestingly, according to a recent paper, probiotic-based restoration of gut permeability in a mouse model of ASD can improve some behavioral abnormalities, but not social behaviors (Hsiao et al., 2013). Given that we identified a different probiotic candidate, L. reuteri, that rescues social behavior (Figures 4 and 5), but not other behavioral endophenotypes associated with ASD (Figure S6) in MHFD mice, we propose that a carefully selected combination of probiotics may be useful as a potential non-invasive treatment for patients suffering from neurodevelopmental disorders including ASD.
EXPERIMENTAL PROCEDURES
Mice and Maternal Diet
C57Bl6/J mice were obtained from Jackson Laboratories (#000-664) and were kept on a 12h light/dark cycle and had access to food and water_ad libitum_. Females were placed on either a regular diet (RD) consisting of 13.4% kcal from fat, 30% kcal from protein, and 57% kcal from carbohydrates (Lab Diets, #5001) or high fat diet (HFD) consisting of 60% kcal from fat, 20% kcal from protein, and 40% kcal from carbohydrates (Research Diets, #D12492). Maternal weight was measured weekly. Maternal total and fat mass were measured using an mq7.5 Minispec NMR body composition analyzer. After eight weeks on diet, females were paired with C57Bl6/J adult males to produce subject offspring. Resulting offspring were weaned at three weeks of age and all placed on RD, regardless of maternal diet (MRD or MHFD). Germ-free mice (C57Bl6/J) were maintained in a flexible isolator fed with HEPA-filtered air and provided with irradiated food and water. Germ-free offspring were weaned at four weeks of age. All behavioral tests were performed on 7–12-week-old male mice. Animal care and experimental procedures were approved by Baylor College of Medicine’s Institutional Animal Care and Use Committee in accordance with all guidelines set forth by the U.S. National Institutes of Health.
Reciprocal Social Interaction
Mice were placed in a 25×25×25cm Plexiglass arena, to which they had not been previously habituated, with either a familiar (cage-mate) or stranger age- and sex-matched conspecific. In all experiments, paired mice were matched for maternal diet, colonization source, and/or treatment. We recorded the time a pair of mice engaged in social interaction (close following, touching, nose-to-nose sniffing, nose-to-anus sniffing, and/or crawling over/under each other). The human observer was blind to maternal diet and/or treatment group. Social behavior was analyzed with AnyMaze automated software.
Three-Chamber Social Test
Crawley’s three-chamber test for sociability and preference for social novelty was performed as described (Silverman et al., 2010). In brief, the mouse first experienced a 10-minute period of habituation during which it was allowed to freely explore a 60×40×23cm Plexiglass arena divided into three equally sized, interconnected chambers (Left, Center, Right). Sociability was measured during a second ten-minute period in which the subject could interact either with an empty wire cup (Empty) or a wire cup containing an age and sex-matched stranger conspecific (Mouse 1). Time spent interacting (sniffing, crawling upon) with either the empty cup or the stranger mouse contained in the other cup as well as time spent in each chamber was recorded using the AnyMaze software, by independent observers. Empty cup placement in the Left or Right chamber during the Sociability period was counterbalanced between trials. Finally, preference for social novelty was assayed by introducing a second stranger mouse (Mouse 2) into the previously the empty wire cup. Time spent in each chamber as well as time spent interacting with either Mouse 1 or Mouse 2 was recorded using the automated AnyMaze software by independent observers.
Marble Burying
Marble burying was performed as previously described (Thomas et al., 2009). Briefly, mice were placed in a standard-sized cage containing 20 regularly-spaced black marbles sitting on fine-wood chipped bedding 5 cm in depth. After 20 minutes, the mouse was removed and marbles with at least two-thirds of their depth obscured by wood chips were counted as buried.
Open Field
Mice were placed in an open arena (40×40×20 cm) and allowed to explore freely for 10 minutes while their position was continually monitored using tracking software (ANY-Maze). Tracking allowed for measurement of distance traveled, speed, and position in the arena throughout the task. Time spent in the center of the arena, defined as the interior 20×20 cm, was recorded.
16s rRNA Gene Sequencing
Methods were adapted from protocols developed for the NIH-Human Microbiome Project (Human Microbiome Project, 2012a, b). For a detailed protocol see Supplemental Experimental Procedures.
Whole Genome Shotgun Sequencing
Individual libraries constructed from each sample were loaded onto the HiSeq platform (Illumina) and sequenced using the 2×100 bp pair-end read protocol. Illumina paired-end libraries were constructed from total genomic DNA isolated from each sample. The DNA was sheared into approximately 400–600 bp fragments followed by ligation of Illumina adaptors containing molecular barcodes for downstream de-multiplexing. These products were then amplified through ligation-mediated PCR (LM-PCR) using KAPA HiFi DNA Polymerase (Kapa Biosystems, Wilmington, MA, USA). Following bead purification with Agencourt AMPure XP (Beckman Coulter, Brea, CA, USA), quantification and size distribution of the LM-PCR product was determined using the LabChip GX electrophoresis system (PerkinElmer, Akron, OH, USA). Libraries were pooled in equimolar amounts at 6 samples per pool, and prepared for sequencing with TruSeq PE Cluster Generation Kit (Illumina). Each library pool was loaded onto one lane of a HiSeq 2000 flow cell spiked with 1% PhiX control library. Sequencing files were de-multiplexed with CASAVA version 1.8.3 (Illumina).
Quality filtering, trimming and de-multiplexing was carried out by a custom pipeline containing Trim Galore and cutadapt (Martin, 2011) for adapter and quality trimming, and PRINSEQ (Schmieder and Edwards, 2011) for low-complexity filtering and sequence deduplication. In addition, Bowtie2 v2.2.1 was used to map reads to MetaPhlAn markers for the classification of bacterial species (Segata et al., 2012).
Colonization of Germ-Free Mice by Fecal Microbiota Transplant
Fresh fecal samples were collected from donor mice/microbiome cohort and homogenized on ice in sterile PBS under sterile conditions. The resulting slurry was spun at 1,000_g_ for three minutes at 4°C. The supernatants were isolated and diluted to 5×109 CFU/ml with sterile PBS. Four- or eight-week-old C57Bl6/J germ-free (GF) recipient mice were then immediately colonized by a single gavage with 0.2mL solution. Fecal samples were collected from the colonized GF mice at 24h, 7d, 14d, 28d, and 56d following colonization. Fecal samples were snap frozen and stored at −80°C until prepared for sequencing. Behavioral experiments were initiated at three weeks post-transplant.
Culture and Treatment with L. reuteri and L. johnsonii
Lactobacillus reuteri MM4-1A (ATCC-PTA-6475) and_Lactobacillus johnsonii_ (ATCC 33200) were cultured anaerobically in MRS broth in a 90% N2, 5% CO2, 5% H2 environment. L. reuteri was heat-killed by keeping the bacteria at 80°C for 20 minutes. Bacterial viability was assessed by plating and the efficacy of the heat-kill procedure was confirmed by the absence of colony growth following plating. Cultures were centrifuged, washed, and resuspended in anaerobic solution (PBS) and frozen at −80°C until use. PBS, live or heat-killed L. reuteri were added to the drinking water, which was changed daily to minimize dosage variability. Whereas the experimental group received live bacteria, one control group received identically prepared cultures of heat-killed bacteria. A second group of control mice received water treated with PBS alone. Live and heat-killed L. reuteri were supplied at a dosage of 1×108 organisms/mouse/day continuously in drinking water. Mice consumed the treated water ad libitum over the treatment period. The treated drinking water for each group was replaced daily two hours prior to the onset of the dark cycle to minimize variation in microbial exposure. Behavioral assays were initiated after 4-weeks of L. reuteri or control treatment. The protocol of the L. johnsonii preparation and administration matched the L. reuteri protocol. Fecal samples for sequencing and tissue used in the immunofluorescence studies were collected at the end of the treatment.
Immunofluorescence
Mice were deeply anesthetized by inhalation of isoflurane and perfused transcardially with 10ml 0.9% phosphate-buffered saline followed by 30ml 4% paraformaldehyde in 0.1M phosphate buffer (PFA). Brains were post-fixed in 4% PFA at 4°C overnight, then cryoprotected in 30% sucrose 0.1M PB over three days. Coronal slices (30µm) thick were obtained from frozen tissue using a sliding blade microtome then transferred to ice cold PBS. Slices were blocked with 5% normal goat serum, 0.3% Triton X-100 0.1M PB (PBTgs) for 1h rocking at RT and then incubated in primary antibodies (rabbit anti-oxytocin, ImmunoStar #20068, 1:2000; mouse anti-NeuN, Millipore, #MAB377, 1:2000) diluted in PBTgs rocking at 4°C for 24h. Slices were then washed three times with 0.3% Triton X-100 0.1M PB. Primary antibodies were visualized using secondary goat anti-rabbit Alexa Fluor® 488 (ThermoFisher Scientific, #A-11034) and goat anti-mouse Alexa Fluor® 594 (ThermoFisher Scientific, #A-11032) antibodies (1:1000 dilution). Slices were incubated in secondary antibodies rocking in the dark for 1h at RT. Five minute final washes with each of PBTgs, 0.1M PB, and 0.05M PB preceded mounting onto 2% gelatin (Sigma-Aldrich, #G9391)-coated coverslips. Nuclei were visualized using Vectashield H-1200 with DAPI (Vector Labs, #H-1200).
Fluorescent imaging and data acquisition was performed on a Zeiss AxioImager.Z2 microscope (Car Zeiss MicroImaging) mounted with an AxioCam digital camera (Carl Zeiss MicroImaging). Images were captured using AxioVision acquisition software (Carl Zeiss Microimaging). All images within a given data set were acquired at identical exposure times, within a given channel, to allow comparison of signal intensity. In some images, contrast and brightness were linearly adjusted using Photoshop (Adobe). Image processing was applied uniformly across all images within a given data set. Fluorescence intensity was measured in ImageJ (NIH) by selecting regions of interest (Oxytocin- and NeuN-positive hypothalamic cell bodies). Hypothalamic oxytocin-expressing neuronal population and NeuN+ cell number was assessed in ImageJ using the following operational sequence: (1) open image file, (2) subtract background, (3) adjust threshold, (4) convert to mask, (5) watershed, (6) analyze particles. Automatic identification of cell boundaries was validated against the source image by an experimenter blind to group allocation.
Oxytocin Administration
Oxytocin was obtained from Tocris Bioscience (product 1910) and solubilized in 10% dimethyl sulfoxide (DMSO) in PBS. 10% DMSO in PBS was used as the vehicle control. Mice received oxytocin intranasally (at approximately 200µg/kg) thirty minutes prior to behavior. 1.25µl of oxytocin or vehicle solution was injected into each nostril from P10 pipette. Oxytocin dose was selected according to dosages reported to rescue social behavior in genetic models phenotypically expressing ASD-like behaviors (Penagarikano et al., 2015).
Electrophysiology
Recordings were performed as recently described (Huang et al., 2016) and the investigators were kept blind to treatment conditions. Briefly, mice were anesthetized with a mixture of ketamine (100 mg/kg), xylazine (10 mg/kg) and acepromazine (3 mg/kg). Horizontal slices (225 – 300 µm thick) containing the VTA were cut from the brains of C57BL/6J mice. Mice were transcardially perfused with an icecold, oxygenated solution containing (in mM) NaCl, 87; NaHCO3, 25; KCl, 2.5; NaH2PO4, 1.25; MgCl2, 7; CaCl2, 0.5; dextrose, 25; sucrose, 75. Horizontal slices were cut with a vibrating tissue slicer (VF-100 Compresstome, Precisionary Instruments, San Jose, CA, or Leica VT 1000S, Leica Microsystems, Buffalo Grove, IL), incubated at 34°C for 40 min, kept at room temperature for at least 30 min before their transfer to a recording chamber continuously perfused with artificial cerebrospinal fluid (ACSF) at 32°C and a flow rate of 2–3 ml/min. The recording ACSF contained in mM: 120 NaCl, 3.3 KCl, 1.25 NaH2PO4, 25 NaHCO3, 10 Dextrose, 1 MgCl2 and 2 CaCl2. Recording pipettes were made from thin-walled borosilicate glass (TW150F-4, WPI, Sarasota, FL). After filling with intracellular solution (in mM): 117 CsMeSO3; 0.4 EGTA; 20 HEPES; 2.8 NaCl, 2.5 ATP-Mg 2.0; 0.25 GTP-Na; 5 TEA-Cl, adjusted to pH 7.3 with CsOH and 290 mosmol/l, they had a resistance of 3–5 MΩ. Data were obtained with a MultiClamp 700B amplifier, digitized at 20 kHz with a Digidata 1440A, recorded by Clampex 10 and analyzed with Clampfit 10 software (Molecular Devices). Recordings were filtered online at 3 kHz with a Bessel low-pass filter. A 2 mV hyperpolarizing pulse was applied before each EPSC to evaluate the input and access resistance (Ra). Data were discarded when Ra was either unstable or greater than 25MΩ, holding current was > 200 pA, input resistance dropped > 20% during the recording, or EPSCs baseline changed by > 10%. After establishing a gigaohm seal (> 2GΩ) and recording stable spontaneous firing in cell-attached, voltage clamp mode (−70 mV holding potential), cell phenotype was determined by measuring the width of the action potential and the presence of an Ih current. AMPAR/NMDAR ratios were calculated as previously described (Huang et al., 2016). Briefly, neurons were voltage-clamped at +40 mV until the holding current stabilized (at < 200 pA). Monosynaptic EPSCs were evoked at 0.05 Hz with a bipolar stimulating electrode placed 50–150 µm rostral to the lateral VTA. Picrotoxin (100 µM) was added to the recording ACSF to block GABARA-mediated IPSCs. After recording the dual-component EPSC, DL-AP5 (100 µM) was bath-applied for 10 min to remove the NMDAR component, which was then obtained by offline subtraction of the remaining AMPAR component from the original EPSC. The peak amplitudes of the isolated components were used to calculate the AMPAR/NMDAR ratios. Picrotoxin and DL-AP5 were purchased from Tocris Bioscience and all other reagents were obtained from Sigma-Aldrich.
Statistical Analysis
Data are presented as mean ± SEM. For behavioral experiments, statistics were based on the two-sided unpaired Student’s_t_-tests, one- or two-way ANOVA with Bonferroni post-hoc analysis to correct for multiple comparisons, unless otherwise indicated.P, t, and F values are presented in the figure legends,n values are provided in the figures.P<0.05 was considered significant. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. GraphPad’s Prism 6 (La Jolla, CA) software was used to perform statistical analyses and for generating graphical representations of data. For 16S rRNA gene sequencing, analysis and visualization of microbiome communities was conducted in RStudio 0.99.292 [http://www.R-project.org/(2014)], utilizing the phyloseq package (McMurdie and Holmes, 2013) to import sample data and calculate alpha- and beta-diversity metrics. 16S rRNA gene sequencing data was analyzed using Silva 115 and 123 (Quast et al., 2013). Analyses were performed on datasets that were rarefied 1,000×, then averaged and rounded. Amplicon sequences were deposited to the NCBI Sequence Read Archive under accession number TBD. Dirichlet Multinomial Mixture modeling was performed in mother (Schloss et al., 2009). Significance of categorical variables were determined using the non-parametric Mann-Whitney test for two category comparisons or the Kruskal-Wallis test when comparing three or more categories. One-way ANOVA followed by Dunnett’s multiple comparisons test was performed using GraphPad’s Prism 6. Correlation between two continuous variables was determined by linear regressions, where _P_-values indicate the probability that the slope of the regression line is zero. Principal coordinate plots employed the Monte Carlo permutation test to estimate _P_-values. All _P_-values were adjusted for multiple comparisons with the FDR algorithm (Benjamini et al., 2001). No animals or data points were excluded from analyses.
Supplementary Material
1
2 revised
3
4 revised
5
6
7
8
9
Acknowledgments
We thank members of the Costa-Mattioli laboratory, K. Krnjević, M. Beauchamp, and H. Dierick for comments on the manuscript; A. Swennes, J. Garcia, D. Quach and J. Auchtung for technical advice. This work was supported by funding from the National Institutes of Health (NIMH 096816, NINDS 076708) to M.C.-M and the Alkek Foundation and Baylor College of Medicine to the Alkek Center for Metagenomics and Microbiome Research to J.F.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization and Methodology, S.A.B. and M.C.-M.; Investigation, S.A.B. and G.V.D.P.; Analysis, S.A.B., G.V.D.P., T.A.A., N.J.A., J.F.P., and M.C.-M.; Writing – Original Draft, S.A.B. and M.C.-M.; Writing – Review & Editing, S.A.B., G.V.D.P., T.A.A., N.J.A., J.F.P, and M.C.-M.; Visualization, S.A.B.; Funding Acquisition, M.C.-M.; Supervision, M.C.-M.
REFERENCES
- Aye IL, Rosario FJ, Powell TL, Jansson T. Adiponectin supplementation in pregnant mice prevents the adverse effects of maternal obesity on placental function and fetal growth. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:12858–12863. doi: 10.1073/pnas.1515484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Bilder DA, Bakian AV, Viskochil J, Clark EA, Botts EL, Smith KR, Pimentel R, McMahon WM, Coon H. Maternal prenatal weight gain and autism spectrum disorders. Pediatrics. 2013;132:e1276–e1283. doi: 10.1542/peds.2013-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton JL, Bilbo SD. Developmental programming of brain and behavior by perinatal diet: focus on inflammatory mechanisms. Dialogues in clinical neuroscience. 2014;16:307–320. doi: 10.31887/DCNS.2014.16.3/jbolton. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan M, Hornig M, Schultz AF, Gunnes N, Hirtz D, Lie KK, Magnus P, Reichborn-Kjennerud T, Roth C, Schjolberg S, et al. Association of maternal report of infant and toddler gastrointestinal symptoms with autism: evidence from a prospective birth cohort. JAMA psychiatry. 2015;72:466–474. doi: 10.1001/jamapsychiatry.2014.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly N, Anixt J, Manning P, Ping ILD, Marsolo KA, Bowers K. Maternal metabolic risk factors for autism spectrum disorder-An analysis of electronic medical records and linked birth data. Autism research : official journal of the International Society for Autism Research. 2016 doi: 10.1002/aur.1586. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nature reviews Neuroscience. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Davari S, Talaei SA, Alaei H, Salami M. Probiotics treatment improves diabetes-induced impairment of synaptic activity and cognitive function: behavioral and electrophysiological proofs for microbiome-gut-brain axis. Neuroscience. 2013;240:287–296. doi: 10.1016/j.neuroscience.2013.02.055. [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Molecular psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Darvishzadeh A, Huang KW, Malenka RC. Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature. 2013;501:179–184. doi: 10.1038/nature12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322:900–904. doi: 10.1126/science.1158668. [DOI] [PubMed] [Google Scholar]
- Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PloS one. 2014;9:e113026. doi: 10.1371/journal.pone.0113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe SE, Gossen A, Rademacher L, Hahn A, Westphal L, Grunder G, Spreckelmeyer KN. Oxytocin influences processing of socially relevant cues in the ventral tegmental area of the human brain. Biological psychiatry. 2013;74:172–179. doi: 10.1016/j.biopsych.2012.12.023. [DOI] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of general psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Placzek AN, Viana Di Prisco G, Khatiwada S, Sidrauski C, Krnjevic K, Walter P, Dani JA, Costa-Mattioli M. Translational control by eIF2alpha phosphorylation regulates vulnerability to the synaptic and behavioral effects of cocaine. eLife. 2016;5 doi: 10.7554/eLife.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, Hessler NA. Social modulation during songbird courtship potentiates midbrain dopaminergic neurons. PloS one. 2008;3:e3281. doi: 10.1371/journal.pone.0003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project, C. A framework for human microbiome research. Nature. 2012a;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012b;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC. Maternal obesity, metabolism, and pregnancy outcomes. Annual review of nutrition. 2006;26:271–291. doi: 10.1146/annurev.nutr.24.012003.132249. [DOI] [PubMed] [Google Scholar]
- Krakowiak P, Walker CK, Bremer AA, Baker AS, Ozonoff S, Hansen RL, Hertz-Picciotto I. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics. 2012;129:e1121–e1128. doi: 10.1542/peds.2011-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, Ebstein RP. Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Molecular psychiatry. 2008;13:980–988. doi: 10.1038/sj.mp.4002087. [DOI] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nature communications. 2014;5:3889. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adaptor sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- Mathers JC, McKay JA. Epigenetics - potential contribution to fetal programming. Advances in experimental medicine and biology. 2009;646:119–123. doi: 10.1007/978-1-4020-9173-5_13. [DOI] [PubMed] [Google Scholar]
- Mayer EA, Padua D, Tillisch K. Altered brain-gut axis in autism: comorbidity or causative mechanisms? BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36:933–939. doi: 10.1002/bies.201400075. [DOI] [PubMed] [Google Scholar]
- Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. The Journal of clinical investigation. 2015;125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Batshaw ML, Hoffman EP. Genomics, intellectual disability, and autism. N Engl J Med. 2012;366:733–743. doi: 10.1056/NEJMra1114194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis MR, Melis T, Cocco C, Succu S, Sanna F, Pillolla G, Boi A, Ferri GL, Argiolas A. Oxytocin injected into the ventral tegmental area induces penile erection and increases extracellular dopamine in the nucleus accumbens and paraventricular nucleus of the hypothalamus of male rats. The European journal of neuroscience. 2007;26:1026–1035. doi: 10.1111/j.1460-9568.2007.05721.x. [DOI] [PubMed] [Google Scholar]
- Modi ME, Young LJ. The oxytocin system in drug discovery for autism: animal models and novel therapeutic strategies. Hormones and behavior. 2012;61:340–350. doi: 10.1016/j.yhbeh.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss BG, Chugani DC. Increased risk of very low birth weight, rapid postnatal growth, and autism in underweight and obese mothers. American journal of health promotion : AJHP. 2014;28:181–188. doi: 10.4278/ajhp.120705-QUAN-325. [DOI] [PubMed] [Google Scholar]
- Parracho HM, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. Journal of medical microbiology. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- Pedersen CA, Caldwell JD, Walker C, Ayers G, Mason GA. Oxytocin activates the postpartum onset of rat maternal behavior in the ventral tegmental and medial preoptic areas. Behavioral neuroscience. 1994;108:1163–1171. doi: 10.1037//0735-7044.108.6.1163. [DOI] [PubMed] [Google Scholar]
- Penagarikano O, Lazaro MT, Lu XH, Gordon A, Dong H, Lam HA, Peles E, Maidment NT, Murphy NP, Yang XW, et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Science translational medicine. 2015;7:271ra278. doi: 10.1126/scitranslmed.3010257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutahidis T, Kearney SM, Levkovich T, Qi P, Varian BJ, Lakritz JR, Ibrahim YM, Chatzigiagkos A, Alm EJ, Erdman SE. Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PloS one. 2013;8:e78898. doi: 10.1371/journal.pone.0078898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier N, Leng G, Menzies J. Oxytocin, feeding, and satiety. Frontiers in endocrinology. 2013;4:35. doi: 10.3389/fendo.2013.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nature reviews Neuroscience. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner AC, Skelton JA. Prevalence and trends in obesity and severe obesity among children in the United States, 1999–2012. JAMA pediatrics. 2014;168:561–566. doi: 10.1001/jamapediatrics.2014.21. [DOI] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, et al. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EL, Nousen EK, Chamlou KA. Maternal high fat diet consumption during the perinatal period programs offspring behavior. Physiology & behavior. 2014;123:236–242. doi: 10.1016/j.physbeh.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Chen Z, Tao H, Li C, Zhang X, Tang A, Liu Y. Oxytocin activation of neurons in ventral tegmental area and interfascicular nucleus of mouse midbrain. Neuropharmacology. 2014;77:277–284. doi: 10.1016/j.neuropharm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology. 2009;204:361–373. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Uvnas-Moberg K, Handlin L, Petersson M. Self-soothing behaviors with particular reference to oxytocin release induced by non-noxious sensory stimulation. Frontiers in psychology. 2014;5:1529. doi: 10.3389/fpsyg.2014.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang BR, Zhang XJ, Xu Z, Ding YQ, Ju G. Evidences for vagus nerve in maintenance of immune balance and transmission of immune information from gut to brain in STM-infected rats. World journal of gastroenterology. 2002;8:540–545. doi: 10.3748/wjg.v8.i3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, Gong X, Zhang Y, Yang X, Zhang D. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biological psychiatry. 2005;58:74–77. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2 revised
3
4 revised
5
6
7
8
9