Writ large: Genomic Dissection of the Effect of Cellular Environment on Immune Response (original) (raw)
. Author manuscript; available in PMC: 2016 Nov 16.
Published in final edited form as: Science. 2016 Oct 7;354(6308):64–68. doi: 10.1126/science.aaf5453
Abstract
Cells of the immune system routinely respond to cues from their local environment and feedback to their surrounding through transient responses, choice of differentiation trajectories, plastic changes in cell state, and malleable adaptation to their tissue of residence. Genomic approaches have opened the way for comprehensive interrogation of such orchestrated responses. Focusing on genomic profiling of transcriptional and epigenetic cell state, we discuss how they are applied to investigate immune cells faced with various environmental cues. We highlight some of the emerging principles, on the role of dense regulatory circuitry, epigenetic memory, cell type fluidity, and reuse of regulatory modules, in achieving and maintaining appropriate responses to a changing environment. These provide a first step toward a systematic understanding of molecular circuits in complex tissues.
Introduction
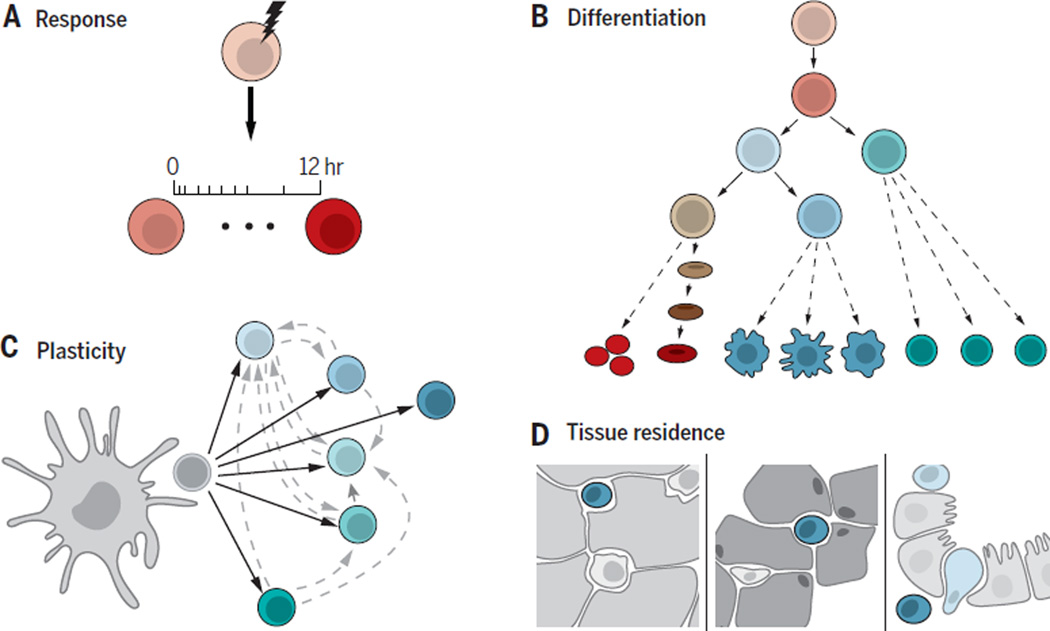
Homeostasis is a hallmark of biological systems that actively maintain a near-constant function in the face of a changing environment. In most animals, multiple systems, from the cellular to the organismal level, including the immune system, the nervous system and fibroblasts in connective tissue, play crucial homeostatic roles, as they sense, respond and adapt to an ever-changing environment – both external and intra-organismal – in different tissues in the body. In particular, the immune system achieves tunability, plasticity and adaptability to the environment at several levels (Figure 1). First, immune cells have transient responses to diverse factors, such as microbes, vaccines, tissue damage, or cancer cells (1). Second, controlled differentiation from progenitor cells generates different cell type balances (2). Furthermore, cells exhibit plasticity, such that certain immune cells can change their identity in the context of new signals (3, 4). Finally, cells can locate and relocate throughout the body, adapting their identity to their locale (1, 5).
Figure 1. Key modes of immune-environment interaction.
A) Transient responses to signals. (B) Balanced differentiation along hematopoiesis; (C) Stable yet plastic cell type polarization; (D) Malleable adaptation of tissue resident cells.
These abilities are controlled by a complex molecular circuitry, both intra-cellular (within immune cells) and through interaction amongst immune cells, or between immune cells and other cell types, including cells of the nervous system or fibroblasts. Malfunction in each of these mechanisms can contribute and give rise to disease. Manipulating them, in turn, provides important avenues for therapies, as has been the case in autoimmune disease and cancer. However, given the diversity of molecules, cell types and tissues, as well as the inherent uncertainties and noise in both molecular systems and measurement techniques, systematic dissection of these intra- and inter- cellular circuitries is remarkably challenging.
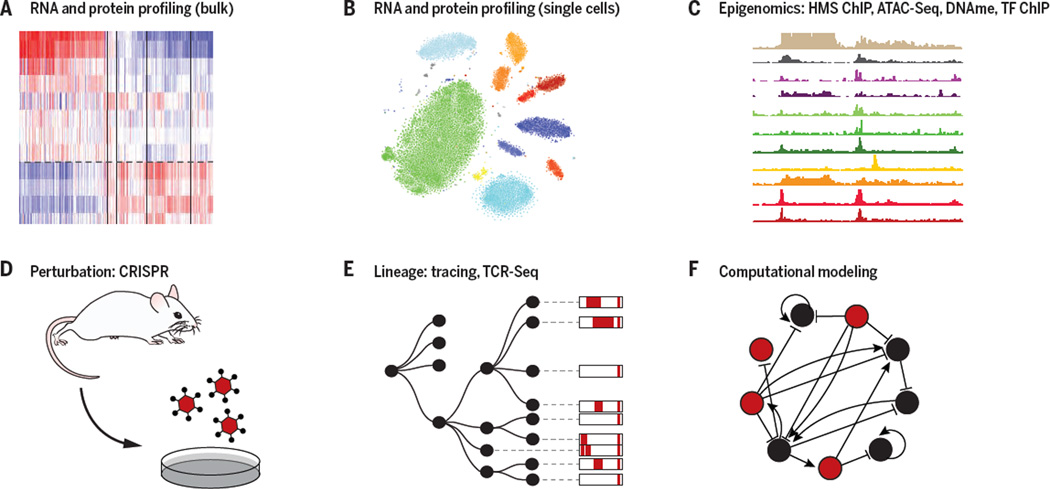
Genomics approaches have opened unique opportunities to address this challenge (Figure 2). Profiling of the genome, epigenome, transcriptome, proteome, and metabolome has been instrumental in defining cell types and states and characterizing the molecular changes that occur as cells respond to their surroundings. Recently, single cell genomics can distinguish these with remarkable resolution, even when the types and states of immune cells are not necessarily known (6–8), and when they are embedded in complex tissues (6, 9, 10) with spatial resolution (11–13). Profiling assays, especially of molecular interactions with ChIP-Seq (14) and interaction proteomics help identify key aspects of the underlying molecular mechanisms – such as key transcription factors (TFs) and regulatory regions. To determine causality, large-scale perturbations, either engineered with RNAi and CRISPR-based genome editing (15), or natural variation between individuals in a population (16–19), provide a systematic mean to assess the causal role of different circuit components, including the context of disease in vivo.
Figure 2. Genomic tools for dissecting immune-environment interactions.
Shown are key components of the current genomic tool box, including profiling of RNA, protein and protein modification levels in bulk samples (A) and single cells (B), epigenomic measurements of TF binding, histone modification, DNA methylation, and chromatin accessibility (C), the ability to systematically perturb genes through genome engineering (D) or natural variation, tracing of cells with engineered molecular barcodes or TcR and BcR clonality (E), and computational algorithms that use profiling and perturbation data to infer genetic causality and molecular mechanisms (F).
While these assays can be applied in principle to many systems, analysis of immune cell responses has been at the forefront, providing a paradigm for other systems. First, the identity of many immune cell sub-types is known, and they can be isolated for analysis from both humans and mice; this has been critical, especially prior to the advent of single cell genomics. Furthermore, many primary immune cells can be studied both ex vivo and in vivo, including adoptive transfer of cells, bone marrow transplants and lineage tracing in animal models (6, 20, 21). Finally, immune cells are present throughout the body, differentiate continuously, and are implicated in many diseases, thus providing a broad lens into organismal physiology.
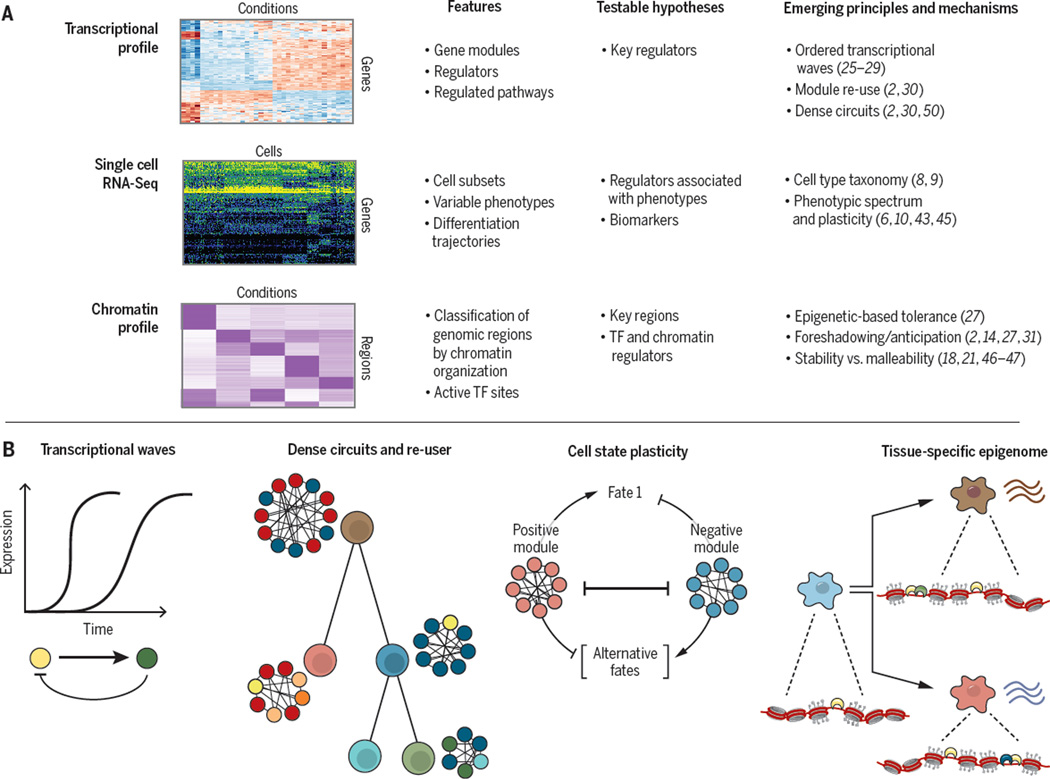
Here, we highlight the power of genomics to dissect environmental responses in immune cells. We focus on assays related to the regulation of mRNA expression, especially transcriptional and epigenetic profiling. Using macrophages and T cells as case studies, we illustrate the types of features that are underscored with different profiles, the resulting testable hypotheses that can be followed up in dedicated low-throughput experiments, and the emerging organizing principles (Figure 3). Finally, we discuss how these lessons, learned in immune cells, can be extended to develop approaches to dissect the overall function of diverse cells in maintaining homeostasis.
Figure 3. Key features, hypotheses and principles revealed by genomic studies of immune-environment interactions.
Three key genomic tools used to analyze transcription and epigenetic mechanisms that participate in immune cell responses. Bottom: The main features characterized by each tool, testable hypotheses derived by computational analysis; and current emerging principles from such studies.
Molecular responses of the immune system through transient intracellular circuits
Macrophages, innate immune cells of the myeloid lineage serve key immune defense functions through phagocytosis, and by communicating with adaptive immune cells through antigen presentation and secretion of and response to cytokines and chemokines (22–24). Mononuclear phagocyte subpopulations are located in the circulatory system and spleen and can differentiate into macrophages, but macrophages also reside in other tissues where they acquire specific characteristics and contribute to local hemostasis (1, 22, 23).
In their role as immune sensors, macrophages express pattern recognition molecules (e.g., Toll like receptors (TLR)), which detect conserved pathogen-associated or tissue damage-associated molecular patterns, and mount the appropriate response. Genomic analysis of macrophages provided key insights on how these transient responses (Figure 1a) are carried out. For instance, transcriptional profiling along a time course following TLR4 activation with bacterial lipopolysaccharide (LPS), an inducer of inflammation, showed that genes are induced in several consecutive “waves” (25), a phenomenon observed in many other response systems (26) (Figure 3). The response waves are regulated through successive activation of transcriptional regulators (27, 28), whose identity can be predicted from DNA-binding motifs enriched in the 5’ regions. This approach helped identify activating TF 3 (ATF3) as an early regulator of the LPS response in macrophages (25). ATF3 was then shown to recruit histone deacetylase to repress its target genes – thus forming a negative feedback. This mechanism may be essential for controlling the extent and duration of TLR-induced inflammation during infection, avoiding rampant inflammation and tissue damage.
The importance of epigenetic regulation as a way of controlling macrophage activation was further demonstrated by investigating the formation of “memory” in antigen-exposed macrophages (Figure 3) – where macrophages repeatedly exposed to a specific component (e.g., LPS) become tolerant, and selectively produce anti-microbial, but not pro-inflammatory signals, to avoid tissue damage (27, 28). Profiling of gene expression and histone modifications (Figure 3) during the macrophage response to repeated stimulation reveals that tolerant genes that are not re-induced in repeated exposure to LPS are enriched for pro-inflammatory functions, and are transiently silenced by loss of activating histone marks. Conversely, non-tolerant genes are enriched for anti-microbial functions and are associated with a faster and stronger transcriptional response upon LPS re-stimulation (compared to the primary stimulation), through persistence or rapid acquisition of activating histone marks (H3 trimethylation and H4 acetylation respectively) and recruitment of RNA polymerase II (Pol II) and chromatin remodeling complexes. These distinct epigenetic mechanisms depend on the protein products generated during the first exposure to LPS, emphasizing the common role of negative feedback in controlling innate immune response (26, 29) (Figure 3).
Depending on their tissue of origin and stimulus, macrophages can acquire distinct functional states. Two well-studied states are pro-inflammatory _M_1 cells, derived in the presence of interferon gamma, and immuno-suppressive _M_2 cells that can be induced with interleukin 4 or 13 (30). Profiling the transcriptional response of monocyte-derived macrophages to a more diverse set of stimuli suggests that macrophages can mount diverse transcriptional programs beyond these two states, depending on the metabolites, cytokines, and ligands to which they are exposed (30). Computational analyses of gene modules that are co-regulated across programs suggested that diversity between programs is generated by different combinations of active transcriptional regulators (Figure 3). Some of these regulators are “re-used” across all programs (e.g., the lineage specifying factor PU.1), whereas others are important only in certain contexts (e.g., STAT1 and STAT6, in the interferon gamma and interleukin 4 responses, respectively). Mapping the diverse activation programs also provided a way to decompose bulk samples into constituent responses, for instance, proposing that alveolar macrophages from chronic obstructive pulmonary disease patients are depleted in the inflammatory (M1) state, which may explain their poor response to anti-inflammatory therapeutics. Such analysis could further benefit from application of single-cell genomics (Figure 3).
Systematic perturbations have helped establish the causality of these observations. Causal loci were discovered either by associating natural genetic variation with variation in the transcriptional response across human individuals (16, 18, 19) or mouse strains (17), or by perturbing genes and measuring the effect on the transcriptome (15, 29).
Balanced differentiation from progenitor cells
The diverse cell types of the hematopoietic system are organized in a taxonomy along different lineages, and are produced daily from a small pool of stem cells (Figure 1b). The composition of hematopoietic cell subsets is tightly controlled, ensuring both homeostatic control and responsiveness to environmental cues. As in studies of transient immune responses, genomic and epigenomic profiling have shed light on the transcriptional shifts during hematopoiesis and the regulatory programs that govern them (Figure 3), primarily focusing on unperturbed, homeostatic conditions in humans (2, 31) and mice (14, 32). Transcriptional profiling revealed substantial expression changes between hematopoietic cell subtypes, comparable to those between different tissues (2). Computational analysis of these data, focused on “modules” of co-regulated genes and the regulators associated with them, has identified global organizing principles in hematopoiesis that may also apply more generally (Figure 3).
First, a large set of predicted transcriptional regulators (across all hematopoietic lineages) form a dense inter-connected network of regulatory interactions in each cell type, and with the same regulator used in multiple hematopoietic subsets. This organization may confer robustness, but could also be liable to dysregulation and cancer (2). This model challenged and expanded an earlier hierarchal model of hematopoiesis controlled by a small number of TFs, expressed sequentially (33).
Furthermore, there is no simple partitioning of regulatory activity at different lineages. Instead, entire modules of co-regulated genes, along with their upstream regulators are re-used across distinct lineages, either because of shared functional needs in otherwise different cells (e.g., mitochondrial and oxidative phosphorylation in erythroid progenitors, granulocytes and monocytes (2)), or due to shared developmental descent. This latter pattern is often reflected in “transitional” cases (Figure 3), with a gradual onset and offset of programs along the hematopoietic cell hierarchy (2). The transitional gene modules could be explained by either the presence of cells at different phases of development within seemingly pure populations of progenitor cells, or because regulatory programs of a more differentiated state are foreshadowed by pre-existing programs at earlier stages. Both models are plausible, and non-exclusive, the second model is strongly supported by profiling of TF binding in humans (2) and of chromatin organization in hematopoiesis in mice (14) and humans (31), where a large portion of the enhancers exhibited a “transitional” behavior – already established in the precursor cells, possibly in a poised (and transcriptionally inactive) state. Single cell RNA-Seq studies can help further address how transition to multiple lineages is concomitantly encoded in progenitor cells. To date, some studies suggested there may be distinct subsets within myeloid progenitor cells that are partially skewed towards distinct functional fates by the expression of key sub-lineage regulators (34), whereas others emphasized evidence for obligatory mixed-lineage states within the same single progenitor cell (35).
While it is tempting to think of hematopoiesis as stereotypic, differentiation is affected and driven by the environment, including not only the stromal niche and other immune cells, but also stress and pathogens (27, 36). For example, stress signals can lead to production of more innate immune myeloid cells at the expense of other lineages, especially lymphoid cells (36). Furthermore, distinct subpopulations of stem and progenitor cells can be activated (37) to produce cytokines that affect core immune responses. Genomic analysis, including at the single cell level, will shed more light on the regulation of hematopoiesis by such signals.
Plasticity of cell differentiation
As immune cells become more committed, differentiation and balancing between sub-lineages become even more intertwined with environmental responses. For example, naïve T helper (Th) cells can differentiate into multiple specialized cell types, including conventional (Tconv) Th cells (e.g., Th1, Th2, Th17, Th9 cells) and regulatory Th cells (e.g., Treg, Tr1) (4). Given the diverse, and partially opposing functions of different Th cells (4), it is critical to maintain their correct blend, in a manner sensitive to and controlled by environmental signals. First, the relative proportion of Th subtypes that will develop from a limited pool of naive Th cells is regulated by the blend of cytokines to which a naive cell is exposed, often produced by antigen presenting cells (e.g., to Treg cells in the presence of TGF-s, but skewed to Th17 cells if interleukin 6 is also present). Second, while differentiated Th cell sub-types can be maintained stably over time, including in the memory pool (38), some can also transition into other, parallel sub-types (Figure 1c), depending on extra-cellular signals, from cytokines, to oxygen or nutrients (39), to components of the microbiome (3). The process leading to these diverse types is often called polarization, rather than terminal differentiation, and the change between the types is referred to as plasticity (1, 4), and can play critical physiological roles. For example, plastic polarization of tissue-resident macrophages helps fulfill changing functional demands from the tissue in which they reside (1, 40); Th cell plasticity could allow an organism to respond to a changing environment even if cells were originally committed to the memory pool in a different state (20).
The distinction between a permanent and plastic state can be defined in principle, but challenging to identify in practice (41) because it can be hard to determine whether a stable state is permanent, and whether a cell’s expression of a marker of one type and functions of another is not mere noise (41). Genomic profiling coupled with lineage tracing and functional studies were instrumental in addressing these questions by defining the spectrum of cell states that can be attained by Th cells, the transition between states through differentiation and polarization, and the underlying regulatory mechanism (Figure 3).
Using the RNA profile of the cell as it’s functional identity, and coupled it to lineage tracing has helped identify both how a cell state it stably maintained and when it shifts plastically ((20, 38, 42), reviewed in (4)). For example, Th17 cells can begin to express both cytokines and seminal TFs of other Th cells (20), but these could reflect either transition to another type, or a transient functional deviation. Lineage tracking of Th17 cells in the gut followed by RNA-Seq, showed that they can adopt a transcriptional signature of regulatory T cells and anti-inflammatory capacity. Conversely, tracing Th17 cells in a melanoma mouse model showed that while they can acquire transcriptional features of Th1 cells, they remain distinct from similarly traced Th1 cells, acquiring a stem-cell like signatures and longevity, with increased tumor eradication capacity (38).
While these studies profiled cell populations defined by cell surface, cytokine or TF expression, recent single cell genomic studies (6, 43) have increased the resolution at which we characterize cellular populations and their fluidity. For example, Th17 cells were shown to span a continuum of states, from higher expression of a program associated with pathogenic effect to one characteristic of regulatory cells, with distinct regulators for each program (6). Single cell RNA-Seq also provides a way for lineage tracing, by capturing the sequence of the T cell receptor transcript (10, 44, 45).
Profiling of chromatin organization, especially histone marks, across different Th cell types highlighted how epigenetic memory maintains cell state stably, while remaining sufficiently malleable to allow for plasticity (Figure 3). While signature cytokines often have a chromatin pattern congruent with strict cell type definitions, chromatin at other key signature genes of one lineage is not always repressed in other Th lineages, offering a possible basis for future plasticity. Indeed, chromatin marks and accessibility can change even for signature cytokines or TFs following stimulation (46). Conversely, DNA de-methylation and stable chromatin organization, with contribution from chromatin regulators and long non-coding RNAs, play an important role in stability. For instance, de-methylation of specific regulatory elements in a CpG island in the locus of Foxp3, a key regulator of Treg cells, helps stabilizing the cells’ identity, further reinforced by a transcriptional positive feedback loop (47). The ability of chromatin organization to function as a malleable memory device, is reflected by the preponderance of DNA variants associated with autoimmune disease that map to enhancer and other regulatory regions in Th cells (48).
Finally, RNA and TF ChIP-seq profiles, combined with genetic perturbations, have shed light on the intricate intracellular circuits controlling these processes in Treg (49) and inflammatory Th17 cells (50, 51). For example, in Th17 cells they revealed a “Yin-Yang” network of TFs, with two densely connected self-reinforcing, but mutually antagonistic, modules: a larger module promotes the Th17 cell fate and suppresses alternative fates, and a smaller module has an opposite function. The dense, interconnected positive module provides stability; as has also been proposed for other Th lineages (42, 49). The smaller negative module could promote alternative plastic fates.
Malleable cell states mirror tissue location
Immune cells sense, adapt, respond to and affect their environment in the context of the tissue (Figure 1d). Tissue-resident immune cells, sometimes life-long sessile inhabitants, play critical roles in homeostasis and pathology, well beyond responses to pathogens. Tissue-resident macrophages perform unique functions as “accessory cell types” (1) that serve “client” primary cells defining the respective tissue. For example (1, 23), alveolar macrophages are critical for surfactant homeostasis in the lungs, microglia are essential for synaptic pruning in the brain, osteoclasts are critical for the dynamic balance of bone, and splenic red-pulp macrophages help manage heme and iron from aging red blood cells. Tissue-resident Tregs (5) have been identified in visceral adipose tissue (VAT) (52, 53), the intestine (54), muscle (55), and lung (56), with roles from metabolic homeostasis to tissue repair and regeneration.
Genomic analysis has played a critical role in identifying tissue-resident immune cells, characterizing their unique features, determining their tissue-specific functions, and inferring the principles and mechanisms by which they adapt to the diversity of tissues in the body and their changing local conditions.
RNA profiling has identified the level at which immune cells of a single “type” vary given their tissue-of-residence (Figure 3). In addition to a core set of macrophage-associated genes, tissue resident macrophages (18, 21, 40, 57) express distinct gene modules in each tissue type. For example, brain-resident microglia (which are deposited prenatally), develop lock-step with the rest of the CNS during brain development and are susceptible to environmental signals pre-natally (58). Tregs isolated from different tissues have shown a similar distinctions (52–55). The profiles and derived signatures then become the fingerprint of the cell’s identity, and – when coupled to transfer, chimera or lineage tracing experiments – can establish if a cell is stably resident in a tissue (52). They also allow us to follow their clonal expansion in response to stimulus. It is possible that other immune lineages may also follow such principles; single cell profiling of entire tissues (9, 10) will help determine this.
Individual genes expressed in these tissue-specific modules – including TFs, cytokines, chemokines and receptors – provide critical starting points to determine the cells’ functions (e.g., lipid metabolism in Tregs in VAT (53), regulatory mechanisms (e.g., Gata6 in peritoneal macrophages (57)), and interaction with other tissue cell (e.g., Treg-adipocyte interaction through IL10 (53)). The exquisite tunability reflected by these programs, led in turn to the exploration of how they are diversely yet stably imprinted on a cell type based on its local environment (1). Two distinct mechanisms (or a blend thereof) can in principle underlie this phenomenon: a pre-programmed set of states, both preceding and succeeding tissue residence; and/or an environmentally-directed process, either permanent (tissue-resident differentiation) or signal-dependent (plasticity).
Epigenomic analysis of macrophages from different tissues strongly supports the signal-dependent model. Tissue resident macrophages have distinct enhancer landscapes compared to promoters, with a large number of unique, tissue-specific, poised and active enhancers. These enhancers can be substantially – albeit not completely – reprogrammed by environment-specific signals in either bone marrow transplant, tissue transfer experiments or ex vivo manipulations (18, 21). As for Th cell plasticity, differential enhancer usage may underlie the preponderance of genetic variants associated with human immune disease in enhancer regions (48).
Combinatorial regulation by TFs helps in turn to establish transcriptional programs for tissue resident macrophages. During differentiation, a first layer of lineage determining “pioneer” factors delineates cell-type specific enhancers through nucleosome repositioning and recruitment of histone modifying enzymes; after differentiation, a second layer of signal-dependent factors binds in those pre-existing loci. Some enhancers are shared across all tissue resident macrophages (18, 21), but are only poised, and signal-dependent factors modulate the activity of this pre-existing enhancer repertoire to achieve context-dependent gene expression. Other enhancers are formed “de novo” to create epigenetic memory of tissue-residence. Thus, signal-dependent (40), tissue-specific TFs can either work cooperatively with the macrophage pioneer factor PU.1 to form new enhancers or can activate poised enhancers that have been formed and pre-bound by Pu.1. This mechanism can also account for transient tissue-resident programs.
Tissue-specific Tregs also exhibit cooperation between tissue-specific and lineage-specific factors (18, 21, 40, 53). For example, PPARγ, the master regulator of adipocyte differentiation, is the key regulator of the tissue specific program in visceral adipose tissue (VAT) Tregs (53). PPARγ in adipose tissue Tregs responds to the tissue’s signal of insulin and orchestrates the relevant metabolic response, mediated through the same biochemical and molecular mechanisms as in adipocytes. It is tempting to speculate that this intracellular “molecular mirroring” between two different cell types in a single tissue, could help synchronize not only their response to signal but also how this signal is precisely processed and affects the same output modules, beyond what could be achieved by inter-cellular communication alone. Such mirroring could exist between other cell types in tissue, immune and non-immune.
Perspective: Towards a tissue circuit
The cellular environment is interwoven into a single integrated whole in tissues, bringing together diverse cell types – epithelial, immune, neurons, stromal and more – as they differentiate and respond to each other, microbes, nutrients, and other stimuli. These responses can be transient, permanent or plastic, and include migration. Every aspect of this “tissue circuit”, including the proportion of cell types, and their states, function and interactions, changes as the local environment varies.
Dissecting how cells interact to maintain tissue function requires knowing the census of cell types and states, their biological roles in the tissue, the signals received and emitted by each cell and their effects, and the cascade of events underlying dynamic tissue processes. Addressing these questions requires the ability to profile the individual cells that comprise the tissue, consider their spatial position and physical interactions in faithful biological samples, have computational means to reconstruct molecular-level quantitative models that combine intra-cellular with inter-cellular circuits, and functional means to test them. Recent technological breakthroughs – in single cell genomics (59) spatially resolved profiling (12, 13), systematic genetic perturbations (15), and access to tissue biopsies and orgnaoids provide promising steps in that direction.
Analysis at a whole-tissue level should provide an unprecedented view into the cellular and molecular composition of tissues and an understanding of the molecular and functional interactions by which cells cooperate to fulfill tissue function and maintain homeostasis. Ultimately, such an understanding will have the potential to be translated to exceptionally effective new therapies that can restore tissue function and human health.
Acknowledgments
AR was supported by the Howard Hughes Medical Institute. NY was supported by NIH grants U01MH105979 and U01HG007910.
References
- 1.Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nature immunology. 2016;17:9–17. doi: 10.1038/ni.3320. published online EpubJan. [DOI] [PubMed] [Google Scholar]
- 2.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. published online EpubJan 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonelli M, Shih HY, Hirahara K, Singelton K, Laurence A, Poholek A, Hand T, Mikami Y, Vahedi G, Kanno Y, O'Shea JJ. Helper T cell plasticity: impact of extrinsic and intrinsic signals on transcriptomes and epigenomes. Current topics in microbiology and immunology. 2014;381:279–326. doi: 10.1007/82_2014_371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nature reviews. Immunology. 2016;16:149–163. doi: 10.1038/nri.2015.18. published online EpubMar. [DOI] [PubMed] [Google Scholar]
- 5.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annual review of immunology. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. published online EpubMay 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, Kuchroo VK, Park H, Regev A. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. published online EpubDec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JJ, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. published online EpubJun 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, Regev A. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. published online EpubJun 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. published online EpubFeb 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. published online EpubApr 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. published online EpubApr 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Gerner MY, Van Panhuys N, Levine AG, Rudensky AY, Germain RN. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature. 2015;528:225–230. doi: 10.1038/nature16169. published online EpubDec 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, Friedman N, Amit I. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. published online EpubAug 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye C, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, Shalem O, Satija R, Raychowdhury R, Mertins P, Carr SA, Zhang F, Hacohen N, Regev A. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015 doi: 10.1016/j.cell.2015.06.059. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, Knight JC. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. published online EpubMar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gat-Viks I, Chevrier N, Wilentzik R, Eisenhaure T, Raychowdhury R, Steuerman Y, Shalek AK, Hacohen N, Amit I, Regev A. Deciphering molecular circuits from genetic variation underlying transcriptional responsiveness to stimuli. Nature biotechnology. 2013;31:342–349. doi: 10.1038/nbt.2519. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. published online EpubDec 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, Ward LD, Raddassi K, McCabe C, Lee MH, Frohlich IY, Hafler DA, Kellis M, Raychaudhuri S, Zhang F, Stranger BE, Benoist CO, De Jager PL, Regev A, Hacohen N. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. published online EpubMar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limon P, Paiva RS, Ching T, Weaver C, Zi X, Pan X, Fan R, Garmire LX, Cotton MJ, Drier Y, Bernstein B, Geginat J, Stockinger B, Esplugues E, Huber S, Flavell RA. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015;523:221–225. doi: 10.1038/nature14452. published online EpubJul 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. published online EpubDec 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nature immunology. 2016;17:18–25. doi: 10.1038/ni.3325. published online EpubJan. [DOI] [PubMed] [Google Scholar]
- 23.Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nature reviews. Immunology. 2015;15:731–744. doi: 10.1038/nri3920. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. published online EpubApr 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. published online EpubMay 11. [DOI] [PubMed] [Google Scholar]
- 26.Yosef N, Regev A. Impulse control: temporal dynamics in gene transcription. Cell. 2011;144:886–896. doi: 10.1016/j.cell.2011.02.015. published online EpubMar 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. published online EpubJun 21. [DOI] [PubMed] [Google Scholar]
- 28.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. published online EpubJul 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. published online EpubOct 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. published online EpubFeb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, Ravindra M, Chang HY. Lineage-specific and single cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature genetics. 2016 doi: 10.1038/ng.3646. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, Immunological Genome Project C, Best AJ, Knell J, Goldrath A, Joic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemarte-Pelletier A, Malhotra D, Turley S. Identification of transcriptional regulators in the mouse immune system. Nature immunology. 2013;14:633–643. doi: 10.1038/ni.2587. published online EpubJun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. published online EpubJun. [DOI] [PubMed] [Google Scholar]
- 34.Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, David E, Cohen N, Lauridsen FK, Haas S, Schlitzer A, Mildner A, Ginhoux F, Jung S, Trumpp A, Porse BT, Tanay A, Amit I. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. published online EpubDec 17. [DOI] [PubMed] [Google Scholar]
- 35.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016 doi: 10.1038/nature19348. published online EpubAug 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao JL, Ma C, O'Connell RM, Mehta A, DiLoreto R, Heath JR, Baltimore D. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell. 2014;14:445–459. doi: 10.1016/j.stem.2014.01.007. published online EpubApr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldridge MT, King KY, Goodell MA. Inflammatory signals regulate hematopoietic stem cells. Trends in immunology. 2011;32:57–65. doi: 10.1016/j.it.2010.12.003. published online EpubFeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, Roychoudhuri R, Ferreyra GA, Shen W, Durum SK, Feigenbaum L, Palmer DC, Antony PA, Chan CC, Laurence A, Danner RL, Gattinoni L, Restifo NP. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–985. doi: 10.1016/j.immuni.2011.09.019. published online EpubDec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Collins M, Kuchroo VK. Effector T cell differentiation: are master regulators of effector T cells still the masters? Current opinion in immunology. 2015;37:6–10. doi: 10.1016/j.coi.2015.08.001. published online EpubDec. [DOI] [PubMed] [Google Scholar]
- 40.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. published online EpubMay 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakaguchi S, Vignali DA, Rudensky AY, Niec RE, Waldmann H. The plasticity and stability of regulatory T cells. Nature reviews. Immunology. 2013;13:461–467. doi: 10.1038/nri3464. published online EpubJun. [DOI] [PubMed] [Google Scholar]
- 42.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C. A multiply redundant genetic switch 'locks in' the transcriptional signature of regulatory T cells. Nature immunology. 2012;13:972–980. doi: 10.1038/ni.2420. published online EpubOct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proserpio V, Piccolo A, Haim-Vilmovsky L, Kar G, Lonnberg T, Svensson V, Pramanik J, Natarajan KN, Zhai W, Zhang X, Donati G, Kayikci M, Kotar J, McKenzie AN, Montandon R, Billker O, Woodhouse S, Cicuta P, Nicodemi M, Teichmann SA. Single-cell analysis of CD4+ T-cell differentiation reveals three major cell states and progressive acceleration of proliferation. Genome biology. 2016;17:103. doi: 10.1186/s13059-016-0957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stubbington MJ, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nature methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afik S, Yates KB, Bi K, Darko S, Godec J, Gerdemann U, Swadling L, Douek DC, Klenerman P, Barnes EJ, Sharpe AH, Haining WN, Yosef N. Targeted reconstruction of T cell receptor sequence from single cell RNA-sequencing links CDR3 length to T cell differentiation state. bioRxiv. 2016 doi: 10.1093/nar/gkx615. http://dxdoiorg/101101/072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. published online EpubMay 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. published online EpubAug 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. published online EpubFeb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, Leslie C, Shaffer SA, Goodlett DR, Rudensky AY. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nature immunology. 2012;13:1010–1019. doi: 10.1038/ni.2402. published online EpubOct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, Shakya A, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK, Regev A. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. published online EpubApr 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. published online EpubOct 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolodin D, van Panhuys N, Li C, Magnuson AM, Cipolletta D, Miller CM, Wagers A, Germain RN, Benoist C, Mathis D. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell metabolism. 2015;21:543–557. doi: 10.1016/j.cmet.2015.03.005. published online EpubApr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. published online EpubJun 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, Harrison OJ, Owens BM, Lohning M, Belkaid Y, Fallon PG, Powrie F. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–568. doi: 10.1038/nature13577. published online EpubSep 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. published online EpubDec 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, Treuting PM, Rudensky AY. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. published online EpubAug 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma'ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ. C. Immunological Genome, Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. published online EpubNov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, Elinav E, Sieweke MH, Schwartz M, Amit I. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016 doi: 10.1126/science.aad8670. published online EpubJun 23. [DOI] [PubMed] [Google Scholar]
- 59.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. published online EpubMay 21. [DOI] [PMC free article] [PubMed] [Google Scholar]