Defining the mode of tumour growth by clonal analysis (original) (raw)
. Author manuscript; available in PMC: 2017 Aug 11.
Published in final edited form as: Nature. 2012 Aug 23;488(7412):527–530. doi: 10.1038/nature11344
Abstract
Recent studies using the isolation of a subpopulation of tumour cells followed by their transplantation into immunodeficient mice provide evidence that certain tumours1,2, including squamous skin tumours3–5, contain cells with high clonogenic potential that have been referred to as cancer stem cells (CSCs). Until now, CSC properties have only been investigated by transplantation assays, and their existence in unperturbed tumour growth is unproven. Here we make use of clonal analysis of squamous skin tumours using genetic lineage tracing to unravel the mode of tumour growth in vivo in its native environment. To this end, we used a genetic labelling strategy that allows individual tumour cells to be marked and traced over time at different stages of tumour progression. Surprisingly, we found that the majority of labelled tumour cells in benign papilloma have only limited proliferative potential, whereas a fraction has the capacity to persist long term, giving rise to progeny that occupy a significant part of the tumour. As well as confirming the presence of two distinct proliferative cell compartments within the papilloma, mirroring the composition, hierarchy and fate behaviour of normal tissue, quantitative analysis of clonal fate data indicates that the more persistent population has stemcell-like characteristics and cycles twice per day, whereas the second represents a slower cycling transient population that gives rise to terminally differentiated tumour cells. Such behaviour is shown to be consistent with double-labelling experiments and detailed clonal fate characteristics. By contrast, measurements of clone size and proliferative potential in invasive squamous cell carcinoma show a different pattern of behaviour, consistent with geometric expansion of a single CSC population with limited potential for terminal differentiation. This study presents the first experimental evidence for the existence of CSCs during unperturbed solid tumour growth.
Different models have been proposed to explain tumour heterogeneity and growth1,6,7. In the stochastic model, all tumour cells are considered equipotent and only a fraction (due to genetic and/or epigenetic changes) will contribute to tumour growth. By contrast, in the CSC model, only a fraction of tumour cells have high clonogenic potential, and give rise to a predictable hierarchical mode of tumour growth. These two models of tumour growth are not mutually exclusive and competition may still arise in the hierarchical model. Until now, CSC properties have been demonstrated by the ability of a subpopulation of tumour cells to reform a tumour identical to the parental tumour on transplantation into immunodeficient mice. This ‘gold standard’ assay has some caveats as tumour cells are usually transplanted at a heterotopic site, embedded in Matrigel containing a cocktail of growth factors, into an allogenic or xenogenic immunocompromised host8. These studies clearly demonstrate the long-term renewal potential of cancer cells in these experimental conditions, but they do not necessarily reflect the physiological fate of these cells in their native environment1.
Here we sought to investigate the fate of individual tumour cells within their natural microenvironment in the early stage of squamous skin tumours. Chemical-induced carcinogenesis is the most commonly used tumour model in mice9,10. Although this model may only imperfectly mimic the occurrence of human skin cancers, it has proved to be extremely useful in dissecting the different steps of tumour initiation, promotion and malignant progression, and has been instrumental in defining many paradigms in cancer biology relevant to human pathology9,11,12. In this protocol, mice are first treated with the carcinogen DMBA (7,12-dimethylbenz[α]anthracene) followed by continuous administration of the mitogen TPA (12-_O_-tetradecanoylphorbol-13-acetate). During the course of TPA administration, benign tumours, known as papilloma, appear (initiation) and grow steadily (promotion), with a fraction eventually progressing into invasive tumours. Papilloma are characterized by a skin excrescence that expands slowly and presents a stereotypical architecture similar to normal epidermis with a basal layer of proliferative tumour cells expressing K5/K14 and suprabasal layers of non-proliferative cells expressing K1/K10, which finally enucleate and form a stratum corneum structure (Fig. 1a). In contrast with normal homeostasis, in which the rate of cell production matches the rate of loss, during tumour growth this balance is shifted towards proliferation, with the diameter of the papilloma doubling in size every month (Supplementary Fig. 1). Although these tumours contain a mass of amorphous material arising from the terminal differentiation of tumour cells, the proportion of differentiated cells remains relatively constant over time (Supplementary Fig. 1), indicating that tumours grow mostly through basal epithelial cell expansion.
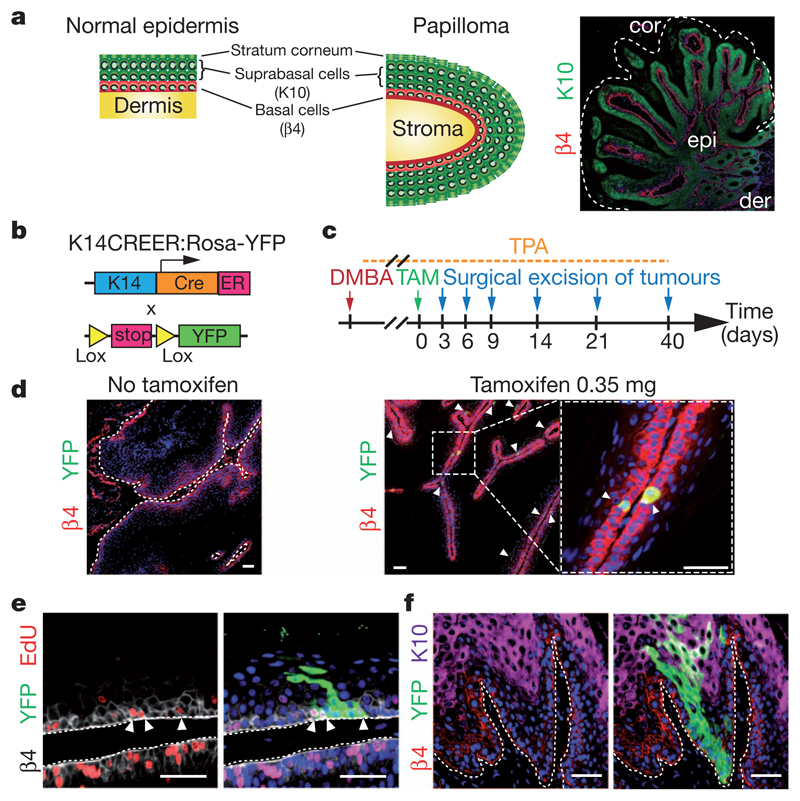
Figure 1. Genetic tracing of skin papilloma at clonal density.
a, Scheme of the stereotypical architecture of papilloma similar to normal epidermis. cor, stratum corneum; der, dermis; epi, epithelial cells. b, Scheme of genetic strategy to induce YFP expression in papilloma. ER, tamoxifen-responsive hormone-binding domain of the oestrogen receptor; K14, human keratin 14 promoter. c, Protocol of DMBA/TPA and tamoxifen (TAM) administration. d, Immunostainings for β4-integrin (β4) and YFP in papilloma with or without TAM. e, Immunostaining for β4-integrin, YFP and 5-ethynyl-2′-deoxyuridine (EdU) in a papilloma 6 days post-labelling. f, Immunostaining for β4 integrin, YFP and K10 in a papilloma 14 days post-labelling. Scale bars, 50 μm.
These observations raise several important questions. Why is the growth of the papilloma relatively slow? Classical theories of tumour development often involve unbridled replication of the tumour-maintaining cell population, leading to geometric expansion of the tumour mass13. However, papilloma arise sporadically following DMBA/TPA treatment, after which they grow slowly, giving rise to a heterogeneous size distribution (Supplementary Fig. 1). Is the characteristic stem/progenitor cell hierarchy of normal tissue recapitulated in the tumour? Are proliferative tumour cells equipotent, or is the papilloma sustained by a distinct population of tumour cells with stem-cell-like properties? To address these questions, we used a quantitative lineage-tracing strategy that has proved successful in resolving cellular hierarchy and stem-cell fate in normal epithelial tissues14–19.
To mark individual tumour cells, we made use of an inducible genetic lineage tracing system (Fig. 1b). Following the administration of tamoxifen (TAM) to K14CREER/Rosa-YFP mice bearing skin tumours induced by DMBA/TPA treatment, we were able to label cells at clonal density within the papilloma (Fig. 1c). Titrating down the dose of TAM to 0.35 mg generated yellow fluorescent protein (YFP) expression in around 1% of basal tumour epithelial cells (Fig. 1d and Supplementary Fig. 2), a density that allows the fate of individual labelled cells to be traced over time. In the weeks following TAM administration, some YFP-labelled basal cells actively divided (Fig. 1e), and formed clones containing several basal cells as well as suprabasal cells that reach the top of the cornified layer (Fig. 1f), demonstrating that labelled tumour cells are capable of generating all cell types that comprise the tumour. Interestingly, the initial shape of the marked tumour clones is reminiscent of the column of marked cells (sometimes termed epidermal proliferative units) observed in the interfollicular epidermis during normal homeostasis14,15,20–22.
To gain further insight into the proliferative potential of labelled tumour cells, we first quantified the size and persistence of clones across a range of time points from 3 days up to 7 weeks post-labelling (Fig. 2a–c). Analysis of the junction between normal epidermis and papilloma excluded the possibility that tumour clones arise from cells coming from normal epidermis (Supplementary Fig. 3). Whereas the frequency of labelled tumour clones (number per sectional area) remained relatively constant during the first 2 weeks after TAM administration, this number had dropped by around 3 weeks, and by 7 weeks involved only 20% of the initial clones (Fig. 2b), indicating that only a fraction of proliferative tumour cells present long-term self-renewal potential. The near-absence of active caspase 3+ cells in these tumours (0.28 ± 0.03% active caspase 3+ tumour epithelial cells) indicates that clones were lost through differentiation rather than by apoptosis. (Supplementary Fig. 4). Of the clones that did survive at 7 weeks post-labelling, many were enormous, containing several hundreds to thousands of cells, and sometimes comprising an entire lobe of the tumour (Fig. 2a, c). These data indicate heterogeneity in the proliferative potential of tumour cells, and indicate that a fraction may conform to the CSC paradigm in vivo by massively contributing at the clonal level to tumour growth.
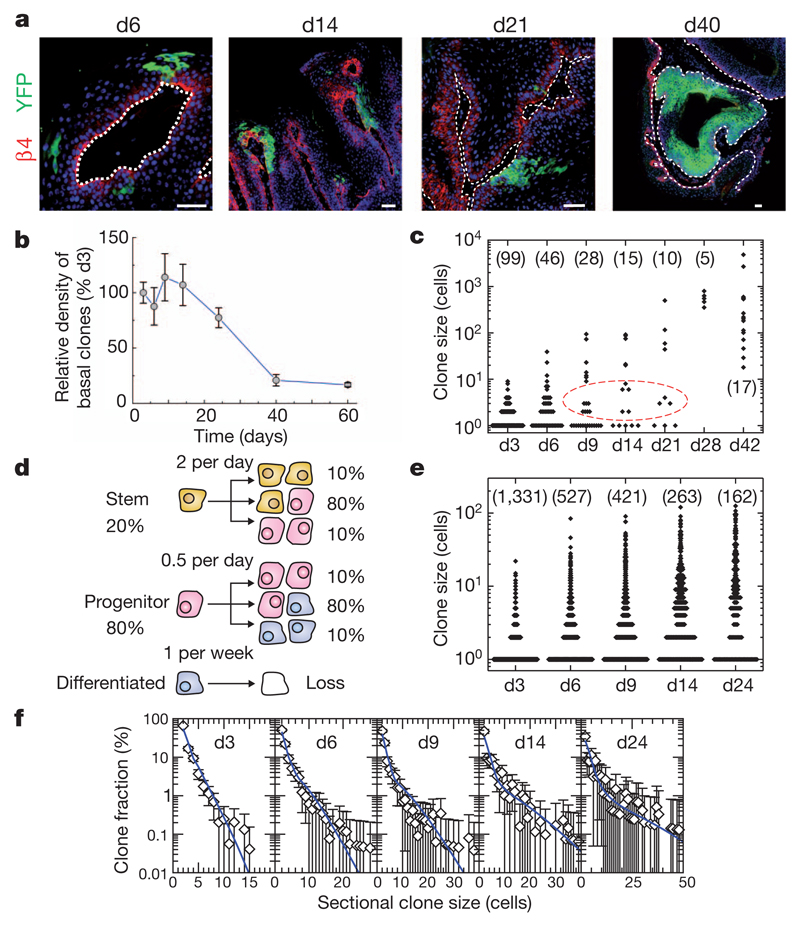
Figure 2. Quantitative clonal analysis in skin papilloma.
a, Immunostainings for β4-integrin and YFP in papilloma after TAM administration. b, Quantification of the relative density of basal clones over time (n = 666, 383, 328, 194, 162, 35 and 33 clones from at least 5 different tumours at day 3, 6, 9, 14, 24, 40 and 60, respectively). c, Total size of clones derived from 3D serial reconstruction over time. The n value is shown. d, Tumour growth model showing the proliferative stem/progenitor cells. e, Clone sizes derived from single sections over time. The n value is shown. f, Fit of the hierarchical model (d) to the corresponding cumulative sectional clonal size distribution. Points show data (e) and the blue lines show the model prediction. Scale bars, 50 μm. Error bars denote s.e.m.
Although the stratified architecture is conserved in papilloma, its folded structure does not facilitate whole-mounting. Instead, clones can only be visualized through the acquisition of sectional data. As a result, total clone size could only be scored at single-cell resolution for individual clones by three-dimensional (3D) reconstruction using confocal analyses of serial (25 µm) sections (Supplementary Fig. 5). Following this procedure, we scored a limited number of clones over a 7-week time course (Fig. 2c), from which several qualitative features emerged. First, single-cell clones, which account for some 50% of the total YFP-labelled clones at 3 days post-labelling, showed a decline only around day 9, indicating that the loss rate of terminally differentiated cells is relatively slow in comparison to the cell division rate. Second, from the size distribution (Fig. 2c), there is further evidence of proliferative heterogeneity. Although the spectrum of clone sizes is broad and continuous at 3 days post-labelling (with clones of ten cells or more coexisting with clones of only two cells), by 9 days the distribution becomes bimodal (Fig. 2c). Of the clones involving multiple cells (and therefore derived from proliferative cells), a minority continue to grow and expand rapidly over the time course, whereas others generate clones with fewer than ten cells that become progressively lost. At 7 weeks post-labelling, clones still present significant heterogeneity in size ranging from dozens of cells to several thousands of cells (Fig. 2c).
Although data from the 3D reconstruction provides qualitative insight into the proliferative cell dynamics, to further resolve the properties of these populations, we turned to a quantitative analysis. Because acquisition of a large statistical ensemble of clone sizes from serial reconstruction is impractical, we used data from independent sections, which could be obtained in larger numbers. Fortunately, analysis of data from the 3D reconstruction revealed a robust quadratic relationship between sectional clone size, _n_2D, and total clone size,_n_3D, which allows the statistical distribution of the latter to be reliably reconstructed from the former (Supplementary Fig. 5 and Supplementary Information). This quadratic dependence, _n_3D≈ _n_2D2/3, follows from the architecture of the tissue that constrains clones to grow laterally as cohesive ‘pancake-like’ structures once the clone has spanned the suprabasal cell layers.
Data from the serial reconstruction indicate that papilloma are sustained by a cellular hierarchy in which a minority population of tumour cells with stem-cell-like properties gives rise to a more transient progenitor cell pool. Motivated by lineage-tracing studies of normal epidermis14,15,18, we propose that the progenitor cell population follows a stochastic pattern of fate in which cell division can lead to all three fate outcomes: two dividing cells, two non-dividing, and one dividing and one non-dividing (Fig. 2d). Further, we propose that the stem-cell-like population divides rapidly, leading to a similar pattern of stochastic fate involving self-renewal and the generation of progenitor cell progeny (Fig. 2d). With this model, we made use of the sectional clone size data (Fig. 2e) to fit the experimental parameters (for further details of the modelling scheme and fitting procedure, see Supplementary Information). This procedure is simplified considerably by the divergence in size of the progenitor-and stem-cell-derived clones, with the former dominating the distribution for smaller clone sizes and early time points, and the latter contributing substantially to the larger clone sizes and later time points. From the predicted dynamics, we obtain an excellent fit to the clone size distributions over the wide time course (Fig. 2f) if we suppose that the stem-cell-like population accounts for just 20% of the labelled tumour proliferative cells, and take both the progenitor and stem cell fates to be approximately balanced with some 80% of divisions leading to asymmetric fate outcome while the remaining 20% lead to either symmetrical duplication or differentiation, with equal probability (Fig. 2d). Moreover, the inferred stem-cell-like tumour cell division rate of twice per day is approximately four times larger than the progenitor cell division rate in the tumour, and an order of magnitude larger than the progenitor cell division rate in normal tissue. Finally, as expected from the analysis of the serial reconstruction, the loss rate of terminally differentiated cells is slow, at around once per week.
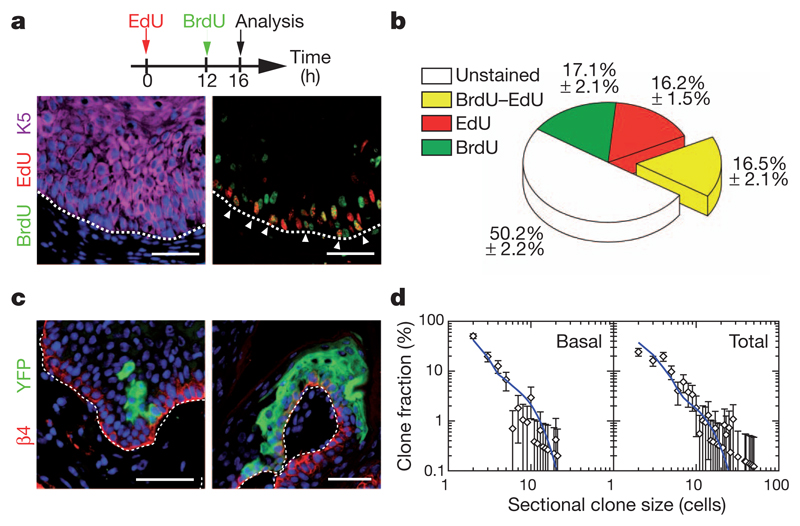
To test the predictions of the model, we performed a double-labelling nucleotide pulse-chase assay to estimate the proportion of cells that re-enter the cell cycle after 12 h. To this end, we first administrated a pulse of 5-ethynyl-2′-deoxyuridine (EdU) to mice bearing papilloma, and administrated 5-bromo-2-deoxyuridine (BrdU) 12 h later, and analysed double-labelled basal cells 4 h after that (Fig. 3a). Significantly, whereas virtually no double-positive cells can be found in normal epidermis on the edge of the tumour (Supplementary Fig. 6), around 17% of the basal tumour epithelial cells were BrdU–EdU double-positive (Fig. 3b), consistent with the predictions made by the analysis of the clonal fate data (Fig. 2d and Supplementary Information). To challenge the model further, we looked for additional clonal fate characteristics that can be matched against the predicted dynamics. To fit the parameters of the model, we make use of the total clone size data without reference to the relative contribution of basal and suprabasal cells within the clones. To test the model, at 9 days post-labelling, we explored the clone size distribution disaggregated into basal and total cell contributions, focusing on the ensemble of basal clones (those with at least one cell that retains an attachment to the basal lamina) (Fig. 3c and Supplementary Fig. 7). With the same parameters, we found good agreement with the measured size distributions (Fig. 3d), lending further support to the findings.
Figure 3. Challenging the stem/progenitor cell model in papilloma.
a, Immunostaining for BrdU, EdU and K5 in papilloma from mice treated following the protocol schemed above. b, Quantification of the proportion of unmarked cells, BrdU, EdU and EdU–BrdU double-labelled cells within the basal layer of papilloma (n = 5,512 cells from 8 papilloma). c, Immunostaining for YFP and β4-integrin on papilloma 9 days after TAM administration. d, Comparison between the model prediction (blue lines), with parameters defined in Fig. 2d and in the text, and the clone size distribution (n = 105 clones from 7 tumours). Scale bars, 50 μm. Error bars denote s.e.m.
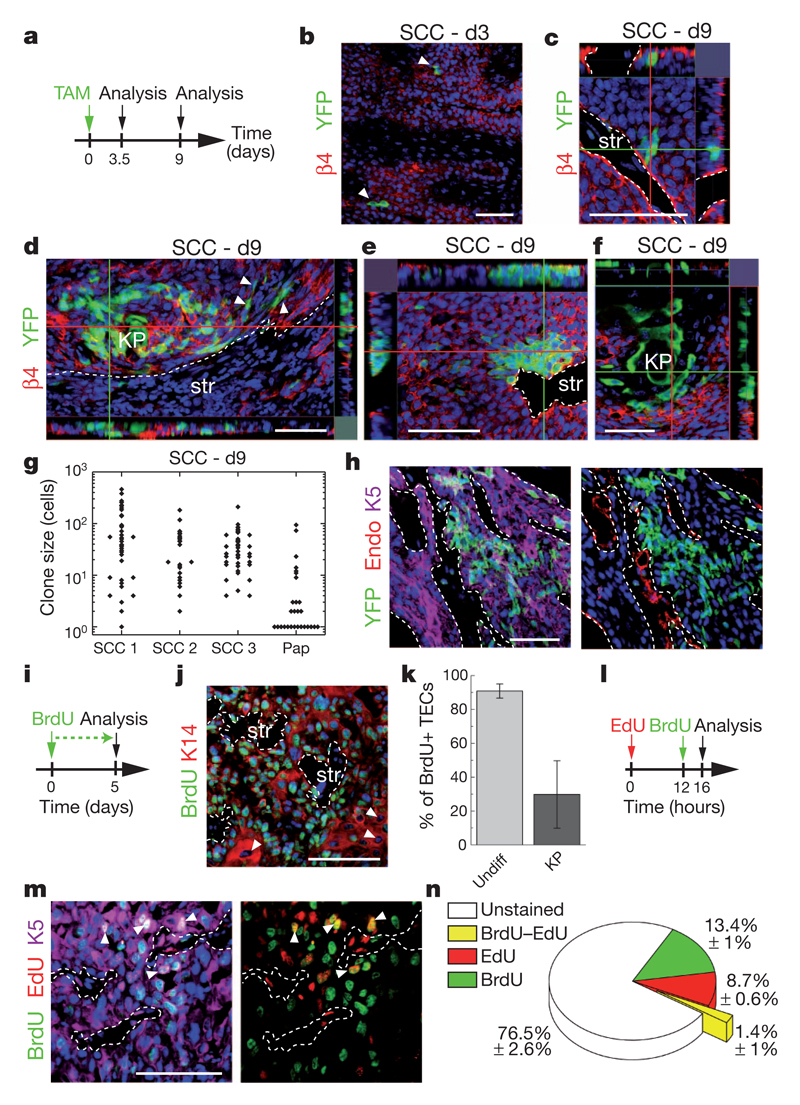
To determine whether the transition from benign papilloma to malignant invasive squamous cell carcinoma (SCC) is accompanied by a change in the cellular hierarchy and proliferation dynamics, we combined a study of clonal fate with proliferation kinetics in invasive SCC arising from the spontaneous malignant transformation of papilloma in DMBA/TPA-treated mice. Whereas SCCs do not express keratins associated with epidermal differentiation (such as K10), evidence of terminal differentiation is visible in cell clusters known as keratin pearls (one of the pathognomonic features of skin SCCs), with a frequency that varies from tumour to tumour (Supplementary Fig. 8). Administration of 1 mg TAM to K14CREER/Rosa-YFP mice presenting invasive SCC induced YFP expression at clonal density (0.9 ± 0.3 clone per mm2) (Fig. 4a, b). From a confocal analysis of serial thick sections (Supplementary Fig. 9), we were able to score the total clone size at 9 days post-labelling, the time point for which we have the most comprehensive clonal data set in papilloma. The proportion of clones presenting signs of differentiation through the presence of keratin pearls is similar to the overall frequency of differentiated cells within tumours, indicating that the targeted clones are representative (Supplementary Fig. 8). Whereas clones showed significant heterogeneity in size (Fig. 4c–g), in contrast with papilloma, SCC also showed tumour-to-tumour variability, and the incidence of small clones was significantly diminished. Interestingly, all of the largest clones contained cells that contact the stroma and endothelial cells as well as cells that have lost their cohesion with the rest of the clone, and present signs of epithelial to mesenchymal transition with a fibroblastic-like morphology (Fig. 4d, e, h) Most cells within these clones present no sign of terminal differentiation despite having undergone many rounds of division (Fig. 4e), indicating that these clones are comprised of proliferative cells with a very low probability of terminal differentiation.
Figure 4. Quantitative clonal analysis of squamous cell carcinoma.
a, Protocol of TAM administration and clonal analysis in SCC. b, Immunostaining for YFP and β4-integrin in SCC 3 days after TAM administration. c–f, Same as in b 9 days after TAM administration. KP, keratin pearl; str, stroma. g, Total size of clones quantified from serial 30 μm sections 9 days after TAM administration (n = 37, 25, 42 and 28 clones in SCC 1, 2, 3 and papilloma (Pap), respectively). h, Immunostaining for YFP, K5 and endoglin (Endo) in SCC 9 days after TAM administration. i, Protocol of BrdU administration. j, Immunostaining for BrdU and K14 in SCC. k, Quantification of the BrdU+ tumour epithelial cells (TECs) within SCC after 5 days of continuous BrdU administration (n = 676 undifferentiated tumour cells and 282 KP cells). Error bars denote s.d. Undiff, undifferentiated. l, Protocol of BrdU and EdU administration. m, Immunostaining for BrdU, EdU and K5 in SCC from mice treated following the protocol shown in l. n, Quantification of the proportion of unmarked cells, BrdU, EdU and EdU–BrdU double-labelled cells within SCC (n = 9,010 cells from 2 SCC). Scale bars, 50 μm.
Consistent with the notion that most cells in invasive SCCs are proliferative, about 70% of cells in SCC are Ki67+ and continuous administration of BrdU for 5 days labelled about 90% of undifferentiated cells in invasive SCCs (Fig. 4i–k and Supplementary Fig. 8). Although heterogeneity of clone size could reflect tumour cells with a range of proliferative potential, the exponential form of the cumulative clone size distribution of individual tumours (Supplementary Fig. 9) is equally consistent with a single equipotent proliferative cell population, biased towards symmetric self-renewal, with a broad distribution of cell cycle times (Supplementary Information). Within this paradigm, we estimate an average cell division time that varies between 1 and 2 days (according to the tumour). This figure is consistent with the results of a BrdU–EdU double-labelling assay, which revealed that, whereas 23% of SCC cells acquire BrdU or EdU expression following a short pulse, the frequency of double-labelled cells after 16 h chase was below 2% (Fig. 4l–n and Supplementary Information).
In summary, our data present the first attempt to dissect the mode of tumour growth using clonal analysis by lineage tracing in mice, and demonstrate for the first time the existence of tumour cells with stem-cell-like properties in an unperturbed solid tumour, consistent with the CSC paradigm. The neutral competition of tumour cells uncovered here in skin papilloma is reminiscent of the dynamics found in normal tissue and indicates that this hierarchical mode of tumour growth is mediated at the cellular level by stochastic cell fate decisions. This mode of tumour growth, involving the chance expansion and extinction of neighbouring clones, may explain the clonal heterogeneity found in many different tumours23. In future studies, it will be important to define the mechanisms regulating the balance between proliferation and differentiation during tumour progression, the influence of the external environment and other physical constrains on tumour expansion, and whether this stochastic mode of tumour progression also operates in other types of cancer.
Methods Summary
Squamous skin tumours were induced by treating K14CREER/Rosa-YFP mice with the two-step DMBA/TPA chemical carcinogenesis protocol10. Clonal YFP expression in skin tumours was performed by administrating 0.35 mg or 1 mg TAM to K14CREER/Rosa-YFP mice bearing benign or malignant skin tumours. Immunostainings were performed as described24. Quantification of the total clone size was determined by counting the number of YFP-positive cells in 3D reconstruction of the whole clone using serial thick sections analysed by confocal microscopy. Two-dimensional clone size was determined by counting the number of YFP cells in a single tumour thin section and its relation to the total clone size was inferred using the algorithm defined in the main text and Supplementary Information. BrdU–EdU double-labelling approach was performed as described25. For further details see Supplementary Methods.
Methods
Skin tumour induction and clonal YFP expression
K14CREER26 and Rosa-YFP mice were obtained from JAX. Mouse colonies were maintained in a certified animal facility in accordance with European guidelines. Mice were treated with DMBA and TPA as previously described4,9. Briefly, mice were treated with DMBA at postnatal day 23, 25 and 27 with DMBA (9,10-dimethyl-1,2-benzanthracene) and then treated weekly for several weeks with TPA (12-_O_-tetradecanoylphorbol-13-acetate). 7-week-old mice were treated again twice with DMBA. 0.35 mg Tamoxifen (TAM) was administered intraperitoneally to K14CREER/Rosa-YFP mice bearing papilloma and 1 mg to mice bearing SCC. The tumours were surgically excised at different time points after TAM injection.
Measurement of tumour growth
Skin tumours were measured using a precision calliper allowing to discriminate size modifications >0.1 mm. Tumour surface was measured every week with the formula A = π(dD/2), where d is the minor tumour axis and D is the major tumour axis.
Histology, immunostaining and imaging
Tissues were pre-fixed for 2–8 h in 4% paraformaldehyde (PAF) and washed in sucrose 30% solution to remove the excess of PAF before being embedded in OCT. Samples were sectioned serially in thin 6–7 µm or thick 25–30 µm sections using a CM3050S Leica cryostat (Leica Microsystems GmbH).
The following primary antibodies were used: anti-K5 (rabbit, 1:1,000, Covance), anti-β4 (rat, 1:200, BD), anti-K14 (chicken, 1:1,000, Covance), anti-GFP (rabbit, 1:1,000, Molecular Probes), anti-K10 (rabbit, 1:2,000, Covance), anti-active caspase 3 (rabbit, 1:600, R&D Systems), FITC (fluorescein isothiocyanate)-coupled anti-BrdU (mouse, 1:100, BD), EdU staining was performed following the manufacturer’s instructions (Invitrogen). Sections were incubated in blocking buffer (PBS, normal donkey serum (NDS) 5%, BSA 1%, Triton 0.2% or 0.5% for thin and thick sections, respectively) for 1 h at room temperature. Primary antibodies were incubated in blocking buffer overnight at 4 °C. Sections were rinsed three times in PBS and incubated with secondary antibodies diluted at 1:400 for 1 h at room temperature for thin sections or overnight at 4 °C for thick sections. The following secondary antibodies were used: anti-rabbit, anti-rat, anti-chicken conjugated to AlexaFluor488 (Molecular Probes), to rhodamine Red-X (Jackson ImmunoResearch) or to Cy5 (Jackson ImmunoResearch). Nuclei were stained in Hoechst solution (1:1,000) and slides were mounted in DAKO mounting medium supplemented with 2.5% Dabco (Sigma). In all immunofluorescences, Hoechst nuclear staining is represented in blue.
Pictures were acquired using an Axio Imager M1 Microscope, AxioCamMR3 camera using Axiovision software (Carl Zeiss).
Confocal pictures were acquired using Zeiss LSM 510 or Zeiss LSM 780 confocal microscopes using Zen software. Images were median-filtered (3 to 3) and are represented as orthogonal projections of 20–65 optical sections in 0.3–1 μm increments rendered using Zen software.
Analysis of clone size
Total clone size was determined by counting the number of YFP-positive cells in 3D reconstruction of confocal analysis of the whole clone using serial thick sections.
2D clone size was determined by imaging 30 to 1,330 clones from at least 8 different tumours from different mice at each time point. 6–7 μm sections were analysed to number the cells and the total clone size was inferred after correction as described in the Supplementary Information. The density of basal clones was determined by quantifying the number of basal clones per area of tumour and corrected for the increase in the total surface during tumour growth.
Proliferation assays
To measure the kinetics of tumour cell proliferation, mice were treated with EdU (pulse) and chased for 12 h followed by BrdU administration and another chase of 4 h. Tissues were first stained for K5. Sections were then washed in PBS and fixed again in PAF 4%. After washing in PBS, tissues were incubated for 20 min in HCl 1 N at 37 °C, washed three times and incubated overnight with FITC-coupled anti-BrdU antibody (mouse, 1:100, BD). The tissues were washed in PBS the next day and the EdU staining was performed following the manufacturer’s instructions (Invitrogen). Slides were mounted in DAKO mounting medium supplemented with 2.5% Dabco (Sigma).
FACS quantification of YFP basal tumour cells
3 days after TAM administration to the K14CREER/Rosa-YFP mice bearing skin tumours, tumours were digested in collagenase I (Sigma) for 2 h at 37 °C on a rocking plate. Collagenase I activity was blocked by addition of EDTA (5 mM). After tumour digestion, cells were first incubated in PBS complemented with 30% FCS to block Fc receptors for 15 min at room temperature. Immunostaining was performed using phycoerythrin-conjugated anti-α6-integrin (clone GoH3; BD Pharmingen), allophycocyanin-Cy7-conjugated anti-Epcam (clone G8.8; Biolegend). YFP-positive tumour cells were quantified within the Epcam+/α6-integrin+ population after forward scatter, side scatter selection and Hoechst dye exclusion. Fluorescence-activated cell sorting analysis was performed using BD LSR Fortessa and FACSDiva software (BD Biosciences).
Theory
See Supplementary Information.
Supplementary Material
Supplementary Information
Acknowledgements
C.B. is investigator of WELBIO and chercheur qualifié of the FRS/FNRS. B.B. is chargé de recherche of the FRS/FNRS and G.D. is supported by the Brussels Region. This work was supported by the FNRS, the program d’excellence CIBLES of the Wallonia Region, a research grant from the Fondation Contre le Cancer, the ULB foundation and the fond Gaston Ithier, a starting grant of the European Research Council (ERC), and the EMBO Young Investigator Program. We thank F. Bollet-Quivogne and J.-M. Vanderwinden for their help with confocal imaging.
Footnotes
Author Contributions C.B., G.D., B.B. and B.D.S. designed the experiments and performed data analysis. G.D. and B.B. performed most of the experiments. A.C. provided technical support. C.B. and B.D.S. wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 3.Malanchi I, et al. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 4.Beck B, et al. A vascular niche and a VEGF–Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478:399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- 5.Schober M, Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-β and integrin/focal adhesion kinase (FAK) signaling. Proc Natl Acad Sci USA. 2011;108:10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 7.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nature Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 9.Kemp CJ. Multistep skin cancer in mice as a model to study the evolution of cancer cells. Semin Cancer Biol. 2005;15:460–473. doi: 10.1016/j.semcancer.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nature Protocols. 2009;4:1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owens DM, Watt FM. Contribution of stem cells and differentiated cells to epidermal tumours. Nature Rev Cancer. 2003;3:444–451. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- 12.Perez-Losada J, Balmain A. Stem-cell hierarchy in skin cancer. Nature Rev Cancer. 2003;3:434–443. doi: 10.1038/nrc1095. [DOI] [PubMed] [Google Scholar]
- 13.Klein CA. Parallel progression of primary tumours and metastases. Nature Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 14.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 15.Doupé DP, Klein AM, Simons BD, Jones PH. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell. 2010;18:317–323. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 17.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 20.Ghazizadeh S, Taichman LB. Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. EMBO J. 2001;20:1215–1222. doi: 10.1093/emboj/20.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potten CS. Cell replacement in epidermis (keratopoiesis) via discrete units of proliferation. Int Rev Cytol. 1981;69:271–318. doi: 10.1016/s0074-7696(08)62326-8. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie IC. Retroviral transduction of murine epidermal stem cells demonstrates clonal units of epidermal structure. J Invest Dermatol. 1997;109:377–383. doi: 10.1111/1523-1747.ep12336255. [DOI] [PubMed] [Google Scholar]
- 23.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Keymeulen A, et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J Cell Biol. 2009;187:91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 26.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information