Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein (original) (raw)
Abstract
We purified microtubules from a mammalian mitotic extract and obtained an amino acid sequence from each microtubule-associated protein by using mass spectrometry. Most of these proteins are known spindle-associated components with essential functional roles in spindle organization. We generated antibodies against a protein identified in this collection and refer to it as astrin because of its association with astral microtubule arrays assembled in vitro. Astrin is ≈134 kDa, and except for a large predicted coiled-coil domain in its C-terminal region it lacks any known functional motifs. Astrin associates with spindle microtubules as early as prophase where it concentrates at spindle poles. It localizes throughout the spindle in metaphase and anaphase and associates with midzone microtubules in anaphase and telophase. Astrin also localizes to kinetochores but only on those chromosomes that have congressed. Deletion analysis indicates that astrin's primary spindle-targeting domain is at the C terminus, although a secondary domain in the N terminus can target some of the protein to spindle poles. Thus, we have generated a comprehensive list of major mitotic microtubule-associated proteins, among which is astrin, a nonmotor spindle protein.
The spindle is a complex microtubule-based superstructure responsible for chromosome segregation in mitosis and meiosis (1–6). Spindle microtubules are organized in a complex process that requires both motor and nonmotor microtubule-associated proteins. A great deal of emphasis has been placed on the functional contribution that motor proteins make to spindle organization because of their inherent biological activity. The strong sequence homology of the conserved motor domain in these proteins has facilitated both their rapid identification and characterization during spindle assembly. These investigations have demonstrated that motors of both the kinesin and dynein families play important roles in such varied aspects of spindle organization as spindle bipolarity (6), chromosome movement (7–10), checkpoint regulation (11, 12), spindle pole organization (13–16), and the regulation of microtubule dynamics (17). In contrast, the functional contribution that nonmotor proteins make to spindle organization has not progressed as rapidly. In part, the lag in progress in defining the roles of nonmotor spindle proteins stems from the lack of clearly defined biological activity of nonmotor proteins. A further complication arises from the lack of any clear homology among nonmotor proteins to facilitate their identification. Thus, although some nonmotor spindle proteins have been characterized (18, 19), others are less well understood (20, 21), and many others may await identification.
In this article we use mass spectrometry to identify the proteins associated with microtubules assembled in a cell-free mitotic extract under conditions where >90% of microtubules are organized into astral arrays. Most proteins identified through this strategy have known functional roles for spindle assembly. A protein identified in this collection is referred to as astrin because of its association with microtubule asters assembled under these in vitro conditions. Astrin is a nonmotor coiled coil containing protein that associates with spindles throughout mitosis and localizes to kinetochores of congressed chromosomes. Thus, we have generated a comprehensive list of all major motor and nonmotor mitotic microtubule-associated proteins.
Materials and Methods
Cell Culture.
HeLa cells were maintained in DMEM containing 10% FCS, 50 units/ml penicillin, and 50 μg/ml streptomycin. CFPAC-1 cells were maintained in Iscove's modified Dulbecco's medium containing 10% FCS, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cells were grown at 37°C in a humidified incubator with a 5% C02 atmosphere.
Microtubule Aster Preparation and Enrichment.
Mitotic extracts and microtubule asters were prepared as described (22). Latrunculin B (5 μg/ml) was added to the final extract to reduce actin polymerization and the contamination of microtubule pellets with actin and actin-associated proteins. Extracts corresponding to ≈3 × 107 cells were layered onto a 50% (wt/vol) sucrose cushion in KHM buffer (78 mM KCL/50 mM Hepes, pH 7.0/4 mM MgCl2/2 mM EGTA/1 mM DTT), and microtubules were collected by sedimentation at 100,000 × g for 2 h at 4°C. The insoluble pellet was collected directly in SDS/PAGE sample buffer. The inclusion of Latrunculin B and the use of 50% sucrose cushions significantly reduced contamination of the insoluble pellet by nonmicrotubule-binding proteins relative to the initial aster enrichment reported previously (23).
Peptide sequencing was performed at the Harvard Microchemistry Facility (Cambridge, MA) by microcapillary reverse-phase high pressure liquid chromatography nanoelectrospray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer.
Antibodies.
The N-terminal 609 aa of astrin was expressed in bacteria as a fusion protein with glutathione _S_-transferase by inserting a 1,827-nt segment of astrin cDNA into the _Sma_I and_Eco_RI sites of pGEX-5X-2 (Amersham Pharmacia). The C-terminal 586 aa of astrin was expressed in bacteria as a fusion protein with glutathione _S_-transferase by inserting a 1,951-nt segment of astrin cDNA into the _Eco_RI site of pGEX-5X-1. Glutathione _S_-transferase-astrin fusion proteins were induced with 1 mM isopropyl β-D-thiogalactoside in liquid cultures, and recombinant proteins were purified by electroelution from SDS/PAGE and injected into rabbits (Covance Research Products, Denver, PA).
Antibodies against NuMA (22), HSET (24), Eg5 (24), Arp1 (25), tubulin (DM1A, Sigma–Aldrich), dynein (26), and TOGp (23) have all been described previously. Antibodies against KIF4 and MCAK both were generated by immunizing rabbits with recombinant proteins expressed in bacteria (A. Levesque and D.A.C., unpublished results). Antibodies against TACC3, MAP4, and centromeres were generous gifts from Jordan Raff, Chloe Bulinski, and Kevin Sullivan, respectively.
Immunologic Techniques.
Indirect immunofluorescence microscopy of HeLa and CFPAC-1 cells was performed as described (27). Immunoblotting of cell extracts or total cell protein was performed as described (27).
Astrin cDNA Analysis and Transfection Constructs.
Full-length astrin cDNA in the vector pCMVSPORT 6 (GenBank accession no. AL530294) was purchased from Invitrogen and designated pSPORT-Astrin. Protein sequence alignment and identity shading was preformed by using MEGALIGN of DNAstar (Madison, WI).
Expression constructs were created in the vector series pEGFP-C (CLONTECH) with the exception of construct H, which used the vector pEGFP-N1. Fragment A was amplified from pSPORT-Astrin by PCR using_Pfu_ polymerase (Stratagene) and the primers AstF (5′-ATGTGGCGAGTGAAAAAACTGAGCC-3′) and AstR (5′-GCTCAGAAATTCCAGCAATCCC-3′) and inserted into the_Ecl136_II site of pEGFP-C3. Fragment B was amplified by using the primers AstF and AstR2 (5′-GTTTGTCCACCTCCTGAGAGAGCC-3′) and inserted into the _Ecl136_II site of pEGFP-C3. Fragment C was amplified by using the primers AstF and AstR and digested with_Eco_RI. The resulting 1,827-nt fragment was inserted into the_Ecl136_II/_Eco_RI site of pEGFP-C3. Fragment D was amplified by using the primers AstF and AstR and digested with_Pvu_II. The resulting 1,374-nt fragment was inserted into the_Ecl136_II site of pEGFP-C1. Fragment E was generated by digesting pSPORT-Astrin with _Dra_I and inserting the resulting 3,207-nt fragment into the _Ecl136_II site of pEGFP-C1. Fragment F was generated by digestion of pSPORT-Astrin with_Eco_RI and _Dra_I and inserting the resulting 1,865-nt fragment into the _Eco_RI/_Sma_I site of pEGFP-C3. Fragment G was generated by digesting pSPORT-Astrin with_Sst_I and _Dra_I and inserting the resulting 492-nt fragment into the _Sac_I/_Sma_I site of pEGFP-C3. Fragment H was generated by digestion of pSPORT-Astrin with_Sac_I and inserting the resulting 1,810-nt fragment into the_Sac_I site of pEGFP-N1.
Transient Transfection.
HeLa cells were seeded onto glass coverslips and grown to 60–80% confluency. Cells were transfected with 2 μg of plasmid DNA and 8 μl of Lipofectamine reagent (GIBCO/BRL) in 1 ml of Opti-MEM (GIBCO/BRL) according to manufacturer recommendations. The transfection reagent was removed after 3 h, replaced with complete growth medium, and processed for indirect immunofluorescence by using anti-green fluorescent protein (GFP) antibody after 24 h (Molecular Probes).
Results
Identification of the Major Mitotic Microtubule-Associated Proteins Using Mass Spectrometry.
Microtubules were polymerized in a cell-free mitotic extract under conditions where >90% of microtubules organized into astral arrays (22). Microtubule complexes were separated from soluble extract components by sedimentation through 50% sucrose. The microtubule pellet was solubilized in SDS, and the proteins were identified by Coomassie blue staining after size separation on 7.5% SDS/PAGE (Fig.1A). As a control for the microtubule-dependent enrichment of proteins, an equivalent amount of mitotic extract was incubated in the absence of taxol, and a proportional quantity of that insoluble pellet was run in parallel. Immunoblotting for NuMA and Eg5, two proteins known to associate with microtubules under these conditions, verified the enrichment of microtubules relative to the total extract by using this technique (Fig. 1A).
Figure 1.
Identification of mitotic microtubule-associated proteins. (A) Equivalent quantities of the total extract and the insoluble fractions obtained after incubation with (+) and without (−) taxol were separated by size on 7.5% SDS/PAGE and stained with Coomassie blue. Protein identities revealed by mass spectrometry are indicated on the right of each protein band. The lower panels show immunoblots of each of these three samples for NuMA and Eg5 to verify the enrichment of microtubule-associated proteins by using this technique. Molecular masses in kDa are indicated on the left. (B) Immunoblots of various microtubule-associated proteins (as indicated) identified by mass spectrometry of soluble (S) and insoluble (P) fractions obtained after microtubule aster assembly in the mitotic extract.
Sixteen protein bands were enriched in the extract after microtubule polymerization compared with the control sample in which microtubules were not polymerized because of the absence of taxol. Each protein band was excised from the polyacrylamide gel and amino acid sequence obtained from each band by using mass spectrometry of tryptic peptides. Although some stained bands contained more than one major protein of equal size, all these proteins were represented in the genome database, and we classified them into five categories (Fig.1A). The first category includes NuMA, TOGp, p150Glued, Eg5, HSET, and tubulin and are proteins with previously identified roles in both spindle organization in vivo and microtubule aster organization in vitro. The second category includes KIAA0622 (the human homologue of the fruit fly gene orbit), KIF4, TACC3, TPX2, and MCAK and are proteins with established spindle functions in other systems but are untested for a functional role in the organization of microtubules into asters in this system. Immunodepletion of KIF4 or MCAK did not alter microtubule aster organization in this system in any detectable manner (A. Levesque and D.A.C., unpublished results), substantiating our previous conclusion that cytoplasmic dynein, Eg5, and HSET may be the primary microtubule motors responsible for organizing microtubule asters in this system (24). The third category includes the microtubule-associated proteins MAP4 and MAP7, which were expected to have been enriched given the isolation strategy used. The fourth category includes plectin, clathrin, SMC1 and -3, and HSP70 and -71, which have no documented role as microtubule-associated proteins and may represent contaminants. The final category includes a protein of ≈140 kDa that we refer to as astrin on the basis of its enrichment with microtubules under conditions where most microtubules assemble into asters.
To verify the mass spectrometry data we performed immunoblot analysis of the 10,000 × _g_-soluble and -insoluble protein fractions obtained after microtubule assembly in these mitotic extracts (Fig. 1B). All the proteins identified by mass spectrometry for which we had access to antibodies displayed enrichment in the microtubule pellet fraction with some (e.g., TOGp) being highly enriched, others (e.g., Eg5) moderately enriched, and yet others [e.g., dynactin(Arp1)] weakly enriched. Cytoplasmic dynein provides an example of a protein that remains largely soluble and shows very little enrichment on microtubules under these conditions.
Astrin Is a Spindle-Associated Protein.
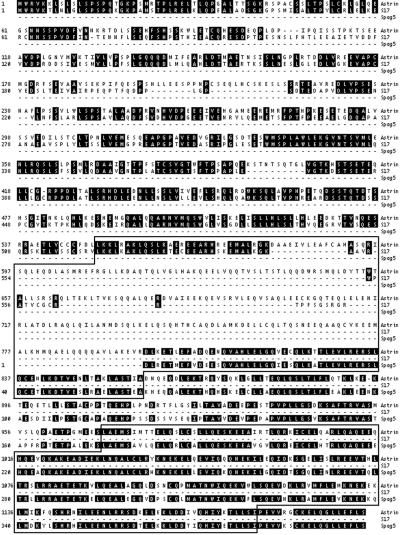
To characterize astrin, the only previously unidentified protein identified in this collection, we obtained a full-length cDNA encoding this protein from Invitrogen. Sequence analysis indicates that the cDNA is 3,843-nt long and encodes a protein of 1,193 aa with a predicted molecular mass of 134,400 Da (Fig.2; accession no. AF399910). With the exception of two predicted coiled-coil domains in the C-terminal half of the protein and numerous predicted phosphorylation sites, astrin contains no identifiable functional motifs. BLAST searches identified numerous expressed sequence tag sequences and cDNA clones encoding partial segments of astrin, only one of which had the correct ORF (accession no. BC000322). Astrin also showed strong homology to proteins from mouse and rat (Fig. 2). A partial sequence of a mouse gene of unknown function called S17 (28) showed 56.4% amino acid identity to the N-terminal 711 aa of astrin. The C-terminal 397 aa of astrin were 72.8% identical to the rat protein Spag5, which was identified in a yeast two-hybrid screen for proteins that interact with Odf1, an outer dense filament protein from sperm flagella (29).
Figure 2.
Alignment of the human astrin protein sequence with mouse S17 and rat Spag5 protein sequences. Identical amino acids residues are shaded, and predicted coiled-coil domains spanning amino acids 550–857 and 970–1,175 are in boxes.
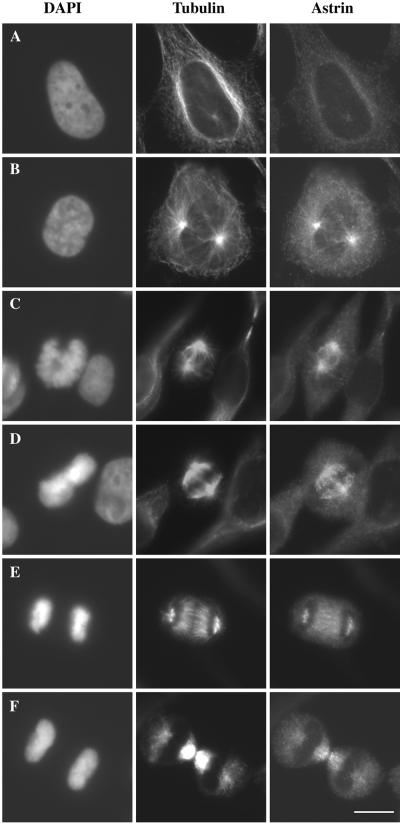
We raised rabbit polyclonal antibodies against the N-terminal 609 aa and the C-terminal 586 aa of astrin. Both antibodies were species-specific and reacted specifically with a protein of ≈140 kDa on immunoblots of total HeLa cell protein (Fig. 1B and data not shown). Both antibodies showed similar staining patterns in human cells under various fixation conditions (Fig.3). Astrin was diffuse in the cytoplasm of interphase cells with some concentrated near the centrosome (Fig.3A). In prophase, astrin associated with microtubules and concentrated at the vertices of the developing spindle poles (Fig.3B). Astrin localized to spindle microtubules throughout prometaphase, metaphase, anaphase, and telophase (Fig. 3 C–F). In metaphase, astrin localized throughout the central spindle in a pattern that mirrored microtubules. It also associated with midzone microtubules in anaphase and telophase. The localization of astrin to the spindle during mitosis was microtubule-dependent as the staining pattern was abolished by nocodazole treatment.
Figure 3.
Cell cycle distribution of astrin. HeLa cells were fixed in cold methanol and stained with antibodies against astrin, tubulin, and the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI) as indicated. Cells were classified as being in interphase (A), prophase (B), prometaphase (C), metaphase (D), anaphase (E), and telophase (F) on the basis of the morphology and alignment of chromosomes and spindle organization. (Bar, 10 μm.)
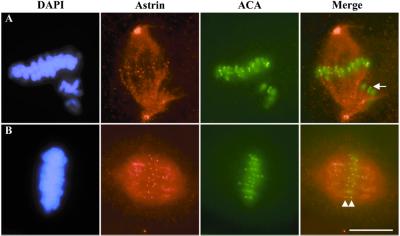
We also observed a punctate staining pattern for astrin adjacent to the chromosomes during metaphase reminiscent of kinetochores, and double labeling with an anti-centromere antibody verified that astrin concentrates at kinetochores during metaphase (Fig.4B). Surprisingly, in prometaphase, kinetochores of chromosomes that were unaligned did not stain for astrin, whereas those chromosomes that were aligned at the metaphase plate displayed kinetochore staining (Fig.4A). Astrin required intact microtubules for both its establishment and maintenance at kinetochores, because nocodazole treatment abolished all kinetochore staining.
Figure 4.
Localization of astrin to kinetochores. CFPAC-1 cells were fixed in glutaraldehyde and stained with antibodies against astrin, anti-centromere antibodies (ACA), and the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI) as indicated. Cells were classified as being in prometaphase (A) and metaphase (B) on the basis of chromosome alignment. Arrowheads highlight sister kinetochores in metaphase that stain for astrin, and the arrow highlights the kinetochore of an unaligned chromosome in prometaphase that lacks astrin staining. (Bar, 10 μm.)
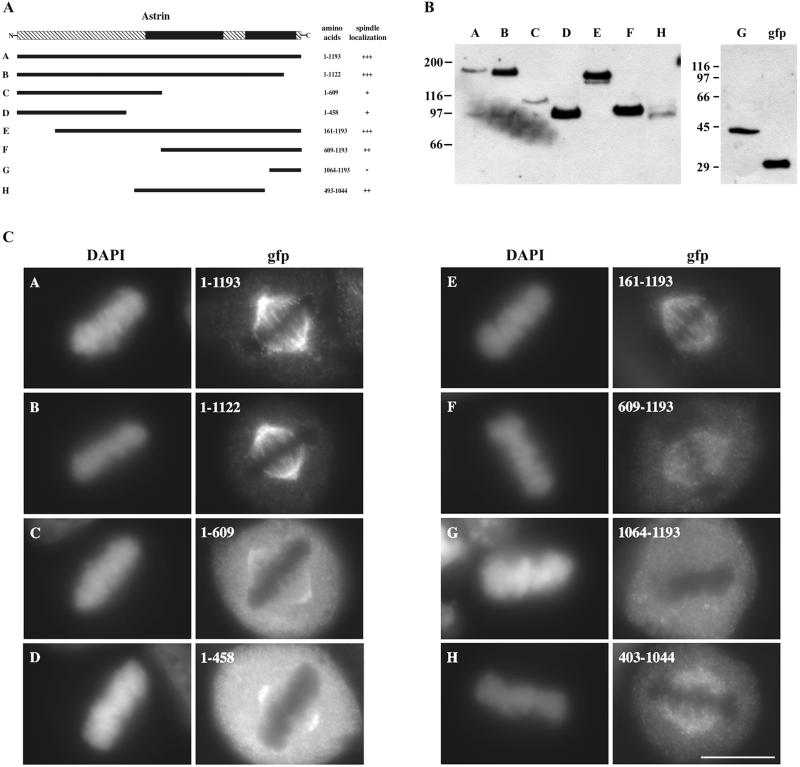
To map the domain of astrin required for spindle targeting, we expressed various segments of astrin in HeLa cells and determined the localization of each segment during mitosis (Fig.5A). Immunoblot analysis of cell extracts obtained after transient expression of each domain confirmed that a protein of the expected size was expressed from each plasmid (Fig. 5B). Full-length astrin and segments of astrin lacking short parts of either the N or C termini localized throughout the mitotic spindle in metaphase in a pattern indistinguishable from the endogenous protein (Fig. 5C, A,B, and E). Central segments of astrin containing the predicted coiled-coil regions displayed spindle localization similar to the full-length protein, although the spindle staining intensity was diminished and there was higher cytoplasmic background (Fig. 5C, F and H). The extreme C-terminal 129 aa failed to localize to the spindle and was localized diffusely in mitotic cells (Fig. 5C, G). Finally, expression of the N-terminal region lacking the predicted coiled-coil domains showed diffuse cytoplasmic staining in addition to some spindle staining (Fig. 5C, C and D). The spindle staining observed with these N-terminal fragments was dissimilar to the full-length protein, because they were restricted to spindle poles and did not extend throughout the spindle. Kinetochore staining was not observed consistently under conditions of transient expression, thus we were unable to define unequivocally a domain of astrin responsible for kinetochore accumulation.
Figure 5.
Expression of GFP-astrin fusion proteins in HeLa cells. (A) Schematic diagram of astrin fragments that were tested for their ability to bind the mitotic spindle. +++, strong binding; ++, moderate binding; +, weak binding; −, no binding; shaded boxes, regions of astrin predicted to form a coiled-coil structure. (B) Immunoblot of transfected HeLa cell lysates expressing the various GFP-astrin fusion proteins. Molecular masses in kDa are indicated on the left. (C) Immunofluorescent images of metaphase mitotic cells expressing the various GFP-astrin fusion proteins. Cells were fixed in cold methanol, and GFP was detected with a rabbit anti-GFP antibody. (Bar, 10 μm.)
Discussion
We report the identification of all major proteins associated with microtubules assembled in a mammalian mitotic extract. The use of mass spectrometry to sequence each protein is unbiased to the specific biological function of any given protein and succeeded in identification of both motor and nonmotor microtubule-associated proteins, most of which have known functional roles in spindle organization. These results suggest that these proteins represent a comprehensive catalogue of all major noncentrosomal and nonchromosomal spindle-associated proteins. Additional experimental approaches will be needed to identify those proteins that associate only transiently with asters/spindles (such as cytoplasmic dynein) and those proteins that are minor components of asters/spindles.
Among this collection of mitotic microtubule-associated proteins is astrin, a coiled coil-containing, nonmotor protein that localizes throughout the spindle during mitosis. Astrin is unique in that although it is expressed throughout the cell cycle, it is localized only to microtubules during mitosis. The C terminus is the primary determinant for targeting astrin to spindles; however, the association of astrin with spindles may involve more complex intermolecular interactions, because the N-terminal domain is sufficient to target some of the protein to spindle poles. Another unusual aspect of astrin is its association with kinetochores. It is the only protein to our knowledge that associates with kinetochores of only those chromosomes that have aligned at the metaphase plate. Thus, although astrin requires intact microtubules to localize to kinetochores and is thus not a constitutive kinetochore component, it can serve as a useful marker for kinetochore maturation and chromosome alignment.
Currently, the function of astrin is unknown. Astrin's specific association with spindles would suggest a functional role in spindle organization; however, we have been unable to confirm/refute that suggestion, because our antibodies do not react efficiently with the native protein, making immunodepletion of mitotic extracts and microinjection experiments uninformative. We speculate that astrin may play a structural role within the spindle based on two observations. First, the targeting of astrin to spindles is complex and involves both the N and C termini, indicating that astrin may bind to multiple spindle components through different domains. Second, astrin is homologous to rat Spag5, a gene product that was identified as interacting with odf1 (outer dense filament protein 1) from sperm (29). The outer dense filament is postulated to play a role in sperm motility or elastic recoil through cross-links with axonemal microtubules (30). Another component of the outer dense filament (odf2) has been shown recently to localize preferentially to the mother centriole in centrosomes, where it was speculated to play a role in microtubule anchoring to the centrosome (31). Thus, by analogy astrin may play a structural role in the spindle by cross-linking microtubules to other spindle components.
Acknowledgments
We thank Kevin Sullivan (Scripps Research Institute), Chloe Bulinski (Columbia University) and Jordan Raff (Wellcome/Cancer Research Campaign Institute) for generously providing antibodies. This work was supported by National Institutes of Health Grant GM51542.
Abbreviation
GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF399910).
References
- 1.Compton D A. Annu Rev Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh J R, Koonce M P. Science. 1989;246:622–628. doi: 10.1126/science.2683078. [DOI] [PubMed] [Google Scholar]
- 3.Mitchison T J. Curr Opin Cell Biol. 1989;1:69–74. doi: 10.1016/s0955-0674(89)80039-0. [DOI] [PubMed] [Google Scholar]
- 4.Rieder C L. Curr Opin Cell Biol. 1991;3:59–66. doi: 10.1016/0955-0674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- 5.Hyman A A, Karsenti E. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- 6.Sharp D J, Rogers G C, Scholey J M. Nature (London) 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 7.Savoian M S, Goldberg M L, Rieder C L. Nat Cell Biol. 2000;2:948–952. doi: 10.1038/35046605. [DOI] [PubMed] [Google Scholar]
- 8.Sharp D J, Rogers G C, Scholey J M. Nat Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- 9.Lombillo V A, Nislow C, Yen T J, Gelfand V I, McIntosh J R. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon M G, Howard L, Compton D A. J Cell Biol. 2001;152:425–434. doi: 10.1083/jcb.152.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan G K, Jablonski S A, Sudakin V, Hittle J C, Yen T J. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrieu A, Kahana J, Wood K W, Cleveland D W. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 13.Compton D A. J Cell Sci. 1998;111:1477–1481. doi: 10.1242/jcs.111.11.1477. [DOI] [PubMed] [Google Scholar]
- 14.Verde F, Berrez J-M, Antony C, Karsenti E. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heald R, Tournebize R, Blank T, Sadaltzopoulos R, Beker P, Karsenti E. Nature (London) 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 16.Gaglio T, Saredi A, Bingham J B, Hasbani M J, Gill S R, Schroer T A, Compton D A. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai A, Verma S, Mitchison T J. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 18.Compton D A, Cleveland D W. Curr Opin Cell Biol. 1994;6:343–346. doi: 10.1016/0955-0674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 19.Wittmann T, Wilm M, Karsenti E, Vernos I. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff J W. Proc Natl Acad Sci USA. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue Y H, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover D M. J Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaglio T, Saredi A, Compton D A. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dionne M A, Sanchez A, Compton D A. J Biol Chem. 2000;275:12346–12352. doi: 10.1074/jbc.275.16.12346. [DOI] [PubMed] [Google Scholar]
- 24.Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton D A. J Cell Biol. 1999;147:351–365. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafer D A, Gill S R, Cooper J A, Heuser J E, Schroer T A. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steuer E R, Schroer T A, Wordeman L, Sheetz M P. Nature (London) 1991;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- 27.Compton D A, Cleveland D W. J Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nehls M, Luno K, Schorpp M, Pfeifer D, Krause S, Matysiak-Scholze U, Dierbach H, Boehm T. Mamm Genome. 1995;6:321–331. doi: 10.1007/BF00364794. [DOI] [PubMed] [Google Scholar]
- 29.Shao X, Tarnasky H A, Lee J P, Oko R, van der Hoorn F A. Dev Biol. 1999;211:109–123. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- 30.Baltz J M, Wiliams P O, Cone R A. Biol Reprod. 1990;43:485–491. doi: 10.1095/biolreprod43.3.485. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Mol Biol Cell. 2001;12:1687–1697. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]