Disc1 Point Mutations in Mice Affect Development of the Cerebral Cortex (original) (raw)
Abstract
Disrupted-in-Schizophrenia 1 (DISC1) is a strong candidate gene for schizophrenia and other mental disorders. DISC1 regulates neurodevelopmental processes including neurogenesis, neuronal migration, neurite outgrowth, and neurotransmitter signaling. Abnormal neuronal morphology and cortical architecture are seen in human postmortem brain from patients with schizophrenia. However, the etiology and development of these histological abnormalities remain unclear. We analyzed the histology of two Disc1 mutant mice with point mutations (Q31L and L100P) and found a relative reduction in neuron number, decreased neurogenesis, and altered neuron distribution compared to wild-type littermates. Frontal cortical neurons have shorter dendrites and decreased surface area and spine density. Overall, the histology of Disc1 mutant mouse cortex is reminiscent of the findings in schizophrenia. These results provide further evidence that Disc1 participates in cortical development, including neurogenesis and neuron migration.
Introduction
Schizophrenia (SZ) is a common psychiatric disorder characterized by reduced hippocampal and cortical volume (Ross et al., 2006), abnormal cytoarchitecture (Kovalenko et al., 2003), reduced neuronal density in superficial cortical layers (Akbarian et al., 1993), decreased neuron size (Sweet et al., 2003), and reduced dendritic arborization (Young et al., 1998) and dendritic spine density (Garey et al., 1998). Many potential schizophrenia susceptibility genes have been identified (Wong and Van Tol, 2003; Ross et al., 2006), including Disrupted-in-Schizophrenia 1 (DISC1), first identified in a large Scottish family carrying a balanced (1q42.1:11q14.3) translocation cosegregating with major mental illnesses including SZ, bipolar disorder, and major depression (Millar et al., 2000). The DISC1 locus shows genetic linkage with SZ, and DISC1 variants show genetic association with SZ (Nakata et al., 2009; Rastogi et al., 2009; Schumacher et al., 2009). DISC1 acts as a regulatory scaffold protein, interacting with multiple binding partners including cytoskeletal and signaling proteins that link DISC1 to important brain developmental functions such as neurogenesis, neuron migration, neurite outgrowth, spine development, and neurotransmitter signaling (Camargo et al., 2007; Brandon et al., 2009).
The cortical histology of transgenic mice expressing various truncated mouse or human DISC1 fragments is similar to that seen in postmortem brain samples from patients with SZ (Hikida et al., 2007; Li et al., 2007; Pletnikov et al., 2008). Mice engineered to express truncated DISC1 have morphological alterations in medial prefrontal cortex and hippocampus (Kvajo et al., 2008). Because the truncated DISC1 gene is unique to the original translocation family and because common disease-associated DISC1 variants are single-nucleotide polymorphisms (SNPs) (Chubb et al., 2008), we sought to characterize brain histology in mice with Disc1 SNPs. Our group described previously two mutant Disc1 mice, each with a different SNP: Q31L (127A/T) and L100P (334T/C) (Clapcote et al., 2007). Both mutants have reduced brain volume, deficits in spatial working memory, and decreased prepulse inhibition. In addition, the Q31L mutants have abnormalities in social behavior and the forced swim test, while the L100P mice have increased open field activity (Clapcote et al., 2007). While the point mutations in our mice are not identical to human disease-associated variants, they may still provide important mechanistic insights into Disc1 SNP effects in contrast to the more drastic translocation mutations.
We undertook a comprehensive histological analysis of the cerebral cortex of Disc1 Q31L and L100P mutant mice. Our mutants have fewer neurons, decreased neuronal proliferation, and altered cortical layer positioning compared to wild-type (WT) littermates. Golgi staining showed shorter pyramidal neuron dendrite length in frontal cortex and reduced spine density in both frontal cortex and hippocampus. Our findings are similar to the abnormalities seen in postmortem human studies of SZ and with transgenic Disc1 mutant mouse models. Our results provide evidence for the effects of DISC1 SNPs on neurodevelopment and cortical structure that may be useful for interpreting DISC1 genetics in the general SZ patient population, and represent a starting point for further investigations of molecular disease mechanisms in SZ.
Materials and Methods
Mice.
_N_-ethyl-_N_-nitrosurea-mutagenized Disc1 mutant mouse lines on a C57BL/6 background (Q31L and L100P homozygous −/−) were generated as described previously (Clapcote et al., 2007), and additional mice were bred for histological analysis at the Toronto Centre for Phenogenomics (TCP) (Toronto, Canada). WT littermates from both Q31L and L100P groups were combined and used as controls. All mouse protocols were approved by the TCP Animal Care Committee.
Bromodeoxyuridine labeling.
Timed pregnant female mice were injected with bromodeoxyuridine (BrdU) (i.p., 50 mg/kg) at embryonic day 14 (E14) for proliferation experiments and at E12, E15, or E18 for investigation of neuronal positioning. Embryonic brains were harvested 24 h after BrdU injection at E15 and embedded in paraffin. Brains at postnatal day 21 (P21) were fixed in 4% paraformaldehyde overnight, cryoprotected in 30% sucrose, and frozen at −80°C before further processing.
Immunohistochemistry.
Paraffin-embedded and frozen coronal sections of 5 and 10 μm thickness, respectively, were cut using a microtome cryostat system (Bright Instruments 5030). All sections were initially incubated in blocking solution (0.1 m PBS, 1% Triton X-100, 0.5% Tween 20, 2% skim milk) or serum-free protein block (DakoCytomation) for 1 h at room temperature to reduce nonspecific background and then incubated with primary and secondary antibodies overnight at 4°C. The following primary antibodies were used: anti-NeuN (1:200; Millipore), anti-BrdU (1:200; BD Biosciences or Abcam), anti-Ki67 (1:200; Neomarkers), anti-Cux1 (1:200; Santa Cruz Biotechnology), and anti-Brn2 (1:200; Santa Cruz Biotechnology). Fluorescent secondary antibodies conjugated to Alexa 488 or Rhodamine Red-X (1:200; Invitrogen) or Cy3 (1:100; Jackson ImmunoResearch Laboratories) were used for detection of primary antibodies.
Golgi–Cox staining.
Golgi–Cox staining was performed as described previously (Gibb and Kolb, 1998). In brief, adult mice (age 6–8 weeks) were anesthetized with xylazene/ketamine (10 ml/kg) and intracardially perfused with 0.9% saline. Brains were removed and immersed in Golgi-Cox solution in the dark for 14 d before transfer to 30% sucrose solution for 5 d. Sections of 200 μm were sliced using a microtome (Leica VT1000S) and placed on 2% gelatinized microscope slides. The slides were stored in a humidified chamber for 3 d before further staining and fixation.
Analysis of immunohistochemistry: neuron number, distribution within the cortex, and neurogenesis.
Immunohistochemistry (IHC) images of the whole cortex were captured using a confocal microscope (Zeiss LSM510 Meta) at 5× or 25× magnification. All fluorescently labeled images were converted to gray values and normalized to background staining. Sections chosen for analysis were anatomically matched along the rostral-caudal axis for all samples. Regions of interest (ROIs) were positioned over the cortex as sampling windows. A two-dimensional cell counting approach of random sampling was used to provide accurate estimates of cell densities (Benes and Lange, 2001). For all IHC images, neurons were counted using the ITCN plugin for ImageJ (http://rsb.info.nih.gov/ij/) (ICTN parameters: width, 10 pixels; minimum distance, 2 pixels; threshold, 1 pixel). Specific procedures for defining areas of analysis differed slightly for each antibody, since each was chosen to address different questions. These procedures are described in detail below.
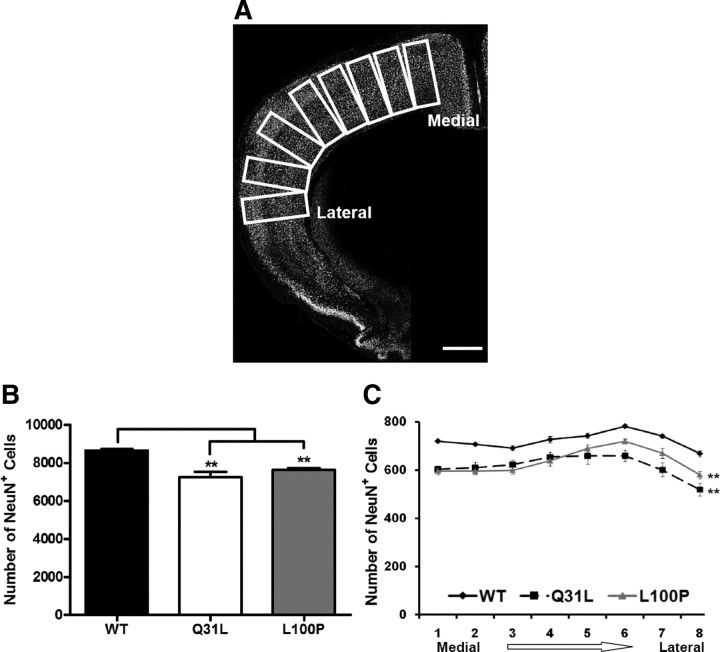
NeuN antibody-labeled images were used to examine overall neuron numbers in the cortex. Eight rectangular ROIs of fixed size (500 μm high × 250 μm wide), with the long axis perpendicular to the pial surface, were outlined throughout the neocortex from medial to lateral (see Fig. 1A). Each ROI was straightened with ImageJ software, and the number of neurons in the ROI was counted.
Figure 1.
Relative reduction in cortical neuron density in Disc1 Q31L and L100P −/− mutant mice. A, NeuN-immunostained confocal images were converted to gray scale, and eight equal ROIs were delimited along the medial to lateral axis. Scale bar, 250 μm. B, C, Quantification of NeuN+ cells is shown in B for all eight ROIs, and in C each ROI showed significantly fewer neurons in both Q31L and L100P mutants (n = 14) when compared to WT littermates (n = 30; two-way ANOVA, p < 0.01). All data are presented as mean ± SEM; **p < 0.01 versus WT.
BrdU and Ki67 were used to assess neurogenesis in both the embryonic and the postnatal cerebral cortex. For neuronal progenitor proliferation, fluorescently labeled cells in the ventricular zone (VZ) and subventricular zone (SVZ) were counted in a fixed area ROI of 100 × 120 μm. Similarly with P21 brains, BrdU-labeled cells in the frontal cortex were counted in an ROI of fixed width (500 μm) but of variable length, corresponding to the thickness of the cortex. Frontal cortical regions were defined according to the Golgi Atlas of the Postnatal Mouse Brain (Valverde, 1998). These BrdU ROIs were subdivided into 10 deciles along the axis perpendicular to the pia to assess the distribution of each wave of newly born neurons within the layers of cortex.
Cux1 and Brn2 antibodies were used to investigate whether neurons normally destined for the superficial cortical layers II and III would instead be seen in deeper layers (IV-VI) in Disc1 mutant mice. In both Cux1- and Brn2-labeled sections, two ROIs with a fixed width of 500 μm and of variable length spanning the thickness of the cortex were delineated. Each ROI was subdivided into eight equal regions from the pia to the inner border of the cortex to assess neuron distribution across the layers of the cortex.
Neuron morphology and dendritic spines.
For morphometric analyses of individual neurons, Golgi images at 40× magnification were captured under brightfield illumination with a Nikon Eclipse E600 microscope. Neurons were chosen based on the following criteria: (1) fully visible and characterized by clear, distinct morphology; (2) all dendrites seen within the 40× magnification field; and (3) only pyramidal neurons in layers III and V of the frontal cortex and CA1 area of the hippocampus, as shown in the Golgi Atlas of the Postnatal Mouse Brain (Valverde, 1998). A z-stack of different focal lengths for each individual neuron was generated to capture the three-dimensional dendritic branching tree in different planes. Acquisition parameters were kept the same for all images. The neurites of each neuron were traced, and the length and surface area were estimated using Neuromantic software (http://www.rdg.ac.uk/neuromantic). All parameters were further normalized to soma surface area for comparison.
Sholl analysis provides a quantitative measure of the radial distribution of neuronal dendritic arborization (Sholl, 1953). Using ImageJ, we created 15 concentric and equidistant circles (8 μm separation of each radius) centered at the perikaryon and then counted the number of dendritic intersections at each circle of increasing radius. The log of the number of intersections per circle area versus the circle radius was plotted (the semilog Sholl method). The slope of the regression line (κ = Sholl regression coefficient) is a measure of the decay rate of the number of branches with distance from the soma (Sholl, 1953). The Schoenen ramification index (maximum number of intersections/number of primary dendrites), a measure of the ramification richness for each neuron (Schoenen, 1982), and the number of dendritic bifurcations provide important information on the degree of dendritic branching complexity.
Spine density was measured with Golgi-stained images captured at 100× magnification (Nikon Eclipse E600). Spines were counted only on the apical dendrites of pyramidal neurons in layers III and V of frontal cortex and CA1 of hippocampus. Spine density was expressed as the number of spines per dendritic length (micrometer). All images for quantification were blinded before analysis.
Statistical analysis.
Statistical differences among different mutant lines and across genotypic groups against various measured parameters were determined using one-way or two-way ANOVA (SPSS 13.0), followed by Bonferroni's correction for multiple testing. To further confirm significance, Student's two-tailed t test was performed in comparing two sets of data. Data are expressed as mean ± SEM. A significance level of p < 0.05 was used for all analyses.
Results
Fewer NeuN-positive neurons in Q31L and L100P (−/−) mutant mice
Experimental evidence is accumulating for the role of DISC1 in neuronal proliferation and neuronal migration (Kamiya et al., 2005; Mao et al., 2009; Singh et al., 2010). We therefore examined the number of neurons throughout the neocortex along the medial-lateral axis. Neurons were labeled with antibodies to NeuN, a neuronal marker, and then counted in WT, Q31L, and L100P mice (Fig. 1A). Although NeuN does not label some types of neuron such as Purkinje cells, the high specificity and dense labeling of cortical neurons and interneurons provide a suitable measure of neuronal density (Wolf et al., 1996). We observed significantly fewer NeuN-labeled neurons in both Q31L (7260 ± 863; p < 0.001) and L100P mutant mice (7638 ± 346; p < 0.001) across the neocortex when compared with WT mice (8636 ± 522) (Fig. 1B). This observation of fewer NeuN-labeled neurons in Q31L and L100P compared to WT was also seen within each individual ROI, spanning from medial to lateral (Fig. 1C).
Reduced neuronal proliferation in Q31L and L100P compared to WT
We next determined whether the differences in the number of NeuN-positive neurons could be related to differences in neurogenesis between WT, Q31L, and L100P mice. First, BrdU was injected into pregnant dams at E14 and embryonic brains were collected at E15 to analyze neural progenitor proliferation in SVZ/VZ of the embryonic cortex (Fig. 2A). Disc1 mutants had fewer BrdU+ cells when compared to WT controls (Q31L: 70 ± 18 and L100P: 66 ± 24 vs WT: 102 ± 33; p < 0.001) (Fig. 2B, C). A similar pattern was observed with Ki67 immunostaining, further confirming a decrease in proliferation (Fig. 2D). As previous studies have shown that DISC1 knockdown causes premature neuronal differentiation (Mao et al., 2009), we investigated whether our mutations have similar effects. We identified cells that had left the cell cycle as BrdU positive and Ki67 negative. The percentage of BrdU+/Ki67− cells was only slightly increased in L100P mutants (97.41 ± 2.91%; p = 0.018) but not Q31L (96.75 ± 3.04%) compared to WT (95.26 ± 3.00%) (Fig. 2E). These subtle effects suggest that our Disc1 mutations may not have strong effects on the timing of neuronal differentiation.
Figure 2.
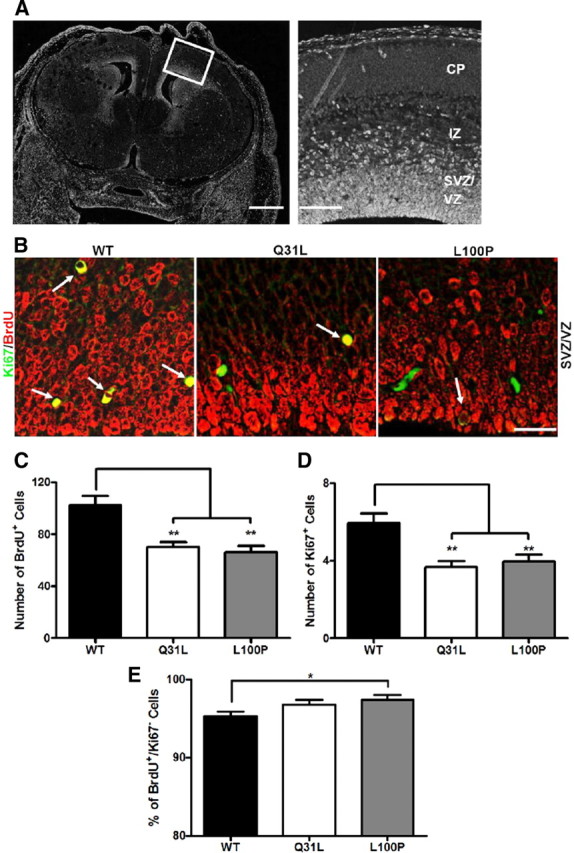
Decreased neuronal progenitor proliferation in Disc1 mutant mice. A, Coronal section of an E15 brain section with a higher magnification image of the different layers in the embryonic cortex (boxed area in the left image) shown in the right image. Scale bars, 250 μm (left) and 50 μm (right). BrdU was injected into pregnant dams at E14, and embryonic brains were collected at E15 for BrdU and Ki67 immunohistochemistry. CP, Cortical plate; IZ, intermediate zone. B, BrdU (red) and Ki67 (green) fluorescently labeled images of the SVZ/VZ region in WT, Q31L, and L100P mutants (left to right). White arrows indicate double-labeled (BrdU+ and Ki67+) cells. Scale bar, 30 μm. C, D, Quantification of BrdU+ (C) and Ki67+ (D) cells in a fixed ROI (100 μm × 120 μm) within the SVZ/VZ region revealed fewer BrdU- and Ki67-labeled cells in both Q31L and L100P mutants when compared to WT (n = 24; t test, p < 0.01). E, The cell cycle exit index is calculated as the fraction of cells labeled only with BrdU and no longer dividing (BrdU+ and Ki67−) divided by the total number of BrdU+ cells. Only a small significant increase in the percentage of BrdU+/Ki67− cells was observed with the L100P mutants, but not the Q31L mutants, when compared to WT. All data are presented as mean ± SEM; *p < 0.05, **p < 0.01 vs WT.
To observe the numbers and eventual location of neurons born at different times during embryonic corticogenesis, BrdU was injected at three different time points, E12, E15, and E18, and analyzed at P21. Coimmunostaining with NeuN confirmed the neuronal identity of BrdU-labeled cells (Fig. 3A). The average total number of BrdU-positive cells was significantly lower in Q31L and L100P cortexes compared to WT at E12 (Q31L: 61 ± 14 and L100P: 64 ± 14 vs WT: 149 ± 25; p < 0.001) and at E15 (Q31L: 185 ± 21 and L100P: 199 ± 40 vs WT: 254 ± 31; p < 0.001) (Fig. 3B). Interestingly, E18-injected BrdU+ neurons were only significantly decreased in Q31L (Q31L: 37 ± 17 vs WT: 54 ± 7; p < 0.05) but not in L100P mutants (42 ± 21) (Fig. 3B). Thus our results suggest that these single point mutations within Disc1 are associated with decreased neuronal proliferation.
Figure 3.
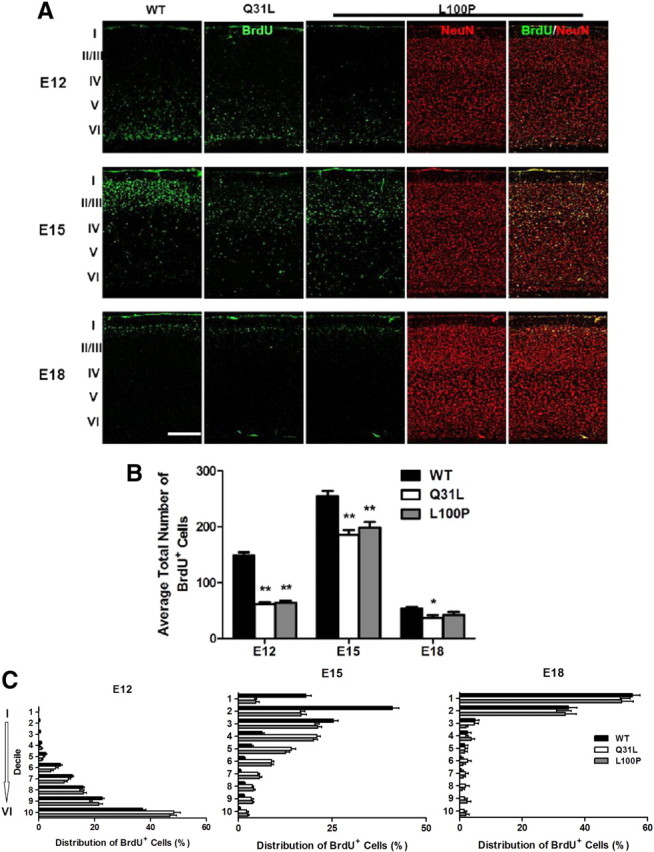
Reduced BrdU incorporation and mispositioning of cortical neurons in Disc1 mutant mice. A, BrdU was injected into E12, E15, and E18 pregnant females of WT, Q31L, and L100P mutants. Mice were killed at P21 for BrdU and NeuN immunostaining. Scale bar, 250 μm. B, The total number of BrdU+ cells was significantly lower in both E12 and E15 BrdU-injected Q31L and L100P mutants, but only Q31L showed fewer labeled cells with BrdU injection at E18 when compared to WT (n = 7–20; t test, p < 0.01). C, The distribution of BrdU-labeled cells was analyzed across cortical layers in all BrdU-injected time points. A rectangular ROI of fixed width (500 μm), spanning the thickness of the frontal cortex, was defined in each slice. This rectangle was then divided into 10 equal deciles along the axis perpendicular to the pial surface. The number of BrdU-positive cells in each decile was counted, and is shown as a percentage of the number in entire ROI. There were more BrdU-positive cells in deeper layers for E12 and E15 time points in both mutants when compared to WT (n = 7–12; two-way ANOVA, p < 0.01). In contrast, the distribution of E18-injected BrdU-labeled cells was similar in all cortical layers between WT and mutants, with only a small number of BrdU-positive cells observed in both mutants in deep cortical layers but not in control mice. All data are presented as mean ± SEM; *p < 0.05, **p < 0.01 versus WT. I—VI, Cortical layers I-VI.
Because neurons destined for more superficial layers are born later and because there is evidence that DISC1 may affect neuronal migration (Marín and Rubenstein, 2003; Kamiya et al., 2005; Young-Pearse et al., 2010), we next examined the cortical distribution and position of BrdU-labeled cells for each time point of BrdU injection. Confocal imaging and quantification revealed that BrdU+ cells were located in deeper cortical layers for E12- and E15-injected Q31L and L100P mice compared with WT (Fig. 3C). At E18, the distribution of BrdU-labeled cells was similar in all cortical layers and all groups, with a small number of BrdU+ cells observed in deeper layers of both Q31L and L100P lines but not in WT (Fig. 3C). Together, these results suggest that point mutations in the Disc1 gene affect neuronal proliferation and the location of neurons within the cortex.
Differential Cux1 and Brn2 staining in Q31L and L100P mice compared to WT
To further examine the relationship between DISC1 and neuronal positioning, we performed IHC with two layer II/III-specific protein markers, Cux1 and Brn2 (Molyneaux et al., 2007). When compared to WT, both Cux1- and Brn2-labeled neurons in Q31L and L100P animals were further away from the pia (Fig. 4A). To quantify the positions of Cux1- and Brn2-labeled cells, two rectangles were positioned over the neocortical region of fluorescent staining, and each was divided into eight equal octants (spanning superficial to deep layers) (Fig. 4B). The percentage of Cux1-labeled cells was significantly higher in both octant 1 and octant 3 of WT littermates (octant 1: 12.83 ± 3.55%; octant 3: 37.37 ± 4.52%) compared to Q31L (octant 1: 5.99 ± 1.76%; octant 3: 31.42 ± 4.07%, p < 0.01) and L100P (octant 1: 7.43 ± 2.61%, p < 0.01; octant 3: 35.11 ± 4.81%, p < 0.05) mice. In contrast, WT controls (12.85 ± 4.84%) displayed a significantly lower proportion of Cux1+ cells in octant 4 than in Q31L (22.53 ± 5.98%, p < 0.01) and L100P (22.04 ± 9.1%, p < 0.01) (Fig. 4B). For Brn2, WT mice showed a significantly higher proportion of Brn2+ cells in superficial cortical layers (octants 1 and 2), and significantly fewer such cells in the deep layers of octants 4, 5, and 6 when compared to both mutants (Fig. 4C). Moreover, we measured the ratio of distance between the inner edge of Cux1fluorescently labeled cells and pia mater versus total cortical thickness and saw a higher ratio in Q31L (0.433 ± 0.039, p < 0.01) and L100P (0.432 ± 0.041, p < 0.01) mutants versus WT (0.417 ± 0.031; p < 0.01) (Fig. 4D). These data suggest that the localization of cortical neurons in Q31L and L100P mice is altered compared to WT mice.
Figure 4.
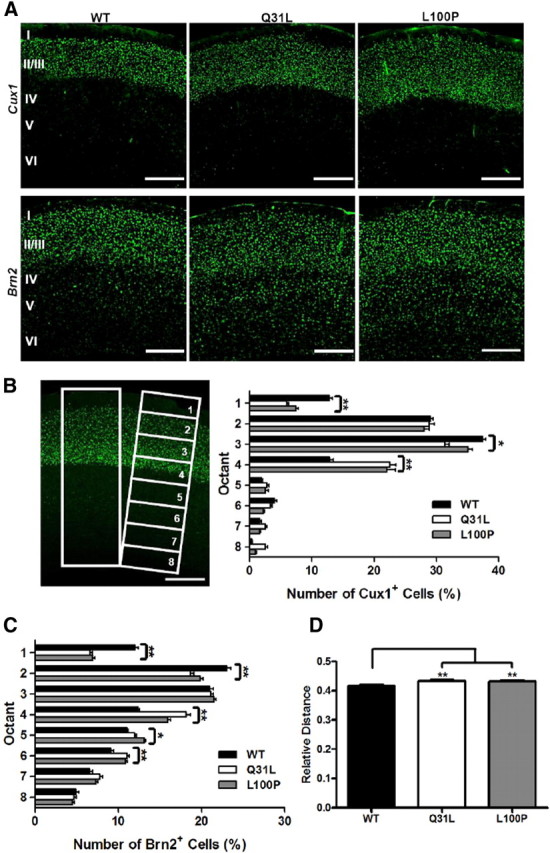
Altered cortical neuronal distribution in Disc1 mutant mice. A, Cux1-labeled (top) and Brn2-labeled (bottom row) fluorescent images of WT, Q31L, and L100P (left to right) brains showed a slight shift of distribution of neurons toward deeper cortical layers in the mutants. B, Two equal ROIs across the cortex of both the Cux1-labeled and Brn2-labeled neurons were outlined. Each ROI was subdivided into eight equal octants in which the number of Cux1+ and Brn2+ cells were counted and expressed as a percentage of total cells in each ROI. A significantly higher proportion of Cux1+ cells was observed in WT (n = 62) than in both Q31L (n = 36) and L100P mutants (n = 44) in the first and third octant. Conversely, the proportion of Cux1+ cells in the fourth octant was significantly lower in WT than the two mutants (two-way ANOVA, t test, p < 0.01). C, Similarly, more Brn2+ cells were present in superficial cortical layers (octants 1 and 2) in WT than in mutants, while fewer Brn2+ cells were seen in deeper cortical layers (octants 4, 5, and 6) in WT (n = 52; two-way ANOVA, t test, p < 0.01). Together, these results indicate that more Cux1-labeled and Brn2-labeled cells are positioned in deeper cortical layers in Disc1 mutants vs WT. D, The distance from the deep edge of the Cux1 fluorescent neurons to the pia versus total cortical thickness within each ROI was also significantly higher in both mutants (t test, p < 0.01). Scale bar, 250 μm. All data are presented as mean ± SEM; *p < 0.05, **p < 0.01 versus WT. I–VI, Cortical layers I-VI.
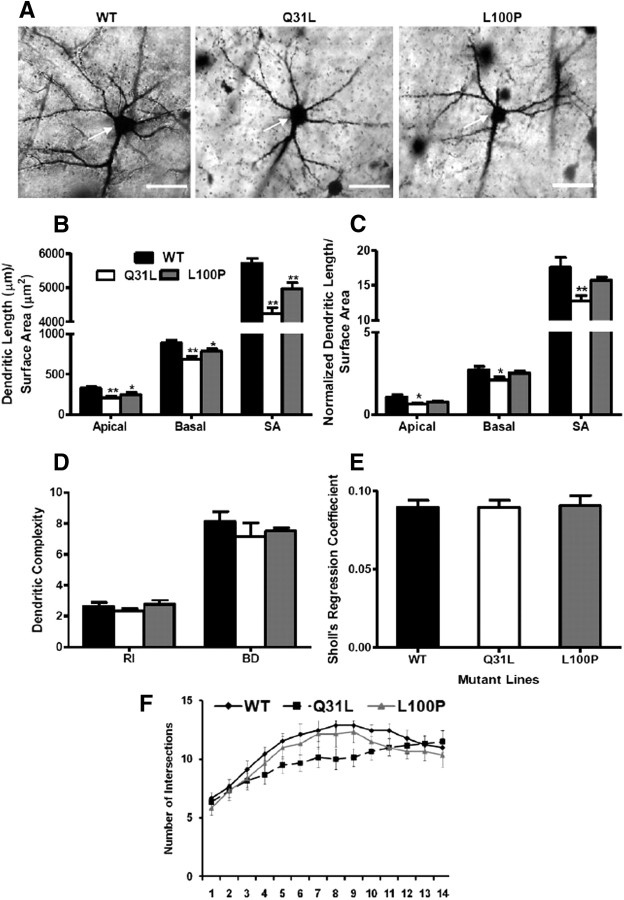
Differences in frontal cortical dendritic morphology in Q31L and L100P mice
There is strong evidence that DISC1 regulates neurite outgrowth and dendritic arborization; therefore, we performed a detailed morphological analysis of dendritic trees for individual neurons in Disc1 Q31L and L100P mutants and WT mice. Golgi staining provides a clear and complete image for a subgroup of neurons without interference by neighboring neurons. Representative neurons from WT, Q31L, and L100P mice are shown in Figure 5A. We observed a significantly shorter apical dendritic length (ADL) in Q31L (208.9 ± 68.9 μm, p < 0.01) and L100P mutants (242.4 ± 94.9 μm, p = 0.03) when compared to WT (328.3 ± 55.5 μm). Consistently, basal dendritic length (BDL) showed a similar trend with significant differences in Q31L (690.6 ± 100.5 μm, p < 0.01) and L100P (786.8 ± 90.1 μm, p = 0.021) versus WT (884 ± 109.6 μm) (Fig. 5B). We also found a significantly lower total dendritic surface area (DSA) of Q31L (4250.9 ± 514.1 μm2, p < 0.001) and L100P mutants (4966.3 ± 550.7 μm2, p = 0.005) when compared to WT (5708.5 ± 439.5 μm2) (Fig. 5B).
Figure 5.
Morphology of neurons from the frontal cortex in Q31L and L100P −/− mutants. A, Golgi-stained images of an individual neuron at 40× magnification from WT, Q31L, and L100P mutant mice (left to right). Scale bar, 50 μm. The white arrows indicate pyramidal neurons with clear, distinct dendrites. B, Both Q31L and L100P mutants had shorter dendrites (apical and basal) and smaller surface area (SA) compared to WT neurons (n = 12; t test, p < 0.05). C, After normalizing to soma SA, only the Q31L mutants had significant differences in all three parameters, while no significant effects were observed with L100P mutants. However, a similar trend was still visible across all groups. D, No significant difference in dendritic branching complexity was observed when measured by Schoenen's ramification index (RI) and the number of bifurcations of dendrites (BD). E, Sholl analysis revealed no significant difference in Disc1 mutant mice versus WT on Sholl's regression coefficient (n = 12). F, Quantification of the number of dendritic intersections with increasing circular radius from the cell body was similar between WT and mutants. All data are presented as mean ± SEM; *p < 0.05, **p < 0.01 versus WT.
Cell soma size has been reported to correlate with dendritic structure (Somogyi and Klausberger, 2005). Thus, we normalized the measured parameters described above to soma surface area. Interestingly, neurite outgrowth was significantly lower by 40.5% (p = 0.014), 23% (p = 0.042), and 27.7% (p = 0.007) in ADL, BDL, and DSA, respectively, within Q31L mutants only, while L100P mice showed a similar but nonsignificant trend (Fig. 5C).
We evaluated dendritic arbor complexity in our Disc1 mutant mice versus WT via Sholl analysis. There were no significant differences in the ramification index (Q31L: 2.3 ± 0.2, p = 0.37; L100P: 2.8 ± 0.3, p = 0.73; vs WT: 2.6 ± 0.3), the dendritic bifurcations (Q31L: 7.2 ± 2.1, p = 0.39; L100P: 7.5 ± 0.5, p = 0.48; vs WT: 8.1 ± 2.0), Sholl's regression coefficient (Q31L: 0.089 ± 0.005, p = 0.97; L100P: 0.091 ± 0.007, p = 0.92; vs WT: 0.090 ± 0.004), or the number of dendritic intersections per radial segments between Q31L and L100P and WT controls (Fig. 5D–F).
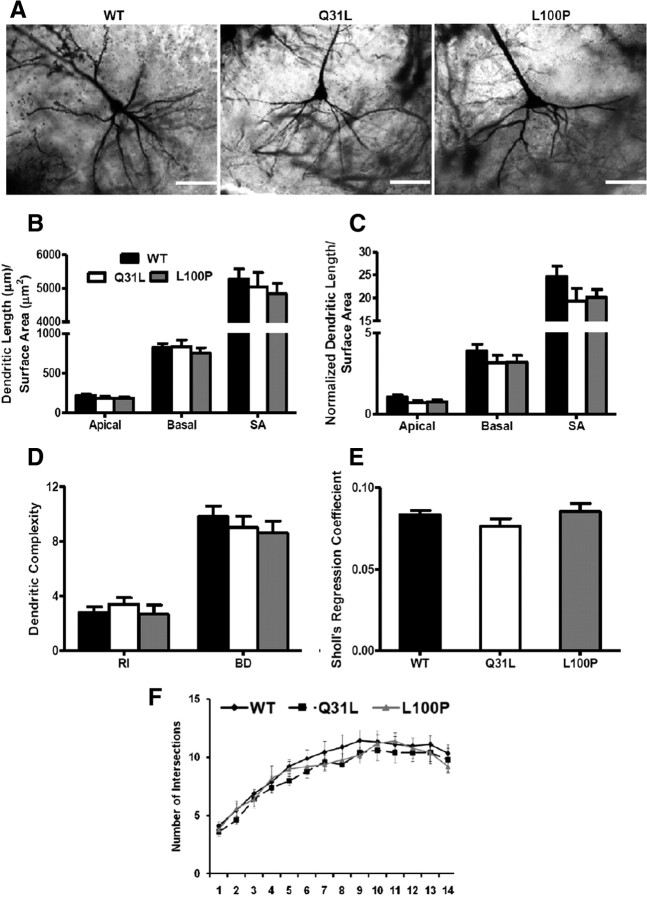
In addition to the frontal cortex, the hippocampus is another important brain area of interest in SZ (Harrison, 2004). Hence, we extended our morphological analyses to pyramidal neurons in hippocampal CA1 regions. In our study, we did not observe any alterations in dendritic length, surface area, ramification index, number of bifurcations of dendrites, and Sholl's regression coefficient in either mutant line within the hippocampus (Fig. 6).
Figure 6.
No alteration in morphology of hippocampal pyramidal neurons in Disc1 mutants. A, Golgi images of hippocampal neuron morphology in WT, Q31L, and L100P mutants. B, No significant difference was observed in dendritic length and surface area (SA) when compared to control mice (n = 12). C, Similarly, parameters after normalization were not significantly different among the measured groups. A slight decreasing trend was observed that parallels the frontal cortex results. D–F, Hippocampal dendritic branching pattern was not significantly different as measured with Schoenen's ramification index (RI), the number of bifurcations of dendrites (BD), Sholl's regression coefficient analysis, and dendritic intersections with increasing radial segments. (n = 9–12). All data are presented as mean ± SEM. SA, Surface area.
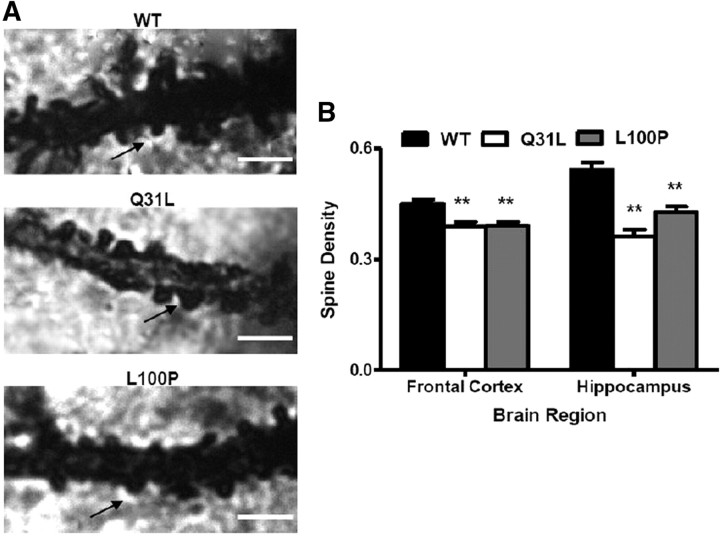
Lower spine density in frontal cortex and hippocampus in Q31L and L100P Disc1 mutant mice
Cognitive deficits associated with SZ have been attributed to altered synaptic transmission and plasticity in which dendritic spines play a critical role (Calabrese et al., 2006). DISC1 is involved in dendritic spine development via Kalirin-7 (Penzes and Jones, 2008; Hayashi-Takagi et al., 2010). To determine whether our Disc1 point mutations affect dendritic spine development, we measured dendritic spine density in the frontal cortex and hippocampus of Q31L, L100P, and WT mice. Golgi images of 100× magnification were captured, and all spine types were counted (Fig. 7A). Both Disc1 mutants had significantly reduced spine density in the frontal cortex with an average spine density of 0.39 ± 0.063 spines/μm in Q31L (p = 0.001) and 0.391 ± 0.057 spines/μm in L100P (p < 0.001) compared to 0.449 ± 0.071 spines/μm in WT (Fig. 7B). Similarly, spine density was also reduced in the hippocampus (Q31L: 0.363 ± 0.087 spines/μm and L100P: 0.427 ± 0.085 spines/μm vs WT: 0.543 ± 0.11 spines/μm; p < 0.001). Thus, DISC1 mutations appear to affect spine density in both the frontal cortex and hippocampus.
Figure 7.
Disc1 mutant mice had lower spine density in frontal cortical and hippocampal pyramidal neurons. A, High magnification (100×) of Golgi-stained images of spine protrusions on apical dendrites. Scale bar, 5 μm. Note the arrows indicating clear dendritic spines. B, Quantification of spine density (number of spines/μm) in all groups showed a significant lower density in mutant mice when compared to WT in frontal cortex and hippocampus (frontal cortex, n = 30; hippocampus, n = 26–33; t test, p < 0.01). All data are presented as mean ± SEM; **p < 0.01 versus WT.
Discussion
There is increasing evidence for a strong association between DISC1 and several major mental illnesses. The mechanism by which DISC1 gene variants produce both cellular and behavioral abnormalities is still unclear. We used two previously described mouse lines with point mutations in Disc1 that have behavioral changes relevant to SZ and depression (Clapcote et al., 2007). In this study, we report a relative decrease in neuron number and decreased neuronal proliferation in the Disc1 mutants compared to WT mice. The mutant mice have differences in neuron positioning and morphology similar to some findings in human SZ histopathological studies (Akbarian et al., 1993; Ross et al., 2006). However, other histological abnormalities observed in postmortem schizophrenia brain, such as interneuron deficits, have not yet been investigated in our Disc1 mutant mice.
We found relatively fewer neurons and decreased neuronal proliferation in Q31L and L100P mutant mice compared to WT. Decreased neuronal density is a common finding of postmortem studies on patients with SZ. Reductions in neuron density in the primary visual cortex (Dorph-Petersen et al., 2007) and reduced glutamatergic neurons in the orbitofrontal cortex have been reported (Garey, 2010). Presently, the DISC1 status of the patients reported in those studies is unknown. Human studies combining genetic markers and histopathological analysis are required.
DISC1 has been well established as a regulator of neurogenesis (Mao et al., 2009). DISC1 participates in a glycogen synthase kinase 3β (GSK3β) signaling pathway involving β-catenin via a direct interaction with GSK3β at two different domains of DISC1, spanning amino acids 1–220 and 356–595. Moreover, DISC1 knockdown results in a reduction of proliferation progenitors likely caused by early cell cycle exit (Mao et al., 2009). However, our Disc1 mutations showed a decrease in neuronal proliferation but not premature neuronal differentiation. It is possible that the Q31L and L100P mutations in DISC1 may only affect part of its interaction with GSK3β and that premature cell cycle exit may not be the sole determinant of neuronal proliferation. Recently, the L100P Disc1 mutant mouse was shown to have reduced interaction with both GSK3α and β (Lipina et al., 2011). Furthermore, genetic and pharmacological inhibition of GSK3 activity rescued DISC1-mediated behavioral effects in these mice.
Examination of neuronal distribution using layer-specific protein markers revealed altered neuron location in Q31L and L100P mice compared to WT. Cortical neuronal positioning can be affected by changes in neurogenesis and neuronal migration. Later-born neurons migrate to more superficial layers of the cortex through radial migration (Marín and Rubenstein, 2003). As DISC1 regulates neurogenesis and neuronal migration, we hypothesized that aberrant neuronal distribution in the cortex may be due to DISC1-mediated effects on both processes (Kamiya et al., 2005; Mao et al., 2009). It was recently shown that DISC1 may participate in neurogenesis and neuronal migration via separate and distinct signaling pathways (Singh et al., 2010). Abnormal cortical cytoarchitecture may also result from malfunctioning of the cytoskeletal machinery mediating neuronal migration. Recent studies have reported several important DISC1-interacting candidates with critical roles in the regulation of neuronal migration, including pericentriolar material 1 (PCM1) (Kamiya et al., 2008), amyloid precursor protein (APP) (Young-Pearse et al., 2010), neuregulin-1/ErbB4 (Jaaro-Peled et al., 2009), and LIS1/NDEL1 (Morris et al., 2003; Wynshaw-Boris, 2007). However, complete details of the mechanisms by which neurogenesis and neuronal migration interact to modulate cortical cytoarchitecture remain to be determined.
The observed differences in frontal cortical neuron morphology in the Disc1 mutants compared to WT may also be mediated by DISC1 interactions with the actin and microtubule cytoskeleton (Ishizuka et al., 2006). Loss of normal DISC1 function or expression of mutant DISC1 disrupts its interaction with NDEL1 and causes abnormal neurite outgrowth in PC12 cells (Ozeki et al., 2003; Kamiya et al., 2005). Consistent with this, transgenic mice expressing truncated Disc1 have an inhibition of neurite outgrowth and a reduction of apical dendritic length (Kvajo et al., 2008; Shen et al., 2008). We reported similar findings of significant reductions in dendritic length and surface area in our Disc1 mutants, further supporting the role of DISC1 in neurodevelopment. Recent studies with transgenic Disc1 mice have shown a disturbance in neuronal arborization both in the developing cerebral cortex and hippocampus (Kamiya et al., 2005; Li et al., 2007; Niwa et al., 2010). In contrast, Kvajo reported no significant changes in dendritic complexity with their mice expressing truncated transgenic Disc1 (Kvajo et al., 2008). Our Disc1 point mutations also showed no alterations in dendritic branching pattern, consistent with Kvajo et al. Dendritic arbor development is a complicated and strictly regulated multistep process involving the following: (1) neurite initiation, outgrowth and guidance; (2) branching and synapse formation; and (3) cytoskeleton stabilization (Urbanska et al., 2008). Proper formation and stabilization of dendritic arbors requires various intrinsic and extrinsic signals (Urbanska et al., 2008).
Hippocampal neurons in SZ have been reported to have subtle morphological changes in size, organization, and perhaps shape (Harrison, 2004). Other Disc1 mutant mouse models show similar features (Li et al., 2007; Shen et al., 2008), and human SNPs in DISC1 have been associated with altered hippocampal structure and cognitive function (Callicott et al., 2005). However, not all SZ postmortem studies are consistent in finding abnormal hippocampal neuronal morphology (Benes et al., 1998). We did not detect any significant changes in hippocampal neuronal morphology with our Disc1 mutations, similar to the findings from Kvajo (Kvajo et al., 2008). Duan et al. (2007) recently demonstrated an acceleration of neuronal integration when downregulating DISC1 in adult hippocampal neurons. As the developmental origin of hippocampus is distinct from the cerebral cortex, DISC1 may modulate different developmental programs in the hippocampus.
Synaptic pathology has been proposed as a cause for cognitive deficits in SZ. Dendritic spines are the postsynaptic targets for synaptic transmission, and decreased spine density in prefrontal cortical and subventricular pyramidal neurons has been reported in human postmortem SZ studies (Garey et al., 1998; Glantz and Lewis, 2000; Rosoklija et al., 2000). Spine density can be used as a measure of neural connectivity (Benes, 2000). DISC1 interacts with Kalirin-7 to modulate Rac1, an important regulator of dendritic spine development and functional plasticity (Penzes and Jones, 2008; Hayashi-Takagi et al., 2010). Long-term suppression of DISC1 can lead to spine shrinkage in primary cortical neurons (Hayashi-Takagi et al., 2010). Our observations are consistent with these previous data, since we see a significant reduction in spine density in both mutants for frontal cortex and hippocampus. Our results further confirm that DISC1 regulates spine development and may represent a possible link between DISC1 genetic variants and cognitive deficits observed in SZ.
Intriguingly, although Q31L and L100P mutant mice have distinct behavioral abnormalities, they have similar histological deficits. Subtle disruption of neuronal architecture and connections can have diverse effects on complex behaviors and on the activity of other brain regions. Q31L and L100P mutants also have different phosphodiesterase 4B (PDE4B) activity and Disc1-PDE4B binding (Clapcote et al., 2007). The discrepancy between histology and behavior may require further histological and biochemical characterization of the effects of these point mutations. Our study provides a general overview of cortical histology, development, and neuronal morphology in two independent Disc1 single-point mutant mouse lines. The function of Disc1 in neuron development has mostly been investigated by observing the effect of drastic reductions in DISC1 expression or by expression of a truncated protein. Our study is novel in characterizing histopathological findings in two mouse lines with Disc1 SNPs. Although the human disease-associated DISC1 SNPs are not the same as the Q31L and L100P mutations in our mice, we argue that our mouse SNPs are more similar to the DISC1 SNPs in the general human population than the truncated Disc1 mutants. Previous studies with truncated DISC1 or severe suppression of DISC1 expression are more relevant to understanding the pathophysiology of the original Scottish translocation pedigree. Ongoing experiments to further understand the molecular mechanisms by which DISC1 regulates brain development are required to understand the causal links between DISC1 variants and susceptibility to schizophrenia.
Footnotes
We thank the Canadian Institutes of Health Research for supporting A.H.C.W. with a Clinician Scientist Phase II Fellowship. M.P.F. is in the Clinician Scientist Program, Department of Psychiatry, University of Toronto. S.J.C. and A.H.C.W. are recipients of National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Awards. We also thank Eric Feng and Carlos Law for blinding of data during analysis.
The authors declare no competing financial interests.
References
- Akbarian S, Bunney WE, Jr, Potkin SG, Wigal SB, Hagman JO, Sandman CA, Jones EG. Altered distribution of nicotinamide-adenine dinucleotide phosphate-diaphorase cells in frontal lobe of schizophrenics implies disturbances of cortical development. Arch Gen Psychiatry. 1993;50:169–177. doi: 10.1001/archpsyc.1993.01820150007001. [DOI] [PubMed] [Google Scholar]
- Benes FM. Emerging principles of altered neural circuitry in schizophrenia. Brain Res Brain Res Rev. 2000;31:251–269. doi: 10.1016/s0165-0173(99)00041-7. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lange N. Two-dimensional versus three-dimensional cell counting: a practical perspective. Trends Neurosci. 2001;24:11–17. doi: 10.1016/s0166-2236(00)01660-x. [DOI] [PubMed] [Google Scholar]
- Benes FM, Kwok EW, Vincent SL, Todtenkopf MS. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biol Psychiatry. 1998;44:88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Millar JK, Korth C, Sive H, Singh KK, Sawa A. Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci. 2009;29:12768–12775. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese B, Wilson MS, Halpain S. Development and regulation of dendritic spine synapses. Physiology (Bethesda) 2006;21:38–47. doi: 10.1152/physiol.00042.2005. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ. Disrupted in Schizophrenia 1 interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, Kaneda H, Shiroishi T, Houslay MD, Henkelman RM, Sled JG, Gondo Y, Porteous DJ, Roder JC. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Dorph-Petersen KA, Pierri JN, Wu Q, Sampson AR, Lewis DA. Primary visual cortex volume and total neuron number are reduced in schizophrenia. J Comp Neurol. 2007;501:290–301. doi: 10.1002/cne.21243. [DOI] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey L. When cortical development goes wrong: schizophrenia as a neurodevelopmental disease of microcircuits. J Anat. 2010;217:324–333. doi: 10.1111/j.1469-7580.2010.01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM, Barnes TR, Hirsch SR. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry. 1998;65:446–453. doi: 10.1136/jnnp.65.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb R, Kolb B. A method for vibratome sectioning of Golgi-Cox stained whole rat brain. J Neurosci Methods. 1998;79:1–4. doi: 10.1016/s0165-0270(97)00163-5. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- Harrison PJ. The hippocampus in schizophrenia: a review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology (Berl) 2004;174:151–162. doi: 10.1007/s00213-003-1761-y. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andradé M, Tankou S, Mori S, Gallagher M, Ishizuka K, Pletnikov M, Kida S, Sawa A. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K, Paek M, Kamiya A, Sawa A. A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol Psychiatry. 2006;59:1189–1197. doi: 10.1016/j.biopsych.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Jaaro-Peled H, Hayashi-Takagi A, Seshadri S, Kamiya A, Brandon NJ, Sawa A. Neurodevelopmental mechanisms of schizophrenia: understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009;32:485–495. doi: 10.1016/j.tins.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Tan PL, Kubo K, Engelhard C, Ishizuka K, Kubo A, Tsukita S, Pulver AE, Nakajima K, Cascella NG, Katsanis N, Sawa A. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko S, Bergmann A, Schneider-Axmann T, Ovary I, Majtenyi K, Havas L, Honer WG, Bogerts B, Falkai P. Regio entorhinalis in schizophrenia: more evidence for migrational disturbances and suggestions for a new biological hypothesis. Pharmacopsychiatry. 2003;36(Suppl 3):S158–S161. doi: 10.1055/s-2003-45124. [DOI] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB, Karayiorgou M, Gogos JA. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci U S A. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, Hennah W, Peltonen L, Lönnqvist J, Huttunen MO, Kaprio J, Trachtenberg JT, Silva AJ, Cannon TD. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U S A. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipina TV, Kaidanovich-Beilin O, Patel S, Wang M, Clapcote SJ, Liu F, Woodgett JR, Roder JC. Genetic and pharmacological evidence for schizophrenia-related DISC1 interaction with GSK-3. Synapse. 2011;65:234–248. doi: 10.1002/syn.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, Biechele T, Petryshen TL, Moon RT, Haggarty SJ, Tsai LH. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, Vakkalanka R, Barenboim M, Sei Y, Weinberger DR, Kleinman JE. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Kamiya A, Murai R, Kubo K, Gruber AJ, Tomita K, Lu L, Tomisato S, Jaaro-Peled H, Seshadri S, Hiyama H, Huang B, Kohda K, Noda Y, O'Donnell P, Nakajima K, Sawa A, Nabeshima T. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65:480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Jones KA. Dendritic spine dynamics–a key role for kalirin-7. Trends Neurosci. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, Mori S, Moran TH, Ross CA. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- Rastogi A, Zai C, Likhodi O, Kennedy JL, Wong AH. Genetic association and post-mortem brain mRNA analysis of DISC1 and related genes in schizophrenia. Schizophr Res. 2009;114:39–49. doi: 10.1016/j.schres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Rosoklija G, Toomayan G, Ellis SP, Keilp J, Mann JJ, Latov N, Hays AP, Dwork AJ. Structural abnormalities of subicular dendrites in subjects with schizophrenia and mood disorders: preliminary findings. Arch Gen Psychiatry. 2000;57:349–356. doi: 10.1001/archpsyc.57.4.349. [DOI] [PubMed] [Google Scholar]
- Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Schoenen J. Dendritic organization of the human spinal cord: the motoneurons. J Comp Neurol. 1982;211:226–247. doi: 10.1002/cne.902110303. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Laje G, Abou Jamra R, Becker T, Mühleisen TW, Vasilescu C, Mattheisen M, Herms S, Hoffmann P, Hillmer AM, Georgi A, Herold C, Schulze TG, Propping P, Rietschel M, McMahon FJ, Nöthen MM, Cichon S. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum Mol Genet. 2009;18:2719–2727. doi: 10.1093/hmg/ddp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, Chatzi C, He S, Mackie I, Brandon NJ, Marquis KL, Day M, Hurko O, McCaig CD, Riedel G, St Clair D. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, Tsai LH. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet RA, Pierri JN, Auh S, Sampson AR, Lewis DA. Reduced pyramidal cell somal volume in auditory association cortex of subjects with schizophrenia. Neuropsychopharmacology. 2003;28:599–609. doi: 10.1038/sj.npp.1300120. [DOI] [PubMed] [Google Scholar]
- Urbanska M, Blazejczyk M, Jaworski J. Molecular basis of dendritic arborization. Acta Neurobiol Exp (Wars) 2008;68:264–288. doi: 10.55782/ane-2008-1695. [DOI] [PubMed] [Google Scholar]
- Valverde F. Golgi atlas of the postnatal mouse brain. Vienna: Springer; 1998. [Google Scholar]
- Wolf HK, Buslei R, Schmidt-Kastner R, Schmidt-Kastner PK, Pietsch T, Wiestler OD, Blümcke I. NeuN: a useful neuronal marker for diagnostic histopathology. J Histochem Cytochem. 1996;44:1167–1171. doi: 10.1177/44.10.8813082. [DOI] [PubMed] [Google Scholar]
- Wong AH, Van Tol HH. Schizophrenia: from phenomenology to neurobiology. Neurosci Biobehav Rev. 2003;27:269–306. doi: 10.1016/s0149-7634(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Young CE, Arima K, Xie J, Hu L, Beach TG, Falkai P, Honer WG. SNAP-25 deficit and hippocampal connectivity in schizophrenia. Cereb Cortex. 1998;8:261–268. doi: 10.1093/cercor/8.3.261. [DOI] [PubMed] [Google Scholar]
- Young-Pearse TL, Suth S, Luth ES, Sawa A, Selkoe DJ. Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J Neurosci. 2010;30:10431–10440. doi: 10.1523/JNEUROSCI.1445-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]