Yotiao, a Novel Protein of Neuromuscular Junction and Brain That Interacts with Specific Splice Variants of NMDA Receptor Subunit NR1 (original) (raw)
Abstract
The molecular machinery underlying neurotransmitter receptor immobilization at postsynaptic sites is poorly understood. The NMDA receptor subunit NR1 can form clusters in heterologous cells via a mechanism dependent on the alternatively spliced C1 exon cassette in its intracellular C-terminal tail, suggesting a functional interaction between NR1 and the cytoskeleton. The yeast two-hybrid screen was used here to identify yotiao, a novel coiled coil protein that interacts with NR1 in a C1 exon-dependent manner. Yotiao mRNA (11 kb) is present modestly in brain and abundantly in skeletal muscle and pancreas. On Western blots, yotiao appears as an ∼230 kDa band that is present in cerebral cortex, hippocampus, and cerebellum. Biochemical studies reveal that yotiao fractionates with cytoskeleton-associated proteins and with the postsynaptic density. With regard to immunohistochemistry, two anti-yotiao antibodies display a somatodendritic staining pattern similar to each other and to the staining pattern of NR1. Yotiao was colocalized by double-label immunocytochemistry with NR1 in rat brain and could be coimmunoprecipitated with NR1 from heterologous cells. Thus yotiao is an NR1-binding protein potentially involved in cytoskeletal attachment of NMDA receptors. Consistent with a general involvement in postsynaptic structure, yotiao was also found to be specifically concentrated at the neuromuscular junction in skeletal muscle.
Keywords: yotiao, NMDA receptor, neuromuscular junction, neuronal synapse, postsynaptic density, cytoskeleton, coiled coil protein, yeast two-hybrid
Proper synaptic function requires accurate localization of appropriate ion channels and neurotransmitter receptors to the postsynaptic site. This localization may occur by means of specific interactions between synaptic membrane proteins and a variety of anchoring or clustering molecules (Froehner, 1993; Hall and Sanes, 1993; Sheng and Kim, 1996). These molecules, via interactions with other proteins, may also bring components of downstream signaling pathways into the proximity of the appropriate channel or receptor, thereby increasing the specificity and efficacy of signal transmission.
The NMDA receptor has been closely scrutinized because it represents a major subclass of glutamate receptor at excitatory synapses in the mammalian brain. NMDA receptors are involved in neural development and excitotoxicity (Constantine-Paton et al., 1990; Choi, 1995) and in mechanisms of activity-dependent synaptic plasticity (Bliss and Collingridge, 1993; Lisman and Harris, 1993). NMDA receptors exist_in vivo_ as heteromultimers composed of the essential NR1 subunit assembled with various members of the NR2 subfamily (NR2A–D) (Ishii et al., 1992; Meguro et al., 1992; Monyer et al., 1992;Kutsuwada et al., 1993; Sheng et al., 1994). Each NR2 subunit confers distinct properties on the heteromeric NMDA receptor complex (Monyer et al., 1994). Further molecular diversity is imparted by alternative splicing at three sites in the mRNA, which generate eight distinct NR1 splice variants (Sugihara et al., 1992; Hollmann et al., 1993).
NMDA receptors were first cloned by functional expression (for review, see Hollmann and Heinemann, 1994), and few NMDA receptor-associated proteins have been identified in brain. An important interaction exists between NR2 subunits and members of the postsynaptic density-95 (PSD-95) family of synaptic proteins (Kornau et al., 1995; Niethammer et al., 1996) (for review, see Sheng, 1996; Sheng and Kim, 1996; Kornau et al., 1997). PSD-95 and its close relative, chapsyn-110, have been shown to cluster NMDA receptors and Shaker K+channels in heterologous cells (Kim et al., 1995, 1996), and the_Drosophila_ PSD-95 homolog Dlg is required for synaptic clustering of Shaker channels in vivo (Tejedor et al., 1997).
Unlike NR2 subunits, the major splice forms of NR1 do not interact with PSD-95. Instead, the C-terminal tail of the most abundant NR1 splice variant (NR1A) interacts with calmodulin (CaM) at two sites: a high-affinity site within the alternatively spliced C1 exon cassette and a lower-affinity site in the C0 membrane-proximal region common to all splice variants of NR1 (Ehlers et al., 1996b). CaM binding to NR1 can inhibit NMDA receptor channel function (Ehlers et al., 1996b). In addition, yeast two-hybrid screens have revealed an interaction between the C0 region of NR1 and α-actinin-2, a protein known to cross-link actin filaments (Wyszynski et al., 1997). This interaction suggests one mechanism by which NMDA receptors can be immobilized via attachment to the postsynaptic actin cytoskeleton. Aside from its potential capacity as an anchoring molecule, α-actinin-2 may also modulate receptor function by virtue of its competitive binding with CaM to the tail of NR1 (Wyszynski et al., 1997).
NR1 subunits, in the absence of NR2, can cluster in heterologous cells via a mechanism dependent on the C1 exon cassette (Ehlers et al., 1995), suggesting a functional interaction between C1 and the cytoskeleton, the molecular basis of which has not been determined. Here we report the identification and characterization of a novel putative cytoskeletal protein that interacts with the C-terminal tail of NR1 in a C1-dependent manner. Because of its long, coiled coil nature, we have dubbed this protein yotiao, after a popular Chinese breakfast victual consisting of long strands of fried dough. Yotiao is specifically concentrated in the neuromuscular junction (NMJ) as well as in neuronal synapses, suggesting that it may have a general function in organizing postsynaptic specializations.
MATERIALS AND METHODS
Yeast two-hybrid screening and analysis of NR1–yotiao interaction. Yeast two-hybrid screening and assays were performed as described previously using the L40 yeast strain harboring HIS3 and β-galactosidase (β-gal) as reporter genes (Kim et al., 1995;Niethammer et al., 1996; Wyszynski et al., 1997). Approximately 2 × 106 clones were screened using a human brain cDNA library (Clontech, Palo Alto, CA) constructed in the Gal4 activation domain vector pGAD10. The C-terminal constructs of various NR1 splice variants, Kv1.4, and GluR1, were generated by PCR with specific primers and subcloned in-frame into pBHA to obtain LexA fusion proteins. Clone A1.7 (yotiao), clone A2.10 (α-actinin-2), and clone NAP1 (CaM) were subcloned into pGAD10 to generate Gal4 activation domain fusion proteins. Deletion constructs of yotiao were made by PCR using specific primers and were also used to generate Gal4 activation domain fusion proteins. Deletion constructs were tested for interaction in the yeast two-hybrid assay by using HIS3 and β-gal as reporter genes.
Yotiao cDNA cloning. The original human yotiao cDNA fragment, clone A1.7, was used as a hybridization probe to obtain from a 5′-Stretch Plus human brain cDNA λ phage library (Clontech) further clones (3-1, 9-1, 21-1, 25-1, and 25-2) that encompassed residues 138–1642 of the putative yotiao protein. Other methods were used to obtain sequences encoding residues upstream of amino acid 138. Clones 31-1, 32-3, and 33-2 were obtained from the SuperScript human brain cDNA phagemid library (Life Technologies, Grand Island, NY) by the GeneTrapper system (Life Technologies). Clone 41-2 was obtained by 5′-rapid amplification of cDNA ends (RACE) using Marathon-Ready human brain cDNA (Clontech). All clones were sequenced at an automated sequencing facility and analyzed using the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison WI). The full-length yotiao cDNA was cloned into pBluescript II (Stratagene, La Jolla, CA) by assembling four large overlapping fragments consisting of cDNA clones 33-2, 32-3, 25-1, and 21-1 (see Fig. 2). This full-length nucleotide sequence was deposited in the GenBank database (accession numberAF026245).
Fig. 2.
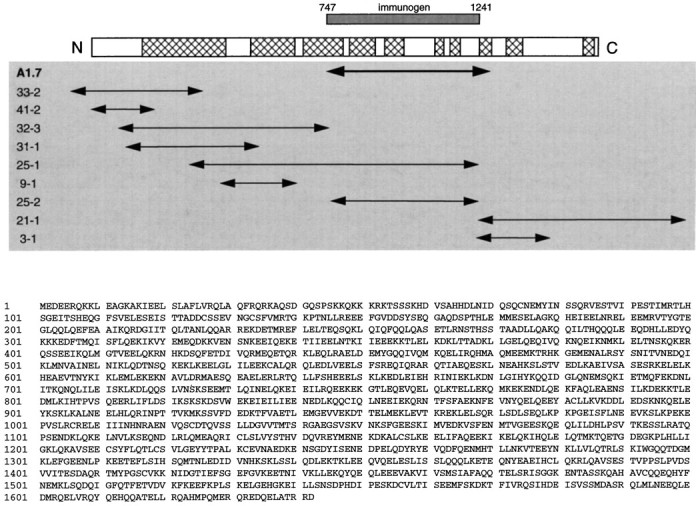
Primary structure of yotiao. A, Aligned below the schematic of yotiao are the overlapping cDNA clones used to determine its full-length sequence. A1.7 is the clone isolated by yeast two-hybrid screen using as bait the NR1A C-terminal tail.Hatched regions within the schematic represent areas of high coiled coil probability as predicted by the Lupas algorithm (Lupas et al., 1991). The region of yotiao prepared as H6-tagged immunogen is shown aligned with full-length yotiao. B, The deduced primary structure of yotiao is shown in single-letter amino acid code.
Northern blots. A human poly(A) mRNA multitissue northern blot (Clontech) was incubated with 32P-labeled probes prepared from clones 21-1 and 25-1, nonoverlapping fragments of the yotiao cDNA (see Fig. 2). Blots were hybridized in ExpressHyb hybridization solution (Clontech) at 68°C for 1 hr, washed under high-stringency conditions (wash 1, 2× SSC and 0.05% SDS at room temperature for 30 min; wash 2, 0.1× SSC and 0.1% SDS at 50°C for 40 min), and exposed 7 hr at −70°C on XAR-5 film (Eastman Kodak, Rochester, NY).
Antibodies. Anti-yotiao antisera (B5843 and B5844) were raised by immunizing rabbits with a hexahistidine (H6)-tagged fusion protein incorporating residues 747–1279 (contained within clone A1.7; see Fig. 2). Yotiao-specific antibodies were then affinity-purified on a Sulfolink column (Pierce, Rockford, IL) coupled to the original H6-tagged immunogen. The rabbit anti-α-actinin-2 antibody was a gift from Alan Beggs (Genetics Division, Children’s Hospital and Harvard Medical School, Boston, MA) and has been described (Wyszynski et al., 1997). The anti-Kv1.4 antibody has been described (Sheng et al., 1992). The widely used anti-NR1 monoclonal antibody 54.1 was obtained from PharMingen (San Diego, CA) (Brose et al., 1993).
Expression constructs and transfections. A yotiao expression construct was created by subcloning the full-length yotiao cDNA into the _Kpn_I site (5′) and the _Bgl_II site (3′) of the mammalian expression vector GW1-CMV (British Biotechnology, Oxford, UK). COS-7 cells were transfected at ∼70% confluency in multiwell plates using the LipofectAMINE method (Life Technologies). Two days after transfection, cells were harvested for preparation of cell extracts.
Immunoblotting, immunoprecipitation, and immunohistochemistry. For immunoblotting, transfected COS-7 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% NP-40, 0.5% deoxycholate, and 0.1% SDS) containing protease inhibitors. After incubation at 4°C for 1 hr, insoluble fractions of cell lysates were pelleted in a microcentrifuge at 14,000 rpm for 15 min. Crude synaptosomal membrane fractions from rat brain were prepared as described previously (Sheng et al., 1992). Immunoblotting with chemiluminescence reagents was performed as described previously (Sheng et al., 1993). All antibodies were used at ∼1 μg/ml in immunoblotting.
For immunoprecipitation, specific antibodies (5–10 μg/ml) were added to RIPA cell lysates (∼150 μg protein) and incubated at 4°C for 2 hr followed by protein A-Sepharose (Pharmacia, Piscataway, NJ) for 2 hr. Immunoprecipitates were washed, eluted, separated by SDS-PAGE, and analyzed by immunoblotting.
Brain immunohistochemistry was performed on Vibratome-cut 50 μm floating brain sections from Sprague Dawley rats (∼6 weeks of age) perfused transcardiacally with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, as described previously (Sheng et al., 1994). For protease treatment, brain sections were subjected to light digestion by 15 μg/ml proteinase K (Boehringer Mannheim, Indianapolis, IN) for 10 min at 37°C. Staining was visualized using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) and diaminobenzidine (DAB) or with Cy3 or FITC-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove PA). In double-labeling experiments, the TSA-Direct kit (DuPont NEN, Boston, MA) was used to enhance NR1 immunostaining signal.
Muscle immunohistochemistry was performed on 15 μm adult rat sternomastoid muscle transverse sections lightly fixed with 0.5% paraformaldehyde. Yotiao was labeled by B5843 at 2 μg/ml, followed by a Texas Red-conjugated secondary antibody (Jackson ImmunoResearch). Acetylcholine receptors (AChRs) were labeled with biotinylated α-bungarotoxin followed by BODIPY-conjugated streptavidin (both from Molecular Probes, Eugene, OR). Desmin was labeled by a 1:100 dilution of a mouse monoclonal antibody (mAb 1403; Porter and Froehner, 1983) followed by Texas Red-conjugated secondary antibody (Jackson ImmunoResearch).
Subcellular and PSD fractionation. Subcellular fractions of whole rat brain were prepared as described (Huttner et al., 1983). Briefly, rat brain Dounce homogenates (H) were centrifuged at 1000 × g to remove nuclei and other large debris (pelleted in P1). The supernatant (S1) was centrifuged at 10,000 ×g to obtain a crude synaptosomal fraction (P2), which was subsequently lysed hypo-osmotically and centrifuged at 25,000 ×g to pellet a synaptosomal membrane fraction (LP1). The supernatant (LS1) was then centrifuged at 165,000 × _g_to obtain a crude synaptic vesicle-enriched fraction (LP2). Concurrently, the supernatant (S2) above the crude synaptosomal fraction pellet (P2) was centrifuged at 165,000 × g to obtain a cytosolic fraction (S3) and a light membrane fraction (P3). The purified PSD fraction was prepared from rat brains as described previously (Carlin et al., 1980). This PSD fraction was then subjected to detergent extraction as described (Cho et al., 1992). Samples were analyzed by SDS-PAGE and immunoblotting as described (Kim et al., 1996).
RESULTS
Yeast two-hybrid screen for NR1 interacting proteins
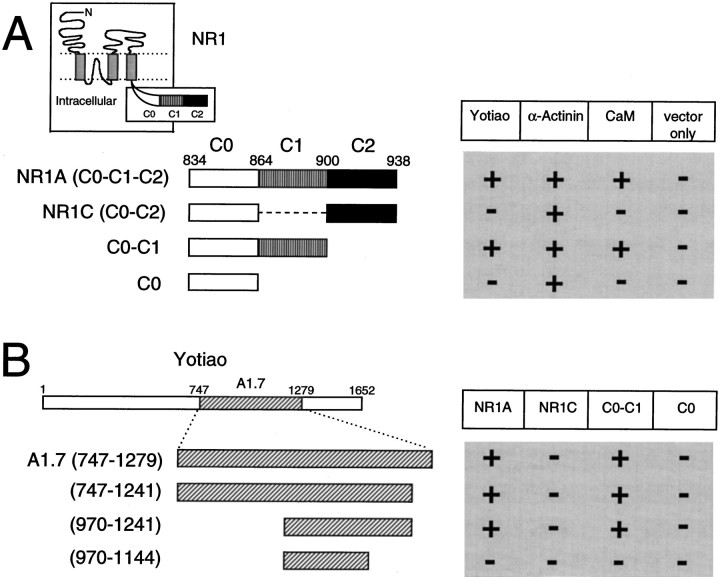
The final 105 amino acids (834–938) constituting the C-terminal tail of the NR1 splice variant NR1A were used as bait to screen ∼2 × 106 clones of a human brain cDNA library (also see Wyszynski et al., 1997). The bait extends from the last transmembrane domain of NR1 to the c-terminus of the protein and contains the C0 membrane proximal region as well as both the C1 and C2 alternatively spliced exon cassettes (Fig.1, inset). The yeast two-hybrid screen yielded multiple isolates of two distinct clones (A1.7 and A2.10) that interacted specifically with NR1A but not with Shaker-type K+ channel subunits or with the AMPA receptor subunit GluR1. Clone A2.10 interacted with both the C0-C1-C2 (NR1A) and C0-C2 (NR1C) splice variants of the NR1 C-terminal tail and was found to encode the C-terminal two-thirds of human α-actinin-2 (described by Wyszynski et al., 1997). In contrast, clone A1.7 interacted with the C0-C1-C2 but not with the C0-C2 variant of the NR1 C-terminal tail, implying that it binds NR1 in a C1 exon-dependent manner. Clone A1.7 contained a 1.6 kb cDNA insert that encoded a fragment of a large novel protein, which we have named yotiao.
Fig. 1.
Yeast two-hybrid analysis of NR1–yotiao interaction. A, C1 exon cassette is necessary for an interaction between yotiao and the NR1 C-terminal tail. NR1 C-terminal tail deletion constructs were subcloned into the bait vector pBHA and assayed by yeast two-hybrid for binding to GAL4 activation domain constructs of yotiao, α-actinin-2, or CaM. Vector only, pGAD10 with no insert. Interaction was determined by activation of the two reporter genes, HIS3 and β-gal.Inset, Membrane topology of NR1. The intracellular tail consists of the membrane proximal segment C0, and alternatively spliced exons C1 and C2. B, A 272 amino acid region of yotiao is sufficient for interaction with the NR1 C-terminal tail in a C1-dependent manner. Clone A1.7 and further deletion constructs of yotiao were tested (as GAL4-activation-domain fusions) by yeast two-hybrid for binding to NR1 C-terminal deletion constructs as in_A_. Numbers refer to the amino acid residues at the boundaries of each construct. The location of the polypeptide fragment encoded by clone A1.7 within the full-length yotiao protein is shown.
Primary structure of yotiao
By using conventional hybridization screening as well as the GeneTrapper system (Life Technologies) and 5′ RACE, we obtained the presumptive full-length coding sequence of yotiao from several human brain cDNA libraries. The open reading frame is 5.1 kb in length and encodes a 1642 amino acid protein (Fig.2). We cannot unambiguously assign the starting methionine of yotiao, because we have not identified an in-frame stop codon in the 5′ untranslated region (UTR). Nevertheless, the veracity of the assigned N terminus of yotiao is supported by the highly GC-rich nature of the 5′ UTR and a six of seven nucleotide match in the Kozak sequence around the putative start codon. Furthermore, this coding sequence can be expressed in heterologous cells to yield a protein of a size roughly consistent with that of endogenous yotiao in rat brain (see Regional and subcellular distribution of yotiao protein). The C terminus of the protein, delineated by an in-frame stop codon, followed by 3′ untranslated region and poly(A) tail, was found in clone 21-1. The accuracy of the yotiao sequence was confirmed in many areas by sequencing numerous overlapping clones (Fig. 2) and by checking the assembled sequence against independent expressed sequence tags in the databases.
Yotiao is a novel protein, and BLAST analysis of the deduced amino acid sequence did not yield significant homologies, except weak similarity to coiled-coil regions of proteins such as myosin heavy chain and intermediate filaments. Yotiao is leucine-rich throughout, and analysis by the Lupas algorithm predicts many long stretches of coiled coil heptad repeats scattered throughout the length of the protein (Fig. 2) (Lupas et al., 1991).
Yotiao interacts with NR1 in a C1 exon cassette-dependent manner
Deletion variants of the NR1 C-terminal tail were constructed in an effort to define the region of NR1 that interacts with yotiao using yeast two-hybrid assays (Fig. 1A). The C1 exon cassette is necessary for binding between NR1 and yotiao. The NR1A C-terminal tail (containing C0-C1-C2) and the C0-C1 constructs both interacted with yotiao, whereas the NR1C C-terminal tail (containing C0-C2) and the C0 constructs did not interact (Fig.1A). In parallel, deletion constructs of clone A1.7 were made to delimit its region of interaction with NR1. A region of ∼270 amino acids in yotiao (970–1241) was sufficient for binding to NR1 in a C1 exon-dependent manner (Fig. 1B). A further deletion of ∼100 amino acids from the C-terminal side abolished the interaction, but whether this deleted fragment (1145–1241) is sufficient as well as necessary for NR1 binding remains to be determined.
Coimmunoprecipitation of yotiao and NR1
We generated rabbit polyclonal antibodies (B5843 and B5844) against yotiao by using as immunogen an H6-tagged fusion protein incorporating residues 747–1241 of yotiao (see Fig. 2). The anti-yotiao antibodies were affinity-purified on a column on which the original H6-tagged immunogen was immobilized.
To confirm the interaction of the full-length yotiao and NR1 proteins within a cellular context, coimmunoprecipitation experiments were performed using COS-7 cells transfected with both yotiao and NR1A expression constructs. A mouse monoclonal antibody against NR1 coimmunoprecipitated the yotiao protein in addition to NR1 (Fig.3). The relative specificity of this coimmunoprecipitation was demonstrated by the inability of yotiao to be coimmunoprecipitated by equal amounts of a control antibody directed against the Shaker potassium channel Kv1.4. Conversely, the yotiao antibodies were able to coimmunoprecipitate NR1, although the efficiency of coimmunoprecipitation was lower than that of the reverse configuration. This difference in efficiency might be attributed to the fact that the yotiao antibodies were raised against the very region of the protein that interacts with NR1. Much of the yotiao protein may therefore have been shielded from the antibody by virtue of its interaction with NR1. We were unable to obtain coimmunoprecipitation of yotiao with NR1 from rat brain membrane extracts, as might be predicted from the differential solubility of these two proteins (see Regional and subcellular distribution of yotiao protein). The harsh detergent conditions needed to extract NR1 from rat brain membrane preparations likely disrupted the interaction between NR1 and the more easily solubilized yotiao protein.
Fig. 3.
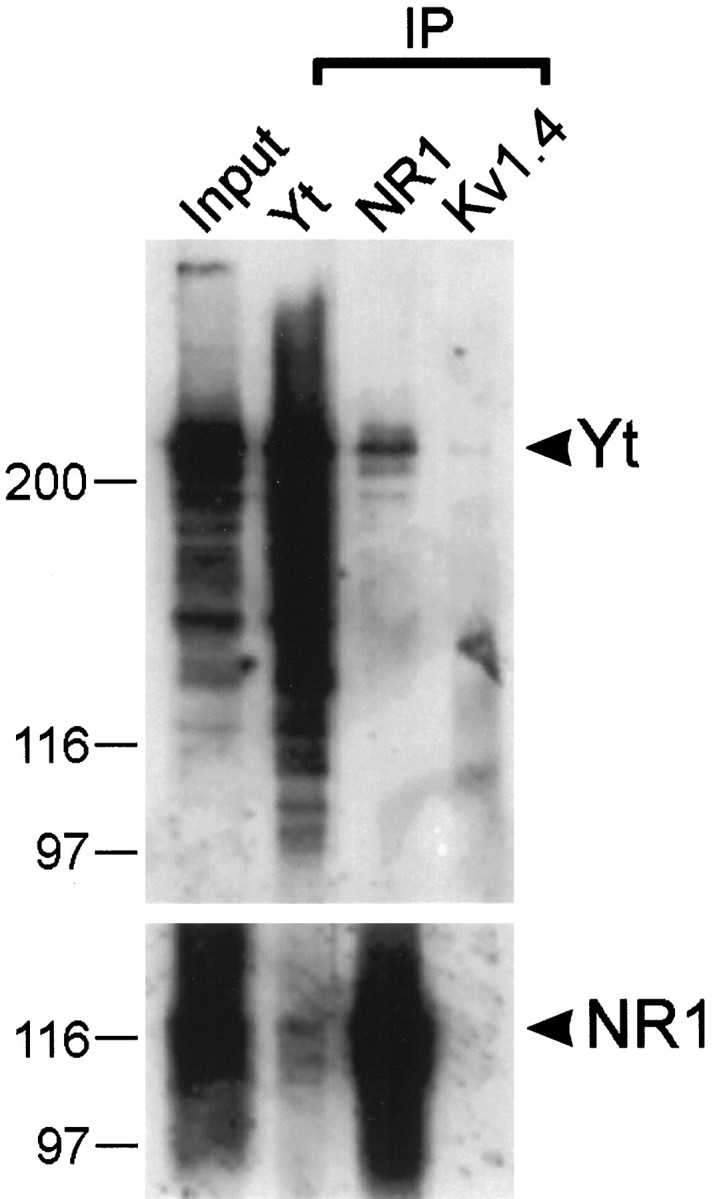
Coimmunoprecipitation of yotiao and NR1. Lysates of COS-7 cells doubly transfected with cDNA expression constructs encoding yotiao and NR1A were immunoprecipitated with antibodies against yotiao, NR1, or Kv1.4. Immunoprecipitates were then immunoblotted for yotiao and NR1 as indicated. Input lane contains 10% of the cell lysate used for the immunoprecipitation reaction. Positions of molecular weight markers are indicated in kilodaltons. Yt, Yotiao.
Tissue distribution of yotiao mRNA
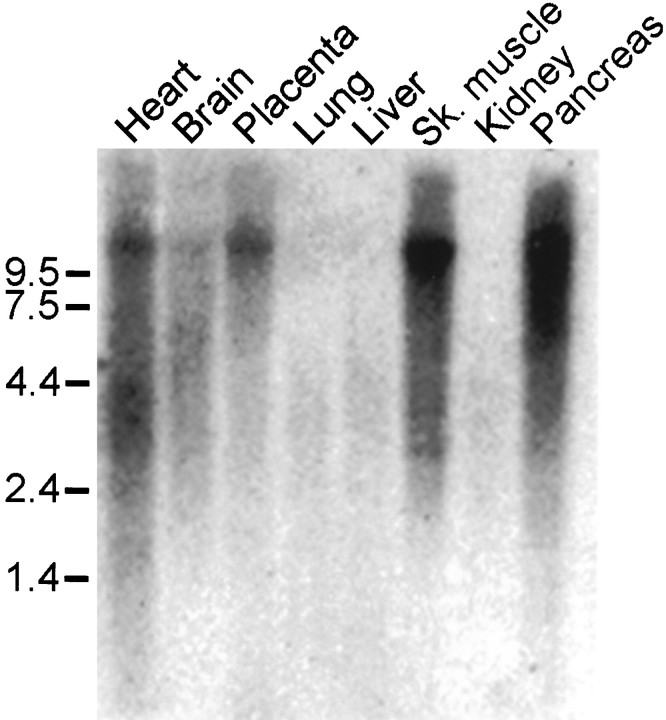
A human multitissue Northern blot hybridized under high stringency conditions revealed an 11 kb yotiao transcript that was present abundantly in skeletal muscle and pancreas, to a lesser degree in heart and placenta, and modestly in brain (Fig.4). Yotiao mRNA was undetectable in lung, liver, and kidney. We were unable to perform _in situ_hybridization experiments to determine the cellular pattern of yotiao mRNA expression in rat brain, because we do not have the rat cDNA homolog of yotiao.
Fig. 4.
Tissue distribution of yotiao mRNA. Poly(A) mRNA multitissue Northern blot was probed with 32P-labeled yotiao cDNA. Positions of RNA molecular size markers are shown in kilobases. Sk., Skeletal.
Regional and subcellular distribution of yotiao protein
Yotiao antibodies were used to study the distribution of yotiao protein in the rat brain. Western blot analysis using rat brain membrane fractions gave similar results with both B5843 and B5844 affinity-purified antibodies. Both antibodies revealed a major band of ∼230 kDa; this compares with the predicted molecular weight of ∼190 kDa based on the amino acid sequence of yotiao (Fig. 5
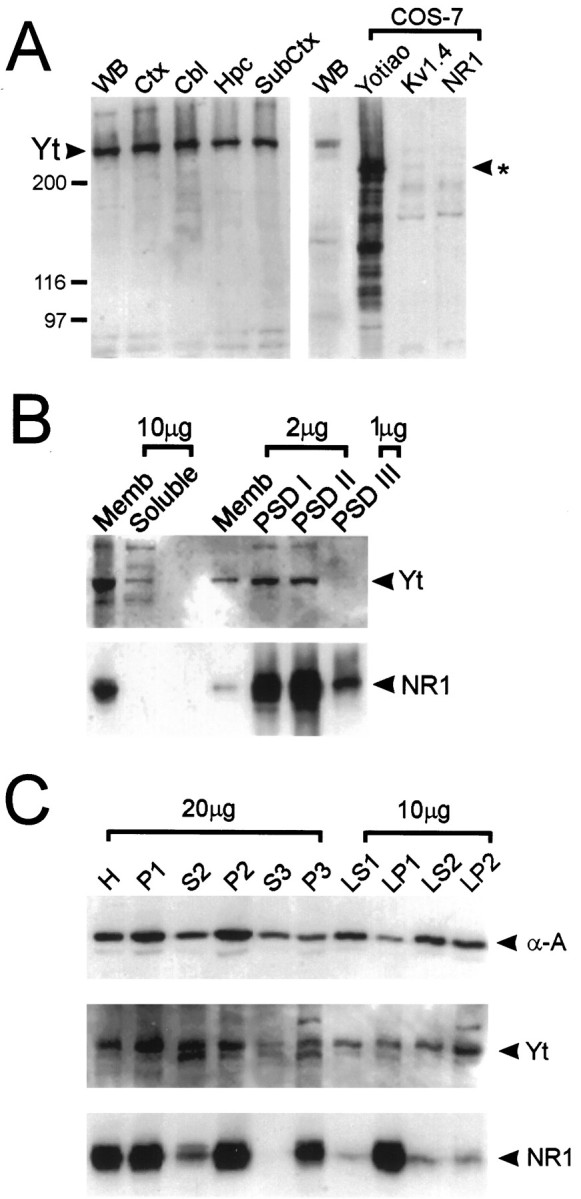
Figure 5. Specificity of yotiao antibody: regional distribution and biochemical fractionation of yotiao in rat brain.A, Synaptosomal membrane preparations from various regions of rat brain were immunoblotted for yotiao along with lysates from COS-7 cells transfected with yotiao, Kv1.4, or NR1. Positions of molecular weight markers are indicated in kilodaltons. The ∼230 kDa band corresponding to endogenous yotiao is indicated. The_asterisk_ indicates the position of the major yotiao band heterologously expressed in COS-7 cells. Yt, Yotiao;WB, whole brain; Ctx, cortex;Cbl, cerebellum; Hpc, hippocampus;SubCtx, subcortical structures including brainstem.B, Membrane association and enrichment of yotiao and NR1 in the PSD, shown by immunoblotting of PSD fractions with yotiao and NR1 antibodies. PSD fractions were extracted with Triton X-100 once (PSD I) or twice (PSD II) or with Triton X-100 followed by sarkosyl (PSD III). Memb, Rat brain crude synaptosomal membrane fraction. Amount of protein (micrograms) in each lane is indicated. C, Yotiao demonstrates a biochemical fractionation profile similar to that of α-actinin-2 and partly overlapping with NR1. The subcellular fractions of rat brain, as described in Materials and Methods, were immunoblotted for α-actinin-2, yotiao, and NR1.
A). To characterize the specificity of yotiao antibodies further, COS-7 cells were transfected with an expression construct containing the full-length yotiao cDNA. Western blotting with both B5843 and B5844 revealed a major band at ∼200 kDa specifically in yotiao-transfected cells but not in control Kv1.4 or NR1-transfected cells (Fig. 5_A_) (data not shown). In addition, multiple specific bands, probably representing yotiao degradation products, were also seen. The discrepancy in size between the major polypeptide expressed in heterologous cells and the endogenous brain protein might be accounted for by post-translational modifications such as phosphorylation or fatty acylation that occur more extensively in vivo than in vitro. We cannot, however, exclude the possibility that our yotiao coding region is incomplete at the 5′ end (see above). Immunoblot analysis of yotiao-transfected COS-7 cells also revealed a prominent band at ∼140 kDa, which may represent a degradation product of the full-length protein (Fig. 5_A_).
Western analysis of soluble and membrane fractions from rat brain indicate that yotiao is predominantly associated with the membrane (Fig. 5_B_). Yotiao is widely expressed in brain; the protein is present at similar levels in crude synaptosomal membrane fractions prepared from cortex, hippocampus, cerebellum, and subcortical structures (Fig. 5_A_). Biochemical studies of rat cerebral cortex revealed a fractionation profile of yotiao that resembles that of a cytoskeleton-associated protein such as α-actinin-2 (Fig. 5_C_). Yotiao is significantly depleted in the cytosolic fraction (S3) and enriched in P2 (crude synaptic fraction), P3 (light membrane pellet), and LP2 (crude synaptic vesicle-enriched fraction). By comparison, however, the integral membrane protein NR1 demonstrates a profile that is more specifically associated with membrane-containing fractions than yotiao. NR1 is highly enriched in P2, P3, and LP1 (lysed synaptosomal membranes) and virtually absent from S3.
The NMDA receptor subunits including NR1 are components of the postsynaptic density (for review, see Kennedy, 1993) and associate tightly with the PSD, resisting extraction by Triton X-100 and sarkosyl detergents (Fig. 5_B_). Yotiao is also enriched in PSD fractions, in which it is resistant to Triton X-100 extraction (PSD I and II) (Fig. 5_B_). However, yotiao apparently differs from NR1 in being extractable by sarkosyl (not enriched in PSD III). Moreover, it is noteworthy that the degree of yotiao enrichment in PSD fractions is not as great as for NR1 (Fig. 5_B_). These differences suggest that yotiao is not as tightly associated with the PSD as NR1 and may not be as exclusively associated with the PSD as NR1. In summary, yotiao is widely expressed in rat brain, and its pattern of subcellular and PSD fractionation is consistent with an interaction between yotiao and NR1 in synapses in vivo. However, this synaptic association may be relatively weak, because yotiao can be stripped from the PSD by sarkosyl.
Immunolocalization of yotiao in rat brain
In immunohistochemical experiments, similar patterns of immunostaining were obtained with both B5843 and B5844 affinity-purified antibodies, supporting the idea that the immunostaining pattern observed corresponds to the true distribution of the yotiao protein (Fig. 6 compare_A_,G and_H_,K). Furthermore, no staining with either antibody was detected when they were preincubated with the H6-tagged immunogen (Fig.6D–F) (data not shown). Yotiao is expressed in a somatodendritic pattern in neurons throughout the rat brain with prominent staining in the cerebral cortex, hippocampus, and cerebellum (Fig. 6).
Fig. 6.
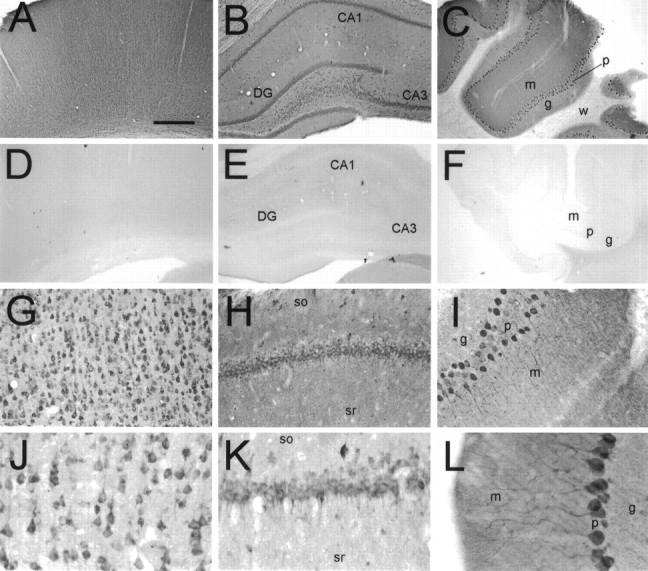
Immunohistochemical localization of yotiao in rat brain. Coronal sections were immunostained with yotiao antibodies B5843 (A, C–F,H, I, L) or B5844 (B, G, J,K) and visualized by DAB. These antibodies give similar staining patterns. A–C, Yotiao immunoreactivity in cerebral cortex (A), hippocampal formation (B), and cerebellum (C), with corresponding controls in D–F (antibody preincubated with immunogen). Note widespread somatodendritic labeling of neurons and diffuse staining of neuropil in neocortex, dentate gyrus, CA1 and CA3 of hippocampal formation, and cerebellum.G, J, Immunostaining of cell soma and apical dendrites of pyramidal neurons in the cerebral cortex. Staining of basal dendrites is also observed. H,K, Labeling of CA1 pyramidal cell layer in a somatodendritic manner. Note scattered interneuron immunoreactivity and neuropil staining in stratum oriens and stratum radiatum.I, L, Staining of Purkinje neurons extending from cell soma throughout major dendritic arborizations.DG, Dentate gyrus; m, molecular layer;g, granular layer; p, Purkinje cell layer; w, white matter; so, stratum oriens; sr, stratum radiatum. Scale bars:A–F, 0.5 mm; G–I, 125 μm;J–L, 62.5 μm.
In the cerebral cortex, yotiao immunoreactivity was found in pyramidal neurons as well as in the surrounding neuropil throughout layers II–VI (Fig. 6A). Pyramidal cell bodies and their proximal apical dendrites were prominently stained. After light pretreatment of brain sections with protease, increased yotiao staining was observed in distal dendrites and in the surrounding neuropil in a punctate pattern suggestive of synaptic localization (Fig.7A). We attempted to colocalize these puncta with synaptophysin in double-labeling experiments, but we were unsuccessful because synaptophysin staining was incompatible with protease pretreatment. The punctate somatodendritic and neuropil staining pattern was specific for yotiao, as evidenced by abolition of staining by competition with excess antigen (Fig. 6) (data not shown).
Fig. 7.
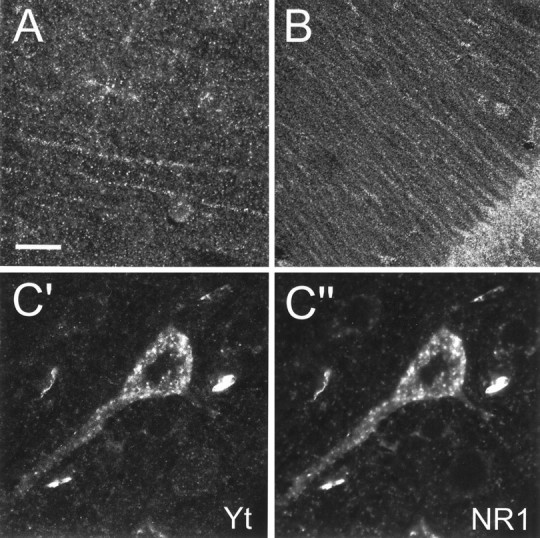
Localization of yotiao in rat cerebral cortex and hippocampus, and colocalization of yotiao and NR1 in pyramidal neurons, by confocal immunofluorescence microscopy.A, B, Specific labeling of distal dendrites of pyramidal cells and the surrounding neuropil in the rat cerebral cortex (A) and the CA1 region of the rat hippocampus (B) in a punctate pattern suggestive of synaptic localization. Brain sections were lightly digested with protease before immunohistochemical processing to enhance distal dendritic and neuropil staining. C′, C", Colocalization of yotiao (C′) and NR1 (C") in immunoreactive puncta in the cell soma and proximal apical dendrites of cortical pyramidal neurons. Brain sections were first stained for yotiao (B5843 visualized by a Cy3-conjugated secondary antibody) followed by immunolabeling for NR1 (54.1 visualized by tyramide amplification system, TSA-green). Scale bar:A, C′, C", 30 μm;B, 75 μm.
All regions of the hippocampus contained immunoreactive neurons (Fig.6B,H,K). The pyramidal cell bodies of CA1, CA2, and CA3 were prominently stained. In all three areas, labeled apical dendrites were observed to extend into the stratum radiatum (Fig. 7B). In CA3, the stratum lucidum was relatively intensely stained compared with the stratum radiatum of CA1 and CA2 (data not shown). Several scattered interneurons were labeled within the stratum oriens as well as the stratum radiatum (Fig.6H,K). In the dentate gyrus, the granule cell soma were positive for yotiao, with weaker immunoreactivity in the stratum moleculare. Scattered neurons in the hilus were stained in a somatodendritic pattern.
The cerebellum showed prominent staining of Purkinje cells in all folia. Yotiao immunoreactivity was observed in the cell bodies and throughout the dendritic arborizations (Fig.6C,I,L). The granule cell layer appeared more lightly stained than the molecular layer.
Immunocolocalization of yotiao and NR1 in rat brain
Colocalization of yotiao and NR1 in brain is a prerequisite for an in vivo association between the two proteins. Double immunofluorescence studies showed that yotiao and NR1 immunoreactivity indeed colocalized in somatodendritic puncta in pyramidal neurons of the cerebral cortex (Fig. 7C′, C′′). The somatodendritic pattern of NR1 staining is similar to that reported previously (Brose et al., 1993; Petralia et al., 1994; Johnson et al., 1996). The immunostaining patterns of yotiao and NR1 did not coincide completely, because a minority of cells demonstrated yotiao staining in the absence of NR1 staining and vice versa (data not shown). This suggests that not all NR1 protein in the brain is associated with yotiao. Because not all NR1 splice variants contain the C1 exon cassette, this result is consistent with the C1 dependence of the interaction between yotiao and NR1. By the same token, not all yotiao protein in the brain is associated with NR1, suggesting that yotiao likely serves functions in neurons other than merely binding NR1.
Immunolocalization of yotiao in rat neuromuscular junction
By Northern analysis, yotiao mRNA is expressed abundantly in skeletal muscle (Fig. 4). Consequently, immunohistochemical localization studies were also directed toward this tissue. Strikingly, yotiao antibodies showed highly concentrated labeling at the NMJ, subjacent to AChRs and extending to the perijunctional membrane (Fig.8A). Outside of the junctional area, yotiao immunoreactivity was also weakly detected in Z lines, the anchoring point of oppositely oriented actin filaments of adjacent sarcomeres (Fig. 8B). Labeling of these structures was eliminated if commercial rabbit IgG was substituted for the anti-yotiao antibodies or if the antibodies were preincubated with the yotiao fusion protein (data not shown). The staining pattern of yotiao is very similar to that of the intermediate filament desmin (described in Sealock et al., 1989), with the exception that unlike desmin, yotiao does not stain the general sarcolemma (Fig.8C). The strand-like pattern of yotiao staining subjacent to the AChR may represent fibrous organization of yotiao (see Fig.8B). However, it is also possible that unstained subjunctional nuclei may be occluding the area of yotiao staining in such a way as to create these strands. In summary, the staining pattern of yotiao in muscle suggests that it is a cytoskeleton-associated protein that may be involved in organization of the NMJ.
Fig. 8.
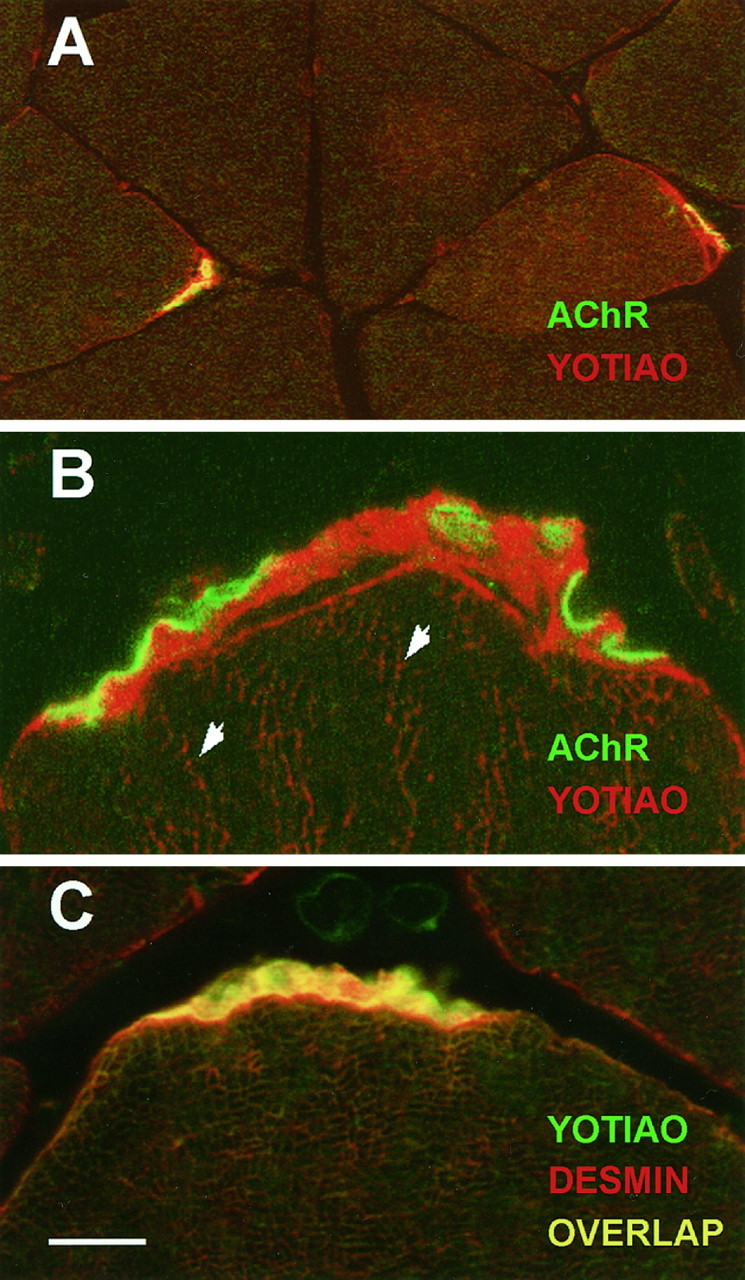
Immunohistochemical localization of yotiao in rat skeletal muscle. Yotiao is predominantly localized subjacent to AChRs in the NMJ and demonstrates a staining pattern similar to the intermediate filament desmin. Yotiao was labeled by B5843 followed by Texas Red-conjugated secondary antibody (A,B; red) or BODIPY-conjugated secondary antibody (C; green). AChR was labeled with biotinylated α-bungarotoxin followed by BODIPY-conjugated streptavidin (A, B;green). Desmin was labeled with mAb 1403 followed by Texas Red-conjugated secondary antibody (C;red). A, Large-field view showing yotiao expression at every NMJ. The sarcolemma is stained only in the perijunctional region but not beyond. B, High-magnification view of the NMJ reveals yotiao staining subjacent to AChR staining. Arrowheads indicate Z lines weakly stained for yotiao. C, Similar pattern of yotiao and desmin localization in the subsynaptic region. However, note absence of yotiao staining, but presence of desmin staining, on the general sarcolemma. As is common with transverse sections, distinct Z lines are not apparent in this view, but Z-line staining by mAb 1403 has been documented (Sealock et al., 1989). Scale bar: A, 25 μm; B, 5 μm; C, 10 μm.
DISCUSSION
The NMDA receptor participates in many important functions and dysfunctions of the nervous system, such as synapse formation, synaptic plasticity, and excitotoxicity. Elucidation of these processes requires an understanding of how NMDA receptors are targeted to particular synapses, how this localization is regulated during development and synaptic activity, and how NMDA receptor activity is transduced into the intracellular signals responsible for particular neuronal responses. In an effort to define the molecular machinery underlying NMDA receptor localization and function, we have identified yotiao, a novel large coiled coil protein that interacts with NR1 in a C1 exon cassette-dependent manner.
The large size and the predicted coiled coil nature of yotiao allows some speculations about this novel protein. First, yotiao can likely assemble itself into homomultimers or form heteromultimers with related proteins, because coiled coil domains are known to be sites of homomultimeric and heteromultimeric interactions (Adamson et al., 1993;Blake et al., 1995). Furthermore, many filamentous cytoskeleton-associated proteins such as myosin heavy chain, tropomyosin, and dystrophin contain extended coiled coil regions (Gimona et al., 1995; Winder et al., 1995; Hoppe and Waterston, 1996). By analogy, therefore, yotiao might also be a filamentous protein that is associated with the cytoskeleton. Supporting this idea, the subcellular fractionation pattern of yotiao is similar to that of α-actinin-2, an actin-binding protein. Furthermore, in skeletal muscle, the immunostaining pattern of yotiao is very similar to that of desmin, a type III intermediate filament; in addition, yotiao staining is associated with Z lines. In its synaptic and Z line localization, yotiao distribution in skeletal muscle is remarkably reminiscent of α-actinin (Bloch and Hall, 1983). Intriguingly, α-actinin also binds NR1 in the brain, although to a different site in the NR1 C-terminal tail than yotiao (Wyszynski et al., 1997). The specific association of yotiao with the NMJ as well as the PSD suggests a possible role for yotiao in the organization of multiple kinds of synapse. As a putative cytoskeleton-associated protein, yotiao may be acting at diverse synapses to help anchor synaptic membrane proteins to the cytoskeleton.
Coincident with this putative structural role, yotiao may also modulate receptor function. NMDA receptors are functionally dependent on their association with the underlying actin cytoskeleton, and α-actinin-2 has been advanced as a candidate protein that links NMDA receptors to actin (Rosenmund and Westbrook, 1993; Krupp et al., 1996; Wyszynski et al., 1997). Yotiao offers an alternative means to connect NMDA receptors to the cytoskeleton. In addition, yotiao may serve as a molecular scaffold that couples NMDA receptors to downstream signaling molecules in a manner similar to PSD-95 (Brenman et al., 1996). Only a small part of yotiao is required for NR1 binding, leaving unoccupied many potential areas of interaction with other proteins. The likelihood of such interactions is bolstered by the fact that coiled coil domains have been identified on many signaling molecules such as G-proteins, protein kinases, and transcription factors (Simonds et al., 1993).
Presently, the evidence for in vivo association between yotiao and NR1 is based on cellular coexpression, subcellular cofractionation, and subcellular colocalization. The inability to coimmunoprecipitate these two proteins in vivo is not surprising, given their differential detergent solubility from synaptic membrane preparations. A robust demonstration of the interaction will come from genetic studies, in which loss of function of the yotiao gene may affect the structure and function of synapses that contain NMDA receptors. On the other hand, binding NMDA receptors is not likely to be the sole function of yotiao in the brain. Based on the incomplete overlap of yotiao and NR1 distribution in brain and the synaptic localization of yotiao at the NMJ (where NMDA receptors do not exist), yotiao can be expected to interact with other receptors or membrane proteins, consistent with a general role in synaptic organization. Additionally, as a putative cytoskeleton-associated protein, yotiao may be involved in protein–protein interactions outside of the synapse, perhaps as part of the transport machinery that targets receptor-containing vesicles to the plasma membrane. We have not definitively localized yotiao to central synapses because of incompatibility of immunolabeling conditions for α-yotiao antibodies and α-synaptophysin antibodies. Therefore, the localization of yotiao at neuronal synapses remains to be confirmed, ideally by electron microscopy.
The dependence of the yotiao–NR1 interaction on the C1 exon cassette is intriguing because of the importance assigned to this alternatively spliced exon cassette by previous studies. This 37 amino acid region has been implicated in aggregation of NR1 in heterologous cells and in modulation of NMDA receptor function in _Xenopus_oocytes (Durand et al., 1992; Ehlers et al., 1995). It has also been identified as a site of phosphorylation by PKC and as a high-affinity calmodulin binding site (Tingley et al., 1993; Ehlers et al., 1996b). As such, the C1 exon cassette has been proposed to serve as a modifiable targeting motif and a regulator of NMDA receptor function (Ehlers et al., 1996a). Our findings indicate that the C1 exon may also function to mediate an interaction with the cytoskeleton through the binding of yotiao.
In light of its C1 exon dependence, the interaction between yotiao and NR1 may be regulated by three distinct mechanisms: (1) alternative splicing of the C1 exon cassette, (2) phosphorylation of the C1 exon cassette, and (3) CaM binding to the C1 exon cassette. Alternative splicing of the NR1 subunit can act through splice-specific interactors such as yotiao to differentially localize or anchor distinct subsets of NMDA receptors. Differential subcellular localization by virtue of subunit-specific or splice variant-specific interactions could be exploited to create glutamatergic synapses with distinct pharmacological and physiological properties within the same neuron (Rubio and Wenthold, 1997). Activity-dependent phosphorylation or CaM binding to C1 could regulate the localization or cytoskeletal attachment of a subset of NR1 by enhancing or disrupting its interaction with proteins such as yotiao. Whether yotiao competes with CaM binding to NR1 and whether the yotiao–NR1 interaction is regulated by PKC phosphorylation of the C1 exon are interesting questions that remain to be answered. Such mechanisms may affect receptor localization and activity and may contribute to the structural and functional plasticity of synapses.
Footnotes
This work was supported by National Institutes of Health Grants NS35050 to M.S. and NS33145 to R.S. and S.C. Froehner, the Muscular Dystrophy Association (R.S.), and the Council for Tobacco Research (R.S.). M.S. is an assistant investigator of the Howard Hughes Medical Institute. We thank Dr. Stanley Froehner for mAb 1403 (anti-desmin).
Correspondence should be addressed to Dr. Morgan Sheng, Howard Hughes Medical Institute/Massachusetts General Hospital, Wellman 423, 50 Blossom Street, Boston, MA 02114.
REFERENCES
- 1.Adamson JG, Zhou NE, Hodges RS. Structure, function and application of the coiled-coil protein folding motif. Curr Opin Biotechnol. 1993;4:428–437. doi: 10.1016/0958-1669(93)90008-k. [DOI] [PubMed] [Google Scholar]
- 2.Blake DJ, Tinsley JM, Davies KE, Knight AE, Winder SJ, Kendrick-Jones J. Coiled-coil regions in the carboxy-terminal domains of dystrophin and related proteins: potentials for protein-protein interactions. Trends Biol Sci. 1995;20:133–135. doi: 10.1016/s0968-0004(00)88986-0. [DOI] [PubMed] [Google Scholar]
- 3.Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 4.Bloch R, Hall Z. Cytoskeletal components of the vertebrate neuromuscular junction: vinculin, alpha-actinin, and filamin. J Cell Biol. 1983;97:217–223. doi: 10.1083/jcb.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 6.Brose N, Gasic GP, Vetter DE, Sullivan JM, Heinemann SF. Protein chemical characterization and imunocytochemical localization of the NMDA receptor subunit NMDA R1. J Biol Chem. 1993;268:22663–22671. [PubMed] [Google Scholar]
- 7.Carlin R, Grab D, Cohen R, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho K-O, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 9.Choi DW. Calcium: still center stage in hypoxic-ischemic neuronal death. Trends Neurosci. 1995;18:58–60. [PubMed] [Google Scholar]
- 10.Constantine-Paton M, Cline HT, Debski E. Patterned activity, synaptic convergence, and the NMDA receptor in developing visual pathways. Annu Rev Neurosci. 1990;13:129–154. doi: 10.1146/annurev.ne.13.030190.001021. [DOI] [PubMed] [Google Scholar]
- 11.Durand GM, Gregor P, Zheng X, Bennett MV, Uhl GR, Zukin RS. Cloning of an apparent splice variant of the rat N-methyl-d-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc Natl Acad Sci USA. 1992;89:9359–9363. doi: 10.1073/pnas.89.19.9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehlers MD, Tingley WG, Huganir RL. Regulated subcellular distribution of the NR1 subunit of the NMDA receptor. Science. 1995;269:1734–1737. doi: 10.1126/science.7569904. [DOI] [PubMed] [Google Scholar]
- 13.Ehlers MD, Mammen AL, Lau L-F, Huganir RL. Synaptic targeting of glutamate receptors. Curr Opin Cell Biol. 1996a;8:484–489. doi: 10.1016/s0955-0674(96)80024-x. [DOI] [PubMed] [Google Scholar]
- 14.Ehlers MD, Zhang S, Bernhardt JP, Huganir RL. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 1996b;84:745–755. doi: 10.1016/s0092-8674(00)81052-1. [DOI] [PubMed] [Google Scholar]
- 15.Froehner SC. Regulation of ion channel distribution at synapses. Annu Rev Neurosci. 1993;16:347–368. doi: 10.1146/annurev.ne.16.030193.002023. [DOI] [PubMed] [Google Scholar]
- 16.Gimona M, Watakabe A, Helfman D. Specificity of dimer formation in tropomyosins: influence of alternatively spliced exons on homodimer and heterodimer assembly. Proc Natl Acad Sci USA. 1995;92:9776–9780. doi: 10.1073/pnas.92.21.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall Z, Sanes JR. Synaptic structure and development: the neuromuscular junction. Neuron. 1993;10:99–122. doi: 10.1016/s0092-8674(05)80031-5. [DOI] [PubMed] [Google Scholar]
- 18.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 19.Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 1993;10:943–954. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- 20.Hoppe P, Waterston R. Hydrophobicity variations along the surface of the coiled-coil rod may mediate striated muscle myosin assembly in Caenorhabditis elegans. J Cell Biol. 1996;135:371–382. doi: 10.1083/jcb.135.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huttner W, Schiebler W, Greengard P, DeCamilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;5:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N, Masu M, Nakanishi S. Molecular characterization of the family of the N-methyl-d-aspartate receptor subunits. J Biol Chem. 1992;268:2836–2843. [PubMed] [Google Scholar]
- 23.Johnson R, Jiang X, Burkhalter A. Regional and laminar differences in synaptic localization of NMDA receptor subunit NR1 splice variants in rat visual cortex and hippocampus. J Comp Neurol. 1996;368:335–355. doi: 10.1002/(SICI)1096-9861(19960506)368:3<335::AID-CNE2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy MB. The postsynaptic density. Curr Opin Neurobiol. 1993;3:732–737. doi: 10.1016/0959-4388(93)90145-o. [DOI] [PubMed] [Google Scholar]
- 25.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 26.Kim E, Cho K-O, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 27.Kornau H-C, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 28.Kornau H, Seeburg P, Kennedy M. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 29.Krupp JJ, Vissel B, Heinemann SF, Westbrook GL. Calcium-dependent inactivation of recombinant N-methyl-d-aspartate receptors is NR2 subunit specific. Mol Pharmacol. 1996;50:1680–1688. [PubMed] [Google Scholar]
- 30.Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T, Arakawa M, Mishina M. Molecular diversity of the NMDA receptor channel. Nature. 1993;358:36–41. doi: 10.1038/358036a0. [DOI] [PubMed] [Google Scholar]
- 31.Lisman JE, Harris KM. Quantal analysis and synaptic anatomy–integrating two views of hippocampal plasticity. Trends Neurosci. 1993;16:141–147. doi: 10.1016/0166-2236(93)90122-3. [DOI] [PubMed] [Google Scholar]
- 32.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 33.Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–73. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- 34.Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 35.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 36.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petralia RS, Yokotani N, Wenthold RJ. Light and electron microscope distribution of the NMDA receptor subunit NMDAR1 in the rat nervous system using a selective anti-peptide antibody. J Neurosci. 1994;14:667–696. doi: 10.1523/JNEUROSCI.14-02-00667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter S, Froehner SC. Characterization and localization of the Mr = 43,000 proteins associated with acetylcholine receptor-rich membranes. J Biol Chem. 1983;258:10034–10040. [PubMed] [Google Scholar]
- 39.Rosenmund C, Westbrook GL. Calcium-induced actin depolymerization reduces NMDA channel activity. Neuron. 1993;10:805–814. doi: 10.1016/0896-6273(93)90197-y. [DOI] [PubMed] [Google Scholar]
- 40.Rubio ME, Wenthold RJ. Glutamate receptors are selectively targeted to postsynaptic sites in neurons. Neuron. 1997;18:939–950. doi: 10.1016/s0896-6273(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 41.Sealock R, Murnane AA, Paulin D, Froehner SC. Immunochemical identification of desmin in Torpedo postsynaptic membranes and at the rat neuromuscular junction. Synapse. 1989;3:315–324. doi: 10.1002/syn.890030404. [DOI] [PubMed] [Google Scholar]
- 42.Sheng M. PDZs and receptor/channel clustering: rounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 43.Sheng M, Kim E. Ion channel associated proteins. Curr Opin Neurobiol. 1996;6:602–608. doi: 10.1016/s0959-4388(96)80091-2. [DOI] [PubMed] [Google Scholar]
- 44.Sheng M, Tsaur M-L, Jan YN, Jan LY. Subcellular segregation of two A-type K+ channel proteins in rat central neurons. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- 45.Sheng M, Liao JY, Jan YN, Jan LY. Presynaptic A-current based on heteromultimeric K+ channels detected in vivo. Nature. 1993;365:72–75. doi: 10.1038/365072a0. [DOI] [PubMed] [Google Scholar]
- 46.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 47.Simonds WF, Husseini K, Garritsen M, Garritsen A, Lupas AN. G proteins and dlg in synaptic clustering of shaker K+ channels in vivo. J Neurosci. 1993;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugihara H, Moriyoshi K, Ishii T, Masu M, Nakanishi S. Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem Biophys Res Commun. 1992;185:826–832. doi: 10.1016/0006-291x(92)91701-q. [DOI] [PubMed] [Google Scholar]
- 49.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tingley WG, Roche KW, Thompson AK, Huganir RL. Regulation of NMDA receptor phosphorylation by alternative splicing of the C-terminal domain. Nature. 1993;364:70–73. doi: 10.1038/364070a0. [DOI] [PubMed] [Google Scholar]
- 51.Winder S, Gibson T, Kendrick-Jones J. Dystrophin and utrophin: the missing links! FEBS Lett. 1995;369:27–33. doi: 10.1016/0014-5793(95)00398-s. [DOI] [PubMed] [Google Scholar]
- 52.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]