Biomolecular condensates and cancer (original) (raw)
. Author manuscript; available in PMC: 2022 Feb 8.
SUMMARY
Malignant transformation is characterized by dysregulation of diverse cellular processes that have been the subject of detailed genetic, biochemical, and structural studies, but only recently has evidence emerged that many of these processes occur in the context of biomolecular condensates. Condensates are membraneless bodies, often formed by liquid-liquid phase separation, that compartmentalize protein and RNA molecules with related functions. New insights from condensate studies portend a profound transformation in our understanding of cellular dysregulation in cancer. Here we summarize key features of biomolecular condensates, note where they have been implicated—or will likely be implicated—in oncogenesis, describe evidence that the pharmacodynamics of cancer therapeutics can be greatly influenced by condensates, and discuss some of the questions that must be addressed to further advance our understanding and treatment of cancer.
INTRODUCTION
Decades of investigation into cancer pathophysiology have revealed that diverse cellular processes become dysregulated in the malignant state. This includes maintenance of genome integrity, chromatin structure, transcription, RNA processing, proliferative signaling, and others (Bradner et al., 2017; Hanahan and Weinberg, 2000, 2011; Jeggo et al., 2016; Labbé and Brown, 2018; Morgan and Shilatifard, 2015; Negrini et al., 2010; Stehelin et al., 1976; Wang and Aifantis, 2020). These processes occur throughout the cellular space, involving protein, DNA, and RNA molecules interacting with spatial and temporal precision. These cellular processes have been the subject of much study, producing a deep mechanistic understanding of cellular regulation in both normal and transformed cells and providing therapeutic hypotheses that have yielded advances in medicine (Chmielecki and Meyerson, 2014; Sawyers, 2004; Vogelstein et al., 2013). Recent studies, however, have revealed that most cellular processes are compartmentalized in biomolecular condensates, which have physicochemical properties that contribute to regulatory mechanisms beyond those anticipated by conventional molecular biology (Banani et al., 2017; Choi et al., 2020; Forman-Kay et al., 2018; Hyman et al., 2014; Jiang et al., 2020; Shin and Brangwynne, 2017). This new understanding has compelled us and others to examine how condensate biology contributes to oncogenesis and consider new therapeutic hypotheses that might be exploited to benefit cancer patients.
Biomolecular condensates are non-membrane-bound organelles that compartmentalize and concentrate components involved in similar cellular processes. In contrast to classical membrane-bound organelles like the nucleus, mitochondria, and Golgi apparatus, these structures are not constrained by a lipid bilayer and are not constitutive stable features of the cell. Rather, they often form reversibly and dynamically by virtue of phase separation. Biological phase separation is governed, in part, by weak, multivalent and dynamic interactions among proteins and nucleic acid polymers (Alberti, 2017; Boeynaems et al., 2018; Riback et al., 2020). At a threshold of concentration and affinity, these biomolecules can coalesce into liquid-like droplets that compartmentalize and regulate biochemical reactions. Many of the cellular processes dysregulated in cancer have recently been shown to occur in biomolecular condensates. This has prompted investigators to begin asking how oncogenic alterations influence condensate biology and contribute to the malignant state. In addition, recent evidence that condensates influence the pharmacodynamic behavior of small-molecule drugs suggests novel therapeutic approaches for cancer (Klein et al., 2020).
In this review, we summarize how the study of condensates is advancing our understanding of cancer and leading to novel therapeutic hypotheses. We survey the broad array of cellular condensates that have been described thus far and describe their shared features. We then discuss the various ways in which condensates are altered in malignancy, with a focus on how condensate physicochemical properties can contribute to dysregulated cellular processes. Condensates influence antineoplastic drug pharmacodynamics, and we suggest how this can be exploited to develop a new generation of cancer therapies. Finally, we highlight key areas for future investigation and speculate on how condensates might contribute to furthering new discoveries in cancer biology.
BIOMOLECULAR CONDENSATES
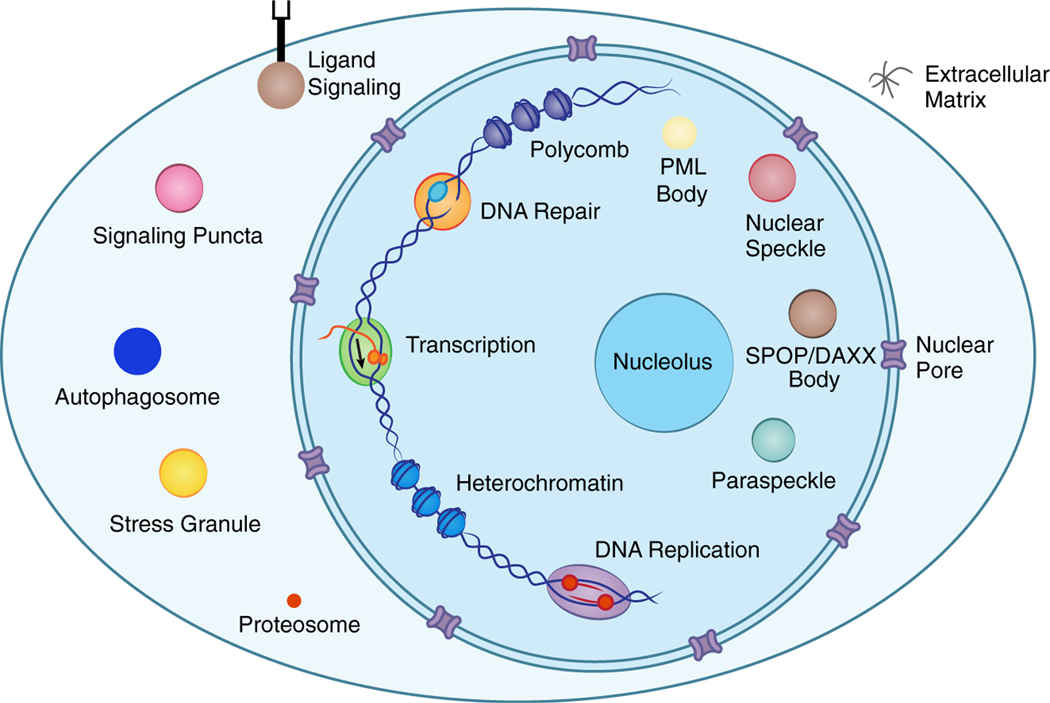
The cell is organized into diverse membrane-bound compartments such as the nucleus and mitochondria, as well as dozens of non-membrane condensates located throughout the nucleus and cytoplasm (Figure 1) (Banani et al., 2017). Whereas classical biochemical complexes have a defined stoichiometry, condensates are non-stoichiometric assemblies composed of biomolecules with weak multivalent interactions. Thus, they form a local concentration of molecules that continuously exchange with the surrounding bulk phase (Kato et al., 2012; Li et al., 2012; Sanders et al., 2020).
Figure 1. Biomolecular condensates located throughout the nucleus and cytoplasm.
Cartoon depicting the cell composed of various condensates that compartmentalize biomolecules involved in shared processes. PML, promyelocytic leukemia protein; SPOP, Speckle-type POZ protein.
The behavior of polymers in solution can be described by simple thermodynamic models that illuminate the behaviors of condensates and their constituent components. The Flory-Huggins theory describes the free energy of mixing polymers within solvent, and has been useful in explaining some of these behaviors (Brangwynne et al., 2015; Chen and Kriwacki, 2018; Flory, 1942). At thresholds of concentration and affinity, the net attraction between polymers drives phase separation into polymer-rich and polymer-poor phases. Alterations to the polymer structure or composition, the affinity between polymers, and the environment can change the point at which a condensate forms; thus, modest perturbations can result in fundamental shifts in a phase-separating system (Banani et al., 2016; Dignon et al., 2019; Pak et al., 2016; Riback et al., 2017, 2020; Yoo et al., 2019). Applying such principles to polymer-like biomolecules and their resulting condensates can help explain their functional behaviors as well as their dysfunction in diverse disease states.
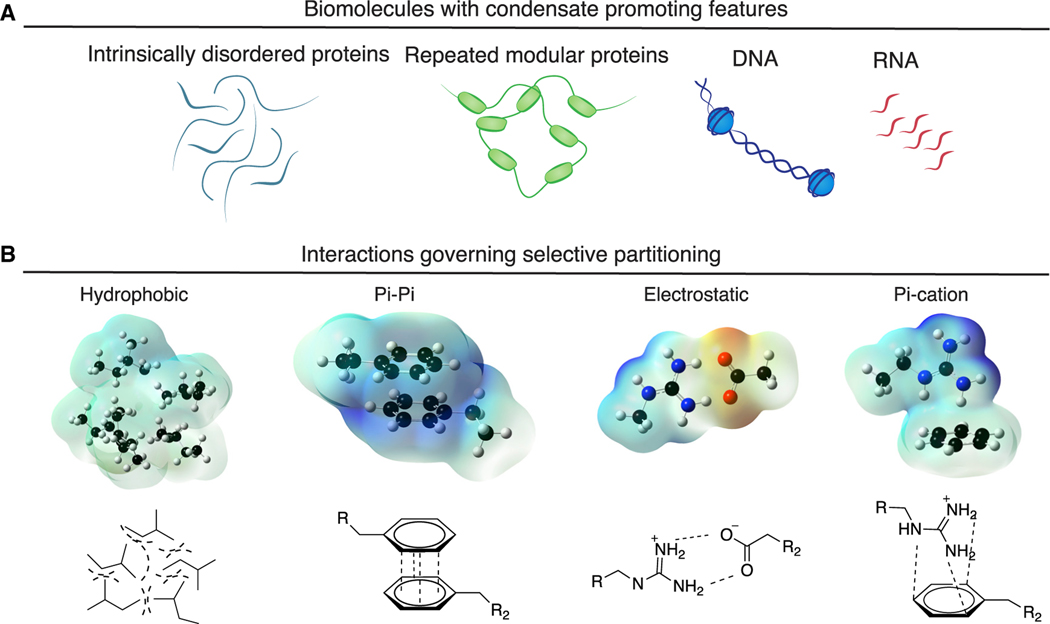
Proteins with intrinsically disordered protein regions (IDRs), repeats of short oligomerizing motifs, and nucleic acid chains are biopolymers that have been observed to promote condensate formation (Banani et al., 2016; Bienz, 2020; Gomes and Shorter, 2019; Harmon et al., 2017; Jain and Vale, 2017; Pak et al., 2016; Shrinivas et al., 2019) (Figure 2A). The formation of condensates, and selective partitioning of biomolecules into condensates, has been attributed to specific weak and dynamic interactions among the molecules, including salt bridges, pi-pi, pi-cation, and hydrophobic interactions (Brangwynne et al., 2015; Lin et al., 2017; Vernon et al., 2018; Wang et al., 2018) (Figure 2B). Condensates are formed when the concentration of and the interaction strength between biopolymers reach a threshold level where the interactions favoring assembly overcome opposing forces.
Figure 2. Condensate-promoting features of biomolecules.
(A) Condensate-promoting features of biomolecules include intrinsically disordered regions and repeated modular domains as well as DNA and RNA.
(B) Electrostatic surface potential plots of interactions governing partitioning of biomolecules into condensates, including hydrophobic, pi-pi, electrostatic, and pi-cation interactions.
Biomolecular condensation involves multivalent interactions between an indefinite number of components that undergo self-assembly via clustering, which is distinct from the formation of smaller stoichiometric protein complexes with defined numbers of subunits, such as a 12-subunit RNA polymerase II complex or a viral capsid (Banani et al., 2017). Biomolecular condensates are thought to arise by either liquid-liquid or liquid-solid phase transition (Boeynaems et al., 2018; Hyman et al., 2014; Peran and Mittag, 2020; Posey et al., 2018; Woodruff et al., 2018). Thus, condensates can have properties of liquids, gels, or solids, and liquid condensates can “age” to take on properties of gels or solids. These properties can produce condensates with various shapes, dimensions, and behaviors (Molliex et al., 2015; Patel et al., 2015). Classical references to these non-membrane assemblies have used diverse descriptive terms such as bodies, puncta, dots, granules, inclusions, aggregates, etc. It is likely that all of these assemblies are governed by physicochemical properties that are now under intense study in the fields of soft matter physics, chemistry, and biology.
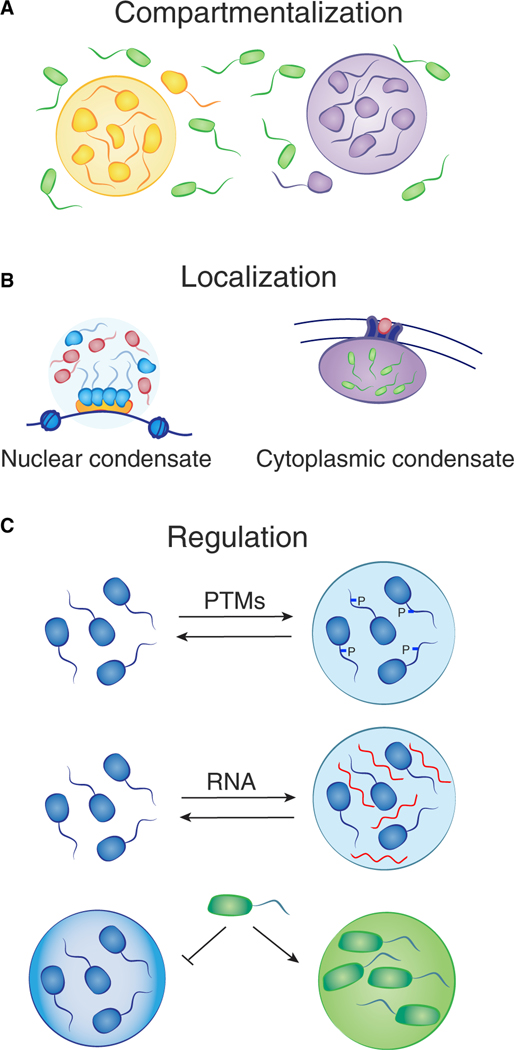
Condensates have properties distinct from the surrounding milieu that allow for novel functions. Condensates can compartmentalize large numbers of biomolecules with related functions, thus concentrating components and accelerating biochemical reactions (Alberti et al., 2019; Banjade and Rosen, 2014; Case et al., 2019a; Huang et al., 2019; Li et al., 2012; Sheu-Gruttadauria and MacRae, 2018; Su et al., 2016; Woodruff et al., 2017; Zhang et al., 2020c). Condensates can be localized to specific sites in cells anchored, for example, by components that interact with DNA sequences or membranes (Boija et al., 2018; Case et al., 2019a; Feng et al., 2018; Huang et al., 2019; Sabari, 2020; Sabari et al., 2020; Shrinivas et al., 2019; Zeng et al., 2018). Condensate regulation can occur through modification of concentration or affinity of components, as well as through their selective partitioning. The attributes of condensates that contribute to cellular function—compartmentalization, localization, and regulation—are discussed further below.
Compartmentalization
Condensates provide a means to organize the 5–10 billion protein molecules of the cell into distinct cellular compartments with specific functions. Cellular processes such as transcription and DNA damage repair typically involve dozens of different biomolecules that must engage with temporal and spatial precision. Compartmentalization allows for high local concentrations of biomolecules and their substrates and exclusion of other molecules that are not functionally relevant (Figure 3A) (Gibson et al., 2019; Li et al., 2020). In addition, condensates are non-stoichiometric assemblies of factors involved in shared processes, so, for example, a condensate at the promoter of a gene can assemble multiple RNA polymerase molecules, thereby producing a burst of transcription (Cho et al., 2018; Guo et al., 2019; Kwon et al., 2013; Sabari et al., 2018; Wei et al., 2020). Condensates can form and dissolve in short time frames, which provides the cell with a means to produce transient compartments, thereby releasing biomolecules for use elsewhere when they are no longer needed at a specific location (Guillén-Boixet et al., 2020). Compartmentalization in condensates also serves to stabilize protein concentrations in cells by buffering the inherent stochasticity in gene expression (Klosin et al., 2020). Furthermore, condensates can be organized in multiple phases, one surrounding another, to enable spatiotemporal regulation of a process. For example, nucleoli consist of distinct liquid phases where RNA polymerase I and FIB1 occur within a larger NPM1 condensate, allowing for the synthesis of rRNA molecules and spatiotemporal regulation of pre-ribosome assembly (Feric et al., 2016; Riback et al., 2020; Mitrea et al., 2018).
Figure 3. Compartmentalization, localization, and regulation are common features of condensates.
(A) Compartmentalization allows for high local concentrations of biomolecules and their substrates, as well as exclusion of other molecules.
(B) Localization of nuclear condensates can be mediated by proteins that bind to specific DNA or RNA sequences (left), and cytoplasmic condensates can form at sites on the plasma membrane (right).
(C) Regulation of condensates can occur at many levels, for example, post-translational modifications (PTMs) of molecules or the presence of RNA may change the properties that influence formation. The chemical environment of condensates dictates selective partitioning, e.g., BRD4 is preferentially concentrated in euchromatin condensates versus heterochromatin condensates.
Localization
Condensates often contain components that can anchor the body to a specific location in the cell. For example, nuclear condensates can form with proteins that bind to specific DNA or RNA sequences, and cytoplasmic condensates can form at sites on the plasma membrane (Figure 3B) (Boija et al., 2018; Case et al., 2019a; Guillén-Boixet et al., 2020; Huang et al., 2019; Shrinivas et al., 2019; Su et al., 2016; Wei et al., 2020). Transcriptional condensates form at specific enhancer and promoter elements by virtue of selective transcription factor (TF) binding (Boija et al., 2018; Shrinivas et al., 2019). TFs are bifunctional proteins that contain both a structured DNA binding domain and an IDR that can condense with coactivator proteins (Sabari et al., 2020). Enhancer and promoter elements contain multiple TF binding sites, thus crowding TFs and driving assembly of TFs and coactivators past the threshold for phase separation (Shrinivas et al., 2019). Constitutive heterochromatin condensates form at methylated satellite repeats, due in part to the binding of methylated DNA by MeCP2 and methylated histone H3K9 by HP1 proteins (Larson et al., 2017; Li et al., 2020; Strom et al., 2017). Facultative heterochromatin condensates form at sites of trimethylated H3K27 by virtue of Polycomb-repressive-complex phase separation (Plys et al., 2019; Tatavosian et al., 2019). Similarly, ligand binding by cell-surface signaling receptors can elevate signaling component concentration at the plasma membrane and thereby stimulate formation of condensates with signaling molecules (Case et al., 2019a; Huang et al., 2019; Su et al., 2016).
Regulation
Condensates can be regulated at many levels, as anything that changes the properties that influence formation, dissolution, viscoelasticity, and other physicochemical properties can modify condensate function (Figure 3C). We discuss specific features that play prominent roles in condensate regulation—concentration, chemical modification, non-coding RNAs (ncRNAs), and selective partitioning—here, because they are known, or likely, to be dysregulated in cancer cells.
The concentration of biomolecules is a key parameter in condensate formation and dissolution. Condensation occurs at a threshold concentration, which can be achieved through increased biosynthesis, reduced degradation, transport into a membrane-bound compartment, or binding to a substrate with multiple binding sites. The loss of the nuclear membrane during mitosis reduces the concentration of nuclear components and is associated with dissolution of nuclear condensates (Dammermann and Merdes, 2002; Rai et al., 2018; Sivan et al., 2007; Spector and Smith, 1986). The nuclear condensates re-form when the nuclear envelope is re-established, which may in part be due to the higher concentration of components enabled by transport of proteins into the nucleus.
Chemical modification, such as the post-translational modification (PTM) of histone proteins, alters the physicochemical properties of proteins and thus the condensates with which they associate. Chromatin can occur in phase-separated condensates, and the behavior of chromatin in unmodified and modified states provides an example of this type of regulation. Repressed genes are generally associated with unacetylated nucleosomes, whereas active genes are associated with acetylated nucleosomes (Bradner et al., 2017). Organized into separate subdomains within the nucleus, these two types of chromatin are highly compact yet dynamically accessible to regulation by diverse modifying enzymes and proteins that bind the modified nucleosomes (Larson et al., 2017; Li et al., 2020; Sabari et al., 2018; Strom et al., 2017). Reconstituted unmodified chromatin can undergo liquid-liquid phase separation due to the IDRs of histone tails, producing dense and dynamic droplets. Acetylation of this chromatin in the presence of BRD4, a protein that binds acetylated nucleosomes at active genes, produces a different phase-separated state, with droplets that exhibit distinct physical properties. The acetylated chromatin becomes less miscible with unmodified chromatin droplets, mimicking the separation of chromatin subdomains observed in cells (Gibson et al., 2019). Histone tails are subjected to diverse chemical modifications that alter the physicochemical properties of chromatin, and thus each modification has the potential to modulate chromatin condensate behavior.
RNA molecules play regulatory roles in diverse biomolecular condensates, including the nucleolus, transcriptional condensates, cotranscriptional splicing condensates, nuclear speckles, paraspeckles, and stress granules (Fay and Anderson, 2018; Guo et al., 2019; Henninger et al., 2021; Roden and Gladfelter, 2020; Sabari, 2020; Strom and Brangwynne, 2019). Condensates are formed by an ensemble of low-affinity molecular interactions, including electrostatic interactions, and RNA can be a powerful regulator of condensates that are formed and maintained by these forces (Banani et al., 2017; Elbaum-Garfinkle et al., 2015; Henninger et al., 2021; Maharana et al., 2018; Peran and Mittag, 2020; Shin and Brangwynne, 2017; Zhang et al., 2015a). The functions of the vast majority of ncRNA species expressed in cells are not known, and it seems likely that many will be found to play regulatory roles in diverse condensates.
Selective partitioning of biomolecules into specific condensates allows for high local concentrations of functionally related molecules. The behaviors of proteins involved in chromatin and transcriptional condensates provide instructive examples of selective biomolecular partitioning. Some euchromatic and heterochromatic proteins selectively partition into condensates formed by other components of euchromatin and heterochromatin, and this partitioning behavior may contribute to the separation of these two compartments in the nucleus (Fasciani et al., 2020; Li et al., 2020). Phosphorylation of RNA polymerase II during transcription initiation causes the polymerase to switch between transcriptional and splicing condensates, illustrating how IDR modification can change the partitioning behavior of a macromolecule (Guo et al., 2019; Kwon et al., 2013).
CONDENSATE DYSREGULATION IN CANCER
Cancer cells acquire mutations that affect diverse condensate-mediated cellular processes, including transcription, chromatin structure, proliferative signaling, and others. The study of condensates in cancer cells is in its infancy, but there are already notable examples of dysregulated condensates that have been reported, and the known effects of cancer mutations on the concentration and modification of regulatory biomolecules predict that condensate dysregulation is a common feature of cancer cells.
Dysregulated compartmentalization
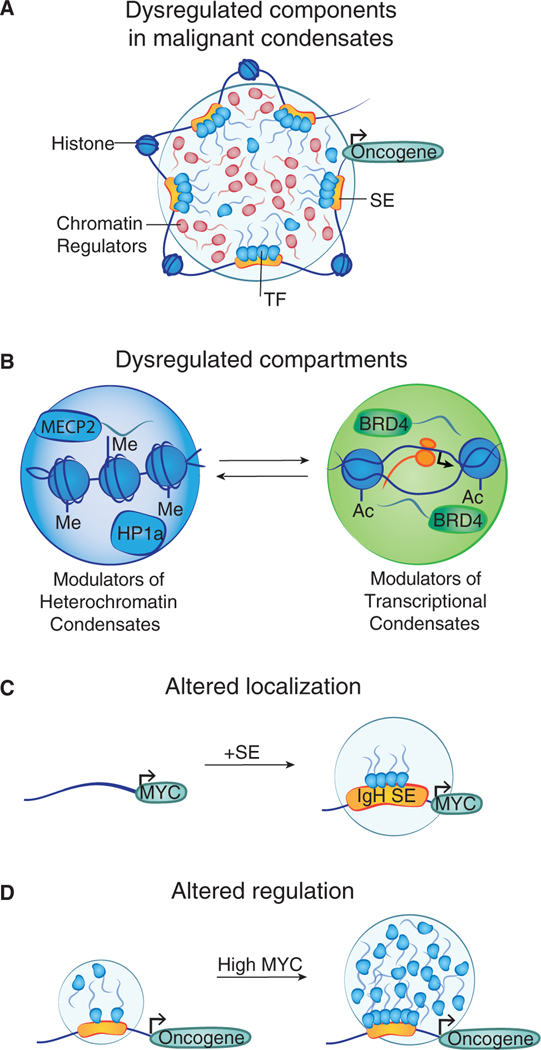
Cancer cells acquire genomic alterations that activate oncogenes and inactivate tumor suppressor genes (Kastenhuber and Lowe, 2017; Prior et al., 2012). Both oncogene activation and tumor suppressor inactivation involve condensate compartments that can become dysregulated in malignant cells (Figure 4A and Table 1).
Figure 4. Condensate compartments dysregulated in malignant cells.
(A) Cartoon depicting a transcriptional condensate with various components that have been reported to be altered in cancer.
(B) Recurrent mutations affecting histones, chromatin modifiers, and proteins that interact with modified biomolecules can alter chromatin condensates.
(C) Translocation of the IgH super-enhancer (SE) to the MYC locus is an oncogenic event in aggressive B cell lymphomas that is likely to result in the formation of a membrane less compartment at the MYC gene.
(D) Elevated levels of the oncogenic MYC protein in metastatic tumor cells may alter the behavior of transcriptional condensates in these cells.
Table 1.
Cellular processes dysregulated in cancer associated with condensates
| Dysregulated process | Protein | Biological role | Reference |
|---|---|---|---|
| Transcription | MED1 | coactivator overexpressed and modified in cancer | (Cho et al., 2018; Nagalingam et al., 2012; Russo et al., 2019; Sabari et al., 2018) |
| CDK7 | kinase overexpressed and targeted in cancer | (Klein et al., 2020; Kwiatkowski et al., 2014) | |
| P-TEFb | kinase implicated in ovarian cancer | (Guo et al., 2019; Kohoutek, 2009) | |
| YAP/TAZ | coregulator with increased activity in various cancers | (Cai et al., 2019a; Zanconato et al., 2018) | |
| EWS | fused to FLI in Ewing’s sarcoma | (Boulay et al., 2017; Chong et al., 2018) | |
| OCT4 | master TF regulator of cell identity | (Boija et al., 2018) | |
| HSF1 | TF overexpressed in cancer | (Carpenter and Gö kmen-Polar, 2018; Gaglia et al., 2020) | |
| eRNP | implicated in breast cancer | (Nair et al., 2019) | |
| FUS | translocated in sarcoma | (Crozat et al., 1993; Kato et al., 2012) | |
| MYC | overexpressed in cancer | (Boija et al., 2018; Dang, 2012) | |
| p53 | tumor suppressor misregulated in cancer | (Kamagata et al., 2020; Lemos et al., 2020; Safari et al., 2019) | |
| TAF15 | cofactor implicated in cancer | (Altmeyer et al., 2015; Kwon et al., 2013; Shin et al., 2018; Wei et al., 2020) | |
| ENL | cofactor translocated in leukemia | (Wan et al., 2017, 2020) | |
| Epigenetic regulation | BRD4 | chromatin factor upregulated and fused in cancer | (Filippakopoulos et al., 2010; Sabari et al., 2020; Shin et al., 2018) |
| HP1a | chromatin factor downregulated in cancer | (Larson et al., 2017; Strom et al., 2017; Vad-Nielsen and Nielsen, 2015) | |
| m6a | DNA modification decreased in cancer | (He et al., 2019; Ries et al., 2019) | |
| Polycomb | gene silencing complex altered in cancer | (Plys et al., 2019; Tatavosian et al., 2019) | |
| MeCP2 | chromatin factor amplified in breast cancer | (Li et al., 2020; Neupane et al., 2016) | |
| MALAT1 | lncRNA dysregulated in cancer | (Greig et al., 2020; Tripathi et al., 2010) | |
| Cell signaling | B-catenin | Wnt factor driving colon cancer | (Nusse and Clevers, 2017; Zamudio et al., 2019) |
| ER (estrogen receptor) | TF and hormone receptor mutated in breast cancer | (Boija et al., 2018; Nair et al., 2019) | |
| PML | fused to RARa in APML | (Banani et al., 2016; Salomoni and Pandolfi, 2002) | |
| PKA | involved in oncogenic signaling | (Sapio et al., 2014; Zhang et al., 2020b) | |
| TCR | mediator of tumor immunity | (June et al., 2018; Su et al., 2016) | |
| SOS | cofactor involved in RAS signaling | (Cai et al., 2019b; Huang et al., 2019) | |
| Immune signaling | cGAS | involved in cancer immunity | (Du and Chen, 2018) |
| Ribosome biosynthesis | NPM1 | nucleolar factor mutated in leukemia | (Feric et al., 2016; Mitrea et al., 2016, 2018) |
| Degradation | SPOP | tumor suppressor mutated in prostate cancer | (Bouchard et al., 2018) |
| DNA repair | RAD52 | HR factor and tumor suppressor | (Oshidari et al., 2020) |
| 53BP1 | DNA repair factor and tumor suppressor | (Kilic et al., 2019; Mirza-Aghazadeh-Attari et al., 2019; Pryde et al., 2005) | |
| FUS | effector of transcription-coupled repair | (Singatulina et al., 2019) | |
| PARP | enzyme targeted for cancer therapy | (Rouleau et al., 2010; Singatulina et al., 2019) | |
| Splicing | SRSF2 | mutated in myelodysplasia | (Guo et al., 2019; Sperling et al., 2017) |
| Nuclear transport | NUPs | nucleoporin dysregulated in cancer | (Schmidt and Gö rlich, 2015, 2016; Simon and Rout, 2014) |
| DNA replication | ORC/CDC6/CDT1 | dysregulated replication | (Gaillard et al., 2015; Parker et al., 2019) |
| Storage | NEAT1 | ncRNA of paraspeckles implicated in cancer | (Shin et al., 2019; Yamazaki et al., 2018) |
| Autophagy | Atg1 | implicated in macromolecule recycling in cancer | (Fujioka et al., 2020; Mulcahy Levy and Thorburn, 2020) |
| Proteasome | Multiple proteins | target of multiple myeloma therapies | (Anderson, 2016; Manasanch and Orlowski, 2017; Yasuda et al., 2020) |
| Telomeres | Multiple proteins | prevents replicative senescence | (Min et al., 2019; Zhang et al., 2020a) |
| Stress response | FUS, hnRPNA1, G3NP1/2 | stress granules are antiapoptotic | (Arimoto et al., 2008; Molliex et al., 2015; Protter and Parker, 2016) |
Oncogene activation is often accomplished by the formation of super-enhancers, which are clusters of enhancers occupied by exceptionally high densities of transcriptional components that drive high-level expression of genes (Bradner et al., 2017; Hnisz et al., 2013; Lovén et al., 2013). Super-enhancers promote the formation of phase-separated condensates that draw together the clustered DNA elements into a non-membrane bound compartment (Figure 4A) (Cho et al., 2018; Sabari et al., 2018). These condensates are nucleated by the binding of multiple TF molecules to genomic regulatory elements where TF activation domains, many of which are IDRs, condense with transcriptional coactivators (Boija et al., 2018; Chong et al., 2018; Nair et al., 2019; Shrinivas et al., 2019). These compartments can then recruit hundreds of molecules of RNA polymerase II to effect transcription of target genes (Cho et al., 2018; Cisse et al., 2013). Thus, oncogene activation often involves condensate-mediated compartmentalization of high densities of transcription apparatus at driver oncogenes. Mutations have been observed to affect many components of these transcriptional condensates in cancer cells, and these are almost certain to cause condensate dysregulation. These mutations alter the functional levels of lineage-specific master TFs, MYC, signaling TFs, transcriptional cofactors, and RNA polymerase II itself (Bhagwat and Vakoc, 2015; Bradner et al., 2017).
Cancer cell sequencing has revealed recurrent mutations that affect both histones and their regulatory proteins, and these are likely to alter the landscape of chromatin condensates (Figure 4B). For example, the oncogenic histone mutation H3K27M frequently occurs in childhood brain-stem gliomas and changes the chromatin landscape from poised to active at bivalent promoters, resulting in dysregulated gene expression (Larson et al., 2019; Schwartzentruber et al., 2012; Wu et al., 2014). The histone mutation H3K36M, which is recurrent in chondroblastoma and drives sarcoma development, inhibits the activity of histone methyltransferases and causes global changes in gene expression (Lu et al., 2016). Diverse chromatin regulators, including histone-modifying enzymes (writers and erasers), proteins that bind selectively to modified histones (readers), and proteins involved in nucleosome mobilization, are mutated in specific cancers (Bradner et al., 2017; Valencia and Kadoch, 2019). Thus, oncogenic mutations that alter chromatin condensates are likely to be a common theme in cancer cells.
Dysregulated condensate compartmentalization has also been observed with tumor suppressors. The tumor repressor SPOP (Speckle-type POZ protein) is an E3 ligase that has been implicated in a wide range of solid tumors (Kim et al., 2013). SPOP forms condensates in a substrate-dependent fashion, and cancer-causing mutations in SPOP prevent the protein from condensing with its substrate and diminish its enzymatic activity (Bouchard et al., 2018). Similarly, the tumor suppressor promyelocytic leukemia protein (PML) forms nuclear bodies that lack membranes and are thought to be biomolecular condensates (Banani et al., 2016). PML bodies compartmentalize a wide variety of proteins, including p53 and DNA repair factors, and have various roles in regulating cell function, including cell death and genome stability (Lallemand-Breitenbach and de Thé, 2010). PML loss-of-function mutations are associated with increased tumor formation and poor prognosis, and these phenotypes are coincident with alterations to the PML bodies (Zhu et al., 2018). Such alterations might compromise the ability of PML body condensates to partition clients and degrade oncoproteins.
Condensate mislocalization
Cancer cells acquire several different types of mutations that alter transcriptional condensate localization, including insertions or deletions (indels), translocations, and other chromosome rearrangements. Small indels have been shown to create oncogenic super-enhancers at genomic loci that do not normally harbor those elements. Recurrent small indels near the TAL1 gene in T cell acute lymphoblastic leukemia were found to form a binding site for a single molecule of the MYB TF, whose binding was found to initiate formation of a large super-enhancer that drives oncogenic TAL1 expression in those cells (Mansour et al., 2014). The ability of a single TF molecule to cause ectopic formation of an apparatus with hundreds of transcriptional components is now understood to be a reflection of the threshold behavior that is characteristic of phase-separated condensates, where the addition of a single TF binding site can create switch-like effects (Shrinivas et al., 2019).
Translocations can effect malignant transformation in leukemias, lymphomas, and solid tumors. This often occurs by juxtaposing a super-enhancer to a proto-oncogene. In aggressive B cell lymphomas, translocation of the IgH super-enhancer to the MYC locus is an oncogenic event that can now be understood as bringing a transcriptional condensate to the MYC gene (Figure 4C) (Klein et al., 2011; Lovén et al., 2013). Chromosome rearrangements and translocations may also create gene fusion events that effect condensate mislocalization. One such example is the fusion oncogenic protein EWS-FLI, composed of the disordered activation domain of the RNA binding protein EWS and the DNA binding domain of the TF FLI1. EWS-FLI has been shown to form condensate-like structures (or hubs) that promote transcription of genes associated with Ewing’s sarcoma (Chong et al., 2018). Diverse oncogenic translocations fuse IDRs to a DNA binding domain, and the ability of the disordered region to promote formation of transcriptional condensates may contribute to the oncogenic function of these fusion onco-TFs (Kumar-Sinha et al., 2015; Winters and Bernt, 2017; Kwon et al., 2013).
Altered regulation
A change in the concentration of any molecule that is contained in a condensate is likely to alter the behavior of that condensate. For example, oncogenic MYC protein accumulates to very high levels in transformed cells—it has been estimated that some tumor cells harbor 500,000 MYC molecules versus 10,000 molecules for a typical TF—and MYC may alter the behavior of transcriptional condensates in these cells (Figure 4D) (Lin et al., 2012).
PTMs can alter the condensation of chromatin. Chromatin is regulated through diverse PTMs, and cancer cells harbor mutations in a broad spectrum of histone-modifying enzymes, histone reader proteins, and nucleosome-mobilizing proteins (Bates, 2020; Mittal and Roberts, 2020; Morgan and Shilatifard, 2015; Pastore and Levine, 2016; Valencia and Kadoch, 2019). As one example, the bromodomain and extraterminal motif protein BRD4, which binds to chromatin through acetylated histones, is overexpressed in some solid tumors (Filippakopoulos et al., 2010; Rhyasen et al., 2018). Although it is possible that the dysregulated transcription in these cells is due simply to canonical binding of excess BRD4 to portions of the genome, evidence that large numbers of BRD4 molecules can assemble into a super-enhancer transcriptional condensate suggests that condensate alterations are in play (Sabari et al., 2018; Shin et al., 2018; Wei et al., 2020).
ncRNA molecules are components of well-studied biomolecular condensates, where they have various regulatory functions (Clemson et al., 2009; Daneshvar et al., 2020; Henninger et al., 2021; Yamazaki et al., 2018). In cancer cells, ncRNAs are overexpressed (MALAT1, HOTAIR, SRA, CCAT2, LincRNAROR, lncRNA-ATB, LncTCF7, SCHLAP1, treRNA, ZEB2-AS1, UCA1), underexpressed (LET, DRAIC, EGOT, GAS5, MEG3, NBAT-1, NKILA, PCAT), and post-transcriptionally modified (Barbieri and Kouzarides, 2020; Wang et al., 2020). The levels of the long non-coding RNA (lncRNA) MALAT1, a component of nuclear speckles, have been shown to be increased in lung, breast, cervical, colorectal, bladder, and liver cancers (Li et al., 2018). Cells that improperly express these ncRNAs exhibit various dysregulated functions, with inconsistent and sometimes conflicting views on the mechanisms of action. It seems likely that altered expression of these ncRNAs will alter the behavior of diverse condensates.
REVISITING MECHANISMS IN COMMON ONCOGENIC EVENTS
The use of condensate models to re-examine the mechanisms involved in common oncogenic events (Figure 5) could lead to new insights and therapeutic approaches. Common events include dysregulated signaling, transcription, DNA damage, metabolism, cellular interactions, immune mechanisms, autophagy, and angiogenesis, and we provide here some examples of new mechanistic models that might be considered in these areas.
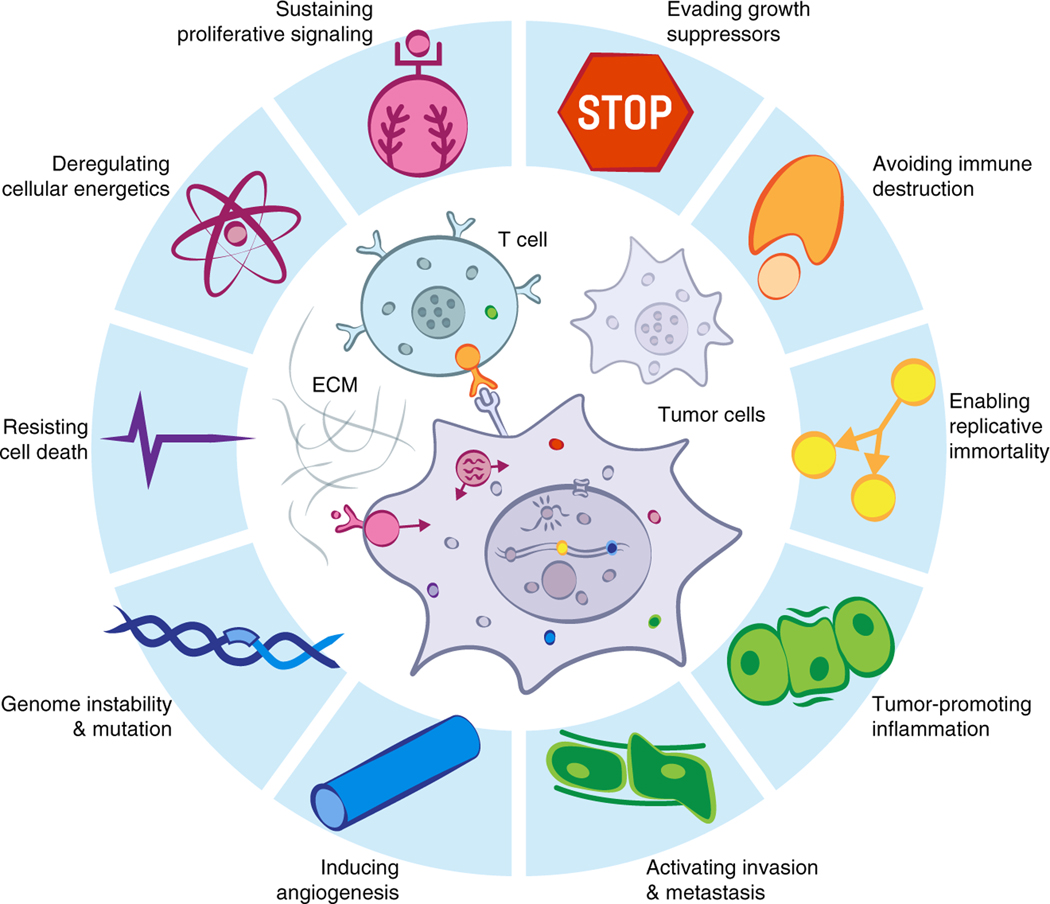
Figure 5. Hallmarks of cancer incorporate processes that involve diverse biomolecular condensates.
Biomolecular condensates are involved in most processes that have been called hallmarks of cancer (modified from Hanahan and Weinberg, 2011).
Diverse signaling pathways controlling cell growth, division, and mobility are altered in cancer; signaling proteins may be overexpressed, mutated, or fused, causing over- or underactivation of the pathway (Klein et al., 2011; Sanchez-Vega et al., 2018; Sever and Brugge, 2015; Vogelstein and Kinzler, 2004; Wong et al., 2010; Zhan et al., 2017). Many of the proteins involved in cancer-associated signaling pathways have been shown to form condensates, thus regulating output of the pathway (Case et al., 2019b; Chong and Forman-Kay, 2016; Su et al., 2016). RAS activation occurs when ligand-bound membrane receptors recruit and modify adaptor proteins, and these adaptor proteins form condensates at the cell membrane that compartmentalize proteins, increasing their “dwell time” and RAS activity (Huang et al., 2019; Zhu et al., 2020). These same adaptor protein condensates also accelerate the rate of actin polymerization (Case et al., 2019a). Cyclic AMP (cAMP)-dependent protein kinase A (PKA) undergoes cAMP-dependent condensate formation, thus compartmentalizing this key signaling molecule, while a PKA fusion oncoprotein disables condensation and causes aberrant signaling (Zhang et al., 2020b). Nuclear signaling proteins such as Wnt, TGF-β, and STAT condense with transcriptional coactivators to activate target genes, accounting for their cell-type-specific effects (Zamudio et al., 2019). Taken together, a novel picture of signaling may be emerging, in which diverse signaling proteins achieve specificity by forming distinct cellular compartments that are disrupted in cancer.
Transcriptional dysregulation is a common feature of cancer cells, and evidence that gene regulation involves formation of transcriptional condensates should prompt new thinking about dysregulated regulatory mechanisms. For example, MYC overexpression is a common event in many metastatic processes (Dang, 2012) and may produce transcriptional condensates at oncogenes that are more long lived. Dysregulated proliferative signaling is a general feature of tumor cells, and evidence that the terminal components of signaling pathways tend to partition into super-enhancer-associated transcriptional condensates (Zamudio et al., 2019) suggests that oncogenes that acquire super-enhancers, such as MYC, are efficiently targeted by dysregulated proliferative signaling through condensate-associated mechanisms. Furthermore, MYC overexpression produces a broad spectrum of cellular effects, including changes in chromatin structure, ribosome biogenesis, metabolic pathways, cell adhesion, cell size, apoptosis, and angiogenesis, among others (Amati et al., 1998; Cole and Cowling, 2008; Cowling and Cole, 2010; Dai and Lu, 2008; Dang, 2012; Eilers and Eisenman, 2008; Facchini and Penn, 1998; Frank et al., 2001; Gallant, 2005; Hanahan and Weinberg, 2011; Herold et al., 2009; Hoffman and Liebermann, 2008; Hurlin and Dezfouli, 2004; Kuttler and Mai, 2006; Lin et al., 2009; Meyer and Penn, 2008; Nieminen et al., 2007; Nilsson and Cleveland, 2003; Peterson and Ayer, 2011; Van Riggelen et al., 2010; Ruggero, 2009; Secombe et al., 2004; Singh and Dalton, 2009). It is possible that most of these cellular effects are secondary consequences of MYC’s increased binding to DNA in the regulatory regions of active genes, with consequent amplification of transcriptional output (Lin et al., 2012; Nie et al., 2012). It is also possible that high levels of MYC cause interference with condensate compartments other than those involved in transcription.
The DNA damage response acts as a barrier to the malignant transformation of pre-neoplastic cells. Most cancer cells display defects in the DNA damage response, and the efficacy of DNA-damaging agents used in cancer treatment is highly influenced by the cellular DNA repair capacity (Gavande et al., 2016). Drugs that inhibit remaining functional DNA repair pathways in cancer can be exploited to selectively kill some malignant cells (Leung, 2020; Oshidari et al., 2020; Pessina et al., 2019). Recent studies have revealed that DNA damage response factors, and RNA produced at sites of DNA damage, promote formation of condensates at the sites of repair and have suggested that selective disruption of these condensates might produce a therapeutic benefit (Altmeyer et al., 2015; Chong et al., 2018; Kilic et al., 2019; Oshidari et al., 2020; Singatulina et al., 2019).
Metabolic reprogramming is a hallmark of malignancy; cancer cells adjust metabolic and nutrient acquisition to support growth and dissemination (Dayton et al., 2016; Vander Heiden, 2011; Hopkins et al., 2016; Martinez-Outschoorn et al., 2017; Pavlova and Thompson, 2016; Schulze and Harris, 2012; Shankaraiah et al., 2018). The metabolic phenotype exhibited by tumor cells—known as the Warburg effect—is characterized by high rates of aerobic glycolysis and has been a topic of special interest in cancer research (Hsu and Sabatini, 2008; Liberti and Locasale, 2016; Warburg, 1956). Transcriptional regulators of mitochondrial biosynthesis, metabolic enzymes, and small metabolites are now thought to function in biomolecular condensates. The level of the transcriptional coactivator PGC-1α, which regulates transcriptional networks associated with mitochondrial biogenesis and function, is altered in diverse cancers (Gravel, 2018); PGC-1α is known to associate with diverse TFs and transcriptional cofactors (Puigserver et al., 1999; Wallberg et al., 2003), and this is now reported to occur via interaction with transcriptional condensates in an RNA-dependent fashion (Pérez-Schindler et al., 2020). Enzymes involved in carbohydrate, nucleotide, fatty acid, and amino acid metabolic pathways have been observed in diverse condensates (An et al., 2008; Jin et al., 2017; O’Connell et al., 2012; Prouteau and Loewith, 2018; Prouteau et al., 2017; Zhang et al., 2020b). Metabolites and the cellular redox state have been shown to have an impact on condensate formation and behavior (Franzmann et al., 2018; Kato et al., 2019; Patel et al., 2017; Riback et al., 2017). Additional insights into dysregulated metabolic processes in cancer will almost certainly come from the study of signaling, transcriptional, and enzymatic condensates.
Altered cell-cell interactions are a feature of the tumor microenvironment and are characteristic of the metastatic state of tumor cells. The extracellular matrix (ECM) is involved in these interactions and both provides physical scaffolding for the cell and mediates responses to biochemical and biomechanical cues that are required for normal morphogenesis, differentiation, and homeostasis (Lu et al., 2011). Deregulation of ECM dynamics promotes cancer cell proliferation, loss of cell differentiation, the epithelial-to-mesenchymal transition, and cancer cell invasion (Henke et al., 2020; Winkler et al., 2020). The multivalent interactions characteristic of ECM components are likely to be involved in diverse condensate behaviors. Elastin is the best characterized ECM protein whose polymeric assembly is initiated by phase separation (Bellingham et al., 2003; Kozel et al., 2006; MacEwan and Chilkoti, 2010; Muiznieks et al., 2018; Reichheld et al., 2017; Vrhovski et al., 1997). There are additional proteins that are thought to contribute to ECM dynamics and may be involved in tumor processes; as an example, galectin3-agglutinated glycosylated molecules in the ECM tumor stroma are thought to undergo phase separation (Chiu et al., 2020). Improved understanding of condensate components and regulation in the ECM might provide novel therapeutic avenues in cancer.
The innate immune response serves as a sensor for oncogenic events and halts malignant transformation (Corrales et al., 2017; Gajewski et al., 2013; Wellenstein and De Visser, 2018; Woo et al., 2015). As a consequence of genomic insults occurring early in the course of oncogenesis, DNA fragments translocate to the cytoplasm, providing a cellular “danger signal” (Dhanwani et al., 2018; Paludan and Bowie, 2013; Won and Bakhoum, 2020). Cytoplasmic DNA sensing and downstream response are critical for anticancer immune responses and have generated excitement as a potential therapeutic opportunity (Chin et al., 2020; Pan et al., 2020). The cytoplasmic DNA sensor is now understood to be a condensate that forms in response to this cellular stress. Upon binding DNA, cGAS forms cytoplasmic condensates to generate cAMP, and this in turn activates STING to induce cytokine production and an appropriate immune response (Du and Chen, 2018; Liu et al., 2018; Sun et al., 2013; Wu et al., 2013; Xie et al., 2019). The understanding that this antineoplastic process occurs by formation of a specialized phase-separated compartment opens the possibility for novel pharmacologic approaches to enhance the activity of this pathway. Consideration should be given to strategies that enhance formation of DNA-sensing condensates or improve the activity of cGAS therein.
Autophagy maintains normal cell homeostasis through the removal of unfolded proteins and damaged organelles (Bento et al., 2016; Dikic and Elazar, 2018; Glick et al., 2010; Levine and Kroemer, 2019). Dysregulation of this process contributes to cancer initiation, and once cancer is established, increased turnover of cell components that provide energy and macromolecular precursors requires autophagy for tumor survival and growth (Levy et al., 2017; Mathew et al., 2007; Mulcahy Levy and Thorburn, 2020). Thus, targeting autophagy holds promise for cancer treatment (Amaravadi et al., 2011). Condensates are now understood to mediate this process. Modification of pre-autophagosomal structural proteins induces phagophore condensate formation, and similar to many other cellular condensates that are anchored to specific membranes, autophagy condensates are localized to vacuoles by specific proteins (Fujioka et al., 2020; Hawkins and Klionsky, 2020; Sun et al., 2018; Yamamoto et al., 2016). A deeper understanding of how autophagy condensates form and function will certainly further illuminate this critical cellular process, and may guide efforts to manipulate autophagy for therapeutic purposes.
Tumors often have an insufficient vascular supply, leading to hypoxia and nutrient deprivation, environmental conditions that lead to stress granule assembly (Ackerman and Simon, 2014; Anderson et al., 2015; Protter and Parker, 2016). Stress granules facilitate high cellular proliferation rates in the face of these environmental conditions by modulating signaling pathways, altering cellular metabolism, and activating the stress response (Buchan and Parker, 2009; Mahboubi and Stochaj, 2017). Phase separation is thought to underlie the formation of stress granules; upon oxidative or osmotic stress, an increase in the concentration of cytoplasmic RNA drives the condensation of RNA binding proteins such as FUS, hnRNPA1, and G3BP1/2 (Guillé N-Boixet et al., 2020; Molliex et al., 2015; Patel et al., 2015; Sanders et al., 2020; Yang et al., 2020). Stress granules compartmentalize antiapoptotic proteins and are associated with resistance to chemotherapeutic drugs (Arimoto et al., 2008; Fournier et al., 2010; Gareau et al., 2011; Kaehler et al., 2014; Thedieck et al., 2013). The understanding that stress granules are condensates may reveal the mechanisms by which cancer cells resist cytotoxic stressors, and impairing stress granule condensation may sensitize tumors to their own harsh environments or antineoplastic drugs.
CONDENSATES AND DRUG ACTION IN CANCER
An understanding of condensate biology in cancer cells presents an opportunity to develop new therapeutic hypotheses. There is now evidence that some drugs concentrate in specific condensates through physicochemical interactions that are independent of the drug’s affinity for its target (Klein et al., 2020). In addition, some drugs appear to selectively disrupt condensates, offering the opportunity to modulate compartments that contribute to disease pathology (Wheeler et al., 2019). Furthermore, drugs that inhibit post-translational modifying enzymes can influence condensate behaviors and might be leveraged to alter oncogenic activities that are compartmentalized in condensates (Monahan et al., 2017; Rai et al., 2018).
Cancer drugs concentrate in condensates
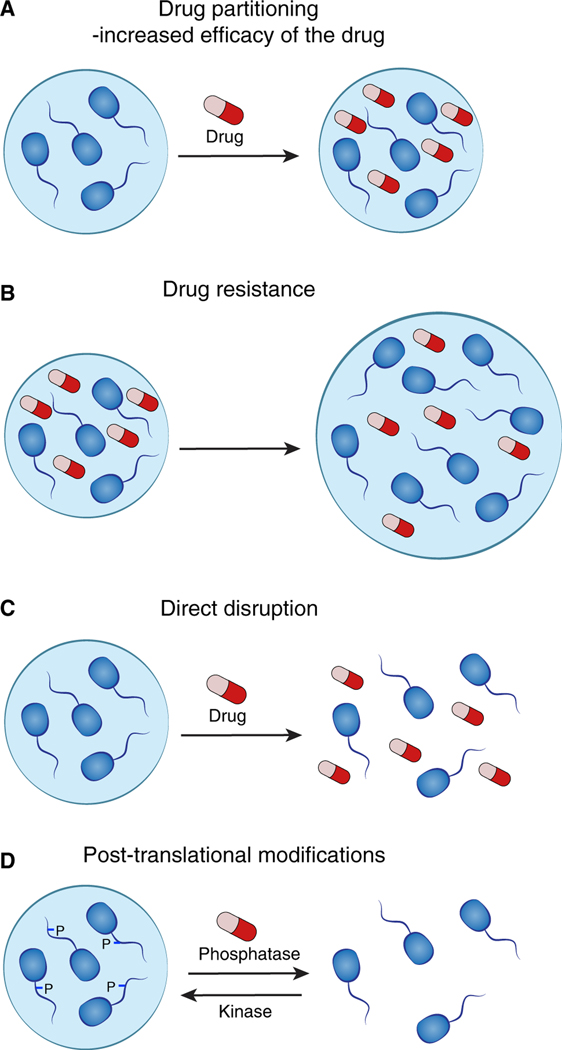
Textbook diagrams of cells generally show membrane-bound organelles, implying an otherwise aqueous environment with free diffusion of macromolecules and proteins. Conventional pharmacological studies do not typically evaluate the intracellular distribution of drugs. Recent evidence, however, indicates that partitioning of drugs into specific non-membrane condensate compartments within cells can play a role in both drug efficacy and drug resistance in cancer (Figures 6A and 6B) (Klein et al., 2020). This new insight suggests the possibility that drugs can be designed to concentrate in specific intracellular compartments where their targets reside, which may provide improved therapeutic index.
Figure 6. Condensates and drug action in cancer.
(A) Partitioning of drugs into specific non-membrane-bound condensate compartments within cells can increase the concentration and efficacy of the drug.
(B) Drug resistance in cancer cells might occur through overexpression of a condensate-promoting protein. This mediates formation of larger condensates with resulting dilution of the drug, rendering the drug less effective.
(C) Certain condensates appear to be sensitive to disruption by small-molecule drugs.
(D) Drugs targeting enzymes mediating post-translational modifications are likely to affect condensate formation.
The targets of many commonly used drugs are now known to occur in condensates, so it might be expected that efficacious drugs can access these compartments to engage their targets. It is possible that drugs become concentrated in condensates due to their selectivity and affinity for the compartmentalized molecules. We now know that drugs can concentrate in the condensates where their targets occur, but they do this through selective partitioning due to the physicochemical properties of the drug and the condensate, independent of their target engagement. Thus, drug molecules can exploit condensate properties, independent of those governing target engagement, to concentrate in the same compartment as their target.
The ability of drugs to partition into specific condensates might be expected to enhance the pharmacological properties of drugs, and indeed, there is evidence that the efficacy of drugs can be influenced by this behavior. Cisplatin, a commonly used antineoplastic intercalating agent, is concentrated up to 600-fold in transcriptional condensates, where it selectively platinates the super-enhancer DNA encompassed in these condensates (Klein et al., 2020). This example shows that condensate partitioning can enhance the pharmacological activity of a drug, but also illustrates how it can enhance target specificity for drugs that would otherwise engage a broader range of substrates. Because some of the largest super-enhancers occur at driver oncogenes, it is possible that cisplatin is especially effective at inactivating the oncogenes embedded in these condensates.
If condensate partitioning properties of drugs play a role in their efficacy, they might also be expected to play a role in drug resistance. Tamoxifen is a highly effective drug in the treatment of estrogen receptor (ER)-positive breast cancer. Tamoxifen resistance can be conferred by ER mutations that reduce drug affinity and MED1 overexpression, which until recently did not have a mechanistic explanation (Fanning et al., 2016; Nagalingam et al., 2012). ER partitions selectively into transcriptional condensates in an estrogen-dependent manner and is evicted from the condensate by tamoxifen, which also partitions selectively into transcriptional condensates and competes for estrogen binding (Klein et al., 2020). MED1 overexpression was found to cause an expansion of the volume of transcriptional condensates, thereby diluting tamoxifen in the condensate and rendering tamoxifen less efficient in evicting ER from the condensate. These results suggest that condensate alterations can contribute to drug resistance in cancer cells.
Drugs that selectively alter condensate properties
Condensates may be selectively disrupted by small-molecule drugs, suggesting a novel therapeutic hypothesis for cancers where dysregulated condensates contribute to the oncogenic state (Figure 6C). FUS and other disordered proteins are common translocation partners to TF DNA binding domains in diverse cancers (Bradner et al., 2017; Crozat et al., 1993; Kumar-Sinha et al., 2015), where they probably contribute to phase-separated transcriptional condensates at key oncogenes (Boulay et al., 2017; Chong et al., 2018). In familial amyotrophic lateral sclerosis, mutations in FUS are characterized by the accumulation of cytoplasmic stress granules containing FUS and other protein and RNA molecules (Patel et al., 2015). A screen for small molecules that selectively affect stress granule formation revealed that lipoamide can selectively dissolve stress granules in cells (Wheeler, 2019). This evidence that a specific condensate can be sensitive to disruption by a specific small-molecule drug suggests a novel therapeutic route for cancers with dysregulated condensates that contribute to disease pathogenesis.
IDR interactions mediate condensate formation, but have long been considered undruggable. This difficulty arises because IDRs lack the stable secondary and tertiary structures generally targeted by traditional medicinal chemistry approaches, and small molecules that do bind IDRs typically do so with low affinity (Csizmok et al., 2016; Dang et al., 2017; van der Lee et al., 2014; Uversky, 2011; Wright and Dyson, 2015). Nonetheless, recent advances in chemistry present an opportunity to drug IDRs, potentially disrupting specific condensates (Chen and Kriwacki, 2018; Metallo, 2010). Small molecules that target the MYC IDR and components of the transcription initiation complex have been identified (Carabet et al., 2019; Zhang et al., 2015b). The ability of the p27 oncogene to interact with the cell-cycle machinery can be disrupted with small molecules that bind its IDR (Ban et al., 2017; Iconaru et al., 2015). Learning how to engineer drugs to selectively concentrate in specific condensates might compensate for the relatively low affinity of drug-IDR interactions.
Modifying condensates by inhibiting post-translational modification
Condensate formation, dissolution, and function can be modified by PTM of constituent proteins; the enzymes that catalyze these modifications are attractive drug targets (Figure 6D). DNA damage repair requires temporal and spatial coordination of diverse effector proteins and is a potent antineoplastic target (Brown et al., 2017; Cleary et al., 2020; Gavande et al., 2016). DNA repair is now understood to take place in specialized condensates, the formation of which is dependent on poly(ADP-ribosyl)ation of proteins and DNA (Kilic et al., 2019; Oshidari et al., 2020; Singatulina et al., 2019). PARP inhibition prevents DNA repair condensate formation and impairs the DNA damage response (Singatulina et al., 2019). Diverse enzymes modify DNA repair components with functional consequences, and those affecting condensation of the DNA repair machinery may be ripe for therapeutic development. Cellular processes that effect high-fidelity mitosis are common and effective antineoplastic targets (Chan et al., 2012). Many cellular condensates are dissolved during mitosis and re-formed after cell division by virtue of DYRK3 kinase activity (Rai et al., 2018). These condensates fail to dissolve upon chemical inhibition of DYRK3, with resulting mitotic defects. Inhibition of the enzymes that regulate cell-cycle-associated condensate formation and dissolution might now be considered for therapeutic discovery.
AREAS FOR FUTURE INVESTIGATION
Condensate regulation and dysregulation
How cancer-associated mutations give rise to the oncogenic state is understood largely from the effects of mutations on the structured regions of individual protein molecules. Understanding how mutations in protein coding sequences affect 3D protein structure has provided mechanistic hypotheses of disease causality and structure-based approaches to medicinal chemistry that have led to valuable therapeutics. However, many proteins contain domains that lack defined, stable 3D structures, and typically contain low amino acid sequence complexity; indeed, more than 1/3 of proteins contain IDRs greater than 30 residues in length (Ward et al., 2004). The emergence of a conceptual framework and new experimental approaches to investigate condensate behaviors presents an opportunity to gain new insights into the dysregulated state of cancer cells and uncover condensate-associated mechanisms that lead to new therapeutic hypotheses.
Now that large databases of recurrent mutations have been generated for diverse cancers, it is possible to identify those that affect DNA, RNA, and protein molecules whose functions are associated with condensates, thus expanding the list of cancer-associated condensate components (Table 1). To the extent that these mutant molecules create pathogenic condensate processes, the mechanisms that are dysregulated and the molecules that are involved become potential targets for therapeutic intervention. For example, it is now possible to explore how diverse modifications of DNA, RNA, and protein molecules contribute to condensate behaviors and how gain- or loss-of-function mutations might affect these compartments.
Drug design and development
The new understanding that the cell is organized into phase separated compartments that concentrate not only cellular components but also therapeutic drugs suggests several approaches that may improve development of novel small-molecule therapeutics.
Novel perturbants of condensates may be discovered using high throughput condensate assays. IDPs are difficult to target by conventional means, and robust assays for small molecules that might disrupt IDR-IDR interactions are lacking. That these domains mediate formation of condensates provides a readout to screen for drugs that perturb their interactions in cells and in vitro. Since condensates can be visualized by using fluorescent-tagged proteins, small molecule screens can be performed to discover perturbants of condensates key for any number of disease processes.
Partitioning of drugs into specific condensates can be optimized in order to enhance target engagement and minimize off-target effects. While our understanding of protein partitioning is still developing, it should be possible to decipher the chemical features of a small molecule - functional groups, lipophilicity, hydrogen bond donor/acceptor count, etc. - that are responsible for selective condensate partitioning and design these into small molecules. These novel design strategies might enable condensate targeting, allowing for partitioning or departitioning from specific cellular compartments. Mapping out the physicochemical rules of each cellular condensate will guide such efforts. Accomplishing this goal could improve the therapeutic index of drugs, allowing patients to be treated at lower doses with fewer side effects.
Based on emergent physicochemical properties of cellular condensates, novel design strategies for therapeutic drugs may be developed. One potential strategy would be bifunctional drugs composed of a condensate addressing module in addition to a target binding molecule. Such bifunctional drugs might allow for generation of higher local drug concentrations or recruitment of a protein of interest, such as an E3 ubiquitin ligase, into a condensate to cause its disruption. Deploying condensate properties as a guiding principle in the design of next generation of drugs may provide a way to drug previously undruggable proteins due to their intrinsically disordered nature - a feature of TFs including the proto-oncoprotein MYC (Liu et al., 2006). The ability to produce high local drug concentrations may allow for the development of lower affinity drugs that are effective in targeting proteins whose features have rendered them “undruggable” (Yan and Higgins, 2013).
With evidence that drugs can selectively partition into specific condensates, revisiting our current understanding of drug mechanisms might provide additional insights valuable for future drug development. Super-enhancer-associated oncogenes can be far more sensitive than other genes to certain drugs that should inhibit transcription generally. For example, treatment of a multiple myeloma cell line with JQ1, an inhibitor of the BRD4 coactivator, leads to selective loss of MYC expression, which is driven by a large super-enhancer (Lovén et al., 2013). Similarly, treatment of various cancer cells with THZ1, a CDK7 inhibitor, produces selective loss of expression of oncogenes that play prominent roles in those cells and that are super-enhancer driven (Chipumuro et al., 2014; Christensen et al., 2014; Kwiatkowski et al., 2014; Wang et al., 2015). These effects have been mysterious, as BRD4 and CDK7 are associated with most active genes, and so it has not been clear why genes with large super-enhancers should be more sensitive than others. Condensate studies have provided a potential solution to this mystery (Hnisz et al., 2017). Larger transcriptional condensates have longer half-lives (Cho et al., 2018) and JQ1 and THZ1 are concentrated in Mediator condensates (Klein et al., 2020), so the concentration of these drugs in long-lived condensates at oncogenes provides an explanation for the gene-selective effects of these drugs.
Drug resistance
Additional condensate-associated mechanisms of drug resistance will almost certainly emerge when various cancer cell resistant states are explored. In prostate cancer, the clinical use of potent androgen receptor (AR) therapies has led to the emergence of a type of castration-resistant prostate cancer (CRPC) called ARL/negative CRPC. This cancer produces abundant AR mRNA but limited AR protein and thus cannot be targeted by hormonal therapies, resulting in poor prognosis. In ARL/negative CRPC cells, the RNA binding protein DDX3 is highly expressed, binds to AR mRNA, and sequesters it in cytoplasmic stress granule condensates, thus limiting its translation. Inhibiting DDX3 was found to be sufficient to restore AR protein expression and sensitivity to AR signaling inhibitors (Vellky et al., 2020). Improved understanding of these mechanisms should lead to more effective strategies to minimize drug resistance for patient benefit.
ACKNOWLEDGMENTS
We thank Henry Kilgore for illustrations depicting chemical interactions and Anthony Hyman, Simon Alberti, Bede Portz, James Shorter, Salman Banani, Charalampos Lazaris, and members of the Young lab for helpful discussions. This work was supported by NIH grants GM123511, CA213333, and CA155258 (R.A.Y.); an American Cancer Society postdoctoral fellowship (I.A.K.); an Ovarian Cancer Research Alliance Mentored Investigator Award (I.A.K.), and a Swedish Research Council postdoctoral fellowship (VR 2017–00372) (A.B.). A.B. is an advisor to Syros Pharmaceuticals. I.A.K. is a shareholder and member of the scientific advisory board of Dewpoint Therapeutics. R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics.
REFERENCES
- Ackerman D, and Simon MC (2014). Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 24, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S. (2017). The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 130, 2789–2796. [DOI] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, and Mittag T. (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates 176, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask MBD, Streicher W, Jungmichel S, Nielsen ML, et al. (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, and White E. (2011). Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 17, 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Alevizopoulos K, and Vlach J. (1998). Myc and the cell cycle. Front. Biosci. 3, d250–68. [DOI] [PubMed] [Google Scholar]
- An S, Kumar R, Sheets ED, and Benkovic SJ (2008). Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106. [DOI] [PubMed] [Google Scholar]
- Anderson KC (2016). Progress and paradigms in multiple myeloma. Clin. Cancer Res. 22, 5419–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, and Ivanov P. (2015). Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 1849, 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, and Takekawa M. (2008). Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324–1332. [DOI] [PubMed] [Google Scholar]
- Ban D, Iconaru LI, Ramanathan A, Zuo J, and Kriwacki RW (2017). A small molecule causes a population shift in the conformational landscape of an intrinsically disordered protein. J. Am. Chem. Soc. 139, 13692–13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional control of phase-separated cellular bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, and Rosen MK (2014). Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife 3, e04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, and Kouzarides T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322. [DOI] [PubMed] [Google Scholar]
- Bates SE (2020). Epigenetic therapies for cancer. N. Engl. J. Med. 383, 650–663. [DOI] [PubMed] [Google Scholar]
- Bellingham CM, Lillie MA, Gosline JM, Wright GM, Starcher BC, Bailey AJ, Woodhouse KA, and Keeley FW (2003). Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers 70, 445–455. [DOI] [PubMed] [Google Scholar]
- Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, and Rubinsztein DC (2016). Mammalian autophagy: how does it work? Annu. Rev. Biochem. 85, 685–713. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, and Vakoc CR (2015). Targeting transcription factors in cancer. Trends Cancer 1, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. (2020). Head-to-tail polymerization in the assembly of biomolecular condensates. Cell 182, 799–811. [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. (2018). Protein Phase Separation: A New Phase in Cell Biology (Elsevier Ltd; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. (2018). Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, et al. (2018). Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. (2017). Cancer-specific retargetingof BAF complexes by a prion-like domain. Cell 171, 163–178.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, Hnisz D, and Young RA (2017). Transcriptional addiction in cancer. Cell 168, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CPP, Tompa P, and Pappu RVV (2015). Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904. [Google Scholar]
- Brown JS, O’Carrigan B, Jackson SP, and Yap TA (2017). Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 7, 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, and Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, and Lippincott-Schwartz J. (2019a). Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 21, 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Choi PS, Gelbard M, and Meyerson M. (2019b). Identification and characterization of oncogenic SOS1 mutations in lung adenocarcinoma. Mol. Cancer Res. 17, 1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabet LA, Rennie PS, and Cherkasov A. (2019). Therapeutic inhibition of myc in cancer. Structural bases and computer-aided drug discovery approaches. Int. J. Mol. Sci. 20, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter RL, and Gökmen-Polar Y. (2018). HSF1 as a cancer biomarker and therapeutic target. Curr. Cancer Drug Targets 19, 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, and Rosen MK (2019a). Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Ditlev JA, and Rosen MK (2019b). Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KS, Koh CG, and Li HY (2012). Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis. 3, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, and Kriwacki RW (2018). Intrinsically disordered proteins: structure, function and therapeutics. J. Mol. Biol. 430, 2275–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin EN, Yu C, Vartabedian VF, Jia Y, Kumar M, Gamo AM, Vernier W, Ali SH, Kissai M, Lazar DC, et al. (2020). Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science 369, 993–999. [DOI] [PubMed] [Google Scholar]
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, et al. (2014). CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YP, Sun YC, Qiu DC, Lin YH, Chen YQ, Kuo JC, and Huang JR (2020). Liquid-liquid phase separation and extracellular multivalent interactions in the tale of galectin-3. Nat. Commun. 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielecki J, and Meyerson M. (2014). DNA sequencing of cancer: what have we learned? Annu. Rev. Med. 65, 63–79. [DOI] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-M, Holehouse AS, and Pappu RV (2020). Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PA, and Forman-Kay JD (2016). Liquid–liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 41, 180–186. [DOI] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. (2014). Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26,909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, and Darzacq X. (2013). Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341, 664–667. [DOI] [PubMed] [Google Scholar]
- Cleary JM, Aguirre AJ, Shapiro GI, and D’Andrea AD (2020). Biomarker-guided development of DNA repair inhibitors. Mol. Cell 78, 1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, and Lawrence JB (2009). An architectural role for a nuclear non-coding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MD, and Cowling VH (2008). Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat. Rev. Mol. Cell Biol. 9, 810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Matson V, Flood B, Spranger S, and Gajewski TF (2017). Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 27, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, and Cole MD (2010). Myc regulation of mRNA cap methylation. Genes Cancer 1, 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Åman P, Mandahl N, and Ron D. (1993). Fusion of CHOP toa novel RNA-binding protein in human myxoid liposarcoma. Nature 363, 640–644. [DOI] [PubMed] [Google Scholar]
- Csizmok V, Follis AV, Kriwacki RW, and Forman-Kay JD (2016). Dynamic protein interaction networks and new structural paradigms in signaling. Chem. Rev. 116, 6424–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M-S, and Lu H. (2008). Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J. Cell. Biochem. 105, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, and Merdes A. (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K, Ardehali MB, Klein IA, Hsieh FK, Kratkiewicz AJ, Mahpour A, Cancelliere SOL, Zhou C, Cook BM, Li W, et al. (2020). lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat. Cell Biol. 22, 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV (2012). MYC on the path to cancer. Cell 149, 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Reddy EP, Shokat KM, and Soucek L. (2017). Drugging the “undruggable” cancer targets. Nat. Rev. Cancer 17, 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton TL, Jacks T, and Vander Heiden MG (2016). PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanwani R, Takahashi M, and Sharma S. (2018). Cytosolic sensing of immuno-stimulatory DNA, the enemy within. Curr. Opin. Immunol. 50, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignon GL, Zheng W, Kim YC, and Mittal J. (2019). Temperature-controlled liquid-liquid phase separation of disordered proteins. ACS Cent. Sci. 5, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, and Elazar Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364. [DOI] [PubMed] [Google Scholar]
- Du M, and Chen ZJ (2018). DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, and Eisenman RN (2008). Myc’s broad reach. Genes Dev. 22, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-HH, Eckmann CR, Myong S, and Brangwynne CP (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U SA 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini LM, and Penn LZ (1998). The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 12, 633–651. [PubMed] [Google Scholar]
- Fanning SW, Mayne CG, Dharmarajan V, Carlson KE, Martin TA, Novick SJ, Toy W, Green B, Panchamukhi S, Katzenellenbogen BS, et al. (2016). Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife 5,e12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciani A, D’Annunzio S, Poli V, Fagnocchi L, Beyes S, Michelatti D, Corazza F, Antonelli L, Gregoretti F, Oliva G, et al. (2020). MLL4-associated condensates counterbalance Polycomb-mediated nuclear mechanical stress in Kabuki syndrome. Nat. Genet. 52, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, and Anderson PJ (2018). The role of RNA in biological phase separations. J. Mol. Biol. 430, 4685–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zeng M, Chen X, and Zhang M. (2018). Neuronal Synapses: microscale signal processing machineries formed by phase separation? Biochemistry 57, 2530–2539. [DOI] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory PJ (1942). Themodynamics of high polymer solutions. J. Chem. Phys. 10, 51–61. [Google Scholar]
- Forman-Kay JD, Kriwacki RW, and Seydoux G. (2018). Phase separation in biology and disease. J. Mol. Biol. 430, 4603–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MJ, Gareau C, and Mazroui R. (2010). The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, and Amati B. (2001). Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. (2018). Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654. [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y, May AI, Knorr RL, Suzuki K, Ohsumi Y, et al. (2020). Phase separation organizes the site of autophagosome formation. Nature 578, 301–305. [DOI] [PubMed] [Google Scholar]
- Gaglia G, Rashid R, Yapp C, Joshi GN, Li CG, Lindquist SL, Sarosiek KA, Whitesell L, Sorger PK, and Santagata S. (2020). HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 22, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, García-Muse T, and Aguilera A. (2015). Replication stress and cancer. Nat. Rev. Cancer 15, 276–280. [DOI] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, and Fu YX (2013). Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P. (2005). Myc, cell competition, and compensatory proliferation. Cancer Res. 65, 6485–6487. [DOI] [PubMed] [Google Scholar]
- Gareau C, Fournier M-J, Filion C, Coudert L, Martel D, Labelle Y, and Mazroui R. (2011). p21WAF1/CIP1 upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One 6, e20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavande NS, Vandervere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, and Turchi JJ (2016). DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol. Ther.160, 65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, and Rosen MK (2019). Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, and Macleod KF (2010). Autophagy: cellular and molecular mechanisms. J. Pathol. 221, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E, and Shorter J. (2019). The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel SP (2018). Deciphering the dichotomous effects of PGC-1α on tumor-igenesis and metastasis. Front. Oncol. 8, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig JA, Nguyen TA, Lee M, Holehouse AS, Posey AE, Pappu RV, and Jedd G. (2020). Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol. Cell 77, 1237–1250.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuilléN-Boixet J, Kopach A, Holehouse AS, Pappu RV, Alberti S, Franzmann Correspondence TM, Wittmann S, Jahnel M, Schlü R, Kim K, et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation article RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D, et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, Spille J-H, Afeyan LK, Zamudio AV, Shrinivas K, et al. (2019). Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2000). The hallmarks of cancer. Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harmon TS, Holehouse AS, Rosen MK, and Pappu RV (2017). Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6, e30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins WD, and Klionsky DJ (2020). A separation that’s for the best: coming together at the PAS. Cell Res. 30, 372–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Li H, Wu A, Peng Y, Shu G, and Yin G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke E, Nandigama R, and Ergün S. (2020). Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 6, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng M, Andrews J, Zamudio A, Lazaris C, Hannett NM, et al. (2021). RNA-mediated feedback control of transcriptional condensates. Cell. 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Herkert B, and Eilers M. (2009). Facilitating replication under stress: an oncogenic function of MYC? Nat. Rev. Cancer 9, 441–444. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, and Young RA (2013). XSuper-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, and Sharp PA (2017). Leading edge perspective A phase separation model for transcriptional control. Cell 169, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B, and Liebermann DA (2008). Apoptotic signaling by c-MYC. Oncogene 27, 6462–6472. [DOI] [PubMed] [Google Scholar]
- Hopkins BD, Goncalves MD, and Cantley LC (2016). Obesity and cancer mechanisms: cancer metabolism. J. Clin. Oncol. 34, 4277–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, and Sabatini DM (2008). Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707. [DOI] [PubMed] [Google Scholar]
- Huang WYC, Alvarez S, Kondo Y, Kwang Lee Y, Chung JK, Monatrice Lam HY, Biswas KH, Kuriyan J, and Groves JT (2019). A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, and Dezfouli S. (2004). Functions of Myc:Max in the control of cell proliferation and tumorigenesis. Int. Rev. Cytol. 238, 183–226. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, and Jülicher F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Iconaru LI, Ban D, Bharatham K, Ramanathan A, Zhang W, Shelat AA, Zuo J, and Kriwacki RW (2015). Discovery of small molecules that inhibit the disordered protein, p27 Kip1. Sci. Rep. 5, 15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, and Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, and Carr AM (2016). DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer 16, 35–42. [DOI] [PubMed] [Google Scholar]
- Jiang S, Fagman JB, Chen C, Alberti S, and Liu B. (2020). Protein phase separation and its role in tumorigenesis. Elife 9, e60264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Han T, Yao Y, Alessi AF, Freeberg MA, Inoki K, Klionsky DJ, Kim JK, Jin M, Klionsky DJ, et al. (2017). Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep. 20, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, O’connor RS, Kawalekar OU, Ghassemi S, and Milone MC (2018). CAR T cell immunotherapy for human cancer. Science 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Kaehler C, Isensee J, Hucho T, Lehrach H, and Krobitsch S. (2014). 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Res. 42, 6436–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata K, Kanbayashi S, Honda M, Itoh Y, Takahashi H, Kameda T, Nagatsugi F, and Takahashi S. (2020). Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci. Rep. 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber ER, and Lowe SW (2017). Putting p53 in context. Cell 170, 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Yang YS, Sutter BM, Wang Y, McKnight SL, and Tu BP (2019). Redox state controls phase separation of the yeast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell 177, 711–721.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, and Altmeyer M. (2019). Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38, e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Je EM, Oh JE, Yoo NJ, and Lee SH (2013). Mutational and expressional analyses of SPOP, A candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS 121, 626–633. [DOI] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. (2011). Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall’Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, et al. (2020). Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosin A, Oltsch F, Harmon T, Honigmann A, Jülicher F, Hyman AA, and Zechner C. (2020). Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468. [DOI] [PubMed] [Google Scholar]
- Kohoutek J. (2009). P-TEFb- the final frontier. Cell Div. 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel BA, Rongish BJ, Czirok A, Zach J, Little CD, Davis EC, Knutsen RH, Wagenseil JE, Levy MA, and Mecham RP (2006). Elastic fiber formation: a dynamic view of extracellular matrix assembly using timer reporters. J. Cell. Physiol. 207, 87–96. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Kalyana-Sundaram S, and Chinnaiyan AM (2015). Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 7, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttler F, and Mai S. (2006). c-Myc, genomic instability and disease. In Genome and Disease, Volff J-N, ed. (KARGER; ), pp. 171–190. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. (2014). Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, and McKnight SL (2013). Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé DP, and Brown M. (2018). Transcriptional regulation in prostate cancer. Cold Spring Harb. Perspect. Med. 8, a030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, and de Thé H. (2010). PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1asuggests a role for phase separation in heterochromatin. Nature 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, Fan Y, Shaw TI, Silveira AB, Qu C, et al. (2019). Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell 35, 140–155.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. (2014). Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos C, Schulze L, Weiske J, Meyer H, Braeuer N, Barak N, Eberspächer U, Werbeck N, Stresemann C, Lange M, et al. (2020). Identification of small molecules that modulate mutant p53 condensation. iScience 23, 101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL (2020). Poly(ADP-ribose): a dynamic trigger for biomolecular condensate formation. Trends Cell Biol. 30, 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, and Kroemer G. (2019). Biological functions of autophagy genes: a disease perspective. Cell 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JMM, Towers CG, and Thorburn A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-CC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, and Zhu YS (2018). MA-LAT1: a potential biomarker in cancer. Cancer Manag. Res. 10, 6757–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Coffey EL, Dall’Agnese A, Hannett NM, Tang X, Henninger JE, Platt JM, Oksuz O, Zamudio AV, Afeyan LK, et al. (2020). MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 586, 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV, and Locasale JW (2016). The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-H, Lin C, Tanaka H, Fero ML, and Eisenman RN (2009). Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS One 4, e7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, and Young RA (2012). Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Currie SL, and Rosen MK (2017). Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs Downloaded from. J. Biol. Chem. 46, 19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, and Dunker AK (2006). Intrinsic disorder in transcription factors †. Biochemistry 45, 6873–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li HB, and Flavell RA (2018). cGAS activation in phased droplets. Cell Res. 28, 967–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Takai K, Weaver VM, and Werb Z. (2011). Extracellular Matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 3, a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR, et al. (2016). Cancer: histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEwan SR, and Chilkoti A. (2010). Elastin-like polypeptides: biomedical applications of tunable biopolymers. Biopolymers 94, 60–77. [DOI] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén-Boixet J, Franzmann TM, et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboubi H, and Stochaj U. (2017). Cytoplasmic stress granules: dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta Mol. Basis Dis.1863, 884–895. [DOI] [PubMed] [Google Scholar]
- Manasanch EE, and Orlowski RZ (2017). Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lee L, Sallan SE, Silverman LB, et al. (2014). An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, and Lisanti MP (2017). Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, and White E. (2007). Role of autophagy in cancer. Nat. Rev. Cancer 7, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo SJ (2010). Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 14, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, and Penn LZ (2008). Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990. [DOI] [PubMed] [Google Scholar]
- Min J, Wright WE, and Shay JW (2019). Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 33, 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza-Aghazadeh-Attari M, Mohammadzadeh A, Yousefi B, Mihanfar A, Karimian A, and Majidinia M. (2019). 53BP1: a key player of DNA damage response with critical functions in cancer. DNA Repair (Amst) 73, 110–119. [DOI] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, and Kriwacki RW (2016). Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5, e13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, Phillips AH, Park C-G, Deniz AA, and Kriwacki RW (2018). Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 9, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal P, and Roberts CWM (2020). The SWI/SNF complex in cancer— biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MA, and Shilatifard A. (2015). Chromatin signatures of cancer. Genes Dev. 29, 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiznieks LD, Sharpe S, Pomès R, and Keeley FW (2018). Role of liquid– liquid phase separation in assembly of elastin and other extracellular matrix proteins. J. Mol. Biol. 430, 4741–4753. [DOI] [PubMed] [Google Scholar]
- Mulcahy Levy JM, and Thorburn A. (2020). Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ.27, 843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam A, Tighiouart M, Ryden L, Joseph L, Landberg G, Saxena NK, and Sharma D. (2012). Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis 33, 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, Wang S, Suter T, Alshareedah I, Gamliel A, et al. (2019). Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 26, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, and Halazonetis TD (2010). Genomic instability an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228. [DOI] [PubMed] [Google Scholar]
- Neupane M, Clark AP, Landini S, Birkbak NJ, Eklund AC, Lim E, Culhane AC, Barry WT, Schumacher SE, Beroukhim R, et al. (2016). MECP2 is a frequently amplified oncogene with a novel epigenetic mechanism that mimics the role of activated RAS in malignancy. Cancer Discov. 6, 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. (2012). c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen AI, Partanen JI, and Klefstrom J. (2007). Cell cycle c-Myc blazing a trail of death: coupling of the mitochondrial and death receptor apoptosis pathways by c-Myc. Cell Cycle 6, 2464–2472. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, and Cleveland JL (2003). Myc pathways provoking cell suicide and cancer. Oncogene 22, 9007–9021. [DOI] [PubMed] [Google Scholar]
- Nusse R, and Clevers H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999. [DOI] [PubMed] [Google Scholar]
- Oshidari R, Huang R, Medghalchi M, Tse EYW, Ashgriz N, Lee HO, Wyatt H, and Mekhail K. (2020). DNA repair by Rad52 liquid droplets. Nat. Commun. 11, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JD, Zhao A, Ellington AD, and Marcotte EM (2012). Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu. Rev. Cell Dev. Biol. 28, 89–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, and Rosen MK (2016). Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, and Bowie AG (2013). Immune sensing of DNA. Immunity 38, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan BS, Perera SA, Piesvaux JA, Presland JP, Schroeder GK, Cumming JN, Wesley Trotter B, Altman MD, Buevich AV, Cash B, et al. (2020). An orally available non-nucleotide STING agonist with antitumor activity. Science 369, eaba6098. [DOI] [PubMed] [Google Scholar]
- Parker MW, Bell M, Mir M, Kao JA, Darzacq X, Botchan MR, and Berger JM (2019). A new class of disordered elements controls DNA replication through initiator self-assembly. Elife 8, e48562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore F, and Levine RL (2016). Epigenetic regulators and their impact on therapy in acute myeloid leukemia. Haematologica 101, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HOO, Jawerth L, Maharana S, Jahnel M, Hein MYY, Stoynov S, Mahamid J, Saha S, Franzmann TMM, et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, and Hyman AA (2017). ATP as a biological hydrotrope. Science 356, 753–756. [DOI] [PubMed] [Google Scholar]
- Pavlova NN, and Thompson CB (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peran I, and Mittag T. (2020). Molecular structure in biomolecular condensates. Curr. Opin. Struct. Biol. 60, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Schindler J, Kohl B, Schneider-Heieck K, Adak V, Delezie J, Maier G, Sakoparnig T, Vargas-Fernández E, Karrer-Cardel B, Ritz D, et al. (2020). RNA-bound PGC-1α controls gene expression in liquid-like nuclear condensates. BioRxiv. 10.1101/2020.09.23.310623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, et al. (2019). Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 21, 1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CW, and Ayer DE (2011). An extended Myc network contributes to glucose homeostasis in cancer and diabetes. Front. Biosci. 16, 2206–2223. [DOI] [PubMed] [Google Scholar]
- Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, and Kingston RE (2019). Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey AE, Holehouse AS, and Pappu RV (2018). Phase separation of intrinsically disordered proteins. Methods Enzymol. 611,1–30, Academic Press Inc. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, and Mattos C. (2012). A comprehensive survey of ras mutations in cancer. Cancer Res. 72, 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouteau M, and Loewith R. (2018). Regulation of cellular metabolism through phase separation of enzymes. Biomolecules 8, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouteau M, Desfosses A, Sieben C, Bourgoint C, Mozaffari NL, Demurtas D, Mitra AK, Guichard P, Manley S, and Loewith R. (2017). TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Nature 550, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie A-M, Melton DW, Jullien D, and Adachi Y. (2005). 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 118, 2043–2055. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, and Spiegelman BM (1999). Activation of PPARgcoactivator-1 through transcription factor docking. Science 286, 1368–1371. [DOI] [PubMed] [Google Scholar]
- Rai AK, Chen JX, Selbach M, and Pelkmans L. (2018). Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216. [DOI] [PubMed] [Google Scholar]
- Reichheld SE, Muiznieks LD, Keeley FW, Sharpe S, and Weitz DA (2017). Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl. Acad. Sci. U S A 114, E4408–E4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyasen GW, Yao Y, Zhang J, Dulak A, Castriotta L, Jacques K, Zhao W, Gharahdaghi F, Hattersley MM, Lyne PD, et al. (2018). BRD4 amplification facilitates an oncogenic gene expression program in high-grade serous ovarian cancer and confers sensitivity to BET inhibitors. PLoS One 13, e0200826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, and Drummond DA (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, and Brangwynne CP (2020). Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, and Jaffrey SR (2019). m6A enhances the phase separation potential of mRNA. Nature 571, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Riggelen J, Yetil A, and Felsher DW (2010). MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309. [DOI] [PubMed] [Google Scholar]
- Roden C, and Gladfelter AS (2020). RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, and Poirier GG (2010). PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. (2009). The role of Myc-induced protein synthesis in cancer. Cancer Res.69, 8839–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JW, Nouri M, and Balk SP (2019). Androgen receptor interaction with mediator complex is enhanced in castration-resistant prostate cancer by cdk7 phosphorylation of med1. Cancer Discov. 9, 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR (2020). Biomolecular condensates and gene activation in development and disease. Dev. Cell 55, 84–96. [DOI] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Co-activator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, and Young RA (2020). Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari MS, Wang Z, Tailor K, Kolomeisky AB, Conrad JC, and Vekilov PG (2019). Anomalous dense liquid condensates host the nucleation of tumor suppressor p53 fibrils. iScience 12, 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, and Pandolfi PP (2002). The role of PML in tumor suppression. Cell 108,165–170. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Ciriello G, Sander C, Schultz N, Chatila WK, Luna A, La KC, Dimitriadoy S, et al. (2018). Oncogenic signaling pathways in the cancer genome atlas article oncogenic signaling pathways in the cancer genome atlas. Cell 173, 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapio L, Di Maiolo F, Illiano M, Esposito A, Chiosi E, Spina A, and Naviglio S. (2014). Targeting protein kinase a in cancer therapy: an update. EXCLI J.13, 843–855. [PMC free article] [PubMed] [Google Scholar]
- Sawyers C. (2004). Targeted cancer therapy. Nature 432, 294–297. [DOI] [PubMed] [Google Scholar]
- Schmidt HB, and Görlich D. (2015). Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like perm-selectivity. Elife 4, e04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, and Görlich D. (2016). Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 41, 46–61. [DOI] [PubMed] [Google Scholar]
- Schulze A, and Harris AL (2012). How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491, 364–373. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DAK, Tönjes M, et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231. [DOI] [PubMed] [Google Scholar]
- Secombe J, Pierce SB, and Eisenman RN (2004). Myc: a weapon of mass destruction. Cell 117, 153–156. [DOI] [PubMed] [Google Scholar]
- Sever R, and Brugge JS (2015). Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 5, a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaraiah RC, Veronese A, Sabbioni S, and Negrini M. (2018). Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. 419, 167–174. [DOI] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, and MacRae IJ (2018). Phase transitions in the assembly and function of human miRISC. Cell 173, 946–957.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, and Brangwynne CP (2018). Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin VY, Chen J, Cheuk IWY, Siu MT, Ho CW, Wang X, Jin H, and Kwong A. (2019). Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis.10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young RA, et al. (2019). Enhancer features that drive formation of transcriptional condensates. Mol. Cell75, 549–561.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, and Rout MP (2014). Cancer and the nuclear pore complex. Adv. Exp. Med. Biol.773, 285–307. [DOI] [PubMed] [Google Scholar]
- Singatulina AS, Hamon L, Sukhanova MV, Bouhss A, Lavrik OI, and Pastré D. (2019). PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 27, 1809–1821.e5. [DOI] [PubMed] [Google Scholar]
- Singh AM, and Dalton S. (2009). The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 5, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Kedersha N, and Elroy-Stein O. (2007). Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 27, 6639–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, and Smith HC (1986). Redistribution of U-snRNPs during mitosis. Exp. Cell Res.163, 87–94. [DOI] [PubMed] [Google Scholar]
- Sperling AS, Gibson CJ, and Ebert BL (2017). The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat. Rev.Cancer 17, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehelin D, Varmus HE, Bishop JM, and Vogt PK (1976). DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260, 170–173. [DOI] [PubMed] [Google Scholar]
- Strom AR, and Brangwynne CP (2019). The liquid nucleome – phase transitions in the nucleus at a glance. J. Cell Sci. 132, jcs235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, and Karpen GH (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, and Vale RD (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Wu R, Zheng J, Li P, and Yu L. (2018). Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, Zhen CY, Ma B, Wang H, and Ren X. (2019). Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 294, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K, Holzwarth B, Prentzell M, Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN, Kremmer E, et al. (2013). Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN (2011). Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 43, 1090–1103. [DOI] [PubMed] [Google Scholar]
- Vad-Nielsen J, and Nielsen AL (2015). Beyond the histone tale: HP1α deregulation in breast cancer epigenetics. Cancer Biol. Ther. 16, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia AM, and Kadoch C. (2019). Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell Biol. 21, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG (2011). Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10, 671–684. [DOI] [PubMed] [Google Scholar]
- Vellky JE, McSweeney ST, Ricke EA, and Ricke WA (2020). RNA-binding protein DDX3 mediates posttranscriptional regulation of androgen receptor: a mechanism of castration resistance. Proc. Natl. Acad. Sci. U S A 117, 28092–28101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon RMC, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, and Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, and Kinzler KW (2004). Cancer genes and the pathways they control. Nat. Med. 10, 789–799. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, and Kinzler KW (2013). Cancer genome landscapes. Science 340, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrhovski B, Jensen S, and Weiss AS (1997). Coacervation characteristics of recombinant human tropoelastin. Eur. J. Biochem. 250, 92–98. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Yamamura S, Malik S, Spiegelman BM, and Roeder RG (2003). Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12, 1137–1149. [DOI] [PubMed] [Google Scholar]
- Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, Joseph JK, Wang X, Loh YHE, Erb MA, et al. (2017). ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, Carroll TS, Li Y, Feng L, Chen G, et al. (2020). Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, and Aifantis I. (2020). RNA splicing and cancer. Trends Cancer 6, 631–644. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, et al. (2015). CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nie H, He X, Liao Z, Zhou Y, Zhou J, and Ou C. (2020). The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics 10, 11049–11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, and Jones DT (2004). Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645. [DOI] [PubMed] [Google Scholar]
- Wei MT, Chang YC, Shimobayashi SF, Shin Y, Strom AR, and Brangwynne CP (2020). Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 22, 1187–1196. [DOI] [PubMed] [Google Scholar]
- Wellenstein MD, and De Visser KE (2018). Review cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity 48, 399–416. [DOI] [PubMed] [Google Scholar]
- Wheeler RJ, Lee HO, Poser I, Pal A, Doeleman T, Kishigami S, Kour S, Anderson EN, Marrone L, Murthy AC, et al. (2019). Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. BioRxiv, 721001, 10.1101/721001. [DOI] [Google Scholar]
- Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, and Werb Z. (2020). Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 11, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters AC, and Bernt KM (2017). MLL-rearranged leukemias-an update on science and clinical approaches. Front. Pediatr. 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won JK, and Bakhoum SF (2020). The cytosolic DNA-sensing cGAS–sting pathway in cancer. Cancer Discov. 10, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Engelman JA, and Cantley LC (2010). Targeting the PI3K signaling pathway in cancer. Curr. Opin. Genet. Dev. 20, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SR, Corrales L, and Gajewski TF (2015). Innate immune recognition of cancer. Annu. Rev. Immunol. 33, 445–474. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Hyman AA, and Boke E. (2018). Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43, 81–94. [DOI] [PubMed] [Google Scholar]
- Wright PE, and Dyson HJ (2015). Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, and Chen ZJ (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. (2014). The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 46, 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Lama L, Adura C, Tomita D, Glickman JF, Tuschl T, and Patel DJ (2019). Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. U S A 116, 11946–11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Fujioka Y, Suzuki SW, Noshiro D, Suzuki H, Kondo-Kakuta C, Kimura Y, Hirano H, Ando T, Noda NN, et al. (2016). The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev. Cell 38, 86–99. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, and Hirose T. (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7. [DOI] [PubMed] [Google Scholar]
- Yan C, and Higgins PJ (2013). Drugging the undruggable: transcription therapy for cancer. Biochim. Biophys. Acta 1835, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, et al. (2020). Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300. [DOI] [PubMed] [Google Scholar]
- Yoo H, Triandafillou C, and Drummond DA (2019). Cellular sensing by phase separation: using the process, not just the products. J. Biol. Chem. 294, 7151–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio AV, Dall’Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, Coffey EL, Li CH, Oksuz O, Sabari BR, et al. (2019). Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell 76, 753–766.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, Quaranta E, Di Biagio D, Sigismondo G, Guzzardo V, et al. (2018). Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat. Med. 24, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Chen X, Guan D, Xu J, Wu H, Tong P, and Zhang M. (2018). Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell 174, 1172–1187.e16. [DOI] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, and Boutros M. (2017). Wnt signaling in cancer. Oncogene 36, 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, and Gladfelter AS (2015a). RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Boskovic Z, Hussain MM, Hu W, Inouye C, Kim HJ, Katherine Abole A, Doud MK, Lewis TA, Koehler AN, et al. (2015b). Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. Elife 4, e07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhao R, Tones J, Liu M, Dilley RL, Chenoweth DM, Greenberg RA, and Lampson MA (2020a). Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 31, 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JFJZJ, Lu TW, Stolerman LM, Tenner B, Yang JR, Zhang JFJZJ, Falcke M, Rangamani P, Taylor SS, Mehta S, et al. (2020b). Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 182, 1531–1544.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Narlikar GJ, and Kutateladze TG (2020c). Enzymatic reactions inside biological condensates. J. Mol. Biol. 10.1016/j.jmb.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H-H, Wu C, Sun Y, and Hu J. (2018). PML mutation in PML-Rarα alters PML nuclear body organization and induces ATRA resistance in acute promyelocytic leukemia. Blood 132, 3923. [Google Scholar]
- Zhu G, Xie J, Kong W, Xie J, Li Y, Du L, Zheng Q, Sun L, Guan M, Li H, et al. (2020). Phase separation of disease-associated SHP2 mutants underlies MAPK hyperactivation. Cell 183, 490–502.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]