Antiviral Actions of Interferons (original) (raw)
Abstract
Tremendous progress has been made in understanding the molecular basis of the antiviral actions of interferons (IFNs), as well as strategies evolved by viruses to antagonize the actions of IFNs. Furthermore, advances made while elucidating the IFN system have contributed significantly to our understanding in multiple areas of virology and molecular cell biology, ranging from pathways of signal transduction to the biochemical mechanisms of transcriptional and translational control to the molecular basis of viral pathogenesis. IFNs are approved therapeutics and have moved from the basic research laboratory to the clinic. Among the IFN-induced proteins important in the antiviral actions of IFNs are the RNA-dependent protein kinase (PKR), the 2′,5′-oligoadenylate synthetase (OAS) and RNase L, and the Mx protein GTPases. Double-stranded RNA plays a central role in modulating protein phosphorylation and RNA degradation catalyzed by the IFN-inducible PKR kinase and the 2′-5′-oligoadenylate-dependent RNase L, respectively, and also in RNA editing by the IFN-inducible RNA-specific adenosine deaminase (ADAR1). IFN also induces a form of inducible nitric oxide synthase (iNOS2) and the major histocompatibility complex class I and II proteins, all of which play important roles in immune response to infections. Several additional genes whose expression profiles are altered in response to IFN treatment and virus infection have been identified by microarray analyses. The availability of cDNA and genomic clones for many of the components of the IFN system, including IFN-α, IFN-β, and IFN-γ, their receptors, Jak and Stat and IRF signal transduction components, and proteins such as PKR, 2′,5′-OAS, Mx, and ADAR, whose expression is regulated by IFNs, has permitted the generation of mutant proteins, cells that overexpress different forms of the proteins, and animals in which their expression has been disrupted by targeted gene disruption. The use of these IFN system reagents, both in cell culture and in whole animals, continues to provide important contributions to our understanding of the virus-host interaction and cellular antiviral response.
INTRODUCTION TO THE INTERFERON SYSTEM
Interferon (IFN) was discovered as an antiviral agent during studies on virus interference (180, 294). Isaacs and Lindenmann reported in 1957 that influenza virus-infected chick cells produced a secreted factor that mediated the transfer of a virus-resistant state active against both homologous and heterologous viruses (180). This seminal observation, along with similar findings described by Nagano and Kojima in 1958 (294), set the stage for subsequent studies that led to the elucidation of the IFN system in exquisite detail.
What is the IFN system? How do IFNs function to inhibit the multiplication of some, but not all, viruses? What strategies are used by viruses to counteract the antiviral actions of IFNs? Considerable progress has been made toward answering these and other questions about IFNs and their effects on the virus-host interaction. Furthermore, IFNs were approved as therapeutics and moved from the basic research laboratory to the clinic. Advances made while elucidating the IFN system contributed significantly to our understanding in multiple areas of mammalian cell biology and biochemistry, ranging from pathways of signal transduction to the biochemical mechanisms of transcriptional and translational control to the molecular basis of viral pathogenesis.
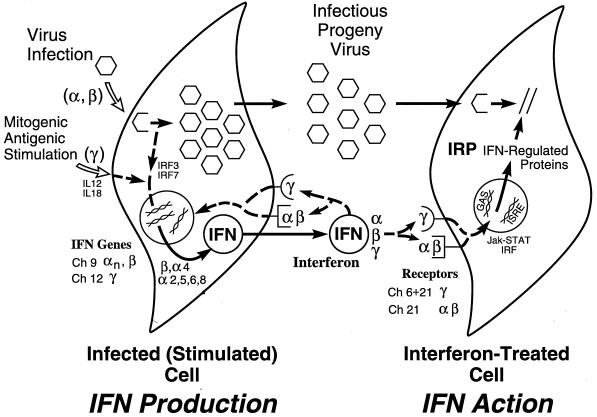
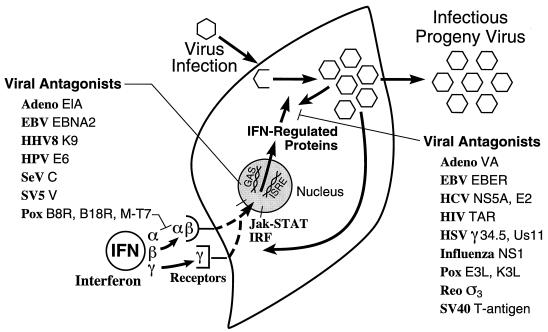
Several of the key features of the human IFN system are summarized in Fig. 1. The IFN system includes cells that synthesize IFN in response to an external stimulus such as viral infection and cells that respond to IFN by establishing an antiviral state (318, 351, 394). Animal viruses are inducers of IFN, and are also sensitive to the antiviral actions of IFNs. Some animal viruses also encode products that antagonize the IFN antiviral response. IFN proteins display autocrine as well as paracrine activities. The IFN response represents an early host defense, one that occurs prior to the onset of the immune response. IFNs possess a wide range of biological activities in addition to the characteristic antiviral activity by which they were discovered (36). This review will focus primarily on the antiviral activities of IFNs. However, IFN cytokines affect a number of other processes including those regulating cell growth, differentiation, and apoptosis, as well as the modulation of the immune response.
FIG. 1.
Schematic summary of the IFN system. Virion particles are illustrated as open hexagons, and IFN proteins are illustrated as open circles. The IFN-producing cell shown on the left depicts a cell induced to synthesize IFN in response to either virus infection (IFN-α and IFN-β) or antigen or mitogen stimulation (IFN-γ). The IFN-treated cell shown on the right depicts paracrine IFN action in a cell induced to synthesize IFN-regulated proteins that collectively constitute the antiviral response that is responsible for the inhibition of virus multiplication. IFN may also act in an autocrine manner on the IFN producer cell. The numbers refer to the chromosome (Ch) assignment of the human genes encoding the IFNs and their receptors. Adapted from reference 354 with permission of the publisher.
INTERFERON GENES AND PROTEINS
IFNs are a multigene family of inducible cytokines (40, 91, 340, 394, 443). They possess antiviral activity (318, 349, 394). Indeed, the biological activity of IFN is most commonly assayed by determining the antiviral activity in cell culture, although radioimmunoassays and enzyme-based immunoassays are also available for IFNs (318). IFNs are commonly grouped into two types (36, 110, 351). Type I IFNs are also known as viral IFNs and include IFN-α (leukocyte), IFN-β, (fibroblast), and IFN-ω. Type II IFN is also known as immune IFN (IFN-γ). The viral IFNs are induced by virus infection, whereas type II IFN is induced by mitogenic or antigenic stimuli. Most types of virally infected cells are capable of synthesizing IFN-α/β in cell culture. By contrast, IFN-γ is synthesized only by certain cells of the immune system including natural killer (NK) cells, CD4 Th1 cells, and CD8 cytotoxic suppressor cells (10, 443). The natural IFN-α-producing cells appear to be precursor dendritic cells (114, 381). Purified CD4+CD11c− type 2 dendritic cell precursors (pDC2s) from human blood produce up to 103 times more IFN in cell culture than do other blood cells following microbial or viral challenge (381). IFN-α genes can be divided into two groups: an immediate-early response gene (IFN-α4), which is induced rapidly and without the need for ongoing protein synthesis, and, a set of IFN-α genes, consisting of IFN-α2, IFN-α5, IFN-α6, and IFN-α8, that display delayed induction and are synthesized more slowly and require protein synthesis (256).
The large number of viral IFN genes in the human include 13 IFN-α genes, 1 IFN-β gene, and 1 IFN-ω gene (340). They all lack introns and are clustered on the short arm of chromosome 9 in the human and chromosome 4 in the mouse. The single IFN-γ gene possesses three introns and maps to the long arm of chromosome 12 in the human, and chromosome 10 in the mouse. Although some IFNs are modified posttranslationally by N- and O-glycosylation, the major human IFN-α subspecies are not glycosylated. The IFN-α gene products appear to function as monomers, whereas IFN-β and IFN-γ appear to function as homodimers (10, 318, 351, 394). It is not known why there are so many IFN-α genes. When the single IFN-β gene is deleted from chromosome 4 by targeted disruption, the resultant mice are highly susceptible to viral infection (82). The IFN-α subspecies do not compensate for the loss of IFN-β, suggesting a unique role for IFN-β that is essential for a fully effective antiviral response.
SIGNAL TRANSDUCTION AND ACTIVATION OF TRANSCRIPTION OF INTERFERON-INDUCIBLE GENES
IFN Receptors
IFNs exert their actions through cognate cell surface receptors that are largely species specific (10, 165, 287, 318). The alpha, beta, and omega IFNs appear to have a common receptor consisting of two subunits, IFNAR-1 and IFNAR-2. Both IFNAR-1 and IFNAR-2 map to chromosome 21 in the human, and chromosome 16 in the mouse. There is a single form of the IFNAR-1 subunit. However, alternative processing of the IFNAR-2 gene transcript produces long (2c), short (2b), and soluble (2a) forms of the encoded subunit (287, 327). IFN-γ binds to a receptor distinct from that used by IFN-α/β. Two kinds of subunits also constitute the IFN-γ receptor complex. The IFN-γ ligand-binding IFNGR-1 subunit and the accessory IFNGR-2 subunit map to chromosomes 6 and 21 in the human and chromosomes 10 and 16 in the mouse, respectively (10). IFN signaling involves an IFN-mediated heterodimerization of the cell surface receptor subunits, IFNAR-1 and IFNAR-2 with IFN-α/β and IFNGR-1 and IFNGR-2 with IFN-γ (10, 12, 287, 379a, 394).
Pathogenesis studies with knockout mice in which the IFN-α/β receptor function has been eliminated by targeted gene disruption illustrate the central importance of the IFN response in virus-host interactions (198). IFN-α/β receptor-null mice are unable to establish an antiviral state, demonstrating that IFN-α/β appears to be of particular importance in the host response to viral pathogens. IFN-α/β receptor-deficient mutant animals are highly susceptible to infection by an array of different viruses exemplified by members of the Poxviridae (vaccinia virus), Arenaviridae (lymphocytic choriomeningitis virus), Rhabdoviridae (vesicular stomatitis virus [VSV]), and Togaviridae (Sindbis virus, Semliki Forest virus, and Venezuelan equine encephalitis virus) despite the presence of an otherwise intact immune system and a normal resistance to the microbial pathogen Listeria monocytogenes (128, 146, 177, 290, 345, 416). By contrast, IFN-γ receptor-deficient mice in which either the IFNGR-1 or IFNGR-2 gene has been disrupted are greatly impaired in their ability to resist a variety of microbial pathogens and some viral pathogens including vaccinia virus and herpes simplex virus (10, 48, 176, 244, 290). Thus, the IFNAR and IFNGR gene knockout studies established that the viral IFN and immune IFN systems are functionally nonredundant. Interestingly, for IFN antiviral responses involving IFN-γ and its receptor, IFNGR knockout mutant mice lacking the receptor are more susceptible to both HSV-1 and vaccinia virus challenge than are mutant mice lacking the gene for the IFN-γ ligand (48). Although animals with IFNGR gene disruptions show no overt defects in embryonic development, they have severe immune system defects (10, 47, 48).
IFN-γ plays an important role in both innate and adaptive immunity (36, 42). It stimulates innate cell-mediated immunity through NK cells; it stimulates specific cytotoxic immunity based on the recognition of cell surface-bound viral antigens expressed in association with major histocompatibility complex (MHC) proteins; and it activates macrophages (10, 42). These immune responses play an important role in the antiviral and antimicrobial actions of IFN-γ. Th cells can be divided into three classes based on the pattern of cytokines produced following activation by antigens and mitogens. In addition to IFN-γ, Th1 cells produce interleukin-2 (IL-2) and tumor necrosis factor beta (TNF-β), Th2 cells produce IL-4 and IL-5, and Tr cells produce IL-10 (56, 260). Cellular immunity mediated by Th1 cells and humoral immunity mediated by Th2 cells are modulated by IFN-γ, which affects the differentiation of naive T cells into either Th1 or Th2 cells (36). IL-12 and IL-18 are IFN-γ-inducing cytokines; IL-12 induction of IFN-γ is dependent on caspase-1 processing of the IL-18 precursor protein (106, 435).
While IFN-γ possesses unique immunoregulatory activities that are especially important in the innate host response to microbial infections, it also plays a role in mediating protection against viral infection, especially long-term control of viral infections (48, 176, 244, 290). Double-knockout mice lacking both the IFN-γ receptor and the IFN-α/β receptor are especially sensitive to viral infection (416). Finally, while studies of mice made deficient in components of the IFN receptors by targeted gene disruption have firmly established the importance of the IFN system in the host antiviral response to a wide range of different RNA and DNA viruses (48, 128, 146, 176, 290, 416), the role of IFNs as inhibitors of the replication do vary with the animal system examined (7, 354, 368). This is illustrated by some members of the Reoviridae, including rotaviruses and orthoreoviruses. Studies of mutant mice deficient in either the IFN-α/β receptor or IFN-γ suggest that neither IFN-α/β nor IFN-γ responses play a major role in the clearance of primary rotavirus infection or affect the duration of rotavirus-induced disease in suckling mice (7). However, treatment of calves with recombinant IFN gave substantial protection from bovine rotavirus-induced diarrhea (368). Differences in IFN-inducing capacity between virus strains may be an important parameter contributing to the host response to viral infection by members of the Reoviridae as well as by other viruses (354). For example, reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than with the generation of infectious virus in cardiac myocytes; it has been speculated that the IFN-inducing capacity of reoviruses may determine in part their potential for causing virus-induced acute myocarditis (376). IFN-γ production can also be induced by reovirus infection, but the capacity of the serotype 1 Lang strain to induce IFN-γ in peripheral lymph node lymphocytes is dependent on the mouse strain; lymphocytes from C3H mice produce significantly higher levels of IFN-γ than do those from BALB/c, C57BL/6, and B10.D2 mice (253). In mouse fibroblasts in culture, serotype 3 is a better inducer of IFN-α/β than is serotype 1 (374); curiously, serotype 3 virus also is more cytopathic in culture than is serotype 1 virus (291).
JAKs, STATs, and IRFs
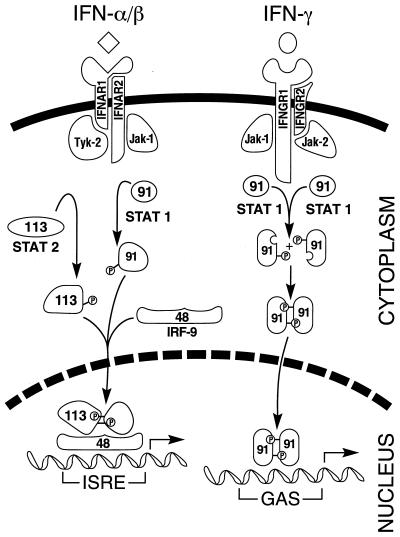
IFN-mediated signaling and transcriptional activation of cellular gene expression are best understood in the context of JAK-STAT pathway proteins (75, 232, 362, 363, 394). The principal components of the pathway are summarized in Fig. 2. The signal transducer and activator of transcription (STAT) family of proteins are latent cytoplasmic transcription factors that become tyrosine phosphorylated by the Janus family of tyrosine kinase (JAK) enzymes in response to cytokine stimulation. There are seven known members of the STAT protein family, Stat-1, Stat-2, Stat-3, Stat-4, Stat-5a, Stat-5b, and Stat-6, and four members of the JAK family, Jak-1, Jak-2, Jak-3, and Tyk-2. Different members of the JAK and STAT families have distinct functions in cytokine signaling. Receptor-associated JAKs are activated following binding of IFNs to their cognate multi-subunit transmembrane receptor. Of the known JAKs and STATs, the Jak-1, Jak-2, and Tyk-2 kinases and the Stat-1 and Stat-2 transcription factors play central roles in mediating IFN-dependent biological responses, including induction of the antiviral state (74, 169, 231, 394).
FIG. 2.
Schematic of IFN signaling by the Jak-Stat pathway. The signaling process is initiated by binding of the IFN ligand to its cognate receptor subunits. IFN-α and IFN-β are depicted by the diamond, and IFN-γ is depicted by the sphere. IFN binding leads to activation of overlapping pairs of Jak and Stat transcription factors by tyrosine phosphorylation. The Jak-1 and Tyk-2 kinases are activated by IFN-α/β, which leads to the phosphorylation and dimerization of the Stat-1 (p91) and Stat-2 (p113) proteins and subsequent translocation, along with IRF-9 (p48), to the nucleus. The complex of these three proteins, known as IFN-stimulated gene factor 3 (ISGF-3), activates the transcription of IFN-α/β–inducible genes through the ISRE. The Jak-1 and Jak-2 kinases are activated by IFN-γ, which leads to the phosphorylation and homodimerization of the Stat-1 protein and subsequent translocation to the nucleus. The Stat-1 dimer complex, known as GAF for gamma activation factor, activates the transcription of IFN-γ inducible genes through the GAS enhancer element.
Overlapping subsets of JAKs are involved in signaling by the two types of IFNs. Jak-1 and Tyk-2 kinases function in IFN-α/β signaling, and the Jak-1 and Jak-2 kinases function in IFN-γ signaling (10, 287). Tyk-2 interacts with the IFNAR-1 receptor subunit, and Jak-1 interacts with the IFNAR-2 subunit of the IFN-α/β receptor. Jak-1 also interacts with the IFNGR-1 receptor subunit, and Jak-2 interacts with the IFNGR-2 subunit of the IFN-γ receptor. Activation of the receptor-associated JAKs leads to the subsequent phosphorylation of latent cytoplasmic STAT transcription factors. For IFN-α/β, the phosphorylated forms of Stat-1α/β and Stat-2, along with an additional non-STAT protein, p48 (also known as IRF-9), translocate to the nucleus and constitute a complex known as ISGF-3. The ISGF-3 trimeric complex binds to a _cis_-acting DNA element, designated ISRE, found in IFN-α/β-inducible genes. For IFN-γ, the phosphorylated Stat-1α factor homodimerizes, translocates to the nucleus, and binds to a different _cis_-acting element, designated the gamma-activated sequence (GAS), that is commonly found in IFN-γ-inducible genes (11, 15, 232).
Targeted disruption of the Jak-1, Jak-2, Stat-1, and Stat-2 genes in mice have firmly established the obligatory physiological roles of the encoded Jak-1 and Jak-2 kinases and the Stat-1 transcription factor in the IFN response (103, 169, 278, 307, 327, 341, 362). Disruption of the mouse Stat-1 gene revealed an unexpected physiologic specificity. Stat-1 null mice show a greatly reduced responsiveness to both IFN-α and IFN-γ and show a compromised innate immunity to viral disease (103, 278). Stat-1-deficient mice are extremely susceptible to viral pathogens. However, if reared in a pathogen-free environment, mice lacking Stat-1 are fertile and show no overt developmental abnormalities. Consistent with the biochemical evidence, Stat-2 null mice are defective primarily in their response to IFN-α/β (327, 362). The Jak-1 gene disruption results in mice that are runted at birth and typically die within 24 h; mutant mouse embryo fibroblasts (MEF) lacking the Jak-1 kinase are unresponsive to either IFN-α or IFN-γ (341). The Jak-2 gene disruption is embryonic lethal on day 12 to 13; mutant MEF lacking the Jak-2 kinase are not responsive to IFN-γ but remain responsive to IFN-α (307).
The IFN regulatory factor (IRF) family of transcriptional regulators, like the STATs, are important regulatory factors in the IFN response (299). The IRF-1 protein was first identified as a regulator of the IFN-α/β gene promoter, as well as the IFN-stimulated response element (ISRE) found in the promoters of some IFN-α/β-regulated genes (363). The IRF family of factors includes nine known members: IRF-1, IRF-2, IRF-3, IRF-4, IRF-5, IRF-6, IRF-7, IRF-8, and IRF-9 (p48, ISGF-3γ). These factors are homologous to each other in the N-terminal region, corresponding to their conserved DNA-binding domain. The IRFs and STATs can function in conjunction with each other to establish the signal transduction and gene regulation events that lead to the induced expression of the proteins that collectively constitute the antiviral state. IRF-9 was initially identified as a component of the trimeric ISGF-3 complex, along with Stat-1 and Stat-2. It is the only component of the ISGF-3 complex for which a nuclear localization signal has been identified (222), and it is the DNA sequence recognition subunit of ISGF-3 (185). The Stat-2 protein appears to form a cytoplasmic complex with IRF-9 that is retained in the cytoplasm in the absence of IFN treatment. The Stat protein subunits are localized in the cytoplasm in untreated cells but rapidly translocate to the nucleus following IFN treatment. IRF-9, on the other hand, is found in both the nucleus and cytoplasm of untreated and IFN-treated cells. Preassociation of IRF-9 with Stat-2 in the cytoplasm has the potential to poise the complex for activation by dimerization with Stat-1, as a rapid response to signaling by IFNs-α/β. IRF-9 also appears to play an ISGF-3-independent role in responses mediated by IFN-γ (195). IRF-9 and IRF-1 play essential but nonredundant roles in the IFN response. IRF-1 binds directly to the ISRE found in the promoters of some IFN-α/β-regulated genes (299) and plays an important role in the antiviral actions of IFN (146, 265, 337).
Most, but not all, of the activities of IFN-γ are mediated through the Stat-1 protein target of the JAK-STAT signaling pathway. IFN-γ treatment leads to the tyrosine phosphorylation of the IFNGR-1 receptor subunit on Tyr-440 and the subsequent interaction and phosphoryation of Stat-1α at Tyr-701. Dimerization of Stat-1α through reciprocal SH2 interactions is followed by nuclear translocation and then DNA binding at GAS elements to activate transcription of IFN-γ-inducible genes (169, 232, 362). Serine phosphorylation of the transcriptional activation domain of Stat-1α at Ser-727, positioned within the C-terminal region of Stat-1, enhances transcription by recruitment of p300/CBP, a ubiquitously expressed global transcriptional coactivator with histone acetyltransferase (HAT) activity (397, 446), and also BRCA1 (306). The BRCA1 tumor suppressor acts in concert with Stat-1 to differentially activate the transcription of a subset of IFN-γ-regulated target genes involved in growth control, including the cyclin-dependent kinase inhibitor p21WAF1. The possible roles of these interactions in the IFN-mediated inhibition of virus multiplication, by affecting responses associated with cell growth and proliferation, are unknown.
STAT factors in addition to Stat-1 and Stat-2, namely, Stat-3, Stat-4, Stat-5, and Stat-6, have also been observed to be activated by IFNs (107, 274, 332). Furthermore, although IFN-mediated signaling and transcriptional activation is presently best understood in the context of the JAK-STAT signal transduction pathway, additional pathways of signal transduction possibly involving the RNA-dependent protein kinase (PKR) (212), mitogen-activated protein kinase (78, 141), and phosphatidylinositol 3-kinase (319) may also be operative under some circumstances. Both IFN-γ and IFN-αβ can regulate the expression of IFN-responsive genes by a Stat1- and PKR-independent alternative signaling mechanism that requires both the IFN-γ receptor and Jak-1 kinase (140a, 330a). Surprisingly, some genes are activated by IFN-γ only when Stat1 is absent, a finding that may be of special significance in those viral infections where Stat1 function is antagonized (330a). Thus, the possibility of an elaborate network that conceivably allows for considerable cross talk between separate pathways involved in IFN signal transduction and the actions of IFNs must be considered.
_cis_-Acting DNA Elements
Two _cis_-acting DNA elements are the known targets of the Stat and IRF transcription factors: ISRE and GAS. ISRE and GAS are largely responsible for the IFN-regulated promoter activity seen for IFN-inducible genes. The ISRE drives the expression of genes inducible by IFN-α/β and has the consensus sequence AGTTTCNNTTTCNPy. It is the binding site for the IFN-stimulated gene factor ISGF-3 and for some of the IRFs (74, 231, 394). Indeed, the crystal structure of an IRF-DNA complex reveals the DNA recognition sequence AANNGAAA and cooperative binding of IRF-2 to a tandem repeat of the GAAA core sequence (118). A novel 15-bp element, designated KCS (for “kinase-conserved sequence”), also is required both for basal and IFN-inducible activity of the promoter for the RNA-dependent protein kinase PKR. The KCS element, GGGAAGGCGGAGTCC, is exactly conserved in sequence and position between the human and mouse Pkr promoters and is the binding site for a protein complex including the Sp1 factor (207, 210).
The GAS element was initially identified as the IFN-γ-responsive DNA element. However, in addition to Stat-1 homodimers that bind DNA at the GAS site, a number of other Stat proteins activated by various cytokines have been shown to bind GAS-like elements as either homo- or heterodimers, including Stat-3, Stat-4, Stat-5, and Stat-6 (74, 169, 231, 394). GAS-like elements have a palindromic core sequence, TTNNNNNAA (10, 362, 394).
Interestingly, the ISRE is not limited to IFN-inducible cellular genes. The Q promoter (Qp) of Epstein-Barr virus (EBV) and the enhancer-1 region of hepatitis B virus (HBV) have an ISRE-like element, vISRE, that is the target of IRF factors (297, 447). Qp is used for the transcription of EBV nuclear antigen 1 (EBNA1) during the highly restricted type I latent infection but is inactive in type III latency. Constitutive activation of EBV EBNA1 gene transcription is mediated in part by IRF-1 activation of Qp. A different IRF factor, IRF-7, binds to the vISRE-like sequence of Qp and represses transcriptional activation by both IFN and IRF-1. Expression of IRF-7 is high in type III latency cells but almost undetectable in type I latency, corresponding to the activity of the Q promoter of EBV in these latency states (447). The EBV latency promoter is positively regulated by Stat factors, and Zta interference with signaling leads to a loss of promoter activity (64). IFN-α treatment suppresses the activity of the hepatitis B virus enhancer-1, which possesses an ISRE-like element that is bound by both IRF-1 and IRF-9. Mutation of the HBV vISRE reduces the suppressive effect of IFN-α, whereas overexpression of IRF-9 enhances the inhibition of enhancer-1 activity in human hepatoma HuH7 cells (297).
Viruses and Double-Stranded RNA as Inducers
Virus infection activates the transcription of a large number of cellular genes (58, 198, 451). The activation may occur either directly through activation of cellular transcription factors such as IRF-3 or indirectly through prior induction of IFN-α/β. IRF-3, a key transcriptional activator affected by viral infection, is a subunit of the double-stranded RNA (dsRNA)-activated transcription factor complex (DRAF) (167, 425). It is constitutively expressed in many cells and tissues. It is directly activated by dsRNA or by virus infection and subsequently plays a role in the transcriptional activation of the IFN-α and IFN-β promoters and IFN-α/β-responsive genes (299, 361, 440). Activation of IRF-3 involves protein serine/threonine phosphorylation, which results in its cytoplasmic-to-nuclear translocation, stimulation of DNA binding, and association with p300/CBP coactivator, leading to increased transcriptional activation (167, 213, 371). Along with IRF-3, IRF-7 probably plays important roles in the regulation of IFN-β synthesis in response to virus infection (424). IRF-7 is also a critical determinant for the induction of IFN-α genes in infected cells and functions in part by a positive feedback induction loop mechanism (256, 438).
Results of analyses of genetically altered cell lines suggest that signal transduction by dsRNA is mediated by ISREs without activation of ISGF-3. The IFN signaling pathway components Jak-1, Tyk-2, IRF-9, and Stat-2, necessary for ISGF-3 activation, are not absolutely required for dsRNA-mediated signal transduction (15). Interestingly, virus infection mediates the induction of the IFN-inducible P56 protein in mutant P2.1 cells even though both dsRNA and IFN signaling are nonfunctional in these cells (155). The biochemical nature of the pathway components implicated in direct induction of IFN-inducible genes by virus in the P2.1 cells in the absense of functional dsRNA and IFN signaling pathways have not yet been defined.
Cellular Negative Regulators of IFN Signaling
Negative regulators of JAK-STAT-mediated signaling and gene activation have been identified. Among these are both viral and cellular proteins which down-regulate JAK-STAT-mediated signals. The family of suppressors of cytokine signaling (SOCS)/STAT-induced STAT inhibitors (SSI)/ cytokine-inducible SH2 protein (CIS) represent cellular proteins that function as negative-feedback regulators of JAK-STAT signaling (295, 395). On the basis of two characteristic conserved domains, a central SH2 domain and the so-called SOCS box at the carboxyl end, there are eight known members (295, 437). SOCS-1 is induced by numerous cytokines including IFN-γ, binds to the kinase domain of all four members of the JAK family of kinases, and inhibits signaling by suppression of Jak activation (104, 295). Consequently, SOCS-1 inhibits the tyrosine phosphorylation and nuclear translocation of Stat-1 in response to both IFN-α and IFN-γ. SOCS-1, but not SOCS-2, inhibits the antiviral and antiproliferative activities of IFNs in cell lines stably expressing the SOCS proteins (388). Targeted gene disruption of SOCS-1 suggests that the most important physiological function of this signaling suppressor is the negative regulation of IFN-γ signaling and Stat-1 function (6, 437).
Two members of the protein inhibitor of activated Stat (PIAS) family of cellular proteins, PIAS-1 and PIAS-3, are also negative regulators of STAT signaling. However, the mechanism is different from that of SOCS, which inactivates the Jak kinases. The PIAS-1 and PIAS-3 proteins directly associate with Stat-1 and Stat-3, respectively, in response to treatment with IFNs or IL-6 (380). PIAS-1 specifically interacts with the Stat-1 dimer, but not monomer, to block the DNA-binding activity and thus the Stat-1-mediated gene activation (234, 380).
Negative regulation of JAK-STAT signal transduction is also achieved by dephosphorylation catalyzed by the protein tyrosine phosphatase SHP-1. SHP-1 suppresses the signal transduction process of a variety of cytokines, including IFN-α (77), by directly interacting with JAKs and catalyzing their dephosphorylation. A dominant negative form of SHP-2 phosphatase suppresses ISGF-3-dependent transcription (79), suggesting that SHP-2 phosphatase, in contrast to SHP-1, plays a role in IFN-α/β-induced gene expression.
Viral Antagonists of IFN Signaling
Both DNA and RNA viruses encode proteins that impair the activity of the JAK-STAT signaling pathway (198). Multiple mechanisms appear to be involved. Among these is mimicry. Several examples exist in which viruses encode products that mimic cellular components of the IFN signal transduction pathway. This molecular mimicry can lead to an antagonism of the IFN signaling process and subsequent impairment of the development of an antiviral state. Poxviruses, for example, encode soluble IFN receptor homologues (vIFN-Rc). These vIFN-Rc homologues are secreted from poxvirus-infected cells and bind IFNs, thereby preventing them from acting through their natural receptors to elicit an antiviral response (385). M-T7, the first poxvirus receptor homologue identified, was found in myxoma virus-infected cells and acts as a decoy to inhibit the biological activity of rabbit IFN-γ. The vIFN-γRc M-T7 gene product is a critical virulence factor for poxvirus pathogenesis. Other poxviruses also encode soluble IFN-γRc homologues, including vaccinia virus, where the B8R gene encodes the IFN-γ receptor homologue. A vIFN-α/βRc protein is secreted by vaccinia virus and several additional orthopoxviruses. The vIFN-α/β receptor homologue, the B18R gene product in the Western Reserve strain and the B19R product in the Copenhagen strain, binds several different IFN-α subspecies as well as IFN-β and blocks IFN-α/β signaling activity (5, 71).
Three additional DNA viruses that affect IFN signaling are adenovirus, papillomavirus, and human herpesvirus 8 (HHV-8). The adenovirus E1A protein blocks IFN-mediated signaling at a point upstream of the activation of ISGF-3. The DNA binding activity of ISGF-3 is inhibited by E1A (230). Overexpression of the IRF-9 subunit of ISGF-3 restores IFN responses and the transcription of ISRE-driven genes in adenovirus-infected cells (230). Sendai virus (SeV), a paramyxovirus that replicates in the cytoplasm of the host, circumvents the IFN-induced antiviral response by interfering with the transcriptional activation of IFN-inducible cellular genes. The C proteins of SeV play an essential role (130); infections with two different C gene mutants of SeV eliminate the ability to prevent the establishment of an antiviral state against vesicular stomatitis virus (131). Impairment of the IFN-induced antiviral response appears to be a key determinant of SeV pathogenicity (93, 131).
Human papillomavirus (HPV) E6 oncoprotein binds selectively to IRF-3 but only very poorly to other cellular IRFs including IRF-2 and IRF-9. Association of E6 with IRF-3 inhibits transactivation, thereby providing HPV with a mechanism to circumvent the antiviral response (343). Adenovirus E1A protein also inhibits IRF-3-mediated transcriptional activation by a mechanism dependent on the ability of E1A to bind p300. CBP/p300 and PCAF histone acetyltransferase (HAT) enzymes are coactivators for several transcription factors including Stat-1α; the viral E1A protein represses HAT activity and inhibits p300-dependent transcription and nucleosomal histone modifications by PCAF (57).
HHV-8, a gammaherpesvirus associated with Kaposi's sarcoma, synthesizes an IRF homologue (vIRF) that functions as a repressor of transcriptional activation induced by IFN-α/β and IFN-γ (127, 167). The HHV-8-encoded vIRF protein also represses IRF-1-mediated transcriptional activation. HHV8 vIRF probably plays an important role in HHV-8 pathogenesis and neoplastic transformation by antagonizing IFN- and IRF-mediated transcriptional control. Expression of vIRF antisense in HHV-8-infected cells increases IFN-mediated transcriptional activation and downregulates the expression of HHV-8 genes. Two other herpesviruses, varicella-zoster virus (VZV) and cytomegalovirus (CMV), also disrupt the function of the JAK-STAT signal transduction pathway (1, 283). VZV inhibits the expression of Stat-1 and Jak-2 proteins but has little effect on Jak-1; the expression of two key transcription factors regulated by IFN-γ signaling, IRF-1 and the MHC class II transactivator (CTIIA), is inhibited in VZV-infected cells, as is the induction of the MHC class II molecules (1). A different strategy of antagonism occurs in CMV-infected cells, where MHC class II expression also is inhibited. There is a specific decrease in the level of Jak-1 due to enhanced protein degradation in CMV-infected fibroblasts (283).
Several nonsegmented negative-strand RNA viruses encode gene products that antagonize IFN receptor-mediated signaling from both type I α/β and type II γ IFN receptors. For example, infection with simian virus 5 or mumps virus leads to an increased proteosome-mediated degradation of Stat-1 (94) whereas in cells infected with parainfluenza virus type 2 there is a degradation of Stat-2 (442). In the case of Sendai virus, the C proteins interfere with IFN action in at least two ways. C proteins prevent the synthesis of Stat-1 and they also induce an increased turnover of Stat-1 (130, 201). The VP35 protein of Ebola virus, a negative-strand RNA virus, functions as a type I IFN antagonist although the precise biochemical mechanism of the antagonism has not yet been defined (21). VP35 inhibits virus induction of the IFN-β promoter and dsRNA- and virus-mediated activation of ISRE-driven gene expression (21).
INTERFERON-INDUCED PROTEINS AND THEIR ANTIVIRAL ACTIVITIES
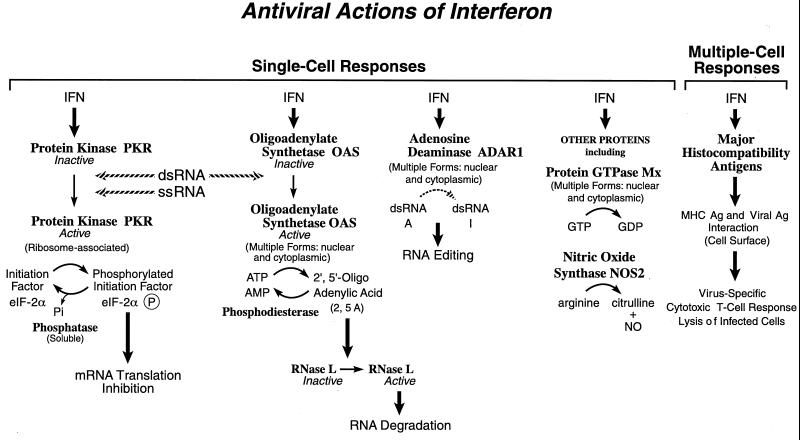
The replication of a wide range of different DNA and RNA animal viruses is inhibited by IFN, both in cell culture and in animals (318, 351, 394). For many of the viruses that are sensitive to the antiviral action of IFN in cell culture systems, the primary step of the virus multiplication cycle inhibited typically is the synthesis of viral polypeptides (36, 351). Exceptions do exist, however. Papovaviruses and certain retroviruses often are inhibited in IFN-sensitive cell systems at early and late steps, respectively, of their multiplication cycles (36, 349, 351), myxoviruses and rhabdoviruses may be inhibited in certain cell types at or before primary transcription (158, 393), and adenoviruses and poxviruses can be comparatively resistant to the antiviral actions of IFN because of virus-encoded antagonists (264, 351). Among the IFN-induced proteins implicated in the antiviral actions of IFNs in virus-infected cells are PKR, the 2′,5′-oligoadenylate synthetase (OAS) and RNase L, the RNA-specific adenosine deaminase (ADAR), and the Mx protein GTPases. Double-stranded RNA plays a central role in modulating protein phosphorylation, RNA degradation, and RNA editing catalyzed by the IFN-inducible enzymes: the PKR kinase, the OAS synthetases, and the ADAR1 deaminase (181, 353). dsRNA interestingly also serves as a template for gene silencing (25). IFN also induces a form of nitric oxide synthase (iNOS2) and the MHC class I and II molecules, all of which play important roles in immune responses to infections.
Protein Kinase PKR
PKR is an IFN-inducible, RNA-dependent protein kinase (68, 352) known in the earlier literature as DAI; dsI; P1 kinase; P1/eIF-2α kinase; p65, p67, or TIK (for the mouse enzyme); and p68 or p69 (for the human enzyme) (69). In IFN-treated cells, PKR is found predominantly in the cytoplasm and associated with ribosomes (318, 352, 412); however, small amounts of PKR have also been localized to the nucleus by cell fractionation and immunofluorescence analyses (183, 412). As summarized in the Fig. 3 schematic, PKR is activated by autophosphorylation, a process mediated by RNA with double-stranded character (68, 328, 352). Following activation, PKR catalyzes the intermolecular phosphorylation of at least six protein substrates: the PKR protein itself (410, 411); the α subunit of protein synthesis initiation factor 2, eIF-2α (348); the transcription factor inhibitor IκB (211, 303); the Tat protein encoded by human immunodeficiency virus (HIV) (272); the 90-kDa NFAT protein (220); and the M-phase specific dsRNA-binding phosphoprotein MPP4 (310).
FIG. 3.
Functions of selected IFN-inducible proteins. Among the IFN-induced proteins believed to affect virus multiplication within single cells are PKR kinase, which inhibits translation initiation through the phosphorylation of protein synthesis initiation factor eIF-2α; the OAS synthetase family and RNase L nuclease, which mediate RNA degradation; the family of Mx protein GTPases, which appear to target viral nucleocapsids and inhibit RNA synthesis; and ADAR, which edits double-stranded RNA by deamination of adenosine to yield inosine. IFN-induced expression of MHC class I and class II antigens and NOS may contribute to the antiviral responses observed within whole animals. Ag, antigen. Adapted from reference 351 with permission of the publisher.
Protein synthesis factor eIF-2 so far is the best characterized of the PKR substrates, in both structural and functional terms. Serine phosphorylation of eIF-2α catalyzed by PKR occurs at Ser-51 (311, 348). This phosphorylation of eIF-2α leads to an inhibition of translation by impairing the eIF-2B-catalyzed guanine nucleotide exchange reaction (68, 125, 352, 366). A variety of physiologic conditions, including IFN treatment and virus infection (351, 352, 355), cause the phosphorylation state of eIF-2α to increase and mRNA translation to subsequently decrease. Activation of PKR and subsequent phosphorylation of eIF-2α change the translational pattern of the host cell.
Human PKR is a 551-amino-acid protein with a molecular mass of about 62 kDa as deduced from the cDNA open reading frame (ORF) (209, 279, 352, 412). By contrast, the mouse and rat PKR proteins are somewhat smaller, 515 amino acids (178, 401) and 514 amino acids (276), respectively. The 11 conserved catalytic subdomains characteristic of protein serine/theonine kinases are, without exception, located in the C-terminal half of PKR (352). Replacement of the subdomain II-invariant Lys-296 with arginine (K296R) eliminates both autocatalytic and eIF-2α protein kinase activities of PKR (17, 409). The N-terminal region of the PKR protein possesses a repeated motif (dsRBM, or R), whose core is about 20 amino acid residues and is responsible for the dsRNA-binding activity (108, 144, 189, 269, 309). Mutational analyses established that the N-terminal proximal copy of dsRBM is both necessary and sufficient for the RNA-binding activity of PKR, although both copies of dsRBM are required for optimal kinase activity (145, 268, 273, 352).
The RNA-dependent autoactivation of PKR involves autophosphorylation of PKR (68, 188, 350, 352, 370, 387). Biochemical and genetic studies suggest that the autophosphorylation can occur by either intramolecular (31, 120) or intermolecular (204, 342, 410, 411) mechanisms. There are multiple sites of autophosphorylation on PKR, predominantly serine residues but also including threonine residues (120, 221, 348). Phosphopeptide analysis suggests a multiple of about four major phosphorylation sites per PKR molecule (411). No PKR phosphorylation on tyrosine is detectable (178, 221).
A number of RNA effectors of PKR function, that is, RNA activators and RNA inhibitors, have been identified. RNA activators include both synthetic and natural dsRNA, for example (rI)_n_-(rC)n and reovirus genome dsRNA, respectively (352). Undistorted A-form dsRNA has its sequence-rich information buried in the major groove, and indeed, no sequence specificity has been observed in interactions between PKR and dsRNA per se (68, 352). Certain highly structured single-stranded viral RNA species are also activators, as exemplified by HIV TAR RNA, reovirus s1 mRNA, and hepatitis delta virus RNA (67, 351). RNA inhibitors of PKR autophosphorylation include dsRNA at high concentration and also three highly structured viral single-stranded RNA (ssRNA) species: adenovirus VAI RNA, EBV EBER RNA, and HIV TAR RNA (68, 264, 351, 352). Synthetic aptamer RNAs selected from a library of ∼1014 RNA sequences containing a randomized region of 50 nucleotides (nt) include both activators and inhibitors of PKR autophosphorylation and eIF-2 phosphorylation (33).
The basis of the RNA selectivity of PKR activation remains an important question. While kinase activation is associated with the formation of a stable PKR dsRNA complex that requires ∼30 to 50 bp of duplex RNA and is optimal with about 80 bp, the PKR protein appears to interact with as little as 11 bp of dsRNA (32, 254). Both the full-length PKR protein and the truncated N-terminal region of the PKR protein containing the two copies of the dsRBM motif bind VAI RNA (267) and an 85-bp dsRNA (365) with comparable affinity, with an apparent KD between 2 and 4 nM. Mutations in PKR that impair RNA binding have similar effects on the binding of both activator and inhibitor RNAs (144, 267, 268). This suggests that the discrimination between activator and inhibitor RNAs presumably takes place after RNA binding. Curiously, the cellular protein activator PACT (308a) and the carbohydrate heparin (136, 170) can substitute for RNA in mediating the autophosphorylation and activation of PKR. Heparin oligosaccharide with 8 sugar residues is nearly as efficient as heparin with 16 residues in activating PKR, whereas heparin with 6 residues is a very poor activator (136). The RNA-binding activity of wild-type PKR is not competed by heparin, and PKR mutants that fail to bind to and be activated by dsRNA can be activated by heparin and by PACT (136, 309).
Southern blot and nucleotide sequence analyses are consistent with a single Pkr gene, and fluorescence in situ hybridization shows that genomic clones colocalize to human chromosome 2p21-22 and mouse chromosome 17E2 (16, 208, 390). The human Pkr gene consists of 17 exons and spans about 50 kb (208), whereas the mouse gene is 16 exons and spans about 28 kb (401). The organization of the regulatory and catalytic subdomains of the PKR protein are remarkably preserved between the human and mouse Pkr genes; the amino acid junction positions for 13 of the 15 protein coding exons are exactly conserved (208).
The TATA-less Pkr promoter possesses an ISRE as well as a novel 15-nt KCS element, which so far has been identified only in the human and mouse Pkr promoters (206, 401). KCS affects basal as well as IFN-inducible expression of PKR (207, 210). IFN-α is an efficient inducer of two Pkr mRNAs in human cells, of ∼2.5 and ∼6.0 kb (279, 412), but IFN-γ is a relatively poor inducer of them (412). Tissue-specific differences in the ratios of the three PKR transcripts are observed in mice; for example, a 2.5-kb mRNA is the predominant species in testes, but in both lung and heart tissues a 4.0-kb species is predominant over the 2.5- and 6.0-kb mRNAs (178). The molecular basis and functional significance of the three forms of PKR mRNA transcripts remain unresolved. For human PKR, alternative exon 2 splice variants with different translational activity and abundance have been found in placental tissue, but their relationship to the 2.5- and 6.0-kb transcripts has not been defined (190). In addition to the regulation of PKR by IFN-inducible transcriptional activation, the synthesis of PKR in transfected mammalian cells is autoregulated primarily at the level of translation by a mechanism dependent on catalytically active PKR (20, 409).
Several studies establish that changes in protein phosphorylation mediated by the IFN-inducible PKR play an important role in the antiviral actions of IFNs as well as the control of cell growth mediated by IFNs (68, 228, 352, 356). Evidence for the involvement of the IFN-inducible PKR in the antiviral actions of IFN and the control of translation in virus-infected cells comes from three types of analyses: (i) the study of virus replication in mammalian cells expressing PKR cDNAs (29, 224, 281, 293); (ii) the analysis of mutant mice and MEF deficient in PKR by targeted disruption of the Pkr gene (2, 13, 436; L. Basu and C. E. Samuel, unpublished data); and (iii) the analysis of virus-encoded inhibitors of the PKR kinase (188, 264, 350, 387). For example, the replication of encephalomyocarditis (EMC) virus (281), HW (29, 293), vaccinia virus (224), and VSV (398; Basu and Samuel, unpublished) is reduced in cell culture by overexpression of the cDNA encoding wild-type PKR but not by expression of the catalytic subdomain II point mutant PKR (1–551)K296R, which lacks kinase activity. Furthermore, in replication-competent HIV-1, chimeric genomes that express the wild-type PKR but not the K296R mutant in place of nef inhibited their own expression in cis and pNL4-3 in trans (29). A systematic analysis designed to identify the precise point(s) of virus replication blocked in transfected cells overexpressing wild-type PKR cDNA, similar to earlier studies carried out with cells treated with purified cloned IFNs (262, 415), has not been reported yet. Mutant Pkr0/0 mice with a targeted disruption in the N-terminal region of PKR that deletes the methionine start codon and RNA-binding motif I show a reduced antiviral response to EMC virus induced by IFN-γ (436). In mutant Pkr0/0 mice with the disruption in the catalytic domain, the antiviral responses to influenza virus and vaccinia virus are normal (2). However, these mutant mice lacking PKR are predisposed to lethal intranasal infection by VSV (13), and MEF derived from these knockout mice treated with recombinant IFN-α display a reduced antiviral state against VSV compared to those from wild-type parental mice (398; Basu and Samuel, unpublished). Tat-mediated activation of transcription factor NF-κB and transcriptional induction of the HIV-1 long terminal repeat LTR also are impaired in mouse cells in which the Pkr gene is knocked out. Both functions are restored by cotransfection of Tat with the cDNA for PKR (81). The HSV ICPγ34.5 gene product mediates neurovirulence in the mouse model by antagonizing the function of PKR. ICPγ34.5 is not required for HSV-1 multiplication in nonneuronal cells in culture but is required for replication in the central nervous system. In mice with the PKR gene disrupted in the N-terminal RNA-binding regions, mutant HSV lacking ICPγ34.5 show wild-type replication and neurovirulence (226).
Adenovirus virus-associated (VA) RNA antagonizes the antiviral action of IFN by preventing the activation of PKR (264, 352). The growth of wild-type adenovirus that produces VA RNA is not inhibited by IFN, but IFN treatment inhibits the growth of adenovirus mutants that are unable to produce VA RNA (197). In the absence of functional VA RNA, adenovirus produces virus-specific RNAs that activate PKR (197, 264), which appears to be responsible for the observed inhibition of translation and the IFN sensitivity of VA mutant virus growth. Additional examples of escape strategies from the antiviral actions of IFN involving antagonism of PKR by virus-encoded gene products will be considered separately (see “Viral antagonists of IFN-induced proteins” below). While adenovirus VA RNA illustrates a well-defined example of how a single viral gene product can drastically affect IFN sensitivity of a virus by targeting a specific IFN-inducible protein (PKR), studies with other viral systems have also shown that the IFN response can differ markedly for a given type of virus. For example, the effect of IFN treatment on the multiplication of reovirus is dependent on the kind of host cell, the type of IFN, and the serotype of reovirus. Reovirus multiplication is sensitive to the antiviral action of IFN in some cell lines but is not significantly inhibited by IFNs in other lines (354). The principal step of reovirus macromolecular synthesis inhibited in reovirus-infected IFN-sensitive mouse fibroblast and monkey kidney cells is the translation of viral mRNA into viral protein. Analysis by two-dimensional isoelectric focusing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting reveals that the phosphorylation of eIF-2α is increased in IFN-treated cells following infection with reovirus (355).
In addition to playing a role in the antiviral actions of IFNs (36, 351), PKR is implicated in the control of cell proliferation (228, 356). For example, stable transformants of NIH 3T3 cells overexpressing catalytically inactive human PKR proteins display a transformed phenotype and are highly tumorigenic when injected into nude mice (19, 203, 280). Further evidence in support of the notion that PKR-mediated perturbation of the homeostatic balance of translation can cause malignant transformation comes from overexpression of either mutated eIF-2 which cannot be phosphorylated (100) or the 58-kDa cellular inhibitor which impairs PKR kinase activity (18). Overexpression of either the PKR inhibitory protein p58 (18) or the nonphosphorylatable mutant Ser51Ala eIF-2α (100) causes malignant transformation of NIH 3T3 cells. The tumor suppressor activity of IRF-1 also appears to be mediated, in part, by PKR (196). And in yeast, the wild-type but not mutant PKR cDNA mediates a growth suppression phenotype (65, 342). Somewhat unexpectedly, however, no evidence of tumor suppressor activity of PKR was observed in two independent mouse mutants devoid of functional PKR by targeted gene disruption (2, 436).
2′,5′-Oligoadenylate Synthetase and RNase L
The IFN-inducible 2-5A response leading to the degradation of RNA requires two enzymes, OAS and RNase L (Fig. 3). OAS catalyzes the synthesis of oligoadenylates of the general structure ppp(A2′p)_n_A, commonly abbreviated 2-5A. As their name implies, they possess a 2′,5′-phosphodiester bond linkage (192). RNase L, a latent endoribonuclease, becomes activated by binding 2-5A oligonucleotides. A third enzymatic activity, that of a phosphodiesterase, also is involved in the metabolism and action of 2-5A oligoadenylates (Fig. 3). The phosphodiesterase catalyzes the hydrolysis of oligonucleotides possessing 2′,5′-phosphodiester bonds, thereby attenuating the 2-5A response. The expression, regulation, and function of the OAS and the 2-5A-dependent RNase L has been characterized extensively in IFN-treated and virus-infected cells (191, 322, 334, 357, 394).
Three size forms of OAS, designated OAS1, OAS2, and OAS3 (334), have been identified in human cells by immunoblotting and by characterization of cDNA and genomic clones. OAS proteins of 40, 46, 69, 71, and 100 kDa are detectable by Western analysis (60, 255, 258). The 40- and 46-kDa forms of OAS, designated OAS1, are identical to each other over their N-terminal 346 amino acids but differ at their C-terminal regions. They are encoded by alternatively spliced 1.6- and 1.8-kb transcripts derived from a single gene (26, 174, 432). The differential splicing of OAS1 occurs between exons 5 and 6 (27, 359). The 69- and 71-kDa medium forms of human OAS, designated OAS2, likewise appear to be generated by differential splicing. The OAS2 isoforms, identical over their N-terminal 683 amino acids, consist of two adjacent domains each homologous to the 40-kDa isoform of OAS1 (255). Several RNA transcripts hybridize to OAS2 cDNAs, with sizes of 2.8, 3.3, 3.9, and 4.5 kb (258). The 100-kDa large isoform of OAS, designated OAS3, is a 1,087-amino acid protein as deduced from cDNA sequence; it is composed of three adjacent repeat domains, each homologous to OAS1 (215, 336). An IFN-inducible transcript of ∼7 kb encodes OAS3. Based on analysis of cDNA and genomic clones, the small (40- and 46-kDa), middle-sized (69-kDa), and large (100-kDa) OAS proteins are products of three distinct genes, clustered over ∼130 kb on human chromosome 12 in the region 12q24.2 (60, 174, 175, 255, 258). Thus, it appears that mammalian OAS genes underwent successive gene duplication events, resulting in the three sizes of enzymes containing one (OAS1), two (OAS2), or three (OAS3) homologous domains (215).
Oligomerization of OAS1 and OAS2 appears necessary for enzymatic activity (137, 358). In their native forms, the small OAS1 isoform exists as a ∼180-kDa tetramer, the middle-sized OAS2 isoform exists as a ∼160-kDa dimer, and the large OAS3 protein exists as a monomer (258). Mutations affecting a tripeptide sequence CFK impair subunit association and are characterized by a reduction in OAS enzymatic activity (137). The three different-sized forms of OAS are associated with different subcellular fractions, including membranes, cytoplasm, and nucleus; they differ in the concentration of dsRNA required for their activation; they differ in the reaction conditions required for their optimal enzymatic activity; and they differ in the size pattern of the 2-5A products produced (172, 173, 179, 334, 433). The full physiological significance of these differences is not yet known. While three forms of OAS are seen in human cells, the cDNAs so far isolated and characterized in other species, including mouse, rat, pig and chicken, appear to correspond most closely to the OAS1 form (334).
Although OAS proteins are activated by dsRNA, there is no obvious structural homology between the dsRNA-binding domains of the OAS proteins and those of PKR or ADAR (139, 269). However, like PKR and ADAR1 (68, 312, 352), the OAS enzymes possess separate subdomain regions responsible for their RNA-binding activity and for their catalytic activity (334, 357). OAS enzymes are activated during viral infection (334). Although in most instances the activator RNA has not been defined precisely, it is presumed to be of viral origin. Conceivably, viral “dsRNA” activators might include single-stranded transcripts which possess significant double-stranded character in addition to the obvious potential sources of dsRNA structures such as RNA duplexes as part of their replicative intermediates, RNA duplexes derived by symmetric transcription from opposing promoters, and RNA duplexes that are genomic dsRNAs released from unstable virions or subviral particles of Reoviridae family members. Two well-characterized viral RNAs that do affect OAS enzymatic activity are the HIV TAR RNA and adenovirus VA RNA. The TAR RNA sequence, present at the 5′-termini of HIV transcripts, forms a stable secondary structure and possesses an intrinsic ability to activate both OAS and PKR (250). By contrast, a mutant form of TAR RNA with disrupted secondary structure does not activate either IFN-induced enzyme. The activation of OAS and PKR by TAR RNA suggests a mechanism for the control of HIV replication by the IFN system (29, 250, 293). Curiously, adenovirus VA RNA that antagonizes PKR has just the opposite effect on the OAS; VA RNA can both bind and activate OAS (86). The ATP-binding domain of recombinant human 40-kDa OAS identified by photoaffinity labeling and sequence analysis includes Lys-K199 that is photolabeled with 8-azido-[α-32P]ATP; Lys-199 is present in a dodecapeptide sequence that is highly conserved among all OAS proteins as deduced from human, mouse, and rat cDNAs (202).
The induction of OAS has been extensively characterized in a variety of different human and mouse cell lines and tissues (36, 351, 394). Early studies revealed that the magnitude of induction is dependent on the type of IFN, the type of cell, and the growth state of the cell. For example, induction levels range from about 10-fold for human HeLa cells that often show a high basal enzyme level (11) to about 10,000-fold for chicken embryo cells that have a low basal enzyme level (14). The three different-sized forms of OAS are all induced by IFN-γ, IFN-α, and IFN-β (60, 433). The 5′-flanking region of the OAS1, OAS2, and OAS3 genes all contain an ISRE (28, 335, 422). However, the induction by IFN-γ, IFN-α, and IFN-β of different isoforms of OAS, as well as total OAS enzymatic activity, differs among human cell lines in vitro and in primary sources, for example among donors of normal peripheral blood mononuclear cells (433). This no doubt reflects differences in the organization of additional regulatory elements between the promoters and the abundance of the _trans_-acting factors in different types of cells and tissues. For example, the OAS3 promoter possesses elements conferring direct inducibility not only by IFNs but also by TNF and retinoic acid (335). Consensus binding sites for IRF family members are present within OAS promoters (63, 335). Ectopic expression of IRF-1 leads to activation of the OAS gene promoter, whereas expression of IRF-2 leads to repression of the OAS promoter (63).
RNase L, the endoribonuclease activated by 2-5A oligonucleotides, is also variously known in the earlier literature as the 2-5A-dependent RNase and RNase F (322). The presence of a functional 2-5A oligomer is required for the conversion of RNase L from an inactive monomeric form to the active dimeric form. The RNase L protein acquires endoribonuclease catalytic activity after binding 2-5A, a process associated with the formation of stable homodimers (97, 98, 160). The ability of 2-5A derivatives to activate RNase L correlates with their ability to mediate dimer formation (97). However, RNase L has also been reported as a dimer of regulatory and catalytic subunits (347), a state which possibly corresponds to a heteromeric complex of the RNase L protein with the RNase L inhibitor protein RLI (38). Activated RNase L catalyzes the degradation of both viral and cellular RNAs, including cellular rRNA, by cleaving on the 3′ side of -UpXp- sequences (113, 434).
The gene for RNase L, designated _RNS_4, maps to human chromosome 1q25 by fluorescence in situ hybridization (389). The mRNA transcript of the human _RNS_4 gene is about 5.0 kb as measured by Northern blot analysis; the protein-coding sequence constitutes only about 40% of the nucleotide sequence of the mRNA (450). The human RNase L protein is 741 amino acids, about 83 kDa as deduced from the cDNA sequence (450). Analysis of aligned human and mouse RNase L sequences suggests several intriguing features of the proteins. RNase L displays a similarity to RNase E, an RNase implicated in the control of mRNA stability in Eschericia coli (403, 450). The N-terminal half of RNase L possesses nine ankyrin-like domains that may be involved in protein-protein interactions (160). A duplicated phosphate-binding loop motif within the N-terminal half of RNase L functions in the binding of the 2-5A oligonucleotides as established from cDNA mutagenesis studies (160, 322). Antagonists that impair 2-5A-dependent RNase L activity have been described. These include a cellular protein of 68 kDa, designated RLI (38), and oligonucleotide derivatives of 2-5A that accumulate in certain types of virus-infected cells (55).
RNase L is constitutively present in most types of cells (112), although treatment with IFN-α/β enhances RNase L activity in some types of cells (111, 182). Subsequent Northern blot analysis established that IFN-α/β increases the steady-state amount of the RNase L transcript in mouse cells by about threefold (450). The RNase L inhibitor, RLI, is not regulated by IFN (38).
The availability of cDNA clones for OAS and for RNase L facilitated studies of the roles of these enzymes in biological processes including virus replication. Among the various families of animal viruses examined, the Picornaviridae shows the best correlation between activation of the 2-5A pathway and inhibition of virus replication (61, 70, 346, 349). Constitutive expression in hamster or mouse cells of cDNA clones encoding the small form of the OAS is sufficient to establish an antiviral state, and this state appears selective for picornaviruses (61, 70, 346). For example, Chinese hamster ovary cells constitutively expressing the 40-kDa form of the human OAS are resistant to infection by Mengo virus but not VSV or herpesviruses (61). Similar results are observed with human and mouse cells transfected to constitutively express a cDNA for OAS1. Human T98G cells that express the cDNA encoding the 40-kDa form of human OAS and mouse NIH 3T3 cells that express the cDNA encoding the 43-kDa form of murine OAS display a resistance to EMC virus replication but not to VSV replication (70, 346). The resistance to EMC virus replication correlates with the expression of OAS enzyme activity (70, 346). Likewise, mouse cells stably expressing either the 69- or 71-kDa isoform of human OAS2 show a partial antiviral response to EMC virus (257). Constitutive expression of the 69-kDa OAS2 in human HT1080 cells likewise causes inhibition of EMC virus replication but not of VSV, SeV, or reovirus replication (138). In agreement with the apparently selective inhibition of picornavirus replication observed for cells overexpressing the OAS cDNAs (61, 70, 346), overexpression of the RNase L inhibitor in stably transfected HeLa cells partially inhibits the antiviral activity of IFN against EMC virus but not against VSV (38). Likewise, overexpression of a dominant negative mutant of RNase L also antagonizes the antiviral activity of IFN against EMC virus (160). Conversely, treatment of cells with 2-5A oligomer molecules provides some protection against picornavirus infection (171, 334).
Targeted disruption of the RNase L gene effectively provides a functional knockout of the 2-5A pathway, because the only known activity of 2-5A is the activation of the latent RNase L protein. Studies carried out with mice with a homozygous RNase L−/− gene disruptions illustrate the importance of the 2-5A pathway in the antiviral actions of IFN. RNase L−/− mice die more rapidly in response to infections with EMC virus than do RNase L+/+ wild-type mice (448, 449). Furthermore, RNase L−/− MEF are defective in apoptotic responses (449). Mice triply deficient in RNase L, PKR, and Mx, generated by combination of the RNase L null with the PKR null in a Mx1−/− background, show an added deficiency in the antiviral response to EMC virus (448). However, a substantial residual antiviral response is observed in MEF lacking all three proteins, RNase L, PKR, and Mx1, further illustrating the redundant nature of pathways that collectively constitute the innate antiviral response of IFN-α/β (448). Somewhat surprising, the level of IFN-induced gene products even may be elevated in the absence of functional RNase L. Comparative kinetic analyses with RNase L−/− mutant and RNase+/+ wildtype MEF suggest an RNase L-dependent destablization of IFN-induced mRNAs, consistent with a possible role for the 2-5A system in the attenuation of the IFN response (233).
RNA-Specific Adenosine Deaminase ADAR1
Posttranscriptional RNA modifications such as deamination of adenosine to yield inosine provide an important mechanism by which the functional activity of viral and cellular RNAs can be altered and, hence, by which biological processes can be affected (22, 51, 252, 344, 384). One important RNA-editing enzyme, ADAR1, is an IFN-inducible RNA-specific adenosine deaminase (134, 312, 313). Several studies implicate the IFN-inducible ADAR1 deaminase in the editing of viral RNA transcripts and cellular pre-mRNAs. The biological importance of RNA editing in animal cells is significant and far ranging (22, 24, 51, 249, 384).
ADAR was first identified as a dsRNA-unwinding activity in Xenopus oocytes (23, 333). ADAR catalyzes the covalent modification of highly structured RNA substrates by hydrolytic C-6 deamination of adenosine to yield inosine. The resultant A-to-I transitions destabilize the dsRNA helix by disrupting base pairing; RNA becomes more single stranded in character because stable AU base pairs are changed to the considerably less stable IU pair (23, 419). Hypoxanthine, the base of the nucleotide inosine generated from adenosine by the deamination, is typically recognized as guanine by the translational and transcriptional machinery (4). Posttranscriptional conversion of adenosine to inosine has been demonstrated in both viral RNAs and nucleus-encoded cellular mRNAs.
The cDNA encoding the human ADAR1 deaminase (K88 cDNA) was isolated in a screen for IFN-regulated cDNAs (312). It hybridizes to a single major transcript of ∼7 kb in human cells (313). Accumulation of ADAR1 transcripts is increased about fivefold by IFN treatment (312, 313). Both IFN-α and IFN-γ induce ADAR1 transcript accumulation (313). As deduced from the cDNA ORF, human ADAR1 is a 1,226-amino-acid protein with a molecular mass of about 136 kDa (194, 312), but the protein migrates at about 150 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (312). Southern blot and sequence analyses are consistent with a single Adar1 gene. Genomic Adar1 clones colocalize to human chromosome 1q21.1-21.2 (423, 427) and mouse chromosome 3F2 (428) as shown by fluorescence in situ hybridization. Sequence analysis demonstrated that the human Adar gene consists of 16 exons and spans about ∼40 kbp (240, 423), including two alternative exon 1 structures (134, 135).
Two immunologically related forms of ADAR are found in human cells: an IFN-inducible ∼150-kDa protein (p150) and a constitutively expressed ∼ 110-kDa protein (p110). Cell fractionation and immunofluorescence studies localized the IFN-inducible p150 protein to both the cytoplasm and nucleus, whereas the constitutively expressed p110 protein is present predominantly if not exclusively in the nucleus (312). The predicted sequence of ADAR includes a putative bipartite nuclear localization signal (194, 302, 312), consistent with immunolocalization and biochemical studies (8, 39, 301). The mechanism of synthesis of the two proteins involves alternative exon 1A and 1B structures that initiate from different promoters, one IFN-inducible and the other not (134, 135). Exons 1A and 1B are spliced to exon 2 at precisely the same junction. The methionine initiation codon for the 1,226-amino-acid ORF specifying the inducible p150 is in exon 1A; the synthesis of exon 1A-containing transcripts is driven by an IFN-inducible promoter that possesses a consensus ISRE (134, 135). Exon 1B does not include an AUG initiation codon; an AUG codon within exon 2 initiates translation of the 931-amino-acid ORF encoding the constitutively expressed p110 protein.
An additional ADAR different from ADAR1 p150/p110, designated ADAR2, has been cloned (216, 275, 286). The 80-kDa ADAR2 protein is not inducible by IFN and does not cross-react immunologically with either the p110 or the p150 protein versions of ADAR1. The ADAR2 gene maps to human chromosome 21q22.3 (286), different from the 1q21 assignment of the inducible ADAR1 (423, 427).
The C-terminal region of the ADAR1 protein constitutes the catalytic domain of the deaminase (217, 237, 248). The nucleic acid-binding domains, for dsRNA and for Z-DNA, are located N-terminal of the catalytic domain. ADAR1 possesses, in the central region of the predicted ORF, three copies of the dsRNA binding motif, designated dsRBMI, dsRBMII, and dsRBMIII (166, 194, 240, 312), that are highly conserved among themselves and with respect to the dsRBM originally identified in the IFN-inducible PKR (144, 269). Mutational analyses show that the dsRBMIII motif of ADAR1 is essential for deaminase activity whereas RII is dispensable (217, 237, 240). Chimeric proteins in which the dsRBM of ADAR1 are replaced with those of PKR retain deaminase activity with synthetic dsRNA substrates but show dramatically reduced editing activity with natural RNA substrates (241). Furthermore, VAI and aptamer RNA antagonists of PKR significantly inhibit the deaminase activity of chimeric PKR-ADAR1 proteins but less so of wild-type ADAR1 (241). These results indicate that the dsRNA-binding motifs are functionally distinct from each other, as measured by binding substrates in a manner recognized by the enzyme catalytic center for deamination. Comparison of cDNA and genomic sequences revealed the exon location of the dsRBD domains within ADAR and PKR. Curiously, the intron phases prior to the three exons of the human Adar1 gene that contain the dsRBM copies, exons 3, 5, and 7, are all phase 2 (240, 423). Even more striking is the finding that this codon phasing observed for the human Adar gene dsRBM-motif exons is conserved exactly for the two R-motif exons of the mouse and human Pkr genes (208, 240, 423). With the IFN-inducible ADAR, dsRNA functions as the substrate for deamination and the dsRBMIII copy is of fundamental importance for catalytic activity (237, 240). With the IFN-inducible PKR kinase, dsRNA functions as an effector that mediates autophosphorylation and kinase activation (68, 188, 352).
ADAR1 also binds Z-DNA in addition to binding highly structured ssRNAs (166, 367). The Z-DNA-binding motif is repeated (Zα, Zβ) within the N-terminal region of ADAR (367). Z-DNA-binding activity is not required for either deaminase activity or dsRNA-binding activity, and, conversely, dsRNA binding activity is not required for Z-DNA-binding activity (235). Although two separate regions of ADAR, the dsRNA-binding motif and the Z-DNA-binding region, display high sequence similarity to the vaccinia virus E3L protein (312), the functional significance of this similarity remains to be established.
RNA editing by adenosine deamination is of major biological significance. RNA A-to-I editing produces RNA transcripts that differ from their template; I is recognized as G (4), not A, by polymerases and ribosomes. Thus, RNA modification has the potential to alter the protein-coding capacity of the edited transcript and the sequence of replicated RNAs. ADAR is implicated in two types of RNA-editing processes that are dependent on double-stranded regions within the substrate RNA (22, 24). First, A-to-I modifications are found at multiple sites in viral RNAs, as exemplified by the biased hypermutations observed in minus-strand RNA virus genomes during lytic and persistent infections. Second, the C-6 adenosine deamination catalyzed by ADAR can be highly site specific, occurring at one or a few sites, as exemplified by hepatitis delta virus (HDV) RNA and the GluR receptor channel and serotonin pre-mRNAs.
Extensive editing, referred to as hypermutation, was first observed in the matrix (M) protein of measles virus recovered from patients with subacute sclerosing panencephalitis (SSPE) and measles inclusion body encephalitis (51–53). Similar hypermutations with clustered U-to-C (A to G) conversions also have been observed for transcripts encoded by other viruses including parainfluenza virus type 3 (292), VSV (304), Borna disease virus (115), avian leukosis virus (157), and polyomavirus (214) in cell culture systems. For the minus-strand RNA viruses, the hypermutation mediated by adenosine deamination is proposed to represent a modification associated with persistence of infection (51, 292). For the dsDNA polyomavirus, the modification is proposed to represent a mechanism by which RNA transcripts expressed early after infection are inactivated after viral replication (214). Recent studies have identified a novel RNase specific for inosine-containing RNA (360), which may affect the stability of I-containing RNAs. A-to-I editing has also been described in the 5′-transactivating response region of HIV RNA transcripts after injection into Xenopus oocytes (372). However, similar editing changes have not yet been observed in HIV transcripts in patients, although other alterations have been seen (43).
In contrast to the extensive A-to-I (G) hypermutation editing observed for several viral RNAs (51–53, 157, 214, 292, 304), both viral and cellular RNA substrates are also known in which the conversion of adenosine to inosine by ADAR occurs with high selectivity at specific A positions (22, 252, 344). For example, in HDV, RNA editing plays an essential role in the production of two HDV proteins from one ORF; the two proteins have different functions in the life cycle of the circular ssRNA HDV. The synthesis of the large form of delta antigen occurs following selective conversion of an amber UAG termination codon to a UIG tryptophan codon, with the editing site present in the self-complementary dsRNA structure (218). The HDV editing occurs in the antigenomic RNA and requires formation of the dsRNA structure near the amber/w site (49, 323). Selective adenosine deamination is also observed with cellular RNAs. The conversion of adenosine to inosine at specific positions in neurotransmitter receptor pre-mRNAs by ADAR leads to specific amino acid substitutions that alter receptor function (248, 344). Adenosine deamination at the Q/R site in exon 11 and the R/G site in exon 13 leads to amino acid substitutions that decrease Ca2+ permeability and alter the kinetics of channel gating, respectively, of the glutamate GluR-B receptor subunit (243, 386). Likewise, for the serotonin 2C receptor pre-mRNA, three amino acid substitutions within exon 3 at the I/V, N/S, and I/V sites caused by selective RNA editing led to a decrease in G-protein coupling (45). A putative RNA duplex structure between the target exon and a complementary sequence in the adjacent downstream intron, referred to as the editing complementary sequence (ECS), is required for the specific editing of the GluR-B and serotonin 2C receptor pre-mRNAs (248, 344). The IFN-inducible ADAR1 catalyzes the A-to-I editing at the R/G site but not the Q/R site of GluR-B (238). For serotonin-2C pre-mRNA, only one of the sites is edited by ADAR1 (239). However, no information is yet available concerning a possible relationship between viral pathogenesis and the editing of the GluR-B and serotonin-2C neurotransmitter receptors. Glutamate neurotransmission involving _N_-methyl-d-aspartate receptors and neuronal NOS (nNOS) activity in part mediates neuronal DNA strand breaks and poly(ADP-ribose) polymerase (PARP) activity (320). PARP transfers ADP-ribose groups from NAD+ to nuclear proteins after activation by DNA strand breaks, and PARP overactivation can cause cell death by depletion of ATP. IFN-mediated increases in ADAR1 levels may potentially contribute to the glutamate toxicity, since A-to-I editing of the _N_-methyl-d-aspartate receptor pre-mRNA yields receptors subunits with amino acid substitutions which affect receptor function (238, 369).
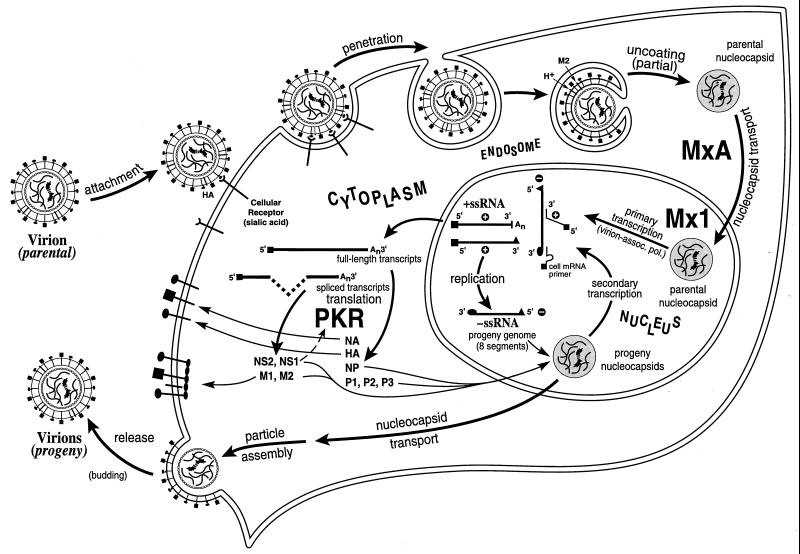
Protein Mx GTPase
Proteins MxA and Mx1 of the Mx family of proteins are possibly the best characterized of the known IFN-inducible gene products with antiviral activity, in the context of direct experimental evidence obtained from animal model studies which establish that Mx alone is sufficient to block the replication of virus in the absence of any other IFN-α/β-inducible proteins (8, 158). Mx proteins are GTPases that belong to the superfamily of dynamin-like GTPases (393, 417). The intrinsic GTPase activity of Mx proteins is required for their antiviral activity (329). The highly conserved tripartite GTP binding motif is present within the N-terminal region of the ∼70- to ∼80-kDa proteins (8, 158, 393). Mx proteins associate with themselves and, importantly, with viral protein complexes. The central and C-terminal regions of Mx play important roles in these protein-protein interactions (199, 325). Mx is inducible by IFN-α and IFN-β but not by IFN-γ (8, 382). A combination of genetic and biochemical evidence establishes that at least some of the Mx proteins, as illustrated by the human MxA protein, possess an intrinsic antiviral activity. The spectrum of antiviral activities of the Mx proteins, and the molecular mechanisms by which they act to inhibit virus replication, are dependent on the specific Mx protein, its subcellular site of localization, and the type of challenge virus examined.
Two members of the Orthomyxoviridae family, influenza virus and Thogoto virus, played pivotal roles in the discovery of IFN (180) and in the subsequent antiviral mechanism studies of the Mx proteins (199, 200, 426). The principal features of the influenza virus multiplication cycle essential to understanding this orthomyxovirus in studies of the antiviral actions of IFN and the biochemistry of Mx proteins are summarized in Fig. 4. Influenza virus virions are enveloped and possess a segmented, minus-strand ssRNA genome (198a). The viral envelope HA glycoprotein mediates binding to cellular sialic acid-containing receptors. Virion penetration occurs via receptor-mediated endocytosis and is followed by partial uncoating and release of the parental viral nucleocapsid into the cytoplasm. Acidification mediated by the viral M2 ion channel is essential for the early stage of infection by the influenza A viruses, whereas the NB and CM2 proteins may be the functional counterparts of M2 in influenza B and C viruses. The parental nucleocapsid is imported into the nucleus, where primary transcription catalyzed by the virion-associated polymerase (P1, P2, P3) occurs using 5′-capped structures derived from cellular mRNAs as primers. Each of the eight minus-strand RNA genome segments is transcribed into ssRNA of the polarity of mRNA. Influenza virus transcripts are processed and transported to the cytoplasm, where they are translated by the host cell protein-synthesizing machinery. Various coding strategies are used by the influenza viruses, including translation of full-length transcripts in a single reading frame, translation of spliced transcripts, and translation of two overlapping reading frames in a transcript. Primary gene expression from parental particles typically accounts for 5% or less of the total viral RNA and protein synthesized in an infected cell. Secondary gene expression from progeny ribonucleoprotein (RNP) particles accounts for the vast majority of the viral mRNA, and thus viral protein, found in influenza virus-infected cells. Newly synthesized progeny nucleocapsids may participate in further transcription or be exported to the cytoplasm, where particle assembly occurs at the plasma membrane, with release of the enveloped virions by a budding process.
FIG. 4.
Schematic diagram of the influenza virus multiplication cycle including sites of action of the IFN-induced Mx and PKR proteins. Enveloped influenza virus virion particles are depicted as spheres, in which the viral HA glycoprotein mediates binding to the cellular receptor. Virus penetration via receptor-mediated endocytosis is followed by partial uncoating and release of the viral nucleocapsid into the cytoplasm. The eight minus-strand RNA genome segments are shown as wavy lines in the nucleocapsid structures. The parental nucleocapsid is imported into the nucleus, where primary transcription is catalyzed by the virion-associated polymerase. Influenza virus transcripts are processed and transported to the cytoplasm, where they are translated by the host cell protein-synthesizing machinery. The IFN-induced PKR protein kinase that inhibits translation by phosphorylation of eIF-2, can be antagonized by the accumulated viral NS1 protein that binds dsRNA. The IFN-induced family of Mx antiviral proteins appear to act by affecting viral nucleocapsid transport and RNA synthesis. Human MxA accumulates in the cytoplasm, whereas mouse Mx1 accumulates in the nucleus of IFN-treated cells. The GTPase activity of Mx proteins is required for their antiviral activity. Newly synthesized progeny nucleocapsids may participate in further transcription or may be exported to the cytoplasm, where particle assembly occurs at the plasma membrane, with release of the enveloped progeny virions by a budding process.
The antiviral activity of IFN-induced Mx proteins is exerted at one of two steps to inhibit myxovirus multiplication. Either viral nucleocapsid transport or viral RNA synthesis is blocked, dependent on whether Mx is present in the cytoplasm or nucleus of the IFN-treated virus-infected host cell (8, 158, 393, 426). The human genome contains two related Mx proteins, MxA and MxB, that are induced by IFN-α/β (3, 158, 393). As deduced from studies with cDNA clones, the human MxA protein is 662 amino acids whereas the MxB protein is 715 amino acids (3). Human MxA and MxB both map to chromosome 21 (132). The MxA protein normally accumulates in the cytoplasm of IFN-treated cells and possesses antiviral activity (3, 158, 393, 426). Stable transformants expressing the MxA protein acquire an antiviral activity against a range of RNA viruses, but MxB so far has not been found to display an antiviral activity (158). The antiviral activity of MxA, which undergoes oligomerization, is dependent upon GTP binding and hydrolysis (325, 338).
Cells that constitutively express the human MxA protein show a high degree of antiviral activity, displaying resistance to several members of the Orthomyxoviridae including influenza A and C viruses and Thogoto virus, but not Dhori virus (8, 117, 158, 259, 315). Interestingly, a number of additional RNA viruses including members of the Bunyaviridae (LaCrosse virus, hantavirus), Paramyxoviridae (measles virus, parainfluenza 3), Rhabdoviridae (VSV), and Togaviridae (Semliki Forest virus) are inhibited by MxA expression either in cultured cells or in transgenic mice (8, 158, 314). The multiplication of members of the Picornaviridae (Mengo virus, EMC virus) is not inhibited. MxA protein protects MxA transgenic mice from lethal virus infections independently of other IFN-α/β-induced proteins (163).
MxA transgenic mice with mutations in the endogenous Mx1 and Mx2 genes have been crossed with IFNAR-1 mutant mice that lack the IFN-α/β receptor subunit as a result of targeted gene disruption. The resultant offspring mice show resistance to Thogoto virus and La Crosse virus, two minus-strand RNA viruses, and also to Semliki Forest virus, a plus-strand RNA virus, thus providing strong evidence for the intrinsic antiviral activity of MxA. The absence of the functional IFN-α/β receptor in this transgene mouse model eliminates the possibility that other IFN-induced proteins might act in conjunction with the ectopic MxA protein to provide protection against virus infection (163).
The mouse genome, like the human genome, encodes two Mx proteins. Mouse Mx1 and Mx2 map to chromosome 16 (8, 158). The mouse Mx1 protein, an IFN-α/β-inducible 631-amino-acid protein, accumulates in the nucleus (102). The Mx2 gene is nonfunctional in all laboratory strains of mice because of a single-nucleotide insertion that alters the reading frame (391). However, the Mx2 mRNA of feral mouse strains NJL and SPR is functional. Feral mouse Mx2 encodes a 656-amino-acid protein, as deduced from the cDNA sequence. The IFN-induced expression of feral mouse Mx2 confers resistance to VSV (184). In contrast to the nuclear localization of mouse Mx1, the mouse Mx2 protein, like the human MxA protein, accumulates in the cytoplasm (184). The mouse Mx1 protein selectively inhibits the replication of members of the Orthomyxoviridae, including influenza virus and Thogoto virus (8, 158, 392). Interestingly, the Mx1-based resistance to Thogoto virus in Mx1+/+ mice is bypassed in tick-mediated virus delivery; the mechanism is unknown but possibly involves an antagonism of IFN production or action by components of the tick saliva (87).
The step of influenza virus multiplication blocked by mouse Mx1 is primary transcription catalyzed by the virion-associated polymerase (205, 315). For human MxA, studies with Thogoto virus have revealed that the IFN-induced MxA GTPase interacts with viral nucleocapsids by binding to the NP nucleoprotein component (199). Thus, the nuclear import of viral nucleocapsids is impaired, which consequently prevents primary transcription (200). Both wild-type MxA which localizes to the cytoplasm, and an engineered nuclear form of MxA that contains the nuclear localization signal of simian virus 40 T antigen, block chloramphenicol acetyltransferase reporter gene expression of reconstituted viral ribonucleoprotein (RNP) complexes produced using minireplicon systems (426). Thus, the available data suggest that MxA recognizes viral RNPs, and that if this interaction occurs in the cytoplasm, nucleocapsid transport to the nucleus is impaired, but if the MxA viral RNP interaction occurs in the nucleus, transcription is impaired.
Major Histocompatibility Complex Proteins
In addition to antiviral effects exerted at the single-cell level that reduce viral macromolecular synthesis and hence virus yield (Fig. 1), both IFN-α/β and IFN-γ modulate a number of immunoregulatory functions involving interactions between cells, for example those of NK cells (37) and Th cells (10, 42) with virus-infected cells. Cytotoxic T lymphocytes play an especially important role in the host response to viral infections. They recognize peptides derived from viral proteins when presented in infected cells in association with MHC proteins, designated HLA in humans. The MHC class I and II molecules present the antigenic peptides, derived by proteolyis of foreign viral protein antigens, to the cytotoxic T cells. Both MHC class I-restricted CD8+ T cells and MHC class II restricted CD4+ T cells are activated during viral infection. Virus-specific, MHC-restricted recognition and killing of infected cells by activated T cells is a key component of the host response to, and recovery from, infection (10, 35, 42, 231, 242, 441).
Elevation of MHC class I and II antigen levels mediated by IFN is believed to increase the efficiency of cellular immune responses to infections in the intact animal. MHC antigen levels are increased in human cells treated with IFN (36, 351). Both IFN-α/β and IFN-γ treatments lead to increased levels of class I (HLA-A, HLA-B, and HLA-C) MHC molecules, but only IFN-γ is an efficient inducer of class II (HLA-D) MHC molecules (42). Differential regulation of MHC class I expression is seen between B and T lymphocytes (223). Loss-of-function studies using knockout mice lacking either IFN receptors or the Stat-1 transcription factor suggest an essential role of IFN signaling and the Stat-1 factor for maintenance of basal expression of the MHC class I antigens (223). IRF-1 plays a key role in the IFN-inducible expression of MHC class I gene expression (337). MHC class II expression is efficiently induced by IFN-γ on many cell types including monocytes, macrophages, microglia, astrocytes, fibroblasts, and endothelial cells (72, 420).
The MHC class II transactivator factor (CIITA) is the master regulator of MHC class II expression. Most cell types do not express basal CIITA. Expression of CIITA and hence MHC class II is inducible by IFN-γ (59, 396). Multiple promoters and alternative exon 1 structures spliced to a shared exon 2 lead to the generation of distinct CIITA mRNAs that determine the cell type specificity and modulation of MHC class II gene expression (289, 321). In addition to MHC class II gene expression, studies of CIITA knockout mice suggest that CIITA is involved in the regulation of IL-4 expression (143). IFN-γ also affects the processing and presentation of antigenic peptides by modulating the expression of cellular components of the proteasomes that generate the peptides and also of cellular components that target the peptides for interaction with MHC class I molecules (42, 441). Viruses have evolved a variety of mechanisms to interfere with the generation of viral peptides, the trafficking of the peptides within the cell, and the surface expression of MHC class I antigens associated with the peptides (439).
The importance of the MHC class II-restricted T-cell response to viral infection also is illustrated by the fact that several viruses, including the herpesviruses CMV and VZV (1, 283), antagonize the IFN-γ-inducible MHC class II molecular expression. This provides a strategy of limiting immune surveillance by T cells. Introduction of mutations in the CTL epitopes of viral proteins represents another strategy used by viruses to escape immune surveillance (305). Mutations may affect the binding to MHC class I proteins or their specific recognition by CTLs, or both. Interestingly, two different mutations in the same CTL epitope of the NP protein of H3N2 influenza A virus, which abrogated MHC class I presentation and allowed escape from recognition by CTLs, were identified in viral isolates at different times that abrogated MHC class I presentation and allowed escape from recognition by CTLs (418). Mutations that affect CTL epitopes that result in escape from CTL-mediated immune surveillance occur for viruses that cause persistent infections including EBV (46), HIV (142, 271), and HCV (430). Conceivably, errors that result from the absence of proofreading during replication by viral RNA polymerases (101) and posttranscriptional RNA editing catalyzed by cellular enzymes such as ADAR1 (24, 312) contribute to the evolution of RNA virus genomes, resulting in sequence variants that give rise to the virus variants that escape from immune surveillance. Although sequence changes in short periods have been described for viruses including influenza virus (109) and HIV (156), examples also exist for relative genetic stability in viral RNA populations possibly due to changes in selective pressures and competition between variants (96).
IFN-γ plays a major immunomodulatory role and is a key mediator of virus-specific cellular immunity (176, 187, 290). IFN-α/β can promote IFN-γ expression in T cells (35). However, in mice, during the endogenous immune response to viral infections when systemic IFN-α/β production is observed, IFN-γ expression is sometimes inhibited. Studies with knockout mice show that the inhibition occurs by a mechanism dependent on both the IFN-α/β receptor and the Stat-1 transcription factor (300). However, in the absence of Stat-1, not only are the IFN-α/β-mediated inhibitory effects completely abrogated but also IFN-α/β themselves can induce IFN-γ expression consistent with the existence of a Stat-1-independent pathway as well as the well-established Stat-1-dependent signaling pathway for IFN-α/β (140a, 330a). Thus, IFN-α/β appear to play a key role in the coordination of innate and adaptive immune responses during viral infection (35, 300).
The host response to HSV infection provides an example of the importance of cell-mediated immunity. IFN-γ can protect mice from fatal HSV-1 encephalitis (47, 48, 133, 444) but has no substantial effect on the efficiency of viral replication or the extent of neuroinvasiveness. In the mouse model of ocular infection, IFN-γ impedes the establishment of HSV-1 latent infection in ganglia of the more susceptible BALB/c mice but not of the less susceptible C57BL/6 mice. IFN-γ has no significant effect on HSV maintenance or UV-mediated reactivation in mice (227). The HSV-1 protein ICP34.5 contributes to the viral neurovirulence seen in the mouse model by a mechanism involving antagonism of PKR function, as revealed from studies using virus deleted in ICP34.5 and mice lacking PKR (12, 226). Studies of Sindbis virus (SV)-induced encephalomyelitis in the antibody knockout mouse model also revealed a specific role for IFN-γ in SV clearance from the central nervous system (34a). T cells used IFN-γ to mediate noncytolytic site-specific clearance of SV from some but not all types of neurons.
The importance of immunological mechanisms in the host response to infection is further illustrated by the effects of IFN-α on Venezuelan equine encephalitis (VEE) virus, a highly infectious arthropod-borne member of the Togaviridae endemic in parts of Central and South America. VEE virus is a plus-strand enveloped RNA virus with a genome of ∼11 kb (198a). In the mouse model for VEE virus infection and pathogenesis, the virus is highly lymphotrophic during the early stages of infection, prior to invasion of the central nervous system. IFN-α and IFN-β play an important role in the pathogenesis of VEE virus (147, 247). Treatment of mice with polyethylene glycol-conjugated recombinant hybrid IFN-αA/D results in enhanced survival after either subcutaneous or aerosol inhalation challenge with VEE virus. The mechanism of protection appears to involve modulation of the host immune response, since the rapid activation of splenic CD4, CD8, and B cells seen in untreated animals was absent in polyethylene glycol-conjugated IFN-α-treated mice which showed increased macrophage activation and decreased TNF-α production (247).
Transgenic mice have been developed as an HBV animal model for the study of viral replication and pathogenesis (150). Adoptive transfer of activated anti-HBV CTLs rapidly eliminates HBV from the liver in this mouse model. The antiviral mechanism mainly involves CTL-mediated induction of cytokine expression, including IFN-γ, rather than target lysis (149). Induction of inflammatory cytokines in HBV transgenic mice occurs following liver infection with unrelated viruses including lymphocytic choriomeningitis virus, adenovirus, and CMV. These infections also lead to a transient inhibition of HBV replication by noncytolytic mechanisms (54, 151). Inactivation of HBV mediated by the CTL-elaborated cytokine expression seems to occur by at least two mechanisms: elimination of HBV nucleocapsids and destablilization of HBV RNAs (152). Intrahepatic HBV replication in the transgenic mouse model is also inhibited by infection with the malarial parasite Plasmodium yoelii. Malaria infection of HBV transgenic mice clears HBV virions from the liver by triggering an inflammatory response; the induction of IFN-γ and IFN-α/β appears essential for the inhibition of HBV (266). Use of an adenovirus vector system to obtain liver-specific expression of the mouse IFN-α2 gene provides protection from induced acute hepatitis following challenge with mouse hepatitis virus type 3 (9). Livers from infected mice treated with IFN-α2 show a substantial reduction in the level of mouse hepatitis virus type 3 mRNAs, along with an increase in transcript levels for OAS and inflammatory cytokines, including TNF-α and IFN-γ.
Inducible Nitric Oxide Synthase
NOS catalyzes NADPH-dependent oxidation of l-arginine to yield nitric oxide (NO) and citrulline (148, 251, 261). NOS is a family of three enzyme isoforms (298): inducible NOS (iNOS, or NOS2), which is inducible by IFN-γ; neuronal NOS (nNOS, or NOS1); and endothelial NOS (eNOS, or NOS3). Maximal expression of mouse iNOS following treatment with IFN-γ and lipopolysaccharide requires the IFN-γ sequence GAS element within the iNOS promoter (126). NO produced by iNOS plays a key role in immunological defenses as an antimicrobial and antiviral agent. iNOS is implicated in the function of activated macrophages as well as the pathogenesis of inflammatory and autoimmune disease (298). Activated macrophages play roles in both innate and adaptive immune responses (73, 176) and IFN-γ, rather than IFN-α/β, mediates the activation of macrophages.
In addition to the NO produced in large amounts by iNOS, further reactive nitrogen intermediates are derived from NO by oxidation, reduction, or adduction. NO and these derived reactive intermediates can greatly affect aspects of mammalian physiology, which include mediating the killing of infected cells (282, 298). Some of the cytotoxic effects of NO are due to its reactivity with iron in the active sites of mitochondrial enzymes, thereby affecting electron transport chain function (250) or cellular regulatory proteins that affect iron metabolism (193). iNOS and NO appear especially important in the antimicrobial mechanisms of macrophages (250, 282, 298, 377). However, for some viruses, including ectromelia virus (mousepox virus), vaccinia virus, and herpesviruses, iNOS plays an important role in the host response to infection and inhibition of virus replication in mouse macrophages (187). iNOS knockout mice are substantially more susceptible to mousepox than are wild-type mice (298). Interestingly, for some antimicrobial responses, loss-of-function studies using knockout mice suggest that the inability to produce reactive nitrogen intermediates by iNOS can be compensated for by phagocyte oxidase-mediated production of reactive oxygen intermediates (377).
Additional Proteins Regulated by IFNs
DNA microarrays or gene chips containing probe sets representing several thousand distinct genes provide a powerful approach to the characterization of transcription profile changes that underlie cellular responses to IFN treatment and viral infection. More than 100 of the ∼6,800 human genes represented on an Affymetrix oligonucleotide array set were identified as candidate genes differentially regulated by IFN treatment of the human fibrosarcoma cell line HT1080. IFN treatment for 6 h with 1,000 U of either IFN-α or IFN-γ showed ∼100 genes in the HT1080 cell line with greater than twofold changes and ∼25 genes with greater than fourfold changes relative to untreated cells (85). Previously, only one gene was known that was selectively induced by IFN-β but not by IFN-α or IFN-γ (331); however, the microarray analysis surprisingly revealed more than 20 candidate genes whose mRNAs were up-regulated by IFN-β but not by IFN-α or IFN-γ (85). IFN-β is an approved therapy for treatment of relapsing forms of multiple sclerosis, and changes in focal areas of relatively severe central nervous system tissue damage detected by MRI in patients with MS are slowed by IFN-β (383). The mechanism is unknown, but the selective effects of IFN-β on gene expression may provide avenues leading to elucidation of the relevant biology. Northern blot or nuclease protection analyses will permit the confirmation and better quantitation of the genes identified as transcriptional targets of IFNs. Conceivably, a temporal analysis of IFN treatment and the examination of additional cell lines or tissues may uncover candidate genes that show even greater levels of induction, or suppression, than that seen after 6 h of IFN treatment of the HT1080 cells. Microarray analyses have also been used to examine global changes in gene expression following virus infection (58, 451). In such studies, IFN-inducible genes are often identified. This is illustrated in studies with human CMV (451, 452) and human papillomavirus (58), where commercial arrays from Affymetrix or Incyte Pharmaceuticals permitted the expression analysis of nearly 7,000 known genes and expressed sequence tags. After human CMV infection of primary human foreskin fibroblasts for 40 min, 8 h, or 24 h, more than 250 genes changed by a factor of 4 or more, with roughly half increasing and half decreasing in steady-state expression level. Among the genes whose expression was increased after 8 or 24 h infection with CMV were more than 20 associated with the IFN system (451). In HPV31 cells, transfected human keratinocytes that stably maintain HPV DNA as an episome, the ∼150 down-regulated genes included those whose expression is normally increased in response to IFN treatment. The basal expression level of at least 10 of the ∼70 most strongly suppressed genes included IFN response genes, among which was the gene for the key transcription factor Stat-1 (58). DNA microarray analysis of liver gene expression during the course of acute resolving HCV infection with genotype 1a virus in a young adult male chimpanzee revealed a dramatically increased expression of several known IFN-inducible genes in a temporally distinct pattern (33a).
In a profile analysis of gene expression using oligonucleotide-based microarray gene chips, the most abundant IFN-induced mRNA identified in HT1080 cells was the 561 mRNA which encodes the IFN-inducible P56 protein (85). In HPV31 cells, the 561 transcript for P56, interestingly, was among the most strongly suppressed (58). The cDNA for P56 was among the earliest IFN-regulated cDNAs cloned (62). Biochemical studies established that protein P56 interacts with the p48 subunit of translation initiation factor eIF-3, also known as the Int-6 protein, and inhibits its function (153, 154). P56 thus has the capacity to alter the translational pattern of IFN-treated cells. However, a role for P56 in the antiviral actions of IFN has not yet been established. The replication of two RNA viruses, VSV and EMC virus, is unaffected in transfected cells that overexpress the P56 protein (155).
Among the increasing number of known IFN-inducible proteins are the P200 family of proteins (219, 229), of which the mouse p202 and p204 proteins are perhaps the best characterized (76, 236, 285, 421). These proteins have the capacity to alter the transcriptional pattern of gene expression in IFN-treated cells. However, like P56, the role of this interesting family of proteins in the antiviral actions of IFN has not yet been established. p202 is a nuclear chromatin-associated protein that acts as a modulator of transcription; p202 inhibits the activities of several factors including NF-κB, c-Fos and c-Jun, E2F1, E2F4, MyoD, and p53 (76, 285). Targeted gene disruption of the Ifi202a gene results in a dosage compensation in the expression of the related Ifi202b gene and no apparent phenotype of the knockout mice (421). p204, another of the IFN-inducible p200 family members, is primarily nucleolar. p204 binds the RNA-specific UBF1 transcription factor and inhibits rRNA transcription (236). Thus, the biochemical mode of action of p202 and p204 IFN-inducible proteins appears similar: they inhibit the activities of distinct sequence-specific transcription factors by binding to them and subsequently inhibiting their sequence-specific binding to DNA. Both p202 and p204 inhibit cell growth when overexpressed (236, 285). Studies of members of the p200 family of proteins from mice and from humans suggest that they are involved in the control of cell proliferation and affect differentiation (89, 429).
Viral Antagonists of IFN-Induced Proteins
The fundamental importance of the IFN system as a host defense against viral infection is further illustrated by the finding that a number of viruses encode gene products that antagonize the IFN-induced antiviral response (Fig. 5). Viruses utilize several different strategies to block the induction and action of IFN-inducible proteins. Members of the Poxviridae family of viruses, including vaccinia virus, occupy a centrally important position in the elucidation of mechanisms by which the IFN-induced antiviral response is impaired by virus-encoded products. The principal features of the vaccinia virus multiplication cycle essential to understanding the use of poxviruses in studies of antagonism of the IFN response are summarized in Fig. 6. Vaccinia virus virions are large, ovoid, enveloped particles with a dumbbell-shaped core. They possess a single linear dsDNA genome of ∼190 kbp with a hairpin loop at each end. Poxviruses replicate in the cytoplasm of their infected host. Following binding to the cell surface, penetration by fusion with the host cell membrane results in the release of viral cores into the cytoplasm. Virion-associated enzymes catalyze the synthesis of 5′-capped and 3′-polyadenylated early viral mRNAs. These mRNAs are translated by the host protein-synthesizing machinery to produce viral enzymes and factors for DNA replication and intermediate transcription (83, 198a). In addition, a number of virus-encoded antagonists of the IFN system are produced (36). These include IFN-γ and IFN-α/β receptor homologues, as discussed already, and two additional proteins, E3L and K3L, which affect the activity of IFN-induced proteins and the induction of IFN-α/β. Following early viral gene expression, uncoating of the subviral core occurs followed by DNA replication. Intermediate gene expression from progeny DNA produces transcription factors required for late-gene expression. The late-gene products include the structural proteins and enzymes necessary for progeny virion assembly. Assembly and maturation of progeny virions occur within the cytoplasm of the infected cell (198a).
FIG. 5.
Antagonism of the antiviral actions of IFN by viral gene products. Virus-encoded products that antagonize the IFN signaling pathway leading to transcriptional activation of IFN-regulated genes include adenovirus E1A; EBV EBNA2; HHV-8 K9; HPV E6; poxvirus B8R, B18R, and M-T7; SV5 V protein; and SeV C proteins. Virus-encoded RNA and protein products that antagonize the biochemical activity of IFN-induced cellular proteins include adenovirus VA RNA, EBV EBER RNA, HCV NS5a and E2 proteins, HSV γ34.5 and Us11 proteins, influenza virus NS1 protein, and SV40 T-antigen.
FIG. 6.
Schematic diagram of the vaccinia virus multiplication cycle including sites of action of poxvirus-encoded antagonists of the IFN response. The enveloped vaccinia virion particle has a large linear dsDNA genome. Virus penetration probably involves fusion between the viral envelope and cellular membranes, with release of the subviral core particle into the cytoplasm, where virus replication occurs. Early viral mRNAs synthesized by the virion-associated RNA polymerase include viral transcripts that encode IFN system antagonists. The E3L RNA-binding protein impairs activation of the PKR kinase and the OAS synthetases, IFN-induced enzymes that are activated by dsRNA. K3L is a substrate homologue of eIF-2α and is believed to antagonize the translation inhibition mediated by PKR. The B8R and B18R proteins are soluble IFN-γ and IFN-α/β receptor homologues, respectively, that serve as decoys to bind the extracellular induced IFNs. Early viral gene products include proteins and enzymes needed for further uncoating and for viral DNA replication and intermediate transcription. Late genes transcribed after DNA replication encode the structural proteins and enzymes necessary for progeny virion formation in the cytoplasm.
The vaccinia virus E3L and K3L gene products stimulate translation through inhibition of the IFN-induced cellular protein kinase PKR (68, 352). E3L and K3L function by different mechanisms. The viral E3L protein is a dsRNA-binding protein that both sequesters dsRNA activators of PKR (379) and interacts directly with the eIF-2α substrate-binding region of PKR (373), thereby down-regulating PKR autoactivation and eIF-2α phosphorylation. The vaccinia virus E3L protein is a potent inhibitor not only of the PKR kinase but also of the OAS (339, 351, 379) and ADAR (Y. Liu, K. Wolff, and C. E. Samuel, unpublished results). E3L acts in part by sequestering activator RNA or PKR and OAS and presumably substrate RNA of ADAR1. Antagonism of PKR autophosphorylation and eIF-2α phosphorylation has been directly linked to inhibition of the IFN-induced antiviral response (68, 121, 263, 352). Deletion mutants of vaccina virus lacking the E3L gene are sensitive to the antiviral effects of IFN. RNA-binding proteins including the reovirus ς3 protein, rotavirus NSP3 and E3L mutants that retain RNA-binding activity complement the E3L gene deletion and reverse the IFN-sensitive phenotype. Vaccinia virus mutants that express E3L proteins that are unable to bind dsRNA are IFN sensitive. The vaccinia virus K3L protein is a virus-encoded homologue of the eIF-2α substrate. The K3L pseudosubstrate inhibits PKR-catalyzed phosphorylation of eIF-2α. Deletion mutants of vaccina virus lacking the K3L gene, which encodes the eIF-2α homologue, are more sensitive to the antiviral effects of IFN than are wild-type virus. Vaccinia virus can rescue IFN-sensitive viruses such as VSV and EMC virus from the antiviral actions of IFN. Complementation studies suggest that the E3L gene product is most probably responsible for the vaccinia virus-mediated rescue of VSV from the antiviral effects of IFN and that the K3L gene product is at least partially responsible for the rescue of EMC virus (378).
The combined use of mutant influenza viruses lacking NS1 and mouse cells and mice genetically deficient in PKR have provided eloquent proof that the NS1 protein of influenza virus can antagonize the IFN-inducible PKR protein kinase in the mouse model. NS1, the only nonstructural protein encoded by influenza A viruses, is abundantly expressed in virus-infected cells and performs several regulatory functions during the infective cycle, including regulation of synthesis, transport, splicing, and translation of mRNAs. NS1 also prevents activation of PKR by binding to dsRNA activator RNAs (246). Temperature-sensitive mutant influenza A viruses with an RNA-binding defective NS1 protein cannot block the activation of PKR in infected cells (161). These temperature-sensitive mutant virus strains exhibit temperature sensitivity in virus protein synthesis at the translational level in a manner that correlates with increased activation of PKR and phosphorylation of eIF-2α. When the NS1 gene is deleted through the use of reverse genetics, the delNS1 virus is able to replicate in IFN-deficient cells and is pathogenic in Stat-1 knockout mice but not in wild-type mice (129). The delNS1 virus does not replicate in wild-type mice expressing PKR but does replicate in mutant knockout mice in which the Pkr gene has been disrupted (30). These results suggest that a major function of the influenza virus NS1 protein is to counteract or prevent the PKR-mediated antiviral response, at least from studies performed with mouse cells and whole mice that are naturally Mx deficient. Curiously, only a very few mouse strains, including A2G and SL/NiA, possess the functional Mx1 gene that confers resistance to influenza in mice (8, 158, 393). However, humans possess a functional Mx gene (MxA) that indeed is a useful marker for IFN-α/β therapy in patients and whose product possesses antiviral activity both in cell culture and in MxA transgenic mice (3, 158). Thus, the relative contribution of the NS1-mediated antagonism of PKR and the MxA-mediated antiviral activity in determining the outcome of influenza virus infections in humans is yet to be fully resolved.
Adenovirus, poliovirus, and SV40 provide three additional examples of viruses that antagonize the IFN-induced PKR kinase by different strategies. Adenovirus VAI RNA antagonizes the antiviral state of IFN by preventing the activation of PKR; deletion mutants that do not express VAI RNA display IFN sensitivity, whereas wild-type virus that produces large amounts of VAI RNA is relatively resistant to IFN-α/β (264). Poliovirus infection leads to a degradation of PKR (39), although the protease responsible for the down-regulation has not yet been identified. Translation of SV40 early RNA is relatively resistant to the antiviral effects of IFN (351). SV40 large-T antigen antagonizes PKR function but does so downstream of the kinase activation step (330). The mechanism is not resolved, but one possibility is a T-antigen-mediated enhancement of the phosphatase activity responsible for the dephosphorylation of eIF-2α similar to that observed in HSV-infected cells. In HSV infection, eIF-2α phosphorylation is reduced even though PKR is activated, due to the interaction of the HSV γ34.5 protein with phosphatase 1α (162).
Studies of the genetic evolution of HCV suggest that IFN-α therapy alters virus-host interactions as a result of changes in the nature of circulating HCV quasi-species (245, 316). HCV genotype 1b resistance to IFN-α therapy is associated with changes in the NS5A gene central region quasi-species (317). The HCV second envelope hypervariable region (HVR1), an 81-nt sequence located at the 5′ end of the E2 gene, shows changes in HVR1 quasi-species major variants during and after IFN therapy in patients who failed to clear HCV RNA. Although the underlying mechanisms are unknown, the quasi-species changes appear to be induced by changes in the host environment that probably result from IFN-induced enhancement and post-IFN attenuation of neutralizing and possibly cytotoxic responses to the envelope hypervariable region HVR1 (316). One intriguing possibility is an IFN-mediated selection for resistant viruses mediated by editing of HCV viral RNA catalyzed by the IFN-inducible RNA-editing enzyme ADAR1 (312). The HCV envelope protein E2 and nonstructural protein NS5A both inhibit PKR under some experimental conditions (122, 404). A potential outcome of the inhibition of PKR by the two HCV-encoded proteins, NS5A and E2, in addition to permitting virus replication in the presence of IFN, might include the promotion of cell growth that may contribute to HCV-associated hepatocellular carcinoma (408). Results obtained with the UHCV-11 human cell line engineered to inducibly express the entire HCV genotype 1a polyprotein have provided further evidence that the expression of HCV proteins interferes with the antiviral action of IFN as measured with EMC virus (116). However, in this system, combined cell localization and biochemical analyses indicate that HCV interferes with the antiviral action of IFN independently of PKR. Neither PKR activation nor eIF-2α phosphorylation was affected by expression of HCV proteins in their biological context (116). Expression of the HCV polyprotein in UHCV-11 cells impairs signal transduction through the JAK-STAT pathway (164), which consequently would affect the IFN-mediated regulation of a large number of genes in addition to PKR.
During virus infection, PKR may become activated by viral RNA products and then, in some cases, later become inhibited, thereby reversing any PKR-mediated translational block that had been established. The mechanism by which PKR-catalyzed phosphorylation of eIF-2 is antagonized differs among viruses and can involve either RNA or protein modulators. Among the best-characterized viral RNA inhibitor of PKR is the adenovirus virus-associated VAI RNA (264). VAI RNA is produced in large amounts at late times after infection and subsequently impairs the activation of PKR and phosphorylation of eIF-2α (263, 264). Virus mutants which lack functional VAI RNA multiply poorly because of virus-specific RNAs which activate PKR, thereby causing an inhibition of late viral protein synthesis. Adenovirus VAI RNA is bound by the dsRBMs of PKR, the same motifs that also bind activator RNAs (267). Modulation of eIF-2α phosphorylation is the primary function of viral VAI RNA, at least in cell culture, because replacement of serine 51 by alanine not only eliminates the phosphorylation site on the eIF-2α substrate but also functionally complements the deletion of VA from the virus genome. Howerver, VAI RNA does antagonize ADAR1 (225, 241) in addition to PKR (197, 264). EBV-encoded EBER RNAs, which functionally can substitute for adenovirus VAI RNA in vivo, also inhibit PKR activation (68).
The fundamental importance of PKR in the modulation of the translational pattern in virus-infected cells is further demonstrated by the large number of viruses that encode proteins that affect PKR function and subsequently the biochemical activity of initiation factor eIF-2 (121, 263, 352). The ς3 major capsid protein of reovirus was among the first animal virus proteins identified that antagonize PKR function by sequestering activator dsRNAs (354). The preferential translation of reovirus mRNAs and the inhibition of cellular protein synthesis appear to be mediated in part by a ς3-dependent mechanism that is dependent upon both the subcellular localization of ς3 (364) and the interaction of ς3 with μ1c, the other major outer capsid protein of reovirus virions (445). Proteolytic cleavage of ς3 results in enhanced dsRNA-binding activity (354), but the association of ς3 with μ1c eliminates dsRNA-binding activity and the ability to antagonize PKR function and stimulate protein synthesis (445). Reovirus mRNAs also can activate PKR. For example, the poorly translated s1 mRNA, which encodes the minor capsid protein ς1, is an efficient RNA activator of PKR, whereas the efficiently translated s4 mRNA, which encodes ς3, is a poor PKR activator (352, 354).
Changes in the translational machinery in influenza virus-infected cells mediated by down-regulation of PKR occur by two different strategies. One involves the viral nonstructural RNA-binding protein NS1 (121, 161), and the other involves the activation of the stress-related cellular P58IPK inhibitor of PKR (121, 277). NS1, found abundantly in influenza virus-infected cells, performs several regulatory functions during the viral infective cycle, including regulation of synthesis, transport, splicing, and translation of mRNAs. NS1 prevents the activation of PKR by binding to dsRNA activator RNAs. Temperature sensitive mutant influenza A viruses with an RNA-binding-defective NS1 protein cannot block the activation of PKR in infected cells. These ts mutant virus strains exhibit temperature sensitivity in virus protein synthesis at the translational level in a manner that correlates with increased activation of PKR and phosphorylation of eIF-2α. P58IPK, a constitutively expressed cellular protein, inhibits PKR activity through direct protein-protein interaction with PKR. Influenza virus infection activates P58IPK by promoting dissociation from its negative regulator, heat shock protein 40 (hsp40); cellular P58IPK is believed to be targeted with hsp70 to PKR, thereby inhibiting kinase function in influenza virus-infected cells (277). The baculovirus PK2 gene product also inhibits PKR by a dimerization mechanism. Baculovirus PK2 protein is a truncated viral kinase that forms a heteromeric complex with the cellular PKR, thereby blocking kinase autophosphorylation and eIF-2α phosphorylation. Insect cells infected with wild-type baculovirus, but not pk2-deleted virus, exhibited reduced phosphorylation and increased translational activity (88).
TAR, a highly structured RNA leader element located at the 5′ end of all HIV transcripts, activates PKR involved in inhibition of viral protein synthesis through eIF-2α phosphorylation (68, 252, 352). HIV uses two proteins to counter this TAR-mediated activation of PKR. PKR function is down-regulated by the cellular TAR-RNA-binding protein (TRBP), which forms an inactive heterodimer with PKR (29), and by the viral trans-activating Tat protein, which inhibits autophosphorylation of PKR and competes with eIF-2 (44). HIV Tat protein also activates transcription factor NF-κB through a process that requires the function of PKR (81). The relative roles during the HIV infective process of the negative (44) and positive (81) effects of Tat protein on PKR activity are not yet fully resolved.
In HSV-infected cells, the viral γ34.5 protein affects phosphatase function, thereby blocking the shutoff of protein synthesis mediated by activated PKR (162, 226). In the absence of the γ34.5 gene, eIF-2α is phosphorylated and protein synthesis is impaired beginning at about 5 h after infection, but in the presence of γ34.5, protein synthesis continues even though PKR is activated. The γ34.5 protein serves as a regulatory subunit of protein phosphatase 1α and redirects the phosphatase to dephosphorylate eIF-2αP (162). The HSV Us11 protein can substitute for the γ34.5 protein if it is present early in infection before PKR is activated. Us11 is an RNA-binding protein that blocks the shutoff of protein synthesis by preventing the activation of PKR and subsequently the phosphorylation of eIF-2α (50, 326).
HCV is the causative agent of the majority of non-A, non-B cases of hepatitis (168). Chronic HCV infection results in liver cirrhosis and hepatocellular carcinoma. There is not yet an effective vaccine for HCV. The current U.S. Food and Drug Administration-approved therapy for hepatitis C is based on systemic administration of recombinant human IFN-α. Although clearance of HCV is seen in only about 20% of patients treated with IFN-α, a combination treatment of IFN-α and ribavirin (Repetron) is efficacious in about 40% of the HCV patients (80, 270). The mechanistic explanation for the low response rate to IFN therapy is not fully resolved but may involve a combination of factors including HCV genotype and functional antagonism of one or more cellular components involved in the signaling or actions of IFN (324, 400, 404). HCV is an enveloped flavivirus with a plus-strand RNA genome of ∼9.5 kb that encodes a precursor protein of ∼3,000 amino acids. Cleavage of this polyprotein precursor generates both viral structural proteins, including the envelope glycoproteins E1 and E2, and nonstructural proteins, including NS5A. HCV, like vaccinia virus, encodes multiple proteins that act synergistically to disrupt PKR function. Two HCV-encoded proteins, the nonstructural protein NS5A (122) and the envelope protein E2 (404), repress PKR through direct interaction with the kinase. NS5A is a phosphoprotein that interacts with a region of the PKR catalytic domain that also serves as a dimerization domain, thereby disrupting PKR dimerization and PKR-mediated eIF-2α phosphorylation (122, 124). The HCV envelope protein E2 also binds to and inhibits PKR; E2 contains a sequence identical to the phosphorylation sites of PKR and the eIF-2α substrate (404). Inhibition of PKR by the NS5A and E2 proteins of HCV may facilitate the survival and proliferation of the infected host, ultimately leading to HCV-associated hepatocellular carcinoma (408). However, the resistance to IFN resulting from the expression of HCV proteins appears in some instances to be independent of PKR-mediated control of protein synthesis (116, 164). Independent of the IFN sensitivity-determining region (ISDR) of protein NS5A, IFN-α treatment inhibits HCV RNA replication in human hepatoma cells in culture. A deletion of 47 amino acids encompassing the ISDR of NS5A is among the adaptive mutations clustered in NS5A that confer an increased efficiency of HCV RNA replication in culture; this and other adaptive HCV mutants were IFN sensitive (41). The NS5A protein also interacts with cellular proteins in addition to PKR, including growth factor receptor protein Grb2 (400) and karyopherin β3, which is implicated in nucleoplasmic transport (66). The binding of Grb2 by NS5A in a SH3 domain ligand-dependent manner perturbs mitogenic signaling (400). The binding of human karyopherin β3 by NS5A blocks function of the β3 protein in the yeast model system (66). It is conceivable that the antagonism of the antiviral actions of IFN attributed to the HCV NS5A protein are the consequence of multiple different NS5A-host cell protein interactions, including PKR, Grb2, karyopherin β3, and possibly other cellular proteins as well, which collectively contribute to enhancing virus growth and pathogenesis by disruption of multiple cellular functions (41, 116, 164).
Dengue virus (DV) is a flavivirus that causes dengue hemorrhagic fever and shock syndrome, an arthropod-borne infectious disease in humans that is prevalent in the tropical and subtropical regions of the world (288). Like HCV (168), there is no effective vaccine at present for DV (288). DV infection, both antibody dependent and independent, is significantly inhibited by pretreatment of cells in culture with IFN-α and IFN-β (90). The mechanism is not resolved, but indications are that IFN inhibits DV infection of hepatoma HepG2 cells by impairing the production or stability of minus-strand DV RNA. Such an effect could result from a block in translation of the parental DV plus-strand genome RNA that encodes the viral polymerase or, alternatively, from a block in the transcription or increased degradation of minus-strand viral RNA. The importance of the IFN system in DV infection and pathogenesis of humans is supported by studies in a mouse model. Mutant mice deficient in IFN receptors through targeted gene disruption are killed by intraperitoneal infection with mouse-adapted DV, but wild-type control mice survive infection (186).
INTERFERON ACTION AND APOPTOSIS
Apoptosis, or controlled cell death, can serve as a defense mechanism for the host cell to combat viral infection. The apoptotic response involves a cascade of intracellular signals initiated in response to a wide variety of stimuli, including viral infection. Morphologic changes associated with apoptosis of virus-infected cells include condensation of chromatin and vacuolization of the infected-cell cytoplasm. Biochemical changes include activation of cellular proteases and nucleases and degradation of cell DNA to nucleosome-sized fragments. The fundamental importance of apoptosis as a component of the host response to viral infection is illustrated by the fact that a number of viruses encode gene products that either induce or inhibit apoptosis (105, 159, 375, 405). Untimely early destruction of infected host cells may greatly reduce virus yields. Hence, viruses have evolved strategies to antagonize the apoptotic response by encoding viral gene products that target effector components in the apoptotic cascade. Suppression or delay of apoptosis can extend the time available following infection to produce new virions prior to destruction of the host biosynthetic pathways utilized by the virus. The opposite also occurs. Viral gene products have been identified that function as inducers of apoptosis, possibly to facilitate the release and spread of progeny virions from the infected host (105, 159, 375, 405).
A wide variety of strategies are used by viruses to affect the balance between growth and death of the infected host cell. The adenovirus E1A gene product stabilizes p53 and induces p53-dependent apoptosis (375, 405). This effect of E1A is countered by the adenovirus E1B-19K, E1B-55K, and E3 proteins, which all antagonize the apoptotic process. The adenovirus E3 gene product promotes the degradation of Fas, which prevents apoptosis (413). The E1B-55K-mediated block of apoptosis involves antagonism of p53 function (406), whereas the E1A-19K protein is a Bcl-2 homologue that forms heterodimers with proteins such as Bax (431). Bcl-2 is an apoptosis suppressor that prolongs cell survival (34). Among additional viral Bcl-2 homologues identified are the LMW5-HL protein of African swine fever virus, which functions to antagonize apoptotic cell death, and the BHRF1 protein of EBV, which protects human B cells from programmed death (159). The E6 protein of HPV and SV40 large-T antigen also impair p53-mediated apoptosis (159, 375, 405).
Cowpox virus blocks apoptosis by encoding CrmA, a serpin family protease inhibitor that blocks the IL-1β-converting enzyme (ICE) protease (119). ICE, a cysteine protease, is a key component of the apoptotic response in mammalian cells. The vaccinia virus B13R gene product is homologous to CrmA and inhibits apoptosis induced by Fas or TNF-α (95). The baculovirus p35 protein, which prevents apoptosis during infection of insect cells, likewise inhibits the ICE family of proteases (284). Apoptosis via the Fas and TNF death receptors involving the death domain (FADD) signaling cascade includes the recruitment of several proteins such as the procaspase-8 protease FLICE. Lymphotropic gammaherpesviruses including HHV-8, herpesvirus saimiri, equine herpesvirus -2, and bovine herpesvirus 4, encode FLICE-inhibitory proteins known as vFLIPS that are recruited to the activated receptor but that lack proteolytic activity and thus do not activate death signals (414).
IFN-α/β are essential mediators of apoptosis. Primary MEF undergo apoptosis when infected with EMC virus, VSV, and HSV, but apoptosis induced by these viruses is inhibited by anti-IFN-α/β antibodies and in homozygous null cells lacking either the IFN receptor or the Stat-1 signaling factor (402). IFN alone does not induce apoptosis unless the treatment is combined with dsRNA. Two important mediators of the IFN-induced antiviral response, PKR (12, 399) and the 2-5A-dependent RNase L (92, 449), play key roles as effectors of apoptosis. Overexpression of wild-type PKR but not catalytically inactive PKR causes apoptosis in the presence of dsRNA (12). Induction of apoptosis by PKR involves eIF-2α and NF-κB, because overexpression of the S51A mutant of eIF-2α or the S32,36A mutant of IκBα leads to an inhibition of apoptosis (140, 407). Interestingly, up-regulation of Fas receptor mRNA accumulation correlates with PKR activation and subsequent apoptosis (99, 140). Apoptosis is substantially suppressed following treatment with dsRNA, TNF-α, or lipopolysaccharide of cells from mice with a targeted disruption of either the PKR gene (84) or the RNase L gene (449). Suppression of apoptosis in the PKR null cells was associated with defects in activation of IRF-1 and in Fas mRNA induction (84). Several viral antagonists of PKR function have been described (121, 352); presumably repression of PKR by these viral products would impair the apoptotic response. For example, the antiapoptotic and oncogenic potentials of HCV are linked to IFN resistance by viral repression of PKR (123). Resistance to apoptosis is attributed to an HCV NS5A-mediated block in eIF-2α phosphorylation; furthermore, cells expressing NS5A exhibit a transformed phenotype.
INTERFERON THERAPY FOR DISEASES OF KNOWN VIRAL ORIGIN
Success in the basic research laboratory led to the subsequent utilization of IFNs as therapeutics in the clinic. Three kinds of human IFNs, IFN-α, IFN-β, and IFN-γ, have been approved for clinical use by the Food and Drug Administration (http://www.fda.gov/cber/products.htm). Diseases of known viral origin for which IFN-α species are most widely used are hepatitis C and hepatitis B (Table 1). Indications for which IFN-α has been approved, in addition to chronic hepatitis C and chronic hepatitis B, are Kaposi's sarcoma, genital warts (condylomata acuminata), hairy cell leukemia, chronic myelogenous leukemia, and malignant melanoma. IFN-β is approved for treatment of relapsing forms of multiple sclerosis, and IFN-γ is approved for treatment of the chronic hereditary immune disorder chronic granulomatous disease). IFNs for clinical use are the products of several biotechnology and pharmaceutical companies. These include IFN-α2a as Roferon A (Hoffman-La Roche), IFN-α2b as Intron A (Schering Corp), consensus IFN-αcon as Infergen (Amgen), human leukocyte-derived IFN-αn3 as Alferon N (Interferon Sciences), and human lymphoblastoid-derived IFN-αn1 as Wellferon (Glaxo Wellcome). Avonex (Biogen) and Betaseron (Chiron, Berlex) are IFN-β, and Actimmune (Genentech) is IFN-γ. In January 2001, the Food and Drug Administration approved a pegylated form of IFN-α2b (PEG-Intron; Schering) for treatment of chronic hepatitis C in patients not previously treated with IFN-α. Roche is also developing pegylated IFN-α2a (Pegasys) for use in treatment of chronic hepatitis C.
TABLE 1.
IFNs approved by the Food and Drug Administration for the treatment of viral hepatitis
| IFN | Trade name | Manufacturer | Disease |
|---|---|---|---|
| IFN-α2a | Roferon A | Hoffman LaRoche | Hepatitis C |
| IFN-α2b | Intron A | Schering | Hepatitis B, Hepatitis C |
| IFN-αnl (lymphoblastoid) | Wellferon | Glaxo Wellcome | Hepatitis C |
| IFN-αcon | Infergen | Amgen | Hepatitis C |
| Peg-IFN-α2b | PEG-Intron | Schering | Hepatitis C |
CONCLUSIONS.
Tremendous progress has been made in understanding the molecular basis of the antiviral actions of IFN, and the strategies that viruses have evolved to antagonize these actions. The actions of IFN are pleiotropic and affect many biological processes in addition to the multiplication of viruses. The characteristics of the IFN-induced antiviral response are dependent on the specific combination of the three key components: the virus, the host cell, and the IFN. Responses, both IFN induction and IFN action, differ quantitatively if not qualitatively and are dependent on the virus strain, the kind of infected host cell, and type of IFN. PKR, OAS, RNase L, and protein Mx are among the best-characterized contributors to the IFN-induced antiviral state displayed at the level of the single virus-infected cell. However, a multitude of additional genes have been identified by microarray analyses whose expression is altered in response to IFN treatment and virus infection. In the whole animal infected with virus, elevated cellular immune responses mediated by enhanced expression of MHC class I and II antigens induced by IFN-α/β and IFN-γ may contribute significantly to the overall antiviral response. Structured RNA is centrally involved in the function of at least three IFN-induced enzymes: protein phosphorylation by PKR, RNA editing by ADAR, and RNA degradation by the 2-5A pathway all involve dsRNA either as an effector or as a substrate. The availability of cDNA and genomic clones of many of the components of the IFN system including the IFNs, their receptors, Jak and Stat, IRF signal transduction components, and proteins such as PKR, OAS, Mx, and ADAR, whose expression is regulated by IFNs, has permitted the generation of mutant proteins, cells that overexpress different forms of the proteins, and animals in which the expression of these and other proteins has been disrupted by targeted gene disruption. The use of these IFN system reagents in studies of the virus-host interaction, both in cell culture and in intact animals, continues to provide seminal contributions to our understanding not only of mechanisms of viral pathogenesis but also of mechanisms of signal transduction and the transcriptional and translational control of macromolecular synthesis that is so important in many areas of mammalian cell biology and virology. The emergence of new tools and approaches for study of the virus-host interaction, including functional genomics and proteomics, together with advances in the areas of structural biology and combinatorial chemistry for the identification of novel molecules that affect the function of viral and cellular targets, should lead to further insights into the structure-function relationships for the IFN system components.
ACKNOWLEDGMENTS
I thank the members of my laboratory for their helpful discussions and the many investigators within the international IFN community for their basic and clinical research contributions that made this review possible.
Work from my laboratory was supported in part by research grants from the National Institute of Allergy and Infectious Diseases (AI-12520 and AI-20611).
REFERENCES
- 1.Abendroth A, Slobedman B, Lee E, Mellins E, Wallace M, Arvin A M. Modulation of major histocompatibility class II protein expression by varicella-zoster virus. J Virol. 2000;74:1900–1907. doi: 10.1128/jvi.74.4.1900-1907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham N, Stojdl D F, Duncan P I, Méthot N, Ishii T, Dubé M, Vanderhyden B C, Atkins H L, Gray D A, McBurney M W, Koromilas A E, Brown E G, Sonenberg N, Bell J C. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 3.Aebi M, Fäh J, Hurt N, Samuel C E, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA Structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1989;9:5062–5072. doi: 10.1128/mcb.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing; 1994. [Google Scholar]
- 5.Alcami A, Symons J, Smith G L. The vaccinia virus soluble alpha/beta interferon receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol. 2000;74:11230–11239. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander W S, Starr R, Fenner J F, Scott C L, Handman E, Sprigg N S, Corbin J E, Cornish A L, Darwiche R, Owczarek C M, Kay T W, Nicola N A, Hertzog P J, Metcalf D, Hilton D J. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 7.Angel J, Franco M A, Greenberg H B, Bass D. Lack of a role for type I and type II interferons in the resolution of rotavirus-induced diarrhea and infection in mice. J Interferon Res. 1999;19:655–659. doi: 10.1089/107999099313802. [DOI] [PubMed] [Google Scholar]
- 8.Arnheiter H, Frese M, Kamadur R, Meier E, Haller O. Mx transgenic mice-animal models of helath. Curr Top Microbiol. 1996;206:119–147. doi: 10.1007/978-3-642-85208-4_8. [DOI] [PubMed] [Google Scholar]
- 9.Aurisicchio L, Delmastro P, Salucci V, Paz O G, Rovere P, Ciliberto G, La Monica N, Palombo F. Liver-specific alpha 2 interferon gene expression results in protection from induced hepatitis. J Virol. 2000;74:4816–4823. doi: 10.1128/jvi.74.10.4816-4823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bach E A, Aguet M, Schreiber R D. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 11.Baglioni C, Maroney P A, West D K. 2′,5′ Oligo(A) polymerase activity and inhibition of viral RNA synthesis in interferon treated HeLa cells. Biochemistry. 1979;18:1765–1770. doi: 10.1021/bi00576a020. [DOI] [PubMed] [Google Scholar]
- 12.Balachandran S, Kim C N, Yeh W-C, Mak T W, et al. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;23:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balachandran S, Roberts P C, Brown L E, Truong H, Pattnaik A K, Archer D R, Barber G N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 14.Ball L A. Induction of 2′5′-oligoadenylate synthetase activity and a new protein by chick interferon. Virology. 1979;94:282–296. doi: 10.1016/0042-6822(79)90462-8. [DOI] [PubMed] [Google Scholar]
- 15.Bandyopadhyay S K, Leonard G T, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-inducible response elements withouth activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 16.Barber G N, Edelhoff S, Katze M G, Disteche C M. Chromosomal assignment of the interferon-inducible double-stranded RNA-dependent protein kinase (PRKR) to human chromosome 2p21–p22 and mouse chromosome. Genomics. 1993;16:765–767. doi: 10.1006/geno.1993.1262. [DOI] [PubMed] [Google Scholar]
- 17.Barber G N, Tomita J, Hovanessian A G, Meurs E, Katze M G. Functional expression and characterization of the interferon-induced double-stranded RNA activated p68 protein kinase from E. coli. Biochemistry. 1991;30:10356–10361. doi: 10.1021/bi00106a038. [DOI] [PubMed] [Google Scholar]
- 18.Barber G N, Thompson S, Lee T G, Strom T, Jagus R, Darveau A, Katze M G. The 58-kilodalton inhibitor of the interferon induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber G N, Wambach M, Wong M L, Dever T E, Hinnebusch A G, Katze M G. Translational regulation by the interferon-induced double-stranded-RNA-activated 68-kDa protein kinase. Proc Natl Acad Sci USA. 1993;90:4621–4625. doi: 10.1073/pnas.90.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basler C F, Wang X Y, Muhlberger E, Volchkov V, Paragas J, Klenk H D, Garcia-Sastre A, Palese P. The Ebola virus VP35 protein functions as a type IIFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bass B L. RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 23.Bass B L, Weintraub H. An unwinding activity that covalently modified its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 24.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O'Connell M A, Samuel C E, Herbert A. A standardized nomenclature for adenosine deaminases that act in RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 25.Bass B L. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 26.Benech P, Mory Y, Revel M, Chebath J. Structure of two forms of the interferon-induced (2′-5′) oligo A synthetase of human cells based on cDNAs and gene sequences. EMBO J. 1985;4:2249–2256. doi: 10.1002/j.1460-2075.1985.tb03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benech P, Merlin G, Revel M, Chebath J. The 3′-end structure of the human (2′-5′) oligo A synthetase gene: prediction of two distinct proteins with cell type-specific expression. Nucleic Acids Res. 1985;13:1267–1281. doi: 10.1093/nar/13.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benech P, Vigneron M, Peretz D, Revel M, Chebath J. Interferon-responsive regulatory elements in the promoter of the human 2′,5′-oligo(A) synthetase gene. Mol Cell Biol. 1987;7:4498–4504. doi: 10.1128/mcb.7.12.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K T. Oncogenic potential of TAR RNA-binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergman M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster, T T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berry M J, Knutson G S, Lasky S R, Munemutsu S M, Samuel C E. Mechanism of interferon action: purification and substrate specificites of the double-stranded RNA-dependent protein kinase from untreated and interferon-treated mouse fibroblasts. J Biol Chem. 1985;260:11240–11247. [PubMed] [Google Scholar]
- 32.Bevilacqua P C, Cech T. Minor-groove recognition of double-stranded RNA of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 33.Bevilacqua P C, George C X, Samuel C E, Cech T R. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–6316. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- 33a.Bigger C B, Brasky K M, Lanford R E. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilbao G, Contreras J L, Zhang H, et al. Adenovirus-mediated gene expression in vivo is enhanced by the antapoptotic Bcl-2 gene. J Virol. 1999;73:6992–7000. doi: 10.1128/jvi.73.8.6992-7000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Binder G K, Griffin D E. Interferon-γ-mediated site-specific clearance of alphavirus from CNS neurons. Science. 2001;293:303–306. doi: 10.1126/science.1059742. [DOI] [PubMed] [Google Scholar]
- 35.Biron C A. Role of early cytokines, including α and β interferons (IFN-α/β), in innate and adaptive immune respones to viral infections. Semin Immunol. 1998;5:383–390. doi: 10.1006/smim.1998.0138. [DOI] [PubMed] [Google Scholar]
- 36.Biron C A, Sen G C. Interferons and other cytokines. In: Knipe D M, Howley P M, Griffin D E, Martin M, Roizman B, Straus S E, editors. Fields virology. 4th ed. Philadelphia, Pa: Lippincott-Raven; 2001. pp. 321–351. [Google Scholar]
- 37.Biron C A, Nguyen K B, Pien G C, Cousens L P, Salazar-Mather T P. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 38.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 39.Black T L, Barber G N, Katze M G. Degradation of the interferon-induced 68,000 Mr protein kinase by poliovirus requires RNA. J Virol. 1993;67:791–800. doi: 10.1128/jvi.67.2.791-800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blatt L M, Davis J M, Klein S B, Taylor M W. The biologic activity and molecular characterization of a novel synthetic interferon-alpha species, consensus interferon. J Interferon Cytokine Res. 1996;16:489–499. doi: 10.1089/jir.1996.16.489. [DOI] [PubMed] [Google Scholar]
- 41.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 42.Boehm U, Klamp T, Groot M, Howard J C. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 43.Bourara K, Litvak S, Araya A. Generation of G-to-A and C-to-U changes in HIV-1 transcripts by RNA editing. Science. 2000;289:1564–1566. doi: 10.1126/science.289.5484.1564. [DOI] [PubMed] [Google Scholar]
- 44.Brand S R, Kobayashi R, Mathews M B. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase PKR. J Biol Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 45.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Regulation of serotonin 2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 46.Burrows J M, Burrows S R, Poulsen L M, Sculley T B, Moss D J, Khanna R. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. J Virol. 1996;70:2490–2496. doi: 10.1128/jvi.70.4.2490-2496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantin E, Tanamachi B, Openshaw H, Mann J, Clarke K. Gamma interferon (IFN-gamma) receptor null-mutant mice are more susceptible to herpes simplex virus type 1 infection than IFN-gamma ligand null-mutant mice. J Virol. 1999;73:5196–5200. doi: 10.1128/jvi.73.6.5196-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cantin E, Tanamachi B, Openshaw H. Role for gamma interferon in control of herpes simplex virus type 1 reactivation. J Virol. 1999;73:3418–3423. doi: 10.1128/jvi.73.4.3418-3423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casey J L, Gerin J L. HDV RNA-editing: specific modification of adenosine in the antigenomic RNA. J Virol. 1995;69:7593–7600. doi: 10.1128/jvi.69.12.7593-7600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassady K A, Gross M, Roizman B. The herpes simplex virus Us11 protein effectively compensates for the γ34.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J Virol. 1998;72:8620–8626. doi: 10.1128/jvi.72.11.8620-8626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cattaneo R. Biased A → I hypermutation of animal RNA virus genomes. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 52.Cattaneo R, Billeter M A. Mutations and A/I hypermutations in measles virus persistent infections. Curr Top Microbiol Immunol. 1992;176:73–74. doi: 10.1007/978-3-642-77011-1_5. [DOI] [PubMed] [Google Scholar]
- 53.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter M A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavanaugh V J, Guidotti L G, Chisari F V. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cayley P J, Davies J A, McCullagh K G, Kerr I M. Activation of the ppp(A2′p)nA system in interferon-treated herpes simplex virus infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent Rnase. Eur J Biochem. 1984;143:165–174. doi: 10.1111/j.1432-1033.1984.tb08355.x. [DOI] [PubMed] [Google Scholar]
- 56.Chakraborty N G, Li L, Sporn J R, Kurtzman S H, Ergin M T, Mukherji B. Emergence of regulatory CD4+ T cell response to repetitive stimulation with antigen-presenting cells in vitro: implications in designing antigen-presenting cell-based tumor vaccines. J Immunol. 1999;162:5576–5583. [PubMed] [Google Scholar]
- 57.Chakravarti D, Ogryzko V, Kao H Y, et al. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 58.Chang Y J E, Laimins L A. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chebath J, Benech P, Hovanessian A, Galabru J, et al. Four different forms of interferon-treated 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J Biol Chem. 1987;262:3852–3857. [PubMed] [Google Scholar]
- 61.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of 2′5′ oligo A synthetase conferes resisteance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 62.Chebath J, Merlin G, Metz R, Benech P, Revel M. Interferon-induced 56,000 Mr protein and its mRNA in human cells; molecular cloning and partial sequence of the cDNA. Nucleic Acids Res. 1983;11:1213–1226. doi: 10.1093/nar/11.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chek-Coccia E M, Del Russo N, Stellacci E, Orsatti R, Benedetti E, Marziali G, Hiscott J, Battistini A. Activation and repression of the 2-5A synthetase and p21 gene promoters by IRF-1 and IRF-2. Oncogene. 1999;18:2129–2137. doi: 10.1038/sj.onc.1202536. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Lee J M, Wang Y, Huang D P, et al. The Epstein-Barr virus latency BamHI-Q promotes is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc Natl Acad Sci USA. 1999;96:9339–9344. doi: 10.1073/pnas.96.16.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong K L, Feng L, Schappert K, Meurs E, Donahue T, Friesen J D, Hovanessian A G, Williams B R. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chung K M, Lee J, Kim J E, Song O K, Cho S, Lim J, Seedorf M, Hahm B, Jang J K. Nonstructural protein 5A of hepatitis C virus inhibits the function of karyopherin beta3. J Virol. 2000;74:5233–5241. doi: 10.1128/jvi.74.11.5233-5241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Circle D A, Neel O D, Robertson H D, Clarke P A, Mathews M B. Surprising specificity of PKR binding to delta agent genomic RNA. RNA. 1997;3:438–448. [PMC free article] [PubMed] [Google Scholar]
- 68.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 69.Clemens M J, Hershey J W B, Hovanessian A C, Jacobs B L, Katze M G, Kaufman R J, Lengyel P, Samuel C E, Sen G C, Williams B R G. PKR: proposed nomenclature for the RNA-dependent protein kinase induced by interferon. J Interferon Res. 1993;13:241. doi: 10.1089/jir.1993.13.241. [DOI] [PubMed] [Google Scholar]
- 70.Coccia E M, Romeo G, Nissim A, Marziali G, et al. A full-length murine 2-5A synthetase cDNA transfected in NIH-3T3 cells impairs EMCV but not VSV replication. Virology. 1990;179:228–233. doi: 10.1016/0042-6822(90)90292-y. [DOI] [PubMed] [Google Scholar]
- 71.Colamonici O R, Domanski P, Sweitzer S M, Larner A, Buller R M L. Vaccinia virus B18R gene encodes a typ3e I interferon binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 72.Collins T, Korman A J, Wake C T, Boss J M, Kappes D J, Fiers W, Ault K A, Gimbrone M A, Jr, Strominger J L, Pober J S. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc Natl Acad Sci USA. 1984;81:4917–4921. doi: 10.1073/pnas.81.15.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 74.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 75.Darnell J E, Jr, Kerr I M, Stark G M. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 76.Datta B, Min W, Burma W, Lengyel P. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Mol Cell Biol. 1998;18:1074–1083. doi: 10.1128/mcb.18.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.David M, Chen H E, Goelz S, Larner A C, Neel B G. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.David M, Petricoin E, Benjamin C, Pine R, Weber M J, Larner A C. Requirement for MAP kinase (ERK2) activity in interferon alpha-and interferon beta-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 79.David M, Zhou G, Pin R, Dixon J E, Larner A C. The SH2 domain-containing tyrosine phosphatase PTP1D is required for interferon α/β-induced gene expression. J Biol Chem. 1996;271:15862–15865. doi: 10.1074/jbc.271.27.15862. [DOI] [PubMed] [Google Scholar]
- 80.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 81.Demarchi F, Gutierrez M I, Giacca M. Human immunodeficiency virus type 1 tat protein activates transcription factor NF-κB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J Virol. 1999;73:7080–7086. doi: 10.1128/jvi.73.8.7080-7086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deonarain R, Alcamí A, Alexiou M, Dallman M J, Gewert D R, Porter A C. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J Virol. 2000;74:3404–3409. doi: 10.1128/jvi.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DePamphilis M. DNA replication in eukaryotic cells. Cold Spring Harbor, New York: Cold Spring Harbor Press; 1996. pp. 751–773. , [Google Scholar]
- 84.Der S D, Yang Y-L, Weissmann C, Williams B R G. A double-stranded RNA-activated protein kinase dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Der S D, Zhou A, Williams B R, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Desai S Y, Patel R C, Sen G C, Malhotra P, Ghadge G D, Thimmapaya B. Activation of interferon-inducible 2′-5′ oligoadenylate synthetase by adenoviral RNA. J Biol Chem. 1995;270:3454–3461. doi: 10.1074/jbc.270.7.3454. [DOI] [PubMed] [Google Scholar]
- 87.Dessens J T, Nuttall P A. Mx1-based resistance to thogoto virus in A2G mice is bypassed in tick-mediated virus delivery. J Virol. 1998;72:8362–8364. doi: 10.1128/jvi.72.10.8362-8364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dever T E, Sripriya R, McLachlin J R, Lu J, et al. Disruption of cellular translational control by viral truncated eukaryotic translation initiation factor 2alpha kinase homolog. Proc Natl Acad Sci USA. 1998;95:4164–4169. doi: 10.1073/pnas.95.8.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DeYoung K I, Ray M E, Suy Y A, Anzick S I, Johnstone R W, Trapani J A, Meltzer P S, Trent J M. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 90.Diamond M S, Roberts T G, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diaz M O, Bohlander S, Allen G. Nomenclature of the human interferon genes. J Interferon Cytokine Res. 1996;16:179–180. doi: 10.1089/jir.1996.16.179. [DOI] [PubMed] [Google Scholar]
- 92.Diaz-Guerra M, Rivas C, Esteban M. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology. 1997;236:354–363. doi: 10.1006/viro.1997.8719. [DOI] [PubMed] [Google Scholar]
- 93.Didcock L, Young D F, Goobourn S, Randall R E. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dideock L, Young D F, Goodburn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dobbelstein M, Shenk T. Protection against apoptosis by the vaccinia virus SP!-2 (B13R) gene product. J Virol. 1996;70:6479–6485. doi: 10.1128/jvi.70.9.6479-6485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 97.Dong B, Silverman R H. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 98.Dong B, Xu L, Zhou A, Hassel B A, et al. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J Biol Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 99.Donze O, Dostie J, Sonenberg N. Regulatable expression of the interferon-induced double stranded RNA dependent protein kinase PKR induces apoptosis and Fas receptor expression. Virology. 1999;256:322–329. doi: 10.1006/viro.1999.9618. [DOI] [PubMed] [Google Scholar]
- 100.Donze O, Jagus R, Koromilas A E, Hershey J W B, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Drake J W, Holland J J. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dreiding P, Staeheli P, Haller O. Interferon-induced protein Mx accumulates in nuclei of mouse cells expressing resistance to influenza viruses. Virology. 1985;140:192–196. doi: 10.1016/0042-6822(85)90460-x. [DOI] [PubMed] [Google Scholar]
- 103.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 104.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;386:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 105.Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7:160–165. doi: 10.1016/s0966-842x(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 106.Fantuzzi G, Reed D A, Dinarello C A. IL-12-induced IFN-gamma is dependent on caspase-1 processing of the IL-18 precursor. J Clin Investig. 1999;104:761–767. doi: 10.1172/JCI7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fasler-Kan E, Pansky A, Wiederkehr M, Battegary M, Heim M H. Interferon-alpha activates signal transducers and activators of transcription 5 and 6 in Daudi cells. Eur J Biochem. 1998;254:514–519. doi: 10.1046/j.1432-1327.1998.2540514.x. [DOI] [PubMed] [Google Scholar]
- 108.Feng G-S, Chong K, Kumar A, Williams B R G. Identification of double-stranded RNA-binding domains in the interferon-induced double-stranded RNA-activated p68 kinase. Proc Natl Acad Sci USA. 1992;89:5447–5451. doi: 10.1073/pnas.89.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fields S, Winter G. Nucleotide-sequence heterogeneity and sequence rearrangements in influenza virus cDNA. Gene. 1981;15:207–214. doi: 10.1016/0378-1119(81)90130-x. [DOI] [PubMed] [Google Scholar]
- 110.Finter N B. The naming of cats—and alpha-interferons. Lancet. 1996;348:348–349. doi: 10.1016/s0140-6736(05)64985-9. [DOI] [PubMed] [Google Scholar]
- 111.Floyd-Smith G. 2′,5′ An-dependent endoribonuclease: enzyme levels are regulated by IFN-β, IFN-γ, and cell culture conditions. J Cell Biochem. 1988;38:13–21. doi: 10.1002/jcb.240380103. [DOI] [PubMed] [Google Scholar]
- 112.Floyd-Smith G, Denton J S. A 2′,5′-dependent endonuclease: tissue distribution in BALB/c mice and the effects of IFN-β treatment and anti-IFN immunoglobulin on the levels of the enzyme. J Interferon Res. 1988;8:517–525. doi: 10.1089/jir.1988.8.517. [DOI] [PubMed] [Google Scholar]
- 113.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a 2′-5′ oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 114.Foster G R, Germain C, Jones M, Lechler R I, Lombardi G. Human T cells elicit IFN-alpha secretion from dendritic cells following cell to cell interationis. Eur J Immunol. 2000;30:3228–3235. doi: 10.1002/1521-4141(200011)30:11<3228::AID-IMMU3228>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 115.Formella S, Jehle C, Sauder C, Staeheli P, Schwemmle M. Sequence variability of Borna disease virus: resistance to superinfection may contribute to high genome stability in persistently infected cells. J Virol. 2000;74:7878–7783. doi: 10.1128/jvi.74.17.7878-7883.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.François C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum H E, Gatignol A, Wychowski C, Moradpour D, Meurs E F. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J Virol. 2000;74:5587–5596. doi: 10.1128/jvi.74.12.5587-5596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Frese M, Kochs G, Meier-Dieter U, Siebler J, Haller O. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J Virol. 1995;69:3904–3909. doi: 10.1128/jvi.69.6.3904-3909.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gagliardini V, Fernandez P-A, Lee R K K, et al. Prevention of vertebrate neuronal death by the crmA gene. Science. 1994;263:826–828. doi: 10.1126/science.8303301. [DOI] [PubMed] [Google Scholar]
- 120.Galabru J, Hovanessian A G. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 121.Gale M, Katze M G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 122.Gale M, Jr, Blakely C M, Kwieciszewski G, Tan S L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gale M, Kwieciszewski B, Dossett M, et al. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gale M, Jr, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 125.Gale M, Jr, Tan S L, Katze M G. Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gao J, Morrison D C, Parmely T J, Russell S W, Murphy W J. An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J Biol Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 127.Gao S J, Boshoff C, Jayachandra C, et al. KSHV ORF K9 (vIRF) is an oncogene which inhibit the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 128.Garcia-Sastre A, Durbin R K, Zheng H Y, Palese P, et al. The role of interferon in influenza virus tissue tropism. J Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 130.Garcin D, Curran J, Kolakofsky D. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J Virol. 2000;74:8823–8830. doi: 10.1128/jvi.74.19.8823-8830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Garcin D, Latorre P, Kolakofsy D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gardiner K, Horisberger M, Kraus J, Tantravahi U, Korenberg J, Rao V, Reddy S, Patterson D. Analysis of human chromosome 21: correlation of physical and cytogenetic maps: gene and CpG island distributions. EMBO J. 1990;9:25–34. doi: 10.1002/j.1460-2075.1990.tb08076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Geiger K D, Nash T C, Sawyer S, Krahl T, Patstone G, Reed J C, Krajewski S, Dalton D, Buchmeier M J, Sarvetnick N. Interferon-gamma protects against herpes simplex virus type 1-mediated neuronal death. Virology. 1997;238:189–197. doi: 10.1006/viro.1997.8841. [DOI] [PubMed] [Google Scholar]
- 134.George C X, Samuel C E. Human RNA specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon-inducible. Proc Natl Acad Sci USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.George C X, Samuel C E. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene. 1999;229:203–213. doi: 10.1016/s0378-1119(99)00017-7. [DOI] [PubMed] [Google Scholar]
- 136.George C X, Thomis D C, McCormack S J, Svahn C M, Samuel C E. Characterization of the heparin-mediated activation of PKR, the interferon-induced RNA-dependent protein kinase. Virology. 1996;221:180–188. doi: 10.1006/viro.1996.0364. [DOI] [PubMed] [Google Scholar]
- 137.Ghosh A, Sarkar S N, Guo W D, Bandyopadhyay S, Sen G C. Enzymatic activity of 2′-5′-oligoadenylate synthetase is impaired by specific mutations that affect oligomerization of the protein. J Biol Chem. 1997;272:33220–33226. doi: 10.1074/jbc.272.52.33220. [DOI] [PubMed] [Google Scholar]
- 138.Ghosh A, Sarkar S N, Sen G C. Cell growth regulatory and antiviral effects of the P69 isozyme of 2-5 (A) synthetase. Virology. 2000;266:319–328. doi: 10.1006/viro.1999.0085. [DOI] [PubMed] [Google Scholar]
- 139.Ghosh S K, Kusari J, Bandyopadhyay S K, et al. Cloning sequencing and expression of two murine 2′-5′-oligoadenylate synthetases. Structure-function relationships. J Biol Chem. 1991;266:15293–15299. [PubMed] [Google Scholar]
- 140.Gil J, Alcami J, Esteban M. Induction of apoptosis by double-stranded RNA-dependent protein kinase PKR involves the α subunit of eukaryotic translation initiation factor 2 and NF-κB. Mol Cell Biol. 1999;19:4653–4663. doi: 10.1128/mcb.19.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140a.Gil M P, Bohn E, O'Guin A K, Ramana C V, Levine B, Stark G R, Virgin H V, Schreiber R D. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goh K C, Haque S J, Williams B R G. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation by interferons. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Goulder P J, Phillips R E, Colbert R A, McAdam S, Ogg G, Nowak M A, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;2:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 143.Gourley T, Roys S, Lukacs N W, Kunkel S L, Flavell R A, Chang C H. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–386. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- 144.Green S R, Mathews M B. Two RNA binding motifs in the double-stranded RNA activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 145.Green S R, Manche L, Mathews M B. Two functionally distinct RNA binding motifs in the regulatory domain of the protein kinase, DAI. Mol Cell Biol. 1995;15:358–364. doi: 10.1128/mcb.15.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Grieder F B, Vogel S N. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology. 1999;257:106–118. doi: 10.1006/viro.1999.9662. [DOI] [PubMed] [Google Scholar]
- 147.Grieder F B, Davis B K, Zhou X D, Chen S J, Finkelman F C, Gause W C. Kinetics of cytokine expression and regulation of host protection following infection with molecularly cloned Venezuelan equine encephalitis virus. Virology. 1997;233:302–312. doi: 10.1006/viro.1997.8617. [DOI] [PubMed] [Google Scholar]
- 148.Griffith O W, Stuehr D J. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 149.Guidotti L G, Chisari F V. Cytokine-mediated control of viral infections. Virology. 2000;273:221–227. doi: 10.1006/viro.2000.0442. [DOI] [PubMed] [Google Scholar]
- 150.Guidotti L G, Ando K, Hobbs M V, Ishikawa T, Runkel L, Schreiber R D, Chisari F W. Cytotoxix T lymphocyes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B A, Chisari F W. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F W. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 153.Guo J, Sen G C. Characterization of the interaction between the interferon-induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumor virus. J Virol. 2000;74:1892–1899. doi: 10.1128/jvi.74.4.1892-1899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guo J, Hui D J, Merrick W C, Sen G C. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guo J, Peters K L, Sen G C. Induction of human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 156.Hahn B H, Shaw G M, Taylor M E, Redfield R R, Markham P D, Salahuddin S Z, Wong- Staal F, Gallo R C, Parks E S, Parks W P. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 157.Hajjar A M, Linial M L. Modifications of retroviral RNA by double-stranded RNA adenosine deaminase. J Virol. 1995;69:5878–5882. doi: 10.1128/jvi.69.9.5878-5882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haller O, Frese M, Kochs G. Mx proteins: mediators of innate resistance to RNA viruses. Rev Sci Technol Off Int Epizootol. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 159.Hardwick J M. Viral interference with apoptosis. Cell Dev Biol. 1998;9:339–349. doi: 10.1006/scdb.1998.0243. [DOI] [PubMed] [Google Scholar]
- 160.Hassel B A, Zhou A, Sotomayor C, Maran A, et al. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hatada E, Saito S, Fukuda R. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol. 1999;73:2425–33. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.He B, Gross M, Roizman R. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hefti HP, Frese M, Landis H, Di Paolo C, Aguzzi A, Haller O, Pavlovic J. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J Virol. 1999;73:6984–6991. doi: 10.1128/jvi.73.8.6984-6991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Heim M H, Moradpour D, Blum H E. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heim M H. The Jak-STAT pathway: cytokine signalling from the receptor to the nucleus. J Receptor Signal Transduction Res. 1999;19:75–120. doi: 10.3109/10799899909036638. [DOI] [PubMed] [Google Scholar]
- 166.Herbert A, Lowenhaupt K, Spitzer J, Rich A. Chicken double-stranded RNA adenosine deaminase has apparent specificity for Z-DNA. Proc Natl Acad Sci USA. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 168.Hoofnagle J H. Therapy of acute and chronic viral hepatitis. Adv Intern Med. 1994;39:241–275. [PubMed] [Google Scholar]
- 169.Horvath C M. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 170.Hovanessian A G, Galabru J. The double- stranded RNA-dependent protein kinase is also activated by heparin. Eur J Biochem. 1987;167:467–473. doi: 10.1111/j.1432-1033.1987.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 171.Hovanessian A G, Wood J N. Anticellular and antiviral effects of pppA(2′p5′A)n. Virology. 1980;101:81–90. doi: 10.1016/0042-6822(80)90485-7. [DOI] [PubMed] [Google Scholar]
- 172.Hovanessian A G, Laurent A G, Chebath J, Galabru J, et al. Identification of 69-kd and 100-kd forms of 2-5A synthetase in interferon-treated human cells by specific monoclonal antibodies. EMBO. 1987;6:1273–1280. doi: 10.1002/j.1460-2075.1987.tb02364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hovanessian A G, Svab J, Marie I, Robert N, et al. Characterization of 69- and 100-kDa forms of 2-5A-synthetase from interferon-treated human cells. J Biol Chem. 1988;263:4945–4949. [PubMed] [Google Scholar]
- 174.Hovnanian A, Rebouillat D, Mattei M G, Levy E R, Marié I, Monaco A P, Hovanessian A G. The human 2′,5′-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics. 1998;52:267–277. doi: 10.1006/geno.1998.5443. [DOI] [PubMed] [Google Scholar]
- 175.Hovnanian A, Rebouillat D, Levy E R, Mattei M G, Hovanessian A G. The human 2′,5′-oligoadenylate synthetase-like gene (OASL) encoding the interferon-induced 56-kDa protein maps to chromosome 12q24.2 in the proximity of the 2′,5′-OAS locus. Genomics. 1999;56:362–363. doi: 10.1006/geno.1998.5737. [DOI] [PubMed] [Google Scholar]
- 176.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 177.Hwang S Y, Hertzog P J, Holland K A, Sumarsono S H, Tymms M J, Hamilton J A, Whitty G, Bertoncello I, Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Icely PL, Gross P, Bergeron J J M, Devault A, Afar D E H, Bell J C. TIK, a novel serine/threonine kinase, is recognized by antibodies directed against phosphotyrosine. J Biol Chem. 1991;266:16073–16077. [PubMed] [Google Scholar]
- 179.Ilson D H, Torrence P F, Vilcek J. Two molecular weight forms of human 2′5′-oligoadenylate synthetase have different activation requirements. J Interferon Res. 1986;6:5–12. doi: 10.1089/jir.1986.6.5. [DOI] [PubMed] [Google Scholar]
- 180.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc London Ser B. 1957;147:258–267. [PubMed] [Google Scholar]
- 181.Jacobs B L, Langland J O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 182.Jacobsen H, Czarniecki C W, Krause D, Friedman R M, et al. Interferon-induced synthesis of 2-5A-dependent Rnase in mouse JLS-V9R cells. Virology. 1983;125:496–501. doi: 10.1016/0042-6822(83)90222-2. [DOI] [PubMed] [Google Scholar]
- 183.Jimenez-Garcia L F, Green S R, Mathews M B, Spector D L. Organization of the double-stranded RNA-activated protein kinase DAI and virus-associated VA RNA1 in adenovirus-2-infected He-La cells. J Cell Sci. 1993;106:11–22. doi: 10.1242/jcs.106.1.11. [DOI] [PubMed] [Google Scholar]
- 184.Jin H K, Takada A, Kon Y, Haller O, Watanabe T. Identification of the murine Mx2 gene: interferon-induced expression of the Mx2 protein from the feral mouse gene confers resistance to vesicular stomatitis virus. J Virol. 1999;73:4925–4930. doi: 10.1128/jvi.73.6.4925-4930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.John J, McKendry R, Pellegrini S, Flavell D, Kerr L M, Stark G R. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol Cell Biol. 1991;11:4189–4195. doi: 10.1128/mcb.11.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Johnson A J, Roehrig J T. New mouse model for dengue virus vaccine testing. J Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Karupiah G, Xie Q W, Buller R M, Nathan C, Duarte C, MacMicking J D. Inhibition of viral replication by interferon-gamma-induced nitric oxide synthase. Science. 1993;261:1445–1448. doi: 10.1126/science.7690156. [DOI] [PubMed] [Google Scholar]
- 188.Katze M G. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 189.Katze M G, Wambach M, Wong M L, Garfinkel M, Meurs E, Chong K, Williams B R G, Hovanessian A G, Barber G N. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated 68,000-Mr protein kinase in a cell-free system. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kawakubo K, Kuhen K L, Vessey J W, George C X, Samuel C E. Alternative splice variants of the human PKR protein kinase possessing different 5′-untranslated regions: expression in untreated and interferon-treated cells and translational activity. Virology. 1999;264:106–114. doi: 10.1006/viro.1999.9995. [DOI] [PubMed] [Google Scholar]
- 191.Kerr I M. The 2-5A system: a personal view. J Interferon Res. 1987;7:505–510. doi: 10.1089/jir.1987.7.505. [DOI] [PubMed] [Google Scholar]
- 192.Kerr I M, Brown R E. pppA2′ p5′ A2′ p5′ A: an inhibitor of protein synthesis syntheized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim S, Ponka P. Effects of interferon-gamma and lipopolysaccharide on macrophage iron metabolism are mediated by nitric oxide-induced degradation of iron regulatory protein 2. J Biol Chem. 2000;275:6220–6226. doi: 10.1074/jbc.275.9.6220. [DOI] [PubMed] [Google Scholar]
- 194.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan R S, Takasugi T, Matsuyama T, Mak T W, Noguchi S, Taniguchi T. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 196.Kirchhoff S, Koromilas A E, Schaper F, Grashoff M, Sonenberg N, Hauser H. IRF-1 induced cell growth inhibition and interferon induction requires the activity of the protein kinase PKR. Oncogene. 1995;11:439–445. [PubMed] [Google Scholar]
- 197.Kitajewski J, Schneider R, Safer B, Munemutsu S, Samuel C E, Thimmapaya B, Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 198.Knipe D M, Samuel C E, Palese P. Virus-host cell interactions. In: Knipe D M, Howley P M, Griffin D E, Martin M, Roizman B, Straus S E, editors. Fields virology. 4th ed. Philadelphia, Pa: Lippincott-Raven; 2001. pp. 133–170. [Google Scholar]
- 198a.Knipe D M, Howley P M, Griffin D E, Martin M, Roizman B, Straus S E, editors. Fields virology. 4th ed. Philadelphia, Pa: Lippincott-Raven; 2001. [Google Scholar]
- 199.Kochs G, Haller O. GTP-bound human MxA protein interacts with the nucleocapsids of Thogoto virus (Orthomyxoviridae) J Biol Chem. 1999;274:4370–4376. doi: 10.1074/jbc.274.7.4370. [DOI] [PubMed] [Google Scholar]
- 200.Kochs G, Haller O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci USA. 1999;96:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kon N, Suhadolnik R J. Identification of the ATP binding domain of recombinant human 40-kDa 2′,5′ -oligoadenylate synthetase by photoaffinity labeling with 8-azido-[α-32P]ATP. J Biol Chem. 1996;271:19983–19990. doi: 10.1074/jbc.271.33.19983. [DOI] [PubMed] [Google Scholar]
- 203.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 204.Kostura M, Mathews M B. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Krug R M, Shaw M, Broni B, Shapiro G, Haller O. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. J Virol. 1985;56:201–206. doi: 10.1128/jvi.56.1.201-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kuhen K L, Samuel C E. Isolation of the interferon-inducible RNA-dependent protein kinase Pkr promoter and identification of a novel DNA element within the 5′-flanking region of the human and mouse Pkr genes. Virology. 1997;227:119–130. doi: 10.1006/viro.1996.8306. [DOI] [PubMed] [Google Scholar]
- 207.Kuhen K L, Samuel C E. Mechanism of interferon action: functional characterization of positive and negative regulatory domains that modulate transcriptional activation of the human RNA-dependent protein kinase Pkr promoter. Virology. 1999;254:182–195. doi: 10.1006/viro.1998.9536. [DOI] [PubMed] [Google Scholar]
- 208.Kuhen K L, Shen X, Carlisle E R, Richardson A L, Weier H U G, Tanaka H, Samuel C E. Structural organization of the human gene PKR encoding an interferon-inducible RNA-dependent protein kinase and differences from its mouse homolog. Genomics. 1996;36:197–201. doi: 10.1006/geno.1996.0446. [DOI] [PubMed] [Google Scholar]
- 209.Kuhen K L, Shen X, Samuel C E. Mechanism of interferon action. Sequence of the human interferon-inducible RNA-dependent protein kinase (PKR) deduced from genomic clones. Gene. 1996;178:191–193. doi: 10.1016/0378-1119(96)00314-9. [DOI] [PubMed] [Google Scholar]
- 210.Kuhen K L, Vessey J W, Samuel C E. Mechanism of interferon action: identification of essential positions within the novel 15-base-pair KCS element required for transcriptional activation of the RNA-dependent protein kinase pkr gene. J Virol. 1998;72:9934–9939. doi: 10.1128/jvi.72.12.9934-9939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion of the PKR gene: role of IRF1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kumar K P, McBride K M, Weaver B K, Dingwall C, Reich N C. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kumar M, Carmichael G G. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcript. Proc Natl Acad Sci USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kumar S, Laneback C, Valente G, Floyd-Smith G. Expansion and molecular evolution of the interferon-induced 2′-5′ oligoadenylate synthetase gene family. Mol Biol Evol. 2000;17:738–750. doi: 10.1093/oxfordjournals.molbev.a026352. [DOI] [PubMed] [Google Scholar]
- 216.Lai F, Chen C X, Carter K C, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 218.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 219.Landolfo S, Gribaudo G, Lembo D. The Ifi2000 genes: an emerging family of IFN-inducible genes. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 220.Langland J O, Kao P N, Jacobs B L. Nuclear factor-90 of activated T-cells: a double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry. 1999;38:6361–6368. doi: 10.1021/bi982410u. [DOI] [PubMed] [Google Scholar]
- 221.Lasky S R, Jacobs B L, Samuel C E. Mechanism of interferon action: characterization of the sites of phosphorylation in the interferon-induced phosphoprotein P1 from mouse fibroblasts: evidence for two forms of P1. J Biol Chem. 1982;257:11087–11093. [PubMed] [Google Scholar]
- 222.Lau J F, Parisien J-P, Horvath C M. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc Natl Acad Sci USA. 2000;97:7278–7283. doi: 10.1073/pnas.97.13.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lee C-K, Gimeno R, Levy D E. Differential regulation of constitutive major histocompatibility complex class I expression in T and B lymphocytes. J Exp Med. 1999;190:1451–1464. doi: 10.1084/jem.190.10.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated human p68 protein kinase inhibits the replication of vaccinia virus. Virology. 1993;193:1037–1041. doi: 10.1006/viro.1993.1223. [DOI] [PubMed] [Google Scholar]
- 225.Lei M, Liu Y, Samuel C E. Mechanism of Interferon Action. Adenovirus VAI RNA antagonizes the RNA-editing activity of the ADAR adenosine deaminase. Virology. 1998;245:188–196. doi: 10.1006/viro.1998.9162. [DOI] [PubMed] [Google Scholar]
- 226.Leib D A, Machalek M A, Williams B R G, Silverman R H, Virgin H W. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci USA. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lekstrom-Himes J A, LeBlanc R A, Pesnicak L, Godleski M, Straus S E. Gamma interferon impedes the establishment of herpes simplex virus type 1 latent infection but has no impact on its maintenance or reactivation in mice. J Virol. 2000;74:6680–6683. doi: 10.1128/jvi.74.14.6680-6683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc Natl Acad Sci USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lengyel P, Choubey D, Li S-J, Datta B. The interferon-activatable gene 200 cluster: from structure toward function. Semin Virol. 1995;6:203–213. [Google Scholar]
- 230.Leonard G T, Sen G C. Restoration of interferon responses of adenovirus E1A-expressing HT1080 ell lines by overexpression of p48 protein. J Virol. 1997;71:5095–5101. doi: 10.1128/jvi.71.7.5095-5101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Leonard W J, O'Shea J J. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 232.Levy D E. Physiological significance of STAT proteins: investigations through gene disruption in vivo. Cell Mol Life Sci. 1999;55:1559–1567. doi: 10.1007/s000180050395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Li X-L, Blackfor J A, Judge C S, Liu M, Xiao W, Kalvakolanu D V, Hassel B A. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2-5A system in attenuation of the interferon response. J Biol Chem. 2000;275:8880–8888. doi: 10.1074/jbc.275.12.8880. [DOI] [PubMed] [Google Scholar]
- 234.Liao J, Fu Y, Shuai K. Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1-Stat1 interaction. Proc Natl Acad Sci USA. 2000;97:5267–5272. doi: 10.1073/pnas.97.10.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu Y, Herbert A, Rich A, Samuel C E. Double-stranded RNA-specific adenosine deaminase: nucleic acid binding properties. Methods. 1998;15:199–205. doi: 10.1006/meth.1998.0624. [DOI] [PubMed] [Google Scholar]
- 236.Liu C-J, Wang H, Lengyel P. The interferon-inducible nucleolar p204 protein binds the ribosomal RNA-specific UBF1 transcription factor and inhibits ribosomal RNA transcription. EMBO J. 1999;18:2845–2854. doi: 10.1093/emboj/18.10.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu Y, Samuel C E. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liu Y, Samuel C E. Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274:5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- 239.Liu Y, Emeson R B, Samuel C E. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274:18351–18358. doi: 10.1074/jbc.274.26.18351. [DOI] [PubMed] [Google Scholar]
- 240.Liu Y, George C X, Patterson J B, Samuel C E. Functionally distinct dsRNA binding domains associated with alternative splice variants of the IFN-inducible ADAR. J Biol Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 241.Liu Y, Lei M, Samuel C E. Chimeric dsRNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of dsRNA-binding domains from ADAR1 and the protein kinase PKR. Proc Natl Acad Sci USA. 2000;97:12541–12546. doi: 10.1073/pnas.97.23.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu T, Chambers T J. Yellow fever virus encephalitis: properties of the brain-associated T-cell response during virus clearance in normal and gamma interferon-deficient mice and requirement for CD4+ lymphocytes. J Virol. 2001;75:2107–2118. doi: 10.1128/JVI.75.5.2107-2118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lomeli H, Mosbacher J, Melcher T, Höger T, Geiger J R P, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;226:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 244.Lu B, Epensperger C, Dembic Z, Wang Y L, Kvatyuk M, Lu T H, Coffman R L, Pestka S, Rothman R B. Targeted disruption of the interferon gamma receptor 2 gene results in severe immune defects in mice. Proc Natl Acad Sci USA. 1998;95:8233–8238. doi: 10.1073/pnas.95.14.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lu M, Wiese M, Roggendorf M. Selection of genetic variants of the 5′ noncoding region of hepatitis C virus occurs only in patients responding to interferon alpha therapy. J Med Virol. 1999;59:146–153. doi: 10.1002/(sici)1096-9071(199910)59:2<146::aid-jmv4>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 246.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 247.Lukaszewski R A, Brooks T J G. Pegylated alpha interferon is an effective treatment for virulent venezuelan equine encephalitis virus and has profound effects on the host immune response to infection. J Virol. 2000;74:5006–5015. doi: 10.1128/jvi.74.11.5006-5015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Maas S, Melcher T, Herb A, Seeburg P H, Keller W, Krause S, Higuchi M, O'Connell M A. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 249.Maas S, Rich A. Changing genetic information through RNA editing. Bioessays. 2000;22:790–802. doi: 10.1002/1521-1878(200009)22:9<790::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 250.MacMicking J D, North R J, LaCourse R, Mudgett J S, Shah S K, Nathan C F. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.MacMicking J D, Xie Q-W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 252.Maitra R K, McMillan N A J, Desai S, McSwiggen J, et al. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 253.Major A S, Cuff C F. Effects of the route of infection on immunoglobulin G subclasses and specificity of the reovirus-specific humoral immune response. J Virol. 1996;70:5968–5974. doi: 10.1128/jvi.70.9.5968-5974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Manchie L, Green S R, Schmedt C, Mathews M B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Marie I, Hovanessian A G. The 69-kDa 2-5A synthetase is composed of two homologous and adjacent functional domains. J Biol Chem. 1992;267:9933–9939. [PubMed] [Google Scholar]
- 256.Marie I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Marie I, Rebouillat D, Hovanessian A G. The expression of both domains of the 69/71 kDa 2′,5′ oligoadenylate synthetase generates a catalytically active enzyme and mediates an anti-viral response. Eur J Biochem. 1999;262:155–165. doi: 10.1046/j.1432-1327.1999.00361.x. [DOI] [PubMed] [Google Scholar]
- 258.Marie I, Svab J, Robert N, Galabru J, Hovanessian A G. Differential expression and distinct structure of 69- and 100-kDa forms of 2-5A synthetase in human cells treated with interferon. J Biol Chem. 1990;265:18601–18607. [PubMed] [Google Scholar]
- 259.Marschall M, Zach A, Hechtfischer A, Foerst G, Meier-Ewert H, Haller O. Inhibition of influenza C viruses by human MxA protein. Virus Res. 2000;67:179–188. doi: 10.1016/s0168-1702(00)00140-4. [DOI] [PubMed] [Google Scholar]
- 260.Mason D, Powrie F. Control of immune pathology by regulatory T cells. Curr Opin Immunol. 1998;10:649–655. doi: 10.1016/s0952-7915(98)80084-8. [DOI] [PubMed] [Google Scholar]
- 261.Masters B S, McMillan K, Sheta E A, Nishimura J S, Roman L J, Martasek P. Neuronal nitric oxide synthase, a modular enzyme formed by convergent evolution: structure studies of a cysteine thiolate-liganded heme protein that hydroxylates L-arginine to produce NO as a cellular signal. FASEB J. 1996;10:552–558. doi: 10.1096/fasebj.10.5.8621055. [DOI] [PubMed] [Google Scholar]
- 262.Masters P S, Samuel C E. Mechanism of interferon action: inhibition of vesicular stomatitis virus replication in human amnion U cells by cloned human eukocyte interferon. II. Effect on viral macromolecular synthesis. J Biol Chem. 1983;258:12026–12033. [PubMed] [Google Scholar]
- 263.Mathews M B. Viral evasion of cellular defense mechanisms: regulation of the protein kinase DA by RNA effectors. Semin Virol. 1993;4:247–257. [Google Scholar]
- 264.Mathews M B, Shenk T. Adenovirus virus-associated RNA and translational control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kündig T M, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 266.McClary H, Koch R, Chisari F V, Guidotti L G. Inhibition of hepatitis B virus replication during schistosoma mansoni infection in transgenic mice. J Exp Med. 2000;192:289–294. doi: 10.1084/jem.192.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.McCormack S J, Samuel C E. Mechanism of interferon action. RNA-binding activity of full-length and R-domain forms of the RNA-dependent protein kinase (PKR): determination of the KD values for VAI and TAR RNAs. Virology. 1995;188:47–56. doi: 10.1016/s0042-6822(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 268.McCormack S J, Ortega L G, Doohan J R, Samuel C E. Mechanism of interferon action. Motif I of the interferon-induced, RNA-dependent protein kinase (PKR) is sufficient to mediate RNA binding activity. Virology. 1994;198:92–99. doi: 10.1006/viro.1994.1011. [DOI] [PubMed] [Google Scholar]
- 269.McCormack SJ, Thomis D C, Samuel C E. Mechanism of interferon action. Identification of an RNA-binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2α protein kinase. Virology. 1992;188:47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- 270.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 271.McMichael A J, Phillips R E. Escape of human immunodeficiency virus from immune control. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 272.McMillan N A J, Chun R F, Siderovski D P, Galabru J, Toone W, Samuel C E, Mak T, Hovanessian A, Jeang K, Williams B R G. HIV-1 Tat directly interacts with the interferon-induced, double-stranded-RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 273.McMillan N A J, Carpick B W, Hollis B, Toone M, Zamanian M, Williams B R G. Mutational analysis of the double-stranded RNA binding domain of the dsRNA-activated protein kinase, PKR. J Biol Chem. 1995;270:2601–2606. doi: 10.1074/jbc.270.6.2601. [DOI] [PubMed] [Google Scholar]
- 274.Meinke A, Barahmandpour F, Wohrl S, Stoiber D, Decker T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol Cell Biol. 1996;16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 276.Mellor H, Flowers K M, Kimball S R, Jefferson L S. Cloning and characterization of a cDNA encoding rat PKR, the double-stranded RNA-dependent eukaryotic initiation factor-2 kinase. Biochim Biophys Acta. 1994;1219:693–696. doi: 10.1016/0167-4781(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 277.Melville M W, Tan S L, Wambach M, et al. The cellular inhibitor of the PKR protein kinase, P58(IPK), is an influenza virus-activated co-chaperone that modulates heat shockprotein 70 activity. J Biol Chem. 1999;274:3797–3803. doi: 10.1074/jbc.274.6.3797. [DOI] [PubMed] [Google Scholar]
- 278.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 279.Meurs E, Chang K, Galabru J, Thomas N S, Kerr I M, Williams B R G, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 280.Meurs E F, Galabru J, Barker G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Meurs E F, Wantanabe Y, Kadereit S, Barber G N, Katze M G, Chong K, Williams B R G, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5805–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Michel T, Feron O. Nitric oxide synthases: which, where, how, and why? J Clin Investig. 1997;100:2146–21552. doi: 10.1172/JCI119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Miller D M, Rahill B M, Boss J M, Lairmore M D, Durbin J E, Waldman J W, Sedmak D D. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Miller L K. Baculovirus interaction with host apoptotic pathways. J Cell Physiol. 1997;173:178–18. doi: 10.1002/(SICI)1097-4652(199711)173:2<178::AID-JCP17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 285.Min W, Ghosh S, Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16:359–368. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Mittaz L, Scott H, Rossier C, Seeburg P, Higuchi M, Antonarakis S. Cloning of a human RNA editing deaminase (ADARB1) of glutamate receptors that maps to chromosome 21q22.3. Genomics. 1997;41:210–217. doi: 10.1006/geno.1997.4655. [DOI] [PubMed] [Google Scholar]
- 287.Mogensen K E, Lewerenz M, Reboul J, Lutfalla G, Uzé G. The type I interferon receptor: structure, function, and evolution of a family business. J Interferon Cytokine Res. 1999;19:1069–1098. doi: 10.1089/107999099313019. [DOI] [PubMed] [Google Scholar]
- 288.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Muhlethaler-Mottet A, Otten L A, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Muller U, Steinhoff U, Reis L F, Hemmi S, Pavlovic J, Zinkernagel R M, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 291.Munemitsu S M, Samuel C E. Biosynthesis of reovirus-specified polypeptides. Multiplication rate but not yield of reovirus serotypes 1 and 3 correlates with the level of virus-mediated inhibition of cellular protein synthesis. Virology. 1984;136:133–143. doi: 10.1016/0042-6822(84)90254-x. [DOI] [PubMed] [Google Scholar]
- 292.Murphy D G, Dimock K, Kang C Y. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology. 1991;181:760–763. doi: 10.1016/0042-6822(91)90913-v. [DOI] [PubMed] [Google Scholar]
- 293.Muto N F, Martinand-Mari C, Adelson M E, Suhadolnik R J. Inhibition of replication of reactivated human immunodeficiency virus type 1 (HIV-1) in latently infected U1 cells transduced with an HIV-1 long terminal repeat-driven PKR cDNA construct. J Virol. 1999;73:9021–9028. doi: 10.1128/jvi.73.11.9021-9028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Nagano Y, Kojima Y. Inhibition de I'infection vaccinale par le virus homologue. C R Seances Soc Biol Fil. 1958;152:1627–1630. [PubMed] [Google Scholar]
- 295.Naka T, Fujimoto M, Kishimoto T. Negative regulation of cytokine signaling: STAT-induced STAT inhibitor. Trends Biochem Sci. 1999;24:394–398. doi: 10.1016/s0968-0004(99)01454-1. [DOI] [PubMed] [Google Scholar]
- 296.Nakajima K, Desselberger U, Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978;247:334–339. doi: 10.1038/274334a0. [DOI] [PubMed] [Google Scholar]
- 297.Nakao K, Yamashita M, Tamada Y, Hamasaki K, Ishikawa H, Kato Y, Eguchi K, Ishii N. p48 (ISGF-3γ) is involved in interferon-α-induced suppression of hepatitis B virus enhancer-1 activity. J Biol Chem. 1999;274:28075–28078. doi: 10.1074/jbc.274.40.28075. [DOI] [PubMed] [Google Scholar]
- 298.Nathan C. Inducible nitric oxide synthase: what difference does it make? J Clin Investig. 1997;100:2417–2423. doi: 10.1172/JCI119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 300.Nguyen K B, Cousens L P, Doughty L A, Pien G C, Durbin J E, Biron C A. Interferon α/β-mediated inhibition and promotion of interferon γ: STAT1 resolves a paradox Nat. Immunol. 2000;1:70–76. doi: 10.1038/76940. [DOI] [PubMed] [Google Scholar]
- 301.O'Connell M A, Keller W. Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc Natl Acad Sci USA. 1994;91:10596–10600. doi: 10.1073/pnas.91.22.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.O'Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Offermann M K, Zimring J, Mellits K H, Mathews M B. Activation of the double-stranded-RNA-activated protein kinase and induction of vascular cell adhesion molecule-1 by poly(I).poly(C) in endothelial cells. Eur J Biochem. 1995;232:28–36. doi: 10.1111/j.1432-1033.1995.tb20777.x. [DOI] [PubMed] [Google Scholar]
- 304.O'Hara P J, Nichol S T, Horodyski F M, Holland J J. Vesicular stomatitis virus defective interfering particles can contain extensive genome sequence rearrangements and base substitutions. Cell. 1984;36:915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- 305.Oldstone M B A. How viruses escape from cytotoxic lymphocyes: molecular parameters and players. Virology. 1997;234:179–185. doi: 10.1006/viro.1997.8674. [DOI] [PubMed] [Google Scholar]
- 306.Ouchi T, Lee S W, Ouchi M, Aaronson S A, Horvath C M. Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc Natl Acad Sci. 2000;97:5208–5213. doi: 10.1073/pnas.080469697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Parganas E, Wang D, Stravopodis D, Topham D J, Marine J C, Teglund S, Vanin E F, Bodner S, Colamonici O R, vanDeursen J M, Grosveld G, Ihle J N. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 308.Patel R C, Sen G C. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J Biol Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- 308a.Patel R C, Sen G C. PACT, a protein activator of the interferon-induced protein kinase. PKR EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Patel R C, Stanton P, Sen G C. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by dsRNA and heparin. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 310.Patel R C, Vestal D J, Xu Z, Bandyopadhyay S, Guo W, Erme S M, Williams B R, Sen G C. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 311.Pathak V K, Schindler D, Hershey J W B. Generation of a mutant form of protein synthesis initiation factor eIF-2 lacking the site of phosphorylation of eIF-2 kinases. Mol Cell Biol. 1988;8:993–995. doi: 10.1128/mcb.8.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Patterson J B, Samuel C E. Expression and regulation by interferon of a double-stranded RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Patterson J B, Thomis D C, Hans S L, Samuel C E. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- 314.Pavlovic J, Arzet H A, Hefti H P, Frese M, Rost D, Ernst B, Kolb E, Staeheli P, Haller O. Enhanced virus resistance of transgenic mice expressing the human MxA protein. J Virol. 1995;69:4506–4510. doi: 10.1128/jvi.69.7.4506-4510.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Pawlotsky J M, Germanidis G, Frainais P O, et al. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J Virol. 1999;73:6490–6499. doi: 10.1128/jvi.73.8.6490-6499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Pawlotsky J M, Germanidis G, Neumann A U, et al. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Pestka S, Langer J A, Zoon K C, Samuel C E. Interferons and their actions. Annu Rev Biochem. 1987;56:317–332. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 319.Pfeffer L M, Mullersman J E, Pfeffer S R, Murti A, Shi W, Yang C H. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 320.Pieper A A, Blackshaw S, Clements E E, Brat D J, Krug D K, White A J, Pinto-Garcia P, Favit A, Conover J R, Snyder S H, Verma A. Poly(ADP-ribosyl)ation basally activated by DNA strand breaks reflects glutamate-nitric oxide neurotransmission. Proc Natl Acad Sci USA. 2000;97:1845–1850. doi: 10.1073/pnas.97.4.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Piskurich J F, Wang Y, Linhoff M W, White L C, Ting J P. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- 322.Player M R, Torrence P F. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Polson A G, Bass B L, Casey J L. RNA editing of HDV antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 324.Polyak S J, Paschal D M, McArdle S, Gale M J, Jr, Moradpour D, Gretch D R. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262–1271. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- 325.Ponten A, Sick C, Weeber M, Haller O, Kochs G. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J Virol. 1997;71:2591–2599. doi: 10.1128/jvi.71.4.2591-2599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Poppers J, Mulvey M, Khoo D, Mohr I. Inhibition of PKR activation by a proline-reich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J Virol. 2000;74:11215–11221. doi: 10.1128/jvi.74.23.11215-11221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Prejean C, Colamonici O R. Role of the cytoplasmic domains of the type I interferon receptor subunits in signaling. Semin Cancer Biol. 2000;10:83–92. doi: 10.1006/scbi.2000.0311. [DOI] [PubMed] [Google Scholar]
- 328.Proud C G. PKR: a new name and new roles. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 329.Ptossi F, Blank A, Schröder A, Schwarz A, Hüssi P, Schwemmle M, Pavlovic J, Staeheli P. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J Virol. 1993;67:6726–6732. doi: 10.1128/jvi.67.11.6726-6732.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Rajan P, Swaminathan S, Zhu J, Cole C N, et al. A novel translational regulation function for the simian virus 40 large-T antigen gene. J Virol. 1995;69:785–795. doi: 10.1128/jvi.69.2.785-795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330a.Ramana C V, Gil M P, Han Y, Ransohoff R M, Schreiber R D, Stark G R. Stat1-independent regulation of gene expression in response to IFN-γ. Proc Natl Acad Sci USA. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Rani M R S, Foster G R, Leung S, Leaman D, Stark G R, Ransohoff R M. Characterization of β-R1, a gene ihat is selectively induced by interferon beta (IFN-β) compared with IFN-α. J Biol Chem. 1996;271:22878–22884. doi: 10.1074/jbc.271.37.22878. [DOI] [PubMed] [Google Scholar]
- 332.Raz R, Durbin J E, Levy D E. Acute phase response factor and additional members of the interferon-stimulated gene factor 3 family integrate diverse signals from cytokines, interferons, and growth factors. J Biol Chem. 1994;269:24391–24395. [PubMed] [Google Scholar]
- 333.Rebagliati M R, Melton D A. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 334.Rebouillat D, Hovanessian A G. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J Interferon Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- 335.Rebouillat D, Hovnanian A, David G, Hovanessian A G, Williams B R G. Characterization of the gene encoding the 100-kDa form of human 2′,5′ oligoadenylate synthetase. Genomics. 2000;70:232–240. doi: 10.1006/geno.2000.6382. [DOI] [PubMed] [Google Scholar]
- 336.Rebouillat D, Hovnanian A, Marie I, Hovanessian A G. The 100-kDa 2′,5′-oligoadenylate synthetase catalyzing preferentially the synthesis of dimeric pppA2′p5′A molecules is composed of three homologous domains. J Biol Chem. 1999;274:1557–1565. doi: 10.1074/jbc.274.3.1557. [DOI] [PubMed] [Google Scholar]
- 337.Reis L F L, Harada H, Wolchok J D, Taniguchi T, Vilcek J. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Richter M F, Schwemmle M, Herrmann C, Wittinghofer A, Staeheli P. Interferon-induced MxA protein. GTP binding and GTP hydrolysis properties. J Biol Chem. 1995;270:13512–13517. [PubMed] [Google Scholar]
- 339.Rivas C, Gil J, Melková Z, et al. Vaccinia virus E3L protein is an inhibitor of the interferon (IFN)-induced 2-5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 340.Roberts R M, Liu L, Guo Q, Leaman D, Bixby J. The evolution of the type I interferons. J Interferon Res. 1998;18:805–816. doi: 10.1089/jir.1998.18.805. [DOI] [PubMed] [Google Scholar]
- 341.Rodig S J, Meraz M A, White J M, Lampe P A, Riley J K, Arthur C D, King K L, Sheehan K C F, Pennica D, Johnson E M, Schreiber R D. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 342.Romano P R, Green S R, Barker G N, Mathews M B, Hinnebusch A G. Structural requirements for double-stranded RNA binding, dimerization, and activation of the human eIF-2α kinase DAI in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:365–368. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor 3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Rueter S M, Emeson R B. Adenosine to inosine conversion in mRNA. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998. pp. 343–361. [Google Scholar]
- 345.Ryman K D, Klimstra W B, Nguyen K B, Biron C A, Johnston R E. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J Virol. 2000;74:3366–3378. doi: 10.1128/jvi.74.7.3366-3378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Rysiecki G, Gewert D R, Williams B R G. Constitutive expression of a 2′,5′-oligoadenylate synthetase cDNA results in increased antiviral activity and growth suppression. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 347.Salehzada T, Silhol M, Steff A M, Lebleu B, et al. 2′,5′-Oligoadenylate-dependent Rnase L is a dimer of regulatory and catalytic subunits. J Biol Chem. 1993;268:7733–7740. [PubMed] [Google Scholar]
- 348.Samuel C E. Mechanism of interferon action. Phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated protein kinase possessing site-specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc Natl Acad Sci USA. 1979;76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Samuel C E. Mechanisms of the Antiviral Actions of IFN. Prog Nucleic Acid Res Mol Biol. 1988;35:27–72. doi: 10.1016/s0079-6603(08)60609-1. [DOI] [PubMed] [Google Scholar]
- 350.Samuel C E. Role of the RNA-dependent protein kinase in the regulated expression of genes in transfected cells. Pharmacol Ther. 1992;54:307–317. doi: 10.1016/0163-7258(92)90005-k. [DOI] [PubMed] [Google Scholar]
- 351.Samuel C E. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 352.Samuel C E. The eIF-2α protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 353.Samuel C E. Protein-nucleic acid interactions and cellular responses to interferon. Methods J. 1998;15:161–165. doi: 10.1006/meth.1998.0620. [DOI] [PubMed] [Google Scholar]
- 354.Samuel C E. Reoviruses and the interferon system. Curr Top Microbiol Immunol. 1998;233:125–145. doi: 10.1007/978-3-642-72095-6_7. [DOI] [PubMed] [Google Scholar]
- 355.Samuel C E, Duncan R, Knutson G S, Hershey J W. Mechanism of interferon action. Increased phosphorylation of protein synthesis initiation factor eIF-2 alpha in interferon-treated, reovirus-infected mouse L929 fibroblasts in vitro and in vivo. J Biol Chem. 1984;259:13451–13457. [PubMed] [Google Scholar]
- 356.Samuel C E, Kuhen K L, George C X, Ortega L G, Rende-Fournier R, Tanaka H. The PKR protein kinase—an interferon-inducible regulator of cell growth and differentiation. Int J Hematol. 1997;65:227–237. doi: 10.1016/s0925-5710(96)00544-0. [DOI] [PubMed] [Google Scholar]
- 357.Sarkar S N, Sen G C. Production, purification, and characterization of recombinant 2′,5′-oligoadenylate synthetases. Methods. 1998;15:233–242. doi: 10.1006/meth.1998.0627. [DOI] [PubMed] [Google Scholar]
- 358.Sarkar S N, Ghosh A, Wang H-W, Sung S-S, Sen G C. The nature of the catalytic domain of 2′-5′-oligoadenylate synthetases. J Biol Chem. 1999;274:25535–25542. doi: 10.1074/jbc.274.36.25535. [DOI] [PubMed] [Google Scholar]
- 359.Saunders M E, Gewert D R, Tugwell M E, McMahon M, et al. Human 2-5A synthetase: characterization of a novel cDNA and corresponding gene structure. EMBO J. 1985;4:1761–1768. doi: 10.1002/j.1460-2075.1985.tb03848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Scadden A D J, Smith C W J. A ribonuclease specific for inosine-containing RNA: a potential role in antiviral defense. EMBO J. 1997;16:2140–2149. doi: 10.1093/emboj/16.8.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 362.Schindler C. Cytokines and JAK-STAT signaling. Exp Cell Res. 1999;253:7–14. doi: 10.1006/excr.1999.4670. [DOI] [PubMed] [Google Scholar]
- 363.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 364.Schmechel S, Chute M, Skinner P, et al. Preferential translation of reovirus mRNA by a s3-dependent mechanism. Virology. 1997;232:62–73. doi: 10.1006/viro.1997.8531. [DOI] [PubMed] [Google Scholar]
- 365.Schmedt C, Green S R, Manchi L, Taylor D R, Ma Y, Mathews M B. Mechanism of interferon action. RNA-binding activity of full-length and R-domain forms of the RNA-dependent protein kinase (PKR): determination of the KD values for VAI and TAR RNAs. J Mol Biol. 1995;249:29–44. [Google Scholar]
- 366.Schneider R J, Shenk T. Impact of virus infection on host cell protein synthesis. Annu Rev Biochem. 1987;56:317–332. doi: 10.1146/annurev.bi.56.070187.001533. [DOI] [PubMed] [Google Scholar]
- 367.Schwart T, Lowenhaupt K, Kim Y G, Li L Y, Brown B A, Herbert A, Rich A. Protein dissection of Zab, the Z-DNA-binding domain of human ADAR1. J Biol Chem. 1999;274:2899–2906. doi: 10.1074/jbc.274.5.2899. [DOI] [PubMed] [Google Scholar]
- 368.Schwers A, Vanden B C, Mainhoudt M, Beduin J M, Werenne J, Pastoret P P. Experimental rotavirus diarrhoea in colostrum-deprived newborn calves: assay of treatment by administration of bacterially producted human interferon (Hu-IFN alpha 2) Ann Rech Vet. 1985;16:213–218. [PubMed] [Google Scholar]
- 369.Seeburg P H, Higuchi M, Sprengel R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 370.Sen G C, Lengyel P. The interferon system: a bird's eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 371.Servant M J, ten Oever B, LePage C, Conti L, Gessani S, Julkunen I, Lin R, Hiscott J. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J Biol Chem. 2000;276:355–363. doi: 10.1074/jbc.M007790200. [DOI] [PubMed] [Google Scholar]
- 372.Sharmeen L, Bass B, Sonenberg N, Weintraub H, Groudine M. Tat-dependent adenosine-to-inosine modification of wild-type transactivation response RNA. Proc Natl Acad Sci USA. 1991;88:8096–8100. doi: 10.1073/pnas.88.18.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Sharp T V, Moonan F, Romashko A, et al. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology. 1998;25:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 374.Sharpe A H, Fields B N. Pathogenesis of reovirus infection. In: Joklik W K, editor. The Reoviridae. New York, N.Y: Plenum Press; 1983. pp. 229–285. [Google Scholar]
- 375.Shen Y, Shenk T E. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 376.Sherry B, Baty C J, Blum M A. Reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than generation of infectious virus in cardiac myocytes. J Virol. 1996;70:6709–6715. doi: 10.1128/jvi.70.10.6709-6715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Shiloh M U, MacMicking J D, Nicholson S, Brause J E, Potter S, Marino M, Fang F, Dinauer M, Nathan C. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 378.Shors S T, Beattie E, Paoletti E, et al. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-α. J Interferon Cytokine Res. 1998;18:721–729. doi: 10.1089/jir.1998.18.721. [DOI] [PubMed] [Google Scholar]
- 379.Shors T, Kibler K V, Perkins K B, et al. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology. 1997;239:269–276. doi: 10.1006/viro.1997.8881. [DOI] [PubMed] [Google Scholar]
- 379a.Shtrichman R, Samuel C E. Role of gamma interferon in antimicrobial immunity. Curr Opin Microbiol. 2001;4:251–259. doi: 10.1016/s1369-5274(00)00199-5. [DOI] [PubMed] [Google Scholar]
- 380.Shuai K. The STAT family of proteins in cytokine signaling. Prog Biophys Mol Biol. 1999;71:405–422. doi: 10.1016/s0079-6107(98)00051-0. [DOI] [PubMed] [Google Scholar]
- 381.Siegal F P, Kadowaki N, Shodell M, Fitzgerald-Bocarsly P A, Shah K, Ho S, Antonenko S, Liu Y J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 382.Simon A, Fäh J, Haller O, Staeheli P. Interferon-regulated Mx genes are not responsive to interleukin-1, tumor necrosis factor, and other cytokines. J Virol. 1991;65:968–971. doi: 10.1128/jvi.65.2.968-971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Simon J H, Lull J, Jacobs L D, Rudick R A, Cookfair D L, Herndon R M, Richert J R, Salazar A M, Sheeder J, Miller D, McCabe K, Serra A, Campion M K, Fischer J D, Goodkin D E, Simonian N, Lajaunie M, Wende K, Martens-Davidson A, Kinkel R P, Munschauer F E., III A longitudinal study of T1 hypointense lesions in relapsing MS: MSCRG trial of interferon beta-1a. Multiple Sclerosis Collaborative Research Group. Neurology. 2000;55:185–192. doi: 10.1212/wnl.55.2.185. [DOI] [PubMed] [Google Scholar]
- 384.Simpson L, Emeson R B. RNA editing. Annu Rev Neurosci. 1996;19:27–52. doi: 10.1146/annurev.ne.19.030196.000331. [DOI] [PubMed] [Google Scholar]
- 385.Smith G L, Symons J A, Alcami A. Poxviruses: interfering with interferon. Semin Virol. 1998;8:409–418. [Google Scholar]
- 386.Sommer B, Köhler M, Sprengel R, Seeburg P H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 387.Sonenberg N. Measures and countermeasures in the modulation of initiation factor activities by viruses. New Biol. 1990;2:402–409. [PubMed] [Google Scholar]
- 388.Song M M, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 389.Squire J, Zhou A, Hassel B A, Nie H, et al. Localization of the interferon-induced, 2-5A-dependent Rnase Gene (RNS4) to human chromosome 1q25. Genomics. 1994;19:174–175. doi: 10.1006/geno.1994.1033. [DOI] [PubMed] [Google Scholar]
- 390.Squire J, Meurs E G, Chong K L, McMillan N A J, Hovanessian A G, Williams B R G. Localization of the human interferon-induced, ds-RNA activated p68 kinase gene (PRKR)to chromosome 2p21–p22. Genomics. 1993;16:768–770. doi: 10.1006/geno.1993.1263. [DOI] [PubMed] [Google Scholar]
- 391.Staeheli P, Sutcliffe J G. Identification of a second interferon-regulated murine Mx gene. Mol Cell Biol. 1988;8:4524–4528. doi: 10.1128/mcb.8.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 393.Staeheli P, Pitossi F, Pavlovic J. Mx proteins: GTPases with antiviral activity. Trends Cell Biol. 1993;3:268–272. doi: 10.1016/0962-8924(93)90055-6. [DOI] [PubMed] [Google Scholar]
- 394.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 395.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 396.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 397.Sterner D E, Berger S L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Stojdal D F, Abraham N, Knowles S, Marius R, Brasey A, Lichty B D, Brown E G, Sonenberg N, Bell J C. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J Virol. 2000;74:9580–9585. doi: 10.1128/jvi.74.20.9580-9585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Tan S L, Katze M G. The emerging role of the interferon-induced PKR protein kinase as a apoptotic effector: a new face of death. J Interferon Cytokine Res. 1999;19:543–554. doi: 10.1089/107999099313677. [DOI] [PubMed] [Google Scholar]
- 400.Tan S L, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Tanaka H, Samuel C E. Mechanism of interferon action. Structure of the gene encoding mouse PKR, the interferon-inducible RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:7995–7999. doi: 10.1073/pnas.91.17.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Tanaka N, Sato M, Lamphier M, Nozawa H, et al. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells. 1998;3:29–37. doi: 10.1046/j.1365-2443.1998.00164.x. [DOI] [PubMed] [Google Scholar]
- 403.Taraseviciene L, Miczak A, Apirion D. The gene specifying RNAase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- 404.Taylor DR, Shi S T, Romano P R, Barber G N, Lai M M. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 405.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Teodoro J G, Branton P E. Regulation of p53-dependent apoptosis, transcriptional repression and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J Virol. 1997;71:3620–3627. doi: 10.1128/jvi.71.5.3620-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 408.Thomas H C, Török M E, Foster G R. Hepatitis C virus dynamics in vivo and the antiviral efficacy of interferon alfa therapy. Hepatology. 1999;29:1333–1334. doi: 10.1002/hep.510290453. [DOI] [PubMed] [Google Scholar]
- 409.Thomis D C, Samuel C E. Mechanism of interferon action: autoregulation of RNA-dependent P1/eIF-2α protein kinase (PKR) expression in transfected mammalian cells. Proc Natl Acad Sci USA. 1992;89:10837–10841. doi: 10.1073/pnas.89.22.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Thomis D C, Samuel C E. Mechanism of interferon action: evidence for intermolecular autophosphorylation and autoactivation of the interferon-induced, RNA-dependent protein kinase PKR. J Virol. 1993;67:7695–7700. doi: 10.1128/jvi.67.12.7695-7700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Thomis D C, Samuel C E. Mechanism of interferon action: characterization of the intermolecular autophosphorylation of PKR, the interferon-induced, RNA-dependent protein kinase. J Virol. 1995;69:5195–5198. doi: 10.1128/jvi.69.8.5195-5198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Thomis D C, Doohan J D, Samuel C E. Mechanism of interferon action: cDNA structure, expression and regulation of the interferon-induced, RNA-dependent PI/eIF-2α protein kinase from human cells. Virology. 1992;188:33–46. doi: 10.1016/0042-6822(92)90732-5. [DOI] [PubMed] [Google Scholar]
- 413.Tollefson A E, Hermiston T W, Lichtenstein D L, et al. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature. 1998;392:726–730. doi: 10.1038/33712. [DOI] [PubMed] [Google Scholar]
- 414.Tschopp J, Thome M, Hofmann K, Meinl E. The fight of viruses against apoptosis. Curr Opin Genet Dev. 1998;8:82–87. doi: 10.1016/s0959-437x(98)80066-x. [DOI] [PubMed] [Google Scholar]
- 415.Ulker N, Samuel C E. Mechanism of interferon action: inhibition of vesicular stomatitis virus replication in human amnion U cells by cloned human gamma-interferon. II. Effect on viral macromolecular synthesis. J Biol Chem. 1985;260:4324–4330. [PubMed] [Google Scholar]
- 416.Van den Broek M F, Müller U, Huang S, et al. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Van der Bliek A M. Functional diversity in the dynamin family. Trends Cell Biol. 1999;3:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 418.Voeten J T M, Bestebroer T M, Nieuwkoop N J, Fouchier F A M, Osterhaus A D, Rimmelzwaan G F. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J Virol. 2000;74:6800–6807. doi: 10.1128/jvi.74.15.6800-6807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Wagner R W, Smith J E, Cooperman B S, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Waldburger J M, Masternak K, Muhlethaler-Mottet A, Villard J, Peretti M, Landmann S, Reith W. Lessons from the bare lymphocyte syndrome: molecular mechanisms regulating MHC class II expression. Immunol Rev. 2000;178:148–165. doi: 10.1034/j.1600-065x.2000.17813.x. [DOI] [PubMed] [Google Scholar]
- 421.Wang H, Chatterjee G, Meyer J J, Liu C-J, Manjunath N A, Bray-Ward P, Lengyel P. Characteristics of three homologous 202 genes (ifi202a, ifi202b, ifi202c) from the murine interferon-activatable gene 200 cluster. Genomics. 1999;60:281–294. doi: 10.1006/geno.1999.5923. [DOI] [PubMed] [Google Scholar]
- 422.Wang Q, Floyd-Smith G. The p69/71 2-5A synthetase promoter contains multiple regulatory elements required for interferon-alpha-induced expression. DNA Cell Biol. 1997;16:1385–1394. doi: 10.1089/dna.1997.16.1385. [DOI] [PubMed] [Google Scholar]
- 423.Wang Y, Zeng Y, Murray J M, Nishikura K. Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene. J Mol Biol. 1995;254:184–195. doi: 10.1006/jmbi.1995.0610. [DOI] [PubMed] [Google Scholar]
- 424.Wathelet M G, Linc C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 425.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Weber F, Haller O, Kochs G. MxA GTPase blocks reporter gene expression of reconstituted Thogoto virus ribonucleoprotein complexes. J Virol. 2000;74:560–563. doi: 10.1128/jvi.74.1.560-563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Weier H U G, George C X, Greulich K M, Samuel C E. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1-21.2. Genomics. 1995;30:372–375. doi: 10.1006/geno.1995.0034. [DOI] [PubMed] [Google Scholar]
- 428.Weier H U G, George C X, Lersch R A, Breitweser S, Cheng J F, Samuel C E. Assignment of the RNA-specific adenosine deaminase gene (Adar) to mouse chromosome 3F2 by in situ hybridization. Cytogenet Cell Genet. 2000;89:214–215. doi: 10.1159/000015615. [DOI] [PubMed] [Google Scholar]
- 429.Weiler S R, Gooya J B, Ortiz M, Tsai S, Collins S J, Keller R. D3: a gene induced during myeloid cell differentiation of Linlo c-Kit+Sca-1(+) progenitor cells. Blood. 1999;93:527–536. [PubMed] [Google Scholar]
- 430.Weiner A, Erickson A L, Kansopon J, Crawford J, Muchmore E, Hughes A L, Houghton M, Walker C M. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Nat Acad Sci USA. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 432.Williams B R G, Saunders M E, Willard W F. Interferon-regulated human 2-5A synthetase gene maps to chromosome 12. Somatic Cell Mol Genet. 1986;12:403–408. doi: 10.1007/BF01570735. [DOI] [PubMed] [Google Scholar]
- 433.Witt P L, Marie I, Robert N, Irizarry A, et al. Isoforms p69 and p100 of 2′,5′-oligoadenylate synthetase induced differentially by interferons in vivo and in vitro. J Interferon Res. 1993;13:17–23. doi: 10.1089/jir.1993.13.17. [DOI] [PubMed] [Google Scholar]
- 434.Wreschner D H, McCaule J W, Skehel J J, Kerr I M. Interferon action-sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 435.Xing Z, Zganiacz A, Wang J, Divangahi M, Nawaz F. IL-12 independent Th1-type immune responses to respiratory viral infection: requirement of IL-18 for IFN-gamma release in the lung but not for the differentiation of viral-reactive Th1-type lymphocytes. J Immunol. 2000;164:2575–2584. doi: 10.4049/jimmunol.164.5.2575. [DOI] [PubMed] [Google Scholar]
- 436.Yang Y L, Reis L F L, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R G, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Yasukawa H, Sasaki A, Yoshimura A. Negative regulators of cytokine signaling pathways. Annu Rev Immunol. 2000;18:143–164. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- 438.Yeow W-S, Au W-C, Juang Y-T, Fields C D, Dent C L, Gewert D R, Pitha P. Reconstitution of virus-mediated expression of interferon a genes in human fibroblast cells by ectopic interferon regulatory factor-7. J Biol Chem. 2000;275:6313–6320. doi: 10.1074/jbc.275.9.6313. [DOI] [PubMed] [Google Scholar]
- 439.Yewdell J W, Bennink J R. Mechanisms of viral interference with MHC class I antigen processing and presentation. Annu Rev Cell Dev Biol. 1999;15:579–606. doi: 10.1146/annurev.cellbio.15.1.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.York I A, Rock K L. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 442.Young D F, Didcock L, Goobourn S, Randall R E. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology. 2000;269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]
- 443.Young H A. Regulation of interferon-γ gene expression. J Interferon Cytokine Res. 1996;16:563–568. doi: 10.1089/jir.1996.16.563. [DOI] [PubMed] [Google Scholar]
- 444.Yu Z, Manickan E, Rouse B T. Role of interferon-gamma in immunity to herpes simplex virus. J Leukoc Biol. 1996;60:528–532. doi: 10.1002/jlb.60.4.528. [DOI] [PubMed] [Google Scholar]
- 445.Yue Z, Shatkin A J. Double-stranded RNA-dependent protein kinase PKR is regulated by reovirus structural proteins. Virology. 1997;234:364–371. doi: 10.1006/viro.1997.8664. [DOI] [PubMed] [Google Scholar]
- 446.Zhang J J, Zhao Y, Chait B T, Lathem W W, Ritzi M, Knippers R, Darnell J E., Jr Ser727-dependent recruitment of MCM5 by Stat1alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Zhou A, Paranjape J M, Der S D, Williams B R G, Silverman R H. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]
- 449.Zhou A, Paranjape J, Brown T L, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman R H. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Zhou A, Hassel B A, Silverman R H. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 451.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Zhu H, Cong J P, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]