Sit down, relax and unwind: structural insights into RecQ helicase mechanisms (original) (raw)
Abstract
Helicases are specialized molecular motors that separate duplex nucleic acids into single strands. The RecQ family of helicases functions at the interface of DNA replication, recombination and repair in bacterial and eukaryotic cells. They are key, multifunctional enzymes that have been linked to three human diseases: Bloom's, Werner's and Rothmund–Thomson's syndromes. This review summarizes recent studies that relate the structures of RecQ proteins to their biochemical activities.
INTRODUCTION
Helicases are molecular motors that couple the energy of nucleoside triphosphate (NTP) hydrolysis to the unwinding and remodeling of double-stranded (ds) DNA and RNA. They are essential components of nearly every cellular nucleic acid metabolic process including replication, recombination, repair, transcription and translation. Helicases have historically been categorized by their conservation of characteristic amino acid sequences (helicase motifs) and by their direction of translocation along nucleic acid substrates (1). The addition of high-resolution structures of several helicases gathered over the past decade have resolved molecular details of helicase function and have provided important insights into the biochemical mechanisms utilized by different helicase families (2). This review focuses on recent structural and functional studies of individual domains from the RecQ family of DNA helicases. We have arranged the review to first describe the three-dimensional structures of domains found in RecQ proteins and then to relate these structures to each domain's biochemical and cellular roles.
RecQ DNA HELICASES
RecQ DNA helicases are wide spread in nature and have been identified in organisms ranging from bacteria to humans. The first RecQ helicase was identified over 20 years ago in Escherichia coli through a screening for mutants resistant to thymineless-death (3). recQ genes have since been identified in every eubacterial and eukaryotic species sequenced to date. The importance of the RecQ family is illustrated by three diseases that are linked to defects in different human RecQ paralogs (4–6). These diseases, Bloom's, Werner's and Rothmund–Thomson syndromes, are caused by mutations in the BLM, WRN and RECQ4 genes, respectively. Although these autosomal-recessive disorders are distinct, they all share a predisposition to cancer, increased chromosomal aberrations and sensitivity to DNA-damaging agents (7). The cellular phenotypes of RecQ-deficiency syndromes underscore the caretaking role that RecQ helicases are thought to play in higher eukaryotes in maintaining genomic integrity. The functions of RecQ proteins have been the focus of numerous investigations and there have been several recent reviews that highlight biochemical, cellular and clinical aspects of these proteins (7–10). We will therefore only briefly describe the activities of RecQ proteins here as a backdrop for the subject of this review.
In E.coli, RecQ is a component of the RecF pathway of recombinational repair (3). The RecF pathway processes gapped DNA and can also act on dsDNA ends in cells when the RecBCD repair pathway has been inactivated (11). In its dsDNA break (DSB) repair function, RecQ is thought to aid recombination by generating a single-stranded (ss) DNA through its helicase activity; this ssDNA is processed by RecJ exonuclease, another component of the RecF pathway, which degrades the 5′ ssDNA. RecFOR then mediates RecA loading on to the remaining 3′ ssDNA to form a filament that is competent for recombination (12–14). RecQ helicase activity has also been shown to function in suppressing illegitimate recombination, rescuing stalled replication forks and promoting induction of the SOS response in E.coli (15–17). Thus, E.coli RecQ is important for cellular genome maintenance and its activities are linked by RecQ's DNA helicase function.
As is the case with their bacterial cousin, eukaryotic RecQ proteins are also broadly important for genome maintenance. Several diverse cellular functions have been ascribed to eukaryotic RecQ proteins, including roles in replication fork maintenance, DNA-damage checkpoint signaling, recombination regulation and telomere stability [reviewed in (7,10,18–20)]. As with bacterial RecQ proteins, most eukaryotic RecQ family members are helicases and this activity is the key to their functions in cells. In addition, recent findings have demonstrated that several human RecQ helicases (BLM, WRN, RecQ1 and RecQ5β) are capable of annealing complimentary ssDNA (21–25). This activity has been hypothesized to contribute to RecQ's ability to aid strand-invasion and to promote formation of structures such as regressed replication forks (23). Under cellular conditions, however, it is likely that these RecQs primarily act to unwind DNA, due to the presence of ssDNA-binding protein and high concentrations of ATP, which favor unwinding over annealing (22–25). Although a satisfying unified model linking the apparently diverse roles of RecQ proteins, especially those in eukaryotic cells, is currently lacking, it is the notion of this review that structural and functional analyses of common structural elements in RecQ proteins will be informative as to the conserved mechanisms employed by the RecQ family and that this approach is key to understanding their cellular roles.
MOTORING AT HIGH RESOLUTION: THE STRUCTURES OF RecQ FUNCTIONAL DOMAINS
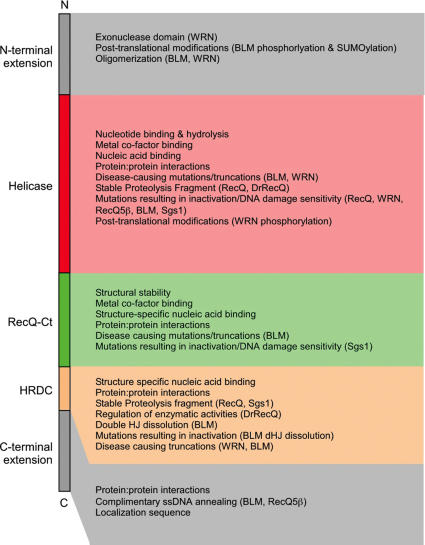
Three conserved sequence elements are commonly found in RecQ helicases [Helicase, RecQ-C-terminal (RecQ-Ct) and Helicase-and-RNaseD-like-C-terminal (HRDC) domains] (26) (Figure 1). The helicase domain is conserved in all RecQ proteins whereas the RecQ-Ct and HRDC domains are found in most RecQs but are missing in a small subset of family members (e.g. human RecQ4 lacks recognizable RecQ-Ct and HRDC domains and RecQ5β lacks an HRDC domain). In addition to these elements, eukaryotic RecQ proteins often encode N- and C-terminal extensions that confer additional enzymatic activities, such as the exonuclease domain in WRN, sequences that are important for binding to heterologous proteins and motifs that facilitate proper subcellular localization (7) (Figure 1).
Figure 1.
Schematic illustration of the structural or functional properties of RecQ protein domains. The N- and C-terminal extensions (gray) from the conserved core domains and the Helicase (red), RecQ-Ct (green) and HRDC (peach) domains are shown on the left-hand side. Boxes outlining contributions or deficiencies associated with each region are shown on the right-hand side in corresponding colors.
Limited proteolysis studies demonstrated that the prototypical E.coli RecQ protein is composed of a modular structure that can be divided into two structural domains (27). The first includes the Helicase and RecQ-Ct elements, which combine to form what has been termed the RecQ ‘catalytic core’ owing to its retention of ATPase and helicase activities. The HRDC domain forms the second structural domain in E.coli RecQ in which it is important for structure-specific DNA binding. Dissection of domains from BLM (28), WRN (29) and Saccharomyces cerevisiae Sgs1 (30) are consistent with conservation of a similar modular domain structure throughout the RecQ family. Crystal structures of both the structural domains of E.coli RecQ have been determined as has an NMR structure of the S.cerevisiae Sgs1 HRDC domain (Figure 2) (30–32). The combination of these structures presents a high-resolution view of the conserved elements found in a typical RecQ helicase and has helped to explain the functions of RecQ domain as well as providing a rationale for why mutations in the conserved regions of eukaryotic RecQ family members cause disease.
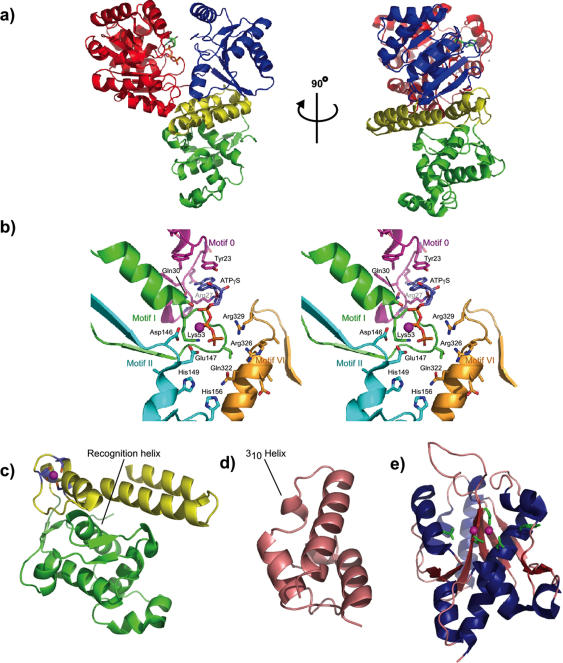
Figure 2.
Structural features of RecQ DNA helicases. (a) Orthogonal views of a ribbon diagram of RecQ catalytic core crystal structure (31). The catalytic core domain is composed of the helicase domain shown in red and blue, and the RecQ-Ct domain shown in yellow and green. The ATPγS moiety is colored according to atom identity where carbon = slate, nitrogen = blue, oxygen = red, and phosphorus or sulfur = orange. (b) Stereo diagram of the ATP-binding site and residues predicted to be involved in hydrolysis. The main chain of the conserved motifs and their relevant residues are colored as follows: Motif 0 in purple, Motif I in green, Motif II in cyan and Motif VI in gold. An Mn2+ ion is shown as a purple sphere above Lys53. (c) Subdomains of the RecQ-Ct domain. The zinc-binding domain is shown in yellow, with the four Zn2+ coordinating cysteine residues shown in blue. The winged-helix domain is shown in green and contains the ‘recognition helix’. (d) The HRDC domain (32) is shown in salmon and its 310 helix is labeled. (e) Structure of the WRN exonuclease domain bound to Mg2+ (74). Helices (blue), sheets (red) and loops (salmon) are shown with active site residue sidechains as green sticks and Mg2+ ions as magenta spheres. Structure figures created using PyMol (79).
Helicase domain
The RecQ helicase domain is typical of other Superfamily (SF) 1 and 2 helicases in that it contains the seven commonly conserved helicase motifs (I, Ia, II, III, IV, V, VI) involved in coupling the energy of NTP hydrolysis to the separation of nucleic acid duplexes (33). The first high-resolution structures of helicases demonstrated that a subset of these conserved motifs folds to create an NTP-binding site and that the structures of helicases share significant similarity to that of RecA (34–36). It now appears that all helicases contain RecA-like modules that form their core motor domains, and that many such enzymes use two RecA-like domains to facilitate DNA unwinding (Figure 2a, shown in blue and red). The helicase domain of RecQ (an SF2 helicase), with some exceptions noted below, is quite similar to other SF1 and SF2 helicases of known structure.
ATP binding and hydrolysis
In addition to the classic helicase motifs, RecQ family members possess a sequence element N-terminal to motif I called ‘motif 0’ that is composed of the sequence Lx3(F/Y/W)Gx3F(R/K)x2Q (27). A similar element called a ‘Q motif’ has also been characterized in DEAD-box RNA helicases where it is important for NTP binding and hydrolysis (37). The crystal structure of the E.coli RecQ catalytic core domain bound to ATPγS (an ATP analog) showed that motif 0 forms a loop connecting two helices that creates a pocket to accommodate the adenine base of the ATPγS molecule (31) (Figure 2b). The C-terminal Gln residue (Gln30) of the motif forms hydrogen bonds with the N6 and N7 atoms of the adenine; in this way motif 0 binds ATP specifically, which is E.coli RecQ's preferred NTP cofactor (38). Two other motif 0 residues, Tyr23 and Arg27, are positioned along either side of the adenine base and stabilize its association with the enzyme (Figure 2b, shown in purple). This three-dimensional arrangement is remarkably similar to that of the Q motif in RNA helicases (37) and could indicate an evolutionary linkage between RecQ and DEAD-box helicases.
The effects of motif 0 mutations on RecQ activity show that it is critical for enzyme function. First, a naturally occurring BLM missense mutation that causes Bloom's syndrome maps to motif 0 (4). This mutation substitutes an Arg for the Gln in motif 0, which, when integrated in murine BLM, inactivates the variant as an ATPase and helicase (39). An analogous mutation in S.cerevisiae Sgs1 renders the cells as sensitive to DNA-damaging agents as an sgs1 deletion strain (40), whereas its mutation in human RecQ5β reduces the ATPase activity of the enzyme by half (25). These studies demonstrate the importance of motif 0 to RecQ helicase function and indicate that RecQ motor function is the key to its cellular activities.
Helicase motifs I and II, also known as the Walker A and B motifs, share similarity to NTP-binding sequences of the E.coli ATP synthase (41). These motifs serve as sites of interaction with ATP and Mg2+ in many enzymes, and function by stabilizing the transition-state intermediate formed between a water nucleophile and the γ-phosphate of ATP (42). Motif I forms the phosphate-binding loop (or ‘P loop’) and has the consensus sequence Gx4GK(T/S). In E.coli RecQ, the ‘GKS’ residues lie at the end of a loop connecting its β1 strand and α3 helix (31). The last residue of motif I, Ser54, helps in positioning a Mg2+ ion near the ATP phosphate groups where it is likely to stabilize the hydrolysis transition state (Figure 2b, shown in green) (43). The penultimate residue, Lys53, is thought to bind the γ-phosphate during ATP hydrolysis. Similar to the results from motif 0 mutations, alterations of motif I sequence in RecQ family members produce dire consequences: mutation of the motif I lysine in E.coli RecQ (44), human WRN (45), human RecQ5β (25) or S.cerevisiae Sgs1 (46) results in the loss of ATPase and helicase activity. Moreover, introduction of the human WRN lysine mutant into mice induces a Werner's syndrome phenotype in tail-derived fibroblasts (47).
Motif II is located in the loop connecting the β5 strand and α8 helix of E.coli RecQ, and is composed of the conserved sequence DExH. The carboxyl side chain of Asp146 helps to coordinate the catalytic Mg2+ ion, whereas Glu147 probably acts as the catalytic base that polarizes a water molecule for attack on the ATP γ-phosphate. The structures of other SF2 helicases have revealed a conserved interaction between the histidine residue of motif II and a glutamine residue from motif VI (48,49). E.coli RecQ encodes two histidines in this portion of the molecule that are in close proximity to Gln322 from its motif VI: His149 and His156. His149 is part of motif II whereas His156 is in an aromatic-rich loop (or ‘ARL’) located between motifs II and III that is similar to an ARL involved in DNA binding in PcrA (Figure 2b, shown in cyan) (50). Thus, it is presently unclear how motifs II and VI might interact during ATP hydrolysis in RecQ but the structure of the RecQ catalytic core has identified candidates that could facilitate coordination between the motifs.
Helicase domain dynamics
As is the case with other helicases, RecQ proteins are almost certainly dynamic enzymes that have different structures when bound to different reaction components (e.g. ATP, ADP and/or DNA). A consideration of domain mobility in RecQ proteins is important since it will be linked to activity in the enzyme. The aforementioned proximity of residues from motifs II and VI may be important for such motions. It is possible, for example, that the position of His149 or His156 could be sensitive to the presence of ATP, ADP or DNA and that this difference is relayed to the second RecA-like module in the RecQ helicase domain through an interaction with Gln322. Interestingly, residues in the E.coli RecQ ARL, which contains His156, have been recently shown to be critical for coupling ATP hydrolysis to DNA unwinding (51). Mutation of Trp154, Phe158 or Arg159 in E.coli RecQ reduces its DNA-dependent ATP hydrolysis and helicase activity without affecting DNA binding. Mutation of the equivalent Trp and Arg residues in S.cerevisiae Sgs1 makes cells as sensitive to DNA-damaging agents as sgs1 strain (52), indicating their importance in vivo. This may point toward the ARL as an important link among ATP hydrolysis, DNA binding and DNA unwinding as has been demonstrated in PcrA and Rep helicases when similar residues in their ARLs have been mutated (53,54), although the ARLs in these SF1 enzymes form part of motif III.
Another frequent means by which the RecA-like domains of helicases communicate is through an ‘arginine finger’ that protrudes into the ATP-binding pocket from the second RecA-like module (2). Once an NTP is bound, this arginine is displaced and the position of the second domain is subsequently altered. In E.coli RecQ, Arg326 and Arg329 are both near the ATP-binding site of the first domain, though it is unknown which, if either, of these two residues might serve as an arginine finger (Figure 2b). Arg329 is a conserved residue in the middle of motif VI (Qx2GRx2R) and its position relative to the ATP-binding lobe in RecQ is similar to that of the Arg-finger in PcrA (Figure 2b, shown in gold color) (50). Although such studies are only beginning to take shape, an understanding of RecQ helicase domain dynamics will be essential for an appreciation of the mechanisms underlying its motor functions.
Addition features of the RecQ helicase domain
The roles of conserved helicase motifs Ia, IV and V are less well defined for RecQ than for its other motifs. These motifs typically comprise aromatic and charged residues and are involved in binding to the phosphate backbone or bases of oligonucleotides in SF1 helicases such as Rep, PcrA and NS3 (35,50,55), and the SF2 helicase HCV (48). Precisely how RecQ binds DNA is presently unclear, though it may combine strategies from both SF1 and SF2 helicases. Based on its sequence, RecQ is an SF2 helicase, but the presence of aromatic and charged regions similar to the ssDNA-binding residues in SF1 helicases (51) and the diminished ability of the WRN and BLM proteins to bind DNA substrates with restricted backbone flexibility (56) suggest that RecQs may also utilize a base-flipping mechanism proposed for SF1 helicases. Further examination of the roles of potential DNA-binding residues in RecQ proteins should provide useful insights into its mechanism of DNA binding and unwinding.
Several disease-causing and other debilitating mutations map to the Helicase domain of eukaryotic RecQ proteins. All of the known disease-linked WRN and RECQ4 mutations, along with most of the known BLM disease-linked mutations, result in the production of truncated proteins that are mislocalized due to a failure to express a C-terminal localization sequence (4–6). However, there are also seven known BLM missense mutations that cause Bloom's syndrome; five map to the helicase domain whereas the remaining two map to the RecQ-Ct domain (4). Besides the motif 0 Gln-to-Arg substitution described above, the four remaining mutations change the polarity or size of residues within the helicase domains and are predicted to lead to destabilization of the helicase domain (31). In addition, a recent study on the effects of polymorphisms in WRN that do not cause Werner's syndrome highlighted an Arg-to-Cys mutation near motif VI that reduced both its helicase and exonuclease activity (57). These findings indicate that proper helicase activity is an essential part of RecQ cellular activity.
RecQ-Ct domain
The RecQ-Ct domain forms the second largest conserved domain in the RecQ family. It is unique to RecQ family members and is best defined at the sequence level by the conservation of four cysteine residues in a zinc-binding motif, C(R/H)(R/H)x_n_CxxCDxC. In the E.coli RecQ catalytic core structure, Cys380 and Cys403 are encoded within a pair of helices whereas Cys397 and Cys400 lie within a loop connecting the helices (31). Together, they form a zinc-binding scaffold that is integral to the structural stability of the protein, as is evident by the insolubility and degradation of E.coli RecQ and BLM when mutations in these conserved cysteines are made (Figure 2c, shown in blue) (28,31,58,59). Mutations in either of the two of these cysteines in BLM are sufficient to cause Bloom's syndrome (4), probably due to misfolding of the protein when this region is disrupted. Similar missense mutations of S.cerevisiae Sgs1 Cys residues result in enhanced sensitivity to DNA-damaging agents (40,52). Additional activities of RecQ's zinc-binding motif beyond protein stability, such as DNA binding or protein binding, are certainly possible but are likely to have been masked in mutagenesis and deletion analyses due to destabilization of the domain through either approach.
In addition to the zinc-binding region, the RecQ-Ct domain contains a winged-helix (WH) subdomain. RecQ's WH sequence is not as well conserved as the helicase domain and zinc-binding motif in RecQ helicases, but the E.coli RecQ catalytic core crystal structure and a recent NMR structure of the WH region from human WRN showed that the WH fold is present in both proteins despite their sharing only 17% identity (31,60). The WH element forms a helix–turn–helix fold; one of these helices has been termed a ‘recognition helix’ and is predicted to be important for dsDNA binding (Figure 2c, shown in green) (31). Consistent with this idea, the RecQ-Ct domain in E.coli RecQ, BLM and WRN bind several distinct DNA structures such as Holliday Junction (HJ) DNA, D-loops and G4 DNA (29,61,62). Similar WH folds have been identified in the structures of transcription factors CAP (63) and hRFX1 (64) where they bind along the major groove to dsDNA, and more recently in the human DNA repair protein AGT where it binds dsDNA through a minor groove interaction (65). Interestingly, the WH domain in WRN is also an important site for binding to heterologous genome maintenance proteins in cells [reviewed in (7,66)]. Therefore, it appears that the RecQ-Ct domain is remarkably multi-faceted, with important roles in DNA binding, heterologous protein binding and protein stability.
HRDC domain
The C-terminal-most conserved domain in RecQ family members is the HRDC domain. The E.coli RecQ and S.cerevisiae Sgs1 HRDC domains form independently folded structural domains that are capable of binding DNA (27,30–32). The NMR structure of the S.cerevisiae Sgs1 HRDC domain and crystal structure of the E.coli RecQ HRDC domain have been solved and both form globular helical domains (Figure 2d) (30,32). The E.coli RecQ HRDC domain is distinguished from the Sgs1 domain by virtue of an additional 310 helix connecting the α1 and α2 helices. This fold exposes a hydrophobic patch consisting of residues 552–556 in E.coli RecQ, of which Tyr555 is important for binding to both ssDNA and partial duplex DNA with a 3′-overhang (32). Surprisingly, mutation of this Tyr to Ala increases the affinity of E.coli RecQ for a synthetic HJ structure, indicating that mutations in this domain can dramatically alter the structure-specific DNA-binding properties of the enzyme. Additional basic residues help to form an electropositive region involved in DNA binding along with the Tyr in the E.coli RecQ HRDC domain (32). In contrast, while the HRDC domain of S.cerevisiae Sgs1 also utilizes an electropositive surface for interactions with ssDNA, it appears to do so using residues from a different face of the domain to form its binding site (30). Since the HRDC domain is the most variable of the conserved RecQ domains, the presence of species-specific DNA-binding modes suggest that the domain can be adapted to fit specific needs of individual RecQ family members.
Interestingly, there are RecQ proteins that do not include an HRDC domain at all as well as a handful in which multiple HRDC domains are present. Three human RecQ homologs, RecQ1, RecQ4 and all isoforms of RecQ5, lack identifiable HRDC domains altogether. In contrast, two HRDC domains are present in Rhodobacter sphaeroides RecQ and three are found in RecQ from Deinococcus radiodurans, Neisseria meningitidis and Neisseria gonorrhea. Biochemical analysis of the D.radiodurans RecQ showed that the additional C-terminal HRDC domains attenuate DNA binding, unwinding and ATP hydrolysis in the enzyme, whereas the N-terminal-most HRDC domain enhances these activities (67). This is another example of how HRDC domains can be specialized to carry out specific roles in regulating RecQ's activities.
The importance of the HRDC domain and other C-terminal elements has been demonstrated in higher eukaryotic RecQ proteins as well. First, the HRDC domain is a critical determinant for BLM-catalyzed double-HJ (dHJ) unwinding and dissolution (along with topoisomerase III) but not for unwinding forked substrates (68). The HRDC domain of E.coli RecQ is also required for efficient unwinding of dHJs, indicating conservation of HRDC domain function, although E.coli RecQ does not stimulate dHJ dissolution. Second, a C-terminal segment of BLM that contains the RecQ-Ct and HRDC domains interacts with the telomere-associated protein TRF2, which stimulates unwinding of telomeric 3′OH and D-loop structures (69). Third, a C-terminal domain of the WRN including its HRDC domain specifically binds HJ and forked DNA substrates (70). The latter two observations are somewhat complicated by the use of protein fragments that include sequences outside of the defined HRDC domain elements; however, the accumulated evidence suggests that the HRDC domain plays an important role in controlling the activities of RecQ helicases by virtue of its structure-specific DNA binding and possible protein-binding properties.
WRN exonuclease domain
The domain architecture of the WRN protein is an exception to other RecQ family members since it encodes an N-terminal exonuclease domain (71–73). Crystal structures of a protease-resistant fragment of this domain (residues 38–236) with several different cofactors have recently been solved (Figure 2e) (74). The structures reveal a similar overall fold to the DnaQ family of proteins, most notably the exonuclease domain of E.coli DNA polymerase I or ‘Klenow fragment’ (75) and the Arabidopsis thaliana protein At5G06450 (www.pdb.org) (76). Alignment of these structural homologs shows a strong conservation of the active site residues that are involved in coordinating two catalytic metal ions (Figure 2e, shown in green). Two of these residues, Glu84 and Tyr212, have been shown to be critical for efficient WRN nuclease activity in vitro (74). In addition, the identity of the active site metal is important for biochemical activity: magnesium and manganese facilitate efficient nucleic acid digestion whereas europium inhibits this activity, possibly due to misalignment of the DNA backbone across the active site.
WRN function in the non-homologous end-joining pathway has been inferred from its interaction with the DNA-PK subunit Ku70/80 (77,78). This interaction stimulates activity of the isolated WRN exonuclease domain comparably to that observed in the full-length WRN protein, consistent with direct interaction between the WRN exonuclease module and Ku70/80 (74). Moreover, when examined in human WS fibroblast cells, a plasmid-based end-joining assay indicated that both the exonuclease and helicase activities of WRN are required to fully complement WS cells (74). These studies, together with the WRN exonuclease domain structure, offer important insights into the role of the exonuclease domain in WRN function.
CONCLUSION
High-resolution structures of RecQ helicase domains have provided an important primer for analyzing the structural basis for RecQ function. RecQ's structural features suggest a modular design in which the mechanism of ATP hydrolysis is conserved with other helicases and its substrate specificity and targeting are provided by the RecQ-Ct and HRDC domains. Recent studies have also begun to highlight the structural mechanisms of the nuclease domain found in WRN. Mutations in each of these regions affect RecQ's catalytic function and/or its interactions with other proteins and DNA structures in the cell. Even in E.coli, however, aspects of RecQ function remain unclear, such as the specific mechanism used by RecQ to bind and unwind nucleic acids, the identity of protein partners that modulate this activity and how these factors may contribute to each of its proposed cellular roles. Future experiments coupled with the growing body of structural information from various RecQ family members should help refine an integrated model of RecQ function.
Acknowledgments
Studies in the Keck laboratory are supported by a grant from the National Institutes of Health (GM068061). J.L.K. is a Shaw Scientist. M.P.K. is a Cremer Scholar and is supported in part by an NIH training grant in Molecular Biophysics. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM068061 to J.L.K.
Conflict of interest statement. None declared.
REFERENCES
- 1.Singleton M.R., Wigley D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama H., Nakayama K., Nakayama R., Irino N., Nakayama Y., Hanawalt P.C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 4.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 5.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 6.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 7.Bachrati C.Z., Hickson I.D. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko H., Fukao T., Kondo N. The function of RecQ helicase gene family (especially BLM) in DNA recombination and joining. Adv. Biophys. 2004;38:45–64. [PubMed] [Google Scholar]
- 9.Furuichi Y. Premature aging and predisposition to cancers caused by mutations in RecQ family helicases. Ann. N. Y. Acad. Sci. 2001;928:121–131. doi: 10.1111/j.1749-6632.2001.tb05642.x. [DOI] [PubMed] [Google Scholar]
- 10.Bennett R.J., Keck J.L. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 11.Kowalczykowski S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 12.Umezu K., Kolodner R.D. Protein interactions in genetic recombination in Escherichia coli interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 13.Kantake N., Madiraju M.V., Sugiyama T., Kowalczykowski S.C. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: a common step in genetic recombination. Proc. Natl Acad. Sci. USA. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morimatsu K., Kowalczykowski S.C. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 15.Hanada K., Ukita T., Kohno Y., Saito K., Kato J., Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hishida T., Han Y.W., Shibata T., Kubota Y., Ishino Y., Iwasaki H., Shinagawa H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courcelle J., Hanawalt P.C. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 18.Wu L., Hickson I.D. RecQ helicases and cellular responses to DNA damage. Mutat. Res. 2002;509:35–47. doi: 10.1016/s0027-5107(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama H. RecQ family helicases: roles as tumor suppressor proteins. Oncogene. 2002;21:9008–9021. doi: 10.1038/sj.onc.1205959. [DOI] [PubMed] [Google Scholar]
- 20.Hickson I.D. RecQ helicases: caretakers of the genome. Nature Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 21.Machwe A., Xiao L., Groden J., Matson S.W., Orren D.K. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 22.Cheok C.F., Wu L., Garcia P.L., Janscak P., Hickson I.D. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machwe A., Lozada E.M., Xiao L., Orren D.K. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol. Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S., Sommers J.A., Choudhary S., Faulkner J.K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R.M., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 25.Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morozov V., Mushegian A.R., Koonin E.V., Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein D.A., Keck J.L. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003;31:2778–2785. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janscak P., Garcia P.L., Hamburger F., Makuta Y., Shiraishi K., Imai Y., Ikeda H., Bickle T.A. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 29.von Kobbe C., Thoma N.H., Czyzewski B.K., Pavletich N.P., Bohr V.A. Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z., Macias M.J., Bottomley M.J., Stier G., Linge J.P., Nilges M., Bork P., Sattler M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure. 1999;7:1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein D.A., Zittel M.C., Keck J.L. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein D.A., Keck J.L. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 34.Subramanya H.S., Bird L.E., Brannigan J.A., Wigley D.B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 35.Korolev S., Hsieh J., Gauss G.H., Lohman T.M., Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E.coli. Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 36.Story R.M., Weber I.T., Steitz T.A. The structure of the E.coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 37.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 38.Umezu K., Nakayama K., Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc. Natl Acad. Sci. USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahr A., De Graeve F., Kedinger C., Chatton B. Point mutations causing Bloom's syndrome abolish ATPase and DNA helicase activities of the BLM protein. Oncogene. 1998;17:2565–2571. doi: 10.1038/sj.onc.1202389. [DOI] [PubMed] [Google Scholar]
- 40.Onoda F., Seki M., Miyajima A., Enomoto T. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 2000;459:203–209. doi: 10.1016/s0921-8777(99)00071-3. [DOI] [PubMed] [Google Scholar]
- 41.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye J., Osborne A.R., Groll M., Rapoport T.A. RecA-like motor ATPases—lessons from structures. Biochim. Biophys. Acta. 2004;1659:1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Smirnova I.N., Kasho V.N., Faller L.D. Inferences about the catalytic domain of P-type ATPases from the tertiary structures of enzymes that catalyze the same elementary reaction. FEBS. Lett. 1998;431:309–314. doi: 10.1016/s0014-5793(98)00760-1. [DOI] [PubMed] [Google Scholar]
- 44.Xu H.Q., Deprez E., Zhang A.H., Tauc P., Ladjimi M.M., Brochon J.C., Auclair C., Xi X.G. The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem. 2003;278:34925–34933. doi: 10.1074/jbc.M303581200. [DOI] [PubMed] [Google Scholar]
- 45.Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshima J., Loeb L.A. The Werner syndrome protein is a DNA helicase. Nature Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 46.Lu J., Mullen J.R., Brill S.J., Kleff S., Romeo A.M., Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 47.Wang L., Ogburn C.E., Ware C.B., Ladiges W.C., Youssoufian H., Martin G.M., Oshima J. Cellular Werner phenotypes in mice expressing a putative dominant-negative human WRN gene. Genetics. 2000;154:357–362. doi: 10.1093/genetics/154.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao N., Hesson T., Cable M., Hong Z., Kwong A.D., Le H.V., Weber P.C. Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa N., Sugahara M., Masui R., Kato R., Fukuyama K., Kuramitsu S. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem. (Tokyo) 1999;126:986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- 50.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 51.Zittel M.C., Keck J.L. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33:6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ui A., Satoh Y., Onoda F., Miyajima A., Seki M., Enomoto T. The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Genomics. 2001;265:837–850. doi: 10.1007/s004380100479. [DOI] [PubMed] [Google Scholar]
- 53.Dillingham M.S., Soultanas P., Wigley D.B. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brosh R.M., Jr, Matson S.W. A partially functional DNA helicase II mutant defective in forming stable binary complexes with ATP and DNA. A role for helicase motif III. J. Biol. Chem. 1996;271:25360–25368. doi: 10.1074/jbc.271.41.25360. [DOI] [PubMed] [Google Scholar]
- 55.Kim J.L., Morgenstern K.A., Griffith J.P., Dwyer M.D., Thomson J.A., Murcko M.A., Lin C., Caron P.R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 56.Garcia P.L., Bradley G., Hayes C.J., Krintel S., Soultanas P., Janscak P. RPA alleviates the inhibitory effect of vinylphosphonate internucleotide linkages on DNA unwinding by BLM and WRN helicases. Nucleic Acids Res. 2004;32:3771–3778. doi: 10.1093/nar/gkh709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamath-Loeb A.S., Welcsh P., Waite M., Adman E.T., Loeb L.A. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J. Biol. Chem. 2004;279:55499–55505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- 58.Liu J.L., Rigolet P., Dou S.X., Wang P.Y., Xi X.G. The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J. Biol. Chem. 2004;279:42794–42802. doi: 10.1074/jbc.M405008200. [DOI] [PubMed] [Google Scholar]
- 59.Guo R.B., Rigolet P., Zargarian L., Fermandjian S., Xi X.G. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom's syndrome helicase. Nucleic Acids Res. 2005;33:3109–3124. doi: 10.1093/nar/gki619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu J.S., Feng H., Zeng W., Lin G.X., Xi X.G. Solution structure of a multifunctional DNA- and protein-binding motif of human Werner syndrome protein. Proc. Natl Acad. Sci. USA. 2005;102:18379–18384. doi: 10.1073/pnas.0509380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Brabant A.J., Ye T., Sanz M., German I.J., Ellis N.A., Holloman W.K. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 62.Huber M.D., Duquette M.L., Shiels J.C., Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 63.Schultz S.C., Shields G.C., Steitz T.A. Crystal structure of a CAP–DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 64.Gajiwala K.S., Chen H., Cornille F., Roques B.P., Reith W., Mach B., Burley S.K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 65.Daniels D.S., Woo T.T., Luu K.X., Noll D.M., Clarke N.D., Pegg A.E., Tainer J.A. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nature Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 66.Lee J.W., Harrigan J., Opresko P.L., Bohr V.A. Pathways and functions of the Werner syndrome protein. Mech. Ageing Dev. 2005;126:79–86. doi: 10.1016/j.mad.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 67.Killoran M.P., Keck J.L. Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J. Biol. Chem. 2006;281:12849–12857. doi: 10.1074/jbc.M600097200. [DOI] [PubMed] [Google Scholar]
- 68.Wu L., Chan K.L., Ralf C., Bernstein D.A., Garcia P.L., Bohr V.A., Vindigni A., Janscak P., Keck J.L., Hickson I.D. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 70.Cheng W.H., von Kobbe C., Opresko P.L., Fields K.M., Ren J., Kufe D., Bohr V.A. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol. Cell. Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mushegian A.R., Bassett D.E., Jr, Boguski M.S., Bork P., Koonin E.V. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc. Natl Acad. Sci. USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang S., Li B., Gray M.D., Oshima J., Mian I.S., Campisi J. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nature Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen J.C., Gray M.D., Oshima J., Kamath-Loeb A.S., Fry M., Loeb L.A. Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide. J. Biol. Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 74.Perry J.J., Yannone S.M., Holden L.G., Hitomi C., Asaithamby A., Han S., Cooper P.K., Chen D.J., Tainer J.A. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nature Struct. Mol. Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 75.Beese L.S., Steitz T.A. Structural basis for the 3′→5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wesenberg G.E., Smith D.W., Phillips G.N.,Jr, Johnson K.A., Bingman C.A., Center for Eukaryotic Structural Genomics (CESG) X-ray structure of gene product from Arabidopsis thaliana At5g06450. 2004.
- 77.Cooper M.P., Machwe A., Orren D.K., Brosh R.M., Ramsden D., Bohr V.A. Ku complex interacts with and stimulates the Werner protein. Genes. Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 78.Li B., Conway N., Navarro S., Comai L. A conserved and species-specific functional interaction between the Werner syndrome-like exonuclease atWEX and the Ku heterodimer in Arabidopsis. Nucleic Acids Res. 2005;33:6861–6867. doi: 10.1093/nar/gki984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Delano W.L. The PyMol Molecular Graphics System. San Carolos, CA: DeLano Scientific; 2002. [Google Scholar]