Microtubule depolymerization can drive poleward chromosome motion in fission yeast (original) (raw)
Abstract
Prometaphase kinetochores interact with spindle microtubules (MTs) to establish chromosome bi-orientation. Before becoming bi-oriented, chromosomes frequently exhibit poleward movements (P-movements), which are commonly attributed to minus end-directed, MT-dependent motors. In fission yeast there are three such motors: dynein and two kinesin-14s, Pkl1p and Klp2p. None of these enzymes is essential for viability, and even the triple deletion grows well. This might be due to the fact that yeasts kinetochores are normally juxtapolar at mitosis onset, removing the need for poleward chromosome movement during prometaphase. Anaphase P-movement might also be dispensable in a spindle that elongates significantly. To test this supposition, we have analyzed kinetochore dynamics in cells whose kinetochore–pole connections have been dispersed. In cells recovering from this condition, the maximum rate of poleward kinetochore movement was unaffected by the deletion of any or all of these motors, strongly suggesting that other factors, like MT depolymerization, can cause such movements in vivo. However, Klp2p, which localizes to kinetochores, contributed to the effectiveness of P-movement by promoting the shortening of kinetochore fibers.
Keywords: β-Tubulin mutant, dynein, electron tomography, kinetochore, kinesin-14
Introduction
During mitotic division, sister chromatids must segregate equally to ensure healthy daughter cells. The fidelity of segregation is dependent in part upon different factors that drive chromosome movements during various mitotic stages (McIntosh et al, 2002; Maiato et al, 2004). At the onset of mitosis in vertebrate cells, the kinetochores lie at a variety of distances from the spindle poles. When they first contact pole-originated microtubules (MTs), kinetochores often interact with the lateral MT surface (Rieder and Alexander, 1990). Subsequent pole-directed movement (P-movement) occurs along the MT wall and is likely to be driven by ‘minus end' motors, like dynein (Savoian et al, 2000; Sharp et al, 2000), although contributions from motors of the kinesin-14 family are also possible (Tanaka et al, 2005). Later, kinetochore-associated MTs rearrange so their plus ends become embedded in a kinetochore. Thereafter, kinetochore movements must be accompanied by kinetochore MT shortening or elongation, so MT depolymerization may also contribute force for kinetochores P-movement (Koshland et al, 1988; Lombillo et al, 1995; Grishchuk et al, 2005).
The complexity of kinetochore–MTs associations during spindle formation and the large number of the contributing players has made it hard to evaluate the roles of each possible factor during the division of multicellular eukaryotes. Recently, pole-directed chromosome movements were studied in the budding yeast Saccharomyces cerevisiae by creating an artificial condition in which one kinetochore was positioned far from the mitotic spindle (Tanaka et al, 2005). In this system, just as in vertebrate mitosis, a distant kinetochore moved towards the pole by sliding on the MT surface. From the six kinesins and one dynein that are present in S. cerevisiae only Kar3p, a kinesin-14, contributed to the observed movement. Even in its absence, however, the movement persisted, but it was less processive (Tanaka et al, 2005).
In the fission yeast Schizosaccharomyces pombe there are two members of the kinesin-14 family, either of which could affect P-movement. Both have been shown to contribute to MT organization, but their exact mitotic functions are unknown (Pidoux et al, 1996; Troxell et al, 2001; Carazo-Salas et al, 2005). Dynein, Dhclp, is another candidate for this role, although it is not known to contribute to vegetative growth in fission yeasts (Yamamoto et al, 1999). To study the contributions of each minus-end motor enzyme in kinetochore P-movement, we have taken advantage of a cold-sensitive allele of the S. pombe gene for β-tubulin, nda3-KM311 (Hiraoka et al, 1984; Kanbe et al, 1990). As in budding yeast, S. pombe segregates its chromosomes within a nuclear envelope, and the kinetochores of its three interphase chromosomes are normally associated with the centrosome, or ‘spindle pole body' (SPB) (Funabiki et al, 1993). When nda3-KM311 cells are grown at 18°C, MTs are reversibly disrupted, and the kinetochores frequently lose their attachment to SPBs. Here, we show that these detached kinetochores can be retrieved, that is, they re-establish an association with a pole, and then bi-orient so sister kinetochores bind to MTs growing from sister poles. During retrieval, P-motions can be fast and robust, even in the absence of each or all of these motors. These experiments provide the first evidence that such motions can occur in vivo without the help of minus end-directed motors, leading us to propose that they can be driven by MT depolymerization.
Results
Mitotic kinetochores of S. pombe frequently detach from SPBs during temporary MT disruption
During 10 h incubation at 20°C, nda3 cells lose visible MTs and accumulate at prometaphase with highly condensed chromosomes (Hiraoka et al, 1984; Kanbe et al, 1990). Sometimes, the chromosomes remain clustered near the SPBs (Supplementary Figure 1a and b), but in about half the cells condensed chromosomes become scattered and appear as distinct, DAPI-staining objects of various sizes (Figure 1A; Supplementary Figure 1c–g). The duplicated SPBs too may dissociate, albeit infrequently; some retain their associations with kinetochores, but others lose them. The detached chromosomes display a single dot of staining with the kinetochore-specific marker, Mis12p (Supplementary Figure 1d and e), or, less frequently, two dimmer dots, which correspond to sister kinetochores (Supplementary Figure 1f). We have confirmed that such chromosomes have scattered a-mitotically and represent unsegregated sister chromatids by staining a centromere-linked marker on chromosome 2. As expected, this marker associates with only one of the three Mis12p dots (Supplementary Figure 1g). Consistent with published results (Hiraoka et al, 1984; Kanbe et al, 1990), these cells contained no MTs, as seen by either antitubulin immunofluorescence, live cell imaging of tubulin-green fluorescent protein (GFP) or electron microscopy (EM) (Supplement). Thus, it is likely that when MTs are absent, chromosomes scatter by thermal motions. Occasionally, such motions lead to a fragmented nuclear envelope. The ensuing mitosis is then prone to errors (Supplement); we therefore focused our study of kinetochore P-movement on cells in which all three chromosomes remained close to the SPB, although their kinetochore(s) had dispersed during the arrest.
Figure 1.
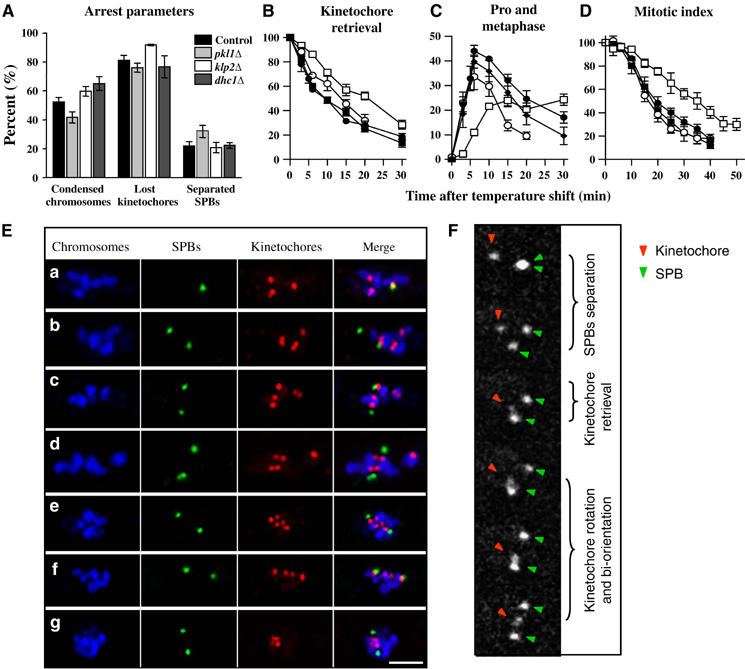
‘Lost' kinetochores are successfully retrieved and bi-orientated. (A) At 18°C cells arrested with condensed chromosomes that remained clustered together (‘condensed chromosomes'). In many such cells at least one kinetochore lost its connection with SPBs (‘lost kinetochores'), and some had separated SPBs. The error bars are standard errors of the mean (s.e.m.) with 95% confidence everywhere in the manuscript unless stated otherwise. (B–D) Progress though mitosis was monitored by fixing cultures sampled at the indicated times after the shift to 32°C and immunostaining. Control—black circles, _pkl1_Δ—white circles, _klp2_Δ—white squares, _dhc1_Δ—black squares. Numbers in (B) and (D) are normalized to the percent at time 0. (B) Percent of cells in which at least one of the kinetochores is neither close to the SPB(s), nor is it positioned on the line between the separated SPBs (see E (a–d)). (C) The percent of cells in which the SPBs are separated and all kinetochores are found on the line between the SPBs, that is, either mono- or bi-oriented (see E, e–g). (D) Percent of cells with condensed chromosomes that are clustered together. The deviations between control and either _klp2_Δ or _pkl1_Δ cells are statistically significant (P<0.05), but _dhc1_Δ and control cells cannot be distinguished. (E) Representative images from synchronized cultures fixed 3–10 min after the shift. Bar: 2 μm. (F) Selected images of an nda3 cell as it recovers from a cold-block, showing kinetochore retrieval and bi-orientation.
Upon MT polymerization, the detached kinetochores return to the poles and become bi-oriented
When arrested nda3 cells are returned to 32°C, their MTs polymerize within a few minutes; the SPBs then separate, and the cells proceed through mitosis (Hiraoka et al, 1984; Kanbe et al, 1990). To examine the process by which ‘lost' kinetochores are ‘retrieved' by spindle MTs, we analyzed the progression of these cells through mitosis by immunostaining (Figure 1). The percent of lost kinetochores decreased fast (τ1/2∼10 min) from the initial 81% (Figure 1A and B). Concomitant with kinetochore–SPB re-association, there was a transient increase in the percent of kinetochores positioned between the SPBs (Figure 1C and E). Approximately 6 min after the shift to 32°C, cells began to exit mitosis (τ1/2∼19 min), as judged by a decrease in the percent of cells with condensed, clustered chromosomes (Figure 1D). By 40 min more than 80% of the cells had exited mitosis having succeeded in retrieving the detached kinetochores. Thus, after initiation of MT polymerization, the lost kinetochores are first drawn to the separated poles (Figure 1E (b–d)); they then bi-orient and go on to anaphase (Figure 1E (e–g)), behaviors that are strikingly similar to those found in normal vertebrate mitosis.
This conclusion was tested by visualizing the poles and the kinetochores of chromosome 2 in living cells with GFP-marked centrosomes and centromeres (see Materials and methods). At restrictive temperature, the detached SPBs and kinetochore underwent seemingly random motions (Supplementary Video 1). The kinetochore frequently split into two, indicating a transient separation of sister chromatids, presumably due to thermal motions and chromatin visco-elasticity. Following the shift to permissive temperatures, the SPBs separated in 2.9±0.3 min (_N_=41). About 7.0±1.7 min later (_N_=30), the tagged kinetochore was drawn towards one of the poles (Figure 1F). Bi-orientation (defined as the first time when the kinetochore was roughly midway between SPBs) occurred 6.9±0.7 min (_N_=17) after the kinetochore's arrival at the SPB. It was frequently preceded by a rotation of the kinetochore from its orientation during the approach to the pole, allowing it now to face the opposite pole. By 30 min after the shift anaphase had occurred in ∼2/3 of the cells. Thus, the results from live imaging and from immunofluorescence were fully consistent. Together they demonstrate that a lost kinetochore can move poleward and bi-orient successfully. Presumably, similar motions occur during normal mitosis, albeit over shorter distances.
The efficiency of kinetochore retrieval is enhanced by the stability of kinetochore–MT attachment and the processivity of the ensuing movements
We then asked about the effectiveness of this kinetochore retrieval process. With increasing distance between the detached kinetochore and poles, the time for retrieval increased slightly (Supplementary Figure 2A, red curve); at distances comparable to the diameter of a fission yeast nucleus, fewer kinetochores were recovered (Supplementary Figure 2B). Nonetheless, ∼80% of the ‘lost' kinetochores were pulled to the pole within 15 min, so long as they had scattered <1.6 μm from the poles. Thus, the MT capture and kinetochore retrieval was effective at these distances.
The shortness of the MTs that connected SPBs with retrievable kinetochores made their visualization difficult, whether by immunofluorescence with antitubulin or in live cells with tubulin-GFP (Supplement). To circumvent these difficulties and gain more information about the kinetics of kinetochore retrieval, we have collected the three-dimensional (3D) coordinates of both poles and the kinetochore. The resulting data were analyzed with custom computer programs (see Materials and methods). For example, Figure 2A shows consecutive kinetochore positions (red X's connected with lines) relative to the poles (green circles) as a 2D image, displayed in the plane that contains these three objects. In the cold (red line) kinetochore movements appeared random (Supplementary Video 2); its position fluctuated around a mean with a standard deviation (s.d.) of 0.318 μm. About 6 min after the shift to 32°C, the kinetochore's movement became directed towards one of the poles (Figure 2A, blue line). The s.d. for a line fitted to this part of the trajectory was 0.077 μm, significantly less than the s.d. determined from the same data fit to the hypothesis of continued random movement (0.544 μm); this phase of movement was essentially linear.
Figure 2.
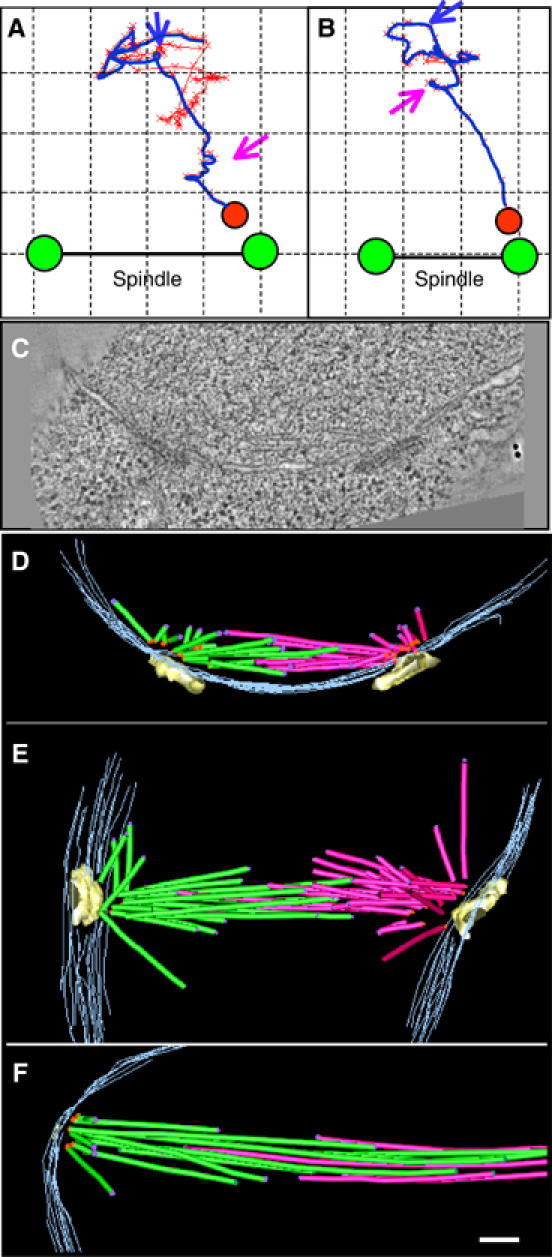
Kinetics and structure of initial chromosome–MT interaction. (A, B) Trajectories of kinetochore movements prior to (red line) and after (blue line) the temperature shift to 32°C in two control cells. The images show movements in the plane that contained the kinetochore (red X's) and both spindle poles (see Materials and methods). Blue arrows point to the onset of the final movements that brought these kinetochores to the pole. Purple arrows point to pauses. Final positions for the kinetochores and poles are shown as red and green circles, respectively. The grid squares are 0.5 μm. (C) A slice from a tomogram, built from nda3 cell frozen during early stage of spindle formation. (D–F) Reconstructions of three spindles (frozen 3–7 min after the shift to 32°C), one of which is shown in (C). MTs nucleated from the different poles are colored green and red, spindle poles are yellow, and the nuclear envelope is gray. Minus ends are shown with orange dots, plus ends with purple dots; ends that lie between the sections are not marked. Occasionally, there were MTs that continued beyond the reconstructed sections; their most distal (plus) ends were marked with green dots. Bar: 0.1 μm.
We have used this data format to analyze both the persistence of kinetochore–MT attachments and the processivity of kinetochore P-movement. If a kinetochore frequently captured and lost MTs that grew from opposite poles, one would expect the kinetochore's trajectory to consist of segmented movements toward different poles. However, all persistent motions were directed towards only one of the poles (Figure 2B). They occurred at average rate of 0.6±0.1 μm/min (range 0.2–2.8 μm/min, _N_=8), and they were quite processive. The kinetochore moved without interruption for 1.0±0.1 μm (_N_=12), with a longest uninterrupted run of 1.5 μm. The pauses (Figure 2A and B; purple arrows) lasted for 124±30 s, which could have resulted either from a stalled movements or from kinetochore–MT detachment, followed by recapture.
P-movement in S. pombe occurrs via pole-originated MTs and is driven by kinetochore-dependent mechanisms
We measured the angles between the direction of the kinetochore's final trajectory and the spindle axis. Their range was broad (82±25°; M±s.d., _N_=28), suggesting that during prometaphase the SPBs nucleated MTs in a wide range of directions, broad enough to cover most pole-proximal regions in the nucleus. This view differs from the published description of more mature, metaphase spindles, which comprised a narrow bundle of MTs (Ding et al, 1993). We therefore used electron tomography (ET) to examine the MT distributions in the forming spindles. Early in prometaphase the duplicated SPBs enter a fenestra in the nuclear membrane (Ding et al, 1997; Uzawa et al, 2004). As the spindle grew in nda3 cells recovering from the cold block, the SPBs initially lay almost parallel to each other (Figure 2C and D). Their separation was probably driven by the few overlapping MTs that ran oblique to the SPB's nuclear surface (Supplementary Video 3). Other MTs projected in diverse directions into the nucleoplasm, which should maximize the chance of their encountering kinetochores (Sagolla et al, 2003). As the spindle matured and its length increased, the poles turned to face each other, and the number of overlapping MTs grew. There were still, however, many MTs of various lengths that grew in a variety of directions, reminiscent of a vertebrate spindle (Figure 2E; Supplementary Video 4). We found no examples in which MTs from both poles pointed towards the same, pole-distant region of the nucleus, as one might expect during merotelic chromosome attachment (Maiato et al, 2004). This is consistent with the suggestion that a lost kinetochore attaches initially to only one pole (in either a monotelic or syntelic configuration). The exact mode of attachment cannot, however, be determined, due to insufficient contrasting of yeast kinetochores in the EM. Although some regions near the MTs plus ends had structural features that made them candidates for kinetochores, these features were difficult to quantify (Supplementary Figure 3A). All such areas were at the ends of highly splayed MT plus ends, implying that the kinetochores interact with the polymer end, rather than with the MT's lateral surface.
In all spindles reconstructed by ET (_N_=24), every MT minus end lay in the vicinity of the SPBs (average distance 86±5 nm, _N_=70) (Supplementary Figure 3B). We found no evidence for MTs initiated at kinetochores, as happens in some systems (Khodjakov et al, 2003). At least 91% of the minus ends (_N_=483) were capped by a roughly conical structure (Supplementary Figure 3C), which resembled the γ-tubulin ring complex in both budding yeast and nematode spindles (O'Toole et al, 1999, 2003). These results suggest a stable attachment of the minus MT end to the SPB, consistent with the apparent lack of MT dynamics at this end (Mallavarapu et al, 1999). Thus, it is unlikely that pole-associated kinesins in S. pombe contribute to P-movements by destabilizing the MT minus ends (Rogers et al, 2004). We conclude that kinetochore P-movement in fission yeast are caused by MT-dependent, kinetochore-associated, minus-end directed motor(s) and/or by forces generated by depolymerization at the MT's plus end.
Neither dynein nor Pkl1p is required for normal kinetochore retrieval
We have used this experimental system to examine the contributions of different minus-end directed motors. Strains lacking the gene for Dhc1p, Pkl1p or Klp2p (all in the nda3 background) were synchronized as described above. By immunofluorescence of fixed cells, the overall parameters of the arrest were similar for all strains, indicating that none of these deletions interfered with the cell cycle block or kinetochore scattering (Figure 1A). In the dynein deletion strain, the kinetics of kinetochore retrieval and bi-orientation were not statistically different from those of the control strain (Figure 1B–D). Live cell imaging has also revealed normal kinetochore retrieval in this strain (_N_=23), though the mean retrieval time was slightly longer than in control cells (Supplementary Figure 4A).
Pkl1p is a S. pombe motor of the kinesin-14 family; it is normally found along the spindle and is enriched at the SPBs (Pidoux et al, 1996). During the initial stages of mitotic progression in _pkl1_Δ cultures (<5 min), the kinetics of kinetochore retrieval, bi-orientation and exit from mitosis were indistinguishable from those of the control strain (Figure 1B–D). However, in _pkl1_Δ the fraction of cells in prometaphase and metaphase decreased prematurely (Figure 1C), which led to a correspondingly higher percent of unretrieved kinetochores at the later times (5–15 min; Figure 1B). The slight advancement in mitotic exit suggested a precocious anaphase onset (Figure 1D); apparently kinetochores are retrieved normally in _pkl1_Δ cells, but there is a defect later in mitosis (Grishchuk and McIntosh, manuscript in preparation).
We have traced the 3D coordinates for kinetochores and poles in 5 _pkl1_Δ and 6 _dhc1_Δ cells. The trajectories of pole-directed kinetochore movements showed no significant deviations from those of control cells (Supplementary Figure 4B and C). The variations in the onset of kinetochore movement to a pole and its speed are almost as great within each of these genotypes as they are between the genotypes (Figure 3A). We conclude that normal P-movements can occur in the absence of either dynein or Pkl1p.
Figure 3.
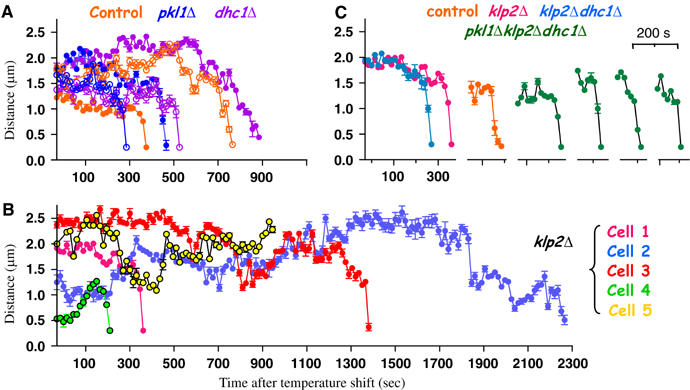
Movements during chromosome retrieval in live cells. (A, B) Example traces of the kinetochore-to-pole distances versus time after the shift to 32°C (see also Supplementary Figure 6). (C) Comparison of the slopes of final kinetochore approaches for the _klp2_Δ cell 1 (Supplementary Video 5), _klp2_Δ_dhc1_Δ cell (Supplementary Video 12), one control cell and 4 _pkl1_Δ _klp2_Δ _dhc1_Δ cells.
Deletion of Klp2 leads to a delay in kinetochore retrieval, due to an increased heterogeneity in the kinetics of kinetochore movements
S. pombe's Klp2p is another member of the kinesin-14 family; it is the only minus-end directed motor that is known to localize to mitotic kinetochores (Troxell et al, 2001). Nda3 cells lacking Klp2p had a reproducible increase in the percent of lost kinetochores, suggesting that Klp2p contributes to normal kinetochore–pole attachments (Figure 1A). Following the return of these cells to 32°C, their kinetochores re-established association with SPBs significantly more slowly than cells of the three other genotypes (τ1/2 20 min; Figure 1B). There was also a delay in the accumulation of kinetochores in prometaphase and metaphase (Figure 1C) and a slower mitotic exit (τ1/2 35 min; Figure 1D), indicating normal operation of the mitotic checkpoint.
By following live mitosis in _klp2_Δ cells (_N_=30), we identified two factors that contribute to slower kinetochore retrieval. First, _klp2_Δ cells retrieve fewer distant kinetochores (⩾1.2 μm from the poles) than control cells (Supplementary Figure 2B). Second, the average time required for kinetochore recovery from shorter distances was increased: 11.8±2.9 min in _klp2_Δ cells versus 6.8±1.1 min in control (Supplementary Figure 4A). Since the Klp2 motor is localized at kinetochores, it might contribute directly to the P-movement. One would then expect that the slower average rate of kinetochore recovery was due to a slower kinetochore movement. However, direct visualization of kinetochore motion showed that this was not so: some of the _klp2_Δ kinetochores were recovered efficiently and moved with the highest speed we observed (∼3 μm/min; Supplementary Video 5). Movements of other kinetochores were greatly delayed, even though they were initially close to the poles (Supplementary Video 6). Thus, in the absence of Klp2p kinetochores can still move rapidly, but there is an increased probability of slower recovery. Since the klp2 + gene is deleted from these cells (Troxell et al, 2001), this behavior could not be due to variations in the levels of Klp2p. We have therefore explored other possibilities.
Klp2p promotes processivity of kinetochore movement by provoking kinetochore fiber shortening and/or preventing its elongation
To analyze mechanisms that led to movement heterogeneity in _klp2_Δ cells, we traced kinetochore–pole distances over time. Some records showed periods when the distance was unchanged or even increased (e.g. cells 2–5 in Figure 3B). This suggested that _klp2_Δ kinetochores were either defective in MT attachment and moved randomly until a new capture could occur, or that their P-movement stalled frequently and even reversed. To distinguish between these possibilities, we examined the 2D kinetochore trajectories. For example, the retrieval path for the kinetochore in cell 3 can be divided into two segments (Figure 4A). Initially, the trajectory resembles a circular arc with its center at the right pole. This model can be evaluated by the s.d. of the data fit to this hypothesis (0.141 μm), while the s.d. for a model based on random movement is 0.410. Such motions could arise in part if the kinetochore's movement were restricted by its proximity to the nuclear envelope. Alternatively, the kinetochore might have attached to one or more stable MTs originating from the right pole. The later segments of the path fit well to a line pointing directly toward that pole (s.d. 0.128 μm, s.d. for random fit 0.422), consistent with the latter model. These movements were not, however, processive; the kinetochore moved forward and back before finally arriving at that pole (Supplementary Video 7). The backward movement could have resulted from the chromatin recoiling after its kinetochore detached from an associated MT(s), but the conspicuous absence of random kinetochore motions, which one would expect for a detached chromosome, suggests that during retrieval this _klp2_Δ kinetochore maintained a stable attachment to a kinetochore MT. This prediction was tested by performing ET on 4 prometaphase spindles from _nda3 klp2_Δ cells delayed in mitotic progression. All of them contained 1–2 MT bundles that diverged from the spindle axis, each consisting of 2–3 MTs (a pombe kinetochore binds 2–4 MTs (Ding et al, 1993; Figure 4B and C; Supplementary Video 8). Although individual kinetochores cannot be located in these tomograms, the positions of the MT plus ends in these bundles (identified through ET) were similar to the positions of kinetochores during their retrieval in _klp2_Δ cells, as seen via live imaging. Together, these results suggest that _klp2_Δ kinetochores capture MTs early after the shift to 32°C, but they pause in their P-movement and frequently move away from that pole.
Figure 4.
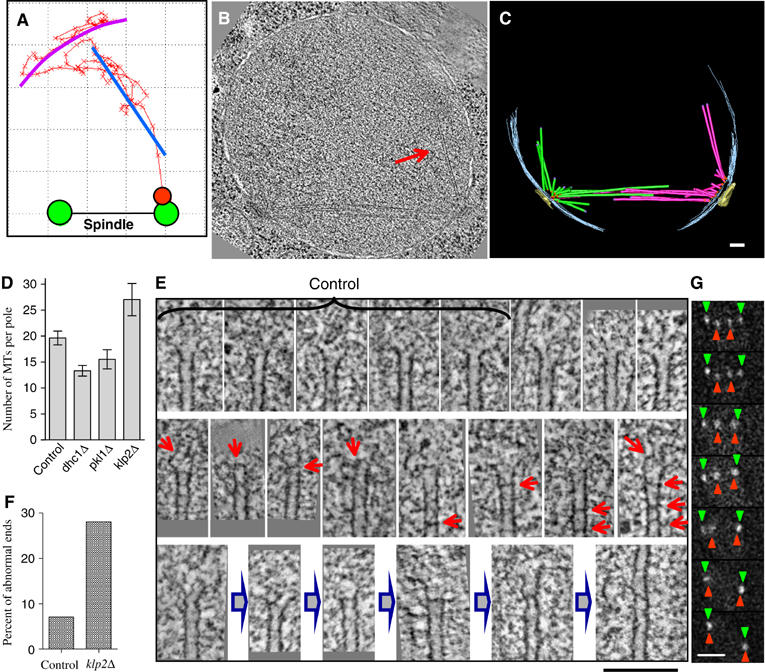
Klp2p facilitates kinetochore retrieval. (A) 2D trajectory of kinetochore retrieval in cell 3. The best fitting circular arc (magenta) for frames 1–50 (Supplementary Video 7); a best linear fit (blue) for frames 51–96. Tomographic slice (B) and 3D model (C) of _nda3klp2_Δ cell delayed in prometaphase (frozen 15 min after the shift to 32°C). Arrow in (B) points to one of the MTs in a two-MTs bundle growing from the right pole. Bar: 100 nm. (D) Average numbers of MTs per spindle pole for control nda3 cells (4 poles), _nda3dhc1_Δ (10), _nda3pkl1_Δ (6) and delayed _nda3klp2_Δ cells (6). (E) ET slices of the MTs plus ends. The first five images were collected from a control nda3 cell; all the rest are from six _klp2_Δ cells. There is a great variability in the appearance of the plus ends in both normal and mutant cells, which makes it difficult to quantify differences. Usually, the plus MT ends are splayed open (upper row), but in _klp2_Δ cells they frequently contain occlusions (red arrows). The images in middle row may represent intermediates that lead to ‘scarred' MTs with altered polymerization behavior; the bottom row suggests a possible pathway for the rescue of rapid shortening at an MT abnormality, followed by renewed MT polymerization. Bar: 100 nm. (F) Percent of MTs plus ends that show darker ‘bars' inside the MT lumen. Number of ends examined: control 68, _klp2_Δ 239. (G) Selected live images (30 s apart) from an _nda3 klp2_Δ _dhc1_Δ cell undergoing anaphase A. Bar: 2 μm.
The inability of these cells to sustain P-movements and the tendency of their chromosomes to switch to movements away from the pole appear to be the primary causes of their delayed chromosome retrieval. For example, the kinetochore in cell 4 (Figure 3B) was initially close to the unseparated poles. Immediately after the shift to 32°C, this kinetochore began moving away from the pole at 0.5 μm/min before moving back again and bi-orienting (Supplementary Video 9). In cell 5 (Figure 3B) the kinetochore began with movement towards the pole, then abruptly switched its direction and moved away (Supplementary Video 10). Our ultrastructural analysis of spindles in delayed _klp2_Δ cells revealed a small increase in the number of MTs growing from each pole relative to wild type (Figure 4D). This difference might result from a longer time spent in mitosis, or the MTs in these cells may show altered MT dynamics (Troxell et al, 2001). Close examination of the MT ends in this strain showed that their protofilaments tended to be less splayed than in other strains (Figure 4E). Many MTs showed occlusions, for example, dark ‘bars' across their lumens (Figure 4F). These structures probably represent proteinaceous material, perhaps protofilament pieces, trapped in a growing MT (Figure 4E, gray arrows). Such irregularities may arise during normal MT dynamics, forming ‘scarred' MTs whose depolymerization is partially impeded. Perhaps Klp2p is required for MT shortening when depolymerization encounters such abnormalities; in its absence, depolymerization rescue and subsequent MT elongation at the kinetochore may be more common.
The maximum rate of P-movement is maintained in the absence of both Klp2p and dynein, as well as in a triple deletion mutant
As described above, the Pkl1 and dynein motors were not required for normal kinetochore recovery in our test system. Further examination of the _pkl1_Δ phenotype revealed that this motor might play an important role in SPB organization (Grishchuk and McIntosh, manuscript in preparation). Dhc1p, on the other hand, is expressed in vegetative cells at such low levels that it has not been detected during mitosis (Yamamoto et al, 1999). It has been suggested that this motor plays no role in fission yeast mitosis, but we found a small increase in kinetochore recovery times in the absence of dynein. Moreover, Northern analysis showed a low level of dhc1 mRNA in vegetative cells (West and McIntosh, unpublished observation). Thus, we have asked if kinetochore movement in the absence of Klp2p might require the activity of dynein. Live cell imaging showed that kinetochores in this double deletion strain were still recovered, although there was a noticeable heterogeneity in the movements. Some of the _klp2_Δ _dhc1_Δ kinetochores failed to move to the pole, even though they stayed close for a long time (70 min, Supplementary Video 11); others were eventually retrieved (Supplementary Figure 2A). Strikingly, fast and processive P-movement could still be seen in this strain (Figure 3C; Supplementary Video 12). Likewise, the P-movement of chromosomes during anaphase A was not abolished (Figure 4G; Supplementary Video 12). Similar results were seen in a strain that lacked all three of these motors (Figure 3C; Supplementary Video 13 and Supplementary Figure 2A). Since S. pombe has no other known minus-end directed motors (and its genome is fully sequenced), the most likely explanation for these movements is that depolymerization of the plus ends of kinetochore-associated MTs can drive poleward chromosome movement in vivo.
Discussion
Determining the relative contributions of MT-dependent motors and of MT depolymerization in the generation of force for chromosome motion is a long-standing goal in cell biology (Inoue and Salmon, 1995). The task has turned out to be difficult because mitosis depends at some level on many MT-dependent motors. With the exception of the kinesin-5s, however, the mitotic phenotype of removing a given motor, either by genetic deletion, RNAi, or antibody-mediated inhibition, is relatively mild, and no motor can so far be described as the sole contributor to a given chromosome movement (reviewed in McIntosh et al, 2002; Maiato et al, 2004). Although there has been no direct evidence that MT depolymerization drives poleward chromosome movement in vivo, experiments in vitro have shown that shortening MTs can generate forces large enough to cause such motions (Grishchuk et al, 2005). The fact that chromosome retrieval in S. pombe continues with runs of rapid pole-directed movement in the absence of Klp2p, dynein and Pkl1p suggests that this significant part of the retrieval process (and of anaphase A) can be motor-independent. In the absence of other reasonable candidates for this motility, we propose that P-movement can be driven by the depolymerization of kinetochore MTs from their plus ends.
It is difficult to see, however, how this mechanism alone could provide the versatility and flexibility that would ensure high fidelity chromosome segregation. Kinetochore capture is inherently random, but mitosis must be accurate and expedient in the face of many variables, for example, initial chromosome position, the number of MTs that make contact with one or both sister kinetochores and the pole from which they came, and finally, in the mode of initial MT–kinetochore interaction (lateral or otherwise). An emerging view of mitotic mechanism is that although motors can and in some systems do affect the rates of some mitotic movements, their more important role may lie in helping to resolve difficult situations that arise by chance, thanks to the random initial arrangements of some key mitotic components.
By combining information from live imaging, fluorescent microscopy, ET and a mathematical analysis of kinetochore trajectories, we have characterized several important aspects of MT–kinetochore interactions in fission yeast following recovery from an MT depolymerization block. Although the genetic background we used to impose such a block might have altered some aspects of MT dynamics, the overall features of kinetochore–MT interactions are likely to be normal at permissive temperature. When all mitotic motors are present, search for and capture of kinetochores, even those that lie at a larger-than-normal distance, can be effective in S. pombe. Successful capture is ensured in part by the spread of orientations in the MTs nucleated by SPBs early in mitosis. The distant kinetochores become stably attached to one of the poles via 1–3 MTs. The subsequent kinetochore-dependent movement is generally robust and processive but variable in speed, and it may contain pauses. Such variability suggests that different strategies and mechanisms might be required to move distant kinetochores to the pole.
Capture and bi-orientation of SPB-proximal kinetochores can proceed without any of the three, minus-end directed motors (Troxell et al, 2001). However, when kinetochores are at some distance from the SPBs, there is an additional stress on the system, and the input from motors becomes more important for successful chromosome segregation. Since the absence of either dynein or Pklp1p does not affect the measurable characteristics of P-movement, these motors are unlikely to contribute directly to chromosome motions during retrieval. In the absence of the kinetochore-associated motor Klp2p, on the other hand, a distant kinetochore appears to remain stably attached to its MTs, but the average rate of the P-movement is decreased. Our data do not exclude the possibility that Klp2p contributes to kinetochore movement along the lateral MT surface, as has been suggested for its S. cerevisiae homolog Kar3p (Tanaka et al, 2005). They do, however, reveal an unexpected and novel role of inhibiting chromosome motions away from the pole. Kar3p has been shown to work as a plus-end localized, MT disassembly factor (Maddox et al, 2003; Sproul et al, 2005; Molk et al, 2006). If Klp2p plays a similar role at the plus ends of kinetochore-associated MTs in fission yeast, its absence might lead to their excessive growth. The abnormal structural features at or near the ends of the _klp2_Δ MTs (Figure 4E) may lead to an increased frequency of MT depolymerization rescue events, processes that would promote movements away from the pole. We propose that Klp2p normally contributes to the processivity of kinetochore movement by promoting and/or sustaining depolymerization at the plus ends of kinetochore MTs. This function may be of little importance during normal mitosis, but it becomes valuable when the chromosomes are positioned unusually far from spindle poles (Figure 5).
Figure 5.
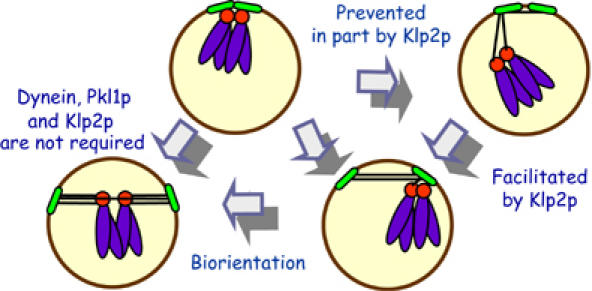
P-movement and bi-orientation of chromosomes in S. pombe. As normal mitosis begins, the SPBs (green ovals) are close to the kinetochores (red circles; only one chromosome (blue) is shown); this situation may explain why all three minus-end directed motors are not required for mitotic progression. In the absence of Klp2p, or during transient MT depolymerization, the kinetochores can move some distance away from the poles. Their subsequent movement to the pole is driven by MT depolymerization, facilitated by Klp2.
Materials and methods
Cell cycle synchronization and immunofluorescence microscopy
S. pombe cells were grown using standard techniques and reagents (Moreno et al, 1991). Strains were constructed from random spores followed by a PCR screen with deletion-specific primers. Complete genotypes of the strains with null alleles pkl1_Δ, klp2_Δ and _dhc1_Δ are described in Supplementary Table 1. Cells carrying the nda3-KM311 mutation (hereafter referred to as nda3) were grown in YPD (1% yeast extract, 2% proteose peptone, 2% dextrose) at 32°C and incubated for 8 h at 18°C. For immunostaining and for chromosome staining with DAPI cells were fixed in 4% paraformaldehyde. Primary antibodies were provided by I Hagan (anti-Sad1, a SPB marker); by P Silver and J Kahana (anti-GFP for _cen2_-GFP), and by K Gull (TAT1 antibody against tubulin). Antibodies to the HA epitope (for kinetochore marker, Mis12-HA, provided by M Yanagida) and to a nucleoporin (Mab414) were purchased from BAbCO. Secondary antibodies, conjugated to Alexa-488 or -594, were from Molecular Probes. Images were acquired with a × 100 apochromatic objective, NA 1.4 (Zeiss Axioplan II light microscope) as _z_-stacks (0.3 μm step) and deconvolved using software from Intelligent Imaging Innovations, Inc. (Denver, CO). Maximum intensity projections were contrasted and arranged into figures using Adobe Photoshop.
Live imaging
The centromere of chromosome 2 was visualized using the cen2-GFP construct (Supplementary Table 1), kindly provided by A Yamamoto (Yamamoto and Hiraoka, 2003). The immunolocalizations of the cen2-GFP marker and of the kinetochore component Mis12-HA are always either co-localized or immediately adjacent (e.g. see Supplementary Figure 1g). Spindle poles were visualized with Pcp1p-GFP (Flory et al, 2002). The microscope stage was precooled to 14°C with a custom made water jacket of metal tubing wrapped around an oil-immersion × 100 NeoFluar objective and oil-immersion condenser. Temperature shift occurred within 1–2 min after switching the tubing of the water jacket from a 14°C to a 32°C bath. Images were acquired as stacks of 7–12 planes (0.25 μm step, 150 ms exposure) every 15 s with a Photometrics Cascade 650 CCD camera (Roper Scientific) controlled via the Metamorph (Universal Imaging). The videos are the averaged, depixelated XY-projections manually aligned to minimize the image drift brought about by the abrupt temperature changes. In all videos, except Video 1, the temperature was shifted to 32°C after the third frame. Since S. pombe nuclei/spindles frequently rotate around randomly oriented axes, the changes in apparent distance between objects in these 2D projections do not necessarily reflect actual changes in separation, and additional image processing was necessary to obtain accurate distances between objects in the images.
Image analysis
All computer programs were custom written using MatLab 6.5. Briefly, the XYZ coordinates of the labeled kinetochore and of the two poles at each time point (collected manually by selecting the apparent centers of the brightest signals in each stack) were transformed, so one of the poles was always at the center of a Cartesian coordinate system. The direction of the _X_-axis was chosen to run through the second pole; the _Y_-axis lay in the plane that contained all three objects. The resulting images display the true 3D distances. This program also eliminates movements caused by stage drift or spindle rotations. In the trajectories the points are moving averages of four consecutive points.
EM
Strains were grown at 18°C, shifted to 32°C for 2–15 min, quickly collected by filtration, high-pressure frozen and processed as described (Ding et al, 1997). After freeze-substitution in 2% gluteraldehyde and 0.1% uranyl acetate, they were embedded in Lowicryl HM20 and cut into 200 nm sections. Dual axes tilt series were acquired (tilt increment 1°; tilt range±60°) using an F30 electron microscope (FEI Corp, Hillsboro, OR), and tomograms were generated using IMOD (Kremer et al, 1996). Data were collected from three independent experiments. The numbers of full and partial reconstructions for spindles from each genotype are, respectively, control: 2,4; _klp1_Δ: 3,7; _klp2_Δ: 7,3; and _dhc1_Δ: 5,0. Full reconstruction means that all MT ends were found within the reconstructed thick sections, with the exception of 1–2 ends, which are marked in the resulting models with green dots.
Supplementary Material
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Acknowledgments
The ET was carried out in the Boulder Laboratory for 3D EM of cells, and was supported by RR000592 to JRM. M Morphew prepared the samples for EM and acquired TEM images of nda3 cells at 18°C; E O'Toole and other members of this lab helped with data acquisition and reconstruction. The temperature control for light microscope was built by V Sarbash and F Ataullakhanov. Strains and reagent were kindly provided by Drs I Hagan, P Silver, K Gull, Y Hiraoka, M Yanagida and T Davis. We are grateful to I Spiridonov for technical assistance, J Becerril for help with collecting 3D coordinates, to M Molodtsov and F Ataullakhanov for the software used to analyze kinetochore trajectories, and to M Winey for helpful discussions. This work was supported in part by GM33787 to JRM.
References
- Carazo-Salas RE, Antony C, Nurse P (2005) The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science 309: 297–300 [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR (1993) Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol 120: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR (1997) The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell 8: 1461–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Morphew M, Joseph JD, Means AR, Davis TN (2002) Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ 13: 47–58 [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121: 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR (2005) Force production by disassembling microtubules. Nature 438: 384–388 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M (1984) The NDA3 gene of fission yeast encodes β-tubulin: a cold- sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39: 349–358 [DOI] [PubMed] [Google Scholar]
- Inoue S, Salmon ED (1995) Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell 6: 1619–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe T, Hiraoka Y, Tanaka K, Yanagida M (1990) The transition of cells of the fission yeast β-tubulin mutant nda3-311 as seen by freeze-substitution electron microscopy requirement of functional tubulin for spindle pole body duplication. J Cell Sci 96: 275–282 [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM (2003) Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol 160: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW (1988) Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 331: 499–504 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR (1995) Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature 373: 161–164 [DOI] [PubMed] [Google Scholar]
- Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K (2003) The minus end-directed motor kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr Biol 13: 1423–1428 [DOI] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T (1999) A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol 9: 1423–1426 [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, West RR (2002) Chromosome–microtubule interactions during mitosis. Annu Rev Cell Dev Biol 18: 193–219 [DOI] [PubMed] [Google Scholar]
- Molk JN, Salmon ED, Bloom K (2006) Nuclear congression is driven by cytoplasmic microtubule plus end interactions in S. cerevisiae. J Cell Biol 172: 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- O'Toole ET, McDonald KL, Mantler J, McIntosh JR, Hyman AA, Muller-Reichert T (2003) Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J Cell Biol 163: 451–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR (1999) High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell 10: 2017–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, LeDizet M, Cande WZ (1996) Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol Biol Cell 7: 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP (1990) Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol 110: 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ (2004) Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427: 364–370 [DOI] [PubMed] [Google Scholar]
- Sagolla MJ, Uzawa S, Cande WZ (2003) Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J Cell Sci 116: 4891–4903 [DOI] [PubMed] [Google Scholar]
- Savoian MS, Goldberg ML, Rieder CL (2000) The rate of poleward chromosome motion is attenuated in Drosophila ZW10 and rod mutants. Nat Cell Biol 2: 948–952 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM (2000) Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol 2: 922–930 [DOI] [PubMed] [Google Scholar]
- Sproul LR, Anderson A, Mackey T, Saunders WS, Gilbert SP (2005) Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol 15: 1420–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU (2005) Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434: 987–994 [DOI] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR (2001) pkl1+ and klp2+: two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell 12: 3476–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa S, Li F, Jin Y, McDonald KL, Braunfeld MB, Agard DA, Cande WZ (2004) Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of Cdc10. Mol Biol Cell 15: 5219–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y (2003) Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J 22: 2284–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol 145: 1233–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7