Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling (original) (raw)
Short abstract
Genome-wide expression profiling of mouse and human leukocytes reveal conserved transcriptional programs of plasmacytoid or conventional dendritic cell subsets.
Abstract
Background
Dendritic cells (DCs) are a complex group of cells that play a critical role in vertebrate immunity. Lymph-node resident DCs (LN-DCs) are subdivided into conventional DC (cDC) subsets (CD11b and CD8α in mouse; BDCA1 and BDCA3 in human) and plasmacytoid DCs (pDCs). It is currently unclear if these various DC populations belong to a unique hematopoietic lineage and if the subsets identified in the mouse and human systems are evolutionary homologs. To gain novel insights into these questions, we sought conserved genetic signatures for LN-DCs and in vitro derived granulocyte-macrophage colony stimulating factor (GM-CSF) DCs through the analysis of a compendium of genome-wide expression profiles of mouse or human leukocytes.
Results
We show through clustering analysis that all LN-DC subsets form a distinct branch within the leukocyte family tree, and reveal a transcriptomal signature evolutionarily conserved in all LN-DC subsets. Moreover, we identify a large gene expression program shared between mouse and human pDCs, and smaller conserved profiles shared between mouse and human LN-cDC subsets. Importantly, most of these genes have not been previously associated with DC function and many have unknown functions. Finally, we use compendium analysis to re-evaluate the classification of interferon-producing killer DCs, lin-CD16+HLA-DR+ cells and in vitro derived GM-CSF DCs, and show that these cells are more closely linked to natural killer and myeloid cells, respectively.
Conclusion
Our study provides a unique database resource for future investigation of the evolutionarily conserved molecular pathways governing the ontogeny and functions of leukocyte subsets, especially DCs.
Background
Dendritic cells (DCs) were initially identified by their unique ability to present antigen for the priming of naïve CD4 and CD8 T lymphocytes [1]. DCs have more recently been shown to be key sentinel immune cells able to sense, and respond to, danger very early in the course of an infection due to their expression of a broad array of pattern recognition receptors [2]. Indeed, DCs have been shown to play a major role in the early production of effector antimicrobial molecules such as interferon (IFN)-α and IFN-β [3] or inducible nitric oxide synthase [4] and it has been demonstrated that DCs can also activate other innate effector cells such as natural killer (NK) cells [5]. In light of these properties, it has been clearly established that DCs are critical for defense against infections, as they are specially suited for the early detection of pathogens, the rapid development of effector functions, and the triggering of downstream responses in other innate and adaptive immune cells.
DCs can be divided into several subsets that differ in their tissue distribution, their phenotype, their functions and their ontogeny [6]. Lymph node-resident DCs (LN-DCs) encompass conventional DCs (cDCs) and plasmacytoid DCs (pDCs) in both humans and mice. LN-cDCs can be subdivided into two populations in both mouse (CD8α and CD11b cDCs) [6] and in human (BDCA1 and BDCA3 cDCs) [7]. In mouse, CD8α cDCs express many scavenger receptors and may be especially efficient for cross-presenting antigen to CD8 T cells [8] whereas CD11b cDCs have been suggested [9,10], and recently shown [11], to be specialized in the activation of CD4 T cells. As human cDC functions are generally studied with cells derived in vitro from monocytes or from CD34+ hematopoietic progenitors, which may differ considerably from the naturally occurring DCs present in vivo, much less is known of the eventual functional specialization of human cDC subsets. Due to differences in the markers used for identifying DC subsets between human and mouse and to differences in the expression of pattern recognition receptors between DC subsets, it has been extremely difficult to address whether there are functional equivalences between mouse and human cDC subsets [6].
pDCs, a cell type discovered recently in both human and mouse, appear broadly different from the other DC subsets to the point that their place within the DC family is debated [3]. Some common characteristics between human and mouse pDCs that distinguish them from cDCs [3] include: their ability to produce very large amounts of IFN-α/β upon activation, their limited ability to prime naïve CD4 and CD8 T cells under steady state conditions, and their expression of several genes generally associated with the lymphocyte lineage and not found in cDCs [12]. Several differences have also been reported between human and mouse pDCs, which include the unique ability of mouse pDCs to produce high levels of IL-12 upon triggering of various toll-like receptors (TLRs) or stimulation with viruses [13,14]. Adding to the complexity of accurately classifying pDCs within leukocyte subsets are recent reports describing cell types bearing mixed phenotypic and functional characteristics of NK cells and pDCs in the mouse [15,16]. Collectively, these findings raise the question of how closely related human and mouse pDCs are to one another or to cDCs as compared to other leukocyte populations.
Global transcriptomic analysis has recently been shown to be a powerful approach to yield new insights into the biology of specific cellular subsets or tissues through their specific gene expression programs [17-21]. Likewise, genome-wide comparative gene expression profiling between mouse and man has recently been demonstrated as a powerful approach to uncover conserved molecular pathways involved in the development of various cancers [22-27]. However, to the best of our knowledge, this approach has not yet been applied to study normal leukocyte subsets. Moreover, DC subsets have not yet been scrutinized through the prism of gene expression patterns within the context of other leukocyte populations. In this report, we assembled compendia comprising various DC and other leukocyte subtypes, both from mouse and man. Using intra- and inter-species comparisons, we define the common and specific core genetic programs of DC subsets.
Results
Generation/assembly and validation of the datasets for the gene expression profiling of LN-DC subsets
We used pan-genomic Affymetrix Mouse Genome 430 2.0 arrays to generate gene expression profiles of murine splenic CD8α (n = 2) and CD11b (n = 2) cDCs, pDCs (n = 2), B cells (n = 3), NK cells (n = 2), and CD8 T cells (n = 2). To generate a compendium of 18 mouse leukocyte profiles, these data were complemented with published data retrieved from public databases, for conventional CD4 T cells (n = 2) [28] and splenic macrophages (n = 3) [29]. We used Affymetrix Human Genome U133 Plus 2.0 arrays to generate gene expression profiles of blood monocytes, neutrophils, B cells, NK cells, and CD4 or CD8 T cells [30]. These data were complemented with published data on human blood DC subsets (pDCs, BDCA1 cDCs, BDCA3 cDCs, and lin-CD16+HLA-DR+ cells) retrieved from public databases [31]. All of the human samples were done in independent triplicates. Information regarding the original sources and the public accessibility of the datasets analyzed in the paper are given in Table 1.
Table 1.
Information on the sources and public access for the datasets analyzed in the paper
| Figures‡ | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | Population* | Laboratory† | Public repository | Accession number | 1a,c; 2a | 1b,d; 2b | 1e | 3 | 4a | 4b | 5a | 5b |
| Affymetrix Mouse Genome 430 2.0 data | ||||||||||||
| Spleen CD8 DCs (2) | MD/SCPK | GEO [95] | GSE9810 | X | X | X | X | X | ||||
| Spleen CD11b DCs (2) | MD/SCPK | GEO | GSE9810 | X | X | X | X | X | ||||
| Spleen pDCs (2) | MD/SCPK | GEO | GSE9810 | X | X | X | X | X | ||||
| Spleen NK cells (2) | MD/SCPK | GEO | GSE9810 | X | X | X | ||||||
| Spleen CD8 T cells (2) | MD/SCPK | GEO | GSE9810 | X | X | |||||||
| Spleen B cells (3) | MD/SCPK | GEO | GSE9810 | X | X | X | ||||||
| Spleen CD4 T cells (2) | AYR | GEO | GSM44979; GSM44982 | X | X | X | ||||||
| Spleen monocytes (3) | SB | NCI caArray [96] | NA | X | X | X | ||||||
| Spleen monocytes (2) | BP | GEO | GSM224733; GSM224735 | X | ||||||||
| Peritoneal MΦ (1) | SA | GEO | GSM218300 | X | ||||||||
| BM-MΦ (2) | RM | GEO | GSM177078; GSM177081 | X | ||||||||
| BM-MΦ (1) | CK | GEO | GSM232005 | X | ||||||||
| BM-DCs (2) | RM | GEO | GSM40053; GSM40056 | X | ||||||||
| BM-DCs (2) | MH | GEO | GSM101418; GSM101419 | X | ||||||||
| Affymetrix Mouse U74Av2 data | ||||||||||||
| Spleen CD4 T cells (3) | CB/DM | GEO | GSM66901; GSM66902; GSM66903 | X | ||||||||
| Spleen B2 cells (2) | CB/DM | GEO | GSM66913; GSM66914 | X | ||||||||
| Spleen B1 cells (2) | CB/DM | GEO | GSM66915; GSM66916 | X | ||||||||
| Spleen NK cells (2) | FT | EBI ArrayExpress [97] | E-MEXP-354 | X | ||||||||
| Spleen CD4 DCs (2) | CRES | GEO | GSM4697; GSM4707 | X | ||||||||
| Spleen CD8 DCs (2) | CRES | GEO | GSM4708; GSM4709 | X | ||||||||
| Spleen DN DCs (2) | CRES | GEO | GSM4710; GSM4711 | X | ||||||||
| Spleen IKDCs (2) | FH | GEO | GSM85329; GSM85330 | X | ||||||||
| Spleen cDCs (2) | FH | GEO | GSM85331; GSM85332 | X | ||||||||
| Spleen pDCs (2) | FH | GEO | GSM85333; GSM85334 | X | ||||||||
| Affymetrix Human Genome U133 Plus 2.0 data | ||||||||||||
| Blood monocytes (3) | FRS | Authors' webpage [86] | NA | X | X | X | X | |||||
| Blood CD4 T cells (3) | FRS | Authors' webpage | NA | X | X | X | ||||||
| Blood CD8 T cells (3) | FRS | Authors' webpage | NA | X | X | X | ||||||
| Blood B cells (3) | FRS | Authors' webpage | NA | X | X | X | ||||||
| Blood NK cells (3) | FRS | Authors' webpage | NA | X | X | X | ||||||
| Blood neutrophils (3) | FRS | Authors' webpage | NA | X | X | X | ||||||
| Blood pDCs (3) | CAKB | EBI ArrayExpress | E-TABM-34 | X | X | X | X | X | ||||
| Blood BDCA1 DCs (3) | CAKB | EBI ArrayExpress | E-TABM-34 | X | X | X | X | X | ||||
| Blood BDCA3 DCs (3) | CAKB | EBI ArrayExpress | E-TABM-34 | X | X | X | X | X | ||||
| Blood CD16 DCs (3) | CAKB | EBI ArrayExpress | E-TABM-34 | X | X | |||||||
| PBMC-derived MΦ (2) | SYH | GEO | GSM109788; GSM109789 | X | ||||||||
| Monocyte-derived MΦ | LZH | GEO | GSM213500 | X | ||||||||
| Monocyte-derived DCs (3) | MVD | GEO | GSM181931; GSM181933; GSM181971 | X |
To verify the quality of the datasets mentioned above, we analyzed signal intensities for control genes whose expression profiles are well documented across the cell populations under consideration. Expression of signature markers were confirmed to be detected only in each corresponding population (see Table 2 for mouse data and Table 3 for human data). For example, Cd3 genes were detected primarily in T cells and often to a lower extent in NK cells; the mouse Klrb1c (nk1.1) gene or the human KIR genes in NK cells; Cd19 in B cells; the mouse Siglech and Bst2 genes or the human LILRA4 (ILT7) and IL3RA (CD123) genes in pDCs; and Cd14 in myeloid cells. As expected, many markers were expressed in more than a single cell population. For example, in the mouse, Itgax (Cd11c) was found expressed to high levels in NK cells and all DC subsets; Itgam (Cd11b) in myeloid cells, NK cells, and CD11b cDCs; Ly6c at the highest level in pDCs but also strongly in many other leukocyte populations; and Cd8a in pDCs and CD8α cDCs. However, the analysis of combinations of these markers confirmed the lack of detectable cross-contaminations between DC subsets: only pDCs expressed high levels of Klra17 (Ly49q) and Ly6c together, while Cd8a, ly75 (Dec205, Cd205), and Tlr3 were expressed together at high levels only in CD8α cDCs, and Itgam (Cd11b) with Tlr1 and high levels of Itgax (Cd11c) only in CD11b cDCs. Thus, each cell sample studied harbors the expected pattern of expression of control genes and our data will truly reflect the gene expression profile of each population analyzed, without any detectable cross-contamination.
Table 2.
Expression of control genes in mouse cells
| Dendritic cells | Lymphocytes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe set ID | Gene | Myeloid cells | pDC | CD8α DC | CD11b DC | NK | CD8 T | CD4 T | B |
| 1419178_at | Cd3g | 40 ± 10 | <20 | <20 | <20 | 97 ± 31 | 2,074 ± 287 | 1,974 ± 478 | 22 ± 3 |
| 1422828_at | Cd3d | 111 ± 14 | <20 | <20 | <20 | 214 ± 16 | 2,815 ± 11 | 4,520 ± 1,414 | 21 ± 2 |
| 1422105_at | Cd3e | 115 ± 30 | 27 ± 10 | 22 ± 2 | 23 ± 5 | 26 ± 9 | 387 ± 58 | 522 ± 210 | 26 ± 10 |
| 1426396_at | Cd3z | <20 | <20 | <20 | <20 | 1,147 ± 81 | 1,545 ± 10 | 2,117 ± 482 | 25 ± 9 |
| 1426113_x_at | Tcra | 83 ± 8 | <20 | 23 ± 4 | <20 | 116 ± 39 | 2,517 ± 42 | 5,601 ± 1,818 | 34 ± 13 |
| 1419696_at | Cd4 | 24 ± 2 | 1,233 ± 144 | <20 | 369 ± 49 | <20 | <20 | 1,052 ± 73 | <20 |
| 1450570_a_at | Cd19 | 190 ± 44 | <20 | <20 | <20 | <20 | <20 | 23 ± 5 | 2,259 ± 292 |
| 1449570_at | Klrb1c (NK1.1) | <20 | <20 | <20 | <20 | 2,328 ± 112 | <20 | 25 ± 7 | <20 |
| 1425436_x_at | Klra3 (Ly49C) | 130 ± 11 | 24 ± 3 | 156 ± 0 | 242 ± 31 | 9,186 ± 479 | 170 ± 61 | 70 ± 42 | <20 |
| 1450648_s_at | H2-Ab1 | 6,887 ± 84 | 7,339 ± 5 | 9,101 ± 100 | 9,056 ± 277 | 81 ± 6 | 83 ± 56 | 978 ± 11 | 7,028 ± 239 |
| 1419128_at | Itgax (CD11c) | 454 ± 5 | 1,928 ± 169 | 2,827 ± 454 | 4,701 ± 56 | 3,403 ± 45 | 108 ± 44 | 22 ± 2 | <20 |
| 1457786_at | Siglech | 31 ± 4 | 3,454 ± 536 | 24 ± 5 | <20 | <20 | <20 | 33 ± 13 | <20 |
| 1425888_at | Klra17 (Ly49Q) | 98 ± 4 | 3,413 ± 116 | 30 ± 14 | 163 ± 2 | 28 ± 11 | 24 ± 6 | 38 ± 10 | <20 |
| 1424921_at | Bst2 (120G8) | 2,364 ± 149 | 5,571 ± 718 | 237 ± 30 | 196 ± 44 | 61 ± 24 | 162 ± 12 | 90 ± 3 | 88 ± 32 |
| 1421571_a_at | Ly6c | 4,420 ± 261 | 8,255 ± 151 | 98 ± 5 | 30 ± 8 | 2,082 ± 365 | 4,530 ± 229 | 1,789 ± 1,242 | 302 ± 303 |
| 1422010_at | Tlr7 | 439 ± 13 | 846 ± 40 | <20 | 322 ± 45 | <20 | <20 | 22 ± 2 | 118 ± 83 |
| 1440811_x_at | Cd8a | <20 | 337 ± 134 | 825 ± 44 | <20 | <20 | 1,235 ± 227 | 22 ± 2 | <20 |
| 1449328_at | Ly75 (Dec205) | 249 ± 27 | <20 | 159 ± 4 | 22 ± 3 | 24 ± 6 | 170 ± 29 | 79 ± 1 | 21 ± 1 |
| 1422782_s_at | Tlr3 | 27 ± 2 | 25 ± 3 | 3,376 ± 159 | 287 ± 14 | <20 | <20 | <20 | 52 ± 45 |
| 1422046_at | Itgam (CD11b) | 956 ± 57 | <20 | <20 | 162 ± 1 | 188 ± 38 | <20 | <20 | 21 ± 1 |
| 1449049_at | Tlr1 | 1,218 ± 54 | 31 ± 15 | 101 ± 4 | 1,601 ± 92 | <20 | 889 ± 109 | 498 ± 103 | 1,141 ± 484 |
| 1417268_at | Cd14 | 7,649 ± 169 | 187 ± 52 | 107 ± 0 | 115 ± 34 | <20 | <20 | 31 ± 8 | 27 ± 12 |
| 1449498_at | Marco | 174 ± 19 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| 1460282_at | Trem1 | 415 ± 19 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
Table 3.
Expression of control genes in human cells
| Lymphocytes | Dendritic cells | Myeloid cells | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probe set ID | Genes | NK | CD8 T | CD4 T | B | pDC | BDCA1 | BDCA3 | Mono | Neu |
| 206804_at | CD3G | 858 ± 71 | 1,760 ± 241 | 1,975 ± 132 | 53 ± 6 | <50 | <50 | <50 | <50 | 52 ± 4 |
| 213539_at | CD3D | 5,413 ± 238 | 7,134 ± 635 | 6,291 ± 285 | 276 ± 24 | <50 | <50 | 51 ± 2 | 112 ± 9 | 276 ± 4 |
| 205456_at | CD3E | 247 ± 21 | 569 ± 67 | 679 ± 91 | <50 | <50 | <50 | <50 | <50 | <50 |
| 210031_at | CD3Z | 8,688 ± 181 | 5,223 ± 218 | 4,749 ± 123 | 2,996 ± 217 | 56 ± 10 | 60 ± 17 | 54 ± 7 | 914 ± 96 | 132 ± 15 |
| 209671_x_at | TCR@ | 147 ± 16 | 3,127 ± 260 | 3,462 ± 170 | 71 ± 7 | <50 | <50 | <50 | <50 | 111 ± 16 |
| 205758_at | CD8A | 911 ± 26 | 5,259 ± 217 | 67 ± 10 | 79 ± 16 | <50 | <50 | <50 | <50 | 99 ± 7 |
| 207979_s_at | CD8B | 77 ± 9 | 3,596 ± 299 | <50 | <50 | <50 | <50 | <50 | <50 | 53 ± 5 |
| 203547_at | CD4 | <50 | <50 | 391 ± 20 | 83 ± 20 | 1,301 ± 119 | 1,004 ± 74 | 278 ± 61 | 205 ± 34 | <50 |
| 206398_s_at | CD19 | <50 | 51 ± 1 | <50 | 1,726 ± 115 | <50 | <50 | <50 | 57 ± 12 | <50 |
| 212843_at | NCAM1 (CD56) | 2,074 ± 96 | 144 ± 14 | 65 ± 2 | 135 ± 9 | <50 | <50 | 82 ± 17 | 52 ± 3 | <50 |
| 207314_x_at | KIR3DL2 | 3,131 ± 172 | 454 ± 14 | 227 ± 18 | 265 ± 16 | <50 | <50 | <50 | 59 ± 8 | <50 |
| 208203_x_at | KIR2DS5 | 3,472 ± 140 | 444 ± 7 | 236 ± 10 | 284 ± 14 | <50 | <50 | <50 | <50 | <50 |
| 239975_at | HLA-DPB2 | <50 | <50 | <50 | 63 ± 22 | 777 ± 701 | 1,565 ± 519 | 2,056 ± 577 | <50 | <50 |
| 210184_at | ITGAX (CD11c) | 1,017 ± 50 | 112 ± 37 | 166 ± 17 | 752 ± 45 | 74 ± 21 | 2,151 ± 430 | 729 ± 98 | 1,284 ± 115 | 2,133 ± 196 |
| 210313_at | LILRA4 (ILT7) | 226 ± 10 | 117 ± 13 | 346 ± 42 | 1,109 ± 76 | 7,916 ± 612 | 230 ± 16 | 1,659 ± 1,183 | 524 ± 41 | <50 |
| 206148_at | IL3RA (CD123) | 84 ± 3 | 59 ± 8 | 91 ± 2 | 324 ± 9 | 4,728 ± 365 | 61 ± 10 | 116 ± 110 | 120 ± 3 | 74 ± 12 |
| 1552552_s_at | CLEC4C (BDCA2) | 93 ± 6 | 61 ± 5 | 99 ± 4 | 408 ± 9 | 6,789 ± 737 | 76 ± 39 | 859 ± 434 | 217 ± 8 | 175 ± 25 |
| 205987_at | CD1C (BDCA1) | 76 ± 8 | 61 ± 12 | 159 ± 8 | 1,715 ± 85 | 64 ± 23 | 8,313 ± 272 | 722 ± 845 | 560 ± 59 | <50 |
| 204007_at | FCGR3B (CD16) | 459 ± 54 | 115 ± 24 | 65 ± 5 | 322 ± 46 | 63 ± 23 | <50 | 51 ± 1 | 160 ± 11 | 5,554 ± 57 |
| 201743_at | CD14 | 94 ± 3 | 139 ± 5 | 343 ± 5 | 1,274 ± 113 | <50 | 202 ± 183 | <50 | 7,638 ± 446 | 4,621 ± 374 |
| 205786_s_at | ITGAM (CD11b) | 5,688 ± 116 | 1,980 ± 147 | 1,161 ± 71 | 2,513 ± 117 | 360 ± 184 | 703 ± 28 | 86 ± 63 | 5,541 ± 193 | 5,232 ± 576 |
| 208982_at | PECAM1 (CD31) | 2,232 ± 48 | 2,144 ± 91 | 1,487 ± 58 | 4,644 ± 102 | 3,834 ± 601 | 2,825 ± 290 | 2,680 ± 363 | 5,479 ± 219 | 7,699 ± 853 |
| 205898_at | CX3CR1 | 10,056 ± 53 | 6,633 ± 232 | 4,351 ± 170 | 6,055 ± 263 | 262 ± 45 | 1,296 ± 84 | 362 ± 419 | 5,717 ± 451 | 616 ± 21 |
| 39402_at | IL1B | 69 ± 6 | 72 ± 7 | 52 ± 3 | 209 ± 27 | <50 | 195 ± 131 | 69 ± 27 | 198 ± 9 | 2,920 ± 183 |
| 202859_x_at | IL8 | 95 ± 7 | 77 ± 6 | 72 ± 5 | 385 ± 26 | 218 ± 185 | 90 ± 9 | 680 ± 561 | 310 ± 17 | 8,685 ± 776 |
| 207094_at | IL8RA | 199 ± 30 | 74 ± 8 | 81 ± 12 | 82 ± 2 | <50 | 61 ± 9 | 67 ± 1 | 90 ± 1 | 4,784 ± 521 |
LN-DCs constitute a specific leukocyte family that includes pDCs in both the human and the mouse
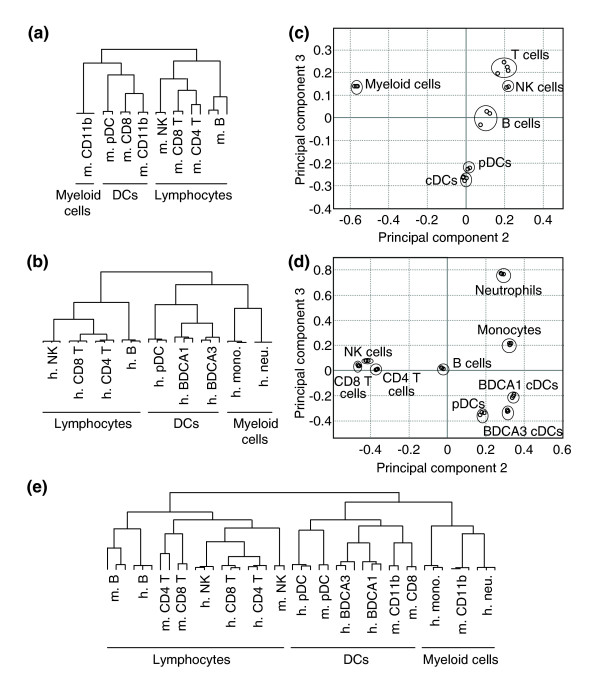
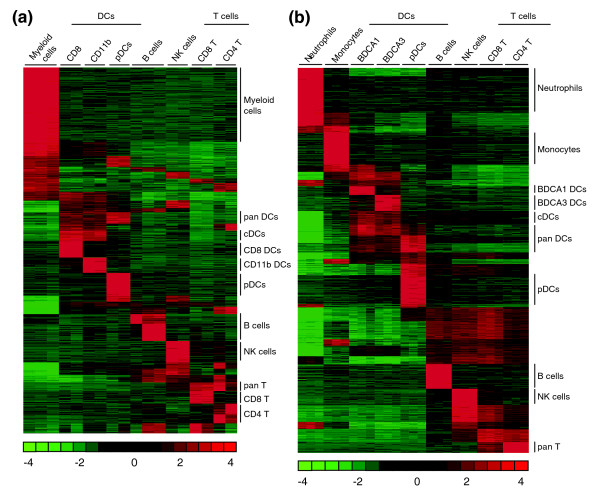
To determine whether LN-DCs may constitute a specific leukocyte family, we first evaluated the overall proximity between LN-DC subsets as compared to lymphoid or myeloid cell types, based on the analysis of their global gene expression program. For this, we used hierarchical clustering with complete linkage [32], principal component analysis (PCA) [33], as well as fuzzy c-means (FCM) partitional clustering approaches [34]. Hierarchical clustering clearly showed that the three LN-DC subsets studied clustered together, both in mouse (7,298 genes analyzed; Figure 1a) and human (11,507 genes analyzed; Figure 1b), apart from lymphocytes and myeloid cells. The close relationship between all the DC subsets in each species was also revealed by PCA for mouse (Figure 1c) and human (Figure 1d). Finally, FCM clustering also allowed clear visualization of a large group of genes with high and specific expression levels in all DC subtypes (Figure 2, 'pan DC' clusters). These analyses, which are based on very different mathematical methods, thus highlight the unity of the LN-DC family. To investigate the existence of a core genetic program common to the LN-DC subsets and conserved in mammals, clustering of mouse and human data together was next performed. We identified 2,227 orthologous genes that showed significant variation of expression in both the mouse and human datasets. After normalization (as described in Materials and methods), the two datasets were pooled and a complete linkage clustering was performed. As shown in Figure 1e, the three major cell clusters, lymphocytes, LN-DCs, and myeloid cells, were obtained as observed above when clustering the mouse or human data alone. Thus, this analysis shows that DC subsets constitute a specific cell family distinct from the classic lymphoid and myeloid cells and that pDCs belong to this family in both mice and humans. All the LN-DC subsets studied therefore share a common and conserved genetic signature, which must determine their ontogenic and functional specificities as compared to other leukocytes, including other antigen-presenting cells.
Figure 1.
Clustering of mouse and human leukocyte subsets. Hierarchical clustering with complete linkage was performed on the indicated cell populations isolated from: (a) mouse, (b) human, and (e) mouse and human. PCA was performed on the indicated cell populations isolated from: (c) mouse and (d) human. Mono, monocytes; neu, neutrophils.
Figure 2.
FCM partitional clustering. FCM partitional clustering was performed on the mouse and human gene chip datasets. (a) FCM partitional clustering for mouse data. (b) FCM partitional clustering for human data. The color scale for relative expression values as obtained after log10 transformation and median centering of the values across cell samples for each gene is given below the heat map.
Identification and functional annotation of the conserved transcriptional signatures of mouse and human leukocyte subsets
Genes that are selectively expressed in a given subset of leukocytes in a conserved manner between mouse and human were identified and are presented in Table 4. Our data analysis is validated by the recovery of all the genes already known to contribute to the characteristic pathways of development or to the specific functions for the leukocyte subsets studied, as indicated in bold in Table 4. These include, for example, Cd19 and Pax5 for B cells [35], Cd3e-g and Lat for T cells [36], as well as Ncr1 [37] and Tbx21 (T-bet) [38] for NK cells. Similarly, all the main molecules involved in major histocompatibility (MHC) class II antigen processing and presentation are found selectively expressed in antigen-presenting cells (APCs). Indeed, a relatively high proportion of the genes selectively expressed in lymphocytes or in APCs has been known for a long time to be involved in the biology of these cells. However, we also found genes identified only recently as important in these cells, such as March1 [39] or Unc93b1 [40,41] for APCs, and Edg8 for NK cells [42]. Interestingly, we also identified genes that were not yet known to be involved in the biology of these cells, to the best of our knowledge, such as the E430004N04Rik expressed sequence tag in T cells, the Klhl14 gene in B cells, or the Osbpl5 gene in NK cells.
Table 4.
Specific transcriptomic signatures identified in the leukocyte populations studied
| Expression ratio (log2) of specific genes* | ||||
|---|---|---|---|---|
| Cell type | 3-4 | 2-3 | 1-2 | 0,4-1 |
| Myeloid cells | - | Steap4; Clec4d; Clec4e; Fpr1 | Nfe2; Mpp1; Snca; Ccr1; Slc40a1; S100a9; Cd14; Tlr4; F5; Fcgr3; Fpr-rs2; Tlr2; Abhd5; Gca; Atp6v1b2; Ier3; Sod2; Pilra; Slc11a1 | Sepx1; Ninj1; Hp; Sdcbp; Bst1; Ifit1; S100a8; Adipor1; Bach1; Marcks; Pira2; Wdfy3; Ifrd1; Fcho2; Csf3r ; C5ar1; Cd93; Snap23; Cebpb; Clec7a; Yipf4; Hmgcr; Slc31a2; Fbxl5 |
| Pan-DC | Flt3 | Sh3tc1 | Trit1; Bri3bp; Prkra; Etv6; Tmed3; Bahcc1; Scarb1 | |
| cDC | - | - | Arhgap22; Btbd4; Slamf8; 9130211I03Rik; Nav1 | C2ta ; Avpi1; Spint1; Cs |
| pDC | Epha2 ; Pacsin1; Zfp521; Sh3bgr | Tex2; Runx2; Atp13a2; Maged1; Tm7sf2; Tcf4; Gpm6b; Cybasc3 | Nucb2; Alg2; Pcyox1; LOC637870; Scarb2; Dnajc7; Trp53i13; Plac8 ; Pls3; Tlr7 ; Ptprs; Bcl11a | |
| B cells | Ebf1 ; Cd19 ; Klhl14 | Bank1 ; Pax5 | Blr1 ; Ralgps2; Cd79b ; Pou2af1 ; Fcer2a; Cr2; Cd79a ; Fcrla | Ms4a1; Blk ; Cd72 ; Syvn1; BC065085; Fcrl1; Phtf2; Tmed8; Grap; Pip5k3; Pou2f2 |
| NK cells | - | Ncr1 | Tbx21 ; Osbpl5 | Rgs3; 1700025G04Rik; Plekhf1; Fasl ; Zfpm1; Edg8 ; Cd160 ; Klrd1 ; Il2rb ; Il18rap ; Ctsw; Ifng ; Prf1 ; Sh2d2a; Llgl2; Gpr178; Prkx; Gab3; Nkg7 ; Cst7; Sntb2; Runx3; Myo6; F2r; Vps37b; Dnajc1; Gfi1 |
| Pan-T cells | - | Camk4 ; E430004N04Rik; Trat1 | Cxcr6 ; Tnfrsf25; Ccdc64; Plcg1 | Cd3e ; Cd5 ; Lrig1; Cd3g ; Ubash3a; Cd6 ; Lat ; Bcl11b; Tcf7 ; Icos |
| CD8 T cells | - | - | - | Gzmk |
| CD4 T cells | - | Ctla4 | - | Icos ; Tnfrsf25; Cd5 ; Cd28 ; Trat1 |
| Lymphocytes | - | - | Ablim1; Lax1; D230007K08Rik; Rasgrp1; Bcl2 | Spnb2; Cdc25b; Ets1 ; Sh2d2a; Ppp3cc; Cnot6l |
| Myeloid, B, DC | - | H2-DMb2 ; H2-DMb1 | C2ta ; March1 ; Aldh2; Bcl11a; Btk | Ctsh ; H2-Eb1 ; Cd74 ; Ctsz ; Clic4; Kynu ; 5031439G07Rik; Nfkbie; Unc93b1 |
| Non-DC | Gimap4 | - | Vps37b | Lck ; Pde3b |
In contrast to the high proportion of documented genes selectively expressed in the cell types mentioned above, most of the genes specifically expressed in LN-DCs have not been previously associated with these cells and many have unknown functions. Noticeable exceptions are Flt3, which has been recently shown to drive the differentiation of all mouse [43-45] and human [46] LN-DC subsets [47], and Ciita (C2ta), which is known to specifically regulate the transcription of MHC class II molecules in cDCs [48]. Interestingly, mouse or human LN-DCs were found to lack expression of several transcripts present in all the other leukocytes studied here, including members of the gimap family, especially gimap4, which have been very recently shown to be expressed to high levels in T cells and to regulate their development and survival [49-51].
Thus, the identity of the gene signatures specific for the various leukocyte subsets studied highlights the sharp contrast between our advanced understanding of the molecular bases that govern the biology of lymphocytes or the function of antigen presentation and our overall ignorance of the genetic programs that specifically regulate DC biology. This contrast is enforced upon annotation of each of the gene signatures found with Gene Ontology terms for biological processes, molecular functions, or cellular components, and with pathways, or with interprotein domain names, using DAVID bioinformatics tools [52,53] (Table 5). Indeed, many significant annotations pertaining directly to the specific function of myeloid cells, lymphocyte subsets or APCs are recovered, as indicated in bold in Table 5. In contrast, only very few significant annotations are found for LN-DCs, most of which may not appear to yield informative knowledge regarding the specific functions of these cells.
Table 5.
Selected annotations for the conserved transcriptomic signatures identified for the cell types studied
| Cell type* | Annotation | Genes |
|---|---|---|
| Myeloid cells | Defense response/response to pest, pathogen or parasite/inflammatory response | C5ar1, Sod2, Fcgr3, Tlr2, Ccr1, Ifrd1, Csf3r, Clec7a, Bst1, Ifit1, Clec4e, Tlr4, Clec4d, Cd14, Cebpb, Hp |
| Response to bacteria or fungi/pattern recognition receptor activity/C-type lectin | SLC11A1, TLR2, TLR4, CLEC7A, Clec4e, Clec4d | |
| H_tollpathway: Toll-like receptor pathway | CD14, TLR2, TLR4 | |
| Regulation of cytokine biosynthesis/positive regulation of TNF-α or IL-6 biosynthesis | Fcgr3, Tlr2, Tlr4, Cebpb, Clec7a | |
| Macrophage activation/mast cell activation/neutrophil chemotaxis | CD93, TLR4, Fcgr3, Csf3r | |
| Pan-DC | Binding | ETV6, PRKRA, FLT3, SCARB1, TRIT1, BAHCC1, SH3TC1 |
| cDC | Nucleobase, nucleoside, nucleotide and nucleic acid metabolism | NAV1, BTBD4, CIITA, SNFT |
| Molecular function unknown | Btbd4, Avpi1, Arhgap22 | |
| pDC | Transcription cofactor activity | Maged1, Bcl11a, Tcf4 |
| Integral to membrane | TLR7, EPHA2, TMEPAI, SCARB2, ATP13A2, ALG2, CYBASC3, TM7SF2, GPM6B, PTPRS | |
| Cellular component unknown | Maged1, Sh3bgr, Cybasc3, Alg2, Plac8 | |
| B cells | MMU04662: B cell receptor signaling pathway/B cell activation | Cr2, Cd79a, Cd79b, Cd72, Cd19, Blr1, Ms4a1 |
| MMU04640: hematopoietic cell lineage | Cr2, Fcer2a, Ms4a1, Cd19 | |
| Defense response/response to pest, pathogen or parasite/humoral immune response | PAX5, POU2F2, CR2, MS4A1, CD72, CD19, POU2AF1, BLR1, CD79A, CD79B, FCER2 | |
| NK cells | MMU04650: natural killer cell mediated cytotoxicity/apotosis | Klrd1, Ifng, Ncr1, Fasl, Prf1, Prf1, Plekhf1 |
| Defense response | IL18RAP, CTSW, IFNG, FASLG, CD160, NCR1, PRF1, KLRD1, CST7 | |
| Pan-T cells | HSA04660: T cell receptor signaling pathway/immunological synapse | CD3E, ICOS, PLCG1, LAT, CD3G, Trat1 |
| Defense response/immune response | Cd5, Icos, Cd3e, Ubash3a, Lat, Trat1, Cd3g | |
| HSA04640: hematopoietic cell lineage | CD3E, CD3G, CD5 | |
| CD8 T cells | No annotations | - |
| CD4 T cells | Defense response/immune response | Cd28, Icos, Cd5, Ctla4, Trat1 |
| M_ctla4pathway: the co-stimulatory signal during T-cell activation | Cd28, Icos, Ctla4 | |
| Lymphocytes | Immune response | BCL2, LAX1, ETS1 |
| Myeloid, B, DC | Antigen presentation, exogenous antigen via MHC class II | H2-Eb1, H2-DMb2, H2-DMb1, Cd74 |
| HSA04612: antigen processing and presentation | HLA-DRB1, CIITA, CD74, HLA-DMB | |
| Defense response/immune response | H2-Eb1, H2-DMb2, H2-DMb1, Bcl11a, Cd74 | |
| Non-DC | Phosphoric ester hydrolase activity | LCK, PDE3B |
Thus, when taken together, our data show that LN-DC subsets constitute a specific family of leukocytes, sharing selective expression of several genes, most of which are still of unknown function. We believe that the identification of these genes selectively expressed in LN-DC subsets in a conserved manner between mouse and human will be very helpful for future investigation of the mechanisms regulating LN-DC biology by the generation and study of novel genetically manipulated animal models.
Search for a genetic equivalence between mouse and human LN-DC subsets
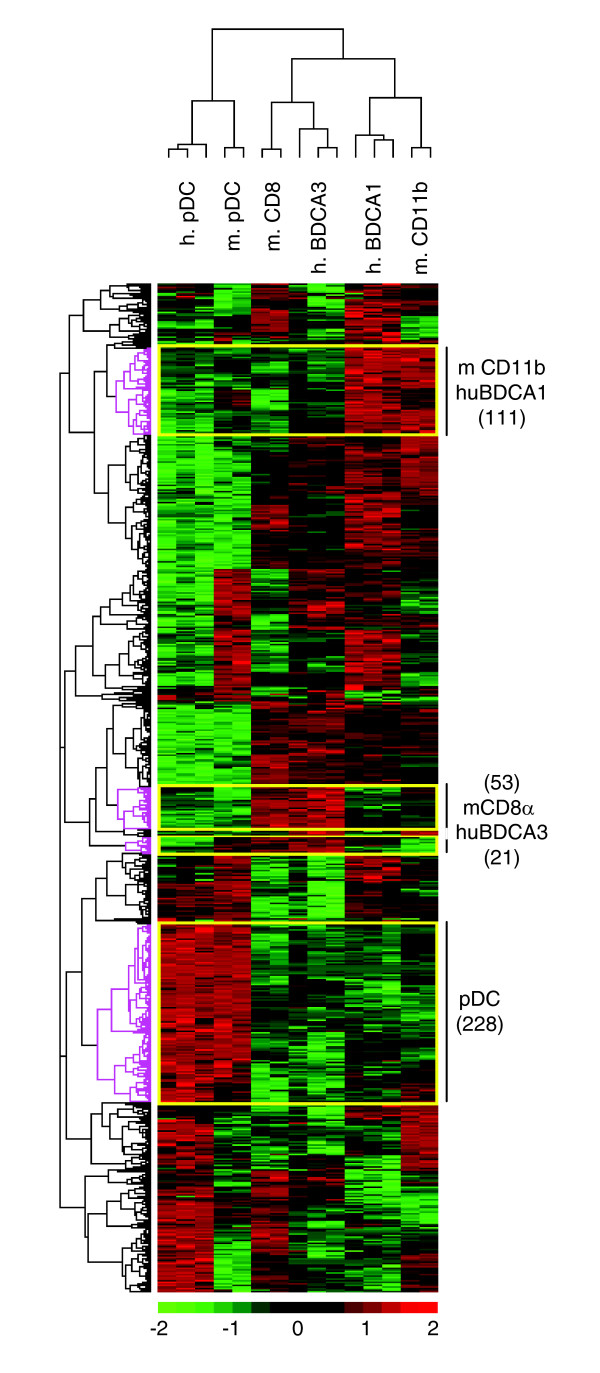
To search for equivalence between mouse and human LN-DC subsets, we examined their genetic relationships in the hierarchical clustering depicted in Figure 1e. Two observations can be made. First and remarkably, mouse and human pDCs clustered together. This result indicates a high conservation in their genetic program and establishes these two cell types as homologs. Indeed, human and mouse pDCs share a large and specific transcriptional signature (Table 4), with a number of genes comparable to those of the transcriptional signature of NK or T cells. To the best of our knowledge, most of these genes had not been reported to be selectively expressed in pDCs, with the exception of Tlr7 [31,54] and Plac8 (C15) [55]. Second, although mouse and human cDCs clustered together, the two cDC subsets of each species appeared closer to one another than to the subsets of the other species. Thus, no clear homology could be drawn between human and mouse cDC subsets in this analysis. However, it should be noted that known homologous human and mouse lymphoid cell types also failed to cluster together in this analysis and were closer to the other cell populations from the same species within the same leukocyte family. This is clearly illustrated for the T cell populations as mouse CD4 and CD8 T cells cluster together and not with their human CD4 or CD8 T cell counterparts (Figure 1e). Therefore, to further address the issue of the relationships between human and mouse cDC subsets, we used a second approach. We performed hierarchical clustering with complete linkage on the mouse and human LN-DC datasets alone (1,295 orthologous LN-DC genes), without taking into account the pattern of expression of each gene in the other leukocyte subsets as it may have hidden some degree of similarity between subsets clustering in the same branch. The results of the analysis of gene expression focused on DCs confirmed that mouse and human pDCs cluster together and apart from cDCs (Figure 3). Importantly, when analyzing the DC datasets alone, mouse CD8α and human BDCA3 cDCs on the one hand, and mouse CD11b and human BDCA1 cDCs on the other hand, clustered together and shared a conserved genetic signature (Figure 3 and Table 6). Thus, although a higher genetic distance is observed between mouse and human conventional DC subsets as opposed to pDCs, a partial functional equivalence is suggested between these cell types. The majority of the genes conserved between mouse CD8α and human BDCA3 cDCs versus mouse CD11b and human BDCA1 cDCs have unknown functions and have not been previously described to exhibit a conserved pattern of expression between these mouse and human cell types. Notable exceptions are Tlr3 [31,56] and the adhesion molecule Nectin-like protein 2 (Cadm1, also called Igsf4) [57], which have been previously described to be conserved between mouse CD8α and human BDCA3 cDCs. When comparing cDC to pDCs, a few genes already known to reflect certain functional specificities of these cells when compared to one another are identified. Tlr7 and Irf7 are found preferentially expressed in pDCs over cDCs, consistent with previous reports that have documented their implication in the exquisite ability of these cells to produce high levels of IFN-α/β in response to viruses [58-60]. Ciita, H2-Ob, Cd83 and Cd86 are found preferentially expressed in cDCs over pDCs, which is consistent with their higher efficiency for MHC class II antigen presentation and T cell priming [61].
Figure 3.
Conserved genetic signatures between mouse and human DC subsets. Hierarchical with complete linkage clustering was performed on the indicated DC populations isolated from mouse and human.
Table 6.
Conserved specific transcriptomic signatures of DC subsets compared to one another
| Expression ratio (log2) of specific genes* | |||||
|---|---|---|---|---|---|
| Cell type | >4 | 3-4 | 2-3 | 1-2 | 0,4-1 |
| pDC | Pacsin1; Sla2; 2210020M01Rik | - | Epha2; Sh3bgr; Ets1; Cobll1; Blnk; Myb; Sit1; Zfp521; Nucb2; Igj; Stambpl1; Ptprcap; Spib; Glcci1; Syne2; Ahi1; Atp13a2; Tcf4; Lair1 | Runx2; LOC637870; Hs3st1; Asph; L3mbtl3; Tex2; Nrp1; Npc1; Maged1; Tm7sf2; Igh-6 ; Csf2rb2; Ccr2; Cdk5r1; Fcrla; Rnasel; Arid3a; Rassf8; Tgfbr3; Tlr7 ; Trp53i11; Ltb4dh; Arhgap24; Creb3l2; Itpr2; Bcl11a; Usp11; Gpm6b; Snx9; Hivep1; Irf7 ; Cnp1; Cybasc3; Pcyox1; Aacs | Ifnar2; Ugcg; Kmo; Tspan31; Xbp1; Alg2; Txndc5; Abca5; Carhsp1; Ptp4a3; Lypla3; Cxxc5; Sema4c; Vamp1; Klhl9; BC031353; Cybb; Scarb2; Card11; Cdkn2d; 4931406C07Rik; Gimap8; Plxdc1; Lman1; 4631426J05Rik; Tcta; Mgat5; Ern1; Atp8b2; Lrrc16; Cln5; Rexo2; Atp2a3; Tspyl4; Anks3; Slc23a2; Gata2; Trp53i13; Slc44a2; Tmem63a; Dnajc7; Rhoh; Daam1; Lancl1; Aff3; Chst12; Unc5cl; Rwdd2; Armcx3; Vps13a; Mcoln2; Tm7sf3; Stch; Glt8d1; Pscd4; Ormdl3; 1110028C15Rik; Snag1; Prkcbp1; Klhl6; Cbx4; Pcmtd1; Bet1; Ccs; Tceal8; Dpy19l3; Pcnx; LOC672274; Sec11l3; Ctsb; Slc38a1; Ostm1; Acad11; Zbtb20; 1110032A03Rik; Ralgps2; Dtx3; Pls3; Ptprs; Zdhhc8; Rdh11; Bcl7a; Tbc1d2b |
| cDC | - | 9130211I03Rik; Hnrpll; Fgl2; Id2; Slamf8 | Chn2; Ddef1; Havcr2; A530088I07Rik; Rab32; Adam8; 2610034B18Rik; Dusp2; Btbd4; Pak1; Bzrap1; Anpep; Apob48r; Aif1 | Arrb1; H2-Ob ; Arhgap22; Aytl1; 2810417H13Rik; Pik3cb; Nav1; Acp2; Tnfaip2; Tspan33; Ralb; Marcks; Epb4.1l2; Rab31; Aim1; Cias1; Cd86 ; Cdca7; Rin3; Hk2; Actn1; Snx8; Cd1d1; Cxcl9; Sestd1; Anxa1; Il15; Ahr; Myo1f; Avpi1; Pde8a; Stom; Spint1; Kit; 1100001H23Rik; Specc1; Bcl6; Tpi1; Kcnk6; Efhd2; Cxcl16; Ddb2; C2ta ; Tgif; Pfkfb3; Ptpn12; Pitpnm1; Rtn1; Maff; Sgk; BB220380; Tes; Elmo1; Tm6sf1; Mast2; Stx11; Dhrs3; Tlr2 | Il18; Vasp; Ppfibp2; Itfg3; Wdfy3; Atad2; Hck; Cnn2; BC039210; Lima1; Fhod1; Klhl5; Flna; Egr1; Mrps27; Gas2l3; Atp2b1; Gypc; Lst1; 8430427H17Rik; Lmnb1; Junb; Irf2; Soat1; Cd83 ; Spg21; Nab2; Rbpsuh; Tiam1; Spfh1; Gemin6; Entpd1; Lzp-s; Lyzs; Slc8a1; Dusp16; Plscr1; Ptcd2; Slc19a2; Mthfd1l; Copg2; Dym; Limd2; Bag3; Csrp1; Ppa1; Nr4a2; Snx10; Hmgb3; Plekhq1; Oat; Rgs12; Numb; Hars2; Pacs1; Gtdc1; Ezh2; Swap70; Rasgrp4; Asahl; Susd3; Lrrk2; Sec14l1; Asb2; Txnrd2; E330036I19Rik; Sla; Fscn1; Nr4a1; Inpp1; Tdrd7; 4933406E20Rik; Usp6nl |
| mCD8 and hBDCA3 | - | Clnk | Gcet2; BC028528; Igsf4a | sept3; Sema4f; Fkbp1b; Tlr3 ; Lima1; Dbn1; Plekha5; Fuca1; Fgd6; Snx22; Gfod1 | Rasgrp3; Btla; Asahl; 4930506M07Rik; Lrrc1; 1700025G04Rik; Tspan33; Fnbp1; Itga6; Zbed3; 9030625A04Rik; Rab32; Ptcd2; Gas2l3; Rab11a; Ptplb; Cbr3; Pqlc2; Slamf8; St3gal5; 4930431B09Rik; Dock7; Stx3; Csrp1; Nbeal2; Gnpnat1; Slc9a9; Ncoa7 |
| mCD11b and hBDCA1 | - | - | Il1rn; Papss2; Pram1 | Il1r2; Oas3; Rin2; Ptgs2; Csf1r; Tlr5; Centa1; Pygl; Igsf6; Csf3r; Tesc; Ncf2; S100a4; Rtn1; Cst7; Car2; Ifitm1; 1810033B17Rik; Lrp1; Dennd3; Ifitm3 | Gbp2; Oas2; Ccl5; Pilra; Sirpa; Pla2g7; Ifitm2; Ms4a7; Cdcp1; Nfam1; BC013672; Slc7a7; Ripk2; Map3k3; Ripk5; Lactb; Rsad2; Parp14; D930015E06Rik; Gyk; Ank; Atp8b4; Emilin2; Arrdc2; Slc16a3; Fcgr3; Clec4a2; Ksr1; Itgax; Sqrdl; Hdac4; Rel; Pou2f2; Chka; Lyst; Ubxd5; Jak2; Cd300a; Lst1; Ssh1; Casp1; D12Ertd553e; Ogfrl1; Rin3; Cd302; Pira2 |
The functional annotations associated with the genes selectively expressed in specific DC subsets when compared to the others are listed in Table 7. The most significant clusters of functional annotations in pDCs point to the specific expression in these cells of many genes expressed at the cell surface or in intracellular compartments, including the endoplasmic reticulum, the Golgi stack, and the lysosome. A cluster of genes involved in endocytosis/vesicle-mediated transport is also observed. This suggests that pDCs have developed an exquisitely complex set of molecules to sense, and interact with, their environment and to regulate the intracellular trafficking of endocytosed molecules, which may be consistent with the recent reports describing different intracellular localization and retention time of endocytosed CpG oligonucleotides in pDCs compared to cDCs [62,63]. The most significant clusters of functional annotations in cDCs concerns the response to pest, pathogens or parasites and the activation of lymphocytes, which include genes encoding TLR2, costimulatory molecules (CD83, CD86), proinflammatory cytokines (IL15, IL18), and chemokines (CXCL9, CXCL16), consistent with the specialization of cDCs in T cell priming and recruitment. Clusters of genes involved in inflammatory responses are found in both pDCs and cDCs. However, their precise analysis highlights the differences in the class of pathogens recognized, and in the nature of the cytokines produced, by these two cell types: IFN-α/β production in response to viruses by pDCs through mechanisms involving IRF7 and eventually TLR7; and recognition and killing of bacteria and production of IL15 or IL18 by cDCs through mechanisms eventually involving TLR2 or lysozymes. Many genes selectively expressed in cDCs are involved in cell organization and biogenesis, cell motility, or cytoskeleton/actin binding, consistent with the particular morphology of DCs linked to the development of a high membrane surface for sampling of their antigenic environment and for the establishment of interactions with lymphocytes. pDCs and cDCs also appear to express different arrays of genes involved in signal transduction/cell communication, transcription regulation and apotosis. A statistically significant association with lupus erythematosus highlights the proposed harmful role of pDCs in this autoimmune disease [64].
Table 7.
Selected annotations for the conserved transcriptomic signatures identified for DC subsets when compared to one another
| Cell type | Annotation | Genes |
|---|---|---|
| pDC | Endoplasmic reticulum | Ern1, Lman1, Txndc5, Rdh11, Tm7sf2, Asph, Ormdl3, Stch, Nucb2, Ugcg, Itpr2, Bet1, Sec11l3, Atp2a3 |
| Golgi stack | BET1, HS3ST1, CHST12, SNAG1, LMAN1, MGAT5, GLCCI1, Pacsin1 | |
| Lysosome | Lypla3, Npc1, Scarb2, Ctsb, Pcyox1, Cln5 | |
| Endocytosis/vesicle-mediated transport | Bet1; Gata2; Igh-6; Lman1; Npc1; Pacsin1; Vamp1 | |
| Integral to plasma membrane | EPHA2, SCARB2, CSF2RB, SIT1, ATP2A3, IFNAR2, VAMP1, PTPRS, SLC23A2, PTPRCAP, LANCL1, TM7SF2, CCR2, TSPAN31 | |
| Inflammatory response | TLR7, CYBB, IRF7, CCR2, BLNK | |
| Intracellular signaling cascade/I-κB kinase/NF-κB cascade | SNAG1, SLC44A2, TMEPAI, CARD11, ERN1, SLA2, IFNAR2, CARHSP1, SNX9, RALGPS2, CXXC5, CCR2, BLNK, RHOH | |
| Regulation of transcription, DNA-dependent/DNA binding/transcription regulator activity/RNA polymerase II transcription factor activity/IPR004827: Basic-leucine zipper (bzip) transcription factor | 1110028C15Rik; Aff3; Anks3; Arid3a; Bcl11a; Carhsp1; Cbx4; Cdkn2d; Creb3l2; Cxxc5; Ern1; Ets1; Gata2; Hivep1; Ifnar2; Irf7; Maged1; Myb; Nucb2; Prkcbp1; Runx2; Sla2; Spib; Tcf4; Tspyl4; Xbp1; Zbtb20 | |
| Systemic lupus erythematosus | LMAN1, CCR2, ETS1 | |
| Regulation of apoptosis | CDK5R1, CARD11, ERN1, CBX4, TXNDC5, CTSB | |
| cDC | Response to pest, pathogen or parasite/defense response/immune response/response to stress/inflammatory response/cytokine biosynthesis/response to bacteria/lymphocyte activation | ANXA1; NR4A2; CIAS1; TLR2; CD83; CD86; IL18; CXCL16; MAST2; AIF1; CIITA; SNFT; Lzp-s, Lyzs; ENTPD1; CXCL9; PLSCR1; BCL6; SGK; TXNRD2; DDB2; AHR; IRF2; LST1; SOAT1; HLA-DOB; CD1D; IL15; Rbpsuh; Swap70; Hmgb3; Egr1 |
| Cytoskeleton/actin binding/filopodium/cell motility | FLNA; FHOD1; CNN2; MYO1F; ACTN1; VASP; EPB41L2; FSCN1; KLHL5; MARCKS; Epb4,1l2; Mast2; Aif1; Csrp1; Elmo1; LIMA1; LMNB1; STOM; Nav1, CXCL16, ANXA1 | |
| Morphogenesis/cell organization and biogenesis/neurogenesis | Rasgrp4; Myo1f; Aif1; Pak1; Pacs1; Vasp; Tiam1; Lst1; Cnn2; Numb; Csrp1; Fhod1; Nav1; Rab32; Stx11; Ezh2; Epb4,1l2; Flna; Acp2; Elmo1; Ralb; Rab31; Id2; Tnfaip2; Txnrd2; Anpep; Il18; Rbpsuh, Nr4a2; Spint1 | |
| Signal transduction/cell communication/MMU04010:MAPK signaling pathway/regulation of MAPK activity/GTPase regulator activity/small GTPase mediated signal transduction/IPR003579:Ras small GTPase, Rab type | ADAM8; AHR; ANXA1; ARRB1; Asb2; Avpi1; CD83; CD86; Chn2; CIAS1; CXCL9; Dusp16; DUSP2; Elmo1; ENTPD1; FLNA; Hck; IL15; IL18; INPP1; Kit; Lrrk2; Mast2; NR4A1; NR4A2; PAK1; PDE8A; PIK3CB; PPFIBP2; Rab31; Rab32; Ralb; Rasgrp4; RBPSUH; RGS12; Rin3; RTN1; Sla; SLC8A1; Snx10; Snx8; Tiam1; TLR2; Arhgap22; Ddef1; Rgs12; Usp6nl | |
| Transcription regulator activity | Junb, Id2, Asb2, Ddef1, Irf2, Nr4a2, C2ta, Nab2, Egr1, Nr4a1, Ahr, 9130211I03Rik, Tgif, Rbpsuh, Bcl6 | |
| Apoptosis | Ahr, Nr4a1, Il18, Bag3, Cias1, Elmo1, Cd1d1, Sgk, Bcl6 | |
| mCD8 and hBDCA3 | Cell organization and biogenesis | DBN1, RAB32, ITGA6, FGD6, RAB11A, SEMA4F |
| Intracellular signaling cascade/small GTPase mediated signal | MIST, TLR3, SNX22; DOCK7; FGD6; RAB11A; RAB32; RASGRP3; sep3 | |
| mCD11b and hBDCA1 | Immune response/defense response/inflammatory response/positive regulation of cytokine production/response to pest, pathogen or parasite/antimicrobial humoral response/IPR006117:2-5-oligoadenylate synthetase | IFITM3, PTGS2, POU2F2, LST1, GBP2, CCL5, OAS2, FCGR2A, NCF2, CSF1R, TLR5, CSF3R, IL1R2, CST7, IL1RN, NFAM1, IFITM2, IFITM1, LILRB2, OAS3, LYST, CLEC4A, IGSF6, HDAC4, PLA2G7, RIPK2, OAS2, OAS3; Rel; Fcgr3 |
| Signal transduction/cell communication/signal transducer activity/positive regulation of I-κB kinase/NF-κB cascade/protein-tyrosine kinase activity/IPR003123:Vacuolar sorting protein 9; vesicle-mediated transport; endocytosis | CASP1; CCL5; CD300A; CD302; CENTA1; CHKA; CLEC4A; CSF1R; CSF3R; FCGR2A; IFITM1; IGSF6; IL1R2; IL1RN; ITGAX; JAK2; KSR1; LILRB2; LRP1; LYST; MAP3K3; MS4A7; NFAM1; OGFRL1; REL; RIN2; RIN3; RIPK2; RIPK5; RTN1; TLR5; Fcgr3 | |
| Chemotaxis/cell adhesion | ITGAX, CD300A, CSF3R, EMILIN2, CLEC4A, CCL5, Fcgr3 | |
| HSA04640:hematopoietic cell lineage | CSF1R, CSF3R, IL1R2 | |
| Asthma. Atopy | PLA2G7, CCL5, |
The mCD11b/hBDCA1 cDC cluster of genes comprises many genes involved in inflammatory responses and the positive regulation of the I-kappaB kinase/NF-kappaB cascade. A statistically significant association with asthma also highlights the proinflammatory potential of this cell type. Recently, it has been reported that the mouse CD11b cDC subset is specialized in MHC class II mediated antigen presentation in vivo [11]. In support of our findings here that mouse CD11b cDCs are equivalent to human BDCA1 cDCs, we found that many of the genes involved in the MHC class II antigen presentation pathway that were reported to be expressed to higher levels in mouse CD11b cDCs over CD8α cDCs [11] are also preferentially expressed in the human BDCA1 cDC subset over the BDCA3 one. These genes include five members of the cathepsin family (Ctsb, Ctsd, Ctsh, Ctss, and Ctsw) as well as Ifi30 and Lamp1 and Lamp2 (see Additional data file 2 for expression values). Thus, it is possible that, like the mouse CD11b cDC subset, human BDCA1 cDCs serve as a subset of DCs that are specialized in presenting antigen via MHC class II molecules. It is also noteworthy that mCD11b and hBDCA1 cDCs express high constitutive levels of genes that are known to be induced by IFN-α/β and that can contribute to cellular antiviral defense (Oas2, Oas3, Ifitm1, Ifitm2, Ifitm3).
No significant informative functional annotations are found for the mCD8α/hBDCA3 cDC gene cluster. However, groups of genes involved in cell organization and biogenesis or in small GTPase regulator activity are found and the study of these genes may increase our understanding of the specific functions of these cells. Mouse CD8α cDCs have been proposed to be specialized for a default tolerogenic function but to be endowed with the unique ability to cross-present antigen for the activation of naïve CD8 T cells within the context of viral infection [65]. It will be important to determine whether this is also the case for hBDCA3 cDCs. From this point of view, it is noteworthy that hBDCA3 cDCs selectively express TLR3, lack TLR7 and TLR9, and exhibit the highest ratio of IRF8 (ICSBP)/TYROBP (DAP12) expression, all of which have been shown to participate in the regulation of the balance between tolerance and cross-presentation by mouse CD8α cDCs [65,66].
Use of leukocyte gene expression compendia to classify cell types of ambiguous phenotype or function
Interferon-producing killer dendritic cells
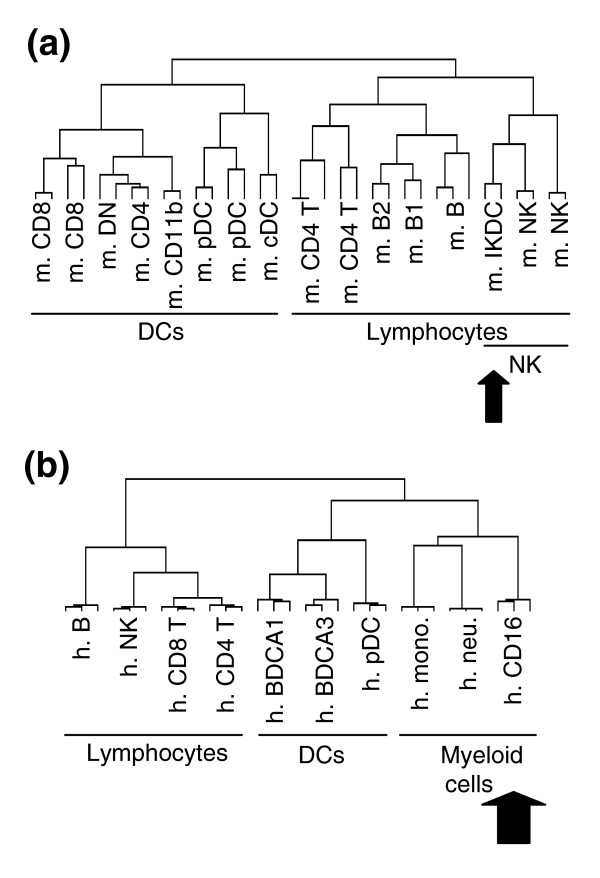
A novel cell type has been recently reported in the mouse that presents mixed phenotypic and functional characteristics of pDCs and NK cells, IKDCs [15,16]. A strong genetic relationship between IKDCs and other DC populations was suggested. However, this analysis was based solely on comparison of the transcriptional profile of IKDCs to DCs and not to other cell populations [15]. As IKDCs were also reported to be endowed with antigen presentation capabilities [15] and to be present in mice deficient for the expression of RAG2 and the common γ chain of the cytokine receptors [16], they have been proposed to belong to the DC family rather than to be a subset of NK cells in a particular state of differentiation or activation. However, IKDCs have been reported to express many mRNA specific for NK cells and many of their phenotypic characteristics that were claimed to discriminate IKDCs from NK cells [16] are in fact consistent with classical NK cell features as recently reviewed [67], including the expression of B220 [68] and CD11c [69,70] (BD/Pharmingen technical datasheet of the CD11c antibody) [71]. To clarify the genetic nature of IKDCs, we reanalyzed the published gene chip data on the comparison of these cells with other DC subsets [15], together with available datasets on other leukocyte populations. We thus assembled published data generated on the same type of microarrays (Affymetrix U74Av2 chips) to build a second mouse compendium, allowing us to compare the transcriptomic profile published for the IKDCs (n = 2) with that of pDCs (n = 2), cDCs (n = 2) [15], CD8α+ (n = 2), CD4+ (n = 2) or double-negative (n = 2) cDC subsets [56], NK cells [72], CD4 T cells (n = 2), and B1 (n = 2) and B2 (n = 2) cells [18]. Information regarding the original sources and the public accessibility of the corresponding datasets are given in Table 1. As depicted in Figure 4a, the hierarchical clustering with complete linkage results of these data sets, together with our novel 430 2.0 data, clearly show that IKDCs cluster with NK cells, close to other lymphocytes, and not with DCs. Indeed, IKDCs express the conserved genetic signature of NK cells but not of DCs (Table 8 and Additional data file 4). Thus, these results strongly support the hypothesis that the cells described as IKDCs feature a specific subset of mouse NK cells that are in a particular differentiation or activation status, rather than a new DC subset.
Figure 4.
Clustering of mouse IKDCs and human CD16 cells. Hierarchical clustering with complete linkage was performed on the indicated cell populations isolated from: (a) mouse and (b) human. Mono, monocytes; neu, neutrophils.
Table 8.
Expression of APC, DC and NK signature genes in IKDCs
| Ratio | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe set ID | Gene | CD8 DC | DN DC | CD4 DC | pDC | cDC | IKDC | NK | IKDC/DC | NK/DC | IKDC/NK |
| APC signature genes | |||||||||||
| 98035_g_at | H2-DMb1 | 2,701* | 3,416 | 4,281 | 1,105 | 2,722 | 179 | 36 | 0.2 | <0.1 | 5 |
| 92668_at | Btk | 454 | 259 | 331 | 252 | 277 | 91 | 20 | 0.4 | <0.1 | 5 |
| 94834_at | Ctsh | 1,606 | 2,650 | 2,862 | 2,993 | 1,653 | 129 | 20 | 0.1 | <0.1 | 6 |
| 94285_at | H2-Eb1 | 8,183 | 7,761 | 7,201 | 5,285 | 14,120 | 1,018 | 74 | 0.2 | <0.1 | 14 |
| 101054_at | Cd74 | 9,094 | 7,810 | 7,313 | 5,158 | 12,258 | 1,031 | 55 | 0.2 | <0.1 | 19 |
| 92633_at | Ctsz | 520 | 1,246 | 1,171 | 887 | 750 | 117 | 44 | 0.2 | <0.1 | 3 |
| 94256_at | Clic4 | 1,668 | 1,067 | 1,234 | 739 | 717 | 440 | 295 | 0.6 | 0.4 | 1 |
| 160781_r_at | Unc93b1 | 683 | 710 | 789 | 301 | 138 | 36 | 22 | 0.3 | 0.2 | 2 |
| Pan-DC signature genes | |||||||||||
| 95295_s_at | Flt3 | 2,769 | 2,004 | 2,231 | 2,069 | 2,547 | 270 | 45 | 0.1 | <0.1 | 6 |
| 100095_at | Scarb1 | 716 | 405 | 333 | 297 | 398 | 125 | 73 | 0.4 | 0.2 | 2 |
| Non-DC signature genes | |||||||||||
| 96172_at | Gimap4 | 29 | 62 | 20 | 314 | 319 | 5,274 | 982 | 263 | 49 | 5 |
| 92398_at | Vps37b | 111 | 139 | 44 | 76 | 56 | 462 | 159 | 11 | 4 | 3 |
| 161265_f_at | Lck | 99 | 80 | 105 | 235 | 199 | 1,991 | 366 | 25 | 5 | 5 |
| NK signature genes | |||||||||||
| 97781_at | Ncr1 | 20 | 20 | 20 | 73 | 39 | 1,483 | 120 | 20 | 2 | 12 |
| 97113_at | Fasl | 20 | 28 | 20 | 22 | 30 | 440 | 263 | 15 | 9 | 2 |
| 102272_at | Cd160 | 75 | 107 | 62 | 82 | 58 | 780 | 246 | 7 | 2 | 3 |
| 100764_at | Il2rb | 26 | 45 | 40 | 50 | 65 | 84 | 501 | 1 | 8 | 0.2 |
| 99334_at | Ifng | 20 | 20 | 20 | 29 | 38 | 203 | 109 | 5 | 3 | 2 |
| 93931_at | Prf1 | 33 | 21 | 35 | 94 | 86 | 839 | 1,287 | 9 | 14 | 1 |
| 92398_at | Vps37b | 111 | 139 | 44 | 76 | 56 | 462 | 159 | 11 | 4 | 3 |
Lineage-CD16+HLA-DR+ cells
A subset of leukocytes characterized as lineage-CD16+HLA-DR+ (hereafter referred to as CD16 cells) has been reported in human blood, and claimed to be a subpopulation of DCs based on their antigen-presentation capabilities. This subset segregates apart from BDCA1 and BDCA3 DCs and pDCs upon gene expression profiling [31]. It is not found in significant amounts in secondary lymphoid organs of healthy donors, contrary to pDCs and BDCA1 or BDCA3 cDCs. It expresses specific pattern recognition receptors, such as TLR4 and TLR8, and chemokine receptors, such as CX3CR1 and CMKOR1 [31], which were initially described to be preferentially expressed by monocytes in humans [73]. As the transcriptional relationship of CD16 cells with other known DC populations was originally established based solely on the transcriptional profile of DCs, we sought to better understand the nature of these cells. For this, we reanalyzed the global gene expression profile of CD16 cells in comparison to not only DC subsets but also to monocytes, neutrophils, and lymphocytes. The results depicted in Figure 4b clearly show that the CD16 cells cluster with neutrophils and monocytes and not with LN-DCs. Indeed, we find many genes that are expressed to much higher levels in monocytes or neutrophils and CD16 cells than in LN-DC subsets (Table 9 and Additional data file 2). Interestingly, MAFB, which has been described to inhibit the differentiation of DCs but to promote that of macrophages from hematopoeitic precursors [74], is expressed to much higher levels in CD16 cells and monocytes compared to DCs (average signal intensity of 6,263 in CD16 cells compared to 3,479 in monocytes, 65 in pDCs, 309 in BDCA1 DCs and <50 in BDCA3 DCs). CD16 cells also express to high levels many genes that are absent or only expressed to very low levels in LN-DCs compared to both lymphoid and myeloid cells, in particular many members of the gimap family. Reciprocally, many of the genes characterized above as specifically expressed in human and mouse LN-DCs are absent or expressed only to low levels in CD16 cells, in particular FLT3 and SCARB1. Thus, CD16 cells likely differentiate along the canonical myeloid lineage rather than belong to the LN-DC family. However, many genes are also specifically expressed to much higher levels in LN-DC subsets and CD16 cells than in monocytes, neutrophils and lymphocytes, attesting to the existence of biological functions common, and specific, to DC subsets and CD16 cells. Thus, these results strongly suggest that CD16 cells represent a particular subset of monocytes endowed with DC-like properties. One possibility is that CD16 cells are the naturally occurring equivalents of the 'monocyte-derived DCs' generated in vitro.
Table 9.
Expression of APC, DC and myeloid signature genes in CD16 cells
| Dendritic cells | Myeloid cells | Ratio to DC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probe set ID | Gene | BDCA1 | BDCA3 | pDC | Mono | Neu | CD16 cells | CD16 | Mono | Neu |
| APC signature genes | ||||||||||
| 203932_at | HLA-DMB | 8,636* | 7,929 | 5,894 | 5,194 | 173 | 2,581 | 0.3 | 0.6 | <0.1 |
| 205101_at | CIITA | 2,803 | 2,354 | 724 | 531 | 50 | 226 | <0.1 | 0.2 | <0.1 |
| 219574_at | MARCH1 | 587 | 777 | 544 | 1,214 | 58 | 810 | 1 | 2 | <0.1 |
| 201425_at | ALDH2 | 9,279 | 7,841 | 6,034 | 8,504 | 706 | 1,760 | 0.2 | 0.9 | <0.1 |
| 222891_s_at | BCL11A | 569 | 747 | 4,502 | 310 | 50 | 213 | <0.1 | <0.1 | <0.1 |
| 205504_at | BTK | 1,120 | 822 | 1,132 | 1,409 | 281 | 1,786 | 2 | 1 | 0.3 |
| 202295_s_at | CTSH | 6,197 | 2,528 | 1,211 | 3,949 | 75 | 2,440 | 0.39 | 0.6 | <0.1 |
| 213831_at | DQA1 | 11,535 | 7,503 | 5,919 | 4,701 | 50 | 252 | <0.1 | 0.4 | <0.1 |
| 215536_at | DQB2 | 432 | 391 | 157 | 180 | 81 | 52 | 0.1 | 0.4 | 0.2 |
| 209312_x_at | DRB1 | 14,608 | 14,477 | 13,250 | 11,915 | 228 | 14,007 | 1 | 0.8 | <0.1 |
| 209619_at | CD74 | 12,533 | 12,210 | 10,498 | 9,020 | 867 | 7,383 | 0.6 | 0.7 | <0.1 |
| 210042_s_at | CTSZ | 906 | 848 | 692 | 370 | 153 | 673 | 0.7 | 0.4 | 0.2 |
| 201560_at | CLIC4 | 920 | 305 | 663 | 3,023 | 165 | 354 | 0.4 | 3 | 0.2 |
| 217388_s_at | KYNU | 2,414 | 1,059 | 2,204 | 3,516 | 50 | 3,738 | 2 | 1 | <0.1 |
| 203927_at | NFKBIE | 529 | 272 | 232 | 197 | 63 | 290 | 0.5 | 0.4 | 0.1 |
| 220998_s_at | UNC93B1 | 966 | 850 | 1,938 | 862 | 449 | 1,235 | 0.6 | 0.4 | 0.2 |
| Pan-DC signature genes | ||||||||||
| 206674_at | FLT3 | 3,032 | 5,883 | 2,169 | 208 | <50 | <50 | <0.1 | <0.1 | <0.1 |
| 219256_s_at | SH3TC1 | 1,263 | 899 | 1,128 | 392 | 166 | 858 | 0.7 | 0.3 | 0.1 |
| 218617_at | TRIT1 | 1,159 | 1,246 | 1,851 | 509 | <50 | 339 | 0.2 | 0.3 | <0.1 |
| 231810_at | BRI3BP | 691 | 735 | 836 | 298 | 146 | 279 | 0.3 | 0.4 | 0.2 |
| 209139_s_at | PRKRA | 846 | 1,067 | 1,440 | 316 | 74 | 497 | 0.3 | 0.2 | <0.1 |
| 225764_at | ETV6 | 2,172 | 2,432 | 1,726 | 1,143 | 938 | 941 | 0.4 | 0.5 | 0.4 |
| 208837_at | TMED3 | 1,317 | 1,852 | 1,859 | 665 | <50 | 1,022 | 0.6 | 0.4 | <0.1 |
| 219218_at | BAHCC1 | 87 | 86 | 250 | <50 | <50 | <50 | 0.2 | 0.2 | 0.2 |
| 1552256_a_at | SCARB1 | 325 | 425 | 942 | 165 | 128 | 59 | <0.1 | 0.2 | 0.1 |
| Non-DC signature genes | ||||||||||
| 219243_at | GIMAP4 | 68 | <50 | <50 | 4,404 | 3,504 | 1,334 | 20 | 65 | 52 |
| 221704_s_at | VPS37B | 54 | <50 | <50 | 593 | 962 | 487 | 9 | 11 | 18 |
| 204891_s_at | LCK | <50 | <50 | <50 | 92 | 181 | 65 | - | - | - |
| 214582_at | PDE3B | 78 | <50 | <50 | 129 | 625 | 114 | 1 | 2 | 8 |
| Myeloid signature genes | ||||||||||
| 225987_at | STEAP4 | <50 | <50 | <50 | 877 | 6,090 | <50 | - | - | - |
| 1552773_at | CLEC4D | <50 | <50 | <50 | 452 | 520 | <50 | - | - | - |
| 222934_s_at | CLEC4E | 214 | 124 | 133 | 2,837 | 5,885 | 229 | 1 | 13 | 28 |
| 202974_at | MPP1 | 591 | 281 | 377 | 3,721 | 2,408 | 1,341 | 2 | 6 | 4 |
| 205098_at | CCR1 | 93 | <50 | 115 | 3,712 | 3,627 | 106 | 1 | 32 | 31 |
| 223044_at | SLC40A1 | 769 | 276 | 321 | 5,018 | 3,444 | <50 | - | 6 | 4 |
| 224341_x_at | TLR4 | 94 | <50 | <50 | 1,411 | 2,869 | 540 | 6 | 15 | 31 |
| 204714_s_at | F5 | <50 | <50 | <50 | 1,392 | 2,313 | <50 | - | - | - |
| 203561_at | FCGR2A | 1,010 | 44 | 51 | 2,985 | 7,151 | 2,857 | 3 | 3 | 7 |
| 210772_at | FPRL1 | <50 | <50 | <50 | 389 | 3,454 | 70 | 3 | - | - |
| 204924_at | TLR2 | 904 | 211 | 57 | 2,870 | 5,548 | 1,606 | 2 | 3 | 6 |
| 215223_s_at | SOD2 | 1,474 | 946 | 528 | 3,528 | 7,599 | 4,236 | 3 | 2 | 5 |
| 222218_s_at | PILRA | 1,168 | 150 | 136 | 2,899 | 4,035 | 3,982 | 3 | 2 | 3 |
| 210423_s_at | SLC11A1 | 81 | 60 | 38 | 1,767 | 2,930 | 3,334 | 41 | 22 | 36 |
| 203045_at | NINJ1 | 357 | 66 | 71 | 1,104 | 3,129 | 1,934 | 5 | 3 | 9 |
| 201669_s_at | MARCKS | 521 | 389 | <50 | 2,449 | 3,224 | 1,730 | 3 | 5 | 6 |
| 207697_x_at | LILRB2 | 1,271 | 78 | 774 | 3,353 | 3,711 | 4,903 | 4 | 3 | 3 |
| 1553297_a_at | CSF3R | 1,902 | 409 | 156 | 3,433 | 6,687 | 282 | 0.2 | 2 | 4 |
| 220088_at | C5AR1 | 56 | 34 | 93 | 2,316 | 5,099 | 3,824 | 41 | 25 | 55 |
| 221698_s_at | CLEC7A | 3,229 | 4,295 | 79 | 6,642 | 7,061 | 5,680 | 1 | 2 | 2 |
| 204204_at | SLC31A2 | 442 | 187 | <50 | 1,579 | 2,047 | 1,671 | 4 | 4 | 5 |
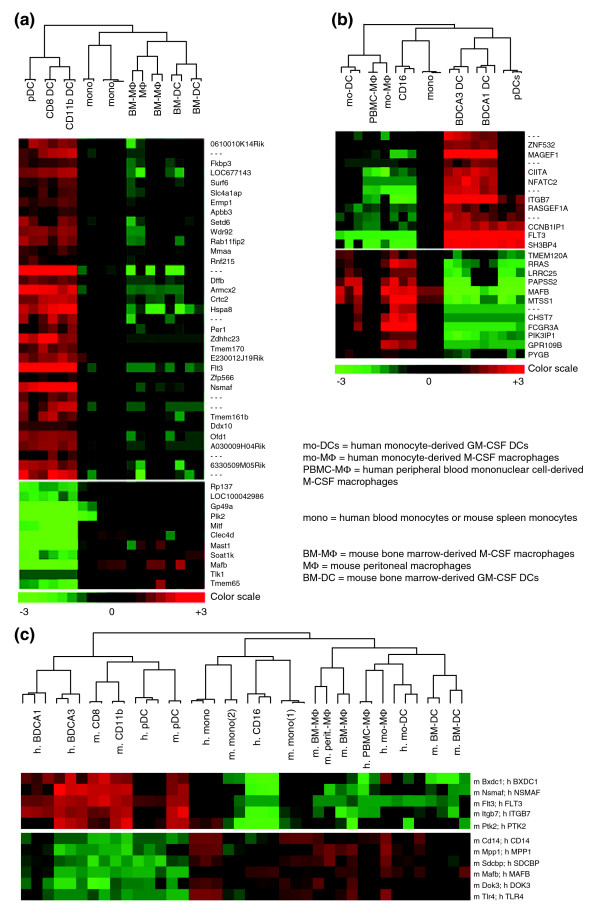
In vitro GM-CSF derived DCs
In vitro derived GM-CSF DCs are the most commonly used model to analyze DC biology. They are often used to investigate the interaction between DCs and other cell types or with pathogens, both in mouse (bone marrow (BM)-derived GM-CSF DCs) and human (monocyte-derived GM-CSF DCs). However, the relationship between these in vitro GM-CSF-derived DCs and the LN-DC subsets present in vivo in the steady state is not clear. A very recent publication suggests that in vitro derived GM-CSF mouse DCs may correspond to the DCs that differentiate from Ly6C+ monocytes in vivo only under inflammatory conditions and appear specialized in the production of high levels of tumor necrosis factor-α and inducible nitric oxide synthase in response to intracellular bacteria, therefore differing from LN-DCs according to both ontogenic and functional criteria [75]. To gain further insights into the relationship between monocytes, macrophages, LN-DCs, and in vitro derived GM-CSF DCs, we thus compared their global gene expression profiling in both human and mouse, using publicly available gene chip data. Information regarding the original sources and the public accessibility of the corresponding datasets are given in Table 1. The results depicted in Figure 5 clearly show that the in vitro derived GM-CSF DCs cluster with monocytes and macrophages and not with the LN-DCs. This result was further confirmed by PCA, which also showed that both mouse and human GM-CSF DCs are close to macrophages, and distant from LN-DCs (Additional data file 6). Indeed, we found many genes that are expressed to much higher levels in monocytes, macrophages and in vitro derived GM-CSF DCs than in LN-DC subsets (Tables 10 and 11). As for human CD16 cells, these genes include the transcription factor Mafb. Reciprocally, some of the genes identified in this study as specific to LN-cDCs are expressed only to much lower levels in GM-CSF DCs. However and interestingly, compared to monocytes, in vitro derived GM-CSF DCs harbor stronger levels of other lymph node resident cDC-specific genes, including scarb1, snft/9130211l03Rik, spint1, ctsh, C22ORF9/5031439G07Rik, and bri3bp. Thus, in vitro derived GM-CSF DCs seem to harbor a strong myeloid gene signature but also express some of the LN-DC-specific genes, consistent with their myeloid ontogeny and their ability to exert myeloid-type functions but also with their acquisition of DC functional properties. In conclusion, our gene chip data analysis is consistent with a very recent report suggesting that in vitro derived GM-CSF mouse DCs correspond to inflammatory DCs and differ greatly from LN-DCs [75]. Indeed, several papers have recently established that in vitro derived FLT3-L DCs constitute the true equivalent of LN-DCs and constitute the only proper surrogate model currently available for their study [75-77].
Figure 5.
Clustering of in vitro GM-CSF derived DCs with monocytes, macrophages and LN-resident DCs. Hierarchical clustering with complete linkage was performed on the indicated cell populations isolated from: (a) mouse, (b) human, and (c) both. The heat maps used for illustration were selected as the two clusters of genes encompassing either Flt3 or Mafb, with a correlation cut-off for similarity of gene expression within each cluster at 0.8.
Table 10.
Comparison of the transcriptome of human GM-CSF monocyte-derived DCs to that of blood DCs
| Ratio to monocytes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe set ID | Name | Mono | PBMC-MΦ | mo-MΦ | mo-DC | CD16 | BDCA3 | BDCA1 | pDCs |
| Myeloid signature genes | |||||||||
| 222934_s_at | CLEC4E | 2,358 | 0.20 | 0.19 | 0.04 | - | - | 0.05 | - |
| 209930_s_at | NFE2 | 823 | 0.06 | 0.06 | 0.89 | 0.10 | - | 0.06 | - |
| 202974_at | MPP1 | 3,622 | 0.40 | 1.25 | 0.68 | 0.33 | 0.08 | 0.15 | 0.11 |
| 205098_at | CCR1 | 3,528 | 0.76 | 1.63 | 1.83 | 0.03 | - | 0.03 | 0.03 |
| 203535_at | S100A9 | 11,192 | 0.05 | 0.37 | 0.01 | 0.12 | 0.02 | 0.17 | 0.01 |
| 201743_at | CD14 | 8,096 | 0.44 | 1.13 | 0.34 | 0.01 | - | 0.02 | 0.01 |
| 224341_x_at | TLR4 | 1,417 | 0.13 | 1.10 | 0.35 | 0.34 | - | 0.06 | - |
| 203561_at | FCGR2A | 2,946 | 0.18 | 0.80 | 1.36 | 0.85 | - | 0.33 | 0.02 |
| 204924_at | TLR2 | 3,220 | 0.14 | 0.80 | 0.32 | 0.54 | 0.08 | 0.31 | 0.02 |
| 218739_at | ABHD5 | 285 | 0.35 | 0.99 | 0.67 | 0.33 | - | - | - |
| 201089_at | ATP6V1B2 | 3,178 | 2.05 | 2.46 | 1.70 | 0.66 | 0.12 | 0.34 | 0.21 |
| 201631_s_at | IER3 | 2,042 | 0.42 | 1.74 | 0.82 | 0.10 | 0.06 | 0.14 | 0.12 |
| 222218_s_at | PILRA | 2,709 | 0.73 | 1.24 | 1.23 | 1.25 | 0.05 | 0.39 | 0.05 |
| 210423_s_at | SLC11A1 | 1,713 | 0.47 | 0.82 | 0.25 | 1.75 | 0.04 | 0.05 | - |
| 203045_at | NINJ1 | 1,190 | 1.69 | 3.59 | 3.41 | 1.59 | 0.27 | 0.44 | 0.26 |
| 200958_s_at | SDCBP | 11,323 | 0.87 | 1.16 | 0.90 | 0.61 | 0.33 | 0.40 | 0.26 |
| 202917_s_at | S100A8 | 15,661 | 0.02 | 0.41 | 0.01 | 0.11 | 0.01 | 0.27 | 0.03 |
| 217748_at | ADIPOR1 | 2,229 | 0.57 | 0.48 | 1.16 | 0.30 | 0.30 | 0.36 | 0.28 |
| 201669_s_at | MARCKS | 2,340 | 0.84 | 2.57 | 1.57 | 0.65 | 0.16 | 0.20 | - |
| 207697_x_at | LILRB2 | 3,260 | 0.29 | 0.64 | 0.76 | 1.36 | 0.02 | 0.39 | 0.24 |
| 228220_at | FCHO2 | 619 | 4.50 | 4.04 | 3.62 | 0.76 | 0.35 | 0.26 | 0.23 |
| 1553297_a_at | CSF3R | 3,121 | 0.42 | 0.69 | 0.37 | 0.08 | 0.11 | 0.52 | 0.04 |
| 220088_at | C5AR1 | 2,059 | 2.56 | 3.63 | 1.30 | 1.60 | - | 0.03 | 0.04 |
| 212501_at | CEBPB | 3,490 | 3.26 | 3.23 | 3.30 | 1.26 | 0.06 | 0.49 | 0.06 |
| 221698_s_at | CLEC7A | 6,596 | 0.24 | 0.55 | 0.63 | 0.74 | 0.62 | 0.46 | 0.01 |
| 209551_at | YIPF4 | 526 | 0.85 | 1.65 | 1.91 | 0.41 | 0.37 | 0.44 | 0.37 |
| 204204_at | SLC31A2 | 1,933 | 0.94 | 1.14 | 0.69 | 0.76 | 0.10 | 0.22 | 0.03 |
| Pan-DC signature genes | |||||||||
| 206674_at | FLT3 | 221 | - | - | - | - | 24.01 | 12.76 | 9.26 |
| 219256_s_at | SH3TC1 | 395 | 1.02 | 2.73 | 1.12 | 2.01 | 2.22 | 3.01 | 2.86 |
| 218617_at | TRIT1 | 498 | 0.49 | 0.58 | 0.86 | 0.71 | 2.46 | 2.15 | 3.61 |
| 231810_at | BRI3BP | 301 | 0.98 | 1.42 | 1.99 | 0.98 | 2.35 | 2.10 | 2.70 |
| 209139_s_at | PRKRA | 325 | 1.12 | 1.77 | 1.47 | 1.57 | 3.17 | 2.42 | 4.37 |
| 225764_at | ETV6 | 1,097 | 0.43 | 1.13 | 2.00 | 0.75 | 2.04 | 1.78 | 1.48 |
| 208837_at | TMED3 | 595 | 1.50 | 2.81 | 1.64 | 1.46 | 2.91 | 1.98 | 2.94 |
| 219218_at | BAHCC1 | - | - | - | - | - | >1.7 | >1.5 | >4.7 |
| 1552256_a_at | SCARB1 | 151 | 8.98 | 6.58 | 7.21 | - | 2.33 | 1.70 | 5.30 |
| cDC signature genes | |||||||||
| 206298_at | ARHGAP22 | - | >5.8 | >6.5 | >3.1 | - | >6.2 | >4.6 | - |
| 227329_at | BTBD4 | - | >1.6 | >2.8 | >5.8 | - | >9.3 | >8.7 | - |
| 219386_s_at | SLAMF8 | 98 | 24.75 | 38.66 | 23.99 | 0.51 | 15.48 | 5.30 | 0.51 |
| 220358_at | SNFT | 148 | 0.62 | 0.34 | 8.62 | 5.66 | 16.01 | 4.82 | 0.34 |
| 224772_at | NAV1 | 64 | 2.01 | 3.25 | 1.40 | 2.00 | 23.87 | 10.50 | 1.62 |
| 205101_at | CIITA | 481 | 0.29 | 0.12 | 1.09 | 0.48 | 4.51 | 5.28 | 1.43 |
| 218631_at | AVPI1 | - | >18.7 | >31.3 | >64.8 | >1.6 | >3.2 | >7.0 | - |
| 202826_at | SPINT1 | 84 | 4.65 | 7.15 | 8.79 | 0.90 | 2.59 | 2.92 | 0.68 |
| 208660_at | CS | 1,848 | 1.24 | 0.99 | 1.04 | 0.84 | 1.70 | 1.63 | 0.89 |
| APC signature genes | |||||||||
| 203932_at | HLA-DMB | 5,137 | 1.28 | 0.64 | 1.37 | 0.44 | 1.45 | 1.62 | 1.14 |
| 219574_at | MARCH1 | 1,133 | 0.42 | 0.89 | 0.73 | 0.62 | 0.64 | 0.44 | 0.46 |
| 201425_at | ALDH2 | 8,782 | 0.51 | 0.54 | 0.34 | 0.18 | 0.84 | 1.01 | 0.69 |
| 222891_s_at | BCL11A | 310 | 0.98 | 0.34 | 0.50 | 0.74 | 2.40 | 1.73 | 14.23 |
| 205504_at | BTK | 1,372 | 0.29 | 0.47 | 0.64 | 1.13 | 0.58 | 0.75 | 0.81 |
| 202295_s_at | CTSH | 3,755 | 1.76 | 2.37 | 2.09 | 0.56 | 0.63 | 1.57 | 0.31 |
| 209312_x_at | HLA-DRB1 | 12,737 | 1.02 | 0.57 | 1.34 | 1.11 | 1.12 | 1.11 | 1.00 |
| 209619_at | CD74 | 8,540 | 1.49 | 0.86 | 2.12 | 0.73 | 1.33 | 1.34 | 1.11 |
| 210042_s_at | CTSZ | 369 | 0.76 | 1.13 | 17.00 | 1.66 | 2.13 | 2.17 | 1.83 |
| 201560_at | CLIC4 | 2,828 | 0.87 | 0.88 | 1.00 | 0.12 | 0.10 | 0.28 | 0.22 |
| 217388_s_at | KYNU | 3,429 | 1.50 | 1.95 | 0.90 | 0.94 | 0.30 | 0.65 | 0.63 |
| 217118_s_at | C22orf9 | 1,617 | 3.33 | 3.46 | 2.77 | 1.43 | 1.85 | 1.79 | 1.04 |
| 203927_at | NFKBIE | 173 | 3.30 | 9.96 | 3.13 | 1.45 | 1.39 | 2.60 | 1.25 |
| 220998_s_at | UNC93B1 | 847 | 0.60 | 1.31 | 0.97 | 1.31 | 0.99 | 1.06 | 2.27 |
| Non-DC signature genes | |||||||||
| 219243_at | GIMAP4 | 4,384 | 0.15 | 0.11 | 0.19 | 0.27 | - | - | - |
| 221704_s_at | VPS37B | 559 | 0.26 | 0.90 | 0.47 | 0.80 | - | - | - |
| 204891_s_at | LCK | 96 | 1.48 | 0.52 | 0.52 | 0.59 | - | - | - |
| 214582_at | PDE3B | 144 | 2.82 | 2.99 | 2.43 | 0.76 | - | 0.51 | - |
Table 11.
Comparison of the transcriptome of mouse GM-CSF BM-derived DCs to that of spleen DCs
| Ratio to monocytes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe set ID | Name | Mono | Mono(2) | MΦ | BM-MΦ | BM-DC | pDC | CD8 DC | CD11b DC |
| Myeloid signature genes | |||||||||
| 1420804_s_at | Clec4d | 4,934 | 0.65 | 0.49 | 0.75 | 0.41 | - | - | - |
| 1420330_at | Clec4e | 5,511 | 0.11 | 0.22 | 0.23 | 0.11 | - | - | - |
| 1450808_at | Fpr1 | 119 | 1.91 | - | 5.55 | 2.41 | - | - | - |
| 1452001_at | Nfe2 | 139 | 1.44 | - | - | 3.31 | - | - | - |
| 1450919_at | Mpp1 | 1,888 | 0.15 | 2.05 | 1.75 | 0.52 | 0.23 | 0.09 | 0.07 |
| 1419609_at | Ccr1 | 403 | 1.27 | 4.04 | 0.53 | 3.98 | 0.2 | - | - |
| 1417061_at | Slc40a1 | 2,588 | 0.68 | - | 0.56 | 0.07 | 0.01 | 0.01 | 0.02 |
| 1448756_at | S100a9 | 8,664 | 1.2 | - | 0.01 | 0.99 | 0 | 0 | - |
| 1417268_at | Cd14 | 6,745 | 0.1 | 0.3 | 0.6 | 0.19 | 0.02 | 0.01 | 0.01 |
| 1418163_at | Tlr4 | 464 | 0.1 | 0.36 | 0.93 | 0.66 | - | 0.07 | 0.06 |
| 1448620_at | Fcgr3 | 1,471 | 2.02 | 3.56 | 2.15 | 2.46 | - | 0.02 | 0.07 |
| 1422953_at | Fpr-rs2 | 839 | 2.04 | 0.12 | 0.85 | 1.58 | - | - | 0.05 |
| 1419132_at | Tlr2 | 1,763 | 0.11 | 0.42 | 0.24 | 0.48 | 0.04 | 0.1 | 0.14 |
| 1417566_at | Abhd5 | 170 | 0.19 | 0.72 | 0.86 | 2.2 | 0.18 | 0.45 | 0.25 |
| 1415814_at | Atp6v1b2 | 1,556 | 0.22 | 2.75 | 1.57 | 1.43 | 0.18 | 0.27 | 0.24 |
| 1427327_at | Pilra | 434 | 1.53 | 0.16 | 0.47 | 2.29 | 0.1 | - | 0.21 |
| 1418888_a_at | Sepx1 | 4,416 | 0.48 | 0.34 | 0.31 | 0.56 | 0.03 | 0.04 | 0.05 |
| 1438928_x_at | Ninj1 | 5,574 | 0.03 | 1.3 | 0.46 | 0.36 | 0.03 | 0.02 | 0.02 |
| 1448881_at | Hp | 400 | 3.19 | 0.14 | 0.06 | 3.09 | - | - | - |
| 1449453_at | Bst1 | 340 | 1.08 | 4.97 | 0.58 | 1.61 | 0.21 | 0.51 | - |
| 1419394_s_at | S100a8 | 10,190 | 1.37 | 0.01 | 0.01 | 0.66 | - | 0 | - |
| 1437200_at | Fcho2 | 311 | 1.09 | 1.32 | 1.06 | 0.76 | 0.28 | 0.2 | 0.33 |
| 1418806_at | Csf3r | 2,598 | 0.2 | 0.14 | 0.19 | 0.11 | - | - | 0.03 |
| 1439902_at | C5ar1 | 317 | 8.21 | 0.19 | 1.63 | 0.37 | - | - | - |
| 1456046_at | Cd93 | 1,559 | 0.1 | 0.49 | 1.18 | 0.33 | 0.02 | - | - |
| 1418901_at | Cebpb | 3,797 | 0.14 | 0.7 | 0.22 | 0.42 | 0.02 | 0.01 | 0.02 |
| 1420699_at | Clec7a | 2,748 | 0.83 | 2.62 | 0.44 | 1.71 | 0.08 | 0.06 | 0.54 |
| Pan-DC signature genes | |||||||||
| 1419538_at | Flt3 | 51 | 0.74 | - | - | 0.7 | 16.2 | 25.32 | 17.78 |
| 1427619_a_at | Sh3tc1 | - | >1.1 | >6.8 | >2.8 | >4.9 | >5.2 | >6.5 | >4.6 |
| 1424489_a_at | Trit1 | 54 | 7.28 | 0.44 | 0.76 | 1.23 | 9.03 | 11.53 | 8.63 |
| 1428744_s_at | Bri3bp | 161 | 0.84 | 0.6 | 1.44 | 3.28 | 6.09 | 7.24 | 5.98 |
| 1448923_at | Prkra | 72 | 1.28 | 0.77 | 2.89 | 2.57 | 4.45 | 7.88 | 3.63 |
| 1434880_at | Etv6 | 140 | 5.39 | 1.52 | 0.74 | 1.75 | 5.79 | 6.02 | 7.78 |
| 1416108_a_at | Tmed3 | 154 | 0.81 | 3.74 | 2.63 | 4.65 | 10.17 | 4.48 | 3 |
| 1436633_at | Bahcc1 | 41 | 1.77 | - | 0.83 | - | 1.8 | 3.88 | 2.35 |
| 1437378_x_at | Scarb1 | 97 | 5.02 | 1.25 | 2.61 | 3.17 | 7.41 | 8.27 | 4.05 |
| cDC signature genes | |||||||||
| 1435108_at | Arhgap22 | 63 | - | - | 2.37 | 0.57 | 0.59 | 10.65 | 4.43 |
| 1429168_at | Btbd4 | 129 | 0.19 | 0.27 | - | 0.47 | 0.81 | 3.89 | 3.8 |
| 1425294_at | Slamf8 | 146 | 1.06 | 39.89 | 1.83 | 1.77 | 0.39 | 8.48 | 5.27 |
| 1453076_at | 9130211I03Rik | 36 | 1.61 | 2.85 | 1.03 | 13.11 | 0.62 | 30.94 | 25.64 |
| 1436907_at | Nav1 | 102 | 1.59 | 0.74 | 2.63 | 1.96 | 1.21 | 6.08 | 13.14 |
| 1421210_at | C2ta | 125 | 0.17 | 1.79 | 0.19 | 0.93 | 1.46 | 5.94 | 5.43 |
| 1423122_at | Avpi1 | 150 | 0.32 | - | 0.2 | 0.86 | 0.61 | 2.47 | 7.62 |
| 1416627_at | Spint1 | - | >1.5 | >1.1 | - | >22.9 | >1.6 | >25.7 | >30.6 |
| 1450667_a_at | Cs | 396 | 2.47 | 0.9 | 1.19 | 3.54 | 2.83 | 4.64 | 4.5 |
| APC signature genes | |||||||||
| 1419744_at | H2-DMb2 | 451 | 0.12 | 0.1 | 0.08 | 1.47 | 0.45 | 0.48 | 1.69 |
| 1443687_x_at | H2-DMb1 | 547 | 0.56 | 0.13 | 0.11 | 1.56 | 1.06 | 0.82 | 3.13 |
| 1434955_at | March1 | 80 | 32.64 | 0.83 | 1.51 | 3.48 | 3.73 | 13.4 | 8.57 |
| 1448143_at | Aldh2 | 867 | 0.47 | 2.14 | 2.07 | 1.32 | 0.95 | 0.65 | 0.45 |
| 1419406_a_at | Bcl11a | 60 | 1.47 | 0.34 | - | 0.71 | 20.41 | 7.63 | 9.19 |
| 1422755_at | Btk | 416 | 0.56 | 0.76 | 1.3 | 1.15 | 0.88 | 1.45 | 1.17 |
| 1418365_at | Ctsh | 1,393 | 0.81 | 3.9 | 2.19 | 2.15 | 3.69 | 1.24 | 2.16 |
| 1417025_at | H2-Eb1 | 6,385 | 0.13 | 0.39 | 0.04 | 0.8 | 0.9 | 1.31 | 1.33 |
| 1425519_a_at | Cd74 | 8,377 | 0.36 | 0.95 | 0.2 | 0.9 | 0.83 | 0.97 | 0.98 |
| 1417868_a_at | Ctsz | 7,061 | 0.05 | 1.16 | 0.95 | 0.85 | 0.5 | 0.3 | 0.49 |
| 1423393_at | Clic4 | 2,807 | 0.07 | 2.04 | 0.84 | 0.57 | 0.69 | 0.72 | 0.67 |
| 1430570_at | Kynu | 31 | 1.23 | - | - | 3.21 | 12.87 | 5.16 | 11.56 |
| 1435745_at | 5031439G07Rik | 356 | 0.95 | 0.73 | 2.76 | 2.51 | 3.23 | 3.14 | 4.28 |
| 1458299_s_at | Nfkbie | 767 | 0.4 | 0.62 | 0.1 | 0.44 | 1.25 | 0.65 | 1.27 |
| 1423768_at | Unc93b1 | 663 | 0.1 | 2.27 | 2.69 | 1.46 | 1.2 | 0.93 | 0.91 |
| Non-DC signature genes | |||||||||
| 1424375_s_at | Gimap4 | 362 | 0.14 | 0.29 | - | 0.1 | 0.11 | - | 0.11 |
| 1424380_at | Vps37b | 313 | 0.44 | 0.46 | 0.45 | 0.26 | 0.28 | 0.28 | 0.27 |
| 1425396_a_at | Lck | 118 | - | 0.57 | 0.2 | 0.32 | 0.21 | - | 0.17 |
| 1433694_at | Pde3b | 352 | 0.69 | 0.15 | 0.16 | 0.42 | - | 0.65 | 0.35 |
Discussion
By performing meta-analyses of various datasets describing global gene expression of mouse spleen and human blood leukocyte subsets, we have been able to identify for the first time conserved genetic programs common to human and mouse LN-DC subsets. All the LN-DC subsets examined here are shown to share selective expression of several genes, while harboring only low levels of other transcripts present in all other leukocytes. These analyses indicate that LN-DCs, including pDCs, constitute a specific family of leukocytes, distinct from those of classic lymphoid or myeloid cells. Furthermore, we demonstrate a striking genetic proximity between mouse and human pDCs, which are shown for the first time to harbor a very distinct transcriptional signature as large and specific as that observed for NK cells or T cells. In contrast, a higher genetic distance is observed between mouse and human conventional DC subsets, although a partial functional equivalence is suggested between mCD8α and hBDCA3 cDCs on the one hand versus mCD11b and hBDCA1 cDCs on the other hand.
Our finding that LN-DCs constitute a distinct entity within immune cells raises the question of whether these cells form a distinct lineage in terms of ontogeny, or whether their shared gene expression profile (notably that between cDCs and pDCs) reflects a functional rather than a developmental similarity. To date, the place of both cDCs and pDCs in the hematopoietic tree is not clear [78,79]. A BM progenitor, named macrophage and dendritic cell progenitor (MDP), has been recently identified that specifically gives rise to monocytes/macrophages and to cDCs, but not to polymorphonuclear cells or to lymphoïd cells [80,81]. Under the experimental conditions used in the corresponding report, pDCs were not detected in the progeny of MDPs. Here, we show that the transcriptome programs of mouse spleen and human blood cDCs exhibit only a very limited overlap with that of monocytes/macrophages (Figure 2). This is consistent with the recent observation that monocytes can give rise to mucosal, but not splenic, cDCs, suggesting that splenic cDCs develop from MDPs without a monocytic intermediate [81]. While mouse pDCs have been argued to arise from both lymphoid or myeloid progenitors, their gene expression overlaps with lymphoid or myeloid cells are limited. Interestingly, a murine progenitor cell line that exhibits both cDC and pDC differentiation potential has been described recently [82], suggesting that putative pan-DC progenitors might also exist in vivo, which would be consistent with the gene profiling analyses presented here.
Our study identifies transcriptional signatures conserved between mouse and human, common to all LN-DC subsets examined, or specific to pDCs, cDCs, or individual cDC subsets. A genetic equivalence is suggested between mouse CD8α cDCs and human BDCA3 cDCs, and between mouse CD11b cDCs and human BDCA1 cDCs. In contrast to the genes selectively expressed in subsets of myeloid or lymphoid cells in a conserved manner between mouse and human, most of the genes specifically increased in all LN-DC subsets or in individual LN-DC subsets are currently uncharacterized. As a consequence, the functional annotations of the LN-DC transcriptional signatures appear much less informative than those for myeloid cells, lymphocytes or APCs. This highlights how much has already been deciphered regarding the molecular regulation of antigen presentation or lymphocyte biology, as opposed to how little we know about the genetic programs that determine the specific features of LN-DCs. We believe that our study provides a unique database resource for future investigation of the evolutionarily conserved molecular pathways governing specific aspects of the ontogeny and functions of leukocyte subsets, especially DCs.
It should be noted that many genes are found to be expressed to very high levels in specific subsets of either mouse or man while no orthologous gene has been identified in the other species. This could be due to a true absence of orthologous genes between these two vertebrate species, or to a lack of identification of an existing orthology relationship. It is also possible that some of the genes expressed only in mouse DCs or only in human DCs, and not conserved between the two species, might represent functional homologs, similar to what is observed for human KIR and mouse Ly49 NK cell receptors. This may be the case for the human LILRA4 (ILT7) and the mouse SIGLECH molecules, as both of them signal through immunoreceptor tyrosine-based activation motif (ITAM)-bearing adaptors to downmodulate IFN-α/β production by human and mouse pDCs, respectively, upon triggering of TLRs [83,84]. Thus, understanding the role in LN-DCs of genes identified here only in mouse or human might be important. The transcriptional signatures identified for mouse LN-DC subsets in this study have been confirmed by analyses of independent data recently published by others on mouse cDC subsets, B cells and T cells [11] or on cDCs and pDCs [15]. Most of the data for the mouse 430 2.0 compendium were generated in-house, with the exceptions being CD4 T cells and myeloid cells. In humans, we generated the data for non-DC populations, whereas data for DC subsets and CD16 cells were all generated by another group and retrieved from a public database. It is well known that datasets for the same cell type can vary considerably between laboratories. However, many of the genes identified as specific for each mouse LN-DC subset using our own data were confirmed by the analysis of other data independently generated by the groups of M Nussenzweig and R Steinman [11]. These data are given in Additional data file 5.
Our clustering analyses and PCA also showed relatively little dataset-dependent biases, and generally grouped related cell populations together, even if they were from different origins (see, for instance, the PCA clustering of in vitro derived GM-CSF DC samples, which originated from two independent datasets in Additional data file 6). In addition, we analyzed by real-time PCR the expression profile of 27 genes across mouse leukocyte subsets from biological samples independent of those used in the gene chips analysis. All the results were consistent with the gene chip data (Additional data file 7). We also confirmed specific expression of PACSIN1 in human pDCs at both the mRNA and protein levels (Additional data file 8). Finally, we believe that our approach validates the gene expression profile identified for leukocyte subsets in the strongest way possible, by demonstrating the evolutionary conservation between mouse and human. Indeed, the gene signatures that we describe here are based on genes found specifically expressed in putatively homologous subsets of mouse and human leukocytes compared to several other types of leukocytes. This approach does not rely solely on the use of independent biological samples of similar origin and on different techniques for measurement of the expression of mRNA. It actually shows that orthologous genes share the same specific expression pattern in putatively homologous immune cell subsets from two different species, under conditions where the markers used to purify the human and mouse cell populations, and the probes used to check the expression of the orthologous genes, differ considerably. Thus, we believe that the analyses presented here are extremely robust even though they were, in part, performed by creating compendia regrouping data generated by different laboratories for different cell types.
In addition to our discovery of transcriptional signatures specific to all LN-DCs or to LN-DC subsets, we demonstrate that, once identified, the transcriptional signatures of multiple cell types can be effectively used to help determine the nature of newly identified cell types of ambiguous phenotype or functions. In our attempt to appropriately place IKDCs and CD16 cells within the leukocyte family, we used the microarray data from the original reports aimed at characterizing these cells and compared them to the data from several other leukocyte populations. The conclusions of this analysis are in sharp contrast to those originally reported [15,31]. We believe that these opposing conclusions arise from the difference in the contextual framework within which our data and that of the previously mentioned studies were analyzed. Thus, the results of our analysis of the transcriptional signature of both IKDCs and CD16 cells emphasize the need to study the transcriptional signatures of individual cell populations in the context of multiple cell types of various phenotypes and functions. Finally, this approach also allowed us to confirm a very recent report that demonstrated that in vitro derived GM-CSF mouse DCs likely correspond to inflammatory DCs and greatly differ from LN-DCs, based on ontogenic and functional studies [75]. Thus, extrapolation to LN-DCs of the results of the cell biology and functional studies performed with in vitro derived GM-CSF DCs should only be made with extreme caution.
Conclusion
This study comparing whole genome expression profiling of human and mouse leukocytes has identified for the first time conserved genetic programs common to all LN-DCs or specific to the plasmacytoid versus conventional subsets. In depth studies of these genetic signatures should provide novel insights on the developmental program and the specific functions of LN-DC subsets. The study in the mouse of the novel, cDC-specific genes identified here should accelerate the understanding of the mysteries of the biology of these cells in both mouse and human. This should help to more effectively translate fundamental immunological discoveries in the mouse to applied immunology research aimed at improving human health in multiple disease settings.
Materials and methods
Sorting of cell subsets
Duplicates of pDCs (Lin-CD11c+120G8high), CD8α cDCs (Lin-CD11chighCD8α+120G8-/low), CD11b cDCs (Lin-CD11chighCD11b+120G8-/low) and NK cells (NK1.1+TCRβ-) were sorted during two independent experiments from pooled spleens of untreated C57BL/6 mice. Splenic CD19+ B lymphocytes, CD4 T cells and CD8 T cells were sorted in other independent experiments. Purity of sorted cell populations was over 98% as checked by flow cytometry (not shown).
Processing of cell samples for the Affymetrix GeneChip assays
RNA was extracted from between 7.5 × 105 and 1.5 × 106 cells for each leukocyte subset with the Qiagen (Courtaboeuf, France) micro RNAeasy kit, yielding between 200 and 700 ng of total RNA for each sample. Quality and absence of genomic DNA contamination were assessed with a Bioanalyser (Agilent, Massy, France). RNA (100 ng) from each sample was used to synthesize probes, using two successive rounds of cRNA amplification with appropriate quality control to ensure full length synthesis according to standard Affymetrix protocols, and hybridized to mouse 430 2.0 chips (Affymetrix, Santa Clara, CA, USA). Raw data were transformed with the Mas5 algorithm, which yields a normalized expression value, and 'absent' and 'present' calls. Target intensity was set to 100 for all chips.
Individual analysis of the mouse 430 2.0 or human U133 Plus 2.0 compendia
For each compendium, all datasets were normalized with the invariant rank method and only one representative dataset was kept for redundant ProbeSets targeting the same gene. The datasets were further filtered to eliminate genes with similar expression in all samples, by selecting only the genes expressed above 50 (respectively 100) in all the replicates of at least one population for the mouse (respectively human) datasets and whose expression across all samples harbored a coefficient of variation above the median of the coefficient of variation of all ProbeSets. The final dataset consisted of 7,298 (respectively 11,507) ProbeSets for the mouse 430 2.0 compendium (respectively human U133 Plus 2.0), representing individual genes with differential expression between ex vivo isolated cell subsets. The final dataset consisted of 12,857 (respectively 6,724) ProbeSets for the mouse 430 2.0 compendium (respectively human U133 Plus 2.0), representing individual genes with differential expression between LN-DCs, monocytes/macrophages and in vitro derived GM-CSF DCs. These datasets for ex vivo isolated cells are accessible as Excel workbooks in Additional data files 1 and 2. The software Cluster and Treeview were used to classify cell subsets according to the proximity of their gene expression pattern as assessed by hierarchical clustering with complete linkage.
We implemented a function in the Matlab software to perform PCA. This function computes the eigenvalues and eigenvectors of the dataset using the correlation matrix. The eigenvalues were then ordered from highest to lowest, indicating their relative contribution to the structure of the data. For both mouse and human datasets, the first principal component accounted for most of the information (54% and 68% for mouse and human, respectively) and was associated with a similar coordinate for all samples. This component thus reflected the common gene expression among the samples. Second and third components together represented 24% and 21%, respectively, of the information for mouse and human datasets, and thus accounted for a large part of the variability. The projection of each sample on the planes defined by these components was represented as a dot plot to generate the PCA figures.
Partitional clustering was performed using the FCM algorithm, which links each gene to all clusters via a vector of membership indexes, each comprised between 0 and 1 [34]. For both mouse and human datasets, we heuristically set the number of clusters to 30, and the fuzziness parameter m was taken as 1.2 (see [34] for the determination of m). Ten independent runs of the algorithm were performed, and the one minimizing the inertia criterion was selected [34]. A threshold value of 0.9 was taken to select probe sets most closely associated with a given cluster. This selection retained 4,062 and 4,751 probe sets from mouse and human datasets, respectively. Probe set clusters were then manually ordered to provide coherent pictures, which were visualized with Treeview.
Meta-analysis of aggregated mouse and human datasets
We identified 2,227 orthologous genes that showed significant variation of expression in both the mouse 430 2.0 and U133 Plus 2.0 human datasets. This dataset is accessible as an Excel workbook in Additional data file 3. In order to compare the expression patterns of these genes between human and mouse, the log signal values for each of these genes were first normalized to a mean equal to zero and a variance equal to 1, independently in the mouse and human datasets, as previously described for comparing the gene expression program of human and mouse tumors [22,27]. The two normalized datasets were then pooled and a hierarchical clustering with complete linkage was performed. A similar analysis was performed for the comparison of human and mouse LN-DCs, monocytes, macrophages and in vitro derived GM-CSF DCs.
Meta-analysis of mouse 430 2.0 and U74Av2 datasets
In order to classify the IKDCs based on the optimal gene signatures of the different cell subsets examined, with only minimal impact of differences in the experimental protocols used to prepare the cells and to perform the gene chips assays, the clustering of the cell populations was performed as a meta-analysis of our own mouse 430 2.0 dataset together with the published U74Av2 datasets. The Array Comparison support information of the NetAffyx™ analysis center (Affymetrix) was used to identify matched ProbSets between the two types of microarrays. Only one representative dataset was kept for redundant ProbeSets targeting the same gene. This yielded a set of 2,251 genes whose expression could be compared between the two datasets, using the same normalization method as described above. This dataset is accessible as Excel workbooks in Additional data file 4. As expected, this meta-analysis led to co-clustering of all the samples derived from identical cell types whether their gene expression had been measured by us on 430 2.0 microarrays or by others on U74Av2 microarrays, with the exception of the cDC population from [15], which segregated with pDCs rather than with the cDC subsets from the other datasets.
Data mining
Gene lists were analyzed using the DAVID 'functional annotation chart' tool accessible on the NIAID website [52,53]. Different databases were used for these annotations: gene ontology (Amigo), knowledge pathways (KEGG), interactions (BIND), interprotein domains (INTERPRO), and disease (OMIM/OMIA). The annotations shown in Tables 5 and 7 were selected as the most highly significant terms retrieved by performing an over-representation study. To this end, a modified Fisher exact P value called the 'EASE score' was calculated to measure the enrichment in gene-annotation terms between the gene signature specific to the leukocyte subpopulation examined ('List') and the complete set of all the genes selected for the compendium analyzed ('Background'). The significance threshold was set at an EASE score below 0.05 in most instances, or below 0.1 for DC signatures that did not yield many highly significant terms as discussed in Results. Individual significant annotations encompassing many common genes or similar biological processes were regrouped using the 'Functional annotation clustering' tool of the DAVID software. More information on this type of analysis is available on the DAVID website [85].
Public access to the raw data for the datasets analyzed in the paper
Our datasets for mouse DC subsets, NK cells, CD8 T cells, and B lymphocytes have been deposited in the Gene Expression Omnibus (GEO) database under reference number GSE9810. The references for download of the public data used from the original websites where they were first made available are given in Table 1. In addition, all raw transcriptomic data analyzed here have been regrouped on our website [86] and are available for public download.
Abbreviations
APC, antigen-presenting cell; BM, bone marrow; cDC, conventional dendritic cell; CDP, common dendritic progenitor; DC, dendritic cell; FCM, fuzzy c-means; GEO, Gene Expression Omnibus; GM-CSF, granulocyte-macrophage colony stimulating factor; IFN, interferon; IKDC, interferon-producing killer dendritic cell; ITAM, immunoreceptor tyrosine-based activation motif; LN-DC, lymph node-resident DC; M-CSF, macrophage colony-stimulating factor; MDP, macrophage and dendritic cell progenitor; MHC, major histocompatibility; NK, natural killer; PCA, principal component analysis; pDC, plasmacytoid dendritic cell; TLR, toll-like receptor.
Authors' contributions
SHR, TW, SC, PK, and MD designed the research; SHR, TW, CT, HX, MS, GB, AD and MD performed the research; EV and PP contributed new reagents/analytical tools; SHR, TW, CT, HX, DD, MS, FRS, SC, PK, and MD analyzed data; and SHR, TW, and MD wrote the paper.
Note added in proof
During the review process of this paper, two reports were published in Nature Immunology that identified a common progenitor characterized as FLT3+M-CSF+ for mouse LN-DCs (pDCs, CD8α cDCs and CD11b cDCs), devoid of any capability to generate lymphoid cells or monocytes/macrophages, and named common dendritic progenitor (CDP) [87,88]. This observation is thus consistent with our gene profiling analysis of human and mouse leukocytes. The question whether this pathway for LN-DCs is the major one, or just one possibility among others, including differentiation from monocytes, has been raised [89]. Our gene profiling data would suggest that most mouse LN-DCs derive from the recently identified CDP or MDP in vivo, without a monocytic intermediate, consistent with a recent report [81]. It also implies that a similar pathway must exist in humans. The relationship between the CDP and the MDP still remains to be established. Three reports have been published very recently in the Journal of Experimental Medicine that showed that IKDCs are a specific subset of NK cells, based on functional and ontogenic approaches comparing these cells to DCs and NK cells [90-92]. This is consistent with the results of our clustering analysis of IKDCs with other leukocyte subsets. Finally, two recent reports have identified a new transduction pathway in human pDCs involving a B cell receptor-like ITAM-signaling pathway [93,94]. This pathway involves the BLNK transduction molecule, which we have identified here as expressed to very high levels in mouse and human pDCs compared to the other LN-DCs (Table 6) and many other leukocytes. We believe that the conserved transcriptional signatures identified here for mouse and human LN-DC subsets will lead to many more discoveries for the understanding of the specialized functions of these cells.
Additional data files
The following additional data are available. Additional data file 1 is a Microsoft Excel workbook with raw data for the mouse gene chip compendium. Additional data file 2 is a Microsoft Excel workbook with raw data for the human gene chip compendium. Additional data file 3 is a Microsoft Excel workbook with raw data for the human/mouse gene chip compendium. Additional data file 4 is a Microsoft Excel workbook with raw data for the IKDC gene chip compendium. Additional data file 5 is a Microsoft Excel workbook giving the mouse DC subset gene signatures according to our datasets with confirmation from two other independent datasets (one for pDCs and one for cDC subsets). Additional data file 6 is a figure showing the results of PCA for investigation of the relationships between in vitro derived GM-CSF DCs and LN-DCs in mouse and human. Additional data file 7 is a table giving real-time PCR data for the pattern of expression of 27 genes across mouse leukocyte subsets. Additional data file 8 is a figure illustrating PACSIN1 expression in human pDCs versus PBMCs by RT-PCR and western blotting.
Supplementary Material
Additional file 1
Raw data for the mouse gene chip compendium.
Additional file 2
Raw data for the human gene chip compendium.
Additional file 3
Raw data for the human/mouse gene chip compendium.
Additional file 4
Raw data for the IKDC gene chip compendium.
Additional file 5
Mouse DC subset gene signatures according to our datasets with confirmation from two other independent datasets (one for pDCs and one for cDC subsets).
Additional file 6
Results of PCA for investigation of the relationships between in vitro derived GM-CSF DCs and LN-DCs in mouse and human.
Additional file 7
Real-time PCR data for the pattern of expression of 27 genes across mouse leukocyte subsets.
Additional file 8
PACSIN1 expression in human pDCs versus PBMCs by RT-PCR and western blotting.
Contributor Information
Scott H Robbins, Email: SRobbins@gnf.org.
Thierry Walzer, Email: walzer@ciml.univ-mrs.fr.
Doulaye Dembélé, Email: doulaye@igbmc.u-strasbg.fr.
Christelle Thibault, Email: thibault@igbmc.u-strasbg.fr.
Axel Defays, Email: defays@ciml.univ-mrs.fr.
Gilles Bessou, Email: bessou@ciml.univ-mrs.fr.
Huichun Xu, Email: huixu@ucdavis.edu.
Eric Vivier, Email: vivier@ciml.univ-mrs.fr.
MacLean Sellars, Email: msellars@igbmc.u-strasbg.fr.
Philippe Pierre, Email: pierre@ciml.univ-mrs.fr.
Franck R Sharp, Email: frank.sharp@ucdmc.ucdavis.edu.
Susan Chan, Email: chan@igbmc.u-strasbg.fr.
Philippe Kastner, Email: kastner@igbmc.u-strasbg.fr.
Marc Dalod, Email: dalod@ciml.univ-mrs.fr.
Acknowledgements
The authors are indebted to Bertrand Nadel and Jean-Marc Navarro for help with the real-time PCR experiments and to Markus Plomann for the generous gift of the anti-PACSIN1 antibody. The authors also thank the staff of the animal care facilities and the flow cytometry core facility of the CIML for excellent assistance. This work was supported by an ATIP grant from the CNRS, a grant from the Association pour la Recherche sur le Cancer (ARC) and a grant from the Réseau National des Génopoles (RNG) to MD. SHR was supported by the CNRS, the Fondation pour la Recherche Médicale, and the Philippe Foundation. The CIML is supported by institutional grants from the INSERM, the CNRS, and the Université de la Méditerranée. We thank the IPSOGEN company for their advice on the analysis of the data. The authors declare no conflict of interest.
References
- Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/S0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-Producing Dendritic Cells Mediate Innate Immune Defense against Bacterial Infection. Immunity. 2003;19:59–70. doi: 10.1016/S1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: "l'union fait la force". Blood. 2005;106:2252–2258. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nature Rev Immunol. 2002;2:151–161. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- Narbutt J, Lesiak A, Zak-Prelich M, Wozniacka A, Sysa-Jedrzejowska A, Tybura M, Robak T, Smolewski P. The distribution of peripheral blood dendritic cells assayed by a new panel of anti-BDCA monoclonal antibodies in healthy representatives of the Polish population. Cell Mol Biol Lett. 2004;9:497–509. [PubMed] [Google Scholar]
- Heath WR, Belz GT, Behrens GM, Smith CM, Forehan SP, Parish IA, Davey GM, Wilson NS, Carbone FR, Villadangos JA. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol Rev. 2004;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- Kronin V, Fitzmaurice CJ, Caminschi I, Shortman K, Jackson DC, Brown LE. Differential effect of CD8(+) and CD8(-) dendritic cells in the stimulation of secondary CD4(+) T cells. Int Immunol. 2001;13:465–473. doi: 10.1093/intimm/13.4.465. [DOI] [PubMed] [Google Scholar]
- Pooley JL, Heath WR, Shortman K. Cutting edge: intravenous soluble antigen is presented to CD4 T cells by CD8- dendritic cells, but cross-presented to CD8 T cells by CD8+ dendritic cells. J Immunol. 2001;166:5327–5330. doi: 10.4049/jimmunol.166.9.5327. [DOI] [PubMed] [Google Scholar]
- Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, Steinman RM, Nussenzweig MC. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, Traver D, Leder P, Sakaguchi N, Akashi K. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Ito T, Kanzler H, Duramad O, Cao W, Liu YJ. Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood. 2006;107:2423–2431. doi: 10.1182/blood-2005-07-2709. [DOI] [PubMed] [Google Scholar]
- Dalod M, Salazar-Mather TP, Malmgaard L, Lewis C, Asselin-Paturel C, Briere F, Trinchieri G, Biron CA. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J Exp Med. 2002;195:517–528. doi: 10.1084/jem.20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CW, Crafton E, Fan HN, Flook J, Yoshimura K, Skarica M, Brockstedt D, Dubensky TW, Stins MF, Lanier LL, Pardoll DM, Housseau F. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat Med. 2006;12:207–213. doi: 10.1038/nm1352. [DOI] [PubMed] [Google Scholar]
- Taieb J, Chaput N, Menard C, Apetoh L, Ullrich E, Bonmort M, Pequignot M, Casares N, Terme M, Flament C, Opolon P, Lecluse Y, Metivier D, Tomasello E, Vivier E, Ghiringhelli F, Martin F, Klatzmann D, Poynard T, Tursz T, Raposo G, Yagita H, Ryffel B, Kroemer G, Zitvogel L. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12:214–219. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, Goldrath A, Nguyen L, Luckey J, Yamagata T, Herman A, Jacobs J, Mathis D, Benoist C. Gene expression microarrays: glimpses of the immunological genome. Nat Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- Benoist C, Germain RN, Mathis D. A plaidoyer for 'systems immunology'. Immunol Rev. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, Staudt LM. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/S1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- Hutton JJ, Jegga AG, Kong S, Gupta A, Ebert C, Williams S, Katz JD, Aronow BJ. Microarray and comparative genomics-based identification of genes and gene regulatory regions of the mouse immune system. BMC Genomics. 2004;5:82. doi: 10.1186/1471-2164-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/S1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichling T, Goss KH, Carson DJ, Holdcraft RW, Ley-Ebert C, Witte D, Aronow BJ, Groden J. Transcriptional profiles of intestinal tumors in Apc(Min) mice are unique from those of embryonic intestine and identify novel gene targets dysregulated in human colorectal tumors. Cancer Res. 2005;65:166–176. [PubMed] [Google Scholar]
- Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, Kong S, Sakthivel B, Xu H, Reichling T, Azhar M, Boivin GP, Roberts RB, Bissahoyo A, Gonzales F, Bloom GL, Eschrich S, Carter SL, Aronow JE, Kleimeyer J, Kleimeyer M, Ramaswamy V, Settle SH, Boone B, Levy S, Graff JM. et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber TG, Sawyers CL. Cross-species comparisons of cancer signaling. Nat Genet. 2005;37:7–8. doi: 10.1038/ng0105-7. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Chelvarajan RL, Liu Y, Popa D, Getchell ML, Getchell TV, Stromberg AJ, Bondada S. Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages. J Leukoc Biol. 2006;79:1314–1327. doi: 10.1189/jlb.0106024. [DOI] [PubMed] [Google Scholar]
- Du X, Tang Y, Xu H, Lit L, Walker W, Ashwood P, Gregg JP, Sharp FR. Genomic profiles for human peripheral blood T cells, B cells, natural killer cells, monocytes, and polymorphonuclear cells: comparisons to ischemic stroke, migraine, and Tourette syndrome. Genomics. 2006;87:693–703. doi: 10.1016/j.ygeno.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005;175:4839–4846. doi: 10.4049/jimmunol.175.8.4839. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembélé D, Kastner P. Fuzzy C-means method for clustering microarray data. Bioinformatics. 2003;19:973–980. doi: 10.1093/bioinformatics/btg119. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Eberhard D, Horcher M, Rolink AG, Busslinger M. Pax5 determines the identity of B cells from the beginning to the end of B-lymphopoiesis. Int Rev Immunol. 2001;20:65–82. doi: 10.3109/08830180109056723. [DOI] [PubMed] [Google Scholar]
- Malissen B, Aguado E, Malissen M. Role of the LAT adaptor in T-cell development and Th2 differentiation. Adv Immunol. 2005;87:1–25. doi: 10.1016/S0065-2776(05)87001-4. [DOI] [PubMed] [Google Scholar]
- Walzer T, Jaeger S, Chaix J, Vivier E. Natural killer cells: from CD3(-)NKp46(+) to post-genomics meta-analyses. Curr Opin Immunol. 2007;19:365–372. doi: 10.1016/j.coi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- Matsuki Y, Ohmura-Hoshino M, Goto E, Aoki M, Mito-Yoshida M, Uematsu M, Hasegawa T, Koseki H, Ohara O, Nakayama M, Toyooka K, Matsuoka K, Hotta H, Yamamoto A, Ishido S. Novel regulation of MHC class II function in B cells. Embo J. 2007;26:846–854. doi: 10.1038/sj.emboj.7601556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, Alcais A, Picard C, Mahfoufi N, Nicolas N, Lorenzo L, Plancoulaine S, Senechal B, Geissmann F, Tabeta K, Hoebe K, Du X, Miller RL, Heron B, Mignot C, de Villemeur TB, Lebon P, Dulac O, Rozenberg F, Beutler B, Tardieu M, Abel L, Casanova JL. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, Jacques Y, Baratin M, Tomasello E, Vivier E. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol. 2007;8:1337–1344. doi: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SD, Peschon JJ. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Tussiwand R, Lanzavecchia A, Manz MG. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J Exp Med. 2006;203:227–238. doi: 10.1084/jem.20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Onai N, Mazzucchelli L, Manz MG. Inhibition of natural type I IFN-producing and dendritic cell development by a small molecule receptor tyrosine kinase inhibitor with Flt3 affinity. J Immunol. 2005;175:3674–3680. doi: 10.4049/jimmunol.175.6.3674. [DOI] [PubMed] [Google Scholar]
- Maraskovsky E, Daro E, Roux E, Teepe M, Maliszewski CR, Hoek J, Caron D, Lebsack ME, McKenna HJ. In vivo generation of human dendritic cell subsets by Flt3 ligand. Blood. 2000;96:878–884. [PubMed] [Google Scholar]
- Bonasio R, von Andrian UH. Generation, migration and function of circulating dendritic cells. Curr Opin Immunol. 2006;18:503–511. doi: 10.1016/j.coi.2006.05.011. [DOI] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Waldburger JM, Reis e Sousa C, Acha-Orbea H, Reith W. MHC class II expression is differentially regulated in plasmacytoid and conventional dendritic cells. Nat Immunol. 2004;5:899–908. doi: 10.1038/ni1109. [DOI] [PubMed] [Google Scholar]
- Chtanova T, Newton R, Liu SM, Weininger L, Young TR, Silva DG, Bertoni F, Rinaldi A, Chappaz S, Sallusto F, Rolph MS, Mackay CR. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol. 2005;175:7837–7847. doi: 10.4049/jimmunol.175.12.7837. [DOI] [PubMed] [Google Scholar]
- Krücken J, Schroetel RM, Müller IU, Saïdani N, Marinovski P, Benten WP, Stamm O, Wunderlich F. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene. 2004;341:291–304. doi: 10.1016/j.gene.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Nitta T, Nasreen M, Seike T, Goji A, Ohigashi I, Miyazaki T, Ohta T, Kanno M, Takahama Y. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 2006;4:e103. doi: 10.1371/journal.pbio.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AD, Diebold SS, Slack EM, Tomizawa H, Hemmi H, Kaisho T, Akira S, Reis e Sousa C. Toll-like receptor expression in murine DC subsets: lack of TLR7 expression by CD8 alpha+ DC correlates with unresponsiveness to imidazoquinolines. Eur J Immunol. 2003;33:827–833. doi: 10.1002/eji.200323797. [DOI] [PubMed] [Google Scholar]
- Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint Vis B, Briere F, Bates EE. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- Edwards AD, Chaussabel D, Tomlinson S, Schulz O, Sher A, Reis e Sousa C. Relationships among murine CD11c(high) dendritic cell subsets as revealed by baseline gene expression patterns. J Immunol. 2003;171:47–60. doi: 10.4049/jimmunol.171.1.47. [DOI] [PubMed] [Google Scholar]
- Galibert L, Diemer GS, Liu Z, Johnson RS, Smith JL, Walzer T, Comeau MR, Rauch CT, Wolfson MF, Sorensen RA, Van der Vuurst de Vries AR, Branstetter DG, Koelling RM, Scholler J, Fanslow WC, Baum PR, Derry JM, Yan W. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J Biol Chem. 2005;280:21955–21964. doi: 10.1074/jbc.M502095200. [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Ott G, Chan JH, Damon E, Calacsan C, Matray T, Lee KD, Coffman RL, Barrat FJ. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J Exp Med. 2006;203:1999–2008. doi: 10.1084/jem.20060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Schulz O, Diebold SS, Chen M, Naslund TI, Nolte MA, Alexopoulou L, Azuma YT, Flavell RA, Liljestrom P, Reis e Sousa C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- Orabona C, Puccetti P, Vacca C, Bicciato S, Luchini A, Fallarino F, Bianchi R, Velardi E, Perruccio K, Velardi A, Bronte V, Fioretti MC, Grohmann U. Toward the identification of a tolerogenic signature in IDO-competent dendritic cells. Blood. 2006;107:2846–2854. doi: 10.1182/blood-2005-10-4077. [DOI] [PubMed] [Google Scholar]
- Spits H, Lanier LL. Natural Killer or Dendritic: What's in a Name? Immunity. 2007;26:11–16. doi: 10.1016/j.immuni.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Rolink A, ten Boekel E, Melchers F, Fearon DT, Krop I, Andersson J. A subpopulation of B220+ cells in murine bone marrow does not express CD19 and contains natural killer cell progenitors. J Exp Med. 1996;183:187–194. doi: 10.1084/jem.183.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath AT, Sheasby CE, Tough DF. Dendritic cells and NK cells stimulate bystander T cell activation in response to TLR agonists through secretion of IFN-alpha beta and IFN-gamma. J Immunol. 2005;174:767–776. doi: 10.4049/jimmunol.174.2.767. [DOI] [PubMed] [Google Scholar]
- Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- BD/Pharmingen technical datasheet of the CD11c antibody. http://www.bdbiosciences.com/external_files/pm/doc/tds/mouse/live/web_enabled/09709A_550261.pdf http://www.bdbiosciences.com/external_files/pm/doc/tds/mouse/live/web_enabled/09709A_550261.pdf
- Veinotte LL, Greenwood CP, Mohammadi N, Parachoniak CA, Takei F. Expression of rearranged TCRgamma genes in natural killer cells suggests a minor thymus-dependent pathway of lineage commitment. Blood. 2006;107:2673–2679. doi: 10.1182/blood-2005-07-2797. [DOI] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Bakri Y, Sarrazin S, Mayer UP, Tillmanns S, Nerlov C, Boned A, Sieweke MH. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood. 2005;105:2707–2716. doi: 10.1182/blood-2004-04-1448. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential Development of Murine Dendritic Cells by GM-CSF versus Flt3 Ligand Has Implications for Inflammation and Trafficking. J Immunol. 2007;179:7577–7584. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, Lahoud MH, O'Keeffe M, Shao QX, Chen WF, Villadangos JA, Shortman K, Wu L. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174:6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- Lahoud MH, Proietto AI, Gartlan KH, Kitsoulis S, Curtis J, Wettenhall J, Sofi M, Daunt C, O'Keeffe M, Caminschi I, Satterley K, Rizzitelli A, Schnorrer P, Hinohara A, Yamaguchi Y, Wu L, Smyth G, Handman E, Shortman K, Wright MD. Signal regulatory protein molecules are differentially expressed by CD8- dendritic cells. J Immunol. 2006;177:372–382. doi: 10.4049/jimmunol.177.1.372. [DOI] [PubMed] [Google Scholar]
- Zenke M, Hieronymus T. Towards an understanding of the transcription factor network of dendritic cell development. Trends Immunol. 2006;27:140–145. doi: 10.1016/j.it.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, Kalchenko V, Geissmann F, Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C, Sauer M, Ghosh A, Rudolph C, Hegazy A, Schlegelberger B, Welte K, Klein C. Generation and characterization of a novel hematopoietic progenitor cell line with DC differentiation potential. Leukemia. 2006;20:870–876. doi: 10.1038/sj.leu.2404157. [DOI] [PubMed] [Google Scholar]
- Cao W, Rosen DB, Ito T, Bover L, Bao M, Watanabe G, Yao Z, Zhang L, Lanier LL, Liu YJ. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits Toll-like receptor-induced interferon production. J Exp Med. 2006;203:1399–1405. doi: 10.1084/jem.20052454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Cella M, Maldonado J, Takai T, Colonna M. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 2006;107:2474–2476. doi: 10.1182/blood-2005-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVID Bioinformatics Resources National Institute of Allergy and Infectious Diseases (NIAID), NIH. http://david.abcc.ncifcrf.gov/home.jsp http://david.abcc.ncifcrf.gov/home.jsp
- Microarray platform of the IGBMC and Génopole Alsace-Lorraine. http://www-microarrays.u-strasbg.fr/files/datasetsE.php http://www-microarrays.u-strasbg.fr/files/datasetsE.php
- Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O'Keeffe M, Bahlo M, Papenfuss A, Kwak JY, Wu L, Shortman K. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Merad M, Ginhoux F. Dendritic cell genealogy: a new stem or just another branch? Nat Immunol. 2007;8:1199–1201. doi: 10.1038/ni1107-1199. [DOI] [PubMed] [Google Scholar]
- Blasius AL, Barchet W, Cella M, Colonna M. Development and function of murine B220+CD11c+NK1.1+ cells identify them as a subset of NK cells. J Exp Med. 2007;204:2561–2568. doi: 10.1084/jem.20070991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I, Ahmet F, Heger K, Brady J, Nutt SL, Vremec D, Pietersz S, Lahoud MH, Schofield L, Hansen DS, O'Keeffe M, Smyth MJ, Bedoui S, Davey GM, Villadangos JA, Heath WR, Shortman K. Putative IKDCs are functionally and developmentally similar to natural killer cells, but not to dendritic cells. J Exp Med. 2007;204:2579–2590. doi: 10.1084/jem.20071351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshenrich CA, Lesjean-Pottier S, Hasan M, Richard-Le Goff O, Corcuff E, Mandelboim O, Di Santo JP. CD11cloB220+ interferon-producing killer dendritic cells are activated natural killer cells. J Exp Med. 2007;204:2569–2578. doi: 10.1084/jem.20071451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röck J, Schneider E, Grün JR, Grützkau A, Küppers R, Schmitz J, Winkels G. CD303 (BDCA-2) signals in plasmacytoid dendritic cells via a BCR-like signalosome involving Syk, Slp65 and PLCgamma2. Eur J Immunol. 2007;37:3564–3575. doi: 10.1002/eji.200737711. [DOI] [PubMed] [Google Scholar]
- Cao W, Zhang L, Rosen DB, Bover L, Watanabe G, Bao M, Lanier LL, Liu YJ. BDCA2/Fc epsilon RI gamma complex signals through a novel BCR-like pathway in human plasmacytoid dendritic cells. PLoS Biol. 2007;5:e248. doi: 10.1371/journal.pbio.0050248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Expression Omnibus (GEO) Main page. http://www.ncbi.nlm.nih.gov/projects/geo/ http://www.ncbi.nlm.nih.gov/projects/geo/
- National Cancer Institute caArray. https://caarraydb.nci.nih.gov/caarray/publicSearch.do;jsessionid=8933FE3445E9FA5CFDEE9D8EBE52CD30?mode=experiments https://caarraydb.nci.nih.gov/caarray/publicSearch.do;jsessionid=8933FE3445E9FA5CFDEE9D8EBE52CD30?mode=experiments
- ArrayExpress. http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0] http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1
Raw data for the mouse gene chip compendium.
Additional file 2
Raw data for the human gene chip compendium.
Additional file 3
Raw data for the human/mouse gene chip compendium.
Additional file 4
Raw data for the IKDC gene chip compendium.
Additional file 5
Mouse DC subset gene signatures according to our datasets with confirmation from two other independent datasets (one for pDCs and one for cDC subsets).
Additional file 6
Results of PCA for investigation of the relationships between in vitro derived GM-CSF DCs and LN-DCs in mouse and human.
Additional file 7
Real-time PCR data for the pattern of expression of 27 genes across mouse leukocyte subsets.
Additional file 8
PACSIN1 expression in human pDCs versus PBMCs by RT-PCR and western blotting.