The Air Noncoding RNA: An Imprinted cis-silencing Transcript (original) (raw)
. Author manuscript; available in PMC: 2010 Mar 30.
Published in final edited form as: Cold Spring Harb Symp Quant Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55
Genomic imprinting is briefly defined as parental-specific gene expression. It is a feature of organisms with diploid cells that have inherited a complete haploid chromosome set from two parents and, thereby, have a maternal and paternal allele for each autosomal gene. Diploid cells would normally be expected to express both parental alleles equally but imprinted genes are different, because they acquire an epigenetic mark from one parental gamete that results in repression on one parental allele and expression from the other. Genomic imprinting uses an epigenetic mechanism (i.e., a reversible modification to DNA or chromatin) to regulate gene expression in cis that is assumed to be at the level of transcriptional, but this is not often tested.
Genomic imprinting has been so far identified in very diverse organisms: in angiosperm plants, in two types of invertebrates (sciarid flies, mealybugs), and lastly in mammals that have live-born young (marsupial and placental mammals) (Goday and Esteban 2001; Killian et al. 2001; Baroux et al. 2002). These diverse organisms lack a contemporary common ancestor that also shows imprinted gene expression, and this allows the possibility that imprinting arose independently at least three times in distantly related organisms. If this is so, the imprinting mechanism in these diverse organisms is less likely to share common features. The function of genomic imprinting is beginning to be understood following the functional analysis of a large number of imprinted genes in mice (see listing at http://www.mgu.har.mrc.ac.uk/research/imprinting/function.html). There is now sufficient information to suggest that genomic imprinting arose in mammals as a consequence of one parent carrying the total burden of provision for an embryo that carries the genomes of two parents. Thus, imprinted genes in mammals are able to influence embryonic growth in such a manner that imprinted growth suppressor genes are always maternally expressed, while imprinted growth promoter genes are always paternally expressed (Wilkins and Haig 2003). The reason imprinted genes exist in angiosperm plants is not yet clear, although it has been suggested that angiosperm plants resemble mammals because biparental seeds are also grown by one parent (Moore and Haig 1991). The two invertebrates with genomic imprinting appear to use this as a sex-determining mechanism, such that males are produced by inactivating the paternal genome in diploid offspring (Goday and Esteban 2001; Bongiorni and Prantera 2003).
The Harwell Mammalian Genetics Unit has for many years maintained a complete list of the known imprinted mouse genes and also lists mutant phenotypes for those imprinted genes so far analyzed (Beechey et al. 2003). Table 1 was drawn from this information and lists the 70 known imprinted genes by chromosomal subregions (note many imprinted genes have alternative names and these are listed on http://www.mgu.har.mrc.ac.uk/research/imprinting/). The imprinted genes are separated in this list according to parental expression and whether they are coding or noncoding. Several features of mammalian imprinted genes become apparent from this list.
Table 1.
Imprinted Genes Are Mainly Organized into Clusters
| Coding (mRNAs) | Noncoding (ncRNAs) as:antisense | ||||
|---|---|---|---|---|---|
| Chr. | Region | Maternal expression | Paternal expression | Maternal expression | Paternal expression |
| 2 | central2 | Gatm | |||
| distal2 | Nesp, Gnas | Nnat, Gnasxl | Nespas | ||
| 6 | proximal6 | Dlx5, Calcr, Neurabin, Pon3, Pon2, Asb4 | Sgce, Peg10 | ||
| subproximal6 | Asb4, Cop2, | Peg1/Mest, Nap1I5 | Cop2as, Mit1 | ||
| 7 | proximal7 | Zim1, Zim3, | Peg3/Pw1, Usp29, Zfp264, | ||
| central7 | Atp10c, Ube3a, Nap1l4 | Snrpn, Snurf, Magel2, Ndn, Zfp127/Mkrn3, Frat3 | Zfp127/Mkrn3as, IC- Snurf-Snrpn pro- cessed, ncRNA, Ipw, Pwcr1, multiple MB11-snoRNAs, Ube3aas | ||
| distal7a | Obph1, Tssc3, Slc22a1L, Msult, Cdkn1c, Kcnq1, Tssc4, Tapa1, Mash2 | Lit1/Kcnq1ot1(as) | |||
| distal7b | Ins2, Igf2 | H19 | Igf2as | ||
| 9 | distal9 | A19, Rasgrf1 | |||
| 10 | proximal10 | Zac1 | |||
| distal10 | Dcn1 | ||||
| 11 | proximal11 | Grb10/Meg1, Murr1 | U2af1rs1 | ||
| 12 | distal12 | Dlk1, Rtl1, Dio3 | Gtl2, Mir126,136 Rain, C/D SnoRNA cluster | ||
| 14 | distal14 | Htr3a | |||
| 15 | distal15 | Peg13, Slc38a4 | |||
| 17 | proximal17 | Slc22a3, Slc22a2, Igf2r | Air (Igf2ras) | ||
| 18 | proximal18 | Impact | |||
| 19 | Ins1 |
- Only 12 of 19 mouse autosomal chromosomes carry imprinted genes; however, four chromosomes (2, 6, 7, and 10) have multiple subregions carrying imprinted genes.
- Imprinted genes are often clustered. There are 18 regions with imprinted genes and 12 have clusters containing between 2 and 9 imprinted genes.
- Some genes inside the cluster escape imprinting.
- Many imprinted genes are noncoding RNAs (ncRNAs) that show three types of pattern: antisense to one imprinted gene and expressed from the opposite parental allele (e.g., proximal 17: Igf2r/Air; distal 7a: Kcnq1/Lit1); antisense to one imprinted gene and expressed from the same chromosome (distal 7b: Igf2/Igf2as; central 7: Zfp127/Zfp127as); and sense to one imprinted gene and expressed from the opposite parental allele (distal 7b: Igf2/H19; distal 12: Dlk1/Gtl2).
- There are approximately equal numbers of maternally and paternally expressed coding (mRNA) genes, but there are less maternally expressed ncRNAs than paternally expressed ncRNAs (note that the multiple ncRNAs from central 7 and distal 12 may arise from a single long processed ncRNA).
- Reciprocal parental-specific expression between protein-coding genes and an ncRNA is often observed. Two patterns are seen: A simple type where all the protein-coding genes are expressed by one parental chromosome and the ncRNA from the other (e.g., distal 7a, distal 7b, distal 12, proximal 17), and a complex type where multiple protein coding genes are maternally or paternally expressed while the noncoding RNA is expressed from one parental chromosome (e.g., distal 2, subproximal 6, central 7).
The observation that imprinted genes exist in clusters suggests that mechanisms to control imprinted expression act on the chromosomal domain rather than on individual genes (Reik and Walter 2001; Verona et al. 2003). If imprinting control mechanisms are specific to a domain this could explain why moving imprinted genes outside their endogenous locus on transgenes causes them to lose imprinted expression (Lee et al. 1993; Sleutels and Barlow 2001). The observation of reciprocal imprinted expression between the clustered protein-coding genes and the noncoding RNA is fascinating and naturally suggests a link between expression of a noncoding RNA and _cis_-induced silencing of the protein coding RNA. The data that tests the significance of this association between ncRNAs and _cis_-induced silencing is only beginning to appear. There are, however, two significant results so far (for review, see Verona et al. 2003). First, there was a clear demonstration that the H19 ncRNA plays no role in silencing the reciprocally expressed Ins2/Igf2 genes in cis (Jones et al. 1998; Schmidt et al. 1999). Instead a DNA sequence functions as a methyl-sensitive insulator that, when methylated, allows expression of Igf2 and, when unmethlyated, allows expression of H19 (Bell and Felsenfeld 2000; Hark et al. 2000). The second result, and the topic of the remainder of this review, was the demonstration that the Air ncRNA plays a direct role in silencing a cluster of three genes spanning 300 kilobases (kb) in cis (Rougeulle and Heard 2002; Sleutels et al. 2002).
THE MOUSE CHROMOSOME 17 PROXIMAL IMPRINTED CLUSTER
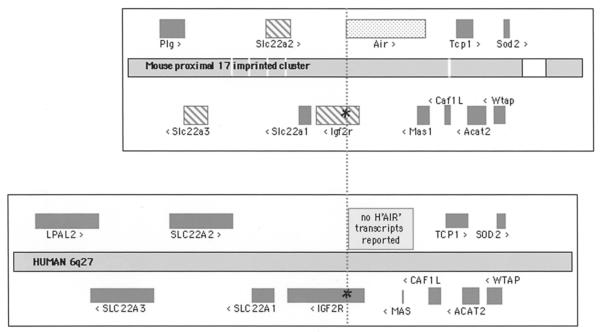
The Air noncoding RNA (ncRNA) is part of an imprinted cluster on proximal mouse chromosome 17/band A2 (http://www.mgu.har.mrc.ac.uk/research/imprinting/). Figure 1 shows a map spanning 500 kb based on the public mouse and human sequence. Genes shown on each side of the sequence line have the same orientation, and it can be seen in the mouse map that the Air ncRNA has an antisense orientation with respect to Igf2r and Mas. The Air ncRNA promoter (marked by asterisks in Fig. 1) lies in the second intron of Igf2r and the transcript overlaps the 5′ part of the Igf2r gene for 29 kb (which includes Igf2r exons 1 and 2) and the 3′ part of the Mas gene (which includes the entire 1-kb coding exon of Mas). Gene overlaps are not rare in the mammalian genome and within this 500-kb region the Acat2 gene overlaps the 3′ coding part of the Tcp1 gene (Shintani et al. 1999). There are three maternally expressed genes indicated by striped boxes: Igf2r (a multifunctional transport receptor with embryonic growth suppressor function [Wang et al. 1994; Ludwig et al. 1996]); Slc22a2 (solute cation transporter family 22a, an organic cation transporter [Jonker et al. 2003]); and Slc22a3 (an organic cation transporter that may function as an extraneuronal monoamine transporter [Zwart et al. 2001b]). Only one paternally expressed gene, the Air ncRNA, indicated in Figure 1 by a dotted box, has been identified (Lyle et al. 2000). The remaining genes show biallelic expression in all tested tissues in embryos and adults.
Figure 1.
The mouse proximal 17 imprinted cluster (top box) and the syntenic human region on 6q27 (bottom box). Genes are positioned above or below the map according to the direction of transcription and their size indicated by the length of the box. Both regions are gene rich and show surprising conservation (n.b., LPAL2 is a human-specific duplication of Plg); the white boxes in the mouse sequence indicate gaps. On the mouse map, the three maternally expressed imprinted genes are shown as striped boxes; the paternally expressed Air ncRNA is shown as a dotted box. The two regions are aligned (vertical dotted line) at the CpG island (*) in Igf2r intron 2 that is common to both mouse and human sequences, which in the mouse is the promoter for the Air ncRNA. The mouse Air ncRNA is 108-kb long and spans from Igf2r intron 2 to the Mas last intron. No human “H”AIR transcripts have been reported in the region between human IGF2R and MAS. Both maps were based on those shown in http://www.ensemble.org/.
The imprinted behavior of genes in this region is fully maintained in transgenes that span 300 kb (from Mas to the intergenic region between Slc22a2 and Slc22a3) when they are integrated into autosomes (Wutz et al. 1997). “Short” transgenes that span 44 kb from 4 kb upstream of the Igf2r promoter to 8 kb downstream of the Air promoter are not reliably imprinted (Sleutels and Barlow 2001). This indicates that all elements and epigenetic marks for the imprinting mechanism lie within a 300-kb region. This rationale has formed the basis for generating epigenetic maps with the aim of identifying all parental-specific epigenetic modifications on the DNA and chromatin, which could contribute to the control of imprinting in this region. An enzyme-based DNA methylation scan that examined a 100-kb region spanning the Igf2r locus, only identified two parental-specific _d_ifferentially _m_ethylated _r_egions (DMR1 and DMR2), the remainder of the analyzed region showed various levels of high and low methylation that were the same on both parental alleles (Stoeger et al. 1993). Both DMRs were large CpG island promoters. DMR1 contained the Igf2r promoter and was specifically methylated on the silent paternal allele, but only after embryonic implantation. DMR2 contained the Air promoter and was specifically methylated on the silent maternal allele from late oocyte stages onward into adulthood (Stoeger et al. 1993). Methylation of DMR2 was also specifically maintained in preimplantation embryos during the process of genome-wide demethylation (Brandeis et al. 1993). While the identification of a DNA methylation imprint on the active maternal Igf2r allele was initially a surprise, it is now clear that the DMR2 methylation mark is the activator of maternal Igf2r expression and also serves as the primary imprint regulator of this cluster. This can be seen from analysis of mouse embryos deficient in genome-wide DNA methylation (through targeted inactivation of the DNA methylation enzyme system) that repress Igf2r on both parental chromosomes (Li 2002). Thus, DNA methylation acts to silence a _cis_-acting repressor of Igf2r but does not act as a direct silencer of Igf2r. Analysis of these methylation-deficient mice showed that the DMR1 methylation mark plays no role in initiating silencing of the paternal Igf2r promoter and, since this methylation mark occurs after embryonic implantation, it may be passively acquired after the promoter is silenced and may fulfill a maintenance role.
In addition to the surprise finding that the maternal Igf2r allele was imprinted by DNA methylation to be active, the lack of widespread DNA methylation on the silent paternal Igf2r allele was unexpected. Textbook models of the mammalian genome often view chromatin as organized into blocks of “euchromatin” containing active genes and blocks of “heterochromatin” containing silent genes. The Igf2r locus spans 86 kb and, although the Air ncRNA overlaps the silent paternal copy at the 5′ end, we had initially anticipated that parts of the Igf2r gene locus downstream of the Air promoter would show euchromatic features on the maternal allele and heterochromatic features on the paternal allele. However, in terms of DNA methylation, no other parental specific methylation marks, not even at the silent Slc22a2/Slc22a3 promoters, were found at the resolution so far used (Stoeger et al. 1993; Zwart et al. 2001a). Histone modifications have been analyzed at specific regions within DMR1 and DMR2 and in the last Igf2r exon (Fournier et al. 2002; Yang et al. 2003). Chromatin immunoprecipitation experiments have shown that histone H3/H4 acetylation and histone H3 K4 methylation specifically distinguish the unmethylated DMRs that contain an active promoter (i.e., the maternal DMR1 and the paternal DMR2) from the methylated DMRs with a silent promoter (i.e., the paternal DMR1 and the maternal DMR2). The methylated DMRs were also marked by histone H3 K9 methylation. However, more detailed analysis of the whole cluster shown in Figure 1 is needed to test if the paternal allele that carries three silenced genes shows more widespread heterochromatin compared to the maternal allele that carries only one silenced gene.
A final chromatin feature associated with this locus is a widespread replication asynchrony that has been reported to cover the 700-kb mouse region shown in Figure 1, and extend approximately 1 Mbp (Mega base pair) upstream and downstream (Kitsberg et al. 1993). Once again this observed replication asynchrony does not follow textbook predictions; instead the early replicating allele is the paternal allele that is silent for Igf2r/Slc22a2/Slc22a3 but active for the Air ncRNA. The early replicating paternal allele also lacks the critical DNA methylation imprint on DMR2; however, replication asynchrony has been shown to be independent of DNA methylation (Gribnau et al. 2003). Our work producing four different targeted alleles at DMR1 and DMR2 has highlighted another phenomenon that could also be related to chromatin structure, which is the preferential targeting in ES cells of the paternal allele (Sleutels et al. 2002, 2003; Wang et al. 1994; P. Latos, unpubl.).
THE _cis_-ACTING SILENCER IS THE Air NONCODING RNA
The analysis of epigenetic marks described above is limited but it already indicates that typical heterochromatic modifications, such as DNA methylation, histone deacetylation, or histone methylation, are not the primary cause of silencing Igf2r, Slc22a2, and Slc22a3 on the paternal allele. Experiments using transgenes and targeted deletion of the endogenous locus, have, however, identified DMR2 as the imprint control element (ICE) for this cluster (Wutz et al. 1997, 2001; Spahn and Barlow 2003). Since DMR2 contained the Air promoter, the next logical step was to test for Air ncRNA involvement in silencing. An experiment was performed that inserted a PolyA signal 3 kb downstream of the Air transcription start (Sleutels et al. 2002). This truncated Air from 108 to 3 kb (in the truncated allele 80% of the transcripts were spliced from a donor at +53 bp to an acceptor in the PolyA signal cassette and only 20% were unspliced and shortened to 3 kb). Expression of the Air promoter from the truncated allele was equal and imprinted exactly as in the wild-type allele. Thus, the truncated allele was predicted to differ from the wild type only in the length of the Air ncRNA. Analysis of _Igf2r_, Slc22a2, and Slc22a3 expression from the paternal chromosome carrying the truncated allele revealed a complete loss of repression of all three genes. This experiment confirms a direct role for the Air ncRNA transcript or its transcription, in the _cis_-silencing of three genes spread over a 300-kb region. A direct role for Air in silencing Igf2r was not unexpected in view of the sense/antisense overlap between the two genes. However, it was unexpected that Air would also silence the Slc22a2 and Slc22a3 genes. In view of this result, it was important to investigate if silencing of Slc22a2 and Slc22a3 arose by a two-step mechanism that initially silenced Igf2r via the sense/antisense overlap with Air and then established a silent chromatin state that spread to the flanking Slc22a2 and Slc22a3 genes. To test the role of the sense/antisense overlap between Igf2r and Air, a mouse was constructed that lacked the Igf2r promoter. In this mouse the Air ncRNA was not overlapped by any known transcript; despite this, the Slc22a2 and Slc22a3 genes, as well as the Air ncRNA itself, maintained their imprinted status (Sleutels et al. 2003). This experiment makes it less likely that a double-stranded RNA-based mechanism, such as RNAi, is involved in imprinting this cluster.
Thus gene-targeting experiments have shown that the Air ncRNA acts as a _cis_-acting silencer of a 300-kb domain of genes. There are still many interesting questions that need to be answered. It is, for example, fascinating why the Air ncRNA does not silence its own promoter yet can silence promoters that lie upstream and downstream. And it is also intriguing to consider why the effects of the Air ncRNA are limited to genes within a 300-kb region and why this region contains some genes that escape imprinting. Some clues to these questions can be obtained by noting that the imprinted expression of the four transcripts in this region shows a developmental and tissuespecific pattern. The Igf2r gene lacks imprinted expression in preimplantation embryos, undifferentiated ES cells, and also in testes, and adult and embryonic brain (Wang et al. 1994; Szabo and Mann 1995a,b; Lerchner and Barlow 1997; Hu et al. 1999; C. Seidl, unpubl.). ES cells do not express Air, and testes and brain express low levels compared to other organs (S. Stricker, unpubl.). However, all tissues that express Air ncRNA also maintain the correct imprinted paternal-specific expression. The Slc22a2 and Slc22a3 genes are not expressed in the embryo proper but both show imprinted expression in the placenta at 11.5 dpc (days postcoitum). However, at 15.5 dpc, while Slc22a2 retains imprinted expression, Slc22a3 shows biallelic expression. Adult tissues such as heart and kidney that retain imprinted expression of Igf2r and Air show biallelic expression of Slc22a2 and Slc22a3. Of the nonimprinted genes displayed in Figure 1, the neighboring genes (Slc22a1 and Mas) are not expressed in the embryo or placenta and show limited tissue-specific expression in adults. Thus, it may not be justified to call these genes “nonimprinted” because they are not expressed when imprinted expression is seen for the Slc22a2 and Slc22a3 genes. In view of the transcriptional overlap between Mas and the Air ncRNA it is of interest that Mas is only expressed in testes and brain, two tissues that lack imprinted Igf2r expression (Schweifer et al. 1997; Hu et al. 1999). Plg, Tcp1, and Sod2 are expressed in the embryo and Tcp1 and Sod2 are also expressed in placenta, but all show biallelic expression (Zwart et al. 2001a). Of the imprinted genes in this cluster, Igf2r shows the strongest imprinted expression pattern and lies 29 kb from the Air promoter, while Slc22a3 shows the weakest imprinted pattern and lies 240 kb distant (Fig. 1). This may indicate that the silencing effects of Air are reduced with distance.
IS THERE HUMAN “H”AIR?
The human IGF2R locus on 6q27 is embedded in a region with a similar organization to the mouse (Fig. 1). All tested human adult tissues appear to show a lack of imprinted IGF2R expression; however, some reports have described imprinted expression of human IGF2R restricted to 50% of the tested samples, in early embryos, in amniotic cell cultures, and in Wilms tumors (Giannoukakis et al. 1993; Kalscheuer et al. 1993; Xu et al. 1997; Oudejans et al. 2001). However, one report showed a fault with the RT reaction that results in a failure to reverse-transcribe one of the polymorphic alleles; thus, reports of imprinted expression from the human IGF2R gene must be confirmed by nonamplification techniques. A more recent report that used different polymorphisms to test the imprinting status of IGF2R in humans and lemurs clearly demonstrated that IGF2R was not imprinted and proposed that imprinted expression of IGF2R was lost in the primate lineage (Killian et al. 2001).
In view of the demonstrated lack of imprinted expression, it is surprising that the human IGF2R gene contains many sequence features and epigenetic marks associated with imprinting of the mouse Igf2r gene. For example, the human IGF2R gene contains a large CpG island in intron 2 in the same relative position to exon 3 as found for the mouse Air promoter (Smrzka et al. 1995; Riesewijk et al. 1996). The human intron 2 CpG island is also a differentially methylated region (DMR) that carries a maternal-specific methylation imprint and it is also contained in a region showing replication asynchrony. However, promoter prediction programs (http://www.gsf.de/biodv/matinspector.html) applied to the mouse and human intron 2 CpG islands predict promoter activity only for the mouse. The 108-kb mouse region spanned by Air contains more than 100 ESTs internally primed from A-rich stretches within the mature Air transcript, in contrast to the region predicted to correspond to a human “H”AIR ncRNA (http://www.ensembl.org/; S. Stricker and I. Yotova, unpubl.). This indicates the human CpG island in intron 2 does not normally act as promoter for an AIR-like transcript. However, the existence of a strong CpG island in intron 2 of the human IGF2R gene that is maintained as a DMR allows the possibility that the human gene could in some circumstances acquire imprinted maternal-specific expression by activating the putative “H”AIR promoter. It remains, therefore, to be tested if expression of a human AIR ncRNA can be found in some situations.
THE MOUSE Air ncRNA IS AN UNUSUAL RNA
The Air ncRNA was shown (Lyle et al. 2000) to span 108 kb from a promoter in Igf2r intron 2, to a PolyA tail in the last intron of Mas. The size of Air cannot be measured by standard Northern blots even in combination with Pulsed Field Gel Electrophoresis, as RNAs above 9 kb were not size-fractionated in formaldehyde gels (Lyle et al. 2000). Instead, the size of Air is assumed from mapping the 5′ and 3′ ends and the demonstration that truncating Air at 3 kb from the promoter removes all transcripts downstream from the truncation site (Sleutels et al. 2002). The major species of Air appears to be unspliced. However, the absence of splicing in the wild-type locus does not indicate that splicing is not possible in some conditions since insertion of a PolyA signal downstream from the Air promoter induced splicing at +53 bp in 80% of the truncated transcripts (Sleutels et al. 2002). In addition, mouse transgenes derived from the Air promoter consistently splice using the same +53-bp donor (Sleutels and Barlow 2001).
Mammalian genomes differ from those of other common experimental organisms (such as yeast, nematodes, fruit flies, and plants) because genes and repeats are comingled; thus, mammalian genes have extremely large introns relative to exons. As a result, unspliced precursor mRNA is rich in interspersed repeats; however, they would not represent a stable population since mRNAs are spliced as they are transcribed (Proudfoot et al. 2002). A recent bioinformatic-based survey indicated that a large number of cDNAs contain interspersed repeats in their 3′UTR (Nagashima et al. 2004); however, this would represent a small percentage of the transcript length. In contrast, the 108-kb Air analyzed by RepeatMasker (http://www.repeatmasker.org/) shows that interspersed repeats occupy 47% of the sequence. Thus the Air ncRNA is unusual because it is repeat rich as a mature transcript.
Seven of the imprinted noncoding RNAs listed in Table 1 show reciprocal parental-specific expression with respect to one or more imprinted protein-coding genes similar to that described for Air and Igf2r, and as a result have the potential to be involved in the imprinting mechanism. Table 2 lists a comparison of the features of these seven imprinted noncoding RNAs with respect to features noted for the Air ncRNA. Two out of seven are maternally expressed ncRNAs (Gtl2 and H19), while five are paternally expressed (Air, Kcnqot1, Nespas, “IC-Snurf-Snurp long processed transcript,” and Cop2as). Of the maternally expressed ncRNAs, the H19 ncRNA has been excluded from a direct role in silencing (Jones et al. 1998; Schmidt et al. 1999), while Gtl2 has not yet been tested. Of the paternally expressed ncRNAs, the Air ncRNA or its transcription has been shown to play a role in the imprinting mechanism (Rougeulle and Heard 2002; Sleutels et al. 2002, 2003). The other four paternally expressed ncRNAs have not yet been tested although deletions of the Kcnq1ot1 and the “_IC-Snurf-Snurp long processed transcript_” promoters allow the possibility that these two ncRNAs may also have a silencing function.
Table 2.
Characteristics of Imprinted Noncoding RNAs
| ImprintedncRNA | Parentalexpression | Size (kb) | Spliced or processed | References in addition toEnsembl build 30 |
|---|---|---|---|---|
| H19 | maternal | 1.8 | 5 exons in 2.2 kb | |
| Gtl2 | maternal | 3.98 | 5 exons over 15.5 kb in ENSEMBL build 32, but 10 exons producing multiply spliced stable RNAs have been reported | Ensembl build 32; Paulsen et al. (2000); Lin et al. (2003) |
| Air | paternal | 108 | 0 intron in 108 kb | Wutz et al. (1997); Lyle et al. (2000) |
| Kcnq1ot1 | paternal | 5′ mapped to Kcnq1 intron10, 3′ end not mapped | not known | Lee et al. (1999) |
| Nespas | paternal | >4.4 possibly longer | not known | Wroe et al. (2000) |
| IC-Snurf-SnrpnncRNA | paternal | 460 kb, _cis_tronic; mature RNA size unknown | >148 exons; long primary transcript possibly processed into multiple RNAs: IPW, PAR, PWCR1, multiple MB11 -snoRNAs, Ube3aas | Runte et al. (2001) |
| Cop2as | paternal | unknown | unknown, overlaps 3′ end of Cop2, possibly contains Mit1 | Lee et al. (2000) |
A comparison of the ncRNAs listed in Table 2 with the features described above for Air does not yet allow clear conclusions to be drawn about similarities and differences. However, it does indicate trends that could be common to imprinted ncRNAs or trends that could be common either to the paternally or maternally expressed imprinted ncRNAs. For example, there is a tendency for both maternal and paternal imprinted ncRNAs to have few or short introns compared to normal mammalian mRNAs that have many and long introns. This tendency for imprinted transcripts per se to have few or short introns was noted before (Pfeifer and Tilghman 1994; Hurst et al. 1996). Interspersed repeats are likely to be present in the five paternally expressed imprinted ncRNAs and it is possible that there is tendency for these ncRNAs to be unusually long, but this will not be clear until more of these transcripts are finely mapped. One feature that may differentiate the maternally and paternally expressed ncRNAs is the presence of tandem direct repeats that map upstream of the transcription start for H19 and Gtl2 (Thorvaldsen et al. 1998; Takada et al. 2000) and downstream of the transcription start for Air and Kcnq1 and thus potentially are present in the ncRNA (Lyle et al. 2000; Mancini-DiNardo et al. 2003).
FISHING FOR Air
RNA FISH (fluorescence in situ hybridization) is a procedure that applies fluorescent DNA hybridization probes to nondenatured cells, which detects singlestranded RNA but not double-stranded DNA (Dirks et al. 2003). This technique identifies a site close to a gene where nascent transcripts are accumulating and RNA processing is occurring. We have here used RNA FISH (Figs. 2-7) to test if the reciprocal parental-specific expression of Air and Igf2r observed in whole organs is also found in single cells.
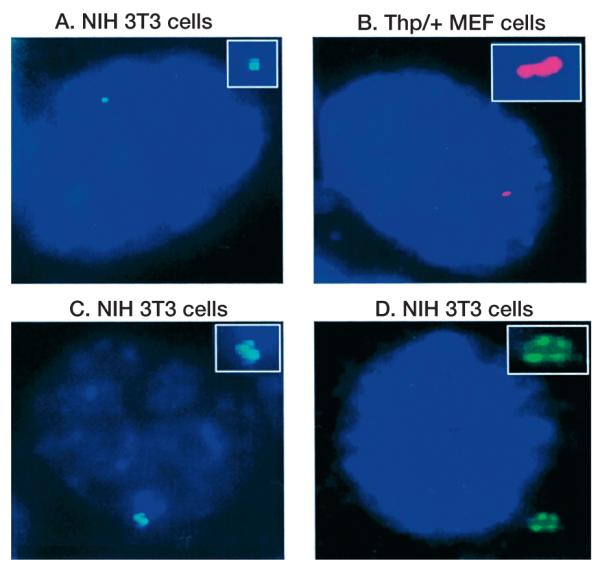
Figure 2.
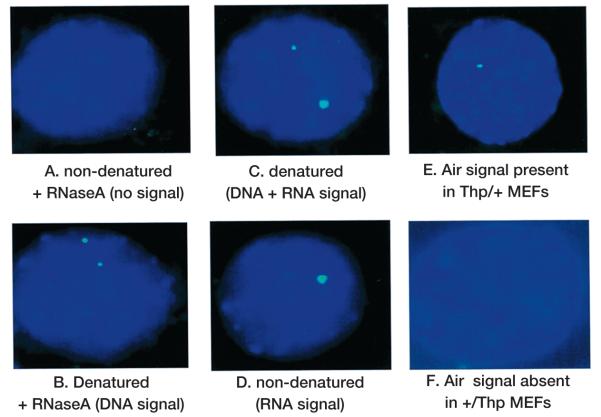
Control experiments for RNA FISH. (A_–_D) Diploid NIH 3T3 cells hybridized with an Igf2r strand-specific probe (green). (E) Thp/+ MEFs hybridized with an Air strand-specific probe (green). (F) +/Thp MEFs hybridized with an Air strand-specific probe. The RNA specific signal from Igf2r is seen in D. The paternal specific expression of Air is seen in E, since Thp/+ cells have a deletion on the maternal chromosome that removes the proximal Chr.17 imprinted cluster. In this and subsequent RNA FISH images, the probes were single stranded and derived from linear PCR. Two _Air_-strand-specific probes (FAS1 and 2) were amplified from cloned genomic DNA from Igf2r intron 1 (TGTCAAGGAGAACTGAGCGTCAT) or intron 2 (GGTAGGATGGTGTCTTACTC). The _Igf2r_-strand-specific probe (FcDNA) was amplified from the full-length mouse cDNA using an exon 28 primer (GTTTTCGAAAGTCAGCTTCTGGC). Probes were labeled with either Digoxigenin or Biotin and detected by anti-Digoxigenin or Avidin antibody conjugated with FITC (green) or Rhodamine (red; see examples in Figs. 3–6). Nuclei were stained with DAPI. The full RNA FISH protocol and image capture has been described (Braidotti 2001).
Figure 7.
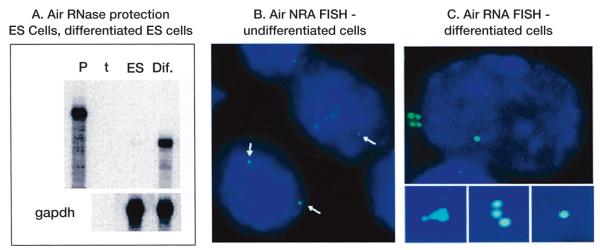
An RNase protection assay for Air (A) shows that Air is not expressed in ES cells but is abundant in ES cells differentiated by exposure for 5 days to retinoic acid. gapdh expression is used as the loading control (P, probe; t, tRNA). (B) The weak Air RNA FISH signal (white arrows) seen in ES cells that were not reproducible enough to count accurately. (C) The strong Air RNA FISH signal seen in 34% of differentiated cells. The three smaller images in C show examples of the Air signal clusters seen in these cells.
Igf2r mRNA IS RESTRICTED TO THE MATERNAL CHROMOSOME IN MOST CELLS
A number of controls were performed to test the specificity of the Igf2r RNA FISH signals (Fig. 2). Note that the cells used in these experiments were not synchronized for cell cycle and the population contained cells with a mixed ploidy number but was dominated by diploid cells. To demonstrate that the observed fluorescent signal arose from RNA, control cells that were not denatured but were treated with Ribonuclease A produced little to no signal (Fig. 2A). Cells that were denatured and treated with RNase A produced two or four signals as expected for specific detection of DNA in diploid and tetraploid cells (Fig. 2B). Cells that were denatured without RNase A treatment showed an enhanced signal from one allele that is assumed to be Igf2r transcription from the maternal allele in addition to the DNA signal (Fig. 2C). These controls demonstrate that the hybridization target in nondenatured cells not treated with RNase A results from RNA (Fig. 2D). Figure 2D also shows that expression of Igf2r mRNA is generally restricted to one chromosome. In RNA FISH experiments with an _Igf2r_-specific probe, only 40–70% of nuclei in the examined population had any RNA FISH signal. The reason for the absence of RNA signals was not determined and may be due to the technical limits of the RNA FISH technique or may arise from stochastic gene expression. However, it should be noted that transcription would not be predicted to occur close to and during M phase. In cells positive for an RNA FISH signal, the majority of nuclei had a single signal (in one experiment with diploid NIH 3T3 cells, 80% of the positive cells had one RNA signal and 16% had two RNA signals). The reason for the presence of two RNA signals in a minority of the cell population was not determined and may be due to tetraploid cells containing two maternal chromosomes, to spontaneous denaturation allowing the probe access to DNA, or to expression of Igf2r from the paternal chromosome. Cells with more than one maternal chromosome, or with more than one maternally inherited transgenic copy of Igf2r, will maintain correct maternal-specific expression, indicating that imprinted expression of Igf2r is not subject to a counting mechanism (Wutz et al. 1997; Vacik and Forejt 2003). It has also been reported that Igf2r can be expressed from the paternal allele in mouse embryos carrying a null Igf2r allele on the maternal chromosome, indicating a leaky repression of the paternal Igf2r promoter in embryos (Wang et al. 1994). In Thp/+ MEFs containing only the paternal Igf2r allele (the maternally inherited Thp chromosome carries a deletion of 6 Mbp that includes this imprinted cluster), a small subset of cells (<5%, data not shown) also produced an RNA signal in agreement with the suggestion that Igf2r mRNA can sometimes be expressed from a paternal allele in embryonic cells.
Air ncRNA IS RESTRICTED TO THE PATERNAL CHROMOSOME IN MOST CELLS
A similar RNA FISH hybridization series using probes for the Air ncRNA showed that Air expression is generally restricted to the paternal chromosome (Fig. 2E,F). In RNA FISH experiments using an _Air_-specific probe, ~50% of nuclei from diploid 3T3 fibroblasts were positive for any RNA signal. The reason for this increased number of nuclei negative for an Air RNA FISH signal was not determined, but may reflect reduced promoter activity or reduced RNA stability for Air compared with Igf2r. Of the 50% of nuclei with a positive signal for Air, 74% showed a single signal while 10% showed two signals (in 265 counted nuclei from NIH 3T3 cells). As with the results obtained with the Igf2r probe, a small proportion of NIH 3T3 cells also showed two Air transcription signals. Although double signals can arise for several reasons, possible Air expression from the maternal allele was seen in a small minority (4% of 71 counted nuclei) of +/Thp MEFs possessing just the maternal Air allele (in +/Thp cells the Thp deletion chromosome is paternally inherited). One difference from the results obtained with Igf2r was that Air showed some cells with a clustered signal pattern containing two to five distinct spots associated together, in ~16% of positive cells (Fig. 3B–D).
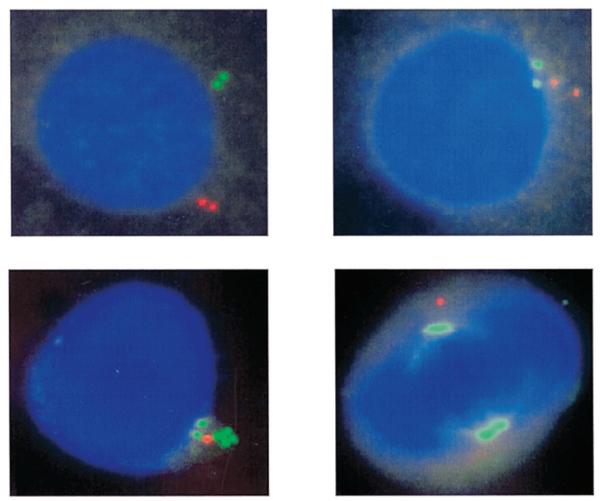
Figure 3.
Four examples of Air RNA FISH showing (A) a single signal and (B_–_D) the clustered spots that appeared in 16% of diploid NIH 3T3 cells or Thp/+ MEFs. An enlarged image for the signals and clusters is shown in the top right of each picture.
Air AND Igf2r ARE NOT COEXPRESSED IN cis
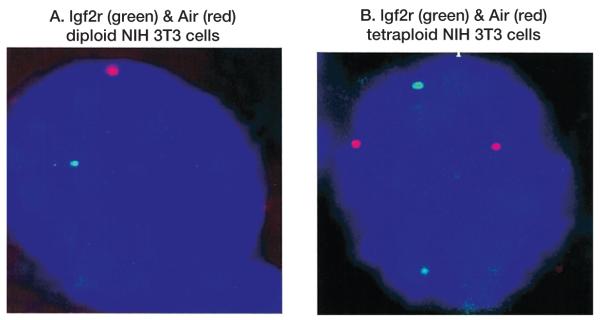
Figure 4A shows cohybridization with two probes, one recognizing the paternally derived Air transcript (red) and the second recognizing the maternally derived Igf2r mRNA (green). All positive cells showed distinct red or distinct green signals, indicating that expression of Igf2r and Air arose on different parental chromosomes. Since the Igf2r and Air probes are separated by ~45 kb, coexpression from the same chromosome would produce a yellow signal and none were observed. This data supports the claim that expression of Igf2r and Air is mutually exclusive in cis. Figure 4B shows a cell assumed to be tetraploid and this confirms other data that multiple copies of imprinted genes are not regulated by a counting mechanism (Wutz et al. 1997; Vacik and Forejt 2003).
Figure 4.
RNA FISH showing cohybridization of diploid and tetraploid NIH 3T3 cells with _Igf2r_-specific (green) and _Air_-specific (red) probes.
THE Air ncRNA FORMS CLUSTERS THAT MIGRATE AWAY FROM CONDENSED CHROMOSOMES
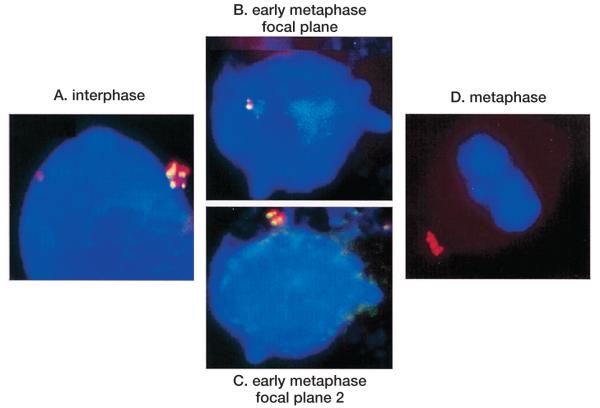
Clustering of the Air ncRNA signal was seen in some cells assumed to be in interphase (Fig. 5A) but most often seen to be separate from the main body of condensing chromosomes in cells assumed from DAPI staining to be in early M phase. In Figure 5B,C, a cell was identified with a single transcription/accumulation signal on one focal plane within the spherical nucleus, plus a cluster of signals on a different focal plane that had migrated away from the condensed chromosomes. We cautiously interpret the behavior of these Air clusters as indicating that the Air transcript does not stay associated with the paternal Igf2r allele as the chromosomes condense in early M phase. In all cases where the Air ncRNA cluster was observed to dissociate from the chromosome, the signals remained grouped together in clusters. The possibility exists that these provisional results indicate that the Air ncRNA may form aggregates either with itself or with other proteins or RNAs. The Air clusters that were separated from metaphase chromosomes were often observed in an asymmetrical distribution relative to the metaphase plate (Fig. 5D), which suggested a link with the centrosome body or the MTOC (microtubule organizing center). However, cohybridization with a gamma tubulin antibody showed that although the Air cluster was often found associated with centrosomes, the cluster was also seen to be separate from the centrosomes (Fig. 6).
Figure 5.
RNA FISH showing Air clustered signals (red) dissociated from chromosomes. Air clusters are seen in interphase nuclei (A), early metaphase nuclei (B and C show two focal planes in the same cell), and in metaphase (D).
Figure 6.
Air RNA FISH (red) combined with gamma tubulin immunofluorescence (green) in NIH 3T3 cells. Gamma tubulin antibody from Dr. Dagmar Ivanyi, NKI, Amsterdam. The top left image shows that Air is localized away from the centrosome, while the three other images show that Air is adjacent to the centrosome.
THE Air CLUSTERS ARE ABSENT IN ES CELLS THAT LACK IMPRINTED EXPRESSION OF Igf2r mRNA
Mouse preimplantation embryos express Igf2r from both parental chromosomes, with imprinted maternalspecific expression occurring between 4.5 and 6.5 dpc (Szabo and Mann 1995a; Lerchner and Barlow 1997). Embryonic stem (ES) cells show the same biallelic expression of Igf2r (Wang et al. 1994). Air ncRNA is weakly detected in the undifferentiated ES cell but is abundant in ES cells induced to differentiate (Fig. 7A). RNA FISH analysis of Air in undifferentiated ES cells identified only a few single very weak signals in ~5% of cells but no clusters were observed (Fig. 7B). Differentiated ES cells, however, produced an array of signal types similar to that observed in NIH 3T3 with a maximum of 34% of cells in the differentiated culture positive for any RNA FISH signal. Figure 7C shows a nucleus of a differentiated ES cell with a pronounced single signal and a cluster located at the edge of the nucleus. Three examples of Air ncRNA clusters in differentiated ES cells are shown magnified below.
The RNA FISH analysis of Igf2r and Air expression in MEFs shows that expression of both transcripts is parental specific. However, there is a possibility that requires further testing—that a minority of cells (<5%) can express Igf2r and Air from the opposite parental chromosome. The cohybridization experiments that assayed expression of Igf2r and Air in the same cell, however, do clearly show that these two genes are not active in cis. This finding supports other data (Sleutels et al. 2002) that shows that Air expression is necessary to silence Igf2r (and the downstream flanking Slc22a2 and Slc22a3 genes) in cis. The RNA FISH analysis has also shown that the Air ncRNA, in contrast to Igf2r, can form clusters of two to five spots. Air clusters were seen in a small number of cells assumed from DAPI staining of chromosomes to be in interphase—the stage when Igf2r silencing must occur. However, in the majority of cases the Air ncRNA clusters were observed dissociated from chromosomes in cells assumed from DAPI staining to be in early metaphase. We cautiously interpret this to indicate that Air clusters form in late interphase and metaphase. The meaning of these Air clusters and why they are also observed dissociated from chromosomes is not yet known. Since expression of Air is known to silence genes spread over 300 kb, it is possible that these clusters are relevant for the silencing function of Air. One speculation is that the clusters that show as multiple distinct bright spots at metaphase are derived from very tight aggregates present at interphase that cannot be resolved into separate spots. However, further work is needed to confirm that these clusters are consistently associated with Air expression or have any functional relevance. It should be noted that an earlier RNA FISH study on the imprinted maternally expressed H19 ncRNA did not find evidence of RNA clusters (Jouvenot et al. 1999). Interestingly, the appearance of the human XIST ncRNA in diploid female cells has been described as an “accumulation of many, very distinct bright clusters” (Clemson et al. 1996). However, in contrast to the results for mouse Air reported here, the human XIST ncRNA clusters were specifically observed in interphase cells associated with the inactive chromosome.
SIMILARITIES BETWEEN GENOMIC IMPRINTING AND X-INACTIVATION
The _cis_-repression of a large subchromosomal domain by the Air ncRNA has clear parallels with the action of the Xist ncRNA in X-chromosome inactivation (Avner and Heard 2001; Wutz 2003). It has also been argued that X-chromosome inactivation arose from a localized form of genomic imprinting that initially affected only part of the chromosome (Graves 1996; Lyon 1999). Similarities between genomic imprinting and X-inactivation have been previously noted (Pfeifer and Tilghman 1994; Lyon 1999; Lee 2003) and the behavior of the Air ncRNA strengthens these similarities. The features in common between the two processes include (for review, see Lyon 1999) the following.
- X-chromosome inactivation is imprinted such that the paternal X is preferentially inactivated in the extraembryonic tissues of placental mammals and in all organs of marsupials.
- The regulatory gene is a _cis_-silencing noncoding RNA that acts over a long range.
- Chromosome-specific methylation regulates expression of the noncoding RNA.
- Some genes escape repression.
The Xist noncoding RNA has some additional features that have not yet been characterized or may be absent for the Air ncRNA. First, experiments indicate that the Xist ncRNA has a sequence-specific function that may lie in the secondary structure (Wutz et al. 2002). Second, the Xist ncRNA has not been observed dissociated from chromosomes; instead it colocalizes with the repressed X chromosome in interphase in human cells (Clemson et al. 1996) and in interphase and metaphase in mouse cells (Sheardown et al. 1997). Third, expression of the Xist transcript is regulated in ES cells by a second antisense noncoding RNA, named Tsix (Lee 2000; Sado et al. 2001). Fourth, Xist recruits repressive chromatin proteins and modifications to the X chromosome (Avner and Heard 2001; Wutz 2003).
The similarities between genomic imprinting and X-inactivation do not imply that these are identical mechanisms. If, as suggested, the latter evolved from the former, it is likely that the Xist ncRNA may have also evolved extra functions when compared to the Air ncRNA, particularly with regard to its ability to spread along the entire X chromosome. While further experiments will be needed to discriminate between the possible modes of actions of the Air ncRNA and to refine the details of the repression mechanism, the high degree of similarity between _cis_-repression on autosomes by imprinted noncoding RNAs and _cis_-repression of the X chromosome by the noncoding Xist ncRNA is striking.
ACKNOWLEDGMENTS
We thank past and present members of the lab for contributing to the work and ideas described here, Dagmar Ivanyi for the gamma tubulin antibody, Roeland Dirks for advice on RNA FISH, and Laura Spahn for reading the manuscript. Research support from GEN-AU—Genomforschung in Österreich (F.P.), FWF—Fonds zur Förderung der wissenschaftlichen Forschung (I.Y. and C.S.), and KWF—the Dutch Cancer Society and the Netherlands Cancer Institute (G.B. and O.S.).
REFERENCES
- Avner P, Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat. Rev. Genet. 2001;2:59. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Baroux C, Spillane C, Grossniklaus U. Genomic imprinting during seed development. Adv. Genet. 2002;46:165. doi: 10.1016/s0065-2660(02)46007-5. [DOI] [PubMed] [Google Scholar]
- Beechey CV, Cattanach BM, Blake A. World Wide Web site—Mouse Imprinting Data and References. 2003. MRC Mammalian Genetics Unit, Harwell, Oxfordshire. ( http://www.mgu.har.mrc.ac.uk/research/imprinting/)
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bongiorni S, Prantera G. Imprinted facultative heterochromatization in mealybugs. Genetica. 2003;117:271. doi: 10.1023/a:1022964700446. [DOI] [PubMed] [Google Scholar]
- Braidotti G. RNA-FISH to analyze allele-specific expression. Methods Mol. Biol. 2001;181:169. doi: 10.1385/1-59259-211-2:169. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Ariel M, Cedar H. Dynamics of DNA methylation during development. Bioessays. 1993;15:709. doi: 10.1002/bies.950151103. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RW, Molenaar C, Tanke HJ. Visualizing RNA molecules inside the nucleus of living cells. Methods. 2003;29:51. doi: 10.1016/s1046-2023(02)00290-6. [DOI] [PubMed] [Google Scholar]
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, Feil R. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukakis N, Deal C, Paquette J, Goodyer CG, Polychronakos C. Parental genomic imprinting of the human IGF2 gene. Nat. Genet. 1993;4:98. doi: 10.1038/ng0593-98. [DOI] [PubMed] [Google Scholar]
- Goday C, Esteban MR. Chromosome elimination in sciarid flies. Bioessays. 2001;23:242. doi: 10.1002/1521-1878(200103)23:3<242::AID-BIES1034>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Graves JA. Mammals that break the rules: Genetics of marsupials and monotremes. Annu. Rev. Genet. 1996;30:233. doi: 10.1146/annurev.genet.30.1.233. [DOI] [PubMed] [Google Scholar]
- Gribnau J, Hochedlinger K, Hata K, Li E, Jaenisch R. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 2003;17:759. doi: 10.1101/gad.1059603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylationsensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hu JF, Balaguru KA, Ivaturi RD, Oruganti H, Li T, Nguyen BT, Vu TH, Hoffman AR. Lack of reciprocal genomic imprinting of sense and antisense RNA of mouse insulin-like growth factor II receptor in the central nervous system. Biochem. Biophys. Res. Commun. 1999;257:604. doi: 10.1006/bbrc.1999.0380. [DOI] [PubMed] [Google Scholar]
- Hurst LD, McVean G, Moore T. Imprinted genes have few and small introns. Nat. Genet. 1996;12:234. doi: 10.1038/ng0396-234. [DOI] [PubMed] [Google Scholar]
- Jones BK, Levorse JM, Tilghman SM. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 1998;12:2200. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker JW, Wagenaar E, Van Eijl S, Schinkel AH. Deficiency in the organic cation transporters 1 and 2 (Oct1/Oct2 [Slc22a1/Slc22a2]) in mice abolishes renal secretion of organic cations. Mol. Cell. Biol. 2003;23:7902. doi: 10.1128/MCB.23.21.7902-7908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenot Y, Poirier F, Jami J, Paldi A. Biallelic transcription of Igf2 and H19 in individual cells suggests a posttranscriptional contribution to genomic imprinting. Curr. Biol. 1999;9:1199. doi: 10.1016/S0960-9822(00)80026-3. [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Mariman EC, Schepens MT, Rehder H, Ropers HH. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat. Genet. 1993;5:74. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, Jirtle RJ. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum. Mol. Genet. 2001;10:1721. doi: 10.1093/hmg/10.17.1721. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Lee JE, Tantravahi U, Boyle AL, Efstratiadis A. Parental imprinting of an Igf-2 transgene. Mol. Reprod. Dev. 1993;35:382. doi: 10.1002/mrd.1080350411. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- — — — Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol. 2003;13:R242. doi: 10.1016/s0960-9822(03)00162-3. [DOI] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. 1999;96:5203. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Park CW, Hahn Y, Park J, Lee J, Yun JH, Hyun B, Chung JH. Mit1/Lb9 and Copg2, new members of mouse imprinted genes closely linked to Peg1/Mest(1) FEBS Lett. 2000;472:230. doi: 10.1016/s0014-5793(00)01461-7. [DOI] [PubMed] [Google Scholar]
- Lerchner W, Barlow DP. Paternal repression of the imprinted mouse Igf2r locus occurs during implantation and is stable in all tissues of the post-implantation mouse embryo. Mech. Dev. 1997;61:141. doi: 10.1016/s0925-4773(96)00630-2. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003;35:97. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Eggenschwiler J, Fisher P, D’Ercole AJ, Davenport ML, Efstratiadis A. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev. Biol. 1996;177:517. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davie C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Imprinting and X-chromosome inactivation. Results Probl. Cell Differ. 1999;25:73. doi: 10.1007/978-3-540-69111-2_4. [DOI] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Ingram RS, Tilghman SM. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum. Mol. Genet. 2003;12:283. doi: 10.1093/hmg/ddg024. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Nagashima T, Matsuda H, Silva DG, Petrovsky N, Konagaya A, Schonbac C, Kasukawa T, Arakawa T, Carninci P, Kawai J, Hayashizaki Y. FREP: A database of functional repeats in mouse cDNAs. Nucleic Acids Res. 2004;32:D471. doi: 10.1093/nar/gkh123. (database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudejans CB, Westerman B, Wouters D, Gooyer S, Leegwater PA, van Wijk IJ, Sleutels F. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics. 2001;73:331. doi: 10.1006/geno.2001.6522. [DOI] [PubMed] [Google Scholar]
- Paulsen M, El-Maarri O, Engemann S, Strodicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum. Mol. Genet. 2000;9:1829. doi: 10.1093/hmg/9.12.1829. [DOI] [PubMed] [Google Scholar]
- Pfeifer K, Tilghman SM. Allele-specific gene expression in mammals: The curious case of the imprinted RNAs. Genes Dev. 1994;8:1867. doi: 10.1101/gad.8.16.1867. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001;2:21. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Riesewijk AM, Schepens MT, Welch TR, van den Berg-Loonen EM, Mariman EM, Ropers HH, Kalscheuer VM. Maternal-specific methylation of the human IGF2R gene is not accompanied by allele-specific transcription. Genomics. 1996;31:158. doi: 10.1006/geno.1996.0027. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Heard E. Antisense RNA in imprinting: Spreading silence through Air. Trends Genet. 2002;18:434. doi: 10.1016/s0168-9525(02)02749-x. [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Levorse JM, Tilghman SM. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl. Acad. Sci. 1999;96:9733. doi: 10.1073/pnas.96.17.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweifer N, Valk PJ, Delwel R, Cox R, Francis F, Meier-Ewert S, Lehrach H, Barlow DP. Characterization of the C3 YAC contig from proximal mouse chromosome 17 and analysis of allelic expression of genes flanking the imprinted Igf2r gene. Genomics. 1997;43:285. doi: 10.1006/geno.1997.4816. [DOI] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AE, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- Shintani S, O’hUigin C, Toyosawa S, Michalova V, Klein J. Origin of gene overlap: The case of TCP1 and ACAT2. Genetics. 1999;152:743. doi: 10.1093/genetics/152.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Barlow DP. Investigation of elements sufficient to imprint the mouse Air promoter. Mol. Cell. Biol. 2001;21:5008. doi: 10.1128/MCB.21.15.5008-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Tjon G, Ludwig T, Barlow DP. Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J. 2003;22:3696. doi: 10.1093/emboj/cdg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrzka OW, Fae I, Stoger R, Kurzbaue R, Fischer GF, Henn T, Weith A, Barlow DP. Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum. Mol. Genet. 1995;4:1945. doi: 10.1093/hmg/4.10.1945. [DOI] [PubMed] [Google Scholar]
- Spahn L, Barlow DP. An ICE pattern crystallizes. Nat. Genet. 2003;35:11. doi: 10.1038/ng0903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: Implications for imprinting mechanisms. Genes Dev. 1995a;9:3097. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- — — — Biallelic expression of imprinted genes in the mouse germ line: Implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995b;9:1857. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr. Biol. 2000;10:1135. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacik T, Forejt J. Quantification of expression and methylation of the Igf2r imprinted gene in segmental trisomic mouse model. Genomics. 2003;82:261. doi: 10.1016/s0888-7543(03)00118-6. [DOI] [PubMed] [Google Scholar]
- Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: Intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Fung MR, Barlow DP, Wagner EF. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature. 1994;372:464. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: The function of parent-specific gene expression. Nat. Rev. Genet. 2003;4:359. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Wroe SF, Kelsey G, Skinner JA, Bodle D, Ball ST, Beechey CV, Peters J, Williamson CM. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl. Acad. Sci. 2000;97:3342. doi: 10.1073/pnas.050015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. RNAs templating chromatin structure for dosage compensation in animals. Bioessays. 2003;25:434. doi: 10.1002/bies.10274. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- Wutz A, Theussl HC, Dausman J, Jaenisch R, Barlow DP, Wagner EF. Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development. 2001;128:1881. doi: 10.1242/dev.128.10.1881. [DOI] [PubMed] [Google Scholar]
- Xu YQ, Grundy P, Polychronakos C. Aberrant imprinting of the insulin-like growth factor II receptor gene in Wilms’ tumor. Oncogene. 1997;14:1041. doi: 10.1038/sj.onc.1200926. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li T, Vu TH, Ulaner GA, Hu JF, Hoffman AR. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology. 2003;144:5658. doi: 10.1210/en.2003-0798. [DOI] [PubMed] [Google Scholar]
- Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001a;15:2361. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart R, Verhaagh S, Buitelaar M, Popp-Snijders C, Barlow DP. Impaired activity of the extraneuronal monoamine transporter system known as uptake-2 in Orct3/Slc22a3-deficient mice. Mol. Cell. Biol. 2001b;21:4188. doi: 10.1128/MCB.21.13.4188-4196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]