RNA Helicases at work: binding and rearranging (original) (raw)
. Author manuscript; available in PMC: 2012 Jan 1.
Published in final edited form as: Trends Biochem Sci. 2011 Jan;36(1):19–29. doi: 10.1016/j.tibs.2010.07.008
Abstract
RNA helicases are ubiquitous, highly conserved enzymes that participate in nearly all aspects of RNA metabolism. These proteins bind or remodel RNA or RNA–protein complexes in an ATP-dependent fashion. How RNA helicases physically perform their cellular tasks has been a longstanding question, but in recent years, intriguing models have started to link structure, mechanism and biological function for some RNA helicases. This review outlines our current view on major structural and mechanistic themes of RNA helicase function, and on emerging physical models for cellular roles of these enzymes.
RNA helicases: ubiquitous and central players in RNA metabolism
RNA helicases are highly conserved enzymes that use ATP to bind or remodel RNA or ribonucleoprotein complexes (RNPs) 1. One of the largest protein classes in RNA metabolism, RNA helicases are found in all kingdoms of life 2. In eukaryotes these enzymes participate in nearly all aspects of RNA metabolism 1. RNA helicases have received significant attention, ever since their identification in the 1980s. Many RNA helicases are essential for viability, and a growing number of these enzymes are known to play major regulatory roles in cells 1, 3. Yet, despite important insights into structural, mechanistic, and cellular aspects of their function, it has remained enigmatic how these enzymes physically perform their cellular tasks. The last few years have now seen a notable increase in the number of cell biological, genetic, molecular biological, biochemical-biophysical, and structural studies on RNA helicases. Although much remains to be learned, intriguing models are emerging that start to link structure, mechanism and biological function for some RNA helicases. In this review, I outline our current view on major structural and mechanistic aspects of RNA helicase function, and how these translate into cellular roles for these enzymes. For space reasons, I will focus mainly on the eukaryotic proteins.
RNA helicase basics: superfamilies, families, and structural themes
RNA helicases are closely related to DNA helicases 4. Both DNA and RNA helicases fall into two categories, those that form oligomeric (mostly hexameric) rings, and those that do not 5. Based on sequence and comparative structural and functional analyses, all helicases are classified into six superfamilies (SFs) 5, 6. The ring-forming helicases comprise SFs 3 to 6, and the non-ring forming ones comprise SFs 1 and 2 5. All eukaryotic RNA helicases belong to SFs 1 and 2 (Fig. 1). Ring-shaped RNA helicases are found in bacteria (e.g. Rho 7 ) and viruses (e.g. Φ29 P4 8). Although these enzymes will not be discussed here, excellent recent reviews on these proteins are available 7, 9.
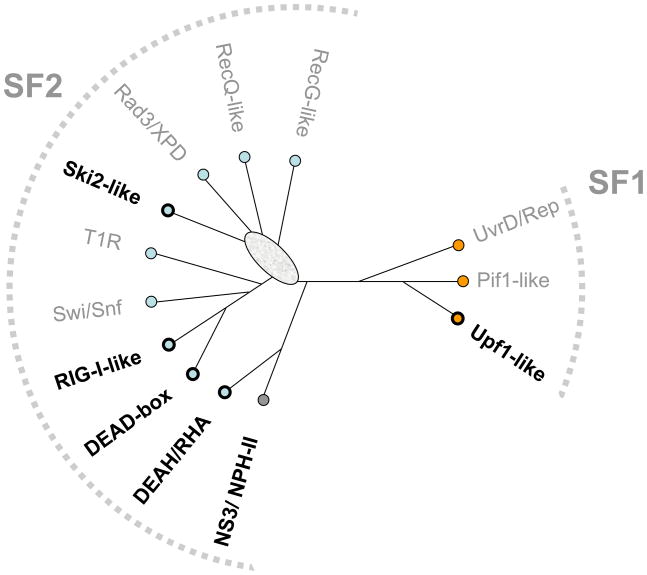
Figure 1. SF1 and SF2 helicase families.
Unrooted cladogram showing the families of the SF1 (right), and the SF2 (left) according to ref. 4. Branch lengths are not to scale. The oval indicates significant uncertainty in cladogram topology in this region. Boldfaced names show families harboring RNA helicases (non-standard abbreviations: T1R – type 1 restriction enzymes, RHA –RNA helicase A).
Sequence comparisons revealed that both SF1 and 2 consist of well-defined helicase families with distinct structural and functional signatures (for a detailed description of the SF1 and SF2 families, see ref. 4). RNA helicases are found in six of these families; the remaining families consist of DNA helicases (Fig. 1). Several helicase families contain both RNA and DNA helicases, and some enzymes, including proteins from the viral NS3/NPH-II group, RNA helicase A (DHX9) and Upf1-like helicases work on both DNA and RNA 10 11, 12. The lack of clear correlation between the helicase families and specificity for RNA or DNA suggests that discrimination between RNA and DNA might not have been a predominant evolutionary force for the differentiation of the helicase families 4.
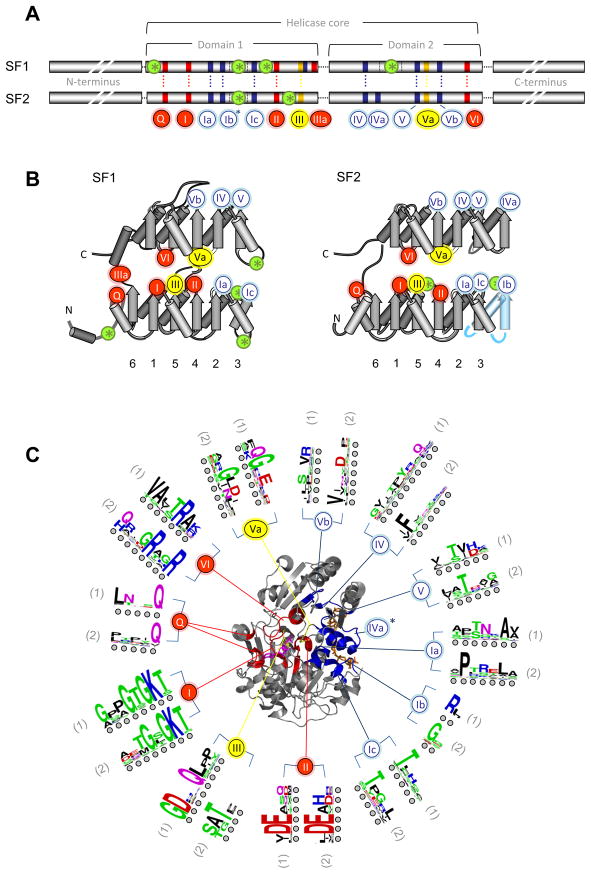
Helicases of SFs 1 and 2 contain a structurally conserved helicase core, formed by two highly similar helicase domains arranged in tandem (Fig. 2). Both SF1 and SF2 helicases contain at least 12 characteristic sequence motifs at defined positions in the helicase core (Fig. 2A, B). However, not all motifs are present in each helicase family 4. The level of sequence conservation in these motifs is high within each family, but decreases between different families 4. Only limited sequence conservation remains across both superfamilies (Fig. 2C).
Figure 2. The helicase core of SF1 and SF2 proteins.
(A) Characteristic sequence motifs of SF1 and SF2 proteins in the helicase core 4. The motifs are colored according to their predominant biochemical function: red, ATP binding and hydrolysis; yellow, coordination between nucleic acid and NTP binding sites; blue, nucleic acid binding. Motif Ib (asterisk) is not present in all SF1 and 2 families. Green circles with asterisks designate insertions of additional domains. Distance between the conserved motifs is not to scale. (B) Location of the helicase motifs in the helicase core fold (arrows: β-strands, cylinders: α-helices). Numbers under the diagrams show connectivity of the β-strands of the first RecA-like domain. Helicase motifs are marked by circles (coloring and numbering as in panel A). Domain insertions are marked by green circles with asterisks. The rightmost β-strand and α-helix in the SF2 (blue) is not present in all SF2 families 4. (C) Location of the characteristic motifs in three-dimensional structure of the helicase core, as represented by SF2 DEAD-box helicase Vasa 49. The bound ATP analog is colored magenta, the RNA wheat. Helicase motifs are colored as in panel A. Corresponding sequence logos indicate conservation within the helicase motifs in SF1 (1) and SF2 (2). Colors mark properties of the amino acids as: green - polar, blue - basic, red – acidic, and black -hydrophobic. Circles under the letters are visual guides.
In addition to the family-typical sequence domains, the Ski2-like, DEAH/RHA and NS3/NPH-II families have a prominent β-hairpin between motifs Va and VI 13–16. This feature is not seen in other RNA helicase families 4. The SF1 Upf1-like family and in the retinoic-acid-inducible gene I (RIG-I)-like family have inserts within or between the helicase core domains 4 (Fig. 2A). The inserts are occasionally large and adopt independent folds, but generally have only minor or no effects on the fold of the helicase core domains 17–19. A small number of individual proteins in other helicase families also feature inserts, such as the DEAD-box protein DDX1 20, but these inserts are not typical for the families.
In essentially all SF1 and SF2 helicases, the structurally conserved helicase core is surrounded by C- and N-terminal domains, which are often larger than the helicase core and frequently contain one or more specific functionalities including nucleases, RNA- or DNA-binding domains 10, 14, 21), protein-binding domains (e.g., CARD domains 22), or oligomerization modules 23. C- and N-terminal domains are thought to be critical for the cellular specificity of helicases by facilitating recruitment of the proteins to specific complexes, either through interactions with other proteins, or as seen in the bacterial DEAD-box protein DbpA or the DEAH/RHA protein RHAU (DHX36), by facilitating recognition of specific nucleic acid regions 24, 25. Except for the C-terminus of the spliceosomal DEAH proteins, C- and N-terminal domains are generally not conserved within or between families 4. However, recent work shows structural conservation in the C-terminal domains of Ski2-like and DEAH/RHA proteins 14, 26.
RNA helicase mechanisms: more than unwinding
Paradoxically, helicases as defined by characteristic sequence motifs are not always helicases as defined by enzymatic function, the ATP-dependent unwinding of nucleic acid duplexes 27, 28. For example, proteins of the Swi/Snf family and the ATP-dependent restriction endonucleases (T1R, Fig. 1) generally display no unwinding activity, even though they hydrolyze ATP in a DNA-dependent fashion, possess all of the helicase motifs, and are built around a helicase core structure.29
RNA helicases generally unwind RNA duplexes in vitro, provided appropriate substrates are used.30 However, RNA helicase activity in vitro does not imply that a given enzyme necessarily unwinds duplexes in the cell. Yet, even for RNA helicases that perform other tasks, RNA helicase activity is an excellent proxy for measuring the ability of the enzymes to remodel RNA structures in an ATP-dependent fashion.
At least two distinct types of RNA helicase activity have been identified: canonical duplex unwinding (Box 1), and unwinding by local strand separation (Box 2). Canonical duplex unwinding refers to the mechanism displayed by many DNA helicases and by several viral RNA helicases of the NS3/NPH-II group 5, 31. The helicase binds to a single stranded region adjacent to the duplex and then translocates along the bound strand with defined directionality, either 3’ to 5’ or 5’ to 3’, thereby displacing the complementary strand (Box 1). As a consequence, most canonically operating helicases require substrates with single stranded regions in a defined orientation (polarity) with respect to the duplex (Box 1). RNA helicases of the Ski2-like, the RIG-I-like, the DEAH/RHA, and the Upf1-like families also display preferred unwinding polarities 10, 12, 32, 33. Based solely on this observation, it is occasionally concluded that these RNA helicases unwind duplexes in the canonical fashion. However, to my knowledge, no eukaryotic RNA helicase has been directly shown to unwind duplexes based on directional translocation, that is, in multiple consecutive unwinding steps. Alternative unwinding scenarios thus remain possible for these helicases.
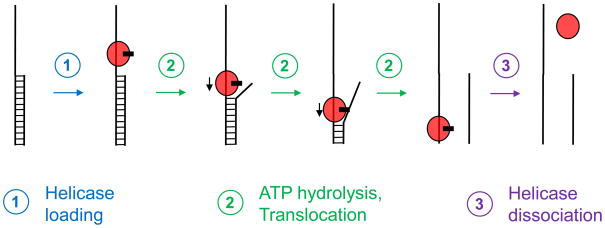
Box 1. Translocation-based duplex unwinding by canonical DNA and RNA helicases.
The helicase binds to the single stranded region and in multiple, ATP-dependent consecutive steps translocates towards the opposite end (Fig. I). In the process, the complementary strand is removed. There are monomeric and oligomeric canonical helicases 27. Each translocation step consists of multiple processes including ATP binding and hydrolysis, a power stroke to produce the forward movement, and dissociation of the products of the ATP hydrolysis. For detailed reviews and discussions on canonical unwinding mechanism, see refs 5, 27, 31, 46.
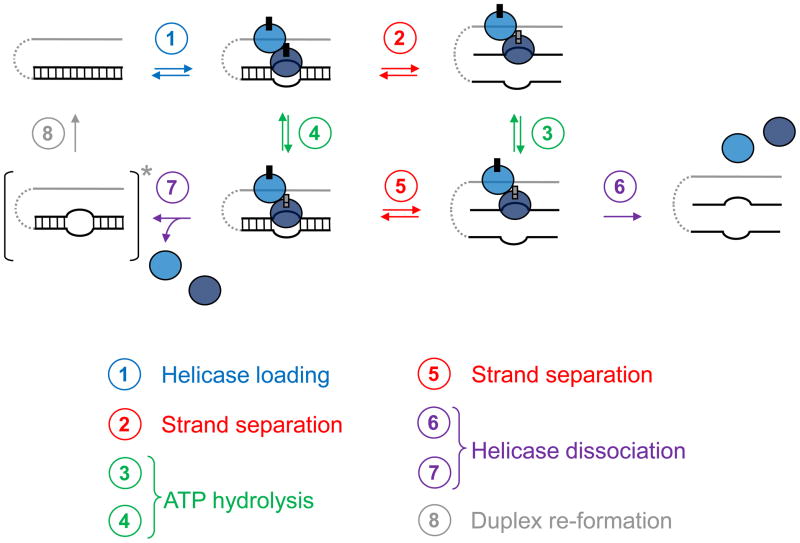
Box 2. Duplex unwinding by local strand separation.
This unwinding mode is employed by DEAD-box helicases 77. The helicase is loaded directly on the duplex region, aided by single stranded or structured nucleic acid regions (Fig. I, step 1). These regions have to be proximal, but no covalent connection to the duplex is necessary. Therefore, DEAD-box proteins unwind tailed substrates without apparent polarity, but often require unpaired regions for efficient unwinding. Duplex loading can involve multiple protomers, but can also be mediated by accessory protein domains 77. Although the exact mechanisms of loading processes are not yet understood, it is known that the loading can occur at any place in the duplex, at an end or internally, and on either strand. Duplex loading is accompanied by ATP binding 34. Upon loading, the DEAD-box protein locally opens the duplex strands (Fig. I, step 1). This step requires ATP, but not ATP hydrolysis, suggesting that ATP binding suffices 39–41. The local helix opening reduces the number of basepairs in the duplex, and the remaining basepairs dissociate without further action from the enzyme (Fig. I, step 2). Unwinding rate constants decrease with duplex length and stability, because more or more stable basepairs dissociate slower. Although ATP hydrolysis is dispensable for duplex unwinding, it is critical for release of the DEAD-box protein from the RNA (Fig. I, step 6), and thus for enzyme recycling 40. Not every ATP-driven local helix opening will lead to complete strand separation. ATP hydrolysis can occur after the helix has been opened by ATP binding, but before the strands have separated (Fig. I, step 4). Although ATP hydrolysis promotes enzyme dissociation, strand separation can take place before the enzyme dissociates (Fig. I, step 5), or the enzyme dissociates before complete helix separation (Fig. I, step 7). In this case, the strands quickly re-anneal (Fig. I, step 8). Such “non-productive” ATP hydrolysis events are more prevalent for longer and more stable duplexes, because unwinding events occur less frequently. Therefore, unwinding of longer or more stable helices involves greater numbers of hydrolyzed ATPs per duplex separated 36, 39.
RNA helicases of the DEAD-box family unwind duplexes by local strand separation (Box 2). This distinct unwinding mechanism is not based on translocation 34–37. Instead, DEAD-box proteins directly load to the duplex region and then pry the strands apart in an ATP-dependent fashion (Box 2).34–37 34, 35 As a result, unwinding occurs without defined polarity, even though single stranded, or in some cases structured RNA extensions, stimulate strand separation by most DEAD-box proteins 35, 37. Unwinding can be accomplished with a single round of ATP binding/hydrolysis, and for several DEAD-box proteins, strand separation does not require ATP hydrolysis, but only ATP binding 38–41. However, ATP hydrolysis is necessary for efficient release of the DEAD-box helicase from the RNA and thus for multiple substrate turnovers 40 (Box 2). Unwinding efficiency greatly decreases with increasing length and stability of the duplex, and most DEAD-box proteins only unwind duplexes containing less than 10–12 basepairs with appreciable activity 36, 39, 42. However, RNAs in eukaryotic cells form few, if any, uninterrupted duplexes exceeding this length 43. The distinct unwinding mode of DEAD-box proteins thus appears uniquely suited for the localized separation of short duplexes in the cell35.
For both canonical and non-canonical RNA helicases, RNA binding and unwinding involves ATP-dependent, coordinated changes in the orientation of the two helicase domains 5, 27, 31, 43. Without ATP, the cleft between the two domains opens, although to varying degrees in different helicase families 43. ATP binding generally promotes closing of the two domains 5, 27, 31, 43. The nucleic acid is bound by all helicases in the same orientation (Fig. 2C). The conserved helicase domains involved in nucleic acid binding make similar contacts in all, canonical and non-canonical RNA helicases; and nearly all contacts are with the sugar-phosphate backbone 5.
For canonically operating helicases (e.g., hepatitis C virus NS3), additional contacts from accessory domains are established with bases 16. These contacts ensure the translocation of the helicase along the RNA/DNA by 1 nt per ATP consumed 44. Structural models now exist for translocation by several canonical RNA and DNA helicases, and all of these models suggest movement by 1 nt per ATP 5, 16, 31. However, the directional movement is accomplished through different base contacts by different enzymes 5, 16, 31. Many translocating helicases appear to move in bursts of several 1 nt steps before repeating a rate limiting step 44–46. The distance translocated during these bursts is often measured as the kinetic step size, reaching up to 18 nt for certain helicases 47, 48. The exact structural bases for the kinetic step sizes are not clear.
Structures with bound RNA are available for only a few DEAD-box proteins 49–53. In all of these structures, the DEAD-box proteins establish contacts to the RNA almost exclusively to the backbone, consistent with their distinct unwinding mode. The conformation of the bound RNA strand in the presence of ATP analogs is characterized by pronounced bends in the backbone, a marked difference to RNA or DNA conformations in most canonical helicases 16, 53. The bends make the bound RNA incompatible with double helical architecture, and probably represent the RNA conformation following strand separation 53. How exactly DEAD-box proteins perform the act of strand separation is not yet understood on a structural level.
In addition to duplex unwinding, RNA helicases display an array of additional activities. Most prominently, several RNA helicases have been directly shown to displace other proteins from RNA in an active, ATP-dependent fashion 54. Protein displacement or RNP remodeling is thought to be central to the physiological function of RNA helicases, because RNAs are generally bound to other proteins in vivo 55. Protein displacement is not necessarily coupled to duplex unwinding, and has also been seen for DEAD-box proteins, indicating that protein removal is not restricted to RNA helicases that unwind duplexes in the translocation-based, canonical fashion 56–58. Nevertheless, in vitro some helicases can only remove a certain spectrum of proteins (e.g., proteins with small RNA binding sites), whereas other helicases displace a more diverse set of proteins 56. The inability of certain helicases to remove a given protein from RNA might spatially regulate helicase activities in larger RNP assemblies 54.
The RNA helicase RIG-I recently was shown to translocate on double stranded RNA in an ATP-dependent fashion, without unwinding the duplex 59. This activity resembles the translocation of type 1 restriction enzymes and some Swi/Snf proteins on dsDNA 29. RIG-I functions in the innate immune system as pattern recognition receptor for the identification of viral RNAs in the cytoplasm, and the translocation is thought to aid the detection of viral RNAs, which can form long dsRNA during viral replication 59. However, translocation on dsRNA appears unlikely to be prevalent among eukaryotic RNA helicases that function on cellular RNAs, given that these RNAs are known to contain only short helical regions.
In addition to the activities listed above, a growing number of RNA helicases are known to facilitate strand annealing, or its intramolecular version, RNA folding 43. Interestingly, pronounced strand annealing activity has also been seen for DNA helicases of the RecQ family 60. Many RNA helicases display a basal annealing activity that enhances the second order rate constant for duplex formation by a factor of 3 to 10 61. However, several DEAD-box helicases including Ded1p (DDX3) and Mss116p are among the strongest known strand annealers. These proteins enhance the second order rate constant for duplex formation by several orders of magnitude up to the diffusion limit, the physically possible ceiling 61, 62. Although most RNA helicases tested do not require ATP to promote strand annealing, some do; however, it is not clear whether ATP hydrolysis is involved 63. Strand annealing activity, in conjunction with duplex unwinding or protein displacement is thought to enable RNA helicases to catalyze RNA or RNP structure conversions that involve both disruption and formation of RNA/RNP structures 64. Indeed, several RNA helicases promote such RNA structure conversions on model RNAs or on physiological substrates 64, 65.
The spectrum of different activities by RNA helicases raised the question of which mechanistic features underlie the different activities. To date, ATP-dependent or ATP-modulated RNA binding appears to be the smallest common denominator for all RNA helicases. Interestingly, the cellular function of at least one RNA helicase, the DEAD-box protein eIF4A-III (DDX48), is based on ATP-dependent RNA binding 66.
RNA helicases in the cell: specific roles for non-specific enzymes
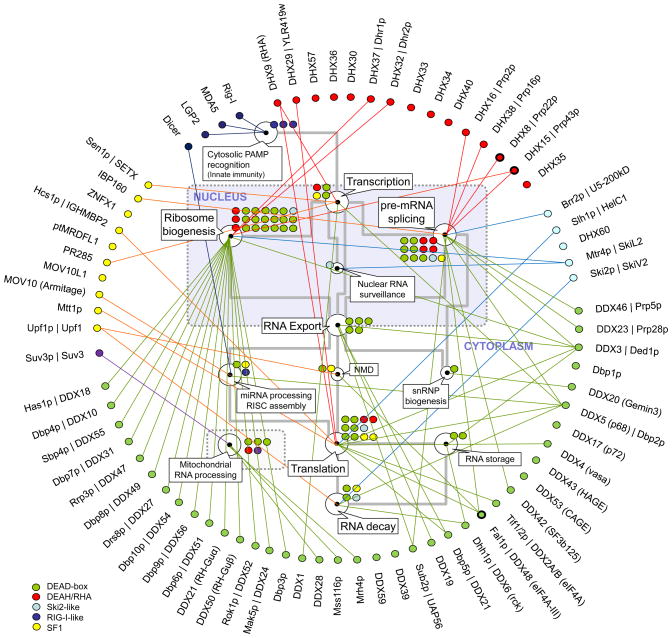
In vitro, the vast majority of RNA helicases do not display sequence or structural preferences, besides the polarity requirements of some enzymes for unwinding RNA duplexes (Box 1). In the cell, however, most RNA helicases function in specific processes such as ribosome biogenesis, pre-mRNA splicing, and translation (Fig. 3). Many RNA helicases appear to participate solely in one process. Several enzymes including Prp43p (DHX15), RNA helicase A (DXH9), eIF4A-III (DDX48), and p68 (DDX5) have been implicated in several processes (Fig. 3).
Figure 3. Cellular roles of eukaryotic RNA helicases.
Selected, basic processes of eukaryotic RNA metabolism are represented by the white circles, as indicated by the callouts (NMD: nonsense mediated decay). The grey lines mark connections between processes. The colored circles represent the number of individual RNA helicases involved in a given process. RNA helicases (yeast and human orthologs) are grouped and color-coded according to their families (see legend at left lower corner). Connectors indicate involvement in one or more processes of RNA metabolism. Clear assignment of Suv3 to either SF is not possible, even though the protein is highly conserved throughout evolution 4. Circles with bold lines emphasize the three RNA helicases (Prp22p, Prp43p, eIF4A-III) for which specific binding site information is available.
Of these proteins, the enzymes functioning in mRNA metabolism are thought to remain bound to a given set of mRNAs, thereby affecting multiple mRNA processing steps 67. Some of these helicases (e.g., DDX3, DHX9, p68) have emerged as key players in the regulation of biological processes, including tumorigenesis 10, 67, 68. Several of these proteins (DHX9, DDX3[SC1]) are also inactivated or co-opted by viruses to enable viral replication 67, 69.
Many RNA helicases have been assigned to specific reaction steps in multi-step processes. For example, it is known for several DEAD-box proteins in which pre-rRNA processing step they participate, and it is comparably well understood at which step in pre-mRNA splicing many spliceosomal RNA helicases function 70, 71. Further roles for the DEAD-box proteins in ribosome biogenesis steps are known for only a few examples, most of which promote the release of small nucleolar RNA (snoRNA) (e.g., refs. 72–74). Many of the spliceosomal RNA helicases appear to provide directionality for inherently reversible transitions during the splicing reaction, most likely by coupling an essentially irreversible ATP turnover to a transition, thereby suppressing reversible steps in the reaction cascade (Box 3).
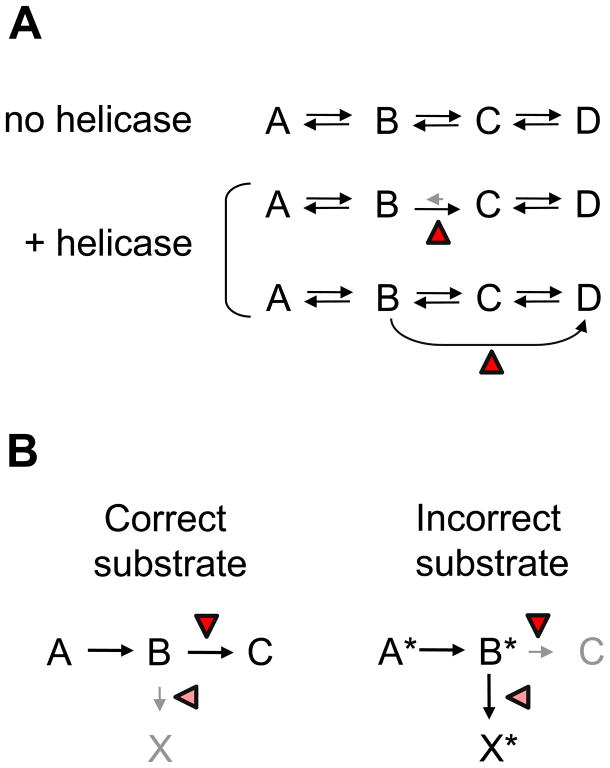
Box 3. Kinetic schemes of RNA helicase functions in RNA metabolic processes.
RNA helicases have been implicated in proofreading processes and in conferring a direction to series of inherently reversible reactions (Fig. I). Without further stimuli (no helicase), a process involving series of inherently reversible reactions would equilibrate, possibly resulting in the continued presence of multiple intermediate species (Fig. IA). Helicases can promote a forward step or inhibit a reverse step, thereby providing direction to the entire process 70. Alternatively, helicases can alter the topology of the reaction mechanism, for example by addition of new reaction pathways. This can be accomplished through remodeling of RNA structures or simply by interactions of the helicase with the RNA 64, 75. Topological alterations of the mechanism changes equilibrium concentrations of final and intermediate species 64. During proofreading, RNA helicases promote the processing pathway for correct substrates or govern the activation of a discard pathway for incorrect substrates, or both 78 (Fig. IB)
From a process point of view, the role of the spliceosomal helicases is similar to the role of RNA helicases that facilitate RNA folding in fungal mitochondria 75, 76 (Box 3). These RNA helicases (Mss116p, CYT-19) function as RNA chaperones that guide RNAs to their native conformation through a series of folding steps 77. The RNA helicases promote forward steps, slow reverse steps or, through remodeling of RNA, open new reactions paths that facilitate correct folding 64, 77 (Box 3). Studies of pre-mRNA splicing also revealed that at least three RNA helicases function in proofreading events at various stages of the splicing reaction 78. As proofreaders, the RNA helicases distinguish between correct and incorrect substrates, and promote further processing of correct substrates while facilitating the discarding of incorrect ones (Box 3).
For the vast majority of RNA helicases, it is not clear which exact physical functions they perform in the cell . Devising physical models for the cellular functions of RNA helicases is the current frontier, but detailed molecular models are only available for very few enzymes. Not surprisingly, attempts to analyze cellular functions of RNA helicases on a detailed molecular level face formidable challenges. Most importantly, RNA targets or RNA binding sites are unknown for the vast majority of RNA helicases, and the lack of sequence or structure specificity greatly complicates target identification. However, without knowing where a helicase binds its substrate, it is essentially impossible to establish physical models for the function of these proteins. Moreover, most, if not all RNA helicases work in the context of large multi-component assemblies and thus interact with many other proteins 43. Because it is known in only few cases whether and how other proteins modulate RNA helicase activities, it is difficult to assess how helicase activities seen in vitro translate into cellular function.
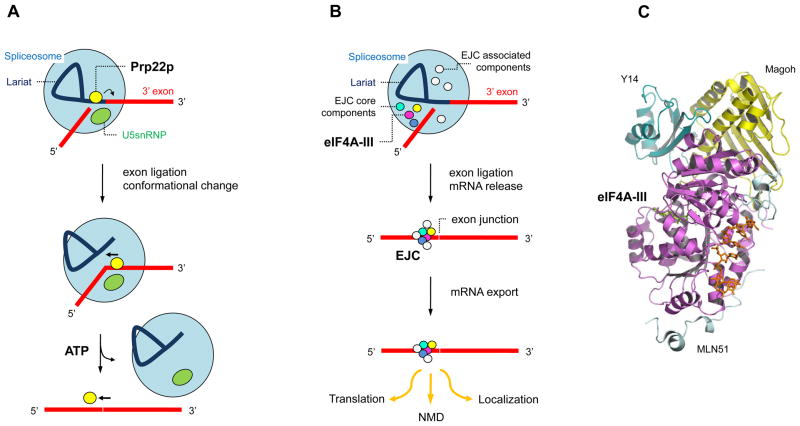
The perhaps most detailed physical models for RNA helicase function in the cell are available for the DEAH/RHA protein Prp22p (DHX8) and the DEAD-box protein eIF4A-III 66, 79 (Fig. 4). Prp22p promotes release of spliced mRNA from the spliceosome and also participates in the exon ligation step 79. During its function, Prp22p changes its position on the RNA several times (Fig. 4A). One of these changes requires ATP and most likely involves directional movement of Prp22p 3’ to 5’ on the mature mRNA, consistent with the unwinding polarity of the protein 79. During this movement, Prp22 breaks several RNA–RNA and RNA–protein contacts, thereby causing the dissociation of the mRNA from the spliceosome 79 (Fig. 4A).
Figure 4. Cellular functions of the DEAH/RHA helicase Prp22p and the DEAD-box protein eIF4A-III.
(A) Prp22p promotes mRNA release from the spliceosome 79. The mRNA exons are marked red; U5snRNP aids in configuring the active site for the second splicing step. Prp22p binds the intron close to the 3' splice site. Upon exon ligation, Prp22p changes it position and binds 3' to the exon junction. Subsequently, Prp22p hydrolyzes ATP and moves towards the 5' end of the spliced mRNA, thereby removing the bound spliceosome. The figure is adapted from ref. 79 (B) eIF4A-III acts as RNA adaptor for the other components of the EJC. The EJC is assembled during mRNA splicing and core components remain stably bound to eIF4A-III. Associated components dissociate at various stages of mRNA metabolism and further associated proteins bind throughout the travel of the EJC bound mRNA 66. The number of associated components shown is not representative. (C) Crystal structure of the core EJC bound to RNA 51. Proteins are labeled and the bound RNA is shown in orange.
eIF4A-III functions as part of the exon junction complex (EJC), a multiprotein complex that is deposited roughly 20 nucleotides upstream of exon–exon junctions during pre-mRNA splicing in higher eukaryotes 66, 80 (Fig. 4B). The EJC remains stably bound to the mRNA after export into the cytoplasm, where it affects several downstream steps of mRNA metabolism including nonsense-mediated decay, translation, and RNA localization 66, 80. Although eIF4A-III can unwind RNA duplexes in vitro, in the cell it functions as adaptor for the other EJC components that ensures their extremely stable association to the RNA 81. Structural studies have shown that the stable binding is accomplished through arrest of the ATP hydrolysis cycle and prevention of hydrolysis product dissociation from the active site of eIF4A-III 50, 51. Trapping of the ATP hydrolysis products is caused by the EJC components Magoh and Y14 82 (Fig. 4B).
The models for both Prp22 and eIF4A-III function provide rationales for their inherent lack of sequence specificity. Both enzymes interact with many different substrates at specific positions. Sequences at these binding sites are probably diverse and pronounced sequence preferences would be detrimental for association of the helicases to such sites.
Binding to various different sites on pre-ribosomal RNA also has been revealed for the DEAH/RHA protein Prp43p 83. Using cross-linking combined with deep sequencing, an approach similar to the crosslinking and immunoprecipitation of RNA–protein complexes (CLIP) technique, binding sites for Prp43p were identified in a genome-wide, unbiased fashion 83. Given the increasing availability of deep sequencing capacity, this approach should become instrumental in locating target sites for RNA helicases. For Prp43p, binding sites on pre-rRNA are now known, and the next challenge will be elucidation of Prp43p function at each position. Suggested roles include snoRNA dissociation, promotion of snoRNA binding, and facilitation of nuclease processing 83.
How Prp43p, Prp22p, eIF4A-III, and most other RNA helicases find their binding sites is not clear. Experimental evidence suggests that several RNA helicases bind co-factors that aid recruitment to complexes such as the spliceosome or to P-bodies 15, 84, 85. Other RNA helicases identify targets on their own, according to a complex code of features on the RNAs. A prominent example is RIG-I, which as part of the innate immune system discriminates cellular from viral RNAs 86. To accomplish this, RIG-I uses a 5’-terminal triphosphate on the viral RNAs, together with RNA secondary structure in the vicinity of the 5’-triphosphate 86.
Although binding to specific sites is most likely critical for the majority of cellular RNA helicases, seemingly indiscriminate action on RNAs by certain RNA helicases might also be important. The DEAD-box helicases Mss116p and CYT-19, RNA chaperones that aid the folding of mitochondrial RNAs in fungi,62, 87, 88 perform their function at least in part by indiscriminate disruption of improperly formed RNA secondary structure that slows RNA folding.64, 89 Sequence and presumably site specificity of these RNA helicases would be incompatible with function on a variety of differently folded substrates 64, 76.
RNA helicases interact constantly with other proteins in the context of larger multi-component complexes 43. An increasing number of studies have focused on identifying interacting proteins (co-factors) and on elucidating their effects on RNA helicase activities. Frequently, co-factors increase unwinding activities or RNA-stimulated ATPase activity of the helicases (e.g., refs.85, 90–92). These stimulations often reflect the ability of the co-factor to increase the RNA affinity of the helicase, which might be a straightforward strategy for recruitment of helicases with inherently poor RNA affinity. A remarkable version of co-factor-mediated stimulation of an RNA helicase was shown for Dbp5p (DDX19/21), a DEAD-box protein that functions in mRNA export. Dbp5p is activated by Gle1p bound to the small molecule inositol hexakisphosphate 93, 94. Gle1p accumulates at the cytosplasmic side of the nuclear pore and thus activates Dbp5p in a localized fashion 93, 94.
Instead of stimulating activities of helicases, co-factors also can inhibit or essentially arrest the ATPase cycle (e.g.eIF4A-III 66). In addition, co-factors can interfere with binding of RNA or prevent association of other proteins. Binding of the cytoplasmic nucleoporin NUP214 blocks the RNA binding site of Dbp5p, and thus its RNA-related functions 52. The tumor suppressor protein programmed cell death 4 (PDCD4), which inhibits translation, binds to the translation initiation factor eIF4A (DDX2), a DEAD-box protein distinct from the EJC component eIF4A-III 95, 96. PDCD4 prevents association of the translation initiation factor eIF4G 97, which is critical for translation initiation and stimulates eIF4A activties 98. Finally, co-factors can bind to RNA helicases without notable effects on activities. This scenario has been observed for the interaction of the DEAD-box protein UAP56 with Aly 99.
Most studies of co-factor-helicase interactions have concentrated on effects on the helicase. Comparably little work has focused on the effect of RNA helicases on other proteins. This aspect has been examined on the interaction between the helicase and the RNA polymerase of hepatitis C virus, and on RNA helicases in the bacterial and the mitochondrial degradosomes 100–102. In all of these cases, functional crosstalk between helicase and other proteins has been observed.
Concluding remarks and future perspectives
This short overview on current topics of research on RNA helicases highlights remarkable progress over the last years. Yet, much remains to be learned about structure, mechanism and physiological function of RNA helicases, before molecular models of cellular functions of these enzymes can be established. On the structural front, the next challenges include obtaining more structures of full length proteins, bound to RNA and ATP analogs, in order to establish detailed structural models of helicase activities for enzymes from different helicase families. On the mechanistic side, focused structure-function studies are needed to verify and complement structural models with dynamic information. In addition, the study of RNA helicases in authentic complexes will probably become a more central theme. With regards to the cellular function of RNA helicases, focus will undoubtedly be on identification of RNA targets, on elucidating means by which the enzymes are recruited to their sites of action, and on devising physical models for RNA helicase function in their physiological environment, building on information from structural and mechanistic studies. Finally, it will be important to examine effects of posttranslational modifications in RNA helicases 103.
Figure I.
Schematic view of the main steps of translocation-based duplex unwinding. Lines represent RNA strands, the oval marks the helicase and the black rectangle indicates the ATP. Only a monomeric enzyme is displayed, but canonical helicases have also been shown to function as oligomers 27. Only selected, main intermediates are shown.
Figure I.
Schematic view of the steps of unwinding by local strand separation. Lines represent RNA strands, the ovals mark the helicase and the small rectangles indicate the ATP/ADP. The different colors of the helicase protomers emphasize their distinct roles in the unwinding process. The asterisk after step 7 highlights the transient nature of the RNA species with a partially opened helix. Adapted, with permission, from ref. 40)
Figure I.
Schemes for RNA helicase functions in the cell. (A) A series of inherently reversible reactions. The red triangle marks the helicase. (B) Proofreading by RNA helicases. The left panel shows processing of the correct substrate, the right panel processing of the incorrect substrate. The triangles mark possible action points of the helicase.
Acknowledgments
Space constraints have limited the depths by which the various aspects of RNA helicase function could be discussed here. I apologize to all colleagues whose recent research was not discussed or directly cited. I thank past and present members of our laboratory for many insightful discussions and for reading the manuscript. Research in our laboratory is funded by the Burroughs Wellcome Fund and by the NIH (GM067700).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tanner NK, Linder P. DExD/H box RNA helicases. From generic motors to specific dissociation functions. Mol Cell. 2001;8:251–261. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 2.Anantharaman V, et al. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Fairman-Williams ME, et al. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singleton MR, et al. Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 6.Gorbalenya AE, Koonin EV. Helicases: amino acid comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 7.Rabhi M, et al. Transcription Termination Factor Rho: A Ring-Shaped RNA Helicase from Bacteria. In: Jankowsky E, editor. RNA helicases. 2010. pp. 243–271. [Google Scholar]; RSC Biomolecular Sciences. Vol. 19 [Google Scholar]
- 8.Kainov DE, et al. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell Mol Life Sci. 2006;63:1095–1105. doi: 10.1007/s00018-005-5450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuma R. Hexameric Viral RNA Helicases. In: Jankowsky E, editor. RNA Helicases. 2010. pp. 213–242. [Google Scholar]; RSC Biomolecular Sciences. Vol. 19 [Google Scholar]
- 10.Zhang S, Grosse F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim Biophys Sin. 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- 11.Taylor SD, et al. The NPH-II helicase displays efficient DNA-RNA helicase activity and a pronounced purine sequence bias. J Biol Chem. 2010;285:11692–11703. doi: 10.1074/jbc.M109.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guenther UP, et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1) Hum Mol Genet. 2009;18:1288–1300. doi: 10.1093/hmg/ddp028. [DOI] [PubMed] [Google Scholar]
- 13.Jackson RN, et al. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, et al. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walbott H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishino T, et al. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Clerici M, et al. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, et al. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godbout R, et al. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J Biol Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- 21.Cui S, et al. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG–I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 22.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Klostermeier D, Rudolph MG. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 2009;37:421–430. doi: 10.1093/nar/gkn947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karginov FV, et al. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J Biol Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 25.Lattmann S, et al. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq372. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Büttner K, et al. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Biol. 2007;14:647–352. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 27.Lohman TM, et al. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 28.Abdel-Monem M, Hoffman-Berling H. Enzymic Unwinding of DNA 2. Chain separtion by an ATP-dependent DNA unwinding enyzme. Eur J Biochem. 1976;65:441–449. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- 29.Durr H, et al. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jankowsky E, Fairman ME. Duplex unwinding and RNP remodeling with RNA helicases. Meth Mol Biol. 2008;488:343–355. doi: 10.1007/978-1-60327-475-3_22. [DOI] [PubMed] [Google Scholar]
- 31.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, et al. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q, et al. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 36.Bizebard T, et al. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 37.Tijerina P, et al. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henn A, et al. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci USA. 2010;107:4046–4050. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, et al. The DEAD-box Protein CYT-19 Uses a Single ATP to Completely Separate a Short RNA Duplex. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, et al. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aregger R, Klostermeier D. The DEAD box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry. 2009;48:10679–10681. doi: 10.1021/bi901278p. [DOI] [PubMed] [Google Scholar]
- 42.Cartier G, et al. Cold adaptation in DEAD-box proteins. Biochemistry. 2010;49:2636–2646. doi: 10.1021/bi902082d. [DOI] [PubMed] [Google Scholar]
- 43.Jankowsky E, Fairman M. RNA helicases - one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Myong S, et al. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumont S, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myong S, Ha T. Stepwise translocation of nucleic acid motors. Curr Opin Struct Biol. 2010;20:121–127. doi: 10.1016/j.sbi.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 48.Serebrov V, et al. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J Biol Chem. 2009;284:2515–2521. doi: 10.1074/jbc.M805460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sengoku T, et al. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 50.Bono F, et al. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 52.von Moeller H, et al. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 53.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linder P. Dead–box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowers HA, et al. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fairman M, et al. Protein displacement by DExH/D RNA helicases without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 58.Tran EJ, et al. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 59.Myong S, et al. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nature Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 61.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 62.Halls C, et al. Involvement of DEAD-box Proteins in Group I and Group II Intron Splicing. Biochemical Characterization of Mss116p, ATP Hydrolysis-dependent and -independent Mechanisms, and General RNA Chaperone Activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chamot D, et al. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J Biol Chem. 2005;280:2036–2044. doi: 10.1074/jbc.M409700200. [DOI] [PubMed] [Google Scholar]
- 64.Bhaskaran H, Russell R. Kinetic Redistribution of Native and Misfolded RNAs by a DEAD-box Chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Q, et al. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol. 2007;368:1087–1100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 67.Tarn WY, Chang TH. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 68.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith DJ, et al. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang XH, Fournier MJ. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kos M, Tollervey D. The Putative RNA Helicase Dbp4p Is Required for Release of the U14 snoRNA from Preribosomes in Saccharomyces cerevisiae. Mol Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 74.Bohnsack MT, et al. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fedorova O, et al. Protein-facilitated folding of group II intron ribozymes. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Del Campo M, et al. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. WIREs: RNA. 2010 doi: 10.1002/wrna.50. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Staley JP, Woolford JL. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tange TO, et al. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 81.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 82.Nielsen KH, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bohnsack MT, et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tritschler F, et al. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka N, et al. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schlee M, et al. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohr S, et al. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohr S, et al. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 89.Del Campo M, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Granneman S, et al. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 2006:3189–3199. doi: 10.1093/nar/gkl419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lebaron S, et al. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009;28:3908–3819. doi: 10.1038/emboj.2009.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maeder C, et al. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alcazar-Roman AR, et al. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 94.Weirich CS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 95.Chang JH, et al. Crystal structure of the eIF4A-PDCD4 complex. Proc Natl Acad Sci U S A. 2009;106:3148–3153. doi: 10.1073/pnas.0808275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loh PG, et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28:274–285. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.LaRonde-LeBlanc N, et al. Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol Cell Biol. 2007;27:147–156. doi: 10.1128/MCB.00867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marintchev A, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen J, et al. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem. 2007;282:22544–22550. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- 100.Carpousis AJ, et al. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 101.Borowski LS, et al. RNA turnover in human mitochondria: More questions than answers? Biochim Biophys Acta. 2010;1797:1066–1070. doi: 10.1016/j.bbabio.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 102.Piccininni S, et al. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J Biol Chem. 2002;277:45670–45679. doi: 10.1074/jbc.M204124200. [DOI] [PubMed] [Google Scholar]
- 103.Kirino Y, et al. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010;285:8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]