A walk-through of the yeast mating pheromone response pathway (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 7.
Abstract
The intracellular signal transduction pathway by which the yeast Saccharomyces cerevisiae responds to the presence of peptide mating pheromone in its surroundings is one of the best understood signaling pathways in eukaryotes, yet continues to generate new surprises and insights. In this review, we take a brief walk down the pathway, focusing on how the signal is transmitted from the cell-surface receptor-coupled G protein, via a MAP kinase cascade, to the nucleus.
Keywords: Yeast mating pheromone, Mitogen-activated protein kinase, Signal transduction, Saccharomyces cerevisiae
1. Introduction
The components of intracellular signaling pathways are dynamically interconnected in a complex network, where the proteins correspond to the nodes of the network and the protein–protein and enzyme–substrate interactions are the links between them. An integrated molecular and systems-level understanding of such networks will require a ‘parts list’ of the nodes, a wiring diagram of the links between them, and experimental understanding of the effects of perturbing individual nodes and links [54,77].
The intracellular signal transduction pathway by which the yeast Saccharomyces cerevisiae responds to the presence of peptide mating pheromone in its surroundings is one of the best understood signaling pathways in eukaryotes; much has been learned from the application of classical and molecular genetics, biochemistry and cell biology. For this pathway, it can be argued that the list of crucial parts is essentially complete, and that the order in which those parts function, particularly with regard to the transmission of the initial signal from outside the cell to the nucleus, is pretty well understood. Furthermore, there is an extensive, though by no means complete, catalog of the links—the protein–protein and enzyme–substrate interactions that connect the parts to each other. The broad challenge for the future, then, is to achieve a detailed understanding of the function of the individual links, and then to synthesize this knowledge into a systems-level understanding of the pathway and the larger network in which it is embedded.
The objectives of this review are to provide a succinct overview of signal transmission through the pathway, with emphasis on recent findings. The focus will be on the pheromone response pathway per se, and not on the fascinating issues concerning how this pathway is integrated with, and insulated from, other pathways within the cell that use similar, or even identical, components. Parallels with more complex eukaryotic cells (mammalian cells in particular) will be highlighted. As this is not intended to be a comprehensive review, I will not attempt to cite a primary reference source for each fact I mention. This information is available in the many excellent reviews of aspects of this pathway that have been published over the last decade [9,32,33,37,56,60,85,115].
2. Overview of the mating process
Saccharomyces cerevisiae (yeast hereafter) is known as bakers or brewer’s yeast for its commercial uses, and as budding yeast for its mode of cell division. The study of the yeast pheromone response pathway began with the isolation of sterile mutants in the laboratories of Mackay and Hartwell in the seventies [59,94,136]. The sterile, or STE, mutants were unable to mate, and those specifically defective in pheromone response did not undergo cell-cycle arrest or change their shape when exposed to purified mating pheromone. Most of the genes in the pathway were cloned in the 1980s and 1990s. Characterization of the gene products continues to the present day, with more recent studies emphasizing functional genomics, aspects of signaling specificity, and detailed characterization of the function of particular protein–protein interactions.
Yeast have two mating types, a and α (genotypes MATa and _MAT_α, respectively). MATa and _MAT_α cells are haploid, and the result of a successful mating will be that two haploid cells of opposite mating type fuse to form a MATa/_MAT_α diploid. _MAT_αcells secrete α-Factor pheromone, a 13 residue peptide (sequence WHWLQLKPGQPMY), and respond to a-Factor. MATa cells secrete a-Factor, a 12 residue peptide (sequence YIIKGVFWDPAC) that is covalently attached to a lipid (farnesyl) group, and respond to α-Factor. When a yeast cell is stimulated by pheromone secreted by a nearby cell of the opposite mating type, it undergoes a series of physiological changes in preparation for mating. These include significant changes in the expression of about 200 genes (about 3% of the genome), arrest in the G1 phase of the cell-cycle, oriented growth toward the mating partner, and, ultimately, the fusion of the plasma membranes of the mating partners, followed shortly thereafter by the fusion of their nuclei. The entire process takes about 4 h.
Many of the same changes also occur when cells of one mating type are exposed to pheromone purified from the opposite mating type. (Since a-Factor is hard to purify, troublesome to synthesize, and sticks to most surfaces, typically MATa cells are treated with synthesized α-Factor peptide.) Cells so treated will arrest their cell-cycle, induce or repress most of the same genes, and even elongate in a default direction determined by the site of their previous bud. These changes can be viewed as the differentiation of vegetatively growing cells into cells with the characteristics of gametes. Cells are not irreversibly committed to this differentiation process, however. Cells that do not successfully mate eventually reenter the cell-cycle and continue vegetative growth as haploids.
The signal transduction pathway that senses the presence of extracellular pheromone and orchestrates the sundry cellular responses to it is known as the yeast mating pheromone response pathway, or mating pathway for short. Several of the components of the mating pathway are also components of distinct signaling pathways that regulate aspects of filamentous invasive growth and the response to certain stresses [91,114,123]. This is not covered here, but has been recently reviewed [18,108,116,142].
3. A walk-through of the mating pathway
3.1. The G-protein-coupled pheromone receptor
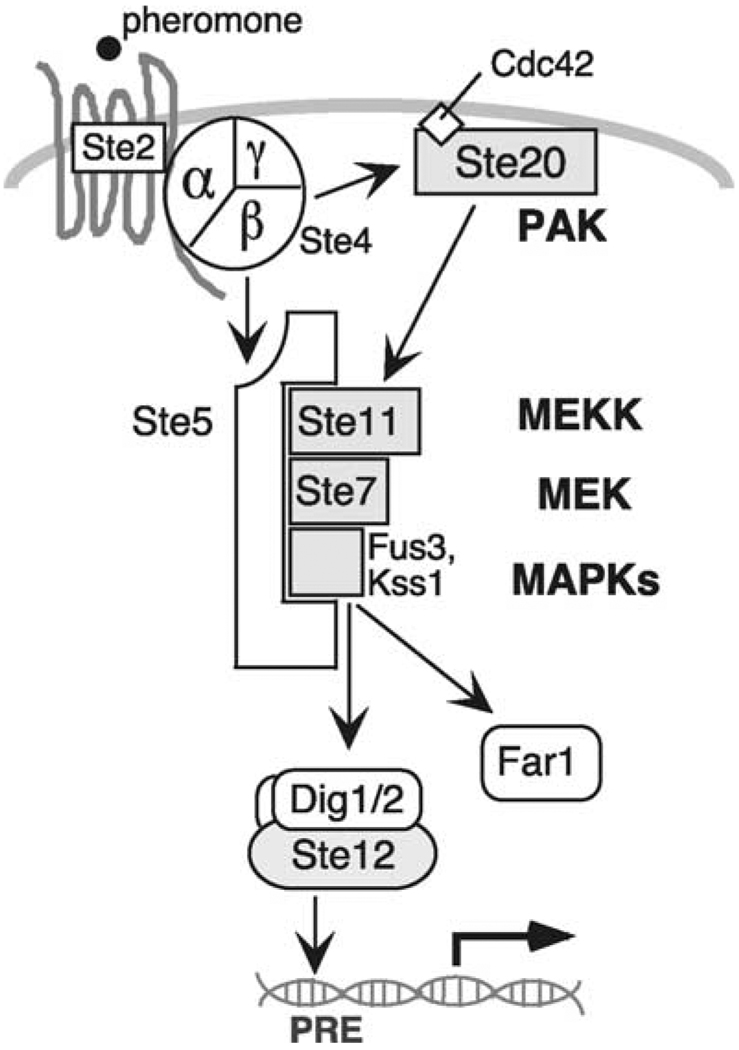
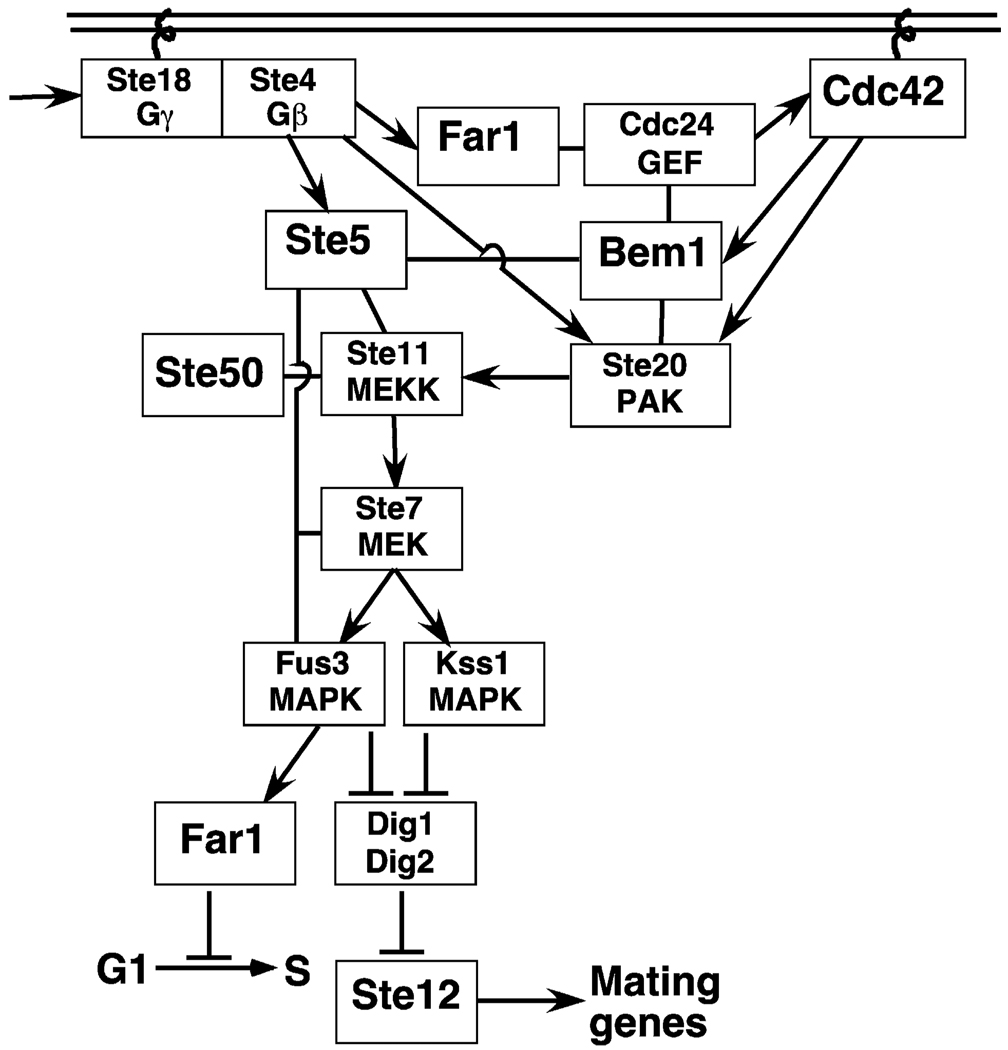
Mating is initiated by the binding of the mating pheromone to a seven-transmembrane, G-protein-coupled receptor (GPCR) on the cell-surface. Receptor-level events are reviewed in much greater detail elsewhere [103a]. As is true for virtually all other GPCR/G-protein modules in eukaryotes, receptor occupancy stimulates the Gα subunit of the G protein to exchange GDP for GTP; GTP-bound Gα then releases the Gβγ heterodimer (see [32] for a recent review of G-protein level events). Gα may also have additional roles in mating besides just regulatingGβγ release [55,102]. Furthermore, Gα may not truly release Gβγ [78]; instead, Gα may remain loosely bound to (and in regulatory communication with) Gβγ and perhaps the receptor as well. The flow of information then proceeds from Gβγ via a four-tiered protein kinase cascade to nuclear transcription factors and other targets. The major components of the pathway and their functions are summarized in Table 1, and a subset of these are depicted in Figs. 1 and 2. Table 2 provides additional information about them, including their closest human homologs. Table 3 explains where some of the names came from.
Table 1.
Some key components of the yeast mating pheromone response pathway
| Protein | Function |
|---|---|
| Ste2/3 | 7-transmembrane-segment, G-protein coupled pheromone receptors |
| Gpa1 | G-protein α subunit |
| Ste4, Ste18 | G-protein βγ subunits |
| Ste5 | Adapter and scaffold, binds Gβ, MAPK cascade kinases, and others |
| Bem1 | Involved in polarity establishment, binds Ste5, Cdc42, Cdc24 and Ste20 |
| Cdc24 | Guanine nucleotide exchange factor (GEF) for Cdc42 |
| Cdc42 | Small rho-like G-protein, binds to Ste20, Bem1, and others |
| Ste20 | PAK (p21-activated protein kinase), activated by Cdc42 |
| Ste11 | MEKK (MEK kinase), activated by Ste20 |
| Ste50 | Binds to N-terminus of Ste11 and aids and/or helps maintain in its activation |
| Ste7 | MEK (MAPK/ERK kinase), activated by Ste11 |
| Kss1, Fus3 | MAP kinases, activated by Ste7 |
| Dig1, Dig2 | MAPK substrates, repressors of Ste12 transcriptional activity |
| Ste12 | MAPK substrate, DNA-binding transcriptional transactivator |
| Far1 | MAPK substrate, inhibits cell-cycle progression, also adapter/scaffold that binds Gβ, Cdc24 and others |
Fig. 1.
Schematic cartoon of selected elements of the yeast mating pheromone response pathway (see text for details).
Fig. 2.
Wiring diagram of selected elements of the yeast mating pheromone response pathway (see text for details).
Table 2.
Size, mass and human homologs of the key players
| Name | Length (aa) | Mass (kDa) | Domains/motifsa | Closest human homologb | ||||
|---|---|---|---|---|---|---|---|---|
| Locus | Name | Identities | E Valuec | Reciprocald | ||||
| Ste2 | 431 | 48 | 7TM (weak) | – | – | – | – | – |
| Ste3 | 470 | 54 | 7TM (weak) | – | – | – | – | – |
| Gpa1 | 472 | 54 | Gα | GNAI2 | Gi alpha 2 | 177/385 (46%) | 1e-67 | No |
| Ste4 | 423 | 47 | WD40 | GNB4 | G beta 4 | 144/386 (37%) | 8e-67 | Yes |
| Ste18 | 110 | 13 | Gγ (weak) | – | – | – | – | – |
| Bem1 | 551 | 62 | SH3 x2, PX, PB1 | SORBS1 | Ponsin | 58/232 (25%) | 4e-09 | Yes |
| Cdc24 | 854 | 97 | CH, RhoGEF, PH, PB1 | VAV3 | Vav3 | 100/461 (21%) | 6e-20 | Yes |
| Cdc42 | 191 | 21 | Rho | CDC42 | Cdc42 | 153/191 (80%) | 2e-88 | Yes |
| Ste5 | 917 | 103 | RING-H2 | – | – | – | – | – |
| Ste50 | 346 | 39 | SAM, RA | – | – | – | – | – |
| Ste20 | 939 | 102 | PBD/CRIB, Kinase | PAK1 | PAK1 | 257/553 (46%) | 1e-123 | Yes |
| Ste11 | 717 | 81 | SAM, Kinase | MAP3K3 | MEKK3 | 128/310 (41%) | 9e-57 | Yes |
| Ste7 | 515 | 58 | Kinase | MAP2K1 | MEK1 | 135/397 (34%) | 5e-56 | No |
| Fus3 | 353 | 41 | Kinase | MAPK1 | ERK2 | 177/346 (51%) | 2e-96 | Yes |
| Kss1 | 368 | 43 | Kinase | MAPK1 | ERK2 | 182/362 (50%) | 7e-96 | No |
| Dig1 | 452 | 49 | – | – | – | – | – | – |
| Dig2 | 323 | 37 | – | – | – | – | – | – |
| Ste12 | 688 | 78 | Homeo (weak) | – | – | – | – | – |
| Far1 | 830 | 94 | RING-H2 | – | – | – | – | – |
| Bar1 | 587 | 64 | Asp-like protease | PGC | Pepsinogen C | 99/369 (26%) | 8e-26 | No |
| Sst2 | 698 | 80 | DEP, RGS | – | – | – | – | – |
| Msg5 | 489 | 54 | Phosphatase | DUSP10 | MKP5 | 44/137 (32%) | 5e-13 | Yes |
| Ptp2 | 750 | 86 | Phosphatase | PTPRC | CD45 | 102/378 (26%) | 5e-21 | No |
| Ptp3 | 928 | 105 | Phosphatase | PTPN6 | SHP-1 | 86/346 (24%) | 2e-16 | No |
Table 3.
What some of the names mean
| Name | Meaning | Why? (phenotype) |
|---|---|---|
| Ste | Sterile | Null mutants cannot mate |
| Gpa1 | G-protein alpha subunit | Named after function |
| Cdc | Cell division control | Cell-cycle arrest at restrictive temperature |
| Fus | Fusion | Null mutants defective for cell fusion during mating |
| Bem | Bud emergence | Budding defect |
| Far | Factor arrest | Null mutants defective for pheromone-imposed cell-cycle arrest |
| Sst | Supersensitive | Null mutants are supersensitive to pheromone |
| Bar | Barrier (to α-factor diffusion) | Null mutants are supersensitive to α-factor pheromone |
| Kss1 | Kinase-suppressor of Sst2 | Multicopy suppressor of sst2 mutant; overproduction of Kss1 inhibits pheromone signaling |
| Dig | Down-regulator of invasive growth | Null mutants exhibit constitutive invasion and derepression of Ste12-regulated genes |
| Ptp | Protein tyrosine phosphatase | Named after function |
| Msg5 | Multicopy suppressor of GPA1 deletion | Overproduction of Mgs5 (a dual-specificity MAPK phosphatase) inhibits pheromone signaling |
3.2. G-protein effectors
Following release from Gα, the membrane-bound Gβγ complex transmits the signal by binding to three different effectors: (1) a Ste5/Ste11 complex; (2) the Ste20 protein kinase, and; (3) a Far1/Cdc24 complex. It is Ste4Gβ that actually binds to each of the effectors, using interaction surfaces that were buried or obscured when it was associated with Gα-GDP; Ste18Gγ anchors the βγ complex to the membrane via covalently attached lipid (farnesyl and palmitoyl) groups. A key result of Gβγ binding to these multiple effectors is that Ste20 and Ste11 are brought near each other; the initial signal is then transmitted further downstream when Ste20 phosphorylates, and thereby activates, Ste11, the first domino in the MAP kinase cascade.
The first Gβγ effector is Ste20. A short conserved motif in the carboxy-terminus of Ste20 binds to Gβγ [81,84]. Ste20 is the founding member of the p21-activated protein kinase (PAK) family [90]. Unactivated, cytoplasmic Ste20PAK is in a low-activity state, because the CRIB domain in its large N-terminal region sterically occludes the active site of the C-terminal kinase domain [80]. In mammalian PAK1, this autoinhibition occurs in trans, in the context of a homodimer [111]. Activation of Ste20 occurs when the CRIB domain binds to a small (21 kD), Rho-like G protein, Cdc42 [3,69]; this interaction antagonizes the ability of Ste20’s CRIB domain to inhibit its kinase domain, thereby permitting autophosphorylation of its now-exposed activation loop [99]. Cdc42, like Ste18Gγ, is permanently tacked to the inner leaflet of the plasma membrane by virtue of a covalently attached lipid (geranylgeranyl) moiety. Hence, another role of Cdc42-Ste20 binding is to localize Ste20 at the membrane. This may also be facilitated by the association of Ste20 with Bem1, which also binds to Cdc42, as well as to two other proteins that are recruited to the membrane in pheromone stimulated cells: Ste5 and Cdc24 (see below) [83,92,103].
The second Gβγ effector is Ste5. An N-terminal region of Ste5, containing a RING-H2 domain, binds to Gβγ near the Ste20 binding site [35,47,66,149]. Ste5 is a large, multifunctional protein that has no catalytic activity, but serves as a binding platform, tugboat, and scaffold for several other proteins. Ste5’s first function is to serve as an adapter, binding to both Gβ and to the Ste11 protein kinase, and thus towing bound Ste11 to the vicinity of the plasma membrane following pheromone stimulation [117]. Here, Ste20 (which is also in the neighborhood by virtue of its association with Cdc42, Gβγ and Bem1) phosphorylates, and thereby activates, Ste11.
The thirdGβγ effector is a complex of the Far1 and Cdc24 proteins [21,105]. A RING-H2 domain in the N-terminal half of Far1 binds to Gβγ; while the C-terminal half of Far1 binds to Cdc24 [21]. Cdc24 is a guanine nucleotide exchange factor (GEF) for Cdc42. Cdc24GEF is complexed tightly to Far1. Similar to how Ste5 functions as an adapter for Ste11 activation (see above), Far1 functions as an adapter for Cdc42 activation. Far1’s adapter function is most analogous to the way Grb2 functions in receptor tyrosine kinase signaling pathways: by binding to the receptor and to Sos, Grb2 brings the Sos exchange factor to the vicinity of the plasma membrane, where Sos’s substrate, Ras, is localized. Analogously, by binding to Gβ and to Cdc24GEF, Far1 brings Cdc24GEF to the plasma membrane, where Cdc24’s substrate, Cdc42, is (literally) hanging. Cdc24 then acts on Cdc42 to promote the exchange of GDP for GTP. GTP-bound Cdc42 binds to several effectors, including Ste20PAK, as detailed above, as well as several other effectors involved in the regulation of cell polarity and the actin cytoskeleton [36,69].
Yeast cells are non motile. They cannot swim, having no cilia or flagella, nor can they crawl; they have a rigid cell wall, and cannot form filopodia like amoeba or mammalian fibroblasts [76]. Rather, although they have ceased dividing, yeast cells elongate by growing asymmetrically in the direction of the nearby mating partner, forming a structure termed a mating projection, and adopting a distended pear-like shape that is termed a ‘shmoo’ [95]. As this shape change, or morphogenesis, is in a particular direction, it is polarized, and as the direction chosen is towards the highest concentration of pheromone, it is chemotropic. The Gβ-Far1-Cdc24-Cdc42 branch of the pathway is crucial for the chemotropic polarized morphogenesis that occurs during mating [21,37,105–107,130,140], as are Cdc42 targets such as Bem1, Bni1, Gic1 and Gic2 [20,24,43]. Cells that crawl use similar regulatory strategies [23]; for example, Gβγ-dependent recruitment of a PAK and a Cdc42 exchange factor also occurs in mammalian chemotaxis [89,101].
Proteins involved in signaling, polarization, cell adhesion, and fusion are localized to the mating projection. As in mammalian cells, this polarized protein localization involves the actin cytoskeleton, cholesterol and sphingolipid-rich lipid rafts, localized exocytosis, and rapid endocytosis to prevent diffusion to equilibrium [4,5,139].
Although the interaction of Gβγ with the Far1/Cdc24 complex is required for pheromone-induced changes in cell polarity, it is not required for initial signal transmission, as shown by the fact that Far1 itself is dispensable for this process [22]. There appears to be enough active Cdc24GEF and Cdc42 constitutively at the membrane to activate the amount of Ste20PAK required for initial signaling [80,117].
3.3. The MAP kinase cascade-overview
Mitogen activated protein kinase (MAPK) cascades are found in all eukaryotes, and are expressed in virtually all tissues. MAPK cascades contribute to the regulation of diverse responses, including, in both yeast and humans, hormone action, cell differentiation, cell-cycle progression, and stress responses [50,88]. The MAPK cascade is a set of three sequentially acting protein kinases. Starting from the bottom and working back up, there is a MAPK (also termed extracellular-signal-regulated kinase, or ERK), which is phosphorylated and thereby activated by a MAPK/ERK kinase (MEK, or MAPKK, or MKK). MEK activity is regulated, in turn, via phosphorylation by the topmost member of the module, a MEK kinase (MEKK). In the yeast mating pathway, the MEKK is Ste11, the MEK is Ste7, and there are two MAPKs, Kss1 and Fus3.
The following is a summary of signal transmission through the MAPK cascade: As a result of Ste5-dependent recruitment to the membrane, the N-terminal regulatory domain of Ste11MEKK is phosphorylated by Ste20PAK. Ste50 is also bound to Ste11, and aids in its activation. Ste11 then activates Ste7MEK by phosphorylating its activation loop, and Ste7MEK, in turn, activates Fus3MAPK and Kss1MAPK, by phosphorylating their activation loops. Distinct regions of Ste5 also bind to Ste7MEK and to the MAPKs. Here, Ste5 is thought to function as a scaffold, co-localizing, sequestering and organizing the component protein kinases of the mating MAPK cascade, thus enhancing signal transmission from MEKK to MEK to MAPK [19,38,49,57,110,118,128,146].
Two very common themes in the regulation of protein kinase activity are: (1) inhibition of the kinase domain by an autoinhibitory domain [132] and (2) regulation of the kinase by phosphorylation of the activation loop, a region of the catalytic domain located between conserved kinase subdomains VII and VIII in the primary structure, just below the catalytic cleft in the tertiary structure [1]. Phosphorylation of the activation loop induces it to refold, causing subtle conformational changes, which reverberate through the rest of the enzyme and increase its catalytic rate by various mechanisms [87]. For example, in MAP kinases, activation loop phosphorylation unblocks the active site and promotes a closure of the upper and lower lobes of the kinase domain that brings the catalytic residues into their correct orientation [70]. Ste20PAK (see above) and Ste11MEKK (see below) are regulated by autoinhibitory domains. In addition, Ste20 (and perhaps Ste11) are also regulated by activation loop phosphorylation. For Ste7MEK and the MAP kinases, activation loop phosphorylation is the primary means of regulation.
3.4. Ste11MEKK
Ste11 consists of an N-terminal regulatory region (comprising roughly half of the protein) and a C-terminal kinase domain. Within the N-terminal regulatory region, three domains have been recognized. First, there is a SAM domain, which binds to the Ste50 protein, followed by a domain that mediates Ste5 binding [67,150], and then a short domain (the catalytic-binding domain, or CBD) that binds to and inhibits the C-terminal catalytic domain [13,137,141]. The CBD is the site of a point mutation (P279S, STE11-1 allele) that constitutively activates Ste11 by weakening the ability of the CBD to bind to and inhibit the kinase domain [133]. The CBD also contains serine and threonine residues that are phosphorylated by Ste20. Ste20-mediated phosphorylation of these residues also antagonizes the ability of the CBD to inhibit the kinase domain, thereby activating Ste11 [141].
Ste50 binds constitutively to the SAM domain of Ste11 via a SAM domain of its own [67,150]. Cells lacking Ste50 are not truly sterile, but are compromised for signaling and mate with a roughly 10–100-fold reduced efficiency, depending upon the strain background. The binding of Ste50 to Ste11 weakens the interaction of the N-terminus of Ste11 with its C-terminus [150]. In so doing, Ste50 may help make the CBD more accessible to Ste20-mediated phosphorylation, or assist in holding phosphorylated Ste11 in a fully open and active conformation, or both.
Ste5 binds to an imprecisely-defined region of Ste11 about 170 residues long that is sandwiched between the SAM domain and the CBD [67]. Ste5–Ste11 binding appears to serve at least three purposes. First, as discussed above, Ste5 serves as an adapter, towing Ste11 to the membrane and near to its activator, Ste20PAK. Second, Ste5, by binding to the N-terminus of Ste11, may, like Ste50, help make the CBD more accessible to Ste20-mediated phosphorylation, and/or assist in holding phosphorylated Ste11 ‘open’. Third, Ste5 also binds to Ste7MEK, and thus may facilitate signal transmission from Ste11MEKK to Ste7MEK.
It is notable that Ste11MEKK has not been reported to bind with measurable affinity to its upstream activator, Ste20PAK, nor to its downstream target, Ste7MEK. Both the Ste20–Ste11 and Ste11–Ste7 interactions, thus, appear to resemble classical, transient enzyme–substrate interactions. As detailed above, however, several other proteins conspire to bring Ste11 and Ste20 to the same region of the membrane, and perhaps to hold them together in a stable multiprotein complex. In addition, Ste5 functions to bring Ste11 and Ste7 together.
There is some confusion in the literature as to whether Ste11 is 717 or 738 residues long. This is because the longest contiguous ORF is 738 residues long, having an extra 21 N-terminal residues. When the transcription start site was mapped by Errede’s lab, however, it was found to be downstream of the first ATG; therefore, translation must start at the second ATG, leading to a 717 residue product [120]. This conclusion is supported by comparison of Ste11 sequences in closely related yeasts [72].
3.5. Ste7MEK and MAPK phosphorylation
Activated Ste11 phosphorylates target residues in the activation loop of Ste7MEK [104,156]. As a result, Ste7 is activated. Activated Ste7 then phosphorylates, and thereby activates, its targets, the MAPKs Kss1 and Fus3 on a threonine and a tyrosine residue in their activation loop [8,41,53,93].
Although Ste7MEK cannot bind stably to Ste11MEKK without help, Ste7 binds directly and with quite high-affinity to its substrates, Kss1MAPK and Fus3MAPK [8]. Ste7-MAPK complexes have a Kd ~5–100 nM, depending on the assay, and a half-life of ~2 min at 30 °C; this is a higher affinity and stability than would be expected for a prototypical enzyme–substrate interaction. Indeed, complex formation does not require the kinase domain of Ste7. Like many other MEKs, Ste7 consists of a highly conserved catalytic domain and a N-terminal extension that exhibits substantially less conservation. It is the first 20 residues of this N-terminal extension that contain the MAPK-binding site, or docking site [7,8]. Similar MAPK-docking sites, or D-sites (consensus sequence (K/R)2–3-X1–6-L/I-X-L/I), are present in the N-terminal extensions of MEKs in organisms representative of many different phyla and even across kingdoms [7,12]. Indeed, the D-sites in mammalian MEK1 [7,151], MEK2 [7], MKK3 and MKK6 [39], and MKK4 [63] have been shown to mediate high-affinity binding to their cognate MAPKs, although the affinity of the mammalian MEK–MAPK interactions (Kd ~5–30 µM [7,63]) is considerably lower than that of the yeast Ste7MEK–MAPK interaction, perhaps because the cellular concentration of the mammalian kinases are higher [48].
It is now widely appreciated that the D-site motif first discovered in Ste7 is found not only in MEKs, but also in transcription factors, phosphatases, scaffolds, other kinases, and other proteins, where it mediates MAPK binding to these substrates and regulators [40,129]. In the yeast mating pathway, putative D-sites are also been found in Gpa1Gα [102], the Ptp3 phosphatase [154], and the Dig1 and Dig2 transcriptional regulators [79]. Hence, D-sites appear to be portable, modular motifs that mediate the interaction of MAPKs with multiple binding partners, contributing to both signal transmission and specificity. Furthermore, the dynamics and specificity of MAPK-mediated signaling is likely to be influenced by the competition between multiple MAPK substrates and regulators for MAPK docking [6,63].
Mutants of Ste7 in which the D-site has been altered or deleted exhibit substantially reduced MAPK binding. When such mutants are introduced into yeast cells in place of wild-type Ste7, however, only a modest defect in pheromone response is observed. This modest defect can be dramatically enhanced, however, by mutations in the Ste5 scaffold that compromise the ability of Ste5 to bind to Ste7 [7]. This observation suggests that scaffolding and docking might have similar, mutually reinforcing roles in achieving efficient signal transmission. In other words, the direct binding of MEK to MAPK, and the binding of both MEK and MAPK to the Ste5 scaffold, may serve much the same purpose. What is this purpose? One possibility is that these stable protein interactions may hold the enzymes together long enough for a relatively slow catalytic phosphotransfer reaction to occur efficiently. Another suggestion is that docking and scaffolding function by making the dual phosphorylation of MAPKs by MEKs processive rather than distributive [19,86]. However, this notion may be inconsistent with evidence that dual phosphorylation cannot occur without prior dissociation of the high-affinity Ste7-MAPK complex, suggesting non-processivity [8]. Regardless of the precise mechanism, it appears that some of the protein–protein interactions in which the MAPKs participate make overlapping, mutually reinforcing contributions to MAPK activation, so that a dramatic phenotype is only observed when multiple links are severed simultaneously.
3.6. MAPK targets
MAPKs, like their cousins, the cyclin-dependent kinases, are proline-directed kinases: they phosphorylate their targets on serine or threonine residues that are immediately followed by a proline. Key substrates of Fus3MAPK and Kss1MAPK are the Ste12/Dig1/Dig2 transcription factor complex and the Far1 protein.
Ste12/Dig1/Dig2
The stimulation of haploid yeast cells with mating pheromone results in the transcriptional induction of at about 200 genes, of which about 100 are induced by at least two-fold [122]. Strains lacking the Ste12 transcription factor are completely defective for these pheromone-induced changes in gene expression [122]. Ste12 is a DNA-binding transcriptional transactivator. Ste12 binds to a DNA motif in the promoters of the genes it regulates, consensus (A/T)GAAACA [58], which is designated the pheromone response element (PRE). Ste12 can also bind combinatorially to composite DNA elements in combination with other transcription factors such as Mcm1 [100] and Tec1 [14,96].
The Dig1 and Dig2 proteins bind to and repress Ste12 [29,135]. In strains lacking Dig1 and Dig2, pheromone-induced genes are constitutively upregulated [10,122,135]. Dig1 and Dig2 display some sequence similarity to each other over a limited region, but appear to repress Ste12 by different mechanisms. Dig2 binds to the DNA-binding domain of Ste12, whereas Dig1 binds to a different region [109].
Fus3MAPK and Kss1MAPK are thought to regulate pheromone-induced gene expression by directly phosphorylating the transcription factors Ste12, Dig1 and Dig2. Fus3 and/or Kss1 must be catalytically active in order for pheromone-induced changes in gene expression to occur [53]. Furthermore, Ste12 [17,65], as well as Dig1 and Dig2 [29,135], are substrates of Fus3 and Kss1. Finally, Dig1 and Dig2 appear to bind Ste12 less tightly following pheromone stimulation [29,135]. These data collectively suggest that MAPK-dependent phosphorylation of Ste12 and/or Dig1/2 alters the ability of Dig1/2 to bind to and repress Ste12. However, it is not known which particular phosphorylation events are crucial, as the target residues have yet to be mapped or mutated.
Ste12-dependent, pheromone-induced genes include positively-acting components of the mating pathway (STE2, FUS3, FAR1), negative feedback regulators of the pathway (SST2, MSG5, GPA1), and genes involved in the process of cell fusion (e.g. FUS1, FUS2, FIG1, FIG2, AGA1) [148]. Ste12 participates in an autoregulatory circuit whereby it binds to its own promoter and upregulates its own expression [82,119]. Ste12 is constitutively bound to some promoters in naive cells, and binds to other promoters only after pheromone stimulation (presumably following Dig2 release) [119,153]. The total number of promoters bound directly by Ste12 seems to be less than 100 [153].
The MAPKs, particularly Kss1, also regulate Ste12 by a novel mechanism: repression of transcription by unactivated MAP kinase [10,11,30,97]. Unphosphorylated Kss1 binds directly to Ste12, and potently represses Ste12-driven transcription [10]. The Dig1 and Dig2 proteins are required cofactors in Kss1-imposed repression of Ste12 [11]; Kss1, by virtue of its ability to bind to both Ste12 and Dig1/2, may help anchor the latter to the former. Fus3 binds much less strongly to Ste12 than Kss1 does [10], and is a correspondingly weaker repressor [30]. Phosphorylation of Kss1 by Ste7 weakens Kss1–Ste12 binding and consequently relieves Kss1-imposed repression, simultaneously activating Kss1 catalytic activity [10]. Repression of transcription by unactivated Kss1 plays a major role in the Kss1-dependent regulation of invasive growth genes. However, mating gene expression is also shaped by this unusual mode of MAPK-dependent regulation [11,31].
Far1
Far1 protein is a multifunctional regulator of the mating process. As detailed above, one function of Far1 is to bind to Gβ and Cdc24GEF, and thereby stimulate the polarized growth of the cell towards its mating partner. A second, apparently independent, function of Far1 is to mediate pheromone-imposed cell-cycle arrest [22]. Mutants of Far1 have been described that separate the arrest and polarity functions [16,52,140]. The mechanism by which Far1 promotes G1 arrest is unclear. It appears to involve the association of Far1 with Cdc28, the cyclin-dependent kinase (CDK) that is the master regulator of the yeast cell-cycle [68,138]. One model proposes that Far1 is a cyclin-dependent kinase inhibitor (CKI) [113], but this is controversial [52]. It is clear, however, that pheromone-induced cell-cycle arrest requires Fus3-mediated phosphorylation of the Far1 protein [52]. Interestingly, relative to Fus3, Kss1 is a poor Far1 kinase [17,112]; this may explain why Kss1 does not support pheromone-imposed arrest as effectively as Fus3.
Microarray studies have shown that about 100 genes are repressed by at least two-fold in pheromone treated cells [122]. Essentially all mating-pheromone-regulated gene repression requires Far1 [122]. Pheromone-regulated gene repression appears, for the most part, to be a consequence of pheromone-imposed cell-cycle arrest; most pheromone-repressed genes are subject to cell-cycle regulation and are expressed preferentially outside G1 phase [122]. On the other hand, pheromone-regulated repression of G1cyclin genes undoubtedly contributes to G1 arrest. Hence, gene repression and cell-cycle arrest are highly interrelated.
Several other ‘Far’ proteins involved in pheromone-imposed arrest have also been identified [26,64,73]. Recent evidence suggests that these may not regulate the initial phase of pheromone-imposed arrest, but are required to prevent premature recovery from arrest [73]. It is not known if any of these proteins are regulated by MAPK phosphorylation.
Other substrates
Other MAPK substrates include several upstream components of the pathway, including Ste5, Ste11 and Ste7; and negative regulators of the pathway including Sst2 and Msg5. With the exception of Sst2 (see next section), the function of these feedback phosphorylations are unclear. The actin-assembly Factor Bni1 is a key Fus3 substrate [99a]. Genetic evidence suggest that there must be other MAPK substrates as well, involved in the regulation of cell-cycle arrest and shmoo formation [27,44].
4. Signal Modulation
In the yeast pheromone response pathway, as in mammalian G-protein-coupled receptor pathways that respond to peptide hormones and other stimuli, negative feedback loops operate at many levels to promote desensitization/adaptation and recovery [33]. This modulation of signal intensity is also crucial for accurate gradient sensing [127]. Some of the negative feedback mechanisms that operate in this pathway are:
- Bar1/Sst1 is an extracellular, pepsin-like protease secreted by MATa cells that degrades α-Factor. BAR1 expression is induced following pheromone stimulation. There is probably not an equivalent activity secreted by _MAT_α cells.
- The pheromone-bound receptor is phosphorylated, mono-ubiquitinated, and then endocytosed [126]. In MATa cells (which express the α-Factor receptor), the kinase responsible for this phosphorylation is probably casein kinase I [45,62], whereas in _MAT_α cells (which express the a-Factor receptor), Fus3MAPK may also participate [46].
- Phosphorylation of the receptor tail further reduces pheromone sensitivity independent of receptor endocytosis [25].
- Sst2 protein, a founding member of the regulator of G protein signaling (RGS) family, accelerates the rate of Gα-mediated GTP hydrolysis by at least 20-fold [2]. The expression of Sst2 is potently induced by pheromone, and Sst2 stability may also be enhanced via phosphorylation by Fus3MAPK [51].
- Following GTP hydrolysis, Gα rebinds to Gβγ, reforming inactive heterotrimer. The expression of Gα is induced by pheromone. Moreover, it has been proposed that Gα may also stimulate desensitization independent of Gβγ sequestration [134].
- There are at least three GTPase-activating proteins (GAPs) for Cdc42, which appear to regulate different subsets of Cdc42 function [131].
- Fus3 controls a negative feedback circuit that limits the magnitude and duration of its own phosphorylation, as well as that of Kss1. This Fus3-dependent feedback circuit plays a crucial role in preventing the mating signal from leaking into other pathways [125]. The relevant target of Fus3 is not yet known.
- Phosphatases operate at every level to reverse the actions of the pathway kinases. For example, the tyrosine phosphatases Ptp2 and Ptp3, and the dual-specificity phosphatase Msg5, act on Fus3MAPK and Kss1MAPK [34,155]. Many of these phosphatase activities are constitutive, but Msg5 is positively regulated at the transcriptional level by pheromone. Dephosphorylation has the potential to eventually reset the pathway to its pre-stimulated state.
- Protein degradation would also eventually lead to the replacement of activated components with newly-synthesized, unactivated ones, thereby resetting the pathway. But in addition, recent studies indicate that the turnover of Ste7 and Ste11 is accelerated by pheromone stimulation [42,145,147].
- As soon as two mating cells fuse, the pheromone response needs to be shut down. Special mechanisms have evolved to accomplish this quickly [74,75,121,124]. A slower, but more permanent solution is then implemented when the transcription of many pathway components is repressed by the a1/α2 diploid-specific heterodimer [61].
5. Where, how fast, and how many?
Where?
As indicated above, the G-protein subunits of the pathway are permanently tacked to membrane via covalently attached lipid groups, and recruit other pathway members, such as Ste20PAK and Ste5, to the membrane when activated. Ste11MEKK and Ste7MEK are predominantly cytoplasmic proteins [143,144], while Ste5 is predominantly found the nucleus, or shuttling between the nucleus and cytoplasm, in resting cells [98,144]. Kss1MAPK is concentrated in the nucleus of resting cells, and this does not change upon pheromone treatment [93]. Fus3MAPK, in contrast, is about equally split between the nucleus and the cytoplasm in unstimulated cells, and concentrates in the nucleus following stimulation [15,28,144]. Ste5, Ste7 and Fus3 localize to tips of mating projections in pheromone-treated cells. Here, Ste5 remains stably bound, but activated Fus3 apparently dissociates from Ste5 and translocates to the nucleus [144].
How fast?
As measured by loss of fluorescence-resonance energy transfer (FRET) between Gα and Gβγ, the G protein is maximally active within 30 s after pheromone addition [152]. Activation of the MAP kinases can be detected within minutes [125]. Changes in gene expression have already begun by 15 min [122].
How many?
There are about 10,000 pheromone receptors on the surface of an unstimulated yeast cell, coupled to about the same number of G-proteins. The amount of Ste5 and Ste11 in the cell is estimated to be between 500 and 1000 molecules [53a]. The same is true of Ste7 [8,53a]. Fus3 and Kss1 are present at about 5000 molecules/cell in resting cells, with Fus3 levels rising about four-fold following pheromone stimulation [8]. The cellular concentration of Dig1, Dig2 and Ste12 is between 1000 and 2000 molecules/cell [53a]. There are only around 100 or so promoters to which Ste12 binds strongly [153]. Some of these have multiple Ste12-binding sites, but it probably takes no more than 1000 Ste12 molecules to occupy all of them.
This counting exercise strongly suggests that substantial amplification does not occur as the signal transits the pathway, except perhaps at the Ste7MEK → MAPK step [48]. Certainly signal amplification could not have been the driving force for the utilization of a four kinase cascade to transmit this signal.
6. Conclusion
The study of the yeast mating pathway played a significant, if not predominant, role in establishing many signaling landmarks and paradigms. A fragmentary and incomplete list of these would include the following: The demonstration that Gβγ subunits transmit the signal to downstream effectors; the combined use of gain and loss-of-function mutants to order gene function in a signaling pathway; insight into how specific extracellular signals regulate cell-cycle progression; the first PAK, MEKK, MEK and MAP kinase cloned from any organism; the discovery of the first MAPK cascade scaffold, and the discovery of the first regulator of G protein signaling. Currently, yeast is one of the lead organisms for functional genomic explorations. In the future, we can anticipate that it will lead us towards an integrated molecular and systems-level understanding of a eukaryotic cell.
Acknowledgements
This work was supported by U.S. National Institute of General Medical Sciences grants GM60366 and GM69013.
References
- 1.Adams JA. Activation loop phosphorylation and catalysis in protein kinases: is there functional evidence for the autoinhibitor model? Biochemistry. 2003;42:601–607. doi: 10.1021/bi020617o. [DOI] [PubMed] [Google Scholar]
- 2.Apanovitch DM, Slep KC, Sigler PB, Dohlman HG. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Galpha protein pair in yeast. Biochemistry. 1998;37:4815–4822. doi: 10.1021/bi9729965. [DOI] [PubMed] [Google Scholar]
- 3.Ash J, Wu C, Larocque R, Jamal M, Stevens W, Osborne M, et al. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics. 2003;163:9–20. doi: 10.1093/genetics/163.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayscough KR, Drubin DG. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr Biol. 1998;8:927–930. doi: 10.1016/s0960-9822(07)00374-0. [DOI] [PubMed] [Google Scholar]
- 5.Bagnat M, Simons K. Cell surface polarization during yeast mating. Proc Natl Acad Sci USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardwell AJ, Abdollahi M, Bardwell L. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell AJ, Flatauer LJ, Matsukuma K, Thorner J, Bardwell L. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J Biol Chem. 2001;276:10374–10386. doi: 10.1074/jbc.M010271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- 10.Bardwell L, Cook JG, Voora D, Baggott DM, Martinez AR, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc Natl Acad Sci USA. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardwell L, Thorner J. A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem Sci. 1996;21:373–374. [PubMed] [Google Scholar]
- 13.Bauman P, Albright CF. Functional analysis of domains in the Byr2 kinase. Biochimie. 1998;80:621–625. doi: 10.1016/s0300-9084(98)80015-1. [DOI] [PubMed] [Google Scholar]
- 14.Baur M, Esch R, Errede B. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol Cell Biol. 1997;17:4330–4337. doi: 10.1128/mcb.17.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackwell E, Halatek IM, Kim HJ, Ellicott AT, Obukhov AA, Stone DE. Effect of the pheromone-responsive G(alpha) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:1135–1150. doi: 10.1128/MCB.23.4.1135-1150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blondel M, Alepuz PM, Huang LS, Shaham S, Ammerer G, Peter M. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 1999;13:2284–2300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitkreutz A, Boucher L, Tyers M. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr Biol. 2001;11:1266–1271. doi: 10.1016/s0960-9822(01)00370-0. [DOI] [PubMed] [Google Scholar]
- 18.Breitkreutz A, Tyers M. MAPK signaling specificity: it takes two to tango. Trends Cell Biol. 2002;12:254–257. doi: 10.1016/s0962-8924(02)02284-5. [DOI] [PubMed] [Google Scholar]
- 19.Burack WR, Shaw AS. Signal transduction: hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211–216. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 20.Butty AC, Perrinjaquet N, Petit A, Jaquenoud M, Segall JE, Hofmann K, et al. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. Embo J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butty AC, Pryciak PM, Huang LS, Herskowitz I, Peter M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 1998;282:1511–1516. doi: 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- 22.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 23.Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- 24.Chen GC, Kim YJ, Chan CS. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Q, Konopka JB. Regulation of the G-protein-coupled alpha-factor pheromone receptor by phosphorylation. Mol Cell Biol. 1996;16:247–257. doi: 10.1128/mcb.16.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherkasova V, Elion EA. far4, far5, and far6 define three genes required for efficient activation of MAPKs Fus3 and Kss1 and accumulation of glycogen. Curr Genet. 2001;40:13–26. doi: 10.1007/s002940100217. [DOI] [PubMed] [Google Scholar]
- 27.Cherkasova V, Lyons DM, Elion EA. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics. 1999;151:989–1004. doi: 10.1093/genetics/151.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi KY, Kranz JE, Mahanty SK, Park KS, Elion EA. Characterization of Fus3 localization: active Fus3 localizes in complexes of varying size and specific activity. Mol Biol Cell. 1999;10:1553–1568. doi: 10.1091/mbc.10.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook JG, Bardwell L, Kron SJ, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 30.Cook JG, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 31.Crosby JA, Konopka JB, Fields S. Constitutive activation of the Saccharomyces cerevisiae transcriptional regulator Ste12p by mutations at the amino-terminus. Yeast. 2000;16:1365–1375. doi: 10.1002/1097-0061(200011)16:15<1365::AID-YEA630>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 32.Dohlman HG. G proteins and pheromone signaling. Annu Rev Physiol. 2002;64:129–152. doi: 10.1146/annurev.physiol.64.081701.133448. [DOI] [PubMed] [Google Scholar]
- 33.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 34.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, et al. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. Embo J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowell SJ, Bishop AL, Dyos SL, Brown AJ, Whiteway MS. Mapping of a yeast G protein betagamma signaling interaction. Genetics. 1998;150:1407–1417. doi: 10.1093/genetics/150.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, et al. A protein interaction map for cell polarity development. J Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elion EA. Pheromone response, mating and cell biology. Curr Opin Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- 38.Elion EA. The Ste5p scaffold. J Cell Sci. 2001;114:3967–3978. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- 39.Enslen H, Brancho DM, Davis RJ. Molecular determinants that mediate selective activation of p38 MAP kinase isoforms. EMBO J. 2000;19:1301–1311. doi: 10.1093/emboj/19.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enslen H, Davis RJ. Regulation of MAP kinases by docking domains. Biol Cell. 2001;93:5–14. doi: 10.1016/s0248-4900(01)01156-x. [DOI] [PubMed] [Google Scholar]
- 41.Errede B, Gartner A, Zhou Z, Naysmith K, Ammerer G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 42.Esch RK, Errede B. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc Natl Acad Sci USA. 2002;99:9160–9165. doi: 10.1073/pnas.142034399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, et al. a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 44.Farley FW, Satterberg B, Goldsmith EJ, Elion EA. Relative dependence of different outputs of the Sacchromyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics. 1999;151:1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y, Davis NG. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol. 2000;20:5350–5359. doi: 10.1128/mcb.20.14.5350-5359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y, Davis NG. Feedback phosphorylation of the yeast a-factor receptor requires activation of the downstream signaling pathway from G protein through mitogen-activated protein kinase. Mol Cell Biol. 2000;20:563–574. doi: 10.1128/mcb.20.2.563-574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Song LY, Kincaid E, Mahanty SK, Elion EA. Functional binding between Gbeta and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- 48.Ferrell JE., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 49.Ferrell JE, Jr, Cimprich KA. Enforced proximity in the function of a famous scaffold. Mol Cell. 2003;11:289–291. doi: 10.1016/s1097-2765(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 50.Garrington T, Johnson G. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 51.Garrison TR, Zhang Y, Pausch M, Apanovitch D, Aebersold R, Dohlman HG. Feedback phosphorylation of an RGS protein by MAP kinase in yeast. J Biol Chem. 1999;274:36387–36391. doi: 10.1074/jbc.274.51.36387. [DOI] [PubMed] [Google Scholar]
- 52.Gartner A, Jovanovic A, Jeoung DI, Bourlat S, Cross FR, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and theonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 53a.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 54.Gilman AG, Taussig R, Ranganathan R, Ross EM. Overview of the Alliance for Cellular Signaling. Nature. 2002;420:703–706. doi: 10.1038/nature01304. [DOI] [PubMed] [Google Scholar]
- 55.Guo M, Aston C, Burchett SA, Dyke C, Fields S, Rajarao SJ, et al. The yeast G protein alpha subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol Cell. 2003;12:517–524. doi: 10.1016/s1097-2765(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 56.Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris K, Lamson RE, Nelson B, Hughes TR, Marton MJ, Roberts CJ, et al. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr Biol. 2001;11:1815–1824. [PubMed] [Google Scholar]
- 58.Harrison R, DeLisi C. Condition specific transcription factor binding site characterization in Saccharomyces cerevisiae. Bioinformatics. 2002;18:1289–1296. doi: 10.1093/bioinformatics/18.10.1289. [DOI] [PubMed] [Google Scholar]
- 59.Hartwell LH. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 61.Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 62.Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ho DT, Bardwell AJ, Abdollahi M, Bardwell L. A docking site in MKK4 mediates high-affinity binding to JNK MAP kinases and competes with similar docking sites in JNK substrates. J Biol Chem. 2003;278:32662–32672. doi: 10.1074/jbc.M304229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horecka J, Sprague GF., Jr Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics. 1996;144:905–921. doi: 10.1093/genetics/144.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hung W, Olson K, Breitkreutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur J Biochem. 1997;245:241–251. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 66.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 67.Jansen G, Buhring F, Hollenberg CP, Ramezani Rad M. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:102–117. doi: 10.1007/s004380000394. [DOI] [PubMed] [Google Scholar]
- 68.Jeoung DI, Oehlen LJ, Cross FR. Cln3-associated kinase activity in Saccharomyces cerevisiae is regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 71.Josefsson LG. Evidence for kinship between diverse G-protein coupled receptors. Gene. 1999;239:333–340. doi: 10.1016/s0378-1119(99)00392-3. [DOI] [PubMed] [Google Scholar]
- 72.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 73.Kemp HA, Sprague GF., Jr Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:1750–1763. doi: 10.1128/MCB.23.5.1750-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J, Bortz E, Zhong H, Leeuw T, Leberer E, Vershon AK, et al. Localization and signaling of G(beta) subunit Ste4p are controlled by a-factor receptor and the a-specific protein Asg7p. Mol Cell Biol. 2000;20:8826–8835. doi: 10.1128/mcb.20.23.8826-8835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim J, Couve A, Hirsch JP. Receptor inhibition of pheromone signaling is mediated by the Ste4p Gbeta subunit. Mol Cell Biol. 1999;19:441–449. doi: 10.1128/mcb.19.1.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimmel AR, Parent CA. The signal to move: D.discoideum go orienteering. Science. 2003;300:1525–1527. doi: 10.1126/science.1085439. [DOI] [PubMed] [Google Scholar]
- 77.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 78.Klein S, Reuveni H, Levitzki A. Signal transduction by a nondissociable heterotrimeric yeast G protein. Proc Natl Acad Sci USA. 2000;97:3219–3223. doi: 10.1073/pnas.050015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kusari AB, Molina DM, Sabbagh W, Jr, Lau CS, Bardwell L. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J Cell Biol. 2004;164:267–277. doi: 10.1083/jcb.200310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamson RE, Winters MJ, Pryciak PM. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leberer E, Dignard D, Thomas DY, Leeuw T. A conserved Gbeta binding (GBB) sequence motif in Ste20p/PAK family protein kinases. Biol Chem. 2000;381:427–431. doi: 10.1515/BC.2000.055. [DOI] [PubMed] [Google Scholar]
- 82.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 83.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, et al. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 84.Leeuw T, Wu C, Schrag JD, Whiteway M, Thomas DY, Leberer E. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 85.Lengeler KB, Davidson RC, D’Souza C, Harashima T, Shen WC, Wang P, et al. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levchenko A, Bruck J, Sternberg P. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lew J. MAP kinases and CDKs: kinetic basis for catalytic activation. Biochemistry. 2003;42:849–856. doi: 10.1021/bi0269761. [DOI] [PubMed] [Google Scholar]
- 88.Lewis TS, Shapiro PS, Ahn NG. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 89.Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- 90.Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 91.Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 92.Lyons DM, Mahanty SK, Choi KY, Manandhar M, Elion EA. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma D, Cook JG, Thorner J. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell. 1995;6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MacKay VL. a’s, α’s and shmoos: mating pheromones and genetics. In: Hall MN, Linder P, editors. The Early Days of Yeast Genetics. Plainview (NJ): Cold Spring Harbor Laboratory Press; 1993. pp. 273–290. [Google Scholar]
- 95.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 96.Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 97.Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 98.Mahanty SK, Wang Y, Farley FW, Elion EA. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- 99.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 99a.Matheos D, Metodiev M, Muller E, Stone D, Rose MD. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the forming Bnilp. J Cell Biol. 2004;165:99–109. doi: 10.1083/jcb.200309089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mead J, Bruning AR, Gill MK, Steiner AM, Acton TB, Vershon AK. Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol Cell Biol. 2002;22:4607–4621. doi: 10.1128/MCB.22.13.4607-4621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meili R, Firtel RA. Two poles and a compass. Cell. 2003;114:153–156. doi: 10.1016/s0092-8674(03)00553-1. [DOI] [PubMed] [Google Scholar]
- 102.Metodiev MV, Matheos D, Rose MD, Stone DE. Regulation of MAPK function by direct interaction with the mating-specific Ga in yeast. Science. 2002;296:1843–1846. doi: 10.1126/science.1070540. [DOI] [PubMed] [Google Scholar]
- 103.Moskow JJ, Gladfelter AS, Lamson RE, Pryciak PM, Lew DJ. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:7559–7571. doi: 10.1128/mcb.20.20.7559-7571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103a.Naider F, Becker JM. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 104.Neiman AM, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: Activation of the yeast MEK homolog STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nern A, Arkowitz RA. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J Cell Biol. 1999;144:1187–1202. doi: 10.1083/jcb.144.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nern A, Arkowitz RA. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol Cell. 2000;5:853–864. doi: 10.1016/s1097-2765(00)80325-1. [DOI] [PubMed] [Google Scholar]
- 107.Nern A, Arkowitz RA. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J Cell Biol. 2000;148:1115–1122. doi: 10.1083/jcb.148.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.O’Rourke SM, Herskowitz I, O’Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- 109.Olson KA, Nelson C, Tai G, Hung W, Yong C, Astell C, et al. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol Cell Biol. 2000;20:4199–4209. doi: 10.1128/mcb.20.12.4199-4209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 111.Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell. 2002;9:73–83. doi: 10.1016/s1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 112.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 113.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 114.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 115.Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- 116.Pryciak PM. MAP kinases bite back. Dev Cell. 2001;1:449–451. doi: 10.1016/s1534-5807(01)00066-1. [DOI] [PubMed] [Google Scholar]
- 117.Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ptashne M, Gann A. Signal transduction. Imposing specificity on kinases. Science. 2003;299:1025–1027. doi: 10.1126/science.1081519. [DOI] [PubMed] [Google Scholar]
- 119.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 120.Rhodes N, Connell L, Errede B. STE11 is a protein kinase required for cell-type-specific transcription and signal transduction in yeast. Genes Dev. 1990;4:1862–1874. doi: 10.1101/gad.4.11.1862. [DOI] [PubMed] [Google Scholar]
- 121.Rivers DM, Sprague GF., Jr Autocrine activation of the pheromone response pathway in matalpha2(™) cells is attenuated by SST2- and ASG7-dependent mechanisms. Mol Genet Genomics. 2003 doi: 10.1007/s00438-003-0914-3. [DOI] [PubMed] [Google Scholar]
- 122.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 123.Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2785. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 124.Roth AF, Nelson B, Boone C, Davis NG. Asg7p-Ste3p inhibition of pheromone signaling: regulation of the zygotic transition to vegetative growth. Mol Cell Biol. 2000;20:8815–8825. doi: 10.1128/mcb.20.23.8815-8825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sabbagh W, Jr, Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schnell JD, Hicke L. Non-traditional Functions of Ubiquitin and Ubiquitin-binding proteins. J Biol Chem. 2003;278:35857–25860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 127.Segall JE. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc Natl Acad Sci USA. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sette C, Inouye CJ, Stroschein SL, Iaquinta PJ, Thorner J. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol Biol Cell. 2000;11:4033–4049. doi: 10.1091/mbc.11.11.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 130.Shimada Y, Gulli MP, Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat Cell Biol. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- 131.Smith GR, Givan SA, Cullen P, Sprague GF., Jr GTPase-activating proteins for Cdc42. Eukaryot Cell. 2002;1:469–480. doi: 10.1128/EC.1.3.469-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Soderling TR. Protein kinases. Regulation by autoinhibitory domains. J Biol Chem. 1990;265:1823–1826. [PubMed] [Google Scholar]
- 133.Stevenson BJ, Rhodes N, Errede B, Sprague GF., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheremone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 134.Stratton HF, Zhou J, Reed SI, Stone DE. The mating-specific G(alpha) protein of Saccharomyces cerevisiae downregulates the mating signal by a mechanism that is dependent on pheromone and independent of G(beta)(gamma) sequestration. Mol Cell Biol. 1996;16:6325–6337. doi: 10.1128/mcb.16.11.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 136.Thorner J. Signal Transduction. In: Linder P, Shore D, Hall MN, editors. Landmark Papers in Yeast Biology. Plainview (NJ): Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 137.Tu H, Barr M, Dong DL, Wigler M. Multiple regulatory domains on the Byr2 protein kinase. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Valdez-Taubas J, Pelham HR. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 140.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van Drogen F, O’Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- 142.van Drogen F, Peter M. MAP kinase cascades: scaffolding signal specificity. Curr Biol. 2002;12:R53–R55. doi: 10.1016/s0960-9822(01)00672-8. [DOI] [PubMed] [Google Scholar]
- 143.van Drogen F, Peter M. MAP kinase dynamics in yeast. Biol Cell. 2001;93:63–70. doi: 10.1016/s0248-4900(01)01123-6. [DOI] [PubMed] [Google Scholar]
- 144.van Drogen F, Stucke VM, Jorritsma G, Peter M. MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- 145.Wang Y, Dohlman HG. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J Biol Chem. 2002;277:15766–15772. doi: 10.1074/jbc.M111733200. [DOI] [PubMed] [Google Scholar]
- 146.Wang Y, Elion EA. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol Biol Cell. 2003;14:2543–2558. doi: 10.1091/mbc.E02-10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang Y, Ge Q, Houston D, Thorner J, Errede B, Dohlman HG. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1-Cullin-F-box complex. J Biol Chem. 2003;278:22284–22289. doi: 10.1074/jbc.M301272200. [DOI] [PubMed] [Google Scholar]
- 148.White JM, Rose MD. Yeast mating: getting close to membrane merger. Curr Biol. 2001;11:16–20. doi: 10.1016/s0960-9822(00)00036-1. [DOI] [PubMed] [Google Scholar]
- 149.Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, et al. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- 150.Wu C, Leberer E, Thomas DY, Whiteway M. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2425–2440. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu B, Stippec S, Robinson FL, Cobb MH. Hydrophobic as well as charged residues in both MEK1 and ERK2 are important for their proper docking. J Biol Chem. 2001;276:26509–26515. doi: 10.1074/jbc.M102769200. [DOI] [PubMed] [Google Scholar]
- 152.Yi TM, Kitano H, Simon MI. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc Natl Acad Sci USA. 2003;100:10764–10769. doi: 10.1073/pnas.1834247100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 154.Zhan X-L, Guan K-L. A specific protein-protein interaction accounts for the in vivo substrate selectivity of Ptp3 towards the Fus3 MAP kinase. Genes Dev. 1999;13:2811–2827. doi: 10.1101/gad.13.21.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhan XL, Deschenes RJ, Guan KL. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- 156.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. Embo J. 1994;13:1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]