Developmental Roles of Pufferfish Hox Clusters and Genome Evolution in Ray-Fin Fish (original) (raw)
Abstract
The pufferfish skeleton lacks ribs and pelvic fins, and has fused bones in the cranium and jaw. It has been hypothesized that this secondarily simplified pufferfish morphology is due to reduced complexity of the pufferfish Hox complexes. To test this hypothesis, we determined the genomic structure of Hox clusters in the Southern pufferfish Spheroides nephelus and interrogated genomic databases for the Japanese pufferfish Takifugu rubripes (fugu). Both species have at least seven Hox clusters, including two copies of Hoxb and Hoxd clusters, a single Hoxc cluster, and at least two Hoxa clusters, with a portion of a third Hoxa cluster in fugu. Results support genome duplication before divergence of zebrafish and pufferfish lineages, followed by loss of a Hoxc cluster in the pufferfish lineage and loss of a Hoxd cluster in the zebrafish lineage. Comparative analysis shows that duplicate genes continued to be lost for hundreds of millions of years, contrary to predictions for the permanent preservation of gene duplicates. Gene expression analysis in fugu embryos by in situ hybridization revealed evolutionary change in gene expression as predicted by the duplication-degeneration-complementation model. These experiments rule out the hypothesis that the simplified pufferfish body plan is due to reduction in Hox cluster complexity, and support the notion that genome duplication contributed to the radiation of teleosts into half of all vertebrate species by increasing developmental diversification of duplicate genes in daughter lineages.
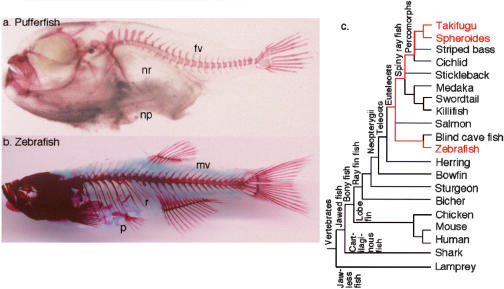
The relationship between genome complexity and phenotypic complexity has yet to be fully understood. In one model, gene number in a taxon correlates with morphological complexity and species diversity (Holland et al. 1994). Alternatively, novel interactions among existing genes may enhance developmental complexity (Wittkopp et al. 2002). Ray-fin fish are species rich and vary in gene number, including numbers of Hox clusters (Amores et al. 1998; Naruse et al. 2000; Malaga-Trillo and Meyer 2001; Scemama et al. 2002; C.H. Chiu, K. Dewar, G. Wagner, K. Takahashi, F. Ruddle, C. Ledje, P. Bartsch, J.L. Scemama, E. Stellwag, C. Fried, et al., in prep.). Some fish are secondarily simplified (Fig. 1a,b). Pufferfish, for example, have fused cranial skeletal elements, lack ribs and pelvis, and have the smallest number of vertebrae among fish (Brainerd and Patek 1998). Skeletal reduction in the pufferfish lineage accompanied genome diminution (Elgar et al. 1999), including seemingly fewer Hox clusters than other teleosts (Aparicio et al. 1997, 2002; Amores et al. 1998; Naruse et al. 2000; Malaga-Trillo and Meyer 2001; Scemama et al. 2002). These data raised the hypothesis that impoverished pufferfish Hox clusters are causally related to simplified morphology (Aparicio et al. 1997; Holland 1997; Meyer and Malaga-Trillo 1999; Snell et al. 1999; Aparicio 2000; Naruse et al. 2000). To test this hypothesis, we constructed a large-insert genomic library for the Southern pufferfish Spheroides nephelus (Amemiya et al. 2001), isolated genomic clones to map pufferfish Hox cluster organization, and compared results with sequences found in genomic databases for the Japanese pufferfish Takifugu rubripes (fugu)(Aparicio et al. 1997; Aparicio et al. 2002).
Figure 1.
Morphology and phylogeny. Pufferfish lack ribs and pelvis and have few vertebrae, as revealed by Alizarin red staining of the skeleton of the pufferfish Takifugu rubripes (a) and the zebrafish Danio rerio (b). (c) A phylogenetic tree for vertebrates (see Nelson 1994), times according to Hedges (2002), Hedges and Kumar (2002), and Santini and Tyler (1999). (fv) Few vertebrae; (mv) many vertebrae;(np) no pelvic appendage; (nr) no ribs; (p) pelvic apparatus; (r) ribs.
Results showed that the organization of pufferfish Hox clusters is similar to that of other teleosts (Amores et al. 1998; Naruse et al. 2000), refuting the hypothesis that morphological simplification is a direct result of the reduction in number of Hox clusters. Pufferfish have duplicate copies of Hox clusters that are present in single copy in tetrapods, including duplicate Hoxa, Hoxb, and Hoxd clusters. Initial analyses suggested that the duplicated pufferfish Hoxa cluster may have arisen in a whole-genome duplication event before the teleost radiation (Aparicio et al. 1997; Amores et al. 1998; Postlethwait et al. 1998, 2002; Wittbrodt et al. 1998; Gates et al. 1999; Meyer and Malaga-Trillo 1999; Meyer and Schartl 1999; Woods et al. 2000; Malaga-Trillo and Meyer 2001), and comparative analysis suggests that this event may have occurred over 300 million years ago (Mya; Taylor et al. 2001).
What processes may have preserved duplicated genes within these duplicated Hox clusters? The classical model for the evolution of gene duplicates suggests that one member of most duplicate pairs should mutate to a pseudogene within a few million generations (Haldane 1933; Ohno 1970; Nei and Roychoudhury 1973; Bailey et al. 1978; Kimura and King 1979; Takahata and Maruyama 1979; Li 1980; Watterson 1983; Ohta 1988; Clark 1994), and investigations of completely sequenced genomes suggests that duplicate genes usually become silenced within about 4 Myr (Lynch and Conery 2000). Many gene duplicates, however, remain for tens of millions of years after the duplication event (Allendorf et al. 1975; Ferris and Whitt 1979; Ahn and Tanksley 1993; Hughes and Hughes 1993; White and Doebley 1998), indicating that other mechanisms must exist to preserve genes, such as the rare evolution of novel positively selected gene functions (Ohno 1970), or the reciprocal sharing of gene subfunctions (Hughes 1994; Force et al. 1999; Stoltzfus 1999). Permanent preservation of gene duplicates is thought to be the outcome of these processes, but the time course of preservation is unclear. The comparison of Hox cluster content in the nested phylogenetic context of two species of pufferfish and zebrafish with tetrapods as outgroup (Fig. 1c) provides the opportunity to test how long this process of permanent preservation can take.
It is presumed that each member of a duplicated pair that has been retained for millions of years must now perform essential and unique functions. To test for function specialization in pufferfish Hox genes, we performed in situ hybridization to pufferfish embryos and compared patterns with those for zebrafish (Prince et al. 1998a,b; McClintock et al. 2002). The results showed evolutionary change in gene expression among teleosts. These experiments support the hypothesis that evolutionary divergence among duplicated genes arising from a genome duplication event in the ray-fin fish lineage contributed to the radiation of teleosts into half of all vertebrate species (Nelson 1994) by increasing developmental diversification of daughter lineages.
RESULTS AND DISCUSSION
Origin and Genomic Organization of Pufferfish Hox Clusters
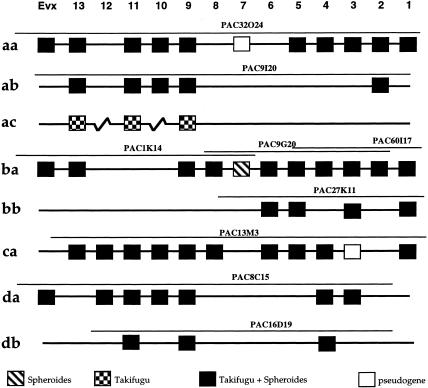
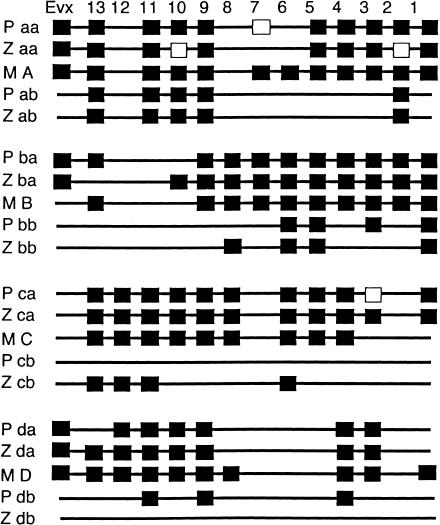
We used degenerate PCR primers (Amores et al. 1998) to identify S. nephalus PACs (Amemiya et al. 2001) that contain Hox cluster genes, and sequenced Hox genes from each PAC to gene-identity Hox cluster gene content. Figure 2 shows PACs isolated and their gene content. We used S. nephalus Hox sequences to query the fugu database (Aparicio et al. 2002) to find fugu orthologs. Results showed that both species have at least seven Hox clusters (Fig. 2). S. nephalus, like medaka (Naruse et al. 2000), has at least two orthologs of mammalian Hoxa, Hoxb, and Hoxd clusters, but a single Hoxc cluster. In contrast, zebrafish (Amores et al. 1998) has two hoxc clusters and just one hoxd cluster (Fig. 3). The finding that pufferfish have the same Hox cluster complement as medaka, which is more closely related to pufferfish than to zebrafish (Fig. 1c), rules out the hypothesis that pufferfish skeletal simplification is due to a grand reduction in Hox cluster number.
Figure 2.
Genomic organization of pufferfish Hox clusters. Hox paralog group shown along the top and cluster designation at left. Plate and well numbers of PACs as well as their genomic extent are sketched. (Filled squares) Genes present in both S. nephalus and T. rubripes. (Squares with diagonal lines) Genes found in S. nephalus butnot T. rubripes. (Checkered squares) Genes found in T. rubripes butnot S. nephalus. (Empty squares) Pseudogenes.
Figure 3.
Comparative genomics of pufferfish (P), zebrafish (Z), and mouse (M) Hox clusters. Paralog group shown along the top and cluster designation at left.
At least three general models can explain duplicate teleost Hox clusters. First, four original clusters may have duplicated in an event before the divergence of the zebrafish lineage and the pufferfish-plus-medaka lineage. Second, all clusters within a lineage may have duplicated in a single event, with one such duplication event occurring in the pufferfish-plus-medaka clade and another event occurring independently in the zebrafish lineage. Third, Hox clusters may have duplicated individually at various times in different lineages. Most data support the first model. First, Hox cluster content is dramatically similar for the two cluster pairs present in both lineages, Hoxa and Hoxb duplicates (Fig. 3). For example, pufferfish and zebrafish Hoxab, Hoxaa, Hoxba, and Hoxbb clusters differ by zero, one, two, and two genes, respectively. What is the likelihood that such a pattern would occur if cluster duplication took place independently in the two lineages and if duplicates were lost at random? Assume that the last common ancestor of zebrafish and pufferfish had a Hoxa cluster with 11 genes (the sum of the Hoxaa plus Hoxab clusters from both species, Evx 13 11 10 9 7 5 4 3 2 1, see Fig. 3). After independent duplication, now assume that six specific genes were lost from one cluster in one of the two lineages to give the content of the Hoxab cluster in the zebrafish lineage. To calculate the probability that the other lineage had the same losses in one of the clusters, note that there is only one way to choose the same six locations, but 11 choose six ways of distributing six losses to 11 sites. So, the probability of this occurrence is 5!6!/11! = .0021645. The caveat to this calculation is that it groups zebrafish Hoxaa with pufferfish Hoxaa, and zebrafish Hoxab with pufferfish Hoxab, and if the original Hoxa cluster duplicated separately in each species, there is no such natural grouping. For the ancestral Hoxb cluster, assuming independent duplication and that the ancestral Hoxb cluster had 12 genes when it duplicated (Evx 13 10 9 8 7 6 5 4 3 2 1; see Fig. 3), then given one loss in the pufferfish lineage and one loss in the zebrafish lineage (the Hoxba clusters), the chance they would overlap is 1/12. Given eight losses in the pufferfish lineage and eight losses in the zebrafish lineage (the Hoxbb clusters), the chance that the losses would overlap seven or more times is [(8 choose 7)(4 choose 1) + 1]/(12 choose 8)= 0.067. These calculations show that the distribution of genes among these duplicated Hox clusters is unlikely to have occurred by chance. The most likely cause is that duplication and gene loss in the a and b copies occurred before lineage divergence, or alternatively, that the probability of Hox cluster gene loss deviates enormously from random between two initially identical duplicated clusters.
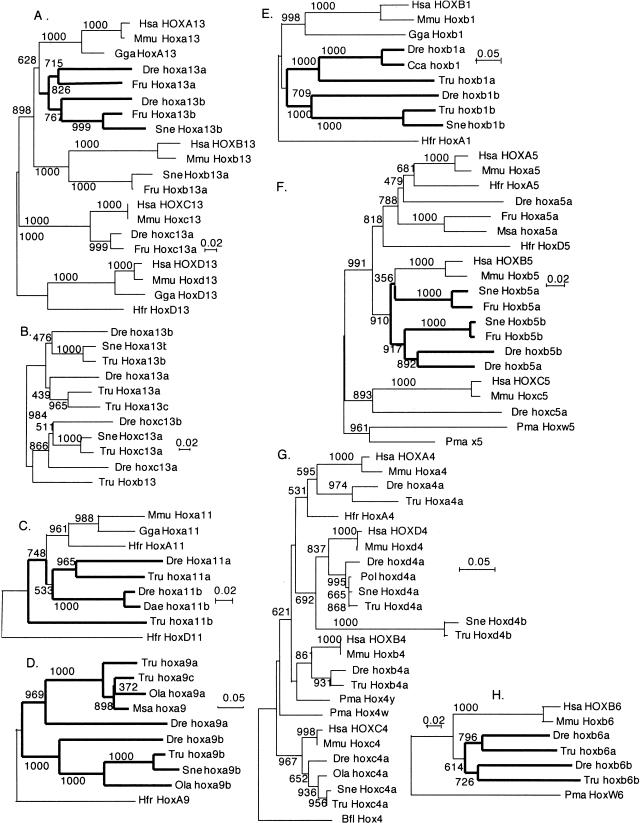
In addition to similar gene content, phylogenetic analysis also supports the duplication-first model. For six original genes, both lineages retain two copies (Fig. 4). Phylogenetic analysis of four of these (Hoxa13, Hoxa9, Hoxb1, and Hoxb6)supports the duplication-first model (Fig. 4A,D,E,H). In the fifth pair (Fig. 4C), Hoxa11a genes group as expected by the duplication-first model, but fugu Hoxa11b is an outgroup, perhaps due to long-branch attraction (Felsenstein 1978). In the final pair (Fig. 4F), zebrafish Hoxb5a and Hoxb5b branch as sisters, contrary to the duplication-first model, but pufferfish Hoxb5a and Hoxb5b do not branch as sisters, thus failing to support the other two models as well. Because most trees rule out the lineage-divergence-first model, but are consistent with the duplication-first model, we conclude that teleost Hox clusters duplicated before divergence of zebrafish and pufferfish lineages. Because these lineages separated at the base of the teleost radiation (Fig. 1b), at least euteleosts share these duplications, which probably occurred in a whole-genome duplication event that occurred sometime between divergence of bichir and Neopterygii and the divergence of pufferfish and zebrafish lineages (Amores et al. 1998; Postlethwait et al. 1998; Taylor et al. 2003; C.H. Chiu, K. Dewar, G. Wagner, K. Takahashi, F. Ruddle, C. Ledje, P. Bartsch, J.L. Scemama, E. Stellwag, C. Fried, et al. in prep.).
Figure 4.
Phylogenetic relationships of pufferfish Hox clusters. Neighbor-joining trees are based on amino acid sequences as described (Amores etal. 1998). (A) Paralog group 13, exon 1, and exon 2. The tree is as expected if Hox cluster duplication occurred before the divergence of pufferfish and zebrafish lineages. (B) Paralog group 13, exon 2 only (due to limited sequence availability). The tree shows that Hoxa13c is the sister group to Hoxa13a. (C) Hoxa11a of pufferfish and zebrafish group as sisters, but Hoxa11b orthologs do not. (D) Hoxa9 tree strongly supports duplication before lineage divergence, and the close similarity of the Hoxaa and Hoxac clusters. (E,F) The Hoxb1 and paralog group-5 trees support duplication before lineage divergence. (G) The paralog group-4 tree shows rapid evolution of Hoxd4b. (H) The Hoxb6 tree supports duplication before lineage divergence. Alignments and accession numbers are available as Supplemental material. Numbers are bootstrap values per 1000 runs. (Cca) Carassius carassius, crucian carp; (Dae) Danio aequipinnatus, giantdanio; (Bfl) Branchiostoma floridae, amphioxus; (Dre) Danio rerio, zebrafish; (Gga) Gallus gallus, chicken; (Hfr) Heterodontus francisci, horned shark; (Hsa) Homo sapiens, human; (Mmu) Mus musculus mouse; (Msa) Morone saxatilis, striped bass; (Ola) Oryzias latipes, medaka; (Pma) Petromyzon marinus, lamprey; (Pol) Paralichthys olivaceus, Japanese flounder; (Sne) Spheroides nephalus, Southern pufferfish; (Tru) Takifugu rubripes, Japanese pufferfish.
Continuing Evolution of Pufferfish Hox Clusters
Although teleost Hox cluster duplicates probably arose in a single event, their structure and contents continue to evolve. Evidence for a large-scale change is found in Scaffolds (S007630, S011648, and S001602, Release 2)we identified in the fugu database that contains copies of Hoxa9, Hoxa11, and Hoxa13, tentatively called here the Hoxac complex (Fig. 2). The phylogenetic relationship of the three Hoxa clusters in fugu shows that Hoxac and Hoxaa clusters group as sisters (Fig. 4B,D). The Hoxac cluster is probably not an allele of Hoxaa because their nucleotide sequences differ by 14.1% in coding regions of Hoxa9, Hoxa11, and Hoxa13, far more than alleles in even widely divergent zebrafish populations, which is ∼0.5% (Stickney et al. 2002). These three coorthologs of the tetrapod Hoxa cluster should probably be called Hoxaaa, Hoxaab, and Hoxab, but that nomenclature seems too unwieldy for now. Assuming that hoxac is absent from zebrafish (Amores et al. 1998), at least a portion of the Hoxaa cluster was duplicated in the pufferfish lineage after it diverged from the zebrafish lineage. The most likely hypothesis is that after the duplication of all four original Hox complexes and the subsequent divergence of zebrafish and pufferfish lineages, a portion of the Hoxaa cluster reduplicated in the pufferfish lineage to produce the Hoxac cluster. We did not find the Hoxac cluster in S. nephalus, although the hox9 tree (Fig. 4D) tentatively suggests that it might have occurred before the divergence of pufferfish, medaka, and striped seabass.
Surprisingly, the Hoxd11b gene in both pufferfish species has a second intron with appropriate splice acceptor and donor sequences in the homeobox, in contrast to other vertebrate Hox cluster genes. Because this intron is absent from tetrapod Hoxd11 and from teleost Hoxd11a, it is a new acquisition after Hox cluster duplication in ray-fin fish.
Further recent evolution in teleost Hox clusters is shown by Hoxa7a, which is a pseuodogene in both pufferfish species, but has normal structure in striped bass and tilapia (Snell et al. 1999; Santini et al. 2003), which are more closely related to pufferfish than to either medaka or zebrafish (see Fig. 1c). The absence of Hoxa7 from zebrafish (Amores et al. 1998) and bichir (C.H. Chiu, K. Dewar, G. Wagner, K. Takahashi, F. Ruddle, C. Ledje, P. Bartsch, J.L. Scemama, E. Stellwag, C. Fried, et al., in prep.) demonstrates multiple independent losses among ray-fin fish.
Teleost Hox clusters show that silencing of duplicated genes can continue far longer than thought previously. Hoxb7a is absent from fugu (Aparicio et al. 1997) but present in S. nephalus (Fig. 2). Both duplicate copies of Hoxb7 must have remained intact from the time of the duplication event about 350 Mya (Taylor et al. 2001) until the divergence of Spheroides and Takifugu lineages only 5-35 Mya (Santini and Tyler 1999). Thus, both duplicate copies were maintained for about 300 Myr before the loss of one copy in the Takifugu lineage after it diverged from the Spheroides lineage. This is hundreds of millions of years longer than the permanent preservation of gene duplicates has been thought to take (Nei and Roychoudhury 1973; Kimura and King 1979; Li 1980; Watterson 1983; Ohta 1988; Clark 1994; Lynch and Conery 2000). In addition, at least four additional duplicates have one intact gene and one detectable pseudogene in teleost Hox clusters (Fig. 3; Aparicio et al. 1997; Amores et al. 1998), and these arose after pufferfish and zebrafish lineages diverged. Because molecular clock estimates suggest that the ray-fin genome duplication occurred about 350 Mya (Taylor et al. 2001), and that zebrafish and pufferfish lineages diverged about 284 Mya (Kumazawa et al. 2000), genes that have recently become pseudogenes must have been retained for tens or hundreds of million years without being lost or permanently preserved. Theory and analysis of sequenced genomes suggest that one copy of a duplicate usually becomes nonfunctional within a few million generations (Li 1980; Lynch and Conery 2000), and that duplicates retained longer probably become essential either by evolving novel, positively selected functions (Ohno 1970)or by partitioning ancestral vital subfunctions (Force et al. 1999). The long persistence of both duplicates, but eventual loss of one copy discovered here suggests either that fully redundant duplicates can be retained far longer than thought previously, or that evolutionary mechanisms that initially preserved both duplicates within a few million years of the duplication event later changed so that one copy secondarily became nonessential.
Expression of Hox Cluster Genes in Fugu Embryonic Development
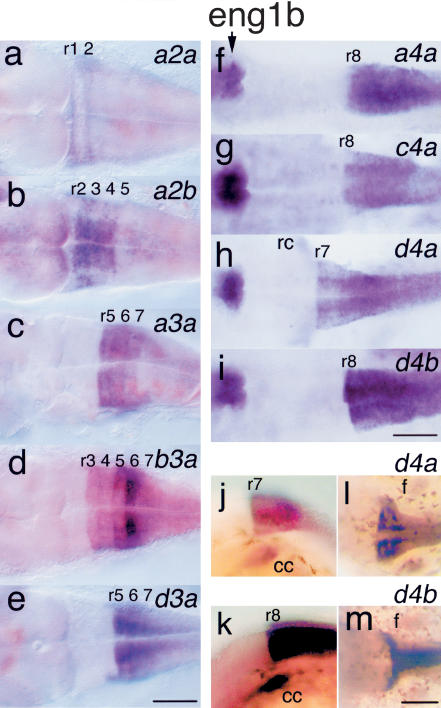
Expression patterns of pufferfish Hox genes support suggestions that gene duplication may facilitate the evolution of new functions and the reciprocal retention of ancestral functions (Ohno 1970; Hughes and Hughes 1993; Force et al. 1999). Our results showed that pufferfish have two orthologs of mammalian HOXD4, but zebrafish has just one (Fig. 3). In fugu, Hoxd4a is expressed with a sharp anterior boundary at hindbrain rhombomeres r6/r7 (Fig. 5h) as it is in tetrapods, zebrafish, and flounder (van der Hoeven et al. 1996; Prince et al. 1998b; Suzuki et al. 1998; Nolte et al. 2003). The duplicate gene Hoxd4b, however, is expressed with a more caudal anterior border (Fig. 5i). The anterior boundary of Hoxd4 expression in mouse is regulated by elements conserved between zebrafish and mouse (Morrison et al. 1996; Nolte et al. 2003). This suggests that fugu Hoxd4b has lost some ancestral subfunctions. Hoxd4a is expressed weakly in hindbrain and neural crest, whereas Hoxd4b is expressed strongly in crest and hindbrain, suggesting quantitative subfunction partitioning (Force et al. 1999) in this pair (Fig. 5j-m). Further analysis is necessary to identify the molecular genetic mechanism for the evolved differences in gene expression for Hoxd4 duplicates, and whether the differences in expression of the single hoxd4 gene in zebrafish and the differently evolved duplicate Hoxd4 genes in pufferfish are related to the evolution of skeletal differences between the species.
Figure 5.
Expression of Hox genes in 5-d fugu embryos. (a) Hoxa2a is expressed in an apparently novel striped pattern in r1 and r2. (b) Hoxa2b is expressed in r2-r5 as in other vertebrates. (c) Hoxa3a has an anterior boundary at the r4/r5 border as in other vertebrates. (d) Hoxb3a has strong expression in r5 and r6 and weak expression in r4 as in zebrafish, but with weak expression extending into r3 in fugu and not in zebrafish (Prince etal. 1998b). (e) Hoxd3a expression mimics that of zebrafish, with an anterior limit at the r5/r6 border, and a small lateral/ventral group of r5 cells (Prince etal. 1998b). Expression of eng1b atthe midbrain/hindbrain border serves as a marker for rhombomere position in f-i. (f) Hoxa4a is expressed with a diffuse anterior expression boundary near the r7/r8 border as in zebrafish (Prince et al. 1998b). (g) Hoxc4a is expressed with a diffuse anterior boundary, and as in zebrafish, the border lies within r7, atleastmedially, and interneurons express eng1b as in zebrafish (Force et al. 1999). (h) Hoxd4a transcript tissue distribution as in other vertebrates. (i) Hoxd4b has a more caudal anterior expression border than Hoxd4 in other vertebrates. (j) Hoxd4a in lateral view. (k) Hoxd4b in lateral view. (l) Hoxd4a in dorsal view showing fin buds. (m) Hoxd4b in dorsal view showing fin buds. Scale bar, 100 μ. (cc) Cranial crest; (f) fin buds.
A novel expression pattern of one Hox gene is tantalizingly related to pufferfish puffing. Fugu Hoxa2b is expressed in hindbrain rhombomeres r2-r5 with sharp borders (Fig. 5b) like hoxa2b in zebrafish and Hoxa2 in mouse (Prince and Lumsden 1994; Prince et al. 1998b; Hunter and Prince 2002). The duplicate Hoxa2a gene, however, is a pseudogene in zebrafish (Amores et al. 1998) but an expressed gene in pufferfish (Fig. 5). Fugu Hoxa2a has a striped expression pattern in r1 and r2 (Fig. 5a), a pattern not reported previously in r1, although in zebrafish, hoxa1a is expressed in small bilateral cell clusters in r1-r3 (McClintock et al. 2001). In fact, ectopic expression of Hoxa2 in r1 of chicken embryos causes motor neuron development in r1, which normally has no motor neurons (Jungbluth et al. 1999). One can speculate that the evolutionary invention of the buccal pump in pufferfish that puffs the stomach with water (Brainerd and Patek 1998) might have involved the origin of new motor neurons stimulated by novel expression of Hoxa2a in r1.
Evolution of the Genomic Organization of Vertebrate Hox Clusters
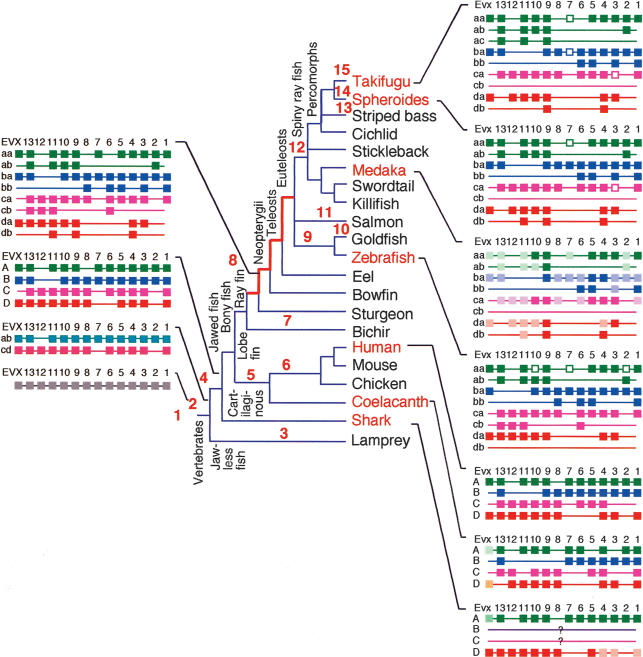
The results of these experiments and those of the accompanying paper (C.H. Chiu, K. Dewar, G. Wagner, K. Takahashi, F. Ruddle, C. Ledje, P. Bartsch, J.L. Scemama, E. Stellwag, C. Fried, et al., in prep.) allow us to paint a more complete picture of vertebrate Hox cluster evolution (Fig. 6). The ancestral vertebrate Hox cluster possessed by the last common ancestor of surviving vertebrates probably consisted of 13 Hox genes and an Evx gene (Fig. 6, #1), even though amphioxus, a cephalochordate representing the sister group of the vertebrates has 14 Hox cluster genes (Ferrier et al. 2000). Evidence from lamprey, a jawless vertebrate, suggests that the ancestral vertebrate Hox cluster probably duplicated once before the lamprey lineage diverged (Fig. 6, #2), and once again after divergence (Fig. 6, #3; Escriva et al. 2002; Force et al. 2002; Irvine et al. 2002). After the initial Hox cluster duplication event (Fig. 6, #2), the paralogy group-12 gene was lost from one copy, which became the proto-ab cluster, and group-7 and group-2 paralogs were lost from the other copy, which became the protocd cluster. These two clusters probably duplicated in an ancestor of jawed fish (Fig. 6, #4), because the horned shark Heterodontus francisci, a cartilaginous fish, has orthologs of mammalian Hoxa and Hoxd clusters (Kim et al. 2000; Chiu et al. 2002). In our phylogenetic trees, the horned shark Hoxa paralogs generally group with the tetrapod clade as a sister to the ray-fin clade (Fig. 4C,D,F, but not G), although bootstrap values are not high. As yet, there is no information about the predicted Hoxb and Hoxc clusters in the shark; they may be missing or more likely have not yet been identified.
Figure 6.
The evolution of vertebrate Hox cluster genomic organization. See text for explanation.
After the second round of Hox cluster duplication in early vertebrates (Fig. 6, #4), but before the divergence of cartilaginous and bony fish, there were cluster-specific gene losses, including the loss of paralogy group-8 from the Hoxa cluster, group-11 from the Hoxb cluster, Evx from the Hoxc cluster, and group-6 from the Hoxd cluster (Fig. 6, #4). The next event was the divergence of ray-fin and lobe-fin fish. The common ancestor of lobe-fin fish had four Hox clusters (Koh et al. 2003), including Hoxc1 (Fig. 6, #5), which is present in the coelacanth but was lost in the tetrapod lineage (Fig. 6, #6). The ancient Evx gene adjacent to the Hoxb cluster was also lost in the lobe-fin, but not the ray-fin lineage.
Among ray-fin fish, the bichir is the only basally diverging lineage that has yet been investigated (Fig. 6, #7), and results show a single Hoxa cluster, suggesting that the last common ancestor of ray-fin fish had just four Hox clusters (C.H. Chiu, K. Dewar, G. Wagner, K. Takahashi, F. Ruddle, C. Ledje, P. Bartsch, J.L. Scemama, E. Stellwag, C. Fried, et al., in prep.). More work needs to be done on this and other basally diverging ray-fin fish, because five group-9 genes have been identified in the bichir (Ledje et al. 2002), which may indicate either tandem duplication or allelic variation of a group-9 gene, or maybe single cluster duplication, or loss of several duplicated clusters in the bichir lineage, leaving a single Hoxa cluster.
Here, we show that two species of pufferfish have at least seven Hox clusters, and because other teleost fish also have duplicates of tetrapod Hox clusters (Misof and Wagner 1996; Amores et al. 1998; Snell et al. 1999; Naruse et al. 2000; Malaga-Trillo and Meyer 2001; Aparicio et al. 2002), there must have been a duplication of all four clusters originally possessed by the last common ancestor of extant bony fishes (Fig. 6, #8). The duplication of at least the Hoxa cluster occurred after the divergence of the bichir and other ray-fin fish (C.H. Chiu, K. Dewar, G. Wagner, K. Takahashi, F. Ruddle, C. Ledje, P. Bartsch, J.L. Scemama, E. Stellwag, C. Fried, et al., in prep.), suggesting that the genome duplication event happened in the time period denoted by the thick red lines in Figure 6.
The time at which Hox clusters duplicated in ray-fin phylogeny has not yet been determined precisely, and the estimate currently spans several hundred million years. It is important to define more accurately the timing of this duplication event, because zebrafish genetic mapping experiments showed that Hox cluster duplication occurred in the context of an ancient whole-genome duplication event (Aparicio et al. 1997; Amores et al. 1998; Postlethwait et al. 1998; Gates et al. 1999; Meyer and Malaga-Trillo 1999; Meyer and Schartl 1999; Woods et al. 2000; Taylor et al. 2001, 2003; Van de Peer et al. 2001, 2002; Postlethwait et al. 2002). It has been hypothesized that the genome duplication event contributed to the vast radiation of teleosts (Amores et al. 1998; Postlethwait et al. 1998). A theoretical reason for thinking that the ray-fin genome duplication could have played a major role in the teleost radiation is the differential resolution of duplicated genes, as evidenced here by the lineage-specific loss of Hox cluster genes and entire Hox clusters, and by lineage-specific gene expression patterns, as with Hoxd4 genes. Such reciprocal loss of duplicated genes or duplicated gene subfunctions in different lineages could act as reproductive isolating mechanisms by decreasing the fitness of hybrids when separately evolving populations come into recontact (Lynch and Conery 2000; Lynch and Force 2000). If future phylogenetic studies show that the ray-fin genome duplication occurred hundreds of millions of years before the teleost radiation, then it is unlikely to have played a major role in spurring lineage divergence among teleost fish. Key lineages to query regarding the timing issue include the Japanese eel, a basally diverging teleost amenable to analysis of embryonic development (Kurokawa et al. 2002), and the bowfin, a nonteleost ray-fin lineage diverging just before the teleost radiation.
Genomic analysis of pufferfish, zebrafish, and medaka Hox clusters permits inferences regarding Hox clusters present in the last common ancestor of euteleosts (Fig. 6, #8). First, it is clear that this organism had two copies of each tetrapod cluster, because the cluster missing in pufferfish and medaka is present in zebrafish, and the cluster missing in zebrafish is present in pufferfish and medaka. Second, at least one gene may have been lost in the ray-fin lineage after the divergence of bichir, and that is Hoxa6, which is present in bichir, but absent from examined teleosts, although this Hoxa6 might also have been lost independently in Hoxaa and Hoxab clusters after duplication Fig. 6, #7 and #8). Third, several additional genes dropped out probably before the ray-fin genome duplication, including Hoxd1, Hoxd5, and Hoxd8.
As teleost lineages diverged, so did their Hox cluster content. In the zebrafish lineage (Fig. 6, #9), the entire hoxdb cluster appears to have been lost. We have identified and mapped to LG6 the zebrafish ESTs orthologous to human loci that directly flank the HOXD cluster, and are currently determining the genomic footprint left by the missing zebrafish cluster. In addition, hoxa10a, hoxa7a, and hoxa2a have all become pseudogenes in the zebrafish lineage, and hoxb13a and hoxb3b have been lost. Some Cyprinids other than zebrafish, including goldfish and some carp species (Risinger and Larhammar 1993; Larhammar and Risinger 1994), have undergone tetraploidization events after divergence from the zebrafish lineage (Fig. 6, #10). Independent additional tetraploidization has occurred in salmonds (Fig. 6, #11)(Allendorf and Thorgaard 1984), catostomids (Uyeno and Smith 1972), and other lineages. These more recent tetraploids we predict to have up to 14 hox clusters, each more depauparate than their single-copy orthologous cluster in zebrafish due to additional reciprocal gene losses between the new duplicates.
Pufferfish and medaka belong to the Acanthopterygii or spiny-ray fish, a group rich in species and morphologically diverse (Nelson 1994). From available data, the loss of the Hoxcb cluster appears to be a shared derived feature of this group (Fig. 6, #12). In the lineage giving rise to pufferfish (Fig. 6, #14 and #15), Hoxa7 become a pseudogene because striped bass has a good Hoxa7 gene (Fig. 6, #13)(Snell et al. 1999). Hoxc3 became a pseudogene sometime after the divergence of medaka and pufferfish lineages, and Hoxd13a dropped out. Finally, several changes occurred after the divergence of the two pufferfish lineages, including the acquisition of an intron in Hoxd11b, and the mutation of Hoxb7a to a pseudogene.
The continuing evolution of Hox cluster genomic organization chronicled in Figure 6 raises the question of the roles of gene and genome duplication in the evolution of novel, lineage-specific morphologies. The pufferfish skeleton is highly derived, and many of these features are adaptations for puffing, the expansion of the gastrointestinal tract with water, which enlarges the fish and makes body spines more formidable defensive armamentation (Brainerd 1994; Wainwright et al. 1995; Wainwright and Turingan 1997). The molecular genetic basis for the evolution of morphologies and behaviors essential for puffing may have now become tractable given the availability of a draft genome sequnce (Aparicio et al. 2002) and the demonstration here that fugu embryos are amenable to developmental genetic investigation.
METHODS
The S. nephalus PAC library was constructed as described (Amemiya et al. 2001). PACs were screened by PCR using a hierarchical pooling protocol (Amores et al. 1998; Amemiya et al. 2001). Degenerate primers for screening the S. nephalus genomic library included posterior Hox forward primers for paralogy groups 9-13 [CGAAAGAAG(C/A)G(I/C)GT(I/C)CC(I/C)TA(T/C)AC], anterior Hox forward primer for paralogy groups 1-9 [GAATTC CACTTCAAC(C/A)(G/A)(C/G)TACCT], and the universal reverse primer (CATCCTGCGGTTTTGGAACCAIAT). PACs shown to contain Hox cluster genes were screened individually by the above primers to amplify _Hox_-containing PCR fragments, which were then sequenced. From the sequenced fragments, gene-specific primers were designed for sequencing portions of each PAC, and in addition, PACs were subjected to shotgun sequencing. For the phylogenetic analyses, neighbor-joining trees were constructed from unambiguously aligned sequences as described (Amores et al. 1998). The alignments are available as Supplemental data available online at www.genome.org. Probes for in situ hybridization experiments were constructed from S. nephalus genomic DNA and used for in situ hybridization experiments on T. rubripes embryos collected as described (Suzuki et al. 2002), and using the zebrafish protocol (Force et al. 1999). To assign rhombomere identity, we used krox20, which is expressed in rhombomeres three and five.
Acknowledgments
We thank Bill Cresko for helpful comments on the manuscript, Elizabeth Housworth for computational help, and NIH grant R01RR10715 and NSF IGERT grant DGE 9972830 for support. Part of this work was a portion of a Clark Honors College Undergraduate Thesis (J.P.) supported by the NSF-IGERT grant Summer Program for Undergraduate Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1717804.
Footnotes
References
- Ahn, S. and Tanksley, S.D. 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. 90**:** 7980-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. and Thorgaard, G. 1984. Tetraploidy and the evolution of salmonid fishes. In Evolutionary genetics of fishes (ed. B.J. Turner), pp. 1-46. Plenum Press, New York.
- Allendorf, F.W., Utter, F.M., and May, B.P. 1975. Gene duplication within the family Salmonidae: II. Detection and determination of the genetic control of duplicate loci through inheritance studies and the examination of populations. In Isozymes (ed. C.L. Markert), pp. 415-432. Academic Press, New York.
- Amemiya, C., Amores, A., Ota, G., Mueller, D., Garraty, J., Postlethwait, J., and Litman, G. 2001. Generation of a P1 artificial chromosome library of the Southern pufferfish. Gene 272**:** 283-289. [DOI] [PubMed] [Google Scholar]
- Amores, A., Force, A., Yan, Y.-L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langel, J., Prince, V., Wang, Y.-L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282**:** 1711-1714. [DOI] [PubMed] [Google Scholar]
- Aparicio, S. 2000. Vertebrate evolution: Recent perspectives from fish. Trends Genet. 16**:** 54-56. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Hawker, K., Cottage, A., Mikawa, Y., Zuo, L., Venkatesh, B., Chen, E., Krumlauf, R., and Brenner, S. 1997. Organization of the Fugu rubripes Hox clusters: Evidence for continuing evolution of vertebrate Hox complexes. Nat. Genet. 16**:** 79-83. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupk, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297**:** 1301-1310. [DOI] [PubMed] [Google Scholar]
- Bailey, G.S., Poulter, R.T.M., and Stockwell, P.A. 1978. Gene duplcation in tetraploid fish: Model for gene silencing at unlinked duplicated loci. Proc. Natl. Acad. Sci. 75**:** 5575-5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainerd, E.L. 1994. Pufferfish inflation: Functional morphology of post cranial structures in Diodon holocanthus (Tetraodontiformes). J. Morphol. 220**:** 243-261. [DOI] [PubMed] [Google Scholar]
- Brainerd, E.L. and Patek, S.N. 1998. Vertebral column morphology, C-start curvature, and the evolution of mechanical defenses in tetraodontiform fishes. Copeia 4**:** 971-984. [Google Scholar]
- Chiu, C.H., Amemiya, C., Dewar, K., Kim, C.B., Ruddle, F.H., and Wagner, G.P. 2002. Molecular evolution of the HoxA cluster in the three major gnathostome lineages. Proc. Natl. Acad. Sci. 99**:** 5492-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A.G. 1994. Invasion and maintenance of a gene duplication. Proc. Natl. Acad. Sci. 91**:** 2950-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgar, G., Clark, M.S., Meek, S., Smith, S., Warner, S., Edwards, Y.J.K., Bouchireb, N., Cottage, A., Yeo, G.S.H., Umrania, Y., et al. 1999. Generation and analysis of 25 Mb of genomic DNA from the pufferfish Fugu rubripes by sequence scanning. Genome Res. 9**:** 960-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriva, H., Manzon, L., Youson, J., and Laudet, V. 2002. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol. Biol. Evol. 19**:** 1440-1450. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology 27**:** 401-410. [Google Scholar]
- Ferrier, D.E., Minguillon, C., Holland, P.W., and Garcia-Fernandez, J. 2000. The amphioxus Hox cluster: Deuterostome posterior flexibility and Hox14. Evol. Dev. 2**:** 284-293. [DOI] [PubMed] [Google Scholar]
- Ferris, S.D. and Whitt, G.S. 1979. Evolution of the differential regulation of duplicate genes after polyploidization. J. Mol. Evol. 12**:** 267-317. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.-L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151**:** 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., Amores, A., and Postlethwait, J.H. 2002. Hox cluster organization in the jawless vertebrate Petromyzon marinus. J. Exp. Zool. 294**:** 30-46. [DOI] [PubMed] [Google Scholar]
- Gates, M.A., Kim, L., Egan, E.S., Cardozo, T., Sirotkin, H.I., Dougan, S.T., Lashkari, D., Abagyan, R., Schier, A.F., and Talbot, W.S. 1999. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 9**:** 334-347. [PubMed] [Google Scholar]
- Haldane, J.B.S. 1933. The part played by recurrent mutation in evolution. Am. Nat. 67**:** 5-9. [Google Scholar]
- Hedges, S.B. 2002. The origin and evolution of model organisms. Nat. Rev. Genet. 3**:** 838-849. [DOI] [PubMed] [Google Scholar]
- Hedges, S.B. and Kumar, S. 2002. Genomics. Vertebrate genomes compared. Science 297**:** 1283-1285. [DOI] [PubMed] [Google Scholar]
- Holland, P.W. 1997. Vertebrate evolution: Something fishy about Hox genes. Curr. Biol. 7**:** 570-572. [DOI] [PubMed] [Google Scholar]
- Holland, P.W., Garcia-Fernàndez, J., Williams, N.A., and Sidow, A. 1994. Gene duplications and the origins of vertebrate development. Dev. Suppl. 125-133. [PubMed]
- Hughes, A.L. 1994. The evolution of functionally novel proteins after gene duplication. Proc. Roy. Soc. Lond. B 256**:** 119-124. [DOI] [PubMed] [Google Scholar]
- Hughes, M.K. and Hughes, A.L. 1993. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 10**:** 1360-1369. [DOI] [PubMed] [Google Scholar]
- Hunter, M.P. and Prince, V.E. 2002. Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev. Biol. 247**:** 367-389. [DOI] [PubMed] [Google Scholar]
- Irvine, S.Q., Carr, J.L., Bailey, W.J., Kawasaki, K., Shimizu, N., Amemiya, C.T., and Ruddle, F.H. 2002. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. J. Exp. Zool. 294**:** 47-62. [DOI] [PubMed] [Google Scholar]
- Jungbluth, S., Bell, E., and Lumsden, A. 1999. Specification of distinct motor neuron identities by the singular activities of individual Hox genes. Development 126**:** 2751-2758. [DOI] [PubMed] [Google Scholar]
- Kim, C.B., Amemiya, C., Bailey, W., Kawasaki, K., Mezey, J., Miller, W., Minoshima, S., Shimizu, N., Wagner, G., and Ruddle, F. 2000. Hox cluster genomics in the horn shark, Heterodontus francisci. Proc. Natl. Acad. Sci. 97**:** 1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. and King, J.L. 1979. Fixation of a deleterious allele at one of two “duplicate” loci by mutation pressure and random drift. Proc. Natl. Acad. Sci. 76**:** 2858-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, E.G.L., Lam, K., Christoffels, A., Erdmann, M.V., Brenner, S., and Venkatesh, B. 2003. Hox gene clusters in the Indonesian coelacanth, Latimeria menadoensis. Proc. Natl. Acad. Sci. 100**:** 1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa, Y., Yamaguchi, M., and Nishida, M. 2000. Mitochondrial molecular clocks and the origin of euteleostean biodiversity: Familial radiation of perciforms may have predated the Cretaceous/tertiary boundary. In The biology of biodiversity (ed. M. Kato,), pp. 35-52. Springer-Verlag, New York.
- Kurokawa, T., Suzuki, T., Ohta, H., Kagawa, H., Tanaka, H., and Unuma, T. 2002. Expression of pancreatic enzyme genes during the early larval stage of Japanese eel Anguilla japonica. Fisheries Sci. 68**:** 736-744. [Google Scholar]
- Larhammar, D. and Risinger, C. 1994. Molecular genetic aspects of tetraploidy in the common carp Cyprinus carpio. Mol. Phylogenet. Evol. 3**:** 59-68. [DOI] [PubMed] [Google Scholar]
- Ledje, C., Kim, C.B., and Ruddle, F.H. 2002. Characterization of Hox genes in the Bichir, Polypterus palmas. J. Exp. Zool. 294**:** 107-111. [DOI] [PubMed] [Google Scholar]
- Li, W.-H. 1980. Rate of gene silencing at duplicate loci: A theoretical study and interpretation of data from tetraploid fishes. Genetics 95**:** 237-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J. 2000. The evolutionary fate and consequences of gene duplication. Science 290**:** 1151-1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Force, A. 2000. The origin of interspecific genomic incompatibility via gene duplication. Amer. Nat. 156**:** 590-605. [DOI] [PubMed] [Google Scholar]
- Malaga-Trillo, E. and Meyer, A. 2001. Genome duplications and accelerated evolution of Hox genes and cluster architecture in teleost fishes. Amer. Zool. 41**:** 676-686. [Google Scholar]
- McClintock, J.M., Carlson, R., Mann, D.M., and Prince, V.E. 2001. Consequences of Hox gene duplication in the vertebrates: An investigation of the zebrafish Hox paralogue group 1 genes. Development 128**:** 2471-2484. [DOI] [PubMed] [Google Scholar]
- McClintock, J.M., Kheirbek, M.A., and Prince, V.E. 2002. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 129**:** 2339-2354. [DOI] [PubMed] [Google Scholar]
- Meyer, A. and Malaga-Trillo, E. 1999. Vertebrate genomics: More fishy tales about Hox genes. Curr. Biol. 9**:** R210-R213. [DOI] [PubMed] [Google Scholar]
- Meyer, A. and Schartl, M. 1999. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish)rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11**:** 699-704. [DOI] [PubMed] [Google Scholar]
- Misof, B.Y. and Wagner, G.P. 1996. Evidence for four Hox clusters in the killifish Fundulus heteroclitus (teleostei). Mol. Phylogenet. Evol. 5**:** 309-322. [DOI] [PubMed] [Google Scholar]
- Morrison, A., Moroni, M.C., Ariza-McNaughton, L., Krumlauf, R., and Mavilio, F. 1996. In vitro and transgenic analysis of a human HOXD4 retinoid-responsive enhancer. Development 122**:** 1895-1907. [DOI] [PubMed] [Google Scholar]
- Naruse, K., Fukamachi, S., Mitani, H., Kondo, M., Matsuoka, T., Kondo, S., Hanamura, N., Morita, Y., Hasegawa, K., Nishigaki, R., et al. 2000. A detailed linkage map of medaka, Oryzias latipes. Comparative genomics and genome evolution. Genetics 154**:** 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. and Roychoudhury, A.K. 1973. Probability of fixation of nonfunctional genes at duplicate loci. Amer. Nat. 107**:** 362-372. [Google Scholar]
- Nelson, J.S. 1994. Fishes of the world. Wiley-Interscience, New York.
- Nolte, C., Amores, A., Nagy Kovacs, E., Postlethwait, J., and Featherstone, M. 2003. The role of retinoic acid response element in establishing the anterior neural expression border of Hoxd4 transgenes. Mech. Dev. 120**:** 325-335. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, New York.
- Ohta, T. 1988. Time for acquiring new gene by duplication. Proc. Natl. Acad. Sci. 85**:** 3509-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait, J., Yan, Y., Gates, M., Horne, S., Amores, A., Brownlie, A., Donovan, A., Egan, E., Force, A., Gong, Z., et al. 1998. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18**:** 345-349. [DOI] [PubMed] [Google Scholar]
- Postlethwait, J., Amores, A., Yan, G., and Austin, C.A. 2002. Duplication of a portion of human chromosome 20q containing Topoisomerase (Top1)and Snail genes provides evidence on genome expansion and the radiation of teleost fish. In: Aquatic genomics: Steps toward a great future (eds. N. Shimizu et al.), pp. 20-31. Springer-Verlag, Tokyo, Japan.
- Prince, V. and Lumsden, A. 1994. Hoxa-2 expression in normal transposed rhombomeres: Independent regulation in the neural tube and neural crest. Development 120**:** 911-923. [DOI] [PubMed] [Google Scholar]
- Prince, V.E., Joly, L., Ekker, M., and Ho, R.K. 1998a. Zebrafish hox genes: Genomic organization and modified colinear expression patterns in the trunk. Development 125**:** 407-420. [DOI] [PubMed] [Google Scholar]
- Prince, V.E., Moens, C.B., Kimmel, C.B., and Ho, R.K. 1998b. Zebrafish hox genes: Expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development 125**:** 393-406. [DOI] [PubMed] [Google Scholar]
- Risinger, C. and Larhammar, D. 1993. Multiple loci for synapse protein SNAP-25 in the tetraploid goldfish. Proc. Natl. Acad. Sci. 90**:** 10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini, F. and Tyler, J.C. 1999. A new phylogenetic hypothesis for the order Tetraodontiformes (Teleostei, Pisces), with placement of the most fossil basal lineages. Amer. Zool. 39**:** 10A. [Google Scholar]
- Santini, S., Boore, J.L., and Meyer, A. 2003. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 13**:** 1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scemama, J.L., Hunter, M., McCallum, J., Prince, V., and Stellwag, E. 2002. Evolutionary divergence of vertebrate Hoxb2 expression patterns and transcriptional regulatory loci. J. Exp. Zool. 294**:** 285-299. [DOI] [PubMed] [Google Scholar]
- Snell, E.A., Scemama, J.L., and Stellwag, E.J. 1999. Genomic organization of the Hoxa4-Hoxa10 region from Morone saxatilis: Implications for Hox gene evolution among vertebrates. J. Exp. Zool. 285**:** 41-49. [PubMed] [Google Scholar]
- Stickney, H.L., Schmutz, J., Woods, I.G., Holtzer, C.C., Dickson, M.C., Kelly, P.D., Myers, R.M., and Talbot, W.S. 2002. Rapid mapping of zebrafish mutations with SNPs and oligonucleotide microarrays. Genome Res. 12**:** 1929-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, A. 1999. On the possibility of constructive neutral evolution. J. Mol. Evol. 49**:** 169-181. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Oohara, I., and Kurokawa, T. 1998. Hoxd-4 expression during pharyngeal arch development in flounder (Paralichthys olivaceus) embryos and effects of retinoic acid on expression. Zool. Sci. 15**:** 57-67. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Kurokawa, T., Hashimoto, H., and Sugiyama, M. 2002. cDNA sequence and tissue expression of Fugu rubripes prion protein-like: A candidate for the teleost orthologue of tetrapod PrPs. Biochem. Biophys. Res. Commun. 294**:** 912-917. [DOI] [PubMed] [Google Scholar]
- Takahata, N. and Maruyama, T. 1979. Polymorphism and loss of duplicate gene expression: A theoretical study with application to tetraploid fish. Proc. Natl. Acad. Sci. 76**:** 4521-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J., Braasch, I., Frickey, T., Meyer, A., and Van De Peer, Y. 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 13**:** 382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.S., van de Peer, Y., Braasch, I., and Meyer, A. 2001. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R Soc. Lond. B 356**:** 1661-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeno, T. and Smith, G.R. 1972. Tetraploid origin of the karyotype of catostomid fishes. Science 175**:** 644-646. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., Taylor, J.S., Braasch, I., and Meyer, A. 2001. The ghost of selection past: Rates of evolution and functional divergence in anciently duplicated genes. J. Mol. Evol. 53**:** 434-444. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., Frickey, T., Taylor, J., and Meyer, A. 2002. Dealing with saturation at the amino acid level: A case study based on anciently duplicated zebrafish genes. Gene 295**:** 205-211. [DOI] [PubMed] [Google Scholar]
- van der Hoeven, F., Sordino, P., Fraudeau, N., Izpisua-Belmonte, J.C., and Duboule, D. 1996. Teleost HoxD and HoxA genes: Comparison with tetrapods and functional evolution of the HOXD complex. Mech. Dev. 54**:** 9-21. [DOI] [PubMed] [Google Scholar]
- Wainwright, P.C. and Turingan, R.G. 1997. Evolution of pufferfish inflation behavior. Evolution 51**:** 506-518. [DOI] [PubMed] [Google Scholar]
- Wainwright, P.C., Turingan, R.G., and Brainerd, E.L. 1995. Functional morphology of pufferfish inflation: Mechanism of the buccal pump. Copeia 1995**:** 614-625. [Google Scholar]
- Watterson, G.A. 1983. On the time for gene silencing at duplicate loci. Genetics 105**:** 745-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S. and Doebley, J. 1998. Of genes and genomes and the origin of maize. Trends Genet. 14**:** 327-332. [DOI] [PubMed] [Google Scholar]
- Wittbrodt, J., Meyer, A., and Schartl, M. 1998. More genes in fish? BioEssays 20**:** 511-515. [Google Scholar]
- Wittkopp, P.J., True, J.R., and Carroll, S.B. 2002. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development 129**:** 1849-1858. [DOI] [PubMed] [Google Scholar]
- Woods, I.G., Kelly, P.D., Chu, F., Ngo-Hazelett, P., Yan, Y.-L., Huang, H., Postlethwait, J.H., and Talbot, W.S. 2000. A comparative map of the zebrafish genome. Genome Res. 10**:** 1903-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/genome/guide/human/Q; Human Genome Resources.
- http://zfin.org/; The Zebrafish Information Network.
- http://fugu.hgmp.mrc.ac.uk/blast/; Fugu BLAST server at the UK Human Genome Mapping Project Resource Centre.
- http://genome.jgi-psf.org/fugu6/fugu6.home.html; Fugu rubripes at the Joint Genome Institute.