The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor (original) (raw)
. Author manuscript; available in PMC: 2013 Sep 1.
Published in final edited form as: Science. 2011 Feb 18;331(6019):881–886. doi: 10.1126/science.1196473
Abstract
Co-translational targeting of membrane and secretory proteins is mediated by the universally conserved Signal Recognition Particle (SRP). Together with its receptor (SR), SRP mediates the GTP-dependent delivery of translating ribosomes bearing signal sequences to translocons on the target membrane. Here we present the crystal structure of the SRP:SR complex at 3.9 Å resolution and biochemical data revealing that the activated SRP:SR GTPase complex bind the distal end of the SRP hairpin RNA where GTP hydrolysis is stimulated. Combined with previous findings, these results suggest that the SRP:SR GTPase complex initially assembles at the tetraloop end of the SRP RNA and then relocalizes to the opposite end of the RNA. This rearrangement provides a mechanism for coupling GTP hydrolysis to the handover of cargo to the translocon.
The signal recognition particle (SRP) is a ubiquitous ribonucleoprotein complex that co-translationally delivers membrane and secretory proteins to the plasma membrane in prokaryotes and to the endoplasmic reticulum in eukaryotes (1, 2). The SRP targeting process starts with recognition of a hydrophobic signal sequence on the ribosome-nascent chain complex (RNC or cargo) by the SRP to yield an RNC:SRP complex. Subsequently, the RNC:SRP complex associates on the membrane with the SRP receptor (SR). The binding between SRP and SR induces conformational changes of yet unknown nature that allow the cargo to be transferred to the protein conducting channel (translocon). The cycle can be resumed following GTP hydrolysis that drives dissociation of the SRP:SR complex (1–3).
Both the SRP and SR include components that are structurally and functionally conserved across the different domains of life (1, 2). In E. coli, SRP comprises of two universally conserved components, the Ffh protein (SRP54 in eukaryotes) and the 4.5S SRP RNA. Ffh contains a methionine-rich (M) domain responsible for high-affinity RNA binding and recognition of signal sequences (4), a helical N-domain responsible for interactions with the ribosome, and a Ras-like GTPase (G) domain. The E. coli SR is a single protein, FtsY, that consists of an N- and a G-domain that are structurally similar to those found in Ffh (5, 6) and an additional A-domain responsible for interactions with the membrane and the translocon (7).
When bound to GTP, SRP and SR form a stable complex through extensive interactions between their NG-domains (5, 6). At the heterodimer interface, a composite active site is formed in which SRP and SR act as reciprocal activating proteins for one another. GTPase activation within the SRP:SR complex is achieved by a set of conformational changes in both proteins that are distinct from those required for their initial complex assembly (8). Mutations that block GTPase activation severely disrupt protein targeting and translocation (8), suggesting that conformational changes leading to GTPase activation play an essential role in the unloading of cargo (5, 9, 10).
The RNA moiety in the SRP system is essential for cell viability in vivo (4, 11) and for protein targeting and translocation in vitro (12, 13). The E. coli 4.5S RNA has two characterized biochemical functions: acceleration of the interaction between Ffh and FtsY by increasing their complex assembly and disassembly rates, and stimulation of GTPase activity once a stable SRP:SR complex is formed (13–15). Additionally, the SRP RNA has been described to act as a platform for conformational changes in Ffh and FtsY following recognition of the signal sequence by the M-domain (15–17).
Despite extensive prior studies of co-translational protein targeting, several fundamental questions remain unanswered: how does the SRP RNA stimulate GTP hydrolysis of the SRP:SR complex, why is this GTPase activation essential for protein targeting, and how is cargo transfer to the translocon coupled to GTP hydrolysis by Ffh and FtsY. To address these questions, we solved the three-dimensional structure of the SRP:SR complex with the non-hydrolysable GTP analog GMP-PCP.
The SRP:SR complex structure
The prokaryotic SRP:SR complexes were crystallized in the pre-GTP hydrolysis state (Fig. 1, A and B) (18, 19). Stable complexes were assembled using E. coli Ffh1-432, full-length 4.5S RNA from E. coli or D. radiodurans, and E. coli FtsY196-497 in the presence of GMP-PCP and the non-ionic detergent C12E8 proposed to mimic a signal peptide (20). Crystals of the homologous E. coli SRP:SR complex and the heterologous complex containing D. radiodurans 4.5S RNA were isomorphous, but the latter diffracted x-rays to higher resolution. After extensive screening, a single crystal of the heterologous complex was identified that diffracted x-rays better than others and permitted recording of a complete dataset to 3.9 Å resolution (table S1). These data were used to produce an atomic-resolution model of the complex. The structure was solved by molecular replacement using high resolution structures of isolated parts of the assembly, including the NG dimer (5) and the 4.5S RNA domain IV in complex with the M-domain (4). Iterative rounds of rebuilding and refinement produced excellent electron density maps at 3.9 Å resolution, which allowed unambiguous tracing of the molecules and placement of side chains and bases for well ordered parts of the structure (Fig. 1C and fig. S1). The missing parts in previous structures, specifically the distal portion of the SRP RNA and the connective linker between the NG and M-domains of Ffh, could be unambiguously identified in the calculated electron density maps (omit map for the linker helix shown in Fig. 1D). The two molecules in the crystallographic asymmetric unit form a head to tail dimer with the N-terminus of the symmetry related Ffh molecule interacting with a groove on the M-domain of the opposing molecule defined by portions of the finger loop (excluding residues 354–368) (21) (fig. S2). Electron density maps for the homologous complex calculated at 7.0 Å resolution, using the coordinates of the heterologous complex for phasing, revealed that their global structures are virtually identical (table S1 and Fig. 1A and fig. S3).
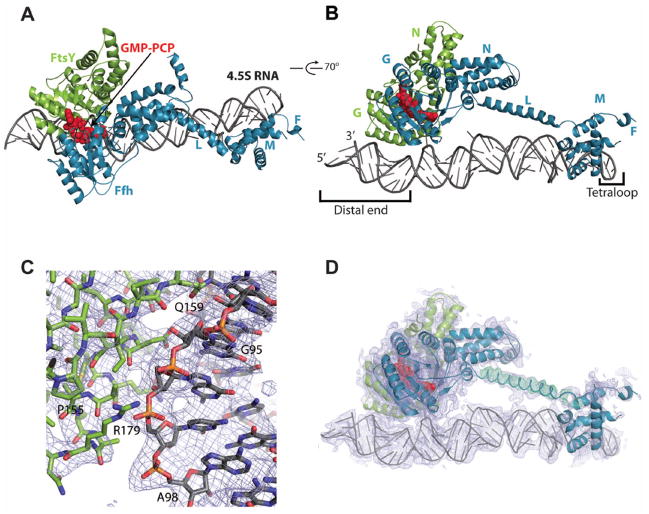
Fig. 1. Structure of Signal Recognition Particle in complex with its Receptor.
(A) Top view of the SRP:SR complex. Ffh is colored in blue, 4.5S RNA in gray, FtsY (SR) is shown in green. The atoms of the two GMP-PCP molecules are displayed as red spheres. (B) Side view of the SRP:SR complex rotated 70° relative to the view in (A). N denotes the N-domain, G denotes the G-domain, M denotes the M-domain, L is the flexible linker and F denotes the finger loop. (C) Visualization of the 2Fo−Fc electron density contoured at 1σ and the stick representation of the SRP:SR complex. (D) Side view of the SRP:SR complex with the contour of a 2Fo−Fc unbiased omit map calculated for linker region residues 300–330 of Ffh. The linker is displayed as a tube together with the difference density Fo−Fc electron density contoured at 3σ, shown as green mesh. The 2Fo−Fc electron density for the entire complex is contoured at 1σ and displayed as gray mesh. The cover radius used for the figure had a cutoff of 2.6 Å for the 2Fo−Fc and Fo−Fc.
The structure shows an unexpected domain arrangement of the Ffh and FtsY proteins relative to the SRP RNA. While the M-domain binds near the tetraloop region (also known as helix 8 or Domain IV) as observed previously (4), the NG heterodimer contacts the opposite end of the 4.5S RNA (also named helix 5) (22). The linker connecting the Ffh NG-and M-domains forms a well defined, 30 amino acids long, helix (Fig. 1, A and D). In previous studies the linker region was either too flexible to be assigned (21) or was modeled as an α-helix and a loop structure (23). There has been no clear evidence for a structural role of the linker helix in previous studies. Interestingly, however, mutations of conserved residues in the linker region abolish the ability of the SRP RNA to stimulate SRP:SR complex formation as well as GTP hydrolysis, indicating that the linker is intimately involved in the function of the SRP RNA (16). In the structure reported here, the linker acts as a spacer placing the activated NG domains of the Ffh and FtsY at the distal end of the 4.5S RNA. This conformation of the SRP:SR complex has not been detected previously (15, 24), possibly because this configuration of the SRP:SR complex is a transient state that was stabilized by intermolecular interactions in the crystal. Nevertheless, the functional importance of this conformational state is strongly supported by the following analyses.
New function of the distal end region of the 4.5S RNA
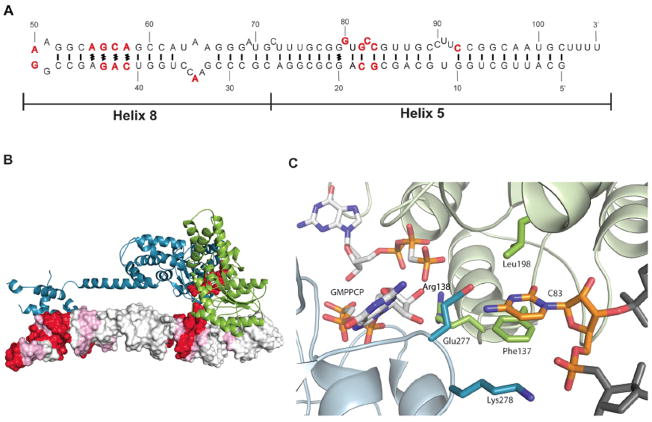
In addition to the structural data presented here, another clue to the importance of the distal RNA region is its sequence conservation (Fig. 2A and fig S4). The sequence motif GUGCCG (bases 83 to 88 in E. coli) can be found in the helix 5 region of most known 4.5S RNAs (Fig. 2A) and is also conserved in the longer prokaryotic 6S SRP RNA (22). The secondary structure prediction for this region of the SRP RNA always features a bulged or unpaired base (Fig. 2A). The conservation of this region is similar to that of the SRP RNA tetraloop and the region recognized by the M domain (Fig. 2, A and B and fig. S4).
Fig. 2. Interaction of the Ffh-SR NG domain with a conserved flipped base at 5′-3′ distal end of the 4.5S RNA.
(A) Secondary structure of the D. radiodurans 4.5S RNA with conserved residues indicated in red (sequence alignment displayed in fig. S4). Base pairings are indicated as (−) and non-canonical interactions are indicated as (#), bulged residues are unpaired. (B) Overall view of the interaction of Ffh and FtsY (displayed as ribbons and colored as in Fig. 1A) with the 4.5S RNA (represented as a contoured surface colored according to the conservation indicated in fig. S4). GMP-PCP molecules are shown as red spheres. (C) Close-up view of the interaction of the conserved flipped C83 with the interface of Ffh and FtsY. 4.5S RNA is displayed as sticks colored in gray with C83 colored in orange. Ffh and FtsY residues that interact directly with C83 are displayed as sticks and colored as in (B). GMP-PCP residues are represented as sticks colored with white carbons, red oxygens, blue nitrogens and orange phosphates.
In our structures, C83 (C86 in E. coli, fig. S5) is positioned close to both GMP-PCP molecules and interacts directly with both the Ffh and FtsY proteins at regions that regulate the GTPase activity (Fig. 2C, 4E and fig. S5) (25–27). Two residues from Ffh can form H-bonds with C83, Lys 278 interacts with the phosphate group of C83 and Glu 277 interacts with the N4 of C83 and the 2′O from GMP-PCP. In contrast, the interaction of C83 with FtsY is hydrophobic: residues Leu 198 and Phe 137 stack with the base; the latter resides in the insertion box domain (IBD) motif previously described as essential for GTP binding and hydrolysis (5, 10, 28). Identical interactions have been described in the Ffh:FtsY NG domains crystal structure (2CNW) containing GDP:AlF4, in which a peripheral nucleotide (GMP) was found to bind in the same position as C83 (27) (fig. S6). The superposition of our NG heterodimer structure with previously determined structures containing GMP-PCP (1OKK), GMP-PNP (1RJ9) and GDP:AlF4 (2CNW) reveals no large differences in the GTPase center, within the resolution limits of our structure (5, 6, 25, 26).
Fig. 4. The SRP RNA distal end specifically stimulates GTP hydrolysis in the SRP:SR complex.
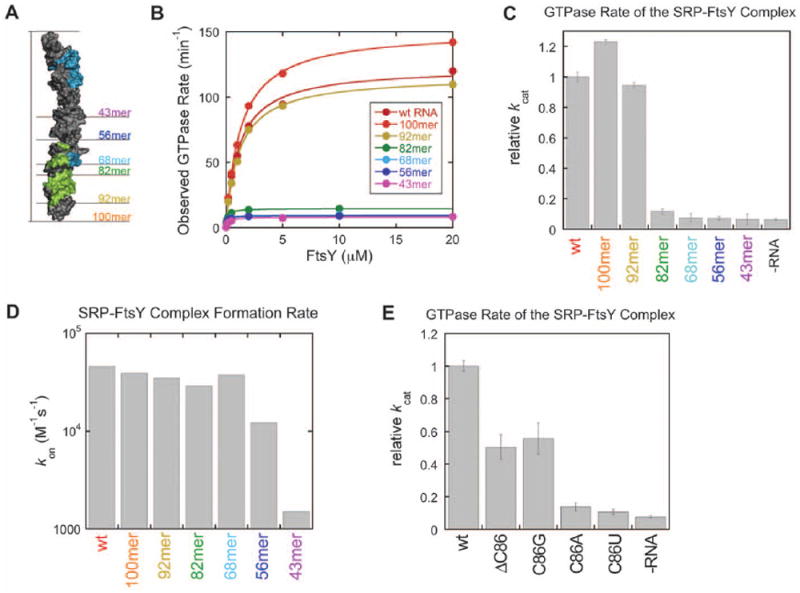
(A) E. coli SRP RNA systematically truncated from the distal end that were used in this study (see fig. S7A for the sequence of the truncated RNA constructs). The FtsY contact area in the RNA is indicated in green and the Ffh contact area in blue, C86 is indicated in blue but contacted by both proteins. (B) Stimulated GTPase activity between SRP and FtsY with the SRP RNA mutants in part A. The data were fit to the Michaelis-Menten equation, and the kinetic constants kcat and kcat/Km are summarized in figure S7B. (C) A significant defect in the GTPase rate of the SRP:FtsY complex was observed upon truncation of the SRP RNA from the 92mer to 82mer. Rate constants were from part B and normalized to that of the wild-type SRP RNA. (D) Truncation of the RNA distal end did not significantly affect SRP-FtsY complex formation rates (_k_on) until the C loop is truncated (43mer). Values of kon were derived from figure S7B. (E) Mutation of the conserved base C86 (C83 in D. radiodurans 4.5S RNA) impairs the ability of SRP RNA to stimulate GTPase hydrolysis of the SRP:FtsY complex. Rate constants were relative to that of the wild type SRP RNA and were derived from the data in figure S7D.
The NG heterodimer exhibits an extensive interaction interface with the 4.5S RNA, burying 890 Å2 (Fig. 3A). FtsY is responsible for the majority of these contacts, whereas Ffh interaction with the distal region of the RNA is limited to two bases, C82 and C83. However, the M-domain of Ffh is also responsible for the interaction with domain IV of the 4.5S RNA. Taken together, the total surface area buried between Ffh and the 4.5S RNA is 780 Å2, which is similar to the extent of interaction between FtsY and the distal portion of the 4.5S RNA (Fig. 3B). These observations explain why FtsY will bind the distal portion of the 4.5 S RNA as part of the GTP-activated twin complex but not in its free state (Fig. 2C and 3B). First, the contact to the RNA is mediated by both FtsY and Ffh. Second, the local concentration of the tethered homodimeric NG domains is very high since they are connected via the linker region to the M-domain, which is tightly bound to the SRP RNA (KD of 5 pM) (29, 30).
Fig. 3. Major contact area of the Ffh:FtsY NG domain with 5′,3′-region of the 4.5S RNA.
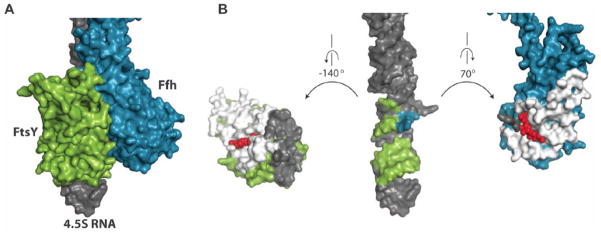
(A) Surface representation of the SRP:SR complex in the 5′,3′-region of the 4.5S RNA. (B) Surface representation of the separated Ffh and FtsY (rotate each to one direction) from the 4.5S RNA (maintained in the same orientation as in A). The interface between Ffh and FtsY is displayed in white in both proteins with GMP-PCP displayed as red spheres. The contact area of each protein to the RNA is indicated in gray. The FtsY contact area in the RNA is indicated in green and the Ffh contact area in blue, C83 is indicated in blue but contacted by both proteins.
The SRP RNA distal end specifically stimulates GTPase activation
Although earlier work implicated the tetraloop region of the SRP RNA in GTP hydolysis (13, 15), interpretation of the observed GTPase activity is complicated by the fact that at subsaturating protein concentrations, the observed GTPase reaction is rate-limited by the assembly of the complex. Recent studies that more rigorously dissected the complex assembly versus GTP hydrolysis steps established that the tetraloop is essential for accelerating SRP:FtsY complex assembly, whereas it affects GTPase activation no more than two-fold once the GTPase complex is formed (12, 31, 32). To define the RNA site(s) responsible for stimulating GTP hydrolysis, we systematically truncated the E. coli 4.5S RNA from the distal end (Fig. 4A, fig. S7, A and B) and tested the effects of these mutations on the stimulated GTPase reaction between SRP and FtsY. The heterologous complex involving D. radiodurans 4.5S RNA was not used for biochemical experiments since it showed significantly lower GTPase activity. This could be either due to small differences in sidechain positions at the interface of the RNA and activated NG domains, beyond the resolution limit of our current data, or due to the possibility that the experimental conditions have been optimized for measuring the activity of the E. coli complex. Truncations of up to ten base pairs from the distal end did not significantly affect the GTPase rate of the SRP:FtsY complex at saturating protein concentrations (Fig. 4, B and C, and fig. S7B, kcat). However, truncation of an additional five base pairs (92mer → 82mer) reduced kcat over eight-fold to values approaching that observed in the absence of RNA (Fig. 4, B and C, and fig. S7B). Therefore, the base-paired region C97–C101:G10–G14 at the distal end plays an important role in the SRP RNA-mediated stimulation of GTPase hydrolysis. Consistent with these findings, this region provides a major site for docking of the SRP:SR NG domain complex at the RNA distal end (Fig. 3, A and B).
In contrast, the values of kcat/Km in the stimulated GTPase reaction were largely unaffected unless the SRP RNA was truncated to less than 56 nucleotides (Figs. 4B and S7B). As the kcat/Km value in this reaction is rate-limited by SRP:FtsY complex formation (18), this strongly argues that the distal end accelerates the actual GTP catalysis step, but not the initial SRP:FtsY complex assembly. To provide independent evidence for this possibility, we directly measured the complex assembly rate constants using acrylodan conjugated at Ffh-C235, a probe that changes fluorescence upon formation of a GTP-dependent SRP:FtsY complex (18). SRP:FtsY complex assembly rates were affected no more than two-fold for RNA truncations up to the 56mer; only with the 43mer a significant defect was detected (Figs. 4D and S7C). Therefore, the distal end of the SRP RNA specifically stimulates GTPase activation in the SRP:FtsY complex without affecting the initial assembly of the complex. Together with previous work (12, 31, 32), these results demonstrate that the SRP RNA contains two separate motifs that regulate distinct stages of the SRP:FtsY interaction: the tetraloop end accelerates the initial assembly of the complex, whereas the distal end facilitates GTPase activation at late stages.
To test the role of the extruded base C86 in GTPase activation, C86 was either removed (ΔC86) or mutated to A, G or U. Deletion of C86 or its mutation to G reduced the GTPase rate constant by two-fold, whereas theC86A and C86U mutations reduced GTPase activity by factors of 7 – 10, to rates approaching that in the absence of the SRP RNA (Figs. 4E and S7D). These data support a role of the extruded base in modulating GTPase activity and explain the conservation of the identity of this base.
Modeling the conformation of the SRP:SR on the ribosome
The data presented here, together with previous structural and biochemical studies, shows that the SRP complex can exist in distinct conformational states depending on the orientation of the linker between the NG and M domains of Ffh. We propose that these conformational states represent different stages of the SRP-mediated protein targeting pathway. Indeed, the large rearrangement of the NG domains from their initial position in the vicinity of the 4.5S RNA tetraloop as observed in EM and biochemical studies (15, 17, 33, 34) to the opposite end of this RNA suggests a plausible mechanism of transferring the RNC from the SRP to the translocon. In order to better understand the implications of the observed conformational changes in the targeting process, we modeled our structure onto the ribosome using available cryo-EM data (33) (Fig. 5A). The SRP bound to the RNC in the absence of the SR is in a conformation in which its NG domain is next to the M-domain with the linker region wrapping around the M-domain (Fig. 5B) (23, 33, 35). We propose that the structure described here represents the cargo release state of the protein targeting cycle. Repositioning the NG domains of Ffh and FtsY to the distal region of 4.5S RNA exposes ribosomal proteins L23 and L29, which constitutes the main translocon binding site. Therefore, this conformational change simultaneously exposes the signal sequence-binding cleft of the M domain and liberates the translocon binding region on the ribosome (35–37).
Fig. 5. Conformation of the SRP and SRP:SR structure on the ribosome showing the large rearrangement between cargo binding and cargo release modes.
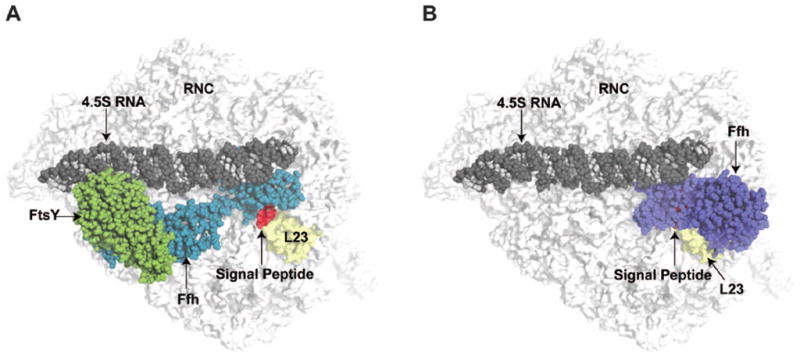
(A) The structure of the SRP:SR is superimposed on the SRP:RNC bound structure from cryo-EM reconstruction (33), with Ffh in the cryo-EM structure omitted. The signal peptide from the SRP:RNC structure was maintained (red surface) as a reference for the exit tunnel. The RNC is displayed as a white surface with the protein L23 highlighted in yellow. SRP:SR are presented as spheres with 4.5S RNA colored with dark gray for phosphate and ribose and light gray for bases, Ffh is blue and FtsY is green. (B) The cargo binding conformation of SRP in the SRP:RNC model structure from cryo-EM is indicated as spheres with Ffh colored in purple.
Since the SRP cycle is evolutionarily conserved, we speculate that many of the molecular features that govern the prokaryotic SRP cycle will also occur in eukaryotes. Upon comparison of the molecular model in Figure 5 with the cryo-EM density of the eukaryotic RNC:SRP:SR complex arrested with the GTP analogue GMP-PNP, certain similarities can be observed (36). Most strikingly, in the eukaryotic complex the density for the NG domains of the SRP54 and SR proteins (homologous to Ffh) is absent from the tetraloop end of the 4.5S RNA, whereas additional density appears in the distal end of the SRP RNA as observed in our complex (fig. S8). It is therefore possible that this density originates from the activated twin NG domains rather than from SR alone, as suggested (36).
The SRP cycle revised
Based on these data, a mechanism can be envisioned that governs the handoff of substrates to the translocon and the role of GTP in this process. The initial interaction of free SRP with the RNC involves binding of the N-domain of Ffh to the ribosomal proteins L23 and L29 (Fig. 6A) as well as interaction of the M-domain of Ffh with the signal sequence, possibly aided by the linker helix that wraps around the M domain in the RNC-bound conformation of the SRP (Fig. 6B) (33, 35). In this “RNC pre-organized” state, the Ffh G-domain is positioned in the vicinity of the tetraloop end of the 4.5S RNA and primed for interaction with the SR (Fig. 6B) (15, 33–35). Receptor binding is initially facilitated by the RNA tetraloop (13, 14, 31, 38), which stabilizes an early conformation of the heterodimeric NG complex through interaction with the SR (Fig. 6C) (5, 10). Subsequent structural rearrangements in the GTPase complex may reduce the affinity of the RNA tetraloop for the SR as well as that of the Ffh N-domain for the ribosome (Fig. 6C) (5, 6, 36). Consequently, the activated NG heterodimer detaches from the ribosome exit site, but stays tethered to the SRP RNA via the tightly bound M-domain, and relocates to the alternative binding site at the distal end of the 4.5S RNA hairpin (Fig. 6D). Since the A-domain of FtsY associates with the translocon (7, 39), repositioning of the NG-domain heterodimer will carry the translocon towards its ribosome binding site (L23), which is now exposed (Fig. 6D). This repositioning could also stabilize the linker domain in the extended helical conformation away from the M domain, thus exposing the signal sequence-binding cleft for peptide release to the translocon. Concurrently, the interaction of the activated NG heterodimer with the distal portion of the 4.5S RNA will stimulate GTPase activity in both Ffh and FtsY, leading to their dissociation and completing the targeting cycle (Fig. 6D).
Fig. 6. Model of the SRP targeting cycle.
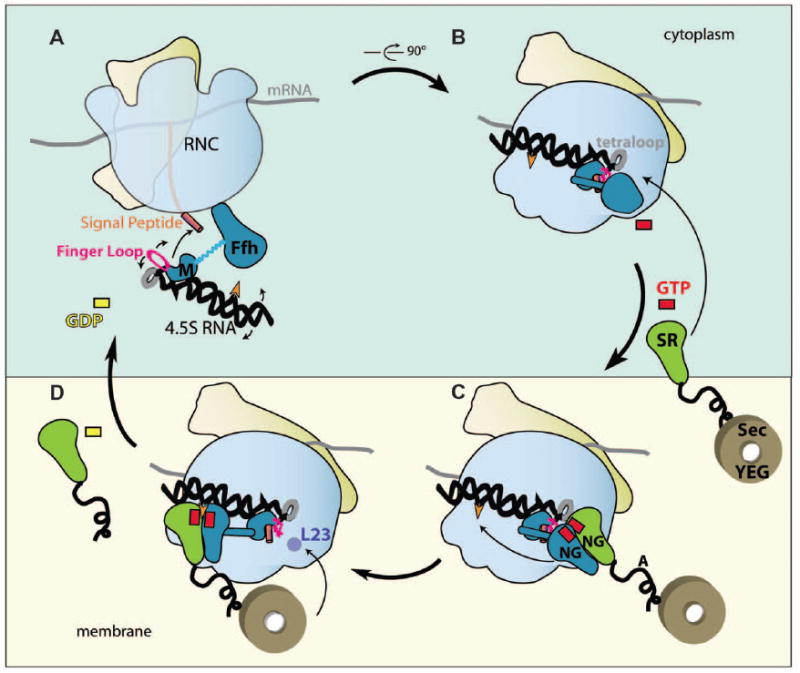
Schematic depiction of the sequence of conformational changes involved in the SRP cycle (viewed in the membrane plane). (A) SRP recognizes the RNC and M-domain interacts with the signal peptide (finger loop is indicated in pink). (B) The N-domain interacts with L23 and the linker region folds on top of the signal peptide covering/shielding the M-domain. (C) SRP bound cargo (RNC) is transferred to the membrane vicinity via SRP interaction with SR. (D) GTP-dependent rearrangements in the SRP:SR complex enables detachment of the Ffh N-domain from L23 and the RNA tetraloop, and the NG domain complex relocates to the 5′,3′-end of the 4.5S RNA sandwiching the C83 (orange arrowhead) at the interface of the two G domains. This repositioning simultaneously exposes the signal sequence bound to the M-domain, and the L23 region of the ribosome for interactions with the translocon (Sec YEG), which is associated with the A-domain of FtsY (black tail). In the final step, signal sequence is transferred from the M-domain to the translocon and the distal region of the 4.5S RNA promotes GTP hydrolysis and subsequent Ffh and FtsY dissociation.
The conformation of the SRP in complex with its receptor observed here also explains many previous observations including the density features observed in the cryo-EM reconstruction of the eukaryotic RNC:SRP:SR complex (36), and the observation that mutations that disrupt the conformational changes leading to GTPase activation block late stages of protein targeting (8). Previous studies also indicated that overexpression of a truncated 4.5S RNA (domain IV) is capable of sustaining cell viability in a 4.5S depleted strain (4). According to the model described here, signal sequence transfer would be possible even in the absence of the distal region of the SRP RNA since the reduced GTPase rate would provide a longer time frame for the less efficient signal sequence transfer to occur.
The results presented here, together with previous biochemical evidence (13–17, 20), define the SRP RNA as a bifunctional molecule acting as a binding platform for the initial receptor interactions on one end of the 142 Å long molecule and for the activated GTPase domains of the Ffh:FtsY complex on the other end. Such conformational change provides a mechanism for the temporal and spatial exchange of large factors that have to access the signal sequence as it emerges from the ribosomal exit tunnel.
Supplementary Material
supplementary
Acknowledgments
We thank Kaihong Zhou for excellent technical assistance and help with crystal preparation during the early stages of the project. Initial crystallographic analysis was performed at beamline 8.2.2 at the Advanced Light Source (ALS), Lawrence Berkeley National Lab; we acknowledge Corie Ralston for outstanding technical assistance at the ALS. Crystallographic data were collected at the beamline X06SA at the Swiss Light Source (SLS). We thank Axel Brunger for the pre-release version of CNS and for helpful comments on the refinement. We are grateful to C. Schulze-Briese and T. Tomizaki for their outstanding support at the SLS. We would like to thank T. Maier and S. Klinge for critical discussion and reading of the manuscript; T. Maier and M. Leibundgut for help and assistance with data collection and solving the structure. SFA was funded initially by the Howard Hughes Medical Institute and currently by an ETH Post-doctoral Fellowship; NS is funded by Boehringer Ingelheim Fonds, and KS is funded by NIH grant GM078024 to S.S. This work was supported in part by the Howard Hughes Medical Institute (J.A.D.) and by the Swiss National Science Foundation (SNSF) and the National Center of Excellence in Research (NCCR) Structural Biology program of the SNSF. Atomic coordinates and structure factors for the SRP:SR crystal structure have been deposited with the Protein Data Bank under accession code 2xxa.
Footnotes
References
- 1.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Batey RT. Structural insights into the signal recognition particle. Annu Rev Biochem. 2004;73:539. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- 3.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995 Jan 16;14:217. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000 Feb 18;287:1232. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 5.Egea PF, et al. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004 Jan 15;427:215. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 6.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004 Jan 16;303:373. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelini S, Deitermann S, Koch HG. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 2005 May;6:476. doi: 10.1038/sj.embor.7400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J Cell Biol. 2007 Aug 13;178:611. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan SO, Walter P. Induced nucleotide specificity in a GTPase. Proc Natl Acad Sci U S A. 2003 Apr 15;100:4480. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004 Oct;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood H, Luirink J, Tollervey D. Evolutionary conserved nucleotides within the E. coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 1992 Nov 25;20:5919. doi: 10.1093/nar/20.22.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Kung S, Shan SO. Demonstration of a multistep mechanism for assembly of the SRP x SRP receptor complex: implications for the catalytic role of SRP RNA. J Mol Biol. 2008 Sep 5;381:581. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siu FY, Spanggord RJ, Doudna JA. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA. 2007 Feb;13:240. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001 Dec 18;40:15224. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 15.Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol. 2005 Dec;12:1116. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 16.Bradshaw N, Walter P. The signal recognition particle (SRP) RNA links conformational changes in the SRP to protein targeting. Mol Biol Cell. 2007 Jul;18:2728. doi: 10.1091/mbc.E07-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neher SB, Bradshaw N, Floor SN, Gross JD, Walter P. SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction. Nat Struct Mol Biol. 2008 Sep;15:916. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci U S A. 2009 Feb 10;106:1754. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material on Science Online
- 20.Bradshaw N, Neher SB, Booth DS, Walter P. Signal sequences activate the catalytic switch of SRP RNA. Science. 2009 Jan 2;323:127. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janda CY, et al. Recognition of a signal peptide by the signal recognition particle. Nature. 2010 May 27;465:507. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwieb C, Larsen N. The signal recognition particle (SRP) database. Nucleic Acids Res. 1992 May 11;20(Suppl):2207. doi: 10.1093/nar/20.suppl.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc Natl Acad Sci U S A. 2003 Dec 9;100:14701. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buskiewicz I, et al. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J Mol Biol. 2005 Aug 12;351:417. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Reyes CL, Rutenber E, Walter P, Stroud RM. X-ray structures of the signal recognition particle receptor reveal targeting cycle intermediates. PLoS One. 2007;2:e607. doi: 10.1371/journal.pone.0000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gawronski-Salerno J, Freymann DM. Structure of the GMPPNP-stabilized NG domain complex of the SRP GTPases Ffh and FtsY. J Struct Biol. 2007 Apr;158:122. doi: 10.1016/j.jsb.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Focia PJ, Gawronski-Salerno J, Coon JSt, Freymann DM. Structure of a GDP:AlF4 complex of the SRP GTPases Ffh and FtsY, and identification of a peripheral nucleotide interaction site. J Mol Biol. 2006 Jul 14;360:631. doi: 10.1016/j.jmb.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan SO, Walter P. Molecular crosstalk between the nucleotide specificity determinant of the SRP GTPase and the SRP receptor. Biochemistry. 2005 Apr 26;44:6214. doi: 10.1021/bi0500980. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz U, et al. NMR studies of the most conserved RNA domain of the mammalian signal recognition particle (SRP) RNA. 1996 Dec;2:1213. [PMC free article] [PubMed] [Google Scholar]
- 30.Batey RT, Doudna JA. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry. 2002 Oct 1;41:11703. doi: 10.1021/bi026163c. [DOI] [PubMed] [Google Scholar]
- 31.Jagath JR, et al. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001 Feb;7:293. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen K, Shan SO. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci U S A. 2010 Apr 27;107:7698. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halic M, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006 Nov 23;444:507. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 34.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004 Feb 26;427:808. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 35.Schaffitzel C, et al. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006 Nov 23;444:503. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 36.Halic M, et al. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006 May 5;312:745. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- 37.Mitra K, et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005 Nov 17;438:318. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jagath JR, Rodnina MV, Wintermeyer W. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J Mol Biol. 2000 Jan 28;295:745. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- 39.Weiche B, et al. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J Mol Biol. 2008 Mar 28;377:761. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary