THE GENOMIC LANDSCAPE OF HYPODIPLOID ACUTE LYMPHOBLASTIC LEUKEMIA (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 10.
Published in final edited form as: Nat Genet. 2013 Jan 20;45(3):242–252. doi: 10.1038/ng.2532
Abstract
The genetic basis of hypodiploid acute lymphoblastic leukemia (ALL), a subtype of ALL characterized by aneuploidy and poor outcome, is unknown. Genomic profiling of 124 hypodiploid ALL cases, including whole genome and exome sequencing of 40 cases, identified two subtypes that differ in severity of aneuploidy, transcriptional profile and submicroscopic genetic alterations. Near haploid cases with 24–31 chromosomes harbor alterations targeting receptor tyrosine kinase- and Ras signaling (71%) and the lymphoid transcription factor IKZF3 (AIOLOS; 13%). In contrast, low hypodiploid ALL with 32–39 chromosomes are characterized by TP53 alterations (91.2%) which are commonly present in non-tumor cells, and alterations of IKZF2 (HELIOS; 53%) and RB1 (41%). Both near haploid and low hypodiploid tumors exhibit activation of Ras- and PI3K signaling pathways, and are sensitive to PI3K inhibitors, indicating that these drugs should be explored as a new therapeutic strategy for this aggressive form of leukemia.
Despite improvements in the outcome of therapy for acute lymphoblastic leukemia (ALL)1, this disease remains the most common cause of cancer-related death in young people. Genomic profiling has identified submicroscopic genetic alterations in ALL that are key determinants of leukemogenesis, notably alterations in B-lymphoid transcription factors in B-progenitor ALL.2,3 Moreover, specific genetic alterations are associated with treatment failure in ALL, including alterations of IKZF1 (Refs.4,5) and CREBBP.6 However, the genetic basis of several subtypes of high-risk ALL remains poorly understood. Notably, hypodiploid ALL with less than 44 chromosomes is characterized by multiple whole chromosomal losses and extremely poor outcome.7–11 Hypodiploid ALL may be subclassified into near haploid cases with 24–31 chromosomes, low hypodiploid cases with 32–39 chromosomes and, less commonly, high hypodiploid cases with 40–43 chromosomes.11 In addition, the hypodiploid genome may undergo reduplication resulting in a hyperdiploid karyotype (“masked” hypodiploid ALL).12 In contrast, near diploid cases with 44–45 chromosomes commonly harbor dicentric chromosomes or other known chromosomal rearrangements (e.g. ETV6-RUNX1)10 and do not share the poor outcome of hypodiploid ALL with more severe aneuploidy. Few hypodiploid ALL cases have been studied in prior genomic analyses and those that have been investigated typically have 45 chromosomes and/or dicentric chromosomes3.
Here we report microarray profiling of gene expression and copy number alterations, Sanger and next-generation sequencing analysis of 124 childhood hypodiploid ALL cases, and have identified multiple recurring genetic alterations that distinguish near haploid and low hypodiploid ALL, and that are rare or absent in other ALL subtypes. Genetic alterations targeting TP53, RB1 and the IKAROS gene family member IKZF2 (HELIOS) are hallmarks of low hypodiploid ALL, while the majority of near haploid ALL cases harbor alterations targeting receptor tyrosine kinase- (RTK-) and Ras signaling, and a high frequency of IKZF3 (AIOLOS) alterations. Notably, the TP53 alterations observed in 91.2% of childhood low hypodiploid ALL are present in non-tumor cells in 43.3% of mutated cases, suggesting that these mutations are inherited and that low hypodiploid ALL represents a manifestation of Li-Fraumeni syndrome13,14 (LFS). Moreover, hypodiploid ALL cells exhibit activation of Ras- and PI3K signaling that is sensitive to the action of PI3K inhibitors, suggesting that these agents represent a new therapeutic strategy in this high-risk disease.
RESULTS
We studied the leukemic blasts from 124 childhood hypodiploid ALL cases with matched remission DNA available for 89 cases (Supplementary Table 1). This cohort included 50 near haploid cases, 18 samples with evidence of doubling of the near haploid clone, either as the sole population (i.e. masked hypodiploidy) or with both non-doubled and a pronounced doubled population, 26 low hypodiploid and 8 masked or partially masked low hypodiploid cases. We also studied 22 near diploid cases, the majority of which had a dicentric chromosome, and a cohort of 117 adult ALL cases including 11 low hypodiploid cases (Supplementary Table 2).
Whole genome-or exome sequencing of tumor and matched remission DNA was performed for 25 pediatric near haploid cases (11 whole genome, 14 exome) and 15 pediatric low hypodiploid cases (9 whole genome, 6 exome; Fig. 1, Supplementary Tables 1 and 3–4 and Supplementary Fig. 1). Validation rates were 83% for identified single nucleotide variations and insertion/deletion mutations, and 86% for structural variations (Supplementary Note, Supplementary Tables 5–8 and Supplementary Figs. 2–3). In contrast to other recently published whole genome sequencing data examining high-risk T-cell leukemia, no complex rearrangements were identified in hypodiploid ALL.15
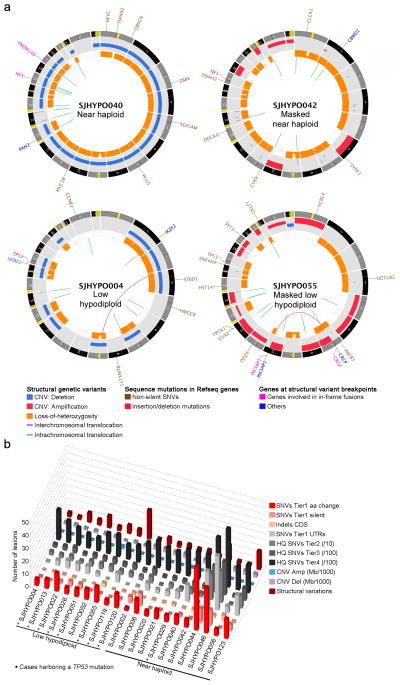
Figure 1. Mutation spectrum of whole genome sequenced hypodiploid ALL.
a, Circos68 plots of representative near haploid (top left), masked near haploid (top right), low hypodiploid (bottom left) and masked low hypodiploid (bottom right) cases. Depicted are structural genetic variants, including DNA copy number alterations, intra- and inter-chromosomal translocations, and sequence alterations. b, Whole genome sequenced cases are depicted from left to right. Colored bars represent number of specific lesions identified in each case as indicated. Masked hypodiploid cases here refer to cases with either a pure doubled hypodiploid clone or cases harboring a doubled clone constituting at least 30%.
Genome-wide profiling of DNA copy number alterations and loss-of-heterozygosity for leukemic and remission samples was performed for all cases using Affymetrix SNP 6.0 microarrays, and gene expression profiling using Affymetrix U133 Plus 2.0 microarrays (Fig. 2). Sanger sequencing was performed for selected exons of 8 genes (CBL, CRLF2, FLT3, JAK1, JAK2, KRAS, NRAS, and PTPN11)or all coding exons for 11 genes (ETV6, IKZF1, IKZF2, IKZF3, KIF2B, MAPK1, NF1, PAG1, PAX5, RB1 and TP53; Supplementary Table 9). The most common targets of aneuploidy were chromosomes 1–7, 9, 11–13, 15–17, 19–20 and 22 in near haploid ALL, and chromosomes 2–4, 7, 9, 12–13, 15–17 and 20 in low hypodiploid ALL(Fig. 2a), as previously shown by cytogenetic analyses.11 Notably, both copies of chromosome 21 were always retained.
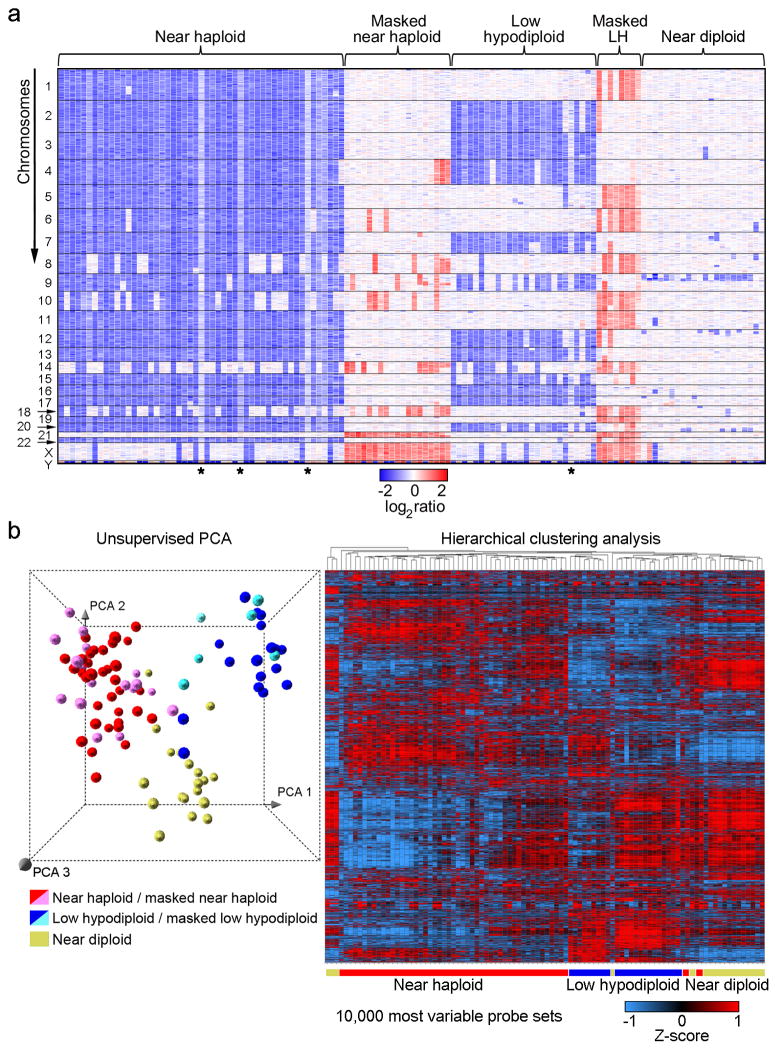
Figure 2. Genome-wide DNA copy number alterations and gene expression profiling of hypodiploid ALL.
a, Log2 ratio DNA copy number heatmap showing SNP 6.0 microarray data for the tumor samples from the pediatric hypodiploid ALL cohort. Chromosomes 1–22, X and Y are depicted from top to bottom, and individual samples are shown from left to right. An asterisk (*) indicates tumor samples with less than 70% blasts. LH, low hypodiploid. Blue indicates loss of genetic material and red, gain. Dicentric chromosomes most frequently involved chromosome arms 9p, and either 12p or 20q, and were predominantly observed in near diploid karyotypes. Copy-neutral loss-of-heterozygosity was observed of diploid chromosomes in masked hypodiploid cases (Supplementary Table 10 and Supplementary Fig. 2) consistent with duplication of the hypodiploid clone. b, Left panel: Unsupervised principal component analysis (PCA) of gene expression data from all hypodiploid ALL cases with available high quality RNA (N=94). PCA analysis using 1000 representative probesets selected by k means distinguishes near haploid/masked near haploid, low hypodiploid/masked low hypodiploid and near diploid subgroups. Right panel: Weighted Pair-Group Method with Arithmetic mean hierarchical clustering analysis performed using the 10,000 most variable probesets. Masked hypodiploid cases here refer to cases with a doubled hypodiploid clone constituting at least 30%.
Subtype specific genetic alterations in hypodiploid ALL
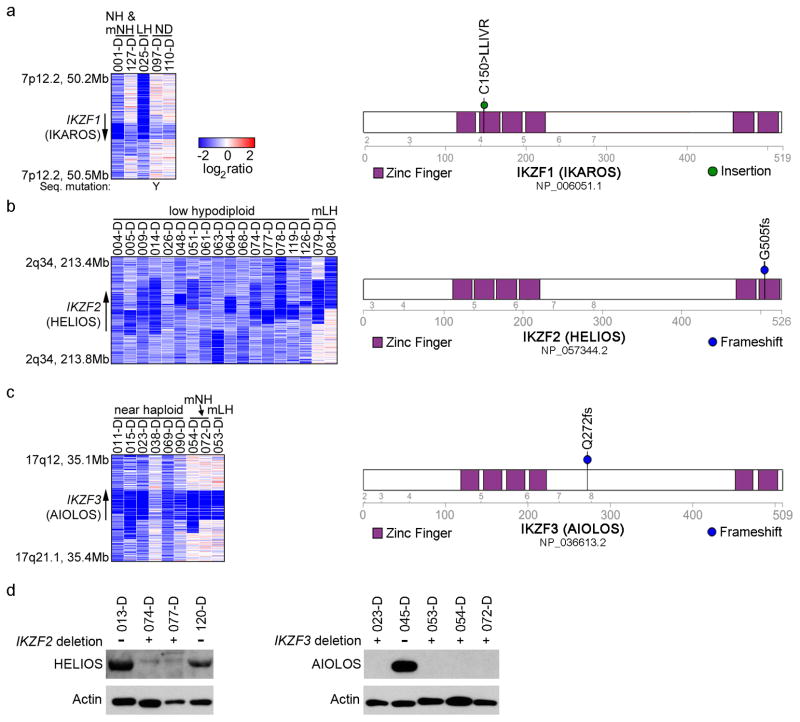
We identified several genes and pathways recurrently targeted by sequence mutations and submicroscopic deletions that were significantly associated with hypodiploid ALL subtype (Table 1 and Supplementary Tables 7–8 and 10–13). There were no significant differences in lesion frequency or transcriptional profiles between non-masked cases and cases that were masked or harbored a clone of at least 30% doubled hypodiploid cells. These cases were grouped together for subsequent analyses (Table 1, Fig. 2b and Supplementary Tables 13–15). Alterations targeting RTK- and Ras signaling were a hallmark of near haploid ALL (70.6% of cases; Table 1). Other recurrent alterations identified in near haploid ALL were deletion of the lymphoid transcription factor IKZF3 (AIOLOS; 13.2%) and deletions of a histone cluster at chromosome 6p22 (19.1%; Table 1).
Table 1. Recurrent copy number alterations and sequence mutations in hypodiploid ALL.
Percentages of cases with the respective alteration are indicated. _P_-values for association between gene alteration and hypodiploid ALL subtype were calculated by an exact chi-square test(2×3 tables) and are given for a comparison between only near haploid and low hypodiploid and between the three subgroups. Masked hypodiploid cases here refer to cases with either a pure doubled hypodiploid clone or cases harboring a doubled clone constituting at least 30%.
| Subgroup | NH | mNH | NH & mNH | LH | mLH | LH & mLH | ND | P value; NH & LH | P value; NH, LH & ND |
|---|---|---|---|---|---|---|---|---|---|
| N= | 50 | 18 | 68 | 26 | 8 | 34 | 22 | ||
| FLT3 | 10 | 5.6 | 8.8 | 0 | 0 | 0 | 0 | 0.074 | 0.075 |
| NF1 | 44 | 44.4 | 44.1 | 7.7 | 12.5 | 8.8 | 4.5 | 8.7×10−5 | 7.3×10−6 |
| KRAS | 4.0 | 0 | 2.9 | 0 | 0 | 0 | 9.1 | 0.31 | 0.17 |
| NRAS | 14 | 16.7 | 14.7 | 0 | 0 | 0 | 18.2 | 0.019 | 0.046 |
| PTPN11 | 2.0 | 0 | 1.5 | 0 | 0 | 0 | 9.1 | 0.48 | 0.072 |
| RTK-/Ras combined | 70 | 72.2 | 70.6 | 7.7 | 12.5 | 8.8 | 31.8 | 7.2×10−10 | 1.7×10−9 |
| IKZF1 | 2.0 | 11.1 | 4.4 | 3.8 | 0 | 2.9 | 9.1 | 1.0 | 0.41 |
| IKZF2 | 0 | 5.6 | 1.5 | 61.5 | 25 | 52.9 | 0 | 3.1×10−10 | 7.9×10−12 |
| IKZF3 | 14 | 11.1 | 13.2 | 0 | 12.5 | 2.9 | 0 | 0.099 | 0.061 |
| TP53 | 2.0 | 5.6 | 2.9 | 96.2 | 75 | 91.2 | 4.5 | 2.1×10−18 | 1.7×10−20 |
| RB1 | 10 | 5.6 | 8.8 | 42.3 | 37.5 | 41.2 | 0 | 1.0×10−4 | 1.2×10−5 |
| Histone, 6p22; * | 18 | 22.2 | 19.1 | 3.8 | 0 | 2.9 | 9.1 | 0.025 | 0.060 |
| CDKN2A/B * | 24 | 16.7 | 22.1 | 11.5 | 62.5 | 23.5 | 77.3 | 0.87 | 4.07×10−6 |
| PAX5 | 10 | 0 | 7.4 | 7.7 | 0 | 5.9 | 59.1 | 0.78 | 1.2×10−8 |
| PAG1 | 12 | 5.6 | 10.3 | 3.8 | 0 | 2.9 | 0 | 0.19 | 0.14 |
In contrast, low hypodiploid ALL was highly enriched for alterations in TP53 (91.2%), RB1 (41.2%) and the IKAROS family gene IKZF2 (HELIOS; 52.9%; Table 1). Adult low hypodiploid ALL cases also had a remarkably high frequency of TP53 mutations (90.9%), indicating that mutations of TP53 are a hallmark of low hypodiploid ALL (Supplementary Table 16).
Mutations targeting RTK- and Ras signaling in near haploid ALL
More than two-thirds (70.6%) of near haploid ALL cases harbored genetic alterations known or predicted to result in activation of RTK- or Ras signaling, including deletion, amplification and/or sequence mutation of NF1, NRAS, KRAS, MAPK1, FLT3 and PTPN11. RTK- and Ras signaling alterations were less frequent in low hypodiploid (8.8%) and near diploid ALL (31.8%; _P_=2.47×10−5; Table 1, Fig. 3 and Supplementary Table 12).
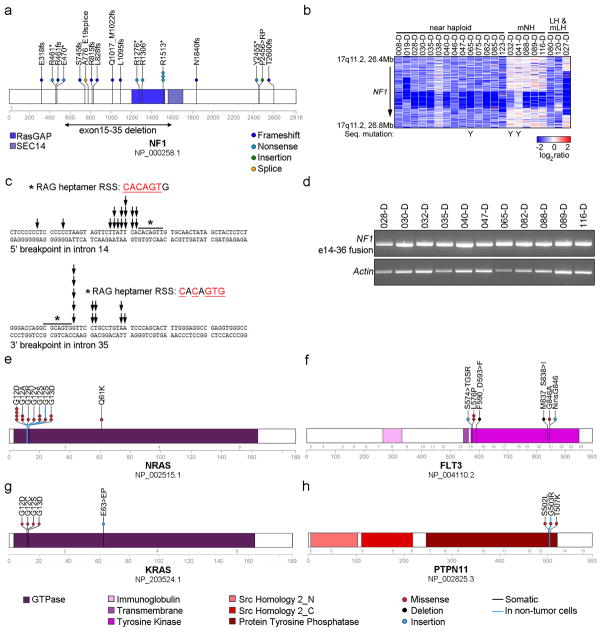
Figure 3. Recurrent alterations of Ras- and RTK signaling in near haploid ALL.
a, Protein domain plot of NF1 depicting identified alterations. The recurrent exon 15–35 deletion in NF1 is indicated by a double-headed arrow. b, SNP 6.0 microarray heatmap showing deletions in NF1. Blue indicates DNA loss. Cases with simultaneous NF1 deletion and sequence mutation are indicated by a Y. Masked hypodiploid cases here include all cases harboring a doubled clone constituting at least 30%. mNH, masked near haploid; LH, low hypodiploid; mLH, masked low hypodiploid. c, The genomic breakpoints for the NF1 deletions cluster in two regions. The individual breakpoints are indicated by arrows. The 5′-break (upper panel) is most frequently present in intron 14, and the 3′-break (lower panel) in intron 35. Immediately internal to the 5′- and 3′-breakpoints are putative, partially conserved RAG heptamer recombination signal sequences (RSS) present, and an insertion of a variable number of non-consensus nucleotides in between the two breakpoints were seen for all these cases. d, RT-PCR on cases harboring an NF1 exon 15–35 deletion, using forward and reverse primers complementary to regions in NF1 exons 14 and 36, respectively, rendering a 648 bp fragment. e–h, Protein domain- and alteration plots for other Ras- and RTK signaling related genes recurrently targeted by sequence mutations in hypodiploid ALL, including NRAS, KRAS, FLT3 and PTPN11 (schematic of MAPK1 can be found in Supplementary Fig. 4). Blue line indicates alterations present also in non-tumor cells.
Thirty of 68 near haploid cases (44.1%) had focal deletions or sequence mutations of NF1 (Fig. 3a and Supplementary Table 12). Due to aneuploidy the NF1 alterations were biallelic in 76.7% of near haploid cases. NF1 encodes Neurofibromin, a tumor suppressor, Ras-GTPase Activating Protein (GAP) and negative regulator of Ras signaling16. Alterations of NF1 result in neurofibromatosis16–19 and juvenile myelomonocytic leukemia (JMML)20. NF1 alterations also occur in T-lineage ALL, acute myeloid leukemia21 and non-hematopoietic malignancies such as glioblastoma.22 Furthermore, neurofibromatosis is associated with an increased risk of developing leukemia.23,24
In contrast to other tumors, in which NF1 deletions usually involve the entire gene25, the focal NF1 deletions in hypodiploid ALL were always intragenic, involving exons 15 through 35 in 17 of 21 cases(Fig. 3b–c, Supplementary Table 12 and Supplementary Fig. 5). This focal deletion has not previously been described, and results in deletion of the GAP domain. Sequencing of the genomic breakpoints suggests that these deletions result from aberrant recombinase-activating gene activity (Fig. 3c and Supplementary Fig. 5b), which has been implicated in the generation of other chromosomal rearrangements and deletions in ALL.26,27
To further characterize the transcriptional consequences of the exon 15–35 NF1 deletion, we performed RT-PCR for NF1 in 17 hypodiploid ALL cases, and mRNA-seq for NALM-1628, a near haploid cell line harboring the same deletion of NF1 exons 15–35. The deletion results in splicing of exon 14 to 36, causing a frame-shift in exon 36 (Fig. 3d and Supplementary Fig. 5). Immunoblotting of NALM-16 indicated that the deletion results in loss of NF1 expression (Supplementary Fig. 6).
We identified RTK/Ras pathway alterations in matched non-tumor DNA from two near haploid cases (Supplementary Fig. 7). A PTPN11 p.Gly503Arg substitution(case SJHYPO036) was heterozygous in remission DNA, and homozygous in the tumor due to aneuploidy. This substitution has been reported previously as a somatic and inherited variant in JMML and Noonan syndrome (NS), and is located in a mutational hotspot region in hematopoietic malignancies and NS.29,30 Exchange of this amino acid results in constitutive activation of the phosphatase.29 An NRAS p.Gly12Ser substitution was identified in remission DNA from case SJHYPO020, and verified by sequencing DNA from normal T-cells purified from bone marrow in this patient (Supplementary Fig. 7). Inherited NRAS mutations are rare in hematopoietic disorders31 and have not previously been described in ALL. The p.Gly12Ser may have a lower transforming potential than other p.Gly12 alterations and thus may be tolerated as inherited.32 These results suggest either an inherited predisposition to leukemia, or acquisition of the RTK/Ras pathway mutations as de novo germline mutations or in a primitive hematopoietic progenitor cell in these patients.
Recurrent alterations of PAG1 in near haploid ALL
PAG1, encoding Phosphoprotein associated with glycosphingolipid microdomains 1, also known as Csk Binding Protein (CBP), was altered in 10.3% of near haploid ALL, but rarely in the other hypodiploid ALL subtypes (Supplementary Table 1). Mutations in this gene have not previously been described in leukemia. The majority of PAG1 deletions was homozygous and involved the upstream region and the first exon, leading to a complete loss of PAG1 expression (Supplementary Fig. 8).
PAG1 is mainly known as an adapter for C-terminal Src kinase in T-cells, and thereby having a role within negative regulation of Src family kinases.33–35 PAG1 has also been identified as a putative Ras signaling inhibitor36, and to have a negative regulatory function in proximal B-cell receptor signaling.37
TP53 mutations in low hypodiploid ALL
A striking finding was TP53 alterations in 91.2% of pediatric low hypodiploid ALL, in comparison to less than 5% of non-low hypodiploid B-ALL38,39(_P_=1.65×10−20; Table 1, Fig. 4 and Supplementary Table 18). Thirty (88.2%) cases had a sequence mutation, the majority of which were homozygous due to aneuploidy. RT-PCR identified an additional case that expressed an aberrantly spliced isoform lacking exons 2–6, including the translational start site, with no expression of the wild-type allele (data not shown). The TP53 mutations included missense, nonsense and insertion/deletion mutations and are in general predicted to be loss-of-function mutations (Fig. 4a and Supplementary Table 18).
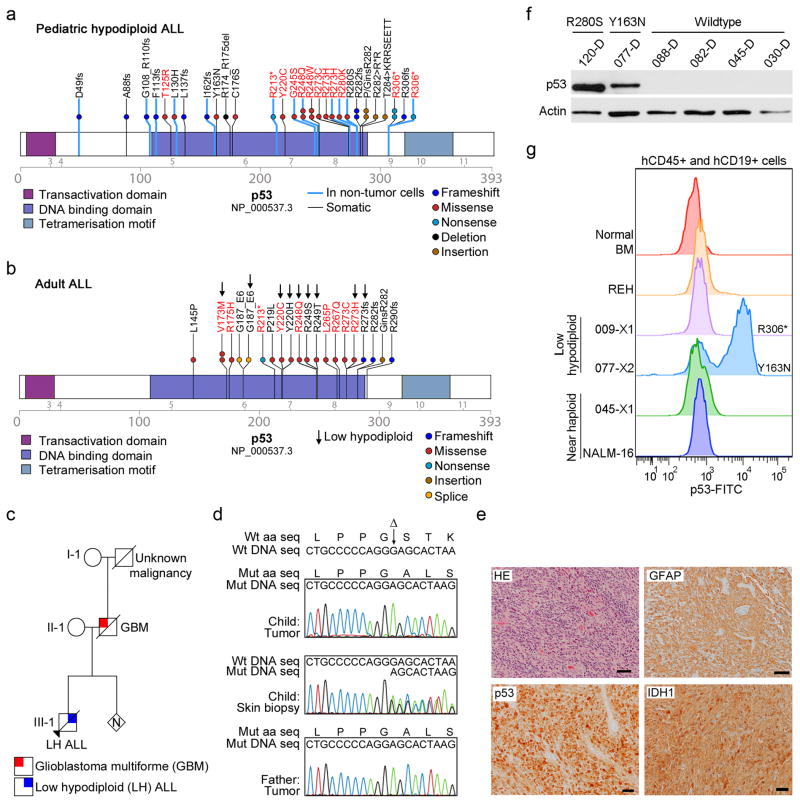
Figure 4. Frequent mutations in TP53 in low hypodiploid ALL.
a–b, Protein domain plots of p53 with alterations identified in (a) pediatric hypodiploid ALL and (b) adult ALL. Known LFS alterations are indicated in red. Alterations present in non-tumor cells are indicated by blue lines. Arrows indicate alterations identified in adult low hypodiploid cases in b. c, Pedigree of a family with an inherited TP53 mutation. N, number of siblings. d, Electropherograms showing the inherited TP53 g.13886delG mutation (resulting in p.G302fs) in the proband (child; leukemic bone marrow (top) and normal skin biopsy(middle)) and father (lower panel). Wild-type (Wt) DNA and amino acid (aa) sequences (seq) are depicted at the top of the panel. Δ indicates the deleted nucleotide. e, Histologic examination of tumor from the proband’s father indicated a diagnosis of glioblastoma. Poorly differentiated glial cells are admixed with sparse cells showing an astrocytic phenotype. Many neoplastic cells are GFAP-immunopositive, and all express the mutant TP53 and IDH1 (p.Arg132His) gene products. Scale bar corresponds to 50 microns. HE, hematoxylin and eosin. f, Immunoblot of p53 on primary hypodiploid ALL samples harboring either wild-type TP53 or a TP53 missense mutation as indicated. g, Flow cytometric analysis of spleen samples from xenografted mice and NALM-16 detecting p53 levels. Only cells positive for human CD45 and CD19 were analyzed. Known nonsense and missense alterations in p53 are indicated. BM, bone marrow.
Almost half (43.3%) of the TP53 mutations in pediatric low hypodiploid ALL were present in non-tumor hematopoietic cells (Supplementary Table 18), suggestive of an inherited origin or acquisition as de novo mutations in the germline or hematopoietic compartment (a comprehensive listing of putative deleterious sequence variants present in non-tumor cells is provided in the Supplementary Note and Supplementary Table 7). Moreover, many of the mutations have previously been reported as LFS associated mutations.40 The most common LFS substitution p.Arg248Trp was present in three low hypodiploid cases, and prevents p53 from binding into the minor groove of its DNA binding site.41 Further, the p.Arg306* alteration present in three cases has previously been identified as both somatic and inherited.40
In support of an inherited origin of the TP53 mutations, we studied a kindred in which a child with low hypodiploid ALL harbored a p53 p.Gly302fs alteration that was homozygous in leukemic cells, and heterozygous in a skin biopsy lacking tumor involvement (Fig. 4c–d and Supplementary Note). The proband’s father developed glioblastoma multiforme, a LFS associated malignancy, and the same homozygous mutation was identified in a tumor sample obtained at diagnosis (Fig. 4d–e).
TP53 mutations were also identified in 10 of 11 adult low hypodiploid cases (90.9%) but only in 8 of 106 (7.5%)non-low hypodiploid ALL cases (_P_=5.03×10−9; Fig. 4b and Supplementary Tables 2 and 16). In contrast to pediatric low hypodiploid ALL, all TP53 mutations observed in adult cases with available remission DNA were somatic. As previously described42,43, TP53 mutations were also identified at relapse (5 of 18 adult ALL relapse cases(27.8%) and one of 5 pediatric hypodiploid ALL cases with available relapse material).
Missense TP53 mutations may result in elevated p53 levels.44 Immunoblotting, immunohistochemistry- and flow cytometry analysis of primary hypodiploid ALL patient material and cells harvested from NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice xenografted with primary hypodiploid ALL cells demonstrated increased levels of p53 in cases harboring missense mutations, but not in cases lacking TP53 mutations (Fig. 4f–g, Supplementary Fig. 9 and data not shown). Expression of CDKN1A, encoding the cyclin-dependent kinase inhibitor p21, is directly regulated by p53.45 The inactivity of one of the p53 mutants (p.Arg280Ser, xenograft SJHYPO120-X) was demonstrated by lack of increase in p21 levels upon treatment of the leukemia cells with etoposide (Supplementary Fig. 9a).
Frequent deletions of IKZF2 and IKZF3 in hypodiploid ALL
Alteration of IKZF1 is a hallmark of high-risk non-hypodiploid B-progenitor ALL5,27,39, but alteration of other IKAROS family genes is rare. In contrast, IKZF1 alterations were infrequent in hypodiploid ALL, while IKZF2 deletion was highly recurrent in low hypodiploid ALL (18 of 34 cases, 52.9%; Fig. 5; Supplementary Table 17). IKZF2 deletions were also common in adult low hypodiploid ALL, with 4 of 11 cases (36.4%) harboring deletions, compared to 2 of 106 adult non-low hypodiploid ALL cases (1.9%; _P_=0.0005; Supplementary Table 19 and Supplementary Fig. 10).
Figure 5. Recurrent deletions of the IKAROS family genes IKZF2 (HELIOS) and IKZF3 (AIOLOS).
a–c, SNP 6.0 microarray heatmaps (left panel) and schematics (right panel) of the alterations identified in the IKAROS family members IKZF1 (IKAROS, a), IKZF2 (HELIOS, b) and IKZF3 (AIOLOS, c). Blue indicates loss of DNA in the heatmaps. One IKZF1 altered case harbors a simultaneous deletion and sequence mutation, indicated by a Y in the left hand panel of a. d, Immunoblot analysis on primary hypodiploid ALL patient samples using the antibodies sc-9866 detecting HELIOS (left), and sc-101982 detecting AIOLOS (right). Deletions in the respective gene are indicated by “+” and lack thereof by “−”.
Alterations of IKZF3 were common in near haploid ALL, with deletions and one frame-shift mutation in 9 of 68 cases (13.2%), but rare in non-near haploid ALL (_P_=0.061; Fig. 5c, Supplementary Table 17). Alterations of IKZF2 and IKZF3 were bi-allelic due to aneuploidy(Supplementary Table 12).
The IKAROS gene family encodes zinc finger transcription factors involved in lymphoid development and differentiation.46 The founding member IKZF1 is essential for lymphopoiesis, and Ikzf1 inactivation leads to complete lack of B-lymphoid cells in mice.47,48 Aiolos has a key role in B-cell differentiation and associates with Ikaros in an Ikaros-NuRD complex49, while Helios has a role in regulatory T-cell development.50–52 During B cell development, Ikzf2 expression is highest in common lymphoid progenitors and pre-pro-B cells, whereas Ikzf3 expression is highest in more mature lymphoid precursors (Supplementary Fig. 11 and refs 53,54), suggesting that the alterations of Ikzf2 and Ikzf3 in low hypodiploid and near haploid ALL, respectively, occur in cells at different stages of lymphoid maturation. Accordingly, the frequency of antigen receptor rearrangements and expression of CD19 are lower in low hypodiploid ALL than non-low hypodiploid B-ALL (Supplementary Fig. 12a–c). Further, low hypodiploid cases have a similar transcriptional profile as immature murine lymphoid progenitors (Hardy stage B; Supplementary Fig. 12d). Together, these observations suggest that low hypodiploid ALL arises from transformation of a less mature lymphoid progenitor than near haploid ALL.
RB1 alterations in low hypodiploid ALL
Alterations of RB1, encoding Retinoblastoma 1, were present in 14 of 34 (41.2%) low hypodiploid cases compared to 6 of 68 (8.8%) near haploid cases and were absent in near diploid ALL (_P_=1.19×10−5; Table 1 and Supplementary Fig. 13a–b). Thirteen (92.9%)_RB1-_altered low hypodiploid cases had concomitant TP53 mutations, while only one (7.1%) had co-occurrence of RB1 and CDKN2A/CDKN2B alterations. A total of 61.8% of low hypodiploid ALL harbored either RB1 or CDKN2A/CDKN2B alteration, suggesting that these are complementary alterations disrupting the RB1 tumor suppressor pathway (Fig. 6a and Supplementary Fig. 14).
Figure 6. Recurring mutations in hypodiploid ALL.
a, Data are shown for the entire hypodiploid ALL cohort split up in the near haploid (including masked near haploid cases), low hypodiploid (and masked low hypodiploid) and near diploid subgroups. b, Data matrix as above, but only including genes involved in histone modification identified in samples sequenced by either whole genome- or exome sequencing. SV, structural variation; Indel, insertion/deletion mutation; SNV, single nucleotide variation; Age, Age at diagnosis; Ind. failure, Induction treatment failure; NGS, next-generation sequencing; WGS, whole genome sequencing; WES, whole exome sequencing; GEP, gene expression profiling.
Similarly, 2 of 11 (18.9%) adult low hypodiploid ALL cases had RB1 deletions, compared to 10 of 106 (9.4%) non-low hypodiploid ALL cases (Supplementary Table 19 and Supplementary Fig. 13c).
High frequency of alterations targeting histones and histone modifiers
Focal deletions of a histone gene cluster at 6p22 were common in near haploid ALL (19.1%; Supplementary Fig. 15a). The minimal region of deletion involved HIST1H2BE, HIST1H4D, HIST1H3D, HIST1H2AD and HIST1H2BF. These deletions have been previously observed in non-hypodiploid ALL, albeit at a lower frequency (8.1% of B-ALL cases5).
Whole genome- and exome sequencing identified a range of mutations in genes encoding proteins involved in histone modifications (Fig. 6b, Supplementary Note and Supplementary Table 20). Twenty-six histone modifier gene mutations were identified in 16 near haploid cases (64% of the 25 next-generation sequenced near haploid cases), and 9 mutations in 9 low hypodiploid ALL samples (60% of 15 cases). The most common target of alteration was the transcriptional co-activator and histone acetyltransferase CREBBP55, with 8 of 25 near haploid cases (32%) harboring a deletion or sequence mutation (Supplementary Fig. 15b). Mutations in CREBBP are uncommon at diagnosis in ALL but enriched at relapse6, and this is the first ALL subtype found to have a high frequency of CREBBP mutations at diagnosis. Two of the CREBBP substitutions(p.Arg1446Cys and p.Gln1500Pro) occur at residues in the histone acetyltransferase domain, important for the histone acetylase activity of CREBBP.6,39 Further, a CREBBPp.Lys389_Met395>Lys alteration overlaps with a deletion in murine Crebbp that enhances Myc-driven lymphomagenesis.56 Three missense mutations with predicted deleterious effect were identified in EZH2, the product of which catalyzes trimethylation of histone 3 lysine 27.57 Two of these mutations appeared in the same relapse sample and were not present at diagnosis (Supplementary Tables 7 and 20).
Gene expression profiling distinguishes subgroups of hypodiploid ALL
The observation of distinct genomic alterations in near haploid and low hypodiploid ALL was recapitulated by gene expression profiling (Table 1 and Supplementary Table 1). Near haploid, low hypodiploid and near diploid cases formed discrete clusters in unsupervised hierarchical clustering and principal component analysis (PCA; Fig. 2b). Further, differential expression analysis between near haploid and low hypodiploid ALL resulted in more than 15,000 differentially expressed probesets (Supplementary Table 21). Similar PCA clustering was observed when restricting the analyses to probes on chromosomes showing identical patterns of aneuploidy between near haploid and low hypodiploid ALL (Supplementary Fig. 16). This indicates that aneuploidy is not responsible for the major differences observed, but that there is a global difference in expression profiles between the hypodiploid ALL subgroups. Furthermore, more than 600 gene sets showed subtype-specific enrichment on gene set enrichment analysis (Supplementary Table 22).
Activation of signaling pathways in hypodiploid ALL
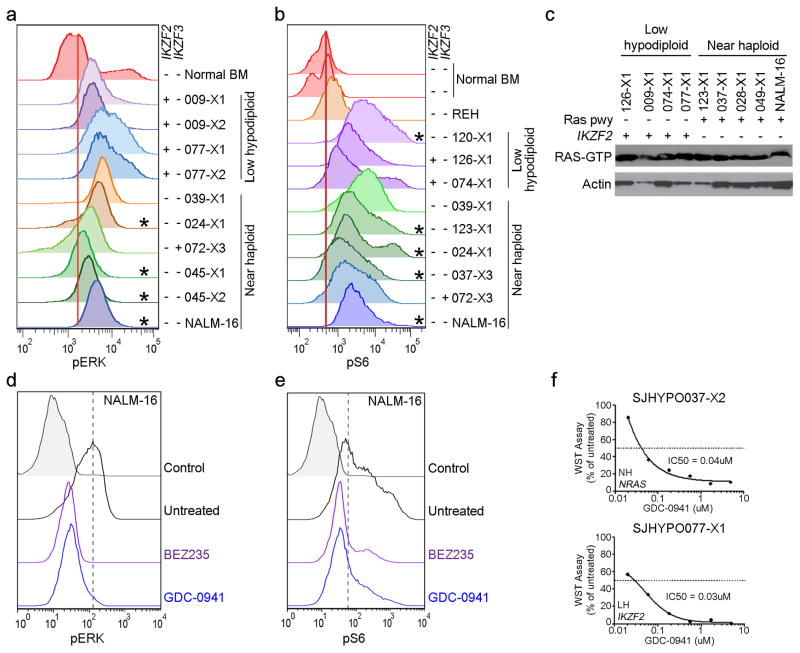
The finding of a high frequency of Ras- and RTK signaling lesions suggested that activation of these pathways is important in the pathogenesis of near haploid ALL. We examined activation of Ras- and related signaling pathways in 8 near haploid and 6 low hypodiploid tumor xenografts. Flow cytometric analysis of pERK, pmTOR and pS6 and assessment of Ras-GTP levels demonstrated activated Ras- and PI3K signaling in tumors with canonical Ras pathway mutations, but also in low hypodiploid tumors lacking mutations in genes known to regulate Ras- and PI3K signaling (Fig. 7 and Supplementary Fig. 17). These findings suggest that alternative mutational mechanisms activate these pathways in low hypodiploid cases lacking Ras/PI3K pathway mutations. Deletions of IKZF2 in low hypodiploid ALL were exclusively observed in cases lacking Ras pathway mutations(Fig. 6a). To investigate the potential role of IKZF2 inactivation in regulation of Ras signaling, we performed shRNA-mediated knockdown of Ikzf2 and Ikzf3 in murine Ba/F3 and Arf−/− pre-B cell lines (Supplementary Fig. 18), which resulted in an increase of both pERK and pS6, suggesting that HELIOS and AIOLOS, in addition to their roles within lymphoid development, may also modulate Ras- and PI3K signaling (Supplementary Fig. 19).
Figure 7. Activation of Ras signaling.
a–b, Flow cytometric analyses of spleen samples from xenografted mice and the near haploid cell line NALM-16 detecting pERK (a) and pS6 (b)levels. Only cells positive for human CD45 were analyzed. *Cases with a known mutation in the Ras signaling pathway. IKZF2 and IKZF3 deletions are indicated by “+” and lack thereof by “−”. c, Immunoblot showing levels of Ras-GTP as assessed by a Raf-Ras binding domain bead pulldown assay in low hypodiploid and near haploid ALL xenograft samples and NALM-16. Presence of Ras pathway (pwy) and IKZF2 alterations are indicated by “+”. d–e, Detection of pERK (d) and pS6 (e) in non-stimulated NALM-16 with and without treatment with the PI3K inhibitor GDC-0941 (0.7uM) and the PI3K/mTOR inhibitor BEZ235 (0.3uM) for 1 hour. The drug concentrations correspond to the respective IC50 value gained from proliferation assays on NALM-16 (Supplementary Table 23). f, ex vivo proliferation assay on near haploid (upper, SJHYPO037-X2) and low hypodiploid (lower, SJHYPO077-X1) xenograft cells harboring the indicated mutations (Supplementary Table 12). BM, bone marrow; NH, near haploid; LH, low hypodiploid.
We next examined the potential for targeting Ras-, PI3K- and mTOR signaling in hypodiploid ALL. The NALM-16 near haploid cell line and a panel of near haploid and low hypodiploid xenograft cells were treated with the MEK inhibitors PD0325901 and MEK162, the PI3K inhibitor GDC-0941 and the dual PI3K/mTOR inhibitor BEZ235. Notably, MEK inhibition did not affect proliferation of the samples tested, but the PI3K- and dual PI3K/mTOR inhibitors substantially inhibited proliferation of all tumors examined, and efficiently attenuated intrinsic PI3K signaling in NALM-16 (Fig. 7d–f and Supplementary Table 23) suggesting that inhibition of the PI3K pathway may serve as a novel treatment alternative in both near haploid and low hypodiploid ALL.
DISCUSSION
Here we report the first genomic analysis of hypodiploid ALL, a leukemic subtype which is associated with poor outcome that hitherto has been poorly understood. These analyses indicate that near haploid and low hypodiploid ALL are two distinct genetic subtypes characterized by distinct focal deletions and sequence mutations. More than 70% of near haploid ALL cases harbor activating Ras- and RTK signaling alterations, while the majority of low hypodiploid ALL cases harbor TP53 alterations (91.2%) and alterations of the lymphoid transcription factor IKZF2 (52.9%), and a high frequency (41.2%) of RB1 alterations (Table 1 and Figs. 3–5). The marked enrichment for Ras pathway, RB1 and TP53 alterations is unusual in non-hypodiploid ALL, and reminiscent of many solid tumors that more frequently also harbor aneuploid genomes.58–60
The remarkably high frequency of TP53 alterations in both pediatric and adult low hypodiploid ALL (91.2% and 90.9%, respectively) suggests that alteration of this gene is an important event in the pathogenesis of low hypodiploid ALL. Furthermore, almost half of the TP53 mutations identified in pediatric low hypodiploid ALL were present as heterozygous mutations in purified normal T-cell populations and the majority of these are known LFS associated mutations40 (Supplementary Table 18). Importantly, we confirmed the inherited origin of the TP53 mutation identified in one pediatric low hypodiploid case by studying non-hematopoietic cells (Fig. 4c–d and Supplementary Note). Further, two of the cases have a known familial history of cancer, while familial cancer history for the other cases is unknown (data not shown). Together, these data strongly suggest that the TP53 mutations are inherited, and suggest that low hypodiploid ALL is a manifestation of LFS.
Catastrophic DNA rearrangements referred to as chromothripsis61 have recently been linked with inherited TP53 mutations in pediatric brain tumors.62 In contrast, there was no evidence of chromothripsis in _TP53-_altered hypodiploid ALL (Fig. 1 and Supplementary Figs. 2–3). TP53 mutations have also been associated with aneuploidy63–65, and may contribute to the development of aneuploidy in low hypodiploid ALL. However, the genetic basis of aneuploidy in near haploid ALL, which lack TP53 mutations, is unknown, and our genome-wide analyses have not identified any other recurrent gene alterations that may contribute to the development of aneuploidy.
RB1 is inactivated in a wide variety of solid tumors60,66 and hematopoietic malignancies, but is uncommonly mutated in B-lineage ALL.39 In contrast, deletion of the CDKN2A/CDKN2B loci is common in ALL and may substitute for the low frequency of RB1 and TP53 alterations seen in ALL.3,39 Low hypodiploid ALL is thus notable for the high frequencies of TP53 and RB1 inactivation seen in both pediatric and adult cases. The majority of RB1 alterations were mutually exclusive with CDKN2A/CDKN2B deletions, while TP53 mutations mostly co-occurred with deletions at these loci (Fig. 6a and Supplementary Fig. 14). This suggests that alteration of p53 provides a survival advantage for the leukemic cells that cannot be compensated for by loss of CDKN2A, whereas RB1 and CDKN2A/CDKN2B loss represent alternative mechanisms of inactivating the RB1 tumor suppressor pathway.
The finding of IKZF2 deletions in 52.9% of low hypodiploid ALL and IKZF3 alterations in 13.2% of near haploid ALL was unexpected and rarely observed in any other hematopoietic or solid tumor. IKZF1 is essential for lymphopoiesis46, and alterations of this gene are associated with poor outcome in B-ALL5, whereas less is known about the roles of IKZF2 and IKZF3 in lymphoid development and leukemogenesis. The distinct mutational patterns of the IKAROS gene family within different ALL subtypes may indicate either that the different gene family members have distinct roles in different ALL subtypes, and/or that the different ALL subtypes have cells of origin from a developmental stage where a certain IKAROS gene family member is expressed. A low frequency of antigen receptor rearrangements, reduced expression of CD19, similarity of the transcriptional profile to immature murine lymphoid progenitors, and expression of murine Ikzf2 in common lymphoid progenitors and pre-pro-B cells, together suggest that low hypodiploid ALL arises from transformation of a less mature lymphoid progenitor than near haploid or non-hypodiploid ALL(Supplementary Figs. 11–12).
In previous studies, murine AIOLOS has been shown to interact with Ras, potentially acting as an inhibitor67. We observed that IKZF2 and Ras pathway alterations are usually mutually exclusive (Fig. 6a), and that there is evidence of activation of downstream Ras- and PI3K signaling in low hypodiploid ALL in the absence of known activating mutations (Fig. 7 and Supplementary Fig. 17). Further, higher levels of pERK and pS6 were observed after knockdown of Ikzf2 and Ikzf3 in cell lines (Supplementary Fig. 19). This raises the possibility that HELIOS and AIOLOS are modulators of Ras/PI3K signaling and that inactivation of these gene products may drive tumor cell proliferation. In addition, PI3K/mTOR inhibitors showed a remarkable potency in ex vivo drug studies on both near haploid and low hypodiploid ALL tumors. Consequently, therapeutic targeting of PI3K/mTOR should be further investigated as a therapeutic modality in the treatment of these high-risk ALL subtypes.
METHODS
Patients and samples
One hundred and twenty-four pediatric patients with hypodiploid ALL treated on St Jude Children’s Research Hospital (SJCRH; N=29) and Children’s Oncology Group (COG; N=95) protocols were studied. Relapse material was available for 5 cases, and 89 cases had a matched remission sample (Supplementary Table 1). This cohort was compared with an adult ALL cohort of 117 cases, including 11 low hypodiploid ALL cases (Supplementary Table 2).
Informed consent was obtained in accordance with the Declaration of Helsinki. Study approval was obtained from the SJCRH and COG institutional review boards (IRB), and the IRBs of the Royal Adelaide Hospital.
Mononuclear cells were purified from bone marrow or peripheral blood by density gradient centrifugation, and cryopreserved in liquid nitrogen. Diagnostic samples were characterized as hypodiploid ALL samples by karyotyping and DNA index measurement.
DNA was directly extracted from tumor samples with more than 70% blasts and remission samples using either phenol/chloroform or a DNA Blood Mini Kit (Qiagen, Valencia, CA), and DNA integrity was assessed by agarose gel electrophoresis. Tumor samples with less than 70% blasts and sufficient material were flow sorted prior to DNA extraction using FACSVantage SE flow cytometers and phycoerythrin (PE) labeled CD19 (BD Biosciences, San Jose, CA) and fluorescein isothiocyanate (FITC) labeled CD45 (Life Technologies, Carlsbad, CA) antibodies. Tumor samples from cases harboring TP53 mutations lacking a matched remission sample were flow sorted using CD45-FITC and allophycocyanin (APC) labeled CD7 (Life Technologies) antibodies to get access to normal T-cells to test the somatic status of the mutations. DNA was whole genome amplified using Qiagen Repli-G mini kit (Qiagen). DNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer or fluorometrically by the Quant-iT BR kit (Life Technologies) using a Synergy HT (BioTek). RNA was extracted from tumor samples using TRIzol (Life Technologies).
Cell lines
Cell lines used were the near haploid cell line NALM-16 (DSMZ #ACC-680), and the murine cell line Ba/F3 and primary Arf−/− pre-B cells (harvested from Arf−/− C57BL/6 mouse bone marrow; e.g. Ref.69). Ba/F3 and Arf−/− pre-B cells were cultured in the presence of 10nM IL-3 and IL-7 (PeproTech), respectively. shRNAs targeting Ikzf2 and Ikzf3 are listed in Supplementary Table 24.
Next generation sequencing
Twenty cases underwent whole genome sequencing (WGS) of tumor and matched non-tumor DNA, and 20 cases whole exome sequencing (WES, Agilent SureSelect Human All Exon 50MB). DNA and RNA library preparations and paired-end sequencing was performed using either Illumina GAIIx or HiSeq 2000 analyzers as previously described.15,70,71
Analysis of sequence and structural variants was performed as previously described.15 Validation was performed on whole genome amplified tumor and matched normal DNA by PCR and either Roche 454 FLX sequencing or Sanger sequencing.
For the mRNA-seq data from the cell line NALM-16, putative variants were compared to a database of variants identified in 254 non-tumor samples sequenced as part of the SJCRH – Washington University Pediatric Cancer Genome Project (PCGP72), and variants present in dbSNP73 (excluding known cancer variants in OMIM and local databases). The remaining variants were also compared with exome variants identified by NHLBI Exome Sequencing Project (Exome Variant Server, NHLBI Exome Sequencing Project, Seattle, WA (http://evs.gs.washington.edu/EVS/)). Data are presented in Supplementary Table 25.
High-risk germline variant analysis
Novel, non-silent germline coding variants were identified as previously described.15 To further identify likely high-risk variants, we selected variants that fit one of the following categories: 1, match to any validated somatic mutation; 2, predicted truncating variants in known cancer genes (using a combined list of CancerGeneCensus genes, downloaded on 2011-03-22, and 120 frequently mutated genes in childhood ALL39); 3, overlap with sites (either chromosome coordinates or amino acid) where confirmed somatic variants have been reported by COSMIC (Catalogue Of Somatic Mutations In Cancer). Variants initially discovered by somatic pipeline but validated as germline were also included in the search. To further filter out common polymorphisms caused by deficiency in current human assembly, we compared the selected germline variants with common germline variants identified by NHLBI Exome Sequencing Project, and an internal database of germline variants collected from PCGP. We further removed variants present in germline but absent in tumor due to aneuploidy.
Single nucleotide polymorphism microarrays
Samples were genotyped using Affymetrix SNP 6.0 microarrays according to the manufacturer’s instructions. CEL files were generated using GeneChip Command Console Software. SNP calls were generated using Genotyping Console (Affymetrix) and the Birdseed v2 algorithm with default parameters with at least 50 arrays in each analysis.
Array normalization and copy number inference were performed according to a previously published workflow.3,27 The importance of correct normalization is clearly exemplified by the highly aneuploid hypodiploid ALL samples (Supplementary Fig. 20). Normalized data were viewed in dChip74 and regions with abnormal copy number identified computationally by circular binary segmentation (CBS)75 and analyzed as previously described.3,27
Recurrent focal deletions were validated by genomic PCR and/or RT-PCR, followed by Sanger sequencing. Oligonucleotide primers for PCR amplification were designed using Primer 3 (Ref. 76). Primers used for mapping the NF1 exon 15–35 deletion are present in Supplementary Table 26.
Targeted gene resequencing
Gene resequencing was performed for all 124 diagnosis samples, 2 relapse cases (SJHYPO056-R and SJHYPO117-R) and the hypodiploid cell line NALM-16 for the genes listed in Supplementary Table 9. Primer sequences are present in Supplementary Table 26. Sequencing was performed by PCR of whole genome amplified DNA, followed by sequencing using 3730xl instruments (Applied Biosystems) as previously described.6 Single nucleotide variations were detected by SNPdetector77 and PolyScan78, and validated by sequencing of both tumor and, where available, matched non-tumor samples. Variations identified in unpaired tumor samples were compared to online databases and to a local database of sequence variation.
Gene expression profiling
Gene expression profiling was performed using Affymetrix GeneChip HT HG-U133+ PM microarrays for human specimens (samples listed in Supplementary Table 1) and MG-430 2.0 microarrays for Hardy Fractions from murine cells (details listed in Supplementary Table 27), according to the manufacturer’s instructions. Arrays with outlier relative log expression metric were excluded from the data depository and downstream analysis. Statistical analyses, principal component analysis and unsupervised hierarchical clustering were performed using R 2.13.0 (Ref. 60), Bioconductor 2.6 (Ref. 61) and Spotfire Decision Site 9.1.1 (Tibco). For HT HG-U133+ arrays, all samples were normalized by the RMA algorithm. Probesets not passing the background signal (twice of the average signal on the control probes with different GC content) across all samples were excluded for differential expression analysis, where limma62 with estimation of false discovery rate (FDR)63 was performed between experimental subgroups. Probesets below a FDR cutoff of 0.05 were deemed differentially expressed. Weighted Pair-Group Method with Arithmetic mean clustering with Euclidean distance was performed using the 10,000 probesets with the highest standard deviation across samples. For MG-430 2.0 arrays, gene expression signals were scaled to a median value of 500 using the Affymetrix MAS 5.0 algorithm. Probesets with absent calls for all samples were excluded, and probeset signals were variance stabilized by adding the number 32 and subsequently log2-transformed. Top 50, 100, 250, and 500 gene signatures for each Hardy Fraction were identified using limma, and Gene Set Enrichment Analysis (GSEA)79 were used to examine the enrichment of the Hardy Fraction genesets for each hypodiploid ALL subgroup. Gene sets with less than 10 or greater 500 genes were excluded, and significantly enriched gene sets after 1000 permutations at a FDR of <0.25 are reported.
Immunohistochemical analyses
Immunohistochemistry of human and mouse tissues were performed using routine techniques, hematoxylin and eosin staining and appropriate primary antibodies.
Generation of xenografted mice
The mouse studies were approved by the SJCRH Institutional Animal Care and Use Committee. Primary leukemic cells were transplanted by tail vein injection into sub-lethally irradiated (250 cGy) immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (2–3 mice per tumor; The Jackson Laboratory). Engraftment was analyzed by bi-weekly retro-orbital bleeds followed by flow cytometry analysis of human CD45-FITC, CD19-PE and CD7-APC, and mouse CD45-PE-Cy7-stained peripheral blood cells. Mice were killed upon signs of disease, and tumors were characterized histologically, immunophenotypically (immunophenotypic markers as above) and by genomic profiling by Affymetrix SNP6.0 microarrays for a subset of samples (Supplementary Note and Supplementary Figs. 21–22).
Fluorescence activated cell sorting analysis
Cell lines were starved in serum free media for 5 hours prior to stimulation with PMA (50 nM; 15min) or treatment with BEZ235 or GDC-0941 (0.3uM and 0.7uM, respectively (IC50 values for NALM-16); 1 hour). Cell lines and splenocytes and bone marrow cells from xenografted mice were fixed with 2% paraformaldehyde and permeabilized with 90–95% methanol prior to staining with antibodies (Supplementary Table 28), and analysis on LSR II or FACS Calibur (Becton Dickinson). Data were collected using DIVA or CellQuest software (Becton Dickinson) and analyzed using FLOWJO (Tree Star) and Cytobank.
Immunoblotting and RAS-GTP assay
Cells were lyzed and protein concentrations determined using the BCA Protein Assay Kit (Pierce). Samples were separated on Criterion polyacrylamide (BioRad) or NuPAGE (Life Technologies) gels and transferred to Immobilon-P (Millipore) or PVDF (Life Technologies) membrane. After staining with the respective antibodies (Supplementary Table 28), target proteins were visualized using ECL (Amersham Biosciences), ECL plus (GE Healthcare) or the Femto Chemiluminescent Kit (Thermo Fisher). Ras-GTP levels were measured using a Raf-Ras binding domain (RBD) bead pulldown assay that has been described previously.80,81
Ex vivo drug studies
Cell viability and proliferation of bone marrow cells obtained from xenografted mice was determined using AO/PI staining (Nexcelom Bioscience LLC, Lawrence, MA) and WST-1 reagent (Roche, Indianapolis, IN) according to manufacturer’s instructions. Cells were treated with BEZ235, GDC-0941, MEK162, or PD0325901 at increasing concentrations as indicated in Supplementary Table 23 for 72 hours. The concentration inhibiting cell proliferation by 50% compared with control cells (IC50) was determined using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA).
Outcome analysis
Event-free survival (EFS) was defined as the time from diagnosis until the date of relapse or until the last follow-up date for all event-free survivors. Death was treated as a competing risk. Associations between genetic variables and EFS were estimated by the methods of Kaplan and Meier. Standard errors were calculated by the methods of Peto et al.82 The Mantel–Haenszel test was used to compare EFS estimates for patients with and without lesion at each locus.83 The proportional hazards model of Fine and Gray was used to adjust for age, presentation leukocyte count, aneuploidy subgroup and levels of minimal residual disease (MRD).84 Analyses were performed using R (www.r-project.org)85, SAS v9.1.2 (SAS Institute) and SPLUS 7.0 (Insightful Corp.). To evaluate associations between genetic alterations and MRD, and hypodiploid ALL subgroup and MRD, MRD data were converted into an ordinal variable (<0.01% (negative) and ≥0.01% (positive)) and association analyses performed using the Chi-Square test (FREQ procedure, SAS) with estimation of false discovery rate (MULTTEST, SAS). Significantly associated variables were adjusted for age, presentation leukocyte count and genetic subtype using logistic regression. Data are presented in Supplementary Note and Supplementary Tables 29–34.
Supplementary Material
Supplementary
Table S1
Table S10
Table S11
Table S12
Table S14
Table S15
Table S2
Table S21
Table S22
Table S25
Table S26
Table S7
Table S8
Acknowledgments
We thank John Morris, Emily Walker and Audra Merriman for performing SNP and gene expression microarrays, and Gerard Zambetti and Paul Brindle for insightful discussions of TP53 and CREBBP mutational data, respectively. We also thank Heather Mulder, Racquel Collins, Michael Barbato, Eileen Stonerock, Emilia Pinto and Megan Ellis for technical assistance, the Tissue Resources Core facility and the Flow Cytometry and Cell Sorting Core facility of SJCRH. This work was supported by The Henry Schueler 41&9 Foundation in conjunction with Partnership4Cures, St. Baldrick’s Foundation, NCI grants RC4CA156329, CA21765 and U01 GM92666, AACR Gertrude B. Elion Cancer Research Award and the American Lebanese and Syrian Associated Charities (ALSAC) of SJCRH. Support was also provided by NCI grants to the Children’s Oncology Group including CA98543, CA98413, and CA114766. L.H. was supported by the Swedish Research Council. S.P.H is the Ergen Family Chair in Pediatric Cancer. M.L.L. is a Clinical Scholar in the Leukemia Lymphoma Society and supported by the Frank A. Campini Foundation. C.G.M. is a Pew Scholar in the Biomedical Sciences and a St. Baldrick’s Scholar. M.L.L. and E.D-F. were supported by the Team Connor Foundation and S.N.P was supported by 5R25CA023944 from the National Cancer Institute. This paper is dedicated to Henry “Hank” Schueler who died from complications of hypodiploid ALL and whose Foundation (www.henryschueler.org) is dedicated to finding a cure for hypodiploid ALL in his memory, and to James B. Nachman M.D. who was instrumental in the genesis of this project and who recently passed away.
Footnotes
Author contributions. L.H., C.G.M., M.L.L. and S.P.H. designed the experiments. L.H. and D.P.-T. prepared patient samples and generated xenografts. L.H., E.D.-F., M.C., K.M., S.N.P. L.A.P. and S.L.H. performed biochemical analyses. C.D. and S.B. performed ex vivo drug studies. L.H. and J.M. analyzed SNP array data. L.H. and J.E. performed transcriptome sequencing. K.B. performed exome sequencing. L.H., D.P.-T. and B.V. performed sequencing validation. L.W., J.Z., L.D., J.B., X.C., C.L. analyzed sequence data. S.-C.C., A.A., G.N. and L.H. analyzed expression microarray data. M.R., E.H., M.P., G.W., R.H. and G.S. provided bioinformatic support. D.P., C.C. and M.D. performed statistical analyses. R.S.F. and L.L.F. supervised whole genome sequencing data generation. D.J.D. supervised the automated analysis pipeline. S.C.R., A.J.C., and N.A.H. performed cytogenetic analyses. M.W., C-H.P., M.M., I.D.L., L.B.T., P.M., G.R., W.S., M.L.L., J.M.G.-F. R.R., B.W., M.J.B. and S.P.H. provided clinical samples and data. D.W.E. performed pathological analyses. R.A.D. and H.G.D. provided important reagents. L.H. and C.G.M. wrote the manuscript. L.H., L.W., J.Z., L.D., D.P-T., M.C., A.A., S-C.C., K.M., J.B., J.M., G.W., G.S., J.E., M.P., X.C., M.R., E.H., C.L., R.S.F., L.L.F., D.J.D., K.O., S.A.S., C-H.P., E.R.M., R.K.W., J.R.D. and C.G.M. are part of the St Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project.
References
- 1.Pui CH, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360:2730–41. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuiper RP, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–66. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 4.Kuiper RP, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–64. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- 5.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–9. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison CJ, et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol. 2004;125:552–9. doi: 10.1111/j.1365-2141.2004.04948.x. [DOI] [PubMed] [Google Scholar]
- 8.Heerema NA, et al. Hypodiploidy with less than 45 chromosomes confers adverse risk in childhood acute lymphoblastic leukemia: a report from the children’s cancer group. Blood. 1999;94:4036–45. [PubMed] [Google Scholar]
- 9.Nachman JB, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood. 2007;110:1112–5. doi: 10.1182/blood-2006-07-038299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raimondi SC, et al. Reassessment of the prognostic significance of hypodiploidy in pediatric patients with acute lymphoblastic leukemia. Cancer. 2003;98:2715–22. doi: 10.1002/cncr.11841. [DOI] [PubMed] [Google Scholar]
- 11.Raimondi SC. Cytogenetics of acute leukemias. In: Pui CH, editor. Childhood Leukemias. Cambridge University Press; 2012. [Google Scholar]
- 12.Carroll AJ, et al. Masked Hypodiploidy: Hypodiploid Acute Lymphoblastic Leukemia (ALL) in Children Mimicking Hyperdiploid ALL: A Report From the Children’s Oncology Group (COG) AALL03B1 Study. Blood: ASH Annual Meeting Abstracts. 2009;114:1580. [Google Scholar]
- 13.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71:747–52. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 14.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2:475–84. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu GF, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 17.Cawthon RM, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 18.Viskochil D, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–92. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 19.Wallace MR, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–6. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 20.Side LE, et al. Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood. 1998;92:267–72. [PubMed] [Google Scholar]
- 21.Balgobind BV, et al. Leukemia-associated NF1 inactivation in patients with pediatric TALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111:4322–8. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- 22.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klopfenstein KJ, Sommer A, Ruymann FB. Neurofibromatosis-Noonan syndrome and acute lymphoblastic leukemia: a report of two cases. J Pediatr Hematol Oncol. 1999;21:158–60. doi: 10.1097/00043426-199903000-00014. [DOI] [PubMed] [Google Scholar]
- 24.Stiller CA, Chessells JM, Fitchett M. Neurofibromatosis and childhood leukaemia/lymphoma: a population-based UKCCSG study. Br J Cancer. 1994;70:969–72. doi: 10.1038/bjc.1994.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messiaen L, et al. Mosaic type-1 NF1 microdeletions as a cause of both generalized and segmental neurofibromatosis type-1 (NF1) Hum Mutat. 2011;32:213–9. doi: 10.1002/humu.21418. [DOI] [PubMed] [Google Scholar]
- 26.Marculescu R, et al. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair (Amst) 2006;5:1246–58. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 28.Kohno S, Minowada J, Sandberg AA. Chromosome evolution of near-haploid clones in an established human acute lymphoblastic leukemia cell line (NALM-16) J Natl Cancer Inst. 1980;64:485–93. [PubMed] [Google Scholar]
- 29.Kratz CP, et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 2005;106:2183–5. doi: 10.1182/blood-2005-02-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tartaglia M, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–90. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Filippi P, et al. Germ-line mutation of the NRAS gene may be responsible for the development of juvenile myelomonocytic leukaemia. Br J Haematol. 2009;147:706–9. doi: 10.1111/j.1365-2141.2009.07894.x. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda K, et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109:5477–80. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- 33.Kawabuchi M, et al. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature. 2000;404:999–1003. doi: 10.1038/35010121. [DOI] [PubMed] [Google Scholar]
- 34.Brdicka T, et al. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J Exp Med. 2000;191:1591–604. doi: 10.1084/jem.191.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidson D, Bakinowski M, Thomas ML, Horejsi V, Veillette A. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol Cell Biol. 2003;23:2017–28. doi: 10.1128/MCB.23.6.2017-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang SQ, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–55. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 37.Kalland ME, Solheim SA, Skanland SS, Tasken K, Berge T. Modulation of proximal signaling in normal and transformed B cells by transmembrane adapter Cbp/PAG. Experimental cell research. 2012;318:1611–9. doi: 10.1016/j.yexcr.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 38.Imamura J, Miyoshi I, Koeffler HP. p53 in hematologic malignancies. Blood. 1994;84:2412–21. [PubMed] [Google Scholar]
- 39.Zhang J, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–7. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 41.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–55. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 42.Hof J, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:3185–93. doi: 10.1200/JCO.2011.34.8144. [DOI] [PubMed] [Google Scholar]
- 43.Wattel E, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–57. [PubMed] [Google Scholar]
- 44.Rovinski B, Benchimol S. Immortalization of rat embryo fibroblasts by the cellular p53 oncogene. Oncogene. 1988;2:445–52. [PubMed] [Google Scholar]
- 45.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 46.Rebollo A, Schmitt C. Ikaros, Aiolos and Helios: transcription regulators and lymphoid malignancies. Immunol Cell Biol. 2003;81:171–5. doi: 10.1046/j.1440-1711.2003.01159.x. [DOI] [PubMed] [Google Scholar]
- 47.Georgopoulos K, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–56. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 48.Wang JH, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–49. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–55. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 50.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–41. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Getnet D, et al. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Mol Immunol. 2010;47:1595–600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugimoto N, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18:1197–209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 53.Hahm K, et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–96. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelley CM, et al. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Current biology: CB. 1998;8:508–15. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 55.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–77. [PubMed] [Google Scholar]
- 56.Kasper LH, et al. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. The EMBO journal. 2005;24:3846–58. doi: 10.1038/sj.emboj.7600846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 60.Lohmann D. Retinoblastoma. Adv Exp Med Biol. 2010;685:220–7. doi: 10.1007/978-1-4419-6448-9_21. [DOI] [PubMed] [Google Scholar]
- 61.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanel W, Moll UM. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113:433–9. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:14188–93. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deshpande A, Hinds PW. The retinoblastoma protein in osteoblast differentiation and osteosarcoma. Curr Mol Med. 2006;6:809–17. doi: 10.2174/1566524010606070809. [DOI] [PubMed] [Google Scholar]
- 67.Romero F, Martinez AC, Camonis J, Rebollo A. Aiolos transcription factor controls cell death in T cells by regulating Bcl-2 expression and its cellular localization. EMBO J. 1999;18:3419–30. doi: 10.1093/emboj/18.12.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boulos N, et al. Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;117:3585–95. doi: 10.1182/blood-2010-08-301267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Downing JR, et al. The Pediatric Cancer Genome Project. Nat Genet. 2012;44:619–22. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin M, et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–40. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 75.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 76.Rozen S, Skaletsky HJ. Bioinformatics Methods and Protocols (Methods in Molecular Biology) Humana Press; 2000. [Google Scholar]
- 77.Zhang J, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen K, et al. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–66. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donovan S, See W, Bonifas J, Stokoe D, Shannon KM. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer Cell. 2002;2:507–14. doi: 10.1016/s1535-6108(02)00214-3. [DOI] [PubMed] [Google Scholar]
- 81.Schubbert S, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–6. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 82.Peto R, et al. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. analysis and examples. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–70. [PubMed] [Google Scholar]
- 84.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 85.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna:Austria: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary
Table S1
Table S10
Table S11
Table S12
Table S14
Table S15
Table S2
Table S21
Table S22
Table S25
Table S26
Table S7
Table S8