DNA targeting specificity of RNA-guided Cas9 nucleases (original) (raw)
. Author manuscript; available in PMC: 2014 Mar 30.
Published in final edited form as: Nat Biotechnol. 2013 Jul 21;31(9):827–832. doi: 10.1038/nbt.2647
Abstract
The Streptococcus pyogenes Cas9 (SpCas9) nuclease can be efficiently targeted to genomic loci by means of singleguide RNAs (sgRNAs) to enable genome editing1–10. Here, we characterize SpCas9 targeting specificity in human cells to inform the selection of target sites and avoid off-target effects. Our study evaluates >700 guide RNA variants and SpCas9-induced indel mutation levels at >100 predicted genomic off-target loci in 293T and 293FT cells. We find that SpCas9 tolerates mismatches between guide RNA and target DNA at different positions in a sequence-dependent manner, sensitive to the number, position and distribution of mismatches. We also show that SpCas9-mediated cleavage is unaffected by DNA methylation and that the dosage of SpCas9 and sgRNA can be titrated to minimize off-target modification. To facilitate mammalian genome engineering applications, we provide a web-based software tool to guide the selection and validation of target sequences as well as off-target analyses.
The bacterial type II clustered, regularly interspaced, short palindromic repeats (CRISPR) system from S. pyogenes can be reconstituted in mammalian cells using three minimal components1: the CRISPR-associated nuclease Cas9 (SpCas9), a specificity-determining CRISPR RNA (crRNA), and an auxiliary trans-activating crRNA (tracrRNA)11. Following crRNA and tracrRNA hybridization, SpCas9 is targeted to genomic loci matching a 20-nt guide sequence within the crRNA, immediately upstream of a required 5′-NGG protospacer adjacent motif (PAM)11. crRNA and tracrRNA duplexes can also be fused to generate a chimeric sgRNA12 that mimics the natural crRNA-tracrRNA hybrid. Both crRNA-tracrRNA duplexes and sgRNAs can be used to target SpCas9 for multiplexed genome editing in eukaryotic cells1,3.
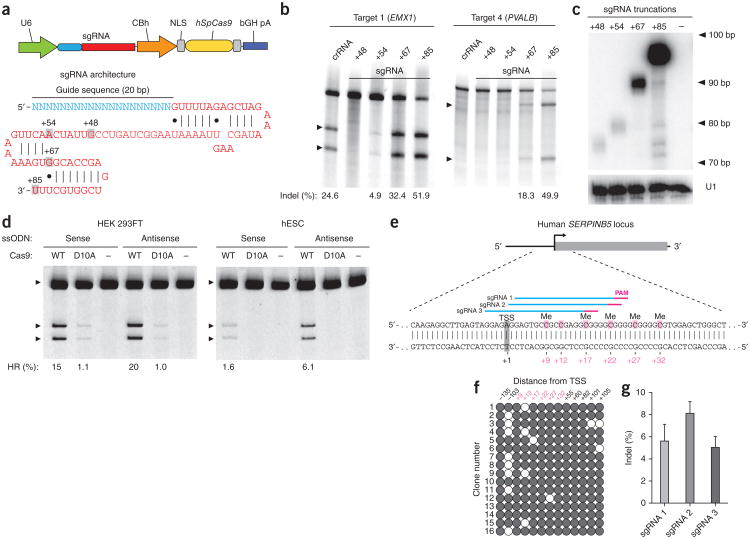
Although an sgRNA design consisting of a truncated crRNA and tracrRNA had been previously shown to mediate efficient cleavage in vitro12, it failed to achieve detectable cleavage at several loci that were efficiently modified by crRNA-tracrRNA duplexes bearing identical guide sequences1. Because the major difference between this sgRNA design and the native crRNA-tracrRNA duplex is the length of the tracrRNA sequence, we tested whether extension of the tracrRNA tail would improve SpCas9 activity.
We generated a set of sgRNAs targeting multiple sites within the human EMX1 and PVALB loci with different tracrRNA 3′ truncations (Fig. 1a). Using the SURVEYOR nuclease assay13, we assessed the ability of each Cas9-sgRNA complex to generate indels in human embryonic kidney (HEK) 293FT cells through the induction of DNA doublestranded breaks (DSBs) and subsequent nonhomologous end joining (NHEJ) DNA damage repair (Online Methods). sgRNAs with +67 or +85 nucleotide (nt) tracrRNA tails mediated DNA cleavage at all target sites tested, with up to fivefold higher levels of indels than the corresponding crRNA-tracrRNA duplexes (Fig. 1b and Supplementary Fig. 1a). Furthermore, both sgRNA designs efficiently modified PVALB loci that were previously not targetable using crRNA-tracrRNA duplexes1 (Fig. 1b and Supplementary Fig. 1b). For all five tested targets, we observed a consistent increase in modification efficiency with increasing tracrRNA length. We performed northern blot analyses for the guide RNA truncations and found increased levels of expression for the longer tracrRNA sequences, suggesting that improved target cleavage was at least partially due to higher sgRNA expression or stability (Fig. 1c). Taken together, these data indicate that the tracrRNA tail is important for optimal SpCas9 expression and activity in vivo.
Figure 1.
Optimization of guide RNA architecture for SpCas9-mediated mammalian genome editing. (a) Schematic of bicistronic expression vector (PX330) for U6 promoter-driven sgRNA and CBh promoter-driven human codon-optimized S. pyogenes Cas9 (hSpCas9) used for all subsequent experiments. The sgRNA consists of a 20-nt guide sequence (blue) and scaffold (red), truncated at various positions as indicated. (b) SURVEYOR assay for SpCas9-mediated indels at the human EMX1 and PVALB loci. Arrowheads indicate the expected SURVEYOR fragments (n = 3). (c) Northern blot analysis for the four sgRNA truncation architectures, with U1 as loading control. (d) Both wild-type (WT) or nickase mutant (D10A) of SpCas9 promoted insertion of a HindIII site into the human EMX1 gene. Single-stranded oligonucleotides, oriented in either the sense or antisense direction relative to genome sequence, were used as homologous recombination templates (Supplementary Fig. 3). (e) Schematic of the human SERPINB5 locus. sgRNAs and PAMs are indicated by colored bars above sequence; methylcytosine (Me) are highlighted (pink) and numbered relative to the transcriptional start site (TSS, +1). (f) Methylation status of SERPINB5 assayed by bisulfite sequencing of 16 clones. Filled circles, methylated CpG; open circles, unmethylated CpG. (g) Modification efficiency by three sgRNAs targeting the methylated region of SERPINB5, assayed by deep sequencing (n = 2). Error bars indicate Wilson intervals (Online Methods).
We further investigated the sgRNA architecture by extending the duplex length from 12 to the 22 nt found in the native crRNA-tracrRNA duplex (Supplementary Fig. 2a). We also mutated the sequence encoding the sgRNAs to abolish any poly-T tracts that could serve as premature transcriptional terminators for U6-driven transcription14. We tested these new sgRNA scaffolds on three targets within the human EMX1 gene (Supplementary Fig. 2b) and observed only modest changes in modification efficiency. Thus, we established sgRNA(+67) as a minimum effective SpCas9 guide RNA architecture and for all subsequent studies we used the most active sgRNA(+85) architecture.
We have previously shown that a catalytic mutant of SpCas9 (D10A nickase) can mediate gene editing by homology-directed repair without detectable indel formation1. Given its higher cleavage efficiency, we tested whether sgRNA(+85), in complex with the Cas9 nickase, can likewise facilitate homology-directed repair without incurring on-target NHEJ. Using single-stranded oligonucleotides as repair templates, we observed that both the wild-type and the D10A SpCas9 mediate homology-directed repair in HEK 293FT cells, whereas only the former does so in human embryonic stem cells (hESCs; Fig. 1d and Supplementary Fig. 3a–c). We further confirmed using SURVEYOR assay that no target indel mutations are induced by the SpCas9 D10A nickase (Supplementary Fig. 3d).
To explore whether the genome targeting ability of sgRNA(+85) is influenced by epigenetic factors15,16 that constrain the alternative transcription activator-like effector nuclease (TALENs)17–21 and potentially also zinc finger nuclease (ZFNs)22–26 technologies, we further tested the ability of SpCas9 to cleave methylated DNA. Using either unmethylated or M. _Sss_I-methylated pUC19 as DNA targets (Supplementary Fig. 4a, b) in a cell-free cleavage assay, we showed that SpCas9 efficiently cleaves pUC19 regardless of CpG methylation status in either the 20-bp target sequence or the PAM (Supplementary Fig. 4c). To test whether this is also true in vivo, we designed sgRNAs to target a highly methylated region of the human SERPINB5 locus (Fig. 1e, f). All three sgRNAs tested were able to mediate indel mutations in endogenously methylated targets (Fig. 1g).
Having established the optimal guide RNA architecture for SpCas9 and having demonstrated its insensitivity to genomic CpG methylation, we sought to conduct a comprehensive characterization of the DNA targeting specificity of SpCas9. Previous studies on SpCas9 cleavage specificity1,2,12 were limited to a small set of single-nucleotide mismatches between the guide sequence and DNA target, suggesting that perfect base-pairing within 10–12 bp directly 5′ of the PAM (PAM-proximal) determines Cas9 specificity, whereas multiple PAM-distal mismatches can be tolerated. In addition, a recent study using catalytically inactive SpCas9 as a transcriptional repressor found no significant off-target effects throughout the Escherichia coli transcriptome27. However, a systematic analysis of Cas9 specificity within the context of a larger mammalian genome has not yet been reported.
To address this, we first evaluated the effect of imperfect complementarity between the guide RNA and its genomic target on SpCas9 activity, and then assessed the cleavage activity resulting from a single sgRNA on multiple genomic off-target loci with sequence similarity. To facilitate large-scale testing of mismatched guide sequences, we developed a simple sgRNA testing assay by generating expression cassettes encoding U6-driven sgRNAs using PCR and transfecting the resulting amplicons (Supplementary Fig. 5). We then performed deep sequencing of the region flanking each target site (Supplementary Fig. 6) for two independent biological replicates. From these data, we applied a binomial model to detect true indel events resulting from SpCas9 cleavage and NHEJ misrepair and calculated 95% confidence intervals for all reported NHEJ frequencies (Online Methods and Supplementary Tables 5–8).
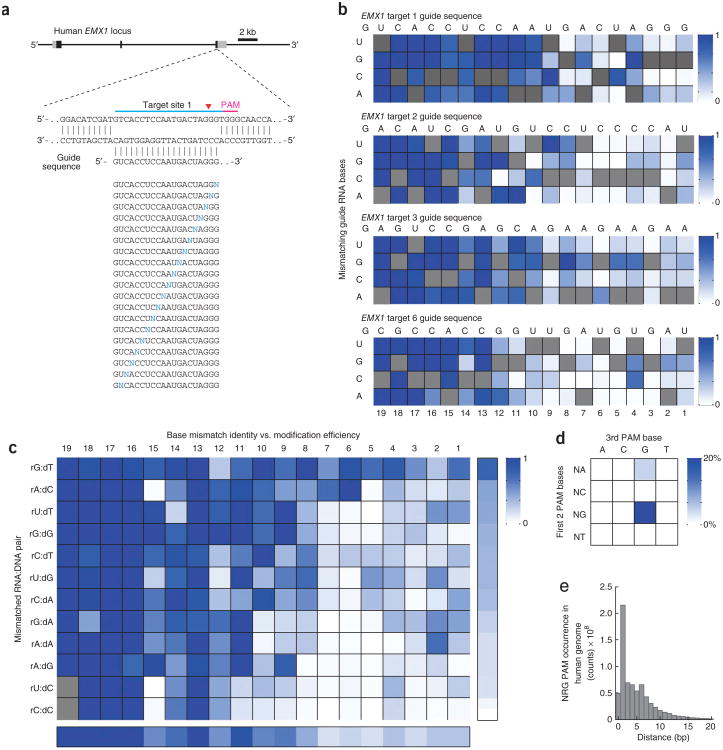
We systematically investigated the effect of base-pairing mismatches between guide RNA sequences and target DNA on target modification efficiency. We chose four target sites within the human EMX1 gene (1, 2, 3 and 6) and, for each, generated a set of 57 different guide RNAs containing all possible single-nucleotide substitutions in positions 1–19 directly 5′ of the requisite NGG PAM (Fig. 2a). The 5′ guanine at position 20 is preserved, given that the U6 promoter requires guanine as the first base of its transcript. These ‘off-target’ guide RNAs were then assessed for cleavage activity at the on-target genomic locus.
Figure 2.
Single-nucleotide specificity of SpCas9. (a) Schematic of the experimental design. sgRNAs carrying all possible single base-pair mismatches (blue Ns) throughout the guide sequence were tested for each EMX1 target site (target site 1 shown as example). (b) Heatmap representation of relative SpCas9 cleavage efficiency by 57 single-mutated and 1 nonmutated sgRNA each for four EMX1 target sites (aggregated from Supplementary Table 5). For each EMX1 target, the identities of single base-pair substitutions are indicated on the left; original guide sequence is shown above and highlighted in the heatmap (gray squares). Modification efficiencies (increasing from white to dark blue) are normalized to the original guide sequence. Sequence logo representation of the same data can be found in Supplementary Figure 7. (c) Heatmap for relative SpCas9 cleavage efficiency for each possible RNA:DNA base pair, compiled from aggregate data from single-mismatch guide RNAs for 15 EMX1 targets (Supplementary Fig. 8). Mean cleavage levels were calculated for the 10 PAM-proximal bases (right bar) and across all substitutions at each position (bottom bar); positions in gray were not covered by the 469 single-mutated and 15 unmutated sgRNAs tested (Supplementary Table 5). (d) SpCas9-mediated indel frequencies at targets with all possible PAM sequences, determined using the SURVEYOR nuclease assay. Two target sites from the EMX1 locus were tested for each PAM (Supplementary Table 4). (e) Histogram of distances between 5′-NRG PAM occurrences within the human genome. Putative targets were identified using both strands of human chromosomal sequences (GRCh37/hg19).
Consistent with previous findings1,2,12, SpCas9 tolerates single-base mismatches in the PAM-distal region to a greater extent than in the PAM-proximal region. In contrast to a model that implies that a prototypical 10–12 bp PAM-proximal seed sequence largely determines target specificity1,2,12, we found that most bases within the 20-bp target site provide varying degrees of specificity. Single-base specificity generally ranges from 8 to 14 bp immediately upstream of the PAM, indicating a sequence-dependent, mismatch-sensitive boundary that varies in length (Fig. 2b, Supplementary Fig. 7 and Supplementary Table 5).
To further investigate the contributions of base identity and position within the guide RNA to SpCas9 specificity, we generated additional sets of mismatched guide RNAs for 11 more target sites within the EMX1 locus (Supplementary Fig. 8), totaling over 400 sgRNAs. These guide RNAs were designed to cover all 12 possible RNA:DNA mismatches for each position in the guide sequence with at least 2× coverage for positions 1–10. Our aggregate single-mismatch data reveal multiple exceptions to the seed sequence model of SpCas9 specificity1,2,6 (Fig. 2c and Supplementary Table 5). Within the PAM-proximal region, the degree of tolerance varied with the identity of a particular mismatch, with rC:dC base-pairing exhibiting the highest level of disruption to SpCas9 cleavage activity (Fig. 2c).
In addition to the target specificity, we also investigated the NGG PAM requirement of SpCas9. To vary the second and third positions of PAM, we selected 32 target sites within the EMX1 locus encompassing all 16 possible alternate PAMs with 2× coverage (Supplementary Table 4). Using the SURVEYOR assay, we showed that SpCas9 also cleaves targets with NAG PAMs, albeit with one-fifth of the efficiency for target sites with 5′-NGG PAMs (Fig. 2d). The tolerance for an NAG PAM is in agreement with previous bacterial studies2 and expands the S. pyogenes Cas9 target space to every 4 bp on average within the human genome, not accounting for constraining factors such as guide RNA secondary structure or certain epigenetic modifications (Fig. 2e). Although we have shown here that methylated DNA sequences can be cleaved, by SpCas9 further characterization of the implications of epigenetic factors on CRISPR editing efficiency are needed.
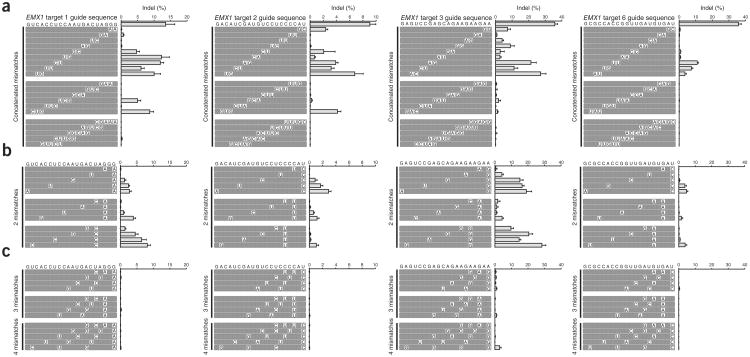
We next explored the effect of multiple base mismatches on SpCas9 target activity. For four targets within the EMX1 gene, we designed sets of guide RNAs that contained varying combinations of mismatches to investigate the effect of mismatch number, position and spacing on SpCas9 target cleavage activity (Fig. 3a, b, and Supplementary Table 6). In general, we observed that the total number of mismatched base-pairs is a key determinant for SpCas9 cleavage efficiency. Two mismatches, particularly those occurring in a PAM-proximal region, considerably reduced SpCas9 activity whether these mismatches are concatenated or interspaced (Fig. 3a, b); this effect is further magnified for three concatenated mismatches (Fig. 3a). Furthermore, three or more interspaced (Fig. 3c) and five concatenated (Fig. 3a) mismatches eliminated detectable SpCas9 cleavage in the vast majority of loci.
Figure 3.
Multiple mismatch specificity of SpCas9. (a–c) SpCas9 cleavage efficiency with guide RNAs containing consecutive mismatches of 2, 3 or 5 bases (a), or multiple mismatches separated by different numbers of unmutated bases for EMX1 targets 1, 2, 3 and 6 (b, c). Rows represent each mutated guide RNA; nucleotide substitutions are shown in white cells; gray cells denote unmutated bases. All indel frequencies are absolute and analyzed by deep sequencing from two biological replicas. Error bars indicate Wilson intervals (Online Methods).
The position of mismatches within the guide sequence also affected the activity of SpCas9. PAM-proximal mismatches are less tolerated than PAM-distal counterparts (Fig. 3a), recapitulating our observations from the single base-pair mismatch data (Fig. 2c). This effect is particularly salient in guide sequences bearing a small number of total mismatches, whether those are consecutive (Fig. 3a) or interspaced (Fig. 3b). Additionally, guide sequences with mismatches spaced four or more bases apart also mediated SpCas9 cleavage in some cases (Fig. 3c). Thus, together with the identity of mismatched base-pairing, we observed that many off-target cleavage effects can be explained by a combination of mismatch number and position.
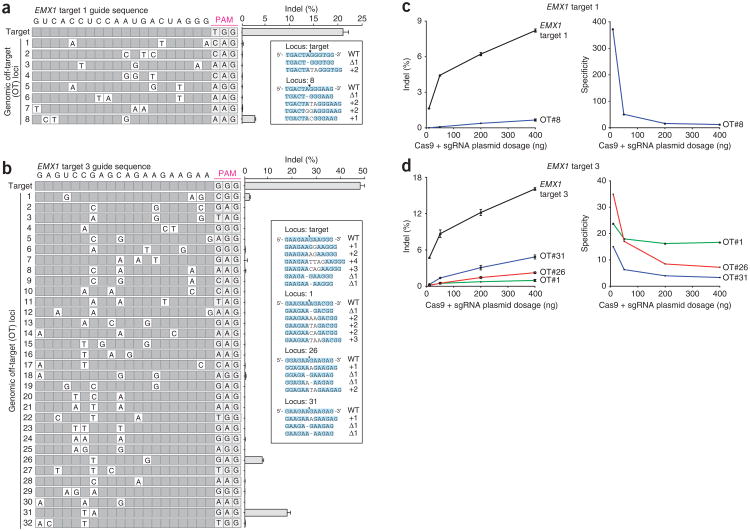
Given these mismatched guide RNA results, we expected that for any particular sgRNA, SpCas9 may cleave genomic loci that contain small numbers of mismatched bases. For the four EMX1 targets described above, we computationally selected 117 candidate off-target sites in the human genome that are followed by a 5′-NRG PAM and meet any of the following additional criteria: (i) up to five mismatches, (ii) short insertions or deletions or (iii) mismatches only in the PAM-distal region. Additionally, we assessed off-target loci of high sequence similarity without the PAM requirement. The majority of off-target sites tested for each sgRNA (30/31, 23/23, 48/51 and 12/12 sites for EMX1 targets 1, 2, 3 and 6, respectively) exhibited modification efficiencies at least 2 magnitudes lower than that of corresponding on-targets (Fig. 4a, b, Supplementary Fig. 9 and Supplementary Tables 7 and 8). Of the four off-target sites that exhibit substantial modification efficiencies, three contained only mismatches in the PAM-distal region, consistent with our multiple mismatch sgRNA observations (Fig. 3). Notably, these three loci were followed by 5′-NAG PAMs, demonstrating that off-target analyses of SpCas9 must include 5′-NAG as well as 5′-NGG candidate loci.
Figure 4.
SpCas9-mediated indel frequencies at predicted genomic off-target loci. (a, b) Cleavage levels at putative genomic off-target loci containing two or three individual mismatches (white cells) for EMX1 target 1 and target 3 are analyzed by deep sequencing. List of off-target sites are ordered by median position of mutations. Putative off-target sites with additional mutations did not have detectable indels (Supplementary Table 8). The Cas9 dosage was 3 × 10−10 nmol/cell, with equimolar sgRNA delivery. Error bars indicate Wilson intervals (Online Methods). (c, d) Indel frequencies for EMX1 targets 1 and 3 and selected off-target loci (OT) as a function of SpCas9 and sgRNA dosage, (n = 2, Wilson intervals). 400 ng to 10 ng of Cas9-sgRNA plasmid corresponds to 7.1 × 10−10 to 1.8 × 10−11 nmol/cell. Cleavage specificity is measured as a ratio of on- to off-target cleavage.
Enzymatic specificity and activity strength are often highly dependent on reaction conditions, which at high enzyme concentration might amplify off-target activity28,29. One potential strategy for minimizing nonspecific cleavage is to limit the enzyme concentration, namely the level of SpCas9-sgRNA complex. Cleavage specificity, measured as the ratio of on- to off-target cleavage, increased dramatically as we decreased the equimolar amounts of SpCas9 and sgRNA transfected into 293FT cells (Fig. 4c, d) from 7.1 × 10−10 to 1.8 × 10−11 nmol/cell (400 ng to 10 ng of Cas9-sgRNA plasmid). qRT-PCR assay confirmed that the level of hSpCas9 mRNA and sgRNA decreased proportionally to the amount of transfected DNA (Supplementary Fig. 10). Whereas specificity increased gradually by nearly fourfold as we decreased the transfected DNA amount from 7.1 × 10−10 to 9.0 × 10−11 nmol/cell (400 ng to 50 ng plasmid), we observed a notable additional sevenfold increase in specificity upon further decreasing transfected DNA from 9.0 × 10−11 to 1.8 × 10−11 nmol/cell (50 ng to 10 ng plasmid; Fig. 4c). These findings suggest that we can minimize the level of off-target activity by titrating the amount of SpCas9 and sgRNA DNA delivered. However, increasing specificity by reducing the amount of transfected DNA also leads to a reduction in on-target cleavage. These measurements enable quantitative integration of specificity and efficiency criteria into dosage choice to optimize SpCas9 activity for different applications. Additional work to explore modifications in SpCas9 and sgRNA design may improve SpCas9-intrinsic specificity without sacrificing cleavage efficiency.
The ability to program SpCas9 to target specific sites in the genome by simply designing a short guide RNA complementary to the desired target site holds enormous potential for applications throughout biology and medicine. Our results demonstrate that the specificity of SpCas9-mediated DNA cleavage is sequence- and locus-dependent and governed by the quantity, position and identity of mismatching bases. Whereas the PAM-proximal 8–12 bp of the guide sequence generally defines specificity, the PAM-distal sequences also contribute to the overall specificity of SpCas9-mediated DNA cleavage. Although there may be off-target cleavage for a given guide sequence, they can be predicted and likely minimized by following general design guidelines.
To maximize SpCas9 specificity for editing a particular gene, one should identify potential ‘off-target’ genomic sequences by considering the following four constraints. First and foremost, they should not be followed by a PAM with either 5′-NGG or 5′-NAG sequences. Second, their global sequence similarity to the target sequence should be minimized, and guide sequences with genomic off-target loci that have fewer than three mismatches should be avoided. Third, at least two mismatches should lie within the PAM-proximal region of the off-target site. Fourth, a maximal number of mismatches should be consecutive or spaced less than four bases apart. Finally, the amount of SpCas9 and sgRNA can be titrated to optimize on- to off-target cleavage ratio.
Using these criteria, we formulated a scoring algorithm to integrate and quantify the contributions of mismatch location, density and identity on SpCas9 on-target and off-target cleavage. We applied the aggregate cleavage efficiencies of single-mismatch guide RNAs to test this scoring scheme separately on genome-wide targets and found that these factors, taken together, accounted for >50% of the variance in cutting-frequency rank among the genome-wide targets studied (Supplementary Fig. 11).
Implementing the guidelines delineated above, we designed a computational tool to facilitate the selection and validation of sgRNAs as well as to predict off-target loci for specificity analyses; this tool can be accessed at http://www.genome-engineering.org/. These results and tools further extend the SpCas9 system as a versatile alternative to ZFNs and TALENs for genome editing applications. Further work examining the thermodynamics and in vivo stability of sgRNA-DNA duplexes will likely yield additional predictive power for off-target activity, whereas exploration of SpCas9 mutants and orthologs may yield novel variants with improved specificity.
Online Methods
Cell culture and transfection
Human embryonic kidney (HEK) cell line 293FT (Life Technologies) was maintained in Dulbecco's modified Eagle's Medium (DMEM) supplemented with 10% FBS (HyClone), 2 mM GlutaMAX (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2 incubation.
293FT cells were seeded onto 6-well plates, 24-well plates or 96-well plates (Corning) 24 h before transfection. Cells were transfected using Lipofectamine 2000 (Life Technologies) at 80–90% confluency following the manufacturer's recommended protocol. For each well of a 6-well plate, a total of 1 μg of Cas9+sgRNA plasmid was used. For each well of a 24-well plate, a total of 500 ng Cas9+sgRNA plasmid was used unless otherwise indicated. For each well of a 96-well plate, 65 ng of Cas9 plasmid was used at a 1:1 molar ratio to the U6-sgRNA PCR product.
Human embryonic stem cell line HUES9 (Harvard Stem Cell Institute core) was maintained in feeder-free conditions on GelTrex (Life Technologies) in mTesR medium (Stemcell Technologies) supplemented with 100 μg/ml Normocin (InvivoGen). HUES9 cells were transfected with Amaxa P3 Primary Cell 4-D Nucleofector Kit (Lonza) following the manufacturer's protocol.
SURVEYOR nuclease assay for genome modification
293FT and HUES9 cells were transfected with DNA as described above. Cells were incubated at 37 °C for 72 h post-transfection before genomic DNA extraction. Genomic DNA was extracted using the QuickExtract DNA Extraction Solution (Epicentre) following the manufacturer's protocol. Briefly, pelleted cells were resuspended in QuickExtract solution and incubated at 65 °C for 15 min, 68 °C for 15 min, and 98 °C for 10 min.
The genomic region flanking the CRISPR target site for each gene was PCR amplified (target sites and primers listed in Supplementary Tables 1 and 2), and products were purified using QiaQuick Spin Column (Qiagen) following the manufacturer's protocol. 400 ng total of the purified PCR products were mixed with 2 μl 10× Taq DNA Polymerase PCR buffer (Enzymatics) and ultrapure water to a final volume of 20 μl, and subjected to a re-annealing process to enable heteroduplex formation: 95 °C for 10 min, 95 °C to 85 °C ramping at −2 °C/s, 85 °C to 25 °C at −0.25 °C/s, and 25 °C hold for 1 min. After re-annealing, products were treated with SURVEYOR nuclease and SURVEYOR enhancer S (Transgenomics) following the manufacturer's recommended protocol, and analyzed on 4–20% Novex TBE polyacrylamide gels (Life Technologies). Gels were stained with SYBR Gold DNA stain (Life Technologies) for 30 min and imaged with a Gel Doc gel imaging system (Bio-rad). Quantification was based on relative band intensities. Indel percentage was determined by the formula, 100 × (1 − (1 − (b + c)/(a + b + c))1/2), where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage product.
Northern blot analysis of tracrRNA expression in human cells
Northern blots were done as previously described1. Briefly, RNAs were extracted using the mirPremier microRNA Isolation Kit (Sigma) and heated to 95 °C for 5 min before loading on 8% denaturing polyacrylamide gels (SequaGel, National Diagnostics). Afterwards, RNA was transferred to a Hybond N+ membrane (GE Healthcare) and crosslinked with Stratagene UV Crosslinker (Stratagene). Probes were labeled with (gamma-32P) ATP (PerkinElmer) with T4 polynucleotide kinase (New England Biolabs). After washing, membrane was exposed to phosphor screen for 1 h and scanned with phosphorimager (Typhoon).
Bisulfite sequencing to assess DNA methylation status
Genomic DNA from 293FT cells was isolated with the DNeasy Blood & Tissue Kit (Qiagen) and bisulfite converted with EZ DNA Methylation-Lightning Kit (Zymo Research). Bisulfite PCR was conducted using KAPA2G Robust HotStart DNA Polymerase (KAPA Biosystems) with primers designed using the Bisulfite Primer Seeker (Zymo Research, Supplementary Table 2). Resulting PCR amplicons were gel-purified, digested with EcoRI and HindIII, and ligated into a pUC19 backbone before transformation. Individual clones were then Sanger sequenced to assess DNA methylation status.
In vitro transcription and cleavage assay
Whole cell lysates from 293FT cells were prepared with lysis buffer (20 mM HEPES, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 0.1% Triton X-100) supplemented with Protease Inhibitor Cocktail (Roche). T7-driven sgRNA was transcribed in vitro using custom oligos (Supplementary Sequences) and HiScribe T7 In vitro Transcription Kit (NEB), following the manufacturer's recommended protocol. To prepare methylated target sites, pUC19 plasmid was methylated by M._Sss_I and tested by digestion with HpaII. Unmethylated and successfully methylated pUC19 plasmids were linearized by NheI. The in vitro cleavage assay was carried out as follows: for a 20 μl cleavage reaction, 10 μl of cell lysate was incubated with 2 μl cleavage buffer (100 mM HEPES, 500 mM KCl, 25 mM MgCl2, 5 mM DTT, 25% glycerol), 1 μg in vitro transcribed RNA and 300 ng pUC19 plasmid DNA.
Deep sequencing to assess targeting specificity
HEK 293FT cells plated in 96-well plates were transfected with Cas9 plasmid DNA and sgRNA PCR cassette 72 h before genomic DNA extraction (Supplementary Fig. 4). The genomic region flanking the CRISPR target site for each gene was amplified (Supplementary Fig. 6, Supplementary Table 5 and Supplementary Sequences) by a fusion PCR method to attach the Illumina P5 adapters as well as unique sample-specific barcodes to the target amplicons (schematic described in Supplementary Fig. 5). PCR products were purified using EconoSpin 96-well Filter Plates (Epoch Life Sciences) following the manufacturer's recommended protocol.
Barcoded and purified DNA samples were quantified by Quant-iT PicoGreen dsDNA Assay Kit or Qubit 2.0 Fluorometer (Life Technologies) and pooled in an equimolar ratio. Sequencing libraries were then sequenced with the Illumina MiSeq Personal Sequencer (Life Technologies).
Sequencing data analysis and indel detection
MiSeq reads were filtered by requiring an average Phred quality (Q score) of at least 23, as well as perfect sequence matches to barcodes and amplicon forward primers. Reads from on- and off-target loci were analyzed by first performing Smith-Waterman alignments against amplicon sequences that included 50 nucleotides upstream and downstream of the target site (a total of 120 bp). Alignments, meanwhile, were analyzed for indels from 5 nucleotides upstream to 5 nucleotides downstream of the target site (a total of 30 bp). Analyzed target regions were discarded if part of their alignment fell outside the MiSeq read itself, or if matched base-pairs comprised less than 85% of their total length.
Negative controls for each sample provided a gauge for the inclusion or exclusion of indels as putative cutting events. For each sample, an indel was counted only if its quality score exceeded μ - σ, where μ was the mean quality-score of the negative control corresponding to that sample and σ was the s.d. of the same. This yielded whole target-region indel rates for both negative controls and their corresponding samples. Using the negative control's per-target-region-per-read error rate, q, the sample's observed indel count n, and its read-count R, a maximum-likelihood estimate for the fraction of reads having target-regions with true-indels, p, was derived by applying a binomial error model, as follows.
Letting the (unknown) number of reads in a sample having target regions incorrectly counted as having at least 1 indel be E, we can write (without making any assumptions about the number of true indels)
Prob(E|p)=(R(1-p)E)qE(1-q)R(1-p)-E
as R(1 − p) is the number of reads having target-regions with no true indels. Meanwhile, because the number of reads observed to have indels is n, n = E + Rp, that is, the number of reads having target-regions with errors but no true indels plus the number of reads whose target-regions correctly have indels. We can then rewrite the above
Prob(E|p)=Prob(n=E+Rp|p)=(R(1-p)n-Rp)qn-Rp(1-q)R-n
Taking all values of the frequency of target-regions with true-indels p to be equally probable a priori, Prob(n|p) ∞ Prob(p|n). The maximum-likelihood estimate (MLE) for the frequency of target regions with true indels was therefore set as the value of p that maximized Prob(n|p). This was evaluated numerically.
In order to place error bounds on the true-indel read frequencies in the sequencing libraries themselves, Wilson score intervals2 were calculated for each sample, given the MLE-estimate for true-indel target-regions, Rp, and the number of reads R. Explicitly, the lower bound l and upper bound u were calculated as
l=(Rp+z22-zRp(1-p)+z2/4)/(R+z2)
u=(Rp+z22+zRp(1-p)+z2/4)/(R+z2)
where z, the standard score for the confidence required in normal distribution of variance 1, was set to 1.96, meaning a confidence of 95%. The maximum upper bounds and minimum lower bounds for each biological replicate are listed in Supplementary Tables 5–8.
qRT-PCR analysis of relative Cas9 and sgRNA expression
72 h post-transfection, total RNA from 293FT cells was harvested with miRNeasy Micro Kit (Qiagen). Reverse-strand synthesis for sgRNAs was performed with qScript Flex cDNA kit (VWR) and custom first-strand synthesis primers (Supplementary Table 2). qPCR analysis was done with Fast SYBR Green Master Mix (Life Technologies) and custom primers (Supplementary Table 2), using GAPDH as an endogenous control. Relative quantification was calculated by the ΔΔCT method.
Supplementary Material
Supp File S1
Supp File S2
Supp File S3
Supp File S4
Supp File S5
Acknowledgments
We thank A. Shalek, E. Stamenova and D. Gray for expert help with DNA sequencing, R. Barretto for genome-wide PAM analysis, as well as D. Altshuler, P.A. Sharp, and the entire Zhang Lab for their support and advice. P.D.H. is a James Mills Pierce Fellow. D.A.S. is an National Science Foundation pre-doctoral fellow and J.A.W. is supported by a Life Science Research Foundation Fellowship. X.W. is a Howard Hughes Medical Institute International Student Research Fellow and is supported by National Institutes of Health (NIH) grants R01-GM34277 and R01-CA133404 to P.A. Sharp, X.W.'s thesis advisor. This work is supported by an NIH Director's Pioneer Award (DP1-MH100706), an NIH Transformative R01 grant (R01-DK097768) to D. Altshuler, the Keck, McKnight, Damon Runyon, Searle Scholars, Klingenstein and Simons Foundations, and Bob Metcalfe and Jane Pauley. The authors wish to dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice. Reagents are available to the academic community through Addgene, and associated protocols, support forums and computational tools are available through the Zhang lab website (http://www.genome-engineering.org/).
Footnotes
Accession codes. All raw reads can be accessed at NCBI BioProject, accession number SRP023129. Indices are described in Supplementary Tables 5–8.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Author Contributions: J.A.W. and F.A.R. contributed equally to this work. P.D.H., D.A.S., F.A.R., S.K. and F.Z. designed and performed the experiments. P.D.H., D.A.S., J.A.W., Y.L., S.K., F.A.R. and F.Z. analyzed the data. V.A. and O.S. contributed computational prediction of CRISPR off-target sites and X.W. performed the northern blot analysis. P.D.H., F.A.R., D.A.S. and F.Z. wrote the manuscript with help from all authors.
Competing Financial Interests: The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 7.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 Jul 2; doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 14.Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 15.Bultmann S, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valton J, et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 19.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu PD, Zhang F. Dissecting neural function using targeted genome engineering technologies. ACS Chem Neurosci. 2012;3:603–610. doi: 10.1021/cn300089k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 23.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 24.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobis-Wozowicz S, Osiak A, Rahman SH, Cathomen T. Targeted genome editing in pluripotent stem cells using zinc-finger nucleases. Methods. 2011;53:339–346. doi: 10.1016/j.ymeth.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaelis, Maud LM. Die kinetik der invertinwirkung. Biochemistry Zeitung. 1913;49:333–369. [Google Scholar]
- 29.Mahfouz MM, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp File S1
Supp File S2
Supp File S3
Supp File S4
Supp File S5