The mammalian dynein/dynactin complex is a strong opponent to kinesin in a tug-of-war competition (original) (raw)
. Author manuscript; available in PMC: 2017 Jan 25.
Published in final edited form as: Nat Cell Biol. 2016 Jul 25;18(9):1018–1024. doi: 10.1038/ncb3393
Abstract
Kinesin and dynein motors transport intracellular cargos bidirectionally by pulling them in opposite directions along microtubules, through a process frequently described as a ‘tug of war’. While kinesin produces a 6 pN force, mammalian dynein was found to be a surprisingly weak motor (0.5–1.5 pN) in vitro, suggesting many dyneins are required to counteract the pull of a single kinesin. Mammalian dynein’s association with dynactin and Bicaudal-D2 (BICD2) activates its processive motility, but how this affects dynein’s force output remained unknown. Here, we show that formation of the dynein-dynactin-BICD2 (DDB) complex increases human dynein’s force production to 4.3 pN. An in vitro tug-of-war assay revealed that a single DDB successfully resists a single kinesin. Contrary to previous reports, the clustering of many dyneins is not required to win the tug-of-war. Our work reveals the key role of dynactin and a cargo adaptor protein in shifting the balance of forces between dynein and kinesin motors during intracellular transport.
Cytoplasmic dynein (“dynein” hereafter) is a minus end-directed microtubule motor responsible for cargo trafficking, organelle positioning, and organization of the mitotic spindle in eukaryotic cells1. The core of the dynein complex comprises a homodimer of two massive heavy chains, each containing a AAA+ motor ring, a microtubule binding domain (MTBD) separated from the ring by a ~15 nm coiled-coil stalk, and a flexible cargo-binding tail which also serves as the dimerization domain and the binding site for dynein light and intermediate chains2,3. Previous studies on mammalian dynein revealed a striking mismatch between the motility of individual motors in vitro and their apparent in vivo functions. Despite high retrograde transport velocities observed in live cells4, single purified mammalian dyneins exhibited diffusive motility or short processive runs in vitro3,5,6 and were found to stall at forces of 0.5–1.5 pN3,7–11, significantly weaker than the 6 pN force production of plus end-directed kinesin-1 motors12. It remained unclear how dynein generates the large forces required for its cellular roles. It has been proposed that multiple (4–7) dynein motors need to be engaged in transport per kinesin-1 in order to balance forces during tug-of-war13, and that the spatial organization of dyneins on the surface of the cargo serves a fundamental regulatory role14. However, due to dynein’s large size, it may not be sterically feasible for multiple dyneins motors to interact with a microtubule when transporting small cargos15. Furthermore, measured dynein to kinesin ratios (~1.5:1) on mouse axonal membranous vesicles16 are inconsistent with the ~5:1 coupling predicted by force-based models.
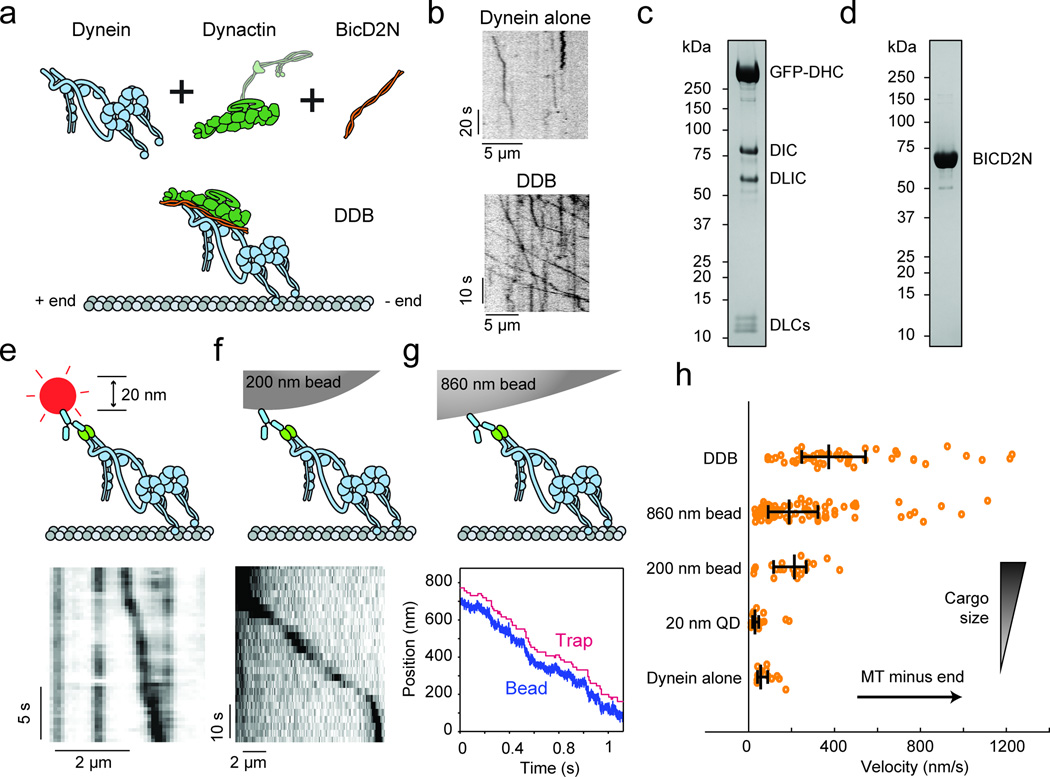
Recent studies with recombinant human dynein have begun to explore the mechanism of its motility. Both velocity and processivity of dynein are dramatically increased by the addition of dynactin, a multi-protein complex that associates with dynein in vivo, together with the N-terminus of the cargo activator Bicaudal-D2 (BICD2N), which increases the affinity of dynactin for dynein17–20 (Fig. 1a–d). In single molecule motility experiments on sea urchin axonemes, we observed that human dynein is poorly recruited to axonemes (0.19 fluorescent spots per µm−1 min−1 at 1 nM motor) and only 16% of dynein spots exhibited slow (79 ± 11 nm s−1, mean ± s.e.m.), processive motility (Supplementary Video 1, Supplementary Fig. 1a). The addition of dynactin and SNAPf-tagged BICD2N (hereafter BICD2N) at a 1:5:2 dynein:dynactin:BICD2N molar ratio increased the recruitment of dynein to 0.66 spots µm−1 min−1 at 1 nM motor, with a substantially higher percentage (52%) of processively moving spots (Supplementary Video 2, Supplementary Fig. 1b), resulting in a tenfold increase in the number of walking molecules. The results are consistent with the enhancement of dynein recruitment and processivity in the presence of dynactin and BICD2N.
Figure 1. Activation of human dynein motility by dynactin, BICD2N, and artificial cargos.
(a) Schematic depiction of the DDB complex on a microtubule. The putative orientation of the p150 subunit of dynactin is shown with a semi-transparent outline and omitted from illustrations elsewhere in the manuscript. (b) Sample kymographs represent the motility of individual dynein alone and DDB motors on axonemes. (c) Denaturing SDS–PAGE gel of purified dynein fractions. Bands corresponding to all dynein subunits can be observed. (d) Denaturing SDS–PAGE gel of purified BICD2N. Full gel scans can be found in Supplementary Fig. 5. (e) Illustration of dynein with a quantum dot attached to the tail (top) and sample kymograph (bottom). The experiment was repeated 4 times. (f) Illustration of dynein with a 200 nm bead attached to the tail (top) and sample kymograph (bottom). The experiment was repeated 3 times. (g) Illustration of dynein with an optically trapped 860 nm bead attached to the tail (top) and sample trajectory of dynein pulling the bead against a constant 0.4 pN hindering force in force feedback mode (bottom). The experiment was repeated 4 times. (h) Effect of cargo size and type on dynein velocity in comparison to the DDB complex. N = 51, 83, 21, 19, 15 runs from 3 independent experiments in order from top to bottom. Vertical bars represent median values and quartiles. Median values, from top to bottom, are 373, 188, 213, 29, and 56 nm/s. Mean ± s.e.m., from top to bottom, are 513 ± 58, 257 ± 26, 200 ± 23, 49 ± 11, and 79 ± 11 nm/s.
The autoinhibition of dynein may be mediated by a large-scale rearrangement of the tail domain, as is the case for some members of myosin21 and kinesin22 families, or by the back-to-back stacking of the motor domains3. The tail-inactivation hypothesis, wherein the tail folds over onto the motor domains and inhibits their activity, has previously been rejected because attaching quantum dots (QDs) to human dynein’s tail did not enhance its motility3. However, QDs are similar in size (~20 nm in diameter) to dynein itself, and may be too small to release inhibition. To test this possibility, we attached a range of artificial cargos to dynein’s tail and tested its motility in vitro. In agreement with Torisawa et al.3, binding a QD to dynein tail did not increase dynein’s velocity as the QD-labeled dynein walked at 49 ± 11 nm/s and the majority of QDs did not exhibit any processive motility along microtubules (N = 19 walking QDs out of 319 observed, Fig. 1e). However, attachment of larger cargos to dynein’s tail resulted in fast processive runs. Single dynein motors carried 200 nm diameter beads at 200 ± 23 nm/s in unloaded conditions (Fig. 1f, Supplementary Video 3) and 860 nm diameter beads at 257 ± 26 nm/s under 0.4 pN constant hindering force (Fig. 1g)23, which was applied to allow for reliable detection of dynein unbinding from the microtubule. This ~4-fold increase in dynein’s velocity indicates that release of dynein from the autoinhibited state can partially be stimulated by attachment of a large cargo to the tail domain, consistent with reports of bead motility driven by nonspecifically adsorbed mammalian dyneins9,23. Importantly, the DDB complex moves ~2-fold faster than dyneins carrying a large bead (Fig. 1h). Therefore, dynactin and BICD2 binding likely leads to additional conformational changes of the heavy chains, such as alignment of the motor domains24 and reorientation of the C-terminus25.
To test whether formation of the DDB complex enhances dynein’s work output, we measured the force generation of dynein, dynein-dynactin, and the DDB complex using an optical trap. We sparsely attached recombinant human dynein motors containing an N-terminal GFP to micron-sized polystyrene beads coated with αGFP antibody (Fig. 2a). With the trap held in a fixed position, the minus-end directed motility of beads driven by individual dynein motors stalled at 2.04 ± 0.02 pN (mean ± s.e.m.) resistive forces, slightly higher than previously reported stall forces of human dynein. Addition of dynactin to human dynein at a five-fold molar excess caused only a modest (p = 0.0025, Welch’s t-test) increase in stall force to 2.48 ± 0.06 pN (Fig. 2b), consistent with previous findings that dynactin alone has little effect on dynein motility17,18.
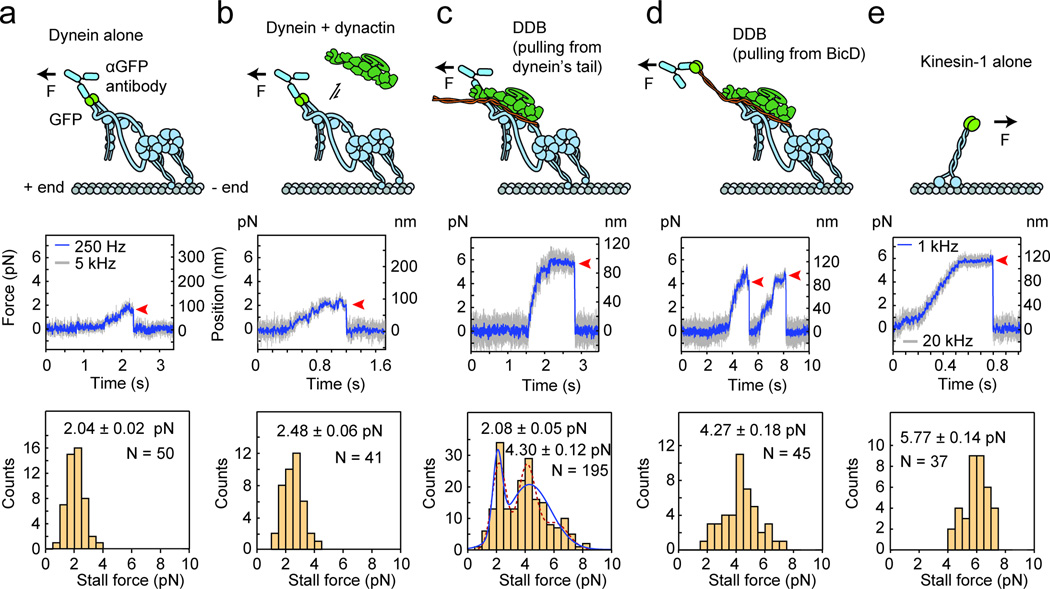
Figure 2. Effect of dynactin and BICD2N on dynein force production.
(a) (Top) The optically trapped bead, represented with a force arrow, is attached to GFP on dynein’s tail via an anti-GFP antibody. (Middle) Trace showing a typical stall of a bead driven by GFP-dynein in a fixed-trap assay. Red arrowhead represents the detachment of the motor from a microtubule after the stall. (Bottom) The histogram of observed stalls reveals the mean stall force (mean ± s.e.m., N = 50 stalls from 12 beads in 4 independent experiments). (b) Stall force of GFP-dynein with the addition of 5× molar excess of dynactin (N = 41 stalls from 10 beads in 4 independent experiments). (c) Stall force measurement of GFP-dynein with the addition of 5× molar excess of dynactin and 2× molar excess of BICD2N. The underlying distribution of the observed stalls is fitted to two Gaussians (blue curve; N = 195 stalls from 47 beads in 19 independent experiments) using a Gaussian Mixture Model. A three-Gaussian fit, shown with a dotted line, is not statistically warranted as determined by the Bayes Information Criterion (see Supplementary Fig. 2). (d) Stall force measurement of dynein with the addition of 5× molar excess of dynactin and 2× molar excess of BICD2N with a C-terminal GFP (BICD2N-GFP). The BICD2N-GFP is the attachment point (N = 45 stalls from 14 beads in 4 independent experiments). (e) Stall force measurement of kinesin-1 with a C-terminal GFP fusion as the attachment point (N = 37 stalls from 15 beads in 4 independent experiments).
We next assembled the DDB complex at a 1:5:2 molar ratio of dynein:dynactin:BICD2N. Single-molecule motility assays showed robust processive motility at this ratio (Fig. 1b). With all three components present, stall forces exhibited a bimodal distribution with a lower peak at 2.1 ± 0.3 pN, comprising 34% of all motors, and a higher peak at 4.4 ± 0.5 pN, comprising 70% of the 195 observed motors (Fig. 2c, Supplementary Fig. 2). A third peak is not warranted in the fit (Fig. 2c, Supplementary Fig. 2). It is likely that the lower peak at 2.1 pN corresponds to incomplete complex formation because in single-molecule motility assays, only 52% of GFP-tagged dynein motors move processively upon addition of dynactin and BICD2N. The stall forces near 4.4 pN only appear when all three components are present, suggesting that they correspond to the force output of DDB. To confirm this, we measured the stall force of DDB by attaching the bead directly to the N-terminal 400 residues of BICD2 via a C-terminal GFP tag (BICD2N-GFP), ensuring that any observed motility is driven by DDB rather than dynein alone. The beads stalled at 4.3 ± 0.2 pN (Fig. 2d), similar to the 4.4 pN peak observed in Figure 2c. A peak in stall events near 2.1 pN was not observed. Thus, we concluded that DDB complexes produce forces above 4 pN. To make a direct comparison with yeast cytoplasmic dynein, whose motility mechanism has been studied in detail26–29, we independently measured the stall force of full-length yeast dynein to be 3.6 ± 0.2 pN (Supplementary Fig. 3). Therefore, force production of mammalian dynein is similar to yeast cytoplasmic dynein28 and less than human kinesin-112 (5.8 ± 0.1 pN) (Fig. 2e).
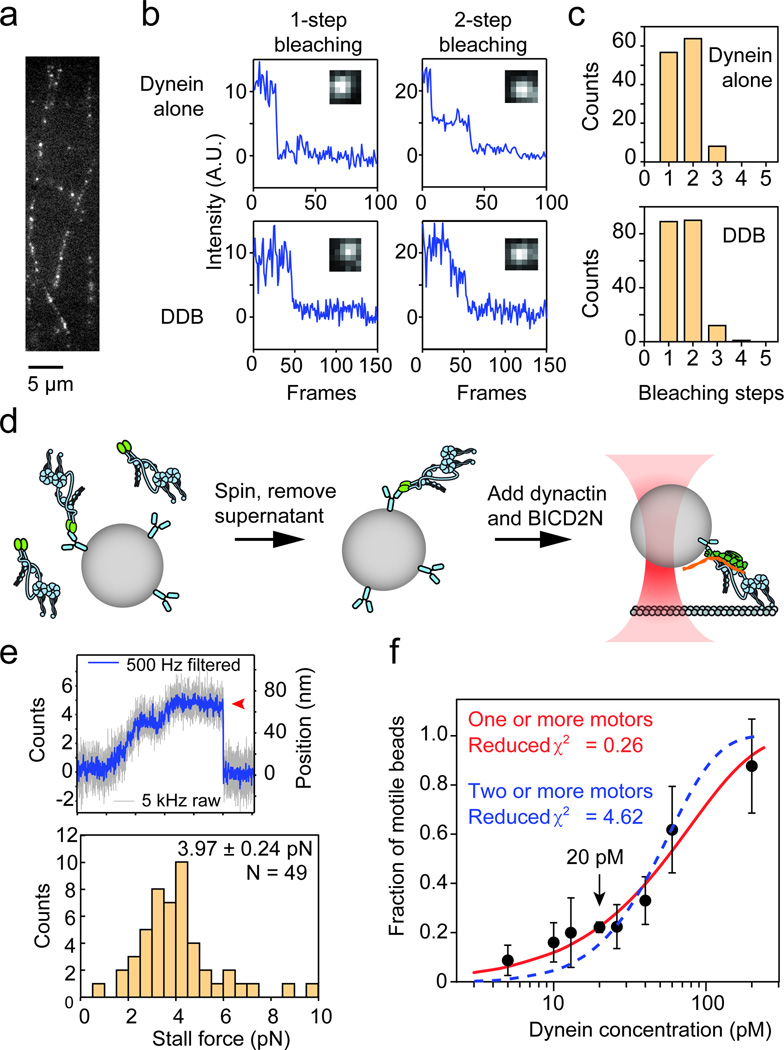
We sought to rule out the possibility that multiple dyneins aggregate in fluorescence or trapping assays. First, we counted the number of photobleaching steps of microtubule-bound dynein dimers tagged with a single GFP on each monomer. 95% of the GFP spots bleached in either one or two steps in the presence and absence of dynactin and BICD2N (Fig. 3a–c), suggesting that they correspond to a single dynein dimer. In the trapping experiments, to prevent the possibility of dynactin- and BICD2N-induced aggregation of dynein on the surface of beads, we pre-bound dynein sparsely to beads, removed the free motors, and then added dynactin and BICD2N (Fig. 3d). Under these conditions, dynein still produced high stall forces, 4.0 ± 0.2 pN, similar to the case in which all three components were mixed prior to bead binding (Fig. 3e). Therefore, the measured increase in stall force of DDB is not an artifact of aggregation, but rather caused by the binding of dynactin and BICD2N to individual dynein motors. Finally, to ensure that trapped beads are driven by a single motor, we quantified the fraction of motile beads as a function of dynein concentration30 (Fig. 3f). The data fit well to the model that each bead is carried by one or more motors and did not fit well to the model that a minimum of two motors is required to carry a bead. All trapping experiments in this work were performed at 20 pM dynein at which > 97% of all beads were driven by single motors.
Figure 3. Processive motility of DDB complexes is driven by single dynein dimers.
(a) GFP-dynein molecules tightly bound to microtubules in the absence of ATP. (b) Intensity traces of GFP-dynein alone in the presence and absence of dynactin and BICD2N show one- and two-step photobleaching. (c) Histograms showing the number of photobleaching steps of dynein in the absence and presence of dynactin and BICD2N. N = 127 for dynein alone and N = 192 for DDB. The experiment was repeated 3 times with dynein from two independent preparations. (d) Schematic depiction of the modified sample preparation for optical trapping. Dynein is mixed with beads and excess dynein is removed by centrifugation. BICD2N and dynactin are added after the removal of free motors to rule out dynactin- and BICD2N-dependent aggregation of dynein motors on beads. (e) Representative stall trace of a DDB complex and distribution of stall forces of the DDB complexes (mean ± s.e.m.). The experiment was repeated 3 times. (f) Fraction of dynein-coated beads moving as a function of dynein concentration. Values are represented as the mean ± the square root of F(1 − F)/N, with N being the number of beads tested. The solid red line represents a fit to the Poisson probability 1 − e−λC that the bead is carried by one or more motors, where C is dynein concentration and λ is the fit parameter (reduced χ2 = 0.26). The dashed blue line represents a fit to the probability 1 − e−λC − (λC)e−λC that the bead is carried by two or more motors (reduced χ2 = 4.62). For each data point, from left to right, N = 23, 25, 10, 482, 28, 35, 20, 24 beads from 3 independent experiments, except for the 20 pM data point, which was obtained from 19 independent experiments. The mean values, from left to right, are 0.087, 0.16, 0.2, 0.22, 0.22, 0.33, 0.61, 0.88.
To investigate the effect of dynein activation by its adaptor proteins on the motor’s ability to transport cargo in competition with motors of opposing polarity, we established an in vitro tug-of-war assay31,32 allowing us to pit one dynein against one human kinesin-1. A dynein construct with an N-terminal SNAPf tag was first labeled sub-stoichiometrically with a 74 nt long single-stranded DNA, and then with an excess of Alexa647 at its tail. This labeling strategy ensured that most dynein dimers were labeled either with one DNA molecule and one Alexa647 fluorophore, or two Alexa647 fluorophores. A similar strategy was used to label kinesin with a complementary DNA and a tetramethylrhodamine (TMR) at its tail using HaloTag (see Methods, Fig. 4a–c). The labeled motors were linked to each other through DNA hybridization, and assayed for motility on microtubules polymerized from pig brain tubulin.
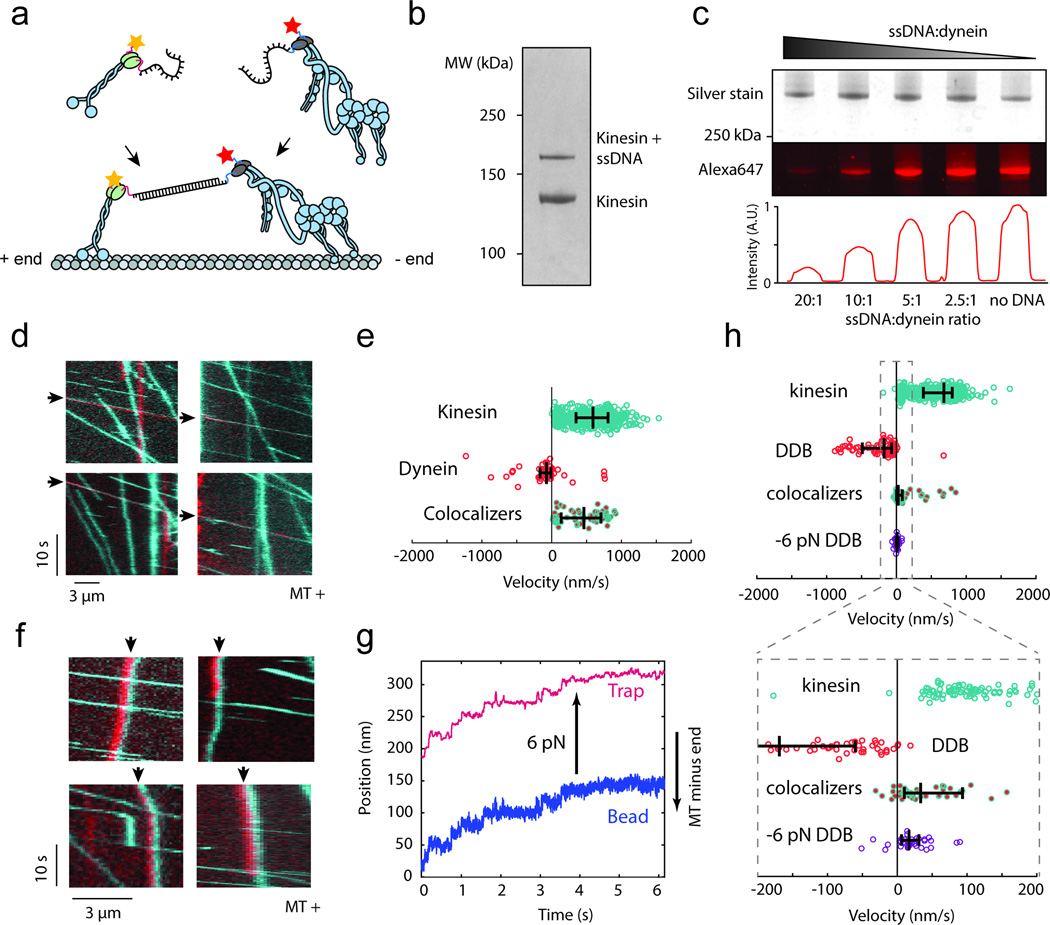
Figure 4. In vitro competition experiment between human dynein and human kinesin.
(a) Kinesin-1 is labeled with a short DNA oligo and TMR, while dynein is separately labeled with a complementary oligo and Alexa647. The two motors are connected through DNA hybridization. (b) The fraction of kinesin motors labeled with DNA was determined from an SDS-PAGE denaturing gel. (c) The fraction of dynein motors labeled with DNA was determined by separately imaging total protein quantity and fraction of DNA-free motors in an SDS-PAGE gel. Higher DNA:dynein ratios resulting in lower Alexa647 fluorescence (bottom), since the DNA occupied larger fractions of SNAPf binding sites, making them unavailable for labeling with Alexa647-BG-GLA. Full gel scans can be found in Supplementary Fig. 5. (d) Kymographs of dynein-Alexa647 (red) and kinesin-TMR (cyan) motility in the absence of dynactin and BICD2N on microtubules. Colocalizers are identified with black arrows. (e) Velocity distribution of kinesin only, dynein only, and kinesin-dynein colocalizers. Positive values correspond to plus end-directed velocities. From top to bottom, N = 655, 44, 59 runs from 3 independent experiments. Vertical bars represent median values and quartiles. Median values, from top to bottom, are 590, −76, and 460 nm/s. (f) Kymographs of dynein and kinesin motility in the presence of dynactin and BICD2N. The black arrows indicate colocalizers. Colocalizers in the top row are walking towards the minus-end of the microtubule. The red and cyan channels are laterally offset by five pixels to enhance the visibility of the colocalizers. The experiment was repeated 4 times. (g) Representative trace of DDB slowly walking towards the plus end in response to a 6 pN pulling force exerted by the optical trap. The experiment was repeated 3 times. (h) Velocity distribution of kinesin only, DDB only, kinesin-DDB colocalizers, and DDB walking against a plus end-directed 6 pN force. From top to bottom, N = 513, 79, 55, 35 runs from 3 independent experiments. Vertical bars represent median values and quartiles. Median values, from top to bottom, are 680, −176, 26, and 10 nm/s.
Simultaneous imaging of TMR-kinesin and Alexa647-dynein on microtubules revealed colocalization and correlated movement of dynein-kinesin complexes. By relying on sub-stoichiometric DNA labeling, we ensured that any observed colocalizers were comprised of one dynein and one kinesin motor. In the absence of dynactin and BICD2N, the velocities of colocalizers were nearly the same as those of kinesins alone (Fig. 4d,e, Supplementary Video 4), revealing that dynein on its own is unable to resist kinesin’s pull. However, the behavior of the colocalizers was markedly different upon the addition of dynactin and BICD2N. The median velocity of DDB-kinesin colocalizers (26 nm/s) was reduced over 20-fold compared to that of dynein-kinesin (464 nm/s) towards the microtubule plus end. Furthermore, 22% of DDB-kinesin colocalizers walked towards the microtubule minus end (Fig. 4f,g, Supplementary Fig. 4, Supplementary Video 5), which was not observed in the absence of dynactin and BICD2N. The velocity distribution of the colocalizers is distinct from those of both DDB alone and kinesin alone, indicating that both motors are contributing to the overall motility rather than one being passively carried by the winning motor. Interestingly, we did not observe frequent reversals of a DDB-kinesin colocalizer’s motility. This agrees with the absence of reversals in the artificial linking of multiple yeast dyneins to multiple human kinesins31. Reversals of cargo motility observed in vivo15 may result from the regulatory factors on the cargo that modulate motor activity, such as JIP133, or transient association of key regulatory proteins such as dynactin and BICD2 in cells15.
Next, we tested whether the dramatic slowing down and reversal of DDB motility can be explained by a purely mechanical response to kinesin’s pulling force towards the plus end. Operating an optical trap in a force-feedback mode, we pulled single DDB complexes towards the plus-end with a force of 6 pN, corresponding to the reported stall force of kinesin-1 (Fig. 2e)12. Under this condition, the median velocity of DDB was 10 nm/s towards the plus end, with 29% of the motors walking towards the minus end (Fig. 4g,h). These values are remarkably similar to the velocities of DDB-kinesin colocalizers, which is consistent with the predictions of the tug-of-war model.
Our results demonstrated that mammalian dynein complexes are strong motors capable of transporting cargos towards the microtubule minus-end against large resistive forces. Contrary to the earlier suggestion that 4–7 dyneins are needed to counteract the force production of a single kinesin-134, the activation of human dynein by dynactin and BICD2N allows it to dramatically slow down and sometimes defeat kinesin-1 in a one-to-one mechanical competition. A large force output of DDB is consistent with high minus end-directed forces exerted on lipid droplets in Drosophila embryos35 and single phagosomes inside mouse macrophage cells8. Furthermore, formation of strong DDB complexes bypasses the previously postulated requirement for a highly ordered spatial organization of dyneins on the surface of cellular cargos. It remains to be seen how diverse cargo adapter proteins other than BICD2 affect the force production of the dynein/dynactin complex and regulate dynein’s activity throughout the cell.
Previous in vivo optical trapping recordings have detected periodic peaks spaced at 1 – 2 pN intervals in the force distributions of dynein-driven cargos. It has been proposed that the peak periodicity represents the force production of single35 or a pair8 of dynein motors, and that the larger collective forces originate from multiple (up to twelve) dyneins simultaneously engaging with the microtubule to transport the cargo. This model relies on the assumption that the measured peak forces in the retrograde direction represent the maximal force production of a single motor multiplied by the motor copy number36. Our results are not fully consistent with this simple mechanical model, because the stall force of single DDB complexes (4.3 pN) is significantly higher than the peak periodicity of these force distributions. We propose that the wide range of cargo stall forces measured during multiple motor transport in vivo can be affected by the modulation of kinesin and dynein activity by cargo adaptor proteins37, premature release of the motors from microtubule before they come to a complete stall, and nonlinear mechanical coupling between multiple motors engaged with the cargo transport38. These possibilities remain to be rigorously tested by characterizing the force production of cargos containing predetermined numbers of motors and their regulators in vitro, such as the artificial cargos employed in this study.
METHODS
Cloning and plasmid construction
Genes for DHC (DYNC1H1, accession number NM_001376.4), DIC (DYNC1I2, IC2C, AF134477), DLIC (DYNC1LI2, LIC2, NM_006141.2), Tctex (DYNLT1, Tctex1, NM_006519.2), LC8 (DYNLL1, LC8-1, NM_003746.2) and Robl (DYNLRB1, Robl1, NM_014183.3) were codon optimized for expression in Sf9 cells, synthesized commercially (Epoch Life Sciences) and assembled into a single plasmid as described previously17. InFusion (Clontech) cloning was used to insert a GFP tag between the His-ZZ-LTLT tag39 and the DHC gene. The BICD2N construct was cloned using the N-terminal 400 amino acids of a codon optimized (Epoch Life Science) version of mouse BICD2 (NM_029791.4) gene. The construct contains a His-ZZ-LTLT tag39 followed by a SNAPf tag (New England Biolabs) at the N-terminus. The C-terminal GFP tagged BICD2N construct (BICD2N-GFP) was made by adding a GFP gene to a construct expressing the His-ZZ-LTLT tag followed by the N-terminal 400 amino acids of a codon optimized mouse BICD239.
Protein expression and purification
Human dynein and BICD2N expression and purification using the baculovirus insect cell system was performed as described previously17. Briefly, constructs for either the full dynein complex or BICD2N were integrated into the EMBac or EMBacY baculovirus genome and p2 baculovirus was produced. For protein expression 500 ml Sf9 cell suspension was infected with 5 ml p2 baculovirus and incubated at 27°C/124 rpm for 70–75 h. A ~2.5 g cell pellet was lysed using a tissue homogenizer (Wheaton), cleared by centrifugation and incubated with 2 to 5 ml IgG Sepharose 6 Fast Flow (GE Healthcare). The protein was removed from the beads with TEV protease and purified by size exclusion chromatography using either a G4000SWXL column (TOSOH Bioscience) or a Superose 6 column (GE Healthcare) on an Ettan LC system (GE Healthcare). The appropriate fractions were concentrated, snap frozen in liquid nitrogen and stored at −80°C. Dynactin was purified from pig brain using the large scale SP-Sepharose purification protocol19. Protein size and purity were confirmed by SDS-PAGE using Novex 4–12% Bis–Tris precast gels using either MOPS or MES buffer (Life Technologies) and stained with Instant Blue (Supplementary Fig. 5). Protein concentration was measured using the Quick Start Bradford kit (Bio-Rad).
Full-length yeast cytoplasmic dynein tagged with GFP at the N-terminal tail and with DHA (Promega) at the C-terminus (GFP-Dyn471kD-DHA)39 was expressed and purified as described previously28. Briefly, frozen yeast pellets were ground in a commercial steel coffee grinder and the resulting powder was thawed in lysis buffer (150mM HEPES, 250mM KAcetate, 10mM MgAcetate, 5mM EGTA, 5mM phenylmethylsulphonyl fluoride (PMSF), 500 mM MgATP, 50% glycerol, pH 7.4). Following centrifugation at 270,000 g for 45 minutes, the supernatant was incubated with IgG sepharose beads (GE Healthcare Life Sciences, 17-0969-01) at 4°C for 1 h. The beads were washed twice and transferred into TEV protease buffer (150mM KCl, 10mM Tris-HCl, 10% glycerol, 1mM TCEP, 1mM PMSF, 100mM ATP, pH 8.0). Dynein was cleaved from the beads by incubation with TEV protease at 16°C for 1 h.
Axonemes were extracted from live sea urchins. Tubulin used for the assembly of microtubules was purified from fresh pig brains through two polymerization-depolymerization cycles in a high molarity buffer40. To assemble microtubules for motility experiments, tubulin was polymerized in the presence of 20 uM taxol in BRB80 buffer at 37°C for 30 minutes and remaining free tubulin dimers were removed by centrifugation.
Functionalization of complementary DNA oligos
Two complementary amine-modified 74bp oligonucleotides (IDT) with the sequences /5AmMC12/TGGTCAATACTAGGAGCAGAGATGGCAGGAGTCAGATGAACAGATAGTGGAGGCAGGGTCAGCGCGAGATCGTC (Strand 1) and /5AmMC12/ATGACGATCTCGCGCTGACCCTGCCTCCACTATCTGTTCATCTGACTCCTGCCATCTCTGCTCCTAGTATTGAC (Strand 2) were designed to minimize potential secondary structures and contain a 2nt overhang on each end, followed by a 12-carbon spacer terminating with an amine group. 25 µM Strand 1 and 2 were separately labeled with 1.25 mM BG-GLA N-Hydroxysuccinimide (NHS) (New England BioLabs, S9151S) and 1 mM alkyl chloride (AC) NHS (Promega, P6751), respectively, in a 50 mM HEPES pH 8.5 buffer containing 50% v/v DMSO. The reaction was allowed to proceed for 30 minutes at room temperature, after which the DNA was de-salted and exchanged into dynein motility buffer (DMB: 30 mM HEPES, 5 mM MgSO4, 1 mM EGTA, pH 7.0 with KOH) through five consecutive spins through 3,000 MWCO spin filters. DNA labeling was confirmed by gel electrophoresis using a 15% TBE-Urea gel, and the DNA concentration and purity were assessed by measuring A230, A260, and A280 absorbances.
Labeling dynein and kinesin with DNA oligos
SNAPf-dynein was mixed with BG-GLA-Strand 1 and kinesin-HaloTag was mixed with AC-Strand 2 for 1h at 4°C. After the reaction, 10-fold molar excess of dye (BG-GLA-Alexa647 for dynein or AC-TMR for kinesin) was added to the reaction mixture for 15 minutes at 4°C to label remaining sites that were not labeled by DNA. The DNA labeling efficiency was highly sensitive to DNA and protein concentrations. In the case of kinesin, labeling efficiency was calculated by comparing the intensities of the labeled and unlabeled bands that were clearly separated on the 4–12% Bis–Tris SDS-PAGE gel (Fig. 5b). For dynein, the labeled and unlabeled bands were virtually indistinguishable on the gel due to its high molecular weight. In this case, the labeling efficiency was estimated by running a band of dynein almost fully labeled with DNA that shows no Alexa647 fluorescence and a band of DNA-free dynein that is strongly fluorescent. The labeling efficiencies of the unknown dynein samples were estimated by comparing the relative Alexa647 fluorescence intensity of the band with that of ~100% labeled and 0% labeled control bands (Fig. 5c). The DNA labeling ratio was optimized to yield a ~30% efficiency for both dynein and kinesin to minimize the likelihood of a single dimeric motor being labeled with two DNA strands to a <9%. Excess DNA and fluorophores were removed from both motors via microtubule bind and release (MTBR).
MTBR purification of human dynein
Human dynein was purified and separated from DNA and Alexa647 using a modification of the MTBR protocol previously established for yeast dynein28. First, ~200 nM dynein was mixed with 2 mg/mL microtubules and allowed to bind at room temperature for 10 min (typical reaction volume was ~300 µL). Then, the dynein/microtubule mixture was centrifuged through 200 µL of sucrose cushion (25% sucrose and 100 µM taxol in DMB buffer: 30 mM HEPES, 5 mM MgSO4, 1 mM EGTA, pH 7.0 with KOH) for 10 min at 22°C and 40,000 rpm in a TLA 120.1 rotor (Beckman Coulter). The pellet was washed twice with DMB supplemented with 100 µM taxol and 1 mM DTT, then resuspended in half of dynein’s original volume of release buffer (DMB supplemented with 150 mM KCl, 2 mM ATP, and 1 mM DTT) and allowed to release at room temperature for 10 min. The solution was then centrifuged for 10 min at 22°C and 40,000 rpm to remove microtubules from free dynein. The supernatant was supplemented with 20% v/w glycerol and stored at −80°C.
Coating beads and quantum dots with αGFP antibodies
860 nm and 200 nm carboxyl latex beads (Life Technologies) were coated with custom-made rabbit polyclonal αGFP antibodies (Covance) with EDC and sulfo-NHS crosslinking (Pierce). 100 µL of 4% v/w bead stock were mixed with ~1 mg of antibody in coupling buffer (100 mM sodium-phosphate buffer, pH 7.4), per) and reacted with shaking at room temperature for 30 min. The labeled beads were then passivated by adding 10 mg/mL BSA and stored in 1× PBS (Phosphate Buffered Saline, pH 7.4) supplemented with 0.1% sodium azide and 0.5 mg/mL BSA at 4°C.
Amine-coated quantum dots emitting at 585 nm (QD585; Invitrogen) were labeled with the same rabbit polyclonal αGFP antibodies (Covance) via sulfo-SMCC crosslinking (Pierce). QD585 were dissolved in 100 mM pH 8.0 borate buffer to a 200 nM final concentration and incubated with a 250 fold excess of sulfo-SMCC for 1 h. Excess crosslinker was removed by two runs through a 30,000 MWCO spin filter and QD585 were transferred into dynein motility buffer (DMB: 30 mM HEPES, 5 mM MgSO4, 1 mM EGTA, pH 7.0 with KOH). Antibodies were reduced with 2 mM DTT for 30 min and residual DTT was removed by three runs through 7,000 MWCO spin de-salting columns. Activated QD585s were then incubated with a 4-fold molar excess of the reduced antibodies for 1 h. The reaction was quenched with the addition of 20 mM Tris pH 8.0 and spin-concentrated to obtain the desired final concentration.
Optical trap assay
Dynein concentration was determined for each batch of protein. The protein was diluted until less than 30% of beads exhibited any activity when brought in contact with an axoneme, ensuring that >95% of observed events can be attributed to the actions of single motors. When BICD2N and/or dynactin were added to the assay, they were mixed with dynein at 1:5:2 molar ratio (dynein:dynactin:BICD2N) and incubated at 4°C for 5 minutes prior to adding the complex to beads. Dynein with any auxiliary proteins was then allowed to bind to 860 nm diameter latex beads for 10 min at 4°C before proceeding with sample preparation. The sample chamber was loaded by first flowing Cy5-labeled axonemes in dynein motility buffer (DMB: 30 mM HEPES, 5 mM MgSO4, 1 mM EGTA, pH 7.0 with KOH), followed by a solution of dynein- or DDB-coated beads in motility/imaging buffer (DMB supplemented with 35 µg/mL PCD, 2.5 mM PCA, 10 mM DTT, 1 mg/mL casein, and 2 mM ATP).
All trapping experiments were performed on a custom-built optical trap microscope, as described previously28. Cy5-labeled axonemes were brought to the center of the field of view with a piezo-driven servo XY stage (M-687, Physik Instrumente). Beads were trapped with a focused 1064 nm beam using a 100× 1.49 N.A. apochromat oil-immersion objective (Nikon). The trap was steered with a pair of perpendicular acousto-optical deflectors (AOD’s, AA Opto-Electronic) and lowered to the surface of the axoneme by moving the trapping objective with a piezo flexure objective scanner (P-721 PIFOC, Physik Instrumente). Position of the bead relatively to the center of the trap was monitored by imaging the back-focal plane of a 1.4 N.A. oil-immersion condenser (Nikon) onto a position-sensitive detector (First Sensor, Inc.). Signals were acquired at 5 kHz, and position feedback was performed at up to 200 Hz. Detector response was calibrated by rapidly raster-scanning the laser across a trapped bead and trap stiffness was obtained from the Lorentzian fit to the power spectrum of a trapped bead. Typical stiffness values used in these assays ranged from 0.008 pN nm−1 to 0.06 pN nm−1. Trap stiffness is adjusted to allow motors to travel 100 nm on average before stall.
Motility and photobleaching assays
Single-molecule motility assays were carried out on a custom-built objective-type TIRF setup, built around the body of a commercial Nikon Ti-E microscope. Fluorophores were visualized with a 100× 1.49 N.A. apochromat oil-immersion objective (Nikon) and imaged onto an EMCCD camera (Andor). Assay preparation was identical to the optical trap sample preparation described in a previous section, except without the addition of latex beads. In the dynein-kinesin crosslinking motility experiments, dynein-DNA and kinesin-DNA were allowed to react with each other for 10 min at 4°C prior to being diluted to the final desired concentration. Photobleaching experiments were performed without the addition of ATP to ensure that motors remain stationary on axonemes. >95% of spots bleached completely over the course of a 200-frame movie.
For dynein-driven bead motility experiments, 200 nm latex beads coated with GFP antibodies were sparsely decorated with dynein (with or without dynactin and BICD2N) and imaged with either brightfield illumination or scattered-light fluorescence. To ensure single-molecule conditions, the concentration of dynein was reduced until >90% of beads did not visibly interact with axonemes upon contact. The trap was used to measure the velocity of individual dynein motors carrying 860 nm diameter beads, because these beads are too large to encounter microtubules with sufficient frequency by diffusion alone.
The dynein-kinesin colocalization motility experiments were performed on microtubules. We observed that using axonemes versus microtubules had a modest effect on dynein recruitment to the tracks and initiation of motility, but not on its velocity or processivity following recruitment. Microtubules were used for the colocalization experiments mainly to obtain longer tracks for processive motility. For surface immobilization of microtubules, 1% of biotinylated tubulin was incorporated into the microtubule polymerization reaction. Biotinylated-microtubules are attached to the coverslips pre-coated with streptavidin and BSA-biotin.
Data analysis
In order to extract run velocities from fluorescence movies, kymographs were created along each individual axoneme using ImageJ. Motile dynein motors were then identified manually from the kymographs. Only molecules that traveled > 530 nm (5 pixels along x) and remained bound for more than 2.5 s (5 pixels along y) were included in the analysis. Diffusive molecules (those that exhibited >530 nm excursions in both directions along the axoneme) were excluded. For molecules that transitioned from static behavior (>30 seconds with < 100 nm displacement) to motile behavior, only the motile segments were analyzed.
To generate dynein stall force histograms, position data from trap recordings were downsampled to 250 Hz or 500 Hz for ease of visualization and stall events were manually selected. To qualify as a stall, the position trace had to reach a plateau and remain stationary (with mean deviations of less than ± 10nm) for at least 100 ms before terminating in a ‘rip’. A ‘rip’, indicating that the motor fully released from the microtubule, had to constitute a rapid (<2 ms) jump towards the trap center of at least 50 nm, larger than the maximum step a dynein molecule can be expected to take. Image analysis was performed in ImageJ.
Statistics and reproducibility
At least three independent repetitions were performed to obtain every published result, and the exact number of repetitions is reported for each experiment. Each statistical analysis method is explicitly stated in the main text and/or figure legend.
Data availability
All data that support the conclusions are available from the authors on request.
Supplementary Material
1
2
3
4
5
6
Acknowledgments
We are grateful to Sinan Can, Luke Ferro, Alexander Chien, and Jigar Bandaria for helpful discussions. This work was supported by NIH (GM094522 (AY)), NSF CAREER Award (MCB-1055017 (AY)), Medical Research Council, UK (MC_UP_A025_1011 (AC)), Wellcome Trust New Investigator Award (WT100387) and EMBO Young Investigator Award (both to AC), a Marie Curie Intra European Fellowship (MS), and an NSF Graduate Research Fellowship [DGE 1106400 (VB)].
Footnotes
SUPPLEMENTARY INFORMATION
Supplementary information is linked to the online version of this paper.
AUTHOR CONTRIBUTIONS
V.B., A.P.C., and A.Y. designed the study. M.A.S. and H.F. prepared the constructs and purified the protein. V.B. and A.E.R. performed the optical trapping experiments. V.B. labeled dynein and kinesin with DNA, performed the fluorescence motility experiments, and analyzed the data. V.B., A.P.C., and A.Y. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 3.Torisawa T, et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat. Cell Biol. 2014;16:1118–1124. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- 4.Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur ELF. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J. Neurosci. 2013;33:13190–13203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Sheetz MP. One-dimensional diffusion on microtubules of particles coated with cytoplasmic dynein and immunoglobulins. Cell Struct. Funct. 1999;24:373–383. doi: 10.1247/csf.24.373. [DOI] [PubMed] [Google Scholar]
- 6.Ayloo S, et al. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat. Commun. 2014;5:4807. doi: 10.1038/ncomms5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rai AKA, Rai AKA, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 9.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 10.Tripathy SK, et al. Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat. Cell Biol. 2014;16:1192–1201. doi: 10.1038/ncb3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nat. Cell Biol. 2010;12:1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 13.Kunwar A, et al. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. PNAS. 2011;108:18960–18965. doi: 10.1073/pnas.1107841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai A, et al. Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell. 2016:722–734. doi: 10.1016/j.cell.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock WO. Bidirectional cargo transport: moving beyond tug of war. Nat. Rev. Mol. Cell Biol. 2014;15:615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendricks AG, et al. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 2010;20:697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014:1–14. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015 doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Splinter D, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thirumurugan K, Sakamoto T, Hammer JA, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaan HYK, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King SJ, Schroer T. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 24.Chowdhury S, Ketcham SA, Schroer TA, Lander GC. Structural organization of the dynein–dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 2015;22:345–347. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas MP, et al. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat. Commun. 2015;6:6206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleary FB, et al. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nat. Commun. 2014;5:4587. doi: 10.1038/ncomms5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belyy V, Hendel NL, Chien A, Yildiz A. Cytoplasmic dynein transports cargos via load-sharing between the heads. Nat. Commun. 2014;5:5544. doi: 10.1038/ncomms6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 31.Derr ND, et al. Tug-of-War in Motor Protein Ensembles Revealed with a Programmable DNA Origami Scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehl MR, Zhang K, Lee HJ, Tirrell DA. Engineering Cooperativity in Biomotor-Protein Assemblies. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 33.Fu MM, Holzbaur ELF. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol. 2013;202:495–508. doi: 10.1083/jcb.201302078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19381–19386. doi: 10.1073/pnas.0906524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leidel C, Longoria Ra, Gutierrez FM, Shubeita GT. Measuring molecular motor forces in vivo: Implications for tug-of-war models of bidirectional transport. Biophys. J. 2012;103:492–500. doi: 10.1016/j.bpj.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shubeita GT, et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu M, Holzbaur ELF. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24:564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klumpp S, Lipowsky R. Cooperative Cargo Transport by Several Molecular Motors. PNAS. 2005;102 doi: 10.1073/pnas.0507363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 39.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization- depolymerization in a high-molarity buffer. Protein Expr. Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
Data Availability Statement
All data that support the conclusions are available from the authors on request.