Targeting and Readout Strategies for Fast Optical Neural Control In Vitro and In Vivo (original) (raw)
Introduction
Major obstacles faced by neuroscientists in attempting to unravel the complexity of brain function include both the heterogeneity of brain tissue (with a multitude of cell types present in vivo) and the high speed of brain information processing (with behaviorally relevant millisecond-scale electrical activity patterns). To address different aspects of these technical constraints, genetically targetable neural modulation tools have been developed by a number of groups (Zemelman et al., 2002; Banghart et al., 2004; Karpova et al., 2005; Lima and Miesenbock, 2005; Thompson et al., 2005; Chambers et al., 2006; Tan et al., 2006; Gorostiza et al., 2007; Lerchner et al., 2007; Szobota et al., 2007). One approach recently brought to neurobiology, combining both high speed and genetic targeting, is based on a family of fast light-responsive microbial opsins including halorhodopsins (e.g., NpHR) and channelrhodopsins (e.g., ChR2) (for review, see Zhang et al., 2007b). These microbial opsins are single-component transmembrane conductance regulators encompassing light sensitivity and fast membrane potential control within a single open reading frame, which can be used to achieve fast bidirectional control of specific cell types even in freely moving animals (Adamantidis et al., 2007; Zhang et al., 2007a). Although the basic functioning of these tools has been reviewed previously (Zhang et al., 2007b), here we describe a collection of targeting and readout strategies designed for rapid and flexible application of the microbial opsin system, and provide pointers to the relevant literature. Combinations of these multiple levels of targeting and readout define an evolving toolbox that may open up new possibilities for basic neuroscience investigation.
Channelrhodopsin-2 (ChR2) and halorhodopsin (NpHR)
Although it had been known that microbial opsins can control membrane potential in their native settings and various reduced systems (e.g., Ehrlich et al., 1984; Kalaidzidis et al., 1998; Nagel et al., 2002, 2003), the surprising ability of these proteins when heterologously expressed to precisely control neurons and the technology for applying these tools in freely behaving animals have been demonstrated only over the past two years (for review, see Zhang et al., 2007b). ChR2 was the first microbial opsin brought to neurobiology, where it was initially found in hippocampal neurons that ChR2-expressing neurons can fire blue light-triggered action potentials with millisecond precision, as a result of depolarizing cation flux, without addition of chemical cofactors (Boyden et al., 2005). This approach has since proven versatile across a variety of preparations (Li et al., 2005; Nagel et al., 2005; Bi et al., 2006; Deisseroth et al., 2006; Ishizuka et al., 2006; Schroll et al., 2006; Zhang et al., 2006, 2007a; Arenkiel et al., 2007; Petreanu et al., 2007; H. Wang et al., 2007; Zhang and Oertner, 2007). Second, an inhibitory member of the microbial opsin family (Ehrlich et al., 1984; Hegemann et al., 1985; Duschl et al., 1988; Bamberg et al., 1993; Kalaidzidis et al., 1998; Kolbe et al., 2000) was brought to neurobiology; in work stimulated by the finding that the all-trans retinal chromophore required by microbial opsins appears already present within mammalian brains (Zhang et al., 2006), it was found that neurons targeted to express the light-activated chloride pump halorhodopsin from Natronomonas pharaonis (NpHR) can be hyperpolarized and inhibited from firing action potentials when exposed to yellow light in intact tissue and behaving animals (Zhang et al., 2007a); because of the excitation wavelength difference, the two optical gates can be simultaneously used in the same cells even in vivo (Zhang et al., 2007a) and may modulate aspects of neural synchrony with high temporal precision (Han and Boyden, 2007) (for review, see Zhang et al., 2007b). Third, genetic targeting tools have been developed for versatile use of microbial opsins with existing resources including cell type-specific promotor fragments or Cre-LoxP mouse driver lines suitable for a wide variety of neuroscience investigations (Adamantidis et al., 2007; Zhang et al., 2007a,c; described below). Finally, integrated fiber-optic and solid-state optical approaches have provided the complementary technology to allow specific cell types, even deep within the brain, to be controlled in freely behaving mammals (Adamantidis et al., 2007; Aravanis et al., 2007) (for review, see Zhang et al., 2007b).
What factors limit opsin efficacy? One potential limitation is that for invertebrates like Caenorhabditis elegans and Drosophila, the all-trans retinal must be delivered, but this turns out to be straightforward to provide in food or in the environment (Nagel et al., 2005; Schroll et al., 2006; Zhang et al., 2007a). In general, sufficient levels of functional expression (which may depend on gene delivery method, copy number, promoter strength, and time after initiation of gene expression) must be obtained to either potentiate or inhibit spiking. However, care must be taken not to express the opsins at excessive levels, because overexpression of membrane proteins can lead to toxicity and loss of membrane integrity in a wide variety of systems. We and our colleagues have generated healthy ChR2-EYFP transgenic mice lines with expression throughout the brain using the Thy1 promoter; under these conditions, ChR2 can be used to map synaptically connected neurons in anesthetized mice and in slices (Arenkiel et al., 2007; H. Wang et al., 2007). We note that visualization of NpHR-transduced neurons indicates that intracellular halorhodopsin accumulation is particularly pronounced (Zhang et al., 2007a). Indeed, as we recently have explored the regime of very high levels of expression (strong promoters and high viral-vector multiplicity of infection or high gene copy number) to determine upper bounds of effect sizes, we have seen intracellular accumulation and even membrane blebbing toxicity that can be prevented by returning to lower, effective expression levels (Zhang et al., 2007a); this first-generation solution soon may be enhanced by second-generation molecular modifications to modulate membrane targeting. NpHR is well tolerated at expression levels that subserve its inhibitory function both in behaving C. elegans and in intact mammalian brain tissue (Zhang et al., 2007a), and NpHR can be introduced developmentally or in adulthood at suitable titers (Fig. 1A) (Zhang et al., 2007a).
Figure 1.
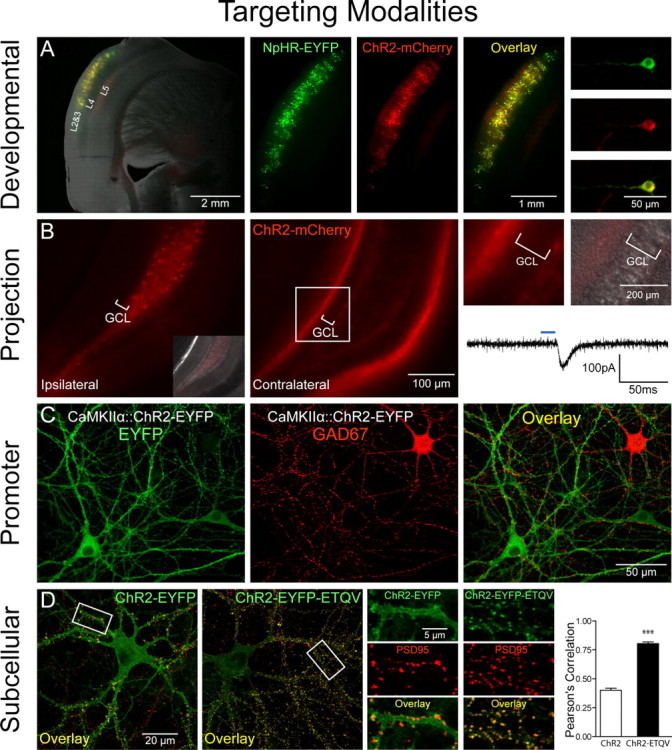
Targeting modalities. A, Developmental targeting: NpHR-EYFP (green) and ChR2-mCherry (red) expression in layers 2 and 3 of the mouse cortex via timed in utero electroporation (E16 surgery, P27 coronal slicing). A mixture of CMV::NpHR-EYFP and CMV::ChR2-mCherry DNA (1 μg/μl each) was pressure injected unilaterally into the ventricle and electroporated [5 50 ms pulses, 1 Hz, 40 V (see also Petreanu et al., 2007)]. Shown are pictures of acute slices taken on a Leica (Bannockburn, IL) MZ16FA stereomicroscope. B, Anterograde projection-based targeting: unilateral delivery of CaMKIIα::ChR2-mCherry lentivirus (1.5 μl, 109 pfu/ml) to the adult mouse dentate hilus/CA3 (left, and inset showing differential interference contrast image superposition) results in axonal ChR2-mCherry expression in the contralateral hippocampus (middle), with no expression in the granule cell layer (GCL) on either side of the brain (right). Note contralateral ChR2-mCherry-positive axonal projections to the molecular layer and subgranular zone. Optical stimulation with 15 ms pulses of blue light (HQ470 nm/40×; Chroma, Brattleboro, VT) of ChR2-positive axonal projections in acute slices from the contralateral hippocampus evoked inward synaptic currents in ChR2-negative dentate granule cells. A Lambda DG-4 optical switch (Sutter Instruments, Novato, CA) and 300 W xenon lamp were used for light delivery. Two injection sites were used at the following coordinates: anteroposterior (AP), 2 mm; mediolateral (ML), 1.5 mm; dorsoventral (DV), −2 mm; and AP, 3 mm; ML, 3 mm; DV, −3 mm. C, Promoter-based targeting: ChR2-EYFP (green) was targeted to excitatory hippocampal neurons in culture via CaMKIIα::ChR2-EYFP lentivirus [infected 2 d in vitro (div)]; immunostaining for GAD67 (red; 1:250; Millipore, Billerica, MA) in 14 div cultures shows exclusion of ChR2 expression from inhibitory GAD67+ neurons. D, Subcellular targeting: ChR2-EYFP (green) is expressed ubiquitously throughout the cell membrane in 16 div cultures. Adding the PDZ-domain binding motif ETQV to the C terminus by in-frame cloning led to concentration of ChR2-EYFP at postsynaptic sites, indicated by colocalization with PSD-95 staining (red; 1:200; Affinity Bioreagents, Golden, CO). Pearson's correlation (Manders et al., 1993) was r = 0.78 for ChR2-EYFP-ETQV and r = 0.40 for untargeted ChR2-EYFP; unpaired t test, ***p < 0.0001; n = 22 and 24 dendrites, respectively. Single confocal sections through 50 μm segments of proximal dendrites were thresholded and correlation coefficients determined using Volocity software.
Putting opsins in their place: beyond genetic targeting in networks
The rate-limiting step in using these tools is now directing their activity to the specific locations where electrical control is needed, by controlling either localization of the light or localization of protein expression (e.g., via cell type-specific promoters like the excitatory neuron-specific CaMKIIα promoter) (Fig. 1C) (Aravanis et al., 2007). To rapidly capitalize on the many available mouse lines driving (for example) Cre recombinase under the control of cell type-specific promoters, AAV vectors have been developed (Zhang et al., 2007c) carrying microbial opsins driven by a strong promoter followed by a floxed-stop sequence that can be injected into Cre-expressing mice. The cell type of interest efficiently recombines the stop sequence out of the genome and allow opsin expression under the strong promoter; the cell type of interest is then susceptible to control by fiber-optic light delivery registered to the virus injection sites using published methods for reliably achieving this crucial coregistration (Adamantidis et al., 2007; Aravanis et al., 2007). These promoter-based approaches are well suited to distinguish the roles played by specific neural types in the physiological or pathological functioning of the nervous system.
A potential limitation is that specific promoter-based targeting strategies or mouse Cre lines are not always immediately available or suitable for some experiments. However, in many cases, projection patterns can be used to achieve optical control over the desired neurons. Contralateral projections in the brain have been targeted by labeling cortical neurons in vivo with ChR2, followed by high-speed circuit mapping with light exposure delivered to brain slices from the contralateral hemisphere (Petreanu et al., 2007). In this experiment, contralateral axonal projections showed similar target preference in comparison with the ipsilateral projections. These experiments illustrate a targeting principle that may be generalizable. For example, Figure 1B shows selective labeling of hippocampal contralateral projections, in which axons of ChR2-positive hippocampal cells from the virus-injected hemisphere synapse onto ChR2-negative granule cells in the contralateral dentate gyrus. Optical stimulation with blue light of the ChR2-positive axonal projections contralateral to the site of injection can activate postsynaptic ChR2-negative dentate granule cells (Fig. 1B, right). In this way the selective impact of fibers traveling from one brain region to another, spatially separated, brain target region can be studied simply by illuminating the target brain region, a principle that could extend to many other circuits (e.g., corticothalamic and nigrostriatal projections) and to species in which promoters are less well defined (e.g., primates). However, a limitation of this method is that in some cases insufficient opsin accumulation in the presynaptic terminal may prevent robust spiking responses, which may be overcome with molecular targeting strategies. Another non-promoter-based targeting method would be to use transsynaptic viral methods to introduce opsins; for example, transsynaptic tracers based on rabies virus will spread only to directly connected cells, thereby allowing tests of the functional role of neurons one synapse removed from the initially infected cells (Wickersham et al., 2007).
Cell types also can be targeted developmentally. One example involves the targeting of specific cortical layers with in utero electroporation, as demonstrated for ChR2 by Petreanu et al. (2007). Indeed, in utero electroporation at precisely timed embryonic days in mouse can be used to target both ChR2 and NpHR together to individual cortical layers such as layers II and III [embryonic day 15.5 (E15.5)] (Fig. 1A), layer IV (E13.5), or layers V and VI (E12.5). In utero electroporation also could be used to express ChR2/NpHR in the inhibitory neurons of the striatum or in the hippocampus (Borrell et al., 2005; Navarro-Quiroga et al., 2007). A key advantage of in utero electroporation is that, unlike viral delivery methods, there are no DNA size restrictions, which allows for expression of opsins under control of a wider array of promoters. Promoter-based strategies could also be used to target specific cortical layers, because emerging knowledge of layer-specific markers (Lein et al., 2007) may lead to highly specific targeting strategies, and layer V of cortex already has been targeted by generating transgenic mice expressing ChR2-EYFP under control of the Thy1 promoter (Arenkiel et al., 2007; H. Wang et al., 2007). Of course, these spatial targeting strategies could be complemented with traditional promoter-based strategies, for example, with promoter fragments, BAC (bacterial artificial chromosome) transgenics (Heintz, 2001), Cre/ROSA26-loxP mice (Srinivas et al., 2001), or Gal4-UAS enhancer expression systems (Perrimon, 1998). For example, a combined developmental/promoter-based strategy could allow selective targeting of excitatory (CaMKIIα-expressing) neurons only in layer 2/3 of cortex.
ChR2 and NpHR are ubiquitously localized membrane proteins when heterologously expressed, and full-field illumination will generate membrane potential changes synchronously across the entire cell. This pattern of activity modulation is suitable for most classes of circuit-level experiments in which neurons simply need to be activated or inhibited in specific temporal patterns. However, subcellular targeting of opsins could be used to modulate activity in a localized manner even with full-field flash illumination; for example, dendritic targeting of ChR2 could preserve physiological information flow from synapse to soma to axon for studies of subcellular signaling (synapse-to-nucleus signaling, dendritic integration rules, spike timing-dependent plasticity, and other backpropagating action potential effects). To achieve precise optical control within neurons, we have explored the use of intracellular targeting motifs that can be attached to the opsin nucleotide sequences. As one example of this approach, we have attached the PDZ-domain binding sequence ETQV, a motif involved in NMDA receptor clustering at postsynaptic sites, to the C terminus of ChR2-EYFP (Zito et al., 1999; Guerrero et al., 2005). When expressed in hippocampal neurons, ChR2-EYFP-ETQV was indeed concentrated at the sites of postsynaptic densities, as indicated by selective colocalization with PSD-95, in contrast to the nonselective localization of ChR2-EYFP (Fig. 1D). The postsynaptically recruited protein maintained its ability to induce precisely timed depolarization and action potential firing in hippocampal neurons (data not shown). If this synaptic targeting were combined with precise optical guidance strategies, the causal role of neural population codes, in which members of the population are individual synapses rather than individual neurons, could be studied. Other targeting strategies (for example, to presynaptic terminals, to axon hillocks and nodes, or to intracellular membranes) should also be possible and will facilitate temporally precise studies of subcellular signaling events as well as network-level optogenetic experiments.
Integrating multimodal readout technologies with optical control
The fully equipped neuroscience optical toolbox would include not only ChR2/NpHR optical control, but also multiple reliable readout technologies for answering a broad range of experimental questions, in vitro and in vivo. Indeed, simultaneous optical control and electrophysiological recording of neuronal activity in vivo, even with intraparenchymal stimulation, is now possible with integrated submillimeter scale fiber optics and recording electrodes (Fig. 2A), complementing the capabilities of surface illumination described by Arenkiel et al. (2007). In this way, local-circuit responses can be measured with temporal precision that matches the millisecond-scale precision of ChR2/NpHR-based optical control, even within deep brain structures in vivo. This rapid feedback not only is experimentally valuable, but also could be important for eventual clinical applications of optical stimulation, so that therapeutic stimulation parameters could be rapidly tuned or timed in patients. A major limitation in functional neurosurgery and interventional psychiatry is the fact that electrical artifacts from stimulating electrodes prevent precisely simultaneous recording from local neurons, so that it is not clear whether particular brain stimulation paradigms are activating, inhibiting, or desynchronizing the target tissue. Although care must still be taken with regard to potential artifacts from external electronics, the combined optical stimulation/electrical recording approach allows high-temporal-precision electrical assessment of local responses in the absence of locally generated electrical artifacts, allowing local evoked spikes to be directly visualized (Fig. 2A). Electroencephalography is another electrical readout well suited for simultaneous use with optogenetic stimulation (Adamantidis et al., 2007).
Figure 2.
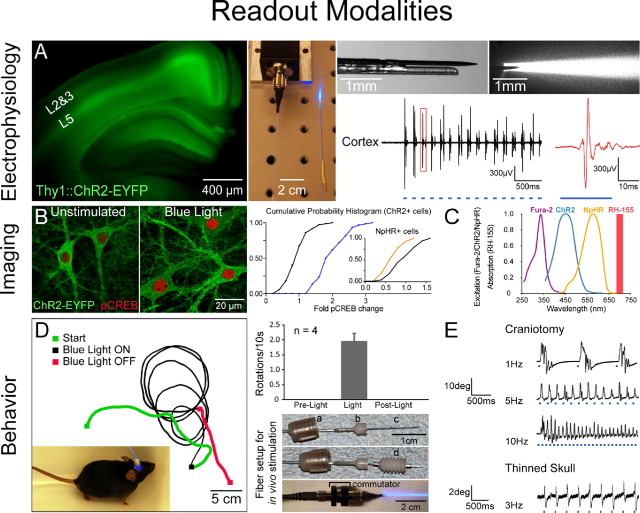
Readout modalities in vitro and in vivo. A, Electrophysiology in vivo: simultaneous optical stimulation and electrical recording from cortex in an intact Thy1::ChR2-EYFP mouse using an extracellular tungsten electrode (1 MΩ, ∼125 μm outer diameter) glued to an optical fiber (∼200 μm outer diameter; ThorLabs, Newton, NJ); the ∼0.5 mm offset (top right) ensures illumination of recorded neurons. The top-right illumination cone picture was generated by placing the electrode and fiber ensemble into a 1-cm-thick block of 1% agar (electrode superficial as shown); the geometrical dispersion is greatly modulated by scattering in brain tissue as described previously (Aravanis et al., 2007). FC/PC-coupled fiber optic is shown (middle) coupled to a 473 nm laser diode from CrystaLaser (Reno, NV) through an FC/PC adapter. Thy1::ChR2-EYFP (line 18) mice (Arenkiel et al., 2007; H. Wang et al., 2007) have strong ChR2 expression in layer 5 of neocortex (left). Recordings in anesthetized mice showed reliable cortex activation with pulsed blue light (10 Hz/25 ms), without stimulation artifacts at either the onset or offset of the stimulus. pClamp 10 and Digidata 1322A board (Molecular Devices, Union City, CA) were used to collect data and generate pulsed light. The recorded signal was bandpass filtered at 300 Hz low/5 kHz high (1800 Microelectrode AC Amplifier, A-M Systems, Sequim, WA). B, Post hoc biochemical imaging modality. Cultured hippocampal neurons [13–14 d in vitro (div), transduced 2 div with ChR2-EYFP and NpHR-EYFP lentiviruses under the CaMKIIα promoter) were preincubated ∼2 h with d-AP5 (25 μm) and NBQX (10 μm) and then stimulated with light in the continued presence of d-AP5/NBQX. Blue light delivery (HQ470 nm/40×; Chroma) for 10 min at 10 Hz/25 ms to ChR2-expressing neurons significantly elevated Ser133 pCREB compared with sham-treated neurons (unpaired t test, p < 0.0001; n = 60 and 72 cells, respectively). Inset, Activation of NpHR with 10 min of continuous amber light (593 nm/20×; Semrock, Rochester, NY) significantly lowered pCREB levels compared with sham-treated neurons (unpaired t test, p < 0.0001; n = 80 and 75 cells, respectively). Cumulative probability histograms show nuclear pCREB intensity normalized to the sham condition mean. For quantification of pCREB immunofluorescence (1:500; Millipore), five confocal images [40×/1.3 numerical aperture (NA) oil] were acquired per stimulated region. Single optical sections through the nucleus were analyzed using Volocity software, and nuclear regions were defined by DAPI labeling. Imaging and quantification were conducted blind to treatment condition. Optical stimulation was conducted on an Olympus (Tokyo, Japan) IX71 inverted scope using a 20×/0.75 NA objective [7 mW/mm2 light intensity measured with Newport (Irvine, CA) 1815-C power meter]. C, Real-time activity imaging modality. ChR2 and NpHR action spectra are shown superimposed with fura-2–Ca2+ indicator excitation spectrum and RH-155 voltage indicator absorption band. Spectral separation allows for simultaneous all-optical control and readout. D, Directional control of mammalian locomotion with fiber-optic light delivery in freely moving animals. Blue light stimulation of the right secondary motor cortex (M2) in Thy1::ChR2-EYFP adult mice induced repetitive rotations to the left that terminated immediately with conclusion of optical stimulation (1.96 ± 0.24 rotations/10 s). Light was delivered from the 473 nm laser via optical fiber through a fiber guide cannula (Plastics One, Roanoke, VA) targeted to the following coordinates: anteroposterior, 1 mm; mediolateral, 0.5 mm; dorsoventral, −0.5 mm. Pulses (30 Hz/15 ms) were generated using a function generator (33220A; Agilent, Palo Alto, CA). Cranioplastic cement was used to stabilize the fiber guide, and a dummy cannula (a) was placed while animals were not being tested. To ensure stability of the fiber during testing in moving animals, an internal cannula adapter (b) was glued to the stripped optical fiber (c), which was inserted into the fiber guide cannula fixed to the skull (d) (see also Adamantidis et al., 2007). A custom aluminum rotating optical commutator (bottom; Doric Lenses, Québec, Canada) was used to release torsion in the fiber caused by the animal's rotation. E, Whisking control with LEDs in sedated Thy1::ChR2-EYFP animals. Optical excitation of vibrissal motor cortex was performed either through a small craniotomy (top traces) or through thinned skull (bottom trace) with a blue LED [LEDtronics, Torrance, CA; skull thinned ∼80% with standard drill as in Yoder and Kleinfeld (2002)]. Whisker deflections were measured as before (Aravanis et al., 2007); a 1 mg rare-earth magnetic particle (Magcraft, Vienna, VA) was attached to the vibrissa, and a magnetoresistive sensor (Honeywell, Morristown, NJ) was used to measure deflections.
However, for some applications, slow or stable readouts are preferable. Slower biochemical markers may be ideal for tracking and validating the outcome of optical neural control, particularly in cases in which simultaneous imaging of controlled cells is not possible but it is still important to quantify the effect of optical control on the stimulated or downstream population. Activation of immediate-early genes like c-fos and Arc can be measured with biochemical methods (e.g., antibodies or GFP-based reporters) 60 min or more after the presumptive optical stimulation, and other transcription factor activation events (e.g., CREB Ser133 phosphorylation) provide sensitive assays that can detect and stably report changes in membrane potential (Fig. 2B). Blue light exposure (470 nm for 10 min at 10 Hz/25 ms) to ChR2-expressing neurons robustly elevated Ser133 pCREB levels (Fig. 2B); in this way, changes in network activity can be tracked using biochemical traces, even after conclusion of optical control of neural activity, although this method is not precise enough for reporting the firing frequency experienced by any one cell during the stimulation protocol. Even in the presence of synaptic activity blockers, optical activation of NpHR (590 nm for 10 min) sufficed to significantly lower Ser133 pCREB levels compared with sham-treated neurons (Fig. 2B, inset); however, if the conditions are not strong enough to separate two populations, the biochemical trace imaging technique is not suitable for precisely determining the fraction of cells controlled by the optical protocol.
Fast optical readouts are needed as well. The action spectra of ChR2 and NpHR are such that even with single-photon imaging, both tools can be used simultaneously together with the fluorescent calcium indicator fura-2 (Fig. 2C) (Zhang et al., 2007a), and each are also compatible in principle with several organic voltage-sensitive dye strategies (Zhang et al., 2006), e.g., the absorbance dye RH-155 for slice physiology (Airan et al., 2007). Moreover, the availability of genetically encoded biosensors that can detect glutamate levels, membrane voltage, protein synthesis, or intracellular calcium in vitro and in vivo (Reiff et al., 2005; Parrish et al., 2006; Garaschuk et al., 2007; Heim et al., 2007) could provide cell type-specific optical readouts for effects of cell type-specific optical control with ChR2/NpHR. A wide range of genetically targetable activity reporters, such as GCaMPs, pericams, or Camgaroos (Nagai et al., 2001; Nakai et al., 2001; Kotlikoff, 2007), could be used together with ChR2/NpHR in vitro and in vivo, but care must be taken in choosing and/or developing the right reporters whose excitation spectra will not overlap with the ChR2/NpHR action spectra. Moreover, the fast troponin C fluorescent resonance energy transfer-based biosensor TN-XL (Mank et al., 2006) is excited at 430 nm and could be a good candidate for pairing with NpHR. The availability of two-photon excitation laser-scanning microscopy (Pettit et al., 1997; Svoboda and Yasuda, 2006) relaxes the need for nonoverlapping spectra, and optical techniques including use of multiple excitation spots, defocused beams, and fast scanning over cells may overcome the fact that two-photon microstimulation will only affect a small number of opsins in a typical excitation volume. Conversely, conventional two-photon imaging of circuit responses could be conducted in parallel with single-photon excitation of large numbers of targeted opsins. Finally, systems imaging modalities like functional magnetic resonance imaging and positron emission tomography will provide compatible in vivo imaging readouts for optogenetic stimulation experiments.
A key readout for systems neuroscience-level questions is behavior of freely moving animals. NpHR and ChR2 have been used in combination to control movement in freely moving C. elegans (Zhang et al., 2007a), and ChR2 has been used to modulate sleep–wake behavior in mice (Adamantidis et al., 2007); here, optical control of locomotion in freely behaving adult mice expressing ChR2 in cortical layer 5 is demonstrated with fiber-optic/laser diode tools (Fig. 2D). Time locked to optical stimulation onset and termination, right motor cortex (area M2)-implanted animals displayed an evoked bias for moving to the left side, a behavior not seen in control (non-ChR2-expressing) mice (Fig. 2D; see supplemental movies for experimental and control animals, available at www.jneurosci.org as supplemental material). The lightweight and thin optical fiber allows unrestricted behavior of the subject over long distances, and a commutator allows even behavior involving rotation to occur, an important feature for some disease models like Parkinson's. In cases in which the targeted cells or their dendrites are superficial, cortical-surface light-emitting diode (LED) illumination through a cortical window and even through thinned skull (Yoder and Kleinfeld, 2002) can also be used to drive temporally precise motor output patterns (Fig. 2E). Light-induced whisker deflections in these intact animals were measured magnetically (Aravanis et al., 2007), and in control experiments blue light illumination in ChR2-negative mice had no effect.
Summary
Interesting integrated applications of these targeting and readout methods could go beyond simply driving specific populations of cells to driving specific cells in different brain regions (e.g., among thalamus, cortex, hippocampus, and striatum) with precise relative timing (synchrony or asynchrony with well defined phase shifts). For example, the motor behavioral studies (Fig. 2D) suggest that specific cortical layers in a region could be turned on or off with high temporal precision in behaving subjects, a possibility unapproachable with electrode-based, pharmacological, or genetic methods. These experiments are within reach now, but there are also many refinements of microbial opsin technology still pending that could enhance the power of the approach. For example, the independence (in mammals) from exogenous cofactors is a positive aspect, but it remains possible that some mammalian cells will not have sufficient retinoids to fully support opsin function; in these cases, cointroduction of enzymes governing rate-limiting steps in retinoid uptake and metabolism (T. Wang et al., 2007; Yang and O'Tousa, 2007) (Tracey, personal communication) might be helpful in driving maximally efficacious opsin function.
The subcellular-targeting approach (Fig. 1D) suggests additional strategies to target axons, somata, and synaptic terminals. Other molecular modifications may result in usefully shifted spectral properties [e.g. redshifted opsins for better tissue penetration and multicolor optical activation experiments (Luecke et al., 2001; Shimono et al., 2003)], greater or lesser Ca2+ permeance, or improved conductance properties (Nagel et al., 2005). We find in hippocampal neurons that H134R mutation in ChR2 expressed under the synapsin I promoter gives rise to a 2.37 ± 0.07- (p < 0.005) fold enhancement in steady state current and 1.73 ± 0.10 (p < 0.005) fold enhancement in peak current (n = 5; comparison with wild-type ChR2). It is important to remember that in principle, ChR2 and NpHR will contribute to small, brief, and reversible elevations in intracellular Ca2+ and Cl−, respectively; likewise, native excitatory transmission also elevates intracellular Ca2+ via Ca2+-permeable glutamate receptors and voltage-activated Ca2+ channels, and native inhibitory transmission typically also elevates intracellular Cl−. Because elevations in Ca2+ can drive intracellular biochemical changes, and elevations in Cl− can modulate the effects of endogenous GABAergic neurotransmission, these factors should be considered in experimental design.
Findings obtained from microbial opsin work in animal models may ultimately inform novel clinical approaches, even without introduction of opsins into patients. Clinically compatible, noninvasive interventions that are likely to modulate neural activity, including radiation, ultrasound, and magnetic methods, all can be depth targeted in various ways (for example by using stereotactically guided accumulation of energy at subcentimeter resolution, delivered from multiple calculated trajectories as with the radiation-based CyberKnife/Gamma Knife, or depth focused as with high-intensity focused ultrasound). Energy can be depth targeted in other ways as well, for example with fiber optics or implanted transducers (Adamantidis et al., 2007). In principle, many focusing methods could be used to directly transduce energy, or even to activate custom stereotactically or endovascularly delivered micro-“antennas” that locally modulate radiation sensitivity, ultrasound response, and magnetic susceptibility, at specific circuit nodes in the clinical setting that had previously been implicated with optogenetic animal models (Zhang et al., 2007b). However, work in animal models will be a primary focus of the field, and in itself can promote understanding of the circuit basis of neuropsychiatric diseases like narcolepsy (Adamantidis et al., 2007).
Looking toward the future, plants and many fungi and microbes are highly dependent on light and therefore have developed highly specialized light-sensing proteins that will likely continue to provide novel and powerful tools for perturbing and interrogating the intact nervous system. Optical activation of intracellular signaling via modified opsins (Kim et al., 2005) and blue-light-activated adenylate cyclases (PACs) from the flagellate Euglena gracilis (Iseki et al., 2002; Ntefidou et al., 2003) can trigger rapid and reversible increases in cAMP levels in vitro and modulate behavior in Drosophila in vivo (Schroder-Lang et al., 2007). Phototropins, photo-regulated protein kinases (Huala et al., 1997; Briggs et al., 2001), might also provide useful tools for biochemistry studies of phosphorylation events within a cell that could complement ChR2/NpHR modulation of biochemical signaling, along with other light-activated compounds yet to be identified. Perhaps only a small fraction of the potential opsins and other tools identified in macrogenetic approaches (e.g., Venter et al., 2004) will be immediately useful, as the experience with the comparatively unstable Halobacterium salinarum halorhodopsin (HsHR) revealed (Zhang et al., 2007a), and the broad action spectra of these naturally occurring proteins may limit the number of independently addressable optical control channels achievable, but identification of new players may refine the microbial opsin approach, and molecular optimizations have the potential to incrementally further unlock latent utility. Together, these novel tools, targeting techniques, and readout technologies will extend the utility of the microbial opsins in bidirectional, reversible, and spatiotemporally precise control of activity in targeted neurons.
Footnotes
Editor's Note: Toolboxes are intended to briefly highlight a new method or a resource of general use in neuroscience or to critically analyze existing approaches or methods. For more information, see http://www.jneurosci.org/misc/itoa.shtml.
K.D. is supported by National Alliance for Research on Schizophrenia and Depression, American Psychiatric Institute for Research and Education, and the Snyder, Culpeper, Coulter, Klingenstein, Whitehall, McKnight, Kinetics, and Albert Yu and Mary Bechmann Foundations, as well as by National Institute of Mental Health, National Institute on Drug Abuse, the National Institutes of Health (NIH) Director's Pioneer Award Program, and the National Science Foundation. V.G. is supported by a Stanford Graduate Fellowship. F.Z. is supported by an NIH National Research Service Award. Special thanks to the Deisseroth laboratory, Russ Altman, Matthew Scott, Dan Tracey, Peter Hegemann, Georg Nagel, Alexander Gottschalk, Guoping Feng, George Augustine, and Jaimie Henderson for discussions. The materials and methods described herein are freely distributed and supported by the authors (www.stanford.edu/group/dlab).
References
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airan RD, Hu E, Vijaykumar R, Roy M, Meltzer LA, Deisseroth K. Integration of light-controlled neuronal firing and fast circuit imaging. Curr Opin Neurobiol. 2007 doi: 10.1016/j.conb.2007.11.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, Schneider MB, Deisseroth K. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberg E, Tittor J, Oesterhelt D. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci USA. 1993;90:639–643. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi A, Cui J, Ma YP, Olshevskaya E, Pu M, Dizhoor AM, Pan ZH. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Yoshimura Y, Callaway EM. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 2005;143:151–158. doi: 10.1016/j.jneumeth.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A, Okada K, Salomon M, Rudiger W, Sakai T, Takano M, Wada M, Watson JC. The phototropin family of photoreceptors. Plant Cell. 2001;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JJ, Banghart MR, Trauner D, Kramer RH. Light-induced depolarization of neurons using a modified Shaker K(+) channel and a molecular photoswitch. J Neurophysiol. 2006;96:2792–2796. doi: 10.1152/jn.00318.2006. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska A, Ting AE, Miesenböck G, Schnitzer MJ. Next-generation optical technologies for illuminating genetically-targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duschl A, McCloskey MA, Lanyi JK. Functional reconstitution of halorhodopsin. Properties of halorhodopsin-containing proteoliposomes. J Biol Chem. 1988;263:17016–17022. [PubMed] [Google Scholar]
- Ehrlich BE, Schen CR, Spudich JL. Bacterial rhodopsins monitored with fluorescent dyes in vesicles and in vivo. J Membr Biol. 1984;82:89–94. doi: 10.1007/BF01870735. [DOI] [PubMed] [Google Scholar]
- Garaschuk O, Griesbeck O, Konnerth A. Troponin C-based biosensors: a new family of genetically encoded indicators for in vivo calcium imaging in the nervous system. Cell Calcium. 2007;42:351–361. doi: 10.1016/j.ceca.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Gorostiza P, Volgraf M, Numano R, Szobota S, Trauner D, Isacoff EY. Mechanisms of photoswitch conjugation and light activation of an ionotropic glutamate receptor. Proc Natl Acad Sci USA. 2007;104:10865–10870. doi: 10.1073/pnas.0701274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero G, Reiff DF, Agarwal G, Ball RW, Borst A, Goodman CS, Isacoff EY. Heterogeneity in synaptic transmission along a Drosophila larval motor axon. Nat Neurosci. 2005;8:1188–1196. doi: 10.1038/nn1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann P, Oesterhelt D, Bamberg E. The transport activity of the light-driven chloride pump halorhodopsin is regulated by green and blue light. Biochim Biophys Acta Biomemb. 1985;819:195–205. [Google Scholar]
- Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, Kovalchuk Y, Konnerth A, Griesbeck O. Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nat Methods. 2007;4:127–129. doi: 10.1038/nmeth1009. [DOI] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- Kalaidzidis IV, Kalaidzidis YL, Kaulen AD. Flash-induced voltage changes in halorhodopsin from Natronobacterium pharaonis. FEBS Lett. 1998;427:59–63. doi: 10.1016/s0014-5793(98)00394-9. [DOI] [PubMed] [Google Scholar]
- Karpova AY, Tervo DG, Gray NW, Svoboda K. Rapid and reversible chemical inactivation of synaptic transmission in genetically targeted neurons. Neuron. 2005;48:727–735. doi: 10.1016/j.neuron.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Kim JM, Hwa J, Garriga P, Reeves PJ, RajBhandary UL, Khorana HG. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- Kotlikoff MI. Genetically encoded Ca2+ indicators: using genetics and molecular design to understand complex physiology. J Physiol (Lond) 2007;578:55–67. doi: 10.1113/jphysiol.2006.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lerchner W, Xiao C, Nashmi R, Slimko EM, van Trigt L, Lester HA, Anderson DJ. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron. 2007;54:35–49. doi: 10.1016/j.neuron.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Crystal structure of sensory rhodopsin II at 2.4 angstroms: insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-color confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Mank M, Reiff DF, Heim N, Friedrich MW, Borst A, Griesbeck O. A FRET-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys J. 2006;90:1790–1796. doi: 10.1529/biophysj.105.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Navarro-Quiroga I, Chittajallu R, Gallo V, Haydar TF. Long-term, selective gene expression in developing and adult hippocampal pyramidal neurons using focal in utero electroporation. J Neurosci. 2007;27:5007–5011. doi: 10.1523/JNEUROSCI.0867-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntefidou M, Iseki M, Watanabe M, Lebert M, Hader DP. Photoactivated adenylyl cyclase controls phototaxis in the flagellate Euglena gracilis. Plant Physiol. 2003;133:1517–1521. doi: 10.1104/pp.103.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish AR, Wang W, Wang L. Manipulating proteins for neuroscience. Curr Opin Neurobiol. 2006;16:585–592. doi: 10.1016/j.conb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Perrimon N. New advances in Drosophila provide opportunities to study gene functions. Proc Natl Acad Sci USA. 1998;95:9716–9717. doi: 10.1073/pnas.95.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Pettit DL, Wang SS, Gee KR, Augustine GJ. Chemical two-photon uncaging: a novel approach to mapping glutamate receptors. Neuron. 1997;19:465–471. doi: 10.1016/s0896-6273(00)80361-x. [DOI] [PubMed] [Google Scholar]
- Reiff DF, Ihring A, Guerrero G, Isacoff EY, Joesch M, Nakai J, Borst A. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder-Lang S, Schwarzel M, Seifert R, Strunker T, Kateriya S, Looser J, Watanabe M, Kaupp UB, Hegemann P, Nagel G. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods. 2007;4:39–42. doi: 10.1038/nmeth975. [DOI] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, Fiala A. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Shimono K, Hayashi T, Ikeura Y, Sudo Y, Iwamoto M, Kamo N. Importance of the broad regional interaction for spectral tuning in Natronobacterium pharaonis phoborhodopsin (sensory rhodopsin II) J Biol Chem. 2003;278:23882–23889. doi: 10.1074/jbc.M301200200. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, Kolstad KD, Tulyathan O, Volgraf M, Numano R, Aaron HL, Scott EK, Kramer RH, Flannery J, Baier H, Trauner D, Isacoff EY. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Tan EM, Yamaguchi Y, Horwitz GD, Gosgnach S, Lein ES, Goulding M, Albright TD, Callaway EM. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51:157–170. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Thompson SM, Kao JP, Kramer RH, Poskanzer KE, Silver RA, Digregorio D, Wang SS. Flashy science: controlling neural function with light. J Neurosci. 2005;25:10358–10365. doi: 10.1523/JNEUROSCI.3515-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, Wang D, Zhang F, Boyden E, Deisseroth K, Kasai H, Hall WC, Feng G, Augustine GJ. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci USA. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Jiao Y, Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, O'Tousa JE. Cellular sites of Drosophila NinaB and NinaD activity in vitamin A metabolism. Mol Cell Neurosci. 2007;35:49–56. doi: 10.1016/j.mcn.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Yoder EJ, Kleinfeld D. Cortical imaging through the intact mouse skull using two-photon excitation laser scanning microscopy. Microsc Res Tech. 2002;56:304–305. doi: 10.1002/jemt.10002. [DOI] [PubMed] [Google Scholar]
- Zemelman BV, Lee GA, Ng M, Miesenbock G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007a;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007b;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Wang LP, Deisseroth K. Genetically-targeted optical neural modulation: development of tools for probing disease models. Soc Neurosci Abstr. 2007c;33:318–11. [Google Scholar]
- Zhang YP, Oertner TG. Optical induction of synaptic plasticity using a light-sensitive channel. Nat Methods. 2007;4:139–141. doi: 10.1038/nmeth988. [DOI] [PubMed] [Google Scholar]
- Zito K, Parnas D, Fetter RD, Isacoff EY, Goodman CS. Watching a synapse grow: noninvasive confocal imaging of synaptic growth in Drosophila. Neuron. 1999;22:719–729. doi: 10.1016/s0896-6273(00)80731-x. [DOI] [PubMed] [Google Scholar]