miR-10 in development and cancer (original) (raw)
- Review
- Published: 22 May 2009
Cell Death & Differentiation volume 17, pages 209–214 (2010)Cite this article
- 5454 Accesses
- 126 Citations
- 6 Altmetric
- Metrics details
Abstract
The microRNA (miRNA) miR-10 family has attracted attention because of its conservation and the position of the miR-10 genes within the Hox clusters of developmental regulators. In several species, miR-10 is coexpressed with a set of Hox genes and has been found to regulate the translation of Hox transcripts. In addition, members of the miR-10 family are de-regulated in several cancer forms. Aside from acting in translational repression, miR-10 was recently found to bind a group of transcripts containing a terminal oligo-pyrimidine (TOP) motif and to induce their translation, thereby adding a new function to the miRNA repertoire.
Similar content being viewed by others
Main
With the discovery of small ncRNAs, such as microRNAs (miRNAs), a new group of gene expression regulators came into play. Over the last years, scientists from very diverse fields of biomedical and life sciences have undertaken the huge effort to elucidate the functions of these thousands of new regulatory molecules and place them into cellular pathways. The miR-10 family constitutes a particularly fascinating case because the miR-10 genes have been retained within the Hox cluster of developmental regulators over the course of evolution. As miR-10 has also been found to target Hox transcripts in several species, it is likely that miR-10 family members play important roles during the development. In addition, miR-10 family members are de-regulated in several types of cancer, but it is still largely unclear if miR-10 is causally related to cancer initiation or progression. Although repression of HOXD10 has been suggested to account for a role for miR-10b in breast cancer metastasis,1 an alternative model suggests that miR-10 may facilitate cancer by regulating ribosome biogenesis.2 In this review, I summarise the knowledge of miR-10 with respect to Hox gene regulation and cancer and discuss some of the outstanding issues to be addressed in the future.
miR-10 and Hox gene regulation
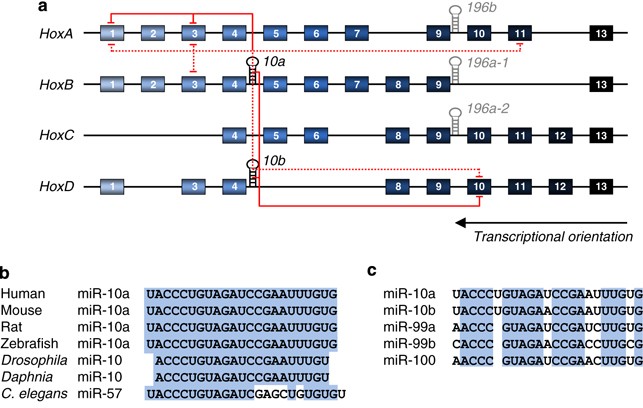
Hox genes encode highly conserved transcription factors characterized by the presence of a homeobox domain capable of binding to DNA.3 The Hox genes play crucial roles during development and are key determinants for correct anterior–posterior patterning of the body axis.3 Hox genes reside in genomic clusters and the temporal and spatial pattern of Hox gene expression is correlated to the genomic position within the cluster, a property coined colinearity.3 The Hox clusters produce several types of ncRNAs,4, 5 including miRNAs, exemplified by miR-10 and miR-196 in mammals.6 Whereas a common bilaterian ancestor is thought to have had a single Hox cluster (as is still the case in the fly), two duplication events over the course of evolution have resulted in the presence of four paralogous Hox clusters in mammals, containing five miRNA genes: miR-10a, miR-10b, miR-196a-1, miR-196a-2 and miR-196b7 (Figure 1a). Consequently, the miR-10 family is widely conserved with orthologs present in both the fly8 and the worm9 (Figure 1b). In mammals, miR-10a resides upstream from Hoxb4 and miR-10b upstream from Hoxd4, relative to the transcriptional orientation of the cluster. In addition, three miRNA encoded outside the Hox clusters, miR-99a/b and miR-100, are highly homologous to miR-10a/b and may have overlapping targets despite a difference within the seed region (Figure 1c). In nematodes, the Hox cluster has disintegrated over the course of evolution10 and although the Caenorhabditis elegans ortholog of miR-10, miR-57, has the same seed sequence as miR-10, it is not predicted to target Hox transcripts.11 The Drosophila miR-10 constitutes a particularly interesting example of Hox gene regulation by miR-10. The fly miR-10 is located in the Antennapedia Hox cluster and the mature miR-10 strand is predicted to target the neighbouring Hox gene, Sex combs reduced (Scr).12, 13 However, the fly miR-10 hairpin produces two mature miRNAs and the miR-10*, which is expressed to much higher levels than miR-10, is also predicted to target Hox cluster transcripts,14, 15 thereby reinforcing the notion of miR-10 as an integral regulator of Hox expression. The conserved position of miR-10 within the Hox gene clusters is intriguing and indicates a requirement for the same _cis_-regulatory elements that regulate the Hox genes. This notion is sustained by an observed pattern of miR-10 and Hox gene coexpression during the development, where the miR-10 genes are transcribed in the same orientation as the Hox genes and display expression boundaries typical for Hox gene expression.16, 17
Figure 1
(a) microRNAs encoded by the Hox cluster. The mammalian Hox cluster encodes miR-10 and miR-196 family members. The arrows indicate predicted Hox targets based on TargetScan and Pictar predictions (dotted line) and experimentally verified Hox targets for miR-10 (solid line) in mammals. See text for details and references. (b) Alignment of miR-10 representatives from seven species. (c) Homology among mammalian miR-10 family members
There is good evidence from several species that miR-10 targets Hox gene transcripts. Bioinformatics algorithms predict 3–4 Hox gene targets for the miR-10 family18, 19 (Figure 1a). An analysis based on TargetScan predictions of miRNAs targeting Hox genes showed that when considering all the predicted miRNA-binding sites in Hox genes, belonging to the 73 highly conserved miRNA families, miR-10 ranks third and the miR-10-related miRNA family miR-99/100 ranks second only surpassed by miR-196 – the other miRNA family encoded in the Hox clusters.11 In accordance with the bioinformatics predictions, several human HOX transcripts have been experimentally validated as miR-10 targets, including HOXA1, HOXA3 and HOXD101, 20, 21 (Figure 1a). Aside from Hox transcripts, miR-10 has also been shown to regulate USF2,22 Ran,2 Pbp12 and a set of ribosomal protein mRNAs,2 as discussed below. In zebrafish, miR-10 is represented by five paralogs and they have been shown to target HoxB1a and HoxB3a.16 Furthermore, overexpression of miR-10 in zebrafish embryos induce phenotypic changes similar to the loss of HoxB1a and HoxB3a.16
miR-10 deregulation in cancer
Several papers have reported of deregulation of miR-10 family members in human cancers, and miR-10a resides in a region amplified in melanoma and breast cancer23 (Table 1). In the haematopoietic system, miR-10a is normally expressed in CD34+ cells,24 and in vitro differentiation of CD34+ cells into megakaryocytes is accompanied by a marked decrease in the levels of both miR-10a and miR-10b.20 In accordance, levels of miR-10a are markedly higher in haematopoietic stem cells than in peripheral blood lymphocytes,25 perhaps reflecting a role for miR-10a in stem or progenitor cells. In haematological cancer cell lines as a group, miR-10a is found downregulated,26 and miR-10a was reported to be downregulated in chronic myeloid leukaemia22 and acute myeloid leukaemia,27 in comparison to CD34+ cells. Although this apparent deregulation of miR-10a in chronic myeloid leukaemia and acute myeloid leukaemia may represent cancer-specific changes, it could also reflect a general downregulation of miR-10 during differentiation.
Table 1 Deregulation of miR-10 family members in cancer
Conversely, miR-10b is upregulated in B-cell chronic lymphocytic leukaemia relative to normal CD5+ cells.28 In addition, noteworthy is the marked overexpression of both miR-10a, miR-10b and miR-100 in the subset of acute myeloid leukaemias carrying mutations in the nucleophosmin gene, NPM1.29 In fact, the three miR-10 family members come out as the top three upregulated miRNAs in this tumour subset,29 suggesting a causal relation to NPM1 mutations. Nucleophosmin is a ubiquitously expressed, multifunctional protein that shuttles between the nucleus and cytoplasm, and the gene is mutated in ∼30% of adult acute myeloid leukaemias because of mutations or translocations.30, 31 Interestingly, amongst its many functions, nucleophosmin is involved in the ribosome biogenesis.30, 32 Given the function proposed for miR-10a in translational regulation of ribosomal proteins (RPs) and ribosome biogenesis (discussed below), it is tempting to speculate that overexpression of miR-10 family members constitutes a compensatory mechanism in acute myeloid leukaemias carrying NPM1 mutations.
Both miR-10a and miR-10b have been found to be upregulated in glioblastoma and anaplastic astrocytomas, in some cases reaching more than a 100-fold overexpression.26, 33, 34 In primary hepatocellular carcinomas both miR-10a and miR-100 are overexpressed, when compared with the levels in normal liver,35 and both miR-10b, miR-99 and miR-100 are upregulated in pancreatic cancer as compared with normal pancreas.36 Furthermore, miR-10a was found overexpressed in colon cancer as compared with normal colon tissue.37 In a debated paper, miR-10b was found to be highly expressed in metastatic breast cancer cell lines relative to non-metastatic cell lines, and ectopic overexpression of miR-10b in non-metastatic cell lines was shown to promote tumour invasion and metastasis in a xenograft mouse model.1 The mechanism suggested involves translational repression of HOXD10 by miR-10b, resulting in upregulation of the invasion-promoting GTPase RHOC. The paper also reported of a moderate overexpression of miR-10b in a subset of primary tumour samples from patients with metastatic breast cancer, and the authors1 concluded that miR-10b plays a part in the processes of metastasis, rather than in cancer initiation per se. This paper has since been challenged by a study analysing a large cohort of patients with breast cancer, in which no correlation could be found between the miR-10b expression levels and the development of distance metastasis.38
miR-10 on TOP
An unexpected role for miR-10 family members was shown when miR-10a was identified as a regulator of ribosome biogenesis and consequently global protein production.2 The involvement of miR-10a in the translational regulation of RPs was discovered in an unbiased forward analysis of mRNAs associating with miR-10a. Although most validated miRNA targets have been identified using miRNA overexpression or inhibition followed by expression array analyses,39, 40, 41 or have been cherry picked from lists of predicted targets,18, 19, 42, 43 both of these approaches have critical drawbacks. As an unknown number of mRNA targets are regulated exclusively at the level of translational repression without affecting mRNA stability, these are often missed in expression array-based approaches. Bioinformatics predictions of miRNA targets, based primarily on evolutionary conservation of sequence motifs in the 3′-UTR matching the miRNA seed regions, have to a large extent driven the miRNA research field forward by providing testable hypotheses for the experimentalists.18, 19, 42, 43 However, the algorithms generate a high rate of false positives and false negatives and are still founded on relatively small sets of experimental data focusing either on highly expressed miRNAs or employing miRNA overexpression.
We devised an affinity purification method, in which miRNA targets are identified from miRNA/mRNA complexes isolated on streptavidin beads after transfection of cells with a biotin-labelled miRNA.44 For miR-10a, a specific set of transcripts was isolated from mouse embryonic stem cells, of which more than 60% of the mRNAs encoded factors involved in translation or translational regulation, mostly RPs.2 Interestingly, the binding site for miR-10 was mapped biochemically and genetically to the 5′-UTR, just downstream from the 5′-terminal oligo-pyrimidine (TOP) motif found in many RPs and translational regulators. Where most mRNAs have an A residue immediately after the cap structure, TOP mRNAs initiate with a C followed by a stretch of 4–14 uninterrupted pyrimidines (reviewed in Hamilton et al.45 and Meyuhas et al.46). TOP mRNAs comprise of RPs, elongation factors and other proteins associated with the translational apparatus and they form part of a cellular sensory mechanism by responding to a range of physiological stimuli. In the event of stresses, such as amino acid starvation, TOP mRNAs are selectively repressed at the level of translation.45, 46 The upstream regulatory pathways regulating TOP translation involves the phosphatidylinositol 3-kinase (PI3K) and mTOR pathways,47, 48 and several RNA-binding proteins have been proposed to regulate the translation of TOP mRNAs.49, 50, 51 However, the exact mechanism and wiring of upstream pathways is still largely unresolved.
We showed that the regulation of several TOP mRNAs is dependent on miR-10a and that ectopic expression of miR-10a can alleviate TOP repression in the face of amino acid starvation, thereby providing a novel link between miRNAs and translational regulation.2 The binding of miR-10a to the 5′-UTR of TOP mRNAs is atypical, in that it does not involve complete base pairing of the seed region. Although this has been noted for several other miRNAs,13, 52, 53 the bulk amount of available data sustain the notion that base pairing of the miRNA seed region is a major determinant for target recognition and repression.54 However, the ability of miR-10 to induce translation is not dependent on the absence of complete seed match, as an Rps16 5′-UTR vector engineered to have a perfect seed match is still translationally induced (UA Ørom and AH Lund, unpublished observation).
Surprisingly, the binding of miR-10a resulted in a modest increase in the translation of RPs, as evident from both analysis of newly synthesised endogenous RPs and heterologous reporter assays.2 Whereas the vast majority of miRNA research papers have shown translational repression, only few examples of miRNA-mediated induction of translation have been reported. In hepatitis C virus, the liver-specific miR-122 binds the viral 5′-UTR and plays an important role in viral replication.55 In addition, a recent report documented that binding of miR-122 also stimulates translation of the viral mRNA.56 Furthermore, translational activation was reported for both natural and artificial miRNAs during cell cycle arrest.57, 58 In the latter case, translational upregulation was dependent on Ago2 and the RNA-binding protein FXR1,57 but so far little mechanistic insight has been provided for miRNA-mediated translational stimulation. Although miR-10a efficiently precipitates Ago2 from mouse embryonic stem cells, it is unclear to what extend Ago2 associates with TOP mRNAs.2 It is, however, interesting to note that Argonaute proteins were originally described as enhancers of translation.59, 60 Hence, miRNA-mediated translational induction may involve an alternative translational mechanism.
Importantly, the miR-10a-mediated increase in the translation of RPs (and presumably other TOP mRNAs) resulted in an increase in the synthesis of mature rRNA and in a 30% overall enhanced protein production. Conversely, inhibition of miR-10a resulted in a 40% drop in global protein production.2 This effect of miR-10a on global protein production could explain the upregulation of miR-10 family members in several forms of cancer. Towards establishing a causal link between miR-10a and cancer, we showed that miR-10a can influence the capacity of NIH-3T3 cells overexpressing oncogenic RAS-V12 to undergo transformation.2 In these experiments, cells transfected with miR-10a transformed more readily, as measured by their ability to form colonies in soft agar assays, than mock transfected cells and introduction of a miR-10a inhibitor significantly reduced the number of transformed colonies. Similar results were obtained using a human glioblastoma cell line and in primary human astrocytes artificially transformed by the introduction of hTERT/RAS-V12/E6 and E7, indicating a broader, conserved mechanism (UA Ørom & AH Lund, unpublished data).
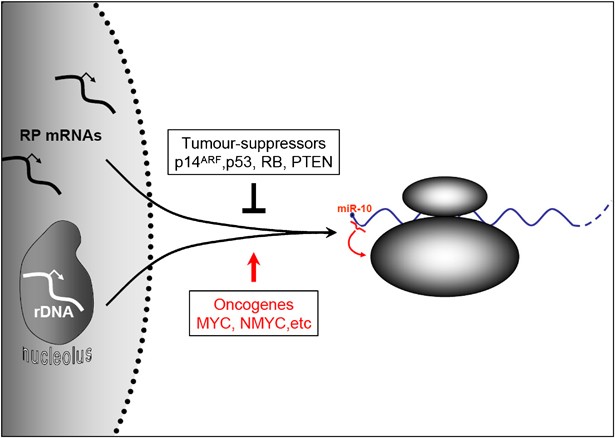
Hence, the level of miR-10a affects the capacity of cells to undergo oncogenic transformation. In normal cells, the translational machinery is tightly controlled, in part through controlled translation of RPs, and mechanisms to hijack the translational apparatus are likely required for cancer to develop.61 Accordingly, RPs are found deregulated in cancer,62, 63 and individual translation factors may directly promote cellular transformation.64, 65 The importance of translational control is also underlined by the fact that major tumour suppressors, such as p14ARF p53, RB and PTEN, negatively regulate ribosome biogenesis,66, 67, 68, 69 whereas oncogenes, such as MYC and NMYC, bind and activate a large group of genes involved in translation, including genes for rRNA, RPs and other translation cofactors.70, 71, 72 Interestingly, the effect of p14ARF on ribosome biogenesis depends, at least in part, on the direct binding of p14ARF to nucleophosmin.69 As mentioned above, patients with acute myeloid leukaemias containing nucleophosmin mutations have upregulated several miR-10 family members,29 suggesting that miR-10 can rescue defective ribosome biogenesis in cells harbouring mutated nucleophosmin. Interestingly, the nucleophosmin transcript is also believed to be TOP regulated.46 On the basis of the identification of RPs as targets for miR-10a-mediated translational stimulation, a role for miR-10 in cancer can be envisioned, in which miR-10 act as an important regulator of ribosome biogenesis to facilitate transformation (Figure 2). According to this model, miR-10, or a functionally similar mechanism, is required to grant the cancer control of the translational apparatus without acting as a classical oncogene (pro-proliferative, anti-apoptotic and so on). miR-10 would thereby constitute a target for anticancer therapy.
Figure 2
Control of global translation. Ribosome biogenesis is under transcriptional, post-transcriptional and translational control through several mechanisms including major oncogene and tumour suppressor pathways. Through regulating the translation of RPs, miR-10 affects ribosome biogenesis and may thereby influence cellular transformation
Outlook
Although the first miRNA was identified in the worm already in 1993,73 miRNA have only been intensely studied since their rediscovery in 20018, 9, 74 and much awaits elucidation. For miR-10, no mouse models have been published, which would be pivotal in addressing the role of miR-10 in development and differentiation. Such studies could be complicated by the existence of several miR-10 family members possibly resulting in genetic redundancy. In addition, both knockout and transgenic models will be important to explore the functional importance of miR-10 in cancer. Very little is known about which regulatory networks miR-10 participates in, and so far TWIST1 is the only direct regulator published for miR-10.1 An important future task will therefore be to identify regulatory pathways operating upstream of miR-10 and to obtain a comprehensive picture of miR-10 targets in different cell types. In addition, the mechanism behind miR-10-mediated translational induction of TOP mRNAs is completely unknown and is currently under study in several laboratories.
Abbreviations
miRNA:
microRNA
ncRNA:
non-coding RNA
UTR:
untranslated region
RPs:
ribosomal proteins
5′ TOP:
5′ terminal oligo-pyrimidine
References
- Ma L, Teruya-Feldstein J, Weinberg RA . Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007; 449: 682–688.
Article CAS Google Scholar - Orom UA, Nielsen FC, Lund AH . MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008; 30: 460–471.
Article Google Scholar - Pearson JC, Lemons D, McGinnis W . Modulating Hox gene functions during animal body patterning. Nat Rev Genet 2005; 6: 893–904.
Article CAS Google Scholar - Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129: 1311–1323.
Article CAS Google Scholar - Schmitt S, Paro R . RNA at the steering wheel. Genome Biol 2006; 7: 218.
Article Google Scholar - Tanzer A, Amemiya CT, Kim CB, Stadler PF . Evolution of microRNAs located within Hox gene clusters. J Exp Zoolog B Mol Dev Evol 2005; 304: 75–85.
Article Google Scholar - Lemons D, McGinnis W . Genomic evolution of Hox gene clusters. Science 2006; 313: 1918–1922.
Article CAS Google Scholar - Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T . Identification of novel genes coding for small expressed RNAs. Science 2001; 294: 853–858.
Article CAS Google Scholar - Lau NC, Lim LP, Weinstein EG, Bartel DP . An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001; 294: 858–862.
Article CAS Google Scholar - Aboobaker AA, Blaxter ML . Hox gene loss during dynamic evolution of the nematode cluster. Curr Biol 2003; 13: 37–40.
Article CAS Google Scholar - Yekta S, Tabin CJ, Bartel DP . MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet 2008; 9: 789–796.
Article CAS Google Scholar - Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS . MicroRNA targets in Drosophila. Genome Biol 2003; 5: R1.
Article Google Scholar - Brennecke J, Stark A, Russell RB, Cohen SM . Principles of MicroRNA-target recognition. PLoS Biol 2005; 3: e85.
Article Google Scholar - Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC . Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 2007; 17: 1850–1864.
Article CAS Google Scholar - Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res 2007; 17: 1865–1879.
Article CAS Google Scholar - Woltering JM, Durston AJ . MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE 2008; 3: e1396.
Article Google Scholar - Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 2004; 36: 1079–1083.
Article CAS Google Scholar - Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20.
Article CAS Google Scholar - Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500.
Article CAS Google Scholar - Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA 2006; 103: 5078–5083.
Article CAS Google Scholar - Han L, Witmer PD, Casey E, Valle D, Sukumar S . DNA methylation regulates microRNA expression. Cancer Biol Ther 2007; 6: 1284–1288.
CAS PubMed Google Scholar - Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, Bandres E, Cordeu L et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res 2008; 6: 1830–1840.
Article CAS Google Scholar - Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 2006; 103: 9136–9141.
Article CAS Google Scholar - Georgantas III RW, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA 2007; 104: 2750–2755.
Article CAS Google Scholar - Jin P, Wang E, Ren J, Childs R, Shin JW, Khuu H et al. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. J Transl Med 2008; 6: 39.
Article Google Scholar - Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 2007; 67: 2456–2468.
Article CAS Google Scholar - Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B . MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008; 111: 5078–5085.
Article CAS Google Scholar - Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA 2004; 101: 11755–11760.
Article CAS Google Scholar - Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA 2008; 105: 3945–3950.
Article CAS Google Scholar - Grisendi S, Mecucci C, Falini B, Pandolfi PP . Nucleophosmin and cancer. Nat Rev Cancer 2006; 6: 493–505.
Article CAS Google Scholar - Okuwaki M . The structure and functions of NPM1/nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem 2008; 143: 441–448.
Article CAS Google Scholar - Borer RA, Lehner CF, Eppenberger HM, Nigg EA . Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989; 56: 379–390.
Article CAS Google Scholar - Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008; 6: 14.
Article Google Scholar - Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 2005; 334: 1351–1358.
Article CAS Google Scholar - Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008; 47: 1223–1232.
Article CAS Google Scholar - Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007; 297: 1901–1908.
Article CAS Google Scholar - Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Article CAS Google Scholar - Gee HE, Camps C, Buffa FM, Colella S, Sheldon H, Gleadle JM et al. MicroRNA-10b and breast cancer metastasis. Nature 2008; 455: E8–E9; author reply E9.
Article CAS Google Scholar - Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005; 438: 685–689.
Article Google Scholar - Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH . Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008; 283: 1026–1033.
Article CAS Google Scholar - Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433: 769–773.
Article CAS Google Scholar - Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP . MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007; 27: 91–105.
Article CAS Google Scholar - Betel D, Wilson M, Gabow A, Marks DS, Sander C . The microRNA org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–D153.
Article CAS Google Scholar - Orom UA, Lund AH . Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods 2007; 43: 162–165.
Article CAS Google Scholar - Hamilton TL, Stoneley M, Spriggs KA, Bushell M . TOPs and their regulation. Biochem Soc Trans 2006; 34: 12–16.
Article CAS Google Scholar - Meyuhas O . Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem 2000; 267: 6321–6330.
Article CAS Google Scholar - Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol 2009; 29: 640–649.
Article CAS Google Scholar - Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D et al. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 2001; 21: 8671–8683.
Article CAS Google Scholar - Zhu J, Hayakawa A, Kakegawa T, Kaspar RL . Binding of the La autoantigen to the 5′ untranslated region of a chimeric human translation elongation factor 1A reporter mRNA inhibits translation in vitro. Biochim Biophys Acta 2001; 1521: 19–29.
Article CAS Google Scholar - Pellizzoni L, Lotti F, Rutjes SA, Pierandrei-Amaldi P . Involvement of the Xenopus laevis Ro60 autoantigen in the alternative interaction of La and CNBP proteins with the 5′UTR of L4 ribosomal protein mRNA. J Mol Biol 1998; 281: 593–608.
Article CAS Google Scholar - Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F . La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res 2000; 28: 2927–2934.
Article CAS Google Scholar - Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ . The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev 2004; 18: 132–137.
Article CAS Google Scholar - Didiano D, Hobert O . Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol 2006; 13: 849–851.
Article CAS Google Scholar - Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233.
Article CAS Google Scholar - Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P . Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005; 309: 1577–1581.
Article CAS Google Scholar - Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 2008; 27: 3300–3310.
Article CAS Google Scholar - Vasudevan S, Steitz JA . AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007; 128: 1105–1118.
Article CAS Google Scholar - Vasudevan S, Tong Y, Steitz JA . Switching from repression to activation: microRNAs can up-regulate translation. Science 2007; 318: 1931–1934.
Article CAS Google Scholar - Roy R, Ghosh-Dastidar P, Das A, Yaghmai B, Gupta NK . Protein synthesis in rabbit reticulocytes. Co-eIF-2A reverses mRNA inhibition of ternary complex (Met-tRNAf.eIF-2.GTP) formation by eIF-2. J Biol Chem 1981; 256: 4719–4722.
CAS PubMed Google Scholar - Zou C, Zhang Z, Wu S, Osterman JC . Molecular cloning and characterization of a rabbit eIF2C protein. Gene 1998; 211: 187–194.
Article CAS Google Scholar - Ruggero D, Pandolfi PP . Does the ribosome translate cancer? Nat Rev Cancer 2003; 3: 179–192.
Article CAS Google Scholar - Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR et al. Gene expression profiles in normal and cancer cells. Science 1997; 276: 1268–1272.
Article CAS Google Scholar - Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M . Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res 2001; 21: 2429–2433.
CAS PubMed Google Scholar - Lazaris-Karatzas A, Montine KS, Sonenberg N . Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 1990; 345: 544–547.
Article CAS Google Scholar - Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 2004; 10: 484–486.
Article CAS Google Scholar - Sugimoto M, Kuo ML, Roussel MF, Sherr CJ . Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell 2003; 11: 415–424.
Article CAS Google Scholar - Zhai W, Comai L . Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol 2000; 20: 5930–5938.
Article CAS Google Scholar - White RJ, Trouche D, Martin K, Jackson SP, Kouzarides T . Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature 1996; 382: 88–90.
Article CAS Google Scholar - Bertwistle D, Sugimoto M, Sherr CJ . Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol 2004; 24: 985–996.
Article CAS Google Scholar - Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 2001; 20: 1383–1393.
Article CAS Google Scholar - Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA 2000; 97: 3260–3265.
Article CAS Google Scholar - Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005; 7: 311–318.
Article CAS Google Scholar - Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854.
Article CAS Google Scholar - Lee RC, Ambros V . An extensive class of small RNAs in Caenorhabditis elegans. Science 2001; 294: 862–864.
Article CAS Google Scholar
Acknowledgements
The work in the author's laboratory is supported by EC FP7 funding (ONCOMIRS, Grant agreement number 201102. This publication reflects only author's views. The commission is not liable for any use that may be made of the information herein), The Vilhelm Pedersen and Hustrus Foundation, The Danish National Research Foundation, The Danish Cancer Society, The Lundbeck Foundation and The Association for International Cancer Research.
Author information
Authors and Affiliations
- Biotech Research and Innovation Centre and Center for Epigenetics, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen, Denmark,
A H Lund
Authors
- A H Lund
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toA H Lund.
Additional information
Edited by G Melino
Rights and permissions
About this article
Cite this article
Lund, A. miR-10 in development and cancer.Cell Death Differ 17, 209–214 (2010). https://doi.org/10.1038/cdd.2009.58
- Received: 18 February 2009
- Revised: 21 April 2009
- Accepted: 22 April 2009
- Published: 22 May 2009
- Issue Date: February 2010
- DOI: https://doi.org/10.1038/cdd.2009.58